Abstract
To investigate the determinants of promoter-specific gene regulation by the glucocorticoid receptor (GR), we compared the composition and function of regulatory complexes at two NFκB-responsive genes that are differentially regulated by GR. Transcription of the IL-8 and IκBα genes is stimulated by TNFα in A549 cells, but GR selectively represses IL-8 mRNA synthesis by inhibiting Ser2 phosphorylation of the RNA polymerase II (pol II) C-terminal domain (CTD). The proximal κB elements at these genes differ in sequence by a single base pair, and both recruited RelA and p50. Surprisingly, GR was recruited to both of these elements, despite the fact that GR failed to repress the IκBα promoter. Rather, the regulatory complexes formed at IL-8 and IκBα were distinguished by differential recruitment of the Ser2 CTD kinase, P-TEFb. Disruption of P-TEFb function by the Cdk-inhibitor, DRB, or by small interfering RNA selectively blocked TNFα stimulation of IL-8 mRNA production. GR competed with P-TEFb recruitment to the IL-8 promoter. Strikingly, IL-8 mRNA synthesis was repressed by GR at a post-initiation step, demonstrating that promoter proximal regulatory sequences assemble complexes that impact early and late stages of mRNA synthesis. Thus, GR accomplishes selective repression by targeting promoter-specific components of NFκB regulatory complexes.
Keywords: GR, P-TEFb, repression
Proper control of eukaryotic transcription relies on the assembly of multiprotein regulatory complexes at promoter-proximal genomic response elements. This is accomplished through protein–protein and protein–nucleic acid interactions that occur in a cell- and promoter-specific manner. These myriad interaction surfaces form the infrastructure of a combinatorial control network that regulates tissue- and gene-specific transcription. A major task facing biologists in the post-genomic era is to elucidate the mechanisms and combinatorial code that are required to precisely orchestrate cellular genomic responses.
Transcriptional regulatory factors function at genomic sites through three distinct classes of response elements: simple, composite, and tethering (Yamamoto et al. 1998). Simple response elements recruit a single DNA-binding factor that is necessary and sufficient for regulation. Composite elements also directly bind the regulatory protein, but the function of the regulator is altered by the presence of heterotypic factors. Tethering response elements do not contain a high-affinity binding site for the regulatory factor; rather, factor recruitment is accomplished through stabilizing protein–protein interactions with other DNA-bound factors.
Naturally occurring response elements commonly diverge from the consensus or high-affinity binding sites for regulatory proteins, which are typically defined in vitro. This is due in part to the fact that many response elements are not simply sites for specific localization of transcription factors. Rather, the response elements serve also as effectors of transcription factor function (Tan and Richmond 1990; Miner and Yamamoto 1991; Ikeda et al. 1996; Cleary et al. 1997; Lefstin and Yamamoto 1998); that is, the regulatory surfaces exposed in a given response element context may effect recruitment of specific cofactors or result in the utilization of a general cofactor in functionally distinct ways.
The intricacy of transcriptional networks is clearly illustrated by the mammalian inflammatory response (Jobin and Sartor 2000; Lentsch and Ward 2000; Hawiger 2001; Kracht and Saklatvala 2002). Inflammation is a highly regulated and complex process that occurs in response to injury and invasion. While protective in nature, chronic inflammation has extremely deleterious effects that are associated with a wide variety of human diseases including asthma and rheumatoid arthritis. Early stages in the inflammatory response are regulated by the Rel gene family, which encodes the NFκB transcriptional regulatory proteins. Homo- or heterodimeric complexes of NFκB proteins bind to κB response elements proximal to the promoters of proinflammatory genes, such as IL-8 and ICAM-1, and regulate transcription. In addition to proinflammatory target genes, the NFκB proteins also regulate the expression of the inhibitor of the κB (IκB) gene family. The IκB proteins bind to NFκB dimers and prevent nuclear translocation and DNA binding. Detailed analyses of the IκB-NFκB signaling module indicate that the negative regulators of NFκB comprise a highly complicated sensory system that responds to both the extent and duration of inflammatory signaling (Hoffmann et al. 2002).
Glucocorticoids are the most common therapeutics for treatment of inflammatory diseases. Their anti-inflammatory effects are mediated through the intracellular glucocorticoid receptor (GR), which binds cognate ligands and regulates target gene expression positively or negatively from simple, composite, and tethering response elements. Previous studies suggest that GR can regulate the activities of NFκB via tethering regulatory complexes formed at κB-binding sites. This model is supported by evidence for direct interaction between RelA and GR in vitro and by the observation that NFκB DNA binding is not blocked by GR in vivo (Nissen and Yamamoto 2000). The mechanisms by which GR interferes with transcriptional activation activities in this tethering mode are not well understood, but are likely to depend on both cellular and promoter context (De Bosscher et al. 2000; Rogatsky et al. 2001, 2002).
Nissen and Yamamoto (2000) showed that GR antagonizes RelA activity at the IL-8 gene in A549 human lung carcinoma cells by interfering with Ser2 phosphorylation of the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (pol II), apparently without blocking preinitiation complex (PIC) assembly. These studies also revealed that CTD Ser2 phosphorylation at the IκBα gene is unaffected in the same cells, consistent with the fact that GR does not repress IκBα transcription.
The CTD is important for initiating, sustaining, and terminating the transcription cycle of pol II regulated genes. The human pol II CTD comprises 52 repeats of a consensus heptapeptide sequence, YSPTSPS, which are reversibly phosphorylated on Ser2 and Ser5 during transcription (Dahmus 1995; Palancade and Bensaude 2003). Phosphorylation of Ser5 occurs proximal to the promoter and appears to be important for efficient transition from transcription initiation to elongation, whereas Ser2 phosphorylation stimulates efficient elongation of pol II complexes (Rougvie and Lis 1990; Rasmussen and Lis 1993; O'Brien et al. 1994; Conaway et al. 2000). Moreover, CTD phosphorylation appears to connect transcription with various processing steps in the production of mature mRNA molecules, such as 5′-capping, cap methylation, splicing, polyadenylation, and cleavage (Cho et al. 1997, 1998; McCracken et al. 1997a,b; Ho et al. 1998; Andrulis et al. 2000, 2002; Komarnitsky et al. 2000; Barilla et al. 2001; Fong and Bentley 2001; Fong and Zhou 2001; Ahn et al. 2004; Kornblihtt et al. 2004).
There are several human kinases and phosphatases that appear to modulate pol II activities by affecting CTD phosphorylation (Majello and Napolitano 2001). P-TEFb is a complex of Cdk9 and cyclin T that possesses CTD Ser2 kinase activity and stimulates pol II elongation in vitro (Marshall and Price 1995; Marshall et al. 1996; Peng et al. 1998; Price 2000; Garriga and Grana 2004). Cyclin T interacts with various transcriptional regulators, including CIITA, MyoD, c-Myc, NFκB, and HIV TAT (Napolitano et al. 2000; Eberhardy and Farnham 2001, 2002), which may facilitate recruitment of P-TEFb to regulated promoters. Lis and coworkers demonstrated that recruitment of P-TEFb to the HSP70 gene following heat shock stimulates proper cleavage and polyadenylation of Hsp70 pre-mRNA (Ni et al. 2004).
