Abstract
The Chinese hamster dihydrofolate reductase (DHFR) origin of replication consists of a 55-kb zone of potential initiation sites lying between the convergently transcribed DHFR and 2BE2121 genes. Two subregions within this zone (ori-β/ori-β′ and ori-γ) are preferred. In the DHFR-deficient variant, DR8, which has deleted a 14-kb sequence straddling the 3′ end of the DHFR gene, early-firing origin activity in the downstream ori-β/ori-β′ and ori-γ regions is completely suppressed. We show that the critical deleted sequences reside within a 168-bp segment encompassing the intron 5/exon 6 boundary, exon 6, 54 bp of the 3′ untranslated region (UTR), but not the three natural polyA sites. In wild-type cells, this sequence efficiently arrests transcription in a region a few kilobases downstream, which coincides with the 5′ boundary of the replication initiation zone. In DR8, DHFR-specific transcripts efficiently use an alternative sixth exon (6c) and polyA signals near the middle of the former intergenic region to process primary transcripts. However, transcription proceeds to a position almost 35 kb downstream from these signals, and replication initiation can only be detected beyond this point. When the wild-type 168-bp 3′ element is inserted into DR8 at the same position as alternative exon 6c, transcription is arrested efficiently and initiations occur almost immediately downstream. Thus, the normal 3′ end of the DHFR gene constitutes a boundary element not only for the gene but also for the local origin of replication.
Keywords: DHFR, DR8, E6DO, replication origins, transcription termination
With the completion of genome sequencing projects for several different eukaryotic organisms, it has been possible to identify and localize virtually every transcription unit within each chromosomal complement. However, genes constitute only part of the critical information encoded by the genome, and the challenge now is to determine how to identify the positions of nontranscribed informational elements within the gene scaffolds that have been constructed. Examples of such elements include origins of replication, sister chromatid cohesion sites, matrix attachment regions, and other unknown sequences that may facilitate chromosome segregation, chromatin compaction, nuclear architecture, etc.
Eukaryotic genomes are composed of many individual replicating units (replicons) (Huberman and Riggs 1968) that are tandemly arranged from one end of each linear chromosome to the other. A replicon is defined as a segment of DNA that is synthesized by the forks emerging from a single origin (Huberman and Riggs 1968). With very few exceptions (e.g., during early embryogenesis) (Blumenthal et al. 1974), virtually all eukaryotic origins of replication identified so far are localized in the spacers between active genes (e.g., Vaughn et al. 1990; Biamonti et al. 1992; Berberich and Leffak 1993; Kitsberg et al. 1993; Little et al. 1993; Raghuraman et al. 2001). To understand the significance of this phenomenon, it will be necessary to understand how transcription per se impinges on local origin activity.
In the simple eukaryote Saccharomyces cerevisiae, the positions of origins of replication are fixed by a recognizable ARS consensus sequence, which, when active, nucleates the loading of an origin recognition complex (ORC) (Bell and Stillman 1992) that ultimately effects initiation at that site. Thus, potential origins can be readily positioned vis-a-vis the surrounding genes by analysis of sequence data, and biochemical mapping procedures can be used to assess their activities (Brewer and Fangman 1987; Nawotka and Huberman 1988). To address the relationship between replication timing and gene expression on a global level in S. cerevisiae, DNA synthesized during defined windows of the S period was hybridized to microarrays of sequences distributed at regular intervals throughout the genome (Raghuraman et al. 2001). Although these studies show clearly that origins coincide with a subset of ARS elements that are activated at different times in S phase, there is no detectable correlation in yeast between the time of replication of a gene and its transcriptional activity. Instead, the time of activation of origins seems to depend primarily on other chromosomal context effects (e.g., proximity to centromeres, telomeres, heterochromatin, etc.) (for review, see Weinreich et al. 2004).
In higher eukaryotes, the regulation of chromosomal replication is much more complex. As in S. cerevisiae, the few origins that have been identified by biochemical means reside upstream or downstream from genes. However, they share no recognizable sequence motifs that have been shown to contribute to origin activity. Indeed, with only a few apparent exceptions (Giacca et al. 1994; Aladjem et al. 1998a), most higher eukaryotic origins that have been identified correspond to broad zones of inefficient sites spread throughout intergenic regions (e.g., Vaughn et al. 1990; Little et al. 1993; Shinomiya and Ina 1993, 1994; Dijkwel et al. 2000). Unlike the situation in yeast, there is an array of data suggesting that the activity of genes in many higher eukaryotic organisms can determine the time during S phase when those genes are replicated (e.g., Goldman et al. 1984; Taljanidisz et al. 1989; for review, see Gilbert 2002). Indeed, in a recent genome-wide study performed on cultured Drosophila cells, it was found that >85% of actively expressed genes replicate in the early S period (Schubeler et al. 2002). Thus, the varying developmental constraints imposed on somatic cells in higher eukaryotes apparently involve an interplay between transcription and origin regulation that is not evident in S. cerevisiae.
We are interested in the regulation of chromosome replication in mammalian cells. The model system we and others have studied is a well characterized 240-kb domain encompassing the dihydrofolate reductase (DHFR) gene in Chinese hamster cells (Milbrandt et al. 1981; Looney and Hamlin 1987). Utilizing a variety of replicon mapping techniques, we have shown that replication initiates at any of a large number of inefficient sites scattered throughout the 55-kb spacer between the convergently transcribed DHFR and 2BE2121 genes (Vaughn et al. 1990; Dijkwel et al. 1994, 2002; Dijkwel and Hamlin 1995; Wang et al. 1998). Within this broad zone, two regions (termed ori-β/ori-β′ and ori-γ) are preferred (Anachkova and Hamlin 1989; Pelizon et al. 1996; Kobayashi et al. 1998; Leu and Hamlin 1989). However, using a homologous recombination strategy, we were able to delete the most active initiation sites or, indeed, the central 45-kb region that encompasses >90% of the usual start sites, without a noticeable effect on the time of replication of the locus as a whole (Kalejta et al. 1998; Mesner et al. 2003). Maintenance of replication timing in each case was achieved by an increase in the efficiency of initiation at sites within the truncated spacer that remained.
Therefore, no unique cis-regulatory elements appear to reside within the most active part of the origin itself. To explain these data, we have proposed a simple model in which virtually any sequence in higher eukaryotic genomes can serve as a template for initiation of replication, provided that it is not actively transcribed or otherwise negatively affected by chromatin architecture or other structural constraints. Support for this model was obtained in a recent study in which targeted DHFR promoter deletions that eliminated transcription were shown to allow replication to initiate in the body of the inactive gene (Saha et al. 2004), something that has never been observed in its wild-type, transcriptionally active counterpart (Vaughn et al. 1990; Dijkwel et al. 1994). Thus, as predicted by the model, inhibition of transcription through a template can activate it for initiation of replication. Conversely, the model predicts that transcription through an active origin/initiation zone should inactivate origin function.
In the present study, we tested this latter prediction by examining the relationship between transcription and replication initiation in the DHFR-deficient variant, DR8 (Jin et al. 1995). In this cell line, a 14-kb deletion has removed the 3′ end of the DHFR gene as well as ∼7-kb of the intergenic spacer, and the downstream ori-β/ori-β′ region is completely inactivated (Kalejta et al. 1998). We performed an extensive series of genetic alterations on the endogenous DHFR locus to determine the element(s) in the DR8 deletion whose loss renders the downstream origin inactive. The results of our studies suggest that the normal 3′ ends of genes contain specific signals that not only process the ends of the transcripts efficiently, but also prevent invasion of intergenic origins by the transcription machinery. The results of this and a previous study on the DHFR promoter (Saha et al. 2004) show that well defined elements at the 5′ and 3′ ends of genes are required to set boundaries between transcription units and origins of replication.
Results
The DHFR origin in the DR8 deletion variant is inactivated in early S phase, but can be reactivated by restoration of the wild-type sequence arrangement
The early-firing replication initiation zone (origin) in the wild-type Chinese hamster DHFR domain corresponds to the 55-kb spacer between the DHFR and 2BE2121 genes (Fig. 1B; Dijkwel et al. 1994; Heintz and Hamlin 1982; Leu and Hamlin 1989). The UA21 cell line contains a single wild-type copy of the DHFR domain resulting from a radiation-induced deletion encompassing the second locus (Urlaub et al. 1983). The DHFR-deficient DR8 cell line was derived from UA21 by α-particle irradiation (Jin et al. 1995), and has sustained a 14-kb deletion encompassing ∼4 kb of intron 5 (in5), exon 6 (ex6), the 3′ UTR, and ∼7 kb of the intergenic spacer (Fig. 1C, deletions indicated in yellow; Jin et al. 1995; Kalejta et al. 1998).
