Abstract
Endothelial cells play crucial roles in physiology and are increasingly recognized as therapeutic targets in cardiovascular disease. Here, we analyzed the regulatory landscape of cardiac endothelial cells by assessing chromatin accessibility, histone modifications, and 3D chromatin organization and confirmed the functional relevance of enhancer-promoter interactions by CRISPRi-mediated enhancer silencing. We used this dataset to explore mechanisms of transcriptional regulation in cardiovascular disease and compared six different experimental models of heart failure, hypertension, or diabetes. Enhancers that regulate gene expression in diseased endothelial cells were enriched with binding sites for a distinct set of transcription factors, including the mineralocorticoid receptor (MR), a known drug target in heart failure and hypertension. For proof of concept, we applied endothelial cell–specific MR deletion in mice to confirm MR-dependent gene expression and predicted direct MR target genes. Overall, we have compiled here a comprehensive atlas of cardiac endothelial cell enhancer elements that provides insight into the role of transcription factors in cardiovascular disease.
Enhancer landscape identifies mineralocorticoid receptor as driver of pathological gene expression in cardiac endothelial cells.
INTRODUCTION
While for a long time cardiomyocytes have been in focus of cardiac research, non-myocytes of the heart have received increased attention in recent years. Endothelial cells are among the most abundant cell types of the heart (1, 2), with the greatest proportion of them being capillary endothelial cells (3, 4). The enormous energy demand of the heart goes along with a high blood vessel density of approximately one capillary per each cardiomyocyte to ensure supply with nutrients and oxygen (5). This specialized function of cardiac endothelial cells compared to other organs or vascular beds (6, 7) is also reflected by marked differences in their respective transcriptomes (3, 8). In addition to their classical role in energy supply, endothelial cells are today considered to actively regulate cardiac tissue homeostasis. As they form the inner layer of the vasculature, endothelial cells act as sensors for circulating factors, hemodynamic changes, and other environmental stimuli (9, 10). The endothelial cell barrier regulates influx of circulating cells such as monocytes into the tissue. In addition, endothelial cells form the extracellular matrix and via secreted factors such as nitric oxide interact with other cell types, directly modulating their function in health and disease (9). This makes endothelial cells an interesting therapeutic target in cardiovascular disease.
Promoters and enhancers are key regulatory elements that govern gene expression. Promoters are characterized by histone H3 lysin 4 trimethylation (H3K4me3) and are typically located in close proximity to the transcription start site of a gene. Enhancers, on the other hand, are typically characterized by H3K4me1 and can be located at variable distances from the gene they regulate. The three-dimensional (3D) structure of the chromatin allows enhancers to interact with promoter regions, which may be located megabases away from them (11), and one gene is often contacted by multiple regulatory regions (12). The interplay between enhancers and promoters is crucial for the fine-tuned regulation of gene expression in response to various cellular and environmental cues. A DNA methylation–guided annotation of regulatory elements predicted that approximately 40% of them are specific for one of the major cardiac cell types within the heart (13). Active regulatory elements can be identified by the histone mark H3K27ac (14, 15). Regulatory regions typically contain multiple binding sites for transcription factors that can cooperatively bind to the chromatin (12, 14). While lineage-determining transcription factors have the ability to facilitate the transition from closed to open chromatin, binding of the majority of signal-dependent transcription factors to regulatory elements requires accessible chromatin (14, 16). Similar to other cell types, endothelial cells contain a distinct repertoire of transcription factors (8, 17) that shape cell type–specific gene expression in conjunction with the regulatory landscape and enhancer-promoter interactions.
In this study, we analyzed the regulatory landscape of mouse ventricular cardiac endothelial cells by assessing chromatin accessibility, histone modifications, and 3D chromatin organization to link enhancer elements to gene expression. The functional relevance of enhancer-promoter interactions was confirmed exemplarily by CRISPRi-mediated enhancer silencing. We used the resulting database to explore mechanisms of transcriptional regulation in cardiovascular disease, comparing six different experimental models, including heart failure, hypertension, and diabetes. Enhancer elements that regulate gene expression in diseased endothelial cells were enriched with a distinct set of transcription factor binding sites, including binding sites of the mineralocorticoid receptor (MR), a known drug target in heart failure and hypertension (18). As a proof-of-concept experiment, we used a mouse model of cell-specific MR deletion to confirm MR-dependent gene expression and predict direct MR target genes in endothelial cells.
RESULTS
Isolation of cardiac endothelial cells
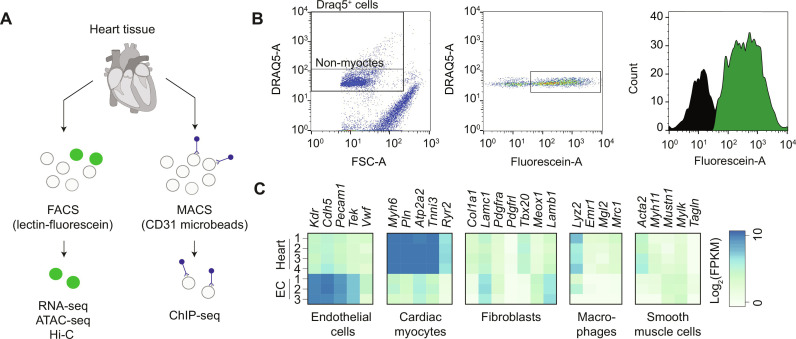
To analyze their regulatory landscape, endothelial cells were isolated from ventricular heart tissue either by fluorescence-activated cell sorting (FACS) or magnetic cell separation (Fig. 1A). Purity of the isolated endothelial cell population was ascertained (Fig. 1B) and validated by comparing mRNA expression levels of cell type–specific marker genes to heart tissue (13) (Fig. 1C). Endothelial marker genes (Kdr, Cdh5, and Pecam1) were found enriched in the isolated endothelial cell population, whereas marker gene expression indicated that other cardiac cell types (1) such as cardiomyocytes (Myh6, Pln, Atp2a2, Tnni3, and Ryr2), fibroblasts (Col1a1, Lamc1, Pdgfra, Pdgfrl, Tbx20, Meox1, and Lamb1), macrophages (Lyz2, Emr1, Mgl2, and Mrc1), or vascular smooth muscle cells (Acta2, Myh11, Mustn1, Mylk, and Tagln) were depleted (Fig. 1C). Expression of marker genes for arterial capillary, venous capillary, large venous, or lymphatic endothelial cell subtypes (4) indicated an enrichment of the capillary arterial endothelial cell subtype (fig. S1).
Fig. 1. Isolation of cardiac endothelial cells.
(A) Endothelial cells were isolated from heart tissue for subsequent transcriptomic and epigenomic analyses via FACS using fluorescein-labeled lectin or via magnetic-assisted cell sorting (MACS) using anti-CD31 magnetic microbeads. (B) Cardiac endothelial cells were separated from other cardiac cell types by DRAQ5 positivity, forward light scatter, and fluorescein intensity. (C) Heatmap showing normalized gene expression of cell type–specific marker genes in heart tissue (13) and isolated cardiac endothelial cells as assessed by RNA sequencing (RNA-seq). FSC, forward scatter; FPKM, fragments per kilobase of transcript per million fragments mapped reads; ATAC-seq, assay for transposase-accessible chromatin with sequencing; ChIP-seq, chromatic sequencing; EC, endothelial cell; FACS, fluorescence-activated cell sorting.
