Abstract
The pathogenetic mechanism of the deafness-associated mitochondrial DNA (mtDNA) T7445C mutation has been investigated in several lymphoblastoid cell lines from members of a New Zealand pedigree exhibiting the mutation in homoplasmic form and from control individuals. We show here that the mutation flanks the 3′ end of the tRNASer(UCN) gene sequence and affects the rate but not the sites of processing of the tRNA precursor. This causes an average reduction of ∼70% in the tRNASer(UCN) level and a decrease of ∼45% in protein synthesis rate in the cell lines analyzed. The data show a sharp threshold in the capacity of tRNASer(UCN) to support the wild-type protein synthesis rate, which corresponds to ∼40% of the control level of this tRNA. Strikingly, a 7445 mutation-associated marked reduction has been observed in the level of the mRNA for the NADH dehydrogenase (complex I) ND6 subunit gene, which is located ∼7 kbp upstream and is cotranscribed with the tRNASer(UCN) gene, with strong evidence pointing to a mechanistic link with the tRNA precursor processing defect. Such reduction significantly affects the rate of synthesis of the ND6 subunit and plays a determinant role in the deafness-associated respiratory phenotype of the mutant cell lines. In particular, it accounts for their specific, very significant decrease in glutamate- or malate-dependent O2 consumption. Furthermore, several homoplasmic mtDNA mutations affecting subunits of NADH dehydrogenase may play a synergistic role in the establishment of the respiratory phenotype of the mutant cells.
Cochlear function depends on a very high rate of ATP production, and mitochondrial DNA (mtDNA)-dependent dysfunctions have often been found to cause hearing defects either in syndromic or nonsyndromic form. mtDNA-linked deafness exhibits in high degree the characteristics of tissue specificity and variable degree of penetrance which are a hallmark of mtDNA mutation-dependent disorders, most typically of Leber’s hereditary optic neuropathy (LHON) (19, 20, 23, 24, 43). Among the identified deafness-causing mtDNA mutations are an A-to-G transition at position 1555 (A1555G) in the 12S rRNA gene (10, 21, 32), a heteroplasmic C-nucleotide insertion at position 7472 in the tRNASer(UCN) gene (39), and a T-to-C transition at position 7445 (T7445C) in the DNA sequence encoding the precursor of this tRNA. The latter mutation has been described in members of a Scottish maternal pedigree with sensorineural bilateral hearing loss (34) and in New Zealand and Japanese pedigrees with sensorineural hearing loss and palmoplantar keratoderma (11, 38). The three pedigrees are unrelated, exhibiting distinct sets of mtDNA polymorphisms (11, 35, 38), besides the identical mutation at position 7445. This mutation changes the stop codon AGA of the heavy-strand (H-strand)-encoded mRNA for subunit COI of cytochrome c oxidase (3, 29, 30) to an equivalent AGG stop codon, and at the same time, causes a U-to-C transition in the light-strand (L-strand)-encoded tRNASer(UCN) precursor (3, 29).
The occurrence of a mutation at position 7445 in three genetically unrelated pedigrees affected by sensorineural deafness and differing considerably in their mtDNA haplotype is clear evidence that this mutation is involved in the pathogenesis of the disorder (11, 33, 38). A recent investigation of a lymphoblastoid cell line derived from a deaf member of the Scottish family carrying the mutation at position 7445 in homoplasmic form compared to a control cell line revealed a marked decrease (60 to 65%) in its tRNASer(UCN) content, which was suggested to result from a decrease in the efficiency of processing of the mutant tRNA precursor. However, no abnormality in mitochondrial protein synthesis nor any significant impairment of respiratory function was reported for this cell line (33).
In order to investigate further the pathogenetic mechanism of the mutation in the New Zealand family, in the present work, lymphoblastoid cell lines derived from four members of this family and from four control individuals have been analyzed. It has been shown that the mutation at position 7445, flanking the 3′ end of the tRNASer(UCN) sequence, affects the rate of processing of the tRNA precursor. This produces a drastic decrease in the steady-state level of this tRNA, with significant effects on protein synthesis. Furthermore, a specific marked reduction in the level of the ND6 mRNA has been shown to be associated and probably mechanistically linked with the mtDNA mutation at position 7445 and to play a determinant role in the respiratory phenotype of the mutant cells.
MATERIALS AND METHODS
Cell lines and media.
Eight human immortalized lymphoblastoid cell lines, derived from four members of the New Zealand family exhibiting the mutation at position 7445 and various degrees of hearing loss (individuals IV-5, IV-7, IV-15, and V-7; the Roman number indicates the generation in the pedigree [11]), and from four genetically unrelated individuals (0913, 0923, 1032, and 0615) lacking the mutation at position 7445 as well as the five additional changes from the Cambridge mtDNA sequence (2) found in the New Zealand family haplotype (11), were used in this work. The cells were grown either in a specially made Dulbecco’s modified Eagle medium (DMEM) containing 1 mg of glucose per ml, 0.11 mg of pyruvate per ml, and 0.36 mM CaCl2 (hereafter referred to as special DMEM-glucose), supplemented with 10% fetal bovine serum (FBS) or in the same medium lacking glucose but containing 0.9 mg of galactose and 0.5 mg of pyruvate per ml (hereafter referred to as special DMEM-galactose), supplemented with 10% dialyzed FBS. The bromodeoxyuridine-resistant 143B.TK− cells (25) were grown in regular DMEM (containing 4.5 mg of glucose and 0.11 mg of pyruvate per ml), supplemented with 100 μg of bromodeoxyuridine per ml and 5% FBS. VA2B cells were grown in DMEM supplemented with 10% bovine serum. The population doubling times (DTs) of the cell lines in special DMEM-glucose or special DMEM-galactose were determined as previously detailed (14).
mtDNA analysis.
Total DNA samples were isolated from the cultured lymphoblastoid cell lines and from 143B.TK− cells with an Applied Biosystems 340A DNA extractor. The A-to-G transition in mtDNA at position 7445 of the Cambridge sequence (3) was detected by taking advantage of the mutation-associated loss of the XbaI site as previously described (11), except for the use of oligodeoxynucleotides corresponding to positions 7396 to 7417 and 7657 to 7676 for the PCR amplification of an appropriate mtDNA fragment.
The quantification of mtDNA was performed by slot blot hybridization, using a 32P-labeled human mtDNA fragment as a specific probe and a 32P-labeled nuclear 28S rRNA gene fragment as a normalization probe, as previously described (14). Quantification of the hybridization was carried out by scanning the fluorograms by laser densitometry or by analyzing the slot blot in a PhosphorImager (Molecular Dynamics). For comparison of the data from different blots, the values obtained for the lymphoblastoid cell lines in each blot were normalized to the values obtained for the 143B.TK− sample in the same blot.
Isolation of mitochondrial and total cell RNA.
Total mitochondrial nucleic acid preparations were obtained by acid phenol-chloroform extraction from mitochondria isolated from lymphoblastoid cell lines (∼3.0 × 108 cells), and from twice EDTA-washed mitochondria of HeLa cells (∼3.0 × 109 cells), as previously described (26). Highly purified total mitochondrial tRNA preparations were obtained from mitochondrial nucleic acids extracted from HeLa cells and from the IV-5 mutant lymphoblastoid cell line by polyacrylamide gel electrophoresis (PAGE), as previously detailed (26). The tRNA fraction isolated by the latter method was used for synthesizing, cloning, and sequencing the cDNA corresponding to tRNASer(UCN), while the total mitochondrial nucleic acid fractions were used for quantification of the mitochondrial tRNAs and 12S rRNA. Total cell RNA was isolated by the RNAzol B procedure (TEL-TEST, Inc., Friendswood, Tex.) which is based on a modification of the single-step method by acid guanidinium thiocyanate-phenol-chloroform extraction (7).
Sequencing of 5′- and 3′-end proximal segments of tRNASer(UCN).
The 5′ and 3′ ends of the mitochondrial tRNASer(UCN) from HeLa cells and from the mutant cell line IV-5 were sequenced after cDNA synthesis, PCR amplification, and cloning, as described elsewhere (46). First, highly purified total mitochondrial tRNA was circularized by incubation in the presence of T4 RNA ligase (Promega) to ligate the 3′ and 5′ ends of the tRNAs. Then, a complementary DNA chain of the tRNASer(UCN) was synthesized by reverse transcriptase after annealing of the circular tRNA with the specific oligodeoxynucleotide SUCN4 (5′-CAAGCCAACCCCATGGCCTC-3′). The second strand of this cDNA was synthesized by using the primer SUCN3 (5′-AAACCAGCTTTGGGGGGTTC-3′), and the artificial tDNA was then amplified by PCR, using both oligodeoxynucleotides SUCN3 and SUCN4. Subsequently, the PCR product was cloned in the TA vector (Invitrogen), and eight clones of HeLa cell tDNA and five clones of IV-5 tDNA were sequenced by the ABI PRISM Dye Terminator Cycle Sequencing Core (Perkin Elmer).
