Abstract
Recently, we identified and classified 926 human endogenous retrovirus H (HERV-H)-like proviruses in the human genome. In this paper, we used the information to, in silico, reconstruct a putative ancestral HERV-H. A calculated consensus sequence was nearly open in all genes. A few manual adjustments resulted in a putative 9-kb HERV-H provirus with open reading frames (ORFs) in gag, pro, pol, and env. Long terminal repeats (LTRs) differed by 1.1%, indicating proximity to an integration event. The gag ORF was extended upstream of the normal myristylation start site. There was a long leader (including a “pre-gag” ORF) region positioned like the N terminus of murine leukemia virus (MLV) “glyco-Gag,” potentially encoding a proline- and serine-rich domain remotely similar to MLV pp12. Another ORF, starting inside the 5′ LTR, had no obvious similarity to known protein domains. Unlike other hitherto described gammaretroviruses, the reconstructed Gag had two zinc finger motifs. Alternative splicing of sequences related to the HERV-H consensus was confirmed using dbEST data. env transcripts were most prevalent in colon tumors, but also in normal testis. We found no evidence for full length env transcripts in the dbEST. HERV-H had a markedly skewed nucleotide composition, disfavoring guanine and favoring cytidine. We conclude that the HERV-H consensus shared a gene arrangement common to gammaretroviruses with gag separated by stop codon from pro-pol in the same reading frame, while env resides in another reading frame. There was also alternative splicing. HERV-H consensus yielded new insights in gammaretroviral evolution and will be useful as a model in studies on expression and function.
About 7% of the human genome has been estimated to consist of retroviruses or retroviral elements (6). Human endogenous retroviruses (HERVs) are currently named after their primer binding sites' (PBS) similarity to one of the human tRNAs. One of the most abundant proviral groups is HERV-H, which uses His-tRNA. It belongs to the genus Gammaretrovirus, previously called the mammalian type C retroviruses. It is represented in about 1,000 copies (62), recently narrowed somewhat to 926 copies (24). It entered the genomes of higher primates about 30 to 35 million years ago, at the time of, but mainly after, the split between old and new world monkeys (1, 35).
The proviral structure consists mainly of 5′ LTR-gag-pro-pol-env-3′ LTR, where the long terminal repeats (LTRs) are identical at the integration event. They are built from untranslated 3′ and 5′ (U3 and U5) sequences separated by a repeat segment (R). The group-specific antigen (Gag) includes the matrix (MA), capsid (CA), and nucleocapsid (NC) proteins. The protease gene (pro) is located between the gag and the polymerase gene (pol), which contains reverse transcriptase (RT), RNaseH, and integrase (IN) domains. The envelope gene (env) consists of the surface unit (SU), with a signal peptide (SP) located at the 5′ end and the downstream transmembrane unit (TM). The PBS is situated between the 5′ LTR and gag, while the polypurine tract (PPT) is located between env and the 3′ LTR. Most HERVs hold defective genomes, but some HERVs have the potential to produce proteins and thus have possible physiological functions. However, HERV proteins have not yet been identified using definite methods like N-terminal sequencing or mass spectrometry. The HERV-W envelope protein (Env), identical to the human protein Syncytin, has the capacity to fuse cytotrophoblast to syncytiotrophoblast in vitro and may thus be important in human placental morphogenesis during pregnancy (40). The TM proteins of many, but not all, gammaretroviruses reportedly have an immunosuppressive unit (ISU) consisting of 17 conserved amino acids (aa) (CKS-17) (for a review, see reference 14). Although the exact mechanism still is not known, gammaretroviral TMs have been shown to promote evasion from anti-tumor cytotoxicity (38, 39). Proviral sequences that do not encode proteins could also have cellular effects if the LTRs serve as promoters for gene expression. Amylase production in human parotid glands became possible after an integration of an HERV-E next to the amylase gene (48, 58). HERVs have also been implicated in several diseases, such as multiple sclerosis (12, 44) and schizophrenia (26), and in different cancers (7, 50, 51). However, this is still speculative. It is of importance to map these proviral structures in order to understand their functions. To study the sequence variability among HERV-H proviruses, we collected a number of representative full-length HERV-H sequences, aligned them, and constructed a consensus HERV-H, likely to be more similar to an “ancestral HERV-H sequence.” Although there may have been several integration events and ancestral HERV-H variants, phylogenetic analysis of the HERV-H group (24) and the computer constructed near open reading frame (ORF) consensus presented here is compatible with the hypothesis of a common ancestral HERV-H. We analyzed the genes and the LTRs in detail with respect to sequence length, position in the viral genome, detectable conserved consensus motifs, and splicing patterns using bioinformatic tools and database searches. Features in the HERV-H consensus were then used in phylogenetic analyses.
MATERIALS AND METHODS
Data collection and processing of the likely HERV-H.
A set of full-length HERV-H sequences were retrieved from GenBank via a BLAT search (http://genome.ucsc.edu/cgi-bin/hgBlat, November 2002 assembly corresponding to human genome build 31 at National Center for Biotechnology Information) using the sequence of HERV-H19, BAC clone AC009495 (positions 9526 through 883) and by a newly developed program, RetroTector (G. O. Sperber and J. Blomberg, unpublished data). Briefly, RetroTector recognizes consensus motifs (Table 1) and constructs putative HERV proteins (“puteins”) from the different reading frames in the gene candidates. The program uses codon statistics, frequency of stop codons, and alignments to known Gammaretrovirus proteins to approximate the original ORF. In this study, RetroTector used the human genome build 29. The criteria for sequence detection by RetroTector was detection of all or some parts of the proviral HERV-H (5′LTR-gag-pro-pol-env-3′LTR). In the BLAT search, sequences with a score (determined by the number of matches versus mismatches in the final alignment of the query to the genome) higher than 4,000 were used. The limit chosen for the study proved to yield hits with almost full-length HERV-H19-like proviruses. BLAT generated HERV-H19-like sequences and RetroTector recognized a more general HERV-H-like sequence when searching the different reading frames of the integration. The nine sequences retrieved by RetroTector and five sequences retrieved by BLAT search were complemented by the previously described RGH2 (D11078), HERV-H/env62 (AJ289709), HERV-H/env60 (AJ289710), and HERV-H/env59 (AJ289711) collected from GenBank. The total of sequences were aligned using ClustalW (version 1.4 in BioEdit using default settings) (57) and edited using BioEdit version 4.8.6 (21). A consensus sequence was generated in the BioEdit program. Already at this stage, the consensus sequence had few stops and shifts in gag, pro, and pol, and the alignment was thus, to a large extent, codon guided (see supplemental data at http://www.kvir.uu.se/supplementary_info/supplementary.html). We finished the construction of a likely HERV-H consensus sequence manually, where stops and frameshifts were adjusted using information from integrations with almost an open reading frame for a gene (e.g., gag, pro, and pol). Nucleotide insertions, one or several nucleotides, present in a single or very few sequences in the alignment, were excluded in order to produce a more likely consensus for a putative original HERV-H. Approximately 1,180 nucleotides (nt), inserted in lengths or as single nucleotides shown by the alignment, were deleted from the sequences in the alignment. Insertions of up to 261 nt were encountered (see supplemental data). These probably represent secondarily inserted repetitive elements or sequences inserted by recombination during reverse transcription. Two separate segments (307 and 165 nucleotides) and additional short segments (one or few nucleotides) that added up to a total of 610 were kept and adjusted manually to fit the sequence prediction of a putative original HERV-H. The consensus construction process can be followed in the supplemental data.
TABLE 1.
Consensus motifs recognized by RetroTectora
| Motif | RetroTector
|
Manually adjusted HERV-H
|
||
|---|---|---|---|---|
| Type | Example | Sequence | Sequence | |
| PBS | C | tRNAHis-RTVLH, ERVfrd | TGGtgccgtgactcggat | tggtgctgtgactcagat |
| MA1 | C | mgnlpp | ||
| CA0 | ABCDELSGO | CA Start NN | kgivkvnapfslsdlsqisqrlgsfssdpt | |
| CA1 | C | BAEV | rtTQGkDESPAaFMERLIEGF | kettqgkdknpaqfmarlaatl |
| CA2 | C | MuLV | QsAPdiGr | qsapdikklqk |
| NC1 | C | S71 | CtyCkqiGHwkkEC | cykcqksghwakec |
| NC2 | B | MMTV | CprCkkgyHwksEC | cpicagphwksdc |
| PR2 | C | MuLV, motif A | lvDTGAqhSv | linteathst |
| PR3 | C | MuLV, motif B | llGRdllt | llgrdiltkls |
| RT1 | C | MLV | wNtPllpVKK | ynspilpvqk |
| RT2 | C | HERVH | svlHLkDaFFtiPL | svldlkhafftipl |
| RT3 | C | MLV | qltWtrLPQGfknSP | qitwavlpqgftdsp |
| RT4 | C | MLV | lqYvDDLlla | iqyiddlllc |
| RT5 | C | MLV | GyrasakKaQ | gyrvspskaq |
| IN2 | B | IAPha | efHkrfHvt | sfhnlfhvg |
| IN3 | B | MPMV | ivkqCpiCvty | itsqcsicyst |
| IN4 | C | MuLV, motif C | hWeidfte | dwqidfth |
| IN5 | C | BAEV | gSDNGPafvSQv | qsdngpaftsqi |
| IN6 | C | HERV-E | AYqPQSsgKVERmnrt | pyhpqssgkvertngl |
| IN7 | C | BAEV | eprWkGPyiVLlttpt | qprwtgpytviystpt |
| SU3 | C | HERVH RGH2 | KRviplitlmvglgl | krvipliplmvglgl |
| TM3 | C | HERV H/ERV9 | IQNhRGLDILTAekGGLClfLE | lqnrrgldlltaekgglcifln |
| TM5 | ABCDELSGO | hydrophobic motif | fllllfgpcifr | |
| PPT | ABCDELSGO | PPT motif | aagaaggcaggaa | |
Amono acids represented by uppercase letters are more conserved and higher scoring than those represented by lowercase letters. Predicted type abbreviations: A, alpha; B, beta; C, gamma; D, delta; E, epsilon; L, lenti; S, spuma-like; G, gypsy; O, copia.
