Abstract
The accumulation of self-renewed polarized microglia in the penumbra is a critical neuroinflammatory process after ischemic stroke, leading to secondary demyelination and neuronal loss. Although known to regulate tumor cell proliferation and neuroinflammation, HDAC3’s role in microgliosis and microglial polarization remains unclear. We demonstrated that microglial HDAC3 knockout (HDAC3-miKO) ameliorated poststroke long-term functional and histological outcomes. RNA-seq analysis revealed mitosis as the primary process affected in HDAC3-deficent microglia following stroke. Notably, HDAC3-miKO specifically inhibited proliferation of proinflammatory microglia without affecting anti-inflammatory microglia, preventing microglial transition to a proinflammatory state. Moreover, ATAC-seq showed that HDAC3-miKO induced closing of accessible regions enriched with PU.1 motifs. Overexpressing microglial PU.1 via an AAV approach reversed HDAC3-miKO–induced proliferation inhibition and protective effects on ischemic stroke, indicating PU.1 as a downstream molecule that mediates HDAC3’s effects on stroke. These findings uncovered that HDAC3/PU.1 axis, which mediated differential proliferation-related reprogramming in different microglia populations, drove poststroke inflammatory state transition, and contributed to pathophysiology of ischemic stroke.
HDAC3/PU.1 axis regulates mitosis of proinflammatory microglia in ischemic stroke.
INTRODUCTION
Upon ischemic stroke, microglia, the major brain resident immune cell, rapidly shift to an “activated” state from a surveillance state within minutes after ischemia onset. In the subacute phase of ischemic stroke, massive self-renewed microglia occupy the ischemic penumbra that retains compromised blood flow, serving as the first nonneuron cell that responds to injury. Pharmacological inhibition of microglial proliferation attenuates neuropathic pain hypersensitivities (1) and prevents progression of Alzheimer’s disease–like pathology (2), while the cell cycle of microglia in stroke has been hardly observed. Notably, in the context of ischemic stroke, previous reports involving microglia have often focused on the immune-related role of microglia, such as phagocytosis or inflammation over microglia proliferation (3). Although a large portion of these reports also documented impaired amplification of the microglia population following beneficial interventions to stroke (4), the exact role of these activated microglia remains controversial. Depleting microglia using colony-stimulating factor 1 receptor (CSF-1R) inhibitors exacerbates stroke-induced neuronal excitotoxicity, leukocyte infiltration, and brain injury (5). Another microglia-depleting system using CXCR1creER:iDTR transgenic mice and diphtheria toxin inhibits neuronal degeneration and increases the expression level of anti-inflammatory factors instead (6). Considering the heterogeneity of microglia, it seems reasonable that these controversial conclusions may result from inhibited proliferation of certain specific subsets of microglia that are either beneficial or detrimental. Therefore, we propose that the proliferative microglia are an essential target in stroke progression.
By removing acetyl groups from lysine residues on the N-terminal tails of histones, histone deacetylases (HDACs) play a crucial role in chromatin modification and the consequent transcriptional regulation of genes associated with inflammation, metabolism, and proliferation (7). Among various class I HDACs, only HDAC3 is a stoichiometry component of both the nuclear receptor corepressor 1 (NCoR1 or NCoR) and silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) (8). Recently, HDAC3 has been found to be a target for neuroinflammation in some neurological diseases (9, 10). It was shown that microglia-specific HDAC3 deletion attenuated stroke-induced neuroinflammation by regulating the microglial cGAS-STING pathway (9). In a mouse model of traumatic brain injury (TBI), HDAC3 knockout also drove microglial turnover from a proinflammatory phenotype to an inflammation-resolving phenotype (10). Intriguingly, these findings, together with a few related studies using pan-HDAC inhibitors, also showed that depletion of HDAC3 caused a remarkable reduction of microglia number in the acute phase of diseases (11), suggesting its simultaneous involvement in microgliosis, strongly reminiscent of its initial role as an antiproliferative target for cancer therapy (12).
As reported in previous studies, PU.1, whose expression is limited to microglia in the central nervous system, controls the development and function of microglia. Early microgliogenesis is dependent on PU.1 level (13). It was also verified to be a master regulator of multiple cell cycle genes in hematopoietic stem cells from which macrophages develop (14). Notably, knockdown or overexpression of PU.1 in the BV2 rodent microglia cell line identified a hub of neuroinflammation-related differentially expressed genes (DEGs) (15). In various neurological disorders, attenuating PU.1 can potentially restrain microglia-mediated neuroinflammation (16).
Here, we have demonstrated that, in addition to restoring microglia polarization as in previous reports, depleting HDAC3 impaired microglial acute proliferation after stroke by inducing cell cycle arrest at the G2-M phase. Notably, while HDAC3 knockout brought CD16+ microglia to a halt, Arg1+ microglia did not show such change, suggesting selective implication of HDAC3 in the proliferation of different microglia populations. Furthermore, combining RNA sequencing (RNA-seq) with assay for transposase-accessible chromatin sequencing (ATAC-seq), we exploited the underlying epigenetic mechanism that PU.1 might be the critical transcriptional factor contributing to HDAC3-regulated microglia proliferation and polarization. Therefore, we used a recently developed adeno-associated virus (AAV) approach, which is able to specifically target microglia (17), and revealed that overexpressing microglial PU.1 reversed microglial HDAC3 knockout (HDAC3-miKO)–induced proliferation inhibition and protective effects on ischemic stroke, suggesting the contribution of PU.1 to HDAC3’s impact. Overall, our results unveiled the complex effects of microglial HDAC3 and highlighted the notable contributions of blocking microglial cell cycle that was seriously underestimated in previous studies. This may broaden our understanding of the multifaceted nature of microglia and expand our knowledge of inflammatory activation in the context of ischemic stroke, thereby opening a window of promising therapies and even for the use of some old anti-proliferative medications.
RESULTS
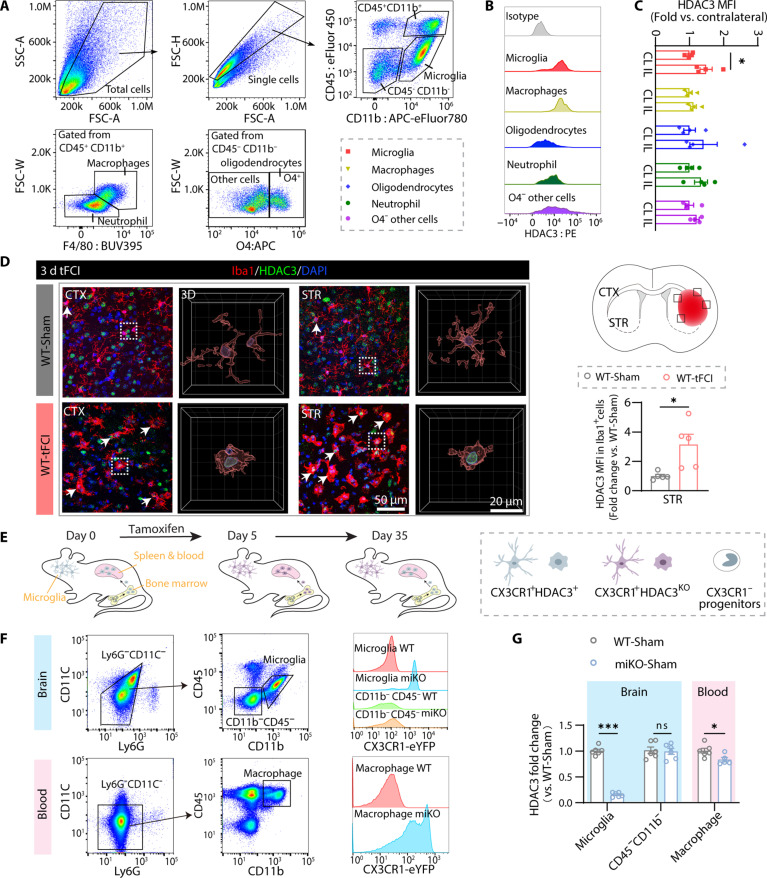
Microglial HDAC3 expression increases after ischemia and the engineering of HDAC3-miKO mice
Pharmacologically inhibiting HDAC3 has been reported to protect blood-brain barrier and prevent neuronal loss and down-regulate inflammasome in experimental stroke, suggesting an important role of HDAC3 in brain cells that contributes to the stroke pathology. In this case, to determine the cell type that was most susceptible to HDAC3 inhibition and thereby served as the key HDAC3-deficient protector for ischemic stroke, we first used flow cytometry to assess HDAC3 expression after transient focal cerebral ischemia (tFCI) in different brain cells (CD45intCD11b+ microglia, CD45+CD11b+F4/80+ macrophages, CD45+CD11b+F4/80− neutrophils, CD45+CD11b−O4+ oligodendrocytes, and CD45−CD11b−O4− other cells) 3 days following tFCI (Fig. 1A). As an essential epigenetic modifier, HDAC3 was expressed in all the detected cell types (Fig. 1B), consistent with a previous study (18). However, compared to the contralateral hemisphere, the injury side had higher expression of HDAC3 exclusively in microglia (Fig. 1C). Immunofluorescence staining confirmed that microglia had their processes contracted and cell body expanded. HDAC3 was colabeled with more microglia and remained mainly localized in the microglial nucleus in the acute phase of ischemic stroke (Fig. 1D). Meanwhile, we observed that neurons and oligodendrocytes both exhibited high levels of HDAC3 expression but remained unchanged following tFCI, while astrocytes exhibited low levels of HDAC3 expression and did not show an increase following tFCI (fig. S1, C and D). These findings suggested that HDAC3 might regulate multiple biological processes in microglia, which served as initial responses to ischemia. This also aligns with findings from a recent study, where HDAC3 was observed to colocalize with microglia/macrophages during the acute phase of retinal ischemic injury, further supporting the pivotal role of HDAC3 in ischemic contexts (19).
Fig. 1. Microglial HDAC3 expression increases after ischemia and the engineering of HDAC3-miKO mice.
(A) Flow cytometry gating strategy for microglia (CD45intCD11b+), macrophages (CD45+CD11b+F4/80+), neutrophils (CD45+CD11b+F4/80−), oligodendrocytes (CD45−CD11b−O4+), and the other cells (CD45−CD11b−O4−). (B) Representative flow cytometry plot showing HDAC3 level in microglia, macrophages, oligodendrocytes, neutrophil, and O4− other cells. (C) Mean fluorescence intensity (MFI) of HDAC3 in different cells by flow cytometry. n = 4 per group. IL, ipsilesional; CL, contralateral. (D) Schematic depicting where the Iba1/HDAC3 immunofluorescence images were taken from in the peri-infarct region of CTX and STR 3 days after tFCI for representation and quantification. Representative images demonstrating the location of HDAC3 in microglia [three dimensional (3D) reconstruction was performed on the cell indicated by white boxes] and the increase of HDAC3 number and MFI in microglia after tFCI (HDAC3+Iba1+, white arrows). n = 5 per group. (E) Diagram illustrating the strategy for microglia-specific knockout of HDAC3. (F) Gating strategy of flow sorting for microglia (CD45intCD11b+) and other brain cells (CD45−CD11b−) in brain (top left) or macrophages (Ly6G−CD11C−CD45+CD11b+) in blood (bottom left). Representative flow plot showing CX3CR1Cre-eYFP fluorescence intensity in the brain (top right) or in blood (bottom right). (G) Quantification of HDAC3 mRNA detected by quantitative reverse transcription polymerase chain reaction (qRT-PCR) at 30 days after tamoxifen treatment on FACS-sorted cells to verify the deletion of HDAC3 in microglia. n = 5 to 6 per group. All data are presented as means ± SEM. Data were analyzed using unpaired two-tailed Student’s t test or Mann-Whitney test (C, Oligodendrocytes panel). *P < 0.05 and ***P < 0.001; ns, no significance.
Therefore, to further study the de novo role of HDAC3 in microglia, we engineered HDAC3flox/floxCX3CR1creER/wt mice (HDAC3-miKO) by mating HDAC3flox/flox mice with CX3CR1creER/wt mice (fig. S1, A and B). In the HDAC3-miKO mice, HDAC3 would be deleted only after tamoxifen administration, excluding the potential effects of HDAC3 deletion on developing brain. Specifically, tamoxifen induced ablation of HDAC3 in CX3CR1-expressing cells, including microglia and also peripheral myeloid cells. However, peripheral myeloid cells were continuously replaced by CX3CR1− progenitors in the bone marrow so that the original HDAC3-knockout macrophage population was almost completely renewed within 30 days, whereas microglia underwent self-renewal and retained CX3CR1 expression during the lifetime (20, 21). Hence, we conducted tFCI on day 30 after tamoxifen administration in all the following experiments to delete HDAC3 in microglia exclusively but not in peripheral myeloid cells, and therefore, we were able to ascertain the definite role of microglial HDAC3 (Fig. 1E). To verify the efficiency of deleting microglial HDAC3, on day 30 after tamoxifen administration, we used fluorescence-activated cell sorting (FACS) to isolate cerebral microglia (Ly6G−CD11C−CD45intCD11b+), other brain cells (Ly6G−CD11C−CD45−CD11b−), and also macrophages (Ly6G−CD11C−CD45+CD11b+) in the peripheral blood (Fig. 1F, left). The signal emitted from enhanced yellow fluorescent protein (eYFP) was only detected in miKO microglia and macrophages, indicating the expression of Cre recombinase in these cells (Fig. 1F, right). As expected, quantification of Hdac3 mRNA expression in these FACS-isolated microglia revealed that Hdac3 was almost eliminated in microglia, while Hdac3 expression in other brain cells was not affected (Fig. 1G, blue background). Consistent with a previous report (10), we also observed a significant but much milder reduction of Hdac3 in peripheral blood macrophage in miKO mice (Fig. 1G, pink background).
HDAC3-miKO ameliorates long-term stroke outcome
The mouse tFCI model mimics the middle cerebral artery occlusion (MCAO) in clinical patients, so the decrease of middle cerebral artery blood flow caused by surgery in mice is the key to the model’s success. First, we used laser Doppler and laser speckle to monitor regional cerebral blood flow (rCBF) of the parietal cortex in mice before, during, and after tFCI. No significant difference in rCBF decrease was observed between genotypes [miKO-tFCI and wild-type (WT)–tFCI; fig. S1, E and F], suggesting that the mice suffered the same ischemia.
