Abstract
Alzheimer's disease (AD) is a neurodegenerative disease that severely affects the activities of daily living in aged individuals, which typically needs to be diagnosed at an early stage. Generative adversarial networks (GANs) provide a new deep learning method that show good performance in image processing, while it remains to be verified whether a GAN brings benefit in AD diagnosis. The purpose of this research is to systematically review psychoradiological studies on the application of a GAN in the diagnosis of AD from the aspects of classification of AD state and AD-related image processing compared with other methods. In addition, we evaluated the research methodology and provided suggestions from the perspective of clinical application. Compared with other methods, a GAN has higher accuracy in the classification of AD state and better performance in AD-related image processing (e.g. image denoising and segmentation). Most studies used data from public databases but lacked clinical validation, and the process of quantitative assessment and comparison in these studies lacked clinicians' participation, which may have an impact on the improvement of generation effect and generalization ability of the GAN model. The application value of GANs in the classification of AD state and AD-related image processing has been confirmed in reviewed studies. Improvement methods toward better GAN architecture were also discussed in this paper. In sum, the present study demonstrated advancing diagnostic performance and clinical applicability of GAN for AD, and suggested that the future researchers should consider recruiting clinicians to compare the algorithm with clinician manual methods and evaluate the clinical effect of the algorithm.
Keywords: generative adversarial network (GAN), Alzheimer's disease (AD), mild cognitive impairment (MCI), deep learning, computational psychoradiology, classification, magnetic resonance imaging (MRI), positron emission tomography (PET)
Introduction
Alzheimer's disease (AD) is a neurodegenerative disease that mainly affects elderly individuals. It is characterized by a decline in memory, cognitive function, and behavioral function. AD severely affects the activities of daily living of patients (Kimura et al., 2020). It is estimated that the global prevalence of AD will increase 4-fold by 2050, and that the total number of patients will exceed 100 million (Brookmeyer et al., 2007). Mild cognitive impairment (MCI) is a cognitive state that lies between that of normal aging and early AD (Petersen, 2004). It can be divided into two categories: stable MCI (sMCI) and progressive MCI (pMCI). The pMCI patients progress to AD, while sMCI patients remain stable and may even return to a healthy state (Chong and Sahadevan, 2005; Davis et al., 2018).
At present, there is no effective drug for AD in clinical practice. The focus of treatment has shifted to diagnosing patients in the early stage of AD (Chong and Sahadevan, 2005; Davis et al., 2018). Identifying whether the patient is in a normal state of cognitive decline or has AD, sMCI, or pMCI (classification of AD state) can help to identify high-risk individuals and take targeted treatment measures to delay disease progression. In addition, the processing of AD-related images (such as image denoising and image segmentation) is also helpful for the early diagnosis of AD.
In recent years, psychoradiological research has provided clinical evidence for the identification of diagnostic and therapeutic neuroimaging biomarkers in patients with psychiatric disorders (Lui et al., 2016). Computational psychoradiology with artificial intelligence has been widely used in the diagnosis of AD. The diagnosis requires the input of more high-quality, processed images. However, there are still many disadvantages to image processing with existing deep learning methods (Sorin et al., 2020). Therefore, a deep learning method that can process images well is greatly needed.
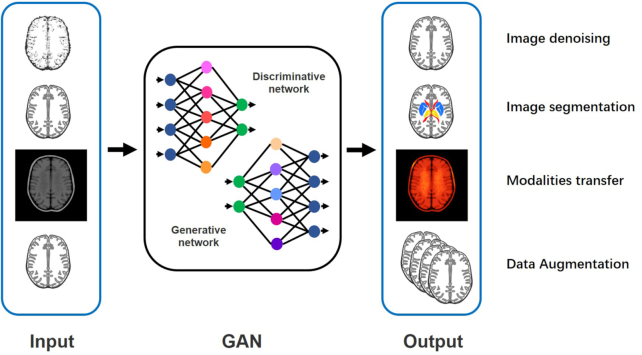
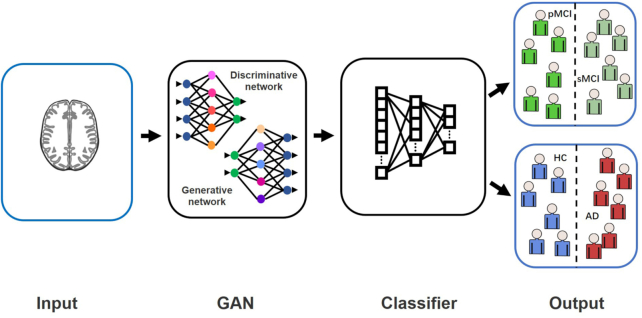
A generative adversarial network (GAN) was proposed by Goodfellow et al. in 2014. This is a deep learning generative model mainly used to process images (Goodfellow et al., 2014). Its main principle is the game between the generator and the discriminator (Sorin et al., 2020). The deep learning classification framework based on a GAN can classify AD state (Bowles et al., 2018; Pan et al., 2018; Yan et al., 2018; Wegmayr et al., 2019; Islam and Zhang, 2020; Kim et al., 2020). GANs also have a wide range of applications in AD-related image processing (shown in Fig. 1). For example, GANs can denoise low-dose positron emission tomography (PET) to obtain high-quality images (Wang et al., 2018; Ouyang et al., 2019; Wang et al., 2019). Accurate segmentation of brain images by a GAN is conducive to feature location (Choi et al., 2018; Shi et al., 2019; Kang et al., 2020). A GAN can convert different image modalities (Choi et al., 2018; Kang et al., 2018; Kang et al., 2020). These applications related to image processing can provide high-quality processed data for the feature extraction step in the AD state classification framework and improve the effect of the classification algorithm.
Figure 1:
Schematic diagram of AD-related image processing by a GAN.
At present, some reviews have reported the role of GANs in medical fields (Lan et al., 2020; Sorin et al., 2020). Sorin et al. introduced the application of GANs in radiology (Sorin et al., 2020). Lan et al. reported the application of different types of GANs in biomedical informatics (Lan et al., 2020). However, previous articles have little significance for the application of a GAN in AD. The aim of this article is to systematically review psychoradiological studies on the application of a GAN in the diagnosis of AD from the aspects of the classification of AD state and AD-related image processing compared with other methods. In addition, we evaluated the research methodology (data, GAN architecture, quantitative assessment, and comparison methods) from the perspective of clinical practitioners and made suggestions for future research.
Materials and Methods
This systematic review was conducted in accordance with the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guide.
PICOS
P (participants): Patients with AD or MCI (no restrictions on age or gender).
I (interventions): GAN-based algorithm.
C (comparison): Other computer algorithms or manual methods.
O (outcome): Outcomes of classification (e.g. classification accuracy), outcomes of image processing [e.g. PSNR, Dice similarity coefficient (DSC)].
S (study design): Psychoradiological studies on algorithm design and application.
Literature search
Two researchers independently searched PubMed, Cochrane Library, EMBASE, Web of Science, and IEEE Xplore to find literature in English on the application of a GAN in the diagnosis of AD before August 2020. The following keywords were used for the search strategy: Alzheimer, AD, dementia, mild cognitive, F-18-FDG, FDG-PET, amyloid, Tau-PET, generative model, and generative adversarial network.
Inclusion and exclusion criteria
Researchers selected the documents required for this systematic review based on the following inclusion and exclusion criteria:
Inclusion criteria
The inclusion criteria followed were: (i) the application of a GAN in human AD; (ii) training and validation data including AD or MCI patients; and (iii) studies including quantitative assessment of the algorithm and comparison with other methods.
Exclusion criteria
The exclusion criteria followed were: (i) no access to the full text; (ii) reviews, conference abstracts, comments, etc.; (iii) literature not in English; (iv) studies using animal models; and (v) studies using a generative model but not a GAN.
Two researchers independently screened the articles according to the inclusion and exclusion criteria and extracted information as follows: general information (title, author, year, journal, etc.), data source, data type, modality of data, quantitative assessment indicator, quantitative assessment result, method compared, and comparison result. The information mentioned was organized into a basic information sheet of literature. After completion, two evaluators cross-checked the information, and differing opinions were resolved through negotiation.
Inclusion and exclusion process
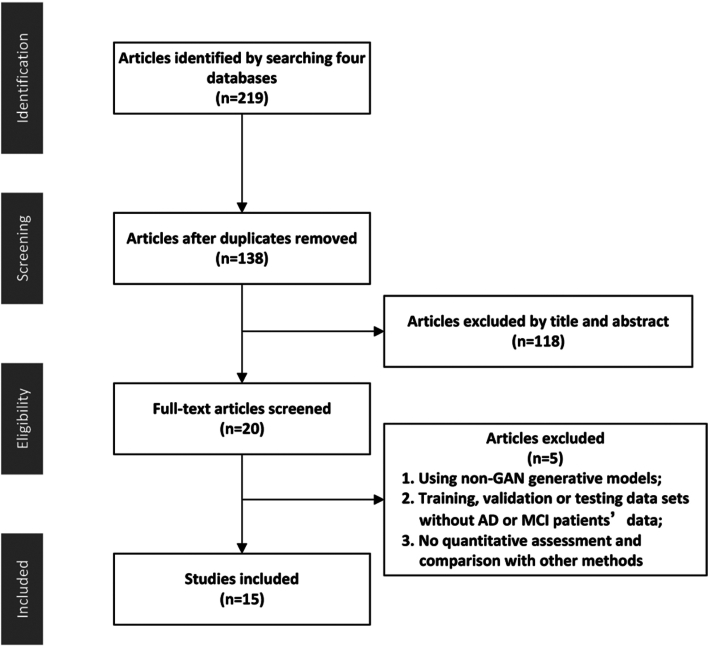
Through the initial search, researchers obtained 225 articles. Twenty-three articles were obtained after removing duplicate and irrelevant documents. The PRISMA flow diagram is shown in Fig. 2. Among these 23 articles, eight articles were excluded after full-text reading analysis. These studies were excluded for the following reasons (details are shown in Table 1): (i) using non-GAN generative models; (ii) training, validation or testing datasets without AD or MCI patients’ data; and (iii) no quantitative assessment and comparison with other methods (Hwang et al., 2018; Armanious et al., 2019; Armanious et al., 2020; Biffi et al., 2020; Kimura et al., 2020).
Figure 2:
The PRISMA flow diagram.
