Abstract
Transcriptional modules of coregulated genes play a key role in regulatory networks. Comparative studies show that modules of coexpressed genes are conserved across taxa. However, little is known about the mechanisms underlying the evolution of module regulation. Here, we explore the evolution of cis-regulatory programs associated with conserved modules by integrating expression profiles for two yeast species and sequence data for a total of 17 fungal genomes. We show that although the cis-elements accompanying certain conserved modules are strictly conserved, those of other conserved modules are remarkably diverged. In particular, we infer the evolutionary history of the regulatory program governing ribosomal modules. We show how a cis-element emerged concurrently in dozens of promoters of ribosomal protein genes, followed by the loss of a more ancient cis-element. We suggest that this formation of an intermediate redundant regulatory program allows conserved transcriptional modules to gradually switch from one regulatory mechanism to another while maintaining their functionality. Our work provides a general framework for the study of the dynamics of promoter evolution at the level of transcriptional modules and may help in understanding the evolvability and increased redundancy of transcriptional regulation in higher organisms.
Keywords: regulatory motifs, transcriptional networks
Transcriptional modules, i.e., groups of coregulated genes, play a central role in the organization and function of regulatory networks (1-3). Comparative studies have demonstrated that various transcriptional modules are highly conserved across a wide variety of organisms from Escherichia coli to humans (4-6). A common tacit assumption is that conserved regulatory mechanisms underlie module conservation, because coregulation imposes tight constraints on the evolution of a module's promoters. Indeed, recent studies showed that orthologous transcriptional modules are often associated with conserved cis-elements (7).
To gain new insights into the evolution of regulation of transcriptional modules, we developed an integrated method for comparative expression and sequence analysis. We applied our method to 17 fully sequenced yeast genomes and identified conserved transcriptional modules and the cis-elements that are associated with them in each species. Although the cis-elements associated with certain modules were conserved in all species, other modules were associated with distinct cis-elements in different species. Divergence of cis-elements in specific promoters has been documented, e.g., in refs. 8 and 9, but it is difficult to explain in a similar way the divergence we observed in a regulatory program associated with dozens of coexpressed genes. In particular, it is not clear how multiple promoters diverge in a coordinated way and how divergence occurs without adversely affecting the coexpression phenotype. To answer this question, we inferred the detailed evolutionary history of modules' regulatory programs and studied different evolutionary events that may have contributed to their divergence, including drift, gain, and loss of cis-elements. Using the paradigmatic example of the ribosomal proteins module, we show that a module can switch from employing one cis-element into another through the formation of redundant intermediate promoters harboring both cis-elements in a tightly coupled configuration. The full spectrum of evolutionary events we discovered, encompassing both conservation and divergence, provides a general framework for the study of the evolution of transcription regulation and highlights the flexibility and evolvability of cis-regulatory programs.
Methods
Sequences Analysis. We used the previously published genomic sequences and annotations for all species used. We reannotated promoters in the Saccharomyces species to ensure short exons are correctly identified. Full details on the data and the procedure are available in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Orthologous Transcriptional Modules. To discover transcriptional modules, we applied the SAMBA algorithm (3) independently to Saccharomyces cerevisiae and Schizosaccharomyces pombe gene expression compendia. We measured the degree of orthology between modules based on the number of orthologous genes shared by them with hypergeometric statistics. Two modules with the lowest reciprocal orthology P values were defined as orthologous and were used in subsequent analysis of conserved transcriptional modules. See Supporting Materials and Methods for details.
Phylogenetic Cis-Profiling. To discover cis-elements enriched in conserved modules in different species, we used the following procedure. Starting with a pair of orthologous transcriptional modules in S. cerevisiae and S. pombe, we first form a projected orthologous module (POM) in each of the other species by taking all of the genes in this species that are orthologous to genes from the S. cerevisiae module. Several alternative definitions of the POMs, such as the union or intersection of the orthologous gene sets from both the S. cerevisiae and S. pombe modules, yielded similar results. Because S. cerevisiae is evolutionarily closer than S. pombe to the additional 15 species we analyzed, we report results based on the simplest procedure, in which we projected all POMs from the S. cerevisiae modules. Next, we apply our cis-element finding algorithm (see Supporting Materials and Methods) to each of the POMs and construct a set of significant position weight matrices (PWMs) in each of them. We now use all of the discovered PWMs (from all species and all modules) as seeds for another iteration of the cis-element finding algorithm on all modules in all species. This final step ensures that the absence of a PWM in one species' POM is not an artifact of the motif finding procedure.