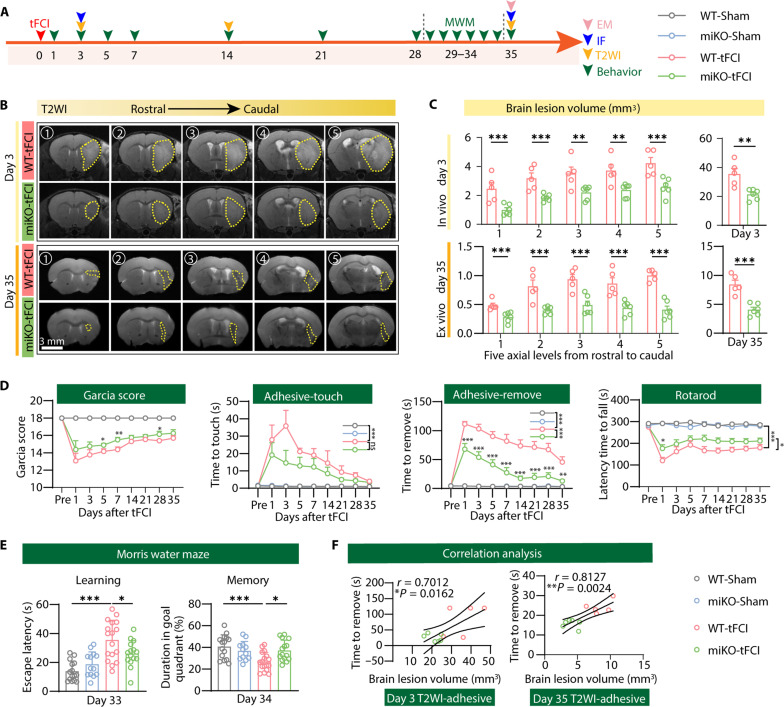
Since assessment of infarct volume via magnetic resonance imaging (MRI) and neurofunctional outcome are the gold standard clinically for the estimation of cerebral ischemic injury (22), we then used T2-weighted MRI (T2WI) to measure brain lesion volume on days 3, 14, and 35 after tFCI (Fig. 2A). miKO-tFCI mice showed significantly smaller infarct volume than WT mice at all the observed time points (Fig. 2, B and C, and fig. S2, A and B).
Fig. 2. HDAC3-miKO ameliorates long-term neurological and histological deficits after ischemic stroke.
(A) Diagram illustrating the experimental design. (B) Representative axial views (five sections from rostral to caudal) of T2WI 3 and 35 days after stroke. Yellow dashed shape indicates lesion areas. (C) Quantification of the brain lesion volume by T2WI at five axial levels from rostral to caudal (left) at day 3 (in vivo) and day 35 (ex vivo) after stroke. Total lesion volume (right) was also evaluated. n = 5/6 per group. (D) Sensorimotor deficits were evaluated before (Pre) and up to 35 days after tFCI or Sham surgery by Garcia score, adhesive touch test, adhesive removal test, and rotarod test. n = 6 to 11 for WT-/miKO-Sham, n = 15 to 17 for WT-/miKO-tFCI. *P < 0.05, **P < 0.01, ***P < 0.001 (miKO-tFCI versus WT-tFCI for individual time points). (E) The escape latency (33 days after tFCI) and the time spent in the target quadrant (34 days after tFCI) were measured in Marris water maze. n = 13 to 17 per group. (F) Correlation analysis between brain lesion volume measured by T2WI and removal time from the adhesive removal test 3 or 35 days after tFCI, respectively. n = 5 to 6 per group. All data are presented as means ± SEM. Generalized estimating equations (GEEs) followed by Tukey’s post hoc test was used to analyze repeated-measures non-normal variables (C, left, and D). Other data were analyzed using unpaired two-tailed Student’s t test (C, right) or one-way analysis of variance (ANOVA) followed by Bonferroni’s post hoc test (E) and Pearson correlation (F). *P < 0.05, **P < 0.01, ***P < 0.001.
Furthermore, to verify the role of microglial HDAC3 in neurofunctional outcomes after ischemic stroke, we performed a battery of neurobehavioral tests for up to 5 weeks. The Garcia score test, the adhesive removal test, and the rotarod test are applied to examine the sensorimotor functions of adult male mice. Mice after tFCI showed remarkable sensorimotor deficits for at least 5 weeks, whereas HDAC3 deletion significantly ameliorated the sensorimotor dysfunction (Fig. 2D). In addition, miKO-tFCI mice also attenuated stroke-induced cognitive impairment, as revealed by the escape latency, especially on the last day of the learning stage and the time spent in the target quadrant in the probe stage in the Morris water maze (Fig. 2E and fig. S2, C and D). Histological damage after stroke often results in behavioral deficits. The correlation analysis between tissue damage and behavioral deficits showed that the increased brain lesion volume detected by MRI was correlated with the poor performance in the adhesive removal test 3 days/35 days after tFCI (Fig. 2F).
Considering the increased risk and severity of stroke in females after menopause, we also used ovariectomized female mice to mimic menopausal females to see whether microglial HDAC3 could be a more general therapeutic target for ischemic stroke (23). Similarly, evaluation of the NeuN+ area of coronal serial sections from anterior to posterior (bregma +1.1 mm to −1.7 mm) revealed that HDAC3 deletion reduced atrophy volume at the late stage in female mice (fig. S3, A and B). However, although female mice also showed impaired spatial learning capability at the late stage of tFCI, miKO-tFCI showed substantial within-group variance and did not ameliorate cognitive deficits, as revealed by the unchanged escape latency and duration in the target quadrant (fig. S3, D and E), probably due to the fact that female microglia exhibit a different phenotype from male microglia in an estrogen-independent manner (24).
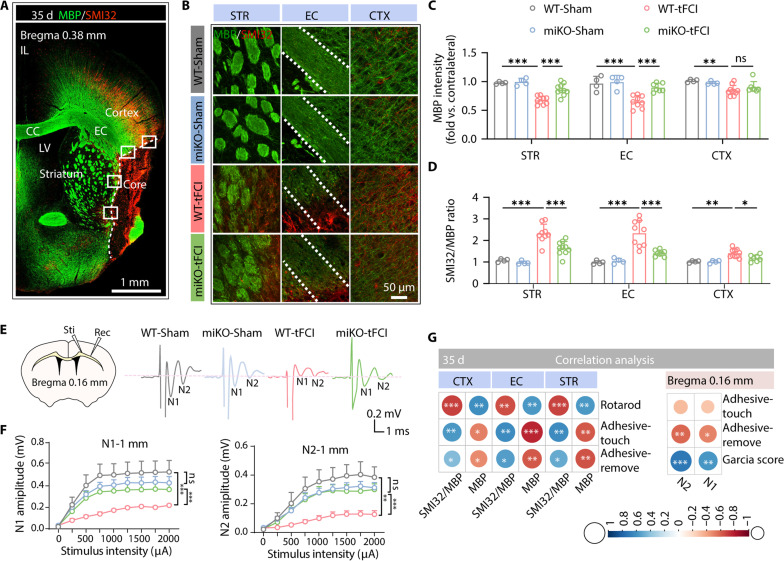
Since structural and functional disruption of white matter often contributes substantially to behavioral dysfunction following stroke (25), we next investigated histological changes of white matter induced by microglial HDAC3 deficiency after stroke. No significant difference was found in the fluorescence intensity of MBP (a major myelin protein) and the ratio of SMI32 (a marker for axon degeneration) relative to MBP among four groups 3 days following tFCI (fig. S4), suggesting no significant structural disruption of white matter at the acute stage of stroke. Nevertheless, the structural disruption of white matter in the peri-infarct regions of the external capsule (EC), striatum, and cortex became significant 35 days after tFCI at the late stage of stroke (Fig. 3, A and B). Of note, HDAC3-miKO remarkably improved myelin loss 35 days after stroke in the striatum and the EC, as well as ameliorated axon degeneration 35 days after stroke in the striatum, EC, and cortex (Fig. 3, B and D), suggesting that deletion of HDAC3 in microglia was beneficial to the structural integrity of white matter after stroke. Either the MBP intensity or the axon degeneration in the two tFCI groups was highly correlated with the sensorimotor behavior outcome assessed on the last day before euthanasia, including performance in the adhesive removal test and the rotarod test (Fig. 3G, left).
Fig. 3. HDAC3-miKO restores structural and functional integrity of white matter after ischemic stroke.
(A) Representative images of MBP/SMI32 immunostaining 35 days after tFCI. (B) The rectangle illustrated where the group-wise images in STR, EC, and CTX were taken. (C) Quantification of the fluorescence intensity of MBP and (D) the ratio of SMI32 to MBP in the STR, EC, and CTX 35 days after tFCI. n = 4 for WT-Sham and miKO-Sham, n = 7 to 10 for WT-/miKO-tFCI. Data were normalized to the intensities of contralateral hemispheres. (E) Schematic illustrating the position of the stimulating (“Sti”) and recording (“Rec”) electrodes for CAP measurements in CC/EC for the slice in bregma 0.16 mm (top left) and representative curves of CAPs in myelinated N1 fibers and unmyelinated N2 fibers (top right). (F) The amplitude of evoked CAPs in myelinated N1 fibers and unmyelinated N2 fibers for bregma 0.16 mm in CC/EC was quantified. n = 4 for WT-Sham, n = 7 for miKO-Sham, n = 6 to 7 for WT-/miKO-tFCI. (G) Correlation analysis between white matter integrity (SMI32/MBP) and behavior (left), and between electrophysiological (N1 and N2) and behavior test (right). The color and the area of circles show the (absolute) value of correlation coefficients. All data are presented as means ± SEM. Data were analyzed using GEE followed by Tukey’s post hoc test (F), one-way ANOVA (C and D), followed by Bonferroni’s post hoc or Pearson correlation (G). *P < 0.05, **P < 0.01, ***P < 0.001.
After demonstrating histological preservation of white matter injury in HDAC3-miKO mice, electrophysiology was then used to examine the long-term functional integrity of white matter manifested as the nerve conduction by evoking and recording the compound action potentials (CAPs) in the corpus callosum (11). The results showed that tFCI decreased the amplitudes of the N1 and N2 segments, representing the fast conduction of myelinated axons and the slow conduction of unmyelinated axons, respectively. Consistent with the histological findings above, miKO-tFCI mice partially restored the decreased amplitude of N1 and N2 segments, further indicating the functional preservation of the white matter at late stage of stroke (Fig. 3, E and F). In addition, both N1 and N2 amplitudes were correlated with the performance in the adhesive removal test and the Garcia score (Fig. 3G, right). These findings extended our understanding of the long-term effects of HDAC3-miKO on neurofunctional deficits and white/gray matter injury induced by ischemic stroke, leading us to exploit the mechanism by which microglial HDAC3 repaired tissue damage or prevented poststroke secondary injury.
RNA-seq highlights hampered inflammatory response and blocked mitosis in HDAC3-deficient microglia after tFCI
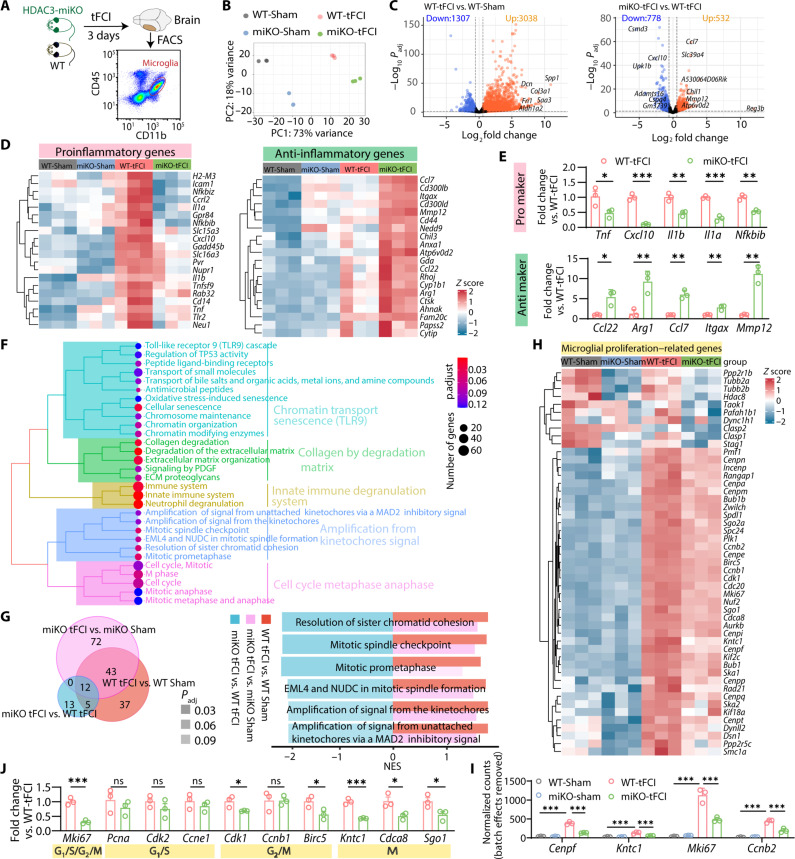
Migrating to the penumbra region soon after ischemia onset, microglia undergo drastic change and are most diverse as early as 3 to 5 days after stroke, which further modulates peri-infarct microenvironment and thereby affects the late outcome of stroke (26). Therefore, to further investigate the mechanism by which HDAC3-miKO preserved ischemic injury, we performed RNA-seq to compare the FACS-purified CD45intCD11b+ microglia isolated from the ipsilateral hemisphere among four groups, all collected 3 days after surgery (Fig. 4A). As shown in the principal components analysis (PCA), microglia from four groups clustered separately in a clearly distinguishable manner. To be specific, microglia isolated from tFCI groups differed significantly from that isolated from Sham groups, as evidenced by the first PC, while the second PC revealed less but still significant distinction between microglia isolated from HDAC3-miKO and WT (Fig. 4B). Massive DEGs were found in both surgery-specific comparison (WT-tFCI versus WT-Sham or miKO-tFCI versus miKO-Sham) and genotype-specific comparison (miKO-tFCI versus WT-tFCI or miKO-Sham versus WT-Sham) (Fig. 4C and fig. S5, A and B). The considerable overlap between different genotypes in terms of tFCI-induced up-regulated and down-regulated genes (fig. S5C) suggested that the surgical procedure itself induced similar effects in both WT and miKO mice. Notably, even in the physiological conditions, a large number of DEGs were also detected in HDAC3-miKO microglia, and the downstream gene set enrichment analysis (GSEA) on the basis of Reactome database highlighted multiple terms for which the genes were enriched. These genes were associated with chromatin modifying enzymes, suggesting that HDAC3-miKO successfully changed the chromatin landscape in physiological conditions (fig. S5D) (27). Comparison between our datasets and M1/M2 markers in public databases (GSE69607) revealed that HDAC3-deficient microglia exhibited a distinct inflammatory expression profile following stroke with decreased proinflammatory gene expression and increased anti-inflammatory gene expression, while these genes were all elevated after tFCI (WT-tFCI versus WT-Sham) (Fig. 4D). As validated by quantitative polymerase chain reaction (qPCR), proinflammatory genes (Tnf, Cxcl10, Il1b, Il1a, and Nfkbib) were down-regulated in miKO-tFCI, while the levels of anti-inflammatory genes (Ccl22, Arg1, Ccl7, Itgax, and Mmp12) were augmented (Fig. 4E). Comparing the expression profiles of the miKO-tFCI microglia and the WT-tFCI microglia, we acquired the DEGs under the threshold (Padj < 0.05) and applied them to the downstream GSEA. The top terms were classified into five categories based on their semantic similarity (Fig. 4F). Several gene sets related to innate immune degranulation system including terms of “Immune System,” “Innate Immune System,” and “Neutrophil degranulation” were enriched in HDAC3-deficient microglia (Fig. 4F, earth-yellow), supporting the expression profile alteration related to microglial polarization in our previous findings (Fig. 4, D and E) (28). These DEGs were also associated with collagen/matrix degradation, as supported by a previous study (29). As expected, like miKO-Sham microglia, the miKO-tFCI microglia suppressed pathways associated with chromatin, such as “chromatin modifying enzymes,” “chromatin organization,” and “chromosome maintenance” (Fig. 4F, turquoise). The gene list was also annotated to several terms associated with various processes in the mitotic phase, including “M phase,” “mitotic anaphase,” and “mitotic metaphase anaphase” (Fig. 4F, pink, and fig. S5E).
