Abstract
We show that reducing the activity of the Drosophila Runx protein Lozenge (Lz) during pupal development causes a decrease in cell death in the eye. We identified Lz-binding sites in introns of argos (aos) and klumpfuss (klu) and demonstrate that these genes are directly activated targets of Lz. Loss of either aos or klu reduces cell death, suggesting that Lz promotes apoptosis at least in part by regulating aos and klu. These results provide novel insights into the control of programmed cell death (PCD) by Lz during Drosophila eye development.
Keywords: Runx, Lozenge, cell death, Drosophila, eye, enhancers
Members of the Runx family of transcription factors are critical for multiple aspects of development, including cell differentiation, proliferation, neurogenesis, and hematopoiesis, and mutations in Runx1 in humans are frequently associated with leukemia (Coffman 2003). The Runx DNA-binding domain (the Runt Domain, RD) is highly conserved throughout the animal kingdom, and the Drosophila RD factor Runt (Run) binds similar sequences as its mammalian homologs (Pepling and Gergen 1995). Moreover, the Drosophila Runx homologs run and lozenge (lz) play analogous roles in development as their mammalian counterparts, indicating that Runx function is also conserved (Canon and Banerjee 2000). There also is evidence that Runx3 may regulate apoptosis in the murine gastric epithelium, suggesting that Runx3 might function as a tumor suppressor by regulating programmed cell death (PCD) (Li et al. 2002).
The Drosophila Runx gene lz is expressed in the eye, an accessible model tissue that is frequently used to study gene function. The fly eye is comprised of ∼800 ommatidia, each of which is a light-sensing unit composed of eight photoreceptors, four lens-secreting cone cells, 11 pigment cells, and three bristle cells (Wolff and Ready 1993). Lz controls the differentiation of a subset of ommatidial cell types, namely photoreceptors R1, R6, R7, and the cone and pigment cells (Daga et al. 1996; Crew et al. 1997). Although the role of lz in promoting cell differentiation during larval stages has been explored in detail, the expression of lz during pupal stages suggests that it may have additional functions in eye development.
To test for a novel function of lz, we used a temperature-sensitive allele of lz (lzts) to reduce Lz activity during pupal stages of eye development. Our results suggest that lz is required for PCD that normally occurs at this stage. In addition, we used an in silico approach to discover novel Lz-regulated enhancers that control the expression of two genes, aos and klu, which have been implicated in PCD in the eye. DNA-binding experiments and mutagenesis of these enhancers suggest that aos and klu are direct Lz targets in cone and pigment cells. Based on these results, we propose a model for Lz-mediated regulation of cell death in the eye.
Results and Discussion
lz is required for cell death in the pupal retina
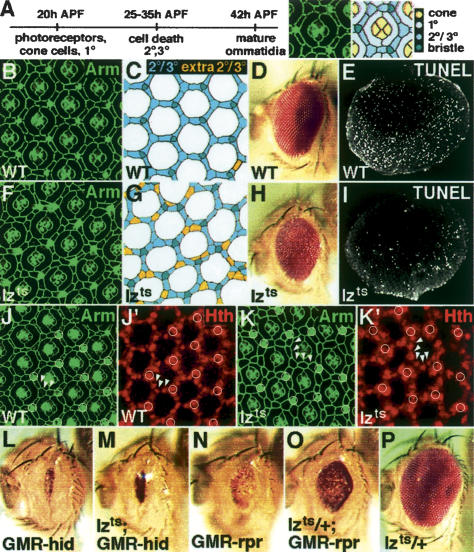
lz is expressed in the Drosophila pupal eye during PCD, suggesting that lz might have a role in this process (Flores et al. 1998). To determine if lz has any effect on apoptosis, we used a lzts allele to reduce Lz activity during PCD, which is later and distinct from the time when Lz promotes cell differentiation and survival in the larval eye disc (Fig. 1A; Daga et al. 1996; Crew et al. 1997; Siddall et al. 2003). Photoreceptor, cone, and 1° pigment cell (1° cell) differentiation is complete by ∼20 h after pupal formation (APF). Excess presumptive 2° and 3° pigment cells (2° and 3° cells, respectively) die between 25 and 35 h APF, and the ommatidia have their full complement of differentiated cells at 42 h APF. We shifted lzts flies to the nonpermissive temperature at 20 h APF and then examined retinas 42 h APF. In temperature-shifted lzts pupal retinas, there was an increased number of 2° and 3° cells, indicating a decrease in cell death (Fig. 1F,G; 2° and 3° cell number is normal in lzts flies raised at 25°C; data not shown). On average, there were an additional two cells/ommatidium (n = 53), which is similar to the number of cells that normally die (two to three cells/ommatidium) (Wolff and Ready 1991). Fifty-five-hour-APF retinas also have additional 2° and 3° cells, indicating that PCD is not delayed (data not shown). Only 2° and 3° cell death is affected; perimeter cell death, which occurs 42 h APF (Lin et al. 2004), is normal in temperature-shifted lzts animals (data not shown). dUTP nick-end labeling (TUNEL) and acridine orange staining, two methods to label dying cells, demonstrated that there is a reduction in apoptosis in temperature-shifted lzts retinas (Fig. 1I; data not shown). Thus, the extra cells in lzts retinas probably result from a decrease in cell death. Moreover, these cells appear to differentiate as 2° and 3° cells because they express homothorax (hth), a marker for these cell types (Fig. 1K; Wernet et al. 2003). In addition, the eyes of temperature-shifted lzts flies have wild-type pigmentation, indicating that the 2° and 3° cells have differentiated normally (Fig. 1H).
