Abstract
Lingonberry (Vaccinium vitis-idaea L.) produces tiny red berries that are tart and nutty in flavor. It grows widely in the circumpolar region, including Scandinavia, northern parts of Eurasia, Alaska, and Canada. Although cultivation is currently limited, the plant has a long history of cultural use among indigenous communities. Given its potential as a food source, genomic resources for lingonberry are significantly lacking. To advance genomic knowledge, the genomes for 2 subspecies of lingonberry (V. vitis-idaea ssp. minus and ssp. vitis-idaea var. ‘Red Candy’) were sequenced and de novo assembled into contig-level assemblies. The assemblies were scaffolded using the bilberry genome (Vaccinium myrtillus) to generate a chromosome-anchored reference genome consisting of 12 chromosomes each with a total length of 548.07 Mb [contig N50 = 1.17 Mb, BUSCO (C%) = 96.5%] for ssp. vitis-idaea and 518.70 Mb [contig N50 = 1.40 Mb, BUSCO (C%) = 96.9%] for ssp. minus. RNA-seq-based gene annotation identified 27,243 and 25,718 genes on the respective assembly, and transposable element detection methods found that 45.82 and 44.58% of the genome were repeats. Phylogenetic analysis confirmed that lingonberry was most closely related to bilberry and was more closely related to blueberries than cranberries. Estimates of past effective population size suggested a continuous decline over the past 1–3 MYA, possibly due to the impacts of repeated glacial cycles during the Pleistocene leading to frequent population fragmentation. The genomic resource created in this study can be used to identify industry-relevant genes (e.g. anthocyanin production), infer phylogeny, and call sequence-level variants (e.g. SNPs) in future research.
Keywords: lingonberry, partridgeberry, mountain cranberry, Vaccinium vitis-idaea ssp. vitis-idaea, V. vitis-idaea ssp. minus, genome assembly
Introduction
Vaccinium vitis-idaea L., commonly known as lingonberry, partridgeberry, or mountain cranberry, is an evergreen dwarf shrub that has cultural, economic, and ecological importance (Debnath and Arigundam 2020). The bright-red colored berries have been consumed among Indigenous communities in northern North America and Scandinavia as a relish and served with meat or fish in traditional meals (Moerman 2010; Vaara et al. 2013). Berry picking has been a cherished cultural practice, and nowadays people commonly preserve berries as jams that are becoming more readily available commercially (e.g. Arctic Lingonberry; https://www.arcticlingonberry.fi/). A growing body of research suggests that lingonberry fruits and leaves have medicinal benefits to human health such as anticancer, cardioprotective, and neuroprotective properties (Ferlemi and Lamari 2016; Kowalska 2021). Despite a long history of utilization as a culturally important food source and its recognized health benefits, the domestication of lingonberry is at its infancy in North America.
Being an evergreen boreal forest understory species, lingonberry propagates vegetatively by forming mat-like clonal communities through rhizomes (Hjalmarsson and Ortiz 1998) or sexually through seeds that are primarily insect pollinated (Jacquemart and Thompson 1996). The species has 2 recognized subspecies (ssp.) based on their geographical origin: V. vitis-idaea ssp. minus and ssp. vitis-idaea, and the species is widely distributed in the circumpolar region (Debnath and Arigundam 2020; Fig. 1a). The European subspecies, ssp. vitis-idaea, currently has active breeding programs with more than a dozen of cultivars available for commercial production, with improved yield and berry size (Penhallegon 2009). The North American ssp. minus, on the other hand, is considered a wild plant with little breeding efforts taken place. The 2 subspecies are distinguishable based on several morphological differences as well as genetic differences (Garkava-Gustavsson et al. 2005; Debnath 2007; Debnath and Arigundam 2020). The extent of genomic differences between the 2 subspecies has not been studied before, and it is somewhat unclear whether they occur sympatrically in the overlapping ranges.
Fig. 1.
a) Worldwide distribution of V. vitis-idaea L. (GBIF 2023). Dots represent occurrence records registered as follows: V. vitis-idaea ssp. minus (blue), V. vitis-idaea ssp. vitis-idaea (red), and V. vitis-idaea L. ssp. unidentified (yellow). b) V. vitis-idaea ssp. vitis-idaea flowers and fruits. c) V. vitis-idaea ssp. minus (left) and ssp. vitis-idaea var. ‘Red Candy’ (right) grown in the greenhouse.
Long-read sequencing technology has fueled exponential growth in the assembly of plant genomes (Marks et al. 2021); there are at least 1,368 unique flowering plant species genomes assembled at higher than scaffold level [NCBI search terms: “Magnoliopsida (flowering plants)” “scaffold+”, by Nov 9th, 2023], and this number is likely underestimated. The use of long reads has been particularly relevant for plant genomes due to their high repeat proportion and propensity for polyploidy. Within Vaccinium, high-quality genomes have been assembled for 9 species (Colle et al. 2019; Diaz-Garcia et al. 2021; Wu et al. 2021; Yu et al. 2021; Cui et al. 2022; Kawash et al. 2022; Yang et al. 2022; Mengist et al. 2023), as well as a pangenome project for cultivated blueberry and cranberry involving 32 cultivars has been completed (Yocca et al. 2023). In contrast, lingonberry's genomics is understudied; only a handful of genetic, chloroplast, or mitochondrial genomic research has been conducted (Garkava-Gustavsson et al. 2005; Debnath 2007; Gailīte et al. 2020; Kim et al. 2020; Tian et al. 2020). This study aimed to provide useful genomic resource to the lingonberry community, through genome assembly of the 2 distinct subspecies: V. vitis-idaea ssp. vitis-idaea and ssp. minus. The resources created from the study will be publicly available, in the hope of furthering our understanding of lingonberry evolution and aiding the future breeding efforts by accelerating the molecular screening of lingonberry cultivars.
Materials and methods
Plant material
The clones of a commercial lingonberry plant (V. vitis-idaea L. ssp. vitis-idaea var. ‘Red Candy’) were obtained from Lochside nursery (Victoria, BC) in September 2021 and July 2022 and kept in the greenhouse, designated as LC1 and LC2, respectively. Since LC1 and LC2 were clones of the same line, they should be genetically identical, but we had used separate identifiers for each. The wild lingonberry clone (V. vitis-idaea L. ssp. minus) designated as LW1, originally collected from Baie-Trinite, Quebec, Canada (latitude: 49°25′N; longitude: 67°18′W; Debnath 2007), was obtained from collaborators at Agriculture and Agri-Food Canada St. John's Research and Development Centre, NL, and kept in the greenhouse. The 3 accessions were vouchered at the University of Victoria herbarium collection: LC1 = UVIC 48749, LC2 = UVIC 48750, LW1 = UVIC 48751, respectively.
High-molecular-weight DNA extraction
Young and mature shoots were excised from each subspecies (LC1, LW1). The leaves (1–2 g dry weight) were collected and wiped with 70% ethanol prior to extractions. The sterilized leaves were flash frozen in liquid nitrogen and ground into fine powder using mortar and pestle (∼5 min). High-molecular-weight (HMW) DNA was extracted using Nucleobond HMW DNA extraction kit (Takara Bio) following the manufacturer's protocol, with double the amount of starting material and the buffers accordingly. The DNA was then size selected using SRE-XS kit or SRE kit (Circulomics) to remove fragments smaller than 10 or 25 kb, respectively.
RNA extraction
Total RNA was extracted for the commercial lingonberry clones, LC1 or LC2 from 5 tissue types: young expanding leaf (LC1), flower (LC2), unripe berry (greenish white; LC2), ripe berry (red; LC2), and rhizome (LC2). Note that the rhizome was technically an underground shoot, but it did not have green leaves. The root-equivalent tissue could not be sampled due to soil contaminations and difficulty in extracting enough root mass without killing the plant. For leaf and flower samples, modified CTAB protocol was used to isolate RNA (Muoki et al. 2012; Yoshida et al. 2015). For rhizome, Spectrum Plant Total RNA Kit (Sigma) was used. For berries, modified CTAB protocol optimized for bilberry was used (Jaakola et al. 2001). Due to low recovery of pure RNA, the unripe and ripe berries were combined to make up 1 berry sample, resulting in the total of 4 RNA samples prepared for sequencing.