One model for glucocorticoid-mediated repression of IL-8 is that GR tethers to promoter proximal NFκB and either inhibits a CTD kinase activity or recruits a CTD phosphatase. However, GR only represses a subset of NFκB-responsive promoters; for example, expression of the IκBα gene is strongly activated by NFκB but is unaffected by dexamethasone (dex). We sought to identify the determinants of promoter-specific glucocorticoid regulation by performing a comparative analysis of the composition and activity of regulatory complexes formed at the IL-8 and IκBα gene regulatory regions in A549 cells.
Results
Repression of NFκB activation activity by GR is gene specific
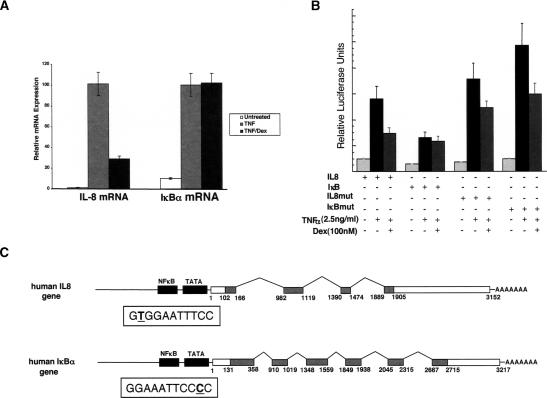
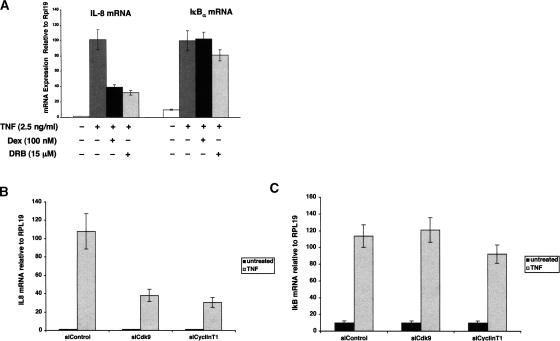
The A549 human lung adenocarcinoma cell line is highly responsive to the proinflammatory cytokine TNFα, which increases transcription of NFκB-responsive genes such as, IL-8, ICAM-1, and GM-CSF, as well as components of the negative regulatory loop of the NFκB pathway including the IκBα gene. A549 cells also contain endogenous GR. Treatment of A549 cells with TNFα for 2 h resulted in a robust increase in the steady-state levels of IL-8 and IκBα mRNA molecules as determined by quantitative real-time PCR (qRT–PCR) analysis. Co-treatment with TNFα and dex for 2 h resulted in a four-fold decrease in IL-8 mRNA accumulation, whereas IκBα mRNA levels were unaffected (Fig. 1A).
Figure 1.
GR differentially regulates NFκB at the IL-8 and IκBα genes in A549 cells. (A) Steady-state IL-8 and IκBα mRNA levels are stimulated in A549 cells treated for 2 h with TNFα (2.5 ng/mL). Cotreatment with TNFα and dex (100 nM) selectively repressed IL-8 mRNA accumulation. (B) A549 cells were transfected with IL-8-Luc, IκB-Luc, IL-8mut-Luc, or IκBmut-Luc (50 ng) reporter genes and RSV-βgal (50 ng) followed by treatment with combinations of TNFα and dex as indicated for 5 h. Luciferase units were normalized to β-galactosidase activity. (C) Diagram of the human IL-8 and IκBα genes. The promoter proximal κB site sequences are boxed.
We were interested in understanding the mechanisms used by GR to selectively target repression to a subset of NFκB response genes within a single cellular context. Previous studies indicate that the IL-8 and IκBα regulatory regions each contain a single κB site that is necessary to confer maximal NFκB responsiveness (Algarte et al. 1999; Warny et al. 2000). First, we tested if the pattern of TNFα and dex regulation could be recapitulated using upstream regulatory fragments from the IL-8 and IκBα genes driving luciferase reporter gene expression. A549 cells were transiently transfected with the reporter plasmids prior to treatment with TNFα or TNFα and dex for 5 h. The IL-8 and IκBα reporters were both induced by treatment with TNFα (five- and fourfold, respectively) and the IL-8 promoter was repressed twofold by dex relative to TNFα treatment alone (Fig. 1B). The TNFα-induced activity of the IκBα reporter construct was not significantly reduced by dex. These experiments indicate that genomic regulatory fragments of the IL-8 and IκBα genes are sufficient to recapitulate the pattern of regulation of endogenous IL-8 and IκBα by TNFα and dex.
We hypothesized that differential GR regulation of the IL-8 and IκBα genes may be the result of differences in core elements that specify formation of distinct regulatory complexes. Interestingly, the κB response elements regulating the IL-8 and IκBα genes differ by a single base pair and in their orientation with respect to the transcription start site (Fig. 1C). The IL-8 κB element is 5′-GTGGAATTTCC-3′ and the element upstream of the IκBα gene is 5′-GGGGAATTTCC-3′. We therefore introduced mutations into the IL-8 and IκBα promoter reporter constructs, which interconvert their respective κB response elements (Fig. 1B). Strikingly, the mutant IκBα reporter was induced by TNFα and repressed by dex to nearly the same extent as the IL-8 reporter. The mutant IL-8 reporter gene is repressed albeit to a lesser extent than the wild-type promoter construct. Together, these results indicate that within a single cellular context two NFκB regulated promoters are differentially regulated by GR. In addition, the degree of promoter regulation by GR is determined in part by the primary sequence of the κB response elements in the promoter proximal regulatory regions of the IL-8 and IκBα genes.
Regulatory complex composition at the IL-8 and IκBα gene in vivo
We were interested in determining whether the IL-8 and IκBα κB response elements recruit compositionally distinct regulatory complexes that could account for the observed differential GR regulation. We assessed the in vivo occupancy of these κB response elements by RelA and p50 using chromatin immunoprecipitation (ChIP). Similar to previous findings at the IL-8 gene in A549 cells, we found that RelA recruitment to IκBα was increased threefold relative to untreated cells and that dex had no effect on RelA occupancy (Fig. 2A). We also performed ChIP assays using an antibody specific to the p50 subunit and the immunoprecipitates were probed with PCR primers specific to the IL-8 and IκBα promoter and downstream genomic regions (Fig. 2B). For both genes, we found that p50 occupancy in the promoter region was enhanced by TNFα treatment, and occupancy was not diminished by dex. In contrast, we did not detect significant binding of p50 to the IκBα or IL-8 downstream regions (Fig. 2B).
Figure 2.
NFκB and GR factor occupancy does not distinguish the IL-8 and IκBα gene regulatory regions. In A, B, and D, ChIP assays were performed on A549 cells treated for 2 h as indicated using polyclonal RelA (A), p50 (B), or GR(N499) (D) antibodies. (A) RelA immunoprecipitates were probed using IκBα promoter primers (–168 to +21) and normalized to U6 snRNA genomic fragment. (B) p50 immunoprecipitates were analyzed for the presence of sequences from the IL-8 and IκBα promoter and downstream regions as indicated; fold enrichment values were normalized to a control region of the HSP70 gene. (C) qRT–PCR analysis of RNA isolated from A549 cells treated with combinations of TNFα and Kamebakaurin for 2 h as indicated. (D) GR immunoprecipitates were probed using primers specific for the IL-8 and IκBα promoter regions and normalized to the U6 control region.
