Abstract
Two distinct genes encode the closely related signal transducer and activator of transcription proteins STAT5A and STAT5B. The molecular mechanisms of gene regulation by STAT5 and, particularly, the requirement for both STAT5 isoforms are still undetermined. Only a few STAT5 target genes, among them the CIS (cytokine-inducible SH2-containing protein) gene, have been identified. We cloned the human CIS gene and studied the human CIS gene promoter. This promoter contains four STAT binding elements organized in two pairs. By electrophoretic mobility shift assay studies using nuclear extracts of UT7 cells stimulated with erythropoietin, we showed that these four sequences bound to STAT5-containing complexes that exhibited different patterns and affinities: the three upstream STAT binding sequences bound to two distinct STAT5-containing complexes (C0 and C1) and the downstream STAT box bound only to the slower-migrating C1 band. Using nuclear extracts from COS-7 cells transfected with expression vectors for the prolactin receptor, STAT5A, and/or STAT5B, we showed that the C1 complex was composed of a STAT5 tetramer and was dependent on the presence of STAT5A. STAT5B lacked this property and bound with a stronger affinity than did STAT5A to the four STAT sequences as a homodimer (C0 complex). This distinct biochemical difference between STAT5A and STAT5B was confirmed with purified activated STAT5 recombinant proteins. Moreover, we showed that the presence on the same side of the DNA helix of a second STAT sequence increased STAT5 binding and that only half of the palindromic STAT binding sequence was sufficient for the formation of a STAT5 tetramer. Again, STAT5A was essential for this cooperative tetrameric association. This property distinguishes STAT5A from STAT5B and could be essential to explain the transcriptional regulation diversity of STAT5.
STAT proteins are latent transcription factors containing a Src homology 2 domain (SH2 domain) that become activated by tyrosine phosphorylation. The binding of the STAT SH2 domains to the phosphorylated cytokine receptors allows their tyrosine phosphorylation by Jak kinases. After dimerization and nuclear translocation, STAT dimers bind to specific DNA sequences, thereby allowing downstream gene regulation. Seven members of the STAT family have been described (STAT1α/β, STAT2, STAT3α/β, STAT4, STAT5A, STAT5B, and STAT6), and specific combinations are involved in the signaling pathways of different cytokines. The association of STAT5 with various coactivators or repressors and the binding of additional transcription factors to the promoter region of target genes, together with various combinations of STAT dimers, could contribute to the diversity and specificity of the transcriptional regulation of target genes (for reviews, see references 4, 11, and 26).
STAT5, originally named mammary gland factor (MGF), has been described as a positive regulator of the β-casein (βCAS) promoter by prolactin (29). In addition to prolactin, STAT5 proteins have been shown to be activated by erythropoietin (Epo), growth hormone, interleukin 2 (IL-2), IL-3, IL-5, IL-7, IL-9, IL-15, granulocyte-macrophage colony-stimulating factor (GM-CSF), thrombopoietin, and several other growth factors. However, only a few STAT5 target sequences have been identified, among them the βCAS promoter (8), the IL-2 receptor alpha (IL-2Rα) chain enhancer (15), p21 (WAF1) (21), oncostatin M (34), the serine protease inhibitor 2.1 (31), and the CIS gene promoter (20). Different MGF boxes located inside these promoters are implicated in transcriptional regulation following STAT5 activation.
Besides positive regulatory pathways, negative regulatory pathways such as phosphatases modulate the response to cytokines. The phosphatase SHP-1 has been shown to bind to both the phosphorylated Epo receptor (Epo-R) and the Jak2 kinase and to dephosphorylate these proteins, thereby leading to inactivation of Epo-R signaling (12). However, under certain conditions, STAT5 itself or carboxy-terminally truncated isoforms of STAT5 act as negative regulators of gene transcription (1, 19, 23, 24, 30). Another negative regulatory mechanism involves the protein CIS. The CIS protein is rapidly induced in hematopoietic cells by IL-2, IL-3, GM-CSF, and Epo. CIS contains an SH2 domain in the central part of the molecule and is associated with the cytoplasmic domains of the tyrosine-phosphorylated Epo-R and the β chain of IL-3R (35). CIS overexpression reduces the activation of promoters regulated by STAT5. Moreover, STAT boxes were shown to be involved in the Epo-dependent promoter activation of the murine CIS promoter in HEK 293 cells (20).
In this report, we identified different nuclear factors which bound to the human CIS gene promoter. This complex combination of transcriptional factors comprised STAT5A, STAT5B, an Sp1-related family protein, and at least three GGAA binding proteins, among which one showed an Epo-dependent DNA binding capacity. Among the four STAT5 boxes of the CIS promoter, only one bound to a STAT5 tetramer. Moreover, we showed that two STAT5A dimers can interact in cooperative binding with two STAT binding sequences of low or undetectable affinity to form a tetramer. This structure involves STAT5A but not STAT5B. The difference in the behavior of the two isoforms of STAT5 could be essential for the differential transcriptional regulation of STAT5 target genes.
MATERIALS AND METHODS
Reagents.
Anti-STAT5 antibodies, directed against the amino terminus of the sheep STAT5A and recognizing both STAT5A and STAT5B, were produced as described previously (7). We produced specific antibodies directed against STAT5A and STAT5B by immunizing rabbits with peptides corresponding to the 12 carboxy-terminal amino acids (AGLFTSARSSLS) of STAT5A or the 8 carboxy-terminal amino acids of STAT5B (QWIPHAQS) coupled to keyhole limpet hemocyanin (data not published). Rabbit polyclonal anti-Elf-1 and anti-Ets-1/2 were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Antibodies directed against the amino-terminal domain of PU.1/Spi-1 were kindly provided by F. Moreau-Gachelin (Institut Curie, Paris, France). Anti-GABPα and -β antibodies were kindly provided by M. Negishi (National Institutes of Health, Bethesda, Md.) (33). Highly purified recombinant human Epo (specific activity, 120,000 U/mg) was a generous gift from M. Brandt (Boehringer Mannheim), and prolactin was purchased from Sigma.
Isolation of human CIS promoter.
The murine CIS cDNA from nucleotides 111 to 1092 (GenBank accession no. MUSSH2DC) (35) was cloned by reverse transcriptase PCR with total RNA from IW32 erythroleukemia cells with a sense primer (5′-CTCCTTCCATCCCGCCGAA-3′) and an antisense primer (5′-CCTGCCTTGTTCTTGCTGGCA-3′).
A human placenta genomic DNA library cloned in the cosmid pWE 15 (Clontech; reference no. HL 1095m) was screened with the murine CIS cDNA PCR probe.
Cell cultures.
The GM-CSF-dependent human megakaryoblastic cell line UT7 (13) was passaged by dilution twice weekly in alpha minimal essential medium containing 10% fetal calf serum containing 2.5 ng of GM-CSF per ml. COS-7 cells (ECCAC no. 87021302) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum.
Preparation of nuclear extracts.