Sequencing
For long-read sequencing with Oxford Nanopore Technologies (ONT), sequencing libraries were prepared with the Ligation Sequencing Kit (SQK-LSK110 or SQK-LSK114) and they were sequenced on MinION Flow Cell R9.4.1 (FLO-MIN106D) or R10.4.1 (FLO-MIN114), respectively, following manufacturer's protocols. For LC1, 1 each of the R9.4.1 flow cell and R10.4.1 flow cell was used. For LW1, 3 R10.4.1 flow cells were used. All the raw output FAST5 reads were then basecalled by the Guppy basecalling software v6.1.2+e0556ff (https://nanoporetech.com/) and minimap2 v2.22-r1101 (Li 2018) using super accurate or “sup” model (-c dna_r9.4.1_450bps_sup.cfg). For reads generated with R10.4.1 flow cells, the reads were further duplex basecalled according to the Guppy Duplex-basecalling pipeline v6.3.8+d9e0f64 (https://nanoporetech.com/). In brief, raw FAST5 files were basecalled using the “fast” model (dna_r10.4_e8.1_fast.cfg), and the duplex candidates were listed as read–pair candidates. Those reads were then duplex basecalled by Guppy-duplex. The remaining reads were identified on the simplex reads already basecalled by “sup” model (dna_r10.4_e8.1_sup.cfg) using a custom perl script (see git repository; “filter_fastq.pl”), and finally the duplex basecalled reads were combined with the duplex-filtered simplex reads. The generated FASTQ files were concatenated as a single raw read output for the downstream procedures. Note that the raw basecalled reads were filtered by the mean >Q10 prior to concatenating. ONT sequencing and basecalling procedures are summarized in Table 1. For short-read sequencing, PCR-free whole-genome sequencing libraries were prepared and sequenced on an Illumina NovaSeq in paired-end mode, targeting 75 M individual reads per sample. The RNA library was prepared by PolyA+ mRNA Library Construction service provided and sequenced on Illumina NovaSeq paired-end mode, targeting 50 M reads per sample. Both RNA and DNA libraries were sequenced using paired-end 150 bp reads. The raw output FASTQ files were visually quality checked with fastqc v0.11.9 (Andrews 2019).
Table 1.
ONT sequencing and basecalling methods used for commercial (LC1) and wild (LW1) lingonberry samples.
| Sample | No. of flow cells used | Flow cell ver. | Flow cell code | Library kit | Simplex/duplex | Basecalling software | Basecalling mode |
|---|---|---|---|---|---|---|---|
| LC1 | 1 | R9.4.1 | FLO-MIN106D | SQK-LSK110 | Simplex | Guppy (v6.1.2+e0556ff)+minimap2 v2.22-r1101 | Super accurate “sup” (dna_r9.4.1_450bps_sup.cfg) |
| 1 | R10.4.1 | FLO-MIN114 | SQK-LSK114 | Simplex | Guppy (v6.1.2+e0556ff)+minimap2 v2.22-r1101 | Super accurate “sup” (dna_r9.4.1_450bps_sup.cfg) | |
| Duplex (∼7%) | Guppy Duplex-basecalling pipeline v6.3.8+d9e0f64 | NA | |||||
| LW1 | 3 | R10.4.1 | FLO-MIN114 | SQK-LSK114 | Simplex | Guppy (v6.1.2+e0556ff)+minimap2 v2.22-r1101 | Super accurate “sup” (dna_r9.4.1_450bps_sup.cfg) |
| Duplex (∼9%) | Guppy Duplex-basecalling pipeline v6.3.8+d9e0f64 | NA |
Assembly and polishing
For LC1 assembly, the filtered ONT reads were used to assemble the initial draft assembly with SmartDenovo v1.4.0 (Liu et al. 2021) with default parameters (smartdenovo.pl -c 1) and was polished 3 times using NextPolish v1.4.0 (Hu et al. 2020). The assembly was further polished with Illumina reads 3 times using Pilon v1.24 (Walker et al. 2014). In brief, the raw FASTQ paired-end reads were first filtered and trimmed using Trimmomatic v0.39 (Bolger et al. 2014; parameters used are ILLUMINACLIP:TruSeq3-PE.fa:2:30:10:2:True SLIDINGWINDOW:4:15 LEADING:3 TRAILING:3 MINLEN:36). The successfully paired reads were aligned to the long-read polished draft genome by BWA mem v0.7.17 (Li 2013), then sorted and indexed with samtools v1.10 (Danecek et al. 2021) prior to polishing with Pilon for a total of 3 rounds with default parameters. Lastly, haplotigs and other redundant contigs were removed using purge_haplotigs v1.1.2 (parameters -l 5 -m 42 -h 95 -j 70 -s 70; Roach et al. 2018). For LW1 assembly, raw ONT reads were corrected and trimmed with Canu v2.2 (Koren et al. 2017) and then assembled by SmartDenovo with default parameters (smartdenovo.pl -c 1). The draft assembly was similarly polished with ONT reads using NextPolish 3 times, with Illumina reads 3 times using Pilon (same parameters as LC1), and haplotigs were removed using purge_haplotigs (parameters -l 5 -m 40 -h 95 -j 70 -s 70). Note that each polishing step was done 3 rounds to ensure the error-prone reads from ONT were corrected while avoiding overpolishing (Chen et al. 2021). The de novo assembled genome was then scaffolded to chromosomes based on mapping contigs to the bilberry genome (Wu et al. 2021), using Ragtag v2.1.0 (Alonge et al. 2019). We did not enable the “correction” mode on Ragtag, meaning it was not looking for potential misassemblies in the de novo assembled contigs because “misassemblies” may represent genome structure differences between bilberry and lingonberry. Importantly, since both subspecies’ genomes were scaffolded from the same reference, structural variation between the 2 genomes may be missed. The final genome assembly was assessed for contiguity (N50, N90 values), per-base accuracy (Quality Value (QV) score or consensus accuracy, error rate), and completeness [Benchmarking sets of Universal Single-Copy Orthologs (BUSCO)] using BBMap v38.86 (Bushnell 2014), Merqury meryl v1.4 (Rhie et al. 2020), and BUSCO v5.1.2 with the following parameters: --lineage_dataset eudicots_odb10, --mode genome (Simão et al. 2015; Manni et al. 2021), respectively.
Gene and transposable element annotation
We performed evidence-based gene annotations following the advice from the unpublished work (Freedman AH, Thomas G, Sackton TB, personal communication from https://github.com/harvardinformatics/GenomeAnnotation), which is particularly relevant for nonmodel species that lacks reliable gene models. After adapter trimming of Illumina RNA-seq reads with Trimmomatic v0.39 with parameters same as DNA (Bolger et al. 2014), the quality of reads was visually checked with fastqc, making sure that there was no sequence bias or decline in read quality throughout. Additionally, published transcriptome data from V. vitis-idaea var. ‘Sunna’ (green, white, and red berries) were added to the data set (Tian et al. 2020). The reads were then aligned to the scaffolded genome including all contigs using Hisat2 v2.2.1 with default parameters (Kim et al. 2019). Following alignment, transcript assembly was performed using StringTie v2.1.5 with default parameters (Pertea et al. 2015), and the transcripts were stored as structural definition file. Gene features [i.e. untranslated regions (UTRs), exons, introns, genes, and mRNAs] were then predicted on the assembled transcripts using TransDecoder v5.5.0 (Haas 2023). The longest ORF prediction (command: TransDecoder.LongOrfs) was run with -S option. A blastp reference library was prepared with Arabidopsis and Vaccinium known proteins from the UniProt database, to retain homologous hits on ORFs even if they did not exceed the coding likelihood scores used to filter ORF candidates in the preceding steps. We used Arabidopsis and Vaccinium protein databases because Arabidopsis is the most well-annotated flowering plant with gene models available in eudicots, and Vaccinium database was the closest published protein gene models to lingonberry, in the hope to discover berry-specific genes. Finally using this information, genes were predicted (command: TransDecoder.Predict) with the parameter --retain_blastp_hits. In cases where there were isoforms (genes of same genomic position, slightly different splicing pattern) or overlapping genes (splicing variants or conflicting candidate gene models), the longest gene hit was chosen as the best candidate sequence. The completeness of the predicted genes was assessed with BUSCO with the following parameters: --lineage_dataset eudicots_odb10, --mode protein (Simão et al. 2015; Manni et al. 2021).
