Abstract
Parasitic nematodes are globally important and place a heavy disease burden on infected humans, crops, and livestock, while commonly administered anthelmintics used for treatment are being rendered ineffective by increasing levels of resistance. It has recently been shown in the model nematode Caenorhabditis elegans that the sensory cilia of the amphid neurons play an important role in resistance toward macrocyclic lactones such as ivermectin (an avermectin) and moxidectin (a milbemycin) either through reduced uptake or intertissue signaling pathways. This study interrogated the extent to which ciliary defects relate to macrocyclic lactone resistance and dye-filling defects using a combination of forward genetics and targeted resistance screening approaches and confirmed the importance of intraflagellar transport in this process. This approach also identified the protein trafficking pathways used by the downstream effectors and the components of the ciliary basal body that are required for effector entry into these nonmotile structures. In total, 24 novel C. elegans anthelmintic survival-associated genes were identified in this study. When combined with previously known resistance genes, there are now 46 resistance-associated genes that are directly involved in amphid, cilia, and intraflagellar transport function.
Keywords: Caenorhabditis elegans, macrocyclic lactone, ivermectin, moxidectin, anthelmintic resistance, xenobiotic resistance, amphids, ciliogenesis, intraflagellar transport
Introduction
Parasitic nematodes place a highly significant and heavy disease burden on infected plants and animals causing annual global yield and productivity losses in excess of $100 billion (Grisi et al. 2014; Singh et al. 2015) and, in addition, requiring over $20 billion annually to treat with anthelmintics (Abongwa et al. 2017). Currently available broad-spectrum anthelmintics are from a limited range of chemical families (Abongwa et al. 2017), and resistance to one or more classes is becoming widespread in field populations (Crook et al. 2016) jeopardizing food security and human health. Therefore, until new anthelmintic classes are developed, it is necessary to prolong the efficacy of existing drugs by finding ways to suppress resistance.
The macrocyclic lactones such as ivermectin (an avermectin) and moxidectin (a milbemycin) are the most commonly administered anthelmintics due to their low cost and high persistent efficacy (McArthur and Reinemeyer 2014); however, the rapid spread of resistance is beginning to render them ineffective (Crook et al. 2016). There has been an observed correlation between macrocyclic lactone resistance caused by reduced uptake and defects in amphid morphology in Caenorhabditis elegans with several causative genes being associated with dye-filling, chemosensation, osmosensation, dauer formation, and mechanosensation defective phenotypes (Dent et al. 2000; Urdaneta-Marquez et al. 2014; Page 2018). Amphid morphology and dye-filling defects have also been noted in field populations of Haemonchus contortus that are resistant to macrocyclic lactones (Freeman et al. 2003; Urdaneta-Marquez et al. 2014). Recently, it has been found that some amphid morphology-associated genes may also influence intestinal anthelmintic exporting P-glycoprotein expression in an NHR-8-dependent manner (Guerrero et al. 2021).
The amphid sensilla consist of 2 pairs of 12–13 neurons (12 in C. elegans), which have nonmotile cilia enriched in G protein-coupled receptors on the dendrites that are exposed to the environment through pores in the cuticle (Perkins et al. 1986; Brear et al. 2014; Vidal et al. 2018; Hong et al. 2019), and function as the primary sensory organ for environmental stimuli (chemical, ion and osmotic gradients, temperature, pheromones, and noxious compounds). Sensory inputs are processed by the nerve ring leading to output motor neuron-mediated responses (Schafer 2016; Cook et al. 2019; Hong et al. 2019) in a 200–300 neuron nervous system whose layout is highly conserved between nematode species. Ciliogenesis of sensory cilia utilizes assembly pathways that are conserved throughout Eukaryota where a centriole-derived basal body anchors to the cell membrane restricting the local diffusion of proteins and lipids and organizes microtubules (Sung and Leroux 2013). These microtubules are then used for the delivery of lipids and proteins to the growing cilia by intraflagellar transport (IFT) complexes that travel along the microtubules using dynein and kinesin motors (Rosenbaum and Witman 2002; Sung and Leroux 2013; Prevo et al. 2017).
Macrocyclic lactones function by paralyzing the central nervous system, which eventually leads to death, through interaction with multiple subunits of the glutamate-gated chloride channel primary target, as well as multiple secondary targets, thereby resulting in constitutive activation (Chen and Kubo 2018). As nematodes have limited capacity for phase I detoxification (functionalization and oxidation) of macrocyclic lactones (Vokřál et al. 2013; Yilmaz et al. 2019), resistance relies on increased phase II conjugation and efflux (James and Davey 2009), target site insensitivity, or reduced drug uptake (Dent et al. 2000). However, all identified and candidate resistance genes that interact directly with macrocyclic lactones or their metabolites function downstream of macrocyclic lactone uptake (Dent et al. 2000; James and Davey 2009; Ménez et al. 2016). The macrocyclic lactones lack the chemical properties that would allow them to spontaneously cross biological membranes (Escher et al. 2008) meaning that uptake is dependent on the ability of the biological systems of the organism to accumulate appropriate concentrations in the target tissues; however, the mechanism and associated genes involved in uptake are still unknown or poorly defined.
This current study uses a mechanistic approach to investigate cellular processes associated with previously discovered resistance genes, in combination with targeted resistance screens in C. elegans, to identify the roles played by anterograde and retrograde IFT in the ciliary distal segment of the amphid neurons in the resistance to macrocyclic lactones (ivermectin and moxidectin). Pathways involved in trafficking ciliary proteins to and from the ciliary gate of the basal body were also investigated, revealing that the UNC-101- and UNC-119-mediated secretion pathways and the polarizers of axon–dendrite protein sorting UNC-33 and UNC-44 are important components involved in macrocyclic lactone resistance, whereas the RAB-35 recycling pathway plays a downstream role. A whole-genome sequencing approach was applied to map candidates from a forward genetic screen for resistance to macrocyclic lactones, and in combination with a targeted resistance screen, 24 novel anthelmintic survival-associated genes were uncovered in C. elegans.
Methods
Chemicals
Suppliers and catalog numbers of all reagents used are listed in the Supplementary Methods.
Nematode strains
Putative orthologs of key basal body genes for which there was no primary literature were chosen using a combination of Protein BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the MARRVEL (Wang et al. 2017) and AceView (Thierry-Mieg and Thierry-Mieg 2006) databases.
TM prefixed strains were obtained from the National BioResource Project, Japan while all other strains used were purchased from the C. elegans Genetics Centre, USA. All strains were maintained on Escherichia coli OP50-1-inoculated Nematode Growth Medium (NGM) plates following standard protocols (http://www.wormbook.org/toc_wormmethods.html). Strains used in this study are listed in the Supplementary Methods.
Anthelmintic resistance assays
Anthelmintic stock solutions were prepared as follows: 10 µM ivermectin stock was made by the serial dilution of a 10 mM stock using DMSO as a solvent for both stocks; 10 µM moxidectin stock was prepared using the same procedure as ivermectin. Stock solutions were dispensed into 1-ml aliquots and stored at −20°C.
NGM plates containing anthelmintics were produced by adding volumes of anthelmintic stock solution to cooled molten NGM agar (50°C) before mixing and pouring onto 3-cm petri dishes. The volume of the anthelmintic stock solution added never exceeded 0.3% of the final volume. Anthelmintic plate concentrations used were 10 nM ivermectin and 5 and 10 nM moxidectin based on resistance threshold criteria used for gastrointestinal nematodes (Kaplan et al. 2007; Crook et al. 2016). Plates were inoculated with 50-µl OP50-1 24 h before starting assays.
To determine ivermectin and moxidectin resistance, survival assays were performed by picking 5 L4 worms of the strain to be tested onto each plate with 2 biological and 2 technical replicates. Growth and mortality were inspected every 48 h using a light microscope. A strain was considered resistant (+) if the F1 generation reached adulthood compared to susceptible strains (−), which showed paralysis and growth arrest and F1s failed to reach adulthood. The wild-type N2 strain was used as a susceptible negative control and DA1316 (ad1305; vu227; pk54) was used as a resistant positive control.
DiI dye-filling assays and microscopy
Worms were washed from populated plates using M9 buffer (3-g KH2PO4, 6-g Na2HPO4, 5-g NaCl, and 1 mM MgSO4/L) and collected in 1.5-ml eppendorfs. Samples were pelleted by centrifugation at 7,000 rpm for 10 s to allow the removal of the supernatant. Two washes with M9 were performed before applying 10-µg/ml DiI (1,1ʹ-dioctadecyl-3,3,3ʹ,3ʹ-tetramethylindocarbocyanine perchlorate) dye in M9 buffer for 30 min. Samples were then washed twice with M9 before incubating at 21°C for 2 h to allow worms to clear their gut of bacteria and dislodge DiI adhered to the cuticle before performing 2 more washes in M9. Worms were pelleted and supernatant removed before transfer to an empty petri dish using a pipette and then picking 20–30 specimens onto prepared microscope slides. Slides were coated with a pad of 2% agar with 1% sodium azide and wet with 10 µl of M9 containing 0.2% sodium azide, and then coverslips were sealed with a thin layer of petroleum jelly.
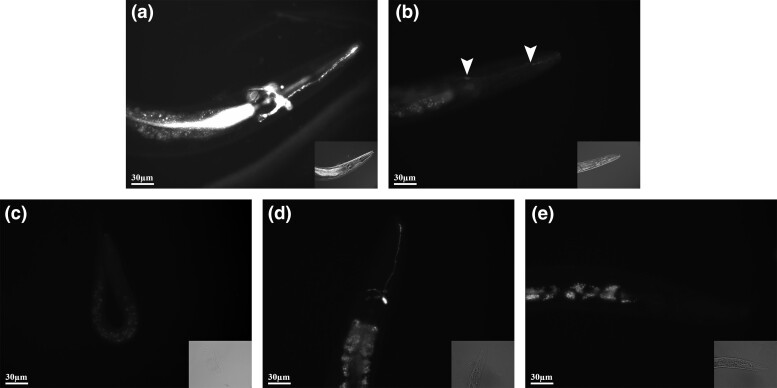
Slides were viewed using a Zeiss Axioskop 2 Plus microscope fitted with a Zeiss Mercury HBO 100 Lamphouse and Zeiss AxioCam camera with images taken using the accompanying AxioVision software. All images were taken at 250× magnification. Control images of worms were taken using a differential interference contrast (DIC) filter, 0.5-s exposure time, and the minimum setting for the internal light source while DiI staining was viewed and imaged using a fluorescein isothiocyanate (FITC) filter, 1-s exposure time, and illumination by the mercury lamp. A minimum of 10 individuals of each strain were observed under FITC conditions to score the average intensity of DiI staining (negative or abnormal dye filling [−] or positive [+]). Representative DIC and FITC images for DiI staining patterns in each category are shown in Fig. 1 while images for individual strains are available upon request.
