Abstract
The phytohormone auxin plays critical roles during plant growth, many of which are mediated by the auxin response transcription factor (ARF) family. MicroRNAs (miRNAs), endogenous 21-nucleotide riboregulators, target several mRNAs implicated in auxin responses. miR160 targets ARF10, ARF16, and ARF17, three of the 23 Arabidopsis thaliana ARF genes. Here, we describe roles of miR160-directed ARF17 posttranscriptional regulation. Plants expressing a miRNA-resistant version of ARF17 have increased ARF17 mRNA levels and altered accumulation of auxin-inducible GH3-like mRNAs, YDK1/GH3.2, GH3.3, GH3.5, and DFL1/GH3.6, which encode auxin-conjugating proteins. These expression changes correlate with dramatic developmental defects, including embryo and emerging leaf symmetry anomalies, leaf shape defects, premature inflorescence development, altered phyllotaxy along the stem, reduced petal size, abnormal stamens, sterility, and root growth defects. These defects demonstrate the importance of miR160-directed ARF17 regulation and implicate ARF17 as a regulator of GH3-like early auxin response genes. Many of these defects resemble phenotypes previously observed in plants expressing viral suppressors of RNA silencing and plants with mutations in genes important for miRNA biogenesis or function, providing a molecular rationale for phenotypes previously associated with more general disruptions of miRNA function.
INTRODUCTION
MicroRNAs (miRNAs) are endogenous 21-nucleotide riboregulators that modulate gene expression in plants and animals (Bartel and Bartel, 2003; Carrington and Ambros, 2003; Bartel, 2004; Mallory and Vaucheret, 2004). In plants, miRNAs are processed from imperfectly complementary stem-loop precursor RNAs (Reinhart et al., 2002), and proper accumulation of some miRNAs depends on the nuclear activity of DICER-LIKE1 (DCL1) as well as HYL1, HEN1, and AGO1 (Park et al., 2002; Reinhart et al., 2002; Boutet et al., 2003; Kasschau et al., 2003; Han et al., 2004; Vaucheret et al., 2004; Vazquez et al., 2004a). In both plants and animals, miRNAs can direct cleavage of target mRNAs and/or inhibition of productive translation, although plant miRNAs appear to act primarily via mRNA cleavage (Lee et al., 1993; Wightman et al., 1993; Olsen and Ambros, 1999; Hutvágner and Zamore, 2002; Llave et al., 2002; Aukerman and Sakai, 2003; Kasschau et al., 2003; Tang et al., 2003; Chen, 2004; Yekta et al., 2004). Studies examining pairing requirements between miR165/166:PHABULOSA and miR171:SCL6 miRNA:target pairs have revealed that mismatches in the miRNA 5′ region disrupt cleavage more effectively than mismatches in the miRNA 3′ region (Mallory et al., 2004b; Parizotto et al., 2004). Many plant and animal miRNAs do not accumulate ubiquitously, but instead are restricted to specific tissue types and developmental stages, suggesting spatial and temporal regulation of miRNA accumulation and, thus, of target expression (Carrington and Ambros, 2003; Ambros, 2004; Bartel, 2004).
Many miRNAs isolated from the dicot Arabidopsis thaliana are conserved in the monocot rice (Oryza sativa) and in other plants, implying conserved evolutionary roles for plant miRNAs (Reinhart et al., 2002; Floyd and Bowman, 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Wang et al., 2004a; Axtell and Bartel, 2005). Plant miRNAs often have extensive, evolutionarily conserved complementarity to plant mRNAs (Rhoades et al., 2002; Axtell and Bartel, 2005). This observation has enabled numerous regulatory targets to be confidently predicted (Park et al., 2002; Rhoades et al., 2002; Xie et al., 2003; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Wang et al., 2004b), more than 50 of which have been experimentally validated in plants (Dugas and Bartel, 2004).
Plant miRNA complementary sites are usually present as single copies in the open reading frame of the target mRNA, although complementary sites in both 5′ and 3′ untranslated regions (UTRs) have been reported. This trend contrasts with the initially characterized animal miRNA complementary sites, which typically are present as multiple sites in the 3′ UTR of target mRNAs (Bartel, 2004). However, recent studies indicate that targeting of single sites and targeting within open reading frames are widespread for mammalian miRNAs (Lewis et al., 2005; Lim et al., 2005).
Many plant miRNA targets encode transcription factors involved in cell fate determination (Rhoades et al., 2002), supporting the idea that miRNAs regulate plant development. In further support of this idea, plants impaired in miRNA accumulation, such as dcl1, ago1, hyl1, and hen1 mutants (Park et al., 2002; Reinhart et al., 2002; Boutet et al., 2003; Kasschau et al., 2003; Han et al., 2004; Vaucheret et al., 2004; Vazquez et al., 2004a), and plants expressing viral suppressors of gene silencing that alter miRNA accumulation (Mallory et al., 2002; Kasschau et al., 2003; Chapman et al., 2004; Chen et al., 2004; Dunoyer et al., 2004) display dramatic anomalies during vegetative and floral development.
The overexpression of miRNAs and the expression of miRNA-resistant targets in vivo have allowed assignment of developmental roles to miR159/miR319/JAW, miR164, miR165/166, and miR172 miRNA families. These miRNAs regulate rosette leaf expansion and curvature (Palatnik et al., 2003), embryonic, vegetative, and floral organ boundary formation (Laufs et al., 2004; Mallory et al., 2004a), radial patterning (Emery et al., 2003; Juarez et al., 2004; Mallory et al., 2004b; McHale and Koning, 2004; Zhong and Ye, 2004), and floral organ identity and flowering time (Aukerman and Sakai, 2003; Chen, 2004). miR162 and miR168 likely influence development through feedback regulation of miRNA pathway components, DCL1 and AGO1, respectively (Xie et al., 2003; Vaucheret et al., 2004). Particular miRNAs accumulate in response to sulfur starvation (Jones-Rhoades and Bartel, 2004), abiotic stresses (Sunkar and Zhu, 2004), or phytohormones (Achard et al., 2004; Sunkar and Zhu, 2004), suggesting that miRNAs are important not only for development but also for responses to environmental stimuli.
The phytohormone auxin influences many aspects of plant development (Rogg and Bartel, 2001; Liscum and Reed, 2002; Swarup et al., 2002; Friml, 2003; Jürgens, 2003), and the identity of several miRNA targets suggests roles for miRNAs in auxin signaling. miR160 is complementary to AUXIN RESPONSE FACTOR10 (ARF10), ARF16, and ARF17 (Rhoades et al., 2002), and miR167 is complementary to ARF6 and ARF8 (Rhoades et al., 2002; Bartel and Bartel, 2003). Thus, at least five of the 23 Arabidopsis ARF transcription factors are potentially miRNA regulated. ARFs are a plant-specific family of DNA binding proteins that control auxin-regulated transcription (Guilfoyle et al., 1998). They bind to auxin-responsive promoter elements (AuxREs), which are found in early auxin response genes, including Auxin/Indole-3-Acetic Acid (Aux/IAA), SAUR, and GH3, and can either enhance or repress transcription (Abel and Theologis, 1996; Ulmasov et al., 1999a, 1999b; Hagen and Guilfoyle, 2002). Most ARFs have a conserved N-terminal DNA binding domain (DBD), a nonconserved middle region conferring transcriptional repression or activation, and a conserved C-terminal dimerization domain (CTD) that can mediate ARF homodimerization and heterodimerization with Aux/IAA repressors (Guilfoyle et al., 1998; Ulmasov et al., 1999a, 1999b; Guilfoyle and Hagen, 2001; Liscum and Reed, 2002; Tiwari et al., 2003).
Aux/IAA proteins, short-lived nuclear proteins, can heterodimerize with activating ARF proteins, preventing early auxin-response gene expression (Kim et al., 1997; Rouse et al., 1998; Reed, 2001; Dharmasiri and Estelle, 2002; Kepinski and Leyser, 2002; Liscum and Reed, 2002; Tiwari et al., 2004). C-terminal domains of Aux/IAA proteins mediate heterodimerization and are conserved with the CTD of most ARF proteins. Increased auxin levels accelerate proteolysis of Aux/IAA proteins, which would allow ARF proteins to homodimerize and impose their regulatory functions on early auxin-response gene expression.
The transcriptional repressors ARF1-ARF4 and ARF9 have Pro-Ser-Thr–rich middle regions, whereas the transcriptional activators ARF5-ARF8 have Gln-Leu-Ser–rich middle regions (Ulmasov et al., 1999b; Tiwari et al., 2003). These observations suggest that ARFs can be classified as activators or repressors based on the amino acid composition of their middle regions. In addition, most tested ARFs require CTD-mediated dimerization to bind stably to AuxREs in vitro (Ulmasov et al., 1997, 1999a); however, ARF1 does not require the CTD to bind AuxREs in vitro (Ulmasov et al., 1997, 1999a), and ARF3 lacks a CTD. ARF10 and ARF16 display typical ARF sequence characteristics: each has a conserved DBD and CTD and a nonconserved middle region. By contrast, ARF17 is unusual because it lacks a conserved CTD, which is present in 21 of 23 Arabidopsis ARF proteins (Ulmasov et al., 1997, 1999a; Hagen and Guilfoyle, 2002).
miR160 and target ARFs are conserved in dicots and monocots (Rhoades et al., 2002; Bartel and Bartel, 2003), but the importance of miR160-directed regulation of ARF17, ARF16, and ARF10 has not been explored. These three ARFs comprise a subgroup of Arabidopsis ARF proteins (Remington et al., 2004). arf10 and arf16 loss-of-function mutants do not display obvious developmental anomalies (Okushima et al., 2005), and ARF17 has not been characterized in vivo. Here, we demonstrate that disrupting miR160 regulation of ARF17 increases ARF17 mRNA levels, leads to severe developmental abnormalities, including defects in embryonic, root, vegetative, and floral development, and alters GH3-like gene expression. These results indicate that miR160-directed regulation is critical for the developmental functions of ARF17 and expose ARF17 as a possible transcriptional regulator of GH3-like early auxin-response genes.