HERV-H consensus analysis.
The HERV-H consensus was analyzed for a broad range of conserved motifs within the different genes by using RetroTector (see above). Transcription factor (TF) binding sites in the LTRs were predicted using the ConSite program made available online at http://mordor.cgb.ki.se/cgi-bin/CONSITE/consite. The LTRs from sequences used to produce the HERV-H consensus were extracted from the original, unedited alignment using RGH2 (D11078) as a template for LTR lengths based on their sequence similarities. TF binding sites were analyzed using the setting for vertebrates, without minimum bit specificity and 85% TF score cutoff. Analysis of nucleic acid folding was conducted using the Mfold web server (http://www.bioinfo.rpi.edu/applications/mfold/) (65). Further, the sequence for the putative HERV-H provirus was subjected to various motif predictions using tools made available online at ExPASy (http://www.expasy.org/).
Phylogenetic analyses of the HERV-H-like sequences used in alignment were conducted in ClustalX (version 1.83) (56) with default settings. An unrooted neighbor-joining (NJ) tree for the HERV-H sequences was produced using the manually adjusted alignment from BioEdit (see above). Bootstrappings of the NJ trees were conducted in 1,000 replications using ClustalX (1.83) and visualized in TreeView (version 1.6.6) (42).
Splice predictions in the putative HERV-H proviral sequence were conducted using NetGene2 at http://www.cbs.dtu.dk/services/NetGene2/ (9) and the NNSPLICE 0.9 at http://www.fruitfly.org/seq_tools/splice.html. The predicted splice donor (SD) and splice acceptor (SA) sites were analyzed in a Gene2EST search (http://woody.embl-heidelberg.de/gene2est/) (20) (with the RepeatMasker disabled) using the putative HERV-H consensus sequence that ranged over the R region of the 5′LTR and over the U3 region in the 3′LTR.
HERV-H env (chromosome 2q24.3, positions 642 through 2396 in AF108843) was used in screening the expressed sequence tag (EST) database at GenBank (dbEST release 061402 containing 4,458,530 sequences) for expression of the entire env gene. A new search in the same database using HERV-H env TM protein (AF108843, positions 1857 through 2396) was performed to decide whether the SU, TM, or both were expressed. Criterium for inclusion was at least 80% sequence identity over at least 100 nucleotides.
Laboratory procedures.
Polyclonal antisera against HERV-H SU (peptide 1437, PPEELIYFLDRSSKTSPDIS, preimmunization serum rabbit 430 and serum VU800) were raised in rabbits. The tetrameric multiantigenic peptide (based on a backbone of β-alanine and two lysines) was purified by reverse-phase high performance liquid chromatography (HPLC) on a C18 column to >95% purity and characterized using mass spectrometry.
Human placenta from an anonymous donor was acquired fresh from the Uppsala Academic Hospital (with ethical committee approval). Samples were homogenized in a denaturing buffer (8 M urea, 50 mM phosphate, 50 mM β-mercaptoethanol, 1% [vol/vol] Tween 20, pH 6.0) using ultra-Turrax 18 (IKA Works Inc. Wilmington, NC). The extract was centrifuged at 4°C and 16,000 ×g for 4 min, and the supernatant was collected. The placenta homogenate supernatant was subjected to cation exchange chromatography (loading buffer, pH 6.0, 8 M urea-50 mM phosphate-50 mM β-mercaptoethanol-0.1% [vol/vol] Tween 20; eluent, pH 6.0, 8 M urea-50 mM phosphate-50 mM β-mercaptoethanol-0.1% [vol/vol] Tween 20-1 M NaCl; column, HiPrep 16/10 SP FF [Amersham Pharmacia Biotech, Uppsala, Sweden]), run with 0 M to 1 M NaCl in 40 min), using a Shimadzu LC8 HPLC.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blots on the protein fractions were run on a Phast System (Pharmacia, Uppsala, Sweden) using dedicated precast gradient gels (10 to 15% acrylamide) and blotted onto polyvinylidene difluoride membranes. Blocking was conducted using 0.20 (vol/vol) Tween 20. Alkaline phosphatase-conjugated goat α-rabbit (Sigma, A3812) antibody (Ab) was used as secondary Ab to detect the primary rabbit α-HERV-H SU and TM Abs.
RESULTS
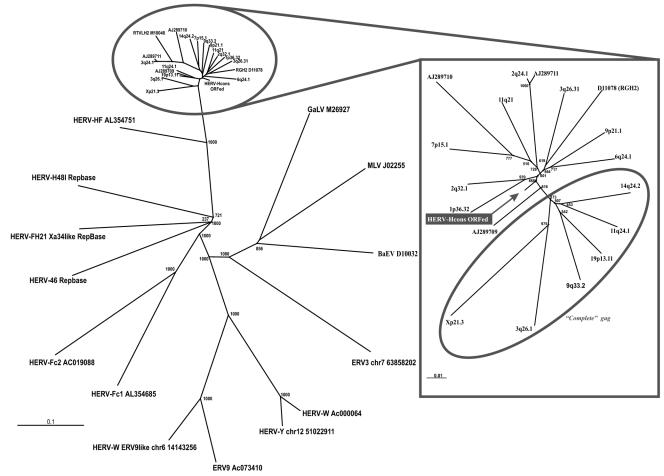
In order to construct and analyze a potential sequence more likely to represent an “original” HERV-H, we collected a representative set of 18 full-length HERV-H provirus sequences (Table 2). Different approaches were used, including the newly developed program RetroTector, a BLAT search and retrieval of previously described sequences in GenBank. The 18 collected sequences were widely dispersed among the HERV-H, based on alignments and the phylogenetic analysis of 1,124 HERV-H-like sequences retrieved with RetroTector (24). Further, the selected HERV-H sequences were separate from adjacent HERV-H-like sequences (24) and other gammaretroviral elements serving as outgroups (Fig. 1). Alignment of the 18 HERV-H sequences with a full set of genes (5′ LTR-gag-pro-pol-env-3′ LTR) gave a primary consensus sequence, where most stops and frameshifts were eliminated (gag, 0 stops/6 shifts; pro, 1/3; pol, 0/13; env, 0/8). Minor manual adjustments of the primary consensus sequence, described in Materials and Methods, yielded ORFs for all proviral genes residing in the first and second reading frames in the HERV-H consensus (Fig. 2). This sequence will be referred to as the “HERV-H consensus” (Fig. 3 and supplemental data). Characteristic motifs (Table 1) for each proviral gene were recognized by RetroTector and were present in frame. Further analyses included in silico identification of probable SD and SA by ESTs and mapping of motifs of special interest in the ORFs of the putative HERV-H consensus. When the nucleotide frequencies of 3,661 pol-containing retroviral chains found by RetroTector version 010 in the human genome version hg15 were analyzed, a markedly skewed distribution was observed in the 926 HERV-H like, and most of the adjacent HERV-H like elements, described recently by us (24). They had over 29% C and below 17% G. The data were compared to nucleotide frequencies of other retroviruses (Table 3). No other retroviral element detected by RetroTector in the human genome had this distribution. Positions described here are relative to the start of the HERV-H consensus provirus. To convert these positions relative to the conventional retroviral start position in the 5′ R region (15), U3 (348 nucleotides) have to be subtracted from the positions presented.
TABLE 2.
Eighteen full-length HERV-H provirus sequences
| Sequence position (synonymous name) | Retrieved by: |
|---|---|
| 1p36.32 | BLAT |
| 2q24.1 | BLAT |
| 3q26.31 | BLAT |
| 7p15.1 | BLAT |
| 14q24.2 | BLAT |
| AJ289709 (HERVH/env62, H19, 2q24.3) | GenBank |
| AJ289710 (HERVH/env60, 3q26.1) | GenBank |
| AJ289711 (HERVH/env59, 2q24.1) | GenBank |
| D11078 (RGH2) | GenBank |
| 2q32.1 | RetroTector |
| 3q26.1 | RetroTector |
| 6q24.1 | RetroTector |
| 9p21.1 | RetroTector |
| 9q33.2 | RetroTector |
| 11q21 | RetroTector |
| 11q24.1 | RetroTector |
| 19p13.11 | RetroTector |
| Xp21.3 | RetroTector |
FIG. 1.