Results
Orthologous Transcriptional Modules in Yeast. To study the evolution of transcriptional programs, we focused on the cis-regulatory mechanisms associated with conserved transcriptional modules. We developed a three-step approach (Fig. 5, which is published as supporting information on the PNAS web site) and applied it to the Ascomycota phylum (sac fungi), the best characterized group of fungi. First, we identified conserved transcriptional modules by using expression data from two distant yeast species, Saccharomyces cerevisiae and Schizosaccharomyces pombe (Fig. 5 a and b). Second, we used sequence information to derive orthologous modules in 15 additional fungal species and identified the cis-regulatory elements associated with each module in each species (Fig. 5 c-e). Third, we reconstructed the evolution of cis-elements associated with each module (Fig. 5f).
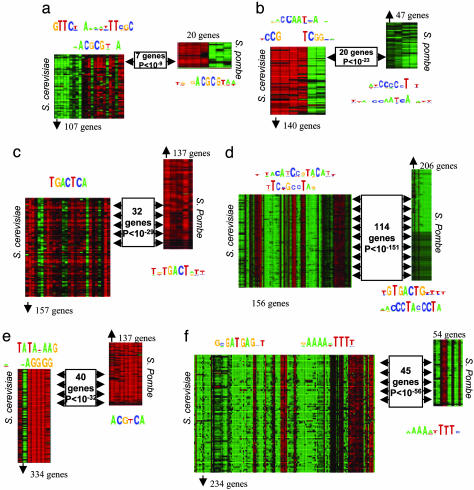
We used expression profiles in the yeasts S. cerevisiae and S. pombe and the SAMBA algorithm (3) (Supporting Materials and Methods) to define transcriptional modules separately in each species. We then identified orthologous transcriptional modules between the two species as pairs of modules that share a significant fraction of orthologous gene pairs (Supporting Materials and Methods). Although existing gene expression data sets only partially cover the relevant biological conditions in each species, we were able to detect several modules that are significantly conserved, encompassing a variety of molecular functions and cellular processes (Fig. 1).
Fig. 1.
Conserved transcriptional modules in S. pombe and S. cerevisiae and their associated cis-elements. Shown are the S. cerevisiae and S. pombe modules for the six key conserved modules we identified, together with the cis-elements enriched in the promoters of these modules' genes. For each module, the profile shows the module genes (rows) induced (red) and repressed (green) across different experiments (columns). Rectangles indicate the orthologous genes, their number, and the P value of their cooccurrence. The enriched cis-elements associated with each module are shown in the sequence logo above or below it. (a) S phase module, associated with the conserved Mlu1 cell cycle box element (ACGCGT, bound by orthologous MBF complexes in both species), and an S. cerevisiae-specific element. (b) Respiration module, associated with the conserved HAP2345 site (CCAATCA, bound by the orthologous Hap2345 and Php2-5 complexes). (c) Amino acid metabolism module, associated with the conserved GCN4 site (TGACTCA; Supporting Materials and Methods, note 2). (d) Ribosomal proteins module associated with RAP1 (TACATCCGTACAT) and IFHL sites (TCCGCCTAG) in S. cerevisiae and with a Homol-D box (TGTGACTG) and a Homol-E site (ACCCTACCCTA) in S. pombe. (e) Stress module associated with the STRE site (AGGGG) in S. cerevisiae and the CRE site (ACGTCA) in S. pombe. (f) Ribosome biogenesis module, associated with the conserved element RRPE (AAAAATTTT) and the S. cerevisiae-specific PAC element (GCGATGAG).
Conserved and Diverged Regulatory Mechanisms. To identify the cis-regulatory mechanisms associated with these conserved modules, we searched for cis-elements enriched in the promoters of each module's genes, separately for S. cerevisiae and S. pombe (Supporting Materials and Methods, note 1). In several cases (Fig. 1), we found similar cis-elements enriched in the orthologous modules of both species, even when the orthologous genes constituted only a small fraction of the modules' genes. The conserved elements were often also known to correspond to binding sites of orthologous transcriptional complexes (Fig. 1 a and b). Surprisingly, we also found several cases where the “phenotypic” conservation of gene expression is not accompanied by a corresponding conservation of the enriched cis-elements. These cases include modules for key molecular functions, such as ribosomal protein synthesis (Fig. 1d) and stress response (Fig. 1e), all of which were demonstrated to be conserved across a wide range of taxa (4, 5). In other cases, such as the ribosome biogenesis module (Fig. 1f) or the S phase module (Fig. 1a), an S. cerevisiae-specific motif is found along with a second, conserved motif. Importantly, this divergence in cis-elements does not stem from the nonorthologous members of these modules because similar results were obtained when considering only the orthologous cores (see Supporting Materials and Methods, note 1), and these cores manifest significant overlap of the modules' genes (Fig. 1 d and e, boxes). It was difficult to envision how such divergence in the mechanisms regulating the expression of highly essential and tightly coordinated modules could take place without deleterious effects.