Table 1:
Reasons for exclusion when 23 full-text articles were screened.
| Study | Reason for exclusion |
|---|---|
| Armanious et al. (2019) | No AD or MCI patients included in the data |
| Hwang et al. (2019) | The generative model but not a GAN |
| Armanious et al. (2020) | No AD or MCI patients included in the data |
| Biffi et al. (2020) | The generative model but not a GAN |
| Kimura et al. (2020) | No quantitative assessment and comparison with other methods |
| Liu et al. (2020) | No comparison with other methods |
| Hu et al. (2020) | No comparison with other methods |
| Roychowdhury et al. (2020) | No quantitative assessment and comparison with other methods |
Fifteen articles met the inclusion criteria and were included in the current systematic review (Baumgartner et al., 2018; Bowles et al., 2018; Choi et al., 2018; Kang et al., 2018; Pan et al., 2018; Wang et al., 2018; Yan et al., 2018; Ouyang et al., 2019; Shi et al., 2019; Wang et al., 2019; Wegmayr et al., 2019; Islam and Zhang, 2020; Kang et al., 2020; Kim et al., 2020; Oh et al., 2020). The relevant information of the 15 articles was extracted. All studies were published between 2017 and 2020.
Results
Among the included studies, nine studies were applied to AD-related image processing (image denoising, image segmentation, modality transfer, and data augmentation), and six studies were applied to the classification of AD state.
AD-related image processing
Image denoising
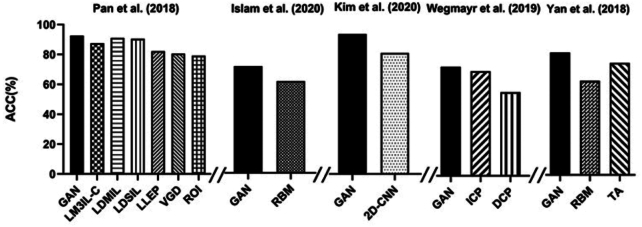
Some studies have been devoted to the development of a denoising framework for low-dose PET images, and images denoised by a GAN achieve a higher peak signal-to-noise ratio (PSNR), which is significantly better than other methods (shown in Fig. 3). Details of the included studies are shown in Table 2. Wang et al. applied a three-dimensional conditional generation adversarial network (3D c-GAN) to generate high-quality full-dose PET images from low-dose PET images (Wang et al., 2018). Compared with the existing convolutional neural network (CNN), mapping-based sparse representation (m-SR), and tripled dictionary learning (t-DL) methods, the GAN method achieved the highest PSNR value, and the lowest normalized mean squared error (NMSE) and standard uptake value (SUV) difference for the normal cognition and the MCI groups. Different from their previous study, Wang et al. then applied a locally adaptive multimode GAN (LA-GAN) model and used an autocontext model to more effectively use context information (Wang et al., 2019). In addition, the generation of high-quality PET images in this study is based on low-dose PET images and corresponding magnetic resonance images. Additionally, compared with the m-SR, t-DL, m-CAA, and autoCNN methods, the GAN method achieved the highest PSNR and SSIM for the normal cognition and the MCI groups. Ouyang et al. combined the perceptual loss in the GAN structure. They trained an additional network to judge the amyloid state, extracted the specific perceptual loss and added it to the GAN structure (Ouyang et al., 2019). This can ensure high visual quality and correct clinical information of amyloid protein. Compared with the networks without perceptual loss and using VGG16 perceptual loss, this research method achieves the highest PSNR and lowest SSIM and RMSE.
Figure 3:
PSNR and DSC of AD-related image processing compared with other methods.
Note: m-CCA, multilevel CCA; AcCNN, autocontext CNN; CCA, canonical correlation analysis; MVPL, models without perceptual loss; MWV, models with perceptual loss computed from VGG16; UGN, UG-net; P2P, Pix2pix_unet; HD, h-dense_unet method; UN, U-net.
Table 2:
Summary of the studies on AD-related image processing.
| Study | Image processing | GAN | Data source | Data type: amount | Modality of data (PET/MRI) | Quantitative assessment indicator | Quantitative assessment result | Method compared | Comparison result |
|---|---|---|---|---|---|---|---|---|---|
| Wang et al. (2018) | Image denoising | 3D c-GAN | Clinical cases | HC: 8MCI: 8 | PET | PSNR; NMSE; SUV | SUV: 0.037 (HC); 0.067 (MCI) | CNN method; m-SR method; t-DL method; 3D U-net-like model (without adversarial training) | The highest PSNR value and the lowest NMSE and SUV difference are achieved by the GAN method. |
| Wang et al. (2019) | Image denoising | LA-GAN | Clinical cases | HC: 8MCI: 8 | MRI; PET; DTI | PSNR; SSIM | PSNR: 24.61 (HC); 15.19 (MCI) SSIM: 0.986 (HC); 0.9843 (MCI) | m-SR method; t-DL method; m-CAA method; autoCNN method; method without autocontext; the generating network (without adversarial training) | The highest PSNR value and SSIM are achieved by the GAN method. |
| Ouyang et al. (2019) | Image denoising | GAN | Clinical cases | Total: 39 | PET | PSNR; SSIM; RMSE; clinical doctors scored image quality | PSNR: compared with low dose PET improved by 4.14 dB;SSIM: compared with low dose PET increased by 7.63%; RMSE: compared with low dose PET reduced by 33.55%; clinical doctors scored image quality = 4.27 | Network without perceptual loss; network using VGG16 perceived loss | This research method achieves the highest PSNR and lowest SSIM and RMSE. |
| Shi et al. (2019) | Image segmentation | GAN | Clinical cases | Total: HC: 21 MCI: 4 AD: 7 training: 5 testing: 27 | MRI | DSC | DSC = 91.6% (overall subfields) | UG-net (without adversarial training); sparse coding and dictionary learning method; CNN; the clinician manual method | This research method has the highest segmentation accuracy in each subfield and overall subfields. |
| Oh et al. (2020) | Image segmentation | cGAN | Clinical cases | Total: 192 training: 154 testing: 19 | MRI: PET | Precision; recall; AUC-PR; DSC; quality scores (scored by observers) | Precision = 0.821; recall = 0.814; AUC-PR = 0.869; DSC = 0.817; quality scores (scored by observers) = 2.6 | pix2pix_unet method; h-dense-Unet method; U-net method | This method achieves the best performance in terms of DSC, AUC-PR, recall, precision and quality scores. |
| Baumgartner et al. (2018) | Image segmentation | WGAN | ADNI | Total: 1288 training: 825 testing: 463 | MRI | NCC | NCC = 0.94 (using synthetic data); NCC = 0.27 (using real data) | Guided backprop; integrated gradients; CAM; additive perturbation | The NCC scores obtained by this study method are significantly higher than other methods. |
| Kang et al. (2020) | Data augmentation | cGAN | Clinical cases | HC: 62 MCI: 99 AD: 137 | PET | MMD; FID;1-NN; Visual Turing Test | MMD = 0.2544; FID = 0.2921; 1-NN = 0.8418; Visual Turing Test = 45.67% | Real data | There is a certain similarity between the generated image and the real image. |
| Kang et al. (2018) | Modalities transfer | GAN | Clinical cases | Training: HC: 338 MCI: 117 AD: 72 testing: HC: 97 MCI: 37 AD: 20 | MRI: PET | Mutual information; mean square error; regional active concentration; SUVR | showed by charts, no specific value | Average template; CAE | The methods based on a GAN and CAE have higher mutual information and smaller mean square error. |
| Choi et al. (2018) | Modalities transfer | GAN | ADNI | Training: HC: 49 MCI: 80 AD: 34 testing: HC: 36 MCI: 41 AD: 21 | MRI: PET | SSIMSUVR | SSIM: 0.91 (AD); 0.92 (MCI); 0.91 (HC); MAE = 0.04 | The method based on PET template; the method based on multiatlas PET template; the method based on PET segmentation; the method based on real MR | The average or MAE of SUVR is smaller than that of other methods. |
Note: HC, healthy control; DTI, diffusion tensor imaging; SSIM, structural similarity index; RMSE, root mean square error; UG-net, a GAN model with the modified U-net; AUC-PR, area under the curve of precision-recall; NCC, normalized cross correlation; CAM, class activation map; MMD, maximum mean discrepancy; FID, Fréchet inception distance; 1-NN, the 1-nearest neighbor classifier; SUVR, SUV ratio; CAE, convolutional autoencoder.
Image segmentation
In terms of image segmentation, two studies using a GAN for segmentation had higher segmentation accuracy than other studies when DSC was the outcome indicator (shown in Fig. 3). Details of the included studies are shown in Table 2. Shi et al. applied a GAN based on an improved U-net generator to segment the hippocampus from magnetic resonance images (Shi et al., 2019). Researchers compared the developed segmentation algorithm using the clinician manual method and showed a good segmentation effect. Compared with the CNN, UG-net and other methods, this research method has the highest segmentation accuracy in each subfield and overall subfields. The overall DSC of all hippocampal subfields reached 91.6%. Oh et al. applied a cGAN based on the pix2pix framework to directly segment white matter in 18F-FDG PET/CT images (Oh et al., 2020). It can be used for quantitative analysis of brain diseases such as AD. Compared with other methods, this method achieved the highest quality score [(2.6 ± 0.7) as scored by five observers]. This method also achieves the best performance in terms of DSC (0.817 ± 0.018), AUC-PR (0.869 ± 0.021), recall (0.814 ± 0.029), and precision (0.821 ± 0.036).
Baumgartner et al. applied the Wasserstein GAN (WGAN) for visual attribution on real 3D neuroimaging data from patients with MCI and AD (Baumgartner et al., 2018). Visual attribution is a process in which the characteristic image areas are labeled depending on the category of the image (AD or MCI). The NCC score of the method using synthetic data was 0.94 ± 0.07, and the score of the method using real data was 0.27 ± 0.15. The accuracy of visual attribution is higher than that of other methods.
Data augmentation and modalities transfer
Details of the included studies are shown in Table 2. Kang et al. used a cGAN to synthesize 18F-florbetaben images (Kang et al., 2020). Researchers performed data augmentation to solve the problem of insufficient data in the development of AD-related deep learning frameworks. Kang et al. applied a GAN to generate individual adaptive PET templates and performed accurate spatial normalization of amyloid PET without using corresponding 3D-MR images (Kang et al., 2018). This helps to conduct objective evaluation and statistical analysis of amyloid PET images. Compared with the method based on the average template, the methods based on a GAN and a CAE have higher mutual information and smaller mean square error. Choi et al. used a GAN to generate realistic structural magnetic resonance images from 18F-florbetapir PET images and applied them to the quantification of cortical amyloid load (Choi et al., 2018). The SSIM values of the AD, MCI, and normal groups were 0.91 ± 0.04, 0.92 ± 0.04, and 0.91 ± 0.04, respectively. Compared with other methods, the average/mean absolute error (MAE) of the SUVR of this method is smaller.
