Abstract
The Ecuadorian brown-headed spider monkey (Ateles fusciceps fusciceps) is currently considered one of the most endangered primates in the world and is classified as critically endangered [International union for conservation of nature (IUCN)]. It faces multiple threats, the most significant one being habitat loss due to deforestation in western Ecuador. Genomic tools are keys for the management of endangered species, but this requires a reference genome, which until now was unavailable for A. f. fusciceps. The present study reports the first whole-genome sequence and assembly of A. f. fusciceps generated using Oxford Nanopore long reads. DNA was extracted from a subadult male, and libraries were prepared for sequencing following the Ligation Sequencing Kit SQK-LSK112 workflow. Sequencing was performed using a MinION Mk1C sequencer. The sequencing reads were processed to generate a genome assembly. Two different assemblers were used to obtain draft genomes using raw reads, of which the Flye assembly was found to be superior. The final assembly has a total length of 2.63 Gb and contains 3,861 contigs, with an N50 of 7,560,531 bp. The assembly was analyzed for annotation completeness based on primate ortholog prediction using a high-resolution database, and was found to be 84.3% complete, with a low number of duplicated genes indicating a precise assembly. The annotation of the assembly predicted 31,417 protein-coding genes, comparable with other mammal assemblies. A reference genome for this critically endangered species will allow researchers to gain insight into the genetics of its populations and thus aid conservation and management efforts of this vulnerable species.
Keywords: whole-genome sequence, genome assembly, reference genome, spider monkey, Ateles fusciceps fusciceps, Oxford Nanopore sequencing
Introduction
The brown-headed spider monkey (Ateles fusciceps fusciceps) is a neotropical primate inhabiting northwestern Ecuador (its presence in Colombia is uncertain). It is most commonly found below 1,200 masl, but its altitudinal range can go as high as 2,300 masl (Gallo-Viracocha et al. 2022). This subspecies plays an important role in the ecosystem as an effective seed disperser; its diet is composed mainly of ripe fruits (70–90%), which is key for the regeneration and maintenance of tree diversity in the forests it inhabits (Calle-Rendón et al. 2016; Morelos-Juárez et al. 2018; Gallo-Viracocha et al. 2022). Female spider monkeys have their first offspring between the ages of 7 and 9, with an interbirth interval of 3–4 years, which means that they have a low reproductive rate compared with other primate species (Eisenberg 1973; Milton 1981; Robinson and Janson 1986; Fedigan and Rose 1995).
A. f. fusciceps is a priority subject for conservation efforts worldwide, currently listed as one of the world's 25 most endangered primates (Tirira et al. 2022) and cataloged as Critically Endangered by the international union for conservation of nature (IUCN) (Moscoso et al. 2021). Anthropogenic factors are the main threats to A. f. fusciceps populations; as a large mammal with slow growth and reproduction rates, it is affected by the subsistence of hunting practices within indigenous communities, as well as poaching of infants for illegal wildlife trade. However, its most important threat is habitat loss. The Chocó region it inhabits in western Ecuador is a biodiversity hotspot (Myers et al. 2000) that requires immediate conservation action, given that it has lost >80% of its original vegetation coverage (Mittermeier et al. 2002; Myers et al. 2000; Critical Ecosystem Partner Fund, Chocó-Darién-Western Ecuador: Chocó-Manabí Conservation Corridor Briefing Book 2005; Sierra et al. 2021). This has led to dramatic population decreases of several species in the region, including the brown-headed spider monkey (Moscoso et al. 2021). Furthermore, spider monkeys are highly frugivorous, devoting ∼80% of their time to the consumption of ripe fruits of different tree species. They are, therefore, extremely dependent on low-availability food resources (Di Fiore et al. 2008), and this makes them more susceptible to local extinction in areas transformed by humans (Garber et al. 2006). The current situation of A. f. fusciceps warrants a stronger focus on its conservation to prevent the extinction of the species.
Reductions in the number of individuals in brown-headed spider monkey populations make them susceptible to inbreeding depression and loss of genetic diversity through drift (Frankham 2003; Rivera Román 2017). These 2 processes reduce the species’ resilience to environmental change, thus increasing its vulnerability (Frankham 2003). Whole-genome sequencing (WGS) has been identified as a key tool to manage threatened species, as genomes from representative numbers of individuals can be used to make inferences on a population's demographic history, inbreeding rates, and past genetic bottlenecks, among other significant events (Taylor et al. 2022). For a critically endangered species like A. f. fusciceps, genomic population studies provide useful information regarding the species’ genetic diversity and population structure, which can assist with the design of adequate management regimes and conservation strategies such as those identified in the Conservation Action Plan for the Ecuadorian Primates (Tirira et al. 2018). Population genomic studies require a reference genome, which was not available for A. f. fusciceps.
Next-generation sequencing has become more accessible in terms of costs and sequencing velocity. Nevertheless, limited resources in developing countries restrict the accessibility for usage and development of genomic tools (Helmy et al. 2016), especially for endangered species in the tropics (regions that harbor at least 50% of the planet's biodiversity; Brancalion et al. 2019). Oxford Nanopore sequencing has facilitated genomic research in developing countries with portable, low-cost sequencers that produce ultra-long reads and allow on-site sequencing (Lin et al. 2021). While only 1% of all threatened species have a published reference genome (Brandies et al. 2019), this could change as access to sequencing technologies increases. Given the overlap of high biodiversity and low accessibility to genomic tools, special emphasis and effort should be placed on genome sequencing projects of endangered species in developing nations.
In the present study, we report the first WGS and assembly of A. f. fusciceps using long reads obtained through Oxford Nanopore Technologies.
Materials and methods
Sampling
The brown-headed spider monkey individual from which the sample was taken was a subadult male named Mishky, born in the Hacienda Jambelí Rescue Center (2°46′30.48″S 79°44′9.51″O) located in the Guayas province in southwestern Ecuador. In 2014, Proyecto Washu started an ex situ conservation program for the rehabilitation and welfare of this species. The Hacienda Jambelí population of A. f. fusciceps is currently considered the largest captive population in Ecuador with a total of 21 individuals: 8 adult males, 1 subadult male, 7 adult females, 1 subadult female, 1 juvenile female, and 3 juvenile males. This population is composed of individuals rescued from the illegal pet trade and others born in the rescue center, as is the case of Mishky.
Mishky was transported to the Tueri Wildlife Hospital (TUERI-USFQ) for medical examination due to injuries sustained while at the Hacienda Jambelí Rescue Center. A 5-ml blood sample was obtained by the TUERI-USFQ veterinarian staff and stored at −80°C in the Laboratorio de Biotecnología Vegetal—USFQ.