Left panel: NJ tree (1,000 bootstraps) of HERV-H-like and reference gammaretrovirus pol sequences. Right panel: outlined NJ tree (1,000 bootstraps) of full-length HERV-H-like elements used in HERV-H consensus construct and the HERV-H consensus which positioned close to the center. Encircled sequences have a 344-nt segment in gag covering the 5′ end of the conserved CA1 consensus motif, which is not present in the other sequences. Sequence comparison for the dendrogram was conducted by neighbor-joining with the Kimura two-parameter model and 500 bootstraps. Sequence comparisons were independent of the different gag lengths through pairwise deletions in the analysis.
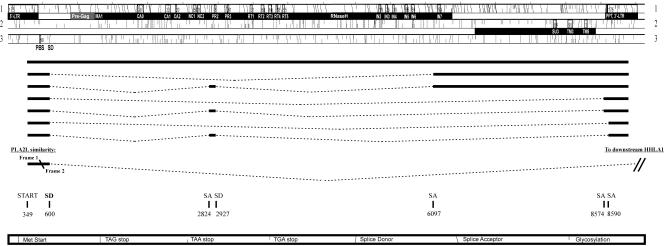
FIG. 2.
Three reading frames and alternative splicing in the HERV-H consensus sequence as interpreted by RetroTector (Sperber and Blomberg, unpublished) and by EST searches. Proviral genes with outlined names of conserved consensus motifs are presented as black bars below each reading frame. Alternative splice patterns are outlined with their corresponding SD and SA sites.
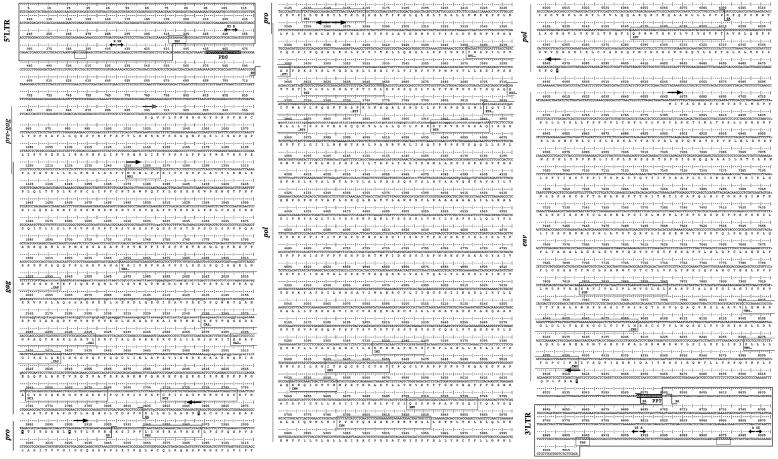
FIG. 3.
The HERV-H consensus sequence likely to represent an “original” provirus. The proviral genes are noted to the left of each panel and with arrows over the nucleotide sequence. Amino acid sequences are presented under the respective gene, and their motifs are presented within boxes. SD and SA are presented over the nucleotide sequence (compare Fig. 2).
TABLE 3.
Nucleotide frequencies of other viruses
| Genus (virus) or DNA type | A | T | G | C |
|---|---|---|---|---|
| Gamma | ||||
| HERV-H consensus | 24 | 28 | 15 | 33 |
| HERV-H like: chr 19_20052799 | 24 | 28 | 15 | 33 |
| HERV-H like: chr11_95227089 | 24 | 28 | 15 | 32 |
| HERV-H like: chr11_122451278 | 24 | 28 | 15 | 32 |
| HERV-H like: chr3_31955536 | 25 | 29 | 15 | 31 |
| HERV-H like: chr5_130938433 | 24 | 28 | 17 | 31 |
| FeLV NC_001940 | 28 | 22 | 23 | 28 |
| MLV NC_001501 | 26 | 21 | 24 | 29 |
| Beta | ||||
| MMTV M15122 | 30 | 26 | 23 | 21 |
| HERV-K(HML2.11_101600013) | 32 | 26 | 21 | 20 |
| SRV-1 M11841 | 31 | 27 | 19 | 24 |
| Delta | ||||
| HTLV-1 Seiki J020209 | 23 | 23 | 19 | 35 |
| BLV NC_0010414 | 22 | 24 | 21 | 33 |
| Lenti | ||||
| HIV-1 HIVMNCG | 36 | 22 | 24 | 18 |
| SIVsykes AY523867 | 35 | 22 | 24 | 19 |
| SIVcpz AF115393 | 35 | 22 | 24 | 18 |
| FIV NC_001482 | 38 | 25 | 22 | 14 |
| EIAV NC_001450 | 36 | 26 | 22 | 16 |
| CAEV NC_001463 | 38 | 21 | 25 | 16 |
| Visna NC_001452 | 37 | 21 | 26 | 15 |
| Spuma like | ||||
| Human Foamy virus NC_001736 | 33 | 29 | 20 | 18 |
| HERV-L chr7_121389860 | 28 | 28 | 23 | 21 |
| HERV-S chr4_78951104 | 25 | 24 | 27 | 24 |
| Randomly Selected Human DNA | ||||
| Human BAC clone AL008733 | 22 | 21 | 28 | 29 |
| Human BAC clone AL023586 | 26 | 26 | 24 | 24 |
| Human BAC clone AL022101 | 28 | 27 | 23 | 22 |
| Selected groups (no. of elements)a | ||||
| Gamma | ||||
| HERV-H (926) | 25.3/1.5 | 28.6/1.1 | 15.5/1.3 | 30.7/2.3 |
| Adjacent HERV-H like (106) | 26.6/2.5 | 27.2/2.2 | 17.1/2.1 | 29.1/3.9 |
| ERV9 and HERV-W like (383) | 29.9/1.8 | 25.0/1.9 | 21.5/1.5 | 23.7/1.7 |
| ERV3 and HERV-E like (145) | 29.1/1.7 | 24.2/1.6 | 23.7/1.7 | 23.0/1.1 |
| Beta: HML 1-10 (672) | 29.8/3.7 | 29.8/4.2 | 20.1/1.6 | 20.3/1.8 |
| Spuma like: HERV-L groups (336) | 28.6/1.7 | 27.9/1.9 | 22.7/1.6 | 20.9/1.3 |
Quantities for nucleotides are given as averages/standard deviations.
LTRs.
The LTRs (Fig. 3) had the characteristic start (TG…) and stop (…CA) described earlier (52). The 5′LTR (1 through 438) and 3′LTR (8585 through 9021) consisted of U3 (1 through 348; 8585 through 8932), R (349 through 409; 8933 through 8992) and U5 (410 through 438; 8993 through 9021). This was based on RetroTector© prediction and EST analysis (Fig. 2). Numerous potential transcription factor binding sites were detected in the LTRs. Progesterone receptor (PR) binding sites were predicted in U3, at positions 194 through 202 and 8778 through 8785. Other sites found in U3 were the TATA-box, TATAAAA (317 through 323; 8901 through 8907), target for the TATA-binding protein (TBP, belonging to the core of transcription factors) (15) and GATA-1 zinc finger transcription factor binding sites (342 through 335; 8926 through 8919). Immediately 3′ of the TATA-box, was a GC/GT box (TATAAAACGGCCCCACCC) required for promoter activity by the DNA bending zinc finger transcription factor SP1 (54). Three SP1 binding sites were predicted (positions 105 through 114, 273 through 282, and 368 through 377). We found four myb binding sites (consensus, AAC[T/G]G) in HERV-H consensus U3, which was consistent with earlier experimental results (16), and located them to positions 76 through 80 (8660 through 8664), 120 through 124 (8704 through 8708), 171 through 175 (8755 through 8759), and 322 through 326 (8906 through 8910) in the HERV-H consensus 5′ LTR (and 3′ LTR). The polyadenylation signal, AATAAA, was found in the R region in the 3′ LTR at positions 8972 through 8977 (and corresponding nucleotide positions 389 through 394 in the 5′ LTR). Among the 18 selected HERV-H-like sequences, there were both “type I” and “type II” LTRs according to the definition of Anderssen et al. (1). We found that 14 among 18 LTR pairs in our data set grouped together with “type I” and subsequently, HERV-H consensus also grouped within “type I” (data not shown). Type I and II LTRs had different tissue specificities in transient transfection experiments (1). The 5′ and 3′ LTR difference in the HERV-H consensus was 1.1%, indicating proximity to a likely ancestral HERV-H provirus, but also representing a certain ambiguity in the consensus.