Starved cells were stimulated at 37°C with 10 U of Epo per ml and quickly chilled in ice-cold phosphate-buffered saline. Cells were pelleted and solubilized with buffer A (buffer A: 20 mM HEPES; 10 mM KCl; 1 mM EDTA; 1 mM dithiothreitol [DTT]; 1 mM phenylmethylsulfonyl fluoride; 0.1 mM Na2VO4; 0.2% Nonidet P-40; 10% glycerol; 1 μg each of aprotinin, pepstatin, and leupeptin per ml, pH 7.9). Cell lysates were centrifuged at 20,000 × g for 2 min, and the pellets were extracted with buffer B (buffer B: 20 mM HEPES; 350 mM NaCl; 10 mM KCl; 1 mM EDTA; 1 mM DTT; 1 mM phenylmethylsulfonyl fluoride; 0.1 mM Na2VO4; 20% glycerol; 1 μg each of aprotinin, pepstatin, and leupeptin per ml, pH 7.9) with 1 ml of buffer B for 5 × 107 cells. The extracts were centrifuged at 20,000 × g for 5 min, and supernatants were quickly frozen and stored at −80°C.
Production of recombinant activated STAT5 protein in Sf9 cells.
Sf9 cells grown in a spinner flask suspension culture were coinfected with baculoviruses encoding either STAT5A and Jak2 or STAT5B and Jak2 with a multiplicity of infection of 10 PFU/cell for each virus. At 60 h after infection, cells were harvested and lysed for 15 min on ice in a buffer containing 10% glycerol, 20 mM HEPES (pH 7.9), 10 mM KCl, 0.2% Nonidet P-40, 1 mM EDTA, 0.1 mM sodium vanadate, 2 mM DTT, 4-(2-aminoethyl)-benzesulfonyl fluoride, phenylmethylsulfonyl fluoride, leupeptin, and aprotinin. Proteins were loaded on a heparin column (Pharmacia) and eluted with an NaCl gradient in lysis buffer.
Electrophoretic mobility shift assay (EMSA).
Two microliters of nuclear extracts was mixed with 20 μl of binding buffer [10 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% Nonidet P-40, 5% glycerol, 1 mg of bovine serum albumin per ml, 2 mg of poly(dI-dC) per ml, pH 7.5] containing 60,000 cpm of end-labelled probe, and the mixture was incubated for 30 min at 4°C. Complexes were separated on 6% nondenaturing polyacrylamide gels in 0.25× Tris-borate-EDTA and detected by autoradiography.
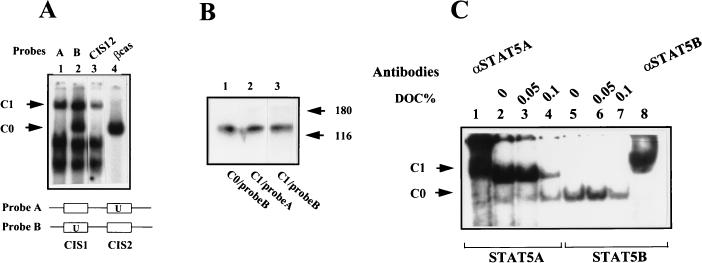
UV cross-linking of CIS12 probe to complexes C0 and C1.
Two probes (A and B) were used in cross-linking experiments. Single-stranded oligonucleotides (25 ng) CIS12 (antisense) and CIS2 (sense) (probe A) or CIS12 (sense) and CIS1 (antisense) (probe B) were annealed. Flanking sequences were transcribed by the Klenow fragment of DNA polymerase under standard conditions in the presence of 250 μM dATP–dGTP–5-bromo-2′-dUTP–80 μCi of [α-32P]dCTP (3,000 Ci/mmol). Probes were purified by Sephadex G-25 chromatography, and 2.5 ng of probes was incubated with UT7 nuclear extracts under EMSA conditions. After separation of the different complexes on a 6% nondenaturing polyacrylamide gel, the gel was irradiated for 15 min with UV light and autoradiographed. The bands were cut out and placed on the top of a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel. After migration of the gel, labelled proteins were detected by autoradiography.
Transfections and luciferase assays.
All plasmids were purified by the Qiagen kit method (Qiagen, Inc.), and two different preparations of plasmid for each construct were tested. Plasmids (1 μg/transfection) of different promoter-luciferase constructs cloned in the pGL2-basic vector (Promega) were introduced in COS-7 cells together with 1 μg of the pCMV-gal plasmid (internal control for transfection efficiency). After 24 h of culture with the appropriate medium, half of the transfected cells were stimulated with 1 μg of prolactin per ml. Total cell extracts were prepared and used for determination of luciferase and β-galactosidase activities according to the manufacturer’s instructions (Promega kit). Final luciferase activity results were obtained after normalization with β-galactosidase activity.
RESULTS
Localization and structural analysis of the human CIS gene 5′-flanking region: constitutive and cytokine-induced nuclear factors bound to the CIS gene promoter.
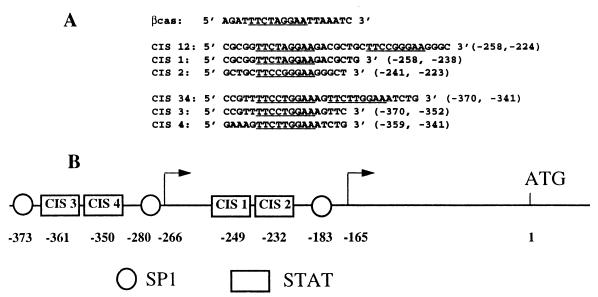
The murine CIS cDNA probe was used to screen a human genomic library as described in Materials and Methods. A cosmid, COS-CIS, that contained the full-length CIS gene was isolated (data not shown). Eight hundred sixty-five base pairs of the murine CIS gene promoter sequences have been previously reported by Matsumoto et al. (20). The nucleotide sequence of the 5′-flanking region of the human CIS gene was compared with this sequence. Computer alignments revealed that regions conserved between the two species began at position −646 of the murine CIS sequence (Fig. 1) and displayed 70% identity. Two blocks of 50 to 60 nucleotides were highly conserved: nucleotides −376 to −324 and −262 to −198 (human sequence) had 94 and 91% identity, respectively. Each conserved region contained two STAT binding consensus sequences (CIS3 and CIS4 and CIS1 and CIS2 at positions −361 and −350 and −249 and −232, respectively [Fig. 1]). These STAT boxes and the distance between CIS3 and CIS4 (2 bases) and between CIS1 and CIS2 (8 bases) were totally conserved between the human and murine species.
FIG. 1.
Nucleotide sequence comparison of human and murine gene 5′-flanking regions. The Kanehisa program was used to compare the human (upper) and murine (lower) 5′-flanking sequences of the CIS gene. The murine sequence corresponds to the sequence published by Matsumoto et al. (20). These sequences are numbered such that +1 refers to the translation initiation codon of the CIS protein. Positions of transcription start sites are indicated by arrows. The consensus binding sites for STAT and SP1 are boxed.