To examine the functional involvement of p50 in regulating IL-8 and IκBα gene transcription, we used kamebakaurin, a diterpene natural product that selectively and covalently modifies p50 and prevents DNA binding by p50 containing dimers (Hwang et al. 2001; Lee et al. 2002). Treatment of A549 cells with kamebakaurin blocked TNFα induction of IL-8 and IκBα reporter genes (data not shown), and reduced the steady-state levels of IL-8 and IκBα mRNA (Fig. 2C). These findings suggest that despite the single base pair sequence difference between the IL-8 and IκBακB core elements, both are regulated in vivo by NFκB heterodimers containing RelA and p50.
GR tethering complexes form at both IL-8 and IκBα promoters
We next tested whether the RelA/p50 complexes at the IL-8 and IκBα promoters differ in their ability to recruit GR. Previous studies indicated that GR interacts directly with RelA in vitro. We demonstrated by ChIP analysis that GR occupancy of the IL-8 promoter region is increased approximately fourfold in vivo upon stimulation with TNFα and dex, and discovered to our surprise that the IκBα promoter region was similarly enriched (Fig. 2D). Thus, it appears that GR is recruited to the IκBα promoter, but fails to repress transcription of the IκBα gene. To gain insight into the mechanism of differential regulation of the IL-8 and IκBα genes, we therefore sought to obtain a more detailed understanding of the mechanism by which GR represses IL-8 mRNA synthesis.
GR represses IL-8 transcription at a step following initiation
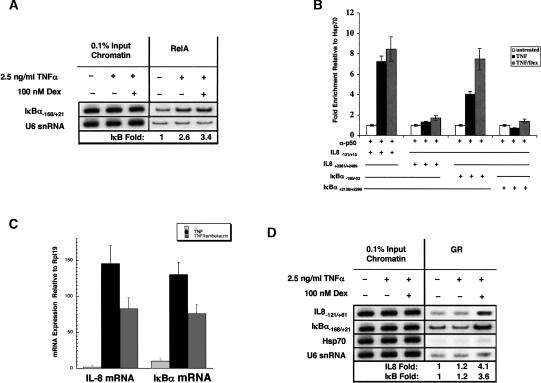
Previous studies indicate that dex treatment does not block PIC assembly at the IL-8 promoter in A549 cells, but instead correlates with a net loss in phosphorylation of the Ser2 position in the pol II CTD heptapeptide repeats (Nissen and Yamamoto 2000). As CTD Ser2 phosphorylation is thought to primarily affect post-initiation functions of pol II, we examined by nuclear run-on assays whether GR affects IL-8 mRNA accumulation without altering the rate of initiation from the IL-8 promoter. A549 cells were treated either with TNFα or cotreated with TNFα and dex for 2 h, nuclei were isolated, and run-on transcription performed. RNA isolated from these nuclei was hybridized to denatured partial cDNA fragments corresponding to the IL-8, IκBα, and GAPDH coding sequences that lack 5′-UTRs (Fig. 3A). These experiments reveal that transcription of the IL-8 and IκBα genes is stimulated by TNFα treatment, and under conditions that repress steady-state IL-8 mRNA levels, there is no apparent decrease in the rate of transcription initiation from the IL-8 promoter. These results suggest that GR interferes with IL-8 gene transcription at a step following initiation.
Figure 3.
IL-8 transcription is repressed by dex at a post-initiation step. (A) Nuclear run-on assays were performed on nuclei isolated from A549 cells treated for 2 h with combinations of TNF and dex as indicated. RNA was hybridized to cDNA fragments of the human IL-8, IκBα, and GAPDH genes. (B) ChIP assays were performed on A549 cells treated for 2 h with combinations of TNFα and dex as indicated using a Pol II polyclonal antibody and probed for sequences located in the IL-8 promoter and 3′-UTR. Fold enrichment values for the IL-8 regions were normalized to a control region from the HSP70 gene.
RNA pol II occupancy of the IL-8 regulatory region in A549 cells was not diminished by dex treatment (Fig. 3B; Nissen and Yamamoto 2000). We performed ChIP analysis in order to assess pol II occupancy of genomic sequences downstream of the IL-8 promoter. Probing total pol II immunoprecipitates with primers specific to the 3′-UTR of the IL-8 gene revealed a twofold decrease in pol II occupancy of this region when cells were treated with TNFα and dex relative to TNFα treatment alone. Taken together, these results suggest that the transcriptional defect introduced by GR impacts a step following initiation and that the defect does not preclude pol II occupancy near the 3′-UTR of the IL-8 gene.
IL-8 and IκBα genes have different requirements for P-TEFb
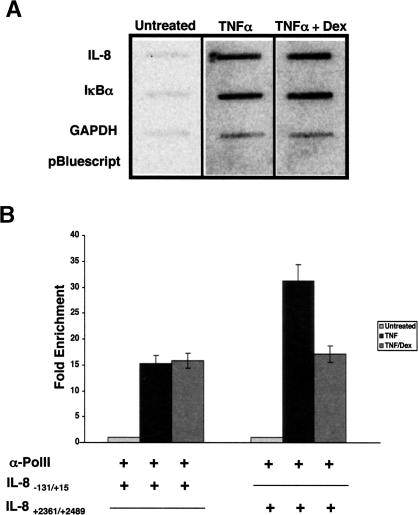
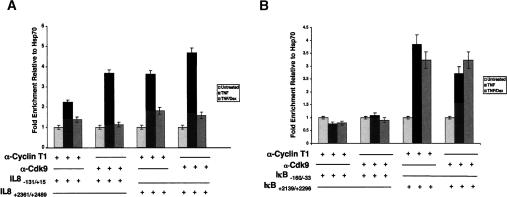
The observations that GR affects a post-initiation event in IL-8 transcription and triggers hypophosphorylation of CTD Ser2 positions on the pol II molecules near the IL-8 promoter led us to pursue P-TEFb. Indeed, previous studies had indicated that RelA can interact directly with P-TEFb through contacts with the cyclin box of cyclin T1 (Barboric et al. 2001). We therefore used ChIP to test the recruitment of P-TEFb to the IL-8 and IκBα gene regulatory regions, and found that TNFα treatment increased occupancy of Cdk9 and cyclin T1 at IL-8 (Fig. 4A) but not at IκBα (Fig. 4B).
Figure 4.
The P-TEFb kinase complex is differentially recruited to and utilized by the IL-8 and IκBα genes in A549 cells. ChIP assays using polyclonal antibodies to Cdk9 (A) or Cyclin T1 (B) were performed on A549 cells treated with combinations of TNF and dex as indicated for 2 h. The immunoprecipitates were probed using qRT–PCR for regions of the IL-8 and IκBα promoter and downstream regions as indicated. The fold enrichment data were normalized to a HSP70 control genomic region.