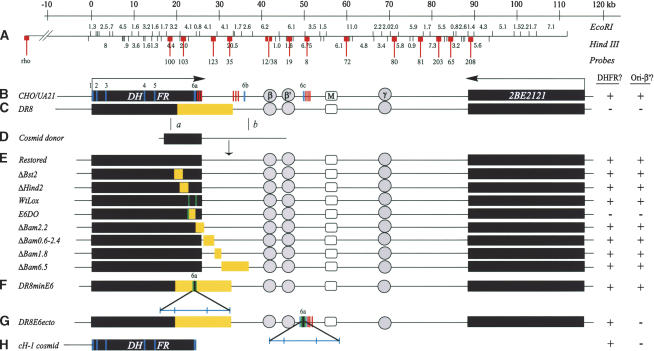
Figure 1.
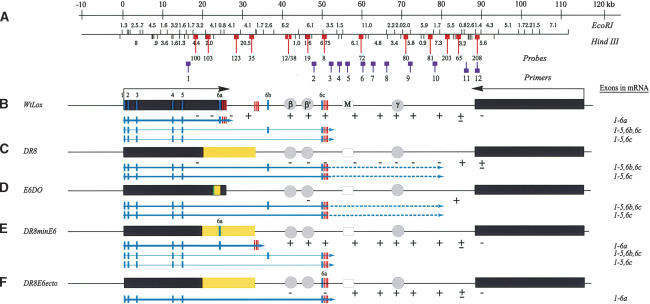
The DHFR locus in wild-type cells, DR8, and engineered variants. The columns to the right indicate whether an active dhfr enzyme is made in each cell line (DHFR?) and whether replication initiation is detected at a diagnostic locus (ori-β′). (A) Scale and EcoRI and HindIII restriction maps of the wild-type locus in CHO and UA21 cells. Probes used to analyze 2D gels of replication intermediates in various cell lines are numbered and indicated with lollipops (see text). (B) Map of the wild-type locus in diploid CHO and haploid UA21 cells, where black rectangles correspond to the DHFR and 2BE2121 genes and arrows indicate the direction of transcription. DHFR exons are indicated with vertical cross-hatches (note the alternate exons 6b and 6c) and polyA signals with a cluster of three vertical bars. The preferred replication initiation sites in the intergenic spacer (ori-β, ori-β′, and ori-γ) are shown in circles, and a centered matrix attachment site is indicated with a square (M). (C) Map of the DHFR locus in the DR8 variant, where the light-yellow rectangle indicates the region deleted. (D) The KZ381 donor cosmid that is used to restore the locus and/or to introduce targeted deletions by homologous recombination events near a and b. (E). Maps of the various targeted deletions constructed via homologous recombination, where yellow rectangles indicate deletions (see text). (F,G) Variants containing the minimal 168-bp 3′ element inserted either at the deletion junction in DR8 or at the site of the alternative ex6c. Brackets indicate (from left to right) 35 bp lying upstream from the splice site; the 78-bp coding region and stop codon; 55 bp of the 3′ UTR. (H) The cosmid, cH-1, which encodes a functional DHFR gene, and which was transfected into DR8 to test the ability of the dhfr enzyme to restore origin activity. In the table on the right, + and – signs indicate whether the particular cell line expressed a functional dhfr enzyme and whether the ori-β′ region was active.
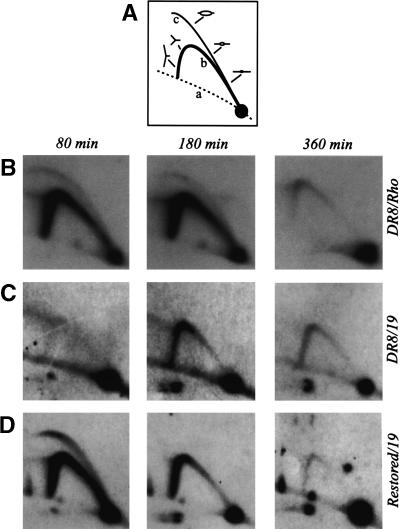
To investigate the properties of the DHFR origin in the DR8 cell line, replication intermediates were isolated 80, 160, and 360 min after release from a G1/S-phase block (i.e., in early, mid-, or late S phase) and were analyzed by a two-dimensional (2D) gel replicon mapping technique (Fig. 2A and legend for principle of the method; Brewer and Fangman 1987). To display the classic pattern exhibited by an early-firing initiation zone such as DHFR, the blot was first hybridized with a radioactive probe specific for the rhodopsin origin, which serves as an internal control on cell synchrony and sample preparation. As shown in Figure 2B, the 80-min sample displays a composite image consisting of a complete bubble arc and a single fork arc. This pattern arises because fragments lying in early-firing initiation zones sometimes serve as templates for internal initiations, but are often replicated passively by forks emanating from starts occurring in nearby fragments in the same zone (Vaughn et al. 1990). The bubble arcs begin to disappear after 180 min, but single fork arcs persist until ∼360 min, since neither the rhodopsin nor DHFR origin fires in every cell cycle, and inactive copies must wait to be replicated by forks arriving from upstream or downstream origins (Dijkwel and Hamlin 1992; Dijkwel et al. 1994).
Figure 2.
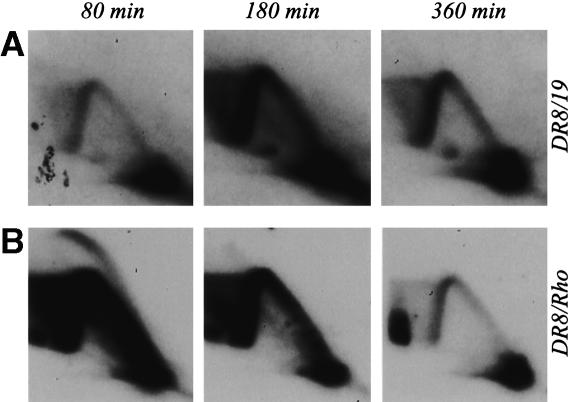
The ori-β′ locus in the DR8 cell line is inactivated in early S phase. (A) Principle of the neutral/neutral 2D gel replicon mapping method (Brewer and Fangman 1987), where curve a denotes the arc of linear fragments, curve b the single fork arc, and curve c the bubble arc. Cells were synchronized at the G1/S boundary by release from an early G1 block into mimosine for 12 h. After drug removal and return to drug-free medium, samples were taken in early, mid-, and late S phase (80, 180, and 360 min, respectively), replication intermediates were prepared using EcoRI to digest the DNA, and the digests were separated on a 2D agarose gel. The DNA was then transferred to a membrane and hybridized with appropriate probes. (B) Replication pattern of the early-firing rhodopsin control origin in DR8 cells. (C) The transfer in B was stripped and rehybridized with a probe specific for the ori-β′ locus. (D) Replication pattern of the cell line obtained by restoring the deletion in DR8 by homologous recombination with cosmid KZ381.
The pattern displayed by the DHFR initiation zone in DR8 is very different. When the transfer shown in Figure 2B was stripped and rehybridized with a probe specific for the ori-β′ locus (probe 19), virtually no intermediates could be detected at 80 min, and only single fork arcs were observed at 180 and 360 min (Fig. 2C). Thus, it appears that the deletion encompassing the 3′ end of the DHFR gene in this cell line has inactivated the downstream origin in early S phase. Note that, although the ori-β′ region is somewhat less active than ori-β when measured by quantitative assays (Kobayashi et al. 1998; Dijkwel et al. 2002), the patterns obtained for the adjacent 6.1- and 6.2-kb fragments that harbor them are indistinguishable on 2D gels (Dijkwel et al. 1994; Dijkwel and Hamlin 1995). Since probe 19 is a larger and more reliable single-copy probe than the 12/38 combination that illuminates ori-β, we used it in most of the studies presented here.
To ensure that the deletion depicted in Figure 1C is responsible for the loss of early-firing origin activity in DR8 (as opposed to an additional α-particle-induced anonymous deletion elsewhere in the genome), we restored the locus to the wild-type arrangement by a homologous recombination approach. DR8 cells were transfected with a cosmid that provided ∼3 kb of homologous overlap with the gene, the missing part of the gene, and ∼6 kb of downstream sequence from the intergenic region (Fig. 1C–E). After selection of DHFR-proficient clones on minimal medium (which does not supply the thymidine, hypoxanthine, and glycine required by DR8; Urlaub and Chasin 1980), faithful restoration of the gene by crossovers near a and b was confirmed by Southern blotting (Fig. 3 and legend). When the ori-β′ locus in one of the resulting restored cell lines was analyzed on a 2D gel, it displayed a pattern very similar to that of the early-firing rhodopsin origin (Fig. 2D). Thus, the deletion illustrated in Figure 1C is solely responsible for the aberrant initiation pattern displayed by DR8.