Double-stranded oligonucleotides corresponding to individual STAT boxes or to a cluster of two STAT boxes were used in gel retardation assays to characterize the nuclear proteins able to bind to these sequences. All the names, sequences, and positions of the oligonucleotides used in this study are indicated in Fig. 2. The four STAT boxes of the CIS gene promoter were compared with the βCAS oligonucleotide sequence: the CIS1 sequence TTCTAGGAA was identical to the βCAS STAT binding site while the three other STAT boxes contained different nucleotides at positions 4 and 5.
FIG. 2.
(A) Synthetic oligonucleotides used in bandshift assays. Double-stranded oligonucleotide probes containing STAT binding sites were end labelled and tested for protein binding by EMSA. Sequences and positions of the upper-strand oligonucleotides used as probes in EMSAs are indicated. The consensus STAT binding sequence TTCNNNGAA is underlined. (B) Schematic representation of the 5′-flanking region of the human CIS gene. Boxes and circles indicate the positions of STAT and SP1 potential binding sequences, respectively. Positions of transcription start sites are indicated by arrows. CIS1 to CIS4 correspond to the names of the four STAT binding oligonucleotides described for panel A.
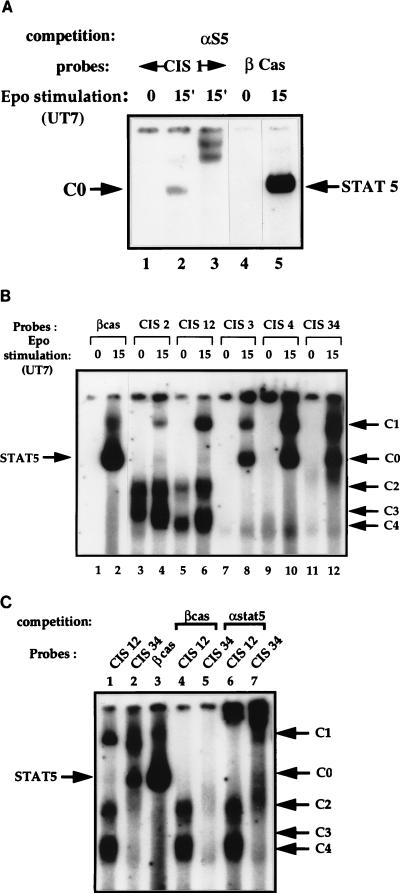
EMSAs were performed with nuclear extracts from resting and Epo-stimulated UT7 cells. The same mobility complex (C0) was obtained with the βCAS and the CIS1 probes. This complex, which is known to contain a dimer of STAT5, was supershifted with antibodies against STAT5. Thus, CIS1 bound specifically to STAT5 but with a weaker affinity than βCAS (Fig. 3A; compare lanes 2 and 5). CIS3, CIS4, and CIS34 oligonucleotides displayed similar Epo-induced retarded bands: a complex migrating like the STAT5-βCAS complex (C0) and a DNA-protein complex of lower mobility (C1) (Fig. 3B, lanes 8, 10, and 12). A different pattern of protein-DNA complexes was detected with the CIS2 and CIS12 probes: the C0 complex was absent, but the Epo-induced complex C1 could be visualized, the CIS12 probe displaying a higher affinity for complex C1 than CIS2 (Fig. 3B, lanes 4 and 6). Two prominent bands of higher mobility were also detected with these two probes. One band contained a constitutive C2 DNA-protein complex, and the lower band contained at least two complexes of similar mobilities, C3 and C4 (Fig. 3B, lanes 3 to 6; see also Fig. 6A, lanes 2 [C4] and 3 [C3+C4]), C3 being detected only in nuclear extracts from Epo-stimulated cells.
FIG. 3.
Gel shift assays of DNA complexes bound to STAT binding oligonucleotides of the human CIS gene promoter. Nuclear extracts from Epo-stimulated UT7 cells were incubated with 32P-labelled probes. DNA-protein complexes were analyzed in nondenaturing acrylamide gels and revealed by autoradiography. Free probes are not visible on these autoradiographs. (A) Identification of DNA-protein complexes which bind to the CIS1 probe (lanes 1 to 3). βCAS probe was used as a control (lanes 4 and 5). An antibody directed against STAT5 (7) was added to nuclear extracts in lane 3. The numbers above the panel show time in minutes. (B) Identification of DNA-protein complexes which bind to the CIS2 (lanes 3 and 4), CIS12 (lanes 5 and 6), CIS3 (lanes 7 and 8), CIS4 (lanes 9 and 10), and CIS34 (lanes 11 and 12) probes. βCAS probe was used as a control (lanes 1 and 2). The position and the name of each specific DNA-protein complex (C) are indicated on the right by an arrow. Numbers above the panel show time in minutes. (C) Nuclear extracts from UT7 cells stimulated with Epo for 15 min were incubated with the clustered STAT binding probes CIS12 (lanes 1, 4, and 6) and CIS34 (lanes 2, 5, and 7). Competition assays with a 50-fold molar excess of unlabelled double-stranded βCAS probe were performed (lanes 4 and 5). For lanes 6 and 7, anti-STAT5 serum was added to the EMSA reaction.
FIG. 6.
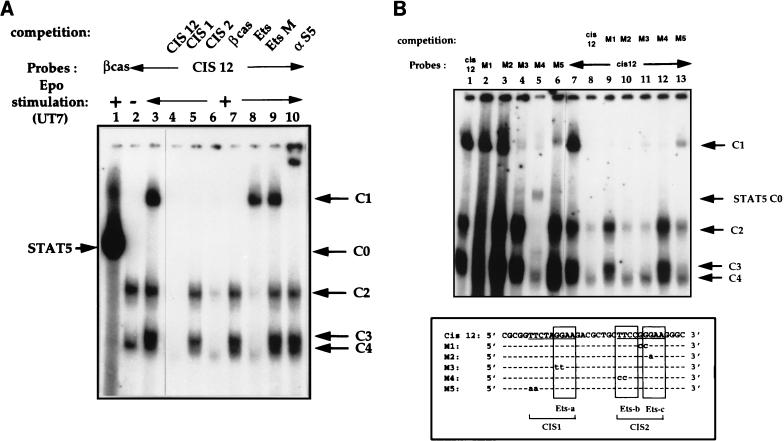
Analysis of DNA-protein complexes which bind to the CIS12 probe. Nuclear extracts from Epo-stimulated UT7 cells were incubated with a 32P-labelled CIS12 probe. (A) Identification of specific DNA-protein complexes by competition experiments. Unlabelled competitors (50-fold molar excess) were added to EMSA mixtures as follows: no competitor (lane 3) and double-stranded oligonucleotides CIS12, CIS1, and CIS2; βCAS; an optimized Ets probe from the Drosophila E74 promoter (3); and EtsM, in which the Ets binding sequence GGAA was mutated to CCAA (lanes 4 to 9, respectively). A βCAS probe was used as a control (lane 1). Lane 2 contains nuclear extracts from unstimulated UT7 cells. (B) Mutated CIS12 oligonucleotide bandshift assays. (Top) Nuclear extracts from Epo-stimulated UT7 cells were incubated with the 32P-labelled CIS12 probe (lanes 1 and 7 to 13) and with 32P-labelled mutated CIS12 M1 to M5 probes (lanes 2 to 6, respectively). Competition assays were performed for lanes 8 to 13 with a 50-fold molar excess of probes as indicated in the figure. (Bottom) Alignment of wild-type CIS12 and mutated M1 to M5 sequences. The consensus STAT binding sequence TTCNNNGAA is underlined, and the Ets domains are boxed.