Sequencing methods and preparation
DNA extraction
For DNA extraction, the DNeasy Blood and Tissue Kit (QIAGEN, Valencia, CA, USA) was used for 16 total reactions with minor modifications. For the final elution, 30 µl of ultrapure water was used to obtain a total elution of 60 µl after 2 elution steps. The final DNA quantification and quality was assessed with Qubit Fluorometric Quantitation and NanoDrop 2000.
Preparation of genomic libraries
The library construction protocol followed the workflow of the Ligation Sequencing Kit SQK-LSK112 (Oxford Nanopore Technologies), which comprises 3 sections. The process started with an average quantity of 2,000 ng per reaction and resulted in a total of 14 libraries. After each section, the DNA concentration was quantified using Qubit Fluorometric Quantitation. The libraries were stored at 4°C awaiting sequencing.
Sequencing
Sequencing was carried out in a MinION Mk1C sequencer using 2 R9.4.1 and 6 R10.4.1 flow cells. The 2 R9.4.1 flow cells were used once each for test runs. Each R10.4.1 flow cell was used for 3–4 runs to generate a total of 21 sequencing runs (>24 h). The libraries that had a high DNA quantity (>800 ng) were used for 2 sequencing runs. Similarly, depending on the final concentration of each library, 6, 7, or 12 µl of the sample was loaded to the flow cell, in order to sequence ∼500 ng of DNA. The real-time base calling was executed with Guppy v5.1.13 (ONT), and the resulting output was raw fastq sequencing reads.
Data processing
Initial processing of reads
The raw sequencing reads (.fastq) were first filtered according to quality scores using NanoFilt v2.3.0 (De Coster et al. 2018). Reads with quality scores <7 were removed from the analysis (Halstead et al. 2021; Feng et al. 2022; Petersen et al. 2022). Adapters from filtered reads were then trimmed in Porechop v0.2.4 (Wick et al. 2017), and sequencing quality was analyzed in Nanoplot v. 1.20.0 (De Coster et al. 2018) for both individual sequencing runs and the complete dataset.
Assembly, mapping, polishing, and scaffolding
Two different assemblers were used to obtain draft genomes using raw reads. First, SMARTdenovo v.1.0.0 (Liu et al. 2021) was used to assemble the obtained reads with the smartdenovo.pl script.
Raw reads were also assembled using Flye v 2.7.1 (Kolmogorov et al. 2019), selecting nano-raw as the type of input reads and with a specified genome size (g) of 2.6 Gb, based on the reported genome size of the closely related species A. geoffroyi (JAKFHY000000000.1) (Shao 2022). The reference genome of A. geoffroyi is part of the Whole Genome Shotgun Sequencing Project. It is a contig-level assembly with a 56.87× genome coverage. The sequencing technology used was PacBio RSII, and the reads were assembled with Wtdb2 v.2 (Shao 2022).
Both de novo assembly drafts were mapped against this reference genome using minimap2 v2.24 (Li 2018) to reorder the contigs generated in the assembly. The resulting mapped assemblies were then polished once using Medaka v1.7.2 (Oxford Nanopore Technologies, 2018). The medaka_consensus program was employed using the r103_fast_g507 model.
Completeness and quality assessment of genome assembly
Genome assembly quality for both assemblies was evaluated with QUAST v5.2.0 (Mikheenko et al. 2018) under default parameters. The reference genome of A. geoffroyi (Shao 2022) was specified as the reference for comparison. BUSCO v5.4.4 (Manni et al. 2021) was then run using the primates_odb10 database with 13,780 genes to evaluate genome completeness based on expected gene content; we provide statistics for complete, single, fragmented, duplicated, and missing BUSCOs.
Genome annotation
The best assembly was selected based on the assembly statistics and BUSCO results, and that assembly was annotated. For genome annotation, a custom repeat library was first created ab initio for the assembled genome of A. f. fusciceps using RepeatModeler v2.0.4 (Flynn et al. 2020). We applied the “LTRStruct” option for long terminal repeat retroelement identification. Repetitive regions of the genome were identified and soft-masked by RepeatMasker v4.0.7 (Smith et al. 2013–2015) in Maker v2.31.9 (Campbell et al. 2014). Contigs were then annotated with Maker v2.31.9 (Campbell et al. 2014) in 3 consecutive rounds. In the first round, ab initio gene prediction algorithms were run with EST and protein evidence using the est2genome and protein2genome functions. Reference proteomes from 4 closely related primate species were gathered from the UniProt database (Bateman et al. 2021) to be used as protein evidence in Maker (Sapajus apella: UP000504640, Callithrix jacchus: UP000008225, Saimiri boliviensis boliviensis: UP000233220, and Aotus nancymaae: UP000233020). EST data were obtained from the NCBI EST database for the most closely related species available (C. jacchus). These initial predictions were then used to train the ab initio gene predictor SNAP (Korf 2004), and a second round of Maker was run using the hidden Markov model from SNAP. Finally, a third round of annotation was run with SNAP. Protein and transcript fasta files and gff files generated along the 3 annotation rounds were then merged. To isolate the best-supported gene models, InterProScan v5.61 (Jones et al. 2014) was first run to identify conserved Pfam domains on the Maker-predicted proteins. Using accessory scripts from Maker, gene models with annotation edit distance (AED) values >0.5 or lacking Pfam domains were then removed from the gff and fasta files. Finally, the agat_sp_statistics.pl script from the Another Gff Analysis Toolkit software was used to obtain the annotation statistics (Dainat 2020).
Foreign contamination screening and elimination
The mapped, polished Flye assembly was screened for foreign contamination using NCBI's FCS-GX tool (Astashyn et al. 2023), which identifies contaminant sequences and removes them from the assembled genome. This clean assembly was evaluated using the parameters described in Completeness and Quality Assessment of Genome Assembly.
Results and discussion
A. f. fusciceps assembly
Oxford Nanopore Sequencing of A. f. fusciceps produced 55.95 Gb from 8.96 million reads with quality scores greater than q7. Reads greater than or equal to q7 were selected due to the fact that various reports of genome assemblies with Oxford Nanopore reads specify q7 as the threshold for acceptable read quality (Halstead et al. 2021; Feng et al. 2022; Petersen et al. 2022). In order to calculate the coverage, we based our predicted genome size on the closely related species, A. geoffroyi, which is 2.6 Gb (Shao 2022). This represents an estimated 21× coverage of the genome. In general, reads had a mean read length of 6.42 kb and a mean read quality score of 10.9 (Table 1).
Table 1.