Figure 1.
Lz promotes pupal cell death. Anti-armadillo (Arm; green) marks the cell boundaries in 42-h-APF pupal retinas. The 2°, 3°, and bristle cells in B and F were traced and are shown to the right of the corresponding photograph (C and G, respectively). The cells in the trace are color-coded: bristle cells are dark green, 2° and 3° cells are blue, and extra 2° and 3° cells are orange; posterior is to the left. (L–P) Adult eyes of the indicated genotypes. (L,M) Male. (N–P) Female. For each genotype, the phenotype is consistent between individuals. (A, left) Timeline of pupal eye development. By 20 h APF the photoreceptors, cone, and 1° cells have differentiated. 2° and 3° cells differentiate shortly after PCD. (Right) One ommatidium from a 42-h-APF retina and corresponding trace with color-coded cells to show the relative location of each cell type. The ommatidia share three 3° cells at the apices of the ommatidial hexagon and six 2° cells between the bristle and 3° cells. (B,C) Wild-type (WT) pupal retina, in which a rare extra 2°/3° cell can be seen in the lower right corner. (D) Wild-type (WT) eye. (E) TUNEL (white) identified dying cells in a whole-mounted 32-h-APF wild-type (WT) retina. (F,G) Temperature-shifted lzts retina. (H) Temperature-shifted lzts adult eyes are only mildly rough and have wild-type pigmentation. (I) TUNEL (white) stain of a whole-mounted 32-h-APF temperature-shifted lzts pupal retina. (J,K) Separated channels of wild-type (WT) (J) and temperature-shifted lzts (K) retinas showing the expression of Hth (red) in the 2° and 3° (arrowheads) and bristle sensory organ, which is comprised of four cells (circle). Apically, Arm marks the cell boundaries, and basally, Hth marks nuclei. (J) There are no bristle cells in the upper left corner because this is a part of the rim of the eye. (L) GMR–hid. (M) Temperature-shifted lzts; GMR–hid. (N) GMR–rpr. (O) Temperature-shifted lzts/+; GMR–rpr. (P) Temperature-shifted lzts/+.
hid, grim, and rpr encode the principal pro-apoptotic factors in Drosophila (Bergmann et al. 2003). In the eye, ectopic expression of hid or rpr under control of the glass multimer reporter (GMR) reduced the overall size and pigmentation of the eye (Fig. 1L,N). GMR–hid- and GMR–rpr-induced eye phenotypes are sensitive to loss- and gain-of-function mutations in genes that affect PCD (Bergmann et al. 1998; Kurada and White 1998). We found that reducing Lz activity using the same temperature-shift protocol described above partially suppresses both the GMR–hid and GMR–rpr adult phenotypes (Fig. 1M,O). The level of suppression is similar to the effect of mutations that stimulate Epidermal Growth Factor (EGF) signaling, which antagonizes cell death by suppressing hid function (Bergmann et al. 1998; Kurada and White 1998). These results suggest that a reduction of Lz activity causes a decrease in cell death and that Lz modulates hid- and rpr-mediated cell death.
Computer-based search for Lz-regulated genes controlling apoptosis
As a transcription factor Lz most likely affects cell death by regulating the expression of gene(s) that have pro- or anti-apoptotic functions. We performed a search for Lz-binding sites in the Drosophila genome to identify genes that Lz might regulate to control cell death. Previous work has shown that Lz regulates gene expression in combination with cofactors (Flores et al. 2000; Xu et al. 2000; Canon and Banerjee 2003). That Lz works with the Ets transcription factor Pointed (Pnt) to regulate both prospero (pros) and D-Pax2 is especially striking given that Runx1 also interacts with Ets factors to regulate transcription (Flores et al. 2000; Wheeler et al. 2000; Xu et al. 2000). Moreover, Pnt is an effector of EGFR signaling, which influences cell death in the Drosophila eye (Bergmann et al. 1998; Kurada and White 1998). Based on these observations, we used the binding site search program Target Explorer to search for clusters of RD- and Ets-binding sites in the Drosophila genome (Sosinsky et al. 2003; see Materials and Methods). In brief, we created positional weight matrices representing RD- and Ets-binding sites (Supplementary Table 1) and searched for clusters of these sites near genes annotated by FlyBase to be expressed or have activity in the eye (so-called eye genes). Using these criteria, we found >500 RD/Ets clusters. Next, we used the cluster score (Sosinsky et al. 2003) to rank the putative enhancers and selected a cut-off score (see Materials and Methods; Supplementary Fig. 1). We found 135 clusters in 112 eye genes that were above this score, including clusters that correspond to the previously identified D-Pax2 and pros enhancers (Supplementary Table 2).