Quantification of the mitochondrial tRNASer(UCN).
To determine the cell content of the mitochondrial tRNASer(UCN), tRNALeu(UUR), tRNALys, tRNAGln, tRNAGlu, and 12S rRNA in all cell lines for use as reference markers, total mitochondrial nucleic acid preparations were electrophoresed through a 5% polyacrylamide–7 M urea gel in Tris-borate-EDTA buffer (TBE) (after heating the sample at 90°C for 5 min) and then electroblotted onto a Zeta-probe membrane (Bio-Rad) for hybridization analysis with specific oligodeoxynucleotide probes. For the detection of tRNASer(UCN), tRNALys, tRNALeu(UUR), tRNAGln, tRNAGlu, and 12S rRNA, the following 5′-end 32P-labeled oligodeoxynucleotides specific for each RNA were used: 5′-CAAGCCAACCCCATGGCCTC-3′ (tRNASer(UCN)); 5′-TCACTGTAAAGAGGTGTTGG-3′ (tRNALys); 5′-TGTTAAGAAGAGGAATTGAA-3′ (tRNALeu(UUR)); 5′-CTAGGACTATGAGAATCGAA-3′ (tRNAGln); 5′-TATTCTCGCACGGACTACAA-3′ (tRNAGlu); and 5′-GAAAGGCTAGGACCAAACCTA-3′ (12S rRNA). The hybridization reactions were carried out in a mixture of 6× SSC (SSC is standard saline citrate; 1× SSC is 150 mM NaCl plus 15 mM trisodium citrate), 5× Denhardt’s solution, 0.1% sodium dodecyl sulfate (SDS), and 200 mg of salmon sperm DNA per ml, for 6 h at 37°C. After hybridization, the samples were washed two times for 10 min in 2× SSC–0.1% SDS at 37 or 50°C. The radioactivity in each band was quantified as detailed above for slot blot hybridization.
mRNA analysis.
For RNA transfer hybridization analysis, 20 μg of total cell RNA was fractionated by electrophoresis through a 1.4% agarose–2.2 M formaldehyde gel (8), transferred to a Zeta-probe membrane (Bio-Rad), and hybridized with a [32P]dCTP-labeled single-stranded (ss) ND6-specific DNA probe (which was synthesized on XbaI-linearized pND6-1 plasmid by using the Klenow fragment of polymerase I and the SP6 primer). After the blot was stripped, the RNA was hybridized with total purified HeLa cell mtDNA, which had been [32P]dCTP labeled by random priming. After the blot was restripped, the RNA was rehybridized with a [32P]CTP-labeled ND6-specific RNA probe synthesized on the pND6-1 plasmid with SP6 RNA polymerase. The plasmid pND6-1 was constructed by amplifying a 275-bp fragment of the human ND6 gene (positions 14343 to 14618) by PCR, cloning it in the TA vector (Invitrogen), and subcloning the 280-bp EcoRI insert into the pGEM-7Zf(+) vector carrying the SP6 and T7 promoters (Promega).
Analysis of mitochondrial protein synthesis.
Pulse-labeling of the cell lines for 30 min with [35S]methionine-[35S]cysteine in the presence of emetine, electrophoretic analysis of the translation products, and quantification of radioactivity in the whole electrophoretic patterns or in individual bands was carried out as detailed previously (14). Labeling of mitochondrial translation products with [3H]serine (30 Ci mmol−1; 50 μCi/ml) for 30 min in serine-free special DMEM-glucose, electrophoretic analysis of the samples, and treatment of the gels were carried out in the same way.
O2 consumption measurements.
Rates of O2 consumption were determined with a Gilson 5/6 oxygraph on samples of 107 cells in 1.5-ml portions of special DMEM-glucose lacking glucose supplemented with 10% dialyzed FBS (25). Polarographic analysis of digitonin-permeabilized cells with different respiratory substrates and inhibitors to test the activity of the various respiratory complexes was carried out as detailed elsewhere (18).
Computer analysis.
Variance analysis was performed by the analysis of variance (ANOVA) test contained in the StatView program for Macintosh (version 4.0) (SAS Institute) and entering individual replicate values. Curve fitting for Fig. 5c was done with the program PSI-Plot (Poly Software International).
FIG. 5.
Quantification of the overall rates of synthesis of the mitochondrial translation products after a 30-min [35S]methionine pulse in different lymphoblastoid cell lines and relationship of the rates of synthesis of the individual polypeptides to their serine (UCN) residue content. (a) Three independent labeling experiments and two electrophoretic analyses of the mitochondrial translation products after each labeling were performed for each cell line. The individual values for the rates of overall mitochondrial protein labeling in the various cell lines, determined as detailed in Materials and Methods, were normalized relative to the value for 143B.TK− cells in each gel, and the mean relative value for each cell line was expressed as a percentage of the average normalized value obtained for the control cell lines, with error bars representing 2 standard errors of the mean. The horizontal dashed lines represent the average value for the control and mutant cell lines, and the vertical arrows represent 2 standard errors of the mean for the two groups. P indicates the significance, according to the ANOVA test, of the difference between mutant and control values. (b) Relationship between the average rate of labeling of the individual mitochondrial translation products in the mutant cell lines, expressed relative to the average value in the control cell lines, and the number of serine (UCN) residues that they contain. CYTB, apocytochrome b. (c) Relationship between average relative rate of synthesis of the individual polypeptides in the mutant cell lines and the proportion of serine (UCN) residues that they contain. The curve shown describes the equation Rx = R0{0.1/[0.1(1 − x) + 1.5x]}, whose parameters have been optimized to make the best fit to the data. In this equation, Rx is the rate of labeling of a polypeptide having x proportion of serine (UCN) residues in mutant cells relative to the rate in wild-type cells, the rates being expressed as reciprocals of the times required for their synthesis (see text for details).
RESULTS
The New Zealand pedigree and derived lymphoblastoid cell lines.
The pedigree of the New Zealand family with maternally inherited deafness has been previously described (11, 38). Immortalized lymphoblastoid cell lines were derived from four members of this family exhibiting the mutation at position 7445, three individuals with moderate to profound sensorineural deafness (IV-5, IV-7, and IV-15; all three between 25 and 31 years old), and one individual with mild conductive deafness (V-7; 11 years old) and from four genetically unrelated individuals lacking the mutation (0913, 0923, 1032, and 0615; all four between 28 and 35 years old) (11). The presence of the mutation in the four chosen lymphoblastoid cell lines from the New Zealand pedigree has been confirmed in the present work, and in all cell lines, the mutation appeared to be homoplasmic.
A complete sequence analysis of the mtDNA from one pedigree member (11) had previously revealed, besides the mutation at position 7445, five additional sequence changes from the Cambridge consensus sequence (3). Of these changes, two, the T4216C transition in the ND1 gene and the G13708A transition in the ND5 gene, have been observed to occur with increased frequency in LHON patients (22). A T-to-C transition at position 10084 in the ND3 gene represents a rare novel mutation, while the G-to-A transition at position 3010 in the 16S rRNA gene and the T-to-C transition at position 14798 in the apocytochrome b gene are polymorphisms commonly found in controls. These five additional sequence changes were absent in the four control cell lines utilized in this study.
Mutation site.
The U-to-C transition at position 7445 in the L-strand-encoded tRNASer(UCN) precursor was originally thought to be located in the 3′-terminal nucleotide of the tRNASer(UCN) (11, 34) on the basis of the proposed structure for this tRNA (2), as deduced from the Cambridge mtDNA sequence (3). However, structural studies on bovine mitochondrial tRNASer(UCN) (47) have led to a reinterpretation of the original sequence, which would place the human nucleotide (nt) 7445 to a position immediately adjacent on the 3′ side to the encoded tRNASer(UCN) (33, 39).
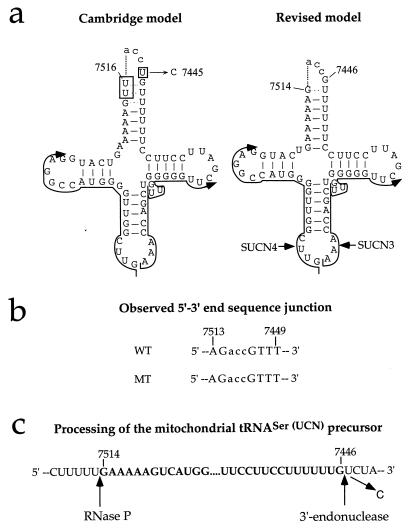
To verify in human mtDNA the position of the 7445 mutation relative to the tRNASer(UCN) coding sequence, as well as to investigate the possible effect of the mutation on the tRNASer(UCN) precursor processing, cDNA clones derived from circularized tRNASer(UCN) (46) from wild-type (HeLa) cells and from a mutant cell line (IV-5) were sequenced. Figure 1a shows the expected structure of the circularized tRNASer(UCN) molecule, as deduced from the original interpretation of the Cambridge mtDNA sequence (2) and from the revised interpretation (47). Figure 1b shows the sequence determined for the 5′-3′ junction sequence in eight cDNA clones from HeLa cell tRNASer(UCN) and five clones from the mutant IV-5 cell tRNASer(UCN). Both sequences lack nt 7445, which is fully consistent with the revised model. Furthermore, no difference was observed between the wild-type and the mutant 5′-3′ junction sequence. Other experiments, involving in vitro processing by partially purified human mitochondrial RNase P of in vitro-transcribed tRNASer(UCN) showed that this transcript was cleaved at a position 3′ to nt 7515, which fully supports the sequencing data (32a). Figure 1c shows the experimentally determined processing site for mitochondrial RNase P in the tRNASer(UCN) precursor and the 3′ endonuclease processing site expected on the basis of the sequencing data.
