Abstract
Although hundreds of distinct animal microRNAs (miRNAs) are known, the specific biological functions of only a handful are understood at present. Here, we demonstrate that three different families of Drosophila miRNAs directly regulate two large families of Notch target genes, including basic helix–loop–helix (bHLH) repressor and Bearded family genes. These miRNAs regulate Notch target gene activity via GY-box (GUCUUCC), Brd-box (AGCUUUA), and K-box (cUGUGAUa) motifs. These are conserved sites in target 3′-untranslated regions (3′-UTRs) that are complementary to the 5′-ends of miRNAs, or “seed” regions. Collectively, these motifs represent >40 miRNA-binding sites in Notch target genes, and we show all three classes of motif to be necessary and sufficient for miRNA-mediated regulation in vivo. Importantly, many of the validated miRNA-binding sites have limited pairing to miRNAs outside of the “box:seed” region. Consistent with this, we find that seed-related miRNAs that are otherwise quite divergent can regulate the same target sequences. Finally, we demonstrate that ectopic expression of several Notch-regulating miRNAs induces mutant phenotypes that are characteristic of Notch pathway loss of function, including loss of wing margin, thickened wing veins, increased bristle density, and tufted bristles. Collectively, these data establish insights into miRNA target recognition and demonstrate that the Notch signaling pathway is a major target of miRNA-mediated regulation in Drosophila.
Keywords: microRNA, Notch signaling, Enhancer of split-Complex, Bearded-Complex
microRNAs (miRNAs) are endogenously encoded 21–22-nucleotide (nt) regulatory RNAs that derive from hairpin precursor transcripts (for review, see Lai 2003; Bartel 2004). Long hidden within the genome, intense efforts over the past 4 years collectively demonstrate miRNAs to constitute a sizable gene family in diverse higher eukaryotes, including both plants and animals. Mature miRNAs reside in a complex known as the RNA-induced silencing complex (RISC) or the micro-ribonucleoprotein complex (miRNP). The RISC/miRNP uses the small RNA as a guide to identify target transcripts for cleavage and/or inhibition of productive translation.
The founding miRNAs, lin-4 and let-7, emerged from forward genetic screens in nematodes (Lee et al. 1993; Reinhart et al. 2000). Mutations in these miRNAs disrupt the timing of developmental transitions, and both miRNAs directly regulate the expression of key timing control factors (Wightman et al. 1993; Moss et al. 1997; Reinhart et al. 2000; Abrahante et al. 2003; Lin et al. 2003). Virtually all of the ∼1000 miRNAs subsequently identified emerged from direct cloning of small RNAs (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001) and/or from genefinding programs that locate miRNAs on the basis of characteristic features of precursor miRNA transcripts (Lai et al. 2003; Lim et al. 2003a,b).
Since miRNA target selection is guided by base-pairing to its target(s), miRNA target-finding should be amenable to computational discovery. This has proven most successful in plants, where plant miRNAs typically display extensive pairing with predicted targets (Rhoades et al. 2002; Jones-Rhoades and Bartel 2004). Biochemical evidence of plant miRNA-directed target cleavage has been readily obtained, and in vivo genetic studies strongly support the biological relevance and importance of miRNA-mediated regulation of specific plant transcripts (Llave et al. 2002; Kasschau et al. 2003; Palatnik et al. 2003; Tang et al. 2003).
The situation has not been as straightforward in animals, since animal miRNAs generally display only modest complementarity to their known targets. Current target-finding informatics has focused on candidates with evolutionarily conserved sites that present strong base-pairing to the 5′-ends of miRNAs (Enright et al. 2003; Lewis et al. 2003, 2005; Stark et al. 2003; Kiriakidou et al. 2004). Although several animal miRNA:target interactions have been validated by various strategies in these studies, it remains generally unclear to what extent miRNA-mediated regulation of any computationally identified target will prove essential for any aspect of animal development or physiology.
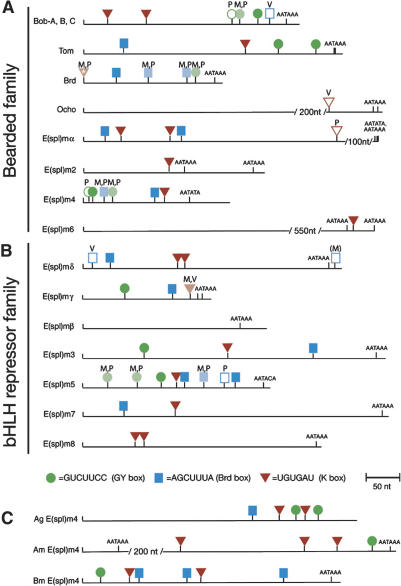
A series of genetic and informatics studies over the past decade cumulatively suggest that the Notch signaling pathway is regulated by miRNAs in Drosophila. The Notch pathway is a signal transduction cascade that mediates local cell–cell communication, and is essential for the proper patterning and development of all metazoan organisms (for review, see Lai 2004a). Genes are directly regulated by Notch signaling via binding sites for a CSL-type transcription factor (Bailey and Posakony 1995; Lecourtois and Schweisguth 1995), and the registry of biologically relevant, direct targets of Notch signaling is perhaps most complete in Drosophila. Although individual Notch target genes are induced only in specific developmental settings, there are two large families of Notch target genes that are collectively deployed across most territories of Notch activity in Drosophila. These genes are clustered in two genomic locations, named the Enhancer of split-Complex [E(spl)-C] and the Bearded-Complex [Brd-C]. Together, these complexes contain seven basic helix–loop–helix (bHLH) repressor-encoding genes and 10 Bearded genes (Fig. 1), all of which regulate Notch signaling (Delidakis and Artavanis-Tsakonas 1992; Knust et al. 1992; Wurmbach et al. 1999; Lai et al. 2000a,b).
Figure 1.
GY-boxes (green circles), Brd-boxes (blue squares), and K-boxes (red triangles) in the 3′-UTRs of Notch target genes of the Brd-Complex and the E(spl)-Complex. These two gene complexes contain 10 members of the Bearded family (A) and seven members of the bHLH repressor family (B). Each 3′-UTR schematic begins with the stop codon and continues to the transcript end as defined by cDNAs and/or consensus polyadenylation signals. Sites are mapped onto D. melanogaster (M) sequences, and those that are conserved in both D. pseudoobscura (P) and D. virilis (V) are dark-filled. Sites that are conserved in only two species are lightly filled, and nonconserved sites are unfilled; species containing these sites are denoted with M, P, or V. The K-box in Brd is starred because it overlaps the stop codon (and thus includes coding sequence), and is present in Dm Brd cDNA but not in Dm Brd genomic DNA (Leviten et al. 1997). The nonconserved Brd-box in E(spl)mδ is in parentheses because it is located ∼10 nt downstream of a utilized polyadenylation site and thus may not formally be located on a 3′-UTR. Note also that there are a limited number of noncanonical box sites in these 3′-UTRs that are not designated in this figure, some of which appear to be functional miRNA-binding sites (see Fig. 6). (C) 3′-UTRs of E(spl)m4 orthologs from the mosquito Anopheles gambiae (Ag), the honeybee Apis mellifera (Am), and the silkmoth Bombyx mori (Bm) also contain multiple GY-boxes, Brd-boxes, and K-boxes.
Gain-of-function alleles of the bHLH repressor gene E(spl)m8 (Knust et al. 1987) and the Bearded family gene Bearded (Brd) (Leviten et al. 1997) are associated with loss of discrete sequence motifs from their 3′-untranslated regions (3′-UTRs). These 6–7-nt motifs, named the GY-box (GUCUUCC), the Brd-box (AGCUUUA), and the K-box (cUGUGAUa), are broadly distributed in the 3′-UTRs of genes in the E(spl)-C and Brd-C (Fig. 1) and mediate negative post-transcriptional regulation (Lai and Posakony 1997, 1998; Lai et al. 1998). The biological importance of these sites was definitively demonstrated by mutation of Brd-boxes and K-boxes in the context of Bearded and E(spl)m8 genomic transgenes, respectively. Such mutant transgenes are hyperactive and induce aberrant peripheral nervous system development, indicating that these sites are required for normal restriction of Notch target gene activity (Lai and Posakony 1997; Lai et al. 1998). Subsequently, GY-box, Brd-box, and K-box motifs were noticed to be perfectly complementary to the 5′-ends of various Drosophila miRNAs, suggesting direct regulatory relationships between miRNAs and these “boxes” (Lai 2002a). Indeed, two E(spl)-C genes with GY-boxes have since been validated as miR-7 targets (Stark et al. 2003).
