Abstract
With the advent of complete genome sequences, large-scale functional analyses are generating new excitement in biology and medicine. To facilitate genomewide functional analyses, we developed a high-density cell array with quantitative and automated readout of cell fitness. Able to print at >×10 higher density on a standard microtiter plate area than currently possible, our cell array allows single-plate screening of the complete set of Saccharomyces cerevisiae gene-deletion library and significantly reduces the amount of small molecules and other materials needed for the study. We used this method to map the relation between genes and cell fitness in response to rapamycin, a medically important natural product that targets the eukaryotic kinase Tor. We discuss the implications for pharmacogenomics and the uncharted complexity in genotype-dependent drug response in molecularly targeted therapies. Our analysis leads to several basic findings, including a class of gene deletions that confer better fitness in the presence of rapamycin. This result provides insights into possible therapeutic uses of rapamycin/CCI-779 in the treatment of neurodegenerative diseases (including Alzheimer's, Parkinson's, and Huntington's diseases), and cautions the possible existence of similar rapamycin-enhanceable mutations in cancer. It is well established in yeast that although TOR2 has a unique rapamycin-insensitive function, TOR1 and TOR2 are interchangeable in the rapamycin-sensitive functions. We show that even the rapamycin-sensitive functions are distinct between TOR1 and TOR2 and map the functional difference to a ≈120-aa region at the N termini of the proteins. Finally, we discuss using cell-based genomic pattern recognition in designing electronic or optical biosensors.
Keywords: live cell array, genetic basis of drug response, molecularly targeted therapy, mitochondrial, vacuolar protein sorting
The standard set of ≈6,000 yeast deletion strains with full-genome coverage created by the Saccharomyces cerevisiae Genome Deletion Project (1, 2) has allowed the systematic investigation of a variety of cellular behaviors, including fitness responses to environmental or chemical agents (reviewed in ref. 3). Two primary methods exist for fitness screening (4): separately as individual strains in multiwell plates and pooled as a mixture of strains grown together. Elegant agar-based wellless “cell arrays” (5) have also been used successfully for fitness screening (6-9). Screening as a pool is efficient and simultaneously assesses competitive fitness in a population with heterogeneous genotypes. Comparatively, screening in the multiplate format (liquid or agar) is a more comprehensive approach, allowing unbiased examination of each and every strain in the library without the complication of intrinsic fitness differences.
Currently existing yeast cell arrays contain from 96 to 786 strains per plate. Screening the whole deletion library thus requires many plates and large volumes (hundreds of milliliters or more) of assay medium dissolved with the small molecule of interest. This setup is generally incompatible with, if not impossible for, the study of molecules that are scarce in quantity. For example, although natural products represent a major source of chemical diversity and therapeutic potential (10), only a tiny fraction of natural products on earth has been studied because of limits in natural sources or cultivable environments (11, 12), and total synthesis of key molecules remains a rather long and low-yielding process. Miniaturization facilitates large-scale genomic analysis, which is of utmost importance for a thorough understanding of the mechanisms of action by natural products and other rare molecules.
To this end, we have developed a high-density cell array strategy (up to 9,600 strains per plate) (Fig. 1a) that relies on nanoliter cell printing by using microarraying pins. This array format greatly reduces the amount needed for a whole genome fitness screen to within submicrograms of natural products (rapamycin as an example). This method is simple and convenient and combines the advantages of both the multiplate method and the pooled method.
Fig. 1.
Chemical genomic screening by using a high-density cell array. (a) A microarraying robot was used to print the full S. cerevisiae single-gene deletion library on yeast extract/peptone/dextrose agar containing ≈0.5 μg of rapamycin (final concentration = 100 nM). Nine thousand six hundred strains can be arrayed on a standard one-well plate area. The yellow arrow points to the rapamycin-resistant fpr1Δ, which is missing the rapamycin receptor FKBP12. (b) The known TOR signaling components (adapted from ref. 20) were colored according to deletion strain phenotypes identified from our screen. Red, rapamycin-resistant when deleted; green, rapamycin-hypersensitive when deleted.
