Abstract
The decay of messenger RNA with a premature termination codon by nonsense-mediated decay (NMD) is an important regulatory pathway for eukaryotes and an essential pathway in mammals. NMD is typically triggered by the ribosome terminating at a stop codon that is aberrantly distant from the poly-A tail. Here, we use a fluorescence screen to identify factors involved in NMD in Saccharomyces cerevisiae. In addition to the known NMD factors, including the entire UPF family (UPF1, UPF2, and UPF3), as well as NMD4 and EBS1, we identify factors known to function in posttermination recycling and characterize their contribution to NMD. These observations in S. cerevisiae expand on data in mammals indicating that the 60S recycling factor ABCE1 is important for NMD by showing that perturbations in factors implicated in 40S recycling also correlate with a loss of NMD.
Keywords: nonsense-mediated decay, ribosome recycling, yeast, translation
Introduction
Nonsense-mediated decay (NMD) is a quality control pathway that targets mRNAs for decay when ribosomes encounter an early or “premature” termination codon (PTC) (Maquat 2004). PTCs can arise from errors in the nucleus such as missplicing events or mutations during DNA replication and transcription, or from errors in translation such as the use of alternative initiation sites or ribosome frameshifting (Popp and Maquat 2013). NMD also plays a broad regulatory role in eukaryotes by targeting both functional, alternatively spliced isoforms, and processed mRNAs that leave the nucleus but that do not encode functional gene products (e.g. long noncoding RNAs) (Smith and Baker 2015). In each scenario, NMD is signaled through ribosome-dependent stop codon recognition and the mRNA is rapidly decayed.
NMD depends on the UPF and SMG-related proteins in all systems (Rebbapragada and Lykke-Andersen 2009; Karousis et al. 2016; Kurosaki and Maquat 2016). These factors are critical for the recognition of terminating/recycling ribosomes at PTCs and for triggering the recruitment of RNA decay machinery. Specifically, the NMD-central RNA helicase Upf1 interacts directly with eRF1 and eRF3 (Czaplinski et al. 1998; Kobayashi et al. 2004), and this interaction is modulated by the phosphorylation status of Upf1 in mammalian systems (Grimson et al. 2004; Kashima et al. 2006). At the same time, eRF3 interacts with the highly conserved NMD factors Upf2 and Upf3 to stabilize a complex of Upf1, Upf2, and Upf3 (Wang et al. 2001). What is essential to understanding the mechanism and specificity of NMD is an understanding of how the ribosome, and in turn release factors and recycling factors, distinguishes between normal termination codon (NTC) and PTC.
The normal processes of translation termination and recycling have been robustly characterized and begin when the ribosome encounters a stop codon (UAA, UAG, or UGA) in the A site. The complex of eRF3:eRF1 recognizes the 3 stop codons and eRF3 then deposits eRF1 into the A site, in a manner analogous to eEF1A loading aminoacyl-tRNAs there during elongation (Hellen 2018; Lawson et al. 2021). Following the release of eRF3, the ATPase Rli1 (or ABCE1 in mammals) binds to the ribosome to promote termination (Shoemaker and Green 2011) and ribosome recycling (Pisarev et al. 2010; Shoemaker and Green 2011) in which the 60S subunit dissociates from the complex composed of the 40S subunit, mRNA, and P-site tRNA. Hcr1, a loosely bound member of the eIF3 initiation complex, has also been implicated in the recruitment of Rli1 to termination sites and in 80S recycling (Khoshnevis et al. 2010; Beznosková et al. 2013). In the final steps of recycling, the 40S subunit is dissociated from the tRNA and mRNA in a reaction promoted by 3 proteins known as Tma64, Tma20, and Tma22 in yeast (or eIF2D, MCT-1, and DENR in mammals) (Skabkin et al. 2010; Lomakin et al. 2017; Weisser et al. 2017; Young et al. 2018).
During any of the steps of termination and recycling, the activity of the ribosome at the stop codon can in principle signal NMD. What then are the features that distinguish between recognition of NTCs and PTCs that lead to NMD? In broad terms, the “context” of a stop codon within an mRNA determines whether the mRNA is targeted for NMD and could include the following: (1) different nucleotide sequences that affect the recruitment of termination and recycling factors, (2) proximity to the poly-A tail, and (3) the composition and context of local RNA-binding proteins. While much is known about how these different models might dictate NMD, we reasoned that important players in the pathway might remain undiscovered and could shed light on molecular mechanism.
Here, using a bidirectional fluorescent reporter in Saccharomyces cerevisiae, we screen for factors that contribute specifically to the decay of the mRNA or its translational repression during NMD. Along with the known NMD regulators in yeast, we identify a group of genes involved in translation termination and recycling. We find that deletion of the known 40S recycling factors TMA20 and TMA22 leads to the stabilization of NMD substrates, and, similar to recent results describing the role of the 60S recycling factor ABCE1 in NMD in mammals (Zhu et al. 2020), we find that HCR1 deletion leads to stabilization of NMD substrates. Taken together with published data demonstrating increased ribosome occupancy in 3′UTRs upon loss of Tma20/22/64 (Young et al. 2018), these data support a model in which perturbations to ribosome recycling disrupt critical signals for NMD.
Materials and methods
Plasmid construction
The OPT reporter plasmid, or pKD065, was constructed as described in D’Orazio et al. (2019). The NMD reporter plasmid (pKD081) was constructed by inserting an in-frame “UAA” stop codon 384-bp upstream of the HIS3-coding region of pKD065.
