Abstract
Circadian clocks are endogenous timekeeping mechanisms that coordinate internal physiological responses with the external environment. EARLY FLOWERING3 (ELF3), PSEUDO RESPONSE REGULATOR (PRR9), and PRR7 are essential components of the plant circadian clock and facilitate entrainment of the clock to internal and external stimuli. Previous studies have highlighted a critical role for ELF3 in repressing the expression of PRR9 and PRR7. However, the functional significance of activity in regulating circadian clock dynamics and plant development is unknown. To explore this regulatory dynamic further, we first employed mathematical modeling to simulate the effect of the prr9/prr7 mutation on the elf3 circadian phenotype. These simulations suggested that simultaneous mutations in prr9/prr7 could rescue the elf3 circadian arrhythmia. Following these simulations, we generated all Arabidopsis elf3/prr9/prr7 mutant combinations and investigated their circadian and developmental phenotypes. Although these assays could not replicate the results from the mathematical modeling, our results have revealed a complex epistatic relationship between ELF3 and PRR9/7 in regulating different aspects of plant development. ELF3 was essential for hypocotyl development under ambient and warm temperatures, while PRR9 was critical for root thermomorphogenesis. Finally, mutations in prr9 and prr7 rescued the photoperiod-insensitive flowering phenotype of the elf3 mutant. Together, our results highlight the importance of investigating the genetic relationship among plant circadian genes.
Keywords: circadian clock, plant development, thermomorphogenesis
Introduction
The daily rotation of Earth generates predictable diel cycles in light and temperature. Circadian clocks are molecular timekeeping mechanisms that anticipate these daily oscillations, allowing physiological responses to be coordinated with the external environment. This anticipatory ability is dependent on the circadian system undergoing daily re-setting in response to stimuli with predictable oscillatory patterns, a process called entrainment. In plants, light at dawn is thought to be a major entrainment signal (Millar 2004). However, temperature, humidity, and sugar availability also function in the entrainment of the plant circadian clock (Webb et al. 2019). The circadian clock has a central role in the life history of plants, regulating germination, vegetative and floral development, metabolism, and the response to biotic and abiotic stress. As such, plants whose circadian clock is not closely aligned with the external environment have reduced fitness (Dodd et al. 2005; Greenham and McClung 2015; Xu et al. 2022).
The plant circadian clock is an interconnected regulatory network of transcriptional, translational, and posttranslational feedback loops (McClung 2019). Genetic and biochemical studies over recent decades have identified more than 20 different components that are involved in the circadian oscillator. Recent mathematical modeling has worked to reduce this complexity, resulting in a compact model that describes 8 genes condensed into 4 components: CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) termed CL, PSEUDO RESPONSE REGULATOR9 (PRR9) and PRR7 termed P97, TIMING OF CAB EXPRESSION1 (TOC1) and PRR5 termed P51, and EARLY FLOWERING4 (ELF4) and LUX ARRYTHMO (LUX) termed EL (De Caluwé et al. 2016). Modifications of this simplified model have been used to understand the effect of external and internal cues on the circadian system and spatial differences in the plant circadian clock (Ohara et al. 2018; Avello et al. 2019; Greenwood et al. 2022).
In these compact models, the EL component describes the evening complex (EC). The EC is a tripartite protein complex composed of EARLY FLOWERING3 (ELF3), ELF4, and LUX (Nusinow et al. 2011; Herrero et al. 2012). LUX is a transcription factor that is necessary for the recruitment of ELF3 and ELF4 onto chromatin (Nusinow et al. 2011). ELF3 recruits chromatin remodeling enzymes including histone deacetylases, histone demethylases, and nucleosome remodeling complexes to repress gene expression (Lee et al. 2019; Park et al. 2019; Tong et al. 2020; Lee and Seo 2021). The role of ELF4 within the EC remains unresolved but may facilitate the nuclear localization of ELF3 and separately facilitate the binding of the EC to DNA (Kolmos et al. 2011; Herrero et al. 2012; Anwer et al. 2014; Silva et al. 2020; Ronald et al. 2021). Mutations in a single ec components result in circadian arrhythmia, although the role and importance of the EC in regulating circadian rhythms in root cells are still uncertain (Covington et al. 2001; Doyle et al. 2002; Onai and Ishiura 2005; Chen et al. 2020; Nimmo et al. 2020). In addition to regulatory activity in the oscillator, ELF3 is also necessary for light and temperature entrainment (Anwer et al. 2020; Zhu et al. 2022). ELF3 functions independently of the other EC components in mediating temperature entrainment (Zhu et al. 2022), while the requirement of the EC in facilitating ELF3-mediated light entrainment remains to be investigated.
A key target of the EC within the circadian clock is PRR9 and PRR7 (Kolmos et al. 2011; Herrero et al. 2012). PRR9 and PRR7 are transcription factors that share partial functional overlap within the circadian clock, with the prr9/prr7 mutant having a longer circadian period than the respective single mutants (Farré et al. 2005; Salomé and McClung 2005). PRR7 and PRR9 along with PRR5 also share genetic redundancy in flowering time and hypocotyl elongation (Nakamichi et al. 2005). Both PRR9 and PRR7 function in the entrainment pathways of the circadian clock. The expression of PRR9 is responsive to red light and this responsiveness is regulated by ELF3 (Farré et al. 2005; Ronald et al. 2022). PRR7 expression is regulated by sugar availability and this regulation underpins sucrose entrainment of the oscillator (Frank et al. 2018). PRR9 and PRR7 also facilitate temperature entrainment, though the nature of this entrainment pathway remains to be investigated (Salomé and McClung 2005). The EC directly regulates PRR9 and PRR7 expression, and accordingly, elf3 mutants have elevated expression of PRR9 and PRR7 (Thines and Harmon 2010; Kolmos et al. 2011; Herrero et al. 2012). At dusk, LUX binds to the promoter of both genes and ELF3 recruits the SWI2/SNF2-RELATED (SWRI) histone-remodeling complex to induce a repressive chromatin state at both loci (Tong et al. 2020). Previous mathematical and genetic analysis indicates a stronger repressive effect of ELF3 on PRR9 than PRR7 (Herrero et al. 2012), although the significance of this remains unclear.
Although the expression of PRR9 and PRR7 is constitutively increased to a highly elevated level in the elf3 mutant background (Kolmos et al. 2011), the importance of this misexpression in contributing to the elf3 circadian and physiological phenotypes has yet to be investigated. Here, through the use of mathematical modeling, molecular assays, and physiological measurements, we sought to test the importance of the individual and high-order mutations in prr9 and prr7 in contributing to the different elf3 phenotypes. Together, our results have highlighted a complex epistatic interaction between ELF3, PRR9, and PRR7.
Materials and methods
Plant lines
All mutant lines described in this work are in the Col-0 background. The Col-0, prr9-1, prr7-3, and prr9-1/prr7-3 mutant lines harboring CCA1::LUC have all been described previously (Farré et al. 2005). The new mutant combinations generated in this work were created by crossing the elf3-1 (Zagotta et al. 1996) CCA1::LUC mutant into the prr9-1/prr7-3 double mutant. The various single, double, and triple mutant combinations were then identified through genotyping.
Modeling
The original De Caluwé model considered PRR9 and PRR7 having similar expression profiles and they were characterized as a single model variable (P97) (De Caluwé et al. 2016). Here, we modified the DC Caluwé model to separate the variable P97 so that 3 models were implemented:
Model 1: The role of each separated component within the network, as described by De Caluwé et al. 2016, was unchanged.
Model 2: The interaction between CCA1/LHY (CL) and PRR9 (P9) was modified; PRR9 is inhibited by CCA1/LHY.
Model 3: The interaction between CCA1/LHY (CL) and PRR9 and PRR7 was modified; PRR9 and PRR7 are now inhibited by CCA1/LHY. Also, a negative autoregulation in CCA1/LHY was introduced as described in Greenwood et al. (2022).
The resulting models consist of 11 ordinary differential equations (ODEs) to reproduce responses to light, rather than 9 as in De Caluwé et al. 2016. Parameter values were taken from the original model. In model 2, where PRR9 is repressed by CCA1/LHY, the mean of the original parameter values that go into P97 inhibition was used for parameterization; that is, the new parameter incorporated into the model is the mean between the Hill function parameters K4 and K5 (De Caluwé et al. 2016) for inhibition of PRR9/PRR7 by PRR5/TOC1 and ELF4/LUX in the original model, respectively. We name this parameter as K11. The added equations are
Additionally, we implemented the U2019.3 model for comparison (Urquiza-García and Millar 2021). This model is governed by the ODEs presented in Pokhilko et al. (2012) but was parameterized using a different approach with the aim to obtain simulated data in absolute units (Urquiza-García and Millar 2021).
Simulations were carried out using experimental conditions, i.e. 8 days of entrainment at 12 hours light and 12h dark in a 24-h cycle. The clock then was released into constant light conditions. Null mutants were simulated by setting the relevant rate constant of transcription to 0 ( and in De Caluwé et al. 2016; n4, n7, n8, n9, and n3 in Urquiza and Millar 2021). All simulations were done in MATLAB.