RESULTS
miR160 Regulates ARF10, ARF16, and ARF17 Expression
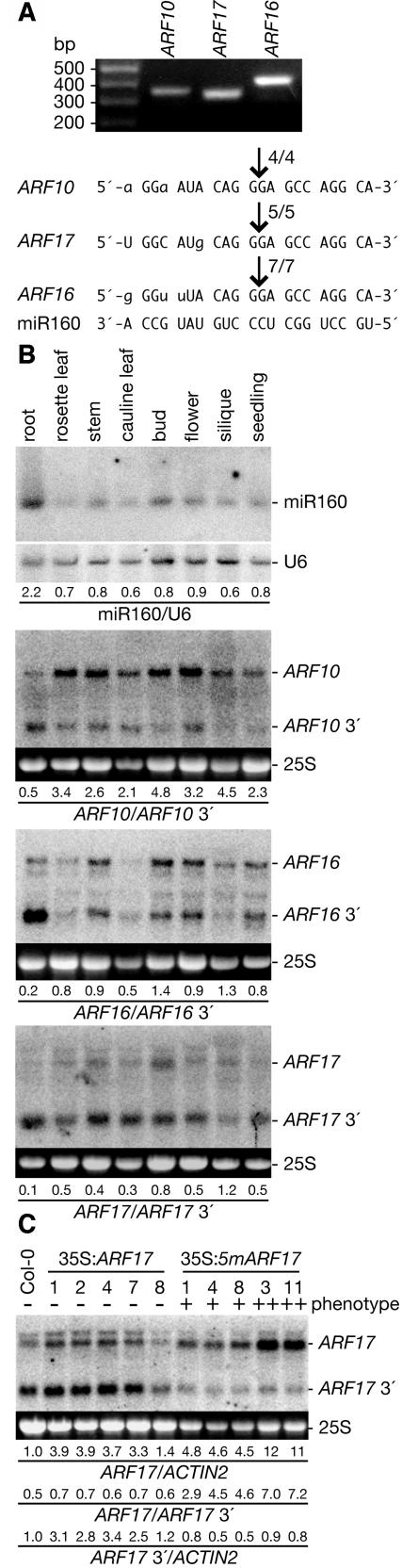
We detected fragments of ARF10, ARF16, and ARF17 mRNAs from Arabidopsis tissues and mapped the end of each fragment to precisely the nucleotide predicted for miR160-directed cleavage (Figure 1A) using a modified form of rapid amplification of cDNA ends (5′ RACE) (Llave et al., 2002). This analysis confirmed what had been reported for ARF10 and ARF17 (Kasschau et al., 2003) and indicated that ARF16 also is posttranscriptionally regulated by miR160. To determine the tissues in which miR160 regulates ARF10, ARF16, and ARF17 mRNAs, we assayed RNA accumulation in a variety of wild-type Arabidopsis tissues using gel blot analysis. We found miR160 and ARF10, ARF16, and ARF17 full-length mRNAs in all tissues analyzed (Figure 1B), indicating that these genes are widely expressed. In addition to full-length transcripts, mRNAs the size of predicted ARF10, ARF16, and ARF17 3′ cleavage products were detected in all tissues analyzed (Figure 1B), suggesting that miR160 is posttranscriptionally regulating these messages in many cells of the plant. The accumulation of miR160 and ARF10, ARF16, and ARF17 3′ cleavage products was highest in roots (Figure 1B), but there was no simple correlation among the level of miR160 and full-length ARF mRNA accumulation in the tissues analyzed (Figure 1B). The relative abundance of ARF17 3′ cleavage product compared with full-length ARF17 mRNA in many tissues (Figure 1B) suggested extensive miRNA-directed cleavage of this transcript and prompted us to explore ARF17 regulation in more detail.
Figure 1.
miR160 Regulates ARF10, ARF16, and ARF17 in Vivo.
(A) miR160 cleavage sites in ARF10, ARF16, and ARF17 mRNAs determined by RNA ligase-mediated 5′ RACE. The agarose gel shows the nested PCR products representing the 3′ cleavage fragments that were cloned and sequenced for each gene. The frequency of 5′ RACE clones corresponding to each cleavage site (arrows) is shown as a fraction, with the number of clones matching the target message in the numerator. As expected for miRNA-directed cleavage (Llave et al., 2002), the clones ended at the nucleotide that pairs with the 10th nucleotide of miR160. Mismatched nucleotides are in lowercase letters.
(B) miR160 and ARF10, ARF16, and ARF17 mRNAs accumulate differentially in Arabidopsis (Col-0) tissues. Shown is an RNA gel blot analysis of 30 μg total RNA prepared from root, rosette leaf, stem, cauline leaf, buds and inflorescence meristems (bud), flower, seedpods and embryos (silique), and 12-d-old seedling tissues with a DNA probe complementary to miR160. The blot was stripped and reprobed with an oligonucleotide complementary to U6 as a loading control. Also shown is RNA gel blot analysis of 7 μg of the same RNA with RNA probes complementary to 3′ ARF10, 3′ ARF16, or 3′ ARF17. The positions of full-length mRNAs and 3′ cleavage products are noted at the right. Ethidium bromide staining of 25S rRNA is shown as a loading control. Ratios of miR160 to U6 RNA or ARF full-length mRNA to 3′ cleavage product are indicated below each lane.
(C) RNA gel blot analysis of 10 μg total RNA from rosette leaves of wild-type Col-0, 35S:ARF17, and 35S:5mARF17 T1 plants with a 3′ ARF17 probe. The absence (−) or presence of a moderate (+) or severe (++) rosette leaf phenotype and individual transformant numbers are indicated above the lanes. The positions of full-length ARF17 mRNA and ARF17 3′ cleavage product are noted at the right. The 25S rRNA is shown as a loading control. The blot was rehybridized with an ACTIN2 probe, and normalized values of ARF17 full-length mRNA and ARF17 3′ cleavage product to ACTIN2 (At3g18780) RNA (with Col-0 levels set at 1.0) and ratios of ARF17 full-length mRNA to 3′ cleavage product are indicated.
35S:ARF17 Plants Overaccumulate ARF17 3′ Cleavage Product
To assess the importance of ARF17 transcriptional regulation during development, we expressed an ARF17 genomic clone in wild-type Arabidopsis under the control of the 35S promoter of Cauliflower mosaic virus (CaMV), which drives strong constitutive expression (Cary et al., 2002). Among 23 35S:ARF17 primary transformants, 21 displayed no obvious developmental defects, whereas two displayed a slight reduction in rosette leaf size and overall stature (data not shown). RNA gel blot analysis revealed a threefold to fourfold increase in full-length ARF17 mRNA accumulation in rosette leaves of four out of five 35S:ARF17 primary transformants, including the two smaller plants (35S:ARF17-1 and 35S:ARF17-4) (Figure 1C). The level of miR160-directed ARF17 3′ cleavage product also was increased approximately threefold in these four 35S:ARF17 plants compared with wild-type Columbia-0 (Col-0) (Figure 1C). The modest increase in ARF17 mRNA expression in 35S:ARF17 Arabidopsis plants prompted us to test the efficacy of our 35S:ARF17 construct with an Agrobacterium tumefaciens–mediated transient expression assay in Nicotiana benthamiana leaves. Although we did not detect endogenous ARF17 mRNA in N. benthamiana leaves, high levels of ARF17 full-length mRNA accumulated after Agrobacterium inoculation with the 35S:ARF17 construct (data not shown), indicating that the 35S:ARF17 construct was capable of driving high level expression. The relatively low level of ARF17 mRNA and the increased level of ARF17 3′ cleavage product in 35S:ARF17 Arabidopsis plants (Figure 1C), along with the wide miR160 expression profile in wild-type plants (Figure 1B), suggested that endogenous miR160 may be sufficiently abundant to direct cleavage of much of the excess ARF17 mRNA that results from 35S driven expression, at least in the relevant tissues.
Plants Expressing miR160-Resistant ARF17 Have Dramatic Developmental Defects
The above results suggested that posttranscriptional regulation limits ARF17 mRNA accumulation. To examine the importance of miR160 in this regulation, we constructed a miR160-resistant version of ARF17. This construct, designated 5mARF17, had five silent mutations within the miR160-complementary domain of an ARF17 genomic clone, thereby increasing the number of mismatches between miR160 and the ARF17 mRNA from one in wild type to six without altering the amino acid sequence of the encoded ARF17 protein (Figure 2A). These substitutions also created an ApaLI restriction site (Figure 2A) that allowed us to distinguish RT-PCR products of 5mARF17 transcripts from those of ARF17.
Figure 2.