Yeast strains and growth conditions
Yeast strains used in this study are described in Supplementary File 5 and are derivatives of BY4741 unless specified otherwise. Yeast strains were constructed using standard lithium acetate transformations. Reporter strains were constructed by linearizing the given plasmids with StuI and integrating them into the ADE2 locus of BY4741. Deletion strains were constructed by replacing the gene of interest with drug resistance cassettes at the given locus; see genotypes in Supplementary File 5.
For synthetic genetic array (SGA) experiments, query strains for the deletion screens were constructed by introducing the RFP–GFP-2A-FLAG-HIS3 cassettes from StuI digested pKD065 or pKD081 at the ADE2 locus in Y7092 (Tong et al. 2001; Tong and Boone 2006) (Supplementary File 5).
For gal-induced growths, overnight cultures were grown in YPAGR media (Supplementary File 5). These overnight cultures were then diluted in the same media to an OD of 0.1 and harvested at an OD of 0.4–0.5.
Flow cytometry
Flow cytometry of individual strains was performed as in D’Orazio et al. (2019). Briefly, cells were harvested in log phase and washed with PBS once and then ran on a Millipore Guava easyCyte flow cytometer for GFP and RFP detection using 488- and 532-nm excitation lasers, respectively. Data for 10,000 cells were collected and gated based on size. Flow cytometry was done in triplicate, with each group of cells taken from individual growths. For triplicate plots, the average of each individual flow cytometry sample was taken and normalized to the indicated strain. The log of the fraction was then plotted for each experiment.
Northern blots
Northern blots were performed as in D’Orazio et al. (2019). Briefly, 25 ml of log-phase cells were harvested. RNA was isolated and 5 μg of RNA was loaded into a 1.2% agarose, formaldehyde denaturing gel and run for 1.5 h. The RNA was vacuum transferred to a nitrocellulose membrane (N + H bond, Amersham). The membrane was UV cross-linked, placed in a prehybridization buffer, and rotated at 42°C for an hour. The indicated DNA oligo listed in Supplementary File 5 was 5′ end-labeled using gamma-ATP and T4 Polynucleotide Kinase radioactive labeling protocol from NEB. Labeled oligos were purified using GE Healthcare illustra ProbeQuant G-50 micro columns, and the membrane was probed overnight, rotating at 42°C. The membrane was washed 3 times in 2× SSC and 0.1% SDS for 20 min at 30°C and then exposed to a phosphoscreen. The phosphoscreen was scanned using a Typhoon FLA 9500.
Western blots
Protein isolation and western blotting were performed as discussed in D’Orazio et al. (2019).
Reporter SGA screens
SGA procedure
SGA screens were performed using a Biomatrix Robot with a few modifications (S&P Robotics Inc.). Briefly, yKD176 and yKD179 query strains (Supplementary File 5) were crossed individually with the yeast nonessential gene deletion library (Tong et al. 2001). The deletion library was arrayed in a 1536-format with 4 colonies per deletion strain. Because we found that our query strains had a slightly lower mating efficiency and a growth defect, incubation times for every step of the SGA protocol were prolonged by 50–75%. Mating and sporulation steps were performed on standard SGA media (Tong et al. 2001; Tong and Boone 2006).
For each deletion screen, diploid strains were selected on DIP media and then haploid double mutant strains were selected for multiple rounds on HAP media listed in Supplementary File 5. Finally, to induce reporter expression, cells were pinned onto haploid double mutant selection medium with raffinose and galactose at a final concentration of 2% (HAPGR media listed in Supplementary File 5). Cells were grown for 26–30 h before scanning on a Typhoon FLA9500 (GE Healthcare) fluorescence scanner equipped with 488- and 532-nm excitation lasers and 520/40 and 610/30 emission filters. Plates were also photographed using a robotic system developed by S&P Robotics Inc. in order to determine colony size.
Reporter screen analysis
Screen analysis was performed as described in previous manuscripts (Kainth et al. 2009; Hendry et al. 2015; D’Orazio et al. 2019). Briefly, GFP and RFP fluorescent intensity data were collected using TIGR Spotfinder microarray software (Saeed et al. 2003). Colony size data were collected using SGATools (Wagih et al. 2013) (http://sgatools.ccbr.utoronto.ca/). After border strains and size outliers (<1,500 or >6,000 pixels) were eliminated, median and mean GFP and RFP values were taken. We then calculated log2(mean GFP/mean RFP) for the replicates and performed a LOESS normalization for each plate. Based on this LOESS-normalized value, Z-scores were calculated without multiple hypothesis testing correction. Strains for the NMD reporter with a Z-score greater than 2.0 or less than −2.0 were considered a hit if their Z-score in the OPT reporter was not also an outlier.
Venn diagrams
The Venn diagram was created using BioVenn, and the input was deletion strains with a Z-score greater than 2.0 or less than −2.0.
GO term analysis
Gene ontology (GO) term analysis was performed with the data from the screen using a list of hits that were in the cutoff of Z > 2.0 or Z < −2.0, and the input gene list is the genes that GFP/RFP data were successfully acquired for in the deletion array screen. The P-value cutoff was set to <10−7 using GOrilla (Eden et al. 2007, 2009). Duplicate strains were removed during analysis.