In this study, we use in vivo assays to demonstrate direct negative regulation of most members of the E(spl)-C and Brd-C by GY-box-, Brd-box-, and/or K-box-class miRNAs. We also demonstrate that GY-boxes, Brd-boxes, and K-boxes are necessary and sufficient for regulation by corresponding miRNAs, and that ectopic expression of Notch target-regulating miRNAs can phenocopy several aspects of Notch pathway loss of function. Collectively, this work establishes key features of miRNA target regulation and demonstrates the breadth of miRNA-mediated negative regulation of Drosophila Notch target gene expression.
Results
Deep conservation of putative miRNA-binding sites in the 3′-UTRs of insect Notch target genes
The E(spl)-C and Brd-C of Drosophila melanogaster (Dm) contain two large families of direct Notch target genes, including seven bHLH repressor-encoding genes and 10 Bearded family genes. With the exception of E(spl)mβ and Ocho, all of these genes contain GY-box (GUCUUCC), Brd-box (AGCUUUA), and/or K-box (UGUGAU) motifs in their 3′-UTRs (Fig. 1), which we propose to be miRNA-binding sites (Lai 2002a). Nine of these genes contain three or more box sites, a density that is especially remarkable when one considers how short their 3′-UTRs are (often <350 nt in length). We systematically assessed the conservation of these sites in their orthologs from Drosophila pseudoobscura (Dp) and Drosophila virilis (Dv), species that are ∼30 million and 60 million years diverged from Dm, respectively. As depicted by darkly filled icons in Figure 1, 33/51 Brd-boxes, GY-boxes, and K-boxes have been perfectly conserved and reside in syntenic locations among all three species; 11 additional sites are identical in two of the three species (denoted with lightly filled icons). This indicates that all three motifs are under strong selective constraint.
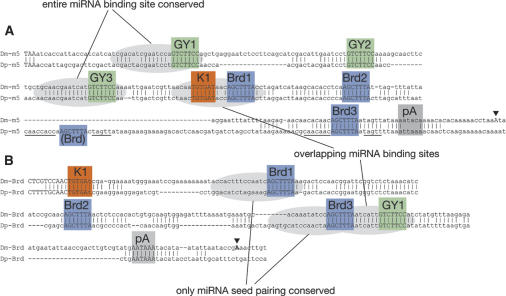
Closer examination of nucleotide divergence surrounding these boxes revealed some unexpected features that are germane to the proposition that these boxes represent miRNA-binding sites. These features are best illustrated by comparing rapidly evolving genes. Notably, Bearded is an unusually rapidly evolving protein, with only 15 residues preserved between Dm and Dv orthologs (out of 81 and 66 amino acids, respectively) (Supplementary Fig. 1), and Dv Bearded has a significantly different arrangement of these 3′-UTR motifs (Fig. 1). The 3′-UTR of Dv E(spl)m5 is also quite different from its counterparts in Dm/Dp. Alignment of Dm/Dp orthologs of Bearded and E(spl)m5 revealed that sequences upstream of most GY-boxes are well conserved (Fig. 2); these regions include most sequences presumed to pair with miR-7 (Fig. 3). Similar patterns are seen for many other GY-boxes in other Notch target genes (data not shown). However, the sequence upstream of many Brd- and K-boxes is strongly diverged, so that only “box”-pairing is often preserved (Figs. 2, 4, 5; data not shown). In fact, many Brd- and K-boxes generally lack extensive pairing to miRNAs outside of the “box” sequence (Figs. 4, 5). These factors likely precluded their identification by various published computational algorithms for miRNA-binding sites (Enright et al. 2003; Stark et al. 2003). Indeed, Brd- and K-boxes in Notch target genes were previously deemed unlikely to represent miRNA-binding sites (Stark et al. 2003). On the other hand, rapid divergence of the upstream portion of miRNA-binding sites is consistent with the idea that pairing between the miRNA “seed” (positions ∼2–8) and the 3′-UTR “box” (approximately the last one-third of the miRNA-binding site) is most critical for miRNA-mediated regulation (Lewis et al. 2003; Doench and Sharp 2004; Brennecke et al. 2005).
Figure 2.
Alignment of the 3′-UTRs of a bHLH repressor gene [E(spl)m5 (m5)] (A) and a Bearded family gene (Bearded, Brd) (B) from D. melanogaster (Dm) and D. pseudoobscura (Dp). Conserved regions of three or more nucleotides are marked. Polyadenylation sites from cDNAs are labeled with triangles, and polyadenylation signals are labeled pA; note that E(spl)m5 appears to use a noncanonical signal. GY-boxes, Brd-boxes, and K-boxes are all well-conserved between these species but display two classes of divergence: those that preserve the entire miRNA-binding site (box + upstream sequence) and those that preserve only miRNA seed-pairing (i.e., only the box); certain miRNA-binding sites are also overlapping. Note that the sequence surrounding a nonconserved Brd-box in Dp E(spl)m5 is highly related to the sequence surrounding the conserved third Brd-box of E(spl)m5 (cf. nucleotide identities between underlined sequences).
Figure 3.
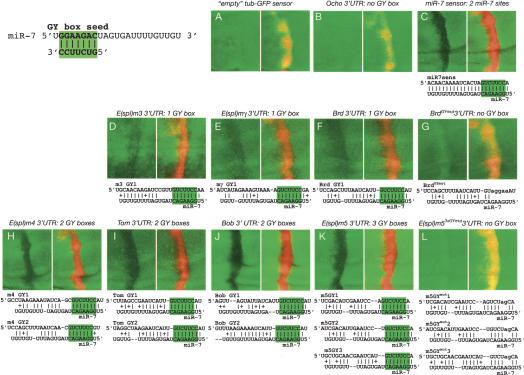
miR-7 directly regulates GY-box-containing Notch target genes. All discs contain ptc-Gal4>UAS-DsRed-miR-7 and one copy of tub>GFP (or arm>YFP in the case of Bearded sensors) attached to the 3′-UTRs as designated above each panel pair. The left panel of each pair depicts GFP expression, and the right panel shows the GFP + DsRed merge; miRNA regulation is inferred by diminution of GFP expression in the DsRed domain. Predicted miR-7:GY-box duplexes are shown below each pair, with the miR-7 seed:GY-box pairing highlighted in green; pairings of two or more adjacent nucleotides are marked. Note that the levels of each sensor are not directly comparable as they have been exposed for varying lengths of time.
Figure 4.
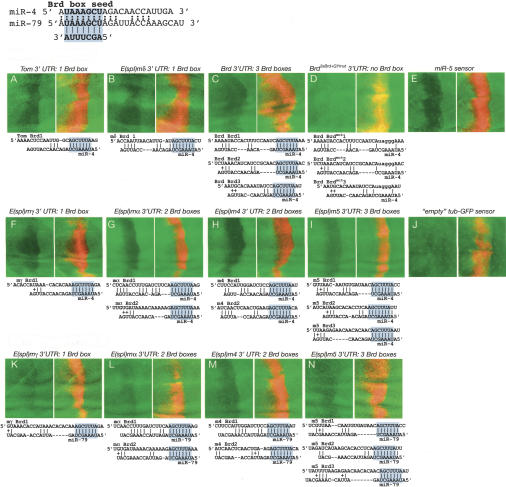
miR-4 and miR-79 directly regulate Brd-box-containing Notch target genes. These discs contain dpp-Gal4>UAS-DsRed-miR-4, dpp-Gal4>UAS-DsRed-miR-5, dpp-Gal4>UAS-DsRed-miR-286 (miR-4) (A–J), or dpp-Gal4>UAS-DsRed-miR-79 (K–N) and one copy of tub>GFP (or arm>YFP in the case of Bearded sensors) attached to the 3′-UTRs designated above each panel pair. Predicted miR4/79:Brd-box duplexes are shown below each pair, with the miR-4/79 seed:Brd-box pairing highlighted in blue; pairings of two or more adjacent nucleotides are marked.
Figure 5.