Fig. 4. RNA-seq reveals that HDAC3-miKO inhibited microglial inflammation and proliferation 3 days after tFCI.
(A) Flow-sorted CD11b+CD45int (microglia) cell population 3 days after tFCI for RNA-seq. n = 3 biological replicates (each includes three to four mice) per group. (B) PCA plot. (C) Differential expression analysis revealed microglia-specific transcriptional changes in response to tFCI and HDAC3-miKO after tFCI. (D) Proinflammatory or anti-inflammatory expression profile. Each column represents a sample. Data were presented as z score, representing the expression levels normalized to the mean and SD of each gene (row). (E) mRNA expression of proinflammatory factors and inflammatory factors for FACS-purified microglia 3 days after tFCI. Data were shown as fold change of WT-tFCI. n = 3 per group (qRT-PCR validation). (F) GSEA using Reactome pathway suggested the role of microglial HDAC3 in response to tFCI (miKO-tFCI versus WT-tFCI). (G) Venn plot showed 97, 36, or 127 terms produced by GSEA analysis (Padj < 0.12) of genes generated by the comparison WT-tFCI versus WT-Sham, miKO-tFCI versus WT-tFCI, or miKO-tFCI versus miKO-Sham (Padj < 0.05), respectively. The overlap referred to terms that gained normalized enrichment score (NES) of different directions in the aforementioned comparison, including six terms all related to mitosis (bar plot). (H) Heatmap showed expression of proliferation-related genes in microglia, while the most prominently affected genes (Cenpf, Kntc1, Mki67, and Ccnb2) were depicted through bar plots for enhanced visualization (I). (J) qRT-PCR validated mRNA expression levels of representative genes involved in different cell cycle phases in FACS-purified microglia 3 days after tFCI. Data were shown as fold change of WT-tFCI controls. n = 3 per group. All data are presented as means ± SEM. Data were analyzed using unpaired two-tailed Student’s t test (E and J) or differential expression analysis via DEseq2 (I). *P < 0.05, **P < 0.01, ***P < 0.001.
To confirm altered pathways in miKO-FCI microglia among all the above pathways, we used comparison analysis between GSEA scores of significantly enriched pathways for WT-tFCI versus WT-Sham and miKO-tFCI versus WT-tFCI as well as those for miKO-tFCI versus WT-tFCI and found out 12 shared pathways. Among these pathways, HDAC3-miKO promoted enrichment scores in six pathways (fig. S5F). We also observed a remarkable reversal of enrichment scores in several pathways associated exclusively with mitosis, specifically related to mitotic spindle checkpoint, resolution of sister chromatid cohesion, and mitotic prometaphase (Fig. 4G), whereas none of these pathways were enriched in the gene list generated from miKO-Sham versus WT-Sham (fig. S5D), suggesting that the mitotic phase was the most critical as well as the most variable process subjected to HDAC3 expression in the context of ischemic stroke. As shown in Fig. 4H, depleting HDAC3 down-regulated numerous highly expressed genes associated with mitosis in microglia under tFCI. Among these genes, Cenpf (centromere protein F), Kntc1 (kinetochore associated 1), Mki67 (marker of proliferation Ki67), and Ccnb2 (cyclin B2) were all included in the top 100 down-regulated genes (miKO-tFCI versus WT-tFCI, overlapped with DEGs generated from WT-tFCI versus WT-Sham) (Fig. 4I and data S2), suggesting that they were the genes mostly associated with the protection against the tFCI model. Overrepresentation analysis of overlapping DEGs between two gene lists (up-regulated genes in WT-tFCI versus WT-Sham and down-regulated genes in miKO-tFCI versus WT-tFCI) confirmed the enrichment of mitotic processes (fig. S5, G to I). Therefore, we proceeded to evaluate microglial mRNA expression of genes present in the cell cycle by qPCR, including the proliferation marker (MKi67) and representative genes associated with G1-S phase (Pcna, Cdk2, and Ccne1), G2-M phase (Cdk1, Ccnb1, and Birc5), and M phase (Kntc1, Cdca8, and Sgo1) (Fig. 4J). Overall, aligned with our RNA-seq results, HDAC3-miKO significantly decreased the expression of most of the genes present in G2-M phase or M phase but did not change the level of genes present in G1-S phase, more directly confirming blockade of G2-M transition induced by HDAC3 loss. Intriguingly, unlike most studies demonstrating that inhibiting HDAC3 regulated cell cycle depending on posttranslational modification to cell cycle–related proteins (30, 31), our findings uncovered instead that HDAC3 was actively engaged in transcriptional regulation of cell cycle–related genes. In summary, our data provided preliminary evidence that ablation of HDAC3 in microglia suppressed the expression of M phase–related genes and thus microglial proliferation.
HDAC3-deficient microglia show mitosis defects
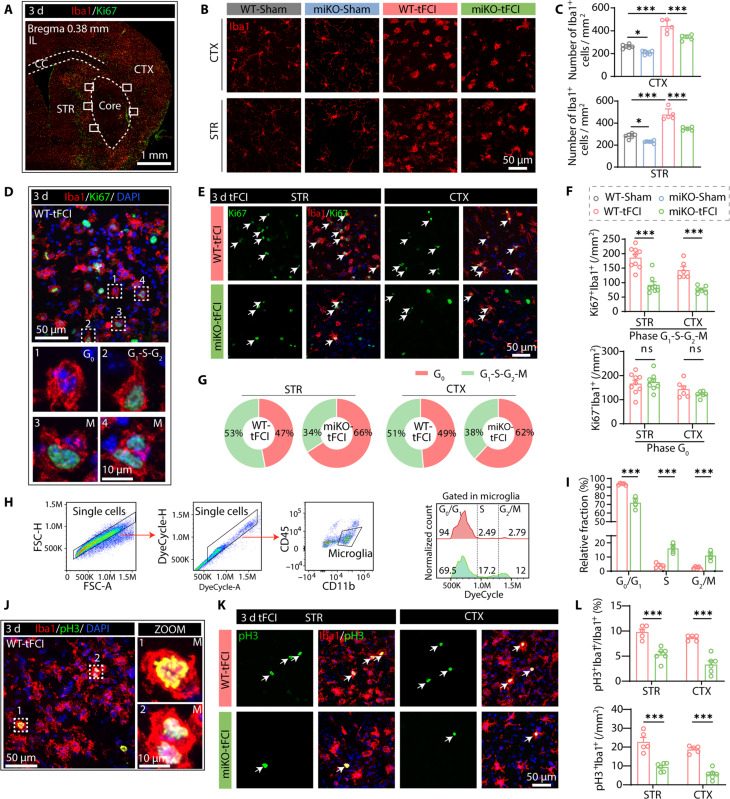
Upon the onset of stroke, microglia undergoing rapid self-renewal aggregate around the infarct region (21). We observed an approximately twofold increase in peri-infarct microglia in the cortex (CTX) or striatum (STR) after tFCI (Fig. 5, A to C). However, this increase was partially inhibited in miKO-tFCI mice (Fig. 5C), suggesting that the proliferation process of HDAC3-deficient microglia was blocked as our RNA-seq data indicated. Therefore, Ki67+ staining was applied to detect proliferating microglia. Microglia in the resting G0 phase were labeled as Ki67−Iba1+, and microglia in the active phase (i.e., G1-S-G2-M phase) were labeled as Ki67+Iba1+. Different cell cycle phases of microglia were all detected in the peri-infarct region by triple immunofluorescence staining of Iba1/Ki67/4′,6-diamidino-2-phenylindole (DAPI) (Fig. 5, D and E). As expected, HDAC3 absence reduced the number of proliferating microglia but did not affect the number of G0-phase microglia after tFCI (Fig. 5F) so that more HDAC3-deficent microglia rested in the G0 phase compared to WT microglia (Fig. 5G).
Fig. 5. HDAC3-miKO inhibits microglial proliferation after tFCI.
(A) Representative image illustrating Iba1/Ki67 immunofluorescence in the ipsilateral hemisphere 3 days following stroke. All the following representative images were taken from where the white boxes indicated. (B) Representative images illustrating the decreased number of Iba1+ cells in CTX/STR in miKO-tFCI compared to WT-tFCI, as reflected by the quantification (C). n = 5 to 6 per group. (D) Representative images of Iba1/Ki67 staining in the peri-infarct region of striatum, indicating microglia in different cell cycle phases, including G0 phase (Ki67−Iba1+) and G1-S-G2-M phase (Ki67+Iba1+). (E) Representative images illustrating the reduction of the number of Ki67+Iba1+ cells (white arrows) in CTX/STR. (F) Quantification of the number of Iba1+Ki67− cells (G0 phase, top) and Iba1+Ki67+ cells (G1-S-G2-M phases, bottom) in peri-infarct STR and CTX 3 days after tFCI. n = 6 to 9 per group. (G) Mean percentage of Iba1+Ki67+/Iba1+ (G1-S-G2-M phase) cells and Iba1+Ki67−/Iba1+ (G0 phase) cells in the peri-infarct region of CTX and STR. (H) Flow gating strategy for single microglia (CD11b+CD45int) (left), identifying microglia in G0-G1, S, or G2-M phases based on the DNA content (right). (I) Quantification of the percentage of microglia in different cell cycles. n = 5 to 6 per group. (J and K) Representative images of Iba1/pH3 immunofluorescence in the peri-infarct region. Cells indicated by white box (J) are microglia undergoing mitosis. Illustrating the reduction of the number of pH3+Iba1+ cells (white arrows) (K) in CTX and STR. (L) Quantification of the number (percentage) of Iba1+pH3+ in peri-infarct STR and CTX 3 days after tFCI. n = 5 to 6 per group. All data are presented as means ± SEM. Data were analyzed using one-way ANOVA followed by Bonferroni’s post hoc (C) or unpaired two-tailed Student’s t test or Mann-Whitney test (F, I, and L). *P < 0.05, ***P < 0.001.
Of note, our behavioral assessment at days 1 and 3 (Fig. 2D) as well as MRI results on day 3 after tFCI (Fig. 2C) also seemed to imply that microglial HDAC3 deficiency potentially reduced lesions at the earlier stage, leading to less microglia loss and thereby the reduction of proliferative response. To exclude this possibility, we increased the occlusion time of HDAC miKO mice to 90 min, resulting in a larger infarct volume (fig. S6, A and B). To detect renewed microglia after tFCI, we conducted intraperitoneal injections of 5-ethynyl-2′-deoxyuridine (EdU) as described in a previous study (21). HDAC3-deficient microglia in mice subjected to 90-min occlusion still showed a severe disability of self-renewal 3 days after tFCI compared to WT microglia in mice subjected to 60-min occlusion (fig. S6, C to E), suggesting that the proliferative potential of microglia was dependent on microglial HDAC3 rather than the initial brain injury volume.
Moreover, to determine whether the reduction of microglial number could also be attributed to exacerbated microglial apoptosis, terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay was used. Our results revealed that HDAC3-miKO significantly decreased the number of TUNEL+ apoptotic microglia instead in the peri-infarct region of the STR (fig. S7), implying that the decreased number of HDAC3-deficient microglia was entirely due to defects in proliferation. Consistently, several prior studies also reported that inhibiting HDAC3 could prevent cell apoptosis (32).
To further confirm the role of HDAC3 in regulating cell cycle process of microglia, we performed cell cycle analysis following DyeCycle staining by flow cytometry in the poststroke brains (Fig. 5H). Compared to WT microglia, HDAC3-deficient microglia showed increased percentage of cells in S phase and G2-M phase but decreased percentage of cells in G0-G1 phase (Fig. 5I), suggesting induction of G2-M arrest and S-phase arrest. To verify the effects of HDAC3 deficiency on M-phase arrest, we also stained microglia with phospho–histone H3 (pH3), which is a mitotic chromosome condensation marker labeling anaphase and telophase (33). Despite the barely observed number of mitotic microglia in the penumbra area (Fig. 5J), a significant decrease of pH3+ microglia number was still detected in miKO-tFCI mice (Fig. 5, K and L), which approximated the extent of the reduction of Ki67+ microglia as demonstrated above (Fig. 5F, top). Together with our prior finding of an increase of cells arrested in G2-M phase in miKO-tFCI mice (Fig. 5, H and I), we concluded that deleting HDAC3 down-regulated mitosis-related genes transcriptionally, which may obstruct mitotic phase entry as suggested by a previous study (34).
HDAC3 deficiency inhibits proliferation of specific microglia populations
Having confirmed that deletion of HDAC3 inhibits microglial proliferation, it remains unknown how much this effect contributes to the peri-infarct microenvironment and thus histological ischemic injury. Notably, previous studies have revealed that blocking microglial proliferation through diphtheria toxin administration in CX3CR1creER:R26iDTR mice significantly decreased inflammatory cells and altered the immune profiles in the context of stroke, suggesting a potential connection between proliferation and inflammatory response in microglia (6). Since the results of RNA-seq suggested the implication of inflammatory responses (Fig. 3, D and E), we hypothesized that deleting HDAC3 not only inhibited microglial proliferation but also changed the expression profile of proliferative microglia.
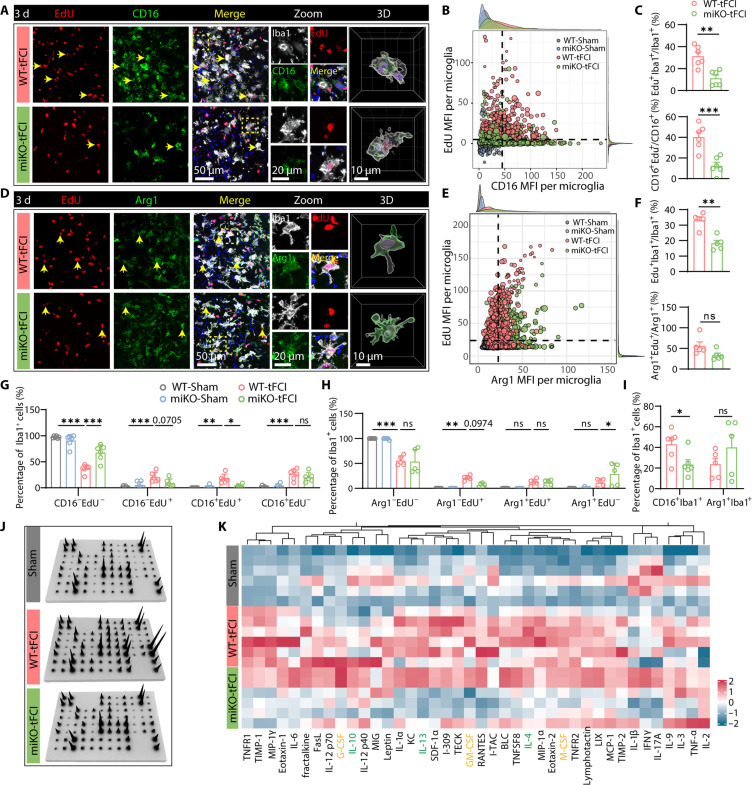
Therefore, we measured the incorporation of 5-Ethynyl-2′-deoxyuridine (EdU) in either CD16-labeling microglia (Fig. 6A) or Arg1-labeling microglia (Fig. 6D), which indicates proinflammatory or anti-inflammatory microglia, respectively. In Sham-operated groups, CD16/Arg1 expression was too low to detect, and no EdU+ microglia were detected (fig. S8, A and B). Like the Ki67 staining results shown in Fig. 5F, the EdU mean fluorescence intensity (MFI) per microglia also decreased in miKO-tFCI mice (fig. S8, C and E). Moreover, the measurements of the MFI in each microglia indicated that CD16 expression in microglia increased 3 days following stroke, while deleting HDAC3 down-regulated CD16 in each microglia (fig. S8D). Conversely, Arg1 expression was augmented in HDAC3-deficient microglia (fig. S8F), consistent with the change of RNA level of Arg1 as evidenced by RNA-seq (Fig. 4D, right). Using CD16 or Arg1 MFI and EdU MFI per microglia in Sham-operated mice as the negative control, we determined the respective thresholds for CD16 or Arg1-postive microglia and EdU-positive microglia (Fig. 6, B and E). Intriguingly, HDAC3-miKO significantly inhibited the proliferation of microglia (Fig. 6C, top), specifically the ones that were positive for CD16 (Fig. 6C, bottom), while the proliferative capacity of Arg1+ microglia did not significantly change (Fig. 6F, bottom). Consistently, while no significant change was observed in the proportion of nonproliferative CD16+ microglia (CD16+EdU−), we observed a significant decrease in the proportion of proliferative CD16+ microglia in miKO-tFCI mice (Fig. 6G), suggesting that HDAC3 deletion selectively inhibited CD16 expression in proliferative microglia, while no significant change was observed in Arg1+EdU+ cells (Fig. 6H). Similar to Arg1+ microglia, CD206+ microglia, which also labels anti-inflammatory microglia/macrophage, also showed constrained proliferative potential (fig. S9).