Using this cell array method, we screened the full collection of yeast deletion strains (including 4,850 nonessential gene MATa haploid deletions and 1,175 essential gene heterozygous diploid deletions) for changes in relative fitness (compared with wild type) in the presence of rapamycin, aiming at identifying all single-gene modifiers of the rapamycin-sensitive Tor pathway. Such genetic modifiers may be Tor pathway components or effectors in parallel pathways. The logic underlying this analysis differs from drug-induced haploinsufficiency screening that aims at identifying direct drug targets (13, 14) but instead facilitates a global understanding of drug effects at the systems level. In the case of rapamycin, its protein targets (FKBP12 and Tor) have been known for more than a decade, but its broad cellular effects, ranging from growth control (15) to possibly lifespan regulation (16, 17), are only beginning to be understood. The ability to profile drug effects at the genomic scale in vivo is fundamental to our understanding of molecular and cellular mechanisms and, as we show here, can shed light on the therapeutic limitations and potential unexpected uses of drugs as well.
Materials and Methods
High-Density Cell Array Printing. A contact microarrayer, Omni-Grid Accent (Genemachines, San Carlos, CA), was used to print cells to agar medium in a one-well plate. Cells that settled on the bottom of the library plates were picked up by using solid quill pins (TeleChem, Sunnyvale, CA) and spotted onto agar. Pins were cleaned between each printing step by sonication (70% ethanol, 1 min), followed by three cycles of rinse (in peristaltic flow of glass distilled water, 15 sec) and dry (vacuum, 15 sec), all in specialized stations on the microarrayer. Using a center-to-center distance of 900 μm between spots, 9,600 strains were printed on the area of a standard microtiter plate (127 × 85 mm). Plates were incubated inverted with a fine mist of distilled water sprayed on the cover to prevent the drying out of the array.
Yeast Deletion Library. A complete yeast deletion strain collection created by the Saccharomyces Genome Deletion Project (http://sequence-www.stanford.edu/group/yeast_deletion_project/deletions3.html) was purchased from Research Genetics (Huntsville, AL). The original library in 96-well plates was reformatted to 384-well plates, which were used as source plates for microarraying. A total of 6,025 strains were screened, including 4,850 haploid (MATa) deletion strains (each deleted of a nonessential gene) and 1,175 heterozygous diploid (MATa/α) deletion strains (each deleted of an essential gene).
Chemical Genomic Screening and Quantitation. A deletion library of 6,025 strains (see above) was arrayed at a density of 6,144 strains per plate on both 10 nM and 30 nM rapamycin and on DMSO as a control (small molecules are dissolved in yeast extract/peptone/dextrose, 2% agar, and 0.1% DMSO). Cell arrays were imaged by using a custom charge-coupled device imaging system, and growth of each strain was measured in the grayscale intensity of the images. To make the three arrays comparable, the intensity values in 10 nM rapamycin were normalized to that in DMSO according to the average intensity (most strains grew in 10 nM rapamycin as well as in DMSO); the intensity values in 30 nM were normalized against DMSO based on the average background value (most strains show little growth in 30 nM rapamycin). The ratios of intensity in rapamycin over that in DMSO were used to score for hypersensitivity or resistance to rapamycin. Only those strains that showed significant growth in DMSO and with 10 nM rapamycin/DMSO ratios <0.5 were accepted as hypersensitive. Only strains that showed significant growth in rapamycin and with 30 nM rapamycin/DMSO ratios >0.5 were accepted as resistant.
Molecular Biology and Yeast Genetics Procedures. Wild-type and rapamycin-resistant TOR1 and TOR2 genes were subcloned into pYES2-1 (Invitrogen) by using standard molecular biological protocols. Domain-swap constructs for Tor2-Tor1 fusions were created through multistep mutagenesis and subcloning procedures (details are available upon request). Yeast transformation and growth assays were performed according to standard protocols (18) with minor modifications.
Results and Discussion
Genomewide Functional Profiling Identifies Major Genetic Modifiers of Rapamycin Sensitivity. Growth of the deletion strains in the presence of rapamycin was measured by using an automated image analysis system and normalized for growth in DMSO (drug carrier). Because wild-type yeast cells are sensitive to rapamycin (i.e., displayed reduced fitness in the presence of rapamycin compared with DMSO control), rapamycin sensitivity of each deletion mutant was defined relative to that of the wild type (relative fitness). Resistant mutants are less sensitive to rapamycin than wild type, whereas hypersensitive mutants are more sensitive than the wild type.