We also addressed the occupancy of P-TEFb at sequences within the IL-8 and IκBα genes. These experiments revealed TNFα-induced P-TEFb recruitment to the 3′-UTR of the IL-8 gene (Fig. 4A), consistent with the idea that P-TEFb is recruited to the IL-8 promoter and associates with elongating pol II complexes. In contrast to the IκBα promoter region, P-TEFb occupancy of the 3′ end of the IκBα gene increased three- to fourfold following TNFα induction (Fig. 4B). Thus, P-TEFb appears to be recruited to the IκBα 3′ end, but this recruitment is not likely mediated by NFκB.
Given the different profiles of P-TEFb recruitment to IL-8 and IκBα, we tested the effect of the Cdk inhibitor, 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), on IL-8 and IκBα mRNA accumulation. A549 cells were treated for 2 h with TNFα or cotreated with TNFα and DRB. The abundance of IL-8 mRNA from cells treated with TNFα and DRB was fourfold lower than in cells treated with TNFα alone (Fig. 5A). In contrast, the amount of IκBα mRNA isolated from DRB treated cells was ∼85% of that isolated from cells treated with TNFα alone.
Figure 5.
IκBα mRNA synthesis is independent of P-TEFb function. (A–C) qRT–PCR analysis of RNA isolated from A549 cells treated with combinations of TNF, dex, and DRB as indicated for 2 h. The abundance of IL-8 and IκBα mRNA were normalized to a RPL19 control. (B,C) A549 cells were transfected with siRNA oligos directed against the Cdk9 or Cyclin T1 subunits of P-TEFb or siRNA control for 26 h. The cells were untreated or treated with TNF (2.5 ng/mL) for 2 h and total RNA was isolated and analyzed by qRT–PCR using primers specific to the IL-8 (B) and IκBα (C) transcripts. These values were normalized to the RPL19 gene.
We also used small interfering RNA (siRNA) directed against Cdk9 and cyclin T1 to test whether the difference in DRB sensitivity of the IL-8 and IκBα mRNA molecules reflected differential requirements for P-TEFb. Transfection of A549 cells with siRNA significantly reduced the amount of Cdk9 and cyclin T1 mRNA and protein (Supplementary Fig. 1). qRT–PCR analysis revealed that IL-8 mRNA stimulation by TNFα was diminished by 60%–70% (Fig. 5B), whereas IκBα mRNA levels were unaffected (Fig. 5C). The extent of Cdk9 and cyclin T1 reduction correlates with the loss of IL-8 mRNA accumulation in TNF-treated cells. Taken together, these results suggest that the IL-8 and IκBα genes have different requirements for P-TEFb, and that recruitment of P-TEFb by RelA appears to be promoter specific.
GR represses IL-8 transcription by competing with P-TEFb
As P-TEFb is required for IL-8 transcription, we tested the effect of dex on recruitment of Cdk9 and cyclin T1 to the IL-8 promoter. ChIP analysis revealed that dex significantly reduced the occupancy of both factors (Fig. 4A); dex treatment also decreased occupancy by P-TEFb of the 3′-UTR of the IL-8 gene. In contrast, P-TEFb recruitment to the 3′ end of the IκBα gene was not significantly affected by dex (Fig. 4B), suggesting that GR successfully antagonizes only P-TEFb that is recruited to promoter proximal κB elements.
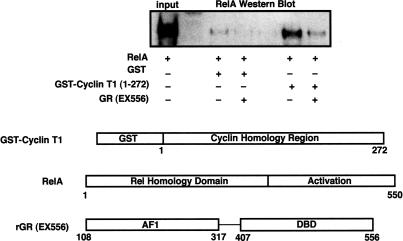
Given that RelA interacts directly with GR and P-TEFb, we tested the possibility that the exclusion of P-TEFb from κB regulatory complexes occurred by direct competition between GR and cyclin T1 for binding to RelA. We performed GST-pulldown experiments using purified recombinant GST-Cyclin T1 (1–272), RelA, and GR (EX556) (Fig. 6). GST-Cyclin T1 was incubated with RelA to allow complexes to form. The first 272 amino acids of human cyclin T1 contain the cyclin boxes required for interaction with RelA (Barboric et al. 2001). The cyclin T1:RelA complexes were isolated following the addition of glutathione agarose beads, washed with binding buffer, and subsequently incubated in the presence or absence of 100 nM purified GR (EX556) for 30 min. The amount of RelA bound to the GST-Cyclin T1 was reduced approximately sixfold in the presence of GR, suggesting that GR and P-TEFb bind to RelA competitively in vitro.
Figure 6.
Interaction between RelA and Cyclin T1 is diminished by GR in vitro. Purified GST-Cyclin T1 (1–272) or GST alone was incubated with purified recombinant RelA. The resulting complexes were isolated and washed on glutathione agarose beads and then were incubated in the presence or absence of 100 nM GR. The amount of RelA bound to the beads was determined by Western blot using sc-109.
Discussion
Modulation of transcription in eukaryotes is accomplished through assembly of regulatory complexes that specify the kinetics and magnitude of mRNA synthesis. Although it is well known that many regulatory proteins can function both in transcriptional activation and repression, no simple rules have been defined that predict factor activity based on genomic response element sequences. One possibility is that regulatory complexes contain independently functioning positive and negative factors, and that the resultant transcriptional output is simply the sum of these components. Another possibility is that the regulatory complexes themselves dictate which protein surfaces are exposed and/or functional; in this case the mechanisms by which a given regulatory factor functions are dependent on the composition of the regulatory complex as a whole and are not intrinsic to the regulator. Exploring these issues requires experimental conditions that allow us to relate the structure or composition of complexes with their function.
In the current study, we have taken a comparative approach that relies upon defining differences in regulatory complex function at two differentially regulated genes within the same cell and correlates those differences with regulatory complex composition. IL-8 and IκBα, two primary NFκB response genes, are differentially antagonized by glucocorticoid signaling in A549 cells. We therefore sought to understand the molecular determinants of differential repression of NFκB by GR. We found that a point mutation in the IκBα regulatory region that altered the κB sequence to match that at IL-8 resulted in a striking increase in TNFα inducibility and in the acquisition of GR repressibility. These experiments suggest that the κB response element sequences specify distinct regulatory functions at these promoters and raise the intriguing possibility that the κB elements function as allosteric modulators of NFκB function.
Similarly, studies of the β-interferon enhancer (Thanos and Maniatis 1995) revealed that substitution of the natural κB response element with other NFκB-binding sites resulted in some cases in the loss of response to viral infection, suggesting that the κB element sequence has evolved to function in a given regulatory complex and promoter context. There is also evidence that single nucleotide changes in κB response elements can alter the requirement for specific coactivator proteins (Leung et al. 2004). These studies underscore that response elements themselves are important components of regulatory complexes that impart specificity on the functions of bound transcriptional regulators.
