Abstract
The ATR-dependent DNA damage response pathway can respond to a diverse group of lesions as well as inhibitors of DNA replication. Using the Xenopus egg extract system, we show that lesions induced by UV irradiation and cis-platinum cause the functional uncoupling of MCM helicase and DNA polymerase activities, an event previously shown for aphidicolin. Inhibition of uncoupling during elongation with inhibitors of MCM7 or Cdc45, a putative helicase cofactor, results in abrogation of Chk1 phosphorylation, indicating that uncoupling is necessary for activation of the checkpoint. However, uncoupling is not sufficient for checkpoint activation, and DNA synthesis by Polα is also required. Finally, using plasmids of varying size, we demonstrate that all of the unwound DNA generated at a stalled replication fork can contribute to the level of Chk1 phosphorylation, suggesting that uncoupling amplifies checkpoint signaling at each individual replication fork. Taken together, these observations indicate that functional uncoupling of MCM helicase and DNA polymerase activities occurs in response to multiple forms of DNA damage and that there is a general mechanism for generation of the checkpoint-activating signal following DNA damage.
Keywords: ATR, DNA damage, Xenopus, checkpoint
The DNA damage response pathway is a cellular surveillance system that senses the presence of damaged DNA and elicits an appropriate and effective response to that damage. First identified as a regulator of cell cycle transitions, the DNA damage response pathway has since been shown to regulate a number of other cellular processes, which include DNA repair, apoptosis, and replication fork stabilization (Zhou and Elledge 2000; Cimprich 2003). The importance of this pathway is demonstrated by the conservation of this response from yeast to humans (Zhou and Elledge 2000; Melo and Toczyski 2002) and by several studies that have shown that loss of checkpoint proteins predisposes affected individuals to cancer (Sherr 2004).
One critical component of the DNA damage response pathway is the ATR–ATRIP complex. ATR is a phosphatidylinositol kinase-related protein kinase that is thought to function as both a sensor and transducer in the DNA damage response. ATR, and its associated protein ATRIP, respond to a broad spectrum of genotoxic agents that includes ultraviolet light (UV), topoisomerase inhibitors, alkylating agents, and cis-platinum, as well as chemicals that disrupt replication, such as aphidicolin and hydroxyurea (HU) (Zhou and Elledge 2000; Cortez et al. 2001; Melo and Toczyski 2002). Following DNA damage, ATR phosphorylates and activates the checkpoint kinase Chk1 (Melo and Toczyski 2002). In higher eukaryotes, the phosphorylation of Chk1 also requires the activities of the Rad9–Rad1–Hus1 (RHR, aka 9–1–1) complex and Claspin (Melo and Toczyski 2002). The RHR complex is a PCNA-related complex that is loaded on to primed DNA in vitro (Ellison and Stillman 2003; Zou et al. 2003) and is recruited to sites of DNA damage in vivo (Kondo et al. 2001; Melo et al. 2001). Claspin was initially identified as a protein that bound the activated form of Chk1, and it has been shown to bind chromatin throughout S phase (Kumagai and Dunphy 2000; Lee et al. 2003).
We and others previously showed that recruitment of ATR and Rad1 to chromatin (Lupardus et al. 2002) and activation of Chk1 (Lupardus et al. 2002; Stokes et al. 2002) requires DNA replication in Xenopus egg extracts following several types of DNA damage. Studies in mammalian cells also indicate that ATR binds UV-damaged chromatin in S phase but not G1 phase (Ward et al. 2004). In addition, other studies show that a replication fork must be established in Saccharomyces cerevisiae for checkpoint activation induced by methylmethane sulfonate (MMS) (Tercero et al. 2003). Taken together, these observations suggest that one or more replication-dependent events are needed to generate the signal that ATR recognizes for many types of DNA damage.
Although the exact nature of the biochemical signal(s) responsible for activating the ATR pathway and the replication-dependent steps necessary for its formation are still unclear, evidence from a number of different systems supports a central role for replication protein A (RPA)-coated single-stranded DNA (ssDNA) in the response. In yeast, certain RPA mutants exhibit a checkpoint defect and also adapt more rapidly to DNA damage (Longhese et al. 1996; Pellicioli et al. 1999). In addition, knock-down of the ssDNA-binding protein RPA results in a significant loss of both Chk1 phosphorylation and ATR foci formation following DNA damage in mammalian cells (Zou and Elledge 2003). In Xenopus egg extracts, RPA is also required for the recruitment of ATR to chromatin following treatment with aphidicolin (You et al. 2002) or etoposide (Costanzo et al. 2003) and for the recruitment of ATR to poly(dA)70 ssDNA (Lee et al. 2003). Importantly, in vitro experiments have shown that RPA is sufficient for the binding of ATRIP to ssDNA (Zou and Elledge 2003) and that RPA also facilitates the association of the RHR complex with DNA (Ellison and Stillman 2003; Zou et al. 2003).
Interestingly, the amount of ssDNA appears to increase following genotoxic stress, as RPA accumulates on chromatin in Xenopus extracts and mammalian cells treated with UV, MMS, HU, or aphidicolin (Michael et al. 2000; Mimura et al. 2000; Walter 2000; Lupardus et al. 2002; Zou and Elledge 2003). Moreover, in budding yeast, increased amounts of ssDNA have been observed by electron microscopy following HU treatment (Sogo et al. 2002). In the case of DNA damage, the mechanism by which this ssDNA accumulates is not known, nor is it clear if ssDNA accumulation is required for checkpoint activation. In principle, a number of DNA repair (e.g., nucleotide excision repair, base excision repair) and recombination processes could lead to the generation of ssDNA following DNA damage at several points in the cell cycle. Alternatively, during DNA replication, ssDNA could be formed if DNA polymerases are slowed by lesions and the replicative helicase continues to unwind DNA. Indeed, uncoupling of helicase and polymerase activities has been previously observed in the presence of aphidicolin (Walter and Newport 2000), and recent studies have shown that this aphidicolin-induced uncoupling is dependent on the MCM helicase (Pacek and Walter 2004).
In this study, we used a cell-free extract system derived from Xenopus eggs (Walter et al. 1998) to examine the mechanism by which ssDNA accumulates following DNA damage. We demonstrate that the appearance and disappearance of a highly unwound form of plasmid DNA that accumulates following aphidicolin treatment (Walter and Newport 2000) correlates with the phosphorylation of Chk1 on Ser 344 (S344). Importantly, this hyperunwound form of DNA was also observed upon replication of plasmid DNA damaged with either UV or cis-platinum. This suggests that DNA damage induces uncoupling of helicase and polymerase activities and that these lesions, as well as aphidicolin, may generate a common checkpoint-activating DNA structure. Moreover, while stalling the replication fork with aphidicolin results in a robust checkpoint response, we find that aphidicolin elicits no checkpoint when the MCM DNA helicase is inactivated. Using plasmids of varying sizes, we also show that functional uncoupling of DNA unwinding and DNA synthesis during S phase may serve to amplify the level of Chk1 phosphorylation that can be achieved at each individual replication fork. Finally, we demonstrate that although DNA unwinding is necessary for checkpoint activation, it is not sufficient and that additional DNA synthesis by Polα is needed. Taken together, these results suggest that functional uncoupling of helicase and polymerase activities is necessary to convert DNA lesions and chemical inhibitors of DNA replication into the signal(s) that activate the ATR-dependent checkpoint.
Results
ATR, Rad1, and Claspin-mediated checkpoint activation with plasmid DNA
We and others have previously shown that addition of aphidicolin, MMS, and UV-treated chromatin to Xenopus egg extracts leads to replication-dependent accumulation of RPA on chromatin (Michael et al. 2000; Mimura et al. 2000; Walter 2000; Lupardus et al. 2002). This observation indicates that ssDNA accumulates following these forms of DNA damage and that some replication-dependent event is necessary for that accumulation. Using the Xenopus egg extract system, we have also shown that aphidicolin treatment can lead to the formation of a highly unwound form of plasmid DNA, suggesting that the activity of the helicase can become uncoupled from the DNA polymerase (Walter and Newport 2000). We wanted to determine if the topological changes that occur in plasmid DNA following aphidicolin treatment are coupled to checkpoint activation. Thus, we first tested the idea that the ATR-mediated checkpoint could be studied using plasmid DNA. For these studies we used a completely soluble system derived from Xenopus egg extracts that allows replication of plasmid DNA (Walter et al. 1998). Efficient replication of plasmid DNA or chromatin in this system requires the initial incubation of the DNA in cytosol to assemble the prereplication complex (pre-RC). Subsequent addition of a concentrated nucleoplasmic extract (NPE) supplies high levels of cdk2 and cdc7 kinase activities, thereby allowing initiation of DNA replication (Walter 2000; Prokhorova et al. 2003).