From our screen, 396 strains were identified that showed altered fitness response to rapamycin (Table 2, which is published as supporting information on the PNAS web site). Among them, 281 were hypersensitive, and 101 were resistant. As expected, all known effectors and downstream targets of Tor (19, 20), such as Gln-3, Ure2, Npr1, and Tip41 (Fig. 1b), were identified, supporting the reliability of our high-density cell-array method. We also obtained identical results with 12 deletion mutants (including controls and random picks) on our array and in two traditional growth conditions (in liquid in 96 wells and on agar plates). Using the Gene Ontology tool in the S. cerevisiae Genome Database (http://www.yeastgenome.org), we found that the majority of the 396 genes map to two biological processes (Fig. 2), “cell growth and/or maintenance” (P value = 1.3 × 10-13) and “metabolism” (P = 0.004), consistent with the known role of Tor in regulating cell growth and nutrient response. Although this pathway appears to be better studied than the genome as a whole (Fig. 2, right set of bars), ≈39% of the identified genes (155 genes) have no identified molecular function and 24% (94 genes) have not been found to participate in any known biological process. Most interestingly, we observed 14 deletion strains (see below) that grew much better in the presence of rapamycin, which we refer to as “rapamycin-enhanced” herein.
Fig. 2.
Gene Ontology (go) mapping of 396 genes that demonstrated altered sensitivity to rapamycin. RsWr, rapamycin-hypersensitive and wortmannin-resistant (when deleted); RrWs, rapamycin-resistant and wortmannin-hypersensitive; R, rapamycin-hypersensitive or rapamycin-resistant.
We previously described the fitness response of the yeast deletion library to wortmannin, a related natural product that inhibits phosphatidylinositol 3-kinase and modulates the phospholipid “signalome” (21). Comparison of the two gene lists of altered drug sensitivity revealed that among the 396 strains of altered rapamycin sensitivity, >72% (284 genes) showed either the opposite response or no response to wortmannin, consistent with the rapamycin specificity of the majority of our identified deletion mutants. Of the 90 genes whose deletions showed similar sensitivity to both rapamycin and wortmannin, some appear to be broad-specificity effectors involved in drug permeability (e.g., Erg4 and Pdr13), and others may indicate common downstream targets or nodes of crosstalk. We performed a quantitative comparison of functional categories affected by rapamycin relative to the genome (Fig. 2). In the list of deletions that are rapamycin-hypersensitive, especially the ones that are strictly rapamycin-hypersensitive (they are either nonresponsive or resistant to wortmannin), genes for the transport process were significantly enriched (×2.7; P < 0.0001). In contrast, in the list of deletions that are specifically resistant to rapamycin, genes for transcription were highly enriched (×3.1; P < 0.01), suggesting that transcription may play a prominent role in modulating rapamycin sensitivity of the cell.
Correlation Between Gene Expression and Drug Sensitivity. The enrichment of genes involved in transcription in the list of rapamycin-resistant deletions prompted us to compare the functional profiling results (Table 2) and whole-genome transcript profiling results (22) from rapamycin-treated cells. Among 396 deletion genes, only 35 (<10%) showed >3-fold changes at the transcript level upon treatment with rapamycin (Fig. 3). Decreases in transcript levels among hypersensitive deletion genes upon rapamycin treatment (e.g., genes 17-33 in Fig. 3) suggests the intriguing possibility that transcriptional down-regulation of these genes may be causal to the cell's sensitivity to rapamycin. Analogously, resistant deletion genes likely have negative roles in TOR signaling; increases in their transcript levels by rapamycin (e.g., genes 10-16) suggests that their up-regulation may be the molecular basis for rapamycin sensitivity in wild-type cells. In contrast, genes 1-9, 34, and 35 (Fig. 3) may reflect cellular effects of rapamycin treatment or defense mechanisms by the cell in an attempt to counter rapamycin toxicity. These changes individually do not appear to alter fitness response to rapamycin. Our combined use of transcript profiling and functional profiling data yields information that would otherwise be inaccessible from each one-dimensional platform alone.
Fig. 3.
