Abstract
The trimeric postfusion structure of the C-terminally truncated fusion protein E of the flavivirus tick-borne encephalitis virus, a class II viral fusion protein, was previously determined (S. Bressanelli, K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey, EMBO J. 23:728-738, 2004). In this study we compared the properties of this truncated form with the full-length trimer and found that the so-called stem-anchor region not only confers additional stability to the full-length molecule but also structurally modifies the protein domain carrying the fusion peptide loop. These data provide experimental evidence to support the model of a fusion process that leads to the interaction of the stem-anchor region with the fusion peptide loop in the postfusion trimer.
The entry of enveloped viruses into cells requires the action of specific surface glycoproteins that are capable of undergoing triggered conformational changes for mediating the fusion of the viral membrane with a cellular membrane. Two classes of such viral fusion proteins have been identified so far that exhibit completely different structural organizations (reviewed in references 5 and 19). Class I fusion proteins form trimeric spikes and are present in orthomyxo-, paramyxo-, retro-, filo-, and coronaviruses. Class II fusion proteins on the other hand are oriented parallel to the viral membrane and form part of icosahedral protein networks in the envelopes of flavi- and alphaviruses (reviewed in references 8 and 13). These viruses enter cells by receptor-mediated endocytosis, and the acidic pH in endosomes triggers irreversible conformational changes in their fusion proteins resulting in fusion from within the endosomes.
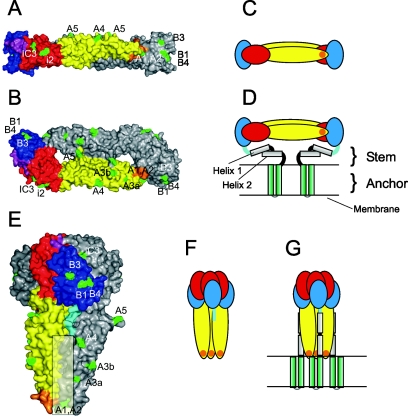
The atomic structures of C-terminally truncated forms of the fusion protein E1 from an alphavirus (Semliki Forest virus [SFV]) and the fusion protein E of two flaviviruses (tick-borne encephalitis virus [TBEV] and dengue virus) have been determined both in their pre- and postfusion conformations (4, 6, 14, 16-18, 24). The corresponding structures of the flavivirus TBEV E protein are shown in Fig. 1. The C-terminally truncated flavivirus E proteins used for X-ray crystallography lack approximately 90 amino acids consisting of the double membrane anchor and the so-called stem region that extends from the C terminus of domain III and contains two α-helices (23) (Fig. 1C, D). In contrast to the class I fusion proteins, the fusion-associated structural rearrangements of the class II proteins do not involve a major refolding of the polypeptide chain but rather a reorganization of the three domains relative to each other (4, 6, 17). Most importantly, domain III changes its position from the end of the rod-like structure and moves to the side of domain II, resulting in a folded-back, hairpin-like structure in a trimeric complex which—in contrast to the native protein—is oriented perpendicular to the membrane (Fig. 1E).
FIG. 1.
Native dimeric and trimeric postfusion structures of the TBEV E protein. (A, B) Surface representations of the X-ray crystal structure of the TBEV sE dimer (18) in a side view (A) and a top view (B). The three domains are color coded in one of the monomeric subunits: red, domain I; yellow, domain II; blue, domain III. The linker between domain I and III is shown in purple, the fusion peptide loop in orange. Positions of amino acid mutations that lead to the loss of MAb binding to virions are indicated in green and designated according to the corresponding MAb. (C, D) Schematic drawings of the E dimer in its truncated form (C) and with the stem-anchor region attached (D). (E, F, G) Representations of the trimeric low-pH form of E (4) analogous to those shown for the native forms in panels A to D. The linker between domain III and helix 1 of the stem is shown in light blue, and the postulated site of interaction of domains II with the stem is indicated by a shaded rectangle in panel E.
The folded-back orientation suggests that the fusion mechanism driven by class II fusion proteins is mechanistically similar to that proposed previously for viruses with class I fusion proteins (4, 6, 17). Although the stem-anchor region was not present in the truncated soluble E trimer (sE trimer) structure, the orientation of domain III suggests that the stem region should run along a groove formed by the domains II of adjacent subunits in the full-length molecule (Fig. 1E, G) (4, 17). In this conformation the fusion peptide loops and the membrane anchors would then be juxtaposed in the same, fused membrane (Fig. 1G). The formation of contacts between the stem-anchor region and domain II in the trimer has been assumed to provide additional energy and to be essential for the fusion process (4, 17). It has also been proposed that in addition to trimer formation, lateral interactions between several adjacent trimers at the fusion site are required for the merger of the membranes (4). So far, these assumptions have not been supported by experimental data.