Sequencing statistics for the A. f. fusciceps genome.
| Generated bases | Read count | Coverage | Mean read length | Mean read quality |
|---|---|---|---|---|
| 55.95 Gb | 8.96 million | 21× | 6.42 kb | 10.9 |
The assembly obtained with SMARTdenovo and later polished by Medaka had a total length of 2.58 Gb and contained 6,856 contigs (Table 2). It had an N50 size of 799,988 bp and an L50 of 985, and its largest contig was 5,164,154 bp. When mapped to the reference genome of the closely related A. geoffroyi, it had 567.9 mismatches per 100 kb. The Flye assembler alongside the Medaka polisher generated a primary assembly for A. f. fusciceps of 2.63 Gb containing 3,861 contigs with an N50 size of 7,560,531 bp (Table 2). The L50 for this assembly was 97, and the largest contig was 44,929,532 bp. In this case, when mapped to A. geoffroyi, the assembly had 539.3 mismatches per 100 kb.
Table 2.
Comparative statistics for the 2 A. f. fusciceps draft assemblies generated using SMARTdenovo and Flye, postpolishing with Medaka.
| Assembly | Total length | Contig number | N50 | Largest contig | L50 | # mismatches per 100 kb |
|---|---|---|---|---|---|---|
| SMARTdenovo | 2,586,824,631 | 6,856 | 799,988 | 5,164,154 | 985 | 567.9 |
| Flye | 2,635,867,907 | 3,861 | 7,560,531 | 44,929,532 | 97 | 539.3 |
The Flye assembly is superior to the SMARTdenovo assembly in all analyzed statistics (Table 2). It has a total length similar to the genome size of the closely related A. geoffroyi (2.68 Gb; Shao 2022) and less mismatches per 100 kb when compared with this genome. It is much less fragmented, with 3,861 contigs compared with 6,856 in the SMARTdenovo assembly. Furthermore, according to the L50, 50% of the A. f. fusciceps genome is represented in 97 contigs in the Flye assembly and in 985 contigs in the SMARTdenovo assembly, proving once again that the SMARTdenovo assembly is less continuous. The Flye assembly also has a much higher N50 and the largest contig size; 50% of the contigs possess a size equal to or longer than 7.56 Mb (Alhakami et al. 2017), which is remarkable, since primate species have very large genomes and first assemblies normally produce contig N50 lengths shorter than 100 kb (Jayakumar et al. 2021). Finally, the largest contig size of the Flye assembly is 44.9 Mb, almost the size of a human chromosome (Brown 2002).
The assemblers employed in this study possess distinct approaches; SMARTdenovo relies on the Overlap-Layout-Consensus (OLC) algorithm, while Flye uses the generalized de Bruijn Graph (DBG; Wang et al. 2021). Primate genomes pose a unique challenge due to their substantial proportion of noncoding regions, rich in repetitive sequences (Ahmad et al. 2020). In the context of contig construction where repeats, sequencing errors, and heterozygosity are influential, OLC usually has the advantage because it tolerates these factors by allowing some mismatches in overlap identification. However, DBG excludes these variations on the k-mer graph, making it particularly suitable for large genome assemblies (Li et al. 2012). Consistent with our results, Wick and Holt (2021) demonstrated the reliability of the Flye assembler, compared with other assemblers. Their research highlighted its superior performance at low read depths and the minimal occurrence of large-scale sequence errors.
Both genome assemblies were analyzed for annotation completeness based on primate ortholog prediction. The gene database used, primates_odb10, comprises 25 primate genomes and 13,780 genes and is categorized as a high-resolution database, which provides a high level of confidence for genome completeness evaluations (Simão et al. 2015; Waterhouse et al. 2018). For the SMARTdenovo assembly, we obtained 10,602 (76.9%) complete BUSCOs, of which 10,384 are single copy (75.4%) and 218 (1.6%) are duplicated (1.58%). There were 2,436 (17.7%) missing BUSCOs and 742 (5.4%) fragmented BUSCOs (Supplementary Fig. 1).
When analyzing the Flye assembly, the BUSCO results improved: we obtained more single-copy complete BUSCOs and less missing or fragmented BUSCOs (Supplementary Fig. 1). Specifically, we obtained 11,604 (84.3%) complete BUSCOs, of which 11,362 (82.5%) are single copy and 242 (1.8%) are duplicated (Supplementary Fig. 1). The high number of complete BUSCOs (84.3%) and the low number of duplicated genes indicate a good level of genome completeness and a precise assembly (Simão et al. 2015; Manni et al. 2021). Regarding the remaining 15.7% of BUSCOs, 564 (4.1%) are fragmented and 1,612 (11.6%) are missing. Technical limitations in gene prediction can inflate the proportions of missing and fragmented BUSCOs, when working with large genomes such as that of A. f. fusciceps (Manni et al. 2021). Additionally, ONT sequences have error rates of 10–30% that are mainly composed of indels (Morisse et al. 2021). However, while the assembly could be improved, the results indicate an overall good quality of the Flye assembly.
Due to the fact that the Flye assembly has better assembly statistics and a more complete annotation, this is the one we selected for further analyses and the one that is reported in this publication. After filtering out foreign contaminations, our A. f. fusciceps assembly was compared with that of the closely related A. geoffroyi (GCA_023783555.1; Table 3, Fig. 1). This contig-level assembly of A. geoffroyi has a total length of 2.68 Gb in 2,732 contigs with a N50 size of 29,212,752 bp and a guanine-cytosine content (GC) content of 40.75%. The values for coverage, contig number, and N50 size for both assemblies were significantly different. However, considering the range of genome size variation among primates (2.09–4.87 Gb; Fantini et al. 2016) and that primate genomes’ GC contents are remarkably consistent (Qi et al. 2016), the similar values for total length and GC (%) clearly show that this primary genome assembly of A. f. fusciceps is adequate, while the differences in coverage, contig number, and N50 suggest there is room for improvement.
Table 3.