Identification of STAT5 nuclear factors.
To identify the nuclear factors bound to the CIS34 and CIS12 probes, we performed EMSA in the presence of an excess of unlabelled oligonucleotides and supershift assays with various antibodies. Competition assays with unlabelled βCAS oligonucleotide showed that C2, C3, and C4 complexes were not affected (Fig. 3C, lane 4). In contrast, Epo-induced C1 and C0 complexes totally disappeared, suggesting that the C1 and C0 complexes both contained STAT5 (Fig. 3C, lanes 4 and 5), as confirmed by supershift experiments with STAT5-specific antibody (Fig. 3C, lanes 6 and 7).
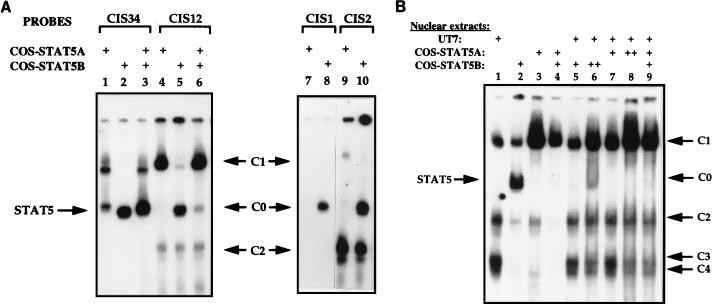
Two highly homologous STAT5 genes encoding STAT5A and STAT5B are expressed in many tissues and cell lines (16, 17, 25). To determine which STAT5 protein bound to the STAT binding sequences of the CIS gene promoter, COS-7 cells were cotransfected with vectors directing the expression of STAT5A or STAT5B and the prolactin receptor. After prolactin stimulation, nuclear extracts (COS-STAT5A and COS-STAT5B) were tested for DNA binding activity. As STAT5A and STAT5B were independently transfected, we verified that both proteins were expressed and phosphorylated at similar levels. When EMSAs were performed with extracts from COS-7 cells transfected with STAT5A, strong C0 and C1 complexes were detected with the CIS34 probe (Fig. 4A, lane 1). In contrast, STAT5B-transfected cell extracts induced a C0-like complex (Fig. 4A, lane 2). When extracts from COS-7 cells transfected with STAT5A and STAT5B were mixed, we detected the same retardation pattern as that with UT7 nuclear extracts (Fig. 4A, lane 3, and 3B, lane 12).
FIG. 4.
Specific binding patterns of STAT5A and STAT5B for the CIS probes. COS-7 cells transfected with expression vectors for the prolactin receptor, STAT5A, or STAT5B were treated with prolactin, and nuclear extracts were prepared (COS-STAT5A and COS-STAT5B, respectively). Bandshift assays were carried out with 32P-labelled CIS34 probe (lanes 1 to 3 [A]), CIS12 probe (lanes 4 to 6 [A] and 1 to 9 [B]), CIS1 probe (lanes 7 and 8 [A]), or CIS2 probe (lanes 9 and 10 [A]) in the presence of different combinations of nuclear extracts (COS-STAT5A, COS-STAT5B, and UT7) as indicated by a + in the figure (++ corresponds to a double quantity of nuclear extracts).
With the CIS12 probe, different results were obtained under the same EMSA conditions (COS-7 reconstituted model): STAT5A bound strongly to the CIS12 probe and appeared as a C1 complex (Fig. 4A, lane 4) whereas a C0-like complex was formed with COS-STAT5B nuclear extracts (Fig. 4A, lane 5). We also observed a faint C1 complex with STAT5B. However, when EMSA was performed with purified recombinant STAT5B, C1 complex was not detected (see below and Fig. 5C, lane 5). COS-7 cells were then tested for endogenous STAT5 expression by Western blot analysis with specific anti-STAT5A and anti-STAT5B antibodies. Both proteins were detected at a low level (data not shown). Therefore, the weak C1 complex observed in COS-7 cells could result from endogenous STAT5A expression.
FIG. 5.
(A and B) UV cross-linking of STAT5 complexes to CIS12 probe. Two double-stranded DNA probes comprising CIS1 and CIS2 were used. The first contained 5-bromo-2′-dUTP in regard to the CIS2 binding site (probe A), and the other contained it in regard to CIS1 (probe B) (see Materials and Methods). (A) EMSAs were carried out with UT7 cell extracts stimulated for 15 min. The complexes C1-probe A, C1-probe B, and C0-probe B were cut out after UV irradiation of the gel. (B) These complexes were electrophoresed on an SDS–10% polyacrylamide gel, and the cross-linked complexes were visualized by autoradiography. Molecular masses of the marker proteins are shown in kilodaltons. (C) Effect of the disruption of STAT5 protein interactions by deoxycholate (DOC). EMSA reactions were performed with the CIS12 probe in the presence of purified activated STAT5A (lanes 1 to 4) or STAT5B (lanes 5 to 8). Specific binding of STAT5 proteins to CIS12 probe was determined by supershift assays with specific antibodies directed against STAT5A (lane 1) and STAT5B (lane 8). Different concentrations of deoxycholate were added in the binding reaction mixtures as indicated. DNA-protein complexes were analyzed in nondenaturing acrylamide gels and revealed by autoradiography.
When COS-STAT5A and COS-STAT5B extracts were mixed, a main C1 complex was detected (Fig. 4A, lane 6), as observed with UT7 nuclear extracts (Fig. 3B, lane 6).
In conclusion, the CIS12 probe preferentially bound to STAT5A and the C1 complex seemed to depend on the presence of STAT5A. With the CIS2 probe, we observed the same complexes, C1 with STAT5A and C0 with STAT5B, although STAT5B showed a stronger affinity for this probe than did STAT5A (Fig. 4A, lanes 9 and 10). In the same conditions, the CIS1 probe bound only to STAT5B (Fig. 4A, lanes 7 and 8).
