Abstract
Transposable elements (TEs), long discounted as “selfish genomic elements,” are increasingly appreciated as drivers of genomic evolution, genome organization and gene regulation. TEs are particularly important in early embryo development, where advances in stem cell technologies, in tandem with improved computational and next-generation sequencing approaches, have provided an unprecedented opportunity to study the contribution of TEs to early mammalian development. Here, we summarize advances in our understanding of TEs in early human development and expand on how new stem cell-based embryo models can be leveraged to augment this understanding.
Keywords: Pluripotency, HERVK, Transposons, Stem Cells, Germ Cells, LTR5Hs
INTRODUCTION
A considerable portion of mammalian genomes are composed of Transposable Elements (TEs); in humans, the most recent assemblies identify ~53% of the genome as being composed of TEs [1,2]. There are two major classes of TE’s: retrotransposons, which involve an RNA intermediate that is reverse transcribed during the process of transposition, and DNA-only TEs, which do not use reverse transcriptase for transposition. The most prolific colonization of mammalian genomes has been achieved by retrotransposons, which can broadly be classified into Long Terminal Repeat (LTR) and non-LTR TEs (Figure 1). Non-LTR TEs include families such as Long interspersed nuclear element (LINE) and short interspersed nuclear elements (SINE), while LTR transposons include the Endogenous Retroviruses (ERVs) families. ERV integrants consist of pro-viral sequences, often flanked by regulatory LTRs, as well as solo-LTRs, which have lost their associated pro-virus through recombination or degradation [3]. In humans, most non-LTR and LTR-family TEs are no longer capable of transposition, while some families such as the LINE1 Human Specific (L1Hs), Alu SINE elements, and the composite retrotransposon family SINE-VNTR-Alu (SVA), a SINE derivative, remain mobile in humans [4].
Figure 1. Schematics of Retroviral Class Elements in Humans:
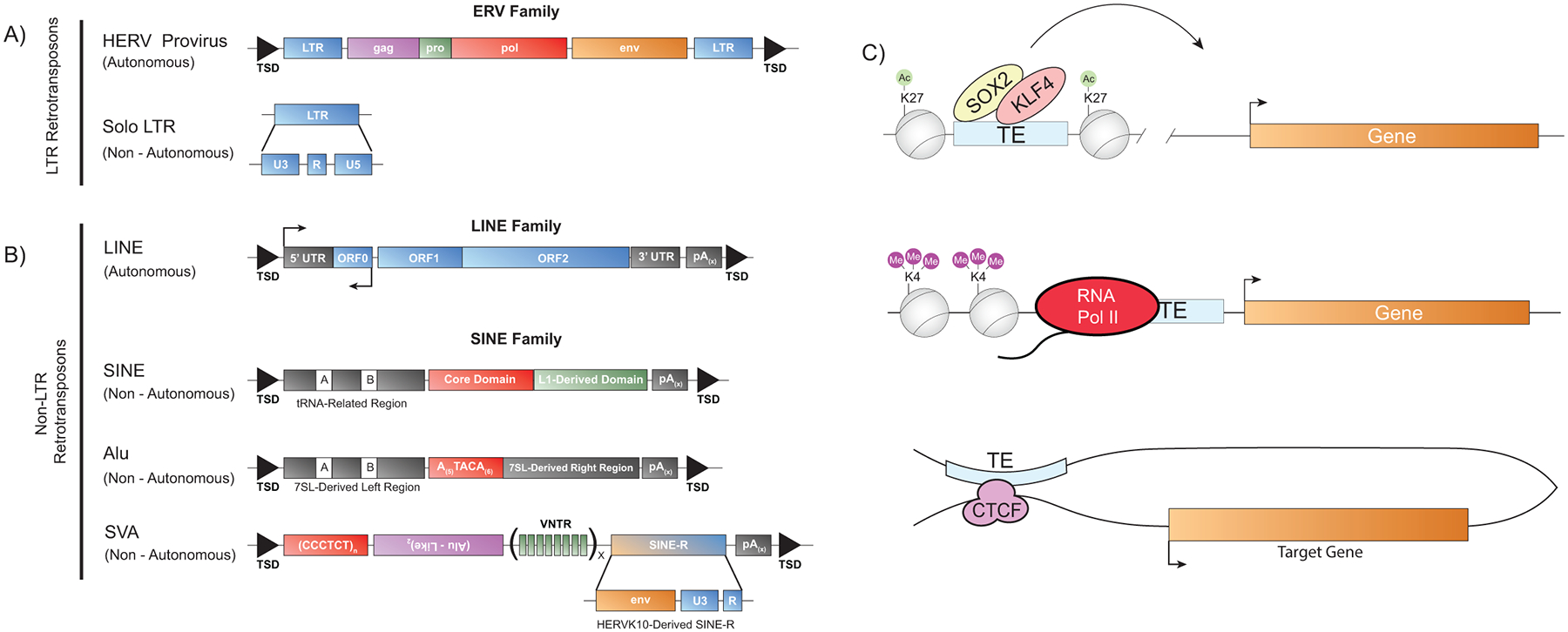
A) (Upper) A typical Human Endogenous Retrovirus (HERV) including (from left to right): Target Site Duplication (TSD), Long Terminal Repeat (LTR), a proviral genome with gag, pro, pol and env genes (Lower) A schematic of the LTR and it’s regulatory regions. Solo LTRs are highly related to those found in the provirus. B) (Top) Long Interspersed Nuclear Element (LINE) containing 2 open reading frames in the sense orientation and one in the anti-sense orientation. (Middle) A Small Interspersed Nuclear Element (SINE), a non-autonomous element which is reliant on LINE activity for mobility. The Alu element is a subfamily of SINEs and is nonautonomous but remains mobile in the human genome. (Bottom) The SINE-VTNR (Variable Number of Tandem Repeat)-Alu (SVA) element, which contains a SINE core repeat, and Alu-like region (antisense to the remainder of the element), a VNTR repetitive region and a SINE-R region, which is derived from HERVK10 and contains homology to LTR5Hs. C) Modes of cis-regulatory TE activity showing (Upper) enhancer activity where H3K27ac-marked TEs are bound by transcription factors (Middle) Promoter-acting TE, where an upstream integrant acts as a promoter for a downstream gene. (Lower) A TE acting as a 3D genome regulatory element.
During early embryogenesis, dynamic changes in RNA expression from TEs is associated with key developmental progressions in totipotent and pluripotent cells including at the time of zygotic genome activation (ZGA), the conversion of totipotent cells in the morula to pluripotent pre-implantation epiblast cells of the blastocyst, as well as during early embryo development including formation of primordial germ cells (PGCs) [5–12]. Studying TE expression and epigenetic regulation in human pre-implantation embryos is possible but limited due to the small number of embryos donated to research following in vitro fertilization (IVF). As described below, most studies on TEs in early human embryos involve tracking RNA expression and chromatin changes. However, a major discovery revealed that pro-viruses originating from ancient HERVK pro-viral genomic integrants are capable of generating viral particles during blastocyst formation when studied in vitro [13].