Fig. 1.
Representative images of DiI phenotypes at 250× magnification. DiI, 1,1ʹ-dioctadecyl-3,3,3ʹ,3ʹ-tetramethylindocarbocyanine perchlorate. a) N2: DiI dye-filling positive, b) ifta-1(nx61): weak DiI dye-filling positive, c) dyf-2(m160): DiI dye-filling negative and c14h10.2(tm10737): novel Dyf mutant that has variable DiI dye filling with d) weak positive individuals in a predominantly e) negative population. Individuals were photographed using a DIC filter (lower right inset image) to highlight the position and orientation of the worm and a FITC filter (main image) to visualize fluorescence. Areas of fluorescence for weak phenotypes are highlighted with arrows.
EMS mutagenesis and whole-genome sequencing
C. elegans L4 stage N2 strain worms were exposed to 50 mM ethyl methanesulfonate (EMS) for 4 h at 20°C following standard mutagenesis procedures (Brenner 1974) and then allowed to recover on OP50-1-seeded NGM plates overnight. Worms were then handled according to Page (2018) selecting for 10 nM moxidectin resistance (see Supplementary Methods for details). Lines were then characterized for DiI dye-filling and ivermectin, albendazole, and levamisole cross-resistance.
From the 14 resulting moxidectin-resistant lines, 5 were selected, and together with uncharacterized ivermectin-resistant lines, TP236(ka30), TP241(ka35), TP272(ka64), and TP274(ka66) from a previous study (Page 2018) were processed for single-nucleotide polymorphism (SNP) mapping. SNP mapping was carried out as described in Doitsidou et al. (2010) using MiModD tools on the public instance of the Galaxy platform (https://usegalaxy.org) (Jalili et al. 2020) (see Supplementary Methods for details). Genomic DNA was extracted using a Gentra Puregene Core Kit A (Qiagen, UK) kit before cleanup and concentration using a Genomic DNA Clean & Concentrator-25 (Zymo Research, USA) kit. Samples were sent for whole-genome sequencing to the Glasgow Polyomics facility, University of Glasgow where libraries were prepared with a TruSeq Nano DNA LT Sample Prep Kit (Illumina), quality controlled on a 2100 Bioanalyzer (Agilent), and run on an Illumina MiSeq platform using 300-bp paired-end reads.
Results
Outcomes of dye-filling and macrocyclic lactone survival assays are listed in Table 1. Strains tested that did not show a phenotype of interest are included in Supplementary Table 1. The relationship between resistance, IFT, protein trafficking, cilia, and dye filling is summarized below.
Table 1.
Many C. elegans mutants for ciliary proteins are resistant to ivermectin and moxidectin.
| Gene (homology) | Strain (allele) | DYF | IVM R | MOX R |
|---|---|---|---|---|
| Transcription factor mutants (2/4) | ||||
| daf-19 (RFX transcription factor) | DR86(m86) | − | + | + |
| hlh-4 (achaete-scute transcription factor) | TM604(tm604) | − | + | + |
| Cell migration/adhesion defect mutants (2/6) | ||||
| dyf-7 (ZP protein) | SP1735(m537) | − | + | + |
| mec-8 (RRM domain/splice factor) | CB398(e398) | + | + | + |
| Amphid channel morphology mutants (1/2) | ||||
| daf-6 (PTCHD-1/4 ortholog) | CB1377(e1377) | − | + | + |
| Protein secretion/trafficking defect mutants (9/52) | ||||
| arl-13 (ARL13B ortholog) | TM1745(tm1745) | + | − | + |
| dyf-5 (map kinase) | SP1745(mn400) | − | + | + |
| dyf-18 (CDK-8/19/20 ortholog) | ET100(ok200) | − | − | + |
| osta-1 (SLC51A ortholog) | TM5255(tm5255) | + | − | + |
| rab-35 (RAB family) | RT206(b1013) | + | + | + |
| unc-33 (CRMP1 ortholog, filamin binding) | CB1193(e1193) | + | + | + |
| unc-44 (ANK2/ANK3 ortholog) | CB1197(e1197) | − | + | + |
| unc-101 (AP1M1 ortholog) | PS529(sy108) | − | + | + |
| unc-119 (HRG4 ortholog) | CB4845(e2498) | − | + | + |
| Ciliary nucleation and region identity mutants (8/44) | ||||
| c14h10.2 (JAKMIP3 ortholog, putative CEP123 ortholog) | TM10737(tm10737) | − | + | + |
| che-10 (rootelin, IFT) | CB3329(e1809) | − | + | + |
| che-12 (TOGARAM1 ortholog, IFT) | CB3332(e1812) | − | + | + |
| dyf-17 (MAGEL2 like) | EG175(ox175) | − | + | + |
| dyf-19 (FBF1 ortholog) | ZP541(jhu455) | − | + | + |
| hyls-1 (hydrolethalus syndrome ortholog) | TM3067(tm3067) | − | + | + |
| nphp-4 (nephrocystin 4 ortholog) | TM925(tm925) | − | + | + |
| yap-1 (WWTR1 ortholog, has CEP164 homology) | TM1416(tm1416) | + | − | + |
| Microtubule mutants (1/2) | ||||
| dyf-10 (α-tubulin homolog) | SP1709(e1383) | − | + | + |
| Dynein and kinesin motor mutants (4/22) | ||||
| che-3 (Dynein HC avr-1, IFT) | CB1124(e1124) | − | + | + |
| dhc-3 (Dynein HC, IFT) | TP239(ka33) | − | + | + |
| osm-3 (kinesin family, IFT) | PR802(p802) | − | + | + |
| xbx-1 (DYNC2LI1 ortholog) | JT11069(ok279) | − | + | + |
| IFT-A complex mutants (5/6) | ||||
| che-11 (IFT140 homolog, IFT) | CB3330(e1810) | − | + | + |
| daf-10 (IFT122A homolog, WD repeat, IFT) | CB1387(e1387) | − | + | + |
| dyf-2 (IFT144 homolog, WRD19, IFT) | SP1234(m160) | − | + | + |
| ift-43 (IFT43 homolog, IFT) | TM8137(tm8137) | + | − | + |
| ifta-1 (IFT122B homolog, WDR35, IFT) | MX124(nx61) | − | + | + |
| IFT-B complex mutants (13/16) | ||||
| che-2 (IFT80 homolog, G-protein, WD repeat) | CB1033(e1033) | − | + | + |
| che-13 (IFT57/Hippi) | CB3323(e1815) | − | + | + |
| dyf-1 (IFT70 homolog, IFT) | SP1205(mn335) | − | + | + |
| dyf-3 (IFT38 homolog, CLUAP protein, IFT) | SP1603(m185) | − | + | + |
| dyf-6 (IFT46 homolog, IFT) | SP1712(m175) | − | + | + |
| dyf-11 (IFT54 homolog, IFT) | SP1713(mn392) | − | + | + |
| dyf-13 (IFT56 homolog, IFT) | SP1678(mn396) | + | + | + |
| ift-20 (IFT20 homolog, IFT) | RB2353(ok3191) | − | + | + |
| ift-74 (IFT72/74 homolog, IFT) | VC2140(ok2866) | + | + | + |
| osm-1 (IFT172 homolog, WD repeat, IFT) | PR808(p808) | − | + | + |
| osm-1 (IFT172 homolog, WD repeat, IFT) | PR816(p816) | − | + | + |
| osm-5 (IFT88 homolog, polaris, IFT) | PR813(p813) | − | + | + |
| osm-6 (IFT52 homolog, IFT) | PR811(p811) | − | + | + |
| rab-28 (RAB family, IFT27 homolog) | RB2484(ok3424) | − | + | + |
| Bardet–Biedl syndrome complex mutants (6/10) | ||||
| bbs-1 (BBS1 ortholog, IFT) | VC837(ok1111) | − | + | + |
| bbs-2 (BBS2 ortholog, IFT) | VC1569(ok2053) | − | + | + |
| bbs-8 (BBS8 ortholog, TPR protein, IFT) | MX52(nx77) | − | + | + |
| bbs-9 (BBS9 ortholog, IFT) | VC1062(gk471) | − | + | + |
| k07c11.10 (BBS10 ortholog) | TM3304(tm3304) | + | + | + |
| osm-12 (bbs7, IFT) | MT3645(n1606) | − | + | + |
| IFT cargo mutants (1/14) | ||||
| osm-9 (TRPV5/6 family) | CX10(ky10) | + | + | + |
| OSM-9 interacting/associated proteins (1/12) | ||||
| npr-1 (NPY1R ortholog) | CX4148(ky13) | + | − | + |
| Other sensory mutants (3/10) | ||||
| inx-19 (innexin homolog) | CX6161(ky634) | − | + | + |
| unc-7 (innexin homolog) | CB5(e5) | + | + | + |
| unc-9 (innexin homolog) | CB101(e101) | + | + | + |
Numbers of positive hits under each category out of the total genes tested are in parentheses.
Underline: previously identified mutants that are resistant to one of the anthelmintics tested.
Dyf, DiI amphid dye filling; IVM R, ivermectin resistance; MOX R, moxidectin resistance; +, dye filling identical to the wild type/resistant (includes weak resistance); −, dye filling defective (includes weak dye filling)/susceptible; IFT, intraflagellar transport component homology.
IFT complex subunits
Of the previously 34 identified ivermectin resistance genes (Dent et al. 2000; Urdaneta-Marquez et al. 2014; Page 2018), 16 encode for proteins of the IFT-A complex, IFT-B complex, and the BBSome, all of which are interacting multiprotein complexes involved in IFT (Rosenbaum and Witman 2002; Prevo et al. 2017). Orthologs of the remaining 14 known, but untested, subunits of these complexes and an ortholog of the chaperone protein BBS10, were investigated for anthelmintic resistance. Out of the 15 genes tested, mutant alleles for 8 showed resistance to ivermectin (Table 1). Within the IFT-A complex mutants, the IFTA-1 dynein-interacting protein mutant was found to be strongly resistant to ivermectin (Table 1), while mutants for the dynein-loading proteins IFT-43 and IFT-139 remained susceptible (Supplementary Table 1). From the IFT-B complex mutants, the Golgi vesicle sorting protein IFT-20 and the tubulin delivery protein IFT-74 mutants were only weakly resistant to ivermectin whereas the IFT27 ortholog RAB-28 mutant was highly resistant (Table 1). Mutants for the core BBSome proteins BBS-2 and BBS-9 and the BBS10 ortholog K07C11.10 all displayed strong ivermectin resistance (Table 1) while those for the cargo adapter subunits BBS-4 and BBS-5 were susceptible (Supplementary Table 1).