From this list of high-scoring clusters, we selected 16 putative enhancers located in 11 eye genes that have pro- or anti-apoptotic functions (Table 1). When available, we used antibodies and lacZ reporter lines to determine if the expression of these genes changed in temperature-shifted lzts pupal retinas. We cloned the remaining putative enhancers into a lacZ reporter gene vector and assayed for lacZ expression in vivo. We found that two enhancers located in two genes, aos and klu, drove expression of lacZ in the pupal retina, suggesting that these genes are Lz targets (Table 1).
Table 1.
High-scoring clusters located in eye genes implicated in cell death
| Putative target | Cluster score | Change in lztsa | DNA size (kb) | Reporter expression (pupal retina) |
|---|---|---|---|---|
| reaper | 3.74 | N.D.b | 1.476 | None detected |
| 3.57 | 1.088 | None detectedc | ||
| roughest | 3.73 | No | — | |
| 3.31 | — | |||
| hid | 3.71 | No | — | |
| Notch | 3.71 | No | — | |
| klu | 3.71 | Yes | 0.560 | R1 R6 R7, cone, 1°, 2°, 3° |
| 3.50 | 1.145 | None detected | ||
| Damm | 3.52 | N.D. | 0.912 | None detected |
| peanut | 3.47 | N.D.b | — | |
| 3.40 | — | |||
| sickle | 3.40 | No | — | |
| aos | 3.38 | N.D. | 1.036 | None detected |
| 3.35 | 0.923 | Cone, 2°, 3° | ||
| pointed | 3.38 | N.D. | 0.424 | None detected |
| Delta | 3.30 | No | — |
(N.D.) not determined; (—) not cloned.
Monitored by antibody stain or lac-Z reporter.
Neither rpr-11-lacZ (Nordstrom et al. 1996) nor Peanut is expressed in wild-type retinas (data not shown).
rpr1088-lacZ is expressed in a subset of glia in the CNS (data not shown).
The aos and klu enhancer expression patterns
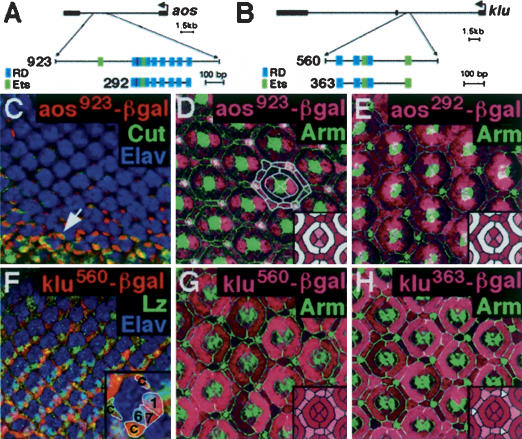
We cloned a 923-bp fragment of the first aos intron containing a cluster of 8 RD sites and two Ets sites to generate aos923–lacZ (Fig. 2A). In larval eye discs aos923–lacZ is expressed in a subset of the cell types that express an aos lacZ enhancer trap, aosw11–lacZ, indicating that this enhancer is responsible for a subset of the aos expression pattern (Fig. 2C). The expression of aosw11–lacZ in photoreceptors is unaffected in lznull larval eye discs, indicating that Lz does not regulate aos during early stages of eye development (data not shown). During pupal stages, aos923–lacZ is expressed in cone, 2°, 3°, and bristle cells (Fig. 2D). The coexpression of aos923–lacZ and lz suggests that aos may be directly activated by Lz in cone and pigment cells. However, aos923–lacZ is also expressed outside the eye, where Lz is not present (data not shown). To focus on the Lz-dependent portion of the enhancer we made a shorter construct (aos292–lacZ) that spans the first and last RD-binding sites (Fig. 2A). aos292–lacZ expression is restricted to cone and pigment cells in the eye and lz-positive cells in the antenna and leg disc (Fig. 2E; data not shown). Thus, aos292 is an aos enhancer whose activity is largely limited to lz-expressing cells.
Figure 2.