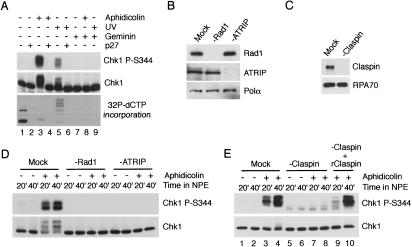
When plasmid DNA was incubated with cytosol and then supplemented with NPE containing aphidicolin, Chk1 underwent robust phosphorylation on S344 (Fig. 1A, lane 3). This residue is phosphorylated in an ATR-dependent manner following UV and aphidicolin treatment (Guo et al. 2000; Hekmat-Nejad et al. 2000; Liu et al. 2000). Incubation of UV-damaged plasmid DNA in cytosol followed by NPE also led to phosphorylation of Chk1 on the same residue (Fig. 1A, lane 5). Inhibition of replication with geminin (McGarry and Kirschner 1998) or p27KIP (Walter and Newport 2000) resulted in a complete loss of Chk1 phosphorylation following both UV and aphidicolin treatment, indicating that both responses are replication-dependent (Fig. 1A, lanes 4,6,8,9).
Figure 1.
Characterization of checkpoint activation using plasmid DNA. (A) Initiation of replication on plasmid DNA is required for Chk1 phosphorylation in response to aphidicolin and UV damage. Plasmid DNA was left untreated or pretreated with UV irradiation (1000 J/m2), incubated in cytosol for 30 min, and then transferred to an equal volume of NPE in the presence or absence of aphidicolin (13 μM) as shown. Geminin was preincubated in cytosol at a concentration of 2 μM for 15 min prior to DNA addition, and p27KIP was added to NPE at a concentration of 10 μM immediately before addition to cytosol. Samples were analyzed for phospho-Chk1 (S344) or total Chk1 by immunoblotting. Replication was analyzed in parallel by incorporation of [α32P]-dCTP into plasmid DNA followed by agarose gel electrophoresis and autoradiography. (B,C) Immunodepletion of Rad1, ATRIP, or Claspin. NPE was incubated with rabbit IgG (mock), α-Rad1, α-ATRIP, or α-Claspin antibodies, and the levels of each protein in depleted extracts were analyzed by immunoblotting. (D,E) Rad1, ATRIP, and Claspin are required for plasmid-mediated checkpoint activation. Depleted extracts were used to replicate plasmid DNA in the presence or absence of aphidicolin as described in A. Samples were taken post-NPE addition at the indicated times and immunoblotted as in A. (Lanes 9,10) Recombinant Claspin (6 nM) was added to cytosol after immunodepletion.
To determine if the ATR complex, RHR complex, and Claspin are required for Chk1 phosphorylation in this plasmid-based system, as they are for chromatin, we immunodepleted ATRIP, Rad1, and Claspin from both cytosol and NPE (Fig. 1B,C). Although aphidicolin induced robust Chk1 phosphorylation in mock-depleted extracts, no detectable Chk1 phosphorylation was observed using extracts immunodepleted of ATRIP, Rad1, or Claspin (Fig. 1D,E). Moreover, addition of recombinant Claspin to Claspin-depleted extracts restored Chk1 phosphorylation (Fig. 1E). These observations demonstrate that the ATR–ATRIP complex, the RHR complex, and Claspin mediate the checkpoint induced by aphidicolin during replication of plasmid DNA.
Chk1 activation follows aphidicolin-induced DNA unwinding
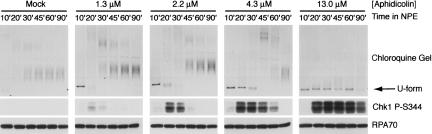
To examine the relationship between DNA unwinding and checkpoint activation, we monitored the topology of plasmid DNA on chloroquine gels in the presence of aphidicolin. Upon protein extraction, plasmids that have been extensively unwound by a DNA helicase are rendered highly negatively supercoiled. Thus, they migrate rapidly on agarose gels even in the presence of high concentrations of chloroquine, which unwinds DNA upon intercalation and causes compensatory positive super-coiling in closed circular plasmids (Walter and Newport 2000). We found that this hyperunwound form of DNA (U form) was generated in a reversible manner at concentrations of aphidicolin as low as 1.3 μM and that increasing the concentration of aphidicolin led to a dose-dependent persistence of this unwound form. Aphidicolin was added to the cytosol prior to the initiation of replication triggered by NPE addition. To determine whether the appearance and disappearance of U-form DNA correlated with activation of Chk1, we examined the relationship between the phosphorylation state of Chk1 and the topology of DNA at these different concentrations of aphidicolin. The phosphorylation of Chk1 occurred within 10 min of the appearance of U-form DNA and was stronger at higher doses of aphidicolin (Fig. 2). Furthermore, the phosphorylation of Chk1 persisted as long as U-form DNA was present, and Chk1 dephosphorylation followed the disappearance of U-form DNA. These observations indicate that there is a temporal relationship between the unwinding of DNA induced by aphidicolin and Chk1 phosphorylation. They are also consistent with a previous study demonstrating a correlation between RPA accumulation on chromatin and Chk1 phosphorylation (Shechter et al. 2004a).
Figure 2.
Chk1 phosphorylation on S344 induced by aphidicolin correlates with DNA unwinding. Plasmid DNA was incubated in cytosol containing the indicated concentration of aphidicolin for 30 min, then added to NPE as in Figure 1A. Parallel samples were removed at the indicated times and analyzed on chloroquine agarose gels and by immunoblotting for the phosphorylation of Chk1. RPA70 was used as a loading control.
Replication of damaged plasmid DNA leads to uncoupling of helicase and polymerase activities
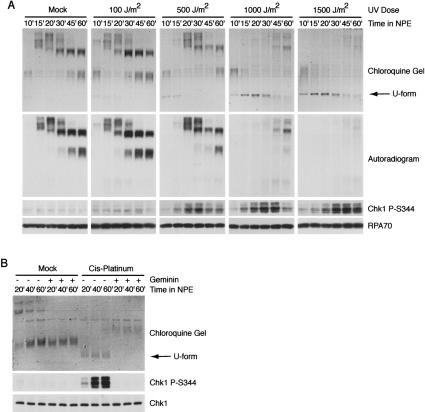
We previously showed that RPA accumulates on chromatin following UV irradiation and MMS treatment (Lupardus et al. 2002). One possibility is that this accumulation is due to the binding of RPA to ssDNA generated during repair of the lesions. However, it is also possible that these lesions cause accumulation of ssDNA by inducing uncoupling of helicase and polymerase activities, as observed for aphidicolin (Walter and Newport 2000). To distinguish between these possibilities, we asked whether U-form DNA was formed upon replication of plasmid DNA damaged by UV irradiation. We observed a dose-dependent formation of U-form DNA upon UV treatment (Fig. 3A). Inhibition of origin firing with geminin completely abrogated the accumulation of U form in response to UV damage (data not shown). Importantly, the disappearance of U-form DNA correlated with increased DNA synthesis, as monitored by radiolabeled dCTP incorporation (Fig. 3A). This indicates that loss of U-form DNA observed at 500 and 1000 J/m2 can be attributed to DNA replication on the unwound strand. Significantly, when the phosphorylation state of Chk1 at S344 was examined in parallel, we found that the phosphorylation of Chk1 consistently followed the appearance of U-form DNA (Fig. 3A). These observations indicate that UV irradiation, like aphidicolin, leads to hyperunwinding of DNA and are consistent with a role for DNA hyperunwinding in activation of the DNA damage checkpoint.
Figure 3.