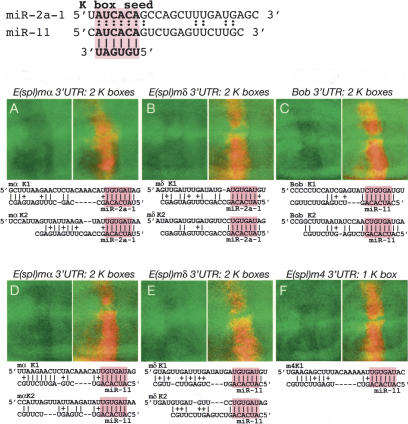
miR-2 and miR-11 directly regulate K-box-containing Notch target genes. These discs contain dpp-Gal4>UAS-DsRed-miR-2a-1, dpp-Gal4>UAS-DsRed-miR-2a-2, dpp-Gal4> UAS-DsRed-miR-2b-2 (miR-2) (A,B) or dpp-Gal4>UAS-DsRed-miR-11 (C–F) and one copy of tub>GFP attached to the 3′-UTRs designated above each panel pair. Predicted miR2/11:K-box duplexes are shown below each pair, with the miR-2/11 seed:K-box pairing highlighted in red; pairings of two or more adjacent nucleotides are marked.
We also note that precise spacing of several motif occurrences that are closely paired is also conserved (Fig. 2; data not shown), even though orthologous 3′-UTRs otherwise display significant insertions and deletions. In these cases, one would presume that simultaneous binding of miRNAs to their respective sites would not be possible unless the 3′-end of the downstream miRNA was unpaired, a configuration that unexpectedly proved functional in vitro (Doench and Sharp 2004). Finally, as shown by the unfilled icons in Figure 1, there are a few nonconserved boxes in these 3′-UTRs (7/51 total sites). In several cases, the nonconserved site is highly related to a neighboring conserved site [i.e., the first and second GY-boxes of Dp E(spl)m4 are equally similar to the first GY-box in Dm E(spl)m4; the third and fourth Brd-boxes in Dp E(spl)m5 are highly related to the third Brd-box in Dm E(spl)m5] (Fig. 2), implying that these nonconserved sites may be functional, newly evolved miRNA-binding sites.
GY-box-, Brd-box-, and K-box-class miRNAs are highly conserved among diverse insects, and many are, indeed, identical (Lai et al. 2003). Therefore, we searched for Brd-boxes, GY-boxes, and K-boxes in the predicted 3′-UTRs of E(spl)bHLH and Brd genes from mosquitoes, bees, and moths; these species cover ∼350 million years of divergence from Drosophila. Impressively, homologs of both E(spl)bHLH and Brd genes in these highly diverged species all contain multiple copies and multiple classes of “box” motifs in their 3′-UTRs (Fig. 1C; data not shown). This strongly suggests that regulation by all three families of miRNAs is an ancient feature of Notch target gene regulation in insects.
Notch target gene 3′-UTRs are directly regulated by multiple families of miRNAs in vivo
To directly test the capacity of miRNAs to regulate the 3′-UTRs of these Notch target genes, we used an in vivo assay developed by the Cohen group (Stark et al. 2003). The target in this assay is a ubiquitously expressed reporter (tub>GFP or arm>lacZ) fused to an endogenous 3′-UTR (a “3′-UTR sensor”). The reporter transgene is introduced into a genetic background in which a UAS-DsRed-miRNA transgene is activated with dpp-Gal4 or ptc-Gal4. This results in ectopic miRNA production in a stripe of red-fluorescing cells at the anterior–posterior boundary of imaginal discs. Inhibition of the green reporter within the red miRNA-misexpressing domain reflects direct miRNA-mediated negative regulation (i.e., Fig. 3C). We focused on the central wing pouch region of the wing imaginal disc; subsequent figures depict sensor expression in this region, by itself and as a merge with DsRed/miRNA expression.
We extensively analyzed the ability of sensor transgenes for most Bearded family genes [Bob, Bearded, Tom, Ocho, E(spl)mα, and E(spl)m4] and most E(spl)bHLH repressor genes [E(spl)mγ, E(spl)mδ, E(spl)m3, E(spl)m5, and E(spl)m8] to be regulated by ectopic GY-box-, Brd-box-, and K-box-class miRNAs. Sensor expression is influenced by the level to which it is negatively regulated by endogenous factors, including miRNAs. In this assay, the disc sensor must be expressed at sufficient levels before one can observe its knock-down by ectopic miRNAs. We observed that 3′-UTR sensor constructs for different Notch target genes accumulate to different levels in vivo, consistent with variable amounts of endogenous miRNA-mediated regulation (data not shown). Nevertheless, we were able to reliably detect expression of all sensors excepting E(spl)m8. As detailed in the following three subsections, we use these sensors to unequivocally demonstrate GY-boxes, Brd-boxes, and K-boxes to be sites of miRNA-mediated negative regulation by corresponding families of complementary miRNAs in vivo.
miR-7 regulates GY-box-containing 3′-UTRs
miR-7 is the only known Drosophila miRNA whose 5′-end is complementary to the GY-box (GUCUUCC) (Lai 2002a; Aravin et al. 2003). Previously, miR-7 was shown to regulate three GY-box targets, including two members of the E(spl)-C, E(spl)m3 and E(spl)m4 (Stark et al. 2003). While these two genes scored well in a genome-wide prediction of miR-7 targets (Stark et al. 2003), many other members of the Brd-C and E(spl)-C also contain between one and three GY-boxes in their 3′-UTRs [Bob, Bearded, Tom, E(spl)mγ, E(spl)m5]. Of these, only Tom was computationally identified as a compelling candidate for miR-7 (Stark et al. 2003).
We established the specificity of the disc sensor assay by showing that neither an empty tub-GFP sensor nor an Ocho sensor were affected by miR-7 (Fig. 3A,B). We also repeated the previous experiments done with E(spl)m3 and E(spl)m4 and observed that both were, indeed, inhibited by ectopic miR-7 (Fig. 3D,H). We then proceeded to use this assay to demonstrate that miR-7 negatively regulates all seven GY-box-containing members of the Brd-C and E(spl)-C, including those with single sites [E(spl)m3, E(spl)mγ, and Bearded] (Fig. 3D–F), those with two sites [E(spl)m4, Tom, Bob] (Fig. 3H–J), and those with three sites [E(spl)m5] (Fig. 3K). These data convincingly support the hypothesis that GY-boxes are general signatures of miR-7-binding sites in Notch target genes, irrespective of the overall amount of pairing between miR-7 and sequences outside of the GY-box. In order to more definitively demonstrate that miR-7-mediated regulation occurs through identified GY-boxes, we tested mutant sensors bearing point mutations in the GY-boxes. As shown in Figure 3G, a Bearded sensor carrying five point mutations in its single GY-box no longer responded to miR-7. In a more stringent test, we generated an E(spl)m5 sensor carrying 2-nt mutations in each of its three GY-boxes. These targeted changes also abolished the ability of miR-7 to negatively regulate E(spl)m5 (Fig. 3L). Therefore, ∼7 continuous base pairs between the “box” motif and its cognate miRNA seed are critical for in vivo target regulation. We also note that when mutant 3′-UTRs are tested, a mild increase in reporter activity in miRNA-misexpressing cells was sometimes observed, the reason for which has not been determined.
Previous work has suggested synergism between miRNA-binding sites on the same transcript (Doench et al. 2003). We observed that multiple GY-box 3′-UTRs were generally subject to greater regulation than single-site 3′-UTRs, even though the amount of miR-7 pairing to individual GY-boxes in multiple-site 3′-UTRs is often less than its pairing with single GY-box 3′-UTRs. Indeed, negative regulation of E(spl)m4, Tom, Bob, and E(spl)m5 by miR-7 (Fig. 3H–K) was qualitatively indistinguishable from an artificial sensor containing two perfectly miR-7-complementary sites (Fig. 3C), even though many sites in these genes display relaxed pairing with miR-7 outside of GY-boxes. This suggests that as little as 7–8 nt of complementarity may suffice for miRNA target recognition, especially where multiple sites are present. However, as all three single GY-box-containing 3′-UTRs were also regulated by miR-7 (Fig. 3D–F), synergism is not required for biologically significant regulation by miRNAs.
miR-4 and miR-79 regulate Brd-box-containing 3′-UTRs
There are two Drosophila miRNAs, miR-4 and miR-79, whose 5′-ends are complementary to the Brd-box (AGCUUUA) (Aravin et al. 2003; Lai et al. 2003). Both miRNAs are resident in miRNA clusters (Lagos-Quintana et al. 2001; Lai et al. 2003), and miR-4 resides in particularly dense clusters containing several unrelated miRNAs. We made use of a UAS-DsRed-miR-286, miR-4, miR-5 transgene that we refer to as “UAS-miR-4” and a UAS-DsRed-miR-79 transgene. miR-4 and miR-79 have only limited similarity outside of their Brd-box seed, and there is little indication from pairwise alignments that these miRNAs are specifically “tuned” to different Brd-box sites in Notch target genes. In fact, all of these Brd-boxes lack the extended complementarity to miRNAs that is typical of miR-7:GY-box pairs, and no Notch target genes were previously predicted computationally as targets of miR-4 or miR-79 (Enright et al. 2003; Stark et al. 2003).