Activity of aos and klu enhancers in the eye. Eye tissue stained for anti-β-gal (red, pink) and Elav (blue). (C,F) The morphogenetic furrow is located at the top of the panel. (D,E,G,H) The cartoons at the bottom right schematize the β-gal expression patterns. (Dark pink) Strong expression; (light pink) weaker expression (D,E), or cells with variable expression (G,H); (white) no expression. (A) The aos923 enhancer. (B) The klu560 enhancer. (C) During larval stages, aos923–lacZ is expressed in Cut-positive (green) cone cells (arrow). (D,E) Forty-two-hour-APF retinas stained with anti-Arm (green). (D) aos923–lacZ is expressed in the cone, 2°, 3°, and bristle cells (the narrow gap in staining between the cone cells and the 2°/3° cells is the 1° cell). The cell boundaries of a single ommatidium are outlined. (E) aos292–lacZ is expressed in cone, 2°, 3°, and bristle cells. aos923–lacZ and aos292–lacZ are expressed in the same cell types, although at different expression levels. (F) klu560–lacZ is initially expressed in a subset of undifferentiated cells (not visible in this confocal plane). klu560–lacZ is coexpressed with lz (green) in photoreceptors R1, R6, and R7 and cone cells (a single ommatidium is enlarged to better show the expression of klu560–lacZ). (G,H) Forty-two-hour-APF retinas stained with anti-Arm (green). (G) klu560–lacZ is expressed in cone, 1°, 2°, 3°, and bristle cells. (H) klu363–lacZ is expressed in cone, 1°, 2°, and 3° cells.
A 560-bp piece of the first klu intron contains a cluster of three RD sites and two Ets sites (klu560–lacZ) (Fig. 2B). During pupal stages, klu560–lacZ is expressed in cone, 1°, 2°, and 3° cells (Fig. 2G). The overlapping expression patterns of klu560–lacZ and lz suggest that klu is also likely to be activated by Lz. A truncated klu560–lacZ construct (klu363–lacZ) that contains only the sequence flanked by the first and last RD sites is expressed in a same pattern as klu560–lacZ (Fig. 2H).
aos292–lacZ and klu363–lacZ are directly activated by Lz
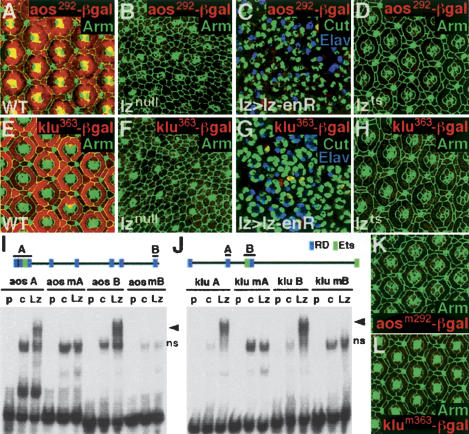
We tested the idea that Lz directly regulates aos and klu by examining the expression of aos292–lacZ and klu363–lacZ in three different genetic backgrounds. First, neither aos292–lacZ nor klu363–lacZ was expressed in lznull pupal retinas (Fig. 3B,F). Second, we expressed a protein that has the Engrailed repressor domain fused to Lz (Lz–enR), which is predicted to repress Lz target genes. Using lz-Gal4, we expressed Lz–enR in the eye and found that aos292–lacZ and klu363–lacZ expression was abolished (Fig. 3C,G). However, cell differentiation was also affected in both the lznull and lz-G4 UAS-lz-enR eyes, raising the possibility that the lack of expression could be due to a change in cell fate. Therefore, we used the lzts allele to reduce lz activity after ommatidial cells have differentiated. Neither aos292–lacZ nor klu363–lacZ was expressed in lzts retinas shifted to the nonpermissive temperature at 20 h APF (Fig. 3D,H). These results suggest that aos and klu are positive targets of lz regulation in cone and pigment cells.
Figure 3.
Lz directly activates aos292–lacZ and klu363–lacZ. Arm (green) delineates cell boundaries (A,B,D,E,F,H,K,L) and Cut (green) marks cone cell nuclei (C,G). The retinas were stained with anti-β-gal (red) and are 42 h APF unless otherwise noted. (A,E) Wild-type expression patterns of aos292–lacZ (A) and klu363–lacZ (E). (B,F) The expression of aos292–lacZ (B) and klu363–lacZ (F) is abolished in lznull pupal retinas. (C,G) In lz-Gal4 UAS-lz-enR retinas the expression of aos292–lacZ (C) and klu363–lacZ (G) is absent (Elav in blue marks the photoreceptors; retinas are <42 h APF). (D,H) Temperature-shifted lzts retinas do not express aos292–lacZ (D) and klu363–lacZ (H). (I,J) EMSAs with radiolabeled oligonucleotide probes containing wild-type or mutant RD sites from the aos292 and the klu363 enhancers. (p) Probe alone, (c) control lysate, (Lz) Lz lysate; (ns) nonspecific band. The arrowhead points to the Lz–DNA complex. Above each EMSA is a cartoon of the aos292 or klu363 enhancers with lines above the binding sites used as probes; (blue boxes) RD sites, (green boxes) Ets sites. (I) Lz forms a complex with two different aos292 probes containing RD sites 1, 2, 3 (aos A) and RD site 8 (aos B). Although aos A contains three putative RD sites, there is no evidence that multiple Lz molecules are bound because the shift is similar to aos B, which has only one RD site. Lz also binds to RD site 4 (data not shown). Lz does not bind to probes with mutant RD sites (mA, mB). (J) Lz binds klu363 probes containing RD sites 2 and 3 (klu A and klu B, respectively). Lz also binds to a probe containing RD site 1 (data not shown). Lz does not bind to probes with mutant RD sites (mA, mB). (K,L) Mutating all the RD sites in each enhancer abolished expression of both reporters: aos292mRD–lacZ (K) and klu363mRD–lacZ (L).