Evaluating the role of TE’s in post-implantation human embryo development poses even more challenges, especially due to highly limited availability of human embryo samples. To fill this gap in knowledge, human pluripotent stem cells, namely human embryonic stem cells (hESCs) and induced PSCs (hiPSCs), are used to model pre- and post-implantation states of pluripotency, emerging somatic lineages, and PGCs. As described below, the field has used relatively simple two-dimensional (2D) models to generate the foundational knowledge regarding expression, epigenetic state and transcription factor binding to TEs in early human development. However, stem cell-based embryo models (embryo models), which recapitulate various three-dimensional aspects of early human embryo development represent the next frontier for evaluating the functional role of TEs in regulating human biology during this key stage of embryo development.
It is now appreciated that some TEs can serve as cis-regulatory elements, and therefore have the potential to drive cell-type specific gene expression. In both mice and humans, extensive binding of transcription factors to ERV-family elements, especially the key regulators of pluripotent stem cell self-renewal (NANOG, OCT4 and KLF4), have driven the hypothesis that TEs contribute to gene regulation in the pluripotent state of mammals [14]. Further supporting this view, many ERV LTRs harbor multiple pluripotency factor binding sites, and Chromatin immunoprecipitation followed by sequencing (ChIP-seq) has shown that ERV LTRs are, in many cases, co-bound by multiple pluripotent transcription factors [14,15]. In addition to being transcription factor-bound, specific families of ERVs are also enriched with chromatin and histone modifications associated with active states of gene expression [11,16–18]. Evidence for a role of TEs in 3D genome organization exists as well, as CTCF binding sites are found in TEs of the LINE, SINE and LTR superfamilies. These sites are thought to be functional, as alterations in 3D genome organization are observed when TEs are altered via CRISPR/cas9 editing [19,20]. These observations support a role for TEs as potential cis-regulatory enhancer/promoter elements in regulating states of pluripotency in the early human embryo.
Epigenetic silencing of ERVs, and therefore the decommissioning of active ERV LTR-family enhancer/promoter elements during early embryo development likely involves Kruppel Associated box Zinc-finger proteins (KRAB-ZFPs), a broad family of DNA binding proteins that bind TEs and work in tandem with TRIM28 (KAP1) to facilitate the deposition of repressive H3K9me3 heterochromatin at the targeted ERVs [21–24]. Enrichment of H3K9me3 at ERVs is highly dynamic during early human embryo development, beginning at the 4-cell stage before ZGA, and through blastocyst formation [7,8]. This is important as the human genome is largely demethylated during these embryonic stages [25], suggesting that H3K9me3 likely serves as the predominant repressive epigenetic modification responsible for dynamically silencing TEs in the early embryo. Supporting this hypothesis, ultra-low input ChIP-seq for H3K9me3 in pre-implantation embryos has revealed that intergenic ERV LTRs are progressively enriched with H3K9me3 from the 4-cell stage to formation of the blastocyst [7,8]. Notably, some primate and hominoid-specific TEs including LTR5Hs, LTR7B and SVA_D are not marked with H3K9me3 by the 8-cell stage and are enriched for pluripotency factor motifs. In particular, SVA_D elements are highly enriched for DUX motifs, suggesting a cis-regulatory role for these elements [7,8]. To evaluate the necessity of TE remodeling at the 8-cell stage, Yu et al. performed CRISPRi-mediated repression of SVA_D elements in human embryos at the 4-cell stage, which resulted in a developmental block at the 8-cell stage, [8] indicating SVA_D expression, or the chromatin state at SVA_D is involved in human ZGA.
Similarly, there is evidence that TEs function in the morula as totipotency is extinguished in order to give rise to trophectoderm and ICM of blastocysts. Specifically, chromatin accessibility of morula cells reveals two distinct chromatin states; 1) cells where regions of high chromatin accessibility are enriched for transcription factor motifs involved in trophectoderm formation (GATA and TEAD families), and 2) cells where regions of high chromatin accessibility are enriched for transcription factor motifs associate with pluripotency (NANOG, SOX2 and KLF4). Interestingly, cells of the putative outer morula (the ones fated to become trophectoderm), acquire H3K9me3 at hominoid-specific ERVs, including those of the ERVK family (MER11B, MER11C LTRs and HERK9-int) and ERV1 family (LTR12). Footprinting analysis of the putative ICM reveals that these ERV LTRs are bound by pluripotency factors OCT4 and SOX2 [8]. Taken together, these observations support the hypothesis that targeted silencing of the aforementioned ERVs safeguards specification of extraembryonic lineages by precluding ectopic activation of the pluripotency program [8]. Collectively, these results suggest that active epigenetic remodeling of TEs is important for early human embryonic development.
Although RNA expression and epigenetic modifications can be mapped to TE families and subfamilies, a caveat exists when studying the role of individual TE integrants and regulation of neighboring gene expression. The repetitive nature of TEs, combined with the short-read lengths acquired using traditional Illumina sequencing precludes accurate mapping to unique genomic sites [26]. Furthermore, in the case of mobile elements, such as L1Hs and Alu, precise mapping of new integrants across individuals using a reference genome is impossible as the reference would likely not include the position of person-specific integrations. Still, computational advances and long-read sequencing (discussed below) are beginning to address these shortfalls, promising better platforms for interrogating the gene regulatory role of TEs in human biology.
In addition to TE regulation at the chromatin level, it is now appreciated the N6-methyladenosine modification to endogenous viral mRNA is also a potent modality of ERV (and LINE1) regulation. Evidence in mouse pluripotent stem cells has shown that ERV RNAs transcribed from the retrotransposition-active IAP family are heavily marked by m6A due to activity of the METTL3/MEETL14 heterodimer. m6A-marked ERV RNAs are then bound by YTHDF1/2/3, m6A readers, which in turn results in clearance of viral mRNAs [27]. While little is known about the role of m6A in regulating ERVs, SVAs or LINE elements in the early human embryo, it is apparent that analysis of m6A at TEs during early human embryo development and human pluripotent stem cells is needed.