Finally, addition of UT7 nuclear extracts which contain phosphorylated STAT5A/B heterodimers to COS-STAT5B extracts led to the disappearance of the C0 complex and to an increase of the C1 complex on the CIS12 probe (Fig. 4B, compare lanes 2 and 5). When COS-STAT5A nuclear extracts were mixed with UT7 nuclear extracts, the intensity of the C1 band decreased while C2, C3, and C4 formation increased (Fig. 4B, lanes 3 and 7). This could result from the high expression in UT7 cells of proteins forming C2, C3, and C4 complexes. These proteins bind to a DNA sequence which overlaps with a STAT binding site and could compete with STAT5 proteins for binding to the CIS12 probe. Increasing the amount of COS-STAT5A and STAT5B nuclear extracts augmented the C1 complex (Fig. 4B, compare lanes 5 with 6 and 7 with 8, respectively). On the other hand, increasing the amount of COS-STAT5B nuclear extracts resulted in the appearance of the C0 complex (Fig. 4B, lane 6). In conclusion, these data suggest that the C0 complex contains STAT5B homodimers whereas the C1 complex is dependent on the binding of STAT5A.
To demonstrate the tetrameric structure of C1 complexes, we first analyzed the nature of the proteins bound to the CIS12 probe by UV cross-linking. Two kinds of CIS12 probes were used, probes A and B, which contained 5-bromo-2′-dUTP in regard to CIS2 or CIS1, respectively (see Materials and Methods and Fig. 5A). EMSAs performed with Epo-stimulated UT7 extracts revealed that probe A bound the same complexes as did the CIS2 probe and that probe B was able to also bind C0. This could be explained by the fact that CIS1 but not CIS2 binds to complex C0 (Fig. 3A and B). The presence of 5-bromo-2′-dUTP in regard to the CIS1 binding site could stabilize C0 complexes on the CIS1 sequence. After UV irradiation of the gel, the complexes C1-probe A, C1-probe B, and C0-probe A were resolved on an SDS-polyacrylamide gel. In all cases, the cross-linked complexes appeared as a single band of about 120 kDa which corresponded to the molecular mass of STAT5 (90 kDa) after subtraction of the mass of the DNA probe (about 30 kDa for 35 bp) (Fig. 5B). These data indicate that the only protein of the C1 complex that binds to CIS12 DNA is STAT5.
To confirm that C1 was composed of a tetramer of STAT5A, we then performed EMSAs with purified recombinant STAT5A and STAT5B proteins. Figure 5C shows that recombinant STAT5A induced C1 complexes with the CIS12 probe, confirming that no additional proteins were necessary for the formation of C1. Only the dimeric C0 complex was detected with recombinant STAT5B. Finally, to demonstrate that the C1 complex depended on interactions between two STAT5A dimers and did not result from the independent binding of two STAT5A dimers, we used deoxycholate, which disrupts protein interactions, and tested this compound on STAT5 complex formation. Increasing the concentration of this detergent reduced the amount of C1 complex (Fig. 5C, lanes 2 to 4). Altogether, these data demonstrate that STAT5 proteins are sufficient for the formation of a C1 complex and that this complex results from an interaction between two STAT5A dimers.
Analysis of C2, C3, and C4 complexes.
The identity of the constitutive C2 and C4 complexes and of the Epo-induced C3 complex was first studied by competition assays with unlabelled oligonucleotides on the CIS12 probe (Fig. 6A). A 50-fold molar excess of CIS12 oligonucleotide resulted in the complete disappearance of all C complexes (Fig. 6A, lane 4); similar results were observed with an excess of CIS2 oligonucleotide, although competition was weaker for C2 and C4 complexes (Fig. 6A, lane 6); in contrast, the same amount of oligonucleotides CIS1 and βCAS competed only for the binding of STAT5-containing complex C1 (Fig. 6A, lanes 5 and 7). The failure of oligonucleotides CIS1 and βCAS to compete for the binding of C2, C3, and C4 to CIS12 confirmed that these three complexes bound to the −240-to-−223 sequence of the human CIS gene promoter (Fig. 2B). The CIS12 sequence contains two 5′-GGAA-3′ or 5′-TTCC-3′ motifs known to be the core binding site of Ets factors. To determine if CIS12 was able to bind Ets proteins, an optimized Ets probe from the Drosophila E74 promoter (3) was used as a competitor. This oligonucleotide competed for the binding of C2, C3, and C4 (Fig. 6A, lane 8); similar results were obtained with the CIS2 probe (data not shown). As a control for specificity, we used as a competitor the oligonucleotide EtsM, in which the Ets binding sequence GGAA was mutated into a CCAA sequence (Fig. 6A, lane 9). In conclusion, the three complexes C2 to C4 recognize the motif 5′-GGAA-3′ and could be related to different members of the Ets family or to GGAA binding proteins. The CIS2 STAT binding sequence overlaps with two Ets sequences (Ets-b and Ets-c [Fig. 6B]) and is also involved in Epo-induced STAT5 C1 complex formation as described above. However, competition with the Ets E74 oligonucleotide did not affect the formation of the C1 complex.
Identification of the C1 to C4 binding sites.
In order to determine the sites required for each protein-DNA interaction, we examined the effects of selected nucleotide modifications in the two STAT binding sequences (CIS1 and CIS2) or in the three Ets sites (Ets-a, -b, and -c). For this purpose, mutated oligonucleotides, shown at the bottom of Fig. 6B, were used as probes or competitors in bandshift assays with the CIS12 probe (Fig. 6B). As a result, C1 binding was not affected by mutations which modified the NNN sequence of STAT5 consensus (M1 mutant) or destroyed the 3′ part of the palindromic proximal CIS2 STAT box and the Ets-c sequence (M2 mutant) (Fig. 6B, lanes 2, 3, 9, and 10). However, the M3 and M5 mutants, containing GG-to-TT and TT-to-AA substitutions that affected either the 3′ binding site of the distal CIS1 STAT box and the Ets-a sequence (M3 mutant) or the 5′ CIS1 STAT-binding sequence (M5 mutant), evidenced a markedly decreased affinity for the C1 complex (Fig. 6B, lanes 4, 6, 11, and 13). This M4 mutant, carrying a TT-to-CC substitution that affected the 5′ binding part of the proximal STAT box (CIS2) and the Ets-b site, was unable to bind C1 (Fig. 6B, lane 5). The faint C0 band detected with this mutant probably corresponded to the binding of STAT5 to the CIS1 sequence (Fig. 6B, lanes 5 and 12). In conclusion, both the CIS1 distal STAT box and the 5′ half of the proximal STAT site seemed to cooperate to bind C1 with high affinity. Moreover, Ets-b was also involved in the binding of C2 and C3: the M4 mutant did not bind C2 or C3 (Fig. 6B, lanes 5 and 12), and the M1 and M2 oligonucleotides, in which the Ets-b environment or the Ets-b site itself was changed, did not compete for the binding of C2 and C3 to the Ets-b sequence (Fig. 6B, lanes 9 and 12). Interestingly, the M2 mutation, in which the second guanine of the Ets-c motif was changed to an adenine, restored a new Ets sequence with increased affinity for C2, C3, and C4 (Fig. 6B, lane 3). Using several anti-Ets antibodies, we identified C2 as GABPα/β-containing complex (data not shown).
A 500-bp human CIS gene promoter shows a basal transcriptional activity and is transactivated by STAT5.