DNA damage induces the functional uncoupling of MCM helicase and DNA polymerase activities. (A) UV damage induces hyperunwinding and Chk1 phosphorylation on S344. A 6-kb plasmid treated with the indicated levels of UV was added to cytosol to a final concentration of 26 μg/mL. After 30 min incubation, an equal volume of NPE was added. Samples were analyzed for phospho-Chk1 (S344) and RPA by immunoblotting at the indicated times as described in Figure 2. Replication was analyzed in parallel by incorporation of [α32P]-dCTP into plasmid DNA followed by analysis on chloroquine agarose gels and autoradiography. (B) Cis-platinum induces replication-dependent hyperunwinding of plasmid DNA and Chk1 phosphorylation on S344. A 6-kb plasmid either mock-treated or treated with cis-platinum was added to cytosol to a final concentration of 15 μg/mL. NPE was added as in A. Geminin was added to cytosol to achieve a final concentration of 2 μM where indicated. Samples were taken at 20, 40, and 60 min post-NPE addition and analyzed as described in Figure 2. Total Chk1 is used as the loading control for the immunoblots.
To determine if other forms of DNA damage can also induce the functional uncoupling of helicase and polymerase activities, we examined the effect of the chemotherapeutic cis-platinum. Cis-platinum has been shown to induce ATR-dependent cell cycle arrest and Chk1 phosphorylation (Cliby et al. 1998; Zhao and Piwnica-Worms 2001). First, we examined whether lesions generated by cis-platinum treatment induced the functional uncoupling of helicase and polymerase activities. Upon replication of plasmid DNA damaged with cis-platinum, significant accumulation of U-form DNA was observed (Fig. 3B). Moreover, when the initiation of replication was blocked with geminin, the formation of U-form DNA was abrogated. No detectable level of U form was observed in samples containing mock-treated plasmid DNA. When we examined the phosphorylation state of Chk1 on S344 in parallel, we observed significant checkpoint activation upon replication of cis-platinum-treated plasmid DNA. No detectable level of Chk1 phosphorylation was observed upon replication of the mock-treated plasmid. It is notable that cis-platinum forms both intra- and interstrand cross-links, the latter of which might be expected to block unwinding and checkpoint activation. However, the vast majority of lesions caused by cis-platinum are intrastrand cross-links (Kartalou and Essigmann 2001), and we expect that the predominance of these intrastrand lesions allows for sufficient uncoupling to induce the checkpoint. Taken together, these results show for the first time that two DNA damaging agents that induce ATR activation, UV and cis-platinum, cause the uncoupling of helicase and polymerase activities that leads to DNA hyperunwinding.
Inhibition of MCM-mediated hyperunwinding during elongation blocks Chk1 phosphorylation
The relationship between U-form DNA and Chk1 phosphorylation suggests that hyperunwinding may be required for checkpoint activation. Because aphidicolin, UV, and cis-platinum-induced hyperunwinding are dependent on the initiation of DNA replication (Walter and Newport 2000; data not shown), they may reflect uncoupling of the replicative DNA helicase from the stalled polymerase. We and others have recently shown that the MCM2–7 complex (Pacek and Walter 2004; Shechter et al. 2004b), as well as the replication factor Cdc45 (Pacek and Walter 2004), are essential for unwinding during both the beginning and middle of S phase. These results strongly support the hypothesis that MCM2–7 functions as the replicative DNA helicase (Labib and Diffley 2001) and that Cdc45 acts as a helicase cofactor (Masuda et al. 2003).
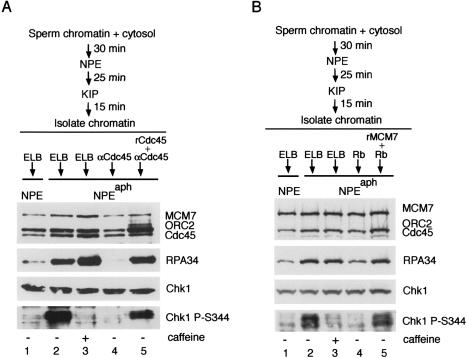
To directly test the hypothesis that MCM-mediated hyperunwinding is required to launch the checkpoint, we allowed replication to initiate, and then stalled the elongating complex with aphidicolin in the presence and absence of helicase inhibitors. Briefly, chromatin was incubated in cytosol to allow pre-RC formation, and it was then mixed with NPE for 25 min at a reduced temperature (19°C) to allow initiation of DNA replication but not completion of elongation (Fig. 4A). Although our previous data indicate that all origins fired during the low-temperature incubation (Pacek and Walter 2004), p27KIP was added here and in all subsequent steps to ensure that no additional origins could fire. The partially replicated chromatin was isolated and incubated with buffer, purified Cdc45-neutralizing antibodies, or antibodies premixed with recombinant Cdc45 protein. The treated chromatin samples were then added to NPE containing aphidicolin. Significant accumulation of RPA on chromatin and caffeine-sensitive Chk1 phosphorylation on S344 were observed with the buffer-treated chromatin when aphidicolin was present, indicating that hyperunwinding and checkpoint activation had occurred (Fig. 4A, lanes 2,3). Importantly, treatment of the isolated chromatin with Cdc45 neutralizing antibodies significantly reduced RPA accumulation and Chk1 phosphorylation (Fig. 4A, lane 4). However, preincubation of the neutralizing antibodies with recombinant Cdc45 protein restored RPA accumulation and Chk1 phosphorylation (Fig. 4A, lane 5). The loading of MCM7, ORC2, and Cdc45 onto chromatin was unaffected by any of these treatments. These observations show for the first time that Cdc45-dependent hyperunwinding during elongation is necessary for activation of Chk1 following aphidicolin treatment.
Figure 4.
MCM7 and Cdc45-mediated hyperunwinding is required for Chk1 phosphorylation. (A) Inhibition of hyperunwinding and checkpoint activation with Cdc45 neutralizing antibodies. Sperm chromatin was incubated with cytosol, NPE, and then p27KIP as indicated. Isolated chromatin was then treated with ELB buffer, Cdc45 neutralizing antibodies, or the same antibodies premixed with recombinant Cdc45 protein. After addition of treated chromatin into NPE, an aliquot was removed for analysis of Chk1 as described in Figure 1A. In parallel, a second aliquot was taken for chromatin isolation. Chromatin-bound proteins (MCM7, ORC2, Cdc45, and RPA34) were analyzed by immunoblotting with indicated antibodies. Aphidicolin (150 μM) was added to NPE where indicated. (B) Inhibition of hyperunwinding and checkpoint activation with Rb1–400. Chromatin was isolated as in A, then treated with ELB, Rb1–400, or Rb1–400 premixed with recombinant MCM7 protein. Aphidicolin (150 μM) and caffeine (5 mM) were added as indicated. Samples were taken and analyzed as described in A.
As an independent means of blocking hyperunwinding, we inhibited the MCM2–7 complex. For these experiments we used an N-terminal fragment of the retinoblastoma protein (Rb1–400) that binds to MCM7 and inhibits DNA replication and aphidicolin-induced unwinding (Sterner et al. 1998; Pacek and Walter 2004). DNA replication forks were synchronized during the elongation phase of DNA replication as described above. Chromatin was isolated and mixed with buffer, Rb1–400, or Rb1–400 that had been preincubated with MCM7 peptide. Finally, NPE-containing aphidicolin was added. As seen in Figure 4B, aphidicolin induced RPA hyperloading (cf. lanes 1 and 2), and caffeine-sensitive Chk1 phosphorylation (cf. lanes 2 and 3). Importantly, Rb1–400 inhibited both RPA hyperloading and Chk1 phosphorylation (Fig. 4B, lane 4). Inhibition was reversed when Rb1–400 was preincubated with MCM7 peptide (Fig. 4B, lane 5). Taken together, these observations strongly suggest that in the context of a stalled DNA replication fork, MCM and Cdc45-dependent DNA hyperunwinding is necessary for the phosphorylation and activation of Chk1.
The amount of DNA unwinding determines the level of Chk1 activation induced with aphidicolin
We previously showed that replication of a 3-kb plasmid in the presence of aphidicolin results in extensive unwinding (Walter and Newport 2000). Although we show that functional uncoupling of DNA unwinding and DNA synthesis is required for checkpoint activation, it is not known if the entire region of hyperunwound DNA contributes to checkpoint signaling. In fact, previous observations in S. cerevisiae suggest that extensive unwinding may not occur. Specifically, EM studies indicate that the amount of additional ssDNA generated at replication forks stalled with HU is ∼100 nucleotides (Sogo et al. 2002). Consistent with this, CHIP studies show also that uncoupling of Cdc45 and RPA from DNA synthesis is minimal in wild-type yeast cells treated with HU (Katou et al. 2003). In addition, it has been suggested that the number of functional replication forks may determine the level of checkpoint activation in S. cerevisiae (Shimada et al. 2002).