Evaluating TEs using Human Pluripotent States in culture
Evaluating TE expression, epigenetic regulation and function in pre-implantation and early post-implantation human embryos is challenging. Therefore, TE analysis in hESCs and hiPSCs has emerged as an important in vitro model to understand the role of TEs in pluripotency. Pluripotency is a spectrum of states from naïve to primed, with three major states of pluripotency (naïve, formative and primed) successfully captured as self-renewing stem cells in vitro. A fourth state of pluripotency, called latent pluripotency, occurs in PGCs and is captured in vitro with the differentiation of PGC-Like cells (PGCLCs) [28–36]. (Figure 2) Culture of human pluripotent stem cells in naïve conditions recapitulates the transcriptome and key epigenomic features of cells from the morula at Carnegie Stage 2 (CS2) and pre-implantation epiblasts at CS3-CS4 (Figure 2). Primed and formative culture conditions recapitulate the post-implantation epiblast at CS4-CS5 [37,38]. The differentiation of hPGCLCs in vitro results in the formation of germ cells equivalent to those found in post-implantation embryos between CS5-CS7 (Figure 2) [10,39,40]. In the following section, we will highlight key studies evaluating TE’s in the four states of human pluripotency.
Figure 2: In Vitro and Stem-Cell Based Embryo Models:
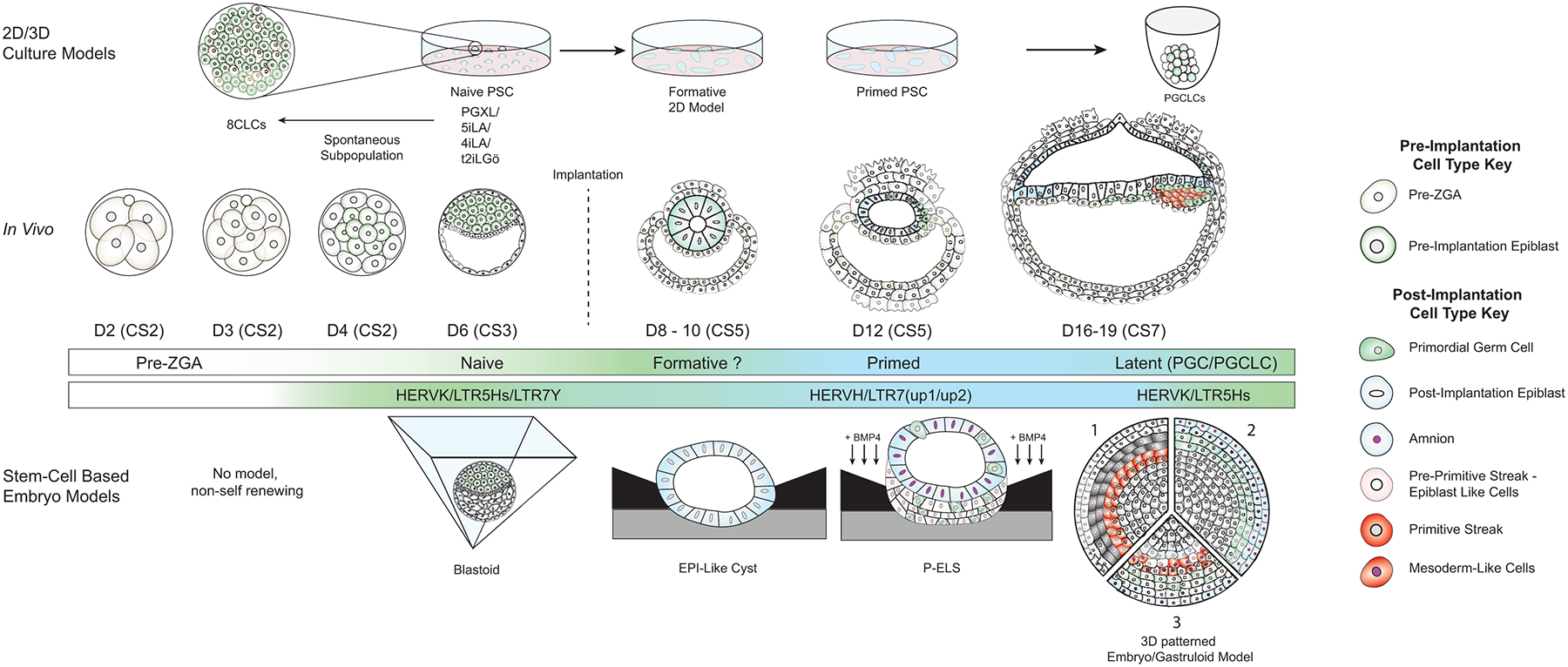
Top) 2D-culture in vitro models representing various states in embryonic development paired with their embryonic time-matched counterparts below (In vivo). From left to right: 8-Cell Like Cells (8CLCs), non-renewing transient subpopulations in naïve human pluripotent stem cell (PSCs) cultures. Naïve PSCs, which represent the inner cell mass of the pre-implantation epiblast. Naïve cells are marked by high expression of HERVK and functional reliance on LTRT5Hs. Formative cells, which can be stably cultured but have not yet been identified in in vivo human embryos, thus the post-implantation time point they may correspond to is unknown (?) . Primed PSCs represent cells of the post-implantation epiblast, and are marked by high expression of HERVH and functional activity of LTR7(up1/2) integrants. Far right is the PGCLC aggregate model, in which PGCLCs occupy a state of latent pluripotency. CS3-CS7 embryos are shown as a lateral cross-section.
Bottom) Stem Cell-based Embryo models (Left to right): Blastoids represent the pre-implantation embryo. The Posteriorized Embryonic-like Sac (P-ELS) model involves gradients of BMP4 exposure driving formation of the amnion, hPGCLCs and expression of T in the pre-streak Epiblast. Two variation of a 3D-printed embryo model and a gastruloid are shown. 1) lineages derived by Warmflash 2014, from outer to inner are trophectoderm-like, Endoderm like, Primitive-streak like, and ectoderm like. 2) Model of Jo et al. 2022, where they observe (from outer to inner) amnion-like cells, PGCLCs, and ectoderm-like cells. 3) A gastruloid model representative of BMP4-treated gastruloids in Minn et al. 2020 (from outer to inner): amnion-Like and trophectoderm-like cells, endoderm and PGCLCs, mesoderm and primitive streak like, epiblast-like and ectoderm-like cells. Pluripotent pre-implantation lineages are noted in the key as well as primitive steak. Other cells represented in the models are shown in grey.