Phylogenetic Cis-Profiles in 17 Yeast Species. To address this question, we analyzed the cis-elements enriched in conserved modules in 15 additional fully sequenced fungal species covering the evolutionary spectrum between S. cerevisiae and S. pombe (Methods). Because genome-wide expression data for these species are scarce (see Supporting Materials and Methods, note 3), we inferred POMs in these species by taking all genes that have an ortholog in the S. cerevisiae-conserved modules (Fig. 5c and Methods). We then searched for enriched cis-elements in the promoters of the projected module's genes (Fig. 5d) and analyzed each motif identified in one species for its presence in all other species. As before, we verified that the motifs we detected were also enriched in modules that were generated by projecting only from the orthologous cores of the S. cerevisiae and S. pombe modules. This step ensured that unique motifs are not simply contributed by the nonorthologous genes (see Supporting Materials and Methods, note 1). The resulting phylogenetic cis-profile (Fig. 5e) associates each module with a set of cis-elements in each species. By examining similarities across the profiles, we can identify conserved mechanisms. Indeed, for modules whose regulatory mechanisms were conserved between S. cerevisiae and S. pombe, the phylogenetic cis-profiles reveal perfect conservation in all intermediate lineages (Fig. 6, which is published as supporting information on the PNAS web site), consistent with recently published results (7). Moreover, by also considering differences between profiles, we can reconstruct the evolutionary scenario that explains the divergence of the regulatory mechanisms associated with conserved transcriptional modules.
The Evolution of the Ribosomal Regulatory Program. A remarkable example of regulatory divergence is the large, tightly regulated and highly conserved ribosomal protein (RP) module. Two elements are associated with the S. cerevisiae module: the well-known RAP1 binding site and an IFHL site TC(C/T)GCCTA (10-12). Two different elements are found in the S. pombe module: the Homol-D box (TGTGACTG) and the homol-E box (CCCTACCCTA), both of which have been shown to regulate the expression of RPs in this species (13). Such disparity can result either from divergence in the DNA binding sequence of the same ancestral transcription factors or from the use of distinct sites bound by distinct transcription factors. The detailed phylogeny of cis-elements in RP promoters (Fig. 2) allows us to infer the evolutionary scenario underlying this divergence.
Fig. 2.
Evolution of the regulatory mechanisms in the highly conserved module of ribosomal proteins. Phylogenetic cis-profile of the RP module. A schematic phylogenetic tree (branches are not drawn to scale) representing the known phylogeny (14) of the 17 analyzed species is shown, together with the sequence logos of the main cis-elements enriched in each module's promoters, grouped into three distinct types (colored boxes): RAP1 (orange), IFHL (blue), and Homol-D (red). The total number of genes in each POM is shown in parentheses, and the number of genes that contain each motif is indicated as well. Although the RP module is phenotypically extremely conserved, the phylogenetic cis-profile reveals a gradual switch from a Homol-D-dominated mechanism to a RAP1-controlled one, beginning before the speciation of A. gossypii. Concomitantly, the IFHL site underwent gradual sequence divergence and possible dimerization or domain duplication of the corresponding transcription factor.
As shown in Fig. 2, the profiles of S. castellii, Candida glabrata, S. kluyveri, Kluyveromyces waltii, and Ashbya gossypii contain both the Homol-D box and the RAP1 site (in addition to a strong IFHL site, discussed below). This apparent redundancy of binding sites in these species has two important implications. First, the presence of intermediate species, in which both the Homol-D and the RAP1 sites appear in RP promoters, suggests that the regulatory mechanism associated with the RP module has “switched” from a Homol-D-based mechanism (in the Ascomycota ancestor and S. pombe) to a RAP1-based one (in S. cerevisiae) and was not modified because of a mere drift in cis-element sequences (Fig. 7, which is published as supporting information on the PNAS web site). Second, it points to a potential process by which such a dramatic change in the regulatory mechanism of an essential module could take place without destroying the coordinated regulation. According to the most parsimonious scenario supported by the data (Fig. 8, which is published as supporting information on the PNAS web site), an ancient homol-D box played the key role in regulating RP transcription. Subsequently, before the divergence of A. gossypii, RAP1 emerged as an additional regulator of this module, whereas the module maintained the functionality of the homol-D box. Because the abundance of RAP1 sites increased, Homol-D lost its central role and was eventually eliminated, possibly after the divergence or loss of the corresponding unknown transcription factor. Thus, a process of infiltration (of RAP1) and loss (of Homol-D) swept through the promoters of the RPs.