Transposable element (TE) annotation was done following the Extensive de novo TE annotator pipeline v2.0.0 (Ou et al. 2019) with sensitive mode. In brief, candidate TEs were identified using LTR-Finder (Xu and Wang 2007; Ou and Jiang 2019), LTRharvest (Ellinghaus et al. 2020), LTR_retriever (Ou and Jiang 2018), TIR-Learner (Su et al. 2019), generic repeat finder (Shi and Liang 2019), and HelitronScanner (Xiong et al. 2014), followed by RepeatModeler (Flynn et al. 2020) to find any missed TEs due to structural-based methods. Finally, the combined repeat libraries were filtered so that coding sequences (CDS) from my transcript-based gene annotation did not get masked by repetitive regions (parameters: --cds, --exclude). Additional filters to effectively remove false positives were also provided at each step of combining multiple independent programs according to EDTA pipeline (Ou et al. 2019). To roughly map the locations of centromeres, centromere regions of the bilberry genome (Vaccinium myrtillus) were transferred to my lingonberry genomes using syntenic positions (Wu et al. 2021; Supplementary Table 1).
Phenolic compound biosynthesis gene expression in different tissues
Phenolic compounds are important berry components for both flavor and health effects. To better understand their biosynthesis in lingonberry, enzymes in select phenolic compound and anthocyanin biosynthesis pathways were identified and then quantified using RNA-seq data in commercial lingonberry genome. Because genes that code for enzymes in anthocyanin production would be of industry and evolutionary interest, we focused our analysis on 20 enzyme-coding genes involved in the anthocyanin biosynthesis pathway, as well as closely connected pathways, in blueberry (Colle et al. 2019; refer to Supplementary Table 2 for the full list of enzymes analyzed). Additionally, we looked for a newly identified structural gene in anthocyanin biosynthesis pathway, glutathione transferase (GST; Eichenberger et al. 2023), in our assembly. We first obtained the protein sequences of structural genes of interest and aligned them against the blueberry genome annotation using BLAST (Altschul et al. 1990) to find which blueberry genes correspond to which enzymes. We then identified gene orthology between lingonberry and other Vaccinium species using OrthoFinder (Emms and Kelly 2019). Note that the tetraploid “Draper” protein sequences were kept as a full set preserving all 4 haplotypes to find a potential match in lingonberry. OrthoFinder places genes into orthogroups representing orthology. Any annotated lingonberry gene found in the same orthogroup as a blueberry gene was a potential enzyme. We then filtered this set to require that the lingonberry gene was ≥95% identical in sequence to its closest blueberry ortholog, and that it was ≥80% of length of the blueberry ortholog. In this way, we enriched for orthologs that were likely to have the same function.
Using the LC1 assembly and gene annotation file produced above as a reference, expression levels of the annotated transcripts/genes were estimated by Hisat2 with -A, -G and -e option (Kim et al. 2019). The abundance estimate from the 7 transcript data sets (i.e. LC1 leaf, LC2 rhizome/flower/berry, and green/white/red berry from Tian et al. 2020) was reported in the units of FPKM for each data set, corresponding to fragments per kilobase of transcript per million mapped fragments (Zhao et al. 2021).
Genomic divergence between subspecies
To calculate pairwise nucleotide divergence between the 2 lingonberry subspecies genomes, the 12 scaffolded chromosomes were aligned using minimap2 v 2.24-r1122 (Li 2018, 2021) with LW1 scaffolded genome as a reference and LC1 scaffolded genome as a query (default parameters: -ax ams5 --cs=long). Following data format conversions (paftools.js sam2paf | view -f maf), the alignment file was filtered to remove duplicate alignments and the pairwise divergence was calculated per 10 kb windows using maffilter v1.3.1 (Dutheil et al. 2014) parameters: Subset(remove_duplicates=yes, keep=no), MinBlockLength(min_length=1000), WindowSplit (preferred_size=10000, align=ragged_left), SequenceStatistics (Pairwise Divergence)). The program computes the number of base pair mismatches based on the alignment file and reports this value as the divergence in % mismatch in the specified window size. Additionally, to explore the presence of structural variations and basic sequence variations, Synteny and Rearrangement Identifier v1.5 (Goel et al. 2019) was used on the aligned chromosomes with default parameters. We note that since both genomes were scaffolded using the same bilberry reference genome, overall synteny was likely inflated and we might not be capturing all structural variation between the species.
Demographic history estimate
In order to investigate the past population history of lingonberry subspecies, we utilized multiple sequentially Markovian coalescent model (MSMC2; Schiffels and Wang 2020) and pairwise sequentially Markovian coalescent model (PSMC; Li and Durbin 2011). MSMC2 requires that the analyzed populations are mapped to the same reference genome. For the purpose of comparing the 2 methods in parallel, we chose to use LW1 as a reference genome for both subspecies because of better contiguity and base pair accuracy than LC1. To first calculate the effective population size (Ne) of each subspecies, the paired Illumina reads were mapped to the LW1 genome using BWA mem v0.7.17 (Li 2013) with default parameters. PCR and optical duplicates were then removed using GATK Picard v2.23.2 “MarkDuplicates” function (Van der Auwera and O’Connor 2020). The mappable heterozygous variant sites were identified separately for each chromosome per subspecies following bamCaller.py in MSMC2 v2.1.3 (Schiffels and Wang 2020). In brief, SNPs were first called using bcftools v1.16 (Danecek et al. 2021) with the command “mpileup” and “call” with the parameters -q 20 -Q 20 -C 50 and -c -V indels, respectively. The results were then filtered and organized based on read coverage (mean coverage set to 38 for LW1, 37 for LC1; filtering applied is the minimum of ×1/2 mean coverage to the maximum of ×2 mean coverage). An additional mappability mask was generated to avoid calling variants from significantly repetitive regions using GenMap v1.3.0 (Pockrandt et al. 2020) with the parameter -K 30 -E 2. For PSMC inputs, SNPs were similarly called using bcftools “mpileup” and “call” with the same parameters as above, and the results were filtered with the minimum of ×1/3 and maximum of ×2 mean coverage, as recommended (Li and Durbin 2011). No repeat mappability mask was considered in PSMC analysis. When running the models, a generation time of 5–10 years was chosen based on a prior experiment observing minimum of 8 years required to consider a seedling fully reproductive (Hjalmarsson and Ortiz 1998) and considering the woody shrub's natural age of first flowering (Ritchie 1955). However, given the potential for reproduction after first maturity, we recognize that this might underestimate the average reproductive age of the natural population. A mutation rate of 3 × 109 substitutions per generation from Arabidopsis thaliana was used (Exposito-Alonso et al. 2018).
Phylogenetic tree construction
Phylogenetic trees were constructed using 2 different approaches. The first approach followed the default pipeline provided using OrthoFinder v2.5.4 (Emms and Kelly 2019). In brief, a total of 11 species protein sequences in amino acid fasta format were collected from published studies: 8 Vaccinium species: 2 V. vitis-idaea subspecies from this study, Vaccinium corymbosum var. ‘Draper’ v1.0 first 12 chromosomes (Colle et al. 2019), Vaccinium macrocarpon var. ‘Stevens’ v1.0, Vaccinium microcarpum v1 (Diaz-Garcia et al. 2021), Vaccinium oxycoccos NJ96-20 v1 (Kawash et al. 2022), V. myrtillus NK2018_v1 (Wu et al. 2021), Vaccinium darrowii v1.2 (Cui et al. 2022), and Vaccinium caesariense W85-20 P0 v2 (Mengist et al. 2023). Kiwi fruit or Actinidia chinensis v3.0 (Tang et al. 2019) and azalea or Rhododendron williamsianum (Soza et al. 2019) were used as outgroups. The species tree was constructed based on the individual gene trees inferred from the orthologous gene groups as per OrthoFinder pipeline (Emms and Kelly 2017, 2018). For further validation using conserved genes only, single-copy BUSCO genes were extracted and aligned to infer species tree. To do that, BUSCO analysis was first performed on the collected genome assembly itself in nucleotide fasta format with --lineage_dataset eudicots_odb10, --mode genome (Simão et al. 2015; Manni et al. 2021). Then the identified single-copy genes were aligned by MAFFT v7.310, and the individual gene trees were inferred with IQ-TREE v1.5.5 (Nguyen et al. 2015). Outlier long branches were trimmed by TreeShrink v1.3.9 (Mai and Mirarab 2018) with default parameters. Finally, the species tree was constructed using the trimmed gene tress in Astral III v5.7.8 (Zhang et al. 2018). For visualization and data interpretation, both species trees were exported in Newick format and then viewed in FigTree. Trees were rooted manually to A. chinensis.