At the RNA level it is now appreciated that the different states of pluripotency in vitro express differing repertoires of TEs, with corresponding ChIP-seq and chromatin accessibility data revealing the epigenetic status of TEs in each state [17,31,32]. For example, naïve pluripotent stem cells exhibit high expression of the HERVK provirus (HERVK-int) and LTR5Hs, a Hominoidea -specific LTR of the HERVK(HML2) family [31]. RNA expression of LTR5Hs in naïve human pluripotent stem cells is coupled with hypomethylation and enrichment of H3K27ac at these TEs. Interestingly, LTR5A and 5B, two older members of the HERVK(HML2) family first identified in Hominoidea (Apes) and Catarrhini (New World Monkeys), respectively, are not highly expressed or marked with H3K27ac in the naïve state. Members of the SVA family, specifically SVA_D, are also expressed by naïve human pluripotent stem cells, and these integrants are correspondingly enriched in H3K27ac [16,31]. This is of particular interest, as the HERVK10-derived SINE-R region of the composite SVA_D element shares significant homology with LTR5Hs (Fig. 1), meaning this region of SVA_D likely recruits the same transcription factors and KRAB-ZFPs as LTR5Hs [16,41]. Functional studies in naïve human pluripotent stem cells using CRISPRi to target LTR5Hs and SVA_D results in wide-spread gene deregulation, especially of genes of involved in 3D genome organization, cell polarity and lineage restriction [16,42]. Thus, naïve human pluripotent stem cells do not simply express LTR5Hs and SVA_D, but are also reliant on chromatin remodeling and/or RNA expression of these TEs for proper maintenance of the naïve state.
Naïve hESCs also have high accessibility and expression of LTR7Y, a regulatory element of the HERVH provirus. While some LTR7B and some LTR7 elements exhibit H3K27ac enrichment in the naïve state, it is the LTR7Y elements that are highly enriched in H3K27ac, and are bound by KLF4, KLF5 and NANOG [16–18]. Notably, different naïve culture conditions lead to differences in LTR7 family expression, with 5i/L/A conditions resulting in higher LTR7Y expression and H3K27ac, while 3i/L naïve conditions lead to higher expression of LTR7. This is likely a result of 5i/L/A conditions and 3i/L naïve conditions capturing different states on the pluripotency spectrum. Indeed, the responsiveness of LTR7, LTR7Y and LTR7B to different naïve culture conditions likely suggests they are highly dynamic and sensitive to developmental progression through the different states of pluripotency in the early embryo.
As the human blastocyst implants during CS4 to CS5, the naïve pluripotent cells of the pre-implantation epiblast convert to a state of pluripotency referred to as formative. This is a state competent to differentiate into somatic cells and PGCs following exposure to appropriate differentiation cues [32]. In the mouse embryo, formative pluripotency corresponds to cells of the post-implantation epiblast at the early egg-cylinder stage. In humans, the location of formative cells in the post-implantation embryo is not yet known, but likely corresponds to post-implantation pluripotent cells somewhere between day 8–12 post-fertilization. Despite this unknown, formative cells have been reported in culture [32]. Formative cells share some TE overlap with naïve human pluripotent stem cells, including expression of HERVK, however formative cells uniquely express LTR6A [32]. Curiously, formative cells share expression of a number of KRAB-ZFPs with naïve cells, including ZNF676, ZNF560, ZNF528. However, much of the formative gene transcriptional profile is shared with the primed state [32]. This similarity in gene expression profiles elevates LTR6A as a key marker of the formative state, and underscores the utility of TEs as markers of developmental populations which are hard to capture in vivo.
Primed hPSCs, which recapitulate post-implantation epiblast cells, express pro-viral integrants of the HERVH and HERVK families [17,43]. Furthermore, the epigenome of HERVH associated LTR regulatory sequence LTR7 are hypomethylated and enriched with H3K27ac in the primed state [17,31,44–47]. In a recently phylogenic analysis of HERVH and LTR7, new subfamilies were identified, and their evolutionary trajectories defined. Of particular interest are Homininae-specific LTR7up1 and LTR7up2, which were previously clustered with LTR7. Re-analysis of LTR7Y, LTR7up1 and LTR7up2 using these updated phylogenies revealed that LTR7Y elements are expressed by naïve human pluripotent stem cells, whereas it is the LTR7up families that are highly expressed in primed cells, with LTR7up1 and LTR7up2 accounting for most of the observed LTR7 transcription in this state. Consistent with these observations, LTR7up1 and LTR7up2 are also enriched with H3K27ac, and are bound by pluripotency factors NANOG, SOX2 and FOXP1 in primed cells [48]. Altogether, this supports the idea that young TEs of the LTR7up families likely acting as cis-regulatory regions in primed state pluripotency.
Latent pluripotency is a feature of PGCs. During embryo implantation and progression of pluripotent cells from the naïve to primed state, hPGCs are specified [49,50]. Based on primate models, this is speculated to occur at CS5, coincident with amnion and extraembryonic mesoderm formation but initiated before the formation of the primitive streak [39,51]. Studying hPGC specification in vivo is extremely challenging due to the rarity of early human post-conceptus tissues donated to research. A recent paper profiling single cells of a CS7 post-implantation human embryo demonstrated that hPGCs exhibit a suite of TEs with similarities to naïve cells [9,10,40]. Specifically, this data set revealed that CS7 hPGCs in vivo express HERVK, LTR5Hs and SVA_D [40].
In order to model the expression and function of TE’s during hPGC specification, the differentiation of hPGCLCs from primed human pluripotent stem cells is used (Figure 2) [33–35,52]. Induction of hPGCLCs in vitro leads to a dynamic change in TE expression relative to the primed state, with one of the most significant changes involving up-regulation of LTR5Hs. At the epigenetic level in hPGCLCs, LTR5Hs sequences become hypomethylated, enriched in H3K27ac, and bound by NANOG, SOX15, SOX17 and TFAP2C transcription factors all of which are necessary for hPGC specification and maintenance [10,49,53,54]. Using CRISPRi to dampen LTR5Hs accessibility before hPGCLC induction results in a significant reduction in the competency human pluripotent stem cells to differentiate into hPGCLCs [10]. This suggests that LTR5Hs is necessary either for acquisition or maintenance of latent pluripotency upon hPGCLC formation.