Assembly statistics for the final, clean A. f. fusciceps genome assembly compared with the closely related A. geoffroyi assembly.
| Genome | Total length | Coverage | Contig number | N50 | GC (%) |
|---|---|---|---|---|---|
| A. fusciceps | 2,639,265,159 | 21× | 3,851 | 7,560,531 | 40.85 |
| A. geoffroyi | 2,683,028,796 | 56.87× | 2,723 | 29,212,752 | 40.75 |
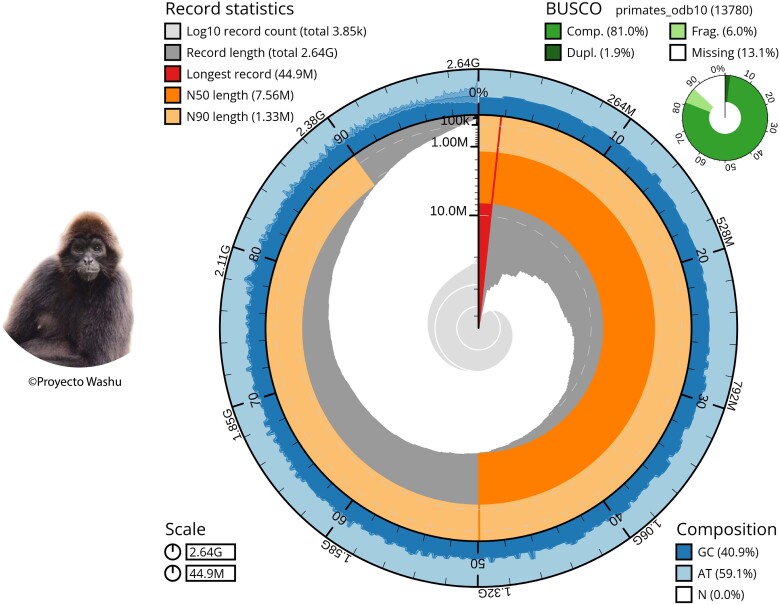
Fig. 1.
The final genome assembly of A. f. fusciceps (Flye assembly) metrics. The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. The plot indicates a total genome size of 2.64 Gb, and a longest obtained contig of 44.9 Mb. The plot also showsthe N50 and N90 values, as well as the GC, AT, and N compositions. A summary of the complete, duplicated, fragmented, and missing BUSCOs (primates_odb10) is represented in the right-hand corner.
Genome annotation
The annotation of the A. f. fusciceps assembly in Maker predicted 35,809 protein-coding genes, 88% (31,417) with an AED value <0.5 (Table 4), indicating good protein and transcript evidence support and reasonable quality of the annotation (Sork et al. 2016; Saenko et al. 2021). AED values closer to 0 generally show greater agreement between the annotation and protein/transcript evidence, while AED values closer to 1 reveal little to no support for the resulting annotation (Eilbeck et al. 2009), which is why all gene models with AED values >0.5 were filtered out of the final annotation.
Table 4.
Summary statistics of the annotated genome (AED < 0.5) of A. f. fusciceps.
| Statistic | Value |
|---|---|
| Number of genes | 31,417 |
| Number of exons | 183,970 |
| Number of introns (in coding sequence [CDS]) | 149,050 |
| Overlapping genes | 715 |
| Mean mRNAs per gene | 1.0 |
| Mean exons per mRNA | 5.9 |
| Mean introns per mRNA | 4.7 |
| Mean gene length (bp) | 16,857 |
| Mean exon length (bp) | 176 |
| Mean intron length (bp) | 3,292 |
| % of genome covered by genes | 20.1 |
| % of genome covered by exons | 1.2 |
| % of genome covered by introns | 18.9 |
The resulting 31,417 protein-coding genes of A. f. fusciceps are comparable with what other mammal genome assemblies have reported like the case of the lowland anoa (Bubalus depressicornis) with 32,393 predicted protein-coding genes (Porrelli et al. 2022). Nonetheless, gene count is slightly higher than expected when compared with the 22,027 protein-coding genes predicted for C. jacchus (GCA_011100555.1; Warren et al. 2009) and the 20,350 protein-coding genes for S. apella (GCF_009761245.1) (Culibrk et al. 2019), both closely related primate species of A. f. fusciceps. In general, eukaryotic genomes have around 15,000–25,000 protein-coding genes (Cantarel et al. 2008) with the human genome (a primate species) reporting ∼19,100 genes (Piovesan et al. 2019). The overestimation of the protein-coding genes could be explained by ONT's long-read accuracy limitations compared with other sequencing technologies (Rang et al. 2018), though the resulting annotation of our genome still shows an accurate prediction. Additionally, since only soft masking was used for repeat masking during MAKER annotation, it is possible that repetitive regions were misconceived as putative genes (Saenko et al. 2021), increasing the predicted number of coding sequences.
Furthermore, the annotation of the A. f. fusciceps genome predicted a mean gene length of 16,857 bp (Table 4), a length comparably smaller to what has been reported for other closely related primate species, with mean gene lengths of ∼40,000 bp (Warren et al. 2009; Culibrk et al. 2019; Harris et al. 2020). The same pattern is evident when we compare mean intron length (3,292 bp) and mean exon length (176 bp). These differences can likely be attributed to the level of fragmentation of our genome and the inaccurate prediction of genomic features in repetitive regions. This is expected since around 50% of a primate genome is covered by repetitive elements (Rogers and Gibbs 2014), making the annotation of other genomic features a challenging task (Okazaki and Hume 2003). Nonetheless, differences in genomic feature predictions between closely related species have been reported in other reference genomes (Jiang et al. 2022; Kaur et al. 2023) and could be attributed to the sequencing technology used and the level of genome fragmentation.
Importance of reference genome
Numerous studies have established the importance of genomic data to understand the evolutionary history of a species and to develop appropriate conservation and management strategies (Kleinman-Ruiz et al. 2017; Saremi et al. 2019; Kenny et al. 2020; Nong et al. 2021; Pfenninger et al. 2021). WGS leads to a better understanding of the biology of a species and provides insights into fundamental processes that shape their evolution (Ryder 2005), and its application can provide important and accurate information about its demographic history, admixture, introgression, recombination, linkage disequilibrium, genomic regions evolving under selective pressures, and other evolutionary processes (Theissinger et al. 2023). For critically endangered species like the brown-headed spider monkey, genomic approaches are even more valuable due to the scarcity of samples for genetic studies; therefore, WGS maximizes the information that researchers can harness from each sample. However, in order to be able to generate and fully take advantage of this information, a reference genome is required (Theissinger et al. 2023).
Species under such conservation threats face a dire need for conservation actions to reverse their declining population trends. Currently, Proyecto Washu is deepening the understanding of the brown-headed spider monkey's behavior and ecology through observational studies of a population of spider monkeys living in a highly fragmented landscape. The sequencing of its genome provides an opportunity to improve its conservation through the development of population-level studies to evaluate its genetic diversity and gene flow. Moreover, genetic population studies may allow us to better differentiate its populations, perform identification of individuals and kinship patterns, evaluate the dispersion and migration of individuals, and identify and prioritize biological corridors through which monkey populations move. Biological corridors prevent the isolation of populations in closed forest fragments, which reduces inbreeding and helps to maintain genetic diversity in the area (Kirchner et al. 2003; Haddad et al. 2015).