Validation screen
We selected NMD reporter genes that gave a Z-score greater than 2.0 or less than −2.0. Then, we removed any hits that also had a Z-score greater than 2.0 or less than −2.0 for the OPT reporter. Strains from the haploid deletion collection were subsequently struck out and transformed with the NMD reporter plasmids (pKD081). Three individual colonies from each transformation were isolated. Strains that did not grow were dropped from the experiment, yielding 100 deletion strains to test (Supplementary File 4). These biological replicates were grown overnight in a flow cytometer plate in YPAGR and put on the flow cytometer the next day. The triplicate flow cytometry data for each strain were analyzed, normalizing to the HIS3 controls for all plates tested.
Results
Developing a reporter system for nonsense-mediated RNA decay
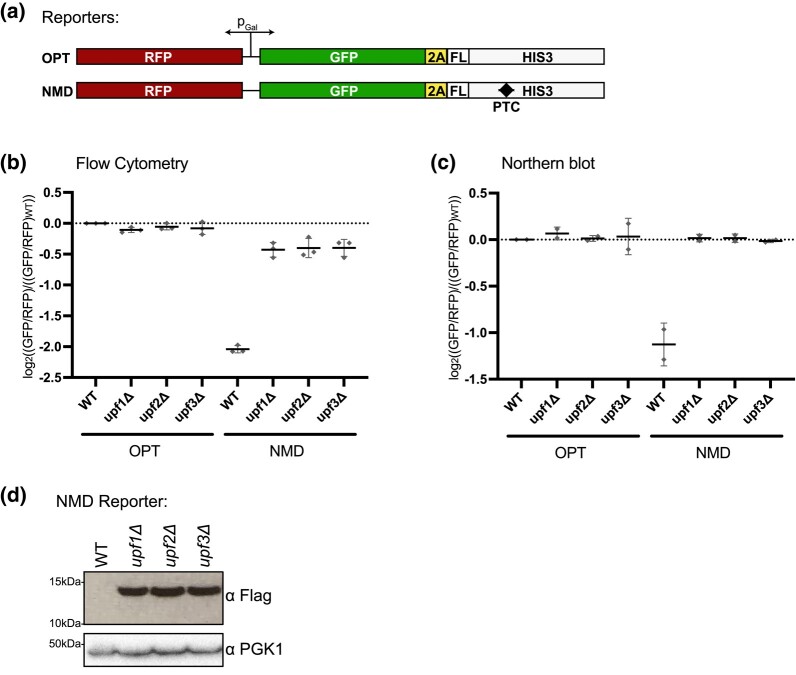
To identify genes necessary for NMD in yeast, we designed gene constructs that would report on both mRNA level and translational repression but not on nascent peptide stability (D’Orazio et al. 2019). To implement this, we used a GFP-His3-conjugated protein and inserted a viral 2A peptide to effectively dissociate the GFP reporter protein from the downstream His3 protein that encodes a PTC. To control for overall expression, we used a bidirectional, inducible galactose promoter that expressed RFP in the opposite direction from GFP-2A-His3, and we normalized to RFP expression. The GFP reporter construct contains a FLAG epitope at the N terminus of His3 for detection of the peptide downstream of the 2A signal (Fig. 1a). In the His3 ORF, we added a PTC to create an NMD signal (the NMD reporter) while the control reporter (the OPT reporter) contained no PTC (Fig. 1a). As anticipated for an NMD reporter construct, we find that insertion of a PTC 384-bp upstream from the normal stop codon of His3 leads to a 3-fold decrease in GFP/RFP levels as determined by flow cytometry and a 2–3-fold decrease in RNA as determined by northern blot analysis (Fig. 1b and c; representative northern blots shown in Supplementary Fig. 2a).
Fig. 1.
Fluorescent reporters reflect mRNA levels of an NMD target. a) Schematic of reporters with a bidirectional, galactose-inducible promoter. RFP is transcribed in one direction and GFP tethered to His3 via the 2A peptide and FLAG tag (FL) is transcribed in the other direction. b) Individual averages of 3 different flow cytometry experiments performed on the indicated strains with the indicated reporters are shown. The GFP/RFP signal for each given strain normalized to the WT strain with the OPT reporter is plotted. c) Northern blot quantification of 2 unique northern blots is shown. The GFP/RFP signal for each strain, normalized to the WT strain with the OPT reporter, is plotted. d) Western blot of the flag-tagged NMD reporter in the given backgrounds is shown with PGK1 as a control.
To confirm that the decreases in RNA levels for the reporter reflect NMD, we deleted the core NMD genes, UPF1, UPF2, and UPF3, and saw a restoration of RNA levels for the NMD reporter by both flow cytometry and northern blots (Fig. 1b and c); as expected, loss of these same factors has no effect on the OPT reporter. Importantly, previous studies have established that NMD impacts the half-life of PTC-containing mRNAs and that deletion of the UPFs results in mRNA stabilization (Leeds et al. 1991, 1992; Cui et al. 1995; Lee and Culbertson 1995; He et al. 1997). These observations lay the groundwork for utilizing GFP output as an indicator for mRNA levels for these reporters in a genetic screen.
Interestingly, in the NMD reporter strains, we saw that GFP RNA levels were fully restored by deletion of UPF1/2/3 but GFP protein levels were only partially restored (compare Fig. 1b and c). These observations invite speculation about potential translational repression of NMD mRNAs as previously documented (Unterholzner and Izaurralde 2004; Loh et al. 2013; Zinshteyn et al. 2021). We next utilized the FLAG epitope to look at the peptide product of the NMD reporter (Fig. 1d). In wild-type (WT) cells, the FLAG epitope was undetectable via western blot as the mRNA was degraded by NMD and very little peptide was made. Reassuringly, in the UPF1/2/3 deletion strains, this peptide construct was stabilized to a similar extent as observed by flow cytometry (Fig. 1d compared with Fig. 1b).