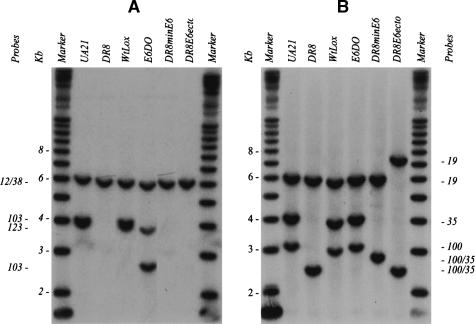
Figure 3.
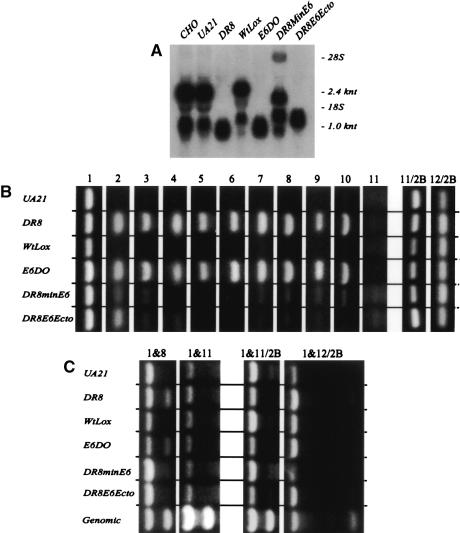
Southern analysis of diagnostic fragments to confirm the success of targeted deletions and insertions into the DHFR locus. Representative Southern transfers containing EcoRI digests of genomic DNA isolated from the indicated cell lines. Transfers were hybridized with radioactive probe mixtures that illuminate diagnostic fragments for each of the targeted modifications, and each fragment is indicated with the probe that detects it (see Fig. 1A for probe positions and the EcoRI restriction map). (A) In the wild-type hemizygote, UA21, a mixture of probes 12/38, 123, and 103 detects a 6.2-kb EcoRI fragment and a doublet at 4.1 kb. In DR8, only the 6.2-kb EcoRI fragment remains. In WtLox, the pattern is indistinguishable from that of UA21, since the two Lox sites add only ∼60 bp of sequence to the wild-type arrangement. In E6DO, probes 12/38 and 123 detect their cognate wild-type fragments at 6.2 and 4.1 kb, but probe 103 detects a variant of ∼2.6 kb predicted by the deletion of the 1.4 kb encompassing part of intron 5 and exon six. DR8minE6 lacks the wild-type 4.1-kb EcoRI fragments recognized by probes 103 and 123. (B) Additional diagnostic EcoRI fragments were hybridized with probes 19, 35, and 100, and yielded the correct spectrum of fragments in each case (see map in Fig. 1A).
Scanning deletion analysis of the 14-kb region missing from DR8
To delineate the sequences in the DR8 deletion whose loss is responsible for inactivating the downstream origin, a nested series of more circumscribed deletions was engineered into the DHFR locus by the same recombination strategy used to restore DR8 to the wild-type arrangement (Fig. 1C–E). The series of deletions indicated in Figure 1E was first engineered into either cosmid KZ381 or a subcloned 21-kb fragment, as described in Materials and Methods. Each donor construct was then transfected into DR8 cells, followed by selection for restoration of dhfr activity by propagation on minimal medium.
With the exception of the sequences removed in the E6DO and ΔBam2.2 variants, none of the deleted sequences outlined in Figure 1E was calculated to compromise dhfr activity, since each normally resides within the fifth intron of the gene or downstream from the 3′ processing signals in the intergenic spacer. To construct the E6DO cell line, which lacks the sixth exon but retains part of the 3′ untranslated region and polyA signals, a cell line was constructed in which this sequence was flanked by lox sites (WtLox). The dhfr-deficient E6DO deletion variant was then derived by introduction of a plasmid encoding the cre recombinase. In the case of the ΔBam2.2 deletion, which encompasses the normal DHFR polyA signals, alternate signals lying further downstream from the deletion were used (see Fig. 1B), and this variant survives on minimal medium. With all cell lines, Southern blotting and hybridization with diagnostic probes were used to confirm that clean homologous recombination events near a and b had occurred, concomitantly introducing the targeted deletion (see Figs. 1, 3 and their legends for details of the analysis).
Identification of a 168-bp element whose loss uniquely elicits the DR8 phenotype
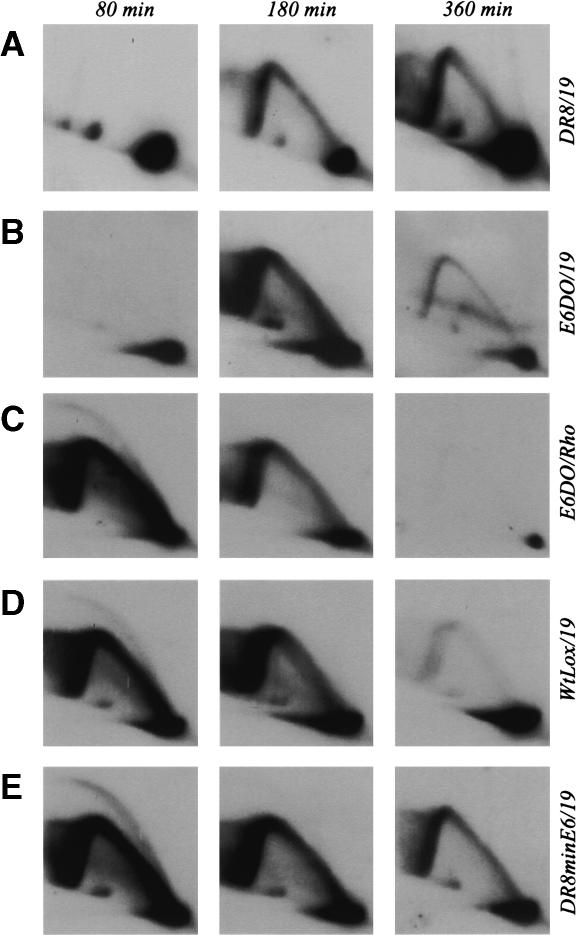
Replication intermediates were prepared from each of the cell lines shown in Figure 1E, and the replication pattern of the ori-β′ locus (one of the most active initiation sites in the spacer) was examined with probe 19. With one exception, none of the deletion variants summarized in Figure 1E showed a pattern of intermediates that differed significantly from that of wild-type UA21 or the restored control pictured in Figure 2 (data not shown). The exception was the E6DO variant, which lacks an ∼1.4-kb StuI/KpnI fragment encompassing a portion of the fifth intron, the sixth exon, and part of the 3′ UTR of the DHFR gene. The replication pattern for E6DO is essentially indistinguishable from that of DR8 itself (Fig. 4A,B; note that the 180-min sample from DR8 is somewhat underexposed relative to the 80- and 360-min time points, while that of E6DO at 180 min is somewhat overexposed). When the E6DO transfer was stripped and rehybridized with a probe for the rhodopsin origin, the classic early-firing pattern was displayed (Fig. 4C). Thus, the synchronizing regimen and sample preparation for E6DO were clearly effective. Importantly, the WtLox cell line, which contains the two lox sites that facilitated the deletion in E6DO, displays the wild-type pattern characteristic of this early-firing origin (Fig. 4D). Thus, we conclude that the 1.4-kb deletion in E6DO, but no other sequences encompassed by the DR8 deletion, must contain an element or elements responsible for maintaining the activity of the downstream origin in the wild-type locus.
Figure 4.
Effects of deletion or restoration of the minimal 3′ element on origin activity at the ori-β′ locus. The indicated cell lines were synchronized as described in Materials and Methods and sampled 80, 180, and 360 min after release from mimosine. Replication intermediates were prepared using EcoRI to digest the DNA, and were separated on a 2D gel. They were then transferred to a membrane and hybridized with the indicated probes.
Targeted restoration of a 168-bp element in the DR8minE6 cell line reactivates the downstream origin in early S phase
To further delineate the elements in the 1.4-kb E6DO deletion that are responsible for maintaining origin activity, lox-flanked subfragments of the 1.4-kb fragment were reintroduced into the single lox site in E6DO by cre-mediated insertion (Fig. 1E). We first tested directly whether reintroduction of the minimal sequence that would reconstitute gene function would also restore origin activity. We found that a 168-bp fragment containing 36 bp of the fifth intron, the 78-bp sixth exon, and 54 bp of the 3′ UTR is able to reconstitute gene activity to E6DO and concomitantly restores the pattern of replication initiation to that of wild-type cells: A complete bubble arc and strong fork arc were detected 80 min after release from mimosine, and the locus was largely finished replicating by 360 min (data not shown).