Luciferase circadian experiments
Seeds were surface sterilized before being sown on 1× MS plates with 3% w/v sucrose, 0.5 g/L MES, and a pH of 5.7. Seeds were then stratified for 3 days. After 3 days, plates were moved to a neutral-day (ND) photoperiod chamber with a set temperature of 22°C and a light intensity of 85 μmol/m-2/s-1 for 5 days. On day 6, seedlings were transferred to a black 96-microwell plate containing the same media as described before. 15-μL 5 mM luciferin was then added superficially to the media. Seedlings were then returned to the same entrainment chamber for 24 h. After 24 h of re-entrainment, seedlings were transferred to the TopCount before subjective dusk. All TopCount experiments were carried out under constant red (3 μmol/m-2/s-1) and blue light (1.1 μmol/m-2/s-1). Data analysis was performed as described previously (Kolmos et al. 2009).
Gene expression analysis
Comparative analysis of selected genes was performed by quantitative real-time PCR (qRT-PCR). Seedings were grown in a plant growth chamber (Percival Scientific, Model CU-36L5) at 22°C under 12:12 long-day (LD) cycles (white light, 100 μmol/m-2/s-1) for 8 days and then transferred into constant light and temperature conditions. Samples were harvested and frozen at 3h intervals. Total RNA was isolated using RNAiso Plus (TaKaRa, Cat. #9109) and reverse transcribed to produce cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Cat. #K1622) following the manufacturer's protocol. The qRT-PCR was performed by using the TB Green Premix Ex Taq (Tli RNase H Plus; TaKaRa, Cat. #RR420A) and CFX Connect Real-Time System (Bio-Rad). Primer sequences used for qRT-PCR are listed in Supplementary Table 1.
Flowering time measurements
Flowering time experiments were carried out as described previously (Kolmos et al. 2011). In brief, seeds were surface sterilized and sown onto 1× MS plates with 0.25% w/v sucrose, 0.5 g/L MES, and a pH of 5.7 and stratified for 3 days at 4°C. Plates were then moved to a ND photoperiod chamber with a set temperature of 22°C and a light intensity of 85 μmol/m-2/s-1 for 14 days. After 14 days, seedlings were transferred to soil and moved to a short-day (SD) or LD photoperiod. Under both photoperiods, the temperature was set to 22°C and light intensity of 85 μmol/m-2/s-1. Flowering was determined as the point at which the inflorescence was ∼1 cm above the rosette. For each genotype, 15–20 plants were analyzed. All experiments were repeated twice with similar results observed each time.
Hypocotyl and root measurements
Seeds were surface sterilized and plated on solid A. thaliana solution medium (Lincoln et al. 1990) with 1% w/v sucrose. Seeds were then stratified for 3 days before being transferred to a SD chamber (8 h of light and 16 h of darkness) with a light intensity of 90 μmol/m-2/s-1. The temperature the seedlings were exposed to is described in the text. Seedlings were allowed to grow for 8 days before scans of the plates were taken. The same scan was used to measure hypocotyl and root growth. Measurements were calculated using ImageJ. A minimum of 42 seedlings were measured per genotype across 2 experimental repeats.
Statistical analysis
To determine the statistical difference for the physiological experiments (Figs. 3–5), an ANOVA with a Tukey honestly significance difference (HSD) post hoc test was performed. The alpha level (P-value) for the post hoc test was set to 0.05 to determine the significant difference between the genotypes and conditions. All statistical analyses were performed in Rstudio (v.2022.07.02) with the version 3.6.1 of R.
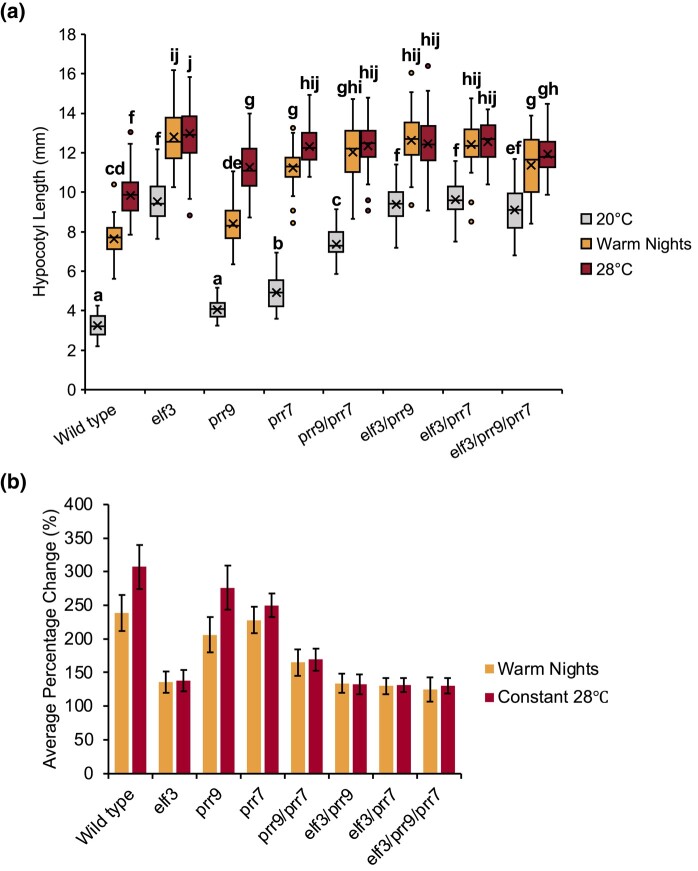
Fig. 3.
ELF3 functions downstream of PRR9/PRR7 in controlling hypocotyl development under ambient and elevated temperatures. a) Hypocotyl length of WT (Col-0) and the different elf3, prr9, and prr7 mutants under SD photoperiods. Seedlings were grown with an ambient temperature of 20°C (gray), 28°C warm nights (orange), or constant 28°C (red) temperatures. b) Average percentage change under warm nights or constant 28°C for the different genotypes. Error bars represent standard deviation. Experiments were repeated twice, with the presented data a combination of the 2 experiments. A minimum of 40 seedlings were analyzed. Letters signify a significant difference of P < 0.05, determined by ANOVA with a Tukey HSD post hoc test.
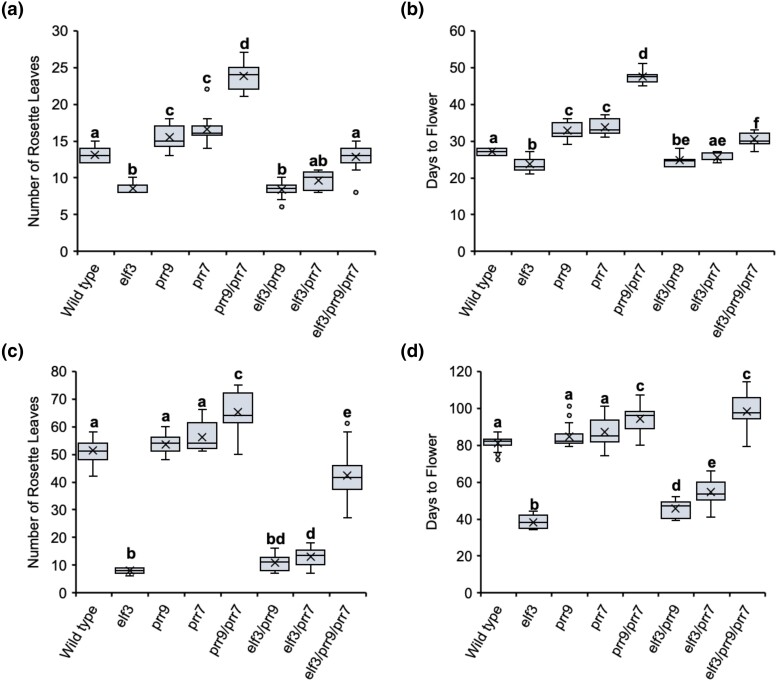
Fig. 5.
Mutations in prr9/prr7 delay the elf3 flowering phenotype under LD and SD photoperiods. Flowering time in the WT (Col-0), elf3, prr, and elf3/prr mutants under LDs a, b) or SDs c, d). Flowering was measured under each photoperiod by the number of rosette leaves a, c) or the days taken to produce a bolt that was ∼1 cm above the rosette b, d). For both photoperiods, plants were grown under a constant temperature of 22°C. Experiments were repeated twice, with a minimum of 8 plants analyzed for each experimental repeat. Letters signify a significant difference of P < 0.05 as determined by ANOVA with a Tukey HSD post hoc test.