Known IFT cargoes
As the primary function of IFT is the delivery of ciliary proteins, genes for known IFT cargo proteins were tested for ivermectin resistance to identify downstream effectors of resistance. Of the 14 cargo protein-encoding genes tested, only the CX10(ky10) mutant of osm-9 was found to exhibit resistance (Table 1); however, this finding was not replicated with the VC1262(ok1677) and JY190(yz6) osm-9 mutant strains (Supplementary Table 1) indicating that perhaps resistance is caused by an unrelated, uncharacterized, mutation in the CX10(ky10) strain. The ciliary membrane protein cargo adaptor tub-1(ok1972) mutant was found to be susceptible to ivermectin (Supplementary Table 1), supporting the hypothesis that the downstream effector for ivermectin resistance must be delivered by another secretion pathway.
Protein trafficking pathways
To gain insight into the trafficking of the downstream effectors for ivermectin resistance, known ciliary protein secretion pathways upstream of the IFT and ciliary membrane protein removal pathways were investigated. The clathrin adapter protein-1 ortholog involved in Golgi vesicle secretion UNC-101, the CRMP1 ortholog involved in polarizing axon–dendrite sorting UNC-33, the ANK2/ANK3 ortholog involved in polarizing axon–dendrite sorting UNC-44, UNC-119 that inserts myristoylated proteins into the cell membrane, and RAB-35 that regulates early endosome recycling were all involved in causing ivermectin resistance when mutated (Table 1). Mutants for the 2 SNAP25 family protein-encoding genes aex-4 and ric-4 were found to be susceptible (Supplementary Table 1), supporting the contention that the downstream effector, which causes ivermectin resistance when absent, must be delivered via vesicle fusion using the essential SNAP-29 protein. All the genes so far tested that are involved in endocytosis, designation to lysosomal degradation, early endosome maturation, extracellular vesicle formation, synaptic vesicle fusion, and other post-Golgi transport complexes did not confer ivermectin resistance (Supplementary Table 1). Intriguingly, mutants for the RAB-8 and RAB-10 exocytosis regulators, which have roles in crossing the ciliary gate, were likewise susceptible to this drug (Supplementary Table 1).
Dyneins and kinesins play an important role in protein trafficking and IFT with osm-3(p802), che-3(e1124), and dhc-3(ka33) already being associated with ivermectin resistance (Dent et al. 2000; Page 2018); therefore, additional members of these families were investigated. Of the 20 genes tested, only mutations in the dynein light-intermediate chain xbx-1 resulted in ivermectin resistance (Table 1). Mutant alleles for all 3 genes encoding the IFT heterotrimeric kinesin (kap-1, klp-11, and klp-20) and the axonal kinesin unc-104 had no impact on ivermectin resistance (Supplementary Table 1).
The ciliary gate
The ciliary gate of the basal body acts as a physical barrier at the base of the cilia that selectively allows the passage of ciliary proteins. Components of the ciliary gate (some putative) were therefore investigated to uncover those required to deliver downstream effectors associated with ivermectin resistance. The MAGEL2-like protein DYF-17, the distal appendage-interacting subunit of the basal body HYLS-1, the FBF1 ortholog DYF-19, the transition fiber subunit NPHP-4, and the JAKMIP3 ortholog with CEP123 homology C14H10.2 were all found to be involved in maintaining ivermectin susceptibility (Table 1) although some nphp-4(tm925) individuals showed incomplete penetrance of the resistance phenotype. Mutants for all other transition fiber genes, putative subdistal appendage proteins, putative ESCRT complex, exocyst vesicle, TRAPP complex, and Rab family-interacting basal body subunits and orthologs of the ARMC9/TOGARAM1 complex were all tested and found to have no impact on ivermectin resistance (Supplementary Table 1).
Cell migration, amphid formation, ciliogenesis, and ciliated neuron-enriched genes tested
As gross morphological defects to amphid neurons, their cilia, and the amphid channel invariably cause ivermectin resistance, some transcription factors that determine amphid neuron cell fate and the proteins involved in axon guidance and lumen formation were assessed for a role in ivermectin resistance. Of the 5 genes tested, only mutant alleles for the ADL neuron determining transcription factor hlh-4 and the lumen endocytosis regulator daf-6 were found to cause resistance to ivermectin (Table 1).
Some genes involved in gap junction formation (unc-7 and unc-9), mechanosensation (mec-1 and mec-8), and osmotic avoidance (osm-1, osm-3, osm-5, osm-6, and osm-12) have been reported to cause ivermectin resistance (Dent et al. 2000; Page 2018), so additional genes in those phenotype categories along with several cilium-enriched membrane proteins (Blacque et al. 2005; Kunitomo et al. 2005) were likewise investigated. Of the genes from this grouping that have been tested, only the gap junction innexin inx-19(ky634) mutant displayed resistance to both ivermectin and moxidectin (Table 1) while no resistance was observed in both mec-1 strains tested (Supplementary Table 1).
Amphidal dye-filling defect correlation with ivermectin resistance
It has previously been found that there is a correlation between ivermectin resistance and dye-filling defects (Page 2018), so the full extent of this relationship was examined. Of previously known ivermectin resistance genes, the mutant alleles daf-19(m86), dyf-7(m537), che-12(e1812), dyf-10(e1383), che-3(e1124), dhc-3(ka33), osm-3(p802), che-11(e1810), daf-10(e1387), dyf-2(m160), che-2(e1033), che-13(e1815), dyf-1(mn335), dyf-3(m185), dyf-6(m175), dyf-7(m537), dyf-10(e1383), dyf-11(mn392), osm-1(p808), osm-1(p816), osm-3(p802), osm-5(p813), and osm-6(p811) were dye-filling negative; bbs-8(nx77), che-10(e1809), bbs-1(ok1111), osm-12(n1606), and dyf-5(mn400) exhibited weak dye filling (Table 1); che-1(p672), che-1(ot75), che-6(e1126), dyf-13(mn396), mec-1(e1066), mec-8(e398), unc-7(e5), and unc-9(e101) were dye-filling positive (Supplementary Table 1); and che-14(e1960) exhibited highly variable degrees of dye filling between individuals. Among the novel ivermectin resistance genes identified in the present study, the mutant alleles unc-101(sy108), daf-6(e1377), ift-20(ok3191), rab-28(ok3424), bbs-2(ok2053), dyf-19(jhu455), and inx-19(ky634) were all dye-filling negative; hlh-4(tm604), unc-119(e2498), hyls-1(tm3067), nphp-4(tm925), ifta-1(nx61), and bbs-9(gk471) displayed weak dye filling (Table 1); rab-35(b1013), unc-33(e1193), ift-74(ok2866), and k07c11.10(tm3304) were dye-filling positive (Table 1); and c14h10.2(tm10737), dyf-17(ox175), unc-44(e1197), and xbx-1(ok279) had highly variable degrees of dye filling between individuals, with c14h10.2(tm10737), dyf-17(ox175), and unc-44(e1197) being predominantly dye-filling negative (Table 1). The tag-278(gk382) mutant also showed highly variable degrees of dye filling between individuals but showed no resistance to any of the tested anthelmintics (Supplementary Table 1).
Processes that are essential for ciliogenesis and cilium maintenance showed a strong correlation between the extent of dye-filling defects and the strength of ivermectin resistance although mec-8(e398), hyls-1(tm3067), ifta-1(nx61), and bbs-9(gk471) defied the trend by showing strong resistance despite having weak dye filling. Mutants for proteins that are involved in trafficking ciliary membrane proteins along the axon such as UNC-33 and proteins that function downstream of IFT, including RAB-35 and helper/regulatory proteins like K07C11.10, showed no correlation. This indicates that although DiI dye filling and ivermectin susceptibility require effector delivery to the cilia through shared pathways, both processes do not necessarily use the same effector.
Observed cross-resistances to moxidectin
Candidate genes were also tested for moxidectin (a milbemycin) resistance to examine possible cross-resistance. Mutants for all genes that were ivermectin resistant were also resistant to moxidectin, indicating as expected, shared mechanisms and also similar levels of resistance to the 2 drugs. Mutants for the kinase DYF-18 that plays a role in ciliogenesis and IFT, a regulator of ciliary protein trafficking OSTA-1, the small GTPase nucleotide exchange factor involved in ciliogenesis ARL-13, a WWTR1 ortholog with CEP164 homology YAP-1, and the IFT-A complex dynein-loading protein IFT-43, however, showed moxidectin resistance but not ivermectin resistance (Table 1).
Whole-genome sequencing of mutants from forward genetic screens
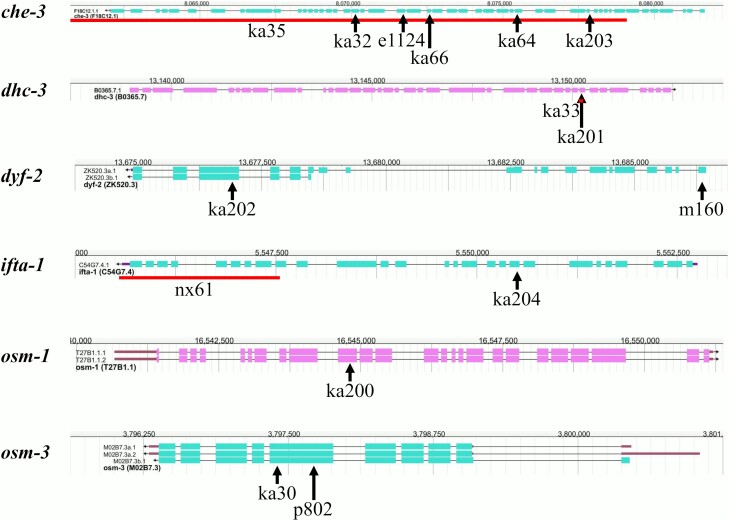
Extensive EMS genetic screens for ivermectin- and abamectin-resistant mutants were carried out previously and mapping of 2 mutants identified 2 IFT-related mutants (che-3(ka32) and dhc-3(ka33)) (Page 2018). In this current study, a new forward genetic screening to identify moxidectin-resistant strains was performed. Together, these screens identified 31 mutants resistant to macrocyclic lactones, which also had their DiI dye-filling phenotypes characterized (Table 2). Based on phenotype, TP236(ka30), TP241(ka35), TP272(ka64), and TP274(ka66) from the previous abamectin screen (Page 2018) along with TP375(ka200), TP378(ka201), TP384(ka202), TP386(ka203), and TP388(ka204) from the current moxidectin screen were selected for backcrossing, whole-genome sequencing, and SNP mapping. Of the selected strains, all were resistant to ivermectin and moxidectin. TP388(ka204) was dye-filling positive while all others were dye-filling negative. The whole-genome sequencing and mapping data (aligned reads available at https://www.ncbi.nlm.nih.gov/sra/PRJNA768320) identified novel alleles of osm-3, che-3 (4 different alleles), osm-1, dhc-3, dyf-2, and ifta-1 (Fig. 2) as the causative genes for resistance to macrocyclic lactones. Details of identified alleles are listed in Supplementary Table 2.