To test if Lz directly regulates aos292–lacZ and klu363–lacZ, we performed electrophoretic mobility shift assays (EMSAs) using oligos containing RD-binding sites found in each enhancer. We tested five of the eight RD-binding sites in aos292 and found that Lz bound to each oligo probe (Fig. 3I,J; data not shown). Lz did not, however, bind to probes that have mutated RD sites, demonstrating that Lz binding is specific (Fig. 3I). Similar experiments showed that Lz binds each of the three RD sites present in klu363 (Fig. 3J; data not shown).
To determine if the RD sites are necessary for aos292–lacZ and klu363–lacZ expression in vivo, we mutated all of the RD sites present in the aos292 and klu363 enhancers (Fig. 3K,L). These reporter genes (aos292mRD–lacZ and klu363mRD–lacZ) were inactive, demonstrating that the RD sites are necessary for the activity of these enhancers in vivo.
EGFR loss of function is epistatic to Lz loss of function
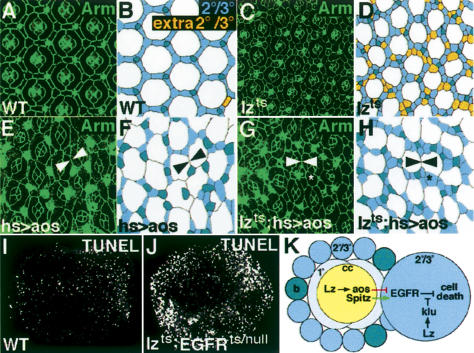
Decreasing EGFR activity results in an increase in cell death in the pupal eye (Brachmann and Cagan 2003). Both aos and klu antagonize EGFR signaling and promote cell death in the pupal retina (Freeman 1994; Sawamoto et al. 1998; Rusconi et al. 2004). If, as we propose, Lz activates these factors, a decrease in EGFR activity should be epistatic to lz loss of function. We tested this by decreasing EGFR activity in two ways. First, we used a heat shock (hs) Gal4 driver to express aos at the same time we reduced Lz activity. As previously reported, ectopic aos caused a decrease in cone, 2°, and 3° cell number (Fig. 4E,F; Freeman 1994; Sawamoto et al. 1998). Ectopic aos produced a similar phenotype in temperature-shifted lzts retinas, indicating that aos normally functions downstream of lz during PCD, albeit we cannot rule out the possibility that aos affects differentiation in these experiments (Fig. 4G,H). Although the overall number of 2° and 3° cells was reduced, a few small clusters of 2°/3° cells remained in the temperature-shifted lzts; hs-Gal4 UAS-aos retinas that were not seen in the hs-Gal4 UAS-aos retinas. These residual pigment cells could be due to a reduction of another Lz-dependent regulator of cell death (such as klu). Nevertheless, ectopic aos is able to at least partially suppress the lz loss-of-function phenotype, supporting the idea that lz promotes cell death by activating aos. In a second test, reducing EGFR signaling (using a temperature-sensitive allele of EGFR) in lzts retinas resulted in an increase in cell death (Fig. 4I,J), also suggesting that EGFR activity is epistatic to Lz.
Figure 4.
Reduction of EGFR activity is epistatic to lz loss of function. Arm (green) marks cell boundaries. To the right of each photograph is a trace of the 2°, 3°, and bristle cells (see Fig. 1). All retinas are 42 h APF. (A,B) Wild-type (WT) retina and corresponding trace (B). (C,D) Temperature-shifted lzts retina. (E,F) hs-Gal4 UAS-aos retina. Because 2° and 3°s are lost, 1°s of neighboring ommatidia contact each other (arrowheads; some cone cells are missing). (G,H) Ectopic expression of aos in temperature-shifted lzts retinas reduced 2° and 3° cell number (some ommatidia also lack cone cells). 1° cells from neighboring ommatidia touch (arrowheads), and there are also small clusters of 2°/3° cells (asterisk). (I,J) TUNEL stain (white) of wild-type (WT) (I) and lzts; EGFRts/EGFRnull (J) pupal retinas. (K) Model of Lz's control of PCD in the pupal retina.