Additionally, divergence times were estimated following (Diaz-Garcia et al. 2021). In brief, single-copy BUSCO gene alignments were used as an input alignment file with RelTime as implemented in MEGA X (Tamura et al. 2012, 2018). Actinidia chinensis was set as an outgroup, and the following calibration time was used based on the average of 16 studies in TimeTree (Kumar et al. 2017): Rhododendron and Vaccinium (45.5–76.9 MYA). Uniform distribution was selected as the calibration density. Due to MEGA X requiring a single sequence alignment file with equal sequence length, only 743 BUSCO genes that were present in all 11 species were selected for analysis. The individually aligned BUSCO genes were concatenated to prepare the input file with seqkit concat function (Shen et al. 2016). Note that 7 ambiguous amino acid “J”s corresponding to isoleucine or leucine in the alignment file were manually replaced with “I”s in order to meet the requirements by MEGA X.
Results and discussion
Sequencing and assembly
Collectively, 35.3 Gb (∼50.0X) of clean (≥Q10) long-read data was generated (read N50 = 20.56 kb), and additional 12.42 Gb (∼37X) of short-read data was generated for the commercial subspecies, LC1. The de novo assembly resulted in 757 contigs of total length 548.004 Mb with BUSCO (Complete) = 96.6%, contig N50 = 1.170 Mb, and per-base accuracy = 99.959%. Similarly, 28.6 Gb (∼46.9X) of clean long-read data (read N50 = 23.16 kb) and 10.9 Gb (∼35X) of short-read data were generated for the wild subspecies, LW1. The final de novo assembly had 518.642 Mb of total assembly length with contig N50 = 1.400 Mb, BUSCO (Complete) = 96.8%, and per-base accuracy = 99.975% (Table 2; Supplementary Tables 3 and 4). The assembled genome sequence lengths were consistent or slightly smaller than flow cytometry estimates that measured a ∼550 Mb genome size (Redpath et al. 2022). Compared to the short-read only assemblies, which generally do not reach N50 of 1 Mb, our ONT-based assemblies were significantly more contiguous (Rhie et al. 2021), and our assembly statistics were comparable to many draft genome assemblies of similar size (Marrano et al. 2020; Wu et al. 2021; Hamilton et al. 2023; Zhang et al. 2023).
Table 2.
Genome assembly statistics.
| De novo assembly | Haploid only | Scaffold assembly | |
|---|---|---|---|
| V. vitis-idaea ssp. vitis-idaea (LC1) | |||
| Total length (Mb) | 614.857 | 548.004 | 548.071 |
| Contig N50 (Mb) | 1.028 | 1.170 | 1.170 |
| Scaffold N50 (Mb) | 43.867 | ||
| No. of fragments/contigs | 1358 | 757 | 757 |
| No. of scaffolds | 92 | ||
| BUSCO (C%) | 96.8 | 96.6 | 96.5 |
| BUSCO (S%) | 84.3 | 87.5 | 88.4 |
| BUSCO (D%) | 12.5 | 9.1 | 8.1 |
| QV score | 33.8254 | ||
| Accuracy (1-error rate) | 99.959% | ||
| Genome anchored to chr (%) | 98.0 | ||
| No. of genes annotated | 27,243 | ||
| Coding gene content (%) | 7.59 | ||
| TE content (%) | 45.82 | ||
| V. vitis-idaea ssp. minus (LW1) | |||
| Total length (Mb) | 545.497 | 518.642 | 518.704 |
| Contig N50 (Mb) | 1.309 | 1.400 | 1.400 |
| Scaffold N50 (Mb) | 42.799 | ||
| No. of fragments/contigs | 1030 | 696 | 696 |
| No. of scaffolds | 76 | ||
| BUSCO (C%) | 96.9 | 96.8 | 96.9 |
| BUSCO (S%) | 89 | 89.7 | 90.5 |
| BUSCO (D%) | 7.9 | 7.1 | 6.4 |
| QV score | 35.9577 | ||
| Accuracy (1-error rate) | 99.975% | ||
| Genome anchored to chr (%) | 98.5 | ||
| No. of genes annotated | 25,718 | ||
| Gene content (%) | 7.37 | ||
| TE content (%) | 44.58 |
Note that the haploid only assembly (for a diploid genome) meant heterozygous alleles were represented as a mixed haplotype from either of the homologous copy, but not both. The allelic sequences with less confidence were purged during assembly correction based on sequence coverage (Roach et al. 2018).
Scaffolding was performed by mapping to the nearest relative with a chromosome-scale genome, bilberry (V. myrtillus; Schlautman et al. 2017; Kim et al. 2020; Fahrenkrog et al. 2022), resulting in the total of 92 and 76 scaffolds, scaffold N50 = 43.867 and 42.799 Mb, and 98.0 and 98.5% of the contigs anchored to chromosomes for LC1 and LW1, respectively (Table 2). We characterized genomic differences between the subspecies using SyRI and found no major translocations, perhaps due to common scaffolding, and low levels of genome-wide divergence in sequence (Supplementary Tables 5 and 6 and Figs. 1 and 2). We recognize that reference-based scaffolding of the genome does not necessarily produce the real genome structure of lingonberry. This is because the true structural variations can be rearranged during scaffolding as the algorithm orients and places contigs based on alignment to the reference genome (Alonge et al. 2019). That being said, a recent study in Eucalyptus scaffolded ONT genomes on congeneric reference genome to study genome structure evolution and found that a very small proportion of synteny breakpoints were at contig joins, as might be expected if scaffolding was inducing false rearrangements (Ferguson et al. 2023). Therefore, the 2 lingonberry genomes created in this study can reasonably serve as a reference genome to identify genes, polymorphic genetic markers, and compare with related species. Future efforts could generate an unbiased scaffolding using Hi-C or optical mapping and additionally test for the amount of bias introduced by scaffolding to a related reference genome.
Annotation
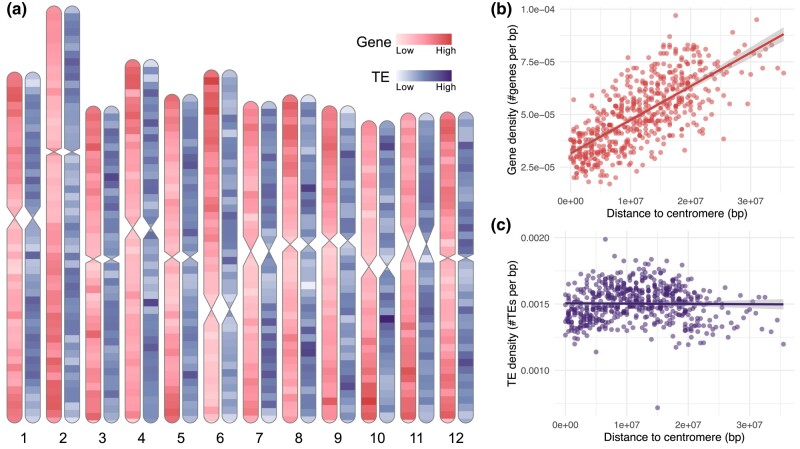
RNA-seq data were produced from leaf sample (∼7.8 Gb), rhizome (∼6.9 Gb), flower (∼11.4 Gb), and berry (∼11.7 Gb) samples in the commercial subspecies, LC1 and LC2. The 2 clones were treated as genetically identical. In addition, transcript data from a published work were added to our analysis (Tian et al. 2020). With the alignment of RNA reads to the assemblies, the total of 27,243 and 25,718 genes were annotated [BUSCO (C): 91.4 and 91.7%]. Excluding non-CDS (introns, UTRs, etc.), the CDS content was 7.59 and 7.37% across the genome, with the average length of 238 and 231 bp for LC1 and LW1, respectively. TEs were also annotated using multiple independent programs and found to cover 45.82 and 44.58% of the genome overall (Table 2). We observed that TE density was fairly even across the genome whereas genes were tended to locate less around putative centromeres and more on distal chromosome positions (Fig. 2). When plotting the TE distributions by different types (Supplementary Fig. 3), some differences in density across the chromosome were observed.
Fig. 2.
a) Gene and TE distributions in lingonberry genome (V. vitis-idaea var. ‘Red Candy’). b) Gene and c) TE densities by distance from centromeres. Centromere positions were approximately mapped from bilberry genome as a range, and distance was calculated to its middle value (Wu et al. 2021). Red shades indicate the gene density, and purple shades indicate the TE density. Genes were filtered to represent only the longest gene in case of isoforms and splicing variants present. All densities are presented as the number of feature counts per 1 Mb, except the terminal windows <1 Mb.