Circular clustergram of the 35 genes that individually contributed to rapamycin sensitivity as well as exhibited significant (>3-fold) changes in gene expression upon rapamycin treatment. The inner circle represents transcript profiling data, decrease is in green, increase in red; the outer circle represents functional profiling data, rapamycin-hypersensitive deletion is in green, rapamycin-resistant deletion in red.
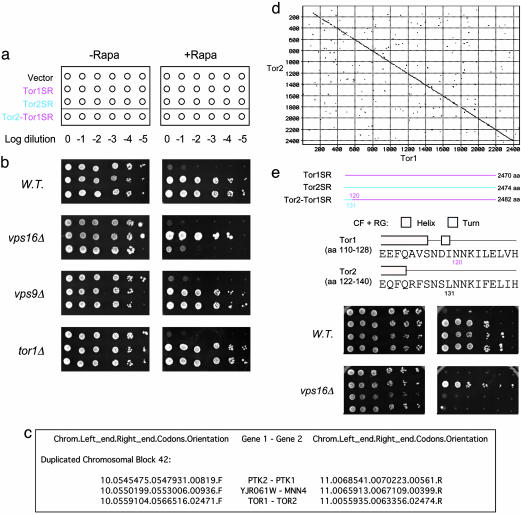
Difference Between TOR1 and TOR2 Concerning the Rapamycin-Sensitive Functions. While most organisms have a single TOR gene, S. cerevisiae has two, which map to the duplicated chromosomal block 42 (23) (Fig. 4c). It is well established that whereas Tor2 has a unique rapamycin-insensitive essential function, Tor1 and Tor2 are identical in their rapamycin-sensitive cell growth (previously also described as the G1 cell cycle) function (24). These two functions have been accounted for biochemically by recent identification of two distinct Tor protein complexes (25), TORC2 and TORC1. Consistent with the notion that the rapamycin-sensitive function is redundant between Tor1 and Tor2, rapamycin sensitivity of wild-type cells (and all mutants reported to date) can be rescued equally by either Tor1SR or Tor2SR, rapamycin-resistant point mutant Tor proteins that are functional but unable to bind FKBP12-rapamycin. However, while testing the specificity of rapamycin-hypersensitive deletions, we found that vps16Δ, vps18Δ, and vps11Δ (thanks to Scott Emr, we corrected the misassignment at the vps18 and vps11 well in the original MATa collection) can be rescued only by Tor1SR but not by Tor2SR (Fig. 4b), indicating a surprising functional distinction even in the rapamycin-sensitive functions of Tor1 and Tor2. Vps16 and Vps18 are subunits of the class C Vps protein complex that regulates vacuolar SNARE pairing and is required for vesicle docking/fusion (26). Interestingly, yeast cells deleted in other rapamycin-sensitive class C complex subunits (27) (e.g., Vps39 and Vps41) do not distinguish between Tor1SR and Tor2SR, possibly suggesting different mechanistic involvements amongst the class C Vps proteins.
Fig. 4.
TOR1 and TOR2 are different in their rapamycin-sensitive function. (a) Schematic representation of plasmids and cell dilutions used for experiments in b and e. (b) Tor1SR is ≈1,000-fold more active than Tor2SR in vps16Δ cells although they are equally active in most (including wild-type) cells. (c) Chromosomal duplication of TOR genes in yeast. (d) Sequence alignment of Tor1 and Tor2 revealed the nonhomologous domain of ≈100 amino acids at the N termini. (e) Sequence junction and activity of the chimeric Tor2-Tor1SR protein, and although it carries only 131 amino acids of Tor2, behaves like Tor2SR and is unable to rescue rapamycin inhibition of either endogenous Tor protein in vps16Δ mutant.
To elucidate the structural basis for the newly uncovered functional difference between Tor1 and Tor2, we performed protein sequence alignments by using Pustell Protein Matrix in macvector (Accelrys, San Diego). Tor1p and Tor2p are known to be highly homologous, as is apparent from the diagonal line across the bulk of the full-length proteins in the alignment matrix map (Fig. 4d). However, in the extreme N termini, Tor1 and Tor2 differ significantly (Fig. 4d). To determine whether the difference in the N termini is responsible for the differential activities for the two Tor proteins in vps16Δ, we swapped the N-terminal domain of Tor2 into Tor1SR (Fig. 4e). Interestingly, the resulting fusion protein, which contains the N-terminal 131 residues of Tor2p and the C-terminal 2,351 residues of Tor1SR, now behaves like Tor2SR and fails to confer rapamycin resistance to vps16Δ cells (Fig. 4e). The fusion protein is folded correctly and is fully functional, because it is able to confer rapamycin resistance to wild-type cells (Fig. 4e). Similar results were obtained in vps18Δ cells (data not shown).