In this report we demonstrate that the full-length trimeric E protein isolated from TBEV in its low-pH conformation has a higher thermostability than the corresponding truncated form consistent with the proposed interactions of the stem-anchor region with domain II in the last stage of the fusion process. We also provide experimental evidence that the membrane anchor and the fusion peptide loop are indeed juxtaposed in the final postfusion form of the molecule as predicted in the model and that interactions with the stem might influence the conformation of domain II—an observation that has important mechanistic implications as discussed below.
Preparation of trimers.
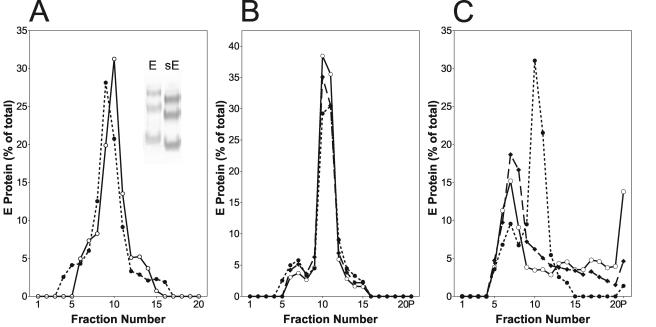
Full-length E trimers were prepared by solubilization of purified low-pH-treated virions with 1% Triton X-100 and sucrose density gradient centrifugation. The solubilized samples were loaded onto 7 to 20% continuous sucrose gradients in TAN buffer, pH 8.0 (0.05 M triethanolamine, 0.1 M NaCl) containing 0.1% Triton X-100, and centrifugation was carried out for 20 h at 38,000 rpm at 15°C in an SW 40 rotor (Beckman). The gradients were fractionated by upward displacement, and E trimer-containing fractions were pooled for their use in further experiments. To prepare the C-terminally truncated forms of E trimers (sE trimers), which were also used for X-ray structure determination (4), we used the procedures described in detail in references 9 and 22. Briefly, purified virions were treated with trypsin to generate the C-terminally truncated form of the soluble E dimer (sE dimer) which was purified by anion exchange chromatography. These sE dimers were then converted into sE trimers by exposure to acidic pH in the presence of liposomes. The membrane-bound trimers were isolated by solubilizing the liposomes with n-octylglucoside and ultrafiltration with Vivaspin20 concentrators as described previously (22). The oligomeric state of full-length and truncated E trimer preparations was verified by sedimentation analysis followed by cross-linking of the corresponding fractions (Fig. 2A).
FIG. 2.
Sedimentation analysis of E trimer preparations. The sedimentation direction is from left to right. (A) Full-length E trimers (○ ____ ○) and truncated sE trimers (• - - - •) were subjected to sedimentation in 7 to 20% sucrose gradients in TAN buffer (pH 8.0) containing 0.1% detergent as described in references 2 and 20. The gradients were fractionated, and the amount of E protein in each fraction was determined by a quantitative four-layer ELISA after denaturation of the samples with 0.4% sodium dodecyl sulfate (10). The inset shows the Western blot analysis after chemical cross-linking of the trimer peak fraction with dimethylsuberimidate as described in reference 2. (B, C) Sedimentation analysis of full-length E trimers (B) and truncated sE trimers (C) after incubation at different temperatures. Trimers were incubated for 10 min at 60°C (• - - - •), 65°C (▴- -▴), or 70°C (○ ____ ○), cooled on ice, and subjected to sedimentation analysis as described for Fig. 3A.
Thermostability of trimers.
To address the issue of whether interactions involving the stem-anchor region provide additional stability to the full-length trimer compared to the sE trimer we examined the effect of increasing temperatures on both forms of the E protein. For this purpose preparations of E trimers and sE trimers were incubated for 10 min at room temperature, 50, 55, 60, 65, and 70°C, and then their oligomeric state was analyzed by rate zonal sucrose density gradient centrifugation in the presence of 0.1% Triton X-100 as described previously (20). Both trimer preparations were found to be more thermostable than the E dimers, which start to denature at 50°C (20). As shown in Fig. 2B, the sedimentation behavior of full-length E trimers was unaffected up to a temperature of 70°C, and almost all of the E protein was detected in the fractions corresponding to a trimer. In contrast, the sE trimers apparently broke down and/or denatured at 65°C and higher temperatures, as indicated by the loss of the trimer peak and the appearance of aggregated material found in the pellet at the bottom of the gradient and more-slowly sedimenting oligomers (Fig. 2C). The latter were shown to be monomers and dimers by dimethyl suberimidate cross-linking and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). These data are consistent with the suggestion that the stem, which is predicted to interact with domain II, stabilizes the final structure by contributing additional trimer contacts (4, 17). They also help to explain the finding that the presence of the first helix of the stem (compare Fig. 1D and G) in recombinant forms of the TBEV E protein is required for the low-pH-induced trimerization in the absence of a lipid membrane (3).