To test whether this sequence is sufficient to restore origin activity to the DR8 cell line, in which the deletion is much larger (Fig. 1C), a lox site was inserted at the DR8 deletion junction, followed by introduction of the lox-flanked 168-bp element by cre-mediated insertion. Selection on minimal medium identified a successful recombinant, which was designated DR8minE6. As shown in Figure 4E, the replication pattern of DR8minE6 is indistinguishable from that of WtLox and also from the rhodopsin internal control (data not shown). Thus, we conclude that this 168-bp element is both necessary and sufficient to restore origin activity to DR8 and to E6DO.
The loss of dhfr enzyme activity per se is not responsible for down-regulation of origin activity in DR8
In only two of the variants that we analyzed was the origin inactivated (DR8 and E6DO). However, these were also the only variants that did not synthesize a functional dhfr enzyme (i.e., could not be selected or propagated on minimal medium). It was therefore important to show that the loss of enzyme activity per se was not responsible for inactivity of the DHFR origin. To test this proposal, a cosmid that encodes a functional dhfr enzyme (cH1, Fig. 1H; Milbrandt et al. 1983) was transfected into DR8, and survivors were selected on minimal medium. A stable cell line was isolated that contains a single copy of the donor cosmid at an anonymous integration site in another chromosome, and which expresses an active enzyme by virtue of growth on minimal medium. When 2D gel analysis was performed on the ori-β′ locus in this cell line (Fig. 5A), the pattern of replication intermediates was essentially the same as that observed for DR8 itself. The rhodopsin control origin in the same cell line still displays the early-firing properties characteristic of wild-type cells (Fig. 5B). Therefore, we conclude that the dhfr enzyme itself cannot restore early-firing activity to the DHFR origin in DR8 cells.
Figure 5.
Restoration of dhfr enzyme activity in trans does not restore activity in the DHFR origin in the DR8 cell line. The cosmid cH1 was transfected into DR8 cells, and a cell line containing a single insert at an anonymous location in the genome was isolated. Synchronized cells were then subjected to 2D gel analysis, and the transfer was hybridized first with probe 19 for the ori-β′ locus and subsequently with the rhodopsin probe.
The pattern of transcription in DR8 and selected engineered variants suggests that read-through transcription down-regulates origin activity
The deletion analysis summarized in Figure 1E implicates the 3′ end of the DHFR gene and its processing signals as necessary for activity of the downstream origin. This suggested that read-through transcription resulting from the lack of termination signals in DR8 and E6DO might repress replication initiation in the region lying between the 3′ end of the gene and ori-β′ (and possibly beyond). In the case of DR8, transcription complexes would have to travel >13 kb beyond the deletion junction before crossing the ori-β′ locus (Fig. 1A,B). To determine whether this actually occurs, we characterized both the steady-state mRNAs and the primary transcripts in wild-type and variant cell lines.
As shown in the Northern blot analysis in Figure 6A, the major processed messages in CHO, UA21, and the restored cell line, WtLox, are 2.4 and 1.0 kDa in length. These two species differ in the position of the polyA site used in each case (Carothers et al. 1983). Surprisingly, however, the major processed mRNAs in DR8 and E6DO are somewhat smaller than the smallest wild-type message. This finding suggested either that transcription terminates earlier than in the wild-type locus or that other aberrant processing events have occurred to yield these novel species.
Figure 6.
RNA polymerase invades the former intergenic region in DR8 cells. (A) Total RNA was isolated from the indicated cell lines, separated on an agarose gel, transferred to a membrane, and hybridized with DHFR exon 1–5. The positions of 28S and 18S rRNAs were assessed from the ethidium bromide staining pattern. (B) RT–PCR analysis of primary transcription products from the indicated cell lines. First-strand cDNA was synthesized from equal amounts of total RNA, using selected oligonucleotides and reverse transcriptase; this was followed by PCR amplification of the products with a second set of primers for 33, 36, 39, 42, or 45 cycles. Primer positions are shown in Figure 7A; products ranged in size from 800 to 2000 bp. The number of cycles selected for presentation for primer sets 2–12 were chosen to normalize the signal in DR8 to that of primer set 1 (see text). Primary data for all cycles available on request. (C) The indicated primer sets were mixed with products of reverse transcriptase reactions, and PCR was carried out for 33–45 cycles to determine the amounts of primary transcripts present at each location relative to the body of the DHFR gene (see Fig. 7A for primer positions).
To understand the 3′ processing events that led to the mRNAs detected in DR8 and E6DO, 3′ RACE was used to amplify and clone the cDNAs for the predominant message species in selected cell lines. The results of sequence analysis of these clones are summarized in Figure 7B–E, where the solid portions of the arrows below each functional map indicate the mRNA species detected by 3′ RACE, and the thickness of the arrows indicates relative abundance (see below for significance of the dashed portions in DR8 and E6DO). The rectangles on the arrows signify the exons included in the final messages, and the set of three hatch marks represents the positions of polyA signals used for termination in each case.
Figure 7.
Transcriptional activity through the template precludes initiation of replication in the DHFR locus. (A) The DHFR locus, showing EcoRI and HindIII maps, as well as relevant probes and primers used in 2D gel and RT–PCR experiments (see text). (B–E) Functional maps of the locus in the different engineered variants used in this study, where light-gray vertical stripes signify exons (note the two alternative exons, 6b and 6c, which are used to a limited extent by wild-type cells but predominantly by DR8 and E6DO). The clusters of three stripes signify polyA addition sites. The thickness of the solid arrows below each map indicates the relative abundance of each transcript; the prevalent processed messages were determined by 3′ RACE, whereas the less abundant species were determined by RT–PCR; the dashed lines in DR8 and E6DO indicate additional downstream sequences transcribed beyond the 3′ processing signals, as determined by RT–PCR (see text). The exons that are included in the final processed transcripts are also summarized in the column to the left. The + and – signs below the maps indicate whether or not that region is used as a template for initiation of replication, as determined by 2D gel analysis.
The predominant message species in WtLox contains the six natural exons and is polyadenylated at the three normal sites just 3′ to the sixth exon (ex6a; Fig. 7B). However, in both DR8 and E6DO, two processed variants were detected that terminate near a novel exon (ex6c; Fig. 7B,C). (As we will show below, these correspond to two minor variants detected by RT–PCR in cells with wild-type arrangements.) The processed messages are smaller than in wild type because the 3′ UTR in E6DO and DR8 is shorter. In DR8minE6, the two major transcripts contain the normal six exons and use cryptic polyA signals at sites near position 35 on the wild-type map, but are somewhat shorter than in WtLox because of their smaller 3′ UTRs (Fig. 7D; 3′ RACE analysis not shown). Not surprisingly, DR8minE6 synthesizes the same two minor transcripts as WtLox, DR8, and E6DO, since these transcripts do not use the 3′ processing signals deleted in DR8 (results of 3′ RACE, data not shown; Fig. 7E).
The DHFR initiation zone is not defined by the position of mRNA 3′ processing events
In cell lines with a wild-type DHFR gene (CHO, UA21, WtLox), the 3′ ends of the major transcription products are processed near map position 26 (Fig. 7B), and replication initiation sites can be detected ∼5 kb downstream (Dijkwel et al. 2002). The same is true for DR8minE6, although the 3′ ends now lie near position 35 on the wild-type map. The 3′ RACE data summarized in Figure 7C and D indicate that in DR8 and E6DO, DHFR messages encompass both ori-β and ori-β′ and are processed near position 50. Thus, the inactivity of these initiation sites could result from read-through transcription (or to some other inhibitory chromosomal ambience related to gene activity).
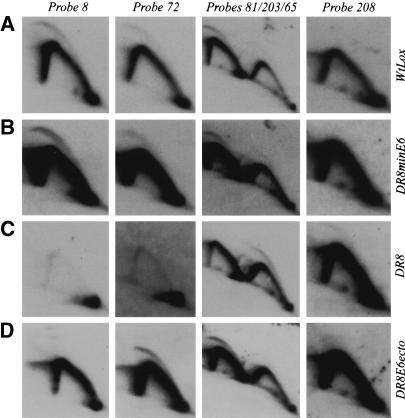
If the DHFR 3′ processing signals normally set the 5′ boundary of the DHFR initiation zone in wild-type cells, then one would predict that replication initiation should be detectable ∼5 kb downstream from the novel processing signals used in DR8 and E6DO. Why, then, are ori-β and ori-β′ so late-replicating, since the nearest initiation sites would be predicted to lie near map position 56, which is only 14 kb and 8 kb away in DR8 and E6DO, respectively? To address this issue, we characterized the distribution of initiation sites in early S phase in WtLox, DR8, E6DO, and DR8minE6 (the relevant hybridization probes are indicated as lollipops on the restriction map in Fig. 7A). The primary data for selected fragments are presented in Figure 8, and data for all the fragments examined are summarized below the functional maps in Figure 7B–E (replication initiation activity indicated with + and – signs; note that only two regions were examined in E6DO).