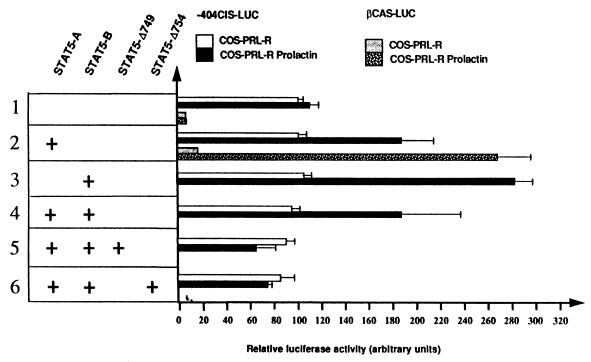
To determine the role of STAT5A and STAT5B on the minimal CIS promoter, the −404CIS-LUC construct was transfected in COS-7 cells in combination with different expression vectors carrying the prolactin receptor, STAT5A, and STAT5B (Fig. 7). We compared the activity of the CIS gene promoter with that of the βCAS gene promoter (βCAS-LUC) (5). Promoter activities were examined in the absence or presence of prolactin stimulation. In the absence of STAT5, the βCAS-LUC construct was inactive; in contrast, the −404CIS-LUC construct evidenced a basal activity which was not increased by prolactin (Fig. 7, lane 1). In the presence of STAT5, prolactin treatment stimulated the luciferase activity of both reporter constructs to approximately the same level; however, if results were expressed in fold induction, the βCAS gene promoter was much more inducible than the CIS gene promoter. This difference results from the basal activity of the −404CIS promoter in the absence of prolactin. Similar results were obtained for UT7 cells grown in the absence or presence of Epo (data not shown). Both STAT5A and STAT5B enhanced reporter gene transcription, and STAT5B alone was more efficient than STAT5A (Fig. 8, lanes 2 and 3). The luciferase activity was identical in cells transfected with STAT5A and STAT5B and in those with STAT5A alone (Fig. 7, lane 4). The specificity of STAT5 transactivation was demonstrated by the expression of STAT5 dominant-negative mutants which lacked the C-terminal transactivation region (STAT5-Δ749 and STAT5-Δ754) (23). As shown in Fig. 7, lanes 5 and 6, basal expression of −404CIS-LUC was still observed in the presence of these STAT5 dominant-negative mutants. Altogether, these data show that the −404-to-−1 CIS promoter region is constitutively active in the absence of cytokine stimulation and that STAT proteins are responsible for the increased activity induced by prolactin or Epo stimulation.
FIG. 7.
STAT5 transactivates the minimal CIS gene promoter in a reconstituted COS-7 model. COS-7 cells were cotransfected with the CIS gene promoter (−404CIS-LUC) or with the βCAS gene promoter-luciferase reporter constructs (βCAS-LUC) (lanes 1 and 2) together with vectors encoding the prolactin receptor and STAT5A, STAT5B, or two dominant-negative forms of STAT5 which lack the C-terminal transactivation region (STAT5-Δ749 and STAT5-Δ754) (23), as indicated in the figure (lanes 1 to 6). To normalize the transfections, a β-galactosidase gene was cotransfected in all cases. Luciferase activities were measured after cell stimulation with (COS-PRL-R Prolactin) or without (COS-PRL-R) prolactin. Values were normalized to the β-galactosidase activities and were the means of three independent transfection assays.
FIG. 8.
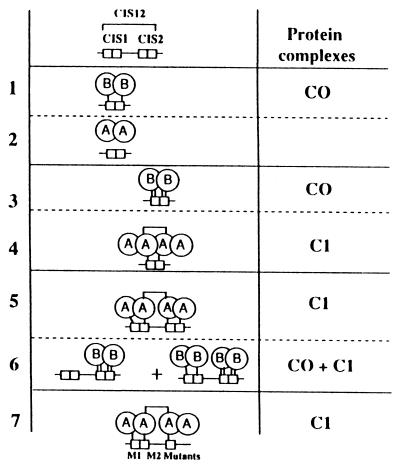
Schematic representation of the binding of STAT5A and STAT5B to the CIS1 and CIS2 proximal STAT sequences of the CIS gene promoter. This figure summarizes the abilities of STAT5A and STAT5B to bind to CIS1, CIS2, or CIS12. Each STAT sequence is composed of the palindromic sequence TTCNNNGAA and is represented by two fused boxes. The affinity of the DNA-protein interactions is represented by small vertical bars, and the level of this affinity is determined as follows: no bar, no interaction; one bar, weak interaction; two bars, strong interaction. The figure represents seven EMSA experiments (lanes 1 to 7) performed with nuclear extracts from COS-7 cells that were transfected with expression vectors for prolactin, STAT5A, or STAT5B and induced with prolactin. The left column represents the DNA complexes with their affinity-binding levels, and the right column indicates the corresponding complexes referenced in previous EMSA figures. STAT5A binding to CIS12 M1 and M2 mutants, in which the 3′ end of the CIS2 palindromic sequence was destroyed, is represented in lane 7. The differences in the DNA complexes observed in the presence of STAT5A and those observed with STAT5B and other information developed in the discussion led us to hypothesize an association between two STAT5A homodimers.
Similar studies were performed with the −292CIS-LUC construct, which contained only the two proximal STAT5 sites (CIS1 and CIS2): in both UT7 and COS-7 cell lines, the level of luciferase activity was reduced by about 50% in comparison with the activity of the −404CIS-LUC construct. We observed a basal activity without hormone stimulation and similar levels of STAT5 transactivation as observed with the −404CIS-LUC construct. Different mutants of this construct in which the two STAT boxes were mutated (separately or together) were also tested. However, no significant difference could be observed due to the low level of activity of these constructs (data not shown).
DISCUSSION
A specific combination of transcriptional factors plays an essential role in the activation of genes induced by cytokines. The study of the CIS gene promoter has revealed the binding of a specific association of ubiquitous and cytokine-induced transcription factors, among which we identified STAT5A and STAT5B in close association with ubiquitous GABPα and -β members (band C2) (data not shown) and an Epo-induced protein (band C3). These GGAA proteins and STAT5 bind to the same DNA region, and their binding sites overlap in the 5′ part of the CIS2 STAT box.