The cardiac endothelial cell epigenome
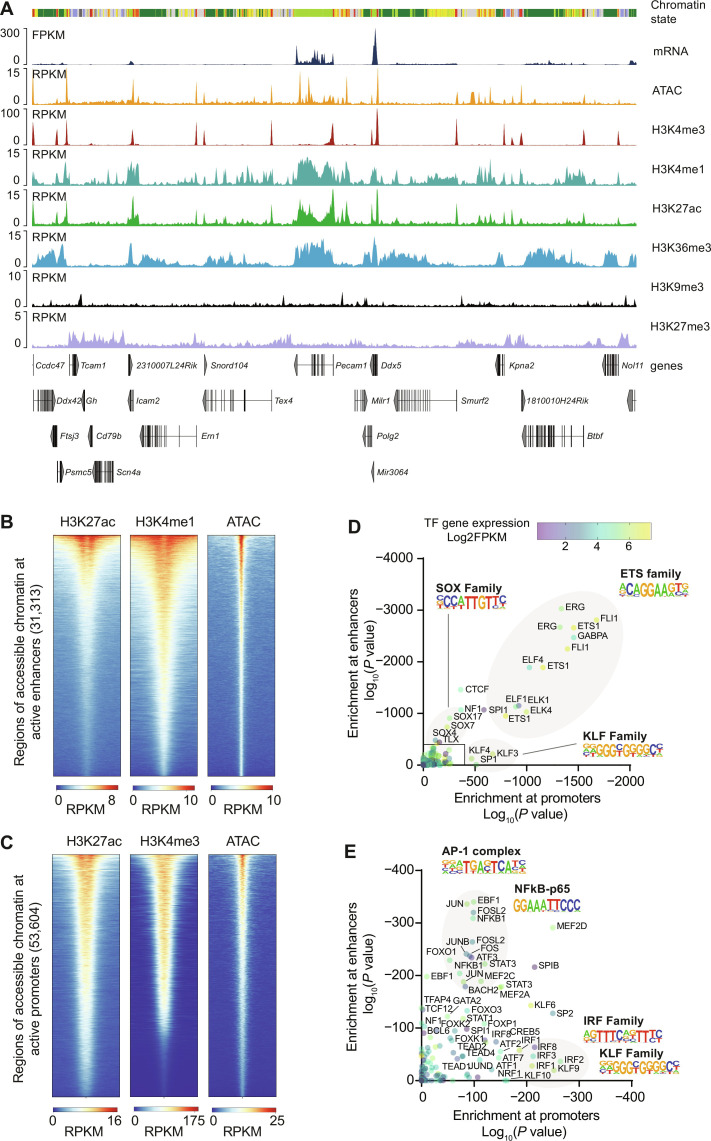
To characterize the epigenome of cardiac endothelial cells, we mapped six different histone modifications (H3K4me3, H3K4me1, H3K27ac, H3K36me3, H3K9me3, and H3K27me3) and profiled chromatin accessibility using assay for transposase-accessible chromatin with sequencing (ATAC-seq) (Fig. 2A), generating more than 900 millions reads in total (table S1). Pecam1, a prototypical endothelial cell gene, shows high expression and an enrichment of the histone modification H3K4me3 at the promoter region (Fig. 2A). As expected, the active mark H3K27ac was enriched at the promoter and active enhancer regions. Furthermore, we observed strong enrichment of H3K36me3 within the gene bodies of actively transcribed genes. In contrast, no relevant mRNA expression, chromatin accessibility, or enrichment of H3K36me3 was detectable at typical cardiomyocyte gene bodies (fig. S2).
Fig. 2. Gene expression and the epigenetic landscape in cardiac endothelial cells.
(A) Representative traces showing mRNA expression, accessible chromatin (ATAC) and histone marks in cardiac endothelial cells at the platelet endothelial cell adhesion molecule 1 (Pecam1) locus. (B) Active enhancer or (C) promoter regions were characterized by chromatin accessibility (ATAC), H3K27ac positivity, and H3K4me1 or H3K4me3 positivity, respectively. (D) Enrichment of transcription factor binding motifs in enhancer and promoter regions was analyzed using HOMER tools and correlated with the expression of the corresponding transcription factor gene. (E) Detailed representation. Further information on chromatin state analysis is provided in fig. S3. RPKM, reads per kilobase per million mapped reads; TF, transcription factor.
On the basis of the combinatorial pattern of generated chromatic sequencing (ChIP-seq) data, we were able to annotate 12 distinct chromatin states (fig. S3) (19, 20). In general, four functional classes could be detected: promoter states, enhancer states, transcribed states, and heterochromatin (fig. S3A). ATAC-seq data analysis revealed 114,741 regions of open chromatin, with the majority of them localized in active regulatory elements annotated as promoters (32%, identified by H3K4me3 and H3K27ac) or enhancers (28%, identified by H3K4me1 and H3K27ac) (fig. S3B). Thirty-four percent of all detected accessible regions were localized in heterochromatin states, reflecting the fact that accessible chromatin could also correspond to insulator proteins or inactive regulatory elements of silenced genes (15, 21).
We next asked which transcription factors could regulate endothelial cell gene expression by binding to accessible enhancer or promoter regions (Fig. 2, B and C). Within the 31,313 sites of open chromatin located at distal enhancer regions, we observed a strong enrichment of binding motifs for several transcription factors that have previously been associated with endothelial cell fate and cell type function such as the ETS (E26 transformation-specific) transcription factor family and the SOX (Sry-related high mobility group box) transcription factor family (17). In addition, we found binding motifs for the AP-1 complex or nuclear factor κB among the most enriched transcription factor binding motifs (Fig. 2, D and E). Within the 53,604 open chromatin sites located at promoter regions, we found an enrichment of Krueppel-like factor family and interferon regulatory factor family motifs (Fig. 2, D and E). Among the top-enriched motifs, only the ETS binding motif was found in both promoter and enhancer regions (Fig. 2D).
Enhancer-promoter interactions in endothelial cells
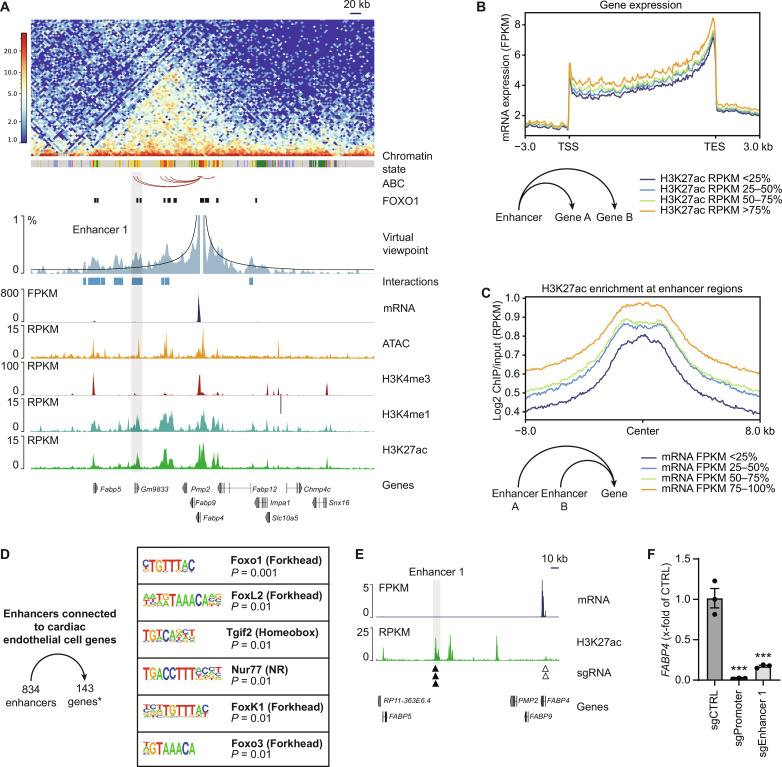
Binding of transcription factors to cis-regulatory elements such as enhancers modulates the expression of their target genes, which are not necessarily located in immediate linear proximity (22). To uncover enhancer-promoter interactions in cardiac endothelial cells, we studied the chromatin organization using in situ Hi-C. In total, we generated 380 million intrachromosomal Hi-C contacts from more than 1.2 billion paired-end sequencing reads from three biological replicates (table S1). A representative heatmap of the resulting interaction data is shown for the Fabp4 (fatty acid binding protein 4) locus (Fig. 3A). To integrate chromatin interaction data for enhancer and promoter regions with their activity, we used the activity-by-contact (ABC) model (23) and predicted in total 75,958 enhancer-promoter interactions. To confirm the validity of this analysis, we representatively generated a virtual 4C viewpoint analysis for the Fabp4 promoter (Fig. 3A). This analysis revealed a highly concordant enhancer-promoter interaction. To further test the impact of enhancer activity, we ranked enhancers or genes according to the strength of their H3K27ac signal or expression level, respectively. As anticipated, genes linked to enhancers with high H3K27ac signals, showed high gene expression (Fig. 3B). Vice versa, enhancers linked to highly expressed genes, showed strong H3K27ac signals (Fig. 3C). These findings confirmed a correlation between H3K27ac enrichment at enhancer sites with gene activity in cardiac endothelial cells.
Fig. 3. Enhancer-promoter interactions in cardiac endothelial cells.
Heatmap of chromatin interactions and representative traces of gene expression, chromatin accessibility, and active histone marks. For Fabp4, a virtual viewpoint analysis and enhancer-promoter interactions annotated by the ABC are shown. (A) Accessible regions of chromatin harboring the FOXO1 binding motif are indicated with one enhancer region (enhancer 1) connected to the Fabp4 promoter highlighted. (B) mRNA expression levels (FPKM) of genes ranked by the activity (H3K27ac level) of enhancers that are connected to those genes. (C) Activity (H3K27ac level) of enhancers ranked by the mRNA expression levels (FPKM) of genes that are connected to those enhancers. Enhancer regions (834) were connected to 143 genes previously defined as specific for cardiac endothelial cells (8). (D) Enriched transcription factor binding motifs within the enhancer regions represent potential regulators of gene expression in cardiac endothelial cell. (E) Representative traces of gene expression and H3K27ac levels at the FABP4 locus in HUVEC as derived from ENCODE datasets ENCFF118SVL and ENCFF656TFQ with target sites for sgRNAs within enhancer 1 (black triangles) and the FABP4 promoter (open triangles). FABP4 mRNA expression in HUVECs after CRISPRi-mediated silencing of enhancer 1 compared to silencing of the FABP4 promoter or nontargeting sgRNA control (sgCTRL) [n = 3 per group, ***P < 0.001 versus sgCTRL, one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparisons test] (F).