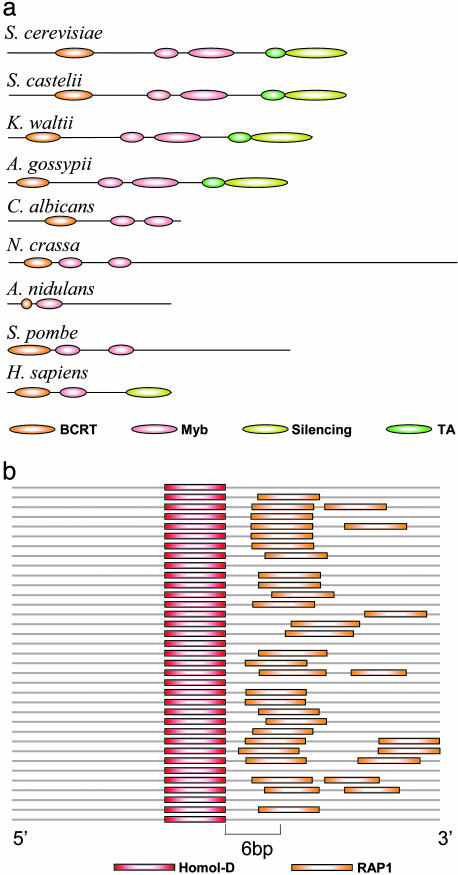
The Basis for Regulator Switching in the Ribosomal Regulatory Program. Several additional lines of evidence support this evolutionary scenario. First, we consider the Rap1p transcription factor. Rap1p's binding specificity is associated with its ancient and conserved function in the regulation of telomere length (15). The RAP1 binding site in RP promoters is a submotif of the telomeric repeat sequence bound by Rap1p in these species (16), including those that do not have a RAP1 site in their RP promoters. Thus, the sequence of the RAP1 motif that emerged in RP promoters matched Rap1p's preexisting DNA binding site. More importantly, analysis of Rap1p's coding sequence in all 17 fungal species, and in mammals, suggests that the invasion of RAP1 sites into RP promoters is associated with the acquisition of a new transactivation (TA) domain by Rap1p after the C. albicans speciation and before the A. gossypii speciation event (Fig. 3a). Thus, whereas the DNA binding domains of Rap1p (Myb-domains) have been conserved in all species, the TA domain, which is responsible for Rap1p's role as an RP transcription factor (17), follows exactly the same evolutionary pattern as the RAP1 binding site in RP promoters and is present only in the clade spanning from A. gossypii to S. cerevisiae. Moreover, Rap1p from species lacking the TA domain, such as C. albicans and S. pombe, cannot functionally complement for the S. cerevisiae Rap1p (18-20), whereas those with the TA domain (e.g., S. castellii) are adequate substitutes (21). Thus, the acquired domain allowed Rap1p to assume a novel role in transcriptional regulation, whereas its conserved DNA binding domain determined the sequence of its corresponding cis-element. Importantly, the evolution of Rap1p's TA domain further supports the parsimonious scenario where a RAP1 site emerged and a Homol-D site was lost from RP promoters (Fig. 8).
Fig. 3.
Mechanisms for evolutionary change in the regulation of ribosomal proteins. (a) Rap1p sequence evolution. A scaled schematic representation of Rap1p sequences is shown for eight species and the human protein. Colored ovals indicate the presence and position of BCRT (orange), Myb (DNA binding, pink), silencing (olive), and TA (dark green) domains. The DNA binding Myb domain is present in all species, but the transactivation domain is apparent only in those species that harbor the RAP1 motif in their RP module genes (S. cerevisiae, S. castellii, K. waltii, A. gossypii, and all of the intermediate species; data not shown). The TA domain is absent from all species lacking the RAP1 element in RP promoters, including C. albicans, N. crassa, A. nidulans, and S. pombe. A Rap1p ortholog cannot be identified in Y. lipolytica, and no significant homology was found to the TA domain for the D. hansenii Rap1p (data not shown). (b) The Homol-D-RAP1 cis-regulatory module. Shown is a scaled schematic representation of the 35 promoters of the A. gossypii RP genes with the highest scoring Homol-D elements. Colored bars indicate the Homol-D (red) and RAP1 (orange) sites. The two sites are extremely close, with the RAP1 trailing the Homol-D site by 2-6 bp, indicating a possible interaction between their corresponding transcription factors.