Because Vps16 and Vps18 are highly conserved in humans (28), it is possible that the N terminus of Tor1 is the ancestor domain, whereas the current N terminus of Tor2 is a result of divergent evolution. The two Tor proteins may have diverged to cope with various nutrient conditions in yeast, although it is unclear at present whether the N terminus of Tor2 has simply degenerated or rather has acquired a new function. Because we have mapped the new functional difference to a small domain (≈120 aa) in the N terminus of the Tor proteins, it should facilitate the use of biochemical and two-hybrid approaches toward a detailed mechanistic understanding of this phenomenon.
Rapamycin-Enhanced Cell Fitness: Therapeutic Opportunities for Neurodegenerative Diseases? As mentioned earlier, we have identified an unexpected “rapamycin-enhanced” phenotype in 14 genes whose deletion each allows cells to grow better in the presence of rapamycin than in its absence (Table 1). Five of these mutants are rapamycin-specific, whereas the other nine are also resistant to wortmannin, suggesting that they may represent common pathways affected by these two natural products.
Table 1. A new class of rapamycin-enhanced gene deletion mutants that exhibits better fitness in the presence of rapamycin than in its absence.
| Gene no. | ORF name | Gene name | DMSO | Rapa | Ratio | Rapa score | Wort score | MMS score | Cellular localization* |
|---|---|---|---|---|---|---|---|---|---|
| 1 | YOR200W | 0.27194 | 5.58246 | 20.53 | + + + + + | 0 | 0 | Unknown | |
| 2 | YOR199W | 0.58382 | 7.57624 | 12.98 | + + + + + | 0 | 0 | Cytoplasm | |
| 3 | YLR288C | MEC3 | 0.50933 | 4.61341 | 9.06 | + + + + + | + + + | - - - | Unknown |
| 4 | YNL005C | MRP7 | 0.90839 | 7.99440 | 8.80 | + + + + + | + + + + | 0 | Mitochondrion |
| 5 | YNR041C | COQ2 | 1.24415 | 9.98004 | 8.02 | + + + + + | 0 | 0 | Mitochondrion |
| 6 | YMR071C | 1.07938 | 8.52237 | 7.90 | + + + + + | + + + + | 0 | Golgi | |
| 7 | YOR187W | TUF1 | 1.09775 | 8.21672 | 7.49 | + + + + + | + + | 0 | Mitochondrion |
| 8 | YPR124W | CTR1 | 1.22578 | 8.24591 | 6.73 | + + + + + | 0 | 0 | Cell periphery; bud |
| 9 | YJR104C | SOD1 | 1.70195 | 10.65302 | 6.26 | + + + + + | + + + + + | - - - | Mitochondrion, etc. |
| 10 | YMR066W | SOV1 | 0.83511 | 5.01229 | 6.00 | + + + + + | + + | 0 | Mitochondrion |
| 11 | YDL198C | YHM1 | 0.53919 | 2.95835 | 5.49 | + + + | 0 | 0 | Mitochondrion |
| 12 | YDL181W | INH1 | 0.75161 | 3.73389 | 4.97 | + + + + + | + + + + | 0 | Mitochondrion |
| 13 | YMR038C | CCS1/LYS7 | 2.75383 | 10.21015 | 3.71 | + + + + + | + + + + + | - - - | Mitochondrion, etc. |
| 14 | YGR076C | MRPL25 | 0.75114 | 2.49108 | 3.32 | + + | + + | 0 | Mitochondrion |
| Control | YNL135C | FPR1 | 3.71285 | 12.16755 | 3.28 | + + + + + | 0 | 0 | Cytoplasm; nucleus |
Jim Broach (personal communication) made the interesting observation that two genes above (YOR200W and YOR199W) and a mitochondrial protein gene PET56 (YOR201C) have overlapping sequences. It is possible that the defect in only one of these genes is responsible for the rapamycin-enhanced phenotype, although the pet56Δ strain in the MATα collection shows equal fitness with or without rapamycin. Complementing either of the two Watson deletions with a vector expressing each of the three genes can be used to ascertain their role (or the lack of) in the rapamycin-enhanced phenotype.