5mARF17 RNA Is Resistant to miR160-Directed Cleavage.
(A) Illustration of the ARF17 genomic clone used to transform Arabidopsis. Locations of the intergenic sequences (solid lines), introns (dashed lines), 5′ and 3′ UTRs (black boxes), DBD, remainder of the ARF17 open reading frame (open boxes), start (ATG) and stop (TAA) codons, miR160 complementary site (asterisk), and the EcoRI and SpeI sites used for cloning are shown. mRNA segments from ARF17 and 5mARF17 are shown paired with miR160, with the ApaLI restriction site unique to 5mARF17 and the protein sequence encoded by the miR160 complementary sites of both mRNAs indicated.
(B) ARF17 RNA cleavage directed by miR160 in wheat germ extracts. The 5′-radiolabeled transcripts prepared from wild-type ARF17 and 5mARF17 constructs were introduced into wheat germ extracts, and the time course of cleavage was examined on a denaturing polyacrylamide gel. The positions of ARF17 RNA and 5′ ARF17 cleavage product are noted at the left.
To determine whether miR160-directed cleavage of 5mARF17 RNA was reduced, we used an in vitro assay that relies on miRNA-programmed silencing complexes endogenously present in wheat germ extract (Tang et al., 2003), which can direct cleavage of wild-type ARF17 RNA (G. Tang, M. Jones-Rhoades, D.P. Bartel, and P. Zamore, unpublished data), consistent with the presence of a miR160 locus in wheat (Triticum aestivum) (Jones-Rhoades and Bartel, 2004) and the cloning of miR160 from wheat germ extract (R. Rajagopalan and D.P. Bartel, unpublished data). For wild-type ARF17 RNA, but not for 5mARF17 RNA, a product of the size expected for miR160-directed cleavage was detected, demonstrating that 5mARF17 mismatches interfered with miR160-directed ARF17 cleavage (Figure 2B).
To assess the in vivo consequences of disrupting miR160 regulation, we transformed wild-type Arabidopsis plants with 5mARF17 and control ARF17 genomic constructs under the control of the native ARF17 5′ and 3′ regulatory sequences. To preserve endogenous ARF17 transcriptional regulation, we included 1.9 kb of 5′ flanking sequence, which extends ∼120 bp into the annotated 3′ UTR of the upstream gene At1g77840 (∼60 bp downstream of the At1g77840 stop codon), and 1.7 kb of 3′ flanking sequence, which stops 170 bp upstream of the stop codon of the downstream, reverse-oriented gene At1g77855 (Figure 2A).
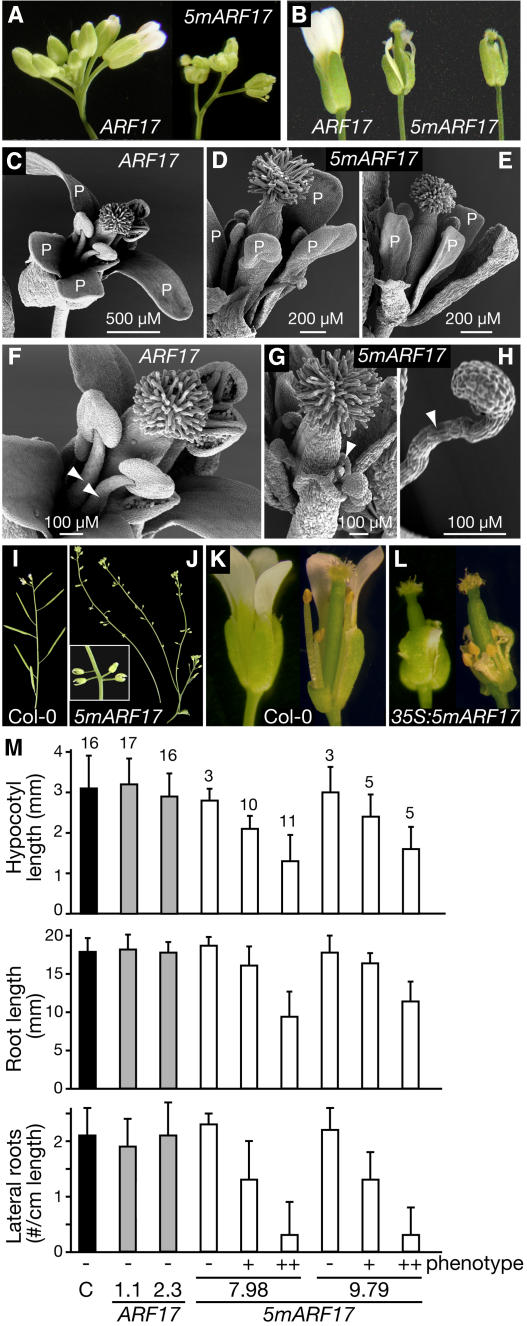
None of the 101 control ARF17 primary transformants displayed obvious or consistent developmental anomalies (Figures 3 and 4). By contrast, 51 of 90 5mARF17 transformants displayed prominent vegetative and floral defects, including rosette and cauline leaf margin serration (Figures 3E to 3I), upward curling of the leaf margins (Figures 3E to 3I), reduced plant size (Figures 3E to 3I), accelerated flowering time (Figures 3G to 3I), altered phyllotaxy along the primary and lateral stems (Figures 4I and 4J), reduced petal size (Figures 4A to 4E), abnormal stamen structure (Figures 4F to 4H), and reduced fertility (Figures 4I and 4J). Eight of the 51 5mARF17 transformants with developmental abnormalities died before the transition to flowering, and five plants flowered successfully but were sterile.
Figure 3.
Embryonic and Vegetative Developmental Defects of 5mARF17 Plants.
(A) Eight-day-old control ARF17 and 5mARF17 T2 seedlings displaying a range of embryonic phenotypes, including lobed cotyledons (bottom left of 5mARF17 panel) and extra cotyledons indicative of trilateral and quadrilateral embryonic symmetry (right 5mARF17 panels).
(B) Scanning electron micrographs of a 6-d-old control ARF17 (left) plant with the normal two cotyledons (asterisks) and two emerging true leaves (arrowheads) and 10-d-old 5mARF17 seedlings (middle and right). The middle plant displays a lobed cotyledon and a downwardly curled cotyledon, and the right plant has three cotyledons (asterisks) and three emerging true leaves (trilateral symmetry).
(C) Scanning electron micrographs of a 10-d-old 5mARF17 seedling (left), 6-d-old 5mARF17 seedling (middle), and magnified view of the 6-d-old 5mARF17 shoot apex (right). Indicative of quadrilaterally symmetric embryos, the 5mARF17 seedlings had four cotyledons (asterisks) and four emerging rosette leaves (arrowheads) rather than two of each.
(D) Scanning electron micrographs of abaxial surfaces of 6-d-old control ARF17 and 5mARF17 cotyledons showing altered epidermal cell shape and organization in the 5mARF17 cotyledon.
(E) Twenty-two-day-old control ARF17 (left) and 5mARF17 (top right) T2 plants and a 30-d-old 5mARF17 T2 plant with severe leaf serration (bottom right).
(F) Rosette leaves from 30-d-old control ARF17 (left) and 5mARF17 (right) T2 plants.
(G) Thirty-two-day-old control ARF17 (left) and 5mARF17 (right) T1 plants. The 5mARF17 plant has bolted.
(H) Magnified view of 5mARF17 plant in (G).
(I) Seventy-two-day-old control ARF17 and three 5mARF17 T2 plants grown in short days (8 h light, 16 h dark). The 5mARF17 developmental defects are increasing in severity from left to right; only the plant with the most severe phenotype has bolted.
(J) Col-0 and 35S:5mARF17 rosette leaves with defects increasing in severity from left to right.
Figure 4.
Floral Organ and Seedling Defects of 5mARF17 Plants.
(A) Side view of the inflorescence bud cluster of ARF17 and 5mARF17 T1 plants.
(B) Mature ARF17 and 5mARF17 T2 flowers with 5mARF17 petal and stamen defects increasing in severity from left to right.
(C) to (E) Scanning electron micrographs of an ARF17 T2 mature flower (C) and 5mARF17 T2 mature flowers with moderate floral organ defects and reduced fertility ([D] and [E]). Petal (P) length and width was reduced in 5mARF17 flowers.
(F) to (H) Scanning electron micrographs of ARF17 T2 (F) and 5mARF17 T2 (G) reproductive structures. Arrowheads point to the stamen. (H) is a magnified view of 5mARF17 stamen. Note the reduced filament length, abnormal stamen orientation, and reduced anther size in 5mARF17 flowers. The 5mARF17 flowers in (G) and (H) were from infertile plants.
(I) Wild-type Col-0 plant floral phyllotaxy along the primary stem.
(J) 5mARF17 floral phyllotaxy along the primary and lateral stems. 5mARF17 plants have reduced fertility, illustrated by the failure to produce filled seedpods. The inset shows a magnified view of altered 5mARF17 floral phyllotaxy.
(K) Mature wild-type Col-0 flower (left) and Col-0 flower with sepals and petals removed to expose reproductive organs (right).
(L) Mature 35S:5mARF17 flower (left) and 35S:5mARF17 flower with sepals and petals removed to expose reproductive organs (right).