While major progress has been made in animal genome sequencing in the last 25 years, significant gaps and biases remain in geographic and taxonomic representation resulting in an improper depiction of the global genetic pool (Hotaling et al. 2021). Ecuador, for instance, has a limited record of genetic and genomic research (Zambrano-Mila et al. 2019) despite its sizable biodiversity (Celi and Villamarín 2020). This is a multifaceted issue resulting from the lack of sequencing platforms and training in genome data analysis and research costs (Hotaling et al. 2021). This makes outsourcing a popular alternative to generate genomic sequences, despite the limitations of using third-party service providers (Helmy et al. 2016). A feasible pathway to democratize sequencing efforts and to involve developing countries is through the usage of portable sequencing devices such as the Oxford Nanopore Technologies MinION, as applied in this study. This is a time-efficient and cost-efficient technology for the assembly of all genome sizes (Wang et al. 2021), which operates on standard computing resources. Its long-read length and portability enable the use of these devices in basic research (e.g. assembly of preliminary nonmodel organism genomes), clinical usage, and on-site applications (Wang et al. 2021). Due to its ease of use and convenience, the current report represents an initial sequencing project, which will be further extended to other underrepresented Ecuadorian mammals. We expect that this and similar efforts will generate critical information for future genomic studies directed toward conservation and management efforts.
Conclusion
The brown-headed spider monkey (A. f. fusciceps) is a critically endangered primate species, facing multiple threats such as habitat loss and hunting, emphasizing the urgent need for conservation efforts. WGS has been identified as a crucial tool for managing threatened species. Here, we present the first WGS and assembly of A. f. fusciceps using long reads obtained through Oxford Nanopore Technologies, which resulted in a good-quality assembly. The genomic insights gained from this study provide valuable information, which can lead to the development of tools for the conservation of A. f. fusciceps. Moreover, the pipelines used in this study can serve as a foundation for sequencing and assembling genomes of other endangered species in developing nations, ultimately aiding in the preservation of global biodiversity.
Supplementary Material
Acknowledgments
We thank Carolina Sáenz and TUERI-USFQ for their valuable assistance in obtaining the sample of the sequenced individual. We also thank the members of the Plant Biotechnology Laboratory (USFQ) for their input and help during this research. Genetic data for the specimen were obtained under the Genetic Resources Permit Number: MAE-DNB-CM-2019-0126 granted to INABIO by Ministerio del Ambiente, Agua y Transición Ecológica in Ecuador, in accordance with Ecuadorian law.
Contributor Information
Gabriela Pozo, Laboratorio de Biotecnología Vegetal, Colegio de Ciencias Biológicas y Ambientales, Universidad San Francisco de Quito (USFQ), Quito 170901, Ecuador; Instituto Nacional de Biodiversidad (INABIO), Quito 170135, Ecuador.
Martina Albuja-Quintana, Laboratorio de Biotecnología Vegetal, Colegio de Ciencias Biológicas y Ambientales, Universidad San Francisco de Quito (USFQ), Quito 170901, Ecuador.
Lizbeth Larreátegui, Laboratorio de Biotecnología Vegetal, Colegio de Ciencias Biológicas y Ambientales, Universidad San Francisco de Quito (USFQ), Quito 170901, Ecuador.
Bernardo Gutiérrez, Laboratorio de Biotecnología Vegetal, Colegio de Ciencias Biológicas y Ambientales, Universidad San Francisco de Quito (USFQ), Quito 170901, Ecuador; Department of Biology, University of Oxford, Oxford OX1 3SZ, UK.
Nathalia Fuentes, Proyecto Washu/Fundación Naturaleza y Arte, Quito 170521, Ecuador.
Felipe Alfonso-Cortés, Proyecto Washu/Fundación Naturaleza y Arte, Quito 170521, Ecuador.
Maria de Lourdes Torres, Laboratorio de Biotecnología Vegetal, Colegio de Ciencias Biológicas y Ambientales, Universidad San Francisco de Quito (USFQ), Quito 170901, Ecuador; Instituto Nacional de Biodiversidad (INABIO), Quito 170135, Ecuador.
Data availability
The raw reads, genome assembly, and annotation can be found at GSA figshare: https://doi.org/10.25387/g3.24076638. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAZHEH000000000. The version described in this paper is version JAZHEH010000000. ONT long-read raw sequences have been deposited in the NCBI Sequence Read Archive database under BioProject PRJNA1009451. The script used for assembly and annotation is described in protocol.io at the following https://doi.org/10.17504/protocols.io.6qpvr3892vmk/v1.
Supplemental material available at G3 online.
Funding
This project was funded by ORG.one, a pilot project that is part of Oxford Nanopore Technologies, and by the Fondos COCIBA grant provided by Universidad San Francisco de Quito (USFQ).
Literature cited
- Ahmad SF, Singchat W, Jehangir M, Suntronpong A, Panthum T, Malaivijitnond S, Srikulnath K. 2020. Dark matter of primate genomes: satellite DNA repeats and their evolutionary dynamics. Cells. 9(12):2714. doi: 10.3390/cells9122714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhakami H, Mirebrahim H, Lonardi S. 2017. A comparative evaluation of genome assembly reconciliation tools. Genome Biol. 18(1):93. doi: 10.1186/s13059-017-1213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astashyn A, Tvedte ES, Sweeney D, Sapojnikov V, Bouk N, Joukov V, Mozes E, Strope PK, Sylla PM, Wagner L, et al. 2023. Rapid and sensitive detection of genome contamination at scale with FCS-GX. bioRxiv 543519. 10.1101/2023.06.02.543519, preprint: not peer reviewed. [DOI]
- Bateman A, Martin MJ, Orchard S, Magrane M, Agivetova R, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R, Bursteinas B, et al. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49(D1):D480–D489. doi: 10.1093/NAR/GKAA1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancalion PHS, Niamir A, Broadbent E, Crouzeilles R, Barros FSM, Zambrano AMA, Baccini A, Aronson J, Goetz S, Reid JL, et al. 2019. Global restoration opportunities in tropical rainforest landscapes. Sci Adv. 5(7):eaav3223. doi: 10.1126/sciadv.aav3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandies P, Peel E, Hogg CJ, Belov K. 2019. The value of reference genomes in the conservation of threatened species. Genes (Basel). 10(11):846. doi: 10.3390/GENES10110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TA. 2002. The human genome. In: Genomes. 2nd ed. Oxford: Wiley-Liss. p. Chapter 1. https://www.ncbi.nlm.nih.gov/books/NBK21134/ [Google Scholar]
- Calle-Rendón BR, Peck M, Bennett SE, Morelos-Juarez C, Alfonso F. 2016. Comparison of forest regeneration in two sites with different primate abundances in Northwestern Ecuador. Rev Biol Trop. 64(2):493–506. doi: 10.15517/rbt.v64i2.18415. [DOI] [PubMed] [Google Scholar]
- Campbell MS, Holt C, Moore B, Yandell M. 2014. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinformatics. 48(1):4.11.1–4.11.39. doi: 10.1002/0471250953.BI0411S48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18(1):188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celi JE, Villamarín F. 2020. Freshwater ecosystems of Mainland Ecuador: diversity, issues and perspectives. Acta Limnol Brasil. 32:e106. doi: 10.1590/S2179-975X3220. [DOI] [Google Scholar]
- Critical Ecosystem Partner Fund, Chocó-Darién-Western Ecuador: Chocó-Manabí Conservation Corridor Briefing Book 2005 . Prepared for: Improving Linkages Between CEPF and World Bank Operations, Latin America Forum, Rio de Janeiro, Brazil—January 24–25, 2005. Available from https://www.cepf.net/sites/default/files/final.chocodarienwesternecuador.chocomanabi.briefingbook.pdf.