Cellular localization data were mined from the yeast GFP fusion localization database (http://yeastgfp.ucsf.edu) and the Saccharomyces Genome Database (www.yeastgenome.org).
Of the 14 gene deletions that produce the rapamycin-enhanced phenotype, 13 genes have human homologs that showed >30% identity (highly significant) at the protein level, and most of them encode mitochondrial proteins (Table 1). Because mitochondrial dysfunction is known to underlie the pathogenesis of a wide range of neurodegenerative disorders due to impaired energy production, increased oxidative damage, and apoptosis (29, 30), our result suggests that rapamycin may be useful in preventing the progression of these diseases, including Alzheimer's, Parkinson's, and Huntington's diseases and brain aging.
It is intriguing that a deficiency in each of the 14 genes is deleterious, as is a deficiency in TOR function, but when both are deficient, the cells exhibit greatly enhanced fitness. One hypothesis is that it is the balance of Tor and each particular protein that is important rather than the absolute amount of each protein. One reason may be that they form a stoichiometric protein complex. It has been shown that mTor is associated with multiple mitochondrial enzymes (ref. 31 and unpublished data) and localizes to the mitochondrial membrane (31). It would be interesting to see whether any of these 14 proteins are directly or indirectly associated with Tor.
Implications for the Genetic Basis of Rapamycin Sensitivity and Resistance in Cancer. Our identification of conserved rapamycin-enhanced gene deletions in yeast raises the possibility that certain tumors may contain similarly rapamycin-enhanceable mutations (or other genetic variation). In such cases, rapamycin would have negative clinical implications as it might actually stimulate tumor growth. Given the importance of rapamycin and its analogue CCI-779 as anti-cancer agents (32), it is important to understand how differences in genotypes, either due to gene mutations or single-nucleotide polymorphisms, affect an individual's response to rapamycin/CCI-779. Although studies of resistance to molecularly targeted drugs have so far focused on the intended drug target while leaving the rest of the genome unexplored, our prediction is that many gene products will affect clinical drug response in a complex manner. There is accumulating evidence that rapamycin sensitivity of cancer cells often correlates with p53 and/or PTEN status and that other genes clearly play important roles as well. However, the nature of these genes remains unknown.
The identity of the many (≈400) genes determined to be important for rapamycin resistance and sensitivity clearly illustrated an important genetic basis of differential rapamycin response. Of the 396 genes isolated from our yeast screen, 102 (≈25%) have significant mammalian homologs (>30% amino acid sequence identity). Varying genotypes at these homologous loci in humans could affect the patients' response to rapamycin. The information may serve to predict human genes involved in determining cellular sensitivity to rapamycin and also possible mechanisms for the emergence of drug resistance from rapamycin/CCI-779-treated tumors. In any case, we expect that a large number of genes will contribute in a complex fashion to determine rapamycin sensitivity of a patient.
Impact of the Technologies. In recent years, there has been tremendous interest and progress in using genomic tools for linking the mechanisms of action of drugs to the genetic roots responsible for their biological effects (9, 13, 14, 21, 33-38). Our cell array technology minimizes both materials and efforts required for such analyses. Because unexpected synergistic activities can coexist (39) in the natural product-producing microorganisms or medicinal plants, our method should also be valuable for whole-genome profiling of complex natural product extracts. Thorough interrogation of the whole genome provides critical information regarding potential drug targets, genetic modifiers of drug sensitivity, and signaling pathways that underlie drug action.
Although not demonstrated here, the cell array technology is readily adaptable to other types of large-scale screens based on growth (e.g., in whole-genome yeast two-hybrid screens; ref. 5), or changes in color/fluorescence or colony morphology. The cell array platform is also applicable to high-throughput investigations of other microbial systems. Furthermore, it may be possible to devise analogous systems to array mammalian cells on appropriate substrates. Such systems will allow high-throughput manipulations of tens of thousands of genetically engineered mammalian cells as an array and complement the use of an elegant mammalian cell array platform developed by the Sabatini laboratory in systematic analyses of gene function and drug mechanisms (40).