We sought to test the contribution of the unwound DNA to Chk1 phosphorylation in the Xenopus system. If the number of origins, and thus the number of replication forks, determines the level of checkpoint activation, comparable levels of Chk1 phosphorylation should be observed with aphidicolin when equal molar quantities of different sized plasmids are examined, assuming an equal number of origins per plasmid. On the other hand, if the unwound DNA contributes to checkpoint activation, then larger plasmids should produce a greater signal with aphidicolin when added on an equal molar basis since these plasmids should have more unwound DNA per mole of plasmid.
To determine whether the unwound DNA contributes to Chk1 phosphorylation, we tested three different sized plasmids and compared the levels of Chk1 phosphorylation generated upon aphidicolin treatment. Plasmids of <10 kb are replicated via a single origin in Xenopus egg extracts (Lucas et al. 2000). Therefore, we initially used equal molar amounts of a 0.8-, 3.0-, and 9 kb plasmid in these experiments. The 0.8-kb plasmid was generated by intramolecular self-ligation of a blunt-ended PCR product, and the plasmid was shown to replicate when incubated sequentially in cytosol then NPE (data not shown). In addition, this plasmid induced Chk1 phosphorylation on S344 following aphidicolin treatment in a geminin-sensitive manner (data not shown).
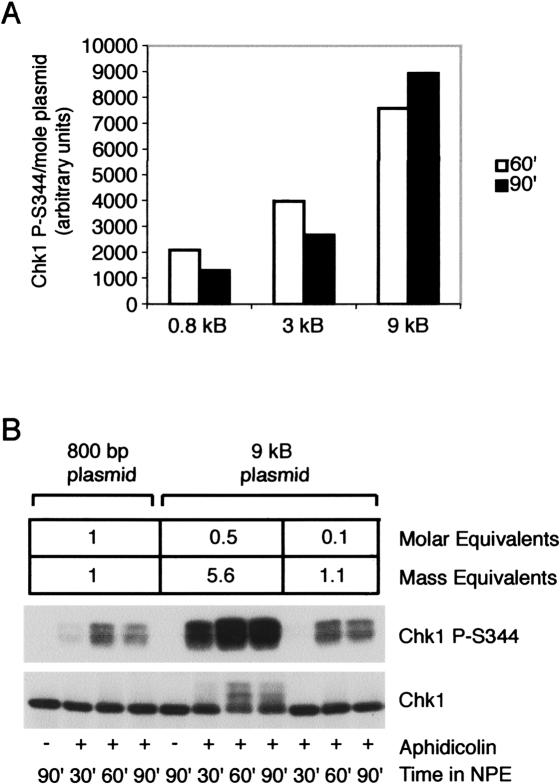
We found that the level of Chk1 phosphorylation increased with increasing plasmid size when the plasmids were present on an equal molar basis (Fig. 5A). Interestingly, we also found that the amount of Chk1 phosphorylation was roughly proportional to the size of the plasmid (Fig. 5A). Consistent with this, when the 9-kb and 0.8-kb plasmids were present on an equal mass basis, the levels of Chk1 phosphorylation observed were comparable (Fig. 5B). These data strongly suggest that it is not the number of origins or replication forks that determines the level of checkpoint activation, but rather the amount of unwound DNA. They also demonstrate that 800 bp of unwound DNA is sufficient to induce the phosphorylation of Chk1 and that additional unwinding contributes to the extent to which Chk1 is phosphorylated.
Figure 5.
The amount of DNA unwinding determines the level of Chk1 activation induced with aphidicolin. (A) Equal molar amounts of larger plasmids induce higher levels of Chk1 phosphorylation than smaller plasmids. Equal molar amounts (∼0.4 nM) of a 9-, 3-, or 0.8-kb plasmid were added to cytosol, then NPE, as described in Figure 1, and aphidicolin was added to a final concentration of 30 μM. Samples were taken at 60 and 90 min and analyzed for phospho-Chk1 (S344) or total Chk1 by immunoblotting. The amount of DNA and level of Chk1 phosphorylation was determined as described in Materials and Methods. The amount of Chk1 phosphorylation achieved per mole of each plasmid was calculated for each plasmid and graphed as shown. (B) Equal mass amounts of 0.8- and 9-kb plasmid DNA generate equivalent levels of Chk1 phosphorylation. Equal mass amounts of a 0.8- or 9-kb plasmid (∼ 0.2 μg/mL) or a fivefold greater amount of the 9-kb plasmid (∼1 μg/mL) were replicated in the presence of aphidicolin (30 μM) as described in A. The amount of DNA was determined as described in Materials and Methods. Samples were taken at 30, 60, and 90 min post-NPE addition and analyzed for phospho-Chk1 (S344) or total Chk1 by immunoblotting. The molar and mass equivalents of each plasmid are shown.
Uncoupling of MCM helicase and DNA polymerase activities is not sufficient for checkpoint activation
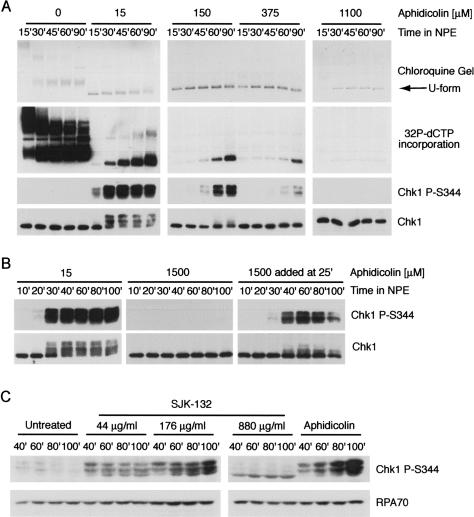
Although we observed a temporal relationship between U-form DNA and checkpoint activation, Chk1 phosphorylation consistently lagged behind U-form DNA by at least 10 min with both UV and aphidicolin (Figs. 2, 3). This indicates that DNA unwinding may be necessary, but not sufficient, to trigger the checkpoint. Since this effect appeared to be more pronounced at higher UV and aphidicolin doses, we examined the effect of further increasing the concentration of aphidicolin on checkpoint activation during replication of plasmid DNA. The addition of 15 μM aphidicolin induced robust Chk1 phosphorylation by the 30-min time point (Fig. 6A). However, when aphidicolin was present at 150 or 375 μM, checkpoint activation was significantly delayed, and at a concentration of 1.1 mM, Chk1 phosphorylation was undetectable. Plasmid hyperunwinding was observed at all concentrations of aphidicolin tested, as determined by the generation of U-form DNA, but replication was progressively inhibited as the concentration of aphidicolin was increased. The appearance of U-form DNA at high concentrations of aphidicolin indicates that hyperunwinding is not sufficient for activation of Chk1 (Fig. 6A). Importantly, however, the phosphorylation of Chk1 strongly correlated with the extent of DNA replication, suggesting that the synthesis of DNA is necessary for checkpoint activation. Moreover, when 1.5 mM aphidicolin was added during S phase, we observed robust checkpoint activation (Fig. 6B). This latter observation demonstrates that checkpoint activation can occur with 1.5 mM aphidicolin if some DNA synthesis has already occurred.
Figure 6.
Uncoupling of MCM helicase and DNA polymerase activities is not sufficient for checkpoint activation. (A) High concentrations of aphidicolin prevent Chk1 phosphorylation. Plasmid DNA was replicated as described in Figure 1A. Aphidicolin was added to achieve a final concentration as shown. Samples were analyzed for phospho-Chk1 (S344) or total Chk1 by immunoblotting at the indicated times. Replication was analyzed in parallel by incorporation of [α32P]-dCTP into plasmid DNA followed by analysis on chloroquine agarose gels and autoradiography. (B) Addition of a high concentration of aphidicolin in S phase induces immediate Chk1 phosphorylation. The experiment was performed as described in A using plasmid DNA except that aphidicolin was added to a concentration of 1.5 mM to cytosol prior to NPE addition or 25 min post-NPE addition. (C) Monoclonal antibody SJK-132 can both activate and block Chk1 phosphorylation. A stock solution of SJK-132 (22 mg/mL stock) was added to crude interphase extract at 0.2%, 0.8%, or 4% v/v prior to chromatin addition. Sperm chromatin was added to a final concentration of 2000 nuclei/μL. Aphidicolin was added to a final concentration of 100 μg/mL. Samples were taken at the times shown and immunoblotted with antibodies to phospho-Chk1 (S344) and RPA70.