Results
Modeling suggests a role of PRR9/PRR7 in the elf3 arrhythmia phenotype
To provide insights into the possible role of PRR9/PRR7 in contributing to the circadian phenotype of elf3, we first simulated the effects of the prr9 and prr7 mutations on the elf3 circadian phenotype using the compact DC2016 model (De Caluwé et al. 2016). ELF3 is not implicitly modeled in the DC2016 model. Instead, the activity of ELF3 is represented by LUX and ELF4 within the EL component. elf4 and lux mutants are similarly arrhythmic to elf3 (Doyle et al. 2002; Onai and Ishiura 2005). Hence, we will use simulated mutations in el as a proxy for mutations in elf3.
In the original compact model (DC2016), the PRR9 (P9) and PRR7 (P7) genes are grouped together in 1 component termed P97 (De Caluwé et al. 2016). Thus, it was firstly necessary to separate the P97 component into 2 components termed P9 and P7 (see Materials and Methods for further details). Three different models were then implemented; in the first model, the individual P9 and P7 components retained the original functions of the P97 component as described in the DC2016 model. In the second model, we introduced a negative regulatory connection from CL (CCA1/LHY) to P9. In the third model, a negative regulatory connection from CL to P9 and P7 was introduced along with a negative autoregulation in CL as described previously (Greenwood et al. 2022) (Fig. 1a). The modifications were made to reflect CL now being described as repressors of PRR expression (Adams et al. 2015).
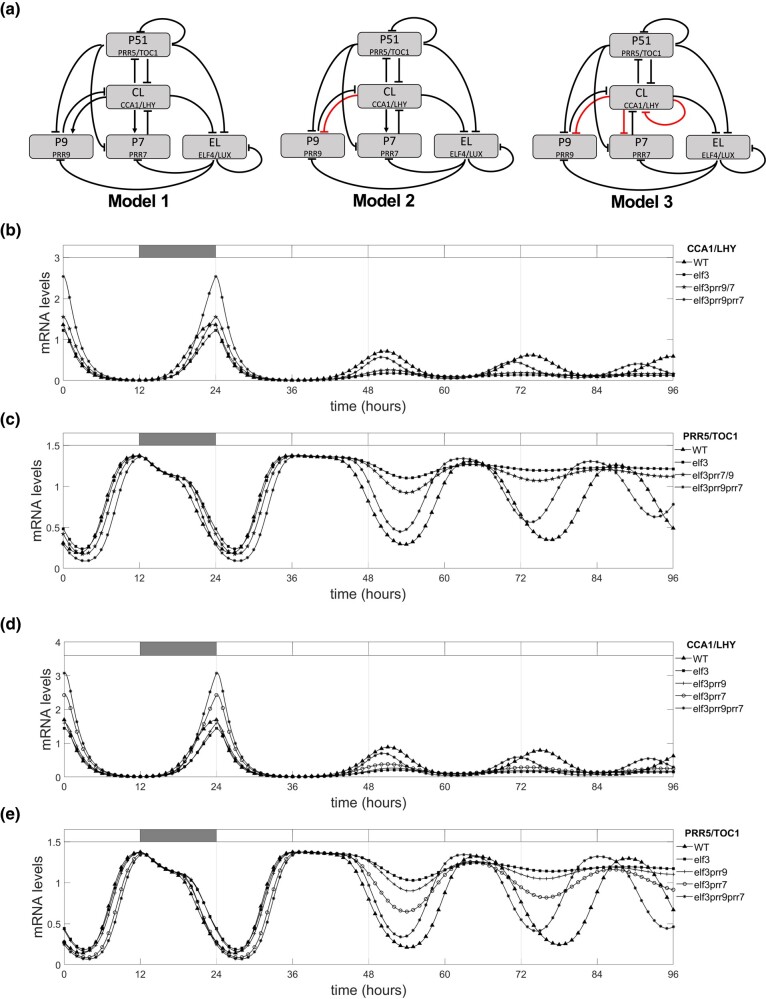
Fig. 1.
Modeling suggests that the prr9/prr7 may contribute to the evening complex circadian arrhythmia. a) Three modifications of the De Caluwé et al. 2016 (DC2016) model presenting PRR9 and PRR7 as separate components. Alterations to the original DC2016 model are highlighted in red. Model 1 is the original layout described in DC2016 but with P9 and P7 split into 2 components. In model 2, a negative interaction is introduced from CL to P9, while in model 3, a negative interaction is introduced from CL to P9 and P7, and negative autoregulation of CL. Outputs from model 1 for the expression of b) CL and c) P51 and outputs from model 2 for the expression of d) CL and e) P51 in the WT and simulated elf3/prr mutant backgrounds are shown. For model 1, the outputs for the elf3/prr9 and elf3/prr7 are represented as a single output (elf3prr9/7) as model 1 keeps the same functions for the respective mutants.
To understand whether the 3 different models could accurately simulate the elf3/prr phenotype, we firstly simulated the expression of CL in wild type (WT) and mutants with well-defined circadian phenotypes. All 3 models were firstly conditioned for 8 days of entrainment under ND photoperiods before being released into constant white light. The simulated expression of CL peaked at dawn for WT in model 1 and model 2, which is consistent with experimental data (Fig. 1b and d) (Kolmos et al. 2011). The output of CL in the simulated elf3, prr9, prr7, and prr9/prr7 mutants also closely replicated the reported expression profile of CCA1/LHY in the respective backgrounds for both model 1 and model 2 (Farré et al. 2005; Kolmos et al. 2011) (Fig. 1b and d; Supplementary Fig. 1a). In contrast, the expression of CL in model 3 did not replicate the reported expression profile of CCA1/LHY in WT or the different mutants (Supplementary Fig. 2a). A similar behavior was also observed for P51 (PRR5/TOC1). Again, the outputs of model 1 and model 2 closely followed the reported expression profile of P51 for WT and the simulated mutants (Fig. 1c and e; Supplementary Fig. 1b) (Kolmos et al. 2011), while model 3 did not accurately reflect the expression of P51 in any instance (Supplementary Fig. 2b). Therefore, we will not further discuss the outputs of model 3 in this work.
We simulated the effect of combining the elf3 and prr9, prr7, prr9/prr7 mutations on the expression of CL and P51 under free-running conditions. As with the elf3 single mutant, the expression of CL was rhythmic in the elf3/prr9 and elf3/prr7 mutants under photocycles but became arrhythmic upon release into free-running conditions (Fig. 1b and d). A similar behavior was observed for the output of P51, with the expression of P51 quickly becoming arrhythmic upon release into free-running conditions for elf3/prr9 and elf3/prr7 (Fig. 1c and e). In contrast, the expression of CL and P51 remained rhythmic under free-running conditions in the simulated elf3/prr9/prr7 mutant. Interestingly, the expression of P51 and CL peaked earlier in the elf3/prr9/prr7 than in WT in model 1 and model 2 (Fig. 1b–d). Previous work has suggested that an extremely early phase in the elf3 mutant may explain the elf3 arrhythmic phenotype (Kim et al. 2005). Together, the output of these models suggests a role of PRR9 and PRR7 in the arrhythmicity of the elf3 mutant.
The prr9/prr7 mutations do not restore rhythmicity in the elf3 mutant background
As the outputs of the models indicate that PRR9/PRR7 may contribute to the arrhythmicity of the elf3 mutant, we generated the different elf3/prr9/prr7 mutant combinations to investigate this possibility. The prr9-1/prr7-3 (prr9/prr7 henceforth) double mutant was crossed into the elf3-1 CCA1::LUC (elf3 henceforth) background to generate all elf3/prr9/prr7 single, double, and triple combinations with the CCA1::LUC reporter gene. This resulted in 8 genotypic comparisons (Fig. 2).
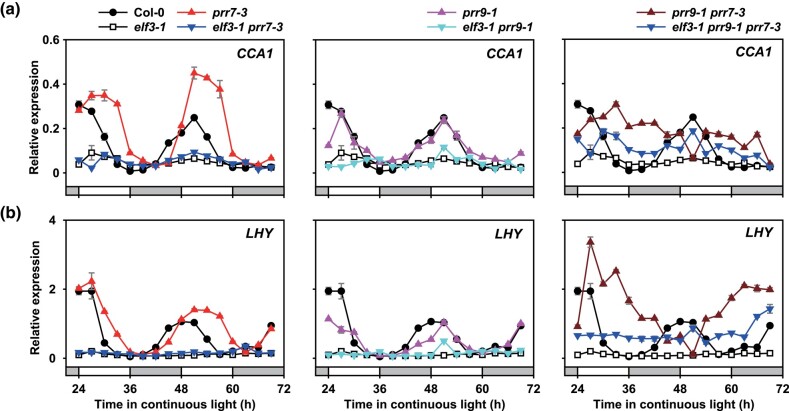
Fig. 2.
Prr9/prr7 does not rescue the arrhythmicity of CCA1 or LHY expression in the elf3 background. The expression of a) CCA1 or b) LHY in WT (Col-0), the elf3, prr, and elf3/prr mutant backgrounds under constant light and constant temperature of 22°C. Seedlings were prior entrained under 12:12 light–dark cycles with a set temperature of 22°C for 8 days. Data are the mean of 3 technical replicas; error bars represent standard error. All experiments were repeated twice, with similar results observed each time. IPP2 was used as a normalization control. White or gray bars represent subjective day and subjective night.