Classification of AD state
Whether a patient is in a normal cognitive decline state or has AD, sMCI, or pMCI is a concern for the classification of AD state. Details of included studies are shown in Table 3. A GAN has significant advantages over other methods in terms of accuracy (shown in Fig. 4). Pan et al. developed a two-stage deep learning framework (Pan et al., 2018). The first step is to impute the PET images according to the corresponding magnetic resonance image by applying a 3D cycle-consistency GAN (3D-cGAN). The second step is to develop a deep multi-instance neural network as a classifier, using paired PET and magnetic resonance images to differentiate between people with and without AD. Pan et al. also classified MCI in their second stage (Pan et al., 2018). The accuracy of Pan's method (AD vs. HC = 92.5% pMCI vs. sMCI = 79.06%) is higher than three methods using hand-crafted features (ROI, VGD, and LLEP), two methods using only magnetic resonance imaging (MRI) data (LDSIL and LDMIL), and one method using real PET and MRI data (LM3IL-C); the overall performance is better than other methods. Islam et al. also developed a two-stage deep learning framework with an accuracy of 71.45%, which is 10% higher than the classification method trained with real PET data (Islam and Zhang, 2020). Kim et al. used the boundary equilibrium GAN (BEGAN) to extract features of AD and normal cognition (Kim et al., 2020). Then, these two disease states were classified. The BEGAN performs better than the 2D-CNN method of Glozman et al. in terms of classification accuracy (94.82%). The deep learning method of Wegmayr et al. can also be divided into two stages (Wegmayr et al., 2019). The first stage simulated the process of brain aging. Researchers input an magnetic resonance image of an MCI patient (xt0), and its corresponding image xt1 at time t1 (t1 = t0 +Δ) was output. In the second stage, researchers input xt1 into the MCI/AD classifier and calculated the probability of AD (pAD). Then, they judged whether this MCI patient had sMCI or pMCI by comparing the pAD with the threshold (γ). The accuracy (73%), precision rate (68%), recall rate (75%), and F1 score (71%) of this method are better than those of the other methods. Yan et al. applied a conditional GAN (cGAN) to generate 18F-florbetapir PET images from corresponding magnetic resonance images, and its MCI classification accuracy was higher than that of the traditional data augmentation method (82 vs. 75%) (Yan et al., 2018).
Table 3:
Summary of the studies on the classification of AD state.
| Study | GAN | Data source | Data type: amount | Modality of data (PET/MRI) | Quantitative assessment indicator | Quantitative assessment result | Method compared | Comparison result |
|---|---|---|---|---|---|---|---|---|
| Pan et al. (2018) | 3D-cGAN | ADNI | Training: AD: 199 HC: 229 pMCI: 167 sMCI: 226 testing: AD: 159 HC: 200 pMCI: 38 sMCI: 239 | MRI; PET | PSNR; accuracy; sensitivity; specificity; F1 score | PSNR = 24.49 ± 3.46 accuracy: 92.50% (AD vs. HC classification); 79.06% (pMCI vs. sMCI classification); sensitivity: 89.94% (AD vs. HC classification); 55.26% (pMCI vs. sMCI classification); specificity: 94.53% (AD vs. HC classification); 82.85% (pMCI vs. sMCI classification); F1 score: 91.37% (AD vs. HC classification); 40.86% (pMCI vs. sMCI classification); | ROI, VGD, LLEP (using hand-crafted features); LDSIL, LDMIL (using only MRI data); LM3IL-C (using real PET and MRI data) | The overall performance (accuracy, sensitivity, etc.) is better than other methods. |
| Islam and Zhang (2020) | Deep CGAN | ADNI | HC: 105 MCI: 208 AD: 98 | PET | PSNR; SSIM; accuracy | PSNR = 32.83; SSIM = 77.48; accuracy = 71.45% | The method using real PET data | The accuracy of the classifier trained with the images generated by GAN is 10% higher than that of the classifier trained with real PET images. |
| Kim et al. (2020) | BEGAN | ADNI; clinical cases | HC: 347 AD: 139 | PET | Accuracy; sensitivity; specificity; AUC | Accuracy = 94.82; sensitivity = 91.78; specificity = 97.06; AUC = 0.98 | 2D-CNN (method of Glozman et al.) | The accuracy is higher than the 2D-CNN method. |
| Wegmayr et al. (2019) | WGAN | ADNI; clinical cases | - | MRI | Accuracy; precision rate; recall rate; F1 score | Accuracy = 73%; precision rate = 68%; recall rate = 75%; F1 score = 71% | Indirect conversion prediction; direct conversion prediction | The WGAN method has higher accuracy, precision rate, recall rate and F1 score. |
| Yan et al. (2018) | cGAN | ADNI | Training: pMCI: 29 sMCI: 50 testing: pMCI: 29 | MRI; PET | SSIM; accuracy; AUC | SSIM = 0.95 ± 0.05; specificity = 82 ± 12%; AUC = 81 ± 7%; | Traditional data augmentation method (images are randomly horizontally and vertically flipped) | The accuracy is higher than that of the traditional data augmentation method. |
| Bowles et al. (2018) | WGAN | ADNI | Total: 1000+ | MRI | The differences between the generated image and the real image | - | Algorithm removed a specific part | The WGAN method without reweighting method has more errors in reconstructing images of severe atrophy or other abnormal cases. |
Note: cGAN: convolutional GAN; ROI, gray matter volume within 90 regions of interest; VGD, voxelwise gray matter density; LLEP, landmark-based local energy patterns; LDSIL, landmark-based deep single-instance learning; LDMIL, landmark-based deep multiinstance learning; LM3IL-C, GAN that use only complete MRI and PET data; AUC, area under curve.
Figure 4:
Accuracy (ACC) of classification of AD state compared with other methods.
Note: LM3IL-C, GAN that uses only complete MRI and PET data; RBM, real image-based method; ICP, indirect conversion prediction; DCP, direct conversion prediction (a CNN classifier); TA, traditional augmentation.
In addition, researchers also made intuitive visual predictions of disease progression. Bowles et al. applied the WGAN to subtract the average potential encoding of a set of magnetic resonance images of healthy participants from that of patients with AD, and separated the potential encodings corresponding to features of AD (Bowles et al., 2018). Researchers can predict the progression of AD by introducing or removing potential encodings from real images.
Data sources and modalities
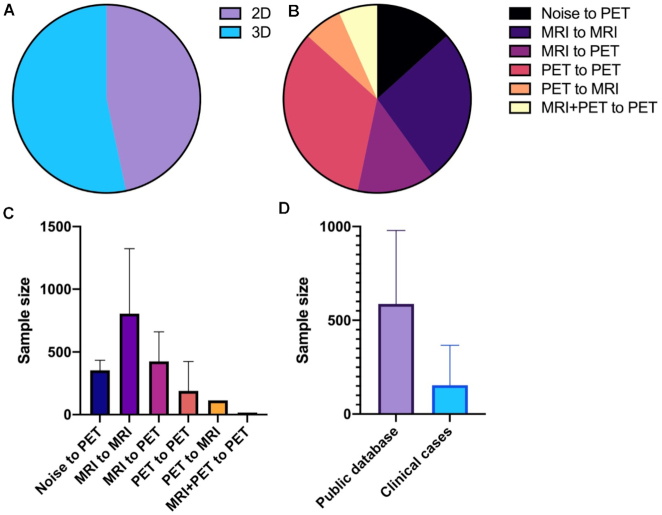
The data sources (public databases or clinical cases) and the amount and mode of data used in the deep learning network were closely related to the training effect; thus, we examined the data used in the included articles (shown in Table 4). We found that among the 15 included studies, eight studies used large public databases (mainly ADNI), six studies used self-collected clinical data as the training and test data of the GAN model, and only Kim et al. trained the GAN model using public databases and verified it with self-collected clinical data. We also note that studies using public datasets usually have a large sample size, while studies using clinical data often have a small sample size. For example, in the study by Wang et al., there were only eight patients in each group (shown in Fig. 5d).
Table 4:
Database, data amount, and modalities in the included studies.
| Database | Data modalities | Data amount | |||||
|---|---|---|---|---|---|---|---|
| Study | Training | Testing | Input | Output | 2D/3D | Training | Testing |
| Islam and Zhang (2020) | Noise | PET | 2D | HC: 105 MCI: 208 AD: 98 | |||
| Bowles et al. (2018) | MRI | MRI | 2D | Total: 1000+ | |||
| Baumgartner et al. (2018) | MRI | MRI | 3D | Total: 1081 | Total: 207 | ||
| Yan et al. (2018) | ADNI | MRI | PET | 3D | pMCI: 29 | pMCI: 21 | |
| Pan et al. (2018) | MRI | PET | 3D | HC: 229 pMCI: 167 sMCI: 226 AD: 199 | HC: 200 pMCI: 38 sMCI: 239 AD: 159 | ||
| Oh et al. (2020) | PET | PET | 2D | Total: 173 | Total: 19 | ||
| Choi et al.(2018) | PET | MRI | 2D | HC: 49 MCI: 80 AD: 34 | HC: 36 MCI: 41 AD: 21 | ||
| Wegmayr et al. (2019) | ADNI and AIBL | MRI | MRI | 2D | HC: 4859 pMCI: 178 sMCI: 232 AD: 700 | ||
| Kang et al. (2020) | Noise | PET | 3D | HC: 62 MCI: 99 AD: 137 | |||
| Shi et al. (2019) | MRI | MRI | 3D | HC: 21 MCI: 4 AD: 7 | |||
| Ouyang et al. (2019) | Clinical cases | PET | PET | 2D | Total: 39 | ||
| Wang et al. (2018) | PET | PET | 3D | HC: 8 MCI: 8 | |||
| Kang et al. (2018) | PET | PET | 3D | HC: 338 MCI: 117 AD: 72 | HC: 97 MCI: 37 AD: 20 | ||
| Wang et al. (2019) | MRI + PET | PET | 3D | HC: 8 MCI: 9 | |||
| Kim et al. (2020) | ADNI | Clinical cases | PET | PET | 2D | HC: 347 AD: 139 | HC: 68 AD: 73 |
Note: AIBL, The Australian Imaging, Biomarker and Lifestyle Flagship Study of Aging.
Figure 5:
Database, data amount, and modalities in the included studies. (A) Dimensions (2D/3D) of data used by included studies; (B) input/output modalities of data used by included studies; (C) sample size of studies using different modalities of data; and (D) sample size of studies using different datasets (public datasets or clinical cases).
We also examined the modalities of data used in different studies and found that most studies used 3D volume data of PET/MR images instead of 2D slices for training (shown in Fig. 5a). In terms of input/output data modes, only Islam et al. and Kang et al. adopted the noise-to-image mode, while most of the remaining studies adopted the image-to-image mode. Most of these studies focus on the conversion of the same modality data (such as MRI to MRI or PET to PET). A small part of the research explores the modality transfer between data with different modalities (mainly MRI to PET). Wang et al. synthesized high-dose PET images based on low-dose PET images and corresponding multimodal magnetic resonance images (T1-weighted MRI and DTI), which is the only study on synthesizing single-mode data based on multimodal data (shown in Fig. 5b).