The LTRs of the 18 HERV-H-like sequences were scanned for potential TF binding sites with the help of the ConSite program (http://mordor.cgb.ki.se/cgi-bin/CONSITE/consite). The search showed numerous potential TF binding sites at an 85% identity cutoff, but no clear conservation, probably due to the poor analysis specificity and the several loci studied. A more detailed analysis of these binding sites was out of scope for this study.
PBS and vicinity.
The PBS (456 through 473) (Fig. 3) had a typical sequence (TGGTGCCGTGACTCGGAT) complementary to His-tRNA. Variation in PBS amounted to 21 substitutions at eight sites (underlined in the HERV-H consensus PBS above) among the 18 collected HERV-H sequences when compared to the HERV-H consensus (supplemental data). According to the manually adjusted alignment and the RetroTector prediction, an insertion (compared to a normal gammaretroviral sequence [15]) was observed just 5′ of gag in the consensus sequence and was present in all of the 18 collected HERV-H-like elements. As shown below, the predicted HERV-H Gag contained hallmarks which identified its extent, from a typical Gag start to the zinc finger motifs in NC, with reasonable certainty. We are therefore confident that this region, an unusually long 5′ leader sequence with an ORF here referred to as “pre-gag” which precedes the traditional gag, is separate from it and is discussed below. The 5′ leader also had an ORF that started within the 5′ LTR region in frame 3 (nucleotides 294 through 1019). There was no known or predicted protein with this sequence in GenBank. However, in a nucleotide BLAST of the nonredundant database of GenBank, the region was found to share similarity with the HERV-H part of an intergenic splice transcript of PLA2L (PLA2 like, accession no. Z14310) that initiates in HERV-H 5′ LTR and splices to genes downstream of the provirus (at chromosome 8q24.1-3) to form a fusion transcript together with the two normally independently expressed HHLA1 (HERV-H LTR associated gene) and Otoconin-90 (18, 30, 60). The similarity was found in frame 1 of the HERV-H consensus ranging over the nucleotides 448 through 504 and continued in frame 2 for nucleotides 506 through 598 (Fig. 2). The intergenic splice transcript most probably uses the major splice donor at corresponding nucleotide position 600 in the HERV-H consensus. This position had a high likelihood score in the NetGene2 and the NNSPLICE 0.9 splice prediction programs (Fig. 2). It has also been found in major HERV-H transcripts (33).
As mentioned above, the 5′ leader region also contained an ORF, “pre-gag” in frame 1, that started at nucleotide 904 and continued into the gag ORF (frame 1 with start at nucleotide 1255) (Fig. 3). This 117-amino acid N-terminal proline- and serine-rich “elongation” of the predicted Gag had three repeated leucine zipper motifs (consensus, L-x(6)-L-x(6)-L-x(6)-L, ranging over the nucleotides 1060 through 1125, 1081 through 1146, and 1102 through 1167) (data not shown). The proline content was 21/117 amino acids in the HERV-H consensus and 18/84 in MLV pp12, whereas the serine content was 19/117 in HERV-H and 8/84 in MLV pp12. Both prolines and serines were repeated in heptads (supplemental data). A BLAST search in GenBank with this sequence did not yield any known or predicted protein, nor were there logically positioned splice donor/acceptor sites or ESTs to indicate a separate “pre-gag” transcript, which would encode such a protein. The HERV-H 5′ leader region was analyzed with respect to secondary structure using the Mfold web server (65) and compared for similarities with the MLV (NC_001501). MLV has earlier been shown to utilize a mechanism involving an internal ribosomal entry site (IRES) situated in the 5′ leader region (4). Although multiple hairpins were predicted in HERV-H “pre-gag,” we could not demonstrate apparent similarities in folding structure and possible IRES formation.
gag.
The gag (nucleotides 1255 through 2850, frame 1) (Fig. 3) was recognized by six motifs with corresponding nucleotide positions in the HERV-H consensus: MA (MA1, 1255 through 1272), CA (CA0, 1852 through 1941; CA1, 2254 through 2319; and CA2, 2389 through 2421), and NC (NC1, 2644 through 2685, and NC2, 2713 through 2751). As mentioned, the N-terminal sequence of Gag MA was predicted to start with “MGNL…” where the G should be a myristylation site, needed for virion assembly (22, 47). The assigned start methionine conforms to the Kozak consensus motif for translational start (32). In the MA-CA region, a PPPY motif, which in several retroviruses encodes a so-called “late” function (63), occurred at nucleotide positions 1729 through 1740, 159 amino acids from the N-terminal of Gag. This “late” motif has been shown to interact with the Nedd4 family of ubiquitine ligases and is required for the budding process in Rous sarcoma virus (RSV p2b) (28). MLV pp12 (NP_955584), which contains the MLV “late” motif, showed a weak similarity to the 5′ extended HERV-H “pre-Gag” in an alignment (supplemental data). The 18 aligned HERV-H sequences could also be divided into two groups on the basis of Gag structure, where one group missed a segment corresponding to the nucleotide positions 1933 through 2277 in the consensus sequence (Fig. 3; see alignment in supplemental data) and thus partly in the 5′ end of the major homology region (MHR) (see below), in the CA1 consensus motif. Interestingly, the sequences with “complete” gag, which lacked the 344-nucleotide deletion, grouped together in a full-length provirus unrooted dendrogram (Fig. 1). Remaining after the deletion was an almost intact C-terminal domain (CTD) of CA. The N-terminal domain (NTD) of the related gammaretroviral MLV CA (also largely similar to human immunodeficiency virus [HIV], Rous sarcoma virus [RSV], and human T-cell leukemia virus [HTLV]) was recently shown by X-ray structure to be involved in Gag assembly (41). Among the 926 HERV-H pol-containing elements in the genome, the HERV-H consensus pol grouped within the RGH2-like subgroup (see supplemental data and Jern et al. [24]). The full HERV-H-like sequence dendrogram was independent of the 344-nucleotide deletion, since pairwise deletions were used in the analysis. Twenty amino acids in the conserved MHR (…TTQGKDKNPAQFMARLAATL…), compared to the consensus of Benit et al. (2) in Fig. 4, were located in the CA 336 amino acids from the N-terminal of Gag and started approximately at nucleotide position 2260 in the HERV-H consensus. The start of the MHR was missing in 12 of the selected HERV-H sequences, since it was located in the 344-nucleotide deletion described above (Fig. 4). The HERV-H elements in hg16 at chromosome 19 position 20068590 and chromosome 10 position 104629785 had the most complete gag genes, each with just 1 shift and 1 stop (supplemental data). In principle, truncated Gag proteins could be expressed from chromosome 19 at position 20068590, chromosome 10 at position 104629785, chromosome 7 at position 106030611, and chromosome 16 at position 9738198.
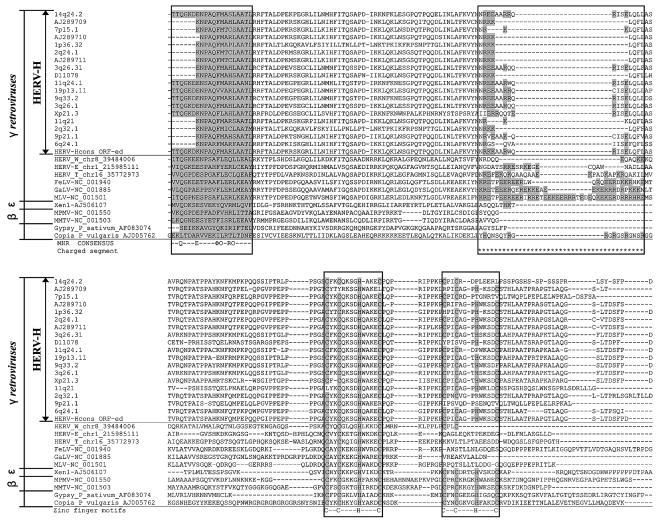
FIG. 4.
Gag alignment ranging from the MHR to the zinc finger motifs for HERV-H consensus and other gamma-HERV-W, -E, -T, -GALV, -MLV, and -FeLV together with two beta-MMTV and -MPMV, one epsilonretrovirus (Xen1), and the more distant retrotransposons (Gypsy and Copia). MHR consensus (Φ, aromatic; O, hydrophobic amino acids; invariants are Q, E, and R) was adapted from Benit et al. (2). The HERV-H sequences share the properties of double zinc finger motifs missing in the other gammaretroviruses that in contrast display an increased density of charged residues in the charged segment.
A highly conserved zinc finger (cys-his motif C-X2-C-X4-H-X4-C) described by Chance et al. (10), was found in NC at corresponding nucleotide positions 2644 through 2685 (…CYKCQKSGHWAKEC…) and another relatively conserved motif probably representing a second zinc finger with an unusual structure, C-X2-C-X3-H-X4-C, ranging from nucleotide positions 2713 through 2751 (…CPICAGPHWKSDC…). An alignment with known gamma- and betaretrovirus Gag proteins indicated similarities both in structure and in length. The second zinc finger could be detected in betaretretrovirus Gag of mouse mammary tumor virus (MMTV) and Mason-Pfizer monkey virus (MPMV), in epsilonretrovirus Gag of Xenopus (Xen1) and in gypsy and copia (delta- and lenti not shown). The other gammaretroviruses MLV, FeLV, GaLV, HERV-E, -W, and -T did not have two zinc finger motifs (Fig. 4).
pro.