We validated seven Brd-box-containing Notch target genes as being regulated by Brd-box-family miRNAs, including those with single sites [Tom, E(spl)mδ, E(spl)mγ] (Fig. 4A,B,F,K) and those with multiple sites [Bearded, E(spl)mα, E(spl)m4, and E(spl)m5] (Fig. 5C,G–I,L–N). Curiously, the negative regulatory effects of miR-4 on E(spl)mγ, E(spl)mα, E(spl)m4, and E(spl)m5 (Fig. 4F–I) were greater than those of miR-79 on these same 3′-UTRs (Fig. 4K–N), even though miR-4 is no more complementary to these sites than is miR-79. Nevertheless, the common ability of miR-4 and miR-79 to down-regulate individual sensors indicates that cross-regulation of individual sites by multiple members of a given miRNA family may occur. Notably, both miRNAs are expressed at high levels during embryonic development (Lagos-Quintana et al. 2001; Lai et al. 2003).
We tested the specificity of miR-4 and miR-79 using two mutant Bearded sensors, one bearing several point mutations in each of its three Brd-boxes and another containing mutations in the Brd-boxes and the GY-box. In both cases, the mutant transgenes accumulate to higher levels, consistent with relief from negative regulation by endogenous Brd-box-class miRNAs in the wing disc (Lai and Posakony 1997). In addition, they are no longer responsive to ectopic Brd-box-class miRNAs, indicating that the observed regulation occurs directly via Brd-boxes (Fig. 4D; data not shown). As well, this experiment demonstrates that regulation by the miR-4 transgene is not attributable to miR-286 and miR-5 carried on this construct. Nevertheless, this miRNA construct efficiently down-regulated a miR-5 sensor containing two miR-5 sites (Fig. 4E), indicating that the other miRNAs carried on this construct are functional. As a final test of the specificity of this assay, we observed that this three-miRNA construct failed to inhibit the expression of an empty tub-GFP sensor (Fig. 4J).
Having demonstrated that Brd-boxes are bona fide miRNA-binding sites, we asked whether regulation of the Bearded 3′-UTR by miR-7 (Fig. 3D) requires the presence of Brd-boxes. This might be the case, for example, if negative regulation of a given 3′-UTR required synergism between different types of miRNA-binding sites. We observed that a Bearded 3′-UTR carrying mutations in each of the three Brd-boxes was still strongly inhibited by miR-7 (Supplementary Fig. 2), indicating that individual types of miRNA-binding sites suffice for regulation in this assay.
miR-2 and miR-11 regulate K-box-containing 3′-UTRs
The largest family of Drosophila miRNAs includes those whose 5′-ends are complementary to the K-box (cUGUGAUa, where the lowercase nucleotides represent positions of strong bias) (Aravin et al. 2003; Lai et al. 2003). The K-box is also the most pervasive motif within these Notch target genes, as it is present in almost every member of the Brd-C and E(spl)-C [excepting E(spl)mβ and Ocho, which lack any box motifs] (Fig. 1). The maximum overall site complementarity of any given K-box site to any K-box family miRNAs is generally modest, and less than that seen with other demonstrated targets of the K-box family miRNA miR-2, namely, the proapoptotic genes grim, reaper, and sickle (Stark et al. 2003). In fact, the sole Notch target gene that was predicted informatically as a target of a K-box family miRNA in any study was E(spl)m8:miR-11, and this pair ranked only 46-th (Stark et al. 2003).
We tested the ability of two quite distinct K-box family miRNAs, those of the miR-2 cluster (miR-2a-1, miR-2a-2, and miR-2b-2) and miR-11, to regulate K-box-containing 3′-UTRs. Given the abundance of K-box complementary miRNAs (as a class, they are among the more frequently cloned fly miRNAs) (Lagos-Quintana et al. 2001), the occupancy of K-box sites by endogenous K-box-class miRNAs may be near-saturating in some cases. In fact, we were unable to convincingly demonstrate negative regulation of E(spl)m8 (data not shown), whose K-boxes mediate 10-fold negative regulation and nearly eliminate expression of this sensor (Lai et al. 1998). In spite of this, we obtained positive evidence that four other K-box-containing 3′-UTRs, E(spl)m4, Bob, E(spl)mα, and E(spl)mδ, are directly regulated by K-box-family miRNAs (Fig. 5), although the amount of regulation observed was weaker than that seen with GY-box- or Brd-box-class miRNAs. As was the case with the two Brd-box-class miRNAs, both miR-2 (Fig. 5A,B; data not shown) and miR-11 (Fig. 5C–F; data not shown) are capable of regulating some of the same K-box-containing targets. This constitutes further evidence for the possibility of cross-regulation of miRNA-binding sites, even where the miRNAs in question display very little similarity outside of their seeds.
Regulation by noncanonical “box” motifs
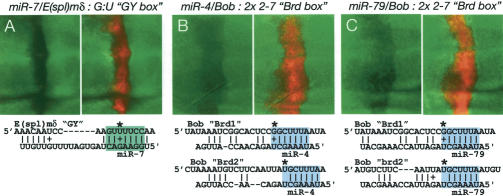
In performing pairwise tests of these miRNAs with Notch target gene sensors, we observed two instances of miRNA-mediated regulation of sensors lacking canonical boxes. First, we observed that the E(spl)mδ sensor was inhibited by miR-7 (Fig. 6A). Although E(spl)mδ lacks a canonical GY-box, it does contain a GY-box-like site that would have a single G:U base pair with the miR-7 seed. The nucleotides that are 5′ and 3′ to the box are also paired with miR-7, and there is a significant region of pairing to the 3′-end of the miRNA. These factors may allow this site to be recognized by miR-7. The 9-mer AGUUUUCCA is found in both Dp and Dv orthologs of E(spl)mδ, indicating that this site is under selection and therefore is likely important for regulation of E(spl)mδ. Second, we observed that the Bob sensor was negatively regulated by both Brd-box-class miRNAs, miR-4 and miR-79 (Fig. 6B,C). Although Bob lacks a canonical Brd-box, it does contain two matches to positions 2–7 of the Brd-box, which would pair to positions 2–7 of the miR-4/79. In this regard, this type of site is reminiscent of the 6-mer K-box, which pairs to positions 2–7 of K-box miRNAs. One of these Brd-box-like sites is conserved in Dp, and the syntenic site in Dv is, in fact, a canonical Brd-box, further indicating a functional relationship between Bob and miRNAs of the Brd-box family.
Figure 6.
Exceptional noncanonical GY-box- and Brd-box-like sites are functional. (A) miR-7 regulates the E(spl)mδ 3′-UTR via a GY-box-like site with G:U seed-pairing (asterisk). (B,C) miR-4 and miR-79 both regulate the Bob 3′-UTR via Brd-box-like sites that match positions 2–7 of the canonical Brd-box (variant first positions are marked with asterisks).