Fig. 6. HDAC3-miKO inhibits the proliferation of proinflammatory microglia after tFCI.
(A) Representative images of Iba1/EdU/CD16 immunofluorescence in the peri-infarct STR 3 days after tFCI and 3D reconstruction for Iba1+CD16+ cells (right). (B) Scatterplot showing MFI of EdU and CD16 per microglia, with density plot drawn on the margin. The black dashed lines indicate the threshold for CD16+ microglia or EdU+ microglia determined by EdU/CD16 expression in the Sham-operated groups. (C) Quantification of the percentage of proliferating microglia (EdU+Iba1+, top) and CD16+ microglia (CD16+EdU+/CD16+, bottom). (D) Representative images of Iba1/EdU/Arg1 immunofluorescence in the peri-infarct STR 3 days after tFCI and 3D reconstruction for Iba1+Arg1+ cells (right). (E) Scatterplot showing MFI of EdU and Arg1 per microglia. (F) Quantification of the percentage of proliferating microglia (EdU+Iba1+, top) and Arg1+ microglia (Arg1+EdU+/Arg1+, bottom). (G) Quantification of the percentage of CD16−EdU−Iba1+, CD16−EdU+Iba1+, CD16+EdU−Iba1+, and CD16+EdU+Iba1+ cells out of the total Iba1+ cells. (H) Quantification of the percentage of microglia with Arg1−EdU−Iba1+, Arg1−EdU+Iba1+, Arg1+EdU−Iba1+, and Arg1+EdU+Iba1+. (I) Quantification of the percentage of CD16+ or Arg1+ microglia. (J) A panel of 40 cytokines was measured in the ipsilesional and contralateral hemispheres via a protein array. (K) Heatmap displaying the relative expression of 40 cytokines. In detail, cytokines in yellow refer to known neurotrophic factors, while cytokines in green refer to anti-inflammatory factors. Other cytokines in black refer to proinflammatory factors. All data are presented as means ± SEM. n = 5 to 6 per group. Data were analyzed using unpaired two-tailed Student’s t test or Mann-Whitney test (C, F, and I) and two-way ANOVA followed by Bonferroni’s post hoc tests (G and H). *P < 0.05, **P < 0.01, ***P < 0.001.
Furthermore, because of the significant infiltration of myeloid cells into the brain, which likely express similar levels of Iba1, CD16, and Arg1 as microglia (35–37), we used P2RY12 as a specific marker to accurately distinguish bona fide microglia (fig. S10, A, D, and G). As expected, HDAC3 deficiency inhibited the proliferation of P2RY12+ microglia (fig. S10, B, E, and H). Furthermore, CD16+P2RY12+ microglia showed a remarkable deficit in the absence of HDAC3 (fig. S10, A and C), while Arg1+P2RY12+ microglia and CD206+P2RY12+ microglia did not show such change (fig. S10, F and I). This consistency did exclude the possibility that contamination of Arg1+ myeloid cells or CD206+ border-associated macrophages (BAMs) masked the suppressed proliferative potential of Arg1+ or CD206+ HDAC3-deficient microglia and further validated that HDAC3 deficiency–induced proliferative effects had cell-specific preference. Collectively, we gained evidence that deleting HDAC3 inhibited the proliferation of specific microglia populations (i.e., CD16+ microglia in our study) sustainedly for at least 3 days after tFCI, while Arg1+ or CD206+ microglia maintained their normal proliferation even in the absence of HDAC3. This may result in the attenuation of specific microglia populations. Deleting HDAC3 decreased the percentage of CD16+ microglia but not affected the percentage of Arg1+ or CD206+ microglia (Fig. 6I and fig. S9E). These findings confirmed the proliferative bias toward different microglia under the regulation of HDAC3. Therefore, we reasoned that after the onset of stroke, microglia developed into different inflammation-associated phenotypes, which simultaneously underwent proliferation that was regulated by HDAC3 in a phenotype-dependent manner, eventually resulting in altered microglial states in miKO-tFCI mice.
Correspondingly, as measured by protein microarray, a remarkable decrease of multiple proinflammatory cytokines was observed in the miKO-tFCI mice (Fig. 6, I and J, and fig. S10D), suggesting that the absence of HDAC3 efficiently ameliorated the peri-infarct inflammatory microenvironment due to the inhibited proliferation of proinflammatory microglia.
Although the initial lesion volume did not show statistical significance between HDAC3-miKO and WT mice, we observed a clear improvement in behavior on day 1 after tFCI (Fig. 2D). This observation led us to question whether there are any preexisting changes in HDAC3-miKO microglia before 3 days after tFCI that could account for these results. Therefore, to gain insight into the microglial landscape at the earlier stage of stroke, we conducted additional analyses. On day 1 after tFCI, there was no evidence of microglial proliferation based on Iba1+EdU+ staining (fig. S11A). This result was aligned with previous studies, which indicates that microglial expansion primarily occurs on day 3 after tFCI, while it remains relatively quiescent on day 1 after tFCI (21). However, through Iba1/CD16 and Iba1/Arg1 staining (fig. S11, B to D), we did observe a reduction in CD16+ microglia but not in Arg1+ microglia in miKO-tFCI on day 1 after tFCI (fig. S11, C and D), which likely accounted for the early-stage trend of mitigated outcomes. This finding suggested that microglia underwent an inflammatory transition before proliferation. It inspired the possibility that different phenotypes, observed during the early stage, may proceed to self-renewal 3 days following tFCI when microglia rapidly expanded themselves. In this context, the inhibited proliferation of CD16+ microglia in miKO mice amplified the overall differences between WT-tFCI and miKO-tFCI 3 days following tFCI.
PU.1 is responsible for selectively inhibited proliferation of microglia in miKO tFCI mice
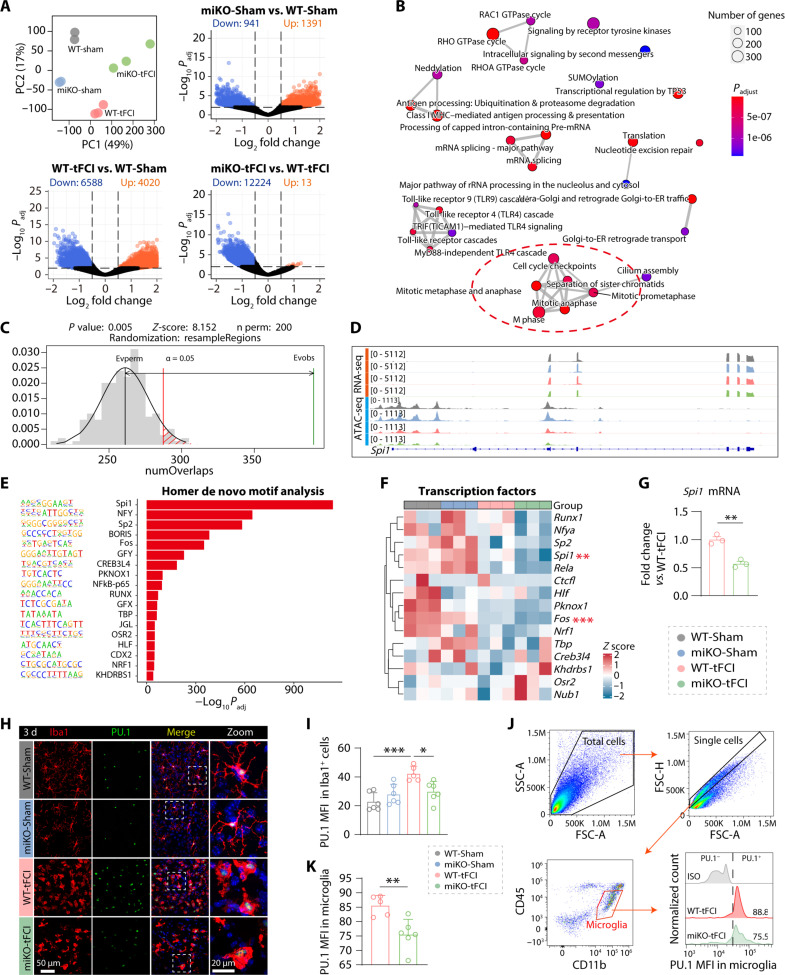
In microglia, the dynamic modulation of the epigenetic landscape guided the reprogramming of the transcriptional profiles (38). To determine the role of HDAC3 as an essential epigenetic modifier, we performed ATAC-seq to identify how HDAC3 deletion reshaped microglial transcriptome profiles and thereby selectively inhibited proliferation of different microglia populations. PCA interpreted large variations among the four groups (Fig. 7A, top left). Replicates of four groups were clearly clustered separately as shown in fig. S12A. With a given false discovery rate (FDR) cutoff of 0.01 and a log2 fold change (log2FC) cutoff of 0.5, we detected tens of thousands of differential accessibility regions (DARs) in two pairwise comparisons among four groups using DiffBind (Fig. 7A, top right and bottom, and fig. S12B), while only 2331 DARs were detected in the comparison between miKO-Sham and WT-Sham (Fig. 7A, top right). Notably, the biggest difference was seen between miKO-tFCI microglia and WT-tFCI microglia, with 12,237 DARs in total. Comparing miKO-tFCI with WT-tFCI, Reactome-based GSEA identified multiple biological processes, specifically including terms associated with mitosis (“M phase,” “Separation of Sister Chromatids,” “Mitotic Metaphase and Anaphase,” and “mitotic prometaphase”) (Fig. 7B, red circle), which were similar to our previous RNA-seq data (Fig. 3, F and G), suggesting epigenetically regulated proliferation in miKO-tFCI microglia. However, unlike DEGs in RNA-seq, the distribution of DARs of miKO-tFCI versus WT-tFCI showed a strong bias, with as many as 12,224 closed DARs that were less accessible but as few as 13 open DARs that were more accessible, while this bias did not exist in the physiological condition (Fig. 7A, top right). This bias toward closed DARs also existed in the comparison between miKO-tFCI and miKO-Sham (fig. S12B), suggesting that in the context of stroke, HDAC3 deficiency led to a pronounced closing of accessible regions. We thus investigated the genomic distribution of differential peaks to identify the main biased regions. Most of the DARs were annotated to introns and promoters. Specifically, for WT-tFCI versus WT Sham, 27.74% of the open DARs in WT-tFCI microglia were located in the promoter region within 3 kb upstream of the transcription start site (TSS) region and 36.15% in the intron region, while 42.41% of the closed DARs were located in the promoter region and 28.81% in the intron region (fig. S12C). For miKO-tFCI versus WT-tFCI, only the distribution of closed DARs was measured because there were too few open DARs; 38.71% of closed DARs were located in the promoter region, and 31.37% were located in the intron region (fig. S12D). For miKO-tFCI versus miKO-Sham, 51% of the closed DARs were located in the promoter region (fig. S12E). The large variability in the percentage of DARs in the promoter region may indicate strong surgery effects and genotype effects. As revealed in GSEA, annotated DARs (miKO-tFCI versus WT-tFCI) (FDR < 0.01) located in the promoter region were enriched for pathways associated with cell cycle (fig. S12F).
Fig. 7. PU.1 may contribute to the selectively inhibited proliferation of microglia in miKO tFCI mice.
(A) PCA showed the separation of ATAC-seq signals of all samples by group. Volcano plots showed the microglia-specific DARs (log2FC >0.5 or <−0.5, FDR < 0.01) in poststroke/Sham-operated brains from HDAC3-miKO and WT mice. (B) Enrichment map visualizing enriched Reactome pathways obtained from GSEA of all annotated DARs (FDR < 0.01) from miKO-tFCI versus WT-tFCI. Mutually overlapping terms clustered together. The red circle indicated the clustered cell cycle–related terms of interest. (C) Permutation test showing the overlap of DEGs between region sets generated from RNA-seq and ATAC-seq. (D) IGV map tracks showing ATAC-seq and RNA-seq signals at Spi1 loci. (E) Homer de novo motif analysis of closed DARs at promoters (<3 kb to TSS) in miKO-tFCI versus WT-tFCI. (F) RNA-seq revealed transcriptional expression of transcriptional factors indicated above in four groups. **P < 0.01, ***P < 0.001 (miKO-tFCI versus WT-tFCI, differential expression analysis via DEseq2). (G) qRT-PCR–validated mRNA expression levels of PU.1 for FACS-purified microglia 3 days after tFCI. Data are shown as fold change of WT-tFCI controls. n = 3 per group. (H) Double-labeled immunofluorescence of Iba1/PU.1 in the peri-infarct region of striatum 3 days after tFCI. (I) MFI of PU.1 in Iba1+ cells in the peri-infarct region of STR 3 days after tFCI. n = 6 per group. (J) Gating strategy for microglia (CD45intCD11b+) and the MFI of PU.1 in microglia. (K) MFI of PU.1 for microglia in the brain by flow cytometry. n = 5 to 6 per group. All data are presented as means ± SEM. Data were analyzed using one-way ANOVA followed by Bonferroni’s post hoc test (I) or unpaired two-tailed Student’s t test (G and K). **P < 0.01. *P < 0.05, ***P < 0.001.
Next, we integrated our ATAC-seq data and previous RNA-seq data generated in the same context to investigate the potential association between peak signal at a gene’s promoter and gene expression (39). Although little overlap was observed between open DARs and up-regulated genes in miKO-tFCI versus WT-tFCI, more than a half of the down-regulated genes had their corresponding promoters less accessible (fig. S12G). Furthermore, little correlation was observed between the differential accessible promoters and the DEGs (fig. S12H). However, the permutation test revealed that the closed promoter-located DARs were significantly associated with down-regulated genes (permutation P < 0.01, Fig. 7C).