(M) Average hypocotyl length, primary root length, and lateral root number (expressed as a fraction of the primary root length) of 7-d-old untransformed control seedlings (C; black bars) and 7-d-old progeny of two ARF17 T3 homozygous lines (gray bars) and two 5mARF17 T3 heterozygous lines (white bars). The 5mARF17 progeny with wild-type phenotypes (−) lacked the transgene; those with mild (+) or severe (++) phenotypes scored positive for the 5mARF17 transgene. The number of plants analyzed is indicated above each bar, and error bars represent standard deviations.
5mARF17 progeny plants displayed presumed embryonic defects, including symmetry anomalies in which the normally bilateral symmetry of germinating seedlings was instead trilateral or quadrilateral, resulting in one or two extra cotyledons (Figures 3A to 3C). Two individual hemizygous 5mARF17 plants segregated 10% (9/89) and 14% (11/81) progeny with three cotyledons and 3% (3/89) and 4% (3/81) progeny with four cotyledons. Plants with three cotyledons had three simultaneously emerging first true leaves (Figure 3B) at a frequency of 6/9 and 7/11, whereas plants with four cotyledons always had four simultaneously emerging first true leaves (Figure 3C). In addition to cotyledon-number defects, 5mARF17 cotyledons were often moderately or deeply lobed (heart shaped) and downwardly curled (Figures 3A to 3C). Abaxial cotyledon epidermal cells were altered in shape, size, and distribution (Figure 3D), perhaps contributing to the downward curling of the cotyledons. By contrast, adaxial epidermal cells of 5mARF17 cotyledons appeared relatively normal (data not shown). 5mARF17 plants displaying mild embryonic phenotypes (moderately lobed cotyledons) had mild vegetative and floral organ defects, whereas plants displaying more severe embryonic phenotypes (heart-shaped or extra cotyledons) had more severe vegetative and floral organ defects. 5mARF17 seedlings also displayed reduced root and hypocotyl lengths and decreased root branching (Figure 4M). Like the primary transformants, 5mARF17 progeny plants flowered slightly earlier than control plants. Five independent 5mARF17 lines with similar rosette leaf defects flowered on day 24.4 ± 1.1 (n = 34), 25.2 ± 1.6 (n = 35), 25.1 ± 1.4 (n = 30), 24.7 ± 1.1 (n = 29), and 24.3 ± 0.7 (n = 30), whereas nontransformed plants flowered on day 27.4 ± 1.7 (n = 44), and four independent ARF17 control lines flowered on day 27.1 ± 1.6 (n = 24), 27.4 ± 1.2 (n = 24), 27.7 ± 1.4 (n = 20), and 27.0 ± 1.5 (n = 20). Despite extensive efforts, we were unable to recover a homozygous 5mARF17 plant with rosette leaf defects that was fertile.
To observe the effects of disrupting both transcriptional and miR160 regulation of ARF17, we transformed wild-type plants with the 5mARF17 construct under the control of the CaMV 35S promoter (35S:5mARF17). Like the 5mARF17 primary transformants, nine out of 19 35S:5mARF17 primary transformants displayed embryonic (data not shown), rosette leaf (Figure 3J), and floral defects (Figures 4K and 4L). Although many of the phenotypes were qualitatively similar to those of 5mARF17 plants, the severity of leaf curling (Figure 3J) and floral organ defects (Figures 4K and 4L) and the frequency of premature death was greater in 35S:5mARF17 plants than 5mARF17 plants; two 35S:5mARF17 plants died as seedlings, four plants never transitioned to flowering but died after producing 8 to 10 upwardly curled and serrated rosette leaves, and three plants produced flowers with stamen and petal defects accompanied by reduced fertility or sterility.
5mARF17 and 35S:5mARF17 Plants Overaccumulate ARF17 mRNA
RNA gel blot analysis revealed that 12- and 16-d-old 5mARF17 seedlings displaying cotyledon phenotypes (Figures 5A and 5C), as well as roots, rosette, and cauline leaves, stems, inflorescence meristems and buds, flowers, and siliques of older 5mARF17 plants with aberrant phenotypes (Figure 5C), all accumulated more ARF17 mRNA than control plants. Moreover, 35S:5mARF17 transformants with aberrant phenotypes accumulated more full-length ARF17 mRNA than 35S:ARF17 or untransformed wild-type Col-0 plants (Figure 1C).
Figure 5.
Phenotypic 5mARF17 Plants Accumulate Increased ARF17 mRNA.
(A) RNA gel blot analysis of 7 μg total RNA prepared from 16-d-old T2 seedlings with a 3′ ARF17 probe. The positions of full-length ARF17 mRNA and ARF17 3′ cleavage product are noted on the right. The 25S rRNA is shown as a loading control. The blot was rehybridized with an ACTIN2 probe, and normalized values of ARF17 full-length mRNA to ACTIN2 mRNA (with Col-0 levels set at 1.0) and ratios of ARF17 full-length mRNA to 3′ cleavage product are indicated.
(B) RNA gel blot analysis of 10 μg total RNA prepared from rosette leaves of 32-d-old T2 plants with a 3′ ARF17 probe. The absence (−) or presence of a mild (+) or severe (++) rosette leaf phenotype is indicated. The positions of full-length ARF17 mRNA and ARF17 3′ cleavage products are noted at the right. Normalization was as in (A).
(C) RNA gel blot analysis of 5 μg total RNA prepared from Col-0 and 5mARF17 root (r), rosette leaf (rl), stem (st), cauline leaf (cl), buds and inflorescence meristems (b), flower (f), silique and embryonic tissues (si), and 12-d-old seedling (se) tissues with a 3′ ARF17 probe. The positions of full-length ARF17 mRNA and ARF17 3′ cleavage product are noted at the right. Two uncharacterized ARF17-hybridizing RNAs, one longer and one shorter than the full-length ARF17 transcript, accumulated in siliques of Col-0 and 5mARF17 plants. Ratios of ARF17 full-length mRNA to 3′ cleavage product are indicated, along with the relative levels of ARF17 mRNA in 5mARF17 plants compared with wild-type Col-0 plants in each tissue (5mARF17/Col-0 ARF17).
(D) Expression of 5mARF17 mRNA in untransformed (untxf) and T2 transgenic plants that were individual progeny of the indicated primary transformant. Rosette leaf RNA was reverse transcribed, and the resulting cDNA was PCR-amplified to completion, then digested with ApaLI, which cuts the 5mARF17 amplicon but not the ARF17 amplicon or heteroduplex molecules resulting from annealing ARF17 and 5mARF17 strands. Agarose gel separation and ethidium bromide staining revealed the full-length PCR product (330 bp) and an ApaLI digestion fragment (250 bp). Each lane is an analysis of an individual plant, and the presence (+) or absence (−) of a rosette leaf phenotype in that plant is indicated. DNA gel blot analysis (data not shown) was used to quantitate the 330- and 250-bp DNAs, and the amount of 5mARF17 relative to ARF17 after correcting for heteroduplex formation is shown below each lane.
The increased ARF17 mRNA levels in 5mARF17 and 35S:5mARF17 plants correlated with the severity of the rosette leaf phenotype (Figures 1C and 5B). The increase was not accompanied by a similar increase in 3′ cleavage product accumulation (Figures 1 and 5), suggesting that the excess ARF17 mRNA did not derive from the endogenous ARF17 gene but instead derived from the 5mARF17 or 35S:5mARF17 transgene, which both produce mRNA refractory to miR160-directed cleavage (Figure 2B). Indeed, the fraction of ARF17 RT-PCR product that corresponded to 5mARF17, as monitored by ApaLI digestion, correlated with the presence of the rosette leaf phenotype (Figure 5D). miR160 levels were unchanged in control ARF17 and 5mARF17 plants (data not shown), indicating that an additional copy of ARF17 or 5mARF17 does not affect miR160 accumulation.
Because developmental defects consistent with those observed in 5mARF17 and 35S:5mARF17 plants were not observed in 101 control ARF17 primary transformants or numerous progeny of these plants or 23 35S:ARF17 primary transformants, we conclude that the developmental phenotypes and increased ARF17 mRNA accumulation in 5mARF17 and 35S:5mARF17 plants resulted from disrupting miR160-directed ARF17 regulation, rather than from expressing an extra copy of ARF17. Together, these results show that miR160 is crucial for the posttranscriptional regulation of ARF17 expression and that this regulation is necessary for the proper growth and development of many Arabidopsis organs.
5mARF17 Plants Respond to Auxin Treatment
5mARF17 plants have fewer lateral roots and shorter hypocotyls than control plants (Figure 4M), traits characteristic of auxin-resistant mutants (Estelle and Somerville, 1987; Hobbie and Estelle, 1995; Monroe-Augustus et al., 2003). 5mARF17 plants also have shorter primary roots than control plants (Figure 4M), whereas auxin-resistant mutants typically have increased root length (Estelle and Somerville, 1987; Hobbie and Estelle, 1995; Monroe-Augustus et al., 2003). To determine if the response to exogenous auxin was altered in 5mARF17 plants, we measured root and hypocotyl length after exposing plants to the auxin IAA. Even though the primary root and hypocotyl lengths of 5mARF17 plants were shorter than those of control plants before IAA treatment, IAA treatment decreased primary root and hypocotyl growth in 5mARF17 plants (Figures 6A and 6B), indicating that 5mARF17 plants were not dramatically impaired in these responses to exogenous IAA.
Figure 6.
IAA Treatment of Wild-Type and 5mARF17 Plants.