The apparent functionality of these noncanonical sites led us to search for other such sites in Notch target 3′-UTRs. Although one might expect to find many-fold more copies of degenerate sites relative to canonical sites, we instead found only a few additional examples of relaxed GY-box-like or Brd-box-like sites. For comparison, there are 28 canonical sites of these classes in Notch target 3′-UTRs (16 Brd-boxes and 12 GY-boxes), but only three additional examples of a 7-mer box-like site with a G:U base-pair to a miRNA seed [all are GY-box-like sites in E(spl)mδ, E(spl)m3, and E(spl)m7]. In addition, there are only five additional examples of sites that match only positions 2–7 of the GY-box or the Brd-box [all of which are Brd-box-like sites: the two in Bob, one in E(spl)m7, one in E(spl)mα, and one in E(spl)mδ]. These considerations strongly suggest that the much more restricted, canonical sites are actively selected for function in these Notch target 3′-UTRs, a conclusion that is bolstered by the patterns of evolutionary conservation of these sites (Figs. 1, 2).
Sufficiency of GY-boxes, Brd-boxes, and K-boxes for miRNA-mediated regulation
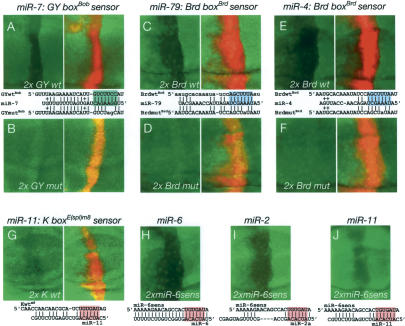
These experiments presented thus far demonstrate that target gene 3′-UTRs harboring sequence elements with Watson-Crick complementarity to the 5′-ends of miRNAs are, indeed, regulated by these miRNAs in vivo, and that such sites are necessary for miRNA-mediated regulation. Are these sites sufficient for regulation by complementary miRNAs? Although a variety of studies of model sites in tissue culture assays indicate site sufficiency, tests in animals suggest that miRNA site context can be less forgiving in vivo. For example, certain reporters containing multimers of six lin-4 or three let-7 sites are not appropriately regulated by lin-4 or let-7 in nematodes (Ha et al. 1996; Vella et al. 2004). In addition, mutation of sequences outside of the let-7-binding sites in lin-41 abolishes regulation by let-7 in vivo (Vella et al. 2004). Therefore, we were interested to test the functionality of GY-boxes, Brd-boxes, and K-boxes when abstracted from endogenous 3′-UTR context.
To do so, we cloned a tandem of isolated GY-box, Brd-box, and K-box elements from Bob, Bearded, and E(spl)m8, respectively, into tub-GFP transgenes. We also cloned mutant versions containing single changes in the Brd-box sites or dual changes in the GY-boxes. We then tested the ability of these “box” sensors to respond to exogenously expressed miRNAs. We found that wild-type GY-box (Fig. 7A), Brd-box (Fig. 7C,E), and K-box (Fig. 7G) sensors were all negatively regulated by corresponding miRNAs. These data directly demonstrate that all three types of box sites are sufficient for miRNA-mediated negative regulation. In contrast, mutant box sensors were nonfunctional in this assay (Fig. 7B,D,F). As the mutant box sensors contain only one or two changes in each site, these data provide strong in vivo support for the idea that Watson-Crick pairing to the 5′-end of the miRNA (the “seed”) is the key essential feature of miRNA target recognition. As a further test of this idea, we assayed the ability of the three different K-box miRNAs, miR-6, miR-2, and miR-11, to down-regulate a miR-6 sensor. As shown in Figure 7H–J, all three inhibited miR-6 sensor expression, consistent with the ability of seed-pairing to mediate regulation by miRNAs.
Figure 7.
Isolated, endogenous GY-boxes, Brd-boxes, and K-boxes are sufficient and necessary for miRNA-mediated regulation. These discs contain ptc-Gal4>UAS-DsRed-miR-7 (A,B), dpp-Gal4>UAS-DsRed-miR-79 (C,D), dpp-Gal4>UAS-DsRed-miR-4, miR-5, miR-286 (E,F), dpp-Gal4>UAS-DsRed-miR-11 (G), along with tub>GFP sensors containing two copies of GY-box, Brd-box, and K-box sites derived from Bob, Bearded, and E(spl)m8, respectively. Note that the GY-box sensor contains both GY-boxes of Bob (Fig. 3J); miRNA pairing is shown only for the more complementary site. (H,J) Expression of a miR-6 sensor in dpp-Gal4>UAS-miR-6-1, dpp-Gal4>UAS-miR-6-2, dpp-Gal4>UAS-miR-6-3 (miR-6) (H), dpp-Gal4>UAS-miR-2a-1, dpp-Gal4>UAS-miR-2a-2, dpp-Gal4>UAS-miR-2b-2 (miR-2) (I), and dpp-Gal4>UAS-DsRed-miR-11 (mir-11) (J) backgrounds.
Ectopic expression of Notch target-regulating miRNAs phenocopies Notch pathway loss of function
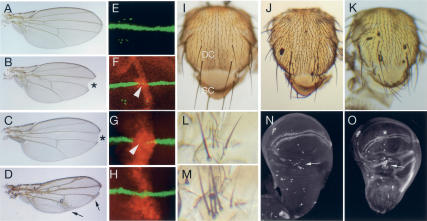
With these UAS-miRNA transgenic lines in hand, we examined the consequences of ectopically expressing miRNAs on Drosophila development. It should be noted that we fully expect Notch target-regulating miRNAs to regulate other functionally unrelated targets in vivo. For example, it has been established that K-box-family miRNAs also negatively regulate the proapoptotic genes reaper, sickle, and grim via K-boxes in their 3′-UTRs, while Brd-box-family miRNAs target the mesodermal determinant bagpipe via a Brd-box in its 3′-UTR (Stark et al. 2003; Brennecke et al. 2005). Therefore, even if ectopic miRNAs were able to affect normal development, we would not necessarily expect them to affect Notch signaling exclusively. Nevertheless, it has been previously reported that ectopic miR-7 induces loss of molecular markers of wing margin development, resulting in wing notching (Stark et al. 2003). This indicates that phenotypic characterization of miRNA misexpression can be informative.
Using an independently derived UAS-miR-7 construct lacking DsRed, we verified that dpp-Gal4>miR-7 wings display notching (Fig. 8A,B) and loss of Cut expression at the developing wing margin of wing imaginal discs (Fig. 8E,F); the size of the L3–L4 intervein domain was also reduced (Stark et al. 2003). We next observed that ectopic K-box miRNAs of the miR-2a-1, miR-2a-2, miR-2b-2 cluster or miR-6-1, miR-6-2, miR-6-3 cluster had similar effects on wing margin development, although two UAS-transgenes were necessary to produce this effect (Fig. 8C,G; data not shown). We also observed loss of anterior crossvein and occasional L3 vein breaks, although these are not indicative of loss of N signaling (Fig. 8C). More generalized expression of miR-7 using bx-Gal4 induced strong thickening of wing veins (Fig. 8D), which is indicative of compromised Notch signaling during lateral inhibition of wing veins. Expression of K-box miRNAs using bx-Gal4 had severe effects on wing development, resulting in tiny, crumpled wings (data not shown). We suspect that this results from misregulation of non-Notch-pathway-related targets. The Brd-box miRNAs miR-4 and miR-79 and the K-box miRNA miR-11 did not affect wing margin development, even when these transgenes were present in two copies, indicating that this phenotype is not generally due to misexpression of miRNAs (Fig. 8H; data not shown). However, miR-79 induced strong wing curling at high levels, potentially due to misregulation of non-Notch-pathway-related targets (data not shown).
Figure 8.