Table 2.
Resistance profiles and causative genes for resistance to macrocyclic lactones in EMS-generated mutant strains.
| Strain (selection screen used for isolation) | Assigned allele | Causal gene | Mutation/effect | DYF | IVM R | MOX R |
|---|---|---|---|---|---|---|
| TP236 (10 nM ivermectin) | ka30 | osm-3 | Substitution/nonsense | − | + | + |
| TP241 (50 nM abamectin) | ka35 | che-3 | Deletion/coding | − | + | + |
| TP272 (10 nM ivermectin) | ka64 | che-3 | Substitution/nonsense | − | + | + |
| TP274 (10 nM ivermectin) | ka66 | che-3 | Substitution/missense | − | + | + |
| TP375 (10 nM moxidectin) | ka200 | osm-1 | Substitution/nonsense | − | + | + |
| TP378 (10 nM moxidectin) | ka201 | dhc-3 | Deletion/frameshift | − | + | + |
| TP384 (10 nM moxidectin) | ka202 | dyf-2 | Substitution/nonsense | − | + | + |
| TP386 (10 nM moxidectin) | ka203 | che-3 | Splice site substitution | − | + | + |
| TP388 (10 nM moxidectin) | ka204 | ifta-1 | Substitution/nonsense | + | + | + |
Dyf, DiI amphid dye filling; IVM R, ivermectin resistance; MOX R, moxidectin resistance; +, dye filling/resistant; −, dye filling defective/susceptible.
Fig. 2.
Position of novel and tested alleles in resistance genes identified by whole-genome sequencing. Transcript structures and positions of genes were obtained from WormBase (https://wormbase.org) (JBrowse version: WS281; genome build WBcel235). Arrows above alleles point to their location in the genomic sequence. Solid lines directly above alleles span the length of deletions. Alleles featured (name = chr-number: position nt-change [aa-change]) are e1124 = I: 8,071,718 G > A (Q > Stop); ka30 = IV: 3,797,404 G > A (Q > Stop); ka32 = I: 8,070,133 C > T (G > R); ka33 = V: 13,150,172–13,150,276 deletion; ka35 = I: 8,058,869–8,079,083 deletion; ka64 = I: 8,075,488 A > T (L > Stop); ka66 = I: 8,072,572 C > T (E > K); ka200 = X: 16,544,813 C > T (Q > Stop); ka201 = V: 13,150,224 AGG > AG frameshift; ka202 = III: 13,676,892 G > A (Q > Stop); ka203 = I: 8,077,873 G > A splice site acceptor change; ka204 = X: 5,550,502 A > T (C > Stop); m160 III: 13,686,367 G > A (R > Stop); nx61 = X: 5,545,532–5,547,540 deletion; p802 = IV: 3,797,722 G > A (Q > Stop); p808 = X: uncharacterized; p816 = X: uncharacterized ∼600-bp deletion.
Discussion
IFT protein resistances and redundancies
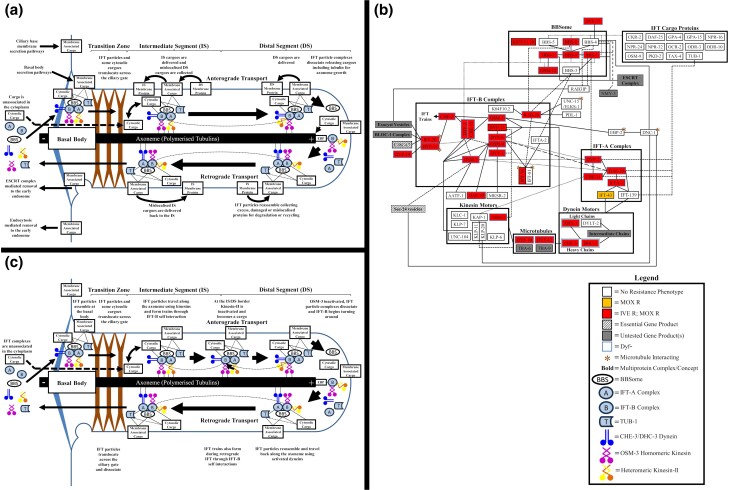
The import and transport of ciliary proteins by IFT are highly conserved throughout Eukaryota (Fig. 3a) with defects impacting cell motility, migration, signaling, and division and the ability to sense environmental stimuli (Boehlke et al. 2015; Taschner and Lorentzen 2016; Prevo et al. 2017; Reiter and Leroux 2017). IFT mutations have also recently been linked to ivermectin resistance in nematodes (Dent et al. 2000; Urdaneta-Marquez et al. 2014; Page 2018). Many of the known intra and inter IFT particle complex protein–protein interactions (Haycraft et al. 2003; Behal et al. 2012; Kubo et al. 2016; Taschner and Lorentzen 2016; Klink et al. 2017; Woodsmith et al. 2017; Zhu et al. 2017; Funabashi et al. 2018; Taschner et al. 2018; Vuong et al. 2018; Nakayama and Katoh 2020) corresponded well with loss of function induced macrocyclic lactone resistance (Fig. 3b). This suggests that IFT particle core subunit structural interactions (BBS-1, BBS-2, BBS-9, CHE-11, DAF-10, DYF-1, DYF-2, DYF-3, DYF-6, IFT-74, IFTA-1, OSM-5, OSM-12, and RAB-28), tubulin import and tubulin microtubule interaction (DTF-10, DYF-11, IFT-20, and IFT-74), IFT train formation (IFT-80/CHE-2), and IFT particle turnaround (Ahmed et al. 2008; Williamson et al. 2012; Ishikawa et al. 2014; Yang and Huang 2020) (Fig. 3c) (DYF-6, DYF-13, and OSM-1) all play key roles in suppressing the macrocyclic lactone resistance mechanism. The importance of the BBSome cargo-interacting subunits (Su et al. 2014) (BBS-1 and BBS-8; Page 2018) implies the effector protein is probably a TUB-1-independent BBSome cargo. There is evidence from the resistance pattern observed in the homomeric kinesin (OSM-3) but not the heterotrimeric kinesin 2 (Snow et al. 2004; Prevo et al. 2015), ciliary dyneins (Signor et al. 1999; Wicks et al. 2000; Hao et al. 2011) (CHE-3, DHC-3, and XBX-1), and ciliary distal segment defect mutants (Burghoorn et al. 2007; Ou et al. 2007; Phirke et al. 2011; Olivier-Mason et al. 2013; Maurya et al. 2019) (dyf-5(mn400), dyf-17(ox175), dyf-18(ok200), osta-1(tm5255), unc-101(sy108), and unc-119(e2498)) that both functional anterograde and retrograde IFT (Fig. 3a) of the effector to the distal segment of the amphid cilia are required to exert an effect. The lack of a role for protein prenylation in macrocyclic lactone susceptibility implies that the resistance observed in rab-28(ok3424) is functioning by a loss in the BBS-8-dependent periciliary membrane interaction and not the BBS-3-mediated interaction with IFT. Resistance may therefore be the result of amphid pore defects (Jensen et al. 2016; Akella et al. 2019, 2020).
Fig. 3.
IFT in C. elegans and resistance patterns in the IFT protein–protein interaction network. a) Summary of ciliary cargo transport in C. elegans during IFT. Line = protein/complex–protein/complex interaction; small arrow = change in protein or complex localization or interaction; large arrow = direction of IFT particle travel. b) A simplified version of predicted IFT protein–protein interaction network in C. elegans showing resistances found in mutants of each node. Box = group of proteins from the same complex or with the same function; line = predicted protein/complex–protein/complex interaction; small arrow = protein self-interaction. c) Summary of IFT complex interactions during IFT C. elegans. Line = protein/complex–protein/complex interaction; small arrow = change in protein or complex localization or interaction; large arrow = direction of IFT particle travel.
The use of single gene loss of function screens has some caveats in that proteins that have functional redundancy will give a greatly reduced phenotype. This was observed in protein dimers of the peripheral subunits of the IFT-B complex (IFT-20-DYF-11 and IFT-74-IFT-81) (Fig. 3b), which have known, but not always equal, redundancies (Kubo et al. 2016; Zhu et al. 2017). Similarly, the IFTA-1-interacting dynein-docking proteins IFT-139 and IFT-43 and the BBS-4 and BBS-5 cargo-interacting proteins of the BBSome are known to display redundancy when interacting with specific proteins (Behal et al. 2012; Xu et al. 2015; Yi et al. 2017; Scheidel and Blacque 2018), suggesting that future work should probe double mutants of these subunits to exclude their role in macrocyclic lactone resistance.
The nonredundant IFT particle subunits with no resistance phenotype association (BBS-3, DCT-14, IFTA-2, and K04F10.2) have niche roles in microtubule stability, receptor subpopulation trafficking, and cell signaling (Schafer et al. 2006; Pinkston-Gosse and Kenyon 2007; Li and Hu 2015; Sanders et al. 2015) suggesting that these processes and downstream effectors are nonessential for the resistance mechanism. Our results also support that DiI dye filling is less prone to disruption by ciliary impairment than the mechanism that induces resistance to macrocyclic lactones, as resistance was associated with the BBSome, dyf-13(mn396), ift-74(ok2866), and ifta-1(nx61) mutants without complete loss of dye-filling capability. Alternatively, these phenotypes could suggest branching in the mechanisms of dye filling and resistance at those subunits. It was surprising that dyf-13(mn396) was found to have no impairment in DiI uptake as that strain was used to first identify the locus with FITC and DiO dyes (Starich et al. 1995); however, Dio and DiI do not always share the same staining pattern (Hong et al. 2019).