TE Expression in Totipotent-like cells in culture
Following fertilization (CS1), the maternal and paternal genomes undergo epigenetic reprogramming to create a totipotent state competent for ZGA, a process where embryonic transcription is activated in the diploid embryo [5,55,56]. In humans, ZGA occurs in the 8-cell embryo, while in the mouse it occurs in the 2-cell embryo. In both mouse and human embryos, ERVL (MERVL/HERVL) family elements are broadly derepressed with ZGA, coincident with high expression of ZSCAN4 and the TRIM family of genes, as well as expression of Dux/DUXA [5,6,57,58]. Using these characteristics, human 8-cell like cells (8CLCs) have been identified sporadically in hPSCs cultured under a variety of naïve culture conditions, including 4 inhibitor (4i), 5i, NHSM (Naïve human stem cell medium), PXGL (PD0325901, Gö6983, XAV939, LIF ), t2iLGö [6] or from naïve hPSCs reverted to a defined media called e4CL (enhanced 4 chemicals + LIF: PD0325901, IWR1, DZNep and TSA), which increases the subpopulation of 8CLCs [5]. Notably, the transcriptomes of 5i and PXGL 8CLCs are closest to those of e4CL, although e4CL was much more potent at generating 8CLC cells [5]. 8CLCs have also been identified in populations of pre-Epiblast-like PSCs (prEpiSCs), which are cultured in media containing inhibitors of MEK (GSK1120212), WNT (XAV939), PKC (Go6983), Src (A419259) and EED/H3K27me3 (DZNep) [12]. In all conditions, 8CLCs are transient and metastable, interconverting between 8CLCs and naïve PSCs in vitro. Therefore, like totipotent cells of the embryo, 8CLCs lack the capacity for self-renewal [56]. Despite these challenges, reliable isolation of 8CLCs and the ability to identify them in single-cell transcriptomic experiments have enabled characterization of their unique TE-repertoire. TE expression in 8CLCs compared to the naïve cells reveals a general enrichment of ERVs including HERVK, but with a marked increase in expression of HERVL and its associated LTRs, MLT2A1 and MLT2A2 [5,6,12]. This is in line with TE expression in the 8-cell human embryo [5,6,12]. Importantly, neither expression nor chromatin accessibility of HERVL, MLT2A1 and MLT2A2 were observed in any other naïve hPSCs in culture, suggesting expression of these LTRs is tightly associated with the 8CLC state and not naïve pluripotency [5,6,12,45,59,60].
Emerging Technologies for Understanding TEs in early Human Embryo Development
Although human pre-implantation embryo development can be studied using IVF embryos, there are limitations in the number of embryos that can be used and, in some jurisdictions, research with human embryos is not allowed [61]. This has led to the emergence of embryo models as critical avatars of human embryo development [62]. New embryo models recapitulate some key aspects of early human development, but not all. The closest representation of an intact embryo are blastoids, resembling pre-implantation blastocysts consisting of spatially organized pre-implantation epiblast, primitive endoderm and trophoblast [63–66]. These emerging models represent a powerful tool in which the function of TE’s can be evaluated using approaches such as CRISRi of SVA_D elements as in [7]. Although powerful, the blastoid model cannot recapitulate the embryonic events in an 8-cell embryo prior to blastocyst formation.
For post implantation stages, embryo models including gastruloids [67,68], post-implantation amniotic sac embryoids (PASE), posteriorized-embryonic-like sacs (P-ELS) [69,70], and micropatterned self-organizing discs [36,65,71] have emerged. These tools enable more thorough interrogation of TEs in lineage specification after embryo implantation, which is of particular importance as the divergence and origin of some post-implantation lineages, such a hPGCs, remains unclear. These models, coupled with advances in sequencing technologies hold promise in understanding the role of TEs throughout development, including a more complex understanding of TE epigenetic reprogramming and its role in properly regulating early embryonic lineage restriction.
In tandem with advancing in vitro models, there has also been a rapid expansion of low-input and single-cell sequencing technologies. Single-cell RNA-seq has become important in examining both rare populations, such as 8CLCs in [5,6], as well as in-depth profiling of the transcriptome of rare clinical samples, such as SMART-seq of the human Carnegie Stage (CS) 7 embryo [40]. Third-generation long-read sequencing technologies also hold promise for better characterization of the RNA expression and mapping of TEs, helping overcome long-standing challenges in mapping to specific genomic sites. For instance, Single-Cell Long-Read RNA-seq (CELLO-seq) [72] can be used to map TE transcripts to their original loci, helping to resolve questions about the cis- and trans-acting roles of TEs. While the higher error rate of third-generation sequencing still imposes some technical limitations on its use, it also harbors distinct advantages, such as the ability to read DNA 5-methycytosine and RNA N6-methyladenosine natively without enzymatic modification to nucleic acids during library preparation [73,74].
Another significant advance in sequencing is the advent of single-cell multi-omics, which are powerful tools to understand how transcriptomes, chromatin accessibility, and epigenetic landscapes are remodeled at the single-cell level. Multi-omic approaches, such as Low-Input Chromatin Accessibility and Transcriptome (Li-CAT-seq) sequencing as in [59], have helped link in vitro observations with in vivo data. These data are of particular importance when studying the expression and epigenetic state of TEs in early development. Here, multi-omic approaches which can map transcriptome, chromatin accessibility and DNA methylome at the single cell level such as scNMT-seq [75] and snmCAT-seq [76], hold promise in better understanding the potential roles of DNA methylation in early human development. These advances are particularly powerful when coupled with low-input chromatin profiling technologies such as CUTnTag and Ultra-Low Input ChIP which have enabled high-quality profiling of chromatin with as few as 100 cells [8]. These advances help understand how epigenetic regulation of TEs, including their activity as enhancers and the dynamics of their silencing during differentiation, contribute to mammalian development.
Conclusions
Here, we summarized major changes in the RNA expression and epigenetic state of TEs across early human embryo development and in pluripotent stem cells in vitro. Importantly, we argue here that distinct states of pluripotency can be defined by a unique repertoire of TEs, their expression profile and epigenetic state forming a molecular fingerprint that can be used to discriminate major transitions in embryo development including ZGA, different states of pluripotency and PGCs. While similar epigenetic profiling of other embryonic stages has not been examined in the same way, such an approach holds promise for improving our understanding of how expression and epigenetic reprogramming at TEs could shape other early embryonic lineage decisions in humans. Disrupting TE expression and chromatin accessibility with CRISPRi provides a glimpse into the functional role of TE’s in early human development, and this functional work is likely to expand when coupled with embryo models. It is important to note, however that confirmation of key observations with embryo models will benefit from access to equivalent stages of human embryo development where possible, and with appropriate regulatory oversight and public engagement as to the use of embryos in research.
Acknowledgements:
A.C and J.D conceived the review, J.D drafted the review, J.D and A.C revised the review.
This work is supported by R01HD079546, R01HD098278 and R01HD058047 to A.C., J.D. is supported by the Whitcome Fellowship and the UCLA Molecular Biology Institute.
References
- 1.Hoyt SJ, Storer JM, Hartley GA, Grady PGS, Gershman A, de Lima LG, Limouse C, Halabian R, Wojenski L, Rodriguez M, et al. : From telomere to telomere: The transcriptional and epigenetic state of human repeat elements. Science 2022, 376:eabk3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, et al. : The complete sequence of a human genome. Science 2022, 376:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalopin D, Naville M, Plard F, Galiana D, Volff JN: Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol 2015, 7:567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Autio MI, Bin Amin T, Perrin A, Wong JY, Foo RS, Prabhakar S: Transposable elements that have recently been mobile in the human genome. BMC Genomics 2021, 22:789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **5.Mazid MA, Ward C, Luo Z, Liu C, Li Y, Lai Y, Wu L, Li J, Jia W, Jiang Y, et al. : Rolling back human pluripotent stem cells to an eight-cell embryo-like stage. Nature 2022, 605:315–324. [DOI] [PubMed] [Google Scholar]; Mazid et al. model the 8C state in culture using enhanced naïve culture conditions. Similar to (6) and (12) they note that 8CLCs are not self-renewing, thought to be a feature of totipotency, and examine distinct controllers of the 8C-state, identifying TPRX1. A similar repetiore of LTR expression to those identified in (6) are also observed in this system as well, in line with observations from 8C in vivo.