The pro (start at nt 2911 extending to approximately nucleotide 3267, frame 1) (Fig. 3) contained two detectable conserved consensus motifs (PR2: 2944 through 2973 and PR3: 3133 through 3165), located in the same reading frame but separated from gag by several stop codons (the first was more conserved than the following two) (see alignment in supplemental data). Thus, an ancestral HERV-H probably had one stop, which is normal for gammaretroviruses (15), despite this data set. The pro structure was similar to that of other gammaretroviral proteases (data not shown). The HERV-H consensus had a noncanonical NTE motif instead of the normal DTG or DSG in the active site (Fig. 3, PR2; see alignment in supplemental data). Consequently, it was present in a majority of the HERV-H Pro puteins (data not shown). However, a small subset of HERV-H had DTG. Thus, HERV-H may have gone through an evolution in this motif. An analysis of betaretroviral Pro puteins resulted in DTD, DTG, and DSG variants (data not shown).
As expected, pro was not separated by stops and frameshifts from the downstream pol. This arrangement of gag and pro-pol ORFs has been described as typical for gammaretroviruses with MLV as a model (15). HERV-H-like proviruses in general had few stops and frameshifts in pro. The pro was found to be the only gene that exists in many completely open forms (see selected alignment in supplemental data). In 48 of 926 HERV-H-like proviruses of the human genome (version hg15), pro had an ORF and was, on average, 300 nucleotides long. However, the pro genes had approximately the same ratio between number of stops and shifts and sequence length as the gag, pol, and env genes (data not shown). This does not support a selection for open pro, nor does it rule out the possibility for pro ORF maintenance selection in a specific locus or loci.
pol.
The pol (nucleotides approximately 3268 through 6369, frame 1) (Fig. 3) was located just 3′ of pro without stops separating the genes in accordance with the typical gammaretroviral genome organization (see above). There were 11 conserved consensus motifs recognized by RetroTector in both RT (RT1, 3457 through 3486; RT2, 3613 through 3654; RT3, 3718 through 3762; RT4, 3826 through 3855; and RT5, 3919 through 3948) and IN (IN2, 5329 through 5355; IN3, 5425 through 5457; IN4, 5527 through 5550; IN5, 5707 through 5742; IN6, 5791 through 5838; and IN7, 6169 through 6216) (Table 1; Fig. 3; supplemental data). An RNaseH motif could also be detected, with the help of alignments and comparison of the RNaseH consensus derived from 1,605 RNaseH sequences in Pfam (DG-(38 aa) E-(20 aa)-DS (64 aa)-N-(3 aa)-D), PF00075, http://www.sanger.ac.uk/Software/Pfam/), just 3′ of RT and ranging approximately over the nucleotide positions 4721 through 4898. The predicted HERV-H consensus Pol protein was similar to other gammaretroviral Pol in its internal RT-RNaseH-IN arrangement and intermotif distances as detected with RetroTector. The characteristic HHCC zinc finger motif and the DD35E catalytic domains (15) were present, as implemented in the RetroTector Pol analysis.
The majority of HERV-H elements had a clear GPY/F domain (explained in discussion), detected as an “IN7” motif by RetroTector (alignment in supplemental data). The deduced GPY/F domain of HERV-H aligned well with the known GPY/F domains of other retroviral elements (29, 36). Despite numerous pol sequences detected by RetroTector, we did not find complete ORFs (see supplemental data).
env.
env (6668 through 8419, frame 2) (Fig. 3) was located 3′ of pol and was preceded by a splice acceptor signal in pol at position 6097 (CCTACTCCAG\ATCCCCAGCC). Further, it contained three detectable motifs: SU3, 7823 through 7867; TM3, 8027 through 8092; and TM5, 8267 through 8302. A von Heijne SP could be recognized in the 5′ of SU (PSNTSTLMKFYSLLLYSLLFSFPFL, using tools at http://www.cbs.dtu.dk/services/SignalP/) and ranged over nucleotide positions 6690 through 6767. Separating the SU from TM, a furin cleavage site, RQKR, was detected at nucleotide positions 7817 through 7828. In exogenous retroviruses, the cleavage (−/−) occurs after R in the consensus sequence RX(R/K)R−/−. The ISU (also noted as the CKS17 motif) with underlined consensus, LQNRRGLDLLTAEKGGLCIF, could be detected in the TM and ranged from nucleotide positions 8027 through 8086. N-Glycosylation sites were found starting at nucleotide positions 6698, 6809, 7334, 7463, 7517, 7724, 7778, 8117, and 8348. The HERV-H consensus aligned well to the three env ORFs (17), extracted with RetroTector from the human genome (supplemental data).
Further, the PPT(AAGAAGGCAGGA) was recognized just upstream of the 3′ LTR and ranged from nucleotide positions 8572 through 8584 (Fig. 3).
Alternative splicing and ESTs.
In the putative HERV-H consensus, numerous SD sites and even more SA sites were predicted. To analyze the predicted SD and SA sites in the putative HERV-H consensus proviral sequence, we performed a search for ESTs using the Gene2EST (20) and the dbEST at National Center for Biotechnology Information. The major SD site was found at the predicted nucleotide position 600, not far downstream of the PBS (Fig. 2; Fig. 3, sequence). Another SD (33, 61) was confirmed at nucleotide position 2927 at the border of gag and pro. An SA site (33, 61) was confirmed at nucleotide position 2824. Further, the major SA for env transcripts (33) was recognized within pol at nucleotide 6097. Splicing in this region was observed in ESTs, but the exact position of this SA could not be confirmed among cDNA in the dbEST. Another previously shown SA was found 9 nucleotides upstream of the 3′ LTR at position 8574 and another SA located 5 nucleotides into the 3′ LTR at position 8590 (61). Neither additional SDs nor additional SAs were confirmed by ESTs. EST confirmations of predicted SD and SA did not always result in identical positions. The observations of ESTs were that splicing occurred in a region plus/minus a few nucleotides from the predicted position.
To analyze the expression of env, we searched the dbEST at GenBank (with HERV-H env AF108843, positions 642 through 2396, which share 98% identity with the HERV-H consensus env). Removal of sequences that were <80% identical to the query sequence over a segment of at least 100 nt resulted in a selection of 129 hits, and in a BLAT search, we noted respective loci for expression (Table 4). As expected, there were generally more hits in a search with the less variable TM than with SU. Colon tumor cDNA libraries generated the most hits (17 hits), exclusively for the TM. These differences may be due to the different cDNA synthesis and cloning strategies in different cDNA libraries. The env-containing ESTs were short, and none of them encompassed the env splice acceptor site, which thus could not be confirmed in this way.
TABLE 4.
Extract of dbEST search results; tissues with deviating expression patterns between SU and TM
| Tissue | No. of HERV-H env hits | HERV-H env TM
|
HERV-H env SU
|
||
|---|---|---|---|---|---|
| No. of hits | Locus (decreasing no. of hits) | No. of hits | Locus (decreasing no. of hits) | ||
| Colon tumor | 17 | 17 | 5q31.1; Xp22.33; 1p32.3; 1q42.2; 2p22.2; 3q22.1; 11q13.3; 13q21.32; 14q12; Xq23 | 0 | |
| Head-neck, tumor | 12 | 8 | 9p21.3; 4q24; 6p21.2; 6q13.3; 8q13.2; 17p12; 19p13.2 | 4 | 14q32.12; 19p13.2 |
| Stomach tumor | 9 | 9 | Xp22.33; 13q14.11; 2p21; 8q23.3; 20p11.23 | 0 | |
| Small intestine, adenocarcinoma | 7 | 7 | 2p24.3; 13q33.3 | 0 | |
| Pooled germ cell tumours, Soares | 7 | 0 | 7 | 4q12; 19p13.2 | |
| Prostate, adenocarcinoma | 6 | 6 | 1p31.2 | 0 | |
| Marrow, tumor | 6 | 5 | 16p12.2; 9q21.33; 16p13.2; Yq11.222 | 1 | 5q23.3 |
| Testis normal | 4 | 1 | 2p14 | 3 | 1p36.11; 3p26.1; 12q24.33 |
| Total no. of hits in dbEST | 129 | 100 | 29 | ||
In an attempt to further investigate the HERV-H env expression, fresh normal human placenta was subjected to cation exchange HPLC. Polyclonal rabbit antiserum was raised against a synthetic peptide derived from HERV-H SU and tested in enzyme-linked immunosorbent assay against the immunogenic peptides (data not shown). Western blots indicated expression of a degraded HERV-H SU fragment of about 23 kDa (supplemental data). However, the band, which bound to the SU antiserum, was not subjected to N-terminal sequencing, for both economic reasons and lack of material.