FIG. 1.
Site of the mutation at position 7445 in the mitochondrial tRNASer(UCN) precursor and effect of the mutation on tRNA processing. (a) Structure of circularized tRNA according to the Cambridge model (2) and the revised model (47). SUCN4 and SUCN3 indicate the two oligonucleotides utilized for the synthesis of the first and second strands of the cDNA, respectively. (b) Sequence of the 5′-3′ end junction determined in eight cDNA clones from wild-type (WT) HeLa cell mitochondrial tRNASer(UCN) and five cDNA clones from the mitochondrial tRNASer(UCN) of the mutant (MT) lymphoblastoid cell line IV-5. (c) Processing sites in the mitochondrial tRNASer(UCN) precursor, experimentally determined for RNase P and predicted from the sequencing data for the 3′ endonuclease.
The fact that the A7445G transition affects the 3′-end nucleotide of the stop codon (AGA) of the COI coding sequence in the H-strand polycistronic transcript (3, 29, 30) raised the possibility of alterations in the processing of this transcript. However, RNA transfer hybridization experiments utilizing total cell RNA and 32P-labeled human mtDNA as a probe failed to show any mobility change or quantitative change in the COI mRNA or its precursor (RNA 6) (29, 30) or the presence of any abnormally migrating band (data not shown), in confirmation of previous findings (33).
Marked decrease in amount of tRNASer(UCN).
It seemed possible that the rate of processing of the tRNASer(UCN) from its precursor is affected by the mutation at position 7445. To test this possibility, the influence of the U7445C transition on the steady-state level of the tRNASer(UCN) was investigated. For this purpose, equal amounts of total mitochondrial nucleic acids (26) from the control and mutant cell lines were run in parallel on a 5% polyacrylamide–7 M urea gel, electroblotted, and hybridized with a 5′-end 32P-labeled oligodeoxynucleotide probe specific for mitochondrial tRNASer(UCN) (9). This tRNA did not show any obvious size change in mutant cells, but the amount decreased markedly from that in control cells (Fig. 2a).
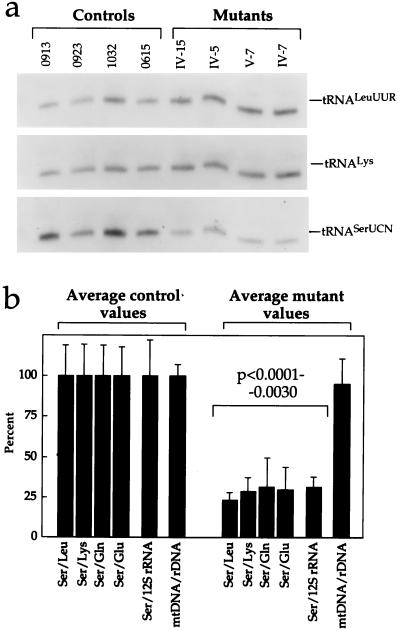
FIG. 2.
Determination of the amount of mitochondrial tRNASer(UCN) and of the mtDNA content in control and mutant lymphoblastoid cell lines. (a) Equal amounts of total mitochondrial nucleic acid samples from the various cell lines were electrophoresed under nonacid conditions, electroblotted, and hybridized with 5′-end 32P-labeled oligonucleotide probes specific for the mitochondrial tRNASer(UCN), tRNALys, and tRNALeu(UUR). (b) Average relative mitochondrial tRNASer(UCN) content per cell, normalized to the average content per cell of mitochondrial tRNALeu(UUR), tRNALys, tRNAGln, tRNAGlu, or 12S rRNA in the four control cell lines and in the four mutant cell lines. The values for the latter are expressed as percentages of the average values for the control cell lines. The calculations were based on one to three independent determinations of tRNASer(UCN) content in each cell line and on one to three determinations of the content of each of the five reference RNA markers in two control cell lines (0913 and 1032) and two mutant cell lines (IV-7 and IV-5) and of the content of tRNALeu(UUR) and 12S rRNA in the other two control cell lines (0615 and 0923) and two mutant cell lines (IV-15 and V-7). The average mtDNA content per cell in the mutant cell lines, normalized relative to the hybridization to a nuclear 28S rRNA probe and expressed as a percentage of the average value determined in the control cell lines, is also shown. Six mtDNA determinations were made for each cell line. See Materials and Methods for details. The error bars indicate two standard errors of the mean; P indicates the significance, according to the ANOVA test, of the difference between mutant and control values for tRNASer(UCN) normalized to the values for each reference marker.
For comparison, the levels of several other mtDNA-coded RNAs derived from the three transcription units of the human mitochondrial genome (27, 28) (Fig. 3) were determined. In particular, the levels of tRNALeu(UUR) and tRNALys (Fig. 2a) (as representatives of the whole H-strand transcription unit [Fig. 3]) and of two tRNAs derived from the L-strand transcription unit [encoded in a region either upstream (tRNAGlu) or downstream (tRNAGln) relative to tRNASer(UCN) (Fig. 3)] were quantified in the control and mutant cell lines on the same blot, after stripping it, using 5′-end 32P-labeled specific oligodeoxynucleotide probes. Similarly, the levels of 12S rRNA, as representative of the H-strand rDNA transcription unit (Fig. 3), in the two groups of cell lines were determined in a blot of total mitochondrial nucleic acids. The average levels of tRNASer(UCN) obtained in the various control or mutant cell lines were then normalized to the average levels determined in the same cell lines for each of the reference RNAs. The average tRNASer(UCN) content values in the different mutant cell lines, normalized to the individual reference RNA levels, when expressed relative to the corresponding average values found in the control cell lines, were remarkably similar, ranging between ∼25% (after normalization to the tRNALeu(UUR) level) and ∼30% (after normalization to the tRNAGln and 12S rRNA levels) (Fig. 2b). These values reflected a very significant decrease in tRNASer(UCN) content in mutant cells relative to the controls (P < 0.0001 to P = 0.0030 by the ANOVA test). The average tRNASer(UCN) content values in the individual mutant cell lines were 28, 38, 27, and 23% of the overall average control value in IV-15, IV-7, IV-5, and V-7, respectively. The average tRNASer(UCN) content values in the individual control cell lines, expressed relative to the same overall average control value, were 101, 107, 114, and 79% in 0913, 0923, 1032, and 0615, respectively.
FIG. 3.
Genetic and transcription maps of the human mitochondrial genome. The two inner circles show the positions of the two rRNA genes, 16S and 12S (black bars), of the reading frames (white bars) and of the tRNA genes (solid circles). The outer portion of the diagram shows the transcripts of the whole H-strand transcription unit (black bars), the H-strand rDNA transcription unit (white bars), and the L-strand transcription unit (hatched bars; in the three longest ones, RNAs 1, 2 and 3, and in ND6 mRNA, the black portion represents the ND6 reading frame). COI, COII, and COIII are subunits I, II, and III of cytochrome c oxidase, respectively; ND1, ND2, ND3, ND4, ND4L, ND5, and ND6 are subunits 1, 2, 3, 4, 4L, 5, and 6 of the respiratory chain NADH dehydrogenase, respectively; and A6 and A8 are subunits 6 and 8 of the H+-ATPase. cyt.b, apocytochrome b; OH and OL, origin of H-strand and L-strand synthesis, respectively, with arrows indicating the direction of synthesis; F-met, formylmethionyl.
An investigation of the mtDNA content of these cell lines by slot blot hybridization, carried out using a 32P-labeled human mtDNA probe and a human 28S rRNA gene probe for normalization purposes, failed to reveal any significant difference between the average mtDNA/ribosomal DNA (rDNA) ratio for the control cell lines and the average (P = 0.4989) (Fig. 2b), as well as the individual values (not shown), for the mutant cell lines. Therefore, the observed decreases in the tRNASer(UCN) levels in the mutants relative to those in the control cell lines do not appear to reflect differences in the amount of mtDNA template.
Mitochondrial protein synthesis defects.