As an alternative method of inhibiting DNA synthesis, we examined the effects of inhibiting Polα with the monoclonal antibody SJK-132. SJK-132 has been shown to specifically inhibit the DNA polymerase activity of Polα (Tanaka et al. 1982) and induce the phosphorylation of Chk1 in Xenopus egg extracts (Michael et al. 2000). If DNA synthesis is required for checkpoint activation, we would expect that, like aphidicolin, partial disruption of DNA replication with SJK-132 would activate the checkpoint by inducing an uncoupling event and allowing an adequate level of DNA synthesis. However, further inhibition would eventually result in loss of the checkpoint, as observed for aphidicolin (Fig. 6A). Consistent with this hypothesis, the addition of SJK-132 to a final concentration of 44 μg/mL or 176 μg/mL induced significant Chk1 phosphorylation (Fig. 6C). However, upon addition of SJK-132 to a final concentration of 880 μg/mL, a dramatic decrease in the level of Chk1 phosphorylation was observed (Fig. 6C). The addition of SJK-132 to a final concentration of 880 μg/mL in crude interphase extracts also abrogated the Chk1 phosphorylation that occurs in response to aphidicolin and UV-damaged chromatin (data not shown). Importantly, significant RPA accumulation on chromatin was observed at all the concentrations of SJK-132 examined, demonstrating that the loss of the checkpoint did not result from a lack of nuclear envelope formation, origin firing, or a failure to functionally uncouple helicase and polymerase activities (data not shown). Taken together, these data demonstrate a requirement for DNA polymerase α in activation of the ATR-dependent checkpoint, consistent with previous observations (Michael et al. 2000). However, since aphidicolin and SJK-132 inhibit DNA polymerase activity and not RNA primer synthesis (Yagura et al. 1983; Sheaff et al. 1991), these results further suggest it is the DNA polymerase activity of Polα that is critical for checkpoint activation.
DNA polymerase activity of Polα is needed for Rad1 chromatin binding
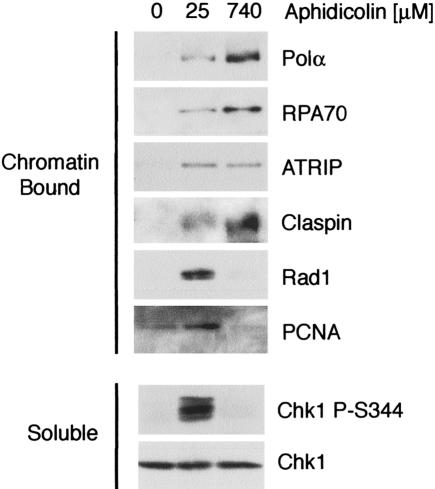
We had previously shown that aphidicolin induces the recruitment of the ATR and Rad1 checkpoint complexes onto chromatin in egg extracts (Lupardus et al. 2002). In order to determine how aphidicolin acts to block checkpoint activation at high concentrations (Fig. 6A), we examined the chromatin binding of both checkpoint protein complexes. When 25 μM aphidicolin was added to extracts, robust Chk1 phosphorylation was observed, and RPA, Polα, PCNA, ATRIP, Claspin, and Rad1 accumulated on chromatin (Fig. 7). However, when the concentration of aphidicolin was increased to 740 μM, Chk1 phosphorylation and chromatin binding of Rad1 was abrogated. This concentration of aphidicolin also blocks PCNA loading (Fig. 7; Michael et al. 2000; Arias and Walter 2005) by preventing adequate elongation of the RNA/DNA primer synthesized by Polα. Importantly, the loading of RPA, Polα, Claspin, and ATRIP were unaffected (Fig. 7). Taken together, these observations suggest that recruitment of RPA, ATRIP, and Polα is not sufficient for Rad1 chromatin binding or Chk1 phosphorylation and that synthesis and elongation of a DNA primer is necessary to recruit the RHR complex onto chromatin for activation of Chk1.
Figure 7.
DNA polymerase activity of Polα is needed for Rad1 chromatin binding. Xenopus sperm chromatin (10,000 nuclei/μL) was preincubated in cytosol containing the indicated concentrations of aphidicolin for 30 min, then mixed with an equal volume of NPE. The chromatin was isolated 50 min later and bound proteins were analyzed by immunoblotting with the indicated antibodies. Prior to chromatin isolation, samples were taken and immunoblotted with antibodies to phospho-Chk1 (S344) and Chk1.
Discussion
Different forms of genotoxic stress can lead to activation of the ATR-dependent DNA damage checkpoint (Melo and Toczyski 2002). This raises the question of whether each distinct lesion is directly recognized by checkpoint proteins or whether these lesions are processed into a common intermediate. Here we provide evidence that functional uncoupling of MCM helicase and DNA polymerase activities occurs in response to multiple forms of DNA damage and that this event is necessary to generate a common DNA intermediate required for activation of Chk1 during S phase. We also show that this uncoupling event serves to increase the amount of checkpoint-activating signal generated at each replication fork.
DNA damage induces uncoupling of helicase and polymerase activities
Previous studies suggest that ssDNA may be a critical intermediate for checkpoint activation and several studies have shown that this structure accumulates following DNA damage (see above). However, it is not known if this accumulation is necessary for checkpoint activation or how the ssDNA is made. To explore the mechanism by which ssDNA is generated following DNA damage, we have analyzed the topology of DNA treated with UV and cis-platinum in Xenopus egg extracts. We find that plasmid DNA damaged with these agents undergoes extensive, replication-dependent unwinding to generate a common DNA intermediate (Fig. 3). This intermediate is also observed upon addition of aphidicolin (Fig. 2; Walter and Newport 2000). These observations indicate for the first time that DNA unwinding and DNA synthesis can become uncoupled in response to DNA damage. From our data, however, we cannot determine if the helicase and polymerase actually become physically uncoupled in the presence of aphidicolin or DNA damage. Instead, this may be only a functional uncoupling whereby the helicase and polymerase remain physically coupled while the helicase continues to unwind DNA.
The observed uncoupling of helicase and polymerase activities induced by UV irradiation and cis-platinum demonstrates that lesions formed by these agents have a greater inhibitory effect on the DNA synthesis machinery than on DNA unwinding mediated by the MCM helicase. Consistent with these results, the T-antigen helicase, which is structurally similar to the MCM helicase (Pape et al. 2003), was shown to display no detectable sensitivity to either UV lesions or N-2-acetylaminofluorene adducts when unwinding double-stranded DNA substrates in vitro (Veaute et al. 2000). In contrast, prokaryotic and eukaryotic replicative polymerases are blocked by lesions on DNA (Moore et al. 1981; Veaute et al. 2000). Our results, along with the previous characterization of the T-antigen helicase, suggest that the ability to efficiently unwind many damaged templates may be a general characteristic of this enzyme class.
Functional uncoupling of MCM helicase and polymerase activities is a determinant of checkpoint activation
Importantly, we provide the first direct evidence that uncoupling of helicase and polymerase activities is necessary for activation of the checkpoint. Inhibition of unwinding by addition of antibodies that neutralize Cdc45, a putative cofactor for MCM activity, or by addition of a fragment of Rb that binds to MCM7 and inhibits DNA replication, prevents activation of the checkpoint (Fig. 4). Importantly, the MCM complex was inhibited during the elongation phase of DNA replication in these experiments. Thus, replication forks have already been assembled and DNA synthesis initiated when the polymerase is stalled by aphidicolin. Since the replication fork is present under these conditions, these results suggest the signal does not emanate from the fork per se. Consistent with this hypothesis, we found that the activation of Chk1 correlated with the amount of unwound DNA and not the number of replication forks assembled in the extract (Fig. 5).
Thus, the checkpoint is a sensor that monitors the product resulting from a difference in helicase and polymerase activities (i.e., unwound DNA) rather than the absolute rate of DNA replication. This strategy has several advantages. First, the checkpoint should be more adaptable to different basal rates of DNA replication. For example, the cell would not be susceptible to activation of the checkpoint when a slowing of all S-phase processes result, such as might occur at low temperature, under conditions of low energy, or when the replication fork progresses through heterochromatin. Instead, the checkpoint would only be activated in what might be the more dangerous situation—when the activities of these two enzymes become uncoordinated at the replication fork.