The phenolic compound biosynthesis pathway in lingonberry
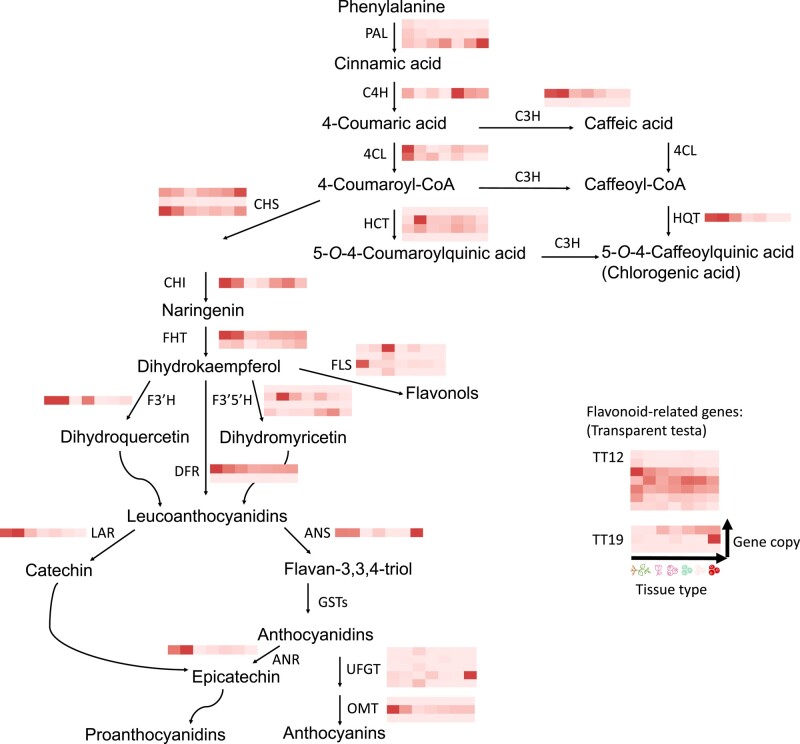
In this study, 49 putative phenolic compound biosynthesis-related genes composed of 20 distinct enzymes/structural gene categories were identified in lingonberry through orthology to the tetraploid commercial blueberry genome (Colle et al. 2019).We did not detect an ortholog of the GST gene in our annotated gene set using OrthoFinder, which uses amino acid similarity for orthology detection. When using BLAST to search our genome nucleotide sequence for the GST gene, we found a single gene (STRG.3821) with ∼96% similarity to the V. corymbosum GST gene, although this gene was more than 7,000 bp long, much longer than the functionally active 700 bp long GST gene (Eichenberger et al. 2023). This suggests significant changes to the lingonberry GST gene or errors in gene annotation. We saw a significantly increased expression of ANS and TT19 in the red berries (Fig. 3), which was consistent with their described roles in anthocyanin production and accumulation (Kitamura et al. 2004; Lin et al. 2018). Although the phenolic-related genes were expected to be highly expressed in berries compared to other tissue types, rhizome and leaf expressed C4H, HCT, HQT, CHI, FHT, F3'H, LAR, and ANS at much higher levels than the berry samples (Fig. 3). Considering various known physiological roles of phenolic secondary metabolites in plants (Albert et al. 2022), abundant expression of genes in vegetative tissues implied that phenolics played roles in stress tolerance.
Fig. 3.
Heatmap of gene abundance related to phenolic compound biosynthesis. Columns represent sample type, and rows represent gene copies on lingonberry genome. Sample types are (from left to right) V. vitis-idaea var. ‘Red Candy’ rhizome, leaf, flower, berry, and var. ‘Sunna’ berries at different ripening stages; green berry, white berry, and red berry (Tian et al. 2020). Abundance was measured by FPKM. Note that the red color gradient was normalized within each heatmap, so comparison cannot be made across heatmaps. The enzyme pathway is based on Colle et al. (2019).
While anthocyanins and the related phenolic compounds are the major targets of breeding due to their health benefits (Edger et al. 2022) and there has been efforts to build QTL maps associating genomic regions to increased anthocyanin production in commercial blueberry and cranberry (Diaz-Garcia et al. 2018; Montanari et al. 2022), the genetic basis for anthocyanin biosynthesis in lingonberry is relatively understudied. The QTL study that specifically targeted the increased anthocyanin production in blueberry suggested candidate genes including BAHD acyltransferase and UFGT to be highly correlated with the increased anthocyanin profile (Montanari et al. 2022). We were able to annotate 5 copies of UFGT in lingonberry genome, 1 of which was highly expressed in red berries (STRG.15162 on chromosome 4; Supplementary Fig. 4). The genomic resource created in this study could be used to find such orthologs and provide a starting point to develop a set of lingonberry-specific markers that could be useful to accelerate the breeding efforts by encouraging marker-assisted selection.
Historical population size and origin of lingonberry
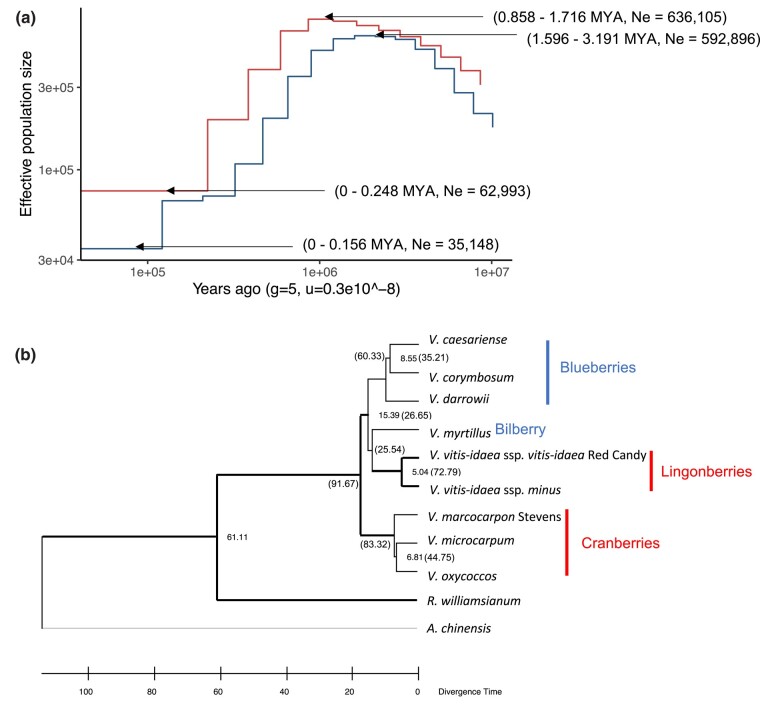
The genetic structures of the contemporary populations can often be shaped by the isolation history, which is especially relevant among subarctic/alpine plants that underwent past population fragmentation due to ice sheets during the Pleistocene (Hewitt 2000; Eidesen et al. 2013). Previous genetic studies in lingonberry revealed the impact of repeated glaciation on its contemporary patterns of genetic diversity (Debnath 2007; Eidesen et al. 2013). Leveraging the genome-wide variant calling along chromosomes, we were able to estimate the historical effective population size (Ne) using PSMC and MSMC2. Despite current range expansions, our result indicated an ongoing population bottleneck for both European (LC1) and North American (LW1) populations. Using a generation time of 5–10 years, we estimated that LC1 and LW1 began declining in Ne around 0.8–1.7 MYA and 1.5–3.2 MYA (Fig. 4a). Lingonberry has likely undergone repetitive range contractions followed by expansion due to ice sheets advancing and receding, which may explain the population size declines over the last 1–2 MYA.
Fig. 4.
a) Past effective population size (Ne) of lingonberry with MSMC2. The Ne of V. vitis-idaea ssp. minus (blue; LW1) and V. vitis-idaea ssp. vitis-idaea var. ‘Red Candy’ (red; LC1) was plotted against years before present. Both x- and y-axes were log scaled. Plots were generated with the generation time of 5 years and mutation rate of 3 × 109 mutations/generation. Note the timings are presented as the range estimate from generation time of 5–10 years. b) Phylogeny of Vaccinium based on 2,226 conserved BUSCO genes. Thick lines indicate nodes supported by >60 STAG support values in OrthoFinder (Emms and Kelly 2018, 2019). The numbers on the selected node represent divergence time in million years (MY), calibrated at the divergence time with Rhododendron (45.5–76.9 MY), and the number in bracket shows the gene concordance factor (0–100) obtained from 2,226 BUSCO genes.