The SDS-PAGE patterns of the mitochondrial translation products from the U7445C mutation-carrying lymphoblastoid cell lines differ from those of the control lymphoblastoid cell lines and from that of the human osteoblastoma-derived cell line 143B.TK− (25) by the faster migration of the ND1 and ND3 polypeptides (Fig. 4a and c). This higher mobility presumably reflects the occurrence of mutations in the reading frames for these polypeptides which was mentioned above. The VA2B cell line, which is related to HeLa cells, shows an even greater electrophoretic mobility of ND3 (Fig. 4a). This abnormal migration of the ND3 polypeptide in HeLa cells and related cell lines has been observed before (45) and shown to be associated with a different mutation of the ND3 reading frame (31). The polypeptide A8 of the control 0923 cell line shows a reduced mobility from that in the other cell lines (Fig. 4b). This change presumably results from the occurrence in this polypeptide of a G-to-A transition at position 8519, producing a Glu-to-Lys change, and a C-to-A transversion at position 8546, producing a Leu-to-Ile change.
FIG. 4.
Electrophoretic patterns of the mitochondrial translation products of the lymphoblastoid cell lines, VA2B cells, and 143B.TK− (143B) cells labeled for 30 min with [35S]methionine in the presence of 100 μg of emetine per ml. Samples containing equal amounts of protein (30 μg), except the 143B.TK− and VA2B samples, which contained 15 μg of protein, were run through SDS–15 to 20% exponential polyacrylamide gradient gels. The three panels represent electrophoretic patterns obtained in separate gel runs, each one including the 143B.TK− control for normalization purposes. The intensities of the bands were quantified by densitometric analysis of appropriate exposures of the fluorograms. The faster migration of the ND1 and ND3 polypeptides is indicated by the arrows in panels a and c. For an explanation of abbreviations, see the legend to Fig. 3.
The cell lines carrying the mutation at position 7445 showed a clear, although variable, decrease in the overall rate of labeling of the translation products relative to the control lymphoblastoid cell lines (Fig. 4). This decrease ranged between 15 (IV-7) and 75% (V-7), with an average of ∼45% (P < 0.0001 by the ANOVA test) (Fig. 5a). No abnormal mitochondrial translation products were observed in the mutant cell lines, as previously found in the cell lines carrying the myoclonic epilepsy and ragged-red fiber (MERRF) tRNALys mutation at position 8344 (9). However, it was possible that alterations in the posttranscriptional modifications of the tRNASer(UCN) related to the reduced rate of processing of the tRNA precursor could cause misincorporation of other amino acids at serine (UCN) codons or inappropriate incorporation of serine residues. To test this possibility, the 0913 and 1032 control cell lines and the IV-5 and IV-7 mutant cell lines were pulse-labeled for 30 min with [3H]serine, and their mitochondrial translation products were analyzed by SDS-PAGE. Protein labeling decreased by 13.5% in IV-7 cells and 46.4% in IV-5 cells relative to the average control labeling; these decreases are comparable to those observed in the same cell lines for the [35S]methionine labeling (15 and 52%, respectively). Moreover, all the products of the mutant cell lines were labeled with [3H]serine in the same relative proportion as in the control cell lines, arguing against either an excess or deficiency of serine incorporation in any of the polypeptides synthesized by mutant cells relative to the expected pattern (data not shown).
The average labeling of each polypeptide in the mutant cell lines, relative to that in the control cell lines, after a 30-min pulse with [35S]methionine, was decreased by 33 to 50%, except that of ND6, which was decreased by ∼59% (Fig. 5b). Interestingly, the reduction in labeling of the various polypeptides did not vary in relationship to the number of serine (UCN) codons in the corresponding mRNAs. In this respect, the behavior of the rate of synthesis versus serine content of the individual polypeptides was dramatically different from the changes in rate of synthesis as related to polypeptide lysine content which had been previously observed in cell lines carrying in nearly homoplasmic form the tRNALys mutation at position 8344 associated with MERRF encephalomyopathy (9). In the latter case, the data indicated clearly an exponential decrease in the rate of synthesis of the individual mitochondrial translation products with increasing lysine content, as a result of a ∼26% probability for a ribosome to fall off the mRNA at or near each lysine codon, releasing the incomplete polypeptide (9). It was therefore hypothesized that the different results obtained in U7445C mutation-carrying cells reflected the fact that the ribosomes did not fall off the mRNA at or near each serine codon but simply stalled for a certain time due to the deficiency of aminoacylated tRNASer(UCN) and then resumed elongation in the proper frame.
As shown in Fig. 5c, the experimentally determined labeling rates of all polypeptides, except ND6, when related to the proportion of serine (UCN) codons in the corresponding mRNAs, conformed well (P < 0.01) to the equation Rx = R0 {0.1/[0.1(1 − x) + 1.5x]} (R0 = 100.8), which describes the transient-pause model mentioned above. In this equation, Rx is the rate of synthesis of a given polypeptide in mutant cells relative to the rate in wild-type cells, both rates being expressed as reciprocals of the times required for their synthesis, and x is the proportion of serine (UCN) codons in the mRNA coding for the polypeptide. According to the equation, amino acids other than those specified by Ser (UCN) codons are assumed to be incorporated into the growing polypeptide chain at an average of one per 0.1 s and serine residues specified by UCN codons are incorporated at the same rate in wild-type cells; by contrast, in U7445C mutation-carrying cells, serine (UCN) residues are assumed to be incorporated at a rate of one per 1.5 s due to the deficiency of aminoacylated tRNASer(UCN) and the consequent pausing of the ribosomes at each serine (UCN) codon. While an average rate of incorporation of one amino acid per 0.1 s is a reasonable one for mitochondrial protein synthesis in human cells (unpublished observations), it should be emphasized that the critical parameter in the equation given above is the ratio of the rate of incorporation of amino acids other than serine residues specified by UCN codons to the rate of incorporation of such serine residues in mutant cells. The value of 1.5 s for the rate of incorporation of serine (UCN) residues in mutant cells was arrived at by trial and error as that which best fitted the data. It should be noted that the curve describing the equation given above extrapolates to about 100% rate of synthesis for a polypeptide lacking serine (UCN) residues. This would imply that, if the model underlying the equation is correct, there are no significant effects on the rate of mitochondrial protein synthesis of factors other than the proportion of serine residues. A very significant observation is that the rate of synthesis of the ND6 polypeptide deviated markedly from the curve describing the equation given above (Fig. 5c); this value was not considered in constructing the curve best fitting the other data. The origin of this deviation is discussed below.
Significant decrease in ND6 mRNA level.
The more marked decrease in the rate of ND6 synthesis in the mutant relative to the control cell lines than predicted from the transient-pause model called attention to a possible decrease in the amount of ND6 mRNA in mutant cells. Both tRNASer(UCN) and the ND6 mRNA are transcribed from the mtDNA L-strand and are derived from the processing of large polycistronic transcripts (29, 30) (Fig. 3). In order to identify and quantify the ND6 mRNA, RNA transfer hybridization experiments were carried out with total cell RNA from three control cell lines (0913, 0923, and 1032) and three mutant cell lines (IV-5, IV-7, and IV-15), using a [32P]dCTP-labeled ND6 mRNA-specific ssDNA probe (synthesized by the Klenow DNA polymerase, using the SP6 primer, from the pND6-1 plasmid; see Materials and Methods). After the blot was stripped, a 32P-labeled total HeLa cell mtDNA probe was used to provide size markers and standards for normalization purposes. These experiments revealed the presence of an RNA species ∼1.1 kb long and of another RNA species ∼2.4 kb long, as shown in Fig. 6a for two representative control cell lines, 0923 and 1032, and two representative mutant cell lines, IV-15 and IV-5. The ∼1.1-kb RNA is equal in size to the ND6 mRNA previously described in mouse (1.15 kb [6]) and rat cells (1.1 kb [42]) and is presumably the human equivalent. As described for the mouse and rat equivalents, the human ND6 mRNA is expected to encompass the 525-nt coding sequence and ∼600 nt of the 3′ untranslated sequence (Fig. 3). The 2.4-kb RNA could conceivably be a precursor of the ND6 mRNA. On the other hand, the ND6 mRNA encoded in the L-strand overlaps the ND5 mRNA encoded in the H-strand (Fig. 3). The identical size of the 2.4-kb RNA species and of the H-strand-encoded ND5 mRNA (29) raised the possibility that mispriming by the SP6 primer within the segment of the ND5 mRNA coding sequence contained in the pND6-1 plasmid or within adjacent vector sequences could produce a probe complementary to the ND5 mRNA. Indeed, when a riboprobe specific for the ND6 mRNA, synthesized on the pND6-1 plasmid template using the SP6 RNA polymerase, was hybridized with the RNA on the stripped blot, only the band corresponding to the ND6 mRNA was labeled (Fig. 6a). This result strongly supported the ND5 identification of the 2.4-kb RNA species.