Secretion pathways used by ciliary proteins
Proteins produced in the soma of ciliated neurons, including those for ciliogenesis, IFT, and any downstream effectors for macrocyclic lactone susceptibility, require delivery to the ciliary gate using one of several secretory pathways (Sato et al. 2005–2018; Nachury et al. 2010; Ding et al. 2017; Monis et al. 2017; Mukhopadhyay et al. 2017; Leitch et al. 2014; Cromm et al. 2019) (summarized in Fig. 4a). The results suggest that the proteins that influence macrocyclic lactone resistance are being secreted via the UNC-101 and UNC-119 secretory pathways before transport along the axon using one or more unidentified axonal kinesins, whose direction of transport along polarized microtubules is dependent on UNC-33 and UNC-44 (Goldstein and Yang 2000; Muresan 2000; Maniar et al. 2011). There is an indication that the effector proteins, along with other proteins for cilia maintenance, are being loaded onto IFT particles that are forming in an ARL-13-dependent manner, making ARL-13 a candidate as one of the effectors of the UNC-119 secretory pathway (Ou et al. 2007; Zhang et al. 2016; Cromm et al. 2019). As some protein trafficking complexes have core subunits that are essential (making them difficult to probe directly), potential roles of the SEC-24(COPII), BLOC-1, and TRAPP complexes/pathways in macrocyclic lactone resistance cannot be entirely excluded. An alternative hypothesis for the resistance seen in unc-33(e1193) and unc-44(e1197) is that the marginally shorter cilia have a smaller area of membrane available for macrocyclic lactone interaction (Hedgecock et al. 1985).
Fig. 4.
Ciliary protein trafficking pathways in C. elegans and resistance patterns in the ciliary gate protein–protein interaction network. a) Protein trafficking pathways used to deliver and remove ciliary proteins. Small arrow = show directionality of protein trafficking between cellular locations or organelles with key proteins and complexes involved in trafficking listed next to the arrow (placed before junctions if merging into a common secretion pathway); large arrow = directionality of axonal transport or passive diffusion. b) A simplified version of predicted basal body protein–protein interaction network in C. elegans showing resistances found in mutants of each node. Box = group of proteins from the same complex or with the same function; line = predicted protein–protein interaction; / = multiple (2–4) candidate genes with homology to a node found in other species (if gene IDs differ only by the last digit, then only the last digit is shown to the right of the candidate with a similar ID); Ce (node name of vertebrate ortholog) = multiple (>4) candidate genes with homology to the node found in other species.
Elements of the ciliary gate important for resistance
IFT particles and other ciliary proteins cross the semi-impermeable ciliary gate (Lambacher et al. 2016; Li, Jensen, Park, et al. 2016; Garcia-Gonzalo and Reiter 2017; Endicott and Brueckner 2018; Blasius et al. 2019). They gain entry to the cilia via the basal body, through the interaction with several proteins from the basal body, distal and subdistal appendages, and protein trafficking complexes (Kilmartin 2003; Bowers et al. 2004; Yoshimura et al. 2007; Babbey et al. 2010; Kaplan et al. 2010; Zhao and Malicki 2011; Joo et al. 2013; Wei et al. 2013; Li, Chen, Fisher, et al. 2016; Mazo et al. 2016; Ojeda Naharros et al. 2017; Zhang et al. 2017) (simplified in Fig. 4b). There is evidence from the results that the effector for the macrocyclic lactone resistance phenotype is gaining entry to the cilia as part of IFT particles in a DYF-19, and potentially C18C4.7, dependent manner (Wei et al. 2013) with other subunits of the distal appendages (C14H10.2 and YAP-1) and linker(s) to the mother centriole (Dammermann et al. 2009; Wei et al. 2016) (HYLS-1) also having key roles. As yap-1 is an ortholog of a Hippo pathway transcription factor (Iwasa et al. 2013; Lee et al. 2018) that also shares homology with the distal appendage subunit CEP164, there will be a need to dissect if resistance in mutants is occurring through the same mechanism as the other distal appendage proteins. The macrocyclic lactone resistance that was observed in the novel dye-filling defective (Dyf) phenotype-associated gene c14h10.2(tm10737) suggests that in addition to being a predicted CEP123 ortholog, it may interact with one or more of the IFT particle complexes.
The interactions between transition fiber proteins are highly redundant in C. elegans (Fig. 4b), meaning that defects in multiple proteins are required to cause the ciliary gate to become permeable, leading to ciliary defects and associated dye-filling phenotypes (Lambacher et al. 2016; Garcia-Gonzalo and Reiter 2017; Warburton-Pitt et al. 2012; Jensen et al. 2016). Consequently, only a single-transition fiber-encoding gene, nphp-4(tm925), was linked to resistance in this study. Among transition fiber-associated proteins and complexes, the TOGARAM1 ortholog CHE-12 has previously been associated with ivermectin resistance (Page 2018; Latour et al. 2019), so the lack of macrocyclic lactone resistance observed in mutants of the periphery subunits was surprising and suggests that unlike vertebrates there is either redundancy or that CHE-12 alone is sufficient for axoneme tubulin modification (Latour et al. 2019). Of the transition fiber-associated proteins, whose specific protein–protein interactions remain to be determined, the resistance observed in osta-1(tm5255) is potentially explained by reduced distal segment surface area (Olivier-Mason et al. 2013) resulting in impaired moxidectin uptake or tethering of cilium-sequestered transcription factors. The role of DYF-17 in distal segment assembly is currently unknown, but orthologs interact with BBS-4 (Lee et al. 2005; Phirke et al. 2011) suggesting a function in facilitating BBSome gating.
Exosomes, recycling, and degradation pathways
Involvement of the UNC-101 (a clathrin adapter protein) secretion pathway and requirement of retrograde IFT for maintaining macrocyclic lactone susceptibility, along with IFT being associated with exosome release phenotypes (Nager et al. 2017; Akella et al. 2019), raised the possibility that endocytosis, secretory vesicles, trafficked endosomes, and the cycling of exosomes (all processes that require membrane folding and targeted fusion; Sato et al. 2005–2018; Mayor et al. 2014; Ni et al. 2020) could be involved in the macrocyclic lactone resistance mechanism. From the results, it can be deduced that the membrane folding is occurring via one or more of the clathrin-independent pathways (Mayor et al. 2014), with only the RAB-35-dependent fast endosome recycling pathway (Sato et al. 2008; Grant and Donaldson 2009) having a role in macrocyclic lactone resistance downstream of IFT. The lack of resistance observed in mutants for the other pathways (Supplementary Table 1) suggests that the slow endosome recycling pathway, lysosomal degradation pathway, and exosomes are not involved in anthelmintic resistance. Loss of RAB-35 might be causing resistance by changes in membrane protein and receptor populations, which could lead to upregulation of resistance gene expression, removal of effectors for anthelmintic uptake, or a restriction in primary target numbers, or by the additional roles that RAB-35 has in cell migration, neurite outgrowth, and cell polarity (Sato et al. 2008; Grant and Donaldson 2009; Overeem et al. 2015; Klinkert and Echard 2016).
The gap junction mutants
Innexins form intercellular channels that function as gap junctions in neurotransmission and allow the exchange of small ions and compounds, including those for nucleotide signaling (Schumacher et al. 2012; Voelker et al. 2019), but might also facilitate the neural distribution of lipophilic dyes and anthelmintics. Comparison of the ivermectin resistance associated innexins unc-7(e5) and unc-9(e101), which are hypothesized to function by modulating the transmission of neurotoxic anthelmintic induced excitations (Dent et al. 2000), with inx-19(ky634) suggests that separate mechanisms are involved as the dye-filling defects indicate structural abnormalities of the ciliated amphid neurons. The macrocyclic lactone resistance and dye-filling phenotypes could be caused by the channel functions of INX-19 or the roles it has in determining neural cell fate (Chuang et al. 2007; Schumacher et al. 2012; Voelker et al. 2019), which could be important for the differentiation into cells involved in dye uptake and anthelmintic resistance.
Whole-genome sequencing of forward genetic screen mutants
The causative genes identified by whole-genome sequencing of the macrocyclic lactone-resistant mutants were all found to be involved in IFT. This is not unexpected, as ciliogenesis and IFT are complex nonredundant processes requiring the interaction of multiple genes. This would make the many genes involved in IFT statistically more likely to undergo mutation than single downstream effectors that rely on functional cilia. The dynein heavy chains che-3 and dhc-3 were overrepresented in the forward genetic screens for macrocyclic lactone resistance, as has been found for screens probing dye-filling defects (Starich et al. 1995; Ou et al. 2007; Page 2018). This finding is linked to the fact that EMS-induced loss-of-function mutations are proportional to gene size (Gengyo-Ando and Mitani 2000), making the long-coding sequences of dynein heavy chains (12,516 and 9,828 nt for che-3 and dhc-3, respectively) more prone to mutation than smaller IFT genes.
Potential mechanisms of resistance
The chemical properties of ivermectin and moxidectin prevent spontaneous crossing of cell membranes (Escher et al. 2008) meaning entry into organisms must be facilitated by either an extracellular membrane-associated carrier protein or transporter or by endocytic pathways (summarized in Fig. 5). The results suggest that the major routes of endocytosis and lysosomal degradation are not a significant mechanism for entry while a capacity for the compounds to induce endosomal escape has never been investigated. This means that uptake is most likely dependent on an elusive protein effector that may localize primarily to the amphid ciliary distal segments of ADL neurons.
Fig. 5.
Potential routes of macrocyclic lactone entry into C. elegans tissues and possibilities eliminated. Summary of routes that would facilitate macrocyclic lactone entry into C. elegans. Red star = macrocyclic lactone molecule; small arrow = show directionality of transport; large arrows = carrier protein cycling between membrane surfaces; gold circle = endosome; red cross = possible entry route that has been eliminated; green question mark = potential entry route remaining.
There is ongoing debate as to whether resistance correlated with defective cilia (represented by osm-3 mutants) is caused by reduced uptake (Dent et al. 2000; Urdaneta-Marquez et al. 2014; Page 2018) or by increased P-glycoprotein-facilitated export in the gut as part of an NHR-8-mediated intertissue signaling pathway (Guerrero et al. 2021). An osm-3;nhr-8 double mutant was recently reported to have higher resistance than wild-type controls (Guerrero et al. 2021), indicating that additional resistance mechanisms are involved. The role of NHR-8 in the macrocyclic lactone-resistant mutants identified in this study could be further investigated using the P-glycoprotein inhibitor verapamil in combination with fluorescently labeled anthelmintics to observe effects on uptake.
Conclusion
The findings of this study not only support strong evidence that the amphid cilia play an important role in responding to xenobiotic challenge by the macrocyclic compounds ivermectin and moxidectin (Page 2018) but also refine the location of the effectors to the distal segment of the cilia. The results from this study suggest that the effectors possess either a transmembrane domain or are anchored via a myristoyl or palmitoyl group. This study also uncovers the pathways used to deliver the effectors and other ciliary proteins in C. elegans and identifies C14H10.2 as a potential CEP123 ortholog. Due to the strong correlation between IFT function with dye-filling defects and resistance to macrocyclic lactones, it may be possible to use resistance phenotypes to identify if novel dye-filling mutants from forward genetic screens are upstream or downstream of IFT. If the resistance-causing genes uncovered in this study have the same functions in other nematode species, then there would be important implications for anthelmintic resistance-monitoring strategies.