Organ specific regulation of gene expression in endothelial cells
Endothelial cells from the heart show distinct gene expression profiles when compared to other organs (3, 8). In a previous study, we had identified a set of 143 genes that are specifically expressed in endothelial cells from heart but not from brain, lung, or kidney (8). These genes show high expression compared to other genes expressed in cardiac endothelial cells [106 ± 29 versus 20.0 ± 0.7 fragments per kilobase of transcript per million fragments mapped reads (FPKM)] and could be connected to 834 active enhancer elements (Fig. 3D). A search for transcription factor binding motifs within the associated enhancer regions revealed a strong enrichment of binding sequences of FOX transcription factor family members such as FOXO1, FOXL2, FOXK1, and FOXO3 (Fig. 3D). In total, 470 enhancer elements contained a FOXO1 binding motif, including enhancer regions connected to the Cd36 (cluster of differentiation 36) or Fabp4 promoter regions, two genes, that are involved in cardiac fatty acid uptake and metabolism and highly expressed in cardiac endothelial cells compared to other organs (8, 24, 25). To validate the functional relevance of enhancer-promoter interactions, we performed CRISPRi-mediated silencing of an enhancer that is linked to the Fabp4 promoter and conserved in mouse and human (Fig. 3E). Enhancer silencing reduced FABP4 mRNA expression by 82.7 ± 1.2% compared to nontargeting single guide RNA (sgRNA) (CTRL). Silencing of the FABP4 gene promoter served as positive control (−97.8 ± 0.4% versus CTRL) (Fig. 3F).
Endothelial cell gene expression in disease
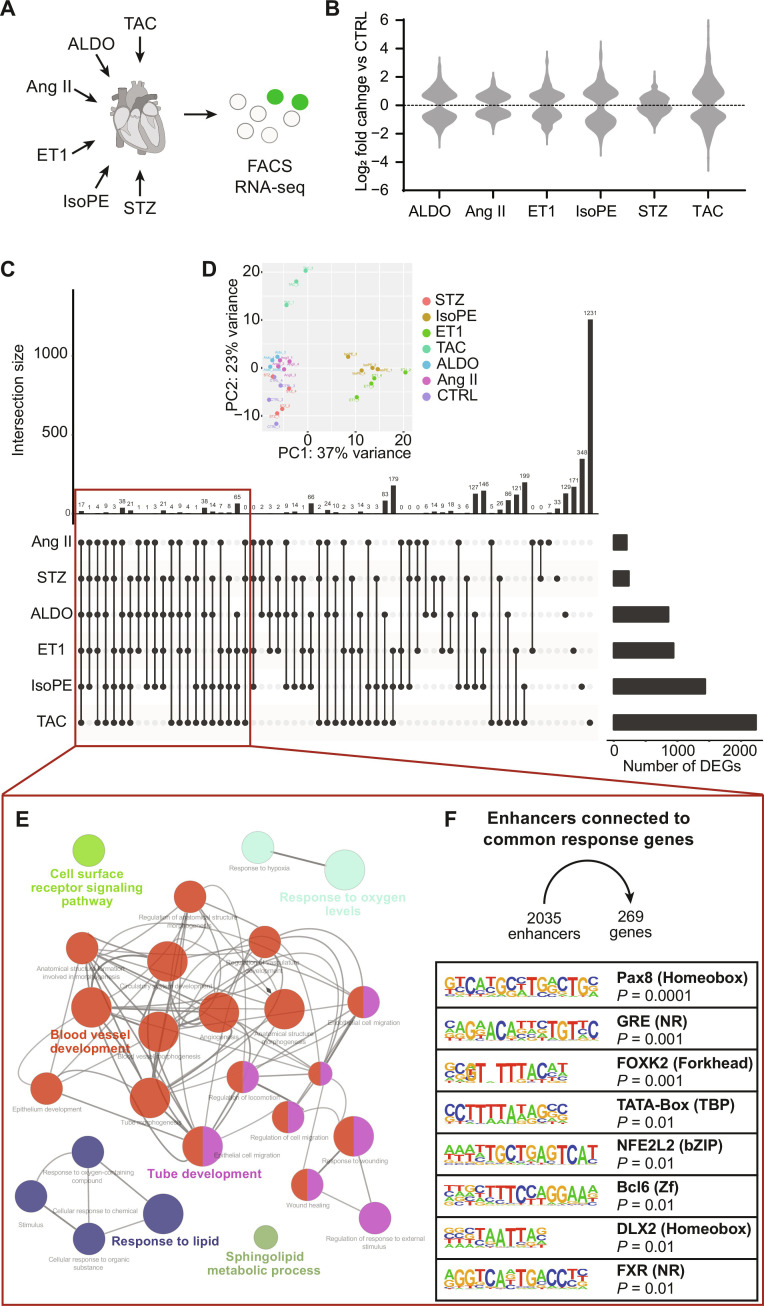
To assess the impact of cardiovascular risk factors or diseases on endothelial cell gene expression, we applied six different animal models, isolated cardiac endothelial cells, and analyzed their transcriptomes (Fig. 4A). The highest number of differentially expressed genes (DEGs; q < 0.05) was observed after transverse aortic constriction (TAC; 2266 DEGs), followed by infusion of isoprenaline and phenylephrine (IsoPE; 1437 DEGs), infusion of endothelin 1 (ET1; 949 DEGs), infusion of aldosterone (ALDO; 863 DEGs), streptozotocin-induced hyperglycemia (STZ; 238 DEGs), and angiotensin II (Ang II) infusion (202 DEGs) (Fig. 4B). Of 3347 genes that were differentially expressed in at least one condition, 1119 genes were not significantly changed in any other condition (Fig. 4C). The heterogeneity in the transcriptional response to different stimuli was also reflected by a principal component analysis that revealed four distinct clusters: cluster 1 including CTRL and STZ samples, cluster 2 including ALDO and Ang II, cluster 3 including IsoPE and ET1, and cluster 4 including only TAC (Fig. 4D). We provide an overview of DEGs per condition, related biological processes from Gene Ontology, and transcription factor binding motifs in linked enhancer regions for each experimental model (fig. S4, table S2, and data S1).
Fig. 4. Gene expression in cardiac endothelial cells in response to stress.
(A) Wild-type mice were subjected to TAC, ALDO infusion, Ang II infusion, ET1 infusion, IsoPE infusion, or streptozotocin (STZ) treatment, and cardiac endothelial cells were isolated by FACS. (B) Violin plots indicating differences in gene expression in endothelial cells from treated hearts compared to untreated CTRL (q < 0.05, n = 3 to 4 per group). Numbers of DEGs in one or more treatment conditions. (C) Genes that are differentially expressed in ≥4 conditions [defined here as common response genes] are highlighted. (D) Principal components analysis of all samples. (E) Enrichment of biological processes among common response genes as derived from gene ontology. (F) Enrichment of transcription factor binding motifs in enhancer regions connected to common response genes.
A proportion of 269 genes showed overlapping regulation in at least four of six treatment conditions and was considered as a common gene response to stress in cardiac endothelial cells (Fig. 4C). Gene Ontology analysis revealed that predominantly genes related to blood vessel development were represented in that common gene set (Fig. 4E). Within the 2035 enhancer regions linked to these genes, we found binding motifs for a set of distinct transcription factors enriched, including forkhead box transcription factor FOXK2 and nuclear factor erythroid 2–related factor 2 (NFE2L2 or NRF-2). Among the top-enriched transcription factor binding motifs was the shared mineralocorticoid and glucocorticoid response element (GRE) (Fig. 4F).
Direct and indirect effects on endothelial cell gene expression
We observed robust expression of the MR (Nr3c2), adrenergic receptor α2B (Adra2b), and endothelin receptor B (Ednrb) in cardiac endothelial cells. In contrast, other receptors from the renin-angiotensin-aldosterone system or adrenergic system were not detectable (fig. S5A). Ang II–induced changes in gene expression showed a strong linear correlation with those induced by aldosterone (fig. S5B). Aldosterone is the main downstream mediator of Ang II in the renin-angiotensin-aldosterone signaling cascade (26) and may be the main effector hormone acting on endothelial cells in absence of Ang II receptors (Agtr1a and Agtr1b).