- **6.Taubenschmid-Stowers J, Rostovskaya M, Santos F, Ljung S, Argelaguet R, Krueger F, Nichols J, Reik W: 8C-like cells capture the human zygotic genome activation program in vitro. Cell Stem Cell 2022, 29:449–459 e446. [DOI] [PMC free article] [PubMed] [Google Scholar]; Taubenschmid and colleagues demonstrate that naïve cells in culture give rise to a spontaneous population of 8C-like cells, marked by expression of DUX, ZSCAN and other markers of the 8C-state.
- *7.Xu R, Li S, Wu Q, Li C, Jiang M, Guo L, Chen M, Yang L, Dong X, Wang H, et al. : Stage-specific H3K9me3 occupancy ensures retrotransposon silencing in human pre-implantation embryos. Cell Stem Cell 2022, 29:1051–1066 e1058. [DOI] [PubMed] [Google Scholar]; Released back-to-back with Yu et al. (8), Xu. And colleagues map the dynamics of H3K9me3 acquisition in early human development. They observe that many TEs silenced by H3K9me3 specifically at the 8-cell stage were more likely to be marked with H3K27ac or H3K4me1 in subsequent stages of development. They also note bivalent domains in TEs which are thought to be regulatory are often silenced in trophectoderm cells, an observation shared by Yu. et al in the morula.
- **8.Yu H, Chen M, Hu Y, Ou S, Yu X, Liang S, Li N, Yang M, Kong X, Sun C, et al. : Dynamic reprogramming of H3K9me3 at hominoid-specific retrotransposons during human preimplantation development. Cell Stem Cell 2022, 29:1031–1050 e1012. [DOI] [PubMed] [Google Scholar]; Yu. et al demonstrate that H3K9me3 is aquired at hominid-specific TEs, often harboring pluripotency factor motifs in the outer cells of the morula, suggesting an early role for TEs in lineage bifuration. Additionally, they observe that SVA elements are necessary during ZGA, and that loss of SVA_D accessibility during ZGA leads to defects in development.
- 9.Ito J, Seita Y, Kojima S, Parrish NF, Sasaki K, Sato K: A hominoid-specific endogenous retrovirus may have rewired the gene regulatory network shared between primordial germ cells and naive pluripotent cells. PLoS Genet 2022, 18:e1009846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *10.Xiang X, Tao Y, DiRusso J, Hsu FM, Zhang J, Xue Z, Pontis J, Trono D, Liu W, Clark AT: Human reproduction is regulated by retrotransposons derived from ancient Hominidae-specific viral infections. Nat Commun 2022, 13:463. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xiang et al. demonstrate that LTR5Hs, a hominidae-specific LTR, is necessaery for PGCLC specification is epigenetically remodeled and bound by SOX15, SOX17, and NANOG, which are necessary for PGC fate. Silencing of LTR5Hs elements reduces PCGLC specification, supporting a functional role of LTR5Hs in germline specification.
- *11.Pontis J, Pulver C, Playfoot CJ, Planet E, Grun D, Offner S, Duc J, Manfrin A, Lutolf MP, Trono D: Primate-specific transposable elements shape transcriptional networks during human development. Nat Commun 2022, 13:7178. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pontis and colleagues demonstrate that early embryonic lineages, including during gastrulation, are marked by compliments of transposable elements and unique KRAB-ZFP expression.
- 12.Yu X, Liang S, Chen M, Yu H, Li R, Qu Y, Kong X, Guo R, Zheng R, Izsvak Z, et al. : Recapitulating early human development with 8C-like cells. Cell Rep 2022, 39:110994. [DOI] [PubMed] [Google Scholar]
- 13.Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. : Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 2015, 522:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G: Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 2010, 42:631–634. [DOI] [PubMed] [Google Scholar]
- 15.Sundaram V, Choudhary MN, Pehrsson E, Xing X, Fiore C, Pandey M, Maricque B, Udawatta M, Ngo D, Chen Y, et al. : Functional cis-regulatory modules encoded by mouse-specific endogenous retrovirus. Nat Commun 2017, 8:14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pontis J, Planet E, Offner S, Turelli P, Duc J, Coudray A, Theunissen TW, Jaenisch R, Trono D: Hominoid-Specific Transposable Elements and KZFPs Facilitate Human Embryonic Genome Activation and Control Transcription in Naive Human ESCs. Cell Stem Cell 2019, 24:724–735 e725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goke J, Lu X, Chan YS, Ng HH, Ly LH, Sachs F, Szczerbinska I: Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 2015, 16:135–141. [DOI] [PubMed] [Google Scholar]
- 18.Ai Z, Xiang X, Xiang Y, Szczerbinska I, Qian Y, Xu X, Ma C, Su Y, Gao B, Shen H, et al. : Kruppel-like factor 5 rewires NANOG regulatory network to activate human naive pluripotency specific LTR7Ys and promote naive pluripotency. Cell Rep 2022, 40:111240. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Li T, Preissl S, Amaral ML, Grinstein JD, Farah EN, Destici E, Qiu Y, Hu R, Lee AY, et al. : Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat Genet 2019, 51:1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhary MNK, Quaid K, Xing X, Schmidt H, Wang T: Widespread contribution of transposable elements to the rewiring of mammalian 3D genomes. Nat Commun 2023, 14:634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coluccio A, Ecco G, Duc J, Offner S, Turelli P, Trono D: Individual retrotransposon integrants are differentially controlled by KZFP/KAP1-dependent histone methylation, DNA methylation and TET-mediated hydroxymethylation in naive embryonic stem cells. Epigenetics Chromatin 2018, 11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D: Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Dev Cell 2016, 36:611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. : KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010, 463:237–240. [DOI] [PubMed] [Google Scholar]
- 24.Imbeault M, Helleboid PY, Trono D: KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 2017, 543:550–554. [DOI] [PubMed] [Google Scholar]
- 25.Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A: DNA methylation dynamics of the human preimplantation embryo. Nature 2014, 511:611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teissandier A, Servant N, Barillot E, Bourc’his D: Tools and best practices for retrotransposon analysis using high-throughput sequencing data. Mob DNA 2019, 10:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chelmicki T, Roger E, Teissandier A, Dura M, Bonneville L, Rucli S, Dossin F, Fouassier C, Lameiras S, Bourc’his D: m(6)A RNA methylation regulates the fate of endogenous retroviruses. Nature 2021, 591:312–316. [DOI] [PubMed] [Google Scholar]
- 28.Kalkan T, Bornelov S, Mulas C, Diamanti E, Lohoff T, Ralser M, Middelkamp S, Lombard P, Nichols J, Smith A: Complementary Activity of ETV5, RBPJ, and TCF3 Drives Formative Transition from Naive Pluripotency. Cell Stem Cell 2019, 24:785–801 e787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalkan T, Olova N, Roode M, Mulas C, Lee HJ, Nett I, Marks H, Walker R, Stunnenberg HG, Lilley KS, et al. : Tracking the embryonic stem cell transition from ground state pluripotency. Development 2017, 144:1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leitch HG, Nichols J, Humphreys P, Mulas C, Martello G, Lee C, Jones K, Surani MA, Smith A: Rebuilding pluripotency from primordial germ cells. Stem Cell Reports 2013, 1:66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **31.Theunissen TW, Friedli M, He Y, Planet E, O’Neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, Imbeault M, et al. : Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell 2016, 19:502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]; Theunissen et al demonstrate that the TE transciptome is signifigantly different between primed and naïve states of pluripotency. These findings demonstrated that HERVK/LTR5Hs and SVA_D transcription are hallmarks of naïve hPSCs in culture, a observation that was later extended to the pre-implantation epiblast.