The promoter sequence at position −258 to −224 (CIS12), which contains two MGF boxes localized in the same DNA helix face (8 bp between each element), is the most interesting because this DNA fragment is able to discriminate between STAT5A and STAT5B binding. Indeed, in a reconstituted COS-7 model, we showed that the distal STAT consensus sequence (CIS1) bound with a low affinity to STAT5B homodimer (C0) but not to STAT5A (Fig. 8, lanes 1 and 2); in contrast, the second element (CIS2) bound to a STAT5B homodimer with good affinity (C0) and weakly to a STAT5A tetramer (C1) (Fig. 8, lanes 3 and 4). We show for the first time that a single STAT box can bind STAT5 only in a tetrameric structure. When EMSAs were performed with an oligonucleotide containing these two clustered STAT elements (CIS12 probe), we observed an increase in STAT5A binding (C1) revealing cooperative binding of two STAT5A dimers (Fig. 8, lane 5). This CIS12 probe bound with high affinity to STAT5A and to STAT5B, and specific shifts were observed with each STAT5 member: there was a low-mobility DNA complex (C1 band) for STAT5A, whereas the fast-migrating C0 protein complex contained STAT5B, demonstrating that STAT5B was in a dimer organization (Fig. 8, lane 6). The formation of C1 with STAT5A strongly suggested that only STAT5A tetramers bound to this probe with high affinity. Competition experiments as well as the use of recombinant STAT5 proteins favored this STAT5A tetramerization. Moreover, disruption of the C1 complex generated with recombinant STAT5A proteins by deoxycholate indicated a direct interaction between two dimers of STAT5A.
To understand how STAT5 interacted with the CIS12 DNA sequence, we determined the sequences required for the formation of the C1 complex with mutated DNA sequences. These studies indicated that the 3′ part of the proximal CIS2 STAT element was not necessary, while the CIS1 STAT box increased STAT5 binding on the CIS12 probe (Fig. 8, lane 7). In conclusion, two neighboring MGF boxes binding (CIS2) or not binding (CIS1) to STAT5A could constitute a high-affinity binding site for a tetramer of STAT5A. The different patterns of STAT5 shifts associated with CIS12, CIS1, or CIS2 probes, indicated in Fig. 8, could be explained by the stronger DNA binding affinity of STAT5B. Thus, the absence of binding of STAT5A to the CIS1 probe, which contains a consensus STAT binding motif, emphasizes the importance of the neighboring sequences of the STAT binding site (10, 14). Because we used an excess of probe, the faintness of the C1 complex observed with the CIS2 probe could result from a competition between STAT5 and Ets-related proteins for binding to the same sequence. STAT5A tetramer formation was strongly increased by the presence of the CIS1 sequence. Cooperative binding has been described for STAT4 and STAT1: Xu et al. (32) and Vinkemeier et al. (28) have shown that the amino-terminal domains of these STATs mediated STAT interaction and led to cooperative binding on clustered nonconsensus STAT boxes. More recently, Meyer et al. (22) described the binding of STAT5A dimers and tetramers on two adjacent STAT boxes of the IL-2Rα enhancer sequence; moreover, by proteolytic clipping of recombinant STAT5A proteins, they demonstrated that the N-terminal domain of STAT5A was required for tetramerization. Our own data indicate that STAT5B did not participate in tetramerization. Alignment of the 190 N-terminal amino acids of STAT5A and STAT5B showed only 17 differences. Creation of chimeric STAT5A-STAT5B proteins would be informative in localizing the amino acids crucial for STAT5A tetramerization.
In this paper, we demonstrate, for the first time, a functional difference in the binding properties of STAT5A and STAT5B. The cooperative binding of STAT5A, leading to a new molecular complex, could contribute to creating a functional diversity of STAT5 regulation: (i) the balance between the activated forms of STAT5A and STAT5B in the cell could modify the combination of the DNA binding complexes on target STAT5 genes, thereby explaining the modulation of cytokine-activated genes during cell differentiation or their tissue-specific expression; and (ii) a low-STAT5-affinity site near a half-STAT consensus site or a STAT sequence which does not bind STAT5 could allow cooperative STAT5 binding to these sites. Among published target STAT5 sequences, either a second STAT sequence or half a sequence (TTC or GAA) is frequently observed, as in the IL-2Rα enhancer (see above and the work of Meyer et al. [22]) and the hepatic serine protease inhibitor 2.1 promoter (2). A TTC motif close to the proximal MGF box of the βCAS promoter is conserved among species (9) and allows the formation of STAT5 tetramer (2).
Individual target disruptions of STAT5A and STAT5B have been realized (6, 18, 27). In EMSAs performed by Feldman et al. (6), disruption of STAT5A led to the disappearance of the C1 tetramer protein complex on the βCAS probe. This observation is in accordance with our own results and implicates STAT5A, but not STAT5B, in C1 complex generation. This change in protein-DNA binding pattern was correlated in vivo with the inhibition of CIS expression in STAT5A-null mice (6). However, our studies with the minimal −404CIS and −292CIS promoter region did not reflect endogenous CIS expression regulation. Indeed, promoter studies showed that the −404CIS or −292CIS promoter was active without STAT activation and that a modest transactivation by STAT5 could be observed (Fig. 7 and data not shown). Therefore, the decrease in CIS expression in STAT5A-null mice cannot be reconstituted in vitro by promoter studies, indicating that other domains could regulate the expression of the CIS gene in vivo: one of these could be a sequence containing four potential STAT boxes found 8 kb downstream of the CIS gene (data not shown). Studies of the CIS promoter and particularly of the CIS12 binding proteins have been worthwhile. However, functional promoter studies with −404CIS-LUC (Fig. 7), −292CIS-LUC, or mutants of these constructions (data not shown) transfected in different cell lines did not clearly demonstrate the functional role of STAT5 tetramerization mediated by STAT5A. Interestingly, however, functional studies of other promoters strongly transactivated by STAT5 have shown that disruption of one of the two STAT5 binding sites led to a decrease or an absence of their transactivation (2, 22). Therefore, these data and ours showing a distinct biochemical difference between STAT5A and STAT5B are in agreement in demonstrating a functional role for STAT5 tetramerization mediated by STAT5A.
ACKNOWLEDGMENTS
This work was supported by grants from the Agence Française du Sang (AFS contract no. 3FS08), from the Association pour la Recherche sur le Cancer (ARC contract no. 1373), and from the Ligue contre le Cancer. Frédérique Verdier is supported by Glaxo Wellcome Laboratories.
We acknowledge Xavier Tatare (ESTBA student) for excellent technical assistance. We are grateful to Emmanuel Gomas and Franck Letourneur for DNA sequencing. We thank M. Negishi for anti-GABPα and -β antibodies.