Figure 8.
Replication patterns suggest that transcription defines the boundaries of the DHFR origin. The indicated cell lines were synchronized as described, and replication intermediates were prepared 80 and 180 min after release from mimosine and were digested with HindIII (only the 80-min samples are shown). After separation on 2D gels and transfer to a membrane, intermediates were hybridized with the indicated probes.
In WtLox, the bubble arcs that indicate internal initiation sites were detected in all of the fragments lying between map positions 32 and 85, the faintest signal arising from the fragment lying just 3′ to the 2BE2121 gene (Figs. 7B, 8A; cf. the bubble arcs in neighboring fragments detected with the mixture of probes 81 and 203 + 65 in Fig. 8A). In DR8minE6, bubbles were also observed in all of the fragments tested, beginning with a region lying ∼6 kb to the right of the inserted 168-bp fragment that contains the 3′ processing elements (data not shown) and extending to map position 85 (probe 65, Figs. 7E, 8B). In contrast, in DR8, initiation signals could not be detected in any of the former intergenic fragments (shown for probes 8, 72, and 81 in Fig. 8C and summarized in Fig. 7C). There was one important exception: The mixture of probes 81 and 203 + 65 detects a relatively pronounced bubble arc in the 3.2-kb HindIII fragment adjacent to the 2BE2121 gene in DR8, whereas no bubbles are observed in the adjacent upstream 7.3-kb HindIII fragment (Fig. 8C). In effect, in DR8 (and by extrapolation, E6DO), the former 55-kb initiation zone appears to have been compressed into a very short region at the extreme 3′ end of the former intergenic spacer, which is normally minimally active as a template for initiation (Fig. 8A, probes 65 and 208; Dijkwel et al. 2002).
Thus, initiation is clearly suppressed within the template for the major elongated messages in DR8 and E6DO (i.e., up to map position 50), but also within the 30-kb region lying downstream from the 3′ processing signals for these messages. A corollary is that the major processing signals present in the wild-type DHFR gene (6a in Fig. 7B) somehow buffer the downstream template for initiation within a distance of ∼5 kb, whereas those used to process the major messages in DR8 and E6DO (6c in Fig. 7B) do not.
Primary transcripts in DR8 and E6DO proceed well beyond their 3′ processing signals
These findings suggested that transcription might be proceeding well beyond the major 3′ processing signals in DR8 and E6DO. To address this possibility, we used RT–PCR to compare the levels of primary unprocessed RNA species at various positions along the template in selected cell lines (see Materials and Methods for details). The same amount of total RNA was used in each case, and primer sets were distributed as shown in Figure 7A. Note that sets 1–11/D are designed to amplify products synthesized from the DHFR template, while sets 11/2B and 12/2B recognize products from the 2BE2121 template. Samples were taken after varying numbers of amplification cycles (33–45), and the products from a given cycle number were run together on an agarose gel. To aid comparison, the cycle numbers chosen for display in Figure 6B were those in which the amount of product detected in DR8 and E6DO with primer sets 2–10 were approximately equal to that obtained with primer set 1 from the DHFR gene in the same sample. With primer set 11, the level of transcripts was too low in any of the samples to detect a reproducible signal (also see below). All of the reactions shown were well below saturation levels (primary data are available on request).
As expected, the levels of primary transcripts in the body of the DHFR gene appear to be approximately equal among all six cell lines when equal amounts of total RNA are subjected to the same number of amplification cycles with primer set 1 (Fig. 6B). However, in the region encompassed by primer sets 2–10, DR8 and E6DO display significantly greater relative numbers of primary transcripts than any of the other cell lines tested. Therefore, although the messages are processed and terminated efficiently at map position 50 between primer sets 2 and 3 in DR8 and E6DO (based on steady-state message levels and 3′ RACE data), the polymerase continues to transcribe efficiently for another 30 kb to the position of primer set 10 (Fig. 7C,D). Importantly, the region encompassed by primer sets 2–10 is very active as a template for initiation of replication in wild-type cells, but appears to be totally inactive for initiation in DR8 (Fig. 7). Primer set 11 lies very near the position of probe 65 (Fig. 7A), which marks the 3′ boundary of the initiation zone in WtLox and the beginning of the small initiation zone in the DR8 cell line. To determine more precisely how the relative levels of primary transcription products in this region compare, PCR reactions were carried out with a mixture of primer sets 1 + 8 or 1 + 11 (Fig. 6C). Samples of genomic DNA were run concurrently to standardize relative primer activities. In agreement with the data in Figure 7B, significant numbers of primary transcripts are detected in DR8 and E6DO with primer set 8 (compare ratio to that of genomic DNA), but essentially no product is detected with primer set 11. Primer sets 11/2B or 12/2B for transcription products from the 2BE2121 template detect a low level of products in all cell lines tested after 45 cycles (Fig. 6B), but when compared directly to DHFR gene transcripts in a mixture with primer set 1, detect only very low levels of primary transcription products, in agreement with earlier studies (Foreman and Hamlin 1989).
Note that in the DR8minE6 cell line, a low but diminishing level of primary transcripts is detected with primer sets 2 and 3, even though the wild-type 168-bp processing element has been reintroduced and a wild-type message is made. This undoubtedly results from the original DR8 deletion, which brings primer sets 2 and 3 ∼10 kb closer to the 3′ processing element, 6a (Fig. 7E), and is consistent with detectable but diminishing levels of primary transcripts between map positions 26 and ∼45 in wild-type cells (A. Pemov and L.D. Mesner, unpubl.). Thus, RNA polymerase appears to cease transcribing the template at random positions well beyond the 3′ processing and polyA addition signals (region indicated with dashed lines in Fig. 7).
The wild-type 3′ processing signals, but not the alternate ones used in DR8 and E6DO, can restore initiation activity 5 kb downstream
In UA21 and WtLox, primary transcripts are efficiently processed at the 3′ element, 6a, and replication bubbles are detected ∼5 kb downstream (Fig. 7). In DR8 and E6DO, primary transcripts are processed efficiently at the novel element, 6c, yet initiation cannot be detected anywhere within the adjacent 30-kb region downstream. Thus, the wild-type element, 6a, appears to be unique in its ability to buffer the immediate downstream region for initiation. To test this directly, the 168-bp element encompassing 6a was inserted into element 6c upstream from the functional polyA addition sites in DR8 to give rise to the DR8E6ecto cell line (Fig. 7F). As shown in Figure 6A, the steady-state message levels in DR8E6ecto are similar to those of DR8 and E6DO and only somewhat less than in the wild-type cell lines. As shown in Figure 6B, the primary transcript levels in DR8E6ecto are similar to those in DR8 and E6DO in the fifth intron of the DHFR gene (primer set 1). However, as with DR8, they are higher than UA21 and WtLox at the position of primer set 2, which is now located within the body of the extended DHFR gene (Fig. 7F). The 3′ RACE analysis showed that the major message species is now processed efficiently at the ectopically situated element 6a, and the same polyA sites are used as in DR8 and E6DO.
When the pattern of replication in DR8E6ecto was examined by the 2D gel replicon mapping method (Fig. 8D), the results confirmed that element 6a, indeed, either acts as a boundary for the origin or plays some other regulatory role in effecting or allowing replication to initiate in the immediate downstream region: As summarized in Figure 7F, initiations can be detected in the 6.1-kb HindIII fragment lying ∼5 kb downstream from the newly positioned element 6a, but not in the 6.75-kb HindIII fragment encompassing the element itself (probe 8) or the fragment immediately to its 5′ side (detected with probes 12 + 38).
Discussion
Since the seminal autoradiographic studies of Huberman and Riggs (1968) on replicating mammalian DNA, the challenge has been to uncover the nature of the bidirectional origins illuminated by those studies and to understand how their activities are regulated. Over the years, it has come to be appreciated that the number of active origins in higher eukaryotic cells can be modulated during development. For example, most expressed genes in a particular somatic cell type are replicated early in the S period (Taljanidisz et al. 1989; Goldsmith et al. 1993), whereas the same genes can become late-replicating in cell types in which they are not expressed, presumably via the inactivation of local origins (Hatton et al. 1988; Leffak and James 1989; Kitsberg et al. 1993; Simon et al. 2001). In addition, in both Drosophila and Xenopus, initiation sites are distributed at very frequent intervals throughout the genome in early cleavage embryos (Blumenthal et al. 1974; Hyrien and Mechali 1993), but are confined to the spacers between rDNA (and presumably other) genes once transcription sets in at the mid-blastula transition (Hyrien et al. 1995). The question, then, is how this relatively plastic modulation of origin activity in different cellular contexts is controlled. Recent work suggests that simple variations of the replicon paradigm established for bacterial genomes may be inadequate to explain regulation of initiation in higher eukaryotes (for review, see Gilbert 2002).