(A) Primary root length of 8-d-old Col-0 and T2 seedlings mock treated (white bars) or treated with 50 nM IAA (black bars). 5mARF17 progeny with wild-type phenotypes (−) lacked the transgene; those with mild (+) or severe (++) phenotypes scored positive for the 5mARF17 transgene. The number of plants analyzed is indicated above each bar, and standard deviations are shown (except in cases where fewer than three plants were analyzed).
(B) Hypocotyl length of 7-d-old Col-0 and T2 seedlings mock treated (white bars) or treated with 0.1 μM IAA (gray bars) and 10 μM IAA (black bars). 5mARF17 progeny with wild-type phenotypes (−) lacked the transgene; those with mild (+) or severe (++) phenotypes scored positive for the 5mARF17 transgene. The number of plants analyzed is indicated above each bar, and standard deviations are shown (except in cases where fewer than three plants were analyzed).
(C) RNA gel blot analysis of 10 μg total RNA prepared from IAA treated (+) or untreated (−) Col-0 seedlings with DNA probes complementary to miR160, miR164, miR167, or U6 (loading control). Blots were stripped between each hybridization. The positions of 32P-labeled RNA oligonucleotides are noted at the right. Also shown are RNA gel blot analyses of 10 μg of the same RNA with RNA probes complementary to IAA1, IAA19, or 3′ ARF17. The positions of each full-length transcript and ARF17 3′ cleavage product are noted at the left. The 25S rRNA is shown as a loading control. Normalized values of miRNA to U6 RNA and IAA1, IAA19, and ARF17 full-length mRNAs to ACTIN2 mRNA (with untreated Col-0 levels set at 1.0) and ratios of ARF17 full-length mRNA to 3′ cleavage product are indicated.
IAA Treatment Does Not Appreciably Alter miR160, miR164, and miR167 Accumulation in Seedlings
In addition to miR160 and miR167, which regulate ARF genes (Figure 1; Rhoades et al., 2002; Kasschau et al., 2003), miR164 regulates NAC-domain genes (Laufs et al., 2004; Mallory et al., 2004a), some of which are also implicated in auxin signaling (Xie et al., 2000; Aida et al., 2002; Furutani et al., 2004). To determine whether levels of miRNAs implicated in auxin signaling are affected by exogenous IAA, we analyzed miR160, miR164, and miR167 accumulation in wild-type seedlings after IAA treatment. Although IAA treatment dramatically increased IAA1 and IAA19 early auxin-response transcript levels, miR160, miR164, and miR167 accumulation was not substantially altered at the times analyzed (Figure 6C), indicating that seedling levels of these miRNAs are not greatly influenced by IAA application. Similarly, ARF17 mRNA and ARF17 3′ cleavage product levels appeared unaffected by IAA treatment (Figure 6C). These results do not provide evidence for a role of auxin in mediating ARF17, miR160, miR164, or miR167 expression in seedlings, suggesting that regulation by these three miRNAs may instead be needed to set components of the auxin-response machinery to proper levels so that tissues can respond appropriately to auxin.
miR160-Directed ARF17 Regulation Is Necessary for Proper Expression of Certain GH3-Like Early Auxin Response Genes
Although no molecular connections between ARF10, ARF16, and ARF17 and auxin responses have been reported, these ARFs all have Pro-Ser-Thr–rich middle regions (see Supplemental Figure 1 online), suggesting that they may repress transcription of early auxin-response genes. To identify molecular changes in 5mARF17 plants, we monitored mRNA accumulation profiles of five GH3-like transcripts and three Aux/IAA transcripts in seedlings and rosette leaves. In 16-d-old seedlings, we found levels of GH3.3 increased approximately fourfold in 5mARF17 seedlings compared with wild-type or control ARF17 seedlings (Figure 7A), whereas levels of IAA1 Aux/IAA mRNA and other GH3-like mRNAs, YDK1/GH3.2, GH3.5, DFL1/GH3.6, and DFL2/GH3.10, were similar in wild-type, control ARF17, and 5mARF17 seedlings (Figure 7). In addition to an approximately fourfold increase in GH3.3 mRNA levels, YDK1/GH3.2 mRNA levels were increased in 30-d-old 5mARF17 rosette leaves (Figure 8A). By contrast, levels of GH3.5 and DFL1/GH3.6 mRNAs were decreased to undetectable levels in rosette leaves of 30-d-old 5mARF17 plants (Figure 8B), whereas levels of DFL2/GH3.10, IAA1, IAA17, and IAA19 mRNAs were similar in rosette leaves of 30-d-old wild-type, control ARF17, and 5mARF17 plants (Figure 8A; data not shown). These results indicate that miR160-directed ARF17 regulation is necessary for proper expression of a subset of GH3-like mRNAs and establish a molecular connection between ARF17 and early auxin responses.
Figure 7.
GH3-Like Expression in 16-d-Old 5mARF17 Seedlings.
(A) RNA gel blot analysis of 7 μg of total RNA from 16-d-old seedlings with RNA probes complementary to YDK1/GH3.2 or GH3.3. The 25S rRNA is shown as a loading control. Blots were rehybridized with an ACTIN2 probe, and normalized values of GH3-like mRNAs to ACTIN2 mRNA are indicated, with mRNA levels in Col-0 control plants set at 1.0.
(B) Similar analysis with a probe complementary to DFL2/GH3.10. Normalized values of DFL2 mRNA to 25S rRNA are indicated, with the mRNA level in Col-0 control plants set at 1.0.
(C) Similar analysis with a probe complementary to GH3.5, DFL1/GH3.6, IAA1, and 3′ ARF17. Normalizations were as in (A).
Figure 8.
GH3-Like Gene Expression Changes in 5mARF17 Rosette Leaves.
(A) RNA gel blot analysis of 7 μg of total RNA from 30-d-old rosette leaves with RNA probes complementary to GH3.3, YDK1/GH3.2, DFL2/GH3.10, IAA1, or 3′ ARF17. The absence (−) or presence of a severe (++) rosette leaf phenotype is indicated. The positions of each full-length transcript and ARF17 3′ cleavage product are noted at the right. The 25S rRNA is shown as a loading control. Normalized values of mRNAs to ACTIN2 mRNA are indicated, with mRNA levels in Col-0 control plants set at 1.0. Because YDK1 transcripts were not detected (ND) in Col-0 or ARF17 control rosette leaves, the approximately fourfold increase in YDK1 signal observed represents a minimum.
(B) Similar analysis with probes complementary to GH3.5 or DFL1/GH3.6. These transcripts were not detected in 5mARF17 rosette leaves, so the fold changes observed represent a minimum.
(C) RNA gel blot analysis of 7 μg of total RNA from 30-d-old rosette leaves with RNA probes complementary to GUS or 3′ ARF17. The absence (−) or presence of a mild (+) or severe (++) rosette leaf phenotype is indicated. The positions of each full-length transcript and ARF17 3′ cleavage product are noted at the right. Normalization is as in (A).
(D) Fifteen-day-old ARF17 DR5-GUS and 5mARF17 DR5-GUS cotyledons.
(E) Twenty-eight-day-old ARF17 DR5-GUS and 5mARF17 DR5-GUS rosette leaves with moderate (top) or more severe (bottom) leaf defects.
YDK1/GH3.2, GH3.3, GH3.5, and DFL1/GH3.6 are group II GH3-like proteins that conjugate IAA to amino acids in vitro (Staswick et al., 2005) and therefore are predicted to decrease active IAA levels in the cell. To determine if responsiveness to endogenous auxin is changed in 5mARF17 plants, we expressed control ARF17 and 5mARF17 in the widely used DR5-GUS reporter line (Guilfoyle, 1999). The DR5-GUS line expresses β-glucuronidase (GUS) under the control of a minimal CaMV 35S promoter fused to multiple copies of the TGTCTC AuxRE, first defined in the auxin responsive promoter of the soybean (Glycine max) GH3 gene. Rosette leaves of control DR5-GUS plants transformed with control ARF17 accumulated similar levels of GUS mRNA as those of the parental line (Figure 8C), and control ARF17 DR5-GUS and DR5-GUS cotyledons and rosette leaves had a similar GUS expression pattern (data not shown). By contrast, phenotypic 5mARF17 DR5-GUS plants with increased ARF17 mRNA levels accumulated approximately twofold more GUS mRNA in rosette leaves (Figure 8C). These increases correlated with more expanded GUS expression in cotyledons and rosette leaves (Figures 8D and 8E), suggesting that auxin responsiveness or perhaps auxin is more widely distributed in cotyledons and rosette leaves of 5mARF17 plants. No change in overall GUS expression was observed in roots (data not shown), suggesting that root auxin responsiveness is not greatly influenced by 5mARF17 expression. Together, these results suggest that miR160-directed regulation of ARF17 is necessary for proper GH3-like gene expression and perhaps auxin distribution.
DISCUSSION
Here, we describe in vivo roles of miR160-directed ARF17 posttranscriptional regulation. Plants expressing a miRNA-resistant version of ARF17 (5mARF17) display increased ARF17 mRNA accumulation and exhibit dramatic developmental defects. These phenotypes correlate with reduced accumulation of GH3.5 and DFL1/GH3.6, two closely related mRNAs (Staswick et al., 2002), and increased accumulation of YDK1/GH3.2 and GH3.3, two other closely related mRNAs (Staswick et al., 2002), as well as DR5-GUS. These results indicate that miR160-directed regulation of ARF17 is critical for proper development, establish a molecular link between ARF17 and the auxin response pathway, and add another posttranscriptional regulatory dimension to ARF-mediated regulation.