Misexpression of Notch target-regulating miRNAs phenocopies many defects associated with loss of Notch signaling. (A) Wild-type adult wing. (B) dpp-Gal4>UAS-miR-7 shows a notched wing and a reduced L3–L4 intervein region. (C) dpp-Gal4>UAS-miR-2a-1, dpp-Gal4> UAS-miR-2a-2, dpp-Gal4>UAS-miR-2b-2 + UAS-miR-6-1, UAS-miR-6-2, UAS-miR-6-3 displays a notched wing, loss of anterior crossvein, and occasional L3 vein breaks. (D) bx-Gal4>UAS-miR-7 wing shows extensive vein thickening. (E) Cut (green) expression at the developing wing margin of a third instar wing imaginal disc. (F) ptc-Gal4>UAS-DsRed-miR-7 disc displays loss of Cut staining (arrowhead) in the miR-7-expressing domain (red). (G) dpp-Gal4> 2×UAS-DsRed-miR-2a-1, dpp-Gal4>2×UAS-DsRed-miR-2a-2, dpp-Gal4>2×UAS-DsRed-miR-2b-2 shows loss of Cut staining in the miR-2-expressing domain (arrowhead). (H) Wing margin expression of Cut is maintained in a dpp-Gal4>2×UAS-DsRed-miR-79 disc. (I) Wild-type adult notum displays a characteristic pattern of microchaetes (small bristles) and macrochaetes (large bristles); (DC) dorsocentral region; (SC) scutellar region. (J) bx-Gal4-UAS-miR-6-1, bx-Gal4-UAS-miR-6-2, bx-Gal4-UAS-miR-6-3 displays a strong increase in microchaete density and ectopic dorsocentral macrochaetes. (K) bx-Gal4>UAS-miR-2a-1, bx-Gal4-UAS-miR-2a-2, bx-Gal4-UAS-miR-2b-2 shows ectopic dorsocentral and scutellar macrochaetes. (L) Wild-type sternopleural bristles. (M) dpp-Gal4>UAS-miR-7 shows tufted sternopleural bristles. (N) Senseless staining of a wild-type imaginal disc highlights sensory organs precursor (SOP) cells; arrow points to SOPs in the dorsal radius. (O) dpp-Gal4>2×UAS-DsRed-miR-7 shows a strong increase in SOPs in the dorsal radius; asterisk denotes a break in the wing margin.
We next focused on development of the adult peripheral nervous system, as exemplified by the bristle sensory organs that decorate the body surface. A classic role for Notch signaling is to restrict the number of sensory organ precursors. We found that misexpression of miR-6 using bx-Gal4 resulted in a strong increase in microchaete bristle density and clustered dorsocentral macrochaetes (Fig. 8I,J), phenotypes that are consistent with loss of Notch signaling during lateral inhibition of sensory organ precursors. Ectopic miR-2 had a similar, but milder, effect and mostly induced clustered dorsocentral and scutellar macrochaetes (Fig. 8K). Therefore, divergent members of the K-box miRNA family have similar effects on sensory organ development, consistent with our data indicating that seed-related miRNAs can regulate overlapping sets of target genes. Ectopic miR-7 also induced macrochaete tufting (Fig. 8L,M), which correlated with the differentiation of supernumerary sensory organ precursors in wing imaginal discs (Fig. 8N,O). Finally, we observed occasional duplication of bristles upon misexpression of the Brd-box miRNA mir-79, although this construct also induced occasional bristle loss (data not shown). Ectopic expression of miRNAs does not in itself induce bristle defects per se, as misexpression of miR-4 or miR-11 did not interfere with bristle development.
Overall, the ability of different classes of Notch-regulating miRNAs to specifically induce phenotypes that are characteristic of Notch pathway loss of function in multiple developmental settings is a strong indication that Notch pathway targets validated in this study are key endogenous targets of these miRNAs.
Discussion
Negative regulation of Notch signaling
The Notch pathway is a fundamental mechanism for determining cell fates and tissue identity throughout the Metazoa (for review, see Lai 2004a). For organisms in which highly detailed phenotypic studies have been performed, such as Drosophila, we may state with confidence that there is scarcely any tissue whose normal development does not depend critically and quite directly on Notch signaling. Because of the profound cellular consequences of Notch activation, it is extremely important for cells to carefully control Notch pathway activity.
It appears, then, that cells go through a significant amount of trouble to actively inhibit Notch signaling. Core components of the Notch pathway are subject to significant negative regulation at every step in their life cycle, including at the transcriptional, post-transcriptional, and post-translational levels. For example, in the absence of activated nuclear Notch, CSL proteins are transcriptional repressors that actively repress Notch target gene activity (Kao et al. 1998; Morel et al. 2001; Barolo et al. 2002). In addition, multiple dedicated ubiquitin ligases promote degradation of Notch pathway components, including the receptor Notch itself (for review, see Lai 2002b). To this list, we may add that transcripts of most direct Notch target genes in Drosophila are negatively regulated by multiple families of miRNAs.
The evidence provided in this study to support this conclusion is that (1) three different classes of miRNA-binding sites (GY-boxes, Brd-boxes, and K-boxes) are pervasive among two major classes of Notch target genes; (2) all three classes of motif are selectively constrained in 3′-UTRs during evolution; (3) transcripts bearing these box sites are negatively regulated by complementary miRNAs in vivo; (4) all three classes of sites are both necessary and sufficient for miRNA-mediated regulation in vivo; and (5) ectopic expression of Notch target-regulating miRNAs phenocopies Notch pathway loss of function during multiple developmental settings. Perhaps most importantly, we have previously shown that genomic transgenes specifically mutated for miRNA-binding sites are sufficiently hyperactive so as to perturb normal development of the peripheral nervous system (Lai and Posakony 1997; Lai et al. 1998). This places these Drosophila Notch target genes in a relatively select group of miRNA targets for which miRNA-mediated regulation is phenotypically essential for normal development.
Implications for miRNA target identification
miRNA target-finding algorithms have been independently developed by many groups (for review, see Lai 2004b). For the most part, these operated with rules gleaned from a very limited set of validated miRNA:target pairs and an incomplete experimental understanding of how miRNAs recognize their targets. Our candidate target list of miRNA:Notch target gene interactions was borne of genetic studies, in which biological indications of phenotypically relevant regulation were observed prior to the recognition of miRNA involvement. Systematic analysis of this target list has allowed us to accumulate a set of newly validated miRNA:target pairs (25) that is comparable to the cumulative number of in vivo validated animal miRNA:target pairs in the literature. The characteristics of this diverse set of miRNA targets are germane to the general pursuit of miRNA target-finding.
While most of the previously characterized in vivo targets of miRNAs are of the “extensive pairing” variety, many of our validated targets display much more limited “box:seed”-pairing to miRNAs. In fact, within the context of the set of Notch target gene 3′-UTRs, the presence of conserved GY-boxes, Brd-boxes, and K-boxes allowed for highly effective prediction of miRNA:target relationships. This is the case even without first taking into account the extent of miRNA-pairing outside of box motifs. Rapid divergence of sequences upstream of box motifs, particularly those of the Brd-box and K-box classes, further indicates that extensive pairing is not selected for in these bona fide target sites. Consistent with this, we have presented multiple lines of evidence that show that divergent seed-related miRNAs can regulate overlapping sets of target in vivo. Conversely, the importance of pairing between 3′-UTR boxes to miRNA seeds was demonstrated by endogenous 3′-UTR and box sufficiency tests, where even single-nucleotide disruption of seed-pairing abolishes regulation by miRNAs in vivo.
Our identification and characterization of miRNA-binding sites in these Notch target 3′-UTRs mesh well with other recent bioinformatics and experimental studies that together help to define the “look” of miRNA-binding sites. The concept of using conserved “boxes” with Watson-Crick complementarity to miRNA seeds to identify miRNA targets is at the heart of the TargetScanS approach (Lewis et al. 2003, 2005). In a recent study, they identify statistically significant signal not only for conserved 3′-UTR sites that match positions 2–8 of the miRNA (as is characteristic of the Brd-box and GY-box), but also for matches to positions 2–7 of the miRNA (as is characteristic of the K-box) (Lewis et al. 2005). In addition, they identify a significant bias for the nucleotide corresponding to position one of the miRNA to be an adenosine in predicted target sites. Interestingly, 27/42 (64%) of GY-boxes, Brd-boxes, and K-boxes in Dm Notch target genes also have an adenosine in this position, consistent with the notion that this feature can help to identify genuine target sites. Our results are also consistent with directed tests of model sites using the imaginal disc sensor assay (Brennecke et al. 2005). Together with the recent observation that miRNAs can down-regulate large numbers of transcripts that contain box:seed matches in their 3′-UTRs (Lim et al. 2005), a current view emerges that conserved 3′-UTR boxes that are 6–7 nt in length and complementary to the 5′-ends of miRNAs need to be considered seriously as functional regulatory sites. While seed-pairings with G:U base pairs are evidently not generally selected for, we have shown evidence that rare sites of this class are functional. This is consistent with other studies that demonstrate that G:U seed-pairing impairs, but does not necessarily abolish target site function (Doench and Sharp 2004; Brennecke et al. 2005).