For example, modern replicon mapping techniques have shown that, while some origins may correspond to well defined genetic elements (e.g., lamin B2 [Giacca et al. 1994]; human β-globin [Aladjem et al. 1998b]), most origins correspond to broad zones of inefficient initiation sites scattered throughout intergenic regions (e.g., the α-polymerase locus in Drosophila [Shinomiya and Ina 1994], the DHFR and rhodopsin domains in Chinese hamster cells [Vaughn et al. 1990; Dijkwel et al. 1994; Dijkwel et al. 2002], and the rDNA locus in human cells [Little et al. 1993]). Since some sites are definitely preferred (e.g., the ori-β, ori-β′, and ori-γ regions in the DHFR origin) (Leu and Hamlin 1989; Kobayashi et al. 1998; Dijkwel et al. 2002), the possibility remained that at least some of these might correspond to classic genetic replicators. Indeed, when a small fragment containing the ori-β region was tested for its ability to initiate at ectopic chromosomal positions, it was found to be active, and small deletions resulted in an apparent reduction in its activity (Altman and Fanning 2001).
However, in studies on the native DHFR locus, deletion of the central 45 kb of the DHFR origin (which encompasses >95% of the usual initiation sites, including ori-β, ori-β′, and ori-γ) had little effect on initiation in the remainder of the spacer nor on the time of replication of the DHFR locus as a whole. Thus, no critical, nonredundant genetic elements reside within the most active portion of this complex origin. Given the clear relationship between gene activity and replication timing illuminated by older studies, we therefore explored the possibility that regulatory elements related to transcription might modulate DHFR origin activity. Indeed, promoter deletions that completely eliminated transcription through the gene had two important consequences: (1) The broad zone of initiation sites that characterizes the wild-type locus expanded to include the body of the inactivated DHFR gene, suggesting that potential sites are distributed throughout the genome; and (2) the overall efficiency of initiation in the expanded origin was significantly reduced, so that the entire S phase was required to replicate what is normally an early-replicating locus (Saha et al. 2004). Based on these data, we suggest that, in effect, active transcription through the body of the DHFR gene sets the upstream boundary of the DHFR origin, but also generally enhances its activity.
In this and a previous study, we showed that the 14-kb deletion in DR8 completely inactivates the ori-β and ori-β′ loci in early S phase (Fig. 2; Kalejta et al. 1998). Scanning deletion analysis eliminated the possibility that a replicator resides within the 7 kb of the intergenic spacer that was deleted in DR8. Rather, we identified a 168-bp element from the 3′ end of the gene whose targeted loss from the wild-type locus in the E6DO cell line inactivates the origin and which fully restores origin activity when reintroduced into DR8 in DR8minE6. This element contains the in5/ex6 boundary of the DHFR gene, part of the 3′ UTR, but not the polyA processing signals. A combination of Northern blot and 3′ RACE analyses revealed that DR8 and E6DO synthesize almost wild-type levels of messages that are terminated efficiently by an alternative exon and adjacent polyA signals positioned near the center of the former intergenic region (ex6c in Fig. 7B). However, RT–PCR analysis of RNA in DR8 and E6DO showed that primary transcripts proceed almost 35 kb beyond ex6c and its neighboring polyA sites, up to but not including the first fragment that displays replication bubbles on 2D gels (Fig. 7C). This is in general agreement with results obtained with wild-type cells and DR8minE6, in which replication initiation can be detected within ∼5 kb of ex6a and the nearby polyA signals. We conclude that the signals defining the ends of most DHFR processed messages at map position 25 in wild-type cells are able to buffer the immediate downstream region for replication initiation, presumably by effecting efficient transcription termination, whereas the signals used by DR8 and E6DO at map position 50 are not.
This conclusion was confirmed by inserting the 168-bp element containing the in5/ex6a boundary and part of the 3′ UTR into the position of alternative in5/ex6c (map position 50) to produce DR8E6ecto. RT–PCR analysis showed that primary transcripts were now efficiently terminated just downstream and replication initiation could be detected within ∼5 kb (Fig. 7F). Importantly, the polyA signals used in the DR8E6ecto processed messages are the same ones used by DR8 and E6DO. Therefore, it must be the in5/ex6a boundary, ex6a, and/or adjacent sequences in the 3′ UTR, but not the particular polyA signals at position 25, which are responsible for efficient termination of transcription and origin boundary definition.
The mechanisms by which RNA polymerase II (pol II) transcription is terminated in higher eukaryotic cells are complex and are not necessarily the same for every gene (for comprehensive reviews, see Hirose and Manley 2000; Proudfoot and O'Sullivan 2002). However, the current general models suggest that transcription is terminated by an initial combinatorial sensing of both the terminal intron/exon junction, a neighboring polyA signal(s), and, in some instances at least, downstream pause sites (Hirose and Manley 2000; Proudfoot and O'Sullivan 2002). It has also been suggested that cleavage at the polyA site may alter the configuration of the polymerase in such a way as to effect its release from the template 700–1000 nt downstream (Proudfoot and O'Sullivan 2002). There is substantial evidence that the proteins involved in both splicing and polyadenylation actually load onto the RNA polymerase via the C-terminal domain (CTD) at the promoter of the gene, thereby coordinating all of the reactions necessary to synthesize and process primary transcripts.
It is also clear from our data on the DHFR gene that both terminal exon definition and efficient polyadenylation are required for termination of transcription. However, there appear to be more constraints on the nature of the sequences that define the terminal exon than on the poly A sites. This conclusion derives from the fact that alternative in5/ex6c, in combination with the neighboring polyA sites at position 50, is very efficient at 3′ end processing, but is very inefficient at evoking transcription termination. Yet when the major in5/ex6a sequence and its immediate sequences are positioned next to the very same polyA site at map position 50, processing and transcription termination are both very efficient. In addition, in the DR8minE6 cell line, in which the major 3′ end signals are restored without the local polyA site, an alternative polyA signal at map position 33 is used to efficiently process the messages and, concomitantly, transcription is terminated efficiently. Therefore, if a combination of specific polyA and neighboring pause sites is required to cause efficient termination, they would have to have been evolutionarily retained at all three of the polyA sites shown in Figure 7B.
We have constructed additional cell lines in an attempt to test the hypothesis that efficient terminal exon splicing, at least in part, mediates transcriptional termination. The most direct approach would be to insert the minimal element minus the splice site into DR8. However, based on results with the E6DO construct (Fig. 7D), the DHFR transcript in such an engineered variant would undoubtedly splice into ex6c, resulting in the DR8 phenotype. Thus, we took an alternative approach in which the 5′-most 40 bp of the minimal element (35 bp of in5 and 5 bp of ex6a) were inserted into DR8 at position 20 with or without a downstream polyA acceptor site (SPAMAZ4; Yonaha and Proudfoot 1999). In both cell lines (as well as in a control cell line that received only the SPAMAZ4 site), the transcription and replication initiation phenotypes were indistinguishable from that of DR8 and E6DO (L.D. Mesner, unpubl.). This finding suggests that sequences in addition to the 3′ splice site in the 168-bp minimal element are required for transcriptional processing and replication initiation.
In combination with previous studies (Dijkwel et al. 2002; Saha et al. 2004), our data reinforce the model that potential initiation sites are scattered throughout genomes, but that active transcription through a region either directly or indirectly precludes its use as a template for initiation (Fig. 7). In fact, it is clear that active transcription through S. cerevisiae ARS elements prevents them from firing, at least in plasmid constructs (Snyder et al. 1988). Calos and coworkers have also presented evidence that cloned genomic DNA fragments that replicate autonomously in mammalian cells are destabilized by transcription from neighboring selectable markers (Haase et al. 1994). Although these examples are artificial, they seem to suggest that the act of transcription per se, rather than some modulation of chromatin architecture resulting from transcription, is responsible for origin inactivation. This could be achieved simply by variations on the model proposed by Gilbert (2002), in which ORCs are suggested to load virtually anywhere in the genome in early G1 phase, but then are removed from the template by the transcription machinery when active transcription begins later in G1. An obvious corollary is that the 3′ ends of genes must be very efficient in unloading RNA polymerase, even if they process the 3′ ends of messages efficiently.