The Developmental Abnormalities of 5mARF17 Plants Overlap with Those of Plants Impaired in miRNA Functioning
In Arabidopsis, mutations in DCL1, AGO1, HYL1, and HEN1 impair the miRNA pathway and lead to developmental defects that overlap with those exhibited by 5mARF17 plants. In particular, hypomorphic ago1 rosette leaves are serrated and ago1, hyl1, and hen1 null mutants exhibit upwardly curled rosette leaves and a dwarfed stature. Indeed, miR160 accumulation is reduced and ARF17 mRNA accumulation is increased in dcl1, ago1, hyl1, and hen1 mutants (Kasschau et al., 2003; Vaucheret et al., 2004; Vazquez et al., 2004a), consistent with the possibility that reduced miR160-directed ARF17 regulation contributes to the developmental abnormalities of these mutants.
Viral proteins can interfere with the miRNA pathway and affect development when expressed in plants (Mallory et al., 2002; Kasschau et al., 2003; Chapman et al., 2004; Dunoyer et al., 2004). For example, Arabidopsis plants expressing viral proteins P1/HC-Pro, p19, p15, and p21 display reduced miRNA-directed mRNA cleavage and developmental abnormalities (Kasschau et al., 2003; Chapman et al., 2004; Dunoyer et al., 2004), whereas plants expressing viral proteins 2b, p38, and p25 are not impaired in miRNA-directed target mRNA cleavage and lack dramatic developmental defects (Chapman et al., 2004; Dunoyer et al., 2004). The small, serrated rosette leaf phenotypes of P1/HC-Pro, p19, p15, and p21 expressing plants (Kasschau et al., 2003; Chapman et al., 2004; Dunoyer et al., 2004) are strikingly similar to those of 5mARF17 plants (Figures 3E to 3I). In addition, P1/HC-Pro plants exhibit reduced stamen size (Kasschau et al., 2003; Chapman et al., 2004; Dunoyer et al., 2004), and p19 and p15 plants have smaller petals (Chapman et al., 2004; Dunoyer et al., 2004), also reminiscent of 5mARF17 plants (Figures 4A to 4H), suggesting that disrupted miR160-directed ARF17 regulation can largely explain many of the developmental defects of plants expressing viral-encoded silencing suppressors.
ARF10, ARF16, and ARF17 Are Similar to Repressing ARFs
ARF proteins can either activate or repress transcription, depending on the nature of the middle domain (Ulmasov et al., 1999b). ARF5, ARF6, ARF7, ARF8, and ARF19 are activating ARFs with Gln-Leu-Ser–rich middle regions, whereas ARF1, ARF2, ARF3, ARF4, and ARF9 are repressing ARFs with Pro-Ser-Thr–rich middle regions (Ulmasov et al., 1999b; Tiwari et al., 2003). ARF10, ARF16, and ARF17 and their four rice homologs have Pro-Ser-Thr–rich middle regions (see Supplemental Figure 1 online), suggesting that they might be repressors. However, the five known repressing ARFs are more closely related to the activating ARFs than either group is to ARF10, ARF16, and ARF17 (Remington et al., 2004), so ARF10, ARF16, and ARF17 may define a specialized ARF class. arf10 and arf16 loss-of-function mutants do not display obvious developmental anomalies (Okushima et al., 2005). In an attempt to identify a loss-of-function arf17 mutant, we searched the Salk Institute Genomic Analysis Laboratory collection (Alonso et al., 2003) for plants with disruptions in ARF17. No mutants were found with insertions in the open reading frame of ARF17, but one mutant, SALK_062511, had a T-DNA inserted ∼210 bp upstream of the ARF17 start codon. Plants homozygous for this insertion did not display obvious developmental defects (data not shown); however, RT-PCR revealed that these plants still accumulated ARF17 mRNA (data not shown), indicating that this mutant was not a null allele. As has been demonstrated for other members of the ARF family (Okushima et al., 2005), functional redundancy among ARF10, ARF16, and ARF17 may preclude informative analyses of single arf mutants in this class.
ARF17 Regulates GH3-Like Expression
There are 20 Arabidopsis GH3 homologs, which fall into three clades. Group II GH3 proteins, including YDK1/GH3.2, GH3.3, GH3.5, and DFL1/GH3.6, conjugate IAA to amino acids in vitro (Staswick et al., 2005). Regulating IAA conjugation is important for maintaining endogenous IAA levels (Ljung et al., 2002); these GH3 proteins likely play an important role in auxin responsiveness by reducing active auxin levels and thus negatively regulating auxin signaling.
We observed decreased GH3.5 and DFL1/GH3.6 mRNA levels in 5mARF17 rosette leaves (Figures 8B). Because the Pro-Ser-Thr–rich middle region of ARF17 is consistent with transcriptional repression, an appealing model is that ARF17 acts directly to repress GH3.5 and DFL1/GH3.6 transcription, and the consequent GH3.5 and DFL1/GH3.6 reduction leads to increased IAA, which is known to stimulate the expression of YDK1/GH3.2, GH3.3, and DR5-GUS. However, because it is currently impossible to discern whether the changes in GH3-like transcript levels were direct or indirect consequences of disrupting miR160-directed ARF17 regulation, there are many other possibilities. For example, ARF17 might indirectly regulate GH3-like and DR5-GUS expression by repressing an activator of GH3.5 and DFL1/GH3.6 or by repressing a repressor of YDK1/GH3.2, GH3.3, and DR5-GUS. Although the exact mechanism by which ARF17 regulates GH3-like gene expression is unclear, the observation that miRNA repression of ARF17 is important for proper GH3-like mRNA accumulation provides an entry point for the study of early auxin-responsive gene expression.
Gain-of-Function ydk1-D Mutants Mimic 5mARF17 Hypocotyl and Root Phenotypes
YDK1/GH3.2 mRNA is increased in 5mARF17 rosette leaves (Figure 8A) but is not obviously changed in 5mARF17 16-d-old seedlings (Figure 7A). Dominant ydk1-D mutants, which overexpress YDK1, have hypocotyl and root phenotypes (Takase et al., 2004) similar to 5mARF17 plants (Figure 4M). Both 5mARF17 and ydk1-D plants display reduced primary root length, lateral root number, hypocotyl length, and stature. ydk1-D plants also exhibit reduced apical dominance, which we did not observe in 5mARF17 plants. Because the hypocotyl and root phenotypes of 5mARF17 and ydk1-D plants are similar, it is possible that increased YDK1 expression contributes to 5mARF17 phenotypes.
Gain-of-function dfl1-D plants are resistant to exogenous auxin and exhibit reduced lateral root number and hypocotyl length and a dwarf stature. Antisense DFL1 plants have increased lateral roots (Nakazawa et al., 2001), whereas 5mARF17 plants, which show reduced DFL1/GH3.6 mRNA accumulation in rosette leaves (Figure 8B) but not in seedlings (Figure 7C), have fewer lateral roots (Figure 4M). The expression of at least two other GH3-like genes, GH3.3 and GH3.5, also is altered in 5mARF17 plants (Figures 7 and 8), but mutants in these genes have not been described. Because the expression of at least four GH3-like genes is altered in 5mARF17 plants, the contribution of individual GH3-like genes to the 5mARF17 developmental defects is difficult to determine.
The expression of GH3.5, DFL1, YDK1, and GH3.3 is altered in 5mARF17 rosette leaves (Figures 8A and 8B), but among the five GH3-like transcripts we monitored, GH3.3 was the only transcript noticeably changed in both 5mARF17 seedlings and rosette leaves (Figures 7A and 8A). Because the increase in ARF17 expression is consistently greater in 5mARF17 rosette leaves than in 5mARF17 seedlings (Figures 5, 7, and 8), it is possible that ARF17 levels in seedlings are not sufficient to detectably change expression of the other GH3-like genes when RNA from entire seedlings is pooled for RNA gel blot analysis. Indeed, we observe expanded DR5-GUS expression domains not only in rosette leaves, but also in 5mARF17 cotyledons (Figures 8D and 8E), and the changes visualized by histochemical staining are obvious, whereas the changes in GUS mRNA levels are modest (approximately twofold).
miR160 and miR167 May Coordinately Modulate GH3-Like Expression
ARF8 appears to negatively regulate free IAA levels by controlling GH3-like gene expression (Tian et al., 2004). Levels of three GH3 mRNAs, GH3.5, DFL1/GH3.6, and GH3.17, are reduced in arf8 loss-of-function mutants and increased in ARF8 overexpressing plants (Tian et al., 2004), suggesting that ARF8 activates GH3-like expression. We found that miR160-directed regulation of ARF17 is also important for proper GH3-like expression (Figures 7 and 8). GH3.3 mRNA levels increase in 5mARF17 seedlings and rosette leaves, YDK1/GH3.2 mRNA levels increase in 5mARF17 rosette leaves, and DFL1/GH3.6 and GH3.5 mRNA levels decrease in 5mARF17 rosette leaves (Figures 7, 8A, and 8B). Only a subset of GH3-like transcripts appears to be regulated by both ARF17 and ARF8; DFL2/GH3.10 mRNA levels are unchanged in 5mARF17 rosette leaves (Figure 8A), and GH3.3, DFL2/GH3.10, and JAR1/FIN219 mRNA levels are unchanged in arf8 mutants and ARF8 overexpressing plants (Tian et al., 2004).