At species level, we generated a phylogeny using all the available Vaccinium whole-genome data. The protein sequence alignment across 8 Vaccinium species and 2 outgroup species resulted in the total number of 377,681 genes analyzed, of which 349,420 were categorized into 31,264 orthogroups by OrthoFinder (Emms and Kelly 2019). The mean orthogroup size was 11.2 genes, and 5,941 orthogroups were shared by all the species, of which 241 were single-copy orthogroups. Additionally, we built species trees based on 2,226 conserved single-copy BUSCO genes to confirm the congruence with the OrthoFinder result using Astral (Zhang et al. 2018). We found that generally there were monophyletic groups for cranberries (V. microcarpum, V. oxycoccos, and V. macrocarpon) and blueberries (V. darrowii, V. caesariense, and V. corymbosum), while bilberry (V. myrtillus) was identified as the closest relative of lingonberry (V. vitis-idaea; Fig. 4b), in agreement with the previous studies (Schlautman et al. 2017; Kim et al. 2020; Fahrenkrog et al. 2022). Interestingly, this suggested that there were multiple color changes of berries in Vaccinium lineage. Adding more species to the current tree, particularly those closely related to lingonberry and bilberry, could address whether the red berry phenotype had convergently evolved in cranberry and lingonberry lineage. Gene concordance values were generally low especially among species in the blueberry, bilberry, and lingonberry (ranging from 25 to 35). Although further analysis is required to fully understand the relationship, it implied that Vaccinium had high levels of incomplete lineage sorting or possibly introgression between species (Coyne and Orr 2004; Beeler et al. 2020).
Our time-calibrated phylogeny suggested that the 2 lingonberry subspecies diverged 5 MYA, which was similar in scale to sister species divergence times in cranberry (6.8 MYA) and blueberry (8.5 MYA). We express some caution in our exact timing because this was based on a single fossil calibration and there was a lack of fossil or geological data in the younger interspecies nodes (Kumar et al. 2017). Compared to previous estimates, our divergence times were consistent (Cui et al. 2022) or overestimated (Diaz-Garcia et al. 2021). Nevertheless, the relative divergence between lingonberry subspecies and other Vaccinium species pairs suggested that the subspecies were near the divergence level expected between species and raised questions about their taxonomic classification. Further work is needed to evaluate where crossability barriers exist between ssp. minus and ssp. vitis-idaea, although high crossability is common between recognized Vaccinium species (Edger et al. 2022). The relatively old divergence time means that the parallel population bottlenecks in both subspecies are not shared but instead are independent events.
Conclusion
This study characterized the genomes of both lingonberry subspecies. Using these genomic resources, we identified genes likely functioning in phenolic compound biosynthesis and clarified the phylogenetic position of lingonberry. The data generated in this study will facilitate future work, such as generation of genetic markers for breeding and analysis of population structure across the species range. Further, the results encouraged scientists in the field to address novel hypotheses regarding not only the evolution of lingonberry but also the evolution of diverse edible berries in the genus Vaccinium.
Supplementary Material
Acknowledgments
The authors acknowledge the collaborators at Agri-Food Canada St. John's Research and Development Centre for collecting, maintaining, and providing the wild lingonberry samples and the Digital Research Alliance of Canada for computational resource and technical support. The authors acknowledge the Michael Smith Genome Sciences Centre at UBC for sequencing. The authors also thank Dr. Ben Koop, Dr. Kris Christensen, and Anne-Marie Flores for sharing their facilities and expertise throughout the project.
Contributor Information
Kaede Hirabayashi, Department of Biology, University of Victoria, 3800 Finnerty Road, Victoria, BC V8W 2Y2, Canada.
Samir C Debnath, Agriculture and Agri-Food Canada, St.John's Research and Development Centre, 204 Brookfield Road, St. John’s, Newfoundland and Labrador L A1E 0B2, Canada.
Gregory L Owens, Department of Biology, University of Victoria, 3800 Finnerty Road, Victoria, BC V8W 2Y2, Canada.
Data availability
This whole-genome sequencing project has been deposited at DDBJ/ENA/GenBank under the accessions JAUYVE000000000 (LC1) and JAUYVF000000000 (LW1). The raw sequences are archived in SRR25468432-50 (ONT) and SRR25477285-90 (Illumina). Full annotations and reference-mapped genome assemblies used in this manuscript can be downloaded from figshare: https://figshare.com/projects/Unveiling_the_evolutionary_history_of_lingonberry_Vaccinium_vitis-idaea_L_through_genome_sequencing_and_assembly_of_European_and_North_American_subspecies/175089. All codes used for assembly pipeline and downstream analysis are available at https://github.com/kaede0e/lingonberry_genomics.
Supplemental material available at G3 online.
Funding
Funding for this project was supplied by a Natural Sciences and Engineering Research Council of Canada Discovery grant to GLO. Computational resources were supplied by grant money supplies to GLO from the Canada Foundation for Innovation and the British Columbia Knowledge Development Fund. Additional computation resources were supplied by the Digital Research Alliance of Canada (alliancecan.ca).
Literature cited
- Albert NW, Lafferty DJ, Moss SMA, Davies KM. 2022. Flavonoids—flowers, fruit, forage and the future. J R Soc N Z. 53(3):304–331. doi: 10.1080/03036758.2022.2034654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, Lippman ZB, Schatz MC. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 20(1):224. doi: 10.1186/s13059-019-1829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2019. A Quality Control Tool for High Throughput Sequence Data. [Accessed 2023 Jan 17]. http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/.
- Beeler RB, Sharples MT, Tripp EA. 2020. Introgression among three western North American bilberries (Vaccinium section Myrtillus). Syst Bot. 45(3):576–584. doi: 10.1600/036364420X15935294613383. [DOI] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B, 2014. BBMap: a fast, accurate, splice-aware aligner. In: Conference: 9th Annual Genomics of Energy & Environment Meeting, Walnut Creek, CA, US.