Analysis of the RP loci that contain a Homol-D site in A. gossypii and K. waltii, two of the species in our collection to also exhibit a RAP1 site, suggests a possible mechanistic model for the process of switching of the transcription factor binding site. In these species, when both RAP1 and Homol-D sites appear in the same promoter, they are usually separated by no more than 2-6 base pairs, in the conserved order 5′-Homol-D-RAP1-3′ (Fig. 3b; see also Fig. 9, which is published as supporting information on the PNAS web site), with Homol-D in a fixed orientation relative to the transcription start site (in contrast to S. pombe, where it has no strand preference). This strong association may indicate a corresponding interaction between the Homol-D binding protein and Rap1p, which may have facilitated Rap1p's infiltration into the RP regulatory program. Taken together, our results for both of the transcription factors and their binding sites propose a coherent view of a process by which RP regulation gradually switched from one transcription factor to another without losing its essential functionality.
Gradual Evolution in the IFHL Box. Additional examination of the phylogenetic cis-profiles (Fig. 2) suggests that the second cis-element in the S. cerevisiae RP module, the IFHL site (TCTGCCTA), has evolved primarily by a different mechanism, involving gradual divergence in DNA binding sequence. First, this element is clearly enriched in the entire Saccharomyces genus as well as S. kluyveri, K. lactis, A. gossypii, and K. waltii. Furthermore, close inspection of motifs enriched in the remaining species, C. albicans, Debaryomyces hansenii, Yarrowia lipolytica, Neurospora crassa, Aspergillus nidulans, and S. pombe, suggests that they also carry related variants of the same motif (albeit not identical ones). In C. albicans, the strongest cis-element in the RP module (AGGGCTATAGCCCT) is a palindrome containing two copies (TAGCCCT and its reverse complemented AGGGCTA) of a variant of the second part of the IFHL motif (GCCTA). A similar complex cis-element is present in D. hansenii and Y. lipolytica. The promoters of RP genes in the evolutionary distant N. crassa and A. nidulans contain an exact match to the second half of the C. albicans motif (GCCCTA), and the S. pombe Homol-E motif (CCCTACCCTA) is a duplicated variant of the same motif (CCCTA). Thus, an ancestral IFHL DNA binding protein may have been associated with the RP module throughout the evolutionary history of the Ascomycota clade. In addition to acquiring smaller-scale mutations causing changes in its DNA recognition site, the IFHL binding protein may have either undergone convergent domain duplication in C. albicans and S. pombe or acquired a dimerization domain in these species. Note that these dimerization or domain duplication events have presumably occurred by different routes in the two species, accounting for the differences in the organization of the respective elements (direct repeats vs. palindromic ones). Additional species-specific motifs are also associated with the RP module, consistent with the evolutionary flexibility of the RP regulatory mechanisms. For example, RP module genes in C. albicans, D. hansenii, and Y. lipolytica are also enriched for the ribosomal RNA processing element (RRPE) motif, which is usually involved in other stress-related modules. Traces of this enrichment can also be found in other species, most notably K. waltii, S. bayanus, and N. crassa.
Conservation of Spatial Configuration in Ribosomal Promoters. To examine the interplay between the three main regulatory elements of the RP module in different species, we analyzed their cooccurrence in the RP genes in each of the species and their relative spatial organization (Fig. 10, which is published as supporting information on the PNAS web site). In A. gossypii and K. waltii, many promoters have “redundant” regulatory mechanisms with three different cis-acting sites, whereas S. cerevisiae promoters are simpler and often contain only a (possibly duplicated) RAP1-binding element. Spatial analysis also reveals that certain features of global promoter organization are conserved across species. For example, we found that IFHL-like sites are typically found 100-200 bp 5′ to the Rap1 site, consistent with the functional constraint imposed by the interaction between Ifh1p, Fhl1p, and Rap1p in the combinatorial regulation of RP genes (10-12). Finally, we asked whether some of the differences in the organization of the regulatory mechanisms may also match phenotypic differences in gene expression. The evidence from S. cerevisiae and S. pombe indicates that switching from a Homol-D to a RAP1 cis-regulatory mechanism does not entail such a change, because RP genes are strictly coregulated in both species and respond similarly to environmental stress. However, some of the organisms, for example C. albicans, employ a regulatory mechanism lacking both RAP1 and Homol-D elements (and using IFHL and RRPE elements). Indeed, a recent expression profiling study (22) indicates that the C. albicans RP module responds much more weakly to environmental stress than either S. cerevisiae or S. pombe (Supporting Materials and Methods, note 3).