Reactivity of truncated and full-length trimers with E protein-specific MAbs.
To obtain additional information about possible conformational differences in the full-length E trimer we made use of a panel of monoclonal antibodies (MAbs) that react with the neutral pH form of the TBEV E protein, whose binding sites have been mapped by the use of neutralization-escape mutants as well as specifically engineered mutations (Fig. 1A, B) (1, 7, 11, 15). In a blocking enzyme-linked immunosorbent assay (ELISA) we measured the capacity of the full-length E trimer and the sE trimer (at a concentration of 1 μg/ml) to inhibit the binding of a predetermined dilution of each of the MAbs to purified virions coated onto the solid phase (22). It is an advantage of this assay format that the antigen under investigation is not subject to denaturation effects that occur during the coating of proteins onto solid phases (12).
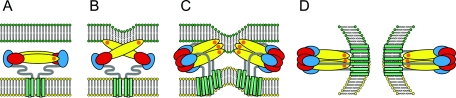
Since the dimer-trimer conversion does not involve a major refolding of the individual domains and a significant proportion of the external surface of the dimer is also exposed in the trimer, most of these antibodies retained their reactivity with the sE trimer (Fig. 1E and Table 1, second column). This includes antibodies to each of the three protein domains with the exception of MAbs i2 and A4 which have lost their reactivity with the sE trimer. The epitope i2 is located in domain I and becomes buried during the dimer-trimer conversion. The lack of binding of MAb A4 on the other hand is probably due to local structural alterations, because the one amino acid (amino acid 233) that has been identified as part of this epitope is still exposed in the sE trimer. Nevertheless, we were still left with a significant panel of MAbs with structurally mapped binding sites in each of the three domains that could be used for determining differences between the full-length E trimer and the sE trimer. As can be seen from the results presented in Table 1, dramatic differences between the two trimers were found with respect to the reactivities of the domain II-specific MAbs A1, A2, and A3, which reacted strongly with the sE trimer but weakly (A3) or not at all (A1 and A2) with the full-length trimer. The binding sites of Mabs A1 and A2 were previously mapped to the region containing the fusion peptide loop at the tip of domain II (1) and were also shown to inhibit the low-pH-induced interaction with target membranes (21). It is probable that their lack of reactivity with the full-length trimer is caused by the shielding of their binding sites by the stem-anchor region, with the stem running down the groove formed between domains II in the trimer (Fig. 1E). These data thus corroborate steps C and D of the proposed flavivirus fusion model (Fig. 3) which emphasize the importance of domain II-stem interactions for the merger of the membranes (Fig. 3C) and the formation of a postfusion structure in which the fusion peptide loops and the stem-anchor region are juxtaposed in the fused membrane (Fig. 3D).
TABLE 1.
Reactivity of E protein-specific MAbs with full-length and truncated E trimers in a blocking ELISAa
| MAb | Full-length E trimer | sE trimer |
|---|---|---|
| Domain I | ||
| C1 | +++ | ++/+++ |
| C2 | +/++ | + |
| C3 | + | −/+ |
| C4 | + | + |
| C5 | ++ | ++ |
| C6 | + | + |
| i2 | − | −b |
| IC3 | + | + |
| Domain II | ||
| A1 | − | +++ |
| A2 | − | +++ |
| A3 | + | +++ |
| A4 | − | −b |
| A5 | +++ | +++ |
| Domain III | ||
| B1 | −/+ | + |
| B2 | ++ | ++ |
| B3 | +++ | +++ |
| B4 | ++ | ++/+++ |
+++, 75-100% blocking; ++, 50-75% blocking; +, 25-50% blocking; −, 0-25% blocking
These MAbs lose their reactivity upon dimer-trimer conversion.
FIG. 3.
Schematic diagram of the proposed membrane fusion process of flaviviruses. E protein: domain I in red, domain II in yellow, domain III in blue, stem region in gray, membrane anchor in green, fusion peptide loop in orange. (A) The E protein homodimer in its native state at the surface of the mature virion. (B) Low-pH-induced dissociation of the E dimer and interaction of the fusion peptide loop with the endosomal membrane. (C) Formation of an E trimer (including the flipping back of domain III) proceeding via still undefined intermediates. (D) Formation of the final postfusion conformation through interactions of the stem-anchor region with domain II, leading to the juxtaposition of the fusion peptide loops and the membrane anchors in the fused membrane.