Architectural design
The included studies all used different GAN modalities, which served as the core of the study. These models had made some improvements to the original GAN to improve its training effectiveness on and adaptability to medical images. We examined the structure and characteristics of the GAN models used in the included studies (shown in Table 5).
Table 5:
GANs used in included studies.
| GANs used | |||||
|---|---|---|---|---|---|
| Study | Main categories | Specific categories | Generator (G) and discriminator (D) | Functions of GANs | Characteristics of GANs |
| Wang et al. (2018) | Conditional GAN (cGAN) | 3D-cGAN | G: 3D U-net based CNND: 3D U-net based CNN | Image denoising (PET to PET) | Adjusting U-net to fit 3D PET dataUsing progressive refinement scheme to improve generating qualityUsing E1 norm estimation error to reduce blurringUsing batch normalization to improve learning efficiency |
| Oh et al. (2020) | 2D-cGAN | G: CNND: CNN | Image segmentation (PET to PET) | Using ReLU for activation function in convolution layer to reduce the vanishing gradient problem | |
| Shi et al.(2019) | 2D-cGAN | G: U-net based CNND: CNN | Image segmentation (MRI to MRI) | Using skip-connection in the U-net to increase the ability of the generator to segment small local regions | |
| Yan et al. (2018) | 2D-cGAN | G: U-net based CNND: Convolutional Markovian discriminator | Modalities transfer (MRI to PET) | Using convolutional Markovian discriminator to improve discrimination performance | |
| Ouyang et al. (2019) | Pix2pix cGAN | G: U-net based CNND: CNN | Image denoising (PET to PET) | Using feature matching to improve training stabilityUsing an extra Amyloid status classifier to make the generated image fit to the patient's real amyloid status | |
| Choi et al. (2018) | Pix2pix cGAN | G: U-net based CNND: CNN | Modalities transfer (PET to MRI) | - | |
| Wang et al. (2019) | “Locality adaptive” multimodality GAN (LA-GAN) | G: 3D U-net based CNND: 3D U-net based CNN | Image denoising (MRI + PET to PET) | Adjusting U-net to fit 3D PET data Using progressive refinement scheme to improve generating quality (autocontext training method) | |
| Baumgartner et al.(2018) | WGAN | WGAN | G: 3D U-net based CNND: CNN | Feature extraction (MRI to MRI) | Using a new map function in generator to generate MRI of AD patients from healthy controls |
| Wegmayr et al. (2019) | WGAN | Same as Baumgartner et al. (2018) | Feature extraction (MRI to MRI) | Same as Baumgartner et al. (2018) | |
| Bowles et al. (2018) | WGAN | - | Feature extraction (MRI to MRI) | Using a training data reweighting schema to improve the generator's ability to produce severely atrophic images | |
| Islam and Zhang (2020) | Deep CGAN | DCGAN | G: CNND: CNN | Data augmentation (noise to PET) | Using BatchNorm to regulate the extracted feature scaleUsing LeakyRelu to prevent the vanishing gradient problem |
| Kang et al. (2020) | DCGAN | G: CNND: CNN | Data augmentation (noise to PET) | Using a regularization term in the Wasserstein loss to improve training stabilityTwo different GAN networks are used to generate Aβ negative and positive images, respectively, to improve the generalization | |
| Kang et al. (2018) | DCGAN | G: CAED: CNN | Modalities transfer (PET to PETSN) | Using the fidelity loss between the MRI-based spatial normalization result and the generated image to generate the template-like image | |
| Pan et al. (2018) | Cycle GAN | 3D Cycle-consistence GAN | Have 2 G & D sets G1 & G2: CNND1 & D2: CNN | Modalities transfer (MRI to PET) | Using two sets of generated countermeasure networks to ensure that the generated image is not only similar to the real image but also corresponding to the input magnetic resonance images |
| Kim et al. (2020) | Boundary Equilibrium GAN (BEGAN) | BEGAN | G: CAE D: CAE | Feature extraction (PET to PET) | The discriminator and generator are trained to maximize and minimize the distance between the real and fake image reconstruction loss rather than the data distribution |
Note: PETSN, PET with spatial normalization; U-net, a modified CNN; ReLU, rectified linear unit.
A total of seven included studies used the conditional GAN (cGAN) model. Wang et al. improved the existing U-net architecture to process 3D PET data and used batch normalization to improve generating efficiency and accuracy. In addition, this study used a training method called the ‘progressive refinement scheme’, which used a series of GANs to input the image generated by the previous GAN into the next GAN to generate a new image; to improve the quality of the generated image as much as possible, their study also used a similar GAN architecture and training method to generate a high-dose PET based on multimodal data (structural MRI, DTI, and low-dose PET). Oh et al. and Shi et al. (2019) used similar 2D-cGAN methods to segment PET and magnetic resonance images. Oh et al. used the residual block based on the rectified linear unit (ReLU) in the generator to reduce the vanishing gradient and improve the speed and stability of training. Shi et al. used skip connections in the U-net to increase the ability of the generator to segment small local regions. Yan et al. also used 2D-cGAN in the modality transfer of MRI to PET. They replaced the discriminator with a convolutional Markovian discriminator so that it could focus on more areas in the image, improve the efficiency of the discriminator, and then improve the efficiency of the whole adversarial network. Ouyang et al. used a pix2pix cGAN for denoising low-dose PET images and used feature matching in the implementation process to reduce the hallucinated structure during training and improve training stability. They also used an extra amyloid state classifier to provide the generator with task-specific perceptual loss to make it generate an image fit to the patient's real amyloid state. Choi et al. completed the transfer from PET to MRI using a similar pix2pix cGAN structure.
A total of three included studies used the WGAN model. Baumgartner et al. and Wegmayer et al. used a similar WGAN model to complete the feature attribute of magnetic resonance images of AD or MCI patients. They added a map generating function to the 3D U-net generator structure to enable it to generate images of another category according to images of one category (such as generating magnetic resonance images of AD patients according to the image of MCI patients or generating images after several years according to the magnetic resonance images at baseline). Bowles et al. also used the WGAN for feature attribution. They used a training data reweighting schema to improve the generator's ability to produce severely atrophic images.
A total of three included studies used the deep convolutional GAN (DCGAN) model. Islam et al. augmented PET data by input random noise. They used the original DCGAN model, which uses BatchNorm to regulate the extracted feature scale, and used LeakyReLU as the activation function to prevent the vanishing gradient problem. Based on this, Kang et al. combined the WGAN model and added a regulation term when calculating the Wasserstein loss to increase training stability. They also trained two different GAN networks to generate both Aβ negative and positive images to improve the generalization of the model. Kang et al. made some improvements to the architecture of DCGAN for the spatial normalization of PET images. First, they used PET images in native space, rather than random noise, as the input of the generator. Second, they used MRI-based spatial normalization results as ‘real’ data to generate template-like images.
Another two included studies used other types of GAN model. Pan et al. used a 3D cycle-consistency GAN for the generation of PET images from magnetic resonance images that have two sets of generators and discriminators to ensure that the generated image is not only similar to the real image but also corresponds to the input magnetic resonance image. Kim et al. used a boundary equilibrium GAN (BEGAN) to extract features from PET images. Different from other studies, the discriminator and generator they used are trained to maximize and minimize the distance between the real and fake image reconstruction loss rather than the data distribution, respectively, which reduces the mode collapse and the training imbalance between the generator and discriminator.
Quantitative assessment and methods compared
To ensure the application value of the research, researchers need to conduct quantitative assessments (setting specific indicators for calculation and evaluation, which is different from simple visual judgment) and compare them with other methods (other algorithms, manual methods, etc.). Therefore, statistics are calculated on quantitative assessment indicators and comparison methods of all included studies (shown in Table 6-8), with particular attention to assessments and comparisons that clinicians were involved in.
Table 6:
Quantitative assessment indicators of image quality and related studies.
| Quantitative assessment indicators of image quality | |||||
|---|---|---|---|---|---|
| Study | PSNR | SSIM | Mean squared error (MSE), NMSE, RMSE | DSC | Manual scoring |
| Wang et al. (2018) | Yes | No | Yes | No | No |
| Wang et al. (2019) | Yes | Yes | No | No | No |
| Ouyang et al. (2019) | Yes | Yes | Yes | No | Yes |
| Shi et al. (2019) | No | No | No | Yes | No |
| Oh et al. (2020) | No | No | No | Yes | Yes |
| Baumgartner et al. (2018) | No | No | No | No | No |
| Kang et al. (2020) | No | No | No | No | No |
| Kang et al. (2018) | No | No | No | No | No |
| Choi et al. (2018) | No | Yes | No | No | No |
| Pan et al. (2018) | Yes | No | No | No | No |
| Islam and Zhang (2020) | Yes | Yes | No | No | No |
| Kim et al. (2020) | No | No | No | No | No |
| Wegmayr et al. (2019) | No | No | No | No | No |
| Yan et al. (2018) | No | Yes | No | No | No |
| Bowles et al. (2018) | No | No | No | No | No |
Note: The PSNR is used to measure the ratio between the maximum possible intensity value and the MSE of the synthetic and real images. SSIM is used to find the similarities within pixels of two images. MSE, NMSE, and RMSE are used to measure the voxelwise intensity differences between the real and estimated images. The DSC is used to measure the voxelwise intensity differences between the real and estimated images.
Table 8:
Methods compared in included studies.
| Comparison method | Study | Detail of method |
|---|---|---|
| The method based on GAN synthesized data vs. the method based on real data | Islam and Zhang (2020) | The method using real PET data |
| Pan et al. (2018) | LM3IL-C (using real PET and MRI data) | |
| Yan et al. (2018) | The method using real data | |
| Kang et al. (2020) | The method using real data | |
| GAN vs. the algorithm removed a specific part | Bowles et al. (2018) | WGAN method without reweighting |
| Wang et al. (2019) | Method without autocontext | |
| Ouyang et al. (2019) | Network without perceptual loss; network using VGG16 perceived loss | |
| Oh et al. (2020) | pix2pix_unet method (u-net replacing residual block) | |
| GAN vs. the generator (removed adversarial training) | Wang et al. (2018) | 3D U-net-like model (without adversarial training) |
| Wang et al. (2019) | Generating network (without adversarial training) | |
| Shi et al. (2019) | The generative network named UG-net (without adversarial training) | |
| Oh et al. (2020) | U-net method (without adversarial training) | |
| GAN vs. other mature algorithms | Kim et al. (2020) | 2D-CNN (method of Glozman et al.) |
| Pan et al. (2018) | ROI; VGD; LLEP (using hand-crafted features); LDSIL; LDMIL (using only MRI data) | |
| Yan et al. (2018) | Traditional data augmentation method (images are randomly horizontally and vertically flipped) | |
| Wegmayr et al. (2019) | WGAN* Conversion prediction; Indirect conversion prediction; Direct conversion prediction | |
| Wang et al. (2018) | CNN method; m-SR method; t-DL method | |
| Wang et al. (2019) | m-SR method; t-DL method; m-CAA method; autoCNN method | |
| Kang et al. (2018) | Average template; CAE | |
| Choi et al. (2018) | The method based on PET template; the method based on multiatlas PET template; the method based on PET segmentation; the method based on real MR | |
| Shi et al. (2019) | Sparse coding and dictionary learning method; CNN | |
| Baumgartner et al. (2018) | Guided backprop; integrated gradients; CAM; additive perturbation | |
| GAN vs. the manual method | Shi et al. (2019) | The clinician manual method |
Note: LM3IL-C, GAN that uses only complete MRI and PET data; UG-net, a GAN model with the modified U-net; VGD, voxelwise GM density.