Regulatory Divergence in the Ribosome Biogenesis Module. Beyond the ribosomal protein genes, a number of other modules exhibit rapid evolution of cis-regulatory motifs. For example, in the ribosomal biogenesis (Fig. 11, which is published as supporting information on the PNAS web site), we detected two elements that were previously associated with the transcription of ribosome biogenesis genes in S. cerevisiae: the RRPE (23) and the polymerase A and C (PAC) element (24). RRPE was detectable in each of the 17 species, whereas we found PAC as a possible innovation of the C. albicans-S. cerevisiae lineage (a GATA-like box in N. crassa may suggest the origin of this innovation). The phylogenetic profile of a third element, TTTCTTTTT, indicates emergence before A. gossypii speciation and loss after the S. kluyveri-K. waltii speciation. Cooccurrence and spatial analysis indicate that, as in the emergence of the RAP1 site in the RP module, the transient TTTCTTTTT site is spatially clustered with the additional binding sites PAC and RRPE (Figs. 12 and 13, which are published as supporting information on the PNAS web site), possibly facilitating its emergence as a regulator of the module.
Our analysis suggests that the transition from one regulatory scheme to the other in a large regulatory module comprised of many genes often occurs through the formation of redundant intermediate regulatory programs. Which forces shape the functional interaction among different cis-elements and affect their evolution? We hypothesize two major potential trends. The first trend (“conservation”) occurs wherever there is a specific regulatory role for each of the cis-elements, and selection conserves a particular combination of cis-elements present in each gene's promoter. The second trend (“buffering”; ref. 25) occurs in cases where two cis-elements have a similar regulatory role and increases the redundancy in the regulatory mechanism. Such buffering could allow stochastic evolutionary changes to occur and would increase the capacity of the system for further evolutionary change (“evolvability”; ref. 26). We characterized the trends affecting specific cis-elements by analyzing their gene-specific patterns across species. For the RP module, such analysis reveals that both conservation and buffering may simultaneously affect different evolving cis-elements in the module (Supporting Materials and Methods, note 4; see also Fig. 14, which is published as supporting information on the PNAS web site).
Discussion
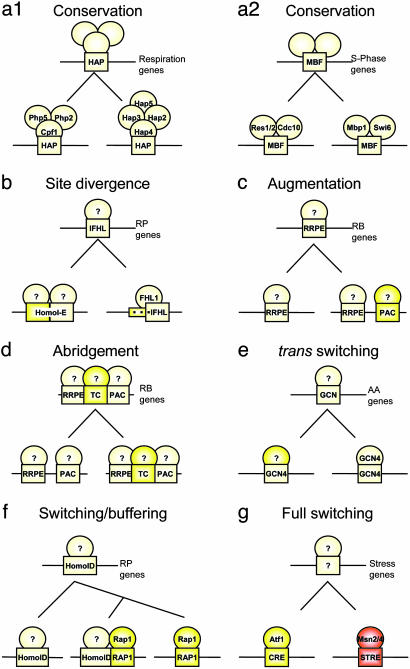
The evolution of transcriptional modules is an important aspect in understanding regulatory networks. Previous studies have suggested that groups of genes that are orthologous to S. cerevisiae expression modules are frequently regulated by conserved cis-elements (7). Our analysis demonstrates that the regulatory mechanisms associated with ancient and tightly conserved transcriptional modules can often be remarkably diverged. Our work suggests a general framework for the study of the evolution of module regulation, including full conservation of binding site and transcription factor, gradual changes in a single DNA binding site, simplification and elaboration of existing programs, and even dramatic events of element infiltration and loss that result in transcription factor switching (Fig. 4). In particular, we suggest that the formation of a redundant intermediate program may explain how a coordinated response may be conserved even though the underlying regulatory mechanisms are changing. This dynamic view of regulatory network evolution is consistent with previous studies on rapid promoter evolution (8, 9, 27) and with the known relative flexibility of cis-regulatory sequences compared with protein coding sequences. Our analysis implies that specific evolutionary processes exploit the dynamic nature of promoters to continuously modify the level of redundancy in regulatory mechanisms. Such redundancy may provide a buffering capacity (25) and may be important for the evolvability (26) of the regulatory program. Additional data and further studies are required to validate our hypotheses and fully elucidate such processes. For example, we still lack experimental evidence demonstrating the redundancy of the various sites in the intermediate programs, and we do not know how a large number of novel binding sites are introduced in a coordinated fashion, whether the coupling of elements we observed within promoters facilitates or constrains the evolution of regulatory programs, and the exact rate of sequence changes necessary to introduce a novel motif.