Having established the importance of the core κB response elements at the IL-8 and IκBα genes, we sought to define the regulatory complexes at the two sites using a candidate approach. We observed that the regulatory complexes formed at the IL-8 and IκBα response elements each contained RelA and p50, despite the fact that the IL-8 κB element has higher intrinsic affinity for RelA homodimers in vitro (data not shown; F.E. Chen et al. 1998; Y.Q. Chen et al. 1998). This underscores the fact that intrinsic DNA-binding affinity does not necessarily correlate with in vivo occupancy and highlights the need to study regulatory complex formation at natural response elements in vivo. We conclude that the differential GR regulation at these response elements does not simply reflect differences in the identity of the NFκB components of the regulatory complexes. Notably, we failed to observe any differences in chromatin modifications at the IL-8 and IκBα promoters that could account for the differential regulation by GR (H.F. Luecke and K.R. Yamamoto, unpubl.). These findings are consistent with those of Saccani and Natoli (2002). Our experiments also revealed that GR was recruited with equal efficiency to the IL-8 and IκBα response elements in vivo. These findings demonstrate that recruitment of GR to NFκB response elements is not sufficient to confer transcriptional regulation in this context.
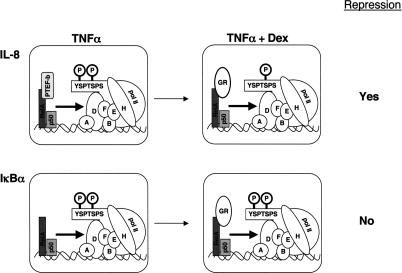
Prior investigations had revealed that the cyclin-dependent kinase P-TEFb can interact directly with the RelA subunit of NFκB, and that P-TEFb is responsible for phosphorylation of the RNA pol II CTD at Ser2. Hence, we examined RelA recruitment of P-TEFb to the IL-8 and IκBα regulatory complexes. Surprisingly, we found that P-TEFb was recruited to the IL-8 but not to the IκBα complex. At IL-8, GR binding to RelA was accompanied by loss of P-TEFb occupancy. These results suggest a model for GR regulation of NFκB in which GR blocks RelA recruitment of P-TEFb to the IL-8 gene, with concomitant diminution of Ser2 phosphorylation of the pol II CTD at IL-8 (Fig. 7). In contrast, at IκBα transcriptional activity and Ser2 phosphorylation of pol II complexes do not require promoter recruitment of P-TEFb, thereby precluding GR-mediated repression at IκBα. Thus, the characteristic that leads to differential GR regulation at IL-8 and IκBα is competition with a promoter-specific NFκB coregulator. It will be interesting to determine if the single nucleotide difference in the κB core sequences regulating these genes is sufficient to specify a difference in P-TEFb recruitment.
Figure 7.
A model for differential regulation of the IL-8 and IκBα genes by the glucocorticoid receptor. Heterodimeric complexes of RelA and p50 occupy both the promoter proximal κB elements of these genes; however, P-TEFb is selectively recruited to the IL-8 gene. Dex treatment results in GR recruitment to both genes, which results in loss of P-TEFb recruitment to the IL-8 promoter, suggesting a direct competition, but has no effect at IκBα.
These studies raise several intriguing issues regarding the role of P-TEFb in transcription of NFκB regulated promoters. Our observation that P-TEFb is differentially recruited to two NFκB regulated promoters indicates that P-TEFb is not a global coregulator of NFκB, but appears to be recruited to only a subset of RelA/p50 response elements. This is consistent with recent results showing that most but not all pol II transcription is sensitive to P-TEFb inhibitors flavopiridol and DRB (Chao and Price 2001; Medlin et al. 2003). It will be important to identify the CTD Ser2 kinase that functions at IκBα and to discover the mechanism of its selective recruitment.
What are the mechanisms by which P-TEFb modulates eukaryotic transcription? The results of our nuclear run-on assays indicate that P-TEFb is required at a post-initiation step in IL-8 transcription. Although we have not identified specific IL-8 mRNA defects that accompany dexamethasone treatment, it is notable that we have characterized a promoter-proximal complex that confers GR repression at a post-initiation step. Recent mechanistic studies on the human U2 snRNA gene and the Drosophila HSP70 gene suggest that one role of P-TEFb is to specify proper control of 3′ end formation and processing of pol II transcripts, presumably mediated through phosphorylation of the pol II CTD (Medlin et al. 2003; Ni et al. 2004). Similar observations have been made for the Saccharomyces cerevisiae Ser2 kinase, Ctk1 (Ahn et al. 2004). These studies suggest that P-TEFb does not simply modulate the rate and processivity of RNA polymerization, but appears to coordinate the events of mRNA synthesis by establishing and maintaining Ser2 phosphorylation patterns on the CTD.
The emerging picture of eukaryotic gene expression is one in which the multiple steps required for eventual emergence of mature mRNA molecules occur in a highly coordinated manner. The intimate coupling of mRNA processing events to pol II transcription has been well documented, especially with respect to capping and co-transcriptional splicing of pre-mRNAs. It has been suggested that coordination of these events, which can function independently in vitro, provides necessary checkpoint mechanisms whereby quality control of mRNA synthesis is accomplished. We provide evidence that promoter-proximal regulatory factors can exploit this coupling to modulate a post-initiation step in mRNA synthesis without affecting the rate of transcription initiation per se. In principle, this implies that all of the events that together give rise to a mature mRNA can be specified at the earliest stages of synthesis, and consequently that virtually any step in mRNA production could be regulated from a promoter-proximal site.
The NFκB signaling module is archetypal of the complexities of gene regulation. In addition to regulating transcription of a host of positive target genes involved in cell division, apoptosis, and inflammation, NFκB also regulates components of its own negative feedback loop. Many important physiological and pharmacological functions of glucocorticoids involve negative regulation of NFκB. To accomplish these, GR selectively represses NFκB activity at positive genes, without blocking expression of components of the negative feedback loop such as IκBα. In this study, we have examined the regulatory complexes formed at canonical positive and negative components of the NFκB signaling module. We found that NFκB differentially utilizes P-TEFb to control expression of the IL-8 and IκBα genes, highlighting the fact that complexity of gene regulatory networks is achieved through combinatorial assembly of multiprotein complexes on genomic response elements. By competing with the P-TEFb selectively recruited to the IL-8 promoter, GR represses IL-8 transcription without blocking expression of IκBα. In principle, these studies suggest that any component of a regulatory complex could serve as a distinguishing determinant, which greatly increases the potential for combinatorial control at the intersection of biological signaling pathways.