Table 7:
Quantitative assessment of classification effect indicators and related studies.
| Quantitative assessment indicators of classification effect | ||||||
|---|---|---|---|---|---|---|
| Study | Accuracy (ACC) | Sensitivity (SEN) | Specificity (SPE) | AUC | F1-score | Recall |
| Pan et al. (2018) | Yes | Yes | Yes | Yes | Yes | No |
| Islam and Zhang (2020) | Yes | No | No | No | No | No |
| Kim et al. (2020) | Yes | Yes | Yes | Yes | No | No |
| Wegmayr et al. (2019) | Yes | No | No | No | Yes | Yes |
| Yan et al. (2018) | Yes | No | No | Yes | No | No |
| Bowles et al. (2018) | No | No | No | No | No | No |
Note: AUC means the area under the receiver operating characteristic curve (ROC) curve; F1-score means the harmonic average of precision and recall.
Most studies commonly used classification and image quality evaluation indicators, such as accuracy (ACC), area under the curve, PSNR, and DSC. Clinicians participated in the evaluation in only a few studies. Ouyang et al. recruited two clinicians to score image quality and judge amyloid status (Ouyang et al., 2019). Oh et al. also used five observers to score the quality of segmentation (Oh et al., 2020).
In terms of comparison methods, the comparison carried out in the included research can be classified into the following categories: (i) comparison with a method based on real data; (ii) comparison with own algorithm removing a specific part; (iii) comparison with the generator but without the adversarial training; (iv) comparison with other mature algorithms; and (v) comparison with the clinician manual method. Only one of these studies compared research methods with clinician manual methods (Shi et al., 2019).
Discussion
The GAN was found to be an emerging deep learning algorithm that has advantages in the diagnosis of AD. In the classification of AD state, the accuracy is significantly better than other algorithms. In the application of AD-related image processing, the image quality after GAN noise reduction and the accuracy of segmentation based on the GAN are higher. The quantitative assessment indicators and comparison methods of GANs are diverse; however, there is a lack of participation by clinicians.
The clinical significance of AD images processing
Images with more details after denoising
The quality of low-dose PET images in the clinical diagnosis of AD are significantly worse than that of full-dose PET images, having more noise and fewer functional details. Wang et al. obtained images with higher PSNR by 3D c-GANs and LA-GANs, improving the quality of low-dose PET images (Wang et al., 2018; Wang et al., 2019). Ouyang et al. ensured the accuracy of amyloid status after image denoising (Ouyang et al., 2019). These images with more details are helpful for clinicians to diagnose AD accurately (Ouyang et al., 2019). The included studies showed that researchers could obtain more accurate classification results by inputting the denoised images.
In addition, AD-related clinical trials are gradually considering the inclusion of young, normal cognitive decline subjects, so it is important to reduce the radiation dose and the risk of radiation exposure (Huang et al., 2009). Therefore, through noise reduction, researchers can not only obtain high-quality pictures and precise diagnostic information, but also can reduce the potential health damage to patients and normal controls..
Located AD state features through segmentation
The precise segmentation of brain images is conducive to locating AD state features. Shi et al. realized the accurate segmentation of hippocampal subfields (CA1, CA2, DG, CA3, Head, Tail, SUB, ERC, and PHG) (Shi et al., 2019). The volume or morphology of these areas are closely related to AD and MCI (Nestor et al., 2013; Hobbs et al., 2016). Oh et al. segmented the white matter compartment of the brain on 18F-FDG PET/CT images using a GAN model (Oh et al., 2020). Quantitative analysis of 18F-FDG PET/CT in the white matter has certain potential for the diagnosis of AD. A classification framework can be established based on processing extracted features in these areas.
Visual attribution is a process in which researchers visualize disease features in an image given the category of diseases. Baumgartner et al. obtained feature maps for different subtypes of AD state using a WGAN (Baumgartner et al., 2018). The feature maps of AD patients by visual attribution contribute to segmentation of structures (Pinheiro and Collobert, 2015; Oquab et al., 2015). For clinicians, changes in the featured area in these images are helpful in assessing AD state progression. For disease research, the generated AD feature map helps to stratify the patient population and prove that AD is composed of multiple subtypes rather than a single disease (Iqbal et al., 2005).
Generating more data and modalities
The main challenge of using deep learning is the lack of sufficient data to train a classification framework (Spasov et al., 2019). Due to the relatively high price of 18F-FDG PET and PET/CT, the problem of lack of data is particularly prominent in AD research (Abdellahi et al., 2018). A GAN can augment 18F-florbetaben image data to solve this problem during the development of AD-related deep learning frameworks (Kang et al., 2020).
Currently, the use of multimodality data is a trend of deep learning to diagnose AD (Liu et al., 2018). MRI contains more structural and textural information, while PET contains metabolic information and the value of quantitative analysis. The combined use of MRIF and PET can provide clinicians with more comprehensive diagnostic information. However, in the clinic, it is common to transfer between modalities to supplement the lack of certain modality data. Choi et al. completed the process of converting PET images into magnetic resonance images by applying a GAN (Choi et al., 2018). AD classification algorithms can be developed and trained based on these multimodal data. More information ensures the accuracy of classification.
The clinical significance of the classification of AD state
At present, no drugs can effectively prevent the development of AD (Neugroschl and Wang, 2011). The clinical trial failure rate of AD drugs is as high as 99.6% (Cummings et al., 2014). Therefore, the current focus of treatments has shifted to diagnosing and intervening patients in the early stages of AD (Chong and Sahadevan, 2005; Davis et al., 2018). In our study, we found that AD patients can be distinguished from normal cognitive decline control groups, and high-risk individuals (pMCI) can be identified among MCI (sMCI and pMCI) patients by the GAN-based classification framework with higher accuracy than other algorithms. This provides an opportunity to delay or even reverse disease progression and to reduce the occurrence of AD patients.
In most studies on AD classification, a two-stage deep learning framework is usually established (shown in Fig. 6). The first stage is to synthesize medical images or extract relevant features, and the second step is to establish a classifier for classification (Rathore et al., 2017). Researchers use GANs to synthesize images and perform feature extraction while using other algorithms (such as CNNs) to build classifiers (Islam and Zhang, 2020; Wegmayr et al., 2019). This structure makes full use of the advantages of GANs in image processing. To obtain higher accuracy, accurate feature extraction is often more important than classification algorithms (Sabuncu et al., 2015). A GAN's processing of AD-related images can better help the extraction of AD-related features in the first stage of this framework, which plays a critical role in the improvement of classification accuracy of the model.
Figure 6:
A general framework for GAN-based AD classification.
Why do GANs outperform other conventional deep learning methods in AD-related tasks?
In this systematic review, GANs outperformed other conventional deep learning methods in AD-related tasks. In feature extraction tasks, GANs can better extract the features of brain images of patients with AD/MCI and complete visual attributes (Baumgartner et al., 2018) to classify AD/HC (Kim et al., 2020) or predict the magnetic resonance images after disease progression (Bowles et al., 2018; Wegmayr et al., 2019). In data augmentation tasks, GANs can generate brain PET images similar to the real data according to random noise to augment the research data and enhance the discrimination and generalization performance of classifiers in the next stage (Islam and Zhang, 2020; Kang et al., 2020). In modality transfer tasks, GANs can generate images for one modality based on another (such as the MRI to PET transfer) and retain more anatomical and functional information (Choi et al., 2018; Kang et al., 2018; Pan et al., 2018; Yan et al., 2018). In image denoising tasks, GANs can better remove noise in low-dose PET images and generate more realistic high-dose PET images (Ouyang et al., 2019; Wang et al., 2018; Wang et al., 2019). In image segmentation tasks, GANs achieve results closer to those of expert manual segmentation when segmenting a specific region (such as the hippocampus) in the brain images of AD patients (Shi et al., 2019; Oh et al., 2020).
In general, in AD-related tasks, GANs show a powerful brain image processing performance that other traditional deep learning methods do not have, which may be related to their better adaptability to the processing of medical imaging data (such as MRI, PET, and other AD-related brain image data). First, medical imaging data are mostly complex high-dimensional data, and the adversarial structure of GANs has advantages over other conventional deep learning methods in processing. Conventional deep learning methods (such as CNNs) often require a very large amount of computation to fit high-dimensional data, resulting in relatively poor image quality, and are often affected by blurring and aliasing artifacts (Chen et al., 2017b; Deng et al., 2020). GANs do not require a preset distribution of data. Theoretically, any differentiable function can be used to construct the generator and discriminator, which means that GANs do not need to customize the complex loss function (Isola et al., 2017), can directly approximate the real probability distribution of high-dimensional data with arbitrary accuracy, and can generate high-quality images (Goodfellow et al., 2014; Radford et al., 2015; ). Second, due to the need for long-term follow-up, professional equipment, and analysis by well-trained medical practitioners, medical imaging data for training are scarce and more likely to have class imbalance problems (Sampath et al., 2021). GANs can learn the potential data distribution from the limited available data for generating high-quality images, which results in less data required for training and an advantage in handling unbalanced data (Kazuhiro et al., 2018), whereas other conventional deep learning methods require a great amount of prior knowledge (such as a very large amount of data) for training and may face more overfitting problems when processing small amounts of data (Shorten and Khoshgoftaar, 2019; Sampath et al., 2021).