- Chen Z, Erickson DL, Meng J. 2021. Polishing the Oxford Nanopore long-read assemblies of bacterial pathogens with Illumina short reads to improve genomic analyses. Genomics 113(3):1366–1377. doi: 10.1016/j.ygeno.2021.03.018. [DOI] [PubMed] [Google Scholar]
- Colle M, Leisner CP, Wai CM, Ou S, Bird KA, Wang J, Wisecaver JH, Yocca AE, Alger EI, Tang H, et al. 2019. Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. Gigascience 8(3):1–15. doi: 10.1093/gigascience/giz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr AH. 2004. Species: reality and concepts. In: Sinauer A, editor. Speciation. Sunderland, MA: Oxford University Press. p. 8–54. [Google Scholar]
- Cui F, Ye X, Li X, Yang Y, Hu Z, Overmyer K, Brosché M, Yu H, Salojärvi J. 2022. Chromosome-level genome assembly of the diploid blueberry Vaccinium darrowii provides insights into its subtropical adaptation and cuticle synthesis. Plant Commun. 3(4):100307. doi: 10.1016/j.xplc.2022.100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10(2):giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath SC. 2007. Inter simple sequence repeat (ISSR) to assess genetic diversity within a collection of wild lingonberry (Vaccinium vitis-idaea L.) clones. Can J Plant Sci. 87(2):337–344. doi: 10.4141/P06-059. [DOI] [Google Scholar]
- Debnath SC, Arigundam U. 2020. In vitro propagation strategies of medicinally important berry crop, lingonberry (Vaccinium vitis-idaea L.). Agronomy 10(5):1–19. doi: 10.3390/agronomy10050744. [DOI] [Google Scholar]
- Diaz-Garcia L, Garcia-Ortega LF, González-Rodríguez M, Delaye L, Iorizzo M, Zalapa J. 2021. Chromosome-level genome assembly of the American cranberry (Vaccinium macrocarpon Ait.) and its wild relative Vaccinium microcarpum. Front Plant Sci. 12:1–12. doi: 10.3389/fpls.2021.633310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Garcia L, Schlautman B, Covarrubias-Pazaran G, Maule A, Johnson-Cicalese J, Grygleski E, Vorsa N, Zalapa J. 2018. Massive phenotyping of multiple cranberry populations reveals novel QTLs for fruit anthocyanin content and other important chemical traits. Mol Genet Genom. 293(6):1379–1392. doi: 10.1007/s00438-018-1464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutheil J, Gaillard S, Stukenbrock E. 2014. MafFilter: a highly flexible and extensible multiple genome alignment files processor. BMC Genom. 15(1):53. doi: 10.1186/1471-2164-15-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Iorizzo M, Bassil NV, Benevenuto J, Ferrão LFV, Giongo L, Hummer K, Lawas LMF, Leisner CP, Li C, et al. 2022. There and back again; historical perspective and future directions for Vaccinium breeding and research studies. Hortic Res. 9:uhac083. doi: 10.1093/hr/uhac083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger M, Schwander T, Hüppi S, Kreuzer J, Mittl PR, Peccati F, Jiménez-Osés G, Naesby M, Buller RM. 2023. The catalytic role of glutathione transferases in heterologous anthocyanin biosynthesis. Nat Catal. 6(10):927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidesen PB, Ehrich D, Bakkestuen V, Alsos IG, Gilg O, Taberlet P, Brochmann C. 2013. Genetic roadmap of the Arctic: plant dispersal highways, traffic barriers and capitals of diversity. New Phytol. 200(3):898–910. doi: 10.1111/nph.12412. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U. 2020. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 9(1):18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2017. STRIDE: species tree root inference from gene duplication events. Mol Biol Evol. 34(12):3267–3278. doi: 10.1093/molbev/msx259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2018. STAG: Species Tree Inference from All Genes. bioRxiv. 267914. doi: 10.1101/267914. [DOI] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Alonso M, Becker C, Schuenemann VJ, Reiter E, Setzer C, Slovak R, Brachi B, Hagmann J, Grimm DG, Chen J, et al. 2018. The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genet. 14(2):e1007155. doi: 10.1371/journal.pgen.1007155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog AM, Matsumoto GO, Toth K, Jokipii-Lukkari S, Salo HM, Häggman H, Benevenuto J, Munoz PR. 2022. Chloroplast genome assemblies and comparative analyses of commercially important Vaccinium berry crops. Sci Rep. 12(1):21600. doi: 10.1038/s41598-022-25434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson S, Jones A, Murray K, Schwessinger B, Borevitz JO. 2023. Interspecies genome divergence is predominantly due to frequent small scale rearrangements in Eucalyptus. Mol Ecol. 32(6):1271–1287. doi: 10.1111/mec.16608. [DOI] [PubMed] [Google Scholar]
- Ferlemi AV, Lamari FN. 2016. Berry leaves: an alternative source of bioactive natural products of nutritional and medicinal value. Antioxidants 5(2):17. doi: 10.3390/antiox5020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci USA. 117(17):9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailīte A, Gaile A, Ruņģis DE. 2020. Genetic diversity and structure of wild Vaccinium populations—V. myrtillus, V. vitis-idaea and V. uliginosum in the Baltic States. Silva Fennica 54(5):10396. doi: 10.14214/sf.10396. [DOI] [Google Scholar]
- Garkava-Gustavsson L, Persson HA, Nybom H, Rumpunen K, Gustavsson BA, Bartish IV. 2005. RAPD-based analysis of genetic diversity and selection of lingonberry (Vaccinium vitis-idaea L.) material for ex situ conservation. Genet Resour Crop Evol. 52(6):723–735. doi: 10.1007/s10722-003-6123-4. [DOI] [Google Scholar]
- GBIF.org (11 August 2023) GBIF Occurrence Download 10.15468/dl.tq9c23. [DOI]
- Goel M, Sun H, Jiao WB, Schneeberger K. 2019. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 20(1):277. doi: 10.1186/s13059-019-1911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ. 2023. https://github.com/TransDecoder/TransDecoder.
- Hamilton JP, Vaillancourt B, Wood JC, Buell CR. 2023. Chromosome-scale assembly of the Verbenaceae species Queen’s wreath (Petrea volubilis L.). BMC Genom Data. 24(1):14. doi: 10.1186/s12863-023-01110-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405(6789):907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hjalmarsson I, Ortiz R. 1998. Effect of genotype and environment on vegetative and reproductive characteristics of lingonberry (Vaccinium vitis-idaea L.). Acta Agric Scand Soil Plant Sci. 48(4):255–262. doi: 10.1080/09064719809362506 [DOI] [Google Scholar]
- Hu J, Fan J, Sun Z, Liu S. 2020. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics 36(7):2253–2255. doi: 10.1093/bioinformatics/btz891. [DOI] [PubMed] [Google Scholar]
- Jaakola L, Pirttilä AM, Halonen M, Hohtola A. 2001. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Appl Biochem Biotechnol—Part B Mol Biotechnol. 19(2):201–203. doi: 10.1385/MB:19:2:201 [DOI] [PubMed] [Google Scholar]
- Jacquemart A-L, Thompson JD. 1996. Floral and pollination biology of three sympatric Vaccinium (Ericaceae) species in the Upper Ardennes, Belgium. Can J Bot. 74(2):210–221. doi: 10.1139/b96-025. [DOI] [Google Scholar]
- Kawash J, Colt K, Hartwick NT, Abramson BW, Vorsa N, Polashock JJ, Michael TP. 2022. Contrasting a reference cranberry genome to a crop wild relative provides insights into adaptation, domestication, and breeding. PLoS One 17(3):e0264966. doi: 10.1371/journal.pone.0264966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Shin J, Oh DR, Kim DW, Lee HS, Choi C. 2020. Complete chloroplast genome sequences of Vaccinium bracteatum Thunb., V. vitis-idaea L., and V. uliginosum L. (Ericaceae). Mitochondrial DNA B Resour. 5(2):1843–1844. doi: 10.1080/23802359.2020.1750318. [DOI] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A. 2004. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 37(1):104–114. doi: 10.1046/j.1365-313X.2003.01943.x. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz B, Berlin K, Miller J, Phillippy A. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalska K. 2021. Lingonberry (Vaccinium vitis-idaea L.) fruit as a source of bioactive compounds with health-promoting effects—a review. Int J Mol Sci. 22(10):5126. doi: 10.3390/ijms22105126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Blair Hedges S. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint. 1303.3997. doi: 10.48550/arXiv.1303.3997. [DOI] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2021. New strategies to improve minimap2 alignment accuracy. Bioinformatics 37(23):4572–4574. doi: 10.1093/bioinformatics/btab705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475(7357):493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wang Y, Li B, Tan H, Li D, Li L, Liu X, Han J, Meng X. 2018. Comparative transcriptome analysis of genes involved in anthocyanin synthesis in blueberry. Plant Physiol Biochem. 127:561–572. doi: 10.1016/j.plaphy.2018.04.034. [DOI] [PubMed] [Google Scholar]
- Liu H, Wu S, Li A, Ruan J. 2021. SMARTdenovo: a de novo assembler using long noisy reads. GigaByte 2021:gigabyte15. doi: 10.46471/gigabyte.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai U, Mirarab S. 2018. TreeShrink: fast and accurate detection of outlier long branches in collections of phylogenetic trees. BMC Genomics 19(S5):272. doi: 10.1186/s12864-018-4620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 38(10):4647–4654. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks RA, Hotaling S, Frandsen PB, Vanburen R. 2021. Representation and participation across 20 years of plant genome sequencing. Nat Plants. 7(12):1571–1578. doi: 10.1038/s41477-021-01031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrano A, Britton M, Zaini PA, Zimin AV, Workman RE, Puiu D, Bianco L, Pierro EAD, Allen BJ, Chakraborty S, et al. 2020. High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. Gigascience 9(5):giaa050. doi: 10.1093/gigascience/giaa050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengist MF, Bostan H, De Paola D, Teresi SJ, Platts AE, Cremona G, Qi X, Mackey T, Bassil NV, Ashrafi H, et al. 2023. Autopolyploid inheritance and a heterozygous reciprocal translocation shape chromosome genetic behavior in tetraploid blueberry (Vaccinium corymbosum). New Phytol. 237(3):1024–1039. doi: 10.1111/nph.18428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman DE. 2010. Native American Food Plants—An Ethnobotanical Dictionary. Portland, London: Timber Press. [Google Scholar]