Screen to identify factors involved in NMD
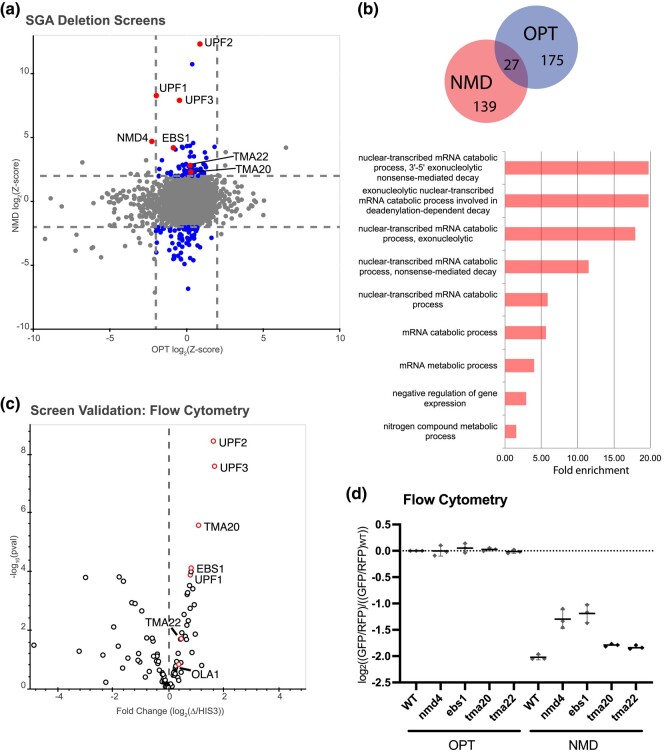
We next performed a high-throughput reverse genetic screen using the S. cerevisiae reporter-SGA (R-SGA) method (Tong et al. 2001; Fillingham et al. 2009). In R-SGA, the reporter haploid strain is crossed with an array of viable haploid budding yeast deletion strains. Then, using a series of selections, an array of haploid strains containing both the reporter gene and a single gene deletion is produced, allowing reporter activity to be scored in each deletion mutant. We performed 2 independent screens using the OPT reporter (screen 1) and NMD reporter (screen 2) strains, where the reporter/deletion arrays were maintained on glucose media and then the cells were shifted to galactose to induce reporter expression. The GFP and RFP signals from the arrays were evaluated by fluorimetry, yielding a readout for both the OPT and NMD reporter (Supplementary Fig. 1a and b) (D’Orazio et al. 2019). For each strain in the plate array, Z-score-normalized ratios of GFP intensity over RFP intensity were calculated. In this setting, Z-scores represent the deviation of the GFP/RFP ratio for a given strain from the mean GFP/RFP ratio for the given array; these data for the OPT and NMD reporter strains are plotted against one another in Fig. 2a (raw data are given in Supplementary Files 1–3). From the NMD screen, there were 76 hits with a Z-score above 2.0 and 94 hits with a Z-score below −2.0 (Fig. 2b; duplicate strains are removed in the Venn diagram). The OPT reporter screen, which has been previously published in D’Orazio et al. (2019), also included 64 hits with a Z-score above 2.0 and 138 hits with a Z-score below −2.0, with 27 genes overlapping as hits in both the OPT and NMD screens that we did not explore further (Fig. 2b) (D’Orazio et al. 2019). GO enrichment data show that the hits in the NMD screen are strongly enriched in “mRNA metabolic process” and in “nuclear-transcribed mRNA catabolic process, NMD,” while there were no GO terms for the hits in the OPT screen with a similar level of enrichment (Fig. 2b). Reassuringly, UPF1/2/3 deletion strains exhibited some of the largest deviations from the mean in the NMD reporter strains, immediately validating the potential of the screen (Fig. 2a). Interestingly, NMD4 and EBS1 deletions are also strong candidates that cause an increase in GFP signal for the NMD reporter (Fig. 2a). Nmd4 and Ebs1 are homologs of the Smg6 and Smg5/7 proteins in mammals that are key players in the nucleolytic decay of mammalian NMD substrates (Eberle et al. 2009; Lykke-Andersen et al. 2014).
Fig. 2.
Yeast SGA screens identify genes important for repression of NMD targets. a) Plot of Z-scores from OPT reporter deletion screen vs the NMD reporter deletion screen. Z-scores reflecting the significance of log2(GFP/RFP) values from each deletion strain are plotted against each other for the 2 different GFP reporters. Dashed lines represent cutoffs at a Z-score greater than 2 or less than −2 for each reporter. Highlighted dots represent deletion strains that have a Z-score value outside the cutoff for the NMD reporter, but not for the OPT reporter. Labeled dots identify the strains known to affect NMD and/or strains of interest in this study. b) (Top) Venn diagram showing the overlap between the OPT screen hits and the NMD screen hits. (Bottom) GO enrichment terms for cellular processes identified from the NMD hits performed using GOrilla software. Note: GO analysis for the OPT reporter hits were insufficient to yield enrichment data. c) Volcano plot showing data from a follow-up screen using newly constructed yeast strains and flow cytometry. The x-axis compares the fold change of individual deletion strains to the control HIS3-deletion strain. Data for each dot were obtained in triplicate and P-values are plotted on the y-axis. Red dots identify the strains known to affect NMD and/or strains of interest in this study. d) Individual averages of 3 different flow cytometry experiments performed on the indicated strains with the indicated reporters are shown. The GFP/RFP signal for each given strain normalized to the WT strain with the OPT reporter is plotted.