We first investigated the expression of CCA1 and LHY in the respective mutants entrained under a ND photoperiod and then released into constant light and constant temperature. As described previously, the expression of CCA1 peaked at dawn in WT before rapidly declining (Fig. 2a). In the elf3 mutant, CCA1 expression was arrhythmic and stayed suppressed below WT levels across the time course. There was no overt effect of the prr9 mutation on CCA1 expression, while in the prr7 background, the expression of CCA1 was elevated and the peak accumulation was shifted from dawn to the early morning (Fig. 2a). The peak expression of CCA1 was further delayed in the prr9/prr7 double mutant until the subjective afternoon. There was no noticeable effect of the prr9 or prr7 mutation on the elf3 phenotype, with the respective double mutants closely resembling the elf3 single and remaining arrhythmic (Fig. 2a). The elf3/prr9/prr7 triple mutant was also arrhythmic, but the expression of CCA1 in the triple mutant was partially elevated relative to elf3. The expression pattern of LHY was comparable to that observed for CCA1 for each respective mutant (Fig. 2b). Neither the prr9, prr7, or prr9/prr7 mutations could rescue the arrhythmicity of LHY in the elf3 background but, as before, there was a partial increase in the expression of LHY in the elf3/prr9/prr7 triple mutant (Fig. 2b). To confirm that neither the prr9, prr7, nor prr9/prr7 mutant could rescue the elf3 arrhythmic phenotype, we analyzed the output of the CCA1::LUC reporter. As with gene expression, all elf3/prr combinations were arrhythmic under free-running conditions (Supplementary Fig. 4). In summary, our data suggest that the misexpression of PRR9 and PRR7 cannot by themselves explain the arrhythmicity of the elf3 mutant.
ELF3 functions downstream of PRR9/7 in controlling ambient and warm temperature-induced hypocotyl elongation
Circadian regulation of hypocotyl elongation primarily occurs through the regulation of PHYTOCHROME-INTERACTING FACTOR4 (PIF4)/PIF5 expression, stability, and transcriptional activity (Favero et al. 2021). Recently, ELF3 and PRR5/TOC1 were described to coordinately regulate the expression of PIF4/5 in controlling hypocotyl elongation under SD and LD photoperiods (Li et al. 2020). Separate studies have also highlighted a role for PRR9/7 in regulating hypocotyl elongation via PIFs (Martín et al. 2018). Therefore, we tested whether there was a similar additive effect of the elf3/prr9/prr7 mutation on hypocotyl elongation as described for the elf3/toc1/prr5 mutant.
Firstly, we analyzed the hypocotyl phenotype of the elf3/prr mutants under SD photoperiods with an ambient temperature of 20°C (Fig. 3; Supplementary Fig. 5). As reported previously, the elf3 mutant had a long hypocotyl phenotype under these conditions compared to WT Col-0 (Fig. 3a). There was no discernible hypocotyl phenotype in the prr9 single mutant, while the prr7 single mutant had a slightly longer hypocotyl compared to WT (Fig. 3a). The hypocotyl of the prr9/prr7 double mutant was further elongated than the prr7 single mutant but remained shorter than the elf3 single mutant. Notably, there was no effect of either prr single or double mutations on the elf3 phenotype, with all mutant combinations closely resembling elf3 (Fig. 3a). Together, this suggests that PRR9 and PRR7 redundantly regulate hypocotyl elongation and function in the same pathway as ELF3 rather than working cooperatively as previously described with ELF3 and TOC1/PRR5.
Next, we investigated the elf3/prr mutant hypocotyl phenotypes in SD photoperiods with either warm nights (28°C exclusively during the dark phase) or constitutive exposure to 28°C. In the WT Col-0 background, exposure to a 28°C warm night was sufficient to promote hypocotyl elongation (Fig. 3a). However, growth under constant temperatures of 28°C had a stronger effect on hypocotyl elongation compared to warm nights only (Fig. 3b). The prr9 and prr7 single mutants had a similar response to WT, although there was a smaller difference between the constant 28°C and warm nights in the prr7 background. Temperature responsiveness was strongly reduced in the prr9/prr7 double mutant and there was no longer an additive effect of constant 28°C compared to warm nights (Fig. 3b). Hypocotyl elongation in the elf3 mutant was only weakly responsive to the elevated temperature, consistent with earlier reports (Jung et al. 2016; Ding et al. 2018). There was also no difference in response between warm nights or constitutive exposure to 28°C in the elf3 background (Fig. 3b). A similar response was also observed in the elf3/prr double and triple mutants, with all combinations closely resembling the response of the elf3 single mutant (Fig. 3a and b). Therefore, as with ambient temperature, ELF3, PRR9, and PRR7 likely function in the same pathway to control warm temperature-induced hypocotyl elongation and ELF3 functions downstream of PRR9/PRR7 in this pathway.
PRR9 is essential for thermomorphogenic root development
Unlike in the hypocotyl, the nature and role of the circadian clock in controlling root development continue to be unclear. Recent work highlighted a role for the EC in controlling lateral root development (Chen et al. 2020), while PRRs also regulate root development (Ruts et al. 2012; Li et al. 2019) and repress the expression of the EC components in root tissue under warm temperatures (Yuan et al. 2021). Therefore, we characterized the elf3/prr primary root phenotypes. As with hypocotyl development, we analyzed root growth under a SD photoperiod with either constant 20°C, 28°C warm nights only, or constant 28°C. Root development was strongly impaired in the elf3, prr9, and prr7 single mutants at 20°C compared to WT, with each respective single mutant having a similar response to each other (Fig. 4a). There was no change in the primary root length of prr9/prr7 double and elf3/prr double combinations compared to the respective single mutants. However, root development was further impaired in the elf3/prr9/prr7 triple mutant compared to the single and double mutants (Fig. 4a). Therefore, at ambient temperatures, ELF3 and PRR9/7 may regulate root development through separate but also partially overlapping pathways.
Fig. 4.
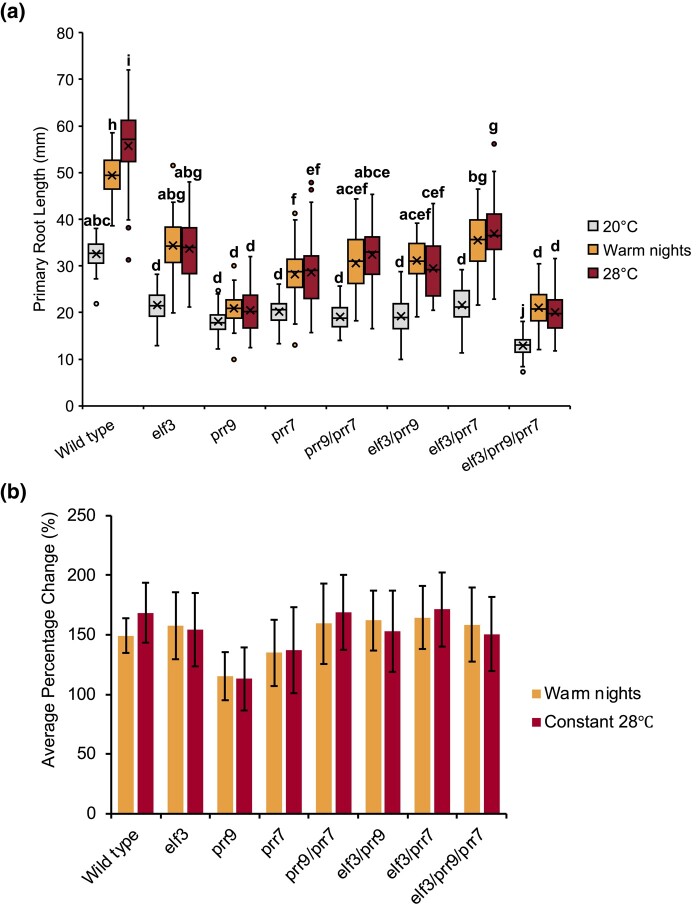
PRR9 is necessary for root thermomorphogenesis. a) Root length of WT (Col-0) and the different elf3, prr9, and prr7 mutants under SD photoperiods. Seedlings were grown with an ambient temperature of 20°C (gray), 28°C warm nights (orange), or constant 28°C (red) temperatures. b) Average percentage change under warm nights or constant 28°C for the different genotypes. Error bars represent standard deviation. Experiments were repeated twice, with the presented data a combination of the 2 experiments. A minimum of 40 seedlings were analyzed. Letters signify a significant difference of P < 0.05, determined by ANOVA with a Tukey HSD post hoc test.