These findings, together with what is known about aos and klu, support the following model: Lz induces aos expression in cone cells, wherefrom Aos diffuses to antagonize EGFR activity in the surrounding 2° and 3° cells (Fig. 4K). The expression pattern of aos923–lacZ indicates that Lz also regulates aos expression in 2° and 3° cells, suggesting that these cells may also send antisurvival signals. Our data further suggest that within the 2° and 3° cells, Lz activates klu, which antagonizes EGFR signaling downstream of the receptor. Lz also activates klu expression in cone and 1° cells, but it is unclear what function klu has in these cells. Although two phases of PCD during retinal development have been proposed (Cordero et al. 2004), our experiments support a role for Lz in promoting only the EGFR-dependent phase. An alternative possibility is that the decrease in cell death in lz mutant retinas is due to an increase in 2° and 3° cell differentiation stimulated by an increase in EGFR signaling. However, given the large body of evidence demonstrating that lz normally functions to promote differentiation, we do not favor a model in which lz acts to suppress differentiation.
The mammalian homolog of Lz, Runx1 (also known as AML1), is also a transcriptional regulator. In humans, translocations that affect Runx1 are associated with acute myelogenous leukemia (AML), which is characterized by the proliferation of undifferentiated hematopoietic cells. Effects on cell cycle regulators have been implicated in contributing to this overproliferation, but it is likely that PCD also plays a role (Alcalay et al. 2001; Bernardin and Friedman 2002). Changes in the amount of the apoptotic regulator Wilms Tumor 1 (WT1) are often found in AML patients (Ostergaard et al. 2004). We have shown here that lz promotes cell death in the Drosophila eye in part by activating the expression of klu, the Drosophila homolog of WT1. We suggest that our findings may be relevant to how Runx1 chimeras lead to the development of AML in humans. Furthermore, they suggest that WT1 may be a direct target of Runx1.
Materials and methods
Search for putative Lz targets
We searched for clusters of RD and Ets sites in the Drosophila genome using Target Explorer (Sosinsky et al. 2003; http://trantor.bioc.columbia.edu/Target_Explorer). Positional weight matrices representing binding sites for each transcription factor family were constructed from training sets of experimentally identified binding sites retrieved from the literature (Supplementary Table 1). We selected the lowest score in each training set as the cut-off score for that matrix. The minimal requirements for defining a putative enhancer were based on the previously identified D-Pax2 and pros enhancers. We searched the noncoding euchromatin fraction of the whole genome with each RD and Ets matrix for clusters of at least two nonoverlapping sites within 500 bp. Additional criteria were that the RD/Ets clusters should: (1) have at least one pair of RD/Ets sites separated by 10 bp or less (based on constraints that limit the cooperative interaction between AML1 and Ets) (Kim et al. 1999); (2) have a cluster score above 3.30 (for a definition of the cluster score, see Supplementary Fig. 1 and Sosinsky et al. 2003); and (3) be located within 15 kb of the transcriptional start site of an “eye” gene. Eye genes were defined as genes annotated by FlyBase as having a loss- or gain-of-function phenotype and/or a known pattern of expression in the eye (511 genes qualified as eye genes). These criteria identified 135 clusters in 112 eye genes. This list was annotated to include information about the expression pattern and mutant phenotypes described in FlyBase and the literature. We selected 16 putative enhancers in genes that were previously shown to have pro- or anti-apoptotic functions (Table 1).
Fly strains
lzr15, a null allele of lz; lzts, and lz-Gal4 were kindly provided by the Banerjee lab. We thank N. Baker (Albert Einstein College of Medicine, New York) for UAS-aos flies. kluP819–lacZ was a gift of T. Klein (Universitat zu Koln, Koln, Germany). H.D. Ryoo (The Rockefeller University, New York) provided GMR–hid and GMR–rpr flies. EGFRf24, a null allele, and EGFRts were obtained from the Bloomington stock center.
Temperature-shift protocols
Oregon-R wild-type and lzts flies were cultured at 25°C. Brown pupae just prior to head eversion that displayed a prominent bubble and were buoyant were considered to be 15 h APF. At 20 h APF, the wild-type and lzts pupae were transferred to 33°C and kept at this temperature until 42 h APF, when the pupae were dissected and fixed. Cone and 1° cell differentiation was not affected in temperature-shifted lzts retinas, and a reduction in Lz immunostaining was observed. Temperature-shifted lzts/lznull retinas also had an increased number of 2° and 3° cells. A similar increase in cell number was also observed in retinas shifted to 31°C at 15 h APF. For the TUNEL assay, temperature-shifted pupae were dissected at 32 h APF. For the aos and EGFR epistasis experiments, flies were cultured at 18°C. For the ectopic aos experiment, staged 15-h-APF pupae were kept at 18°C for 11 h (when they resembled 20-h-APF pupae raised at 25°C), transferred to 35°C, and fixed at 42 h APF. For the EGFR epistasis experiment, staged 15-h-APF pupae were kept at 18°C for 26 h, transferred to 33°C for 5 h, and then fixed.