Genomic sensor pads based on RNA or protein expression patterns have been postulated for drug discovery and development (41). Analogously, our observation of the highly distinct fitness profiles of rapamycin and wortmannin suggests that a “chemical genomic biosensor” based on cell fitness patterns may be created by combining the information from many fitness screens. In the form of a live cell array consisting of representative isogenic yeast deletion strains, such a simple and robust live “pattern-recognition algorithm” can also be integrated with state-of-the-art electronic (42) or optical (43) biosensor platforms in the identification and prediction of drugs or environmental agents.
Supplementary Material
Acknowledgments
We thank Greg Payne, Des Smith, Jon Saxe, William Straub, Kurtis Straub, and our anonymous reviewers for comments on the manuscript; Scott Emr and Jim Broach for sharing their insights; Charlie Boone for communicating results before publication; Fred Zhang for software development; Lisa Zeng and Ron Tongbai for technical assistance; and Tim Cloughesy, Paul Mischel, Hong Wu, Charles Sawyers, Owen Witte, and Mike Phelps for intellectual support. This work was funded, in part, by the General Motors Cancer Research Foundation, the Stein Oppenheimer Endowment Fund, and the Singleton Developmental Grant (to J.H.).
References
- 1.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 2.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- 3.Scherens, B. & Goffeau, A. (2004) Genome Biol. 5, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Que, Q. Q. & Winzeler, E. A. (2002) Funct. Integr. Genomics 2, 193-198. [DOI] [PubMed] [Google Scholar]
- 5.Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. (2000) Nature 403, 623-627. [DOI] [PubMed] [Google Scholar]
- 6.Tong, A. H., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W., Bussey, H., et al. (2001) Science 294, 2364-2368. [DOI] [PubMed] [Google Scholar]
- 7.Chang, M., Bellaoui, M., Boone, C. & Brown, G. W. (2002) Proc. Natl. Acad. Sci. USA 99, 16934-16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begley, T. J., Rosenbach, A. S., Ideker, T. & Samson, L. D. (2002) Mol. Cancer Res. 1, 103-112. [PubMed] [Google Scholar]
- 9.Parsons, A. B., Brost, R. L., Ding, H., Li, Z., Zhang, C., Sheikh, B., Brown, G. W., Kane, P. M., Hughes, T. R. & Boone, C. (2004) Nat. Biotechnol. 22, 62-69. [DOI] [PubMed] [Google Scholar]
- 10.Demain, A. L. (2002) Nat. Biotechnol. 20, 331. [DOI] [PubMed] [Google Scholar]
- 11.Handelsman, J., Rondon, M. R., Brady, S. F., Clardy, J. & Goodman, R. M. (1998) Chem. Biol. 5, R245-R249. [DOI] [PubMed] [Google Scholar]
- 12.Kaeberlein, T., Lewis, K. & Epstein, S. S. (2002) Science 296, 1127-1129. [DOI] [PubMed] [Google Scholar]
- 13.Lum, P. Y., Armour, C. D., Stepaniants, S. B., Cavet, G., Wolf, M. K., Butler, J. S., Hinshaw, J.C., Garnier, P., Prestwich, G. D., et al. (2004) Cell 116, 121-137. [DOI] [PubMed] [Google Scholar]
- 14.Giaever, G., Flaherty, P., Kumm, J., Proctor, M., Nislow, C., Jaramillo, D. F., Chu, A. M., Jordan, M. I., Arkin, A. P. & Davis, R. W. (2004) Proc. Natl. Acad. Sci. USA 101, 793-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fingar, D. C. & Blenis, J. (2004) Oncogene 23, 3151-3171. [DOI] [PubMed] [Google Scholar]
- 16.Vellai, T., Takacs-Vellai, K., Zhang, Y., Kovacs, A. L., Orosz, L. & Muller, F. (2003) Nature 426, 620. [DOI] [PubMed] [Google Scholar]