DISCUSSION
In this study, we have investigated the sequence variation and gene structure of full-length HERV-H proviruses derived from the human genome. Several reported biological functions of ERVs and an implied relevance for disease (7, 38-40, 50, 51) motivate a comprehensive analysis and description of the large HERV-H group. We propose a likely model for the sequence and structure of an original HERV-H based on an alignment of full-length proviruses and few manual adjustments of the consensus to eliminate stops and frameshifts. For this adjustment, we used 18 sequences with almost or totally open reading frame for at least one gene (gag, pro, pol, or env). In support of the consensus construction, the 18 HERV-H like sequences used (Table 2) were reasonably representative of the entire HERV-H group (24) and were widely dispersed in the HERV-H tree (supplemental data). Further, all retrieved sequences formed a distinct clade compared against other gammaretroviruses in pol (Fig. 1). By using different HERV-H proviruses in an alignment of RetroTector suggested proteins (“puteins”), we overcame the effects of random insertions and deletions (indels) from single or few sequences. Save for gene conversion, recombination, and duplication, an indel would unlikely have occurred at the same position in the majority of the 18 sequences that differed in age and integration events indicated by 5′-3′ LTR differences of 2 to 8% and thus have an effect on the “most recent common ancestor” (the HERV-H consensus sequence). This also holds if gene duplication, recombination, and/or gene conversion has not occurred in the majority of sequences used in the alignment. The use of a consensus sequence facilitated structural and functional interpretations of other HERV-H sequences. The full-length or partial HERV-H ORFs observed here were highly similar to the ORFs predicted from the consensus sequence (supplemental data). This supports the validity of the consensus construct. It may be used to guide analyses of other gammaretroviral sequences.
Consensus sequences have been used successfully for other retroelements, e.g., long interspersed nucleotide elements (55). However, that exhaustive analysis was conducted on sequences less than 1 kb. Such a large-scale analysis of full-length retroviruses (approximately 9 kb) in alignments is not amenable. Therefore, we chose full-length sequences scattered over the HERV-H tree so the data set used here is representative (see the cladogram in the supplemental data and Jern et al. [24]). It is separate from other gammaretroviruses (Fig. 1, left panel). A codon-guided alignment is important to judge the effect of selected (preintegrational) and neutral (postintegrational, in absence of physiological or pathogenic function) substitutions. This was to a large degree achieved by the putein-guided minor adjustments of the computer-generated consensus (Fig. 3 and alignment in supplemental data).
The markedly skewed nucleotide frequency in HERV-H disfavoring guanine and favoring cytidine (Table 3) ought to have a functional explanation. In HIV, the G-to-A hypermutation is caused by encapsidation of the host enzyme APOBEC3G, a cytidine deaminase (37), which converts C to U in the antisense strand. This is likely to be a host defense measure against retroviruses (59), and gives HIV an excess of A. In fact, all lentiviruses have an excess of A (Table 3). Skewed nucleotide distributions also occur in the deltaretroviruses (HTLV and bovine leukemia virus), which are rich in C and in betaretroviruses [like HERV-K(HML-2) and MMTV], which, like HIV, are rich in A (3). Gammaretroviruses (like MLV and FeLV) do not have a marked overrepresentation of any nucleotide (Table 3). The mechanisms involved in the skewed distributions of nonlentiviral retroviruses are not known. Our observation indicates that exogenous HERV-H had a hypermutation mechanism which depleted guanine and enriched for cytidine. It is more pronounced than in HTLV. Since guanine is a purine, while cytidine is a pyrimidine, it is difficult to envisage a simple chemical conversion, which leads from one to the other, like the deamination catalyzed by APOBEC3G. However, regardless of mechanism, the skewed nucleotide frequency of HERV-H-related proviruses is an additional and independent tool for the classification of ERVs. The confinement of an extraordinarily low G/C ratio to the 926 HERV-H and most of the 106 adjacent HERV-H proviruses (Table 3) segregates these clades from the rest of the gammaretroviruses.
The HERV-H consensus LTRs differed by 1.1% and thus indicated proximity to the proviral integration event where the LTRs would be identical. Random, postintegrational mutations were most probably evened out in the consensus LTRs. However, this difference may also represent the ambiguity of the sequence and roughly indicates the accuracy of the consensus. The LTR structures were bona fide according to identified TF binding sites. Multiple hits in the dbEST also confirmed the start and end of the R region of the 5′ LTR, which also verified the RetroTector prediction (data not shown).
The 5′ leader region between the PBS and the gag (Fig. 2 and 3) may serve several functions and usually contains the packaging signal ψ downstream of the major splice donor site (for a review, see Coffin et al. [15]). However, the 5′ leader in the HERV-H consensus was rather long and contained additional features. The 117-amino acid 5′ “elongation” of the Gag protein (“pre-Gag”) had heptad repeats of leucine, proline, and serine. Heptad serine repeats in RNA polymerase II CTD have several regulatory functions, e.g., initiation of transcription, which is decreased when CTD is phosphorylated (46). This phosphorylation of the serines also prevents transcription by accidental readthrough of upstream stop codons. However, the function of the “pre-Gag” sequence of HERV-H is uncertain.
MLV has earlier been shown to have an alternative gag start (CUG) upstream of the normal gag start (AUG), thus producing a larger “glyco-Gag” (45). In normal initiation of translation, the ribosome, carrying Met-tRNA and initiation factors, binds to the 5′ CAP and migrates (scans) along the 5′ leader until it encounters an AUG codon. Besides the 5′ CAP initiation of translation, the MLV 5′ leader sequence has an IRES that is involved in the initiation of translation of both Gag and glyco-Gag precursors (4). In a comparison between the HERV-H and the MLV 5′ leaders using the Mfold web server (65), we could not find evidence for a folding similarity. This may be due to the difference in length between the two sequences. However, multiple hairpin structures (δG < −70 kcal/mol) were predicted in both sequences. Kozak showed that a δG of −50 kcal/mol would, in theory, be sufficient to interfere with the ribosome migration in a CAP-initiated translation (31). This is circumstantial evidence for HERV-H sharing the features of MLV with both CAP- and alternative IRES-initiated translation.
Another ORF was found in the HERV-H consensus ranging from nucleotide positions 294 through 1019. The function of this ORF remains to be shown, but additional ORFs upstream of gag have recently been reported for other proviruses (25). The region upstream of HERV-H gag may also have a physiological function. The HERV-H LTR has been demonstrated to initiate a phosholipase A2-related gene, generating the single-copy gene PLA2L (PLA2-like, accession no. Z14310), which is expressed in human teratocarcinoma cells (18, 30, 59). An interpretation is that PLA2L was derived from a HERV-H that suffered a deletion in that particular region of the 5′ leader, resulting in a single ORF instead of separate ORFs in frames 1 and 2 (Fig. 2). The PLA2L intergenic transcript could be a general example of how old retroviruses gain coding potential and how serendipitous generation of physiological functions can take place. In some retroviruses, serine- and proline-rich sequences occur as separate proteins between the MA and CA proteins (MLV, MPMV, MMTV) within the Gag polyprotein and can also contain late domains (19). They are phosphorylated and probably serve regulatory functions (63). HERV-H lacks a pp12, but hypothetically, the proline-rich predicted Pre-Gag protein could serve similar functions.
The HTLV Gag CA has two distinct domains, the NTD and CTD (27, 41), where mutations in the CTD, including the conserved 20 amino acids of MHR, are detrimental for virus assembly and mutations in the NTD produce noninfectious viruses. Interestingly, 12 out of the selected 18 HERV-H-like sequences had deletions in the MHR of CTD (Fig. 4). The uniform pattern of the sequence with the 344-nucleotide gag deletion (located together in Fig. 1), including the CTD and thus the MHR, indicated a single event. In fact, most the 926 HERV have the deletion in CA. The deletion probably arose earlier than the divergence of RTVLH2 and RGH2 subgroups of HERV-H (cladogram in supplemental data). We postulate that these proviruses needed help from RGH2-like “midwife” elements (24) with a complete gag that could provide a functional CA. The lentiviral conserved “AGPI” and HTLV-1 “AGPL” motifs (27), which bind the cellular cyclophilin A (CypA) protein needed in viral replication, could not be found the HERV-H CA. Thus, CypA was probably not needed for the infectivity of HERV-H (for a review, see reference 27).
Like betaretroviruses (MMTV and MPMV), epsilonretrovirus (Xen1), gypsy, copia, deltaretrovirus (not shown), and lentiretrovirus (not shown), the HERV-H consensus Gag had two zinc finger motifs (Fig. 4). Normally, gammaretroviruses (e.g., MLV, GaLV, HERV-E,-W, etc.) have only the first of the two motifs, but remnants of a second zinc finger, or a complete one, were found in all of the 18 representative HERV-H-like sequences. We propose that the original Gammaretrovirus ancestor had two finger motifs in its NC protein and lost one during evolution. The lost finger would have been replaced by a section of charged amino acids in the CA-NC region (…KREETPEER… in MLV [Fig. 4]) (11), sometimes referred to as an “electric wire.” This evolutionary scenario is supported by the presence of two zinc finger motifs in the Epsilonretrovirus Xen1 (AJ506107), from Xenopus laevis, which branches off at the root of Gammaretrovirus (25). Gammaretroviral HERVs other than HERV-H retain a miscellany of a second zinc finger, concomitantly with a more or less dense accumulation of charged amino acids (Fig. 4) upstream of the first zinc finger. The charged amino acid segment (most developed in MLV) upstream of the zinc finger locates in a region overlapped by the Gag interaction (I) domain which is required for virion formation (8) and may have arisen as compensation for the loss of the second zinc finger. Thus, the human genomic record also provides evidence for this sequence of retroviral evolution.