To validate our screen, we independently transformed the NMD reporter into strains from the haploid deletion collection corresponding to the NMD screen hits with a Z-score less than −2.0 or greater than 2.0. In this list, we excluded the 27 genes that were also hits in the OPT screen, ultimately yielding 139 genes to be validated. Strains that did not grow were dropped from the experiment, leaving 100 strains to be tested (Supplementary File 4). We individually analyzed these 100 newly constructed strains by flow cytometry and plotted the fold change of GFP/RFP for each candidate gene deletion relative to a HIS3-deletion control (Fig. 2c and Supplementary File 4). Among the validated hits were genes encoding the UPF proteins, Nmd4, and Ebs1 but also 2 genes encoding factors that were previously implicated in ribosome recycling—TMA20 and TMA22 (Skabkin et al. 2010; Samanfar et al. 2014).
We next decided to focus on several genes previously implicated in ribosome termination and/or recycling. We reconstructed the TMA20 and TMA22 deletions along with deletions for the previously known NMD factors EBS1 and NMD4 in the WT BY4741 strain carrying the OPT or NMD reporter and used these reconstructed strains for the rest of this study. We began by performing flow cytometry on nmd4Δ, ebs1Δ, tma20Δ, and tma22Δ strains in triplicate. As recently reported, deletion of either NMD4 or EBS1 modestly stabilizes the NMD reporter (Fig. 2d) (Dehecq et al. 2018). Deletion of TMA20 and TMA22 even more modestly, but reproducibly, stabilized the NMD reporter relative to the OPT reporter (Fig. 2d), underscoring the capacity of the R-SGA screens to isolate genes with even mild effects on NMD. Although the deletions showed mild effects on NMD, Tma20 and Tma22 have previously been shown to act redundantly, so we reasoned these mild effects might be indicative of a combinatorial role in NMD for this group of proteins (Skabkin et al. 2010; Young et al. 2018).
Perturbations to ribosome recycling correlate with inefficient NMD
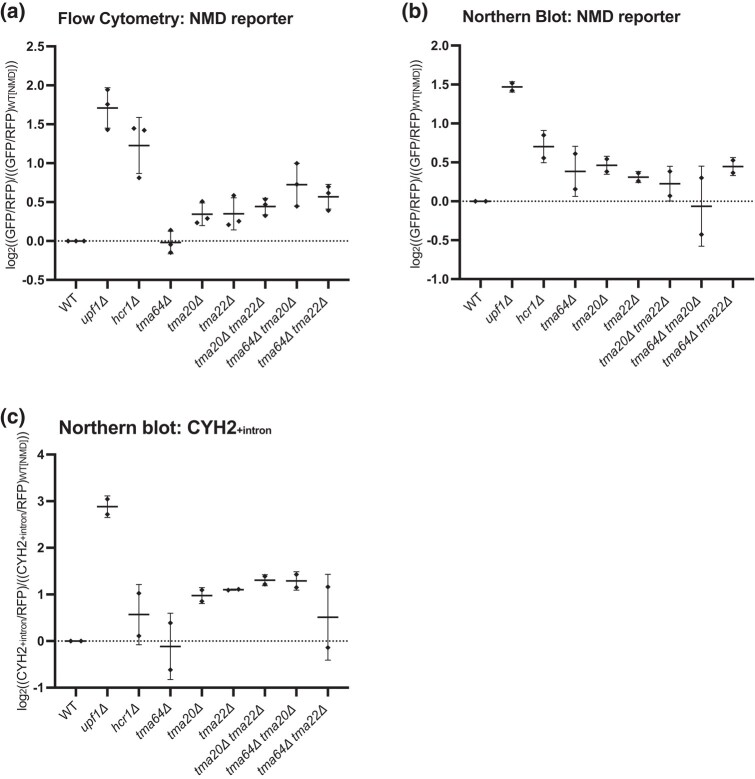
Ribosome profiling and in vitro biochemical experiments previously showed that Tma20/Tma22 and their mammalian homologs MCT-1/DENR, respectively, are involved in the removal of the 40S subunit from mRNA following Rli1/ABCE1-mediated 60S dissociation (Skabkin et al. 2010; Young et al. 2018). These factors are homologous to the N- and C-termini of Tma64 whose human homolog, ligatin, functions redundantly with MCT-1 and DENR in in vitro reconstituted systems to release 40S ribosomes and deacylated tRNA from mRNAs following 60S recycling (Skabkin et al. 2010). Therefore, we asked if deletion of TMA64 similarly increases GFP reporter expression as observed for the tma20Δ and tma22Δ strains. Consistent with the screen data, where TMA64 did not emerge as a candidate, and with recent data showing only a minor role for Tma64 in vivo (Young et al. 2021), deletion of TMA64 alone did not result in an increase in GFP expression from the NMD reporter (Fig. 3a). As a matter of routine, we made the double deletions of TMA64 in combination with deletion of TMA20 or TMA22, though the modest increase the GFP signal from the NMD reporter relative to each of the single deletions was not statistically significant (Fig. 3a). Northern blot data for the NMD reporter similarly revealed a modest (but statistically insignificant) stabilization of the NMD reporter mRNA for the TMA20, TMA22, and TMA64 deletion strains, while the double deletions (i.e. tma20Δ tma64Δ) did not show enhanced effects (Fig. 3b; representative northern blots shown in Supplementary Fig. 2b).