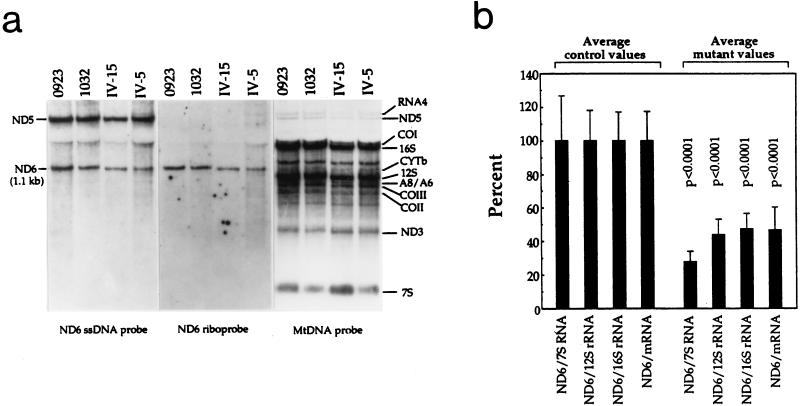
FIG. 6.
Identification and quantification of the ND6 mRNA in control and mutant lymphoblastoid cell lines. (a) Equal amounts (20 μg) of total cell RNA from the mutant cell lines IV-15 and IV-5 and the control cell lines 0923 and 1032 were electrophoresed through a 1.4% agarose-formaldehyde gel, transferred to a Zeta-probe membrane, and hybridized first with a 32P-labeled ND6-specific ssDNA probe, and subsequently (after the blot was stripped) with a human mtDNA probe 32P labeled by random priming. After a further restripping of the blot, the RNA was rehybridized with an ND6-specific RNA probe (riboprobe). See Materials and Methods for details. (b) Average relative ND6 mRNA content per cell normalized to the average content per cell of 7S RNA, 12S rRNA, 16S rRNA, and mRNAs in three control cell lines (0913, 0923, and 1032) and three mutant cell lines (IV-15, IV-7, and IV-5). The values for the latter are expressed as percentages of the average values for the control cell lines. Two independent determinations of ND6 mRNA content and one determination of the content of each of the four RNA reference markers for each cell line were used in the calculations. Graph details and symbols are explained in the legend to Fig. 2b.
In the experiment shown in Fig. 6a, the amount of ND6 mRNA is clearly lower in the two mutant cell lines than in the control cell lines and to a similar extent in both the blot hybridized with the ND6 mRNA-specific ssDNA probe and in the blot hybridized with the ND6 riboprobe. To quantify the ND6 mRNA in the various cell lines, the corresponding bands in the blot hybridized with either of the two probes which is shown in Fig. 6a and in another blot in which RNA preparations from the 0913 and IV-7 cell lines were similarly analyzed were subjected to densitometric analysis. For comparison, the levels of 16S and 12S rRNA, as representatives of the H-strand rDNA transcription unit (Fig. 3), the combined levels of A6 or A8 mRNA, COIII mRNA, COII mRNA, and ND3 mRNA, as representatives of the whole H-strand transcription unit (28) (Fig. 3), and the level of 7S RNA, the leader segment of the L-strand polycistronic transcript (29), as representative of the L-strand transcription unit (Fig. 3), were determined on the same blots stripped and hybridized with the whole mtDNA probe. The average levels of ND6 mRNA in the 0913, 0923, and 1032 control cell lines and in the IV-15, IV-7, and IV-5 mutant cell lines were normalized to the average levels determined in the same cell lines for each of the individual or combined reference RNAs and expressed relative to the corresponding average values obtained in the control cell lines (Fig. 6b). The average relative levels of ND6 mRNA in the three mutant cell lines tested, normalized with respect to the 12S rRNA or 16S rRNA or mRNAs, were fairly similar, ranging between ∼44% of the average control values (after normalization to 12S rRNA) and ∼47% (after normalization to 16S rRNA). These values reflected a very significant decrease of the ND6 mRNA level in the 7445 mutation-carrying cell lines (P < 0.0001 by the ANOVA test). The average relative level of ND6 mRNA in the mutant cell lines was considerably lower when normalized with respect to 7S RNA, i.e., ∼28%. This result may reflect a compensatory stimulation of L-strand transcription in the mutant cell lines (see below). The averages of the ND6 mRNA levels normalized to 12S rRNA or to 16S rRNA or to mRNAs, expressed relative to the overall average control values, were 43, 60, and 35% in the three mutant cell lines IV-15, IV-7, and IV-5, respectively, and 123, 101, and 76% in the control cell lines 0913, 0923, and 1032, respectively.
Specific decrease in complex I-dependent respiration.
The total respiration capacities of the four control and four mutant cell lines were measured by determining the O2 consumption rate in intact cells (Fig. 7a). The rate of total O2 consumption in the four mutant cell lines revealed a variable decrease relative to the mean value measured in the wild-type cell lines; this decrease ranged from ∼10 to ∼45%, with an average reduction of ∼22% (P < 0.0001 by the ANOVA test). The variations in overall respiration among the individual control and mutant cell lines showed a very significant correlation with the corresponding variations in rate of mitochondrial protein synthesis (r = 0.94; P < 0.001).
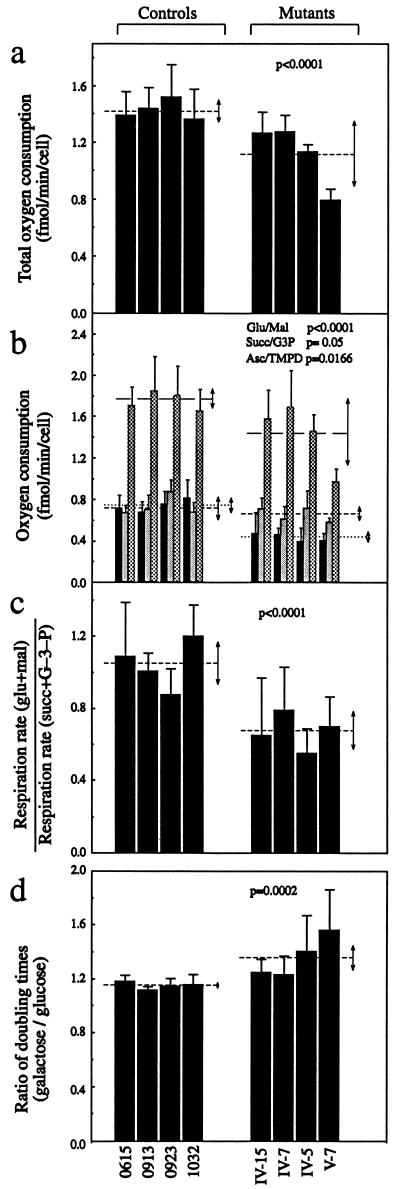
FIG. 7.
Respiration and growth assays. (a) Average rates of total O2 consumption per cell measured in different lymphoblastoid cell lines. Four to eight determinations were carried out for each cell line. (b) Polarographic analysis of O2 consumption in digitonin-permeabilized cells of different cell lines with different substrates. The activities of the various components of the respiratory chain were determined as respiration dependent on glutamate or malate (Glu/Mal) (solid bars; group averages indicated by dotted lines), succinate or glycerol-3-phosphate (G-3-P) (Succ/G3P) (shaded bars; group averages indicated by short-dash lines), or ascorbate plus TMPD (Asc/TMPD) (cross-hatched bars; group averages indicated by long-dash lines). Three to eight determinations were carried out for each cell line. (c) Ratios of glutamate and malate (glu+mal)-driven respiration to succinate and G-3-P (succ+G–3–P)-driven respiration. (d) Ratios of DTs in galactose-containing medium to DTs in glucose-containing medium in different cell lines. Four determinations were carried out for each cell line. Graph details and symbols are explained in the legend to Fig. 5a.
In order to investigate which of the enzyme complexes of the respiratory chain was affected in the mutant cell lines, O2 consumption measurements were carried out on digitonin-permeabilized cells, using different substrates and inhibitors (14). As illustrated in Fig. 7b, in the mutant cell lines, the rate of glutamate- and malate-driven respiration, which depends on the activities of NADH:ubiquinone oxidoreductase (complex I), ubiquinol-cytochrome c reductase (complex III), and cytochrome c oxidase (complex IV) but usually reflects the rate-limiting activity of complex I was very significantly decreased relative to the average rate in the control cell lines, with decreases of 37 to 47% (∼42% on the average; P < 0.0001 by the ANOVA test). By contrast, the rate of succinate- and glycerol-3-phosphate (G-3-P)-driven respiration, which depends on the activities of complex III and complex IV but usually reflects the activity of complex III, showed a decrease in the mutant cell lines relative to the average control value, which was barely significant (P = 0.05). The rate of ascorbate–N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD)-driven respiration, which reflects the activity of complex IV, showed a significant reduction in the mutant cell lines, relative to the average control value (P = 0.0166); here, however, a considerable contribution to the decrease was given by the cell line V-7, which exhibited an ∼46% reduction, for unknown reasons. Figure 7c shows that the glutamate- and malate-driven respiration rates of the various mutant cell lines, normalized to their succinate- and G-3-P-driven respiration rates, were very significantly decreased compared to the mean value in the control cell lines, by an average of ∼36% (P < 0.0001 by the ANOVA test). The results discussed above strongly suggested that the most important effect of the mutation at position 7445 on the respiratory function of the mutant cell lines was a marked specific decrease in the activity of complex I.