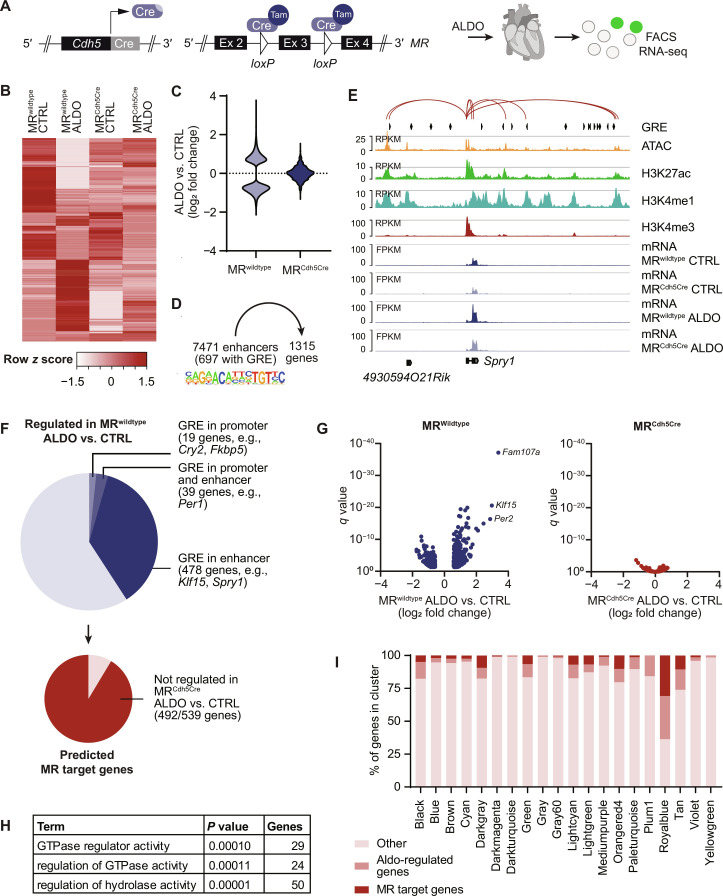
Predicting MR target genes in cardiac endothelial cells
Aldosterone (and cortisol) binds to the MR, a ligand-activated nuclear transcription factor and well-described driver of cardiovascular disease (18). To validate whether aldosterone-induced gene expression was mediated via MR in endothelial cells, mice with cell type–specific deletion of the MR (MRCdh5Cre) and respective controls (MRwildtype) (Fig. 5A) were treated with aldosterone. RNA sequencing (RNA-seq) from isolated endothelial cells revealed 1315 or 214 DEGs upon aldosterone stimulation in MRwildtype or MRCdh5Cre mice, respectively (q < 0.05, ±1.5-fold change; Fig. 5, B and C; table S3; and fig. S6). Hi-C analysis revealed 7471 enhancers linked to genes regulated after aldosterone treatment in MRwildtype mice, 697 of them carrying a GRE (Fig. 5, D and E). In total, we identified 539 genes with a GRE in their promoter (19 genes), connected enhancer (478 genes), or both (39 genes) (Fig. 5F). Four hundred ninety-two of these 539 genes (91.2%) were not differentially expressed upon aldosterone treatment in MR-deficient mice and were thus predicted as direct MR target genes (Fig. 5, F and G). Those predicted direct MR target genes were predominantly associated with guanosine triphosphatase (GTPase) or hydrolase activity functions (Fig. 5H). Using weighted gene network coexpression analysis as another, unbiased approach, we identified a cluster of 286 genes (cluster “royalblue”) that shows a high module-trait correlation (0.87, P < 0.00001) (fig. S7) with aldosterone treatment in MRwildtype and a high proportion of aldosterone-regulated or direct MR target genes (Fig. 5I). GTPase activity and cellular response to steroid hormone stimulus were among the most enriched biological processes among the genes from that cluster (fig. 7D).
Fig. 5. Identification of MR target genes in cardiac endothelial cells.
Deletion of the MR in endothelial cells in mice was achieved using the Cre/loxP system under control of the Cadherin 5 (Cdh5) promoter. (A) Mice with endothelial cell–specific MR deletion (MRCdh5Cre) and Cre-negative controls (MRwildtype) were treated with ALDO from osmotic minipumps, and endothelial cells were isolated for RNA-seq. (B) Heatmap and (C) violin plot indicating 1315 genes that were differentially expressed upon ALDO treatment in MRwildtype mice (q < 0.05, >1.5-fold versus untreated CTRL, n = 4 to 5 per group). (D) Enhancers connecting to ALDO-regulated genes and the proportion of enhancers harboring the shared mineralocorticoid, and GRE were derived from Hi-C data. (E) Representative traces of enhancer connections, mRNA expression, chromatin accessibility (ATAC), and histone marks at the Sprouty 1 (Spry1) locus with genic and intergenic enhancers carrying an GRE. (F) The proportion of aldosterone-regulated genes carrying a GRE in a linked enhancer, promoter, or both that were not regulated in MR-deficient mice was considered as direct MR target genes. (G) Volcano plots indicating aldosterone-induced changes in expression of predicted direct MR target genes in MRwildtype (blue) and MRCdh5Cre mice (red). (H) Gene Ontology analysis revealed enriched molecular functions among predicted MR target genes. (I) Weighted gene coexpression network analysis (WGCNA) revealed clusters of MR target genes and associated other ALDO-regulated genes. GTPase, guanosine triphosphatase; FACS, fluorescence-activated cell sorting.
DISCUSSION
In this study, we analyzed the regulatory landscape of primary isolated cardiac endothelial cells. The interplay between enhancers and promoters with transcription factors binding to these regulatory regions is crucial for the fine-tuned regulation of gene expression in response to various cellular and environmental cues (12, 14, 16). We provide a combined analysis of the histone code, chromatin accessibility, and 3D organization and exemplarily confirm the functional relevance of enhancer-promoter interactions by CRISPRi-mediated enhancer silencing.
ETS, GATA, SOX, and FOX families of transcription factors—among others—have well-defined roles in endothelial cell development, fate, and subspecification, influencing diverse cellular processes such as differentiation, proliferation, migration, and angiogenesis (17, 27). Their binding motifs can be found at the transcription start site (8, 28) and in regulatory elements annotated based on chromatin accessibility (29) or DNA methylation (13). Our data indicate that endothelial cells have a unique set of transcription factor binding sites at active enhancers that is distinct from those found at promoter sites. So far, available data on endothelial cell enhancers mostly derived from cultured cells (30–34), with many of these studies using human umbilical vein endothelial cells as an established work horse model to study endothelial cell biology, while we use primary cardiac endothelial cells. It needs to be acknowledged that our dataset is not suitable to study differences between endothelial cell subtypes within the heart. The cardiac endothelial cell population comprises endothelial cells from different vascular beds, while in our approach, predominantly capillary arterial endothelial cells were analyzed. This is in line with data from single-cell gene expression and ATAC-seq experiments, which indicate that the vast majority of cardiac endothelial cells belongs to the capillary subtype (2–4). However, the differences in gene expression and chromatin accessibility between endothelial cells from vascular beds within the heart were considered to be minor and far exceeded by those observed between endothelial cells from different organs (3, 4). Therefore, this dataset expands our understanding of gene regulation in cardiac endothelial cells beyond traditional views and provides a valuable resource for further analysis.
We applied the dataset provided here to uncover mechanisms of transcriptional control in cardiovascular disease. To mimic the multiple signaling pathways that may contribute to disease progression, we used six different experimental models. We identified a common response gene expression program that was associated with processes involved in blood vessel development. Motif analysis revealed that enhancers connected to this gene program contained a transcription factor subset that was distinct from other endothelial enhancers. For instance, we found an enrichment of NFE2L2 binding sites. NFE2L2 has earlier been found to mediate the response of endothelial cells to oxidative stress and shear stress (35, 36), both crucial in the pathophysiology of hypertension and atherosclerosis. FOXK2 is involved in the development and progression of cancer, e.g., by promoting tumor angiogenesis (37), but little is known about its role in cardiovascular disease (38).
Among the top-enriched transcription factor binding motifs was the MR binding motif. MR antagonists are standard of care for chronic heart failure and an emerging option to prevent cardiovascular events in patients with diabetic kidney disease (18, 39). Several preclinical studies showed that these beneficial effects are related to inhibition of MR in endothelial cells, thereby improving cardiovascular inflammation, fibrosis, and angiogenesis (40–43). Using a stepwise approach, we used our dataset to predict direct MR target genes in endothelial cells. This confirmed previous reports describing the interaction of MR with the promoter region of target genes such as FK506 binding protein 5 (Fkbp5) (44), period circadian clock 1 and 2 (Per1 and Per2) (45), or cryptochrome circadian regulator 2 (Cry2) (45). However, the use of enhancer-gene associations markedly expanded this view by revealing 478 predicted MR target genes carrying a GRE in associated enhancers but not their promoter regions. This is in line with previous studies on kidney epithelial cells, where the majority of MR-bound regions was not located at the transcription start site but at distal enhancers (46). Among the newly predicted MR target genes were multiple regulators of GTPase or hydrolase activity. While GTPases such as Rac1 (Rac Family Small GTPase 1) have previously been described as activators of MR transcriptional activity in cardiac and renal disease (47, 48), our data suggest that they may be involved in the downstream effects of MR activation as well.
In summary, we provide here a comprehensive atlas of cardiac endothelial cell enhancer elements linked to gene expression. This resource can be applied to various datasets and allows to predict regulatory mechanisms of gene expression in health and disease. Our study highlights the role of transcription factors binding to active enhancer elements, particularly MR as an established drug target in cardiovascular disease. These findings not only advance our fundamental knowledge of transcriptional control but also offer potential avenues for the development of innovative therapeutic strategies targeting cardiac endothelial cells (49, 50).