- Culibrk L, Leelakumari S, Tse K, Cheng D, Chuah E, Kirk H, Pandoh P, Troussard A, Zhao Y, Mungall A, et al. 2019. The genome of the tufted capuchin (Sapajus apella). NCBI. [accessed 2023 May 24]. https://www.ncbi.nlm.nih.gov/nuccore/WRPQ00000000.1/ [Google Scholar]
- Dainat J. 2020. Another Gff analysis toolkit to handle annotations in any GTF/GFF format (v0.8.0). Zenodo. [cited 2023 Jun 19]. doi: 10.5281/zenodo.3552717 [DOI] [Google Scholar]
- De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 34(15):2666–2669. doi: 10.1093/BIOINFORMATICS/BTY149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore A, Link A, Dew JL. 2008. Spider Monkeys. In: Christina C, editor. Cambridge: Cambridge University Press. p. 81–137. doi: 10.1017/CBO9780511721915.004. [DOI] [Google Scholar]
- Eilbeck K, Moore B, Holt C, Yandell M. 2009. Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics. 10(1):67. doi: 10.1186/1471-2105-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg JF. 1973. Reproduction in two species of spider monkeys, Ateles fusciceps and Ateles geoffroyi. J Mammal. 54(4):955–957. doi: 10.2307/1379089. [DOI] [PubMed] [Google Scholar]
- Fantini LI, Jeffery NW, Pierossi P, Ryan Gregory T, Nieves M. 2016. Qualitative and quantitative analysis of the genomes and chromosomes of spider monkeys (Primates: Atelidae). [Cited 2023 Jul 12]. Available from www.genomesize.com. [Google Scholar]
- Fedigan LM, Rose LM. 1995. Interbirth interval variation in three sympatric species of neotropical monkey. Am J Primatol. 37(1):9–24. doi: 10.1002/ajp.1350370103. [DOI] [PubMed] [Google Scholar]
- Feng L, Lin H, Kang M, Ren Y, Yu X, Xu Z, Wang S, Li T, Yang W, Hu Q. 2022. A chromosome-level genome assembly of an alpine plant Crucihimalaya lasiocarpa provides insights into high-altitude adaptation. DNA Res. 29(1):dsac004. doi: 10.1093/DNARES/DSAC004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 117(17):9451–9457. doi: 10.1073/PNAS.1921046117/SUPPL_FILE/PNAS.1921046117.SAPP.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. 2003. Genetics and conservation biology. C R Biol. 326(Suppl 1):22–29. doi: 10.1016/S1631-0691(03)00023-4. [DOI] [PubMed] [Google Scholar]
- Gallo-Viracocha F, Urgilés-Verdugo C, Fuentes N, Alfonso-Cortes F, Zurita-Arthos L, Torres TC, Tirira DG. 2022. Distribution, conservation, and vulnerability to climate change of the Ecuadorian Brown-headed Spider Monkey (Primates: Atelidae). Mammalia Aequatorialis. 4:39–52. doi: 10.59763/mam.aeq.v4i.50. [DOI] [Google Scholar]
- Garber PA, Estrada A, Pavelka MSM. 2006. In: Estrada A, Garber PA, Pavelka MSM, Luecke L, editors. New Perspectives in the Study of Mesoamerican Primates. Developments in Primatology: Progress and Prospects. Boston (MA): Springer. p. 563–584. doi: 10.1007/0-387-25872-8_27. [DOI] [Google Scholar]
- Haddad NM, Brudvig LA, Clobert J, Davies KF, Gonzalez A, Holt RD, Lovejoy TE, Sexton JO, Austin MP, Collins CD, et al. 2015. Habitat fragmentation and its lasting impact on Earth's ecosystems. Sci Adv. 1(2):e1500052. doi: 10.1126/sciadv.1500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead MM, Islas-Trejo A, Goszczynski DE, Medrano JF, Zhou H, Ross PJ. 2021. Large-scale multiplexing permits full-length transcriptome annotation of 32 bovine tissues from a single nanopore flow cell. Front Genet. 12:664260. doi: 10.3389/fgene.2021.664260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AR, Raveendran M, Abee C, Williams L, Simmons J, Brady AG, Rui JC, Doddapaneni H, Muzny DM, Meng Q, et al. 2020. Saimiri boliviensis boliviensis breed Bolivian squirrel monkey isolate 100643, whole genome shotgun sequencing project. Nucleotide-NCBI. [Cited 2023 May 24]. Available from https://www.ncbi.nlm.nih.gov/nuccore/1955098031 [Google Scholar]
- Helmy M, Awad M, Mosa KA. 2016. Limited resources of genome sequencing in developing countries: challenges and solutions. Appl Transl Genomics. 9:15–19. doi: 10.1016/J.ATG.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotaling S, Kelley JL, Frandsen PB. 2021. Toward a genome sequence for every animal: where are we now? Proc Natl Acad Sci U S A. 118(52):e2109019118. doi: 10.1073/PNAS.2109019118/-/DCSUPPLEMENTAL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakumar V, Nishimura O, Kadota M, Hirose N, Sano H, Murakawa Y, Yamamoto Y, Nakaya M, Tsukiyama T, Seita Y, et al. 2021. Chromosomal-scale de novo genome assemblies of cynomolgus macaque and common marmoset. Sci Data. 8(1):159. doi: 10.1038/s41597-021-00935-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Wang S, Wang H, Wang A, Xu D, Liu H, Yang B, Yuan L, Lei L, Chen R, et al. 2022. A chromosome-level reference genome of a Convolvulaceae species Ipomoea cairica. G3 (Bethesda). 12(9):jkac187. doi: 10.1093/G3JOURNAL/JKAC187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics. 30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Stinson SA, diCenzo GC. 2023. Whole genome assemblies of Zophobas morio and Tenebrio molitor. G3 (Bethesda). 13(6):jkad079. doi: 10.1093/G3JOURNAL/JKAD079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny NJ, Francis WR, Rivera-Vicéns RE, Juravel K, de Mendoza A, Díez-Vives C, Lister R, Bezares-Calderón LA, Grombacher L, Roller M, et al. 2020. Tracing animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia muelleri. Nat Commun. 11(1):3676. doi: 10.1038/s41467-020-17397-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner F, Ferdy J-B, Andalo C, Colas B, Moret J. 2003. Role of corridors in plant dispersal: an example with the endangered Ranunculus nodiflorus. Conserv Biol. 17(2):401–410. doi: 10.1046/j.1523-1739.2003.01392.x. [DOI] [Google Scholar]