Fig. 4.
Alternative modes for the evolution of the regulation of transcriptional modules. Each panel shows a distinct scenario of the inferred evolution of an ancestral regulatory program (Upper) into programs observed in 2 or more extant species (Lower). For each module, a schematic representative promoter is shown (black line) along with cis-elements (boxes) and transcription factors (ovals). Ancestral conserved sites and proteins are in light yellow, and innovations and divergences are in bright yellow or red. (a) Conservation of both the cis-element and trans-factors [e.g., the S phase (a2) and respiration (a1) modules]. (b) A gradual divergence of binding site sequence (e.g., the IFHL site in the RP module). (c) Augmentation of an existing program by the emergence of a new site along an ancestral one [e.g., the RRPE and PAC sites in the ribosomal biogenesis (RB) module]. (d) Abridgement of an augmented program by binding site loss (e.g., the loss of the TC site in the RB module). (e) Switching of the transcription factor while maintaining the same cis-element (e.g., the AA metabolism module). (f and g) Full switching of a program from one cis-element to another (e.g., the stress and the RP modules). In some cases (f), this can occur by a combination of augmentation and abridgement.
Our results have significant implications for the study of transcription regulation in an evolutionary context. We have shown that computational techniques, merging previously uncharacterized data with well established evolutionary concepts, facilitate improved integration of genomic (sequence) and phenotypic (expression) data and their synthesis into a coherent reconstruction of the evolution of regulatory networks. The evolutionary context is crucial for the exploitation of these data and greatly enhances the potential of comparative methods (28). Whereas previous research in comparative genomics of regulatory networks focused on the identification of conserved cis-elements (29-31), our results emphasize the importance of accounting for changes, both gradual sequence divergence and dramatic innovation processes. Finally, the putative buffering effect of redundant regulatory elements that we report here may be instrumental in enabling rapid evolutionary change of regulatory networks and may play a major role in metazoan eukaryotes. The typical animal promoter is organized into cis-regulatory modules (32) that contain multiple, often redundant, binding sites. It is possible that this organization is a consequence of evolutionary processes similar to those we report here that are essential for the emergence of the increased complexity and evolvability of animals' regulatory networks.
Supplementary Material
Acknowledgments
We thank Nir Friedman, Laura Garwin, Irit Gat-Vicks, Martin Kupiec, Andrew Murray, Dana Pe'er, and Ilan Wapinski for comments on the manuscript. A.T. was supported in part by a scholarship in Complexity Science from the Horvitz Association. R.S. was supported by the Israel Science Foundation. A.R. was supported by the Bauer Center and by the National Institute of General Medical Sciences.
Abbreviations: POM, projected orthologous module; RP, ribosomal protein; RRPE, ribosomal RNA processing element; TA, transactivation.