- **32.Kinoshita M, Barber M, Mansfield W, Cui Y, Spindlow D, Stirparo GG, Dietmann S, Nichols J, Smith A: Capture of Mouse and Human Stem Cells with Features of Formative Pluripotency. Cell Stem Cell 2021, 28:453–471 e458. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kinoshita et al. demonstrate that stable culture of formative mouse PSCs can be achieved, and that similar culture conditions can also induce hPSCs into a stable, self-renewing formative state. They also demonstrate that formative hPSCs are marked by their own compliment of hPSCs, as is common between differing states of pluripotency.
- 33.Irie N, Weinberger L, Tang WW, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, Surani MA: SOX17 is a critical specifier of human primordial germ cell fate. Cell 2015, 160:253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, Tsuchiya H, et al. : Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell 2015, 17:178–194. [DOI] [PubMed] [Google Scholar]
- **35.Jo K, Teague S, Chen B, Khan HA, Freeburne E, Li H, Li B, Ran R, Spence JR, Heemskerk I: Efficient differentiation of human primordial germ cells through geometric control reveals a key role for Nodal signaling. Elife 2022, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jo et al. observe that hPGCLCs can be specified from hPSCs culture on a micropatterned surface after exposure to BMP4 and Nodal. Importantly, they demonstrate that duration and concentration of Nodal and BMP4 require careful titration, and demonstrate that changes in cytokine concentration or exposure time leads to specification of amnion or ectoderm, depending on conditions.
- **36.Minn KT, Fu YC, He S, Dietmann S, George SC, Anastasio MA, Morris SA, Solnica-Krezel L: High-resolution transcriptional and morphogenetic profiling of cells from micropatterned human ESC gastruloid cultures. Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Minn et al. demonstrate tha treatment of micropatterned hPSCs with BPM4 gives rise to a gastruloid with cells represent each of the germ layers, including EPI-like cells and PGCLCs.
- 37.Liu X, Ouyang JF, Rossello FJ, Tan JP, Davidson KC, Valdes DS, Schroder J, Sun YBY, Chen J, Knaupp AS, et al. : Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature 2020, 586:101–107. [DOI] [PubMed] [Google Scholar]
- 38.Guo G, Stirparo GG, Strawbridge SE, Spindlow D, Yang J, Clarke J, Dattani A, Yanagida A, Li MA, Myers S, et al. : Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 2021, 28:1040–1056 e1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niu Y, Sun N, Li C, Lei Y, Huang Z, Wu J, Si C, Dai X, Liu C, Wei J, et al. : Dissecting primate early post-implantation development using long-term in vitro embryo culture. Science 2019, 366. [DOI] [PubMed] [Google Scholar]
- 40.Tyser RCV, Mahammadov E, Nakanoh S, Vallier L, Scialdone A, Srinivas S: Single-cell transcriptomic characterization of a gastrulating human embryo. Nature 2021, 600:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, Lower J, Stratling WH, Lower R, Schumann GG: The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res 2012, 40:1666–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuentes DR, Swigut T, Wysocka J: Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchs NV, Loewer S, Daley GQ, Izsvak Z, Lower J, Lower R: Human endogenous retrovirus K (HML-2) RNA and protein expression is a marker for human embryonic and induced pluripotent stem cells. Retrovirology 2013, 10:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barakat TS, Halbritter F, Zhang M, Rendeiro AF, Perenthaler E, Bock C, Chambers I: Functional Dissection of the Enhancer Repertoire in Human Embryonic Stem Cells. Cell Stem Cell 2018, 23:276–288 e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao L, Wu K, Liu Z, Yao X, Yuan S, Tao W, Yi L, Yu G, Hou Z, Fan D, et al. : Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution. Cell 2018, 173:248–259 e215. [DOI] [PubMed] [Google Scholar]
- 46.Lu X, Sachs F, Ramsay L, Jacques PE, Goke J, Bourque G, Ng HH: The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol 2014, 21:423–425. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Xie G, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al. : Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 2014, 516:405–409. [DOI] [PubMed] [Google Scholar]
- **48.Carter TA, Singh M, Dumbovic G, Chobirko JD, Rinn JL, Feschotte C: Mosaic cis-regulatory evolution drives transcriptional partitioning of HERVH endogenous retrovirus in the human embryo. Elife 2022, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Carter et al. refine the phylogeny of the HERVH LTR, LTR7. In addition to new phylogeny, they demonstrate that LTR7 expression in the post-implantation epiblast is dominated by LTR7up1 and LTR7up2, which are hominidae-specific LTR7s, in line with observation that most regulatory TEs in the early embryo are evolutionarrily young and not shared with lower primates.