Transgenic flies
Genomic DNA isolated from wild-type flies was used to amplify putative enhancers by the PCR. Fragments were cloned into pCaSperR-hs43. Mutant versions of aos292 and klu363 were created using oligos with mutations in the RD-binding sites. UAS-lz-enR was created by substituting the Engrailed Repressor domain (2–297 amino acids of Engrailed) in frame for 542–750 amino acids of Lz (GenBank accession no. AAC47196) and cloning into pUAST.
Immunohistochemistry and cell death assays
Dissected tissue was prepared by standard procedures. The primary antibodies used were rat anti-Elav (1/50; Hybridoma Bank), mouse anti-Arm (1/10; Hybridoma Bank), mouse anti-Cut (1/10; Hybridoma Bank), mouse anti-Lz (1/10; gift of U. Banerjee, University of California at Los Angeles, Los Angeles, CA), rabbit anti-β-gal (1/1000; Cappell), guinea pig anti-Hth (1/1000) (Ryoo and Mann 1999), mouse anti-Dl (1/1000; Hybridoma Bank), mouse anti-N C17.9C6 (1/1000; Hybridoma Bank), mouse anti-Rst (1/10; gift of K. Fischbach, Albert-Ludwigs-Universitat Freiburg, Freiburg, Germany), rabbit anti-Hid (1/1000) (Yoo et al. 2002), anti-Pnut (1/10; Hybridoma Bank), anti-Skl (1/300) (Srinivasula et al. 2002). TUNEL assays were carried out with the ApopTag Fluorescein In Situ Apoptosis Detection kit (Serologicals).
EMSAs
Lz and control lysates were produced using the TNT coupled reticulocyte system (Promega). Lz was transcribed from a full-length lz cDNA in pET3c template (kindly provided by R. Carthew, Northwestern University, Evanston, IL). 32P-end-labeled oligo probes were mixed with 5 μL of either control or Lz lysates in 10 mM Tris (pH 7.5), 50 mM NaCl, 5% glycerol, 67 μg/mL BSA, 13.4 μg/mL poly(dI/dC), and incubated for 20 min at room temperature. The entire 20-μL reaction was run on a 4% polyacrylamide gel.
Acknowledgments
We thank J. Rusconi and R. Cagan for sharing unpublished data and U. Banerjee, N. Baker, R. Carthew, T. Klein, H.D. Ryoo, K. Fischbach, B. Hay, T. Alnemri, and the Developmental Hybridoma Bank for reagents. We also thank H.D. Ryoo, T. Cook, B. Gebelein, L. Johnston, J. Culi, and N. Baker for comments on the manuscript. This work was supported by a grant from the NCI.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1298105.
References
- Alcalay M., Orleth, A., Sebastiani, C., Meani, N., Chiaradonna, F., Casciari, C., Sciurpi, M.T., Gelmetti, V., Riganelli, D., Minucci, S., et al. 2001. Common themes in the pathogenesis of acute myeloid leukemia. Oncogene 20: 5680–5694. [DOI] [PubMed] [Google Scholar]
- Bergmann A., Agapite, J., McCall, K., and Steller, H. 1998. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 95: 331–341. [DOI] [PubMed] [Google Scholar]
- Bergmann A., Yang, A.Y., and Srivastava, M. 2003. Regulators of IAP function: Coming to grips with the grim reaper. Curr. Opin. Cell Biol. 15: 717–724. [DOI] [PubMed] [Google Scholar]
- Bernardin F. and Friedman, A.D. 2002. AML1 stimulates G1 to S progression via its transactivation domain. Oncogene 21: 3247–3252. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B. and Cagan, R.L. 2003. Patterning the fly eye: The role of apoptosis. Trends Genet. 19: 91–96. [DOI] [PubMed] [Google Scholar]
- Canon J. and Banerjee, U. 2000. Runt and Lozenge function in Drosophila development. Semin. Cell Dev. Biol. 11: 327–336. [DOI] [PubMed] [Google Scholar]
- ____. 2003. In vivo analysis of a developmental circuit for direct transcriptional activation and repression in the same cell by a Runx protein. Genes & Dev. 17: 838–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman J.A. 2003. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol. Int. 27: 315–324. [DOI] [PubMed] [Google Scholar]
- Cordero J., Jassim, O., Bao, S., and Cagan, R. 2004. A role for wingless in an early pupal cell death event that contributes to patterning the Drosophila eye. Mech. Dev. 121: 1523–1530. [DOI] [PubMed] [Google Scholar]
- Crew J.R., Batterham, P., and Pollock, J.A. 1997. Developing compound eye in lozenge mutants of Drosophila: Lozenge expression in the R7 equivalence group. Dev. Genes Evol. 206: 481–493. [DOI] [PubMed] [Google Scholar]
- Daga A., Karlovich, C.A., Dumstrei, K., and Banerjee, U. 1996. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes & Dev. 10: 1194–1205. [DOI] [PubMed] [Google Scholar]