Fig. 3.
Deletion of 40S and 60S ribosome recycling factors leads to stabilization of exogenous NMD reporter and an endogenous NMD-targeted gene. a) Individual averages of 3 different flow cytometry experiments performed on the indicated strains with the indicated reporters are shown. The GFP/RFP signal for each strain normalized to the WT strain with the OPT reporter is plotted. b, c) Northern blot quantification of 2 unique northern blots is shown for each graph. The (GFP/RFP) signal in b) and the (CYH2+intron/RFP) in c) are both normalized to the WT strain with the OPT reporter and plotted.
Because TMA20, TMA22, and TMA64 deletions have been shown to function in 40S ribosome recycling, we next asked if NMD was also affected by disrupting 60S ribosome recycling by deleting the Rli1 accessory factor gene HCR1. While the hcr1Δ strain did not emerge from our initial screen due to its slow-growth phenotype, this deletion indeed increased GFP expression and stabilized the NMD reporter (Fig. 3a and b). These data together support the hypothesis that deficiencies in either 40S or 60S ribosome recycling can disrupt NMD.
As final validation of a role for these genes in NMD, we evaluated their effects on a well-characterized endogenous target of NMD, the rare intron-retaining transcript of CYH2 (He et al. 1993), by northern blot analysis. The absence of the 40S recycling factors Tma20 and Tma22 alone again reproducibly stabilized the CYH2+intron transcript (Fig. 3c; representative northern blots shown in Supplementary Fig. 2b). Deletion of the 60S recycling factor HCR1, however, did not have as pronounced an effect on NMD of the intron-containing CYH2 transcript as it did in flow and northern analyses of the NMD reporter. These data provide broad support for both the 40S and 60S recycling factors playing roles in promoting efficient NMD.
Discussion
Translation termination and recycling are inherently linked to NMD signaling through the recognition of a premature stop codon by the ribosome. Many previous studies have worked to define how these ribosome activities, termination and recycling, might be connected to the specification of NMD either directly or through interaction with a host of factors potentially bound to the 3′UTR of the mRNA (Czaplinski et al. 1998; González et al. 2000; Lykke-Andersen et al. 2000; Le Hir et al. 2001; Wang et al. 2001; Kobayashi et al. 2004; Hogg and Goff 2010). Furthermore, there are indications that key NMD factors such as Upf1 may directly influence ribosome recycling (Ghosh et al. 2010).
Here, we used a newly developed genetic screen in yeast with fluorescent reporter constructs to identify key factors that contribute to NMD. We identify 40S ribosome recycling factors Tma20 and Tma22 in the screen and additionally 60S recycling factor Hcr1 in follow-up experiments, as important modulators of NMD for both reporter constructs and an endogenous NMD target (Figs. 2 and 3). These observations are consistent with earlier studies showing that ribosome reinitiation after a PTC abrogates NMD (Ruiz-Echevarria and Peltz 1996; Zhang and Maquat 1997) and several recent studies in mammals implicating ribosome recycling and 3′UTR ribosome activity in NMD specification (Annibaldis et al. 2020; Zhu et al. 2020). These data are broadly consistent with NMD models implicating 3′UTR-bound proteins as key determinants (Peltz et al. 1993; González et al. 2000; Lykke-Andersen et al. 2000; Le Hir et al. 2001; Hogg and Goff 2010).
Rli1, or ABCE1 in mammals, works with partner protein Hcr1 to stimulate termination and recycle 60S subunits (Pisarev et al. 2010; Shoemaker and Green 2011; Beznosková et al. 2013; Young and Guydosh 2019). Our data show that deletion of HCR1 stabilizes NMD substrates (Fig. 3), consistent with recent work done in mammalian cells demonstrating that a loss of ABCE1 stabilizes NMD substrates (Annibaldis et al. 2020; Zhu et al. 2020). Similarly, we found that deletion of 40S ribosome recycling factors, TMA64, TMA20, and TMA22, also stabilizes NMD substrates (Fig. 3). Ribosome profiling experiments in Rli1 depletion, hcr1Δ, tma64Δtma20Δ, and tma64Δtma22Δ strains, reported a transcriptome-wide increase in the abundance of ribosomes in 3′UTRs (Beznosková et al. 2013; Young et al. 2018; Young and Guydosh 2019).
Previous studies have established that the 3′UTR is a critical regulator of NMD. In early studies in yeast, specific “downstream elements” (DSEs) were demonstrated to be important for triggering NMD and these DSEs were later shown to be bound by the RNA-binding protein Hrp1 that was critical for NMD for a subset of DSE-containing targets (Peltz et al. 1993; González et al. 2000). In mammalian cells, the presence of exon junction complexes (EJCs) and the accumulation of Upf1 in the 3′UTR have both been strongly implicated in promoting NMD (Lykke-Andersen et al. 2000; Le Hir et al. 2001; Hogg and Goff 2010). Taken together, these studies lead to simple models invoking positive contributions to NMD by proximal bound proteins (Upf1 and the EJC) that recruit other critical machinery involved in mRNA decay (such as the SMG proteins). The contributions of such proteins in the 3′UTR to NMD signaling provide 1 potential explanation for our results wherein the presence of scanning and/or translating ribosomes in the 3′UTR disrupts NMD. Alternatively, inefficient ribosome clearing at stop codons may interfere with NMD by perturbing other key steps such as recruitment of deadenylation or decapping machinery or by indirectly impairing translation initiation. Further experiments will clearly be needed to define the mechanism by which loss of Tma20 and Tma22 contributes to impaired targeting of NMD substrates for decay.