Supplementary Material
Acknowledgments
Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Some C. elegans strains used in this work were created by the International C. elegans Gene Knockout Consortium. Some strains were provided by the National BioResource Project (Japan). Many thanks to Eric Jorgessen for dyf-17(ox175) and Jinghua Hu for dyf-19(jhu455).
Contributor Information
Robert A Brinzer, School of Biodiversity, One Health and Veterinary Medicine, University of Glasgow, Scotland G61 1QH, UK.
Alan D Winter, School of Biodiversity, One Health and Veterinary Medicine, University of Glasgow, Scotland G61 1QH, UK.
Antony P Page, School of Biodiversity, One Health and Veterinary Medicine, University of Glasgow, Scotland G61 1QH, UK.
Data availability
The strains dyf-17(ox175) and dyf-19(jhu455) can be made available if unable to be obtained from their creators (Eric Jorgessen and Jinghua Hu, respectively). All other strains are available for purchase from the stock centers mentioned in the Methods section. Aligned reads are available at https://www.ncbi.nlm.nih.gov/sra/PRJNA768320. Images of dye-filling assays for individual strains are available upon request. All survival assay data are in the Supplementary Methods.
Supplemental material available at G3 online.
Funding
This study was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) grant no. BB/R00711X/1.
Literature cited
- Abongwa M, Martin RJ, Robertson AP. 2017. A brief review on the mode of action of antinematodal drugs. Acta Vet (Beogr). 67(2):137–152. doi: 10.1515/acve-2017-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. 2008. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol. 183(2):313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akella JS, Carter SP, Nguyen K, Tsiropoulou S, Moran AL, Silva M, Rizvi F, Kennedy BN, Hall DH, Barr MM, et al. 2020. Ciliary Rab28 and the BBSome negatively regulate extracellular vesicle shedding. eLife. 9:e50580. doi: 10.7554/eLife.50580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akella JS, Carter SP, Rizvi F, Nguyen KCQ, Tsiropoulou S, Moran AL, Silva M, Kennedy BN, Hall DH, Barr MM, et al. 2019. A ciliary BBSome-ARL-6-PDE6D pathway trafficks RAB-28, a negative regulator of extracellular vesicle biogenesis. bioRxiv. doi: 10.1101/715730. [DOI]
- Babbey CM, Bacallao RL, Dunn KW. 2010. Rab10 associates with primary cilia and the exocyst complex in renal epithelial cells. Am J Physiol Renal Physiol. 299(3):F495–F506. doi: 10.1152/ajprenal.00198.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behal RH, Miller MS, Qin HM, Lucker BF, Jones A, Cole DG. 2012. Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins. J Biol Chem. 287(15):11689–11703. doi: 10.1074/jbc.M111.287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, Warner A, Khattra J, Holt RA, Ou G, Mah AK, et al. 2005. Functional genomics of the cilium, a sensory organelle. Curr Biol. 15(10):935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Blasius TL, Takao D, Verhey KJ. 2019. NPHP proteins are binding partners of nucleoporins at the base of the primary cilium. PLoS One. 14(9):e0222924. doi: 10.1371/journal.pone.0222924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehlke C, Janusch H, Hamann C, Powelske C, Mergen M, Herbst H, Kotsis F, Nitschke R, Kuehn EW. 2015. A cilia independent role of Ift88/polaris during cell migration. PLoS One. 10:e0140378. doi:ARTN e0140378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. 2004. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 5(3):194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- Brear AG, Yoon J, Wojtyniak M, Sengupta P. 2014. Diverse cell type-specific mechanisms localize G protein-coupled receptors to Caenorhabditis elegans sensory cilia. Genetics. 197(2):667–684. doi: 10.1534/genetics.114.161349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. Genetics of Caenorhabditis elegans. Genetics. 77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghoorn J, Dekkers MP, Rademakers S, de Jong T, Willemsen R, Jansen G. 2007. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 104(17):7157–7162. doi: 10.1073/pnas.0606974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IS, Kubo Y. 2018. Ivermectin and its target molecules: shared and unique modulation mechanisms of ion channels and receptors by ivermectin. J Physiol. 596(10):1833–1845. doi: 10.1113/Jp275236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Vanhoven MK, Fetter RD, Verselis VK, Bargmann CI. 2007. An innexin-dependent cell network establishes left-right neuronal asymmetry in C. elegans. Cell. 129(4):787–799. doi: 10.1016/j.cell.2007.02.052. [DOI] [PubMed] [Google Scholar]
- Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, Yakovlev MA, Nguyen KCQ, Tang LTH, Bayer EA, Duerr JS, et al. 2019. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature. 571(7763):63–71. doi: 10.1038/s41586-019-1352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromm PM, Adihou H, Kapoor S, Vazquez-Chantada M, Davey P, Longmire D, Hennes E, Hofer W, Kuchler P, Chiarparin E, et al. 2019. Lipidated stapled peptides targeting the acyl binding protein UNC119. Chembiochem. 20:2987–2990 . doi: 10.1002/cbic.201900615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook EK, O’Brien DJ, Howell SB, Storey BE, Whitley NC, Burke JM, Kaplan RM. 2016. Prevalence of anthelmintic resistance on sheep and goat farms in the mid-Atlantic region and comparison of in vivo and in vitro detection methods. Small Rumin Res. 143:89–96. doi: 10.1016/j.smallrumres.2016.09.006. [DOI] [Google Scholar]
- Dammermann A, Pemble H, Mitchell BJ, McLeod I, Yates JR 3rd, Kintner C, Desai AB, Oegema K. 2009. The hydrolethalus syndrome protein HYLS-1 links core centriole structure to cilia formation. Genes Dev. 23(17):2046–2059. doi: 10.1101/gad.1810409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent JA, Smith MM, Vassilatis DK, Avery L. 2000. The genetics of ivermectin resistance in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 97(6):2674–2679. doi: 10.1073/pnas.97.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Shao L, Yao Y, Tong X, Liu H, Yue S, Xie L, Cheng SY. 2017. DGKδ triggers endoplasmic reticulum release of IFT88-containing vesicles destined for the assembly of primary cilia. Sci Rep. 7(1):5296. doi: 10.1038/s41598-017-05680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M, Poole RJ, Sarin S, Bigelow H, Hobert O. 2010. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS One. 5(11):e15435. doi: 10.1371/journal.pone.0015435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott SJ, Brueckner M. 2018. NUP98 sets the size-exclusion diffusion limit through the ciliary base. Curr Biol. 28(10):1643–1650.e3. doi: 10.1016/j.cub.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher BI, Berger C, Bramaz N, Kwon J-H, Richter M, Tsinman O, Avdeef A. 2008. Membrane–water partitioning, membrane permeability, and baseline toxicity of the parasiticides ivermectin, albendazole, and morantel. Environ Toxicol Chem. 27(4):909–918. doi: 10.1897/07-427.1. [DOI] [PubMed] [Google Scholar]
- Freeman AS, Nghiem C, Li J, Ashton FT, Guerrero J, Shoop WL, Schad GA. 2003. Amphidial structure of ivermectin-resistant and susceptible laboratory and field strains of Haemonchus contortus. Vet Parasitol. 110(3–4):217–226. doi:Pii S0304-4017(02)00321-7. [DOI] [PubMed] [Google Scholar]
- Funabashi T, Katoh Y, Okazaki M, Sugawa M, Nakayama K. 2018. Interaction of heterotrimeric kinesin-II with IFT-B–connecting tetramer is crucial for ciliogenesis. J Cell Biol. 217(8):2867–2876. doi: 10.1083/jcb.201801039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Reiter JF. 2017. Open sesame: how transition fibers and the transition zone control ciliary composition. Cold Spring Harb Perspect Biol. 9(2):a028134. doi: 10.1101/cshperspect.a028134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K, Mitani S. 2000. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 269(1):64–69. doi: 10.1006/bbrc.2000.2260. [DOI] [PubMed] [Google Scholar]
- Goldstein LS, Yang Z. 2000. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci. 23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. 2009. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 10(9):597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisi L, Leite RC, Martins JR, Barros AT, Andreotti R, Cancado PHD, León AA, Pereira JB, Villela HS. 2014. Reassessment of the potential economic impact of cattle parasites in Brazil. Rev Bras Parasitol Vet. 23(2):150–156. doi: 10.1590/s1984-29612014042. [DOI] [PubMed] [Google Scholar]
- Guerrero GA, Derisbourg MJ, Mayr FAMC, Wester LE, Giorda M, Dinort JE, Hartman MD, Schilling K, Alonso-De Gennaro MJ, Lu RJ, et al. 2021. NHR-8 and P-glycoproteins uncouple xenobiotic resistance from longevity in chemosensory C. elegans mutants. eLife. 10:e53174. doi: 10.7554/eLife.53174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Efimenko E, Swoboda P, Scholey JM. 2011. The retrograde IFT machinery of C. elegans cilia: two IFT dynein complexes? PLoS One. 6(6):e20995. doi: 10.1371/journal.pone.0020995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft CJ, Schafer JC, Zhang Q, Taulman PD, Yoder BK. 2003. Identification of CHE-13, a novel intraflagellar transport protein required for cilia formation. Exp Cell Res. 284(2):251–263. doi: 10.1016/s0014-4827(02)00089-7. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Nichol Thomson J, Perkins LA. 1985. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol. 111(1):158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- Hong RL, Riebesell M, Bumbarger DJ, Cook SJ, Carstensen HR, Sarpolaki T, Cochella L, Castrejon J, Moreno E, Sieriebriennikov B, et al. 2019. Evolution of neuronal anatomy and circuitry in two highly divergent nematode species. eLife. 8:e47155. doi: 10.7554/eLife.47155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ide T, Yagi T, Jiang X, Hirono M, Sasaki H, Yanagisawa H, Wemmer KA, Stainier DYR, Qin HM, et al. 2014. TTC26/DYF13 is an intraflagellar transport protein required for transport of motility-related proteins into flagella. eLife. 3:e01566. doi: 10.7554/eLife.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa H, Maimaiti S, Kuroyanagi H, Kawano S, Inami K, Timalsina S, Ikeda M, Nakagawa K, Hata Y. 2013. Yes-associated protein homolog, YAP-1, is involved in the thermotolerance and aging in the nematode Caenorhabditis elegans. Exp Cell Res. 319(7):931–945. doi: 10.1016/j.yexcr.2013.01.020. [DOI] [PubMed] [Google Scholar]