Materials and methods
Cell lines, plasmids, transient transfection, and treatments
A549 human lung carcinoma cells were cultured in Dulbecco's modified Eagle medium (DMEM, Invitrogen) supplemented with 5% fetal bovine serum (FBS, HyClone Laboratories). Plasmids used for the expression of his-tagged mouse RelA and GST-tagged human Cyclin T1 have been described (Nissen and Yamamoto 2000; Das et al. 2004). The IL-8-Luc reporter gene contains a –1481/+40-bp region of the IL-8 gene driving firefly luciferase expression (Warny et al. 2000). The IκB-Luc reporter construct contains a 400-bp region of the IκBα gene driving firefly luciferase expression (Algarte et al. 1999). The IL-8mut-Luc and IκBmut-Luc reporter genes were constructed by Quick-change mutagenesis using complimentary synthetic oligonucleotides (IL-8, 5′-CATTATGTCAGGGGAAATTCCACGATTTGCAACTG-3′ and IκBα, 5′-GACTGGCTTGGAAATTCCACGAGCCTGACCCCGCC-3′). For transient transfection reporter gene assays, A549 cells were split in DMEM-5% FBS into 24-well plates at 30,000 cells/well and transfected the following day in serum-free DMEM with Lipofectamine-PLUS reagent (Invitrogen) using 0.8 μL/well Lipofectamine and 1.4 μL/well PLUS as per the manufacturer's instructions. Three hours post-transfection, cells were refed with DMEM-5% FBS; 12 h post-transfection, the cells were treated for 5 h with combinations of 2.5 ng/mL TNFα Roche), 100 nM dex (Invitrogen), and 15 μg/mL DRB (Sigma) in DMEM-5% FBS. Cells were lysed in 100 μL/well lysis buffer (Pharmingen) and assayed for luciferase and β-gal activity as previously described. Kamebakaurin was a generous gift from J.J. Lee (Korean Research Institute of Bioscience and Biotechnology, Taejon, Korea).
Antibodies
The human GR rabbit polyclonal antiserum (N499) was generated against a purified polypeptide corresponding to amino acid residues 1–499 of the human GR (R.M. Nissen, B. Darimont, and K.R. Yamamoto, unpubl.). RNA polymerase II (sc-899), RelA (sc-109), p50 (sc-7178), and Cdk9 (sc-484) polyclonal antibodies were purchased from Santa Cruz Biotechnology. The cyclin T1 antibody was a gift from D. Price (University of Iowa, Iowa City, IA).
ChIP
A549 cells grown in 150-mm dishes were treated as indicated in figure legends. Cross-linking was performed by adding 2.5 mL 11× formaldehyde solution (50 mM HEPES-KOH at pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, 11% formaldehyde) to the cells in media for 10 min at 25°C. Formaldehyde was quenched with 125 mM glycine for 5 min at 4°C; cells were rinsed in ice-cold phosphate-buffered saline (PBS), scraped off the dishes, and harvested by centrifugation (600g, 10 min at 4°C). Cell pellets were lysed by nutating at 4°C for 10 min in 10 mL ice-cold lysis buffer (50 mM HEPES-KOH at pH 7.4, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) supplemented with 1 mM phenymethylsulfonyl fluoride (PMSF) and 1 μg/mL each of aprotinin, leupeptin, and pepstatin A. Nuclei were collected by centrifugation (600g for 5 min at 4°C) and washed in 8 mL of wash buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 200 mM NaCl, supplemented with protease inhibitors) for 10 min at room temperature, collected as above, and resuspended in 2 mL of ice-cold RIPA buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 5% glycerol, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 1% Triton X-100, supplemented with protease inhibitors). Samples were sonicated with a Branson Sonifier 250 microtip at power setting 5, in 20-sec bursts separated by 1 min incubation on ice for a total of 3 min per sample. An average DNA fragment size of 200–800 bp was assessed by agarose gel electrophoresis followed by staining with ethidium bromide. Lysates were then cleared by centrifugation (16,000g, 15 min at 4°C) and 20 μL of each sample was saved as “input.” The remaining lysate was used for immunoprecipitation with RelA, p50, pol II, Cdk9, Cyclin T1 (2–3 μg/sample), N499 (24 μg total IgG/sample), or an equivalent amount of normal mouse or normal rabbit serum for 6 h at 4°C; and immune complexes were collected by nutating the lysates for 1 h at 4°C with 30 μL/sample of 50% protein A/G Plus agarose beads (Santa Cruz Biotechnology) preincubated with 100 μg/mL salmon sperm DNA (Invitrogen) in RIPA buffer. The beads were washed once with 0.5 mL of ice-cold RIPA buffer and then five times for 5 min with 1 mL of ice-cold RIPA buffer supplemented with 1 mM PMSF and 100 μg/mL salmon sperm DNA. The beads were then incubated in 100 mL of TE, 0.5% SDS, and 200 μg/mL proteinase K (Sigma) for 3 h at 55°C, and cross-links were reversed for 6 h at 65°C. The DNA was extracted twice with phenol-chloroform and once with chloroform, ethanol-precipitated in the presence of 20 μg glycogen at –20°C overnight, and resuspended in 40 μL TE. The isolated genomic DNA was amplified with primers specific to the human IL-8, IκBα, U6, and Hsp70 sequences. The oligonucleotide sequences used to amplify the IL-8 (–121/+61) region, IκBα (–168/+21), and U6 (–245/+85) were previously reported (Nissen and Yamamoto 2000). The oligonucleotide pairs used to amplify genomic regions of IL-8, IκBα using qPCR were IL-8 (–131/+15), 5′-TGTGATGACTCAGGTTTGC-3′ and 5′-TGT GCCTTATGGAGTGCTCC-3′; IL-8 (+2361/+2489), 5′-ATCTGGC AACCCTAGTCTGC-3′ and 5′-GTGCTTCCACATGTCCTCAC-3′; IκBα (–160/–33), 5′-GCTCAGGGTTTAGGCTTCTT-3′ and 5′-TATAAACGCTGGCTGGGGAT-3′; and IκBα (+2261/+2331), 5′-ACCTGGTGTCACTCCTGTTG-3′ and 5′-CTCTCTGGC AGCATCTGAAG-3′.
RNA isolation and quantitative real-time PCR analysis
Confluent A549 cell monolayers were treated in 100-mm dishes as described in the figure legends. Total RNA was isolated from cells using the guanidium isothiocyanate method according to the manufacturer's protocol (TriReagent, Molecular Research Center). Random-primed cDNA was prepared from 1 μg of total RNA using the ProtoScript first-strand cDNA synthesis kit (New England Biolabs). Five percent of the resultant cDNA was used per 50 μL reaction containing 1.25 units of Taq DNA polymerase (Invitrogen), 1.5 mM MgCl2, 300 nM of each primer, 0.5 mM dNTP mix, and 0.2× SYBR green I dye (Molecular Probes) in 1× Taq buffer. Real-time PCR was performed in an Opticon 2 DNA Engine (MJ Research) and analyzed by using the Ct method (Applied Biosystems Prism 7700 Users Bulletin No. 2 http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf) and ribosomal Rpl19 as a control for data normalization. An IL-8 cDNA fragment was amplified with the 5′-ATGATCTC TTTTGGAATTAAGGAGCAT-3′ and 5′-CATAATTTGGCC CAGGAGGAA-3′ primer pair. An IκBα cDNA fragment was amplified with the 5′-CATGCGCACAAATCCCTTCT-3′ and 5′-CATCTCTGTCGGCAAATTCGT-3′ primer pair.