Second, and perhaps more importantly, by converting lesions or other polymerase-slowing events into a common intermediate, a general solution exists for responding to a broad range of events that threaten genomic stability. These events include multiple types of DNA damage as well as other insults that do not cause DNA damage, at least initially, but which interfere with coordination between helicase and polymerase activities. In accord with this idea, we have found that partial depletion of Polα not only slows the rate of DNA synthesis, but also causes the formation of unwound DNA and activation of the checkpoint (T. Byun and K. Cimprich, unpubl.). This checkpoint activation could result from a discordant synthesis of leading and lagging strands or slower action of the polymerases during DNA replication, but in any case, it shows that other mechanisms of creating an imbalance in the rate of DNA unwinding versus DNA synthesis can generate this common checkpoint-activating signal.
Another consequence of this uncoupling mechanism may be to create a threshold for checkpoint activation. Processes such as DNA repair and lesion by-pass may allow DNA replication to occur efficiently at low levels of DNA damage such that extensive uncoupling of DNA unwinding and DNA synthesis does not occur and the checkpoint is minimally induced. However, when the level of DNA damage exceeds the level that can be repaired or by-passed efficiently, significant stalling of polymerases would occur, leading to functional uncoupling of MCM helicase activity and strong, sustained checkpoint activation. This could provide the cell with the ability to tolerate some level of DNA damage without activating the checkpoint. It might also allow the cell to respond to low but persistent levels of DNA damage or unreplicated DNA.
One important issue to consider is whether this mechanism of generating ssDNA occurs in eukaryotic cells. A study in yeast shows only a modest increase in the amount of ssDNA generated in response to HU (Sogo et al. 2002). This could indicate that only minimal uncoupling is possible and necessary for checkpoint activation in yeast. However, because HU and aphidicolin inhibit DNA synthesis to different degrees and by distinct mechanisms, it is also possible that the difference in the amount of uncoupling may reflect the level of polymerase inhibition. Consistent with the idea that uncoupling does occur in mammalian cells, treatment of human melanoma cells with aphidicolin has been shown to induce the formation of large regions (>20 kb) of ssDNA (Lonn and Lonn 1988). In addition, other studies in mammalian cells have demonstrated the accumulation of ssDNA (Scudiero and Strauss 1974; Stewart and Hristoforidis 1987) or chromatin-bound RPA (Zou and Elledge 2003) in response to various types of genotoxic agents. Although further studies will be required to delineate whether the observed accumulation of RPA and/or ssDNA occurs through the proposed uncoupling model, these observations argue that a similar mechanism is used in eukaryotic cells.
Interestingly, two recent studies have shown a role for the MCM complex in activating the ATR-dependent checkpoint in mammalian cells (Cortez et al. 2004; Tsao et al. 2004). These studies show that depletion of MCM7 prevents Chk1 phosphorylation in response to DNA damage or aphidicolin treatment (Cortez et al. 2004; Tsao et al. 2004) and also causes a defect in the formation of ATR nuclear foci following UV (Tsao et al. 2004). Although a direct role for MCM7 in signaling to Chk1 cannot be ruled out, based on the role of the MCM complex in functional uncoupling following both UV and aphidicolin treatment, these observations could also result from a failure to generate the checkpoint signal.
We also show that while unwinding is necessary for Chk1 phosphorylation, it is not sufficient, and DNA synthesis by the Polα complex is needed. This result is based in part on the observation that high concentrations of DNA polymerase inhibitors, aphidicolin and SJK-132 antibody, blocked Chk1 phosphorylation. Consistent with this idea, only those concentrations of aphidicolin that allowed DNA synthesis led to activation of Chk1, although hyperunwinding was induced by all concentrations of aphidicolin examined (Fig. 6A). Moreover, there was a strong correlation between the timing of DNA synthesis and Chk1 phosphorylation. This model is consistent with findings in Schizosaccharomyces pombe that show a requirement for the DNA polymerase activity of Polα in preventing inappropriate entry into mitosis with no DNA synthesis (Bhaumik and Wang 1998). What is not clear, however, is whether events downstream of Polα DNA polymerase activity, such as PCNA loading, may still be necessary.
At first glance, this result appears to be in conflict with a previous study in Xenopus egg extracts, which concluded that DNA synthesis by the Polα complex is not necessary for checkpoint activation and that RNA primer synthesis is the critical checkpoint-activating event (Michael et al. 2000). However, that conclusion was based on the ability of aphidicolin and SJK-132 to induce Chk1 phosphorylation. Indeed, we found that these agents did induce Chk1 phosphorylation at some concentrations, but that higher concentrations delayed and ultimately inhibited this event. Because RNA primer synthesis is insensitive to aphidicolin (Sheaff et al. 1991) and the monoclonal antibody SJK-132 (Yagura et al. 1983), our results argue that the RNA primer is not sufficient for Chk1 activation and that extension of the RNA primer by the aphidicolin-sensitive polymerase subunit of Polα is necessary. Since even a very low level of replication was sufficient to induce Chk1 phosphorylation (Fig. 6A), it is likely that the potent inhibition of DNA polymerase α needed to block Chk1 phosphorylation was not achieved at the single time point and concentration examined in this previous study.
Finally, we find that Polα DNA polymerase activity is necessary for loading Rad1 but not ATR onto chromatin. These observations strongly suggest that the formation of ssDNA and the chromatin association of RPA and ATR are not sufficient for activation of Chk1. They are also consistent with the observation that depletion of Polα, which blocks all DNA synthesis, prevents RHR loading (You et al. 2002), and they reinforce the idea that ATR and RHR loading are independent events (Kondo et al. 2001; Melo et al. 2001; Zou et al. 2002; Lee et al. 2003).
Functional uncoupling of helicase and polymerase activities amplifies the level of checkpoint activation
Using different sized plasmids, we have also investigated the extent to which DNA unwinding contributes to activation of the checkpoint. Because a plasmid of 800 bp can induce Chk1 phosphorylation, we conclude that extensive accumulation of ssDNA at the replication fork is not necessary to trigger checkpoint activation and that the minimal amount of unwound DNA required for checkpoint activation is 800 bp or less. This is consistent with the idea that ssDNA generated during ongoing DNA replication can regulate the timing of origin firing through ATR (Marheineke and Hyrien 2004; Shechter et al. 2004a). However, we also show that the additional unwound DNA that is formed with larger plasmids can contribute to the strength and duration of Chk1 phosphorylation. Thus, the generation of extensive regions of unwound DNA through this uncoupling event may provide the cell with a mechanism of amplifying the level of Chk1 phosphorylation that can be achieved per replication fork. Interestingly, we found that the amount of Chk1 phosphorylation observed was roughly proportional to the amount of ssDNA that would be formed if full uncoupling occurred with the different sized plasmids. This suggests that unwinding is complete on each of the plasmids used. Indeed, when we examined the extent of unwinding by electron microscopy of a 13-kb plasmid replicated in NPE system in the presence of aphidicolin, we observed that the entire plasmid had become unwound (K. Mowrer and J. Walter, unpubl.). At present it is not clear if the increased level of Chk1 phosphorylation achieved with the larger plasmids is due solely to the longer regions of ssDNA, as the synthesis of additional DNA primers on this unwound DNA is still possible. This model is supported by our previous observation that Polα accumulates on chromatin in response to aphidicolin (Michael et al. 2000; Lupardus et al. 2002) and DNA damage (Lupardus et al. 2002), although the presence of additional primers has not been directly observed.
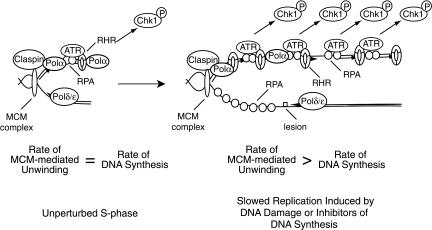
Model for checkpoint activation
Taken together with other findings, our results suggest the following model for checkpoint activation, a major determinant of which is the relationship between DNA helicase and polymerase activities. In the absence of genotoxic stress, the rate of DNA unwinding is less than or equal to the rate of DNA synthesis, likely due to a slower intrinsic rate of unwinding relative to DNA synthesis. This limits the amount of unwound DNA during S phase and keeps checkpoint activation to a minimum. Upon perturbation of DNA synthesis by DNA damage or other inhibitors of DNA replication, however, the rate of unwinding becomes greater than the rate of DNA synthesis, leading to a functional uncoupling of the MCM helicase from the replicative polymerases (Fig. 8). This results in the creation of large regions of unwound ssDNA and leads to the accumulation of RPA, ATRIP, and ATR on chromatin. As unwinding occurs, the Polα–primase complex begins DNA synthesis on the unwound DNA, creating an additional necessary component of the signal that recruits the RHR complex onto chromatin. The induced proximity of the ATR and RHR complexes at primer/template junctions then leads to Chk1 phosphorylation.