MATERIALS AND METHODS
Isolation of cardiac endothelial cells for RNA-seq, ATAC-seq, and Hi-C
For RNA-seq and ATAC-seq, cardiac endothelial cells were isolated by FACS. Briefly, ventricular heart tissue was dissected and incubated in enzyme solution containing 4 mg of collagenase B (Roche), 2.2 mg of hyaluronidase (Sigma-Aldrich), and 10 nM CaCl2 in 5 ml of Hanks’ balanced salt solution (Gibco) for 10 min. Dissociation of the remaining tissue was obtained by repetitive pipetting. The resulting suspension was filtered using a 100-μm cell strainer and centrifuged at 1000g for 5 min at 4°C. Cell pellets were resuspended in phosphate-buffered saline (PBS) containing 2 M EDTA. Cell suspensions were labeled using the far-red fluorescent dye Draq5 (Cell Signaling Technology). Endothelial cells were stained using fluorescein-conjugated Griffonia simplicifolia lectin (Vector Laboratories). Sorting was carried out using a Bio-Rad S3 Cell Sorter. Cardiac endothelial cells were identified by Draq5 positivity, forward light scatter, and fluorescein intensity.
RNA-seq library preparation
For RNA-seq analysis, cells were sorted directly into RLT Plus lysis buffer (QIAGEN). Isolation of RNA was carried out according to the manufacturer’s instruction using the AllPrep Micro Kit (QIAGEN). Subsequently, isolated RNA was used for generation of sequencing libraries with the NuGEN Ovation SoLo Kit (Tecan). Size selection was carried out using AMPure XP Beads (Beckman Coulter, 0.8× beads). Qualitative assessment of the resulting libraries was performed using the Bioanalyzer High Sensitivity Assay (Agilent).
ATAC-seq library preparation
ATAC-seq was performed as described in the Omni-ATAC protocol with minor modifications (51). A total of 50,000 sorted cardiac endothelial cells were used as an input for the transposase reaction. Reactions were cleaned up with the QIAGEN MinElute Reaction Cleanup Kit (QIAGEN). Libraries were generated and amplified using NEBNext High-Fidelity 2X PCR Master Mix (New England Biolabs). Two-sided size selection was carried out using AMPure XP Beads (Beckman Coulter, 1.8× beads).
Isolation of cardiac endothelial cells for ChIP-seq
For ChIP-seq, cardiac endothelial cells were isolated by magnetic-assisted cell sorting (MACS). Fresh heart tissue was gently dissected and enzymatically digested using a solution containing collagenase I (4 mg/ml; Merck Millipore) in Dulbecco’s modified Eagle’s medium (DMEM). Dissociation of remaining minced tissue was achieved by repetitive pipetting. After centrifugation of the obtained cell suspension, cell pellets resuspended with 270 μl of PBS containing 2 M EDTA and 1% bovine serum albumin. Resulting suspensions were incubated with 30 μl of CD31 microbeads (Miltenyi Biotec). CD31-positive cells were isolated by positive selection according to the manufacturer’s protocols using LS columns on a midiMACS separator.
ChIP-seq
Chromatin was isolated from cardiac endothelial cells using the NEXSON protocol (52). Briefly, cells were fixed using 1% formaldehyde in DMEM for 5 min at room temperature. Fixation was quenched by adding glyercin for a final concentration of 0.125 M and incubation for 5 min. Sonication of chromatin was achieved using a bioruptor device (Diagenode) for 3× 10 cycles (30-s on, 30-s off, high-energy settings). ChIP was performed according to the manufacturer’s protocol using the High sensitivity ChIP-IT Kit (Active motif). ChIP experiments were carried out using 300 to 500 ng of isolated chromatin with the following antibodies: H3K4me3 (4 μg; Diagenode, C15410003), H3K27ac (4 μg; Abcam, ab4729), H3K4me1 (4 μg; Abcam, ab8895), H3K36me3 (4 μg; Abcam, ab9050), H3K27me3 (4 μg; Merck, 07-449), and H3K9me3 (4 μg; Merck Millipore, 07-449).
Hi-C
A total of 400,000 to 500,000 endothelial cells per biological replicate were sorted into PBS and centrifuged (1000g, 5 min), and the pellet was snap-frozen. Cell pellets were used for in situ Hi-C using the Arima-HiC Kit according to the user guide for mammalian cells. Fragmentation of DNA to obtain an average fragment size of 400 base pair was performed with the Bioruptor (Diagenode; 30 cycles, 30-s on, 90-s off, “low”). The final Hi-C library was amplified using the KAPA-SYBR Fast DNA amplification reagents (Kapa Biosystems) supplemented with EvaGreen (1×) in technical replicates to monitor library amplification. The final sequencing libraries were constructed using the Swift Bioscience protocol provided by Arima. Technical replicates were pooled and cleaned twice using Ampure Beads XP (Beckman Coulter, 0.9× beads).
Bioinformatics
Libraries were sequenced on Illumina sequencers in paired-end mode. Raw data can be accessed via BioProject IDs PRJNA945592, PRJNA972993, and PRJNA989417. Bioinformatics analyses were performed using the Galaxy platform (53). Quality control was performed with FastQC. Trimming of low read quality reads and adapter contamination was achieved using Trim Galore. RNA-seq data were aligned to the Mus musculus genome (mm9) using RNA-STAR. Subsequently, htseq-count was used to obtain read count data. Differential gene expression analysis was performed with DESeq2 (q < 0.05). Transcript abundance was estimated as FPKM. Data visualization at specific loci was performed using the Integrative Genomics Viewer or pyGenomeTracks (54). Enrichment of biological processes from Gene Ontology (GO) was assessed using Cytoscape with ClueGO (55).
ChIP-seq and ATAC-seq reads were mapped to the mm9 genome assembly by Bowtie2 (56). Polymerase chain reaction (PCR) duplicated were removed using rmdup. MACS2 was used for peak calling over input (q < 0.05). Visualization of sequencing datasets was carried out mainly using deeptools (57): Bigwig tracks for visualization were generated with deeptools bamCoverage. Read coverage was estimated as reads per kilobase per million mapped reads (RPKM). Calculation of log2 reads ratio of ChIP RPKM and input RPKM was achieved using deeptools bamCompare. On the basis of ChIP-seq data for H3K27ac, H3K4me1, H3K4me3, H3K36me3, H3K9me3, and H3K27me3 from isolated cardiac endothelial cells, 12 chromatin states were annotated using ChromHMM using default settings (58). To find enriched transcription factor binding motifs within specific regions relative to the background of all endothelial enhancer regions, HOMER tools (findMotifsGenome.pl) (http://homer.ucsd.edu/homer/) (59) were used.
For Hi-C data analysis, trimmed reads were mapped as single-end reads to the reference genome mm9 with Bowtie2 using the local alignment mode (--local) (56). HiCExplorer (version 3.7.2) (60) was used for further processing Hi-C data. hicBuildMatrix was used to generate Hi-C matrices with a resolution of 1 kb for the predicted restriction cutting sites (GATC and GANTC). Replicates were merged with hicSumMatrices. For visualization of Hi-C interaction heatmaps, the bin size was adjusted to 5 kb with hicMergeMatrixBins and hicCorrectMatrix was used to correct the Hi-C matrix. Viewpoint analysis was performed for the Fabp4 promoter site (±2.5 kb) A background model was created with 10,000 random 5-kb regions using chicViewpointBackgroundModel. To prevent overestimation of chromatin interaction due to sparse background signal in regions far away from the viewpoint, we adjusted the background by replacing bins more than 200 kb distal from the viewpoint with mean background values obtained for the last 10 bins of the 200-kb flanking intervals (61). The matrices used for viewpoint analysis (chicViewpoint) are 1 kb in bin size and were analyzed using a 5-kb smoothing window. Significantly interacting domains were determined using chicSignificantInteractions (P < 0.001 and 1.5-fold over background).
To predict the effect of enhancer on target gene expression, we used the ABC model (62) with ATAC-seq, H3K27ac ChIP-seq, and Hi-C data (1-kb matrix). Data were visualized using pyGenomeTracks (54).
Weighted gene coexpression network analysis
To identify cluster of related genes, a weighted gene coexpression network analysis was carried out as described before (63) with normalized counts of genes retrieved from RNA-seq data. The dataset was filtered for genes (n = 5791) with an expression level of equal or higher than 10 in at least one experimental group to reduce noise. An adjacency matrix was generated by using bicor for correlation calculations, signed hybrid as a network type, and a soft-threshold β = 16 (scale free R2 = 0.9) and transformed into a topological overlap matrix. Hierarchical clustering was used to identify modules with a minimum of 30 genes per cluster (minModuleSize = 30). Eigengenes of modules were calculated, clustered based on correlation, and merged according to a height cut of 0.25 (correlation of 0.75) to combine similar modules. By correlating module eigengenes with treatment groups (traits), gene cluster with significant associations to those traits could be identified (64).