Finally, the presence of multiple classes of miRNA-binding sites in most Notch target gene 3′-UTRs raises the possibility of combinatorial regulation. Although this has been widely suggested as a formal possibility, we have provided extensive evidence that 3′-UTRs can bear multiple classes of functional sites. Phylogenetic considerations indicate that 10 different Notch target genes are likely regulated by multiple classes of miRNAs, and we have provided direct experimental support of this for six Notch target genes. Multiple Brd-box-, K-box-, and GY-box-class miRNAs are present at high levels in the Drosophila embryo, and the Brd-box miRNA miR-4 is co-transcribed with the K-box miRNAs miR-6-1, miR-2, miR-3 (Lagos-Quintana et al. 2001), suggesting that combinatorial control of Notch target genes actually occurs during normal development. Future studies are aimed at examining how different miRNA-binding sites collectively contribute to overall regulation of an individual gene.
`Switch' targets versus `tuning' targets
Of the few animal miRNAs whose in vivo functions and targets are well understood, most act as genetic switches that determine binary, on/off states of target gene activity. For example, lin-4 and let-7 are temporal switches that control progression through nematode larval stages by inhibiting their targets at designated times in development (Lee et al. 1993; Wightman et al. 1993; Reinhart et al. 2000). lsy-6 and miR-273 are spatial switches whose extremely restricted cell-type-specific expression patterns control neuronal identity (Johnston and Hobert 2003; Chang et al. 2004). In these cases, temporally or spatially restricted miRNA expression is central to their control of specific processes, and each of these miRNAs appears to have a small number of key targets.
We propose a different rationale for Brd-box and K-box miRNAs during Drosophila development. Although endogenous territories of GY-box-mediated regulation are not known, negative regulation by Brd-boxes and K-boxes appears spatially and temporally ubiquitous (Lai and Posakony 1997; Lai et al. 1998). Thus, Notch target transcripts of the Brd family and E(spl)bHLH families are subject to modes of miRNA-mediated regulation that operate in all cells, even though the genes themselves display highly restricted patterns of spatial expression. This suggests that these miRNAs are not dedicated to regulating Notch signal transduction, but may “tune” the expression of many target genes (for review, see Bartel and Chen 2004). Indeed, the K-box-family miRNAs miR-2, miR-6, and miR-11 also directly regulate K-box-containing proapoptotic genes (Stark et al. 2003), and the Brd-box-family miRNAs miR-4 and miR-79 regulate the mesodermal determinant bagpipe (Brennecke et al. 2005). One prediction is that even though mutation of Brd-boxes and K-boxes in individual Notch target genes results in specific defects in Notch-mediated cell fate decisions (Lai and Posakony 1997; Lai et al. 1998), mutation of Brd-box and K-box miRNAs would have more general developmental consequences. This is supported by our observation that many, but not all, of the phenotypes induced by ectopic expression of Notch-regulating miRNAs appear to be obviously related to repression of Notch pathway activity.
An important advance of this study is the in vivo validation of a large number of biologically relevant miRNA targets that are minimally paired to miRNAs outside of the “box:seed” region. As we demonstrated that modestly complementary sites are both necessary and sufficient for miRNA-mediated regulation, it might be relatively easy for novel miRNA-binding sites to arise in “tuning” targets. Indeed, a subset of box sites has apparently newly evolved during Drosophilid radiation. In the greater context of insect Notch target genes, it appears to have been important that they be negatively regulated by miRNAs, although the precise numbers and arrangement of different sites is variable (Fig. 1). These features of tuning targets seem to allow for highly customized regulation of individual genes.
The experimental validation of many tuning targets may be challenging or impossible to obtain where quantitative regulation is subtle. Nevertheless, minor changes in gene activity, even of a fraction of a percent, could become highly significant when selecting the fitness of individuals at the population level. Deep evolutionary profiling of related species will therefore be key to revealing the full complement of biologically important miRNA-binding sites. Our data suggest that multiple classes of miRNA-binding sites can be recognized with confidence as highly conserved 3′-UTR “boxes” complementary to miRNA seeds, and this approach has been applied to the analysis of mammalian genomes (Lewis et al. 2005). By mid-2005, 12 Drosophila genomes will be completed, which should enable high-confidence identification of miRNA-binding sites on the genome-wide scale—even in cases in which only 7 nt of the target are paired to a miRNA.
Regulation of Notch target genes by miRNAs in other species?
Recent computational work pointed to regulation of vertebrate Notch and Delta by miR-34 (Lewis et al. 2003); however, no Notch target genes were similarly singled out in various bioinformatics efforts. miR-34 is conserved in flies; however, inspection of fly Notch or its ligands Delta and Serrate failed to reveal “boxes” that might indicate similar regulation by miR-34. Brd-box-, GY-box-, and K-box-complementary miRNAs are likewise conserved between flies and vertebrates. Are any vertebrate Notch target genes predicted to be targeted by these miRNAs by virtue of “boxes”? Although Brd proteins have thus far been found only in insects, E(spl)bHLH proteins are conserved in and are primary effectors of Notch signaling in all vertebrates (for review, see Lai 2004a). We failed to observe enrichment for Brd-boxes, GY-boxes, and K-boxes across the set of vertebrate E(spl)bHLH 3′-UTRs as a whole. However, we did find that members of a specific subset of E(spl)-related repressors, named the Hey genes, contain a preponderance of these boxes in their 3′-UTRs. This appears to be the case in a variety of mammals (human, mouse, and rat) and fish (fugu and zebrafish) (E.C. Lai, unpubl.). Therefore, miRNA-mediated regulation may be a conserved feature of Notch target genes, a scenario that is under current experimental investigation.
Materials and methods
Sensor assay
miRNA expression constructs include miRNA hairpins and ∼150 nt of flanking genomic sequence at both ends; a similar amount of flanking sequence was used in cases of miRNA clusters. These were cloned into either pUAST (Brand and Perrimon 1993) or pUAS-DsRed (Stark et al. 2003) to generate UAS-miR-7, UAS-miR-6-1, UAS-miR-2, UAS-miR-3, UAS-miR-2a-1, UAS-miR-2a-2, UAS-miR-2b-2, and UAS-DsRed-miR-79.3′-UTR sensors for Bob, Tom, Ocho, E(spl)mα, E(spl)mγ, E(spl)mδ, and E(spl)m5 contained the sequence from the stop codon to ∼250 bp downstream of the polyadenylation signal cloned into a tub-GFP transgene (Brennecke et al. 2003). Site mutants of the E(spl)m5 sensor were made using Quikchange (Stratagene). Specific miRNA and 3′-UTR sequences are available upon request. Sequences of box and miRNA sensors cloned into tub-GFP are as follows: GY-box wild type (the two GY-boxes from Bob), gttagtattatcattGTCTTCCattagtttaagaaaatcattGTCTTCCat; GY-box mutant, gttagtattatcattGTCTagCattagtttaagaaaatcattGTCT agCat; Brd-box wild type (two copies of the third Brd-box of Brd), aatgcacaaatatccAGCTTTAataatgcacaaatatccAGCTTTAat; Brd-box mutant, aatgcacaaatatccAGCTTaAataatgcacaaatatccAGCTT aAat; K-box wild type [two copies of the first K-box of E(spl)m8], caaccaacaacgcatcTGTGATagcaaccaacaacgcatcTGTGATag; miR-5 sensor, tCATATCACAACGATCGTTCCTTTtcaacaatcactCAT ATCACAACGATCGTTCCTTTt; miR-6 sensor, cAAAAAGAA CAGCCACTGTGATAtcaacaatcaccAAAAAGAACAGCCACTG TGATAt.
The following transgenic lines were previously described: bx-Gal4, ptc-Gal4, and dpp-Gal4 (FlyBase 2003); wild-type and mutant arm-lacZ-Brd 3′UTR and arm-lacZ-m8 3′UTR lines (Lai and Posakony 1997; Lai et al. 1998); and UAS-DsRed-miR-7, UAS-DsRed-miR-2a-1, UAS-DsRed-miR-2a-2, UAS-DsRed-miR-2b-2, empty tub-GFP sensor, tub-GFP-miR-7 sensor, tub-GFP-m4 3′UTR, and tub-GFP-m3 3′UTR (Stark et al. 2003). UAS-DsRed-miR-286, UAS-DsRed-miR-4, UAS-DsRed-miR-5, and UAS-DsRed-miR-11 were unpublished gifts of Julius Brennecke (EMBL, Heidelberg, Germany) and Stephen Cohen (EMBL, Heidelberg, Germany). Wild-type and mutant arm-YFP-Brd 3′UTR transgenes were unpublished gifts of Garson Tsang (University of California at Berkeley, Howard Hughes Medical Institute, Berkeley, CA) and Adina Bailey (University of California at Berkeley, Howard Hughes Medical Institute, Berkeley, CA).