Growth properties of cell lines in glucose- or galactose-containing medium.
It has been shown that cell lines with impaired oxidative phosphorylation activity have reduced growth capacity in medium containing galactose instead of glucose (14, 15, 17). In order to investigate whether the respiratory defects detected in the lymphoblastoid cell lines carrying the mutation at position 7445 affected their growth capacity, the DTs of the mutant cell lines in special DMEM-galactose and in special DMEM-glucose were compared to those of the control cell lines in the same media. The DTs of the latter cell lines were somewhat higher in galactose-containing medium (average, 52 h) than in glucose-containing medium (average, 45 h). The DTs of the mutant cell lines in galactose-containing medium, though showing considerably variation, exhibited a tendency to be increased relative to the values in glucose-containing medium much more than observed in the control cell lines (from an average of 68 h to an average of 95 h). The ratios of DTs in galactose-containing medium to those in glucose-containing medium showed a clear increase in the mutant versus the control cell lines (from an average of ∼1.15 to an average of ∼1.36; P = 0.0002 by the ANOVA test) (Fig. 7d), confirming the existence of defective oxidative metabolism in the cells carrying the mutation at position 7445. The variations in DT ratio among the individual control and mutant cell lines were inversely correlated with the rate of total O2 consumption (r = 0.96; P < 0.001) and with the rate of mitochondrial protein synthesis (r = 0.91; P < 0.01).
DISCUSSION
Site and primary target of the mutation at position 7445.
The observation made in the present work that the mutant and wild-type circularized mitochondrial tRNASer(UCN) had an identical 5′-3′-end junction sequence lacking nt 7445 and the results of experiments of in vitro processing of a synthetic human tRNASer(UCN) by partially purified mitochondrial RNase P have demonstrated that the mutation flanks the 3′ end of the tRNA. Therefore, the revised structural model originally proposed for bovine tRNA (47) also applies to the human tRNASer(UCN). Furthermore, these findings have excluded the possibility that the mutation alters qualitatively the processing of the primary transcript. Therefore, the most plausible interpretation of the observed reduction in tRNASer(UCN) level in the mutant cell lines is that it is due to an effect of the mutation on the rate of processing of the primary transcripts, in agreement with an earlier suggestion (33).
Pleiotropic secondary effects of the mutation at position 7445.
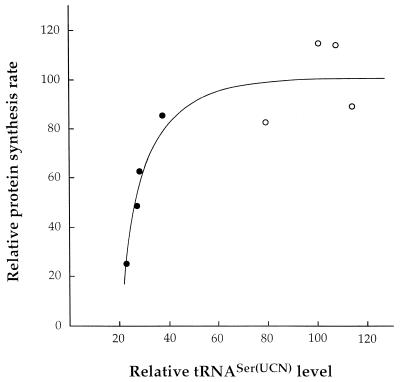
It is reasonable to interpret the overall decrease in rate of organelle-specific protein synthesis in the mutant cell lines analyzed here as being due to the 62 to 77% reduction in the level of total tRNASer(UCN) and to the corresponding expected decrease in the amount of aminoacylated tRNASer(UCN). A plot of the mitochondrial tRNASer(UCN) content in the various control and mutant cell lines versus their rate of organelle-specific translation has revealed a sharp threshold in the capacity of this tRNA to support protein synthesis. As shown in Fig. 8, ∼40% of the control level of the tRNASer(UCN) appeared to be the minimum required to support the wild-type rate of protein synthesis in mitochondria of the lymphoblastoid cell lines. This threshold very probably accounts for the recent report that the single lymphoblastoid cell line analyzed from the Scottish pedigree, which exhibited a 60 to 65% decrease in tRNASer(UCN) level, showed no significant decrease in overall mitochondrial protein synthesis, although it exhibited a slight reduction in growth rate in medium containing galactose versus glucose (33). It is interesting that previous work on cell lines carrying the tRNALys gene A8344G transition associated with the MERRF syndrome had indicated that ∼50% of the control level of aminoacylated tRNALys is the minimum required to support the rate of protein synthesis necessary for full respiratory competence (9). Similarly, the presence of ∼45% of wild-type mtDNA had been found to be the minimum allowing complementation of five missing tRNA genes comprised in the 5-kbp mtDNA deletion associated with chronic progressive external ophthalmoplegia (15). Therefore, the available evidence strongly suggests that, in mammalian mitochondria, there is not a large excess of tRNAs over the levels allowing normal translation.
FIG. 8.
Relationship between the relative level of tRNASer(UCN) and relative rate of mitochondrial protein synthesis in mutant (solid circles) and wild-type (open circles) cell lines. The values of each parameter for the different cell lines are expressed as percentages of the average values for the control cell lines.
The markedly variable decrease in protein synthesis rate, which has been observed in the present work in different mutant cell lines, has clearly shown the important role that the nuclear background plays in determining the severity of the biochemical phenotype of mtDNA-linked diseases. This role has been previously demonstrated for LHON, which is associated with ND gene mutations (19, 20, 43), and for the nonsyndromic deafness associated with the 12S rRNA gene mutation at position 1555 (14).
The present observations on the effects of the mutation at position 7445 on mitochondrial protein synthesis differ from those that have been previously made in mtDNA-less cell transformants carrying in nearly homoplasmic form the tRNALys A8344G mutation associated with the MERRF syndrome (9). In these transformants, a reduction in the level of aminoacylated tRNALys of 50 to 60% was found to cause a decrease in overall rate of protein synthesis of 80 to 90%; furthermore, the synthesis of the higher-molecular-weight translation products was much more affected than that of the smaller products. By contrast, in the cells carrying the mutation at position 7445, an average decrease of ∼45% in protein synthesis rate and no significant difference in the labeling of the larger versus the smaller mitochondrially synthesized polypeptides were observed. In these cells, the lack of any effect of the absolute serine (UCN) content on the rate of synthesis of the individual mitochondrial translation products and the absence of any abnormal translation products have clearly excluded any model involving premature termination of translation at or near each serine (UCN) codon, in contrast to what was previously shown in cells carrying the tRNALys mutation at position 8344 (9). In contrast, the transient-pause model, which assumes that ribosomes stall at each serine UCN codon for a certain time and then resume elongation in the proper frame, has been found to fit well the data on the relationship between reduction in labeling of the various polypeptides and their proportion of serine (UCN) residues.
The dropping-off propensity of mitochondrial tRNALys in human cell lines carrying the MERRF mutation at position 8344 may be related to an unconventional anticodon conformation, which has been associated, in both Escherichia coli and mammalian cytosolic tRNALys species, with the occurrence of hypermodified nucleotides in the wobble position 34 of the anticodon and at position 37 (1, 41). Recently, these hypermodified nucleotides have also been identified in wild-type human mitochondrial tRNALys (16).
As to the explanation for the much more pronounced decrease than expected from the transient-pause model which was observed in the rate of synthesis of the ND6 polypeptide in the cell lines carrying the mutation at position 7445, a crucial observation made in the present work is the significant reduction in these cell lines in the amount of the ND6 mRNA. This reduction does not appear to be due to a decrease in the rate of synthesis of this RNA. In fact, the significantly larger amount of 7S RNA, the leader segment of the L-strand polycistronic transcript (27), in the mutant cell lines relative to those of the control cell lines argues rather in favor of an increase, possibly compensatory, in the rate of L-strand transcription. Therefore, the finding of a strong decrease in ND6 mRNA level in the mutant cell lines points to a reduced stability of the ND6 mRNA precursor. This interpretation provides strong support for the idea that the more marked deficiency in the synthesis of the ND6 subunit compared to the other mtDNA-encoded polypeptides in the mutant cells is related to the processing defect of the tRNASer(UCN) precursor. This idea, as discussed below, is based on the fact that the ND6 mRNA is derived from the same precursor as the tRNASer(UCN) (29, 30).