- Kleinman-Ruiz D, Martínez-Cruz B, Soriano L, Lucena-Perez M, Cruz F, Villanueva B, Fernández J, Godoy JA. 2017. Novel efficient genome-wide SNP panels for the conservation of the highly endangered Iberian lynx. BMC Genomics. 18(1):556. doi: 10.1186/s12864-017-3946-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37(5):540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics. 5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen Y, Mu D, Yuan J, Shi Y, Zhang H, Gan J, Li N, Hu X, Liu B.. 2012. Comparison of the two major classes of assembly algorithms: overlap-layout-consensus and de-bruijn-graph. Briefings in Functional Genomics. 11(1):25–37. 10.1093/bfgp/elr035. [DOI] [PubMed] [Google Scholar]
- Lin B, Hui J, Mao H. 2021. Nanopore technology and its applications in gene sequencing. Biosensors (Basel). 11(7):214. doi: 10.3390/bios11070214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wu S, Li A, Ruan J. 2021. SMARTdenovo: a de novo assembler using long noisy reads. Gigabyte. 2021:gigabyte15. doi: 10.46471/gigabyte.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Zdobnov EM. 2021. BUSCO: assessing genomic data quality and beyond. Curr Protoc. 1(12):e323. doi: 10.1002/CPZ1.323. [DOI] [PubMed] [Google Scholar]
- Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. 2018. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics. 34(13):i142–i150. doi: 10.1093/BIOINFORMATICS/BTY266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton K. 1981. Estimates of reproductive parameters for free-ranging Ateles geoffroyi. Primates. 22(4):574–579. doi: 10.1007/BF02381250. [DOI] [Google Scholar]
- Mittermeier RA, Myers N, Mittermeier CG. 2002. Hotspots: Earth's biologically richest and most endangered terrestrial ecoregions. J. Mammal. 83(2):630–633. doi: 10.1644/1545-1542(2002)0832.0.CO;2. [DOI] [Google Scholar]
- Morelos-Juárez C, Tapia A, Cervera L, Alfonso-Cortes F, Fuetnes N, Araguillin E, Zapata-Ríos G, Spaan D, Peck MR. 2018. Distribución actual, ecología y estrategias para la conservación de un primate críticamente amenazado (Ateles fusciceps fusiceps) en el Ecuador. In: Urbani B, Kowalewski M, Teixeira da Cunha RG, de la Torre S. Cortés-Ortiz L, editors. La Primatología en Latinoamérica 2 - A primatologia na America Latina 2. Caracas, Venezuela: Ediciones IVIC. Instituto Venezolano de Investigaciones Cientìficas (IVIC). p. 441–452. https://www.asp.org/research/PrimLatam%202-%20T.%20II%20-C.R-Vzla.pdf. [Google Scholar]
- Morisse P, Marchet C, Limasset A, Lecroq T, Lefebvre A. 2021. Scalable long read self-correction and assembly polishing with multiple sequence alignment. Sci Rep. 11(1):761. doi: 10.1038/s41598-020-80757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso P, Link A, de la Torra S, Shanee S, Cortes-Ortíz L. 2021. Ateles fusciceps ssp. fusciceps (brown-headed spider monkey). The IUCN Red List of Threatened Species. [cited 2021 Sep 16]. Available from https://www.iucnredlist.org/species/39922/191687911 [Google Scholar]
- Myers N, Mittermeler RA, Mittermeler CG, Da Fonseca GAB, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature. 403(6772):853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Nong W, Qu Z, Li Y, Barton-Owen T, Wong AYP, Yip HY, Lee HT, Narayana S, Baril T, Swale T, et al. 2021. Horseshoe crab genomes reveal the evolution of genes and microRNAs after three rounds of whole genome duplication. Commun Biol. 4(1):83. doi: 10.1038/s42003-020-01637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Hume DA. 2003. A guide to the mammalian genome: figure 1. Genome Res. 13(6b):1267–1272. doi: 10.1101/gr.1445603. [DOI] [Google Scholar]
- Petersen C, Sørensen T, Westphal KR, Fechete LI, Sondergaard TE, Sørensen JL, Nielsen KL. 2022. High molecular weight DNA extraction methods lead to high quality filamentous ascomycete fungal genome assemblies using Oxford Nanopore sequencing. Microb Genom. 8(4):000816. doi: 10.1099/MGEN.0.000816/CITE/REFWORKS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenninger M, Reuss F, Kiebler A, Schönnenbeck P, Caliendo C, Gerber S, Cocchiararo B, Reuter S, Blüthgen N, Mody K, et al. 2021. Genomic basis for drought resistance in European beech forests threatened by climate change. eLife. 10:e65532. doi: 10.7554/eLife.65532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan A, Antonaros F, Vitale L, Strippoli P, Pelleri MC, Caracausi M. 2019. Human protein-coding genes and gene feature statistics in 2019. BMC Res Notes. 12(1):315. doi: 10.1186/S13104-019-4343-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrelli S, Gerbault-Seureau M, Rozzi R, Chikhi R, Curaudeau M, Ropiquet A, Hassanin A. 2022. Draft genome of the lowland anoa (Bubalus depressicornis) and comparison with buffalo genome assemblies (Bovidae, Bubalina). G3 (Bethesda). 12(11):jkac234. doi: 10.1093/g3journal/jkac234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W-H, Yan C, Li W-J, Jiang X-M, Li G-Z, Zhang X-Y, Hu T-Z, Li J, Yue B-S. 2016. Distinct patterns of simple sequence repeats and GC distribution in intragenic and intergenic regions of primate genomes. Aging. 8(11):2635–2654. doi: 10.18632/aging.101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rang FJ, Kloosterman WP, de Ridder J. 2018. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 19(1):90. doi: 10.1186/S13059-018-1462-9/FIGURES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera Román ES. 2017. Filogeografía del mono araña de cabeza café (Ateles fusciceps fusciceps) en el Ecuador. Universidad Central del Ecuador. [cited 2021 Oct 14]. Available from https://www.dspace.uce.edu.ec/bitstream/25000/11043/1/T-UCE-0016-009.pdf [Google Scholar]