In vivo models of cardiovascular disease
Adult wild-type mice on C57BL/6 background were subjected to different models of cardiovascular risk factors or injury. To mimic heart failure, ventricular pressure overload was induced by TAC. Mice were anesthetized with isoflurane (2.0 to 3.0 volume % in oxygen), and after minimal-invasive thoracotomy, a 7.0 silk suture was placed around 27-gauge needle to constrict the transverse aorta (65). After surgery, mice were monitored for 2 weeks. To evaluate the impact of disease-relevant circulating factors, mice received either ALDO (500 μg/kg body weight per day), Ang II (2 mg/kg body weight per day), ET1 (36 μg/kg body weight per day), or isoprenaline and phenylephrine (IsoPE 15 mg + 15 mg/kg body weight per day) (66) for 2 weeks from subcutaneously implanted osmotic minipumps (Alzet). As a model of diabetes, hyperglycemia was induced in mice receiving repetitive low-dose intraperitoneal applications of streptozotocin (60 mg/kg body weight) for six consecutive days with an 8-week follow-up period (13).
Generation of MR mutant mice
Mice with endothelial cell–specific deletion of the MR were generated as described using a tamoxifen-inducible Cre/loxP system (40, 41). Mice expressing Cre under the control of the cadherin 5 promoter (Cdh5Cre-ERT2) (67) were crossed with mice carrying a floxed MR allele (MRflox) (68). Cre-negative (MRwildtype) mice were used as a control. All mice were treated with 2 mg of tamoxigen (20 mg/ml; Sigma-Aldrich) in sunflower oil and 10% ethanol intraperitoneally per day on five consecutive days at least 2 weeks before any further experiments.
All animal procedures were approved by the responsible animal care committees (Regierungspraesidium Freiburg, Germany, G-13/27, G-15/109, G-16/48, and G-16/62), and they conformed to the Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health (2011).
Cell culture
Human umbilical vein endothelial cells (HUVECs) were purchased from PromoCell (C-12203, lot nos. 474Z010, 466Z022, and 471Z011, Heidelberg, Germany) and cultured at 37°C, 5% CO2. Endothelial growth medium, consisting of endothelial basal medium supplemented with human recombinant epidermal growth factor, EndoCGS-Heparin (PeloBiotech, Germany), 8% fetal calf serum (FCS) (F7524, Sigma-Aldrich, Germany), penicillin-streptomycin (15140122, Gibco, USA), was used. For all experiments, three different batches of HUVEC at passage 3 were used.
Human embryonic kidney–293 T (HEK293T) cells were obtained from American Type Culture Collection (Manassas, VA, USA) and cultured at 37°C, 5% CO2 in DMEM high glucose (Gibco, USA) supplemented with FCS (8%) and penicillin-streptomycin (15140122, Gibco, USA).
Construction of the lentiviral CRISPRi plasmid and sgRNA cloning
A lentiviral expression vector was generated using a plasmid gifted by C. Gersbach (pLV hU6-sgRNA-hUbC-dCas9-KRAB-T2a-GFP; Addgene, #71237), in which the human U6, ubiquitin C promoter, and KRAB domain were replaced by synthetic fragments containing the mouse U6 promoter, guide RNA (gRNA) entry site, CAG promoter, and the ZIM3 repressor domain. Cross-species conservation of enhancer regions in mouse (mm9) and human (hg38) genomes was assessed using liftOver (69) and visually confirmed in datasets ENCFF118SVL and ENCFF656TFQ retrieved from the ENCODE portal (www.encodeproject.org/) (70). Two (FABP4 promoter) or three (FABP4 enhancer) sgRNAs were designed for each target site using CRISPOR v5.01 (http://crispor.tefor.net/), ordered as oligonucleotide pairs, annealed, and cloned into the expression vector via golden gate cloning with Esp 3I. A gRNA targeting a region of the lambda phage DNA without homology to mammalian genomes was used as a negative control. The gRNA sequences are provided in table S4.
Transduction of HUVEC and gene expression analysis
For lentiviral packaging, the plasmid coding for the nontargeting gRNA or an equimolar mix of the two (FABP4 promoter) or three (FABP4 enhancer) plasmids coding for the target specific sgRNAs was transfected into HEK293T along with envelope and helper plasmids (gifts from D. Trono; Addgene #12259 and #12260). Viral supernatants were collected after 72 hours, filtered, and added to three batches of HUVEC (P3) for 48 hours before medium change. After 7 days, green fluorescent protein–positive HUVEC cells were sorted into RLT buffer (QIAGEN) using an SH800 cell sorter (Sony). RNA was isolated with the RNeasy Micro Kit (QIAGEN) according to the manufacturer’s instructions, reverse-transcribed into cDNA, and used for quantitative PCR analysis of FABP4 and RPS29 (housekeeping gene) expression with primer pairs indicated in table S5. One-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparisons test was performed for statistical analysis using GraphPad Prism v10.0.2.
Acknowledgments
We acknowledge the support of the Freiburg Galaxy Team: R. Backofen, A. Erxleben, and B. Grüning, Bioinformatics, University of Freiburg (Germany), funded by the German Research Foundation (SFB 992 and SFB 1425) and the German Federal Ministry of Education and Research BMBF grant 031 A538A de.NBI-RBC. We thank the European Molecular Biology Laboratory GeneCore (Heidelberg, Germany) and the Max Planck Institute of Immunobiology and Epigenetics Deep Sequencing Facility (Freiburg, Germany) for providing sequencing services. We acknowledge support by the Open Access Publication Fund of the University of Freiburg.
Funding: This work was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) project ID 422681845–SFB 1425 (to A.L., R.G., and L.H.), project ID 192904750–SFB 992 (to L.H.), and project ID 470188766 (to A.L.); the Else-Kröner-Fresenius-Stiftung project ID 2016_A164 (to A.L.); and the German Heart Foundation/German Foundation of Heart Research, project ID F/50/22 (to A.L.).
Author contributions: Conceptualization: L.D., L.H., R.G., and A.L. Methodology: L.D., L.P., R.B., C.C., P.L., L.W., A.M., C.B., L.H., R.G., and A.L. Resources: L.D., L.P., R.B., C.C., P.L., L.W., A.M., C.B., L.H., R.G., and A.L. Investigation: L.D., L.P., R.B., C.C., P.L., L.W., A.M., C.B., R.G., and A.L. Formal analysis: L.D., L.P., R.B., C.C., P.L., L.W., A.M., C.B., L.H., R.G., and A.L. Data curation: L.D., R.G., and A.L. Visualization: L.D., R.G., and A.L. Supervision: L.H., R.G., and A.L. Writing—original draft: L.D. and A.L. Writing—review and editing: L.D., L.P., R.B., C.C., P.L., L.W., A.M., C.B., L.H., R.G., and A.L. Funding acquisition: L.H., R.G., and A.L. Project administration: L.H., R.G., and A.L.