Supplementary Material
Acknowledgments
We thank Brendan Cormack for his contributions to planning and carrying out experiments.
Contributor Information
Miguel Pacheco, Department of Molecular Biology and Genetics, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Karole N D’Orazio, Department of Molecular Biology and Genetics, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Laura N Lessen, Department of Molecular Biology and Genetics, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Anthony J Veltri, Department of Molecular Biology and Genetics, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Zachary Neiman, Department of Molecular Biology and Genetics, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Raphael Loll-Krippleber, Department of Biochemistry and Donnelly Centre, University of Toronto, Toronto, ON M5S 3E1, Canada.
Grant W Brown, Department of Biochemistry and Donnelly Centre, University of Toronto, Toronto, ON M5S 3E1, Canada.
Rachel Green, Department of Molecular Biology and Genetics, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Data availability
Strains and plasmids are available upon request. The supplementary files contain complete lists of all data and analysis from the screen, including raw data from the synthetic genetic array for the OPT and NMD reporter (Supplementary Files 1 and 2, respectively; a condensed spreadsheet with Z-scores from both reporters is provided in Supplementary File 3), flow cytometry data for each candidate gene deletion relative to a HIS3-deletion control (Supplementary File 4), and a list of yeast strains, plasmids, and DNA oligos used in this study (Supplementary File 5).
Supplemental material available at G3 online.
Funding
RG is supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under a R37 Method to Extend Research in Time (MERIT; 5R37GM059425). KND and MP are supported by the NIGMS under a T32 Training Grant (5T32GM007445-39), as well as LNL and AJV (5T32GM135131-02). RG and MP are supported by the Howard Hughes Medical Institute (HHMI). MP is an HHMI Gilliam Fellow. GWB is supported by the Canadian Institutes of Health Research (FDN-159913). GWB holds a Canada Research Chair in Genome Integrity.
Literature cited
- Annibaldis G, Domanski M, Dreos R, Contu L, Carl S, Kläy N, Mühlemann O. 2020. Readthrough of stop codons under limiting ABCE1 concentration involves frameshifting and inhibits nonsense-mediated mRNA decay. Nucleic Acids Res. 48(18):10259–10279. doi: 10.1093/nar/gkaa758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beznosková P, Cuchalová L, Wagner S, Shoemaker CJ, Gunišová S, von der Haar T, Valášek LS. 2013. Translation initiation factors eIF3 and HCR1 control translation termination and stop codon read-through in yeast cells. PLoS Genet. 9(11):e1003962. doi: 10.1371/journal.pgen.1003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Hagan KW, Zhang S, Peltz SW. 1995. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 9(4):423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 12(11):1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehecq M, Decourty L, Namane A, Proux C, Kanaan J, Le Hir H, Jacquier A, Saveanu C. 2018. Nonsense-mediated mRNA decay involves two distinct Upf1-bound complexes. EMBO J. 37(21):e99278. doi: 10.15252/embj.201899278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazio KN, Wu CC-C, Sinha N, Loll-Krippleber R, Brown GW, Green R. 2019. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during no go decay. Elife. 8:e49117. doi: 10.7554/eLife.49117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 16(1):49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- Eden E, Lipson D, Yogev S, Yakhini Z. 2007. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol. 3(3):e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 10(1):48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J, Kainth P, Lambert J-P, van Bakel H, Tsui K, Peña-Castillo L, Nislow C, Figeys D, Hughes TR, Greenblatt J, et al. 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol Cell. 35(3):340–351. doi: 10.1016/j.molcel.2009.06.023. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Ganesan R, Amrani N, Jacobson A. 2010. Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA. 16(9):1832–1847. doi: 10.1261/rna.1987710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González CI, Ruiz-Echevarría MJ, Vasudevan S, Henry MF, Peltz SW. 2000. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol Cell. 5(3):489–499. doi: 10.1016/s1097-2765(00)80443-8. [DOI] [PubMed] [Google Scholar]
- Grimson A, O’Connor S, Newman CL, Anderson P. 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol Cell Biol. 24(17):7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Brown AH, Jacobson A. 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 17(3):1580–1594. doi: 10.1128/MCB.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc Natl. Acad Sci. 90(15):7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CUT. 2018. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb. Perspect Biol. 10(10):a032656. doi: 10.1101/cshperspect.a032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry JA, Tan G, Ou J, Boone C, Brown GW. 2015. Leveraging DNA damage response signaling to identify yeast genes controlling genome stability. G3 (Bethesda). 5(5):997–1006. doi: 10.1534/g3.115.016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell. 143(3):379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainth P, Sassi HE, Peña-Castillo L, Chua G, Hughes TR, Andrews B. 2009. Comprehensive genetic analysis of transcription factor pathways using a dual reporter gene system in budding yeast. Methods. 48(3):258–264. doi: 10.1016/j.ymeth.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Karousis ED, Nasif S, Mühlemann O. 2016. Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip Rev RNA. 7(5):661–682. doi: 10.1002/wrna.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. 2006. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20(3):355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnevis S, Gross T, Rotte C, Baierlein C, Ficner R, Krebber H. 2010. The iron–sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 11(3):214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Funakoshi Y, Hoshino S, Katada T. 2004. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J Biol Chem. 279(44):45693–45700. doi: 10.1074/jbc.M405163200. [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. 2016. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 129(3):461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson MR, Lessen LN, Wang J, Prabhakar A, Corsepius NC, Green R, Puglisi JD. 2021. Mechanisms that ensure speed and fidelity in eukaryotic translation termination. Science. 373(6557):876–882. doi: 10.1126/science.abi7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BS, Culbertson MR. 1995. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci. 92(22):10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5(12a):2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Leeds P, Wood JM, Lee B-S, Culbertson MR. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 12(5):2165–2177. doi: 10.1128/mcb.12.5.2165-2177.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20(17):4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh B, Jonas S, Izaurralde E. 2013. The SMG5–SMG7 heterodimer directly recruits the CCR4–NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev. 27(19):2125–2138. doi: 10.1101/gad.226951.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Stolboushkina EA, Vaidya AT, Zhao C, Garber MB, Dmitriev SE, Steitz TA. 2017. Crystal structure of the human ribosome in complex with DENR-MCT-1. Cell Rep. 20(3):521–528. doi: 10.1016/j.celrep.2017.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Chen Y, Ardal BR, Lilje B, Waage J, Sandelin A, Jensen TH. 2014. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 28(22):2498–2517. doi: 10.1101/gad.246538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu M-D, Steitz JA. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 103(7):1121–1131. doi: 10.1016/S0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Maquat LE. 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell. Biol. 5(2):89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Peltz SW, Brown AH, Jacobson A. 1993. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 7(9):1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CUT, Pestova TV. 2010. The role of ABCE1 in eukaryotic post-termination ribosomal recycling. Mol Cell. 37(2):196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Maquat LE. 2013. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 47(1):139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. 2009. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol. 21(3):394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, Peltz SW. 1996. Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J. 15(11):2810–2819. doi: 10.1002/j.1460-2075.1996.tb00641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Samanfar B, Tan LH, Shostak K, Chalabian F, Wu Z, Alamgir M, Sunba N, Burnside D, Omidi K, Hooshyar M, et al. 2014. A global investigation of gene deletion strains that affect premature stop codon bypass in yeast, Saccharomyces cerevisiae. Mol Biosyst. 10(4):916–924. doi: 10.1039/C3MB70501C. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. 2011. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci. 108(51):E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CUT, Pestova TV. 2010. Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 24(16):1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE, Baker KE. 2015. Nonsense-mediated RNA decay—a switch and dial for regulating gene expression. BioEssays. 37(6):612–623. doi: 10.1002/bies.201500007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AHY, Boone C. 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 313:171–191. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Tong AHY, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CWV, Bussey H, et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294(5550):2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E. 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 16(4):587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Wagih O, Usaj M, Baryshnikova A, VanderSluis B, Kuzmin E, Costanzo M, Myers CL, Andrews BJ, Boone CM, Parts L. 2013. SGAtools: one-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 41(W1):W591–W596. doi: 10.1093/nar/gkt400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Czaplinski K, Rao Y, Peltz SW. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20(4):880–890. doi: 10.1093/emboj/20.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser M, Schäfer T, Leibundgut M, Böhringer D, Aylett CHS, Ban N. 2017. Structural and functional insights into human re-initiation complexes. Mol. Cell. 67(3):447–456.e7. doi: 10.1016/j.molcel.2017.06.032. [DOI] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR. 2019. Hcr1/eIF3j is a 60S ribosomal subunit recycling accessory factor in vivo. Cell Rep. 28(1):39–50.e4. doi: 10.1016/j.celrep.2019.05.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Makeeva DS, Zhang F, Anisimova AS, Stolboushkina EA, Ghobakhlou F, Shatsky IN, Dmitriev SE, Hinnebusch AG, Guydosh NR. 2018. Tma64 (eIF2D), Tma20 (MCT-1), and Tma22 (DENR) recycle post-termination 40S subunits in vivo. Mol. Cell. 71(5):761–774.e5. doi: 10.1016/j.molcel.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Meydan S, Guydosh NR. 2021. 40S ribosome profiling reveals distinct roles for Tma20/Tma22 (MCT-1/DENR) and Tma64 (eIF2D) in 40S subunit recycling. Nat Commun. 12(1):2976. doi: 10.1038/s41467-021-23223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Maquat LE. 1997. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 16(4):826–833. doi: 10.1093/emboj/16.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhang H, Mendell JT. 2020. Ribosome recycling by ABCE1 links lysosomal function and iron homeostasis to 3ʹ UTR-directed regulation and nonsense-mediated decay. Cell Rep. 32(2):107895. doi: 10.1016/j.celrep.2020.107895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B, Sinha NK, Enam SU, Koleske B, Green R. 2021. Translational repression of NMD targets by GIGYF2 and EIF4E2. PLoS Genet. 17(10):e1009813. doi: 10.1371/journal.pgen.1009813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. The supplementary files contain complete lists of all data and analysis from the screen, including raw data from the synthetic genetic array for the OPT and NMD reporter (Supplementary Files 1 and 2, respectively; a condensed spreadsheet with Z-scores from both reporters is provided in Supplementary File 3), flow cytometry data for each candidate gene deletion relative to a HIS3-deletion control (Supplementary File 4), and a list of yeast strains, plasmids, and DNA oligos used in this study (Supplementary File 5).
Supplemental material available at G3 online.