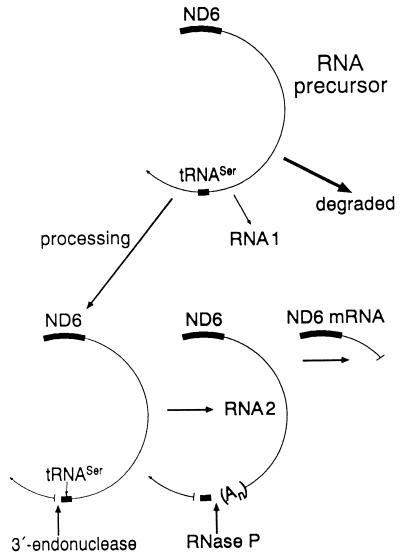
Among the L-strand polycistronic transcripts of HeLa cell mtDNA, three overlapping polyadenylated RNA species, RNAs 1, 2, and 3 (29, 30), had been previously identified, which correspond with their common 5′ end to the 5′ end of the ND6 reading frame, and extend for ∼10,400, ∼7,070, and ∼4,155 nt, respectively (Fig. 3). Of these, RNA 2 is by far the most abundant (4, 13) and extends up to the tRNASer(UCN) without including it (29, 30). The processing of the fast turning-over mtDNA L-strand transcripts is a highly regulated phenomenon, which ensures both the accurate cleavages leading to the formation of eight L-strand-encoded tRNAs and of the ND6 mRNA and the controlled disposal of the large excess of transcripts (4). In this process, the order of the cleavage events may play a determining role. In vitro tRNA processing experiments (36) and in vivo observations (12, 37) have indicated that the order of 5′- and 3′-end cleavages of the tRNA precursors in human mitochondria may vary depending on the tRNA. Other in vitro processing experiments utilizing partially purified mitochondrial RNase P from HeLa cells have shown that a tRNASer(UCN) precursor carrying a 3′-terminal CCA is accurately cleaved by the enzyme at the 5′ end of the tRNA (32a). This result is consistent with the idea that, in the formation of the tRNASer(UCN), the 3′-end cleavage by the mitochondrial 3′ endonuclease precedes the 5′-end cleavage by the mitochondrial RNase P, which is possibly a regulated step (Fig. 9). Furthermore, it is well established that all RNA species other than tRNAs which are derived from the processing of the primary mtDNA transcripts are polyadenylated (30). Therefore, the hypothesis that a failure in tRNASer(UCN) processing and the consequent lack of stabilization by polyadenylation of the upstream L-strand transcript segment (RNA 2), encompassing the ND6 mRNA, would lead to the degradation of the latter RNA is very plausible (Fig. 9). The occurrence, which was mentioned above, of a mechanistic link between the processing defect of the tRNASer(UCN) precursor and the decreased stability of the ND6 mRNA precursor is strongly supported by the observation that the amount of ND6 mRNA in three control and three mutant cell lines was correlated with the tRNASer(UCN) level (r = 0.923; P < 0.01). The results reported here thus represent a striking example of long-distance effects of a posttranscriptional event.
FIG. 9.
Model illustrating the possible role of the tRNASer(UCN) precursor processing on the stabilization by polyadenylation of the L-strand transcript RNA 2. See text for details.
Effects of the mutation at position 7445 on respiration.
In the control and mutant cell lines analyzed here, there was a very significant correlation between the rate of mitochondrial protein synthesis, on one hand, and overall respiratory capacity (P < 0.001) or relative growth rate in galactose- versus glucose-containing medium (P < 0.01), on the other. This correlation is clearly consistent with an important role that the marked decrease in tRNASer(UCN) steady-state level in the mutant cell lines plays in producing their overall respiration and growth defects. On the other hand, a very significant reduction (P < 0.0001) in glutamate- or malate-dependent O2 consumption has been observed in the mutant cell lines compared to the control cell lines. This reduction contrasted with a marginally significant decrease in the same cell lines in complex III- and IV-dependent O2 consumption and pointed to a specific NADH dehydrogenase deficiency. These observations have called attention to the possibility of another factor(s) being involved in the respiratory phenotype of these cell lines. The evidence presented here strongly suggests that one such factor is the significant reduction in the level of ND6 mRNA caused by the mutation, which accentuates the decrease in rate of synthesis of the ND6 subunit beyond the general effect on mitochondrial protein synthesis caused by the tRNASer(UCN) depletion. Recent work on cultured mutant mouse cells carrying different proportions of mtDNA with a nonsense mutation in the gene encoding another subunit of NADH dehydrogenase, ND5, has shown that the glutamate- or malate-dependent respiration declines in these mutants in parallel with the rate of synthesis of ND5, indicating that the synthesis of this subunit is nearly rate-limiting for respiration (5a). The observation made in the present work that an ∼59% average decrease in rate of synthesis of the ND6 subunit in the mutant cell lines is associated with an average reduction of ∼42% in glutamate- or malate-dependent respiration suggests that the above conclusion concerning the ND5 subunit also applies to the ND6 subunit. These findings support the conclusion that, in mammalian mitochondria, there is not a large excess of protein synthesis over the rate necessary to support normal respiration. Other work on mouse cells has shown that the ND6 subunit is essential for the assembly and function of complex I (5). Furthermore, several mutations in the ND6 gene have been shown to cause diseases in humans. Thus, a mutation at position 14,484 (24) has been found to be associated with LHON, and a mutation at position 14,459 (44) or a combination of mutations at positions 4,160 and 14,484 (20) has been found to be associated with LHON and dystonia.
Possible synergistic role of ND gene mutations.
Another factor which may play a synergistic role in the establishment of the respiratory phenotype of the mutant cell lines is the occurrence in these of several homoplasmic mtDNA mutations affecting subunits of NADH dehydrogenase. Among these complex I subunit mutations, that in the ND1 gene at position 4216 (which changes a nonconserved tyrosine to a histidine) and that in the ND5 gene at position 13708 (which changes a moderately conserved alanine to a threonine) belong to the group of so-called secondary LHON mutations (22). These mutations confer upon an individual a lower risk for developing LHON than any of the primary LHON mutations (occurring in the ND4 gene at position 11708, in the ND1 gene at position 3460, in the ND6 gene at position 14459 or 14484, and in the apocytochrome b gene at position 15257) (23, 44), and it has been proposed that they can act synergistically with such primary mutations. The results of a recent haplotype and phylogenetic analysis has indeed suggested that the best candidate for increasing the penetrance of primary LHON mutations is the ancient combination of mutations at positions 4216 and 13708 (40), which has also been found in the present deafness pedigree (11). Another sequence change found in the present work in the ND3 gene of the mutant cell lines at position 10084 (which changes an isoleucine to a threonine) is a previously undescribed rare mutation. In the New Zealand pedigree, the clinical disorder has higher penetrance than in the Scottish pedigree, and it is reasonable to suggest that these coexisting mutations may be responsible.
ACKNOWLEDGMENTS
This study was supported in part by NIH grant GM11726 to G.A., grant 5RO1 DC 0142-04 from the National Institute on Deafness and Other Communication Disorders, NIH, to N.F.-G., and the New Zealand Deafness Research Foundation.
We thank Anne Chomyn for help in the statistical analysis, valuable advice, and useful discussions, and we thank Benneta Keeley, Arger Drew, and Rosario Zedan for technical assistance.
REFERENCES
- 1.Agris P F, Guenther R, Ingram P C, Basti M M, Stuart J W, Sochacka E, Malkiewicz A. Unconventional structure of tRNALysSUU anticodon explains tRNA’s role in bacterial and mammalian ribosomal frameshifting and primer selection by HIV-1. RNA. 1997;3:420–428. [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson S, Bankier A T, Barrell B G, de Bruijn M H L, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Rose B A, Sanger F, Schreier P H, Smith A J H, Staden R, Young I G. Comparison of the human and bovine mitochondrial genomes. In: Slonimski P, Borst P, Attardi G, editors. Mitochondrial genes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 5–43. [Google Scholar]
- 3.Anderson S, Bankier A T, Barrell B G, de Bruijn M H L, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Rose B A, Sanger F, Schreier P H, Smith A J H, Staden R, Young I G. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 4.Attardi G, Chomyn A, King M P, Kruse B, Polosa P L, Murdter N N. Regulation of mitochondrial gene expression in mammalian cells. Biochem Soc Trans. 1990;18:509–513. doi: 10.1042/bst0180509. [DOI] [PubMed] [Google Scholar]
- 5.Bai Y, Attardi G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 1998;17:4848–4858. doi: 10.1093/emboj/17.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Bai, Y., and G. Attardi. Unpublished observations.