Figure 8.
Model for activation of the ATR-mediated checkpoint.
Materials and methods
Xenopus egg extracts
Interphase extracts were prepared as described (Lupardus et al. 2002). Membrane-free egg cytosol and NPE were prepared as described previously (Walter et al. 1998).
Antibodies and recombinant proteins
The 5′ region of xATRIP encoding amino acids 1–107 was cloned by PCR from Image clone 3402491 (GenBank BG020060) with the following primers: 5′-GCGAATTCTATGTCTGC TAACCCCTTG-3′ and 5′-TACTCGAGTTTATGAGCTACT TGTTGTT-3′. The resulting PCR product was digested with EcoRI and XhoI and cloned into pGEX4T3 (Amersham Pharmacia). The GST fusion protein was expressed at 30°C and purified according to the manufacturer's instructions. The production of Geminin and GST-p27 was previously described (Lupardus et al. 2002). Recombinant His6-Claspin was expressed and purified as previously described (Kumagai and Dunphy 2000) using baculovirus provided by William Dunphy (California Institute of Technology, Pasadena, CA). Rabbit polyclonal ATRIP and Claspin antibodies were raised at Josman, LLC. Antibodies for Chk1 (G-4 sc-8408) (Santa Cruz Biotechnology) and phospho Ser 345 Chk1 (Cell Signaling) are commercially available. Antibodies used to immunodeplete and/or immunoblot for Rad1 (Lupardus et al. 2002) and ATR (Hekmat-Nejad et al. 2000) have been previously described. The RPA70 antibody was provided by Peter Jackson (Stanford University, Stanford, CA). The mouse hybridoma used to produce SJK-132 was provided by Teresa Wang (Stanford University, Stanford, CA). SJK-132 was concentrated using YM-10 Microcons (Millipore).
Immunodepletions and chromatin binding
Chromatin binding in NPE was performed as previously described (Edwards et al. 2002). Immunodepletions (three rounds, 1 h each) were carried out at 4°C with Protein A Sepharose beads (Amersham Pharmacia). The depletion of ATRIP and Rad1 were performed using cross-linked antibodies prepared with dimethyl pimelimidate (Pierce) using the manufacturer's suggested protocol. Depletion of Claspin was performed without cross-linking. Specific sera were used at a 1:1 or 1:2 ratio of beads to serum. When depleting cytosol and NPE, the amount of packed beads per volume extract was 0.5:1 to 1:1.
DNA and replication assays
A 6-kb plasmid was used in all experiments, unless otherwise stated, at a final concentration of 13 μg/mL. The 3-kb (pBluescript) and 9-kb plasmids were amplified in DH5α and isolated using the Maxi-Prep kit (Qiagen). For UV treatment, plasmid DNA was diluted to 0.26 mg/mL and treated using a UV Stratalinker (Stratagene) at the specified dosage. For cis-platinum, 0.3 mg/mL stock solution of cis-platinum was prepared in water. The 6-kb plasmid (0.15 mg/mL) was incubated in TE containing 3 μg/mL of cis-platinum and incubated at 37°C for 23 h. The DNA was ethanol precipitated, washed with 70% ethanol, dried, and resuspended in TE to a final concentration of 0.3 mg/mL.
To prepare the 0.8-kb plasmid, a PCR reaction was performed using the following primers and pBS SK (–) as the template: 5′-GCAGAATTCGCGTAATCATGGTCATAGCTGTT-3′ and 5′-TATGAATTCACATACCTCGCTCTGCTAATCCT-3′. The DNA primers were phosphorylated at the 5′ end with T4 polynucleotide kinase prior to use. PCR was performed using Vent polymerase in Thermo Pol Buffer with an annealing temperature of 55°C. After the PCR, the product was diluted 10-fold and ligated using T4 ligase at 16°C overnight. The resulting product was ethanol precipitated and separated in a 1.1% TAE agarose gel in the presence of ethidium bromide (0.5 ng/mL). The band that migrated at ∼600 bp was excised and gel extracted using the Gel Extraction Kit (Qiagen). The eluate was precipitated in the presence of glycogen and resuspended in 10 mM Tris at pH 8.0 and stored at –20°C.
Aphidicolin (Sigma) was diluted in 10 mM PIPES at pH 7.4. Plasmid replication was performed as previously described (Walter and Newport 2000). Samples were analyzed on 0.8% TBE agarose gels containing 4 μM chloroquine (Sigma) when analyzing U-form DNA. Otherwise samples were analyzed on 0.8% TAE agarose gels. DNA was visualized using SybrGold (Molecular Probes).
Method for quantitating molar equivalents of plasmid DNA and levels of Chk1 phosphorylation on S344
The amount of plasmid DNA was determined by replicating plasmid DNA by sequential addition to cytosol and then NPE, then analyzing samples 90 min post-NPE addition. Identical samples containing geminin (2 μM) were also analyzed to subtract incorporation resulting from non-replication-associated events. The samples were separated on a 1.1% TAE agarose gel, dried, and subjected to autoradiography. The level of [α32P]-dCTP incorporation was determined by exposure of the gels to storage phosphor screens (Molecular Dynamics), scanning on Typhoon 9410 (Amersham), and quantitation using Image Quant software (Molecular Dynamics). The level of Chk1 phosphorylation was determined by immunoblotting with the phospho S344 Chk1 antibody and measuring the luminescence on a Lumi-Imager (Boehringer Mannheim), followed by analysis using the Lumi-Analyst software (Boehringer Mannheim).
Treatment of chromatin with Cdc45 neutralizing antibodies and Rb1–400
Procedures for isolation of chromatin and treatment with both Cdc45 neutralizing antibodies and Rb1–400 have been previously described (Pacek and Walter 2004).
Acknowledgments
We thank Drs. William Dunphy, Peter Jackson, and Teresa Wang for generously providing reagents used in this study. This work was supported by NIH grant GM062193 and a Beckman Scholar Award to K.A.C., NIH grant GM62267 to J.C.W., and NIH Training Grant 2 T32 HD07249 and DOD Breast Cancer Research Program grant 04-1-0311 to T.S.B. K.A.C. is a Leukemia and Lymphoma Society Scholar.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1301205.