Particulate fractions from supernatants of multiple sclerosis B cells have been reported to contain HERV-H sequences with fragments of the second zinc finger motif similar to that of RGH2 (13). A database search indicated that they were similar, but not identical, to several HERV-H loci (data not shown). Taken at face value, this could indicate a possibility for RGH2-like “midwife” elements (24) to “break out” (5) under special conditions. A related question is whether the few partial gag ORFs recorded here have any potential for expression of Gag functions. From our data, we do not deduce a packaging-competent HERV-H capsid. However, it should be further investigated.
The stop codons observed in frame between gag and pro (Fig. 3; see alignment in supplemental data) may be a consequence of imperfections during consensus construction, exemplified by the 1.1% error rate estimated through LTR divergence. A consensus sequence will be identical to the ancestral one only if postintegrational mutations are random. If a common inactivating mutation is selected for, it will occur in the consensus. Thus, an ancestral HERV-H probably had one stop, which is normal for gammaretroviruses (15). The NTE protease motif of the consensus may not be representative of the original HERV-H. Its functionality is uncertain. It may be a consequence of the gradual taming of an exogenous HERV-H. (We thank Ronald Swanstrom for pointing this out for us.) The numerous HERV-H-like pro ORFs or near-ORFs in the human genome may simply be due to their shorter sequences (about 300 nucleotides) and thereby a smaller target for random mutations. On the other hand, a separately expressed Pro derived from other proviruses such as the betaretroviral RERV-H was reported (64). We here show the presence of a splice acceptor site in pro (see Lindeskog et al. [33]), raising the possibility of similar mechanisms in HERV-H. However, ESTs supporting a separate HERV-H pro expression were not found. The degree of expression and possible physiological functions of the HERV-H protease should be investigated.
Deletions in pol have been observed in the conserved consensus motifs RT4 and RT5 (Table 1) (see Jern et al. [24]). This was also true for 11 of the 18 representative HERV-H-like sequences used in the consensus construct (supplemental data). The deletion in RT4 that encodes the most conserved YXDD motif used as target in many PCRs may thus cause false-negative results in broadly amplifying HERV-H pol PCRs (for a review, see reference 24). The addition of functional modules in carboxy-terminal IN without disturbing the basic integrase can result in extra features like the conserved GPY/F domain (36). To this domain, another “chromo” (chromatin-binding) domain is sometimes appended (36). The integrase carboxyterminus may interact with chromatin via DNA-binding proteins (49, 53). In the distant relatives gypsy and chromovirus, these two domains together direct the integration to more or less specific genomic positions (53). The functional implications of the broad conservation of the GPY/F motif within retroelements remain to be studied. The large amount of HERV-H integrations may give hints to the possible integration specificity endowed by the HERV-H Pol carboxy terminus.
Several HERV-H proviruses contain env ORFs (17, 23, 34, 38) and a number of HERV-H env polymorphism studies have been conducted (17, 23). However, unlike HERV-W Env (40), the HERV-H Env has hitherto not been detected in human tissues. In this study, we describe the expression of many HERV-H env-containing ESTs. HERV-H RNA expression was found in several malignant tissues. env was detected mostly in colon tumor libraries where TM transcripts dominated over those detected with HERV-H consensus SU. A polyclonal antiserum raised from a HERV-H SU peptide could detect a protein in human normal placenta. The HERV-H env RNA is expressed from several loci (Table 4). Transcripts were not necessarily complete, since in several tissues, we found only TM expression. We found no proof for expression of an entire spliced env product in the dbEST search. This may be explained by a relative incompleteness of the dbEST. Our EST results were also consistent with earlier described data on a HERV-H/F virus (43). The relation of the HERV-H loci identified here, in particular, their env sequences, to various cancers should be studied. Functional, immunological as well as diagnostically useful information may emerge from such a study.
Concluding remarks.
The HERV-H consensus derived from the largest endogenous retrovirus family (constituting nearly one-third of all pol-containing ERVs detected by RetroTector) shared a typical gene arrangement with other gammaretroviruses and may thus be useful as a model in studies on retroviral evolution and in expression and polymorphism studies. Unique features of HERV-H consensus compared to other gammaretroviruses are the pre-gag ORF whose function is obscure, and the two zinc finger motifs in gag, a gene which currently is in the searchlight because of its interaction with cellular factors like cyclophilin, APOBEC3G, and TRIM5α. The two zinc fingers probably existed early in Gammaretrovirus evolution. After the HERV-H branching off, the second motif was lost and was followed by an accumulation of a more or less densely charged section in the CA-NC boundary, which is most marked in MLV, to cover the loss. The rich genetic retroviral sequence record materially helps in the understanding of retroviral evolution.
Acknowledgments
This work was supported by the Swedish research council (grant no. K2004-32X-14252-03A) and Stanley Foundation (grant no. 03R-584).
We thank dr Rüdiger Pipkorn, Deutsche Krebsforschungszentrum, Heidelberg, Germany, for peptide synthesis, purification and characterization.
REFERENCES
- 1.Anderssen, S., E. Sjottem, G. Svineng, and T. Johansen. 1997. Comparative analyses of LTRs of the ERV-H family of primate-specific retrovirus-like elements isolated from marmoset, African green monkey, and man. Virology 234:14-30. [DOI] [PubMed] [Google Scholar]
- 2.Benit, L., N. De Parseval, J. F. Casella, I. Callebaut, A. Cordonnier, and T. Heidmann. 1997. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 71:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkhout, B., A. Grigoriev, M. Bakker, and V. V. Lukashov. 2002. Codon and amino acid usage in retroviral genomes is consistent with virus-specific nucleotide pressure. AIDS Res. Hum. Retrovir. 18:133-141. [DOI] [PubMed] [Google Scholar]
- 4.Berlioz, C., and J. L. Darlix. 1995. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J. Virol. 69:2214-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomberg, J., D. Ushameckis, and P. Jern. 2005. Evolutionary aspects of human endogenous retroviral sequences (HERVs) and disease, p. 227-262. In E. D. Sverdlov (ed.), Retroviruses and primate genome evolution. Eurekah.com/Landes Bioscience, Georgetown, Tex.