- 49.Chen D, Sun N, Hou L, Kim R, Faith J, Aslanyan M, Tao Y, Zheng Y, Fu J, Liu W, et al. : Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors. Cell Rep 2019, 29:4568–4582 e4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang WW, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA: A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell 2015, 161:1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki K, Nakamura T, Okamoto I, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Shiraki N, Takakuwa T, et al. : The Germ Cell Fate of Cynomolgus Monkeys Is Specified in the Nascent Amnion. Dev Cell 2016, 39:169–185. [DOI] [PubMed] [Google Scholar]
- 52.Zheng Y, Yan RZ, Sun S, Kobayashi M, Xiang L, Yang R, Goedel A, Kang Y, Xue X, Esfahani SN, et al. : Single-cell analysis of embryoids reveals lineage diversification roadmaps of early human development. Cell Stem Cell 2022, 29:1402–1419 e1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Veerapandian V, Yang X, Song K, Xu X, Cui M, Yuan W, Huang Y, Xia X, Yao Z, et al. : The chromatin accessibility landscape reveals distinct transcriptional regulation in the induction of human primordial germ cell-like cells from pluripotent stem cells. Stem Cell Reports 2021, 16:1245–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen D, Liu W, Zimmerman J, Pastor WA, Kim R, Hosohama L, Ho J, Aslanyan M, Gell JJ, Jacobsen SE, et al. : The TFAP2C-Regulated OCT4 Naive Enhancer Is Involved in Human Germline Formation. Cell Rep 2018, 25:3591–3602 e3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Posfai E, Schell JP, Janiszewski A, Rovic I, Murray A, Bradshaw B, Yamakawa T, Pardon T, El Bakkali M, Talon I, et al. : Evaluating totipotency using criteria of increasing stringency. Nat Cell Biol 2021, 23:49–60. [DOI] [PubMed] [Google Scholar]
- 56.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL: Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 2012, 487:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hendrickson PG, Dorais JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. : Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 2017, 49:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D: DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 2017, 49:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L, Leng L, Liu C, Lu C, Yuan Y, Wu L, Gong F, Zhang S, Wei X, Wang M, et al. : An integrated chromatin accessibility and transcriptome landscape of human pre-implantation embryos. Nat Commun 2019, 10:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu J, Xu J, Liu B, Yao G, Wang P, Lin Z, Huang B, Wang X, Li T, Shi S, et al. : Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 2018, 557:256–260. [DOI] [PubMed] [Google Scholar]
- 61.Caulfield T, Zarzeczny A, McCormick J, Bubela T, Critchley C, Einsiedel E, Galipeau J, Harmon S, Huynh M, Hyun I, et al. : The stem cell research environment: a patchwork of patchworks. Stem Cell Rev Rep 2009, 5:82–88. [DOI] [PubMed] [Google Scholar]
- 62.Clark AT, Brivanlou A, Fu J, Kato K, Mathews D, Niakan KK, Rivron N, Saitou M, Surani A, Tang F, et al. : Human embryo research, stem cell-derived embryo models and in vitro gametogenesis: Considerations leading to the revised ISSCR guidelines. Stem Cell Reports 2021, 16:1416–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **63.Yu L, Wei Y, Duan J, Schmitz DA, Sakurai M, Wang L, Wang K, Zhao S, Hon GC, Wu J: Blastocyst-like structures generated from human pluripotent stem cells. Nature 2021, 591:620–626. [DOI] [PubMed] [Google Scholar]; Yu et. al introduce model system to derive human blastocyst embryoids, a so-called blastoid. The system devised by Yu. et al. relies on step-wise exposure of developing blastoids to medias which bias naïve hPSCs toward trophectoderm or hypoblast lineages. This step-wise process leads to formation of blastoids.
- **64.Liu X, Tan JP, Schroder J, Aberkane A, Ouyang JF, Mohenska M, Lim SM, Sun YBY, Chen J, Sun G, et al. : Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 2021, 591:627–632. [DOI] [PubMed] [Google Scholar]; Liu. et al demonstrate that reprogramming of fibroblasts under specific minimal conditions followed by 3D culture in an Aggrewell and exposure to a media cocktail including HDAC inhibitors, WNT activation, ALK5/TGFb, and ALK4/NODAL inhibitors gives rise to induced blastoids (iBlastoids) which model the early pre-implantation embryo.
- **65.Kagawa H, Javali A, Khoei HH, Sommer TM, Sestini G, Novatchkova M, Scholte Op Reimer Y, Castel G, Bruneau A, Maenhoudt N, et al. : Human blastoids model blastocyst development and implantation. Nature 2022, 601:600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kagawa et al. demonstrate that naïve hPSCs treated with a cocktail of Hippo, TGFb and ERK inhibitors in hydrogel wells self-assemble into blastoids. These blastoids, like other blastoid models, most closely model the pre-implantation embryo.
- 66.Sozen B, Jorgensen V, Weatherbee BAT, Chen S, Zhu M, Zernicka-Goetz M: Reconstructing aspects of human embryogenesis with pluripotent stem cells. Nat Commun 2021, 12:5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moris N, Anlas K, van den Brink SC, Alemany A, Schroder J, Ghimire S, Balayo T, van Oudenaarden A, Martinez Arias A: An in vitro model of early anteroposterior organization during human development. Nature 2020, 582:410–415. [DOI] [PubMed] [Google Scholar]
- 68.Simunovic M, Metzger JJ, Etoc F, Yoney A, Ruzo A, Martyn I, Croft G, You DS, Brivanlou AH, Siggia ED: A 3D model of a human epiblast reveals BMP4-driven symmetry breaking. Nat Cell Biol 2019, 21:900–910. [DOI] [PubMed] [Google Scholar]
- 69.Shao Y, Taniguchi K, Townshend RF, Miki T, Gumucio DL, Fu J: A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat Commun 2017, 8:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **70.Zheng Y, Xue X, Shao Y, Wang S, Esfahani SN, Li Z, Muncie JM, Lakins JN, Weaver VM, Gumucio DL, et al. : Controlled modelling of human epiblast and amnion development using stem cells. Nature 2019, 573:421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zheng et al. updated the PASE model with the posteriorized embryonic like sac (P-ELS) model and find, upon induction using BMP4, that amnion-like cells and PGCLCs are specified in embyponic micro-patterned culture devices. Change to the compliment of cytokines and inhibitors added results in different lineage biases, enabling its use as a model to understand the interplay between signaling cues in the post-implantation epiblast.
- 71.Warmflash A, Sorre B, Etoc F, Siggia ED, Brivanlou AH: A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat Methods 2014, 11:847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berrens RV, Yang A, Laumer CE, Lun ATL, Bieberich F, Law CT, Lan G, Imaz M, Bowness JS, Brockdorff N, et al. : Locus-specific expression of transposable elements in single cells with CELLO-seq. Nat Biotechnol 2022, 40:546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parker MT, Knop K, Sherwood AV, Schurch NJ, Mackinnon K, Gould PD, Hall AJ, Barton GJ, Simpson GG: Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m(6)A modification. Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W: Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods 2017, 14:407–410. [DOI] [PubMed] [Google Scholar]
- 75.Clark SJ, Argelaguet R, Kapourani CA, Stubbs TM, Lee HJ, Alda-Catalinas C, Krueger F, Sanguinetti G, Kelsey G, Marioni JC, et al. : scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat Commun 2018, 9:781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luo C, Liu H, Xie F, Armand EJ, Siletti K, Bakken TE, Fang R, Doyle WI, Stuart T, Hodge RD, et al. : Single nucleus multi-omics identifies human cortical cell regulatory genome diversity. Cell Genom 2022, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]