References
- Arias E.E. and Walter, J.C. 2005. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes & Dev. 19: 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik D. and Wang, T.S. 1998. Mutational effect of fission yeast pol α on cell cycle events. Mol. Biol. Cell 9: 2107–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K.A. 2003. Fragile sites: Breaking up over a slowdown. Curr. Biol. 18: R231–R233. [DOI] [PubMed] [Google Scholar]
- Cliby W.A., Roberts, C.J., Cimprich, K.A., Stringer, C.M., Lamb, J.R., Schreiber, S.L., and Friend, S.H. 1998. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 17: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Guntuku, S., Qin, J., and Elledge, S.J. 2001. ATR and ATRIP: Partners in checkpoint signaling. Science 294: 1713–1716. [DOI] [PubMed] [Google Scholar]
- Cortez D., Glick, G., and Elledge, S.J. 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. 101: 10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V., Shechter, D., Lupardus, P.J., Cimprich, K.A., Gottesman, M., and Gautier, J. 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 11: 203–213. [DOI] [PubMed] [Google Scholar]
- Edwards M.C., Tutter, A.V., Cvetic, C., Gilbert, C.H., Prokhorova, T.A., and Walter, J.C. 2002. MCM2–7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 277: 33049–33057. [DOI] [PubMed] [Google Scholar]
- Ellison V. and Stillman, B. 2003. Biochemical characterization of DNA damage checkpoint complexes: Clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 1: 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Kumagai, A., Wang, S.X., and Dunphy, W.G. 2000. Requirement for ATR in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes & Dev. 14: 2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Nejad M., You, Z., Yee, M.-C., Newport, J., and Cimprich, K.A. 2000. Xenopus ATR is a replication-dependent chromatin binding protein required for the DNA replication checkpoint. Curr. Biol. 10: 1565–1573. [DOI] [PubMed] [Google Scholar]
- Kartalou M. and Essigmann, J.M. 2001. Recognition of cisplatin adducts by cellular proteins. Mutat. Res. 478: 1–21. [DOI] [PubMed] [Google Scholar]
- Katou Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K., and Shirahige, K. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083. [DOI] [PubMed] [Google Scholar]
- Kondo T., Wakayama, T., Naiki, T., Matsumoto, K., and Sugimoto, K. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294: 867–870. [DOI] [PubMed] [Google Scholar]
- Kumagai A. and Dunphy, W.G. 2000. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell 6: 836–849. [DOI] [PubMed] [Google Scholar]
- Labib K. and Diffley, J.F. 2001. Is the MCM2–7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev. 11: 64–70. [DOI] [PubMed] [Google Scholar]
- Lee J., Kumagai, A., and Dunphy, W.G. 2003. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol. Cell 11: 329–340. [DOI] [PubMed] [Google Scholar]
- Liu Q., Guntuku, S., Cui, X.S., Matsuoka, S., Cortez, D., Tamai, K., Luo, G., Carattini-Rivera, S., DeMayo, F., Bradley, A., et al. 2000. Chk1 is an essential kinase that is regulated by ATR and required for the G(2)/M DNA damage checkpoint. Genes & Dev. 14: 1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Longhese M.P., Neecke, H., Paciotti, V., Lucchini, G., and Plevani, P. 1996. The 70 kDa subunit of replication protein A is required for the G1/S and intra-S DNA damage checkpoints in budding yeast. Nucleic Acids Res. 24: 3533–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonn U. and Lonn, S. 1988. Extensive regions of single-stranded DNA in aphidicolin-treated melanoma cells. Biochemistry 27: 566–570. [DOI] [PubMed] [Google Scholar]
- Lucas I., Chevrier-Miller, M., Sogo, J.M., and Hyrien, O. 2000. Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J. Mol. Biol. 296: 769–786. [DOI] [PubMed] [Google Scholar]
- Lupardus P.J., Byun, T., Yee, M.C., Hekmat-Nejad, M., and Cimprich, K.A. 2002. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes & Dev. 16: 2327–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marheineke K. and Hyrien, O. 2004. Control of replication origin density and firing time in Xenopus egg extracts: Role of a caffeine-sensitive, ATR-dependent checkpoint. J. Biol. Chem. 279: 28071–28081. [DOI] [PubMed] [Google Scholar]
- Masuda T., Mimura, S., and Takisawa, H. 2003. CDK- and Cdc45-dependent priming of the MCM complex on chromatin during S-phase in Xenopus egg extracts: Possible activation of MCM helicase by association with Cdc45. Genes Cells 8: 145–161. [DOI] [PubMed] [Google Scholar]
- McGarry T.J. and Kirschner, M.W. 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93: 1043–1053. [DOI] [PubMed] [Google Scholar]
- Melo J. and Toczyski, D. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14: 237–245. [DOI] [PubMed] [Google Scholar]
- Melo J.A., Cohen, J., and Toczyski, D.P. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes & Dev. 15: 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W.M., Ott, R., Fanning, E., and Newport, J. 2000. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science 289: 2133–2137. [DOI] [PubMed] [Google Scholar]
- Mimura S., Masuda, T., Matsui, T., and Takisawa, H. 2000. Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells 5: 439–452. [DOI] [PubMed] [Google Scholar]
- Moore P.D., Bose, K.K., Rabkin, S.D., and Strauss, B.S. 1981. Sites of termination of in vitro DNA synthesis on ultraviolet- and N-acetylaminofluorene-treated φ X174 templates by prokaryotic and eukaryotic DNA polymerases. Proc. Natl. Acad. Sci. 78: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M. and Walter, J.C. 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 23: 3667–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T., Meka, H., Chen, S., Vicentini, G., van Heel, M., and Onesti, S. 2003. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 4: 1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lucca, C., Liberi, G., Marini, F., Lopes, M., Plevani, P., Romano, A., DiFiore, P.P., and Foiani, M. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18: 6561–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova T.A., Mowrer, K., Gilbert, C.H., and Walter, J.C. 2003. DNA replication of mitotic chromatin in Xenopus egg extracts. Proc. Natl. Acad. Sci. 100: 13241–13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudiero D. and Strauss, B. 1974. Accumulation of single-stranded regions in DNA and the block to replication in a human cell line alkylated with methyl methane sulfonate. J. Mol. Biol. 83: 17–34. [DOI] [PubMed] [Google Scholar]
- Sheaff R., Ilsley, D., and Kuchta, R. 1991. Mechanism of DNA polymerase α inhibition by aphidicolin. Biochemistry 30: 8590–8597. [DOI] [PubMed] [Google Scholar]
- Shechter D., Costanzo, V., and Gautier, J. 2004a. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6: 648–655. [DOI] [PubMed] [Google Scholar]
- Shechter D., Ying, C.Y., and Gautier, J. 2004b. DNA unwinding is an MCM complex-dependent and ATP hydrolysis-dependent process. J. Biol. Chem. 279: 45586–45593. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. 2004. Principles of tumor suppression. Cell 116: 235–246. [DOI] [PubMed] [Google Scholar]
- Shimada K., Pasero, P., and Gasser, S.M. 2002. ORC and the intra-S-phase checkpoint: A threshold regulates Rad53p activation in S phase. Genes & Dev. 16: 3236–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Lopes, M., and Foiani, M. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602. [DOI] [PubMed] [Google Scholar]
- Sterner J.M., Dew-Knight, S., Musahl, C., Kornbluth, S., and Horowitz, J.M. 1998. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol. Cell. Biol. 18: 2748–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart B.W. and Hristoforidis, C. 1987. Persistent structural change in replicating hepatic DNA isolated from diethylnitrosamine-treated rats. Cancer Lett. 35: 263–269. [DOI] [PubMed] [Google Scholar]
- Stokes M.P., Van Hatten, R., Lindsay, H.D., and Michael, W.M. 2002. DNA replication is required for the checkpoint response to damaged DNA in Xenopus egg extracts. J. Cell Biol. 158: 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Hu, S.Z., Wang, T.S., and Korn, D. 1982. Preparation and preliminary characterization of monoclonal antibodies against human DNA polymerase α. J. Biol. Chem. 257: 8386–8390. [PubMed] [Google Scholar]
- Tercero J.A., Longhese, M.P., and Diffley, J.F. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11: 1323–1336. [DOI] [PubMed] [Google Scholar]
- Tsao C.C., Geisen, C., and Abraham, R.T. 2004. Interaction between human MCM7 and Rad17 proteins is required for replication checkpoint signaling. EMBO J. 23: 4660–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X., Mari-Giglia, G., Lawrence, C.W., and Sarasin, A. 2000. UV lesions located on the leading strand inhibit DNA replication but do not inhibit SV40 T-antigen helicase activity. Mutat. Res. 459: 19–28. [DOI] [PubMed] [Google Scholar]
- Walter J.C. 2000. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem. 275: 39773–39778. [DOI] [PubMed] [Google Scholar]
- Walter J. and Newport, J. 2000. Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol. Cell 5: 617–627. [DOI] [PubMed] [Google Scholar]
- Walter J., Sun, L., and Newport, J. 1998. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell 1: 519–529. [DOI] [PubMed] [Google Scholar]
- Ward I.M., Minn, K., and Chen, J. 2004. UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. J. Biol. Chem. 279: 9677–9680. [DOI] [PubMed] [Google Scholar]
- Yagura T., Tanaka, S., Kozu, T., Seno, T., and Korn, D. 1983. Tight association of DNA primase with a subspecies of mouse DNA polymerase α. J. Biol. Chem. 258: 6698–6700. [PubMed] [Google Scholar]
- You Z., Kong, L., and Newport, J. 2002. The role of single-stranded DNA and polymerase α in establishing the ATR, Hus1 DNA replication checkpoint. J. Biol. Chem. 277: 27088–27093. [DOI] [PubMed] [Google Scholar]
- Zhao H. and Piwnica-Worms, H. 2001. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21: 4129–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.B. and Elledge, S.J. 2000. The DNA damage response: Putting checkpoints in perspective. Nature 408: 433–439. [DOI] [PubMed] [Google Scholar]
- Zou L. and Elledge, S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300: 1542–1548. [DOI] [PubMed] [Google Scholar]
- Zou L., Cortez, D., and Elledge, S.J. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes & Dev. 16: 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Liu, D., and Elledge, S.J. 2003. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. 100: 13827–13832. [DOI] [PMC free article] [PubMed] [Google Scholar]