Correspondingly, to uncover the factor that potentially induced these chromatin changes and thus transcriptomic changes, we searched for transcription factor motifs within these less accessible promoters. We observed an apparent enrichment of binding sites for E26 transformation specific (ETS) family transcription factor PU.1 (encoded by Spi1), which is well known for its pivotal role in microglial development and inflammation (Fig. 7, D to F) (15, 40). Notably, we observed a significant increase of PU.1 MFI in peri-infarct microglia compared to Sham-operated microglia (Fig. 7, H and I). Furthermore, in the absence of HDAC3, PU.1 expression was significantly decreased in both mRNA level (Fig. 7G, detected by qPCR) and protein level (Fig. 7, H and I, detected by immunofluorescence staining; Fig. 7, J and K, detected by flow cytometry).
According to previous relevant studies, our ATAC-seq results provided two possible answers to the questions that have permeated our entire study, namely, how HDAC3-miKO altered microglial inflammatory profiles and inhibited expression of cell cycle–related genes. Why were some anti-inflammatory gene signatures increased in the absence of HDAC3 after ischemic stimulation in our study? First, the reason may be that HDAC3 acts at a subset of PU.1-defined macrophage-specific enhancers and thereby restricts histone marks at many interleukin-4 (IL-4)–targeted genes (41). Of note, in our study, many DARs were not located near the DEGs, making it hard to explain why those up-regulated genes had their accessible regions closed, which could be attributed to the epigenetic regulation via long-range interaction so that a DAR can regulate distant genes rather than the closest gene only (42). On the other hand, we noted that PU.1 expression was decreased in miKO-tFCI mice (Fig. 7, F to K), suggesting that HDAC3-miKO selectively inhibited microglial proliferation by suppressing PU.1, given that PU.1 reduction has been reported to result in the down-regulation of massive proinflammatory genes and cell cycle–related genes (14, 43).
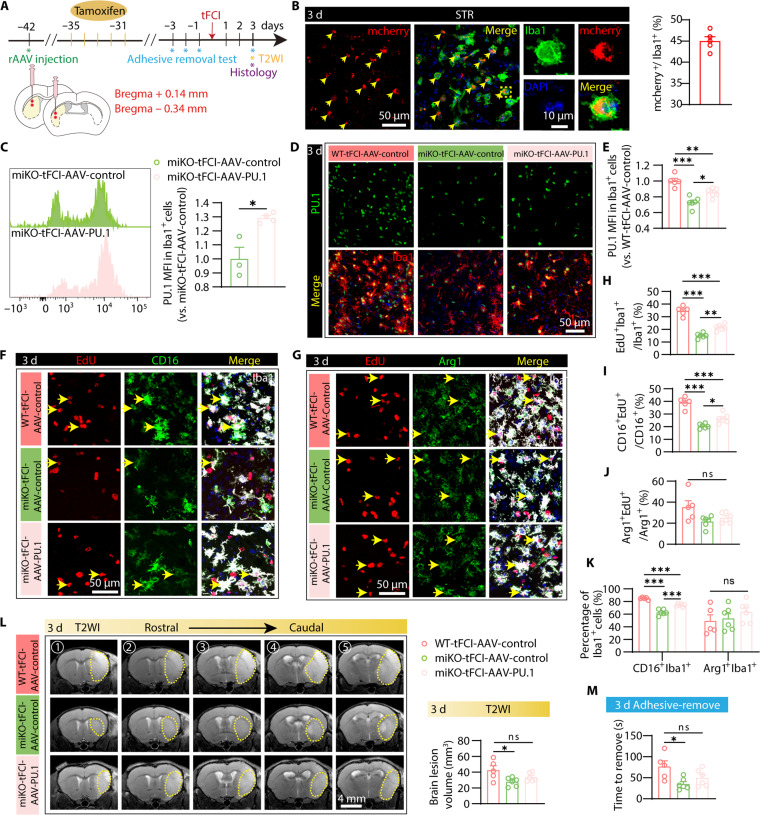
Therefore, to explore the role of PU.1 in the protective effects of HDAC3 ablation, we used a recombinant AAV (rAAV) that was recently developed to specifically target microglia to package rAAV-EF1a-DIO-mPU.1-P2A-mCherry (hereafter referred to as AAV-PU.1), which enables efficient overexpression of PU.1 selectively in Cre-expressing microglia. We performed stereotaxic injection of AAV-PU.1 at four distinct sites within the peri-infarct striatum and injected tamoxifen for five consecutive days a week after AAV injection (Fig. 8A). By immunofluorescence staining of Iba1 and mCherry, we revealed that 45% of microglia in the peri-infarct striatum (Fig. 8B) are positive for mCherry, indicative of efficient AAV infection of microglia. Furthermore, as supported by quantification of PU.1 level via flow cytometry and immunofluorescence, AAV-PU.1 successfully induced significant overexpression of PU.1 in HDAC3-deficient microglia (Fig. 8, C to E). Of note, overexpressing PU.1 significantly reversed inhibited proliferation in microglia deficient in HDAC3 (Fig. 8, F to H), especially the ones that were positive for CD16, while Arg1+ or CD206+ microglia did not show such change (Fig. 8, I and J, and fig. S13, A and B) after AAV-PU.1 injection. As expected, this alteration in the proliferation of different microglia also selectively caused an increase in the proportion of CD16+ microglia but not Arg1+ or CD206+ microglia (Fig. 8K and fig. S13C). Although miKO-tFCI-AAV-PU.1 did not significantly reverse the improved outcomes measured by MRI (Fig. 8L) and the adhesive removal test (Fig. 8M) compared to miKO-tFCI-AAV-control, there were no remarkable change between miKO-tFCI-AAV-PU.1 and WT-tFCI-AAV-control (Fig. 8, L and M) 3 days following tFCI. These results indicated that overexpressing microglial PU.1 partially counteracted the protective effects of microglial HDAC3 ablation. They provided strong support for the involvement of PU.1 as a downstream molecule of HDAC3 in the selective inhibition of microglial proliferation and, thereby, in the protection effects of HDAC3-miKO after tFCI.
Fig. 8. PU.1 partially reversed selective inhibition of CD16+ microglial proliferation and outcome improvement induced by HDAC3-miKO after tFCI.
(A) Diagram illustrating the spatiotemporal strategy for rAAV injection and the following functional and histological measurements. (B) Representative images of the colocalization of mCherry+ and Iba1+ signals in the STR of miKO-Sham mice injected with rAAV-EF1a-DIO-mCherry and the quantification of the infection efficiency indicated by the proportion of mCherry+ microglia. (C) PU.1 MFI in CD45intCD11b+ cells of miKO-tFCI mice with AAV-control/PU.1 injection measured by flow cytometry at 3 days after tFCI. (D and E) PU.1 MFI in Iba1+ cells in the peri-infarct STR measured by immunostaining of Iba1/PU.1 3 days after tFCI. (F and G) Representative images of Iba1/EdU/CD16 (F) and Iba1/EdU/Arg1 (G) staining in the peri-infarct region of STR 3 days after tFCI. (H, I, and J) Quantification of the percentage of different microglia, including all proliferative (EdU+Iba1+), proliferative CD16+ (CD16+EdU+/CD16+), CD16+, proliferative Arg1+ (Arg1+EdU+/Arg1+), and Arg1+ microglia. n = 5 to 6 per group. (K) Quantification of the percentage of CD16+Iba1+ cells or Arg1+Iba1+ of total Iba1+ cells. (L) Representative axial views (five sections from rostral to caudal) of T2WI and quantification of the total lesion volume. n = 5 to 6 per group. (M) Time to remove in adhesive removal test. All data are presented as means ± SEM. Data were analyzed using one-way ANOVA followed by Bonferroni’s post hoc (E, H, and I to M) or unpaired two-tailed Student’s t test (C). **P < 0.01. *P < 0.05, ***P < 0.001.
DISCUSSION
During the past few years, HDAC3 has been reported to regulate the progression of different cell cycle phases in different cells (44). Loss of HDAC3 in hematopoietic progenitor cells blocks only the S phase progression (45), while in Hela cells, knockdown of HDAC3 induces a G1-S transition defect (31). In the nervous system, HDAC3 controls G2-M phase progression in adult neural stem/progenitor cells by posttranslational stabilization of the G2-M–dependent CDK1 (30). In our investigation, an unbiased comparative analysis of RNA-seq has identified the cell cycle M phase as the principal process significantly affected by HDAC3-miKO 3 days after tFCI, which implied a substantial transcript-level down-regulation of genes subsequent to HDAC3 ablation. Notably, Cenpf, Kntc1, Mki67, and Ccnb2 were particularly prominent among these genes, exhibiting the most pronounced alterations. Specifically, Cenpf and Kntc1 both play essential roles in mitotic chromosome segregation; Mki67 is indispensable for maintaining the dispersion of mitotic chromosomes in the cytoplasm, and it is expressed across the cell cycle; cyclin B2 (encoded by Ccnb2) is a critical component of the regulatory machinery governing the cell cycle. Although a recent investigation has indicated that HDAC3 inhibition disrupts the activity of CENP-F (encoded by Cenpf) (46), comprehensive elucidation of how HDAC3 directly modulates the mRNA levels of these cell cycle–related genes remains largely unexplored. Future investigations are warranted to elucidate their intricate interplay. Further cell cycle analysis by flow cytometry revealed that ablating HDAC3 induced both S-phase arrest and G2-M arrest. As expected (47), the number of pH3+Iba1+ and that of Ki67+Iba1+ cells were remarkably reduced as shown in immunostaining. Our study revealed that HDAC3 inhibition intervened in the progressions of microglia cell cycles probably in an elusive way more or less independent of posttranslational modulation of cyclins or CDKs as shown by RNA-seq results and distinct from other cell types (30).
Since HDAC3 can directly regulate cell epigenetic landscape that may subsequently reshape transcriptome profiles (8, 12), we used ATAC-seq to measure microglial chromatin accessibility (48) and uncovered the underlying epigenetic mechanism by which HDAC3 inhibition halted microglia proliferation. Notably, DNA access at transcription factor motif for Spi1 is most remarkably lost in HDAC3-deficient microglia, suggesting its implication in HDAC3-mediated transcriptional regulation, which is validated by the consistent evidence that inhibiting HDAC3 down-regulated microglial PU.1 in both RNA and protein levels. This finding is consistent with previous studies using class I/II HDAC inhibitors, trichostatin A (TSA) (49–52). However, these studies mainly focused on the role of HDAC1 in PU.1 gene transcription, such as its involvement in TAF9 deacetylation and transcript factor IID assembly (51), as well as the increased histone H4 acetylation across the PU.1 promoter region by TSA (52). It remains unclear whether HDAC3 regulates PU.1 mRNA and protein levels through mechanisms similar to HDAC1. In addition, the down-regulation of PU.1 may also involve RUNX1, which is a component of the HDAC3 corepressor complex and has been shown to bind to crucial sites within the PU.1 upstream regulatory element and regulate PU.1 expression (53). Further investigation is necessary to fully elucidate the specific mechanisms by which HDAC3 affects PU.1 regulation.
PU.1 is known to be a vital transcription factor to regulate macrophage proliferation (54) and differentiation (55) as well as microgliogenesis (13) in the developing brain. In the context of Alzheimer’s disease, PU.1 is vital for Aβ clearance (56) and the inflammatory response of microglia (15). The latter role is also found in the model of experimental autoimmune encephalomyelitis (EAE) (16) and optic nerve injury (57). In a mouse model of prion disease, another neurodegeneration model, PU.1 also serves as a key component of the mitogenic pathway driving microglial proliferative response. Similar to our findings, Mullican et al. (41) revealed that HDAC3 in bone marrow–derived macrophages (BMDMs) localizes to a subset of PU.1-defined macrophage enhancers, suggesting that PU.1 likely facilitates adjacent binding of nuclear receptors. In lipopolysaccharide (LPS)–treated BMDMs, the most enriched motif at HDAC3-bound regions also matches the binding specificity of PU.1 (27). High-throughput screening of 1280 US Food and Drug Administration–approved compounds also identified an HDAC inhibitor, vorinostat, as a candidate drug for attenuating PU.1 level, which is then found to mimic partial effects of PU.1 knockdown (43). A recent study found novel AAV variants that are capable of delivering various genetic payloads into microglia in vitro and in vivo without activating microglia with higher efficiency than generating transgene mouse model or conventional rAAVs (20). Therefore, we took advantage of the rAAV and constructed AAV-PU.1 to overexpress PU.1 specifically in microglia. Our findings revealed that overexpressing PU.1 partially reversed selective inhibition of CD16+ microglial proliferation and ameliorated outcomes induced by HDAC3-miKO after tFCI. Together, these previous findings support our hypothesis that HDAC3-PU.1 axis in microglia may mediate microglial proliferation and immune response. Although transcriptomically HDAC3-miKO–induced up-regulated gene counts are comparable to the down-regulated gene counts, ATAC-seq analysis indicated that HDAC3 deletion results in much more down-regulated loci than up-regulated loci. This inconsistency is largely due to the nature that ATAC-seq only provides regulatory insights for phenomena that are probably transcriptionally regulated but miss any posttranscriptional changes such as RNA decay or modification (48). Moreover, since HDAC3 functions as a gene repressor via histone deacetylation, the fact that deleting HDAC3 resulted in a great decrease in the accessibility of a large number of loci seems counterintuitive. In previous studies, loss of HDAC3 impairs the activation of LPS-stimulated genes, which is attributable to the reduction of interferon-β (IFN-β) expression (58). This activation defect is probably independent of HDAC3 deacetylase activity (27). Thus, it could be presumed that in the context of ischemic stroke, it is the noncanonical deacetylase-independent way (27) by which deleting HDAC3 represses gene expression. Nonetheless, how exactly deleting HDAC3 transcriptomically up-regulated genes that may be associated with an alternative phenotype (41) without promoting associated chromatin accessibility is still a complicated issue that needs further investigation.
Our study on microglia apoptosis, proliferation, and associated cell cycle assay provided evidence of why HDAC3 depletion diminishes the expanding microglia population (21) at the acute stage of stroke, a notable feature in common with previous studies (9). We also detected a subtle reduction of microglia number in untreated HDAC3-miKO brain, which is probably due to the fact that even in the healthy adult brain, microglia population also undergoes continuous proliferation for self-renewal (59). It remains elusive whether microgliosis plays a detrimental role in ischemic stroke so that inhibiting microgliosis ameliorates stroke outcomes. Most studies did regard reduced microgliosis as attenuated microglia-based inflammatory responses (21, 60). To answer this question, depleting microglia as far as possible provides the most direct evidence in the context of stroke. In a preventive study, after depleting microglia by blocking CSF-1R for 21 days before tFCI, above 90% of microglia are eliminated, leading to increased brain injury, enhanced excitotoxicity, and altered neuronal activity (61). Similarly, long-term inhibition of CSF-1R before and after stroke exacerbates the production of inflammatory mediators, leukocyte infiltration, and ischemic lesion within the early stage of injury (62), while short-term CSF-1R inhibition resolves inflammation and ameliorates brain damage (63). In addition, it was reported that after inhibiting CSF-1R, the resident microglia display elevated proliferation and inflammation markers (64, 65), which may account for conflicting effects resulting from CSF-1R inhibition. Similarly, direct inhibition of microglial mitosis with AraC fosters microglial development of a proinflammatory phenotype during chronic degeneration (66). Therefore, it remains unclear whether reduction of microglia ameliorates or aggravates stroke outcome. In our study, microglia were not directly depleted but were prevented from rapid proliferation to some extent after stroke. Regardless of the extent, these microglia in two tFCI groups also undergo the similar so-called repopulation, which may confer different characteristics in turn to these renewed microglia (63). A previous study reported that HDAC3-deficent microglia shift from a proinflammatory phenotype to an inflammation-resolving phenotype with a reduction of microglia number in a mouse model of TBI (10).