Exposure to warm temperature promoted root growth in WT, consistent with other reports in the literature (Quint et al. 2005; Hanzawa et al. 2013). As with hypocotyl development, constitutive exposure to 28°C caused a greater response than just warm nights, although the magnitude difference between the 2 growth conditions was smaller than observed for hypocotyl elongation (Figs. 3b and 4b). Root growth in the elf3 single mutant remained responsive to warmth (Fig. 4a) but there was no difference in the magnitude of response between constant exposure to 28°C or warm nights only (Fig. 4b). Root development was also thermoresponsive in the prr7 mutant, but this response was weaker than the response seen in WT or the elf3 mutant (Fig. 4a and b). As with the elf3 mutant, root development in the prr7 mutant also did not show a differing response between the constant 28°C or warm night only (Fig. 4b). In the prr9 mutant, the temperature responsiveness of root growth was largely lost, with only a minimal, nonsignificant, response observed under both warm conditions (Fig. 4a and b). Notably, thermoresponsiveness was restored in the prr9/prr7 double mutant, suggesting a complex regulatory pathway underpinning the prr9 and prr7 phenotype. Root growth in the elf3/prr double and triple mutants was also thermoresponsive and showed a similar response to WT and the elf3 single mutant (Fig. 4a and b). However, as with the elf3 mutant, there was no longer a difference in response between warm nights and constitutive exposure to 28°C (Fig. 4b). Altogether, our results suggest a complex relationship between ELF3 and PRR9/7 in controlling root development in response to warm temperatures.
Elf3 photoperiod insensitivity is rescued by the prr9/7 mutation
ELF3 and PRRs are both critical regulators of the photoperiodic flowering time pathway in Arabidopsis and other plant species (Osnato et al. 2022). In Arabidopsis, mutations in elf3 and prr9/7 lead to opposite effects on flowering time; elf3 mutants are early flowering and photoperiod insensitive (Zagotta et al. 1996), while the prr9/7 retains photoperiod sensitivity but flowers late under LD and SD (Nakamichi et al. 2007). This differing response between the elf3 and prr9/7 mutant led us to explore potential epistatic interactions in flowering time under LD and SD.
Under LD, the elf3 mutant flowered slightly earlier than WT plants, while the prr9 and prr7 single mutants had a moderate late-flowering phenotype (Fig. 5a and b). This late-flowering phenotype was enhanced in the prr9/prr7 double mutant, which flowered much later than respective single mutant (Fig. 5a and b). Introducing the prr9 mutation into the elf3 background resulted in a similar response as to the elf3 single mutant. For the elf3/prr7 mutant, flowering time trended toward a WT response for both measurements of flowering (Fig. 5a and b). However, there was no significant difference between elf3 and elf3/prr7 in the number of leaves produced. In the elf3/prr9/prr7 background, flowering was delayed relative to WT as measured by days to flower but not leaf count (Fig. 5a and b). Together, these results indicate that PRR9/7 and ELF3 likely regulate flowering time under LD through partially overlapping pathways, with PRR9/7 functioning downstream of ELF3.
Under SD, elf3 mutants flowered extremely early (Fig. 5c and d). Neither the prr9 nor the prr7 mutant had a flowering-time phenotype under SD, with both mutants flowering at the same time as WT. Flowering was delayed in the prr9/prr7 double mutant, suggesting genetic redundancy between PRR9/7 in regulating flowering under SD photoperiods (Fig. 5c and d). Neither the prr9 nor prr7 mutation alone was able to fully restore the elf3 phenotype to a WT response under SD photoperiods, with both elf3/prr9 and elf3/prr7 mutants still flowering earlier than WT plants for both days to flower and the number of leaves (Fig. 5c and d). The phenotype for elf3/prr9/prr7 mutant was complex, with a disconnect between leaf number and days to flower (Fig. 5c and d). As measured by days to flower, the elf3/prr9/prr7 triple mutant resembled the prr9/prr7 double mutant. However, when a number of leaves were measured, the elf3/prr9/prr7 had fewer leaves than WT. Therefore, this analysis suggests a wider metabolic or plastochron phenotype in the elf3/prr9/prr7 background.
Discussion
PRR9 and PRR7 are direct targets of ELF3 through the activity of the EC (Herrero et al. 2012; Tong et al. 2020), and as a result, the expression of PRR9 and PRR7 is constitutively upregulated in the elf3 mutant background (Kolmos et al. 2011). However, the consequence of this misexpression has so far not been tested. Here, we utilized mathematical modeling to simulate the effect of the prr9 and prr7 mutations on the circadian phenotype of the elf3 mutant. Our modeling approach was designed to seek qualitative insights on the presence or absence of rhythmicity in the elf3/prr9/prr7 mutant background. Therefore, we chose DC2016 model to build our models' structures as DC2016 model was designed to characterize the plant clock at a qualitative level. This is in line with similar approaches that have been previously published. This is in line with previous work using the DC2016 model (De Caluwé et al. 2017) and other work on minimal models of the plant circadian clock (Avello et al. 2021).
The outputs of the modified DC2016 models used here (Fig. 1a) indicated a possible role for PRR9 and PRR7 in contributing to elf3 arrhythmicity (Fig. 1b and c). However, follow-up experiments analyzing the circadian phenotypes of the different elf3/prr mutants through gene expression analysis (Fig. 2) or luminescence reporter (Supplementary Fig. 4) revealed that all combinations of the elf3/prr9/prr7 mutants were arrhythmic. It is unclear why the mathematical models and our in planta data produced different outputs. PRR9 and PRR7 are transcriptional repressors that are part of a gene family that includes TOC1, PRR3, and PRR5. So far, the EC has been demonstrated to directly repress the expression of TOC1, as well as PRR9 and PRR7 (Herrero et al. 2012; Lee et al. 2019). Whether the EC represses PRR3 or PRR5 expression remains to be investigated. It is possible that the misexpression of additional PRRs in the elf3 background also contributes to the circadian arrhythmia phenotype and further prr mutations are necessary alongside the prr9/prr7 mutations.
Alternatively, limitations within the implemented models may be responsible for the discrepancies we have observed. To understand this possibility, we performed additional modeling with the previously described U2019.3 model (Urquiza-García and Millar 2021) as this model differs in network structure from the compact DC model that we initially used here (Fig. 1; Supplementary Figs. 1 and 2). The U2019.3 model does not incorporate the direct repressive effect of CCA1 and LHY on PRR9 and PRR7 (Adams et al. 2015) and the negative autoregulation effect of TOC1 included in the latest mathematical models of the plant circadian clock (Huang et al. 2012). It also has additional components and a larger number of parameters than the DC2016 model. Simulating the elf3/prr9/prr7 mutation in the U2019.3 model revealed that the elf3/prr9/prr7 triple mutant was arrhythmic for both the CCA1/LHY and TOC1 output (Supplementary Fig. 3), supporting our experimental results (Fig. 3; Supplementary Fig. 4). This paradoxical result highlights the importance of the combined work of modeling and experiments. In future work, it will be necessary to focus further modeling efforts on understanding these discrepancies between models.
Alongside investigating the effect of the prr9/prr7 mutation on the elf3 circadian phenotype, we also characterized the elf3/prr developmental phenotypes. We firstly investigated hypocotyl development, where we found that the elf3/prr combinations closely resembled the elf3 phenotype and there was no enhancement of the elf3 phenotype (Fig. 3). This suggests that PRR9/7 function upstream of ELF3 in the same genetic pathway to control hypocotyl development. Circadian regulation of hypocotyl development primarily occurs through the control of PIF activity. ELF3 regulates the expression of PIF4, PIF5, and PIF7 through the EC, while independently also controlling PIF4 transcriptional activity (Nusinow et al. 2011; Nieto et al. 2015; Jiang et al. 2019). Separately, PRR9 and PRR7 regulate PIF4 expression, while also directly inhibiting PIF4 functional activity (Liu et al. 2016; Martín et al. 2018). Thus, it was unsurprising that there is no further enhancement of the elf3 phenotype in the elf3/prr9/prr7 background as PIF4 expression and activity are already enhanced in the elf3 background.
We also observed that the elf3 and prr single mutants were equally compromised in root development (Fig. 4). This short-root phenotype was not enhanced in the prr9/prr7 or elf3/prr double mutants, with all lines displaying similar phenotypic defects as the respective single mutants. However, the elf3/prr9/prr7 triple mutant had a shorter root than the respective single mutants, indicating that ELF3 and PRR9/7 independently control root development (Fig. 4). Supporting a role for these genes in controlling development, ELF3, PRR9, and PRR7 are all expressed in the seedling root, albeit less than in the hypocotyl or mature leaf (Supplementary Fig. 6; Klepikova et al. 2016). So far, few studies have investigated how the circadian clock controls root development. PRR9/7 may regulate root development via controlling TOR signaling (Li et al. 2019), but further studies are needed to understand how the circadian clock controls root development.