The strongly reduced reactivity of MAb A3, which is believed to bind primarily to the side of domain II, might be caused either by the stem-anchor region physically blocking access to the MAb binding site or by conformational changes within this domain induced by interactions with the stem. Interestingly, the truncated postfusion structures of the alphavirus Semliki forest virus (SFV) and flavivirus fusion protein trimers show differences in conformation at the tips of domain II. In the SFV E1 trimer the fusion peptide loops are widely separated and do not interact with each other as they do in the flavivirus E trimer (4, 6, 17). Bressanelli et al. (4) have attributed this difference to the presence of the s1 sequence element in the crystal structure of the SFV E1 trimer which would roughly correspond to helix 1 of the stem in the flavivirus E protein and suggested that the insertion of the stem between domains II of the trimer would keep the fusion peptide loops apart in the full-length TBEV E trimer as they are in SFV E1. The splayed-apart orientation would allow the fusion peptide loops of adjacent trimers to interact with each other to form higher-order ring-like structures at the fusion site that have been proposed to be involved in the bending of the target membrane (6). Whether the observed differences in MAb A3 reactivity with the full-length and the truncated E trimers of TBEV are related to this proposed conformational difference remains to be determined.
The present report thus provides experimental evidence for important predictions that had been made based on the X-ray crystal structures of the C-terminally truncated postfusion trimers of flaviviruses. This includes the juxtaposition of the fusion peptide loop and the stem-anchor region in the postfusion conformation and stabilizing interactions between the stem-anchor region and domain II of the fusion protein that serve as a driving force for membrane merger.
Acknowledgments
We thank Steven Allison for helpful discussions and for critical reading of the manuscript and Walter Holzer for technical assistance.
This work was supported in part by the Austrian “Fonds zur Förderung der wissenschaftlichen Forschung,” FWF project number P16535-B09.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, S. L., K. Stiasny, K. Stadler, C. W. Mandl, and F. X. Heinz. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimitrov, D. S. 2004. Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol 2:109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibbons, D. L., M. C. Vaney, A. Roussel, A. Vigouroux, B. Reilly, J. Lepault, M. Kielian, and F. A. Rey. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320-325. [DOI] [PubMed] [Google Scholar]
- 7.Guirakhoo, F., F. X. Heinz, and C. Kunz. 1989. Epitope model of tick-borne encephalitis virus envelope glycoprotein E: analysis of structural properties, role of carbohydrate side chain, and conformational changes occurring at acidic pH. Virology 169:90-99. [DOI] [PubMed] [Google Scholar]
- 8.Heinz, F. X., and S. L. Allison. 2003. Flavivirus structure and membrane fusion. Adv. Virus Res. 59:63-97. [DOI] [PubMed] [Google Scholar]
- 9.Heinz, F. X., C. W. Mandl, H. Holzmann, C. Kunz, B. A. Harris, F. Rey, and S. C. Harrison. 1991. The flavivirus envelope protein E: isolation of a soluble form from tick-borne encephalitis virus and its crystallization. J. Virol. 65:5579-5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109-117. [DOI] [PubMed] [Google Scholar]
- 11.Holzmann, H., K. Stiasny, M. Ecker, C. Kunz, and F. X. Heinz. 1997. Characterization of monoclonal antibody-escape mutants of tick-borne encephalitis virus with reduced neuroinvasiveness in mice. J. Gen. Virol. 78:31-37. [DOI] [PubMed] [Google Scholar]
- 12.Holzmann, H., G. Utter, E. Norrby, C. W. Mandl, C. Kunz, and F. X. Heinz. 1993. Assessment of the antigenic structure of tick-borne encephalitis virus by the use of synthetic peptides. J. Gen. Virol. 74:2031-2035. [DOI] [PubMed] [Google Scholar]
- 13.Kielian, M. 2002. Structural surprises from the flaviviruses and alphaviruses. Mol. Cell 9:454-456. [DOI] [PubMed] [Google Scholar]
- 14.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus. An icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 15.Mandl, C. W., F. Guirakhoo, H. Holzmann, F. X. Heinz, and C. Kunz. 1989. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J. Virol. 63:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 18.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 19.Sollner, T. H. 2004. Intracellular and viral membrane fusion: a uniting mechanism. Curr. Opin. Cell Biol. 16:429-435. [DOI] [PubMed] [Google Scholar]
- 20.Stiasny, K., S. L. Allison, C. W. Mandl, and F. X. Heinz. 2001. Role of metastability and acidic pH in membrane fusion by tick-borne encephalitis virus. J. Virol. 75:7392-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stiasny, K., S. L. Allison, J. Schalich, and F. X. Heinz. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 76:3784-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stiasny, K., S. Bressanelli, J. Lepault, F. A. Rey, and F. X. Heinz. 2004. Characterization of a membrane-associated trimeric low-pH-induced form of the class II viral fusion protein E from tick-borne encephalitis virus and its crystallization. J. Virol. 78:3178-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, Y., W. Zhang, S. Ogata, D. Clements, J. H. Strauss, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2004. Conformational changes of the flavivirus E glycoprotein. Structure (Cambridge) 12:1607-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]