Hence, we hypothesized that inflammatory phenotype alteration was at least partially triggered by preferential inhibition of proinflammatory microglia. Our results showed that in the absence of HDAC3, the blockade of proliferation did not occur in Arg1+ or CD206+ microglia, while CD16+ microglia displayed comparable proliferative suppression as in the whole microglia population. Consistently, we also observed that HDAC3 deficiency alleviated neuroinflammation and reduced the number of CD16+ microglia. Because of the channel limitation of immunostaining, we could not directly figure out the exact phenotype of which the proliferation is inhibited by ablation of HDAC3. While a previous study reported that in a model of demyelination, proliferative microglia cluster as a population (Birc5+) independent of other pathological microglia in single-cell RNA sequencing (scRNA-seq) (67), our study revealed the parallel inflammatory properties of proliferative microglia. However, we still do not know the sequence in which microglial proliferation and polarization occur, that is, whether proliferative microglia acquire different inflammatory phenotypes or microglia of different phenotypes proliferate. Besides, since in this study we simply used a limited number of molecules CD16, Arg1, or CD206 to label different states of activated microglia and thereby simplified the true situation of microglia-mediated inflammation, we were not able to uncover the exact connection between microglial proliferation and inflammation, while nowadays, in the single-cell era, evidence from novel unbiased single-cell techniques [e.g., cytometry by time of flight (CyTOF) and scRNA-seq] is accumulating that microglia are context dependent and develop numbers of populations including inflammation/proliferation-associated populations in response to different environments (67, 68). Studies predating the widespread adoption of single-cell techniques commonly used CD206 as a marker for anti-inflammatory microglia/macrophages, without distinguishing whether these cells were bona fide microglia. However, current knowledge, informed by the advancements in single-cell techniques, indicates that CD206, particularly at the RNA level, is highly expressed on macrophages but barely detectable even on activated microglia (69). Contrary to this understanding, our study demonstrated robust expression of CD206 on activated microglia after stroke, indicating a need for further exploration of the presence and role of CD206 on microglia in various diseases (fig. S10D). Arg1 is an enzyme that catalyzes the conversion of arginine to ornithine and urea. It competes with inducible nitric oxide synthase (iNOS) for the substrate of arginine and down-regulates the production of nitric oxide. In this case, Arg1 has been regarded as a marker for beneficial microglia/macrophages and has anti-inflammatory and tissue repair properties under various pathological conditions. Recent investigations have identified Arg1+ microglia as key regulators of postnatal development (70) and reducers of Aβ plaque deposition (71). Stat6/Arg1 axis also orchestrated microglia/macrophage efferocytosis and inflammation resolution after stroke (72). Furthermore, treatment with Arg1 collaborated with ornithine decarboxylase enzyme was found to protect against retinal ischemic injury (19). These discoveries suggest that Arg1 might serve as a more valid marker for anti-inflammatory microglia/macrophages. In line with this, our study revealed high expression of Arg1 in both RNA and protein levels in bona fide stroke-induced microglia. Consequently, we have chosen to use Arg1 as an alternative marker to represent anti-inflammatory microglia in our investigation, allowing us to explore the distinct effects of HDAC3 ablation on different microglial populations. In agreement with our findings, a recent study also reported increased Arg1 expression following HDAC3 deletion in macrophages. Treating LPS-induced macrophages with pegylated arginase 1 (PEG-A1) negatively regulated the expression of HDAC3, highlighting a potential bidirectional regulatory interaction between Arg1 and HDAC3. This underscores the need for future research to understand the intricate relationship between Arg1 and HDAC3 in microglia, particularly in the stroke context (19).
Recently, using scRNA-seq, a proliferative subtype controlled by HDAC3 is also unveiled in the context of spinal cord injury (7). Therefore, future studies using scRNA-seq and scATAC-seq may enable us to understand the association between proliferation and microglia-mediated inflammation and the nature of how depleting HDAC3 affects microglial heterogeneity in the context of ischemic stroke. In addition to discussing the role of HDAC3 for microglia, in the current study, using a singular CX3CR1CreER transgenic mouse line, we cannot exclude the potential impact of HDAC3 deletion in BAMs such as perivascular macrophages and meningeal macrophages, which are also targeted by Cx3cr1CreER (73). While our focus was primarily on microglia, the presence of HDAC3-deficient BAMs raised the possibility of their influence on microglial characteristics through direct contact or paracrine signaling pathways (74) given a degree of heterogeneity of BAMs as reported in recent studies (69). Therefore, future studies using scRNA-seq (69) or binary Cre transgenic approaches (73) could provide deeper insights into the specific roles of HDAC3 in microglia and BAMs and also in their interaction, enabling a comprehensive understanding of their contributions to neuroinflammation and brain function.
MATERIALS AND METHODS
Ethics statement
All animals were housed in facilities with suitable temperature and humidity and had a 12-hour light-dark cycle. Food and water are ad libitum. All animal experiments were approved by the Animal Care and Use Committee of Fudan University (approval numbers 20200306-117 and 20230-004S) and reported following the ARRIVE guidelines (75). All efforts were made to minimize animal suffering and the number of animals used.
Experimental animals
HDAC3flox/flox mice were constructed by Shanghai Model Organisms Center. These mice process LoxP sites flanking exon 3 of the Hdac3 gene. In the presence of Cre, the LoxP sites recombine and delete the floxed region of Hdac3. CX3CR1CreER mice were purchased from The Jackson Laboratory. Two groups of CX3CR1CreER(−/−)HDAC3flox/flox (WT) and CX3CR1CreER(+/−)HDAC3flox/flox (HDAC3-miKO) mice were bred from HDAC3flox/flox and CX3CR1CreER mice. Microglial HDAC3 depletion was induced in HDAC3-miKO mice (8-week-old males and females) by intraperitoneal injection of tamoxifen (100 mg/kg, daily for five consecutive days), which was dissolved in corn oil and absolute ethanol in a ratio of 9:1 (H7904, Sigma-Aldrich, St. Louis, MO, USA).
Of note, male mice were used for most experiments. Female mice were exclusively used for behavioral testing and histological evaluation (i.e., NeuN assessment) to confirm the presence of protective effects similar to those observed in male mice. Apart from these experiments, all the remaining trials were conducted exclusively on male mice for consistency.
Mouse tFCI model
tFCI was induced by MCAO for 60 min or specifically 90 min as indicated. Briefly, mice were anesthetized with 1.5% isoflurane in a 30% O2/70% N2 mixture and placed on a temperature-adjustable heating pad to maintain the mouse rectal temperature at 37 ± 0.5°C during surgery. In tFCI mice, the left carotid artery was separated and exposed. The external carotid artery (ECA) was ligated and cut. The suture (diameter 0.10 mm) wrapped with a silicone tip (0.20 to 0.22 mm, RWD, Shanghai, China) was inserted into the ECA and advanced along the internal carotid arteries until it meets resistance (occluding the middle cerebral artery). Sham-operated mice received the same anesthesia and arterial exposure, except that no MCAO procedure was performed. To ensure the success of the tFCI model, the blood flow of the middle cerebral artery branches (rCBF) was monitored by laser Doppler flowmetry (PeriFlux System 5001, Perimed, Jarfalla, Sweden) and laser speckle (PeriCam PSI System, Perimed, Jarfalla, Sweden). The mice with a drop of less than 75% in cerebral blood flow compared to pre-ischemia levels would be eliminated. Surgeries were performed by investigators blinded to animal genotypes and experimental grouping.
Behavioral tests
Tests assessing Garcia score, rotarod test, and adhesive removal test were performed 1, 3, 5, 7, 14, 21, 28, and 35 days after tFCI as previously described (25). Morris water maze was performed during 29 to 34 days after tFCI for assessment of long-term leaning/cognitive deficits (25). All behavioral tests were performed by an investigator blinded to experimental manipulations (detailed in Supplementary Materials and Methods).
Tissue loss measurement
Mice were euthanized and then perfused transcardially with phosphate-buffered saline (PBS; pH 7.4) followed by 4% (w/v) paraformaldehyde (PFA; dissolved in PBS). Brains were harvested, fixed overnight in 4% PFA, and then cryoprotected in 20% sucrose in PBS for 1 day and in 30% sucrose in PBS for 1 day at 4°C. To measure tissue defects, we performed frozen serial coronal sections of the brain at 25-μm thickness using a frozen sectioning machine (Leica). Immunohistochemical staining of 10 equally spaced coronal brain sections (1.1 to −1.7 mm bregma) was performed with a neuronal marker, rabbit anti-NeuN antibody (1:1000, ab177487, Abcam, Cambridge, MA, USA). Brain tissue loss in each section was measured using ImageJ image analysis software (National Institutes of Health, Bethesda, MD, USA), and the volume of tissue loss was calculated by subtracting the non-infarcted volume of the ipsilateral hemisphere from the volume of the contralateral hemisphere using numerical integration. The volume of brain tissue loss was quantified by a researcher blinded to group allocation. The data were shown as the volume of tissue loss as a percentage of the contralateral hemisphere volume.
Immunofluorescence and image analysis
Immunofluorescence staining was carried out as described previously (25). In brief, 25-μm-thick frozen coronal brain slices were cut, blocked (5% donkey serum), and incubated in primary antibodies (diluted in 0.3% Triton X-100 of PBS containing 1% goat or donkey serum) at 4°C overnight. After washing, slices were incubated in secondary antibodies at room temperature for 2 hours. Brain slices were finally mounted with DAPI Fluoromount-G (36308ES20, Yeasen, Shanghai, China) after washing. ImageJ and Imaris software were used to analyze images that were captured by a Nikon A1 confocal microscope. Source code for immunofluorescence quantitative analysis (Fig. 6 and fig. S8) is available at https://zenodo.org/record/8238087.
CAP measurements
CAPs were recorded in the EC 35 days after tFCI or Sham operation. Mice were anesthetized with isoflurane and decapitated to remove the brain rapidly. Coronal brain sections were made at 350 μm using a vibrating microtome (1200s, Leica). Sections were transferred to artificial cerebrospinal fluid and saturated with a 95% oxygen + 5% CO2 mixture for 0.5 hours at 32°C, followed by 1 hour at room temperature for recovery. A concentric stimulation electrode and a glass microelectrode (5 to 8 megohms) were used for stimulation and recording. A range of stimulation currents (0.25-mA interval from baseline up to 2 mA) was given to elicit CAPs of EC. The amplitudes of the N1 (representing myelinated fibers) and N2 (representing unmyelinated fibers) components of the CAPs were measured by pClamp 10 software (Molecular Devices, San Jose, CA, USA).
Quantitative real-time PCR
Total RNA from brain tissue in the infarcted area was extracted with TRIzol reagent (19201ES60, Yeasen, Shanghai, China). RNA was extracted from flow-sorted microglia using RNeasy Plus Micro Kit (74030, Qiagen, Germany). The RNA was reverse-transcribed into cDNA according to the instructions of the reverse transcription kit (K1622, Thermo Fisher Scientific, Pittsburgh, PA, USA). Real-time qPCR was performed on Hieff QPCR SYBR Green Master Mix (11201ES08, Yeasen, Shanghai, China) as detection dye. The reaction was performed at 95°C for 5 min with 40 cycles of 95°C for 10 s, 55°C for 20 s, and 72°C for 20 s. Values were averaged for triplicate assays, and the relative amounts of mRNA were normalized to Gapdh levels.
MRI measurement
MRI machine model 11.7T (Bruker Biospin, Billerica, MA, USA) was used for scanning using T2WI procedures. In vivo scans were performed at 3 and 14 days after tFCI in mice. The steps were as follows: Mice were anesthetized with 3% isoflurane and placed in the coil of the MRI machine for scanning. The mice were maintained in anesthesia with 1% isoflurane. The body temperature was maintained, and the heartbeat and respiratory rate of the mice were monitored in real time. In vivo T2WI scan parameters were set as follows: repetition time (TR)/echo time (TE) = 3000/26 ms, field of view (FOV) = 18 × 18 mm, acquisition matrix = 256 × 256, 45 slices, slice thickness of 0.3 mm, two mean values, Rapid Acquisition with Relaxation Enhancement (RARE) factor = 8.
The ex vivo scan was performed 35 days after tFCI with the following steps. After perfusing the mice with PBS and PFA (paraformaldehyde), the brains were removed by decapitation and fixed in PFA for 24 hours. After washing PFA with PBS, the brains were air-dried, fixed in 5-ml test tubes containing carbon-free oil, and finally placed in the machine coil for scanning. The ex vivo T2WI scan parameters were set as follows: TR/TE = 4000/17.331636364 ms, FOV = 20 × 20 mm, acquisition matrix = 256 × 256, 28 slices, slice thickness of 0.4 mm, two averages, RARE factor = 8.
EdU injections
EdU (50 mg/kg, ST067, Shanghai Beyotime, China) was injected intraperitoneally every 12 hours for 1 to 3 days after tFCI to label newly generated cells. Microglia proliferation was detected using the BeyoClick EdU Cell Proliferation Kit with Alexa Fluor 594. (C0078, Beyotime Biotechnology, Shanghai, China).
Flow cytometry and FACS
Single-cell suspensions in all flow cytometry/FACS experiments were prepared following procedures as described previously (10). After staining, flow cytometry was performed using a Beckman CytoFLEX flow cytometer operated by CytExpert software. FlowJo software was then used to quantify proportion of positively stained cells or MFI in specific cell populations. In particular, flow-based cell cycle analysis was performed with Vybrant DyeCycle Violet Ready Flow Reagent (R37172, Thermo Fisher Scientific, Pittsburgh, PA, USA) following the manufacturer’s instructions.
For FACS, mononuclear cells were sorted using a Beckman CytoFLEX SRT cell sorter operated by Kaluza analysis software. The flow-sorted cells were then applied to qPCR, ATAC-seq, and RNA-seq.
Protein microarray
Ipsilateral and contralateral brain tissue proteins were extracted 3 days after tFCI. The protein expression of 40 inflammatory factors was detected by kits (RayBio C-Series Mouse Inflammation Antibody Array C1) 3 days after tFCI. Various factors were normalized to levels in the contralateral control brain, and data were expressed as fold change.
RNA sequencing
Isolated microglia were sent to Shanghai Megiddo Biological Pharmaceutical Co. Ltd. for RNA-seq. Total RNA from flow-sorted microglia was extracted using TRIzol reagent according to the manufacturer’s instructions (Invitrogen), and genomic DNA was removed using deoxyribonuclease I (Takara). RNA quality was assessed by 2100 Bioanalyzer (Agilent Technologies) and quantified by ND-2000 (NanoDrop Technologies). The library was then prepared following Clontech-SMART-Seq v4 Ultra Low Input RNA kit for sequencing using 10 ng of total RNA. Paired-end RNA-seq was performed on Illumina NovaSeq 6000 sequencer [2 × 150–base pair (bp) read length].