Exposure to constitutively elevated temperature promotes hypocotyl and root development (Quint et al. 2016; Lee et al. 2021). Here, we observed that elevated temperatures during the night were also sufficient in promoting hypocotyl and root development in WT plants. However, hypocotyl and root development more strongly responded to constant exposure to warm temperatures than night-time only exposure to warm temperature (Figs. 3 and 4). ELF3 was necessary for elongation of the hypocotyl in response to warm temperatures regardless of the duration of temperature exposure (Fig. 3b), supporting previous work (Box et al. 2015; Raschke et al. 2015; Zhang et al. 2021). In contrast, root development remained sensitive to temperature in the elf3 mutant background, but there was no difference between different durations of warmth exposure (Fig. 4b). Together, our results support recent work that highlighted shoot thermosensors may not function as root thermosensors, and thermomorphogenesis in shoots and roots uses different genetic pathways (Lee et al. 2021; Borniego et al. 2022; Ai et al. 2023).
Hypocotyl elongation in prr9 and prr7 remained temperature responsive under both constant conditions and warm nights only, although there was a smaller magnitude difference between the 2 different conditions in the prr7 mutant (Fig. 3b). The temperature responsiveness of the prr9/prr7 double mutant was further impaired, and there was no magnitude difference between the different warm temperature regimes, indicating a redundant role for PRR9/PRR7 in mediating daytime hypocotyl thermomorphogenesis (Fig. 3b). TOC1 regulates the time-of-day response to warm temperature by directly binding to and subsequently inhibiting the activity of PIF4 (Zhu et al. 2016). This mechanism ensures thermomorphogenesis is phased to occur only in the late evening and early morning. Under ambient temperatures, PRR9 and PRR7 directly interact with PIF4 to control the timing of CYCLING DOF FACTOR6 (CDF6) expression, a positive regulator of hypocotyl elongation (Martín et al. 2018). Whether similar mechanisms control hypocotyl thermomorphogenesis remains to be investigated.
In the roots, our results suggest a complex genetic pathway underpins the response to warm temperature response. Root development in the prr9 mutant was insensitive to warm temperature regardless of the length of warmth exposure (Fig. 4b). This response was then fully rescued in the prr9/prr7, elf3/prr9, and elf3/prr9/prr7 mutant combinations. So far, no studies have highlighted a role for PRR9 in controlling root thermomorphogenesis and the pathway(s) regulating root thermomorphogenesis remain unclear. A recent study has revealed that root thermomorphogenesis is mediated by an unidentified regulator that controls auxin-dependent progression of the cell cycle (Ai et al. 2023). This signaling pathway occurs independently of the well-described PIF thermal integration pathway that regulates thermomorphogenesis in aerial tissue (Delker et al. 2022; Ai et al. 2023). Thus, it is unclear how the circadian clock regulates root thermomorphogenesis. TOC1, another PRR within the circadian clock, has been described to regulate cell cycle progression in leaf tissue (Fung-Uceda et al. 2018). Alternatively, all stages of auxin homeostasis and signaling are under circadian control (Covington and Harmer 2007). Further studies are needed to investigate the integration point for the circadian clock in controlling root thermomorphogenesis.
The regulatory connection between ELF3 and PRRs has emerged as a critical node in the photoperiodic control of flowering time in agronomically important crops (Faure et al. 2012; Zhao et al. 2012; Alvarez et al. 2023; Woods et al. 2023). In Arabidopsis, the importance of the PRRs in contributing to the photoperiod-insensitive early flowering of the elf3 mutant has remained untested. Under LD, introducing the prr9/prr7 mutation into the elf3 background rescued the early flowering elf3 phenotype and resulted in a mild late-flowering phenotype when measured by days to flower (Fig. 5b) but only a WT response when measured by leaf count (Fig. 5a). These discrepancies were further enhanced under SD. When measured by days to flower, the elf3/prr9/prr7 triple mutant resembled the late flowering prr9/prr7 double mutant (Fig. 5d). However, by leaf count, the elf3/prr9/prr7 triple mutant had fewer leaves than WT (Fig. 5c), thus technically an early flowering plant. Together, these results may highlight a plastochron phenotype in the elf3/prr9/prr7 mutant. Plastochron regulation is a complex trait, with multiple signaling pathways contributing to the timing of leaf emergence (Werner et al. 2001; Wang et al. 2008; Lohmann et al. 2010; Mimura et al. 2012; Cornet et al. 2021). One regulator of the plastochron is gibberellin (GA) signaling (Mimura et al. 2012; Mimura and Itoh 2014). In barley, elf3 mutants overaccumulate GA and this misregulation contributes to the elf3 early flowering under noninductive SD photoperiods (Boden et al. 2014). It will be of interest to investigate whether GA is a contributory factor to the complex flowering time phenotypes observed in the elf3/prr9/prr7 mutant background.
Conclusion
The elf3 circadian arrhythmicity was first described nearly 30 years ago (Hicks et al. 1996), but we still do not understand the causative factors. In this study, we hypothesized that misregulation of PRR9 and PRR7 may explain the elf3 arrhythmicity. Although our mathematical modeling supports such a hypothesis (Fig. 1), we could not replicate such results in vivo (Fig. 2; Supplementary Fig. 3). Further work is needed to untangle whether further genetic redundancy among the PRRs is responsible for the discrepancy between the simulations and in vivo results or if the differing results are caused by limitations within the model structure used.
Supplementary Material
Contributor Information
Li Yuan, State Key Laboratory of Crop Stress Adaptation and Improvement, School of Life Sciences, Henan University, Kaifeng 475004, China.
Paula Avello, Department of Mathematics, University of York, York, YO10 5DD, UK; School of Biology, Faculty of Biological Sciences, University of Leeds, Leeds LS2 9JT, UK.
Zihao Zhu, Institute of Agricultural and Nutritional Sciences, Martin Luther University Halle-Wittenberg, Halle (Saale) 06108, Germany.
Sarah C L Lock, Department of Biology, University of York, Wentworth Way, York, YO10 5DD, UK.
Kayla McCarthy, Department of Biology, University of York, Wentworth Way, York, YO10 5DD, UK.
Ethan J Redmond, Department of Biology, University of York, Wentworth Way, York, YO10 5DD, UK.
Amanda M Davis, Department of Biology, University of York, Wentworth Way, York, YO10 5DD, UK.
Yang Song, State Key Laboratory of Crop Stress Adaptation and Improvement, School of Life Sciences, Henan University, Kaifeng 475004, China.
Daphne Ezer, Department of Biology, University of York, Wentworth Way, York, YO10 5DD, UK.
Jonathan W Pitchford, Department of Mathematics, University of York, York, YO10 5DD, UK; Department of Biology, University of York, Wentworth Way, York, YO10 5DD, UK.
Marcel Quint, Institute of Agricultural and Nutritional Sciences, Martin Luther University Halle-Wittenberg, Halle (Saale) 06108, Germany.
Qiguang Xie, State Key Laboratory of Crop Stress Adaptation and Improvement, School of Life Sciences, Henan University, Kaifeng 475004, China.
Xiaodong Xu, State Key Laboratory of Crop Stress Adaptation and Improvement, School of Life Sciences, Henan University, Kaifeng 475004, China.
Seth J Davis, State Key Laboratory of Crop Stress Adaptation and Improvement, School of Life Sciences, Henan University, Kaifeng 475004, China; Department of Biology, University of York, Wentworth Way, York, YO10 5DD, UK.
James Ronald, Department of Biology, University of York, Wentworth Way, York, YO10 5DD, UK; School of Molecular Biosciences, College of Medical, Veterinary and Life Sciences, University of Glasgow, Bower Building, University Avenue, Glasgow G12 8QQ, UK.
Data availability
All data associated with this manuscript, including the modeling files, are available in the Zenodo repository: https://doi.org/10.5281/zenodo.10115364.
Supplemental material available at GENETICS online.
Funding
This work was supported by funding from the Biotechnology and Biological Sciences Research Council (BBSRC): SJD (BB/N018540/1), DE (BB/V006665/1), and DE (BB/S506795/1). We also acknowledge BBSRC Whiterose DTP studentship: JR (BB/M011151/1, ref 1792522) and EJR (BB/T007222/1, ref 2444228). DE was also supported by the Royal Society (RGS\R2\212345). This work was also supported by the National Natural Science Foundation of China to QX (32170259) and XX (U1904202 and 32170275) and the National Key Research and Development Program of China to XX (2021YFA1300402). Deutsche Forschungsgemeinschaft (Qu 141/12-1) and the European Social Fund and the Federal State of Saxony-Anhalt (International Graduate School AGRIPOLY—Determinants of Plant Performance, grant no. ZS/2016/08/80644) supported MQ. PF was supported by the CONICYT PFCHA/DOCTORADO BECAS CHILE award (2013–72140562).
Author contributions
Experimental design: PA, AMD, DE, JWP, MQ, QX, XX, SJD, and JR. Data generation: LY, PA, ZZ, SCLL, KM, EJR, AMD, YS, and JR. Data analysis: all authors. Manuscript writing: SJD and JR. Manuscript edits: all authors.