These technical advantages make GANs more suitable for medical image data processing and can explain our findings in this systematic review. The data augmentation task requires that the deep-learning network use only a small amount of data to learn its characteristics to generate very realistic images. Other conventional data augmentation methods (such as fully visible belief networks, recurrent neural networks, and variational autoencoders) require a very large amount of computation when generating high-dimensional data such as images, resulting in slower generation speed and more artifacts of the generated images. The superior characteristics of GANs in processing high-dimensional data make them perfectly capable for this task (Kazuhiro et al., 2018; Sampath et al., 2021); the modality transfer task needs to complete the nonlinear conversion from one modal image to another. Other conventional deep learning methods often need to customize complex loss functions when solving such problems and need perfect alignment between modifications, which is difficult to achieve in medical practice. The good adaptability of GANs (especially cycleGAN) makes it possible to fully learn the corresponding law between the two modal data, to achieve high-quality modalities transfer without the need for perfect alignment between images, which is also one of the biggest advantages of GANs (Isola et al., 2017; Zhu et al., 2017; Largent et al., 2019); In feature extraction, image denoising, and image segmentation tasks, the deep-learning network needs to fully learn the distribution features of the input image to complete the extraction of disease features, the removal of image noise or the segmentation of specific regions. Although other conventional deep learning methods have achieved some success when performing these tasks, they lose some high-frequency structures and textures (Chen et al., 2017a) and may not ensure the spatial consistency between the output image and the real image when applied to continuous three-dimensional data (such as MRI or PET images) (Yi et al., 2019). GANs solve these problems (Kang et al., 2019; Huo et al., 2018) and can generate normal-appearing images from images with abnormal findings to visualize the effects of the disease (i.e. feature attribution) (Sun et al., 2020).
Data and architecture of GANs
Improve data quality and modalities
Data quality has always been a topic of concern in deep learning research. Although a GAN can process image data with high quality, data from different sources will also have effects on AD classification and other clinical applications. We reviewed the training and validation datasets used in the included studies. For most of the included studies, the training data came from the public database Alzheimer's Disease Neuroimaging Initiative (ADNI), which was launched in 2003 as a public–private partnership. The primary goal of the ADNI is to find more biomarkers (such as MRI, PET, etc.) for the early detection and tracking of AD and MCI. After >10 years of development, the ADNI has formulated a strict project plan (which includes patient type, patient age, cognitive status assessment method, subpopulation allocation plan, etc.) to ensure the comparability of the data and has included >1500 AD, MCI or HC subjects from 57 sites across the USA and Canada, which has made it one of the largest neuroimaging databases of AD and MCI (Burton, 2011). Compared with other medical fields, such a large database facilitates the development and training of deep learning frameworks and alleviates the problem of a lack of deep learning data. At the same time, some studies cooperate with medical institutions, using clinically collected data for training. However, in this systematic review, we found that simply using samples from clinical cases may lead to a lower sample size for training, resulting in problems such as poor generation effects and overfitting. The amount of data is a decisive factor in the effectiveness of the deep learning model (Parmar et al., 2018). Therefore, we suggest that researchers using clinically collected data for training try to use a public database (such as ADNI) with a large data volume and high data quality to expand their sample size to improve the generation effect and generalization ability of the GAN model.
For validation data, we noticed that most of the data used to train and validate algorithms were from the same dataset; that is, only the internal validation method was used to evaluate the performance of the GAN model, which can be divided into cross-validation and split-sample validation (Park and Han, 2018; Kim et al., 2020a; Kulkarni et al., 2021). Cross-validation (such as the leave-one-site-out method) takes the previous training data as validation data in iterative training, which can improve training efficiency and the fitting degree of the model to the dataset and can make full use of the dataset. However, it can also result in the ‘leakage’ of the information in the training set to the verification set, which leads to overfitting and overestimation of the model capability (Park and Han, 2018). Split-sample validation uses a small part of the data that is randomly split from the dataset and kept unused for training to evaluate the performance of the algorithm. Although it will not cause the “leakage” problem, spectrum bias and overfitting cannot be avoided (Park and Kressel, 2018; Park and Han, 2018). In general, many studies have suggested that although internal validation can effectively evaluate the technical performance of deep learning models, it may also lead to insufficient generalization performance in real-world, high-volume clinical environments (Zech et al., 2018; Salehinejad et al., 2021). Therefore, using data from clinical collections or external institutions as validation data is important to solve such problems, and researchers can consider using large databases such as ADNI to train deep learning frameworks and use clinically collected data for verification, as Kim et al.
We also checked the input/output modalities of data used by the included studies. Most of the studies only used single modal data (PET or MRI) for AD classification or image processing, while the use of multimodal data was less common. Pan et al. and Yan et al. both use the two-stage deep learning architecture. In the first stage, a GAN is used to generate the missing PET image according to the magnetic resonance images, and then the generated PET image and original magnetic resonance images are input into the CNN classifier for classification. The training effect is better than using MRI data alone, which is similar to the results obtained by Li et al. and proves the good efficiency of the multimodal classification method (Li et al., 2014). In addition, at present, the MRI data used in most PET to MRI modality transfer studies are structural magnetic resonance images (such as T1-weighted MRI) (Hu et al., 2021), which may not be able to synthesize PET images that reflect brain metabolism. Wang et al. creatively combined T1-weighted MRI, which reflects brain structure, with DTI (a kind of functional MRI), which reflects brain function, to synthesize PET images. This modality transfer method based on multimodal MRI has achieved good results and provided insight for subsequent research. However, there were few data samples used in this study, and there is still a lack of follow-up research after expanding the samples.
We also analyzed the training sample size using different modal data. Among the included studies, the sample size of studies using PET images was generally small, which may be due to the expensive cost of obtaining PETs and the relative shortage of MRI–PET paired data in public datasets such as ADNI (Zhang et al., 2012), which suggests that we need to add more of this type of data when building public databases. In addition, nearly half of the included studies used partial 2D slices in MRI or PET images instead of the whole 3D image for training, which may cause the loss of spatial information and discontinuous estimation (Nie et al., 2018). However, a recent study found that using the whole 3D image may increase the scale of the GAN model and then affect the generation efficiency (Yu et al., 2018). Therefore, how to apply 3D image data to train the GAN more efficiently remains to be studied.
Toward better GAN architecture
We checked the characteristics of the GAN architecture used by different image processing tasks. We found that most studies on image-to-image tasks (image denoising, image segmentation, and modality transfer) used the cGAN model, which is a supervised model proposed by Mirza et al. and uses a conditional variable C constraint generator and discriminator to generate the specified target image (Mirza and Osindero, 2014). In the image-to-image task of medical images, the input image is used as the conditional variable C so that cGAN can perform corresponding processing according to the image and obtain the desired output image (Sundar et al., 2020). This GAN model has been proven to achieve good performance in medical imaging denoising (Sundar et al., 2020), segmentation (Yu et al., 2018) and modality transfer (Kawahara and Nagata, 2021), which is also supported by our systematic review. In image feature extraction, most studies use the WGAN model, which was first proposed by Arjovsky et al. This model uses Wasserstein loss instead of Jensen Shannon divergence to avoid modal collapse and makes the training gradient meet the Lipschitz continuity to solve the problem of training difficulty and instability (Radford et al., 2015). This model can minimize the distance between the real and generated distribution so that it can better extract meaningful features in the image and complete the feature attribute task. In the noise-to-image task, most studies choose the DCGAN. This model was proposed by Radford et al. and combines a CNN in supervised learning and a GAN in unsupervised learning, which can improve the stability of training and the quality of generated images (Radford et al., 2015) and is widely used in medical image data augmentation (Kazuhiro et al., 2018). Kang et al. and Islam et al. both used this model to amplify PET data and achieved good results, which confirmed the advantages of this model in data augmentation tasks. In addition, the GAN model used by Pan et al. and Kim et al. also puts forward a new direction for future research. The cycle GAN used by Pan et al. was proposed by Zhu et al. This model creatively uses two sets of generators and discriminators to learn the mapping relationship between the two modified data and then completes the modality transfer without paired data, which has great clinical application potential and research value (Zhu et al., 2017). The BEGAN used by Kim et al. was proposed by Berthelot et al., which generates data by estimating the error of distribution rather than the difference between generated data and real data, which improves the generation stability (Berthelot et al., 2017). However, its application effect in high-resolution images is poor, and its application in medical images is limited, requiring further study.
We also checked the GAN architecture improvement methods used in different studies, most of which aimed to improve training stability and image generation quality (such as improvements to generators, discriminators, and loss functions). However, some studies have improved the GAN model according to the characteristics of AD image data; for example, Ouyang et al. used an extra amyloid status classifier to make the generated image fit to the patient's real amyloid status. Kang et al. used two different GAN modalities to generate Aβ negative and positive images to improve the generalization. This task-specific improvement can make the GAN model better meet the needs of clinical application and has high reference value.
Suggestions on quantitative assessment and method comparison
To ensure that the developed algorithms can be applied in clinical practice, quantitative assessment and comparison methods are worthy of attention. Quantitative assessment can detect factors that reduce generalization performance and evaluate the applicability of training datasets (Kang et al., 2020). Judging from the present results of the included studies, the quantitative assessment indicators of the studies with the same application purpose were not uniform. This is one of the reasons why this article only carried out a systematic review and failed to carry out a meta-analysis. Future research can propose reference assessment indicators for different purposes, such as image segmentation, image denoising, and modality transfer. This can facilitate horizontal comparisons between studies.
Judging from the summary of the comparison methods here, we recommend that researchers consider at least the following three comparison methods: (i) comparison with algorithms removing a specific part; (ii) comparison with other mature algorithms; and (iii) comparison with clinician manual methods. In Wang et al., Shi et al., and Oh et al., the advantages of GANs can be highlighted by comparison with algorithms removing part of the adversarial training (Wang et al., 2018; Shi et al., 2019; Wang et al., 2019; Oh et al., 2020). Therefore, this comparison method is necessary in research based on GANs. Comparison with other published algorithms can help show the advantages and application potential of the algorithm.
From the results of the included studies, the process of evaluation of the algorithm lacks clinician participation. This limits that studies break through the barriers from development to clinical application. The purpose of algorithm training is to reach the level of the clinician and realize automatic diagnosis. The clinician's evaluation reflects the clinical application effect of the algorithm. Whether some key clinical information can be retained in the process of image processing can only be known through the evaluation of clinicians. In addition, it is difficult for clinicians to obtain an intuitive understanding of the application effect of the algorithm from the only objective quantitative assessment indicators (e.g., PSNR, DSC) due to barriers between specialties. This will affect the attitude of clinicians to use algorithms. Therefore, we strongly recommend recruiting clinicians to evaluate the algorithm in future research. Specifically, from the aspect of quantitative assessment, clinicians make rules to score images after image denoising, segmentation and other processing. Then, the scores of different algorithms are compared. Ouyang et al. and Oh et al. reported the advantages of the GAN algorithm by scoring (Ouyang et al., 2019; Oh et al., 2020). From the aspect of method comparison, researchers can consider comparing the effect of clinician manual methods with that of algorithms. For example, Shi et al. showed a good segmentation effect in the hippocampus from magnetic resonance images compared with the manual segmentation method (Shi et al., 2019).