We followed standard protocols for immunofluorescent stainings (Lai and Rubin 2001) using rabbit α-GFP (1:1000; Molecular Probes), rabbit α-β-galactosidase (1:5000; Cappel), guinea pig α-Senseless (1:4000; gift of Hugo Bellen, Baylor College of Medicine, Howard Hughes Medical Institute, Houston, TX), or mouse α-Cut (1:100; from the Developmental Studies Hybridoma Bank), followed by Alexa 488- or 594-conjugated goat secondaries (1:400; Molecular Probes).
Acknowledgments
We thank Julius Brennecke, Steve Cohen, Garson Tsang, and Adina Bailey for generous gifts of unpublished miRNA and sensor lines; James Posakony for Brd and E(spl)m8 sensors; and Hugo Bellen for antibodies. This work was supported by the Howard Hughes Medical Institute. E.C.L. was also supported by a Damon Runyon Cancer Research Foundation Fellowship (DRG 1632) and by a Career Development Award from the Leukemia and Lymphoma Society (LLS #3096-05).
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1291905.
References
- Abrahante J.E., Daul, A.L., Li, M., Volk, M.L., Tennessen, J.M., Miller, E.A., and Rougvie, A.E. 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4: 625–637. [DOI] [PubMed] [Google Scholar]
- Aravin A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337–350. [DOI] [PubMed] [Google Scholar]
- Bailey A.M. and Posakony, J.W. 1995. Suppressor of Hairless directly activates transcription of Enhancer of split Complex genes in response to Notch receptor activity. Genes & Dev. 9: 2609–2622. [DOI] [PubMed] [Google Scholar]
- Barolo S., Stone, T., Bang, A.G., and Posakony, J.W. 2002. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes & Dev. 16: 1964–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs. Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. and Chen, C.Z. 2004. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Genet. 5: 396–400. [DOI] [PubMed] [Google Scholar]
- Brand A.H. and Perrimon, N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark, A., Russell, R.B., and Cohen, S.M. 2005. Principles of microRNA-target recognition. PLoS Biol. 3: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Johnston Jr., R.J., Frokjaer-Jensen, C., Lockery, S., and Hobert, O. 2004. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430: 785–789. [DOI] [PubMed] [Google Scholar]
- Delidakis C. and Artavanis-Tsakonas, S. 1992. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix–loop–helix proteins. Proc. Natl. Acad. Sci. 89: 8731–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G. and Sharp, P.A. 2004. Specificity of microRNA target selection in translational repression. Genes & Dev. 18: 504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J., Petersen, C., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17: 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D.S. 2003. MicroRNA targets in Drosophila. Genome Biol. 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase. 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31: 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha I., Wightman, B., and Ruvkun, G. 1996. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes & Dev. 10: 3041–3050. [DOI] [PubMed] [Google Scholar]
- Johnston R.J. and Hobert, O. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W. and Bartel, D.P. 2004. Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- Kao H.Y., Ordentlich, P., Koyano-Nakagawa, N., Tang, Z., Downes, M., Kintner, C.R., Evans, R.M., and Kadesch, T. 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes & Dev. 12: 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- Kiriakidou M., Nelson, P.T., Kouranov, A., Fitziev, P., Bouyioukos, C., Mourelatos, Z., and Hatzigeorgiou, A.G. 2004. A combined computational–experimental approach predicts human microRNA targets. Genes & Dev. 18: 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust E., Tietze, K., and Campos-Ortega, J.A. 1987. Molecular analysis of the neurogenic locus Enhancer of split of Drosophila melanogaster. EMBO J. 6: 4113–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust E., Schrons, H., Grawe, F., and Campos-Ortega, J.A. 1992. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix–loop–helix proteins. Genetics 132: 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Lai E.C. 2002a. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30: 363–364. [DOI] [PubMed] [Google Scholar]
- ____. 2002b. Protein degradation: Four E3s for the Notch pathway. Curr. Biol. 12: R74–R78. [DOI] [PubMed] [Google Scholar]
- ____. 2003. microRNAs: Runts of the genome assert themselves. Curr. Biol. 13: R925–R936. [DOI] [PubMed] [Google Scholar]
- ____. 2004a. Notch signaling: Control of cell communication and cell fate. Development 131: 965–973. [DOI] [PubMed] [Google Scholar]
- ____. 2004b. Predicting and validating microRNA targets. Genome Biol. 5: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.C. and Posakony, J.W. 1997. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split complex gene expression. Development 124: 4847–4856. [DOI] [PubMed] [Google Scholar]
- ____. 1998. Regulation of Drosophila neurogenesis by RNA:RNA duplexes? Cell 93: 1103–1104. [DOI] [PubMed] [Google Scholar]
- Lai E.C. and Rubin, G.M. 2001. neuralized functions cell-autonomously to regulate a subset of Notch-dependent processes during adult Drosophila development. Dev. Biol. 231: 217–233. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Burks, C., and Posakony, J.W. 1998. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of Enhancer of split complex transcripts. Development 125: 4077–4088. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Bodner, R., Kavaler, J., Freschi, G., and Posakony, J.W. 2000a. Antagonism of Notch signaling activity by members of a novel protein family encoded by the Bearded and Enhancer of split gene complexes. Development 127: 291–306. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Bodner, R., and Posakony, J.W. 2000b. The Enhancer of split complex of Drosophila includes four Notch-regulated members of the Bearded gene family. Development 127: 3441–3455. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Tomancak, P., Williams, R.W., and Rubin, G.M. 2003. Computational identification of Drosophila microRNA genes. Genome Biol. 4: R42.1–R42.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N., Lim, L., Weinstein, E., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lecourtois M. and Schweisguth, F. 1995. The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the Enhancer of split complex genes triggered by Notch signaling. Genes & Dev. 9: 2598–2608. [DOI] [PubMed] [Google Scholar]
- Lee R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with anti-sense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Leviten M.W., Lai, E.C., and Posakony, J.W. 1997. The Drosophila gene Bearded encodes a novel small protein and shares 3′ UTR sequence motifs with multiple Enhancer of split complex genes. Development 124: 4039–4051. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., and Burge, C.B. 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge, C.B., and Bartel, D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Glasner, M.E., Yekta, S., Burge, C.B., and Bartel, D.P. 2003a. Vertebrate microRNA genes. Science 299: 1540. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. 2003b. The microRNAs of Caenorhabditis elegans. Genes & Dev. 17: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., and Johnson, J.M. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773. [DOI] [PubMed] [Google Scholar]
- Lin S.Y., Johnson, S.M., Abraham, M., Vella, M.C., Pasquinelli, A., Gamberi, C., Gottlieb, E., and Slack, F.J. 2003. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 4: 639–650. [DOI] [PubMed] [Google Scholar]
- Llave C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Morel V., Lecourtois, M., Massiani, O., Maier, D., Preiss, A., and Schweisguth, F. 2001. Transcriptional repression by Suppressor of Hairless involves the binding of a Hairless–dCtBP complex in Drosophila. Curr. Biol. 11: 789–792. [DOI] [PubMed] [Google Scholar]
- Moss E.G., Lee, R.C., and Ambros, V. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack, F., Basson, M., Pasquinelli, A., Bettinger, J., Rougvie, A., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. 2002. Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke, J., Russell, R.B., and Cohen, S.M. 2003. Identification of Drosophila microRNA targets. PLoS Biol. 1: E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. 2003. A biochemical framework for RNA silencing in plants. Genes & Dev. 17: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella M.C., Choi, E.Y., Lin, S.Y., Reinert, K., and Slack, F.J. 2004. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes & Dev. 18: 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]
- Wurmbach E., Wech, I., and Preiss, A. 1999. The Enhancer of split complex of Drosophila melanogaster harbors three classes of Notch responsive genes. Mech. Dev. 80: 171–180. [DOI] [PubMed] [Google Scholar]