- Robinson JC, Janson CH. 1986. Capuchins, squirrel monkeys, and atelines: socioecological convergence with old world pritnates. In: Smuts BB, Cheney DL, Seyfarth RM, Wrangham RW, editors. Primate Societies. Chicago: University of Chicago Press. p. 69–82. doi: 10.7208/9780226220468-009. [DOI] [Google Scholar]
- Rogers J, Gibbs RA. 2014. Comparative primate genomics: emerging patterns of genome content and dynamics. Nat Rev Genet. 15(5):347–359. doi: 10.1038/NRG3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder OA. 2005. Conservation genomics: applying whole genome studies to species conservation efforts. Cytogenet Genome Res. 108(1–3):6–15. doi: 10.1159/000080796. [DOI] [PubMed] [Google Scholar]
- Saenko SV, Groenenberg DSJ, Davison A, Schilthuizen M. 2021. The draft genome sequence of the grove snail Cepaea nemoralis. G3 (Bethesda). 11(2):jkaa071. doi: 10.1093/g3journal/jkaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saremi NF, Supple MA, Byrne A, Cahill JA, Coutinho LL, Dalén L, Figueiró HV, Johnson WE, Milne HJ, O’Brien SJ, et al. 2019. Puma genomes from North and South America provide insights into the genomic consequences of inbreeding. Nat Commun. 10(1):4769. doi: 10.1038/s41467-019-12741-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y. 2022. Ateles geoffroyi isolate KIZ-2021_1, whole genome shotgun sequencing project. Nucleotide-NCBI. [cited 2022 Mar 24]. Available from https://www.ncbi.nlm.nih.gov/nuccore/JAKFHY000000000 [Google Scholar]
- Sierra R, Calva O, Guevara A. La Deforestación en el Ecuador, 1990-2018. Factores promotores y tendencias recientes. Ministerio de Ambiente y Agua del Ecuador, Ministerio de Agricultura del Ecuador, en el marco de la implementación del Programa Integral Amazónico de Conservación de Bosques y Producción Sostenible. Quito, Ecuador. p. 216. [Cited 2021 Oct 24]. Available from https://www.proamazonia.org/wp-content/uploads/2021/06/Deforestacio%CC%81n_Ecuador_com2.pdf [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Smith A, Hubley R, Green P. 2013-2015. RepeatMasker Open-4.0. http://www.repeatmasker.org
- Sork VL, Fitz-Gibbon ST, Puiu D, Crepeau M, Gugger PF, Sherman R, Stevens K, Langley CH, Pellegrini M, Salzberg SL. 2016. First draft assembly and annotation of the genome of a California endemic oak Quercus lobata Née (Fagaceae). G3 (Bethesda). 6(11):3485–3495. doi: 10.1534/g3.116.030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RS, Manseau M, Redquest B, Keobouasone S, Gagne P, Martineau C, Wilson PJ. 2022. Whole genome sequences from non-invasively collected caribou faecal samples. Conserv Genet Resour. 14:53–68. doi: 10.1007/s12686-021-01235-2. [DOI] [Google Scholar]
- Theissinger K, Fernandes C, Formenti G, Bista I, Berg PR, Bleidorn C, Bombarely A, Crottini A, Gallo GR, Godoy JA, et al. 2023. How genomics can help biodiversity conservation. Trends Genet. 39(7):545–559. doi: 10.1016/j.tig.2023.01.005. [DOI] [PubMed] [Google Scholar]
- Tirira DG, de la Torre S, Ríos GZ. 2018. Plan de acción para Estudio, de los primates del Ecuador. Ministerio del Ambiente del Ecuador (MAE)/Grupo de estudio de Primates del Ecuador (GEPE)/Asociación Ecuatoriana de Mastozoología (AEM). Quito.
- Tirira DG, Fuentes N, Alfonso-Cortes F, Morelos-Juárez C, Méndez-Carvajal PG, Gutiérrez-Pineda KM, Montilla S, Hernández-Jaramillo A, Morales-Jiménez A, et al. 2022. In: Mittermeier RA, Reuter KA, Rylands AB, Jerusalinsky L, Schwitzer C, Strier KB, Ratsimbazafy J, et al., editors. Primates in Peril: The World's 25 Most Endangered Primates 2022-2023. Washington (DC): IUCN SSC Primate Specialist Group, International Primatological Society. p. 127–132. https://cdn.www.gob.pe/uploads/document/file/3574458/Primates_in_Peril_2022_2023.pdf.pdf. [Google Scholar]
- Wang Y, Zhao Y, Bollas A, Wang Y, Au KF. 2021. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol. 39(11):1348–1365. doi: 10.1038/s41587-021-01108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren W, Ye L, Minx P, Worley K, Gibbs R, Wilson RK. 2009. Proteomes· Callithrix jacchus (White-tufted-ear marmoset). UniProt. [Cited 2022 May 24]. Available from https://www.uniprot.org/proteomes/UP000008225 [Google Scholar]
- Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Holt KE. 2021. Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Res. 8:2138. doi: 10.12688/f1000research.21782.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom. 3(10):e000132. doi: 10.1099/MGEN.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano-Mila MS, Agathos SN, Reichardt JKV. 2019. Human genetics and genomics research in Ecuador: historical survey, current state, and future directions. Hum Genomics. 13(1):64. doi: 10.1186/s40246-019-0249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw reads, genome assembly, and annotation can be found at GSA figshare: https://doi.org/10.25387/g3.24076638. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAZHEH000000000. The version described in this paper is version JAZHEH010000000. ONT long-read raw sequences have been deposited in the NCBI Sequence Read Archive database under BioProject PRJNA1009451. The script used for assembly and annotation is described in protocol.io at the following https://doi.org/10.17504/protocols.io.6qpvr3892vmk/v1.
Supplemental material available at G3 online.