- Flores G.V., Daga, A., Kalhor, H.R., and Banerjee, U. 1998. Lozenge is expressed in pluripotent precursor cells and patterns multiple cell types in the Drosophila eye through the control of cell-specific transcription factors. Development 125: 3681–3687. [DOI] [PubMed] [Google Scholar]
- Flores G.V., Duan, H., Yan, H., Nagaraj, R., Fu, W., Zou, Y., Noll, M., and Banerjee, U. 2000. Combinatorial signaling in the specification of unique cell fates. Cell 103: 75–85. [DOI] [PubMed] [Google Scholar]
- Freeman M. 1994. Misexpression of the Drosophila argos gene, a secreted regulator of cell determination. Development 120: 2297–2304. [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Sieweke, M., Ogawa, E., Wee, H.J., Englmeier, U., Graf, T., and Ito, Y. 1999. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 18: 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurada P. and White, K. 1998. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell 95: 319–329. [DOI] [PubMed] [Google Scholar]
- Li Q.L., Ito, K., Sakakura, C., Fukamachi, H., Inoue, K., Chi, X.Z., Lee, K.Y., Nomura, S., Lee, C.W., Han, S.B., et al. 2002. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109: 113–124. [DOI] [PubMed] [Google Scholar]
- Lin H.V., Rogulja, A., and Cadigan, K.M. 2004. Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development 131: 2409–2418. [DOI] [PubMed] [Google Scholar]
- Nordstrom W., Chen, P., Steller, H., and Abrams, J.M. 1996. Activation of the reaper gene during ectopic cell killing in Drosophila. Dev. Biol. 180: 213–226. [DOI] [PubMed] [Google Scholar]
- Ostergaard M., Olesen, L.H., Hasle, H., Kjeldsen, E., and Hokland, P. 2004. WT1 gene expression: An excellent tool for monitoring minimal residual disease in 70% of acute myeloid leukaemia patients—Results from a single-centre study. Br. J. Haematol. 125: 590–600. [DOI] [PubMed] [Google Scholar]
- Pepling M.E. and Gergen, J.P. 1995. Conservation and function of the transcriptional regulatory protein Runt. Proc. Natl. Acad. Sci. 92: 9087–9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi J.C., Fink, J.L., and Cagan, R. 2004. klumpfuss regulates cell death in the Drosophila retina. Mech. Dev. 121: 537–546. [DOI] [PubMed] [Google Scholar]
- Ryoo H.D. and Mann, R.S. 1999. The control of trunk Hox specificity and activity by Extradenticle. Genes & Dev. 13: 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto K., Taguchi, A., Hirota, Y., Yamada, C., Jin, M.H., and Okano, H. 1998. Argos induces programmed cell death in the developing Drosophila eye by inhibition of the Ras pathway. Cell Death Differ. 5: 262–270. [DOI] [PubMed] [Google Scholar]
- Siddall N.A., Behan, K.J., Crew, J.R., Cheung, T.L., Fair, J.A., Batterham, P., and Pollock, J.A. 2003. Mutations in lozenge and D-Pax2 invoke ectopic patterned cell death in the developing Drosophila eye using distinct mechanisms. Dev. Genes Evol. 213: 107–119. [DOI] [PubMed] [Google Scholar]
- Sosinsky A., Bonin, C.P., Mann, R.S., and Honig, B. 2003. Target Explorer: An automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res. 31: 3589–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula S.M., Datta, P., Kobayashi, M., Wu, J.W., Fujioka, M., Hegde, R., Zhang, Z., Mukattash, R., Fernandes-Alnemri, T., Shi, Y., et al. 2002. sickle, a novel Drosophila death gene in the reaper/hid/grim region, encodes an IAP-inhibitory protein. Curr. Biol. 12: 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet M.F., Labhart, T., Baumann, F., Mazzoni, E.O., Pichaud, F., and Desplan, C. 2003. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell 115: 267–279. [DOI] [PubMed] [Google Scholar]
- Wheeler J.C., Shigesada, K., Gergen, J.P., and Ito, Y. 2000. Mechanisms of transcriptional regulation by Runt domain proteins. Semin. Cell Dev. Biol. 11: 369–375. [DOI] [PubMed] [Google Scholar]
- Wolff T. and Ready, D.F. 1991. Cell death in normal and rough eye mutants of Drosophila. Development 113: 825–839. [DOI] [PubMed] [Google Scholar]
- ____. 1993. Pattern formation in the Drosophila retina. In The development of Drosophila (eds. M. Bate and A. Martinez-Arias), pp. 1277–1326. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Xu C., Kauffmann, R.C., Zhang, J., Kladny, S., and Carthew, R.W. 2000. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell 103: 87–97. [DOI] [PubMed] [Google Scholar]
- Yoo S.J., Huh, J.R., Muro, I., Yu, H., Wang, L., Wang, S.L., Feldman, R.M., Clem, R.J., Muller, H.A., and Hay, B.A. 2002. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 4: 416–424. [DOI] [PubMed] [Google Scholar]