REFERENCES
- 1.Azam M, Lee C, Strehlow I, Schindler C. Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity. 1997;6:691–701. doi: 10.1016/s1074-7613(00)80445-8. [DOI] [PubMed] [Google Scholar]
- 2.Bergad P L, Shih H M, Towle H C, Schwarzenberg S J, Berry S A. Growth hormone induction of hepatic serine protease inhibitor 2.1 transcription is mediated by a Stat5-related factor binding synergistically to two gamma-activated sites. J Biol Chem. 1995;270:24903–24910. doi: 10.1074/jbc.270.42.24903. [DOI] [PubMed] [Google Scholar]
- 3.Bosselut R, Levin J, Adjadj E, Ghysdael J. A single amino-acid substitution in the Ets domain alters core DNA binding specificity of Ets1 to that of the related transcription factors Elf1 and E74. Nucleic Acids Res. 1993;21:5184–5191. doi: 10.1093/nar/21.22.5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 5.Doppler W, Groner B, Ball R K. Prolactin and glucocorticoid hormones synergistically induce expression of transfected rat beta-casein gene promoter constructs in a mammary epithelial cell line. Proc Natl Acad Sci USA. 1989;86:104–108. doi: 10.1073/pnas.86.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldman G M, Rosenthal L A, Liu X, Hayes M P, Wynshaw-Boris A, Leonard W J, Hennighausen L, Finbloom D S. STAT5A-deficient mice demonstrate a defect in granulocyte-macrophage colony-stimulating factor-induced proliferation and gene expression. Blood. 1997;90:1768–1776. [PubMed] [Google Scholar]
- 7.Gouilleux F, Pallard C, Dusanter-Fourt I, Wakao H, Haldosen L A, Norstedt G, Levy D, Groner B. Prolactin, growth hormone, erythropoietin and granulocyte-macrophage colony stimulating factor induce MGF-Stat5 DNA binding activity. EMBO J. 1995;14:2005–2013. doi: 10.1002/j.1460-2075.1995.tb07192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groner B, Gouilleux F. Prolactin-mediated gene activation in mammary epithelial cells. Curr Opin Genet Dev. 1995;5:587–594. doi: 10.1016/0959-437x(95)80027-1. [DOI] [PubMed] [Google Scholar]
- 10.Horvath C M, Wen Z, Darnell J E., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 11.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 12.Klingmuller U, Lorenz U, Cantley L C, Neel B C, Lodish H C. Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu N, Nakauchi H, Miwa A, Ishihara T, Eguchi M, Moroi M, Okada M, Sato Y, Wada H, Yamata Y, Suda T, Miura Y. Establishment and characterization of a human leukemic cell line with megakaryocytic features: dependency on granulocyte-macrophage colony stimulating factor, interleukin 3, or erythropoietin for growth and survival. Cancer Res. 1991;51:341–345. [PubMed] [Google Scholar]
- 14.Lamb P, Seidel H M, Haslam J, Milocco L, Kessler L V, Stein R B, Rosen J. STAT protein complexes activated by interferon-gamma and gp130 signaling molecules differ in their sequence preferences and transcriptional induction properties. Nucleic Acids Res. 1995;23:3283–3289. doi: 10.1093/nar/23.16.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lécine P, Algarté M, Rameil P, Beadling C, Bucher P, Nabholz M, Imbert J. Elf-1 and Stat5 bind to a critical element in a new enhancer of the human interleukin-2 receptor α gene. Mol Cell Biol. 1996;16:6829–6840. doi: 10.1128/mcb.16.12.6829. . (Author’s correction, 17:2351, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin J X, Mietz J, Modi W S, John S, Leonard W J. Cloning of human Stat5B. Reconstitution of interleukin-2-induced Stat5A and Stat5B DNA binding activity in COS-7 cells. J Biol Chem. 1996;271:10738–10744. [PubMed] [Google Scholar]
- 17.Liu X, Robinson G W, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Robinson G W, Wagner K U, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 19.Luo G, Yu-Lee L. Transcriptional inhibition by Stat5. Differential activities at growth-related versus differentiation-specific promoters. J Biol Chem. 1997;272:26841–26849. doi: 10.1074/jbc.272.43.26841. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- 21.Matsumura I, Ishikawa J, Nakajima K, Oritani K, Tomiyama Y, Miyagawa J, Kato T, Miyazaki H, Matsuzawa Y, Kanakura Y. Thrombopoietin-induced differentiation of a human megakaryoblastic leukemia cell line, CMK, involves transcriptional activation of p21 (WAF1/Cip1) by STAT5. Mol Cell Biol. 1997;17:2933–2943. doi: 10.1128/mcb.17.5.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer W K, Reichenbach P, Schindler U, Soldaini E, Nabholz M. Interaction of STAT5 dimers on two low affinity binding sites mediates interleukin 2 (IL-2) stimulation of IL-2 receptor alpha gene transcription. J Biol Chem. 1997;272:31821–31828. doi: 10.1074/jbc.272.50.31821. [DOI] [PubMed] [Google Scholar]
- 23.Moriggi R, Gouilleux-Gruart V, Jahne R, Berchtold S, Gartmann C, Liu X, Hennighausen L, Sotiropoulos A, Groner B, Gouilleux F. Deletion of the carboxyl-terminal transactivation domain of MGF-Stat5 results in sustained DNA binding and a dominant negative phenotype. Mol Cell Biol. 1996;16:5691–5700. doi: 10.1128/mcb.16.10.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mui A L, Wakao H, Kinoshita T, Kitamura T, Miyajima A. Suppression of interleukin-3-induced gene expression by a C-terminal truncated Stat5: role of Stat5 in proliferation. EMBO J. 1996;15:2425–2433. [PMC free article] [PubMed] [Google Scholar]
- 25.Mui A L, Wakao H, O’Farrell A M, Harada N, Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologs. EMBO J. 1995;14:1166–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellegrini S, Dusanter-Fourt I. The structure, regulation and function of the Janus kinases (JAKs) and the signal transducers and activators of transcription (STATs) Eur J Biochem. 1997;248:615–633. doi: 10.1111/j.1432-1033.1997.00615.x. [DOI] [PubMed] [Google Scholar]
- 27.Udy G B, Towers R P, Snell R G, Wilkins R J, Park S H, Ram P A, Waxman D J, Davey H W. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinkemeier U, Cohen S L, Moarefi I, Chait B T, Kuriyan J, Darnell J E., Jr DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 29.Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1994;13:2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle J N. Naturally occurring dominant negative variants of Stat5. Mol Cell Biol. 1996;16:6141–6148. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood T J, Sliva D, Lobie P E, Gouilleux F, Mui A L, Groner B, Norstedt G, Haldosen L A. Specificity of transcription enhancement via the STAT responsive element in the serine protease inhibitor 2.1 promoter. Mol Cell Endocrinol. 1997;130:69–81. doi: 10.1016/s0303-7207(97)00075-0. [DOI] [PubMed] [Google Scholar]
- 32.Xu X, Sun Y L, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 33.Yokomori N, Kobayashi R, Moore R, Sueyoshi T, Negishi M. A DNA methylation site in the male-specific P450 (Cyp 2d-9) promoter and binding of the heteromeric transcription factor GABP. Mol Cell Biol. 1995;15:5355–5362. doi: 10.1128/mcb.15.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshimura A, Ichihara M, Kinjyo I, Moriyama M, Copeland N G, Gilbert D J, Jenkins N A, Hara T, Miyajima A. Mouse oncostatin M: an immediate early gene induced by multiple cytokines through the JAK-STAT5 pathway. EMBO J. 1996;15:1055–1063. [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshimura A, Ohkubo T, Kigushi T, Jenkins N A, Gilbert D J, Copeland N G, Hara T, Miyajima A. A novel cytokine-inducible gene CIS encodes an SH2-containing protein that binds to tyrosine-phosphorylated interleukin 3 and erythropoietin receptors. EMBO J. 1995;14:2816–2826. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]