Limitations of GANs in psychoradiology and AD-related tasks
There are also some common problems with the GAN algorithm itself. For example, GANs are difficult to train. During training, the generator and discriminator often fail to balance well, which may cause problems such as pattern collapse and gradient disappearance and which results in the generator stopping the training after learning only part of the distribution pattern of the data and not converging to global Nash equilibrium (Mertikopoulos et al., 2018; Wiatrak and Albrecht, 2019). Also, the neural network needs good initialization during the training of GANs; otherwise, the learned distribution may still be far from the real distribution, resulting in cyclic, oscillating, or diverting behavior (Goodfellow, 2014; Mertikopoulos et al., 2018; Wiatrak and Albrecht, 2019). In addition, the generator of GANs can only learn an end-to-end mapping function, which does not have explicit expression. Therefore, the interpretability of GANs is poor, and the corresponding relationship between their latent space and the generated image is not clear, which is like a “black box” for researchers (Zhou et al., 2016). Some researchers have proposed optimized GAN models to solve the above problems (such as cGAN, WGAN, and cycleGAN) (Radford et al., 2015; Zhu et al., 2017; Sundar et al., 2020), but GANs still need to be further optimized to fully achieve their optimal generation performance.
The application of GANs in psychoradiology and AD-related tasks still has some limitations. At present, a GAN is mainly used in the processing of AD-related medical images, but its application in other mental illnesses (such as schizophrenia, autism, attention deficit hyperactivity disorder, etc.) is still lacking (Li et al., 2021; Ntelemis et al., 2021). The psychoradiologic data used to study these mental diseases are similar to AD, while more complex and higher-dimensional data (such as functional MRI data) are often used to complete the clustering and classification of those diseases. Deep learning methods (such as CNNs) have been gradually applied to the processing of imaging data of these mental diseases and have achieved some promising results, but their ability to process functional MRI and other high-dimensional psychoradiologic data needs to be improved (Li et al., 2021). Therefore, it is very promising for GANs, which outperform other conventional deep learning methods in processing high-dimensional data, to be applied to these diseases. Meanwhile, the application of a GAN in the field of AD state can be extended to the field of bioinformatics, such as the use of a GAN to analyze AD molecular data (Park et al., 2020). The lack of data in bioinformatics is also a tricky problem. The ability of GANs to amplify data in image processing can be transferred to bioinformatics research (Lan et al., 2020). In addition, in this systematic review we found that researchers paid little attention to the clinical information contained in the image (such as amyloid status) when conducting AD-related research (Sorin et al., 2020). In the future, when studying the application of GANs to AD-related tasks, algorithm researchers should work closely with psychoradiologists to ensure the consistency of clinical information provided by images before and after processing (Yang et al., 2021).
Conclusion
The application value of a GAN in the classification of AD state and AD-related image processing has been confirmed in reviewed studies. Compared with other methods, GAN classification is more accurate, the image quality after denoising is higher, and the image segmentation is more accurate. In the future, researchers need to consider using better data and GAN architecture and comparing algorithms with clinician manual methods and recruiting clinicians to evaluate the effect of the algorithm.
Acknowledgments
This work was supported by grants from National Key Research and Development Project (2018YFC1704605), National Natural Science Foundation of China (81401398), Sichuan Science and Technology Program (2019YJ0049), Sichuan Provincial Health and Family Planning Commission (19PJ080), National College Students' innovation and entrepreneurship training program (C2021116624) and Chinese Postdoctoral Science Foundation (2013M530401). Dr Gong was also supported by the US-China joint grant (Grant No. NSFC81761128023) and NIH/NIMH R01MH112189-01.
Notes
Huaxi MR Research Center (HMRRC), Department of Radiology, West China Hospital of Sichuan University, Chengdu, China.
Contributor Information
Changxing Qu, Huaxi MR Research Center (HMRRC), Department of Radiology, West China Hospital of Sichuan University, Chengdu 610044, China; State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, West China School of Stomatology, Sichuan University, Chengdu 610044, China.
Yinxi Zou, West China School of Medicine, Sichuan University, Chengdu 610044, China.
Qingyi Dai, State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, West China School of Stomatology, Sichuan University, Chengdu 610044, China.
Yingqiao Ma, Huaxi MR Research Center (HMRRC), Department of Radiology, West China Hospital of Sichuan University, Chengdu 610044, China.
Jinbo He, School of Psychology, Central China Normal University, Wuhan 430079, China.
Qihong Liu, College of Biomedical Engineering, Sichuan University, Chengdu 610065, China.
Weihong Kuang, Department of Psychiatry, West China Hospital of Sichuan University, Chengdu 610065, China.
Zhiyun Jia, Huaxi MR Research Center (HMRRC), Department of Radiology, West China Hospital of Sichuan University, Chengdu 610044, China.
Taolin Chen, Huaxi MR Research Center (HMRRC), Department of Radiology, West China Hospital of Sichuan University, Chengdu 610044, China; Research Unit of Psychoradiology, Chinese Academy of Medical Sciences, Chengdu 610041, Sichuan, P.R. China; Functional and Molecular Imaging Key Laboratory of Sichuan Provience, Department of Radiology, West China Hospital of Sichuan University, Chengdu 610041, Sichuan, P.R. China.
Qiyong Gong, Huaxi MR Research Center (HMRRC), Department of Radiology, West China Hospital of Sichuan University, Chengdu 610044, China; Research Unit of Psychoradiology, Chinese Academy of Medical Sciences, Chengdu 610041, Sichuan, P.R. China; Functional and Molecular Imaging Key Laboratory of Sichuan Provience, Department of Radiology, West China Hospital of Sichuan University, Chengdu 610041, Sichuan, P.R. China.
Author Contributions
C.Q., Y.Z., and T.C. reviewed articles and wrote the initial manuscript and proposed an article outline. T.C., Q.G., Z.J., J.H., and Y.M. revised the drafts and gave suggestion for the development of the paper. T.C. and Y.M. contributed to the polish of language. Y.Z. and Q.D. contributed to the making of figures and tables. WK and Q.L. explained the principle of the algorithm. T.C. and Y.Z. approved the final version of the manuscript for its submission to this journal.
Conflict of interest statement
One of the authors, Dr Qiyong Gong, is also the editor-in-chief of Psychoradiology. He was blinded from reviewing or making decisions on the manuscript.
References
- Abdellahi M, Karamian E, Najafinezhad Aet al. (2018) Diopside-magnetite; a novel nanocomposite for hyperthermia applications. J Mech Behav Biomed Mater. 77:534–8. [DOI] [PubMed] [Google Scholar]
- Armanious K, Küstner T, Reimold Met al. (2019) Independent brain F-18-FDG PET attenuation correction using a deep learning approach with Generative Adversarial Networks. Hellenic J Nucl Med. 22:179–86. [DOI] [PubMed] [Google Scholar]
- Armanious K, Hepp T, Küstner Tet al. (2020) Independent attenuation correction of whole body [18F]FDG-PET using a deep learning approach with Generative Adversarial Networks. EJNMMI Res. 10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner CF, Koch LM, Tezcan KCet al. (18-23 June 2018) Visual feature attribution using Wasserstein GANs. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). Salt Lake City, UT, USA:IEEE, 8309–19. [Google Scholar]
- Berthelot D, Schumm T, Metz L (2017) BEGAN: boundary equilibrium generative adversarial networks. arXiv. [Google Scholar]
- Biffi C, Cerrolaza JJ, Tarroni Get al. (2020) Explainable anatomical shape analysis through deep hierarchical generative models. IEEE Trans Med Imaging. 39:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles C, Gunn R, Hammers Aet al. (2018) Modelling the progression of Alzheimer's disease in MRI using generative adversarial networks. Medical Imaging 2018: Image Processing. 10574. [Google Scholar]
- Brookmeyer R, Johnson E, Ziegler-Graham Ket al. (2007) Forecasting the global burden of Alzheimer's disease. Alzheimers & Dementia. 3:186–91. [DOI] [PubMed] [Google Scholar]
- Burton A (2011) Big science for a big problem: ADNI enters its second phase. Lancet Neurol. 10:206–7. [DOI] [PubMed] [Google Scholar]
- Chen Hu, Zhang Yi, Zhang Wet al. (2017a) Low-dose CT via convolutional neural network. Biomed Optics Expr. 8:679–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Hu, Zhang Yi, Kalra MKet al. (2017b) Low-dose CT with a residual encoder-decoder convolutional neural network. IEEE Trans Med Imaging. 36:2524–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Lee DS (2018) Generation of structural MR images from amyloid PET: application to MR-less quantification. J Nucl Med. 59:1111–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MS, Sahadevan S (2005) Preclinical Alzheimer's disease: diagnosis and prediction of progression. Lancet Neurol. 4:576–9. [DOI] [PubMed] [Google Scholar]
- Cummings JL, Morstorf T, Zhong K (2014) Alzheimer's disease drug-development pipeline: few candidates, frequent failures. Alzheimer's Res Ther. 6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M, O`Connell T, Johnson Set al. (2018) Estimating Alzheimer's disease progression rates from normal cognition through mild cognitive impairment and stages of dementia. Curr Alzheimer Res. 15:777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Mo, Goy A, Li Set al. (2020) Probing shallower: perceptual loss trained Phase Extraction Neural Network (PLT-PhENN) for artifact-free reconstruction at low photon budget. Opt Express. 28:2511–35. [DOI] [PubMed] [Google Scholar]
- Goodfellow IJ (2014) On distinguishability criteria for estimating generative models. Statistics. [Google Scholar]
- Goodfellow IJ, Pouget-Abadie J, Mirza Met al. (2014) Generative adversarial nets. Advances in Neural Information Processing Systems 27 (NIPS 2014). 27:2672–80. [Google Scholar]
- Hobbs KH, Zhang P, Shi Bet al. (2016) Quad-mesh based radial distance biomarkers for Alzheimer's disease. 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI). Prague, Czech Republic: IEEE. [Google Scholar]
- Hu S, Yu W,Chen Zet al. (2020) Medical Image Reconstruction Using Generative Adversarial Network for Alzheimer Disease Assessment with Class-Imbalance Problem. 2020 IEEE 6th International Conference on Computer and Communications (ICCC):1323–27. [Google Scholar]
- Hu S, Lei B, Wang Set al. (2021) Bidirectional mapping generative adversarial networks for brain MR to PET synthesis. IEEE Trans Med Imaging. Chengdu, China: IEEE. [DOI] [PubMed] [Google Scholar]
- Huang B, Law MW, Khong PL (2009) Whole-body PET/CT scanning: estimation of radiation dose and cancer risk. Radiology. 251:166–74. [DOI] [PubMed] [Google Scholar]
- Huo Y, Xu Z, Bao Set al. (2018) Splenomegaly Segmentation using Global Convolutional Kernels and Conditional Generative Adversarial Networks. Conference on Medical Imaging - Image Processing 10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SJ, Tao Z, Kim WHet al. (2018) Conditional recurrent flow: conditional generation of longitudinal samples with applications to neuroimaging. University of Wisconsin-Madison; University of Wisconsin-Madison USA; University of Texas at Arlington. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Flory M, Khatoon Set al. (2005) Subgroups of Alzheimer's disease based on cerebrospinal fluid molecular markers. Ann Neurol. 58:748–57. [DOI] [PubMed] [Google Scholar]