FastQC (v0.11.9) was first used for quality control assessment. The low-quality reads (quality score lower than 20) were removed using Trim Galore. Paired-end clean data were then aligned to the reference genome (Mus musculus, UCSC mm10) using HISAT2 (v2.2.1) after being sorted using Samtools. After that, HTseq (v1.99.2) was used to quantify the number of reads mapped to each gene annotated in the GENCODE release M25 (GRCm38.p6). Here, a gene refers to the union of all its exons in any isoforms based on GENCODE annotation. Alignment files were merged by group using Samtools. These merged files were then transformed into normalized bigwig files using BamCoverage from Deeptools and visualized on IGV.
Raw HTseq read counts were imported into R and then transformed and normalized using DEseq2 (v1.32.0). The normalized rlog/vst-transformed counts were adjusted using the removeBatchEffect from the limma package. We visualized PCA on these adjusted values and gene expression in different gene sets as heatmaps using the ComplexHeatmap package.
Differential expression analysis was performed with batch differences controlled. For pairwise comparisons, we compared miKO-Sham to WT-Sham, miKO-tFCI to WT-tFCI, and WT-tFCI to WT-Sham (Padj < 0.05, |log2FC| > 0.58). Shrunk fold change produced by an adaptive shrinkage estimator ashr (76) was used for visualizing and ranking of genes in the following analysis. EnhancedVolcano package was used to visualize differential expression analysis results.
Using clusterProfiler R package (v4.0.5) (77), we applied DEGs produced from different pairwise comparisons to overrepresentation analysis, as well as lists of significantly expressed genes (P < 0.05) that were ranked according to their shrunk fold change to GSEA (default parameters) against Reactome pathway database. We then compared all the significantly enriched pathways generated from WT-tFCI versus WT-Sham to those from miKO-tFCI versus WT-tFCI and visualized all the overlapping pathways via a bar plot. Of note, a positive normalized enrichment scores (NES) indicates that a specific gene set is predominantly represented at the top of the list, while a negative NES suggests representation toward the bottom. For instance, a positive NES for a gene set implies up-regulation in WT-tFCI versus WT-Sham, whereas miKO-tFCI versus WT-tFCI shows a negative NES, indicating down-regulation of genes related to that set. Specifically, in this study, these bars (Fig. 4G) displayed positive NES for “WT-tFCI vs. WT-Sham” and negative NES for “miKO-tFCI vs. WT-tFCI.”
Gene sets, “proinflammatory genes,” or “anti-inflammatory genes” was produced from down-regulated genes or up-regulated genes in miKO-tFCI versus WT-tFCI that overlapped with respective DEGs that were acquired using GEO2R from either LPS and IFN-γ–stimulated BMDMs versus control or IL-4–stimulated BMDMs versus control in a public dataset (GSE69607). “Microglial proliferation-related genes” was the union of all genes enriched for any of the altered pathways as shown in Fig. 3G.
ATAC sequencing
Freshly flow-sorted microglia were also collected for ATAC-seq. ATAC-seq library was prepared as described in a previous study. Microglial nuclei were first extracted with lysis buffer [0.32 M sucrose, 5 mM CaCl2, 3 mM MgAce2, 0.1 mM EDTA, 10 mM tris-HCl (pH 8), and 0.1% NP-40]. Then, ATAC-seq library was constructed using the reagents in TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme, TD501-02). The experimental procedure was as follows: Nuclei were treated with Tn5 at 37°C for 30 min. DNA was then purified with the MinElute Gel Extraction Kit (Qiagen, 28604) and eluted in 27 μl of EB buffer. Twenty-four microliters of DNA samples, 10 μl of 5X TruePrep Amplify Buffer, 5 μl of PCR Primer Mix, 1 μl of TruePrep Amplify Enzyme, and selected aptamers (5 μl of N5XX and 5 μl of N7XX) (Vazyme, TD202) were mixed, followed by PCR amplification. Library DNA was purified and size-selected with SPRIselect beads (Beckman, B23318). Library quality was checked on a Qubit and Agilent 4200 Tapestation and then sent to Novogene in China for paired-end sequencing on Illumina XTen (2 × 150–bp read length).
FastQC and data cleaning were performed as described previously. Paired-end clean data were aligned to the reference genome (M. musculus, UCSC mm10) using Bowtie2 (v2.2.5). Alignment files were merged and visualized as described above. Peaks were called using MACS2 v2.1.4 (--shift -100 --extsize 200–nomodel). Diffbind was used to analyze differential peaks. Read count on peaks was acquired (bUseSummarizeOverlaps = TRUE). Inbuilt DEseq2 was then invoked to perform the normalization and differential analysis, with batch effect considered. ATAC-seq peaks were annotated to their genomic region and proximate gene using ChIPseeker (v1.28.3) (78) and org.Mm.eg.db (v3.13.0) package, respectively. GSEA was performed on DARs (P < 0.01) as previously described. The HOMER2 software suite was used to perform de novo motif enrichment analysis of promoter-located DARs by the program “findMotifs.pl.”
For integrative analysis of bulk RNA-seq and ATAC-seq, up-/down-regulated DEGs (Padj < 0.05 and |log2FC| > 0.58) in RNA-seq and open/closed DARs in ATAC-seq (Padj < 0.01 and |log2FC| > 0.5) generated from the comparison, miKO-tFCI versus WT-tFCI, were exploited. To check whether closed DARs overlap with down-regulated DEGs more than expected, a permutation test was performed with an R package regioneR by the “permTest” function (ntimes = 200, randomize.function = randomizeRegions, evaluate.function = numOverlaps, universe = all.genes); all consensus peaks were assigned to the parameter, “all.genes.”
Intracerebral injection of AAV vector expressing PU.1
The virus infecting microglia was constructed by Braincase Biotechnology Co. Ltd. (Shenzhen, China). In this experiment, we injected the control virus (rAAV-EF1a-DIO-mCherry, AAV-Control) and the virus overexpressing PU.1 (rAAV-EF1a-DIO-mPU.1-P2A-mCherry, AAV-PU.1) with a titer of ≥2.00 × 1012 Vector Genomes per ml. Viruses (500 nl per site) were delivered via stereotaxic injection into four sites [bregma: anterior-posterior (AP) = 0.14 mm, medial-lateral (ML) = 2.0 mm, dorsal-ventral (DV) = 2.5 mm; bregma: AP = 0.14 mm, ML = 2.0 mm, DV = 3.5 mm; bregma: AP = −0.34 mm, ML = 2.5 mm, DV = 3 mm; bregma: AP = −0.34 mm, ML = 2.5 mm, DV = 4 mm]. After the stereotaxic injection of the virus into the mouse brain, a 1-week resting period was allowed. Then, tamoxifen was injected intraperitoneally to induce the expression of Cre enzyme. The virus-infected microglia were overexpressed with PU.1. After 1 month, the mice underwent MCAO surgery. The experiment consisted of three groups: WT-tFCI-AAV-Control, miKO-tFCI-AAV-Control, and miKO-tFCI-AAV-PU.1.
Statistical analysis
Data were expressed as means and SEM. Results were analyzed with GraphPad Prism software (version 8.0, La Jolla, CA, USA). Gaussian distributions were tested using the Kolmogorov-Smirnov test. Data conforming to the Gaussian distribution were analyzed with parametric tests, while data with non-Gaussian distribution were analyzed with nonparametric tests. Two-tailed unpaired Student’s t test was used for the comparison of two sets of Gaussian distributed data. For non-Gaussian distributed data, the Mann-Whitney U rank sum test was used for one-way analysis. Unless otherwise stated, differences in means between multiple groups of Gaussian-distributed data were analyzed by ordinary one-way analysis of variance (ANOVA; F test) followed by Bonferroni’s multiple comparisons, and for non-Gaussian–distributed data, Kruskal-Wallis test was used. Generalized estimation equation was applied to repeated-measures models (behavior tests, MRI, or electrophysiology) with a baseline covariant (e.g., preoperative value) used if available, followed by Tukey’s post hoc test for multiple comparison of treatment effects or interaction effects of treatment and time. Two-way ANOVA followed by Bonferroni’s multiple comparisons was used for two-way analysis. Linear regression analysis was performed using GraphPad Prism software. Pearson correlation analysis was used to test for correlations. In all analyses, P < 0.05 was considered statistically significant.
Acknowledgments
We are indebted to J. J. Li for proofreading the manuscript.
Funding: This work was supported by STI 2030-Major Projects (2022ZD0204704 and 2021ZD0201704) to Y. Gao; National Natural Science Foundation of China (82071311 and 81870971) to Y. Gao; Shanghai Leading Talent Project (3030513405) to Y. Gong; and Shanghai Municipal Science and Technology Major Projects (22ZR1413700 and 2018SHZDZX01), ZJLab, and Shanghai Center for Brain Science and Brain-Inspired Technology to Y. Gao.
Author contributions: Conceptualization: Y. Gao. Methodology: Y. Zhang, J.L., Y. Zhao, Y.H., Z.S., H.W., H.C., C.W., Yana Wang, D.C., S.C., Yangfan Wang, S.M., Y. Zhu, and Y.J. Investigation: Y. Gao and Y. Gong. Visualization: Y. Gao, Y. Zhang, and J.L. Supervision: Y. Gao and Y. Gong. Writing—original draft: J.L., Y. Zhang, and Z.S. Writing—review and editing: Y. Gao and Y. Gong.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Bulk RNA-seq data and ATAC-seq data are available in GEO and can be accessed with GEO accession GSE220043. The script for image quantification analysis is provided in https://zenodo.org/record/8238087.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Supplementary Materials and Methods
Tables S1 to S4
Legends for data S1 and S2
References
Other Supplementary Material for this manuscript includes the following:
Data S1 and S2
REFERENCES AND NOTES
- 1.Gu N., Peng J., Murugan M., Wang X., Eyo U. B., Sun D., Ren Y., DiCicco-Bloom E., Young W., Dong H., Wu L.-J., Spinal microgliosis due to resident microglial proliferation is required for pain hypersensitivity after peripheral nerve injury. Cell Rep. 16, 605–614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olmos-Alonso A., Schetters S. T. T., Sri S., Askew K., Mancuso R., Vargas-Caballero M., Holscher C., Perry V. H., Gomez-Nicola D., Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer's-like pathology. Brain 139, 891–907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia J., Yang L., Chen Y., Zheng L., Chen Y., Xu Y., Zhang M., The role of microglial phagocytosis in ischemic stroke. Front. Immunol. 12, 790201 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sieber M. W., Jaenisch N., Brehm M., Guenther M., Linnartz-Gerlach B., Neumann H., Witte O. W., Frahm C., Attenuated inflammatory response in triggering receptor expressed on myeloid cells 2 (TREM2) knock-out mice following stroke. PLOS ONE 8, e52982 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barca C., Foray C., Hermann S., Herrlinger U., Remory I., Laoui D., Schäfers M., Grauer O. M., Zinnhardt B., Jacobs A. H., The colony stimulating factor-1 receptor (CSF-1R)-mediated regulation of microglia/macrophages as a target for neurological disorders (glioma, stroke). Front. Immunol. 12, 787307 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T., Zhao J., Xie W., Yuan W., Guo J., Pang S., Gan W.-B., Gómez-Nicola D., Zhang S., Specific depletion of resident microglia in the early stage of stroke reduces cerebral ischemic damage. J. Neuroinflammation 18, 81 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wahane S., Zhou X., Zhou X., Guo L., Friedl M.-S., Kluge M., Ramakrishnan A., Shen L., Friedel C. C., Zhang B., Friedel R. H., Zou H., Diversified transcriptional responses of myeloid and glial cells in spinal cord injury shaped by HDAC3 activity. Sci. Adv. 7, eabd8811 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emmett M. J., Lazar M. A., Integrative regulation of physiology by histone deacetylase 3. Nat. Rev. Mol. Cell Biol. 20, 102–115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao Y., Cheng J., Kong X., Li S., Li X., Zhang M., Zhang H., Yang T., Dong Y., Li J., Xu Y., Yuan Z., HDAC3 inhibition ameliorates ischemia/reperfusion-induced brain injury by regulating the microglial cGAS-STING pathway. Theranostics 10, 9644–9662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y., Mu H., Huang Y., Li S., Wang Y., Stetler R. A., Bennett M. V. L., Dixon C. E., Chen J., Shi Y., Microglia-specific deletion of histone deacetylase 3 promotes inflammation resolution, white matter integrity, and functional recovery in a mouse model of traumatic brain injury. J. Neuroinflammation 19, 201 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G., Shi Y., Jiang X., Leak R. K., Hu X., Wu Y., Pu H., Li W.-W., Tang B., Wang Y., Gao Y., Zheng P., Bennett M. V. L., Chen J., HDAC inhibition prevents white matter injury by modulating microglia/macrophage polarization through the GSK3β/PTEN/Akt axis. Proc. Natl. Acad. Sci. U.S.A. 112, 2853–2858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adhikari N., Jha T., Ghosh B., Dissecting histone deacetylase 3 in multiple disease conditions: Selective inhibition as a promising therapeutic strategy. J. Med. Chem. 64, 8827–8869 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Kierdorf K., Erny D., Goldmann T., Sander V., Schulz C., Perdiguero E. G., Wieghofer P., Heinrich A., Riemke P., Hölscher C., Müller D. N., Luckow B., Brocker T., Debowski K., Fritz G., Opdenakker G., Diefenbach A., Biber K., Heikenwalder M., Geissmann F., Rosenbauer F., Prinz M., Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273–280 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Staber P. B., Zhang P., Ye M., Welner R. S., Nombela-Arrieta C., Bach C., Kerenyi M., Bartholdy B. A., Zhang H., Alberich-Jordà M., Lee S., Yang H., Ng F., Zhang J., Leddin M., Silberstein L. E., Hoefler G., Orkin S. H., Göttgens B., Rosenbauer F., Huang G., Tenen D. G., Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol. Cell 49, 934–946 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pimenova A. A., Herbinet M., Gupta I., Machlovi S. I., Bowles K. R., Marcora E., Goate A. M., Alzheimer's-associated PU.1 expression levels regulate microglial inflammatory response. Neurobiol. Dis. 148, 105217 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponomarev E. D., Veremeyko T., Barteneva N., Krichevsky A. M., Weiner H. L., MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nat. Med. 17, 64–70 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin R., Zhou Y., Yan T., Wang R., Li H., Wu Z., Zhang X., Zhou X., Zhao F., Zhang L., Li Y., Luo M., Directed evolution of adeno-associated virus for efficient gene delivery to microglia. Nat. Methods 19, 976–985 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Baltan S., Bachleda A., Morrison R. S., Murphy S. P., Expression of histone deacetylases in cellular compartments of the mouse brain and the effects of ischemia. Transl. Stroke Res. 2, 411–423 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shosha E., Shahror R. A., Morris C. A., Xu Z., Lucas R., McGee-Lawrence M. E., Rusch N. J., Caldwell R. B., Fouda A. Y., The arginase 1/ornithine decarboxylase pathway suppresses HDAC3 to ameliorate the myeloid cell inflammatory response: Implications for retinal ischemic injury. Cell Death Dis. 14, 621 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendes M. S., Le L., Atlas J., Brehm Z., Ladron-de-Guevara A., Matei E., Lamantia C., McCall M. N., Majewska A. K., The role of P2Y12 in the kinetics of microglial self-renewal and maturation in the adult visual cortex in vivo. eLife 10, e61173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T., Pang S., Yu Y., Wu X., Guo J., Zhang S., Proliferation of parenchymal microglia is the main source of microgliosis after ischaemic stroke. Brain 136, 3578–3588 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Birenbaum D., Bancroft L. W., Felsberg G. J., Imaging in acute stroke. West. J. Emerg. Med. 12, 67–76 (2011). [PMC free article] [PubMed] [Google Scholar]