Intriguingly, ARF8 is regulated by miR167 (Rhoades et al., 2002; Kasschau et al., 2003), a miRNA unrelated in sequence to miR160. The miR167 complementary sites of ARF6 and ARF8 are in the conserved CTD (Rhoades et al., 2002; Bartel and Bartel, 2003), whereas the miR160 complementary sites in the ARF10, ARF16, and ARF17 mRNAs (Rhoades et al., 2002) comprise the major block of conservation within the middle regions of these ARFs (see Supplemental Figure 1 online). These differences suggest independent evolutionary origins of two ARF-miRNA regulatory pairings. Although a role for miR167-directed ARF8 regulation in GH3-like expression remains to be examined, it is possible that miR160 and miR167 coordinately modulate GH3-like mRNA expression by regulating expression of repressing and activating ARF proteins encoded by ARF17 and ARF8, thus contributing to the intricate interplay between auxin levels and auxin responses.
miRNAs and Auxin Signaling
In addition to ARF regulation by miR160 and miR167 (Rhoades et al., 2002; Kasschau et al., 2003), miR164 and miR393 also target genes implicated in auxin signaling. miR393 targets mRNAs encoding TIR1 and its three most closely related F-box proteins (Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004). TIR1 is the specificity component of an SCF E3 ubiquitin ligase that targets Aux/IAA proteins for ubiquitin-dependent degradation in response to auxin (Gray et al., 1999, 2001). In addition to targeting CUC1 and CUC2 mRNAs (Rhoades et al., 2002; Kasschau et al., 2003; Laufs et al., 2004; Mallory et al., 2004a), which establish organ boundaries in embryos and flowers (Aida et al., 1997), miR164 targets NAC1 (Rhoades et al., 2002; Mallory et al., 2004a), a putative transcription factor that promotes lateral root development downstream of TIR1 (Xie et al., 2000, 2002). Furthermore, ARF3 and ARF4 mRNAs overaccumulate in zip, sgs3, and rdr6 mutants (Peragine et al., 2004); sgs3 and rdr6 are impaired in the accumulation of trans-acting short interfering RNAs, endogenous small RNAs that, like miRNAs, appear to regulate gene expression by directing mRNA cleavage (Peragine et al., 2004; Vazquez et al., 2004b). The propensity for unrelated miRNAs and possibly trans-acting short interfering RNAs to regulate genes involved in auxin signaling suggests that the severe developmental consequences observed when disrupting miRNA-mediated regulation of ARF17 will be one of numerous examples in which endogenous silencing RNAs are shown to be key players modulating auxin responses during development.
METHODS
DNA Constructs, Transgenic Plants, Agrobacterium tumefaciens–Mediated Transient Expression, and in Vitro Cleavage Assay
The genomic sequence of ARF17 (At1g77850), including ∼1.9 and ∼1.7 kb of putative 5′ and 3′ regulatory sequences, respectively, was cloned as an ∼6.6-kb EcoRI-SpeI fragment into pBluescript II SK+ (Stratagene, La Jolla, CA) from the BAC F28K19. Site-directed mutagenesis using primers ARF17 mutagenesis forward and reverse (Table 1) was performed using PfuUltra polymerase followed by DpnI digestion, as suggested by the manufacturer (Stratagene), to produce the 5mARF17 sequence. After mutagenesis, an ∼1.9-kb SgrAI-SexAI fragment spanning the mutagenized ARF17 miR160 complementary site was subcloned and used to replace the corresponding wild-type sequence of the original ARF17 genomic clone. This 1.9-kb fragment was sequenced to ensure that only the desired silent mutations were present. The control ARF17 and the 5mARF17 ∼6.6-kb EcoRI-SpeI fragments were subcloned into binary vectors pGreenII0129 (hygromycin resistance) and pGreenII0229 (bialaphos resistance) and then electroporated into Agrobacterium tumefaciens strain GV3101:pMP90 (Koncz and Schell, 1986).
Table 1.
Primer Sequences
| Primer | Primer Sequence (5′–3′) |
|---|---|
| 3′ ARF10 for | CAACTCTTCGGATCACCATCTCCGTC |
| 3′ ARF10 rev | GGCCGTAATACGACTCACTATAGGCCGAGAGATCGAGTGTGCGTCC |
| 3′ ARF16 for | GGCCGTAATACGACTCACTATAGGGGCTCCTGATGCATCCCTATAGAG |
| 3′ ARF16 rev | GACAATGGGAACAACACCATGC |
| 3′ ARF17 for | GAGATGATGAACTTTGGCAGTCCG |
| 3′ ARF17 rev | GGCCGTAATACGACTCACTATAGGCCTGCGTTGCTGTTGGAATGTT |
| GH3.5 for | CGTGAAGTTTGCTCCAATTATCGAGC |
| GH3.5 rev | GGCCGTAATACGACTCACTATAGGCATGATAAAAGTCCAAACAATTCCGC |
| GH3.17 for | CAAGCAGAGCATGTACTGTCAGCTTC |
| GH3.17 rev | GGCCGTAATACGACTCACTATAGGGCCATCGAACCAGTCACAATCACC |
| GH3.6 for | GGCCGTAATACGACTCACTATAGGCCTTGGTGTCTTGTACTGATTGATCG |
| GH3.6 rev | GCGCCTCAGTTCAGCTTCATATGC |
| GH3.10 for | GGCCGTAATACGACTCACTATAGGGCATGTGAAACAATAGGCACCAAGG |
| GH3.10 rev | TTTGTCTGAAGACAAAGGGAGTGGTC |
| GH3.2 for | CCATCCTCTGCTCCGACTCGTCC |
| GH3.2 rev | GGCCGTAATACGACTCACTATAGGGCTCCAGTAACAATCACGTCGAGG |
| GH3.3 for | GGCCGTAATACGACTCACTATAGGGACAAGATCATTGACCGGTCACC |
| GH3.3 rev | GCTCTGCGATCTCCGATGATGC |
| IAA1 for | GGAAGTCACCAATGGGCTTAACC |
| IAA1 rev | GGCCGTAATACGACTCACTATAGGGCAGGAGGAGGAGCAGATTCTTCTG |
| IAA17 for | GGCAGTGTCGAGCTGAATCTGAGG |
| IAA17 rev | GGCCGTAATACGACTCACTATAGGGGCCGGAGGTTTGGCTGGATC |
| IAA19 for | GAGAAGGAAGGACTCGGGCTTGAG |
| IAA19 rev | GGCCGTAATACGACTCACTATAGGCGATGCCACGGAAACCGAAGAG |
| GUS for | GCGTATCGTGCTGCGTTTCGATG |
| GUS rev | GGCCGTAATACGACTCACTATAGGGCCTTGTCCAGTTGCAACCACC |
| ARF17 mut for | CGTATTCTACATTTCCTGCTGGAATGCAAGGTGCACGGCAATATGATTTTGGGTCTTTCAATCC |
| ARF17 mut rev | GGATTGAAAGACCCAAAATCATATTGCCGTGCACCTTGCATTCCAGCAGGAAATGTAGAATACG |
| ARF17 wg for | GGCCGTAATACGACTCACTATAGGCCTCTACTTATCAGGAGACCG |
| ARF17 wg rev | CCAGAGGACAGATTAGTGGTG |
| ARF10.1 RACE | CCGAGAGATCGAGTGTGCGTCC |
| ARF10.2 RACE | CTCCTCCGCTTCCGCCTCTTCTTCC |
| ARF16.1 RACE | GGCTCCTGATGCATCCCTATAGAG |
| ARF16.2 RACE | CCAAACCTGATGCATCATGAACTTTCTTAG |
| ARF17.1 RACE | CTTGGGAGCTAGAACCTGCGTTGC |
| ARF17.2 RACE | CCTGCGTTGCTGTTGGAATGTTTAGG |
| ARF17-RT for | GGATGAACCTGAGATTCTGC |
| ARF17-RT rev | TGGTGAATAGCTGGGGAGGATTTC |
| ARF17 screen | CTCTCTGTGGATCACTCTGTCTTGC |
| pGII far SacI | GCTATGACCATGATTACGCC |
| ACTIN2 | AACCCTCGTAGATTGGCACA |
for, forward; rev, reverse; mut, mutagenesis; wg, wheat germ.
To generate 35S:ARF17 and 35S:5mARF17 constructs, 3.3 kb EcoR1-BamH1 fragments of each of the 6.6 kb control ARF17 and 5mARF17 pBluescript clones described above were subcloned into pLBR19 downstream from the duplicated enhancer of CaMV 35S RNA promoter (P70) (Meyermans et al., 2000). 35S:ARF17 and 35S:5mARF17 KpnI-EcoRI fragments were then subcloned into the binary vector pBINPLUS (kanamycin resistance) (van Engelen et al., 1995) and electroporated into Agrobacterium GV3101:pMP90.