- Jalili V, Afgan E, Gu Q, Clements D, Blankenberg D, Goecks J, Taylor J, Nekrutenko A. 2020. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 48(W1):W395–w402. doi: 10.1093/nar/gkaa434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James CE, Davey MW. 2009. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int J Parasitol. 39(2):213–220. doi: 10.1016/j.ijpara.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Jensen VL, Carter S, Sanders AAWM, Li C, Kennedy J, Timbers TA, Cai J, Scheidel N, Kennedy BN, Morin RD, et al. 2016. Whole-organism developmental expression profiling identifies RAB-28 as a novel ciliary GTPase associated with the BBSome and intraflagellar transport. PLoS Genet. 12(12):e1006469. doi: 10.1371/journal.pgen.1006469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo K, Kim CG, Lee M-S, Moon H-Y, Lee S-H, Kim MJ, Kweon H-S, Park W-Y, Kim C-H, Gleeson JG, et al. 2013. CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc Natl Acad Sci U S A. 110(15):5987–5992. doi: 10.1073/pnas.1220927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan OI, Molla-Herman A, Cevik S, Ghossoub R, Kida K, Kimura Y, Jenkins P, Martens JR, Setou M, Benmerah A, et al. 2010. The AP-1 clathrin adaptor facilitates cilium formation and functions with RAB-8 in C. elegans ciliary membrane transport. J Cell Sci. 123(Pt 22):3966–3977. doi: 10.1242/jcs.073908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan RM, Vidyashankar AN, Howell SB, Neiss JM, Williamson LH, Terrill TH. 2007. A novel approach for combining the use of in vitro and in vivo data to measure and detect emerging moxidectin resistance in gastrointestinal nematodes of goats. Intern J Parasitol. 37(7):795–804. doi: 10.1016/j.ijpara.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV. 2003. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol. 162(7):1211–1221. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink BU, Zent E, Juneja P, Kuhlee A, Raunser S, Wittinghofer A. 2017. A recombinant BBSome core complex and how it interacts with ciliary cargo. eLife. 6:e27434. doi: 10.7554/eLife.27434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert K, Echard A. 2016. Rab35 GTPase: a central regulator of phosphoinositides and F-actin in endocytic recycling and beyond. Traffic. 17(10):1063–1077. doi: 10.1111/tra.12422. [DOI] [PubMed] [Google Scholar]
- Kubo T, Brown JM, Bellve K, Craige B, Craft JM, Fogarty K, Lechtreck KF, Witman GB. 2016. Together, the IFT81 and IFT74 N-termini form the main module for intraflagellar transport of tubulin. J Cell Sci. 129(10):2106–2119. doi: 10.1242/jcs.187120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitomo H, Uesugi H, Kohara Y, Iino Y. 2005. Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails. Genome Biol. 6(2):R17. doi: 10.1186/gb-2005-6-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambacher NJ, Bruel AL, van Dam TJP, Szymańska K, Slaats GG, Kuhns S, McManus GJ, Kennedy JE, Gaff K, Wu KM, et al. 2016. TMEM107 recruits ciliopathy proteins to subdomains of the ciliary transition zone and causes Joubert syndrome. Nat Cell Biol. 18(1):122–131. doi: 10.1038/ncb3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latour BL, Van De Weghe JC, Rusterholz TDS, Letteboer SJF, Gomez A, Shaheen R, Gesemann M, Grout ME, van Reeuwijk J, Van Beersum SEC, et al. 2019. ARMC9 and TOGARAM1 define a Joubert syndrome-associated protein module that regulates axonemal post-translational modifications and cilium stability. bioRxiv. doi: 10.1101/817213. [DOI]
- Lee H, Kang J, Lee J. 2018. Involvement of YAP-1, the homolog of yes-associated protein, in the Wnt-mediated neuronal polarization in Caenorhabditis elegans. G3 (Bethesda). 8(8):2595–2602. doi: 10.1534/g3.118.200325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Walker CL, Karten B, Kuny SL, Tennese AA, O'Neill MA, Wevrick R. 2005. Essential role for the Prader–Willi syndrome protein necdin in axonal outgrowth. Hum Mol Genet. 14(5):627–637. doi: 10.1093/hmg/ddi059. [DOI] [PubMed] [Google Scholar]
- Leitch CC, Lodh S, Prieto-Echagüe V, Badano JL, Zaghloul NA. 2014. Basal body proteins regulate Notch signaling through endosomal trafficking. J Cell Sci. 127(Pt 11):2407–2419. doi: 10.1242/jcs.130344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Jensen VL, Park K, Kennedy J, Garcia-Gonzalo FR, Romani M, De Mori R, Bruel AL, Gaillard D, Doray B, et al. 2016. MKS5 and CEP290 dependent assembly pathway of the ciliary transition zone. PLoS Biol. 14(3):e1002416. doi: 10.1371/journal.pbio.1002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FQ, Chen X, Fisher C, Siller SS, Zelikman K, Kuriyama R, Takemaru KI. 2016. BAR domain-containing FAM92 proteins interact with Chibby1 to facilitate ciliogenesis. Mol Cell Biol. 36(21):2668–2680. doi: 10.1128/MCB.00160-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hu J. 2015. Small GTPases act as cellular switches in the context of cilia. In: Schatten H, editors. The Cytoskeleton in Health and Disease. New York, NY: Springer New York. p. 217–237. [Google Scholar]
- Maniar TA, Kaplan M, Wang GJ, Shen K, Wei L, Shaw JE, Koushika SP, Bargmann CI. 2011. UNC-33 (CRMP) and ankyrin organize microtubules and localize kinesin to polarize axon–dendrite sorting. Nat Neurosci. 15(1):48–56. doi: 10.1038/nn.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya AK, Rogers T, Sengupta P. 2019. A CCRK and a MAK kinase modulate cilia branching and length via regulation of axonemal microtubule dynamics in Caenorhabditis elegans. Curr Biol. 29(8):1286–1300 e4. doi: 10.1016/j.cub.2019.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S, Parton RG, Donaldson JG. 2014. Clathrin-independent pathways of endocytosis. Cold Spring Harb Perspect Biol. 6:a016758. doi: 10.1101/cshperspect.a016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo G, Soplop N, Wang WJ, Uryu K, Tsou MFB. 2016. Spatial control of primary ciliogenesis by subdistal appendages alters sensation-associated properties of cilia. Dev Cell. 39(4):424–437. doi: 10.1016/j.devcel.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur MJ, Reinemeyer CR. 2014. Herding the U.S. cattle industry toward a paradigm shift in parasite control. Vet Parasitol. 204(1):34–43. doi: 10.1016/j.vetpar.2013.12.021. [DOI] [PubMed] [Google Scholar]
- Ménez C, Alberich M, Kansoh D, Blanchard A, Lespine A. 2016. Acquired tolerance to ivermectin and moxidectin after drug selection pressure in the nematode Caenorhabditis elegans. Antimicrob Agents Chemother. 60(8):4809–4819. doi: 10.1128/AAC.00713-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monis WJ, Faundez V, Pazour GJ. 2017. BLOC-1 is required for selective membrane protein trafficking from endosomes to primary cilia. J Cell Biol. 216(7):2131–2150. doi: 10.1083/jcb.201611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Badgandi HB, Hwang SH, Somatilaka B, Shimada IS, Pal K. 2017. Trafficking to the primary cilium membrane. Mol Biol Cell. 28(2):233–239. doi: 10.1091/mbc.E16-07-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V. 2000. One axon, many kinesins: what's the logic? J Neurocytol. 29(11–12):799–818. doi: 10.1023/a:1010943424272. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Seeley ES, Jin H. 2010. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nager AR, Goldstein JS, Herranz-Pérez V, Portran D, Ye F, Garcia-Verdugo JM, Nachury MV. 2017. An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell. 168(1–2):252. doi: 10.1016/j.cell.2016.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Katoh Y. 2020. Architecture of the IFT ciliary trafficking machinery and interplay between its components. Crit Rev Biochem Mol Biol. 55(2):179–196. doi: 10.1080/10409238.2020.1768206. [DOI] [PubMed] [Google Scholar]
- Ni K, Wang C, Carnino JM, Jin Y. 2020. The evolving role of caveolin-1: a critical regulator of extracellular vesicles. Med Sci (Basel). 8(4):46. https://www.mdpi.com/2076-3271/8/4/46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeda Naharros I, Gesemann M, Mateos JM, Barmettler G, Forbes A, Ziegler U, Neuhauss SCF, Bachmann-Gagescu R. 2017. Loss-of-function of the ciliopathy protein Cc2d2a disorganizes the vesicle fusion machinery at the periciliary membrane and indirectly affects Rab8-trafficking in zebrafish photoreceptors. PLoS Genet. 13:e1007150. 10.1371/journal.pgen.1007150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier-Mason A, Wojtyniak M, Bowie RV, Nechipurenko IV, Blacque OE, Sengupta P. 2013. Transmembrane protein OSTA-1 shapes sensory cilia morphology via regulation of intracellular membrane trafficking in C. elegans. Development. 140(7):1560–1572. doi: 10.1242/dev.086249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G, Koga M, Blacque OE, Murayama T, Ohshima Y, Schafer JC, Li C, Yoder BK, Leroux MR, Scholey JM. 2007. Sensory ciliogenesis in Caenorhabditis elegans: assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol Biol Cell. 18(5):1554–1569. doi: 10.1091/mbc.e06-09-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overeem AW, Bryant DM, Van IJzendoorn SCD. 2015. Mechanisms of apical-basal axis orientation and epithelial lumen positioning. Trends Cell Biol. 25(8):476–485. doi: 10.1016/j.tcb.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Page AP. 2018. The sensory amphidial structures of Caenorhabditis elegans are involved in macrocyclic lactone uptake and anthelmintic resistance. Int J Parasitol. 48(13):1035–1042. doi: 10.1016/j.ijpara.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 117(2):456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Phirke P, Efimenko E, Mohan S, Burghoorn J, Crona F, Bakhoum MW, Trieb M, Schuske K, Jorgensen EM, Piasecki BP, et al. 2011. Transcriptional profiling of C. elegans DAF-19 uncovers a ciliary base-associated protein and a CDK/CCRK/LF2p-related kinase required for intraflagellar transport. Dev Biol. 357(1):235–247. doi: 10.1016/j.ydbio.2011.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkston-Gosse J, Kenyon C. 2007. DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat Genet. 39(11):1403–1409. doi: 10.1038/ng.2007.1. [DOI] [PubMed] [Google Scholar]