Competing interests: A.L. received fees for lectures and/or serving on advisory boards from Bayer and AstraZeneca, not related to this work. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw sequencing data can be accessed via BioProject ID PRJNA945592, PRJNA972993, and PRJNA989417. HOMER motif database can be assessed via http://homer.ucsd.edu/homer/.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Legends for tables S1 to S3
Tables S4 and S5
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
Data S1
REFERENCES AND NOTES
- 1.Skelly D. A., Squiers G. T., McLellan M. A., Bolisetty M. T., Robson P., Rosenthal N. A., Pinto A. R., Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 22, 600–610 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Litvinukova M., Talavera-Lopez C., Maatz H., Reichart D., Worth C. L., Lindberg E. L., Kanda M., Polanski K., Heinig M., Lee M., Nadelmann E. R., Roberts K., Tuck L., Fasouli E. S., DeLaughter D. M., McDonough B., Wakimoto H., Gorham J. M., Samari S., Mahbubani K. T., Saeb-Parsy K., Patone G., Boyle J. J., Zhang H., Viveiros A., Oudit G. Y., Bayraktar O. A., Seidman J. G., Seidman C. E., Noseda M., Hubner N., Teichmann S. A., Cells of the adult human heart. Nature 588, 466–472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalucka J., de Rooij L., Goveia J., Rohlenova K., Dumas S. J., Meta E., Conchinha N. V., Taverna F., Teuwen L. A., Veys K., Garcia-Caballero M., Khan S., Geldhof V., Sokol L., Chen R., Treps L., Borri M., de Zeeuw P., Dubois C., Karakach T. K., Falkenberg K. D., Parys M., Yin X., Vinckier S., Du Y., Fenton R. A., Schoonjans L., Dewerchin M., Eelen G., Thienpont B., Lin L., Bolund L., Li X., Luo Y., P., Single-cell transcriptome atlas of murine endothelial cells. Cell 180, 764–779.e20 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Wang L., Yang Y., Ma H., Xie Y., Xu J., Near D., Wang H., Garbutt T., Li Y., Liu J., Qian L., Single-cell dual-omics reveals the transcriptomic and epigenomic diversity of cardiac non-myocytes. Cardiovasc. Res. 118, 1548–1563 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakusan K., Flanagan M. F., Geva T., Southern J., Van Praagh R., Morphometry of human coronary capillaries during normal growth and the effect of age in left ventricular pressure-overload hypertrophy. Circulation 86, 38–46 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Potente M., Makinen T., Vascular heterogeneity and specialization in development and disease. Nat. Rev. Mol. Cell Biol. 18, 477–494 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Augustin H. G., Koh G. Y., Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science 357, (2017). [DOI] [PubMed] [Google Scholar]
- 8.Lother A., Bergemann S., Deng L., Moser M., Bode C., Hein L., Cardiac endothelial cell transcriptome. Arterioscler. Thromb. Vasc. Biol. 38, 566–574 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Luxan G., Dimmeler S., The vasculature: A therapeutic target in heart failure? Cardiovasc. Res. 118, 53–64 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lother A., Hein L., Vascular mineralocorticoid receptors. Hypertension 68, 6–10 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Mifsud B., Tavares-Cadete F., Young A. N., Sugar R., Schoenfelder S., Ferreira L., Wingett S. W., Andrews S., Grey W., Ewels P. A., Herman B., Happe S., Higgs A., LeProust E., Follows G. A., Fraser P., Luscombe N. M., Osborne C. S., Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Grosveld F., van Staalduinen J., Stadhouders R., Transcriptional regulation by (Super)enhancers: From discovery to mechanisms. Annu. Rev. Genomics Hum. Genet. 22, 127–146 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Lother A., Bondareva O., Saadatmand A. R., Pollmeier L., Hardtner C., Hilgendorf I., Weichenhan D., Eckstein V., Plass C., Bode C., Backs J., Hein L., Gilsbach R., Diabetes changes gene expression but not DNA methylation in cardiac cells. J. Mol. Cell. Cardiol. 151, 74–87 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Isbel L., Grand R. S., Schubeler D., Generating specificity in genome regulation through transcription factor sensitivity to chromatin. Nat. Rev. Genet. 23, 728–740 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Shlyueva D., Stampfel G., Stark A., Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15, 272–286 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Heinz S., Romanoski C. E., Benner C., Glass C. K., The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park C., Kim T. M., Malik A. B., Transcriptional regulation of endothelial cell and vascular development. Circ. Res. 112, 1380–1400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauersachs J., Lother A., Mineralocorticoid receptor activation and antagonism in cardiovascular disease: Cellular and molecular mechanisms. Kidney Int. Suppl. 12, 19–26 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst J., Kellis M., Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 12, 2478–2492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorkin D. U., Barozzi I., Zhao Y., Zhang Y., Huang H., Lee A. Y., Li B., Chiou J., Wildberg A., Ding B., Zhang B., Wang M., Strattan J. S., Davidson J. M., Qiu Y., Afzal V., Akiyama J. A., Plajzer-Frick I., Novak C. S., Kato M., Garvin T. H., Pham Q. T., Harrington A. N., Mannion B. J., Lee E. A., Fukuda-Yuzawa Y., He Y., Preissl S., Chee S., Han J. Y., Williams B. A., Trout D., Amrhein H., Yang H., Cherry J. M., Wang W., Gaulton K., Ecker J. R., Shen Y., Dickel D. E., Visel A., Pennacchio L. A., Ren B., An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasquali L., Gaulton K. J., Rodriguez-Segui S. A., Mularoni L., Miguel-Escalada I., Akerman I., Tena J. J., Moran I., Gomez-Marin C., van de Bunt M., Ponsa-Cobas J., Castro N., Nammo T., Cebola I., Garcia-Hurtado J., Maestro M. A., Pattou F., Piemonti L., Berney T., Gloyn A. L., Ravassard P., Skarmeta J. L. G., Muller F., McCarthy M. I., Ferrer J., Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 46, 136–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tolhuis B., Palstra R. J., Splinter E., Grosveld F., de Laat W., Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10, 1453–1465 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Fulco C. P., Nasser J., Jones T. R., Munson G., Bergman D. T., Subramanian V., Grossman S. R., Anyoha R., Doughty B. R., Patwardhan T. A., Nguyen T. H., Kane M., Perez E. M., Durand N. C., Lareau C. A., Stamenova E. K., Aiden E. L., Lander E. S., Engreitz J. M., Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng W., Chen L., Nguyen P. K., Wu S. M., Li G., Single cell analysis of endothelial cells identified organ-specific molecular signatures and heart-specific cell populations and molecular features. Front Cardiovasc. Med. 6, 165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coppiello G., Collantes M., Sirerol-Piquer M. S., Vandenwijngaert S., Schoors S., Swinnen M., Vandersmissen I., Herijgers P., Topal B., van Loon J., Goffin J., Prosper F., Carmeliet P., Garcia-Verdugo J. M., Janssens S., Penuelas I., Aranguren X. L., Luttun A., Meox2/Tcf15 heterodimers program the heart capillary endothelium for cardiac fatty acid uptake. Circulation 131, 815–826 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Lother A., Moser M., Bode C., Feldman R. D., Hein L., Mineralocorticoids in the heart and vasculature: new insights for old hormones. Annu. Rev. Pharmacol. Toxicol. 55, 289–312 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Cui Y., Zheng Y., Liu X., Yan L., Fan X., Yong J., Hu Y., Dong J., Li Q., Wu X., Gao S., Li J., Wen L., Qiao J., Tang F., Single-cell transcriptome analysis maps the developmental track of the human heart. Cell reports 26, 1934–1950.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Froese N., Cordero J., Abouissa A., Trogisch F. A., Grein S., Szaroszyk M., Wang Y., Gigina A., Korf-Klingebiel M., Bosnjak B., Davenport C. F., Wiehlmann L., Geffers R., Riechert E., Jurgensen L., Boileau E., Lin Y., Dieterich C., Forster R., Bauersachs J., Ola R., Dobreva G., Volkers M., Heineke J., Analysis of myocardial cellular gene expression during pressure overload reveals matrix based functional intercellular communication., iScience 25, 103965 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hocker J. D., Poirion O. B., Zhu F., Buchanan J., Zhang K., Chiou J., Wang T. M., Zhang Q., Hou X., Li Y. E., Zhang Y., Farah E. N., Wang A., McCulloch A. D., Gaulton K. J., Ren B., Chi N. C., Preissl S., Cardiac cell type-specific gene regulatory programs and disease risk association. Sci. Adv. 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sissaoui S., Yu J., Yan A., Li R., Yukselen O., Kucukural A., Zhu L. J., Lawson N. D., Genomic characterization of endothelial enhancers reveals a multifunctional role for NR2F2 in regulation of arteriovenous gene expression. Circ. Res. 126, 875–888 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalna V., Yang Y., Peghaire C. R., Frudd K., Hannah R., Shah A. V., Osuna Almagro L., Boyle J. J., Gottgens B., Ferrer J., Randi A. M., Birdsey G. M., The transcription factor erg regulates super-enhancers associated with an endothelial-specific gene expression program. Circ. Res. 124, 1337–1349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakato R., Wada Y., Nakaki R., Nagae G., Katou Y., Tsutsumi S., Nakajima N., Fukuhara H., Iguchi A., Kohro T., Kanki Y., Saito Y., Kobayashi M., Izumi-Taguchi A., Osato N., Tatsuno K., Kamio A., Hayashi-Takanaka Y., Wada H., Ohta S., Aikawa M., Nakajima H., Nakamura M., McGee R. C., Heppner K. W., Kawakatsu T., Genno M., Yanase H., Kume H., Senbonmatsu T., Homma Y., Nishimura S., Mitsuyama T., Aburatani H., Kimura H., Shirahige K., Comprehensive epigenome characterization reveals diverse transcriptional regulation across human vascular endothelial cells. Epigenetics Chromatin 12, 77 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mushimiyimana I., Niskanen H., Beter M., Laakkonen J. P., Kaikkonen M. U., Yla-Herttuala S., Laham-Karam N., Characterization of a functional endothelial super-enhancer that regulates ADAMTS18 and angiogenesis. Nucleic Acids Res. 49, 8078–8096 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moonen J. R., Chappell J., Shi M., Shinohara T., Li D., Mumbach M. R., Zhang F., Nair R. V., Nasser J., Mai D. H., Taylor S., Wang L., Metzger R. J., Chang H. Y., Engreitz J. M., Snyder M. P., Rabinovitch M., KLF4 recruits SWI/SNF to increase chromatin accessibility and reprogram the endothelial enhancer landscape under laminar shear stress. Nat. Commun. 13, 4941 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen B., Lu Y., Chen Y., Cheng J., The role of Nrf2 in oxidative stress-induced endothelial injuries. J. Endocrinol. 225, R83–R99 (2015). [DOI] [PubMed] [Google Scholar]