- 6.Bock, M., and J. P. Stoye. 2000. Endogenous retroviruses and the human germline. Curr. Opin. Genet. Dev. 10:651-655. [DOI] [PubMed] [Google Scholar]
- 7.Boller, K., H. Konig, M. Sauter, N. Mueller-Lantzsch, R. Lower, J. Lower, and R. Kurth. 1993. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 196:349-353. [DOI] [PubMed] [Google Scholar]
- 8.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunak, S., J. Engelbrecht, and S. Knudsen. 1991. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 220:49-65. [DOI] [PubMed] [Google Scholar]
- 10.Chance, M. R., I. Sagi, M. D. Wirt, S. M. Frisbie, E. Scheuring, E. Chen, J. W. Bess, Jr., L. E. Henderson, L. O. Arthur, T. L. South, and et al. 1992. Extended x-ray absorption fine structure studies of a retrovirus: equine infectious anemia virus cysteine arrays are coordinated to zinc. Proc. Natl. Acad. Sci. USA 89:10041-10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheslock, S. R., D. T. K. Poon, W. Fu, T. D. Rhodes, L. E. Henderson, K. Nagashima, C. F. McGrath, and W.-S. Hu. 2003. Charged Assembly Helix Motif in Murine Leukemia Virus Capsid: an Important Region for Virus Assembly and Particle Size Determination. J. Virol. 77:7058-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen, T., P. Dissing Sorensen, H. Riemann, H. J. Hansen, and A. Moller-Larsen. 1998. Expression of sequence variants of endogenous retrovirus RGH in particle form in multiple sclerosis. Lancet 352:1033. [DOI] [PubMed] [Google Scholar]
- 13.Christensen, T., P. Dissing Sorensen, H. Riemann, H. J. Hansen, M. Munch, S. Haahr, and A. Moller-Larsen. 2000. Molecular characterization of HERV-H variants associated with multiple sclerosis. Acta Neuro. Scand. 101:229-238. [DOI] [PubMed] [Google Scholar]
- 14.Cianciolo, G. J., T. D. Copeland, S. Oroszlan, and R. Snyderman. 1985. Inhibition of lymphocyte proliferation by a synthetic peptide homologous to retroviral envelope proteins. Science 230:453-455. [DOI] [PubMed] [Google Scholar]
- 15.Coffin, J. M., S. H. Hughes, and H. E. Varmus (ed.). 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 16.de Parseval, N., H. Alkabbani, and T. Heidmann. 1999. The long terminal repeats of the HERV-H human endogenous retrovirus contain binding sites for transcriptional regulation by the Myb protein. J. Gen. Virol. 80:841-5. [DOI] [PubMed] [Google Scholar]
- 17.de Parseval, N., J. Casella, L. Gressin, and T. Heidmann. 2001. Characterization of the three HERV-H proviruses with an open envelope reading frame encompassing the immunosuppressive domain and evolutionary history in primates. Virology 279:558-569. [DOI] [PubMed] [Google Scholar]
- 18.Feuchter-Murthy, A. E., J. D. Freeman, and D. L. Mager. 1993. Splicing of a human endogenous retrovirus to a novel phospholipase A2 related gene. Nucleic Acids Res. 21:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gemund, C., C. Ramu, B. Altenberg-Greulich, and T. J. Gibson. 2001. Gene2EST: a BLAST2 server for searching expressed sequence tag (EST) databases with eukaryotic gene-sized queries. Nucleic Acids Res. 29:1272-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 22.Henderson, L. E., H. C. Krutzsch, and S. Oroszlan. 1983. Myristyl amino-terminal acylation of murine retrovirus proteins: an unusual post-translational proteins modification. Proc. Natl. Acad. Sci. USA 80:339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jern, P., M. Lindeskog, D. Karlsson, and J. Blomberg. 2002. Full-Length HERV-H Elements with env SU Open Reading Frames in the Human Genome. AIDS Res Hum Retroviruses. 18:671-676. [DOI] [PubMed] [Google Scholar]
- 24.Jern, P., G. O. Sperber, and J. Blomberg. 2004. Definition and variation of human endogenous retrovirus H. Virology 327:93-110. [DOI] [PubMed] [Google Scholar]
- 25.Kambol, R., P. Kabat, and M. Tristem. 2003. Complete nucleotide sequence of an endogenous retrovirus from the amphibian, Xenopus laevis. Virology 311:1-6. [DOI] [PubMed] [Google Scholar]
- 26.Karlsson, H., S. Bachmann, J. Schroder, J. McArthur, E. F. Torrey, and R. H. Yolken. 2001. Retroviral RNA identified in the cerebrospinal fluids and brains of individuals with schizophrenia. Proc. Natl. Acad. Sci. USA 98:4634-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khorasanizadeh, S., R. Campos-Olivas, and M. F. Summers. 1999. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J. Mol. Biol. 291:491-505. [DOI] [PubMed] [Google Scholar]
- 28.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kordis, D., and F. Gubensek. 1998. Unusual horizontal transfer of a long interspersed nuclear element between distant vertebrate classes. Proc. Natl. Acad. Sci. USA 95:10704-10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowalski, P. E., J. D. Freeman, and D. L. Mager. 1999. Intergenic splicing between a HERV-H endogenous retrovirus and two adjacent human genes. Genomics 57:371-379. [DOI] [PubMed] [Google Scholar]
- 31.Kozak, M. 1986. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA 83:2850-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak, M. 1989. The scanning model for translation: an update. J. Cell 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindeskog, M., and J. Blomberg. 1997. Spliced human endogenous retroviral HERV-H env transcripts in T-cell leukaemia cell lines and normal leukocytes: alternative splicing pattern of HERV-H transcripts. J. Gen. Virol. 78:2575-85. [DOI] [PubMed] [Google Scholar]
- 34.Lindeskog, M., D. L. Mager, and J. Blomberg. 1999. Isolation of a human endogenous retroviral HERV-H element with an open env reading frame. Virology 258:441-450. [DOI] [PubMed] [Google Scholar]
- 35.Mager, D. L., and J. D. Freeman. 1995. HERV-H endogenous retroviruses: presence in the New World branch but amplification in the Old World primate lineage. Virology 213:395-404. [DOI] [PubMed] [Google Scholar]
- 36.Malik, H. S., and T. H. Eickbush. 1999. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 73:5186-5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 38.Mangeney, M., N. de Parseval, G. Thomas, and T. Heidmann. 2001. The full-length envelope of an HERV-H human endogenous retrovirus has immunosuppressive properties. J. Gen. Virol. 82:2515-2518. [DOI] [PubMed] [Google Scholar]
- 39.Mangeney, M., and T. Heidmann. 1998. Tumor cells expressing a retroviral envelope escape immune rejection in vivo. Proc. Natl. Acad. Sci. USA 95:14920-14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mi, S., X. Lee, X. Li, G. M. Veldman, H. Finnerty, L. Racie, E. LaVallie, X. Y. Tang, P. Edouard, S. Howes, J. C. Keith, Jr., and J. M. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785-789. [DOI] [PubMed] [Google Scholar]
- 41.Mortuza, G. B., L. F. Haire, A. Stevens, S. J. Smerdon, J. P. Stoye, and I. A. Taylor. 2004. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431:481-485. [DOI] [PubMed] [Google Scholar]
- 42.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 43.Patzke, S., M. Lindeskog, E. Munthe, and H. C. Aasheim. 2002. Characterization of a novel human endogenous retrovirus, HERV-H/F, expressed in human leukemia cell lines. Virology 303:164-173. [DOI] [PubMed] [Google Scholar]
- 44.Perron, H., J. A. Garson, F. Bedin, F. Beseme, G. Paranhos-Baccala, F. Komurian-Pradel, F. Mallet, P. W. Tuke, C. Voisset, J. L. Blond, B. Lalande, J. M. Seigneurin, and B. Mandrand. 1997. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. The Collaborative Research Group on Multiple Sclerosis. Proc. Natl. Acad. Sci. USA 94:7583-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prats, A. C., G. De Billy, P. Wang, and J. L. Darlix. 1989. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J. Mol. Biol. 205:363-372. [DOI] [PubMed] [Google Scholar]
- 46.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 47.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samuelson, L. C., K. Wiebauer, C. M. Snow, and M. H. Meisler. 1990. Retroviral and pseudogene insertion sites reveal the lineage of human salivary and pancreatic amylase genes from a single gene during primate evolution. Mol. Cell. Biol. 10:2513-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandmeyer, S. 2003. Integration by design. Proc. Natl. Acad. Sci. USA 100:5586-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sauter, M., K. Roemer, B. Best, M. Afting, S. Schommer, G. Seitz, M. Hartmann, and N. Mueller-Lantzsch. 1996. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res. 56:4362-4365. [PubMed] [Google Scholar]
- 51.Sauter, M., S. Schommer, E. Kremmer, K. Remberger, G. Dolken, I. Lemm, M. Buck, B. Best, D. Neumann-Haefelin, and N. Mueller-Lantzsch. 1995. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 69:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimotohno, K., S. Mizutani, and H. M. Temin. 1980. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature 285:550-554. [DOI] [PubMed] [Google Scholar]
- 53.Singleton, T. L., and H. L. Levin. 2002. A long terminal repeat retrotransposon of fission yeast has strong preferences for specific sites of insertion. Eukaryot. Cell 1:44-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sjottem, E., S. Anderssen, and T. Johansen. 1996. The promoter activity of long terminal repeats of the HERV-H family of human retrovirus-like elements is critically dependent on Sp1 family proteins interacting with a GC/GT box located immediately 3′ to the TATA box. J. Virol. 70:188-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smit, A. F., G. Toth, A. D. Riggs, and J. Jurka. 1995. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 246:401-417. [DOI] [PubMed] [Google Scholar]
- 56.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ting, C. N., M. P. Rosenberg, C. M. Snow, L. C. Samuelson, and M. H. Meisler. 1992. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 6:1457-1465. [DOI] [PubMed] [Google Scholar]
- 59.Vartanian, J. P., P. Sommer, and S. Wain-Hobson. 2003. Death and the retrovirus. Trends Mol. Med. 9:409-413. [DOI] [PubMed] [Google Scholar]
- 60.Wang, Y., P. E. Kowalski, I. Thalmann, D. M. Ornitz, D. L. Mager, and R. Thalmann. 1998. Otoconin-90, the mammalian otoconial matrix protein, contains two domains of homology to secretory phospholipase A2. Proc. Natl. Acad. Sci. USA 95:15345-15350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilkinson, D. A., J. D. Freeman, N. L. Goodchild, C. A. Kelleher, and D. L. Mager. 1990. Autonomous expression of RTVL-H endogenous retroviruslike elements in human cells. J. Virol. 64:2157-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilkinson, D. A., D. L. Mager, and J. C. Leong. 1994. Endogenous human retroviruses, p. 465-535. In J. Levy (ed.), The retroviridae. Plenum Press, New York, N.Y.
- 63.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voisset, C., R. E. Myers, A. Carne, P. Kellam, and D. J. Griffiths. 2003. Rabbit endogenous retrovirus-H encodes a functional protease. J. Gen. Virol. 84:215-225. [DOI] [PubMed] [Google Scholar]
- 65.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]