Transient transfection of siRNA oligos
Confluent A549 cells were transfected with siRNA oligos (100 pmol) directed against hCdk9 or hCyclin T1 using RNAinfect (Qiagen) on 24-well plates according the manufacturer's protocol. After 3 h incubation in the presence of the lipid:siRNA complexes, the cells in each well were trypsinized and split into two clean wells of a 24-well plate. Twenty-four hours later, the cells were left untreated or were treated with TNFα (2.5 ng/mL) for 2 h. Subsequently, the cells were harvested in TriReagent (Molecular Research Center, Inc.) according to the manufacturer's instructions to isolate total RNA. The resulting RNA pellet was resuspended in 50 μL DEPC-treated water. Ten microliters of the RNA suspension was used as template for cDNA synthesis using the ProtoScript first-strand cDNA synthesis kit (New England Biolabs). Five percent of the cDNA template was used for qRT–PCR analysis using IL-8-, IκBα-, or Rpl19-specific PCR primers.
Nuclear run-on transcription assay
Confluent A549 cell monolayers (10-cm dish) were incubated in the presence of TNFα (2.5ng/mL) and/or dex (10–6 M) for 2 h. The media was removed, and the cells were washed once with ice-cold PBS and mechanically detached in 1 mL PBS, transferred to a 1.5-mL tube, and pelleted by centrifugation at 1000g for 2 min. The cell pellets were resuspended in 500 μL lysis buffer (10 mM Tris at pH 7.6, 2 mM MgCl2, 10 mM NaCl, 0.6% Triton X-100, 3 mM CaCl2), incubated for 5 min, and pelleted again. Nuclei were washed once with lysis buffer lacking Triton X-100. The nuclei were stored in freezing buffer (50 mM Tris (8.3), 40% glycerol, 5 mM MgCl2, 0.1 nM EDTA) at –70°C. Nuclei were thawed on ice and incubated in transcription buffer (10 mM Tris at pH 8.0; 0.3 M KCl; 5 mM MgCl2; 5 mM DTT; 1 mM each ATP, CTP, and GTP; 40 units RNasin) with [32P]UTP (0.5 mCi, 3000 Ci/mmol) for 30 min at 30°C. The reactions were terminated by addition of RNase-free DNase (Boehringer Mannheim), followed by treatment with proteinase K for 10 min at 55°C. RNA was extracted with chloroform-phenol-isoamyl alcohol (10:10:1) and precipitated with sodium acetate in EtOH. The pellet was then washed with 90% alcohol and pelleted. A slot-blot apparatus was used to prepare BrighstarPlus (Ambion) membranes containing 20 μg/slot of plasmid DNA, with cDNA insert encoding IL-8 and IκB and 3 μg/slot of cDNA encoding GAPDH. Twenty micrograms of pBluescript vector was used as a control for nonspecific binding. The membranes were hybridized in Ultrahyb for 24 h with equal amounts of labeled RNA (1 × 106 to 2 × 106 cpm/mL) according to the manufacturer's instructions. The filters were washed sequentially at 55°C in 1× SSC, 1% SDS, and 0.2× SSC, 0.2% SDS for 15 min each. This was followed by a wash at 37°C with 2× SSC containing RNase A (10 pg/mL) and RnaseTl (5 units/mL) for 15 min, prior to final washes in 2× SSC. The 32P-labeled RNA bound specifically to the filters was visualized by PhosphorImager.
In vitro binding assays
GST-Cyclin T1 (1–272) fusion protein and GST were expressed in Escherichia coli BL21(DE3) cells and purified as described. Each binding reaction was performed in 100 μL binding buffer (20 mM HEPES at pH 7.9, 1% Triton X-100, 20 mM DTT, 0.5% BSA, 100 mM KCl) for 1 h at 4°C using 50 μg GST fusion proteins and 50 μg purified RelA. After binding, glutathione-conjugated agarose beads were added and nutated for 30 min at 4°C. The complexes were isolated by centrifugation and washed three times with binding buffer lacking BSA. The beads were then resuspended in binding buffer or binding buffer containing 100 nM GR-EX556 and nutated for 30 min at 4°C and then collected by centrifugation. Bound proteins were eluted by boiling in SDS sample buffer and resolved by SDS-PAGE, and RelA was detected by Western blot using sc-109 (Santa Cruz).
Acknowledgments
We thank C. Das, D. Price, and J.J. Lee for their kind gifts of reagents. We are also grateful to members of the Yamamoto laboratory for advice and discussions, and A. Frankel, J. Taunton, G. Narlikar, E. Bolton, S. Meijsing, C. Pantoja, and J.-C. Wang for critical comments on the manuscript. This work was supported in part by an NIH post-doctoral fellowship to H.F.L. and by grants from the National Institutes of Health (R01 CA20535) and National Science Foundation (MCB9604938) to K.R.Y.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1297105.
References
- Ahn S.H., Kim, M., and Buratowski, S. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13: 67–76. [DOI] [PubMed] [Google Scholar]
- Algarte M., Kwon, H., Genin, P., and Hiscott, J. 1999. Identification by in vivo genomic footprinting of a transcriptional switch containing NF-κB and Sp1 that regulates the IκBα promoter. Mol. Cell. Biol. 19: 6140–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E.D., Guzman, E., Doring, P., Werner, J., and Lis, J.T. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes & Dev. 14: 2635–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E.D., Werner, J., Nazarian, A., Erdjument-Bromage, H., Tempst, P., and Lis, J.T. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420: 837–841. [DOI] [PubMed] [Google Scholar]
- Barboric M., Nissen, R.M., Kanazawa, S., Jabrane-Ferrat, N., and Peterlin, B.M. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8: 327–337. [DOI] [PubMed] [Google Scholar]
- Barilla D., Lee, B.A., and Proudfoot, N.J. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 98: 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao S.H. and Price, D.H. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276: 31793–31799. [DOI] [PubMed] [Google Scholar]
- Chen F.E., Huang, D.B., Chen, Y.Q., and Ghosh, G. 1998. Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature 391: 410–413. [DOI] [PubMed] [Google Scholar]
- Chen Y.Q., Ghosh, S., and Ghosh, G. 1998. A novel DNA recognition mode by the NF-κ B p65 homodimer. Nat. Struct. Biol. 5: 67–73. [DOI] [PubMed] [Google Scholar]
- Cho E.J., Takagi, T., Moore, C.R., and Buratowski, S. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 11: 3319–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.J., Rodriguez, C.R., Takagi, T., and Buratowski, S. 1998. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes & Dev. 12: 3482–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary M.A., Pendergrast, P.S., and Herr, W. 1997. Structural flexibility in transcription complex formation revealed by protein–DNA photocrosslinking. Proc. Natl. Acad. Sci. 94: 8450–8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway J.W., Shilatifard, A., Dvir, A., and Conaway, R.C. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25: 375–380. [DOI] [PubMed] [Google Scholar]
- Dahmus M.E. 1995. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim. Biophys. Acta 1261: 171–182. [DOI] [PubMed] [Google Scholar]
- Das C., Edgcomb, S.P., Peteranderl, R., Chen, L., and Frankel, A.D. 2004. Evidence for conformational flexibility in the Tat-TAR recognition motif of cyclin T1. Virology 318: 306–317. [DOI] [PubMed] [Google Scholar]
- De Bosscher K., Vanden Berghe, W., Vermeulen, L., Plaisance, S., Boone, E., and Haegeman, G. 2000. Glucocorticoids repress NF-κB-driven genes by disturbing the interaction of p65 with the basal transcription machinery, irrespective of coactivator levels in the cell. Proc. Natl. Acad. Sci. 97: 3919–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardy S.R. and Farnham, P.J. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 276: 48562–48571. [DOI] [PubMed] [Google Scholar]