Literature cited
- Adams S, Manfield I, Stockley P, Carré IA. 2015. Revised morning loops of the Arabidopsis circadian clock based on analyses of direct regulatory interactions. PLoS One. 10(12):e0143943. doi: 10.1371/journal.pone.0143943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai H, Bellstaedt J, Bartusch KS, Eschen-Lippold L, Babben S, Balcke GU, Tissier A, Hause B, Andersen TG, Delker C, et al. 2023. Auxin-dependent regulation of cell division rates governs root thermomorphogenesis. EMBO J. 42(11):e111926. doi: 10.15252/embj.2022111926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez MA, Li C, Lin H, Joe A, Padilla M, Woods DP, Dubcovsky J. 2023. EARLY FLOWERING 3 interactions with PHYTOCHROME B and PHOTOPERIOD1 are critical for the photoperiodic regulation of wheat heading time. PLoS Genetics. 19(5):e1010655. doi: 10.1371/journal.pgen.1010655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwer MU, Boikoglou E, Herrero E, Hallstein M, Davis AM, Velikkakam James G, Nagy F, Davis SJ. 2014. Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife. 3:e02206. doi: 10.7554/eLife.02206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwer MU, Davis A, Davis SJ, Quint M. 2020. Photoperiod sensing of the circadian clock is controlled by EARLY FLOWERING 3 and GIGANTEA. Plant J. 101(6):1397–1410. doi: 10.1111/tpj.14604. [DOI] [PubMed] [Google Scholar]
- Avello P, Davis SJ, Pitchford JW. 2021. Temperature robustness in Arabidopsis circadian clock models is facilitated by repressive interactions, autoregulation, and three-node feedbacks. J Theor Biol. 509:110495. doi: 10.1016/j.jtbi.2020.110495. [DOI] [PubMed] [Google Scholar]
- Avello PA, Davis SJ, Ronald J, Pitchford JW. 2019. Heat the clock: entrainment and compensation in Arabidopsis circadian rhythms. J Circadian Rhythms. 17(1):5. doi: 10.5334/jcr.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, Swain SM. 2014. EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell. 26(4):1557–1569. doi: 10.1105/tpc.114.123794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borniego MB, Costigliolo-Rojas C, Casal JJ. 2022. Shoot thermosensors do not fulfil the same function in the root. New Phytologist. 236(1):9–14. doi: 10.1111/nph.18332. [DOI] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AAR, et al. 2015. ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol. 25(2):194–199. doi: 10.1016/j.cub.2014.10.076. [DOI] [PubMed] [Google Scholar]
- Chen WW, Takahashi N, Hirata Y, Ronald J, Porco S, Davis SJ, Nusinow DA, Kay SA, Mas P. 2020. A mobile ELF4 delivers circadian temperature information from shoots to roots. Nat Plants. 6(4):416–426. doi: 10.1038/s41477-020-0634-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet F, Pillot J-P, Le Bris P, Pouvreau J-B, Arnaud N, De Saint Germain A, Rameau C. 2021. Strigolactones (SLs) modulate the plastochron by regulating KLUH (KLU) transcript abundance in Arabidopsis. New Phytologist. 232(5):1909–1916. doi: 10.1111/nph.17725. [DOI] [PubMed] [Google Scholar]
- Covington MF, Harmer SL. 2007. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 5(8):e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA. 2001. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell. 13(6):1305–1316. doi: 10.1105/TPC.000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Caluwé J, De Melo JRF, Tosenberger A, Hermans C, Verbruggen N, Leloup J-C, Gonze D. 2017. Modeling the photoperiodic entrainment of the plant circadian clock. J Theor Biol. 420:220–231. doi: 10.1016/j.jtbi.2017.03.005. [DOI] [PubMed] [Google Scholar]
- De Caluwé J, Xiao Q, Hermans C, Verbruggen N, Leloup J-C, Gonze D. 2016. A compact model for the complex plant circadian clock. Front Plant Sci. 7:74. doi: 10.3389/fpls.2016.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Quint M, Wigge PA. 2022. Recent advances in understanding thermomorphogenesis signaling. Curr Opin Plant Biol. 68:102231. doi: 10.1016/j.pbi.2022.102231. [DOI] [PubMed] [Google Scholar]
- Ding L, Wang S, Song Z-T, Jiang Y, Han J-J, Lu S-J, Li L, Liu J-X. 2018. Two B-Box domain proteins, BBX18 and BBX23, interact with ELF3 and regulate thermomorphogenesis in Arabidopsis. Cell Rep. 25(7):1718–1728.e4. doi: 10.1016/j.celrep.2018.10.060. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR. 2005. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 309(5734):630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, Mcwatters HG, Kozma-Bognár L, Nagy F, Millar AJ, Amasino RM. 2002. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature. 419(6902):74–77. doi: 10.1038/nature00954. [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. 2005. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 15(1):47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, Von Korff M, Laurie DA. 2012. Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci U S A. 109(21):8328–8333. doi: 10.1073/pnas.1120496109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favero DS, Lambolez A, Sugimoto K. 2021. Molecular pathways regulating elongation of aerial plant organs: a focus on light, the circadian clock, and temperature. Plant J. 105(2):392–420. doi: 10.1111/tpj.14996. [DOI] [PubMed] [Google Scholar]
- Frank A, Matiolli CC, Viana AJC, Hearn TJ, Kusakina J, Belbin FE, Wells Newman D, Yochikawa A, Cano-Ramirez DL, Chembath A, et al. 2018. Circadian entrainment in arabidopsis by the sugar-responsive transcription factor bZIP63. Curr Biol. 28(16):2597–2606.e6. doi: 10.1016/j.cub.2018.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung-Uceda J, Lee K, Seo PJ, Polyn S, De Veylder L, Mas P. 2018. The circadian clock sets the time of DNA replication licensing to regulate growth in Arabidopsis. Dev Cell. 45(1):101–113.e4. doi: 10.1016/j.devcel.2018.02.022. [DOI] [PubMed] [Google Scholar]
- Greenham K, Mcclung CR. 2015. Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet. 16(10):598–610. doi: 10.1038/nrg3976. [DOI] [PubMed] [Google Scholar]
- Greenwood M, Tokuda IT, Locke JCW. 2022. A spatial model of the plant circadian clock reveals design principles for coordinated timing. Mol Syst Biol. 18(3):e10140. doi: 10.15252/msb.202010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa T, Shibasaki K, Numata T, Kawamura Y, Gaude T, Rahman A. 2013. Cellular auxin homeostasis under high temperature is regulated through a SORTING NEXIN1–dependent endosomal trafficking pathway. Plant Cell. 25(9):3424–3433. doi: 10.1105/tpc.113.115881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. 2012. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell. 24(2):428–443. doi: 10.1105/tpc.111.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA. 1996. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science. 274(5288):790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar A, Antoshechkin I, Riechmann JL, Mas P. 2012. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 336(6077):75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yang C, Huang S, Xie F, Xu Y, Liu C, Li L. 2019. The ELF3-PIF7 interaction mediates the circadian gating of the shade response in Arabidopsis. iScience. 22:288–298. doi: 10.1016/j.isci.2019.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J-H, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. 2016. Phytochromes function as thermosensors in Arabidopsis. Science. 354(6314):886–889. doi: 10.1126/science.aaf6005. [DOI] [PubMed] [Google Scholar]
- Kim WY, Hicks KA, Somers DE. 2005. Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol. 139(3):1557–1569. doi: 10.1104/pp.105.067173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA. 2016. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88(6):1058–1070. doi: 10.1111/tpj.13312. [DOI] [PubMed] [Google Scholar]
- Kolmos E, Herrero E, Bujdoso N, Millar AJ, Tóth R, Gyula P, Nagy F, Davis SJ. 2011. A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell. 23(9):3230–3246. doi: 10.1105/tpc.111.088195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E, Nowak M, Werner M, Fischer K, Schwarz G, Mathews S, Schoof H, Nagy F, Bujnicki JM, Davis SJ. 2009. Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 3(5):350–366. doi: 10.2976/1.3218766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Seo PJ. 2021. The Arabidopsis JMJ29 protein controls circadian oscillation through diurnal histone demethylation at the CCA1 and PRR9 Loci. Genes (Basel). 12(4):529. doi: 10.3390/genes12040529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Mas P, Seo PJ. 2019. The EC-HDA9 complex rhythmically regulates histone acetylation at the TOC1 promoter in Arabidopsis. Commun Biol. 2(1):143. doi: 10.1038/s42003-019-0377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wang W, Huq E. 2021. Spatial regulation of thermomorphogenesis by HY5 and PIF4 in Arabidopsis. Nat Commun. 12(1):3656. doi: 10.1038/s41467-021-24018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Wang Y, Zhang Y, Tian W, Chong K, Jang J-C, Wang L. 2019. PRR5, 7 and 9 positively modulate TOR signaling-mediated root cell proliferation by repressing TANDEM ZINC FINGER 1 in Arabidopsis. Nucleic Acids Res. 47(10):5001–5015. doi: 10.1093/nar/gkz191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang Y, He Y, Wang Y, Wang L. 2020. Pseudo response regulators regulate photoperiodic hypocotyl growth by repressing PIF4/5 transcription. Plant Physiol. 183(2):686–699. doi: 10.1104/pp.19.01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. 1990. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 2(11):1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TL, Newton L, Liu MJ, Shiu SH, Farré EM. 2016. A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol. 170(1):528–539. doi: 10.1104/pp.15.01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann D, Stacey N, Breuninger H, Jikumaru Y, Müller D, Sicard A, Leyser O, Yamaguchi S, Lenhard M. 2010. SLOW MOTION is required for within-plant auxin homeostasis and normal timing of lateral organ initiation at the shoot meristem in Arabidopsis. Plant Cell. 22(2):335–348. doi: 10.1105/tpc.109.071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Rovira A, Veciana N, Soy J, Toledo-Ortiz G, Gommers CMM, Boix M, Henriques R, Minguet EG, Alabadí D, et al. 2018. Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr Biol. 28(2):311–318.e5. doi: 10.1016/j.cub.2017.12.021. [DOI] [PubMed] [Google Scholar]