Materials and methods
Cell culture and cell synchrony
DHFR-deficient cell lines were propagated in Minimal Essential Medium (MEM; Invitrogen) containing 10% fetal clone II (FCII; Hyclone) supplemented with 100 μM hypoxanthine and 16 μM thymidine (Invitrogen). CHO and UA21 cell lines as well as DHFR-expressing variants were cultured in MEM/FCII lacking hypoxanthine and thymidine. For 2D gel experiments, cells were plated at a density of 5 × 106 per 15 cm dish, starved for isoleucine at a density of 4 × 107/dish for 32–36 h, and released into medium containing 200 μM mimosine for 13 h (Dijkwel and Hamlin 1995). The peak of initiation in the wild-type DHFR locus occurs between 70 and 100 min after release from mimosine, and S phase is complete in 8–9 h.
Construction of cosmid and plasmid donors for homologous recombination
Donor clones for modifying the DHFR locus were constructed from the cosmid KZ381 for the “ΔBam” cell lines, and from a subcloned 21-kb HindIII(partial)–XhoI fragment (map position 16–37) for the remaining cell lines (Fig. 1E). The desired deletions were introduced by partial BamHI, BstEII, or HindIII digestion, isolation of the appropriately sized truncated clone on an agarose gel, and self-ligation. The donor used to construct the Wtlox cell line was constructed by first inserting the 1.4-kb StuI–KpnI fragment containing exon6a between lox sites and using this cassette to replace the 1.5-kb HindIII–BamH1 fragment in the 21-kb HindIII(partial)–XhoI fragment. The DR8minE6 donor was constructed by inserting the 168-bp Bsr-GI–NsiI exon6a-containing fragment from the 1.4-kb StuI–KpnI fragment between lox sites, ligating the ∼4-kb SpeI–XhoI fragment (map position [mp] 33–37) immediately downstream from the 3′ lox site, and ligating the ∼3-kb HindIII–MscI fragment (mp 16–19) immediately upstream from the 5′ lox site (note: MscI and SpeI are ∼10 bp from the DR8 deletion junction). The DR8E6ecto donor was engineered by inserting the 168-bp Bsr-GI–NsiI lox cassette into the XhoI site in the 6.75-kb HindIII fragment (note: the XhoI site is in ex6c).
Generating variants by homologous or nonhomologous recombination
The inserts from the various engineered cosmid and plasmid donors (10 μg) were resected with appropriate restriction enzymes and transfected into 5 × 107 DR8 cells by electroporation (1 kV, 25 μF) as described (Saha et al. 2004). Transfectants were plated in MEM containing 10% FCII supplemented with hypoxanthine and thymidine (HT; ∼3-5 × 106 cells/10-cm culture dish). Forty-eight hours later, the medium was replaced with F12-Special medium lacking hypoxanthine, thymidine, and glycine and supplemented with 10% fetal clone I (Invitrogen). After 8–10 d, surviving colonies were expanded and genomic DNAs were analyzed by Southern hybridizations (probes and diagnostic fragments indicated in Fig. 1A). This process yielded all of the recombinants shown in Figure 1E except E6DO. The latter variant was obtained by subsequently transfecting WtLox with a cre expression vector (Sauer and Henderson 1988) and analyzing several colonies for the loss of the lox-flanked sequences. Finally, the cosmid cH1, which contains all but the very 3′ end of the DHFR gene and which expresses a functional dhfr enzyme (Milbrandt et al. 1983), was transfected into DR8 cells, and a cell line was isolated that had integrated the functional gene intact into another chromosome at an anonymous location and survives on minimal medium.
Southern blotting and hybridization analysis of genomic DNAs from homologous recombinants
Genomic DNAs were isolated from an average of 10 colonies for each of the transfections above as described (Gross-Bellard et al. 1973), and a variety of enzyme digests and suitable single-copy probes were used to select variants that had cleanly deleted or inserted the desired sequences at the DHFR locus (see Fig. 1A for maps and probes, Fig. 3 for representative analyses, and text; other mapping data available upon request).
Two-dimensional gel replicon mapping procedure
Isolation of replication intermediates and 2D gel analysis were essentially as described (Brewer and Fangman 1987; Dijkwel et al. 1991; detailed protocol available on request).
Northern and RT–PCR analysis of transcripts
Total RNA from log phase cells was isolated as described (Chomczynski and Sacchi 1987) from ∼6 × 107 cells cultured in MEM/HT containing 10% FCII. Steady-state messages were analyzed by Northern blot analysis as described (Sambrook et al. 1989), using GeneScreen membranes (Dupont) as the transfer medium. To isolate RNAs from early S-phase cells for RT–PCR analysis, replicas of those plated for 2D gel analysis were harvested 80 min after release into S phase. In some experiments, ∼20 μg total RNA was treated with 80 U DNaseI (Roche) in 20 mM Tris-HCl (pH 8.4), 2 mM MgCl2, 50 mM KCl, and 5 mM DTT at 37°C for 10 min (it was later determined that this step was not necessary, as contamination with DNA was insignificant) (L.D. Mesner, unpubl.). The enzyme was inactivated at 75°C for 5 min, and the RNA was precipitated with alcohol in the presence of 250 mM NaCl. cDNA was synthesized using the Thermoscript RT–PCR system (Invitrogen) according to the manufacturer's instructions. DNaseI-treated RNA was resuspended at 1.0 μg/μL, and 5.0 μg RNA was used for cDNA synthesis. Reactions were carried out at 60°C for 60 min. Approximately 10% of each cDNA synthesis reaction was used for PCR reactions, carried out using Platinum Taq DNA polymerase (Invitrogen) in a RoboCycler Gradient 96 instrument (Stratagene). Denaturation at 94°C for 2 min was followed by 33–45 cycles of amplification (denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 3 min). PCR products were analyzed on a 0.7% agarose gel. Primers for cDNA construction and PCR are available upon request.
Acknowledgments
We thank the other members of our laboratory for valuable discussions throughout the course of this work. We are also extremely grateful to Drs. Jim Manley and Nick Proudfoot for valuable discussions and critical reading of the manuscript. This study could not have been undertaken without the gifts of the UA21 and DR8 cell lines by Drs. Lawrence Chasin and Adelaide Carothers (Columbia University), respectively. The project was supported by a grant to J.L.H. from the NIH (RO1 GM26108).
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1307105.