- ____. 2002. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 277: 40156–40162. [DOI] [PubMed] [Google Scholar]
- Fong N. and Bentley, D.L. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes & Dev. 15: 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y.W. and Zhou, Q. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414: 929–933. [DOI] [PubMed] [Google Scholar]
- Garriga J. and Grana, X. 2004. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene 337: 15–23. [DOI] [PubMed] [Google Scholar]
- Hawiger J. 2001. Innate immunity and inflammation: A transcriptional paradigm. Immunol. Res. 23: 99–109. [DOI] [PubMed] [Google Scholar]
- Ho C.K., Sriskanda, V., McCracken, S., Bentley, D., Schwer, B., and Shuman, S. 1998. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 273: 9577–9585. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Levchenko, A., Scott, M.L., and Baltimore, D. 2002. The IκB-NF-κB signaling module: Temporal control and selective gene activation. Science 298: 1241–1245. [DOI] [PubMed] [Google Scholar]
- Hwang B.Y., Lee, J.H., Koo, T.H., Kim, H.S., Hong, Y.S., Ro, J.S., Lee, K.S., and Lee, J.J. 2001. Kaurane diterpenes from Isodon japonicus inhibit nitric oxide and prostaglandin E2 production and NF-κB activation in LPS-stimulated macrophage RAW264.7 cells. Planta Med. 67: 406–410. [DOI] [PubMed] [Google Scholar]
- Ikeda M., Wilcox, E.C., and Chin, W.W. 1996. Different DNA elements can modulate the conformation of thyroid hormone receptor heterodimer and its transcriptional activity. J. Biol. Chem. 271: 23096–23104. [DOI] [PubMed] [Google Scholar]
- Jobin C. and Sartor, R.B. 2000. The I κ B/NF-κ B system: A key determinant of mucosalinflammation and protection. Am. J. Physiol. Cell Physiol. 278: C451–C462. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P., Cho, E.J., and Buratowski, S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 14: 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A.R., de la Mata, M., Fededa, J.P., Munoz, M.J., and Nogues, G. 2004. Multiple links between transcription and splicing. RNA 10: 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracht M. and Saklatvala, J. 2002. Transcriptional and post-transcriptional control of gene expression in inflammation. Cytokine 20: 91–106. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Koo, T.H., Hwang, B.Y., and Lee, J.J. 2002. Kaurane diterpene, kamebakaurin, inhibits NF-κ B by directly targeting the DNA-binding activity of p50 and blocks the expression of antiapoptotic NF-κ B target genes. J. Biol. Chem. 277: 18411–18420. [DOI] [PubMed] [Google Scholar]
- Lefstin J.A. and Yamamoto, K.R. 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392: 885–888. [DOI] [PubMed] [Google Scholar]
- Lentsch A.B. and Ward, P.A. 2000. The NFκBb/IκB system in acute inflammation. Arch. Immunol. Ther. Exp. (Warsz) 48: 59–63. [PubMed] [Google Scholar]
- Leung T.H., Hoffmann, A., and Baltimore, D. 2004. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell 118: 453–464. [DOI] [PubMed] [Google Scholar]
- Majello B. and Napolitano, G. 2001. Control of RNA polymerase II activity by dedicated CTD kinases and phosphatases. Front. Biosci. 6: D1358–D1368. [DOI] [PubMed] [Google Scholar]
- Marshall N.F. and Price, D.H. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270: 12335–12338. [DOI] [PubMed] [Google Scholar]
- Marshall N.F., Peng, J., Xie, Z., and Price, D.H. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271: 27176–27183. [DOI] [PubMed] [Google Scholar]
- McCracken S., Fong, N., Rosonina, E., Yankulov, K., Brothers, G., Siderovski, D., Hessel, A., Foster, S., Shuman, S., and Bentley, D.L. 1997a. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 11: 3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Fong, N., Yankulov, K., Ballantyne, S., Pan, G., Greenblatt, J., Patterson, S.D., Wickens, M., and Bentley, D.L. 1997b. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361. [DOI] [PubMed] [Google Scholar]
- Medlin J.E., Uguen, P., Taylor, A., Bentley, D.L., and Murphy, S. 2003. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 22: 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner J.N. and Yamamoto, K.R. 1991. Regulatory crosstalk at composite response elements. Trends Biochem. Sci. 16: 423–426. [DOI] [PubMed] [Google Scholar]
- Napolitano G., Majello, B., Licciardo, P., Giordano, A., and Lania, L. 2000. Transcriptional activity of positive transcription elongation factor b kinase in vivo requires the C-terminal domain of RNA polymerase II. Gene 254: 139–145. [DOI] [PubMed] [Google Scholar]
- Ni Z., Schwartz, B.E., Werner, J., Suarez, J.R., and Lis, J.T. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 13: 55–65. [DOI] [PubMed] [Google Scholar]
- Nissen R.M. and Yamamoto, K.R. 2000. The glucocorticoid receptor inhibits NFκB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 14: 2314–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T., Hardin, S., Greenleaf, A., and Lis, J.T. 1994. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature 370: 75–77. [DOI] [PubMed] [Google Scholar]
- Palancade B. and Bensaude, O. 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 270: 3859–3870. [DOI] [PubMed] [Google Scholar]
- Peng J., Marshall, N.F., and Price, D.H. 1998. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 273: 13855–13860. [DOI] [PubMed] [Google Scholar]
- Price D.H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell Biol. 20: 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen E.B. and Lis, J.T. 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. 90: 7923–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I., Zarember, K.A., and Yamamoto, K.R. 2001. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. EMBO J. 20: 6071–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I., Luecke, H.F., Leitman, D.C., and Yamamoto, K.R. 2002. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc. Natl. Acad. Sci. 99: 16701–16706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A.E. and Lis, J.T. 1990. Postinitiation transcriptional control in Drosophila melanogaster. Mol. Cell. Biol. 10: 6041–6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S. and Natoli, G. 2002. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes & Dev. 16: 2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. and Richmond, T.J. 1990. DNA binding-induced conformational change of the yeast transcriptional activator PRTF. Cell 62: 367–377. [DOI] [PubMed] [Google Scholar]
- Thanos D. and Maniatis, T. 1995. Identification of the rel family members required for virus induction of the human β interferon gene. Mol. Cell. Biol. 15: 152–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warny M., Keates, A.C., Keates, S., Castagliuolo, I., Zacks, J.K., Aboudola, S., Qamar, A., Pothoulakis, C., LaMont, J.T., and Kelly, C.P. 2000. p38 MAP kinase activation by Clostridium difficile toxin A mediates monocyte necrosis, IL-8 production, and enteritis. J. Clin. Invest. 105: 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K.R., Darimont, B.D., Wagner, R.L., and Iniguez-Lluhi, J.A. 1998. Building transcriptional regulatory complexes: Signals and surfaces. Cold Spring Harb. Symp. Quant. Biol. 63: 587–598. [DOI] [PubMed] [Google Scholar]