- Mcclung CR. 2019. The plant circadian oscillator. Biology (Basel). 8(1):14. doi: 10.3390/biology8010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ. 2004. Input signals to the plant circadian clock. J Exp Bot. 55(395):277–283. doi: 10.1093/jxb/erh034. [DOI] [PubMed] [Google Scholar]
- Mimura M, Itoh J. 2014. Genetic interaction between rice PLASTOCHRON genes and the gibberellin pathway in leaf development. Rice (N Y). 7(1):25. doi: 10.1186/s12284-014-0025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura M, Nagato Y, Itoh J. 2012. Rice PLASTOCHRON genes regulate leaf maturation downstream of the gibberellin signal transduction pathway. Planta. 235(5):1081–1089. doi: 10.1007/s00425-012-1639-5. [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T. 2005. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant and Cell Physiology. 46(5):686–698. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Niinuma K, Ito S, Yamashino T, Mizoguchi T, Mizuno T. 2007. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 48(6):822–832. doi: 10.1093/pcp/pcm056. [DOI] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière JM, Prat S. 2015. ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr Biol. 25(2):187–193. doi: 10.1016/j.cub.2014.10.070. [DOI] [PubMed] [Google Scholar]
- Nimmo HG, Laird J, Bindbeutel R, Nusinow DA. 2020. The evening complex is central to the difference between the circadian clocks of Arabidopsis thaliana shoots and roots. Physiol Plant. 169(3):442–451. doi: 10.1111/ppl.13108. [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA. 2011. The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 475(7356):398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T, Hearn TJ, Webb AAR, Satake A. 2018. Gene regulatory network models in response to sugars in the plant circadian system. J Theor Biol. 457:137–151. doi: 10.1016/j.jtbi.2018.08.020. [DOI] [PubMed] [Google Scholar]
- Onai K, Ishiura M. 2005. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells. 10(10):963–972. doi: 10.1111/j.1365-2443.2005.00892.x. [DOI] [PubMed] [Google Scholar]
- Osnato M, Cota I, Nebhnani P, Cereijo U, Pelaz S. 2022. Photoperiod control of plant growth: flowering time genes beyond flowering. Front Plant Sci. 12:805635. doi: 10.3389/fpls.2021.805635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Baek D, Cha J-Y, Liao X, Kang S-H, Mcclung CR, Lee SY, Yun D-J, Kim W-Y. 2019. HOS15 interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell. 31(1):37–51. doi: 10.1105/tpc.18.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ. 2012. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol. 8(1):574. doi: 10.1038/msb.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, Van Zanten M. 2016. Molecular and genetic control of plant thermomorphogenesis. Nat Plants. 2(1):15190. doi: 10.1038/nplants.2015.190. [DOI] [PubMed] [Google Scholar]
- Quint M, Ito H, Zhang W, Gray WM. 2005. Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J. 43(3):371–383. doi: 10.1111/j.1365-313X.2005.02449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke A, Ibañez C, Ullrich KK, Anwer MU, Becker S, Glöckner A, Trenner J, Denk K, Saal B, Sun X, et al. 2015. Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biol. 15(1):197. doi: 10.1186/s12870-015-0566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald J, Su C, Wang L, Davis SJ. 2022. Cellular localization of Arabidopsis EARLY FLOWERING3 is responsive to light quality. Plant Physiol. 190(2):1024–1036. doi: 10.1093/plphys/kiac072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald J, Wilkinson AJ, Davis SJ. 2021. EARLY FLOWERING3 sub-nuclear localization responds to changes in ambient temperature. Plant Physiol. 187(4):2352–2355. doi: 10.1093/plphys/kiab423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruts T, Matsubara S, Wiese-Klinkenberg A, Walter A. 2012. Aberrant temporal growth pattern and morphology of root and shoot caused by a defective circadian clock in Arabidopsis thaliana. Plant J. 72(1):154–161. doi: 10.1111/j.1365-313X.2012.05073.x. [DOI] [PubMed] [Google Scholar]
- Salomé PA, Mcclung CR. 2005. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 17(3):791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva CS, Nayak A, Lai X, Hutin S, Hugouvieux V, Jung J-H, López-Vidriero I, Franco-Zorrilla JM, Panigrahi KCS, Nanao MH, et al. 2020. Molecular mechanisms of evening complex activity in Arabidopsis. Proc Natl Acad Sci U S A. 117(12):6901–6909. doi: 10.1073/pnas.1920972117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Harmon FG. 2010. Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc Natil Acad Sci U S A. 107(7):3257–3262. doi: 10.1073/pnas.0911006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong M, Lee K, Ezer D, Cortijo S, Jung J, Charoensawan V, Box MS, Jaeger KE, Takahashi N, Mas P, et al. 2020. The evening complex establishes repressive chromatin domains via H2A.Z deposition1. Plant Physiol. 182(1):612–625. doi: 10.1104/pp.19.00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquiza-García U, Millar AJ. 2021. Testing the inferred transcription rates of a dynamic, gene network model in absolute units. In Silico Plants. 3(2):diab022. doi: 10.1093/insilicoplants/diab022. [DOI] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D. 2008. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 20(5):1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AAR, Seki M, Satake A, Caldana C. 2019. Continuous dynamic adjustment of the plant circadian oscillator. Nat Commun. 10(1):550. doi: 10.1038/s41467-019-08398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmülling T. 2001. Regulation of plant growth by cytokinin. Proc Natl Acad Sci U S A. 98(18):10487–10492. doi: 10.1073/pnas.171304098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DP, Li W, Sibout R, Shao M, Laudencia-Chingcuanco D, Vogel JP, Dubcovsky J, Amasino RM. 2023. PHYTOCHROME C regulation of photoperiodic flowering via PHOTOPERIOD1 is mediated by EARLY FLOWERING 3 in Brachypodium distachyon. PLoS Genet. 19(5):e1010706. doi: 10.1371/journal.pgen.1010706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yuan L, Yang X, Zhang X, Wang L, Xie Q. 2022. Circadian clock in plants: linking timing to fitness. J Integr Plant Biol. 64(4):792–811. doi: 10.1111/jipb.13230. [DOI] [PubMed] [Google Scholar]
- Yuan L, Hu Y, Li S, Xie Q, Xu X. 2021. PRR9 and PRR7 negatively regulate the expression of EC components under warm temperature in roots. Plant Signal Behav. 16(2):1855384. doi: 10.1080/15592324.2020.1855384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR. 1996. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10(4):691–702. doi: 10.1046/j.1365-313X.1996.10040691.x. [DOI] [PubMed] [Google Scholar]
- Zhang LL, Shao YJ, Ding L, Wang MJ, Davis SJ, Liu JX. 2021. XBAT31 regulates thermoresponsive hypocotyl growth through mediating degradation of the thermosensor ELF3 in Arabidopsis. Sci Adv. 7(19):eabf4427. doi: 10.1126/sciadv.abf4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Huang X, Ouyang X, Chen W, Du A, Zhu L, Wang S, Deng XW, Li S. 2012. OsELF3-1, an ortholog of Arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS One. 7(8):e43705. doi: 10.1371/journal.pone.0043705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-Y, Oh E, Wang T, Wang Z-Y. 2016. TOC1–PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat Commun. 7(1):13692. doi: 10.1038/ncomms13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Quint M, Anwer MU. 2022. Arabidopsis EARLY FLOWERING 3 controls temperature responsiveness of the circadian clock independently of the evening complex. J Exp Bot. 73(3):1049–1061. doi: 10.1093/jxb/erab473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this manuscript, including the modeling files, are available in the Zenodo repository: https://doi.org/10.5281/zenodo.10115364.
Supplemental material available at GENETICS online.