- 17.Kapahi, P., Zid, B. M., Harper, T., Koslover, D., Sapin, V. & Benzer, S. (2004) Curr. Biol. 14, 885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guthrie, C. & Fink, G. R., eds. (1991) Methods in Enzymology Guide to Yeast Genetics and Molecular Biology (Academic, New York), Vol. 194. [PubMed]
- 19.Chan, T. F., Carvalho, J., Riles, L. & Zheng, X. F. S. (2000) Proc. Natl. Acad. Sci. USA 97, 13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacinto, E. & Hall, M. N. (2003) Nat. Rev. Mol. Cell Biol. 4, 117-126. [DOI] [PubMed] [Google Scholar]
- 21.Zewail, A., Xie, M. W., Xing, Y., Lin, L., Zhang, P. F., Zou, W., Saxe, J. P. & Huang, J. (2003) Proc. Natl. Acad. Sci. USA 100, 3345-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, J., Zhu, H., Haggarty, S. J., Spring, D. R., Hwang, H., Jin, F., Snyder, M. & Schreiber, S. L. (2004) Proc. Natl. Acad. Sci. USA 101, 16594-16599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfe, K. H. & Shields, D. C. (1997) Nature 387, 708-713. [DOI] [PubMed] [Google Scholar]
- 24.Zheng, X. F., Florentino, D., Chen, J., Crabtree, G. R. & Schreiber, S. L. (1995) Cell 82, 121-130. [DOI] [PubMed] [Google Scholar]
- 25.Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J. L., Bonenfant, D., Oppliger, W., Jenoe, P. & Hall, M. N. (2002) Mol. Cell 10, 457-468. [DOI] [PubMed] [Google Scholar]
- 26.Sato, T. K., Rehling, P., Peterson, M. R. & Emr, S. D. (2000) Mol. Cell 6, 661-671. [DOI] [PubMed] [Google Scholar]
- 27.Wurmser, A. E., Sato, T. K. & Emr, S. D. (2000) J. Cell Biol. 151, 551-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huizing, M., Didier, A., Walenta, J., Anikster, Y., Gahl, W. A. & Kramer, H. (2001) Gene 264, 241-247. [DOI] [PubMed] [Google Scholar]
- 29.Beal, M. F. (2000) Trends Neurosci. 23, 298-304. [DOI] [PubMed] [Google Scholar]
- 30.Mattson, M. P. (2000) Nat. Rev. Mol. Cell Biol. 1, 120-129. [DOI] [PubMed] [Google Scholar]
- 31.Desai, B. N., Myers, B. R. & Schreiber, S. L. (2002) Proc. Natl. Acad. Sci. USA 99, 4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bjornsti, M. A. & Houghton, P. J. (2004) Nat. Rev. Cancer 4, 335-348. [DOI] [PubMed] [Google Scholar]
- 33.Giaever, G., Shoemaker, D. D., Jones, T. W., Liang, H., Winzeler, E. A., Astromoff, A. & Davis, R. W. (1999) Nat. Genet. 21, 278-283. [DOI] [PubMed] [Google Scholar]
- 34.Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109-126. [DOI] [PubMed] [Google Scholar]
- 35.Willson, T. M., Jones, S. A., Moore, J. T. & Kliewer, S. A. (2001) Med. Res. Rev. 21, 513-522. [DOI] [PubMed] [Google Scholar]
- 36.Neckers, L. (2003) Curr. Med. Chem. 10, 733-739. [DOI] [PubMed] [Google Scholar]
- 37.Haggarty, S. J., Clemons, P. A. & Schreiber, S. L. (2003) J. Am. Chem. Soc. 125, 10543-10545. [DOI] [PubMed] [Google Scholar]
- 38.Tucker, C. L. & Fields, S. (2004) Comp. Funct. Genomics 5, 216-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang, A., Wong, G. K. & Demain, A. L. (2000) J. Antibiot. 53, 158-162. [DOI] [PubMed] [Google Scholar]
- 40.Ziauddin, J. & Sabatini, D. M. (2001) Nature 411, 107-110. [DOI] [PubMed] [Google Scholar]
- 41.Mihich, E., Feunteun, J. & Friend, S. (2002) Cancer Res. 62, 3883-3887. [PubMed] [Google Scholar]
- 42.Solly, K., Wang, X., Xu, X., Strulovici, B. & Zheng, W. (2004) Assay Drug Dev. Technol. 2, 363-372. [DOI] [PubMed] [Google Scholar]
- 43.Biran, I., Rissin, D. M., Ron, E. Z. & Walt, D. R. (2003) Anal. Biochem. 315, 106-113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.