- 36.McSweeney S. R., Warabi E., Siow R. C., Nrf2 as an Endothelial mechanosensitive transcription factor: Going with the flow. Hypertension 67, 20–29 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Feng H., Jin Z., Liang J., Zhao Q., Zhan L., Yang Z., Yan J., Kuang J., Cheng X., Qiu W., FOXK2 transcriptionally activating VEGFA induces apatinib resistance in anaplastic thyroid cancer through VEGFA/VEGFR1 pathway. Oncogene 40, 6115–6129 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Yu M., Yu H., Mu N., Wang Y., Ma H., Yu L., The function of FoxK transcription factors in diseases. Front. Physiol. 13, 928625 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agarwal R., Filippatos G., Pitt B., Anker S. D., Rossing P., Joseph A., Kolkhof P., Nowack C., Gebel M., Ruilope L. M., Bakris G. L., Cardiovascular and kidney outcomes with finerenone in patients with type 2 diabetes and chronic kidney disease: The FIDELITY pooled analysis. Eur. Heart J. 43, 474–484 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lother A., Deng L., Huck M., Furst D., Kowalski J., Esser J. S., Moser M., Bode C., Hein L., Endothelial cell mineralocorticoid receptors oppose VEGF-induced gene expression and angiogenesis. J. Endocrinol. 240, 15–26 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Lother A., Fürst D., Bergemann S., Gilsbach R., Grahammer F., Huber T. B., Hilgendorf I., Bode C., Moser M., Hein L., Deoxycorticosterone Acetate/Salt-induced cardiac but not renal injury is mediated by endothelial mineralocorticoid receptors independently from blood pressure. Hypertension 67, 130–138 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Rickard A. J., Morgan J., Chrissobolis S., Miller A. A., Sobey C. G., Young M. J., Endothelial cell mineralocorticoid receptors regulate deoxycorticosterone/salt-mediated cardiac remodeling and vascular reactivity but not blood pressure. Hypertension 63, 1033–1040 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Moss M. E., Lu Q., Iyer S. L., Engelbertsen D., Marzolla V., Caprio M., Lichtman A. H., Jaffe I. Z., Endothelial mineralocorticoid receptors contribute to vascular inflammation in atherosclerosis in a sex-specific manner. Arterioscler. Thromb. Vasc. Biol. 39, 1588–1601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartmann J., Bajaj T., Klengel C., Chatzinakos C., Ebert T., Dedic N., McCullough K. M., Lardenoije R., Joels M., Meijer O. C., McCann K. E., Dudek S. M., Sarabdjitsingh R. A., Daskalakis N. P., Klengel T., Gassen N. C., Schmidt M. V., Ressler K. J., Mineralocorticoid receptors dampen glucocorticoid receptor sensitivity to stress via regulation of FKBP5. Cell Rep. 35, 109185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fletcher E. K., Kanki M., Morgan J., Ray D. W., Delbridge L., Fuller P. J., Clyne C. D., Young M. J., Cardiomyocyte transcription is controlled by combined MR and circadian clock signalling. J. Endocrinol. 241, 17–29 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Le Billan F., Khan J. A., Lamribet K., Viengchareun S., Bouligand J., Fagart J., Lombes M., Cistrome of the aldosterone-activated mineralocorticoid receptor in human renal cells. FASEB J. 29, 3977–3989 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Nagase M., Fujita T., Role of Rac1-mineralocorticoid-receptor signalling in renal and cardiac disease. Nat. Rev. Nephrol. 9, 86–98 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Ayuzawa N., Nagase M., Ueda K., Nishimoto M., Kawarazaki W., Marumo T., Aiba A., Sakurai T., Shindo T., Fujita T., Rac1-mediated activation of mineralocorticoid receptor in pressure overload-induced cardiac injury. Hypertension 67, 99–106 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Landmesser U., Poller W., Tsimikas S., Most P., Paneni F., Luscher T. F., From traditional pharmacological towards nucleic acid-based therapies for cardiovascular diseases. Eur. Heart J. 41, 3884–3899 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Clarisse D., Deng L., de Bosscher K., Lother A., Approaches towards tissue-selective pharmacology of the mineralocorticoid receptor. Br. J. Pharmacol. 179, 3235–3249 (2022). [DOI] [PubMed] [Google Scholar]
- 51.Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., Orloff L. A., Risca V. I., Kundaje A., Khavari P. A., Montine T. J., Greenleaf W. J., Chang H. Y., An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arrigoni L., Richter A. S., Betancourt E., Bruder K., Diehl S., Manke T., Bonisch U., Standardizing chromatin research: A simple and universal method for ChIP-seq. Nucleic Acids Res. 44, e67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Community G., The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 50, W345–W351 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez-Delisle L., Rabbani L., Wolff J., Bhardwaj V., Backofen R., Gruning B., Ramirez F., Manke T., pyGenomeTracks: Reproducible plots for multivariate genomic datasets. Bioinformatics 37, 422–423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W. H., Pages F., Trajanoski Z., Galon J., ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramirez F., Dundar F., Diehl S., Gruning B. A., Manke T., deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ernst J., Kheradpour P., Mikkelsen T. S., Shoresh N., Ward L. D., Epstein C. B., Zhang X., Wang L., Issner R., Coyne M., Ku M., Durham T., Kellis M., Bernstein B. E., Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolff J., Rabbani L., Gilsbach R., Richard G., Manke T., Backofen R., Gruning B. A., Galaxy HiCExplorer 3: A web server for reproducible Hi-C, capture Hi-C and single-cell Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 48, W177–W184 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andrey G., Schopflin R., Jerkovic I., Heinrich V., Ibrahim D. M., Paliou C., Hochradel M., Timmermann B., Haas S., Vingron M., Mundlos S., Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding. Genome Res. 27, 223–233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nasser J., Bergman D. T., Fulco C. P., Guckelberger P., Doughty B. R., Patwardhan T. A., Jones T. R., Nguyen T. H., Ulirsch J. C., Lekschas F., Mualim K., Natri H. M., Weeks E. M., Munson G., Kane M., Kang H. Y., Cui A., Ray J. P., Eisenhaure T. M., Collins R. L., Dey K., Pfister H., Price A. L., Epstein C. B., Kundaje A., Xavier R. J., Daly M. J., Huang H., Finucane H. K., Hacohen N., Lander E. S., Engreitz J. M., Genome-wide enhancer maps link risk variants to disease genes. Nature 593, 238–243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Storey J. D., Tibshirani R., Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beetz N., Rommel C., Schnick T., Neumann E., Lother A., Monroy-Ordonez E. B., Zeeb M., Preissl S., Gilsbach R., Melchior-Becker A., Rylski B., Stoll M., Schaefer L., Beyersdorf F., Stiller B., Hein L., Ablation of biglycan attenuates cardiac hypertrophy and fibrosis after left ventricular pressure overload. J. Mol. Cell. Cardiol. 101, 145–155 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Dewenter M., Pan J., Knodler L., Tzschockel N., Henrich J., Cordero J., Dobreva G., Lutz S., Backs J., Wieland T., Vettel C., Chronic isoprenaline/phenylephrine vs. exclusive isoprenaline stimulation in mice: critical contribution of alpha1-adrenoceptors to early cardiac stress responses. Basic Res. Cardiol. 117, 15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y., Nakayama M., Pitulescu M. E., Schmidt T. S., Bochenek M. L., Sakakibara A., Adams S., Davy A., Deutsch U., Luthi U., Barberis A., Benjamin L. E., Makinen T., Nobes C. D., Adams R. H., Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 465, 483–486 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Berger S., Wolfer D. P., Selbach O., Alter H., Erdmann G., Reichardt H. M., Chepkova A. N., Welzl H., Haas H. L., Lipp H. P., Schutz G., Loss of the limbic mineralocorticoid receptor impairs behavioral plasticity. Proc. Natl. Acad. Sci. U.S.A. 103, 195–200 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hinrichs A. S., Karolchik D., Baertsch R., Barber G. P., Bejerano G., Clawson H., Diekhans M., Furey T. S., Harte R. A., Hsu F., Hillman-Jackson J., Kuhn R. M., Pedersen J. S., Pohl A., Raney B. J., Rosenbloom K. R., Siepel A., Smith K. E., Sugnet C. W., Sultan-Qurraie A., Thomas D. J., Trumbower H., Weber R. J., Weirauch M., Zweig A. S., Haussler D., Kent W. J., The UCSC genome browser database: Update 2006. Nucleic Acids Res. 34, D590–D598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo Y., Hitz B. C., Gabdank I., Hilton J. A., Kagda M. S., Lam B., Myers Z., Sud P., Jou J., Lin K., Baymuradov U. K., Graham K., Litton C., Miyasato S. R., Strattan J. S., Jolanki O., Lee J. W., Tanaka F. Y., Adenekan P., O'Neill E., Cherry J. M., New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 48, D882–D889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for tables S1 to S3
Tables S4 and S5
Legend for data S1
Tables S1 to S3
Data S1