- 23.Sohrabji F., Okoreeh A., Panta A., Sex hormones and stroke: Beyond estrogens. Horm. Behav. 111, 87–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villa A., Gelosa P., Castiglioni L., Cimino M., Rizzi N., Pepe G., Lolli F., Marcello E., Sironi L., Vegeto E., Maggi A., Sex-specific features of microglia from adult mice. Cell Rep. 23, 3501–3511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen D., Li J., Huang Y., Wei P., Miao W., Yang Y., Gao Y., Interleukin 13 promotes long-term recovery after ischemic stroke by inhibiting the activation of STAT3. J. Neuroinflammation 19, 112 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X., Li P., Guo Y., Wang H., Leak R. K., Chen S., Gao Y., Chen J., Microglia/macrophage polarization dynamics reveal novel mechanism of injury expansion after focal cerebral ischemia. Stroke 43, 3063–3070 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Nguyen H. C. B., Adlanmerini M., Hauck A. K., Lazar M. A., Dichotomous engagement of HDAC3 activity governs inflammatory responses. Nature 584, 286–290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyce M., Xin Y., Chowdhury O., Shang P., Liu H., Koontz V., Strizhakova A., Nemani M., Hose S., Zigler J. S., Campbell M., Sinha D., Handa J. T., Kaarniranta K., Qian J., Ghosh S., Microglia–neutrophil interactions drive dry AMD-like pathology in a mouse model. Cell 11, 3535 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeda Y., He P., Tachibana I., Zhou B., Miyado K., Kaneko H., Suzuki M., Minami S., Iwasaki T., Goya S., Kijima T., Kumagai T., Yoshida M., Osaki T., Komori T., Mekada E., Kawase I., Double deficiency of tetraspanins CD9 and CD81 alters cell motility and protease production of macrophages and causes chronic obstructive pulmonary disease-like phenotype in mice. J. Biol. Chem. 283, 26089–26097 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y., Hsieh J., HDAC3 controls gap 2/mitosis progression in adult neural stem/progenitor cells by regulating CDK1 levels. Proc. Natl. Acad. Sci. U.S.A. 111, 13541–13546 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vidal-Laliena M., Gallastegui E., Mateo F., Martínez-Balbás M., Pujol M. J., Bachs O., Histone deacetylase 3 regulates cyclin A stability. J. Biol. Chem. 288, 21096–21104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matheson R., Chida K., Lu H., Clendaniel V., Fisher M., Thomas A., Lo E. H., Selim M., Shehadah A., Neuroprotective effects of selective inhibition of histone deacetylase 3 in experimental stroke. Transl. Stroke Res. 11, 1052–1063 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Kim J.-Y., Jeong H. S., Chung T., Kim M., Lee J. H., Jung W. H., Koo J. S., The value of phosphohistone H3 as a proliferation marker for evaluating invasive breast cancers: A comparative study with Ki67. Oncotarget 8, 65064–65076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann M. J., Meneceur S., Hommel K., Schulz W. A., Niegisch G., Downregulation of cell cycle and checkpoint genes by class I HDAC inhibitors limits synergism with G2/M checkpoint inhibitor MK-1775 in bladder cancer cells. Genes 12, 260 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holder G. E., McGary C. M., Johnson E. M., Zheng R., John V. T., Sugimoto C., Kuroda M. J., Kim W. K., Expression of the mannose receptor CD206 in HIV and SIV encephalitis: A phenotypic switch of brain perivascular macrophages with virus infection. J. Neuroimmune Pharmacol. 9, 716–726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faraco G., Park L., Anrather J., Iadecola C., Brain perivascular macrophages: Characterization and functional roles in health and disease. J. Mol. Med. 95, 1143–1152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartalska K., Hübschmann V., Korkut-Demirbaş M., Cubero R. J. A., Venturino A., Rössler K., Czech T., Siegert S., A systematic characterization of microglia-like cell occurrence during retinal organoid differentiation. iScience 25, 104580 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corces M. R., Shcherbina A., Kundu S., Gloudemans M. J., Frésard L., Granja J. M., Louie B. H., Eulalio T., Shams S., Bagdatli S. T., Mumbach M. R., Liu B., Montine K. S., Greenleaf W. J., Kundaje A., Montgomery S. B., Chang H. Y., Montine T. J., Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer's and Parkinson's diseases. Nat. Genet. 52, 1158–1168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Starks R. R., Biswas A., Jain A., Tuteja G., Combined analysis of dissimilar promoter accessibility and gene expression profiles identifies tissue-specific genes and actively repressed networks. Epigenetics Chromatin 12, 16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith A. M., Gibbons H. M., Oldfield R. L., Bergin P. M., Mee E. W., Faull R. L. M., Dragunow M., The transcription factor PU.1 is critical for viability and function of human brain microglia. Glia 61, 929–942 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Mullican S. E., Gaddis C. A., Alenghat T., Nair M. G., Giacomin P. R., Everett L. J., Feng D., Steger D. J., Schug J., Artis D., Lazar M. A., Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 25, 2480–2488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanyal A., Lajoie B. R., Jain G., Dekker J., The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rustenhoven J., Smith A. M., Smyth L. C., Jansson D., Scotter E. L., Swanson M. E. V., Aalderink M., Coppieters N., Narayan P., Handley R., Overall C., Park T. I. H., Schweder P., Heppner P., Curtis M. A., Faull R. L. M., Dragunow M., PU.1 regulates Alzheimer's disease-associated genes in primary human microglia. Mol. Neurodegener. 13, 44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarkar R., Banerjee S., Amin S. A., Adhikari N., Jha T., Histone deacetylase 3 (HDAC3) inhibitors as anticancer agents: A review. Eur. J. Med. Chem. 192, 112171 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Summers A. R., Fischer M. A., Stengel K. R., Zhao Y., Kaiser J. F., Wells C. E., Hunt A., Bhaskara S., Luzwick J. W., Sampathi S., Chen X., Thompson M. A., Cortez D., Hiebert S. W., HDAC3 is essential for DNA replication in hematopoietic progenitor cells. J. Clin. Invest. 123, 3112–3123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh A. K., Bishayee A., Pandey A. K., Targeting histone deacetylases with natural and synthetic agents: An emerging anticancer strategy. Nutrients 10, 731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan W., Su P.-Y. P., Leff J., Gao X., Chen J., Guan A. K., Kalyanasundaram G., Ma A., Guan Z., Distinct phases of adult microglia proliferation: A Myc-mediated early phase and a Tnfaip3-mediated late phase. Cell Discov. 8, 34 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grandi F. C., Modi H., Kampman L., Corces M. R., Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 17, 1518–1552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laribee R. N., Klemsz M. J., Loss of PU.1 expression following inhibition of histone deacetylases. J. Immunol. 167, 5160–5166 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Koyama Y., Adachi M., Sekiya M., Takekawa M., Imai K., Histone deacetylase inhibitors suppress IL-2-mediated gene expression prior to induction of apoptosis. Blood 96, 1490–1495 (2000). [PubMed] [Google Scholar]
- 51.Jian W., Yan B., Huang S., Qiu Y., Histone deacetylase 1 activates PU.1 gene transcription through regulating TAF9 deacetylation and transcription factor IID assembly. FASEB J. 31, 4104–4116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laribee R. N., Klemsz M. J., Histone H4 HDAC activity is necessary for expression of the PU.1 gene. Biochim. Biophys. Acta 1730, 226–234 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Huang G., Zhang P., Hirai H., Elf S., Yan X., Chen Z., Koschmieder S., Okuno Y., Dayaram T., Growney J. D., Shivdasani R. A., Gilliland D. G., Speck N. A., Nimer S. D., Tenen D. G., PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat. Genet. 40, 51–60 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Celada A., Borràs F. E., Soler C., Lloberas J., Klemsz M., van Beveren C., McKercher S., Maki R. A., The transcription factor PU.1 is involved in macrophage proliferation. J. Exp. Med. 184, 61–69 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li G., Hao W., Hu W., Transcription factor PU.1 and immune cell differentiation (Review). Int. J. Mol. Med. 46, 1943–1950 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Lau S.-F., Chen C., Fu W.-Y., Qu J. Y., Cheung T. H., Fu A. K. Y., Ip N. Y., IL-33-PU.1 transcriptome reprogramming drives functional state transition and clearance activity of microglia in Alzheimer's disease. Cell Rep. 31, 107530 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Zhou N., Liu K., Sun Y., Cao Y., Yang J., Transcriptional mechanism of IRF8 and PU.1 governs microglial activation in neurodegenerative condition. Cell 10, 87–103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X., Barozzi I., Termanini A., Prosperini E., Recchiuti A., Dalli J., Mietton F., Matteoli G., Hiebert S., Natoli G., Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc. Natl. Acad. Sci. U.S.A. 109, E2865–E2874 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Askew K., Li K., Olmos-Alonso A., Garcia-Moreno F., Liang Y., Richardson P., Tipton T., Chapman M. A., Riecken K., Beccari S., Sierra A., Molnár Z., Cragg M. S., Garaschuk O., Perry V. H., Gomez-Nicola D., Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Rep. 18, 391–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kodali M. C., Chen H., Liao F.-F., Temporal unsnarling of brain's acute neuroinflammatory transcriptional profiles reveals panendothelitis as the earliest event preceding microgliosis. Mol. Psychiatry 26, 3905–3919 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szalay G., Martinecz B., Lénárt N., Környei Z., Orsolits B., Judák L., Császár E., Fekete R., West B. L., Katona G., Rózsa B., Dénes Á., Microglia protect against brain injury and their selective elimination dysregulates neuronal network activity after stroke. Nat. Commun. 7, 11499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Otxoa-de-Amezaga A., Miró-Mur F., Pedragosa J., Gallizioli M., Justicia C., Gaja-Capdevila N., Ruíz-Jaen F., Salas-Perdomo A., Bosch A., Calvo M., Márquez-Kisinousky L., Denes A., Gunzer M., Planas A. M., Microglial cell loss after ischemic stroke favors brain neutrophil accumulation. Acta Neuropathol. 137, 321–341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willis E. F., MacDonald K. P. A., Nguyen Q. H., Garrido A. L., Gillespie E. R., Harley S. B. R., Bartlett P. F., Schroder W. A., Yates A. G., Anthony D. C., Rose-John S., Ruitenberg M. J., Vukovic J., Repopulating microglia promote brain repair in an IL-6-dependent manner. Cell 180, 833–846.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Jin W.-N., Shi S. X.-Y., Li Z., Li M., Wood K., Gonzales R. J., Liu Q., Depletion of microglia exacerbates postischemic inflammation and brain injury. J. Cereb. Blood Flow Metab. 37, 2224–2236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elmore M. R. P., Najafi A. R., Koike M. A., Dagher N. N., Spangenberg E. E., Rice R. A., Kitazawa M., Matusow B., Nguyen H., West B. L., Green K. N., Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gómez-Nicola D., Fransen N. L., Suzzi S., Perry V. H., Regulation of microglial proliferation during chronic neurodegeneration. J. Neurosci. 33, 2481–2493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hammond T. R., Dufort C., Dissing-Olesen L., Giera S., Young A., Wysoker A., Walker A. J., Gergits F., Segel M., Nemesh J., Marsh S. E., Saunders A., Macosko E., Ginhoux F., Chen J., Franklin R. J. M., Piao X., McCarroll S. A., Stevens B., Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Böttcher C., Schlickeiser S., Sneeboer M. A. M., Kunkel D., Knop A., Paza E., Fidzinski P., Kraus L., Snijders G. J. L., Kahn R. S., Schulz A. R., Mei H. E., Hol E. M., Siegmund B., Glauben R., Spruth E. J., de Witte L. D., Priller J., Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat. Neurosci. 22, 78–90 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Jordão M. J. C., Sankowski R., Brendecke S. M., Sagar G., Locatelli Y. H., Tai T. L., Tay E., Schramm S., Armbruster N., Hagemeyer O., Groß D., Mai Ö., Çiçek T., Falk M., Kerschensteiner D., Grün M. P., Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Stratoulias V., Ruiz R., Kanatani S., Osman A. M., Keane L., Armengol J. A., Rodríguez-Moreno A., Murgoci A. N., García-Domínguez I., Alonso-Bellido I., González Ibáñez F., Picard K., Vázquez-Cabrera G., Posada-Pérez M., Vernoux N., Tejera D., Grabert K., Cheray M., González-Rodríguez P., Pérez-Villegas E. M., Martínez-Gallego I., Lastra-Romero A., Brodin D., Avila-Cariño J., Cao Y., Airavaara M., Uhlén P., Heneka M. T., Tremblay M., Blomgren K., Venero J. L., Joseph B., ARG1-expressing microglia show a distinct molecular signature and modulate postnatal development and function of the mouse brain. Nat. Neurosci. 26, 1008–1020 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cherry J. D., Olschowka J. A., O'Banion M. K., Arginase 1+ microglia reduce Aβ plaque deposition during IL-1β-dependent neuroinflammation. J. Neuroinflammation 12, 203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cai W., Dai X., Chen J., Zhao J., Xu M., Zhang L., Yang B., Zhang W., Rocha M., Nakao T., Kofler J., Shi Y., Stetler R. A., Hu X., Chen J., STAT6/Arg1 promotes microglia/macrophage efferocytosis and inflammation resolution in stroke mice. JCI Insight 4, e131355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim J. S., Kolesnikov M., Peled-Hajaj S., Scheyltjens I., Xia Y., Trzebanski S., Haimon Z., Shemer A., Lubart A., Van Hove H., Chappell-Maor L., Boura-Halfon S., Movahedi K., Blinder P., Jung S., A binary cre transgenic approach dissects microglia and cns border-associated macrophages. Immunity 54, 176–190.e7 (2021). [DOI] [PubMed] [Google Scholar]
- 74.De Schepper S., Ge J. Z., Crowley G., Ferreira L. S. S., Garceau D., Toomey C. E., Sokolova D., Rueda-Carrasco J., Shin S.-H., Kim J.-S., Childs T., Lashley T., Burden J. J., Sasner M., Sala Frigerio C., Jung S., Hong S., Perivascular cells induce microglial phagocytic states and synaptic engulfment via SPP1 in mouse models of Alzheimer's disease. Nat. Neurosci. 26, 406–415 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kilkenny C., Altman D. G., Improving bioscience research reporting: ARRIVE-ing at a solution. Lab. Anim 44, 377–378 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Stephens M., False discovery rates: A new deal. Biostatistics 18, 275–294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu G., Wang L. G., He Q. Y., ChIPseeker: An R/bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Garcia J. H., Wagner S., Liu K. F., Hu X. J., Neurological deficit and extent of neuronal necrosis attributable to middle cerebral artery occlusion in rats. Stroke 26, 627–634 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Supplementary Materials and Methods
Tables S1 to S4
Legends for data S1 and S2
References
Data S1 and S2