- 6.Bhat K S, Bhat N K, Kulkarni G R, Iyengar A, Avadhani N G. Expression of the cytochrome b-URF6-URF5 region of the mouse mitochondrial genome. Biochemistry. 1985;24:5818–5825. doi: 10.1021/bi00342a020. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Chomyn A, Lai S S-A T. cDNA of the 24 kDa subunit of the bovine respiratory chain NADH dehydrogenase: high sequence conservation in mammals and tissue-specific and growth-dependent expression. Curr Genet. 1989;16:117–125. doi: 10.1007/BF00393404. [DOI] [PubMed] [Google Scholar]
- 9.Enriquez J A, Chomyn A, Attardi G. MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNA and premature translation termination. Nat Genet. 1995;10:47–55. doi: 10.1038/ng0595-47. [DOI] [PubMed] [Google Scholar]
- 10.Estivill X, Govea N, Barcelo A, Perello E, Badenas C, Romero E, Moral L, Scozzari R, D’Urbano L, Zeviani M, Torroni A. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment with aminoglycosides. Am J Hum Genet. 1998;62:27–35. doi: 10.1086/301676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischel-Ghodsian N, Prezant T R, Fournier P, Stewart I A, Maw M. Mitochondrial mutation associated with non-syndromic deafness. Am J Otolaryngol. 1995;16:403–408. doi: 10.1016/0196-0709(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 12.Gaines G, Attardi G. Intercalating drugs and low temperatures inhibit synthesis and processing of ribosomal RNA in isolated human mitochondria. J Mol Biol. 1984;172:451–466. doi: 10.1016/s0022-2836(84)80017-0. [DOI] [PubMed] [Google Scholar]
- 13.Gelfand R, Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol Cell Biol. 1981;1:497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan M X, Fischel-Ghodsian N, Attardi G. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet. 1996;6:963–971. doi: 10.1093/hmg/5.7.963. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi J-L, Ohta S, Kikuchi A, Takemitsu M, Goto Y-I, Nonaka I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc Natl Acad Sci USA. 1991;88:10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helm M, Brulé H, Degoul F, Cepanec C, Leroux J-P, Giegé R, Florentz C. The presence of modified nucleotides is required for cloverlead folding of a human mitochondrial tRNA. Nucleic Acids Res. 1998;26:1636–1643. doi: 10.1093/nar/26.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofhaus G, Johns D R, Hurko O, Attardi G, Chomyn A. Respiration and growth defects in transmitochondrial cell lines carrying the 11778 mutation associated with Lebers hereditary optic neuropathy. J Biol Chem. 1996;271:13155–13161. doi: 10.1074/jbc.271.22.13155. [DOI] [PubMed] [Google Scholar]
- 18.Hofhaus G, Shakeley R M, Attardi G. Use of polarography to detect respiration defects in cell cultures. Methods Enzymol. 1996;264:476–483. doi: 10.1016/s0076-6879(96)64043-9. [DOI] [PubMed] [Google Scholar]
- 19.Howell N, Bindoff L A, McCullough D A, Kubacka I, Poulton J, Mackey D, Taylor L, Turnbull D M. Leber hereditary optic neuropathy: identification of the same mitochondrial ND1 mutation in six pedigrees. Am J Hum Genet. 1991;49:939–950. [PMC free article] [PubMed] [Google Scholar]
- 20.Howell N, Kubacka I, Xu M, McCullough D A. Leber hereditary optic neuropathy: involvement of the mitochondrial ND1 gene and evidence for an intragenic suppression mutation. Am J Hum Genet. 1991;48:935–942. [PMC free article] [PubMed] [Google Scholar]
- 21.Hutchin T, Haworth I, Higashi K, Fischel-Ghodsian N, Stoneking M, Saha N, Arnos C, Cortopassi G. A molecular basis for human hypersensitivity to aminoglycoside antibiotics. Nucleic Acids Res. 1993;21:4174–4179. doi: 10.1093/nar/21.18.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johns D R, Berman J. Alternative, simultaneous Complex I mitochondrial DNA mutations in Leber’s hereditary optic neuropathy. Biochem Biophys Res Commun. 1991;174:1324–1330. doi: 10.1016/0006-291x(91)91567-v. [DOI] [PubMed] [Google Scholar]
- 23.Johns D R, Neufeld M J. Cytochrome b mutations in Leber hereditary optic neuropathy. Biochem Biophys Res Commun. 1991;181:1358–1364. doi: 10.1016/0006-291x(91)92088-2. [DOI] [PubMed] [Google Scholar]
- 24.Johns D R, Neufeld M J, Park R D. An ND6 mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Biochem Biophys Res Commun. 1992;187:1551–1557. doi: 10.1016/0006-291x(92)90479-5. [DOI] [PubMed] [Google Scholar]
- 25.King M P, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 26.King M P, Attardi G. Post-transcriptional regulation of the steady-state levels of mitochondrial tRNAs in HeLa cells. J Biol Chem. 1993;268:10228–10237. [PubMed] [Google Scholar]
- 27.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci USA. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montoya J, Gaines G L, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 29.Ojala D, Merkel C, Gelfand R, Attardi G. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell. 1980;22:393–403. doi: 10.1016/0092-8674(80)90350-5. [DOI] [PubMed] [Google Scholar]
- 30.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 31.Oliver N A, Greenberg R D, Wallace D C. Assignment of a polymorphic polypeptide to the human mitochondrial DNA unidentified reading frame 3 gene by a new peptide mapping strategy. J Biol Chem. 1983;258:5834–5839. [PubMed] [Google Scholar]
- 32.Prezant T R, Agapian J V, Bohlman M C, Bu X, Öztas S, Qiu W-Q, Arnos K S, Cortopassi G A, Jaber L, Rotter J I, Shohat M, Fischel-Ghodsian N. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 32a.Puranam, R. S., and G. Attardi. Unpublished data.
- 33.Reid F M, Rovio A, Holt I J, Jacobs H T. Molecular phenotype of a human lymphoblastoid cell-line homoplasmic for the np-7445 deafness-associated mitochondrial mutation. Hum Mol Genet. 1997;6:443–449. doi: 10.1093/hmg/6.3.443. [DOI] [PubMed] [Google Scholar]
- 34.Reid F M, Vernham G A, Jacobs H T. A novel mitochondrial point mutation in a maternal pedigree with sensorineural deafness. Hum Mutat. 1994;3:243–247. doi: 10.1002/humu.1380030311. [DOI] [PubMed] [Google Scholar]
- 35.Reid F M, Vernham G A, Jacobs H T. Complete mtDNA sequence of a patient in a maternal pedigree with sensorineural deafness. Hum Mol Genet. 1994;3:1435–1436. doi: 10.1093/hmg/3.8.1435. [DOI] [PubMed] [Google Scholar]
- 36.Rossmanith W, Tullo A, Potuschak T, Karwan R, Sbisà E. Human mitochondrial tRNA processing. J Biol Chem. 1995;270:12885–12891. doi: 10.1074/jbc.270.21.12885. [DOI] [PubMed] [Google Scholar]
- 37.Sbisà E, Tullo A, Nardelli M, Tanzariello F, Saccone C. Transcription mapping of the Ori L region reveals novel precursors of mature RNA species and antisense RNAs in rat mitochondrial genome. FEBS Lett. 1992;296:311–316. doi: 10.1016/0014-5793(92)80311-4. [DOI] [PubMed] [Google Scholar]
- 38.Sevior K B, Hatamochi A, Stewart I A, Bykhovskaya Y, Allen-Powell D R, Fischel-Ghodsian N, Maw M A. Mitochondrial A7445G mutation in two pedigrees with palmoplantar keratoderma and deafness. Am J Med Genet. 1998;75:179–185. [PubMed] [Google Scholar]
- 39.Tiranti V, Chariot P, Carella F, Toscano A, Soliveri P, Girlanda P, Carrara F, Fratta G M, Reid F M, Mariotti C, Zeviani M. Maternally inherited hearing loss, ataxia and myoclonus associated with a novel point mutation in mitochondrial tRNASer(UCN) gene. Hum Mol Genet. 1995;4:1421–1427. doi: 10.1093/hmg/4.8.1421. [DOI] [PubMed] [Google Scholar]
- 40.Torroni A, Petrozzi M, Durbano L, Sellitto D, Zeviani M, Carrara F, Carducci C, Leuzzi V, Carelli V, Barboni P, De Negri A, Scozzari R. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuchihashi Z, Brown P O. Sequence requirements for efficient translational frameshifting in the Escherichia coli DNAX gene and the role of an unstable interaction between tRNALys and an AAG lysine codon. Genes Dev. 1992;6:511–519. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- 42.Tullo A, Tanzariello F, D’Erchia A M, Nardelli M, Papeo P A, Sbisà E, Saccone C. Transcription of rat mitochondrial NADH-dehydrogenase subunits. Presence of antisense and precursor RNA species. FEBS Lett. 1994;354:30–36. doi: 10.1016/0014-5793(94)01080-3. [DOI] [PubMed] [Google Scholar]
- 43.Wallace D C, Singh G, Lott M T, Hodge J A, Schurr T G, Lezza A M, Elsas L J, Nikoskelainen E K. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 44.Wallace D C, Shoffner J M, Trounce I, Brown M D, Ballinger S W, Corral-Debrinski M, Horton T, Jun A S, Lott M T. Mitochondrial DNA mutations in human degenerative diseases and aging. Biochim Biophys Acta. 1995;1271:141–151. doi: 10.1016/0925-4439(95)00021-u. [DOI] [PubMed] [Google Scholar]
- 45.Yatscoff R W, Goldstein S, Freeman K B. Conservation of genes coding for proteins synthesized in human mitochondria. Somatic Cell Genet. 1978;4:633–645. doi: 10.1007/BF01543155. [DOI] [PubMed] [Google Scholar]
- 46.Yokobori S, Pääbo S. Transfer RNA editing in land snail mitochondria. Proc Natl Acad Sci USA. 1995;92:10432–10435. doi: 10.1073/pnas.92.22.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokogawa T, Watanabe Y, Kumazawa Y, Ueda T, Hirao I, Miura K, Watanabe K. A novel cloverleaf structure found in mammalian mitochondrial transfer RNASer (UCN) Nucleic Acids Res. 1991;19:6101–6105. doi: 10.1093/nar/19.22.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]