- Islam J, Zhang Y (2020) GAN-based synthetic brain PET image generation. Brain Informatics. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isola P, et al. (2017) Image-to-image translation with conditional adversarial networks. 30th IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). Honolulu, HI, USA:IEEE;5967–76. [Google Scholar]
- Kang SK, Seo S, Shin SAet al. (2018) Adaptive template generation for amyloid PET using a deep learning approach. Hum Brain Mapp. 39:3769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E, Koo HJ, Yang DHet al. (2019) Cycle-consistent adversarial denoising network for multiphase coronary CT angiography. Med Phys. 46:550–62. [DOI] [PubMed] [Google Scholar]
- Kang H, Park J-S, Cho Ket al. (2020) Visual and quantitative evaluation of amyloid brain PET image synthesis with generative adversarial network. Appl Sci. 10:2628. [Google Scholar]
- Kawahara D, Nagata Y (2021) T1-weighted and T2-weighted MRI image synthesis with convolutional generative adversarial networks. Rep Prac Oncol Radiother. 26:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuhiro K, Werner RA, Toriumi Fet al. (2018) Generative adversarial networks for the creation of realistic artificial brain magnetic resonance images. Tomography. 4:159–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HW, Lee HaE, Lee Set al. (2020) Slice-selective learning for Alzheimer's disease classification using a generative adversarial network: a feasibility study of external validation. Eur J Nucl Med Mol Imaging. 47:2197–206. [DOI] [PubMed] [Google Scholar]
- Kim DW, Jang HY, Ko Yet al. (2020a) Inconsistency in the use of the term “validation” in studies reporting the performance of deep learning algorithms in providing diagnosis from medical imaging. PLoS ONE. 15:e0238908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Watanabe A, Yamada Tet al. (2020) AI approach of cycle-consistent generative adversarial networks to synthesize PET images to train computer-aided diagnosis algorithm for dementia. Ann Nucl Med. 34:1–4. [DOI] [PubMed] [Google Scholar]
- Kulkarni V, Gawali M, Kharat A (2021; Key technology considerations in developing and deploying machine learning models in clinical radiology practice. JMIR Medical Informatics. 9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, You L, Zhang Zet al. (2020) Generative adversarial networks and its applications in biomedical informatics. Front Pub Health. 8:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Largent A, Barateau A, Nunes J-Cet al. (2019) Comparison of deep learning-based and patch-based methods for pseudo-CT generation in MRI-based prostate dose planning. Int J Radiat Oncol Biol Phys. 105:1137–50. [DOI] [PubMed] [Google Scholar]
- Li F, Sun H, Biswal BBet al. (2021) Artificial intelligence applications in psychoradiology. Psychoradiology. 2:94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhang W, Suk H-Iet al. (2014) Deep learning based imaging data completion for improved brain disease diagnosis. 17th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI). Boston, MA, USA:Springer, Cham, 8675:305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen K, Wu Tet al. (2018) Use of multimodality imaging and artificial intelligence for diagnosis and prognosis of early stages of Alzheimer's disease. Transl Res. 194:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Nai Y-H, Chen Cet al. (2020) Deep Learning-Based Estimation of Non-Specific Uptake in Amyloid-PET Images from Structural MRI for Improved Quantification of Amyloid Load in Alzheimer's Disease. 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS):573–78. [Google Scholar]
- Lui Su, Zhou XJ, Sweeney JAet al. (2016) Psychoradiology: the frontier of neuroimaging in psychiatry. Radiology. 281:357–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertikopoulos P, Papadimitriou C, Piliouras Get al. (2018) Cycles in adversarial regularized learning. 29th Annual ACM-SIAM Symposium on Discrete Algorithms (SODA). New Orleans, LA, USA:Assoc Comp Machinery, SIAM, 2703–17. [Google Scholar]
- Mirza M, Osindero S (2014) Conditional generative adversarial nets. Comput Sci. arXiv. 2672–80. [Google Scholar]
- Nestor SM, Gibson E, Gao Fu-Qet al. (2013) A direct morphometric comparison of five labeling protocols for multi-atlas driven automatic segmentation of the hippocampus in Alzheimer's disease. Neuroimage. 66:50–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugroschl J, Wang S (2011) Alzheimer's disease: diagnosis and treatment across the spectrum of disease severity. Mount Sinai J Med. 78:596–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie D, Trullo R, Lian Jet al. (2018) Medical image synthesis with deep convolutional adversarial networks. IEEE Trans Biomed Eng. 65:2720–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntelemis F, Jin Y, Thomas SA (2021) Image clustering using an augmented generative adversarial network and information maximization. IEEE Trans Neural Netw Learning Syst. arXiv. [DOI] [PubMed] [Google Scholar]
- Oh KT, Lee S, Lee Het al. (2020) Semantic segmentation of white matter in FDG-PET using generative adversarial network. J Digit Imaging. 33:816–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oquab M, Bottou L, Laptev Iet al. (2015) Is object localization for free? Weakly-supervised learning with convolutional neural networks. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Boston, MA, USA:IEEE, 685–94. [Google Scholar]
- Ouyang J, Chen KT, Gong Eet al. (2019) Ultra-low-dose PET reconstruction using generative adversarial network with feature matching and task-specific perceptual loss. Med Phys. 46:3555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YS, Liu M, Lian Cet al. (2018) Synthesizing missing PET from MRI with cycle-consistent generative adversarial networks for Alzheimer's disease diagnosis. Med Image Comput Comput Assist Interv, Pt Iii. 11072:455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SHo, Kressel HY (2018) Connecting technological innovation in artificial intelligence to real-world medical practice through rigorous clinical validation: what peer-reviewed medical journals could do. J Korean Med Sci. 33:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SHo, Han K (2018) Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 286:800–9. [DOI] [PubMed] [Google Scholar]
- Park J, Kim H, Kim Jet al. (2020) A practical application of generative adversarial networks for RNA-seq analysis to predict the molecular progress of Alzheimer's disease. PLoS Comput Biol. 16:e1008099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar C, Barry JD, Hosny Aet al. (2018) Data analysis strategies in medical imaging. Clin Cancer Res. 24:3492–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256:183–94. [DOI] [PubMed] [Google Scholar]
- Pinheiro PO, Collobert R (2015) From image-level to pixel-level labeling with convolutional networks. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Boston, MA, USA: IEEE, 1713–21. [Google Scholar]
- Radford A, Metz L, Chintala S (2015) Unsupervised representation learning with deep convolutional generative adversarial networks. Computer Ence. [Google Scholar]
- Rathore S, Habes M, Iftikhar MAet al. (2017) A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer's disease and its prodromal stages. Neuroimage. 155:530–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury S (2020) A Modular Framework to Predict Alzheimer’s Disease Progression Using Conditional Generative Adversarial Networks. 2020 International Joint Conference on Neural Networks (IJCNN):1–8. [Google Scholar]
- Sabuncu MR, Konukoglu E (2015) Clinical prediction from structural brain MRI scans: a large-scale empirical study. Neuroinformatics. 13:31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehinejad H, Kitamura J, Ditkofsky Net al. (2021) A real-world demonstration of machine learning generalizability in the detection of intracranial hemorrhage on head computerized tomography. Sci Rep. 11:17051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath V, Maurtua I, Aguilar Martín JJet al. (2021) A survey on generative adversarial networks for imbalance problems in computer vision tasks. J Big Data. 8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Cheng K, Liu Z (2019) Hippocampal subfields segmentation in brain MR images using generative adversarial networks. Biomed Eng Online. 18:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorten C, Khoshgoftaar TM (2019) A survey on image data augmentation for deep learning. J Big Data. 6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin V, Barash Y, Konen Eet al. (2020) Creating artificial images for radiology applications using generative adversarial networks (GANs) - a systematic review. Acad Radiol. 27:1175–85. [DOI] [PubMed] [Google Scholar]
- Spasov S, Passamonti L, Duggento Aet al. (2019) A parameter-efficient deep learning approach to predict conversion from mild cognitive impairment to Alzheimer's disease. Neuroimage. 189:276–87. [DOI] [PubMed] [Google Scholar]
- Sun L, Wang J, Huang Yet al. (2020) An adversarial learning approach to medical image synthesis for lesion detection. IEEE J Biomed Health Informatics. 24:2303–14. [DOI] [PubMed] [Google Scholar]
- Sundar S, Iommi LK, Muzik Det al. (2020) Conditional Generative Adversarial Networks (cGANs) aided motion correction of dynamic 18 F-FDG PET brain studies. J Nucl Med. 62:871–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu B, Wang Let al. (2018) 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage. 174:550–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou L, Yu Bet al. (2019) 3D auto-context-based locality adaptive multi-modality GANs for PET synthesis. IEEE Trans Med Imaging. 38:1328–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmayr V, Horold M, Buhmann JM (2019) Generative aging of brain MRI for early prediction of MCI-AD conversion. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 1042–6. [Google Scholar]
- Wiatrak M, Albrecht SV (2019) Stabilizing generative adversarial network training: a survey. arXiv. 13. [Google Scholar]
- Yan Y, Lee H, Somer Eet al. (2018) Generation of amyloid PET images via conditional adversarial training for predicting progression to Alzheimer's disease. Pred Intell Med. 11121:26–33. [Google Scholar]
- Yang Z, Nasrallah IM, Shou Het al. (2021) Disentangling brain heterogeneity via semi-supervised deep-learning and MRI: dimensional representations of Alzheimer's disease. arXiv. [Google Scholar]
- Yi X, Walia E, Babyn P (2019) Generative adversarial network in medical imaging: a review. Med Image Anal. 58:101552. [DOI] [PubMed] [Google Scholar]
- Yu B, Zhou L, Wang Let al. (2018) 3D cgan based cross-modality MR image synthesis for brain tumor segmentation. 15th IEEE International Symposium on Biomedical Imaging (ISBI). Washington, DC, USA:IEEE, 626–30. [Google Scholar]
- Zech JR, Badgeley MA, Liu Met al. (2018) Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 15:e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shen D (2012) Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer's disease. Neuroimage. 59:895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Khosla A, Lapedriza Aet al. (2016) Learning deep features for discriminative localization. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, NV, USA: IEEE, 2921–9. [Google Scholar]
- Zhu J-Y, Park T, Isola Pet al. (2017) Unpaired image-to-image translation using cycle-consistent adversarial networks. 16th IEEE International Conference on Computer Vision (ICCV). Venice, Italy: IEEE, 2242–51. [Google Scholar]