- Prevo B, Mangeol P, Oswald F, Scholey JM, Peterman EJG. 2015. Functional differentiation of cooperating kinesin-2 motors orchestrates cargo import and transport in C. elegans cilia. Nat Cell Biol. 17(12):1536–1545. doi: 10.1038/ncb3263. [DOI] [PubMed] [Google Scholar]
- Prevo B, Scholey JM, Peterman EJG. 2017. Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery. FEBS J. 284(18):2905–2931. doi: 10.1111/febs.14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR. 2017. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 18(9):533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. 2002. Intraflagellar transport. Nat Rev Mol Cell Biol. 3(11):813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Sanders AAWM, de Vrieze E, Alazami AM, Alzahrani F, Malarkey EB, Sorusch N, Tebbe L, Kuhns S, van Dam TJP, Alhashem A, et al. 2015. KIAA0556 is a novel ciliary basal body component mutated in Joubert syndrome. Genome Biol. 16(1):293. doi: 10.1186/s13059-015-0858-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Norris A, Sato M, Barth D, Grant BD. 2005–2018. C. elegans as a model for membrane traffic. In: WormBook. The C. elegans Research Community: WormBook. Pasadena (CA): WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Sato K, Liou W, Pant S, Harada A, Grant BD. 2008. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 27(8):1183–1196. doi: 10.1038/emboj.2008.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer JC, Winkelbauer ME, Williams CL, Haycraft CJ, Desmond RA, Yoder BK. 2006. IFTA-2 is a conserved cilia protein involved in pathways regulating longevity and dauer formation in Caenorhabditis elegans. J Cell Sci. 119(19):4088–4100. doi: 10.1242/jcs.03187. [DOI] [PubMed] [Google Scholar]
- Schafer W. 2016. Nematode nervous systems. Curr Biol. 26(20):R955–R959. doi: 10.1016/j.cub.2016.07.044. [DOI] [PubMed] [Google Scholar]
- Scheidel N, Blacque OE. 2018. Intraflagellar transport complex A genes differentially regulate cilium formation and transition zone gating. Curr Biol. 28(20):3279–3287 e2. doi: 10.1016/j.cub.2018.08.017. [DOI] [PubMed] [Google Scholar]
- Schumacher JA, Hsieh YW, Chen S, Pirri JK, Alkema MJ, Li WH, Chang C, Chuang CF. 2012. Intercellular calcium signaling in a gap junction-coupled cell network establishes asymmetric neuronal fates in C. elegans. Development. 139(22):4191–4201. doi: 10.1242/dev.083428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor D, Wedaman KP, Orozco JT, Dwyer ND, Bargmann CI, Rose LS, Scholey JM. 1999. Role of a class Dhc1b dynein in retrograde transport of ift motors and ift raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J Cell Biol. 147(3):519–530. doi: 10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Singh B, Singh AP. 2015. Nematodes: a threat to sustainability of agriculture. Procedia Environ Sci. 29:215–216. doi: 10.1016/j.proenv.2015.07.270. [DOI] [Google Scholar]
- Snow JJ, Ou G, Gunnarson AL, Walker MRS, Zhou HM, Brust-Mascher I, Scholey JM. 2004. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 6(11):1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- Starich TA, Herman RK, Kari CK, Yeh WH, Schackwitz WS, Schuyler MW, Collet J, Thomas JH, Riddle DL. 1995. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics. 139(1):171–188. doi: 10.1093/genetics/139.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Driscoll K, Yao G, Raed A, Wu M, Beales PL, Zhou J. 2014. Bardet–Biedl syndrome proteins 1 and 3 regulate the ciliary trafficking of polycystic kidney disease 1 protein. Hum Mol Genet. 23(20):5441–5451. doi: 10.1093/hmg/ddu267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Leroux MR. 2013. The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat Cell Biol. 15(12):1387–1397. doi: 10.1038/ncb2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Lorentzen E. 2016. The intraflagellar transport machinery. Cold Spring Harb Perspect Biol. 8:a028092. doi: 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Lorentzen A, Mourao A, Collins T, Freke GM, Moulding D, Basquin J, Jenkins D, Lorentzen E. 2018. Crystal structure of intraflagellar transport protein 80 reveals a homo-dimer required for ciliogenesis. eLife. 7:e33067. doi:ARTN e33067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J. 2006. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 7(1):S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdaneta-Marquez L, Bae SH, Janukavicius P, Beech R, Dent J, Prichard R. 2014. A dyf-7 haplotype causes sensory neuron defects and is associated with macrocyclic lactone resistance worldwide in the nematode parasite Haemonchus contortus. Int J Parasitol. 44(14):1063–1071. doi: 10.1016/j.ijpara.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Vidal B, Aghayeva U, Sun H, Wang C, Glenwinkel L, Bayer EA, Hobert O. 2018. An atlas of Caenorhabditis elegans chemoreceptor expression. PLoS Biol. 16(1):e2004218. doi: 10.1371/journal.pbio.2004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker L, Upadhyaya B, Ferkey DM, Woldemariam S, L’Etoile ND, Rabinowitch I, Bai J. 2019. INX-18 and INX-19 play distinct roles in electrical synapses that modulate aversive behavior in Caenorhabditis elegans. PLoS Genet. 15(10):e1008341. doi: 10.1371/journal.pgen.1008341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokřál I, Jedličková V, Jirásko R, Stuchlíková L, Bártíková H, Skálová L, Lamka J, Holčapek M, Szotáková B. 2013. The metabolic fate of ivermectin in host (Ovis aries) and parasite (Haemonchus contortus). Parasitology. 140(3):361–367. doi: 10.1017/S0031182012001680. [DOI] [PubMed] [Google Scholar]
- Vuong LT, Iomini C, Balmer S, Esposito D, Aaronson SA, Mlodzik M. 2018. Kinesin-2 and IFT-A act as a complex promoting nuclear localization of β-catenin during Wnt signalling. Nat Commun. 9(1):5304. doi: 10.1038/s41467-018-07605-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Al-Ouran R, Hu YH, Kim SY, Wan YW, Wangler MF, Yamamoto S, Chao HT, Comjean A, Mohr SE, et al. 2017. MARRVEL: integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am J Hum Genet. 100(6):843–853. doi: 10.1016/j.ajhg.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton-Pitt SRF, Jauregui AR, Li C, Wang J, Leroux MR, Barr MM. 2012. Ciliogenesis in Caenorhabditis elegans requires genetic interactions between ciliary middle segment localized NPHP-2 (inversin) and transition zone-associated proteins. J Cell Sci. 125(Pt 11):2592–2603. doi: 10.1242/jcs.095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Xu Q, Zhang Y, Li Y, Zhang Q, Hu Z, Harris PC, Torres VE, Ling K, Hu J. 2013. Transition fibre protein FBF1 is required for the ciliary entry of assembled intraflagellar transport complexes. Nat Commun. 4(1):2750. doi: 10.1038/ncomms3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Zhang Y, Schouteden C, Zhang Q, Dong J, Wonesch V, Ling K, Dammermann A, Hu J. 2016. The hydrolethalus syndrome protein HYLS-1 regulates formation of the ciliary gate. Nat Commun. 7:12437. doi: 10.1038/ncomms12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks SR, de Vries CJ, van Luenen HGAM, Plasterk RHA. 2000. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev Biol. 221(2):295–307. doi: 10.1006/dbio.2000.9686. [DOI] [PubMed] [Google Scholar]
- Williamson SM, Silva DA, Richey E, Qin HM. 2012. Probing the role of IFT particle complex A and B in flagellar entry and exit of IFT-dynein in Chlamydomonas. Protoplasma. 249(3):851–856. doi: 10.1007/s00709-011-0311-4. [DOI] [PubMed] [Google Scholar]
- Woodsmith J, Apelt L, Casado-Medrano V, Özkan Z, Timmermann B, Stelzl U. 2017. Protein interaction perturbation profiling at amino-acid resolution. Nat Methods. 14(12):1213–1221. doi: 10.1038/nmeth.4464. [DOI] [PubMed] [Google Scholar]
- Xu Q, Zhang Y, Wei Q, Huang Y, Li Y, Ling K, Hu J. 2015. BBS4 and BBS5 show functional redundancy in the BBSome to regulate the degradative sorting of ciliary sensory receptors. Sci Rep. 5(1):11855. doi: 10.1038/srep11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Huang K. 2020. Dissecting the vesicular trafficking function of IFT subunits. Front Cell Dev Biol. 7:352–352. doi: 10.3389/fcell.2019.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Li W-J, Dong M-Q, Ou G. 2017. Dynein-driven retrograde intraflagellar transport is triphasic in C. elegans sensory cilia. Curr Biol. 27(10):1448–1461.e7. doi: 10.1016/j.cub.2017.04.015. [DOI] [PubMed] [Google Scholar]
- Yilmaz E, Gerst B, McKay-Demeler J, Krücken J. 2019. Minimal modulation of macrocyclic lactone susceptibility in Caenorhabditis elegans following inhibition of cytochrome P450 monooxygenase activity. Exp Parasitol. 200:61–66. doi: 10.1016/j.exppara.2019.03.017. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. 2007. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 178(3):363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BY, Wang G, Xu XW, Yang SS, Zhuang TH, Wang GP, Ren H, Cheng SY, Jiang Q, Zhang CM. 2017. DAZ-interacting protein 1 (Dzip1) phosphorylation by polo-like kinase 1 (Plk1) regulates the centriolar satellite localization of the BBSome protein during the cell cycle. J Biol Chem. 292(4):1351–1360. doi: 10.1074/jbc.M116.765438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li Y, Zhang Y, Torres VE, Harris PC, Ling K, Hu J. 2016. GTP-binding of ARL-3 is activated by ARL-13 as a GEF and stabilized by UNC-119. Sci Rep. 6(1):24534. doi: 10.1038/srep24534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CT, Malicki J. 2011. Nephrocystins and MKS proteins interact with IFT particle and facilitate transport of selected ciliary cargos. Embo J. 30(13):2532–2544. doi: 10.1038/emboj.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Liang Y, Gao F, Pan J. 2017. IFT54 regulates IFT20 stability but is not essential for tubulin transport during ciliogenesis. Cell Mol Life Sci. 74(18):3425–3437. doi: 10.1007/s00018-017-2525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The strains dyf-17(ox175) and dyf-19(jhu455) can be made available if unable to be obtained from their creators (Eric Jorgessen and Jinghua Hu, respectively). All other strains are available for purchase from the stock centers mentioned in the Methods section. Aligned reads are available at https://www.ncbi.nlm.nih.gov/sra/PRJNA768320. Images of dye-filling assays for individual strains are available upon request. All survival assay data are in the Supplementary Methods.
Supplemental material available at G3 online.