References
- Aladjem M.I., Rodewald, L.W., Kolman, J.L., and Wahl, G.M. 1998a. Genetic dissection of a mammalian replicator in the human β-globin locus. Science 281: 1005–1009. [DOI] [PubMed] [Google Scholar]
- Aladjem M.I., Spike, B.T., Rodewald, L.W., Hope, T.J., Klemm, M., Jaenisch, R., and Wahl, G.M. 1998b. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 8: 145–155. [DOI] [PubMed] [Google Scholar]
- Altman A.L. and Fanning, E. 2001. The Chinese hamster dihydrofolate reductase replication origin β is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol. Cell. Biol. 21: 1098–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anachkova B. and Hamlin, J.L. 1989. Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol. Cell. Biol. 9: 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P. and Stillman, B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134. [DOI] [PubMed] [Google Scholar]
- Berberich S. and Leffak, M. 1993. DNase-sensitive chromatin structure near a chromosomal origin of bidirectional replication of the avian α-globin locus. DNA & Cell Biol. 12: 703–714. [DOI] [PubMed] [Google Scholar]
- Biamonti G., Perini, G., Weighardt, F., Riva, S., Giacca, M., Norio, P., Zentilin, L., Diviacco, S., Dimitrova, D., and Falaschi, A. 1992. A human DNA replication origin: Localization and transcriptional characterization. Chromosoma 102: S24–S31. [DOI] [PubMed] [Google Scholar]
- Blumenthal A.B., Kriegstein, H.J., and Hogness, D.S. 1974. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb. Symp. Quant. Biol. 38: 205–223. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman, W.L. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471. [DOI] [PubMed] [Google Scholar]
- Carothers A.M., Urlaub, G., Ellis, N., and Chasin, L.A. 1983. Structure of the dihydrofolate reductase gene in Chinese hamster ovary cells. Nucleic Acids Res. 11: 1997–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P. and Sacchi, N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenolchloroform extraction. Anal. Biochem. 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Dijkwel P.A. and Hamlin, J.L. 1992. Initiation of DNA replication in the dihydrofolate reductase locus is confined to the early S period in CHO cells synchronized with the plant amino acid mimosine. Mol. Cell. Biol. 12: 3715–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1995. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol. Cell. Biol. 15: 3023–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P.A., Vaughn, J.P., and Hamlin, J.L. 1994. Replication initiation sites are distributed widely in the amplified CHO dihydrofolate reductase domain. Nucleic Acids Res. 22: 4989–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P.A., Mesner, L.D., Levenson, V.V., d'Anna, J., and Hamlin, J.L. 2000. Dispersive initiation of replication in the Chinese hamster rhodopsin locus. Exp. Cell Res. 256: 150–157. [DOI] [PubMed] [Google Scholar]
- Dijkwel P.A., Wang, S., and Hamlin, J.L. 2002. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol. Cell Biol. 22: 3053–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman P.K. and Hamlin, J.L. 1989. Identification and characterization of a gene that is coamplified with dihydrofolate reductase in a methotrexate-resistant CHO cell line. Mol. Cell. Biol. 9: 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacca M., Zentilin, L., Norio, P., Diviacco, S., Dimitrova, D., Contreas, G., Biamonti, G., Perini, G., Weighardt, F., Riva, S., et al. 1994. Fine mapping of a replication origin of human DNA. Proc. Natl. Acad. Sci. 91: 7119–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D.M. 2002. Replication timing and transcriptional control: Beyond cause and effect. Curr. Opin. Cell Biol. 14: 377–383. [DOI] [PubMed] [Google Scholar]
- Goldman M.A., Holmquist, G.P., Gray, M.C., Caston, L.A., and Nag, A. 1984. Replication timing of genes and middle repetitive sequences. Science 224: 686–692. [DOI] [PubMed] [Google Scholar]
- Goldsmith K., Bendell, L., and Frappier, L. 1993. Identification of EBNA1 amino acid sequences required for the interaction of the functional elements of the Epstein-Barr virus latent origin of DNA replication. J. Virol. 67: 3418–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet, P., and Chambon, P. 1973. Isolation of high-molecular-weight DNA from mammalian cells. Eur. J. Biochem. 36: 32–38. [DOI] [PubMed] [Google Scholar]
- Haase S.B., Heinzel, S.S., and Calos, M.P. 1994. Transcription inhibits the replication of autonomously replicating plasmids in human cells. Mol. Cell. Biol. 14: 2516–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton K.S., Dhar, V., Brown, E.H., Iqbal, M.A., Stuart, S., Didamo, V.T., and Schildkraut, C.L. 1988. Replication program of active and inactive multigene families in mammalian cells. Mol. Cell. Biol. 8: 2149–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N.H. and Hamlin, J.L. 1982. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc. Natl. Acad. Sci. 79: 4083–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y. and Manley, J.L. 2000. RNA polymerase II and the integration of nuclear events. Genes & Dev. 14: 1415–1429. [PubMed] [Google Scholar]
- Huberman J.A. and Riggs, A.D. 1968. On the mechanism of DNA replication in mammalian chromosomes. J. Mol. Biol. 32: 327–341. [DOI] [PubMed] [Google Scholar]
- Hyrien O. and Mechali, M. 1993. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 12: 4511–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O., Maric, C., and Mechali, M. 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270: 994–997. [DOI] [PubMed] [Google Scholar]
- Jin Y., Yie, T.A., and Carothers, A.M. 1995. Non-random deletions at the dihydrofolate reductase locus of Chinese hamster ovary cells induced by α-particles simulating radon. Carcinogenesis 16: 1981–1991. [DOI] [PubMed] [Google Scholar]
- Kalejta R.F., Li, X., Mesner, L.D., Dijkwel, P.A., Lin, H.B., and Hamlin, J.L. 1998. Distal sequences, but not ori-β/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell 2: 797–806. [DOI] [PubMed] [Google Scholar]
- Kitsberg D., Selig, S., Keshet, I., and Cedar, H. 1993. Replication structure of the human β-globin gene domain. Nature 366: 588–590. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Rein, T., and DePamphilis, M.L. 1998. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 18: 3266–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffak M. and James, C.D. 1989. Opposite replication polarity of the germ line c-myc gene in HeLa cells compared with that of two Burkitt lymphoma cell lines. Mol. Cell. Biol. 9: 586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu T.H. and Hamlin, J.L. 1989. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol. Cell. Biol. 9: 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R.D., Platt, T.H., and Schildkraut, C.L. 1993. Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol. 13: 6600–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney J.E. and Hamlin, J.L. 1987. Isolation of the amplified dihydrofolate reductase domain from methotrexate-resistant Chinese hamster ovary cells. Mol. Cell. Biol. 7: 569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner L.D., Li, X., Dijkwel, P.A., and Hamlin, J.L. 2003. The dihydrofolate reductase origin of replication does not contain any nonredundant genetic elements required for origin activity. Mol. Cell Biol. 23: 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbrandt J.D., Heintz, N.H., White, W.C., Rothman, S.M., and Hamlin, J.L. 1981. Methotrexate-resistant Chinese hamster ovary cells have amplified a 135-kilobase-pair region that includes the dihydrofolate reductase gene. Proc. Natl. Acad. Sci. 78: 6043–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbrandt J.D., Azizkhan, J.C., Greisen, K.S., and Hamlin, J.L. 1983. Organization of a Chinese hamster ovary dihydrofolate reductase gene identified by phenotypic rescue. Mol. Cell. Biol. 3: 1266–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawotka K.A. and Huberman, J.A. 1988. Two-dimensional gel electrophoretic method for mapping DNA replicons. Mol. Cell. Biol. 8: 1408–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelizon C., Diviacco, S., Falaschi, A., and Giacca, M. 1996. High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol. Cell. Biol. 16: 5358–5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. and O'Sullivan, J. 2002. Polyadenylation: A tail of two complexes. Curr. Biol. 12: R855–R857. [DOI] [PubMed] [Google Scholar]
- Raghuraman M.K., Winzeler, E.A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D.J., Davis, R.W., Brewer, B.J., and Fangman, W.L. 2001. Replication dynamics of the yeast genome. Science 294: 115–121. [DOI] [PubMed] [Google Scholar]
- Saha S., Shan, Y., Mesner, L.D., and Hamlin, J.L. 2004. The promoter of the Chinese hamster ovary dihydrofolate reductase gene regulates the activity of the local origin and helps define its boundaries. Genes & Dev. 18: 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sauer B. and Henderson, N. 1988. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. 85: 5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., Scalzo, D., Kooperberg, C., van Steensel, B., Delrow, J., and Groudine, M. 2002. Genome-wide DNA replication profile for Drosophila melanogaster: A link between transcription and replication timing. Nat. Genet. 32: 438–442. [DOI] [PubMed] [Google Scholar]
- Shinomiya T. and Ina, S. 1993. DNA replication of histone gene repeats in Drosophila melanogaster tissue culture cells: Multiple initiation sites and replication pause sites. Mol. Cell. Biol. 13: 4098–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1994. Mapping an initiation region of DNA replication at a single-copy chromosomal locus in Drosophila melanogaster cells by two-dimensional gel methods and PCR-mediated nascent-strand analysis: Multiple replication origins in a broad zone. Mol. Cell. Biol. 14: 7394–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I., Tenzen, T., Mostoslavsky, R., Fibach, E., Lande, L., Milot, E., Gribnau, J., Grosveld, F., Fraser, P., and Cedar, H. 2001. Developmental regulation of DNA replication timing at the human β globin locus. EMBO J. 20: 6150–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Sapolsky, R.J., and Davis, R.W. 1988. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 2184–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taljanidisz J., Popowski, J., and Sarkar, N. 1989. Temporal order of gene replication in Chinese hamster ovary cells. Mol. Cell. Biol. 9: 2881–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G. and Chasin, L.A. 1980. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc. Natl. Acad. Sci. 77: 4216–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Kas, E., Carothers, A.M., and Chasin, L.A. 1983. Deletion of the diploid dihydrofolate reductase locus from cultured mammalian cells. Cell 33: 405–412. [DOI] [PubMed] [Google Scholar]
- Vaughn J.P., Dijkwel, P.A., and Hamlin, J.L. 1990. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell 61: 1075–1087. [DOI] [PubMed] [Google Scholar]
- Wang S., Dijkwel, P.A., and Hamlin, J.L. 1998. Lagging-strand, early-labelling, and two-dimensional gel assays suggest multiple potential initiation sites in the Chinese hamster dihydrofolate reductase origin. Mol. Cell. Biol. 18: 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M., Palacios DeBeer, M.A., and Fox, C.A. 2004. The activities of eukaryotic replication origins in chromatsein. Biochim. Biophys. Acta 1677: 142–157. [DOI] [PubMed] [Google Scholar]
- Yonaha M. and Proudfoot, N.J. 1999. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell. 3: 593–600. [DOI] [PubMed] [Google Scholar]