- Montanari S, Thomson S, Cordiner S, Günther CS, Miller P, Deng CH, McGhie T, Knäbel M, Foster T, Turner J, et al. 2022. High-density linkage map construction in an autotetraploid blueberry population and detection of quantitative trait loci for anthocyanin content. Front Plant Sci. 13:965397. doi: 10.3389/fpls.2022.965397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoki RC, Paul A, Kumari A, Singh K, Kumar S. 2012. An improved protocol for the isolation of RNA from roots of tea (Camellia sinensis (L.) O. Kuntze). Mol Biotechnol. 52(1):82–88. doi: 10.1007/s12033-011-9476-5. [DOI] [PubMed] [Google Scholar]
- Nguyen LS, Schmidt HA, Von Haeseler A, Minh BQ. 2015. Q-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Jiang N. 2018. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176(2):1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Jiang N. 2019. LTR_FINDER_parallel: parallelization of LTR_FINDER enabling rapid identification of long terminal repeat retrotransposons. Mob DNA. 10(1):48. doi: 10.1186/s13100-019-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, Lugo CSB, Elliott TA, Ware D, Peterson T, et al. 2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20(1):275. doi: 10.1186/s13059-019-1905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penhallegon RH. 2009. Lingonberry yields in the Pacific Northwest. Acta Hortic. 810(810):223–228. doi: 10.17660/ActaHortic.2009.810.30. [DOI] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pockrandt C, Alzamel M, Iliopoulos CS, Reinert K. 2020. GenMap: ultra-fast computation of genome mappability. Bioinformatics 36(12):3687–3692. doi: 10.1093/bioinformatics/btaa222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redpath LE, Aryal R, Lynch N, Spencer JA, Hulse-Kemp AM, Ballington JR, Green J, Bassil N, Hummer K, Ranney T, et al. 2022. Nuclear DNA contents and ploidy levels of North American Vaccinium species and interspecific hybrids. Sci Hortic. 297(30):110955. doi: 10.1016/j.scienta.2022.110955. [DOI] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J, et al. 2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592(7856):737–746. doi: 10.1038/s41586-021-03451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM. 2020. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21(1):245. doi: 10.1186/s13059-020-02134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie JC. 1955. Vaccinium vitis-idaea L. J. Ecol. 43(2):701–708. doi: 10.2307/2257030. [DOI] [Google Scholar]
- Roach MJ, Schmidt SA, Borneman AR. 2018. Purge haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinform. 19(1):460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Wang K. 2020. MSMC and MSMC2: the multiple sequentially Markovian coalescent. Methods Mol Biol. 2090:147–166. doi: 10.1007/978-1-0716-0199-0_7. [DOI] [PubMed] [Google Scholar]
- Schlautman B, Covarrubias-Pazaran G, Fajardo D, Steffan S, Zalapa J. 2017. Discriminating power of microsatellites in cranberry organelles for taxonomic studies in Vaccinium and Ericaceae. Genet Resour Crop Evol. 64(3):451–466. doi: 10.1007/s10722-016-0371-6. [DOI] [Google Scholar]
- Shen W, Le S, Li Y, Hu F. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One 11(10):e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Liang C. 2019. Generic repeat finder: a high-sensitivity tool for genome-wide de novo repeat detection. Plant Physiol. 180(4):1803–1815. doi: 10.1104/pp.19.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Soza VL, Lindsley D, Waalkes A, Ramage E, Patwardhan RP, Burton JN, Adey A, Kumar A, Qiu R, Shendure J, et al. 2019. The Rhododendron genome and chromosomal organization provide insight into shared whole-genome duplications across the heath family (Ericaceae). Genome Biol Evol. 11(12):3353–3371. doi: 10.1093/gbe/evz245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Gu X, Peterson T. 2019. TIR-Learner, a new ensemble method for TIR transposable element annotation, provides evidence for abundant new transposable elements in the maize genome. Mol Plant. 12(3):447–460. doi: 10.1016/j.molp.2019.02.008. [DOI] [PubMed] [Google Scholar]
- Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S. 2012. Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci USA. 109(47):19333–19338. doi: 10.1073/pnas.1213199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Tao Q, Kumar S. 2018. Theoretical foundation of the RelTime method for estimating divergence times from variable evolutionary rates. Mol Biol Evol. 35(7):1770–1782. doi: 10.1093/molbev/msy044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Sun X, Yue J, Tang X, Jiao C, Yang Y, Niu X, Miao M, Zhang D, Huang S, et al. 2019. Chromosome-scale genome assembly of kiwifruit Actinidia eriantha with single-molecule sequencing and chromatin interaction mapping. Gigascience 8(4):giz027. doi: 10.1093/gigascience/giz027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Ma Z, Ma H, Gu Y, Li Y, Sun H. 2020. Comparative transcriptome analysis of lingonberry (Vaccinium vitis-idaea) provides insights into genes associated with flavonoids metabolism during fruit development. Biotechnol Biotechnol Equip. 34(1):1252–1264. doi: 10.1080/13102818.2020.1803130. [DOI] [Google Scholar]
- Vaara M, Saastamoinen O, Turtiainen M. 2013. Changes in wild berry picking in Finland between 1997 and 2011. Scand J For Res. 28(6):586–595. doi: 10.1080/02827581.2013.786123. [DOI] [Google Scholar]
- Van der Auwera G, O'Connor B. 2020. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. 1st ed. Sebastopol, California: O'Reilly Media. [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Deng C, Hilario E, Albert NW, Lafferty D, Grierson ERP, Plunkett BJ, Elborough C, Saei A, Günther CS, et al. 2021. A chromosome-scale assembly of the bilberry genome identifies a complex locus controlling berry anthocyanin composition. Mol Ecol Resour. 22(1):345–360. doi: 10.1111/1755-0998.13467. [DOI] [PubMed] [Google Scholar]
- Xiong W, He L, Lai J, Dooner HK, Du C. 2014. HelitronScanner uncovers a large overlooked cache of Helitron transposons in many plant genomes. Proc Natl Acad Sci USA. 111(28):10263–10268. doi: 10.1073/pnas.1410068111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang H. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35(Web Server):265–268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li M, Shen M, Bu S, Zhu B, He F, Zhang X, Gao X, Xiao J. 2022. Chromosome-level genome assembly and annotation of the native Chinese wild blueberry Vaccinium bracteatum. Fruit Res. 2(1):8. doi: 10.48130/FruRes-2022-0008. [DOI] [Google Scholar]
- Yocca AE, Platts A, Alger E, Teresi S, Mengist MF, Benevenuto J, Ferrão LFV, Jacobs M, Babinski M, Magallanes-Lundback M, et al. 2023. Blueberry and cranberry pangenomes as a resource for future genetic studies and breeding efforts. Hortic Res. 10(11):uhad202. doi: 10.1093/hr/uhad202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Ma D, Constabel CP. 2015. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 167(3):693–710. doi: 10.1104/pp.114.253674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hulse-Kemp AM, Babiker E, Staton M. 2021. High-quality reference genome and annotation aids understanding of berry development for evergreen blueberry (Vaccinium darrowii). Hortic Res. 8(1):288. doi: 10.1038/s41438-021-00641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Rabiee M, Sayyari E, Mirarab S. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 19(S6):153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wei Y, Meng J, Wang Y, Nie S, Zhang Z, Wang H, Yang Y, Gao Y, Wu J, et al. 2023. Chromosome-scale de novo genome assembly and annotation of three representative Casuarina species: C. equisetifolia, C. glauca, and C. cunninghamiana. Plant J. 114(6):1490–1505. doi: 10.1111/tpj.16201. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li M-C, Konaté MM, Chen L, Das B, Karlovich C, Williams PM, Evrard YA, Doroshow JH, McShane LM. 2021. TPM, FPKM, or normalized counts? A comparative study of quantification measures for the analysis of RNA-Seq data from the NCI patient-derived models repository. J Transl Med. 19(1):269. doi: 10.1186/s12967-021-02936-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This whole-genome sequencing project has been deposited at DDBJ/ENA/GenBank under the accessions JAUYVE000000000 (LC1) and JAUYVF000000000 (LW1). The raw sequences are archived in SRR25468432-50 (ONT) and SRR25477285-90 (Illumina). Full annotations and reference-mapped genome assemblies used in this manuscript can be downloaded from figshare: https://figshare.com/projects/Unveiling_the_evolutionary_history_of_lingonberry_Vaccinium_vitis-idaea_L_through_genome_sequencing_and_assembly_of_European_and_North_American_subspecies/175089. All codes used for assembly pipeline and downstream analysis are available at https://github.com/kaede0e/lingonberry_genomics.
Supplemental material available at G3 online.