References
- 1.Ihmels, J., Friedlander, G., Bergmann, S., Sarig, O., Ziv, Y. & Barkai, N. (2002) Nat. Genet. 31, 370-377. [DOI] [PubMed] [Google Scholar]
- 2.Segal, E., Shapira, M., Regev, A., Pe'er, D., Botstein, D., Koller, D. & Friedman, N. (2003) Nat. Genet. 34, 166-176. [DOI] [PubMed] [Google Scholar]
- 3.Tanay, A., Sharan, R., Kupiec, M. & Shamir, R. (2004) Proc. Natl. Acad. Sci. USA 101, 2981-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stuart, J. M., Segal, E., Koller, D. & Kim, S. K. (2003) Science 302, 249-255. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann, S., Ihmels, J. & Barkai, N. (2004) PLoS Biol. 2, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarroll, S. A., Murphy, C. T., Zou, S., Pletcher, S. D., Chin, C. S., Jan, Y. N., Kenyon, C., Bargmann, C. I. & Li, H. (2004) Nat. Genet. 36, 197-204. [DOI] [PubMed] [Google Scholar]
- 7.Gasch, A. P., Moses, A. M., Chiang, D. Y., Fraser, H. B., Berardini, M. & Eisen, M. B. (2004) PloS Biol. 2, e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig, M. Z., Bergman, C., Patel, N. H. & Kreitman, M. (2000) Nature 403, 564-567. [DOI] [PubMed] [Google Scholar]
- 9.Ludwig, M. Z., Patel, N. H. & Kreitman, M. (1998) Development (Cambridge, U.K.) 125, 949-958. [DOI] [PubMed] [Google Scholar]
- 10.Schawalder, S. B., Kabani, M., Howald, I., Choudhury, U., Werner, M. & Shore, D. (2004) Nature 432, 1058-1061. [DOI] [PubMed] [Google Scholar]
- 11.Wade, J. T., Hall, D. B. & Struhl, K. (2004) Nature 432, 1054-1058. [DOI] [PubMed] [Google Scholar]
- 12.Martin, D. E., Soulard, A. & Hall, M. N. (2004) Cell 119, 969-979. [DOI] [PubMed] [Google Scholar]
- 13.Witt, I., Kwart, M., Gross, T. & Kaufer, N. F. (1995) Nucleic Acids Res. 23, 4296-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurtzman, C. P. & Robnett, C. J. (2003) FEMS Yeast Res. 3, 417-432. [DOI] [PubMed] [Google Scholar]
- 15.Li, B., Oestreich, S. & de Lange, T. (2000) Cell 101, 471-483. [DOI] [PubMed] [Google Scholar]
- 16.Cohn, M., McEachern, M. J. & Blackburn, E. H. (1998) Curr. Genet. 33, 83-91. [DOI] [PubMed] [Google Scholar]
- 17.Graham, I. R., Haw, R. A., Spink, K. G., Halden, K. A. & Chambers, A. (1999) Mol. Cell. Biol. 19, 7481-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanoh, J. & Ishikawa, F. (2001) Curr. Biol. 11, 1624-1630. [DOI] [PubMed] [Google Scholar]
- 19.Biswas, K., Rieger, K. J. & Morschhauser, J. (2003) Gene 307, 151-158. [DOI] [PubMed] [Google Scholar]
- 20.Uemura, H., Watanabe-Yoshida, M., Ishii, N., Shinzato, T., Haw, R. & Aoki, Y. (2004) Yeast 21, 1-10. [DOI] [PubMed] [Google Scholar]
- 21.Wahlin, J. & Cohn, M. (2002) Yeast 19, 241-256. [DOI] [PubMed] [Google Scholar]
- 22.Enjalbert, B., Nantel, A. & Whiteway, M. (2003) Mol. Biol. Cell 14, 1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes, J. D., Estep, P. W., Tavazoie, S. & Church, G. M. (2000) J. Mol. Biol. 296, 1205-1214. [DOI] [PubMed] [Google Scholar]
- 24.Dequard-Chablat, M., Riva, M., Carles, C. & Sentenac, A. (1991) J. Biol. Chem. 266, 15300-15307. [PubMed] [Google Scholar]
- 25.Hartman, J. L., IV, Garvik, B. & Hartwell, L. (2001) Science 291, 1001-1004. [DOI] [PubMed] [Google Scholar]
- 26.Kirschner, M. & Gerhart, J. (1998) Proc. Natl. Acad. Sci. USA 95, 8420-8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wray, G. A., Hahn, M. W., Abouheif, E., Balhoff, J. P., Pizer, M., Rockman, M. V. & Romano, L. A. (2003) Mol. Biol. Evol. 20, 1377-1419. [DOI] [PubMed] [Google Scholar]
- 28.Tanay, A., Gat-Viks, I. & Shamir, R. (2004) Genome Res. 14, 829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B. A. & Johnston, M. (2003) Science 301, 71-76. [DOI] [PubMed] [Google Scholar]
- 30.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241-254. [DOI] [PubMed] [Google Scholar]
- 31.Pritsker, M., Liu, Y. C., Beer, M. A. & Tavazoie, S. (2004) Genome Res. 14, 99-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markstein, M., Markstein, P., Markstein, V. & Levine, M. S. (2002) Proc. Natl. Acad. Sci. USA 99, 763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.