Arabidopsis thaliana (Col-0 accession) and DR5-GUS Arabidopsis in the Col-0 accession (Guilfoyle, 1999) were transformed using the floral dip method (Clough and Bent, 1998). Collected seeds were surface sterilized and plated on Bouturage 2 medium (Duchefa Biochemie, Haarlem, The Netherlands) containing 30 μg/mL hygromycin or 50 μg/mL kanamycin for selecting Col-0 transformants or 10 μg/mL Glufosinate-ammonium PESTANAL (Sigma-Aldrich, St. Louis, MO) for selecting DR5-GUS Col-0 transformants. Seedlings were grown in long days (16 h light, 8 h dark) at 20°C for ∼14 d before transfer to Metromix 200 soil (Scotts, Maysville, OH), where they were grown at 20°C in either long or short (8 h light, 16 h dark) days. For genotyping, genomic DNA from ARF17 and 5mARF17 plants was extracted and amplified with primers ARF17 screen and pGII far SacI (Table 1).
Agrobacterium infiltration of Nicotiana benthamiana leaves was performed as described (Llave et al., 2000). Bacteria were coinjected each at a final OD600 = 0.50, and the zones of infiltration were harvested 64 h after infiltration for total RNA isolation (Mallory et al., 2001). Expression of 35S:GUS was monitored as an infiltration control.
Arabidopsis ARF17 transcripts containing the miR160 complementary site were generated by PCR amplification of ARF17 or 5mARF17 genomic clones followed by in vitro transcription using T7 RNA polymerase. Primers ARF17 wheat germ forward and reverse (Table 1) were used to generate ARF17 and 5mARF17 templates. Wheat germ lysate preparation, cap labeling, and in vitro cleavage assays were performed as described (Tang et al., 2003).
RNA Isolation, RNA Gel Blot Analysis, and 5′ RACE Analysis
Total RNA was isolated (Mallory et al., 2001) and miRNA gel blot analysis was conducted (Reinhart et al., 2002) as previously described. For mRNA gel blot analysis, RNA was separated on 1.2% agarose gels containing 0.8% formaldehyde and transferred to nylon membranes by capillary action. Blots were hybridized with α-UTP 32P-labeled RNA probes at 68°C in ULTRAhyb buffer as recommended by the manufacturer (Ambion, Austin, TX). Using the ARF17 genomic clone (3′ ARF17 probe), pBI101 vector (GUS probe), Col-0 cDNA (At2g23170 probe), or Col-0 genomic DNA as template, 32P-UTP RNA probes were generated by PCR with primers listed in Table 1 followed by T7-mediated in vitro transcription. mRNA gel blots were rehybridized with an end-labeled ACTIN2 DNA probe. Hybridization signals were quantified using a Fuji phosphor imager (Tokyo, Japan) and normalized to ACTIN2 or 25S rRNA for mRNA gel blot analyses or to U6 for miRNA gel blot analyses.
Poly(A)+ RNA isolation, cDNA synthesis, non-gene-specific 5′ RACE amplifications and gene-specific 5′ RACE amplifications (primers listed in Table 1) were performed as described (Mallory et al., 2004a).
RT-PCR, ApaLI Digestion, and DNA Gel Blot Analysis
Five micrograms of total RNA prepared from rosette leaves of 30-d-old plants as described (Mallory et al., 2001) was used for (dT)20-primed first-strand cDNA synthesis followed by RNase H digestion as recommended by the manufacturer (ThermoScript RT system; Invitrogen, Carlsbad, CA). PCR amplification using 50 ng of cDNA as template was performed to completion using ARF17-RT forward and reverse primers (Table 1). To equalize the possibility of heteroduplex formation in the 5mARF17 samples, the final PCR products were denatured and renatured. ApaLI digestion of the ∼330-bp 5mARF17 PCR product yielded ∼250- and ∼80-bp fragments. To monitor ApaLI digestion efficiency, parallel reactions were spiked with a 2.2-kb DNA fragment containing an ApaLI restriction site, which produced ∼1.75- and ∼0.45-bp fragments after digestion. This control DNA was cleaved to completion, indicating that the undigested fragments in the 5mARF17 RT-PCR lacked the ApaLI site and derived from the endogenous ARF17 gene. DNA gel blot analysis was performed as described (Mallory et al., 2001). Briefly, undigested and ApaLI-digested PCR amplicons were separated on a 2% agarose gel, blotted to a nylon membrane, and hybridized with 32P end-labeled ARF17-RT forward primer (Table 1), which detects both undigested ARF17 and 5mARF17 330-bp PCR products and the 250-bp ApaLI digestion fragment of the 5mARF17 PCR product. Hybridization signals were quantified using a Fuji phosphor imager.
Phenotypic Analyses
Plant tissues were fixed and imaged for scanning electron microscopy as described (Mallory et al., 2004a). For histochemical staining, seedlings were submerged in a solution of 50 mM NaPO4, pH 7.0, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 10 mM EDTA, and 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide cyclohexylammonium salt (Gold Biotechnology, St. Louis, MO), vacuum infiltrated, and incubated 14 to 16 h at 37°C. To remove chlorophyll before photography, seedlings were rinsed repeatedly with 90% ethanol.
For seedling growth analyses, plants were grown at 20°C in long days on Bouturage 2 medium unless otherwise noted. T3 plants were grown under yellow long-pass filters (Stasinopoulos and Hangarter, 1990) to minimize IAA breakdown on horizontal plates for 7 d, after which primary root and hypocotyl lengths were measured and lateral roots were counted.
To monitor IAA-responsive transcripts, Col-0 seedlings were grown on plates for 7 d and then transferred to liquid Bouturage cultures supplemented with 0 or 10 μM IAA. Cultures were grown at 20°C with constant light and shaking (100 rpm) for the specified time, after which seedlings were collected and flash frozen with liquid nitrogen for RNA extraction.
Supplementary Material
Acknowledgments
We thank M. Jones-Rhoades, G. Tang, and P. Zamore for wheat germ extracts, P. Mullineaux and R. Hellens (John Innes Centre and the Biotechnology and Biological Science Research Council, Norwich, UK) for the pGreenII0129 and pGreenII0229 binary vectors, T. Guilfoyle for DR5-GUS transgenic seeds, H. Vaucheret, M. Jones-Rhoades, and D. Dugas for critical comments on the manuscript, the W.M. Keck Foundation Biological Imaging Facility (Whitehead Institute, Cambridge, MA) for scanning electron microscope use, and T. DiCesare for graphics assistance with the cover image. The Arabidopsis Biological Resource Center at The Ohio State University (Columbus, OH) supplied the F28K19 clone and SALK 062511 seeds. This research was supported by the National Institutes of Health (F32-GM071200, A.C.M.; R24-GM069512, B.B. and D.P.B.), the G. Harold and Leila Y. Mathers Charitable Foundation (B.B.), and the Robert A. Welch Foundation (C-1309, B.B.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: David P. Bartel (dbartel@wi.mit.edu) and Bonnie Bartel (bartel@rice.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031716.
References
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131, 3357–3365. [DOI] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida, M., Vernoux, T., Furutani, M., Traas, J., and Tasaka, M. (2002). Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129, 3965–3974. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Ambros, V. (2004). The functions of animal microRNAs. Nature 431, 350–355. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., and Bartel, D.P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17, in press. [DOI] [PMC free article] [PubMed]
- Bartel, B., and Bartel, D.P. (2003). MicroRNAs: At the root of plant development? Plant Physiol. 132, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Boutet, S., Vazquez, F., Liu, J., Béclin, C., Fagard, M., Gratias, A., Morel, J.B., Crété, P., Chen, X., and Vaucheret, H. (2003). Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13, 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301, 336–338. [DOI] [PubMed] [Google Scholar]
- Cary, A.J., Che, P., and Howell, S.H. (2002). Developmental events and shoot apical meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant J. 32, 867–877. [DOI] [PubMed] [Google Scholar]
- Chapman, E.J., Prokhnevsky, A.I., Gopinath, K., Dolja, V.V., and Carrington, J.C. (2004). Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Li, W.X., Xie, D., Peng, J.R., and Ding, S.W. (2004). Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell 16, 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, S., and Estelle, M. (2002). The role of regulated protein degradation in auxin response. Plant Mol. Biol. 49, 401–409. [PubMed] [Google Scholar]
- Dugas, D.V., and Bartel, B. (2004). MicroRNA regulation of gene expression in plants. Curr. Opin. Plant Biol. 7, 512–520. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., Lecellier, C.H., Parizotto, E.A., Himber, C., and Voinnet, O. (2004). Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16, 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. [DOI] [PubMed] [Google Scholar]
- Estelle, M., and Somerville, C. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206, 200–206. [Google Scholar]
- Floyd, S.K., and Bowman, J.L. (2004). Gene regulation: Ancient microRNA target sequences in plants. Nature 428, 485–486. [DOI] [PubMed] [Google Scholar]
- Friml, J. (2003). Auxin transport: Shaping the plant. Curr. Opin. Plant Biol. 6, 7–12. [DOI] [PubMed] [Google Scholar]
- Furutani, M., Vernoux, T., Traas, J., Kato, T., Tasaka, M., and Aida, M. (2004). PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131, 5021–5030. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T.J. (1999). Auxin-regulated genes and promoters. In Biochemistry and Molecular Biology of Plant Hormones, P.J.J. Hooykaas, M.A. Hall, and K.R. Libbenga, eds (Amsterdam: Elsevier), pp. 423–459.
- Guilfoyle, T.J., and Hagen, G. (2001). Auxin response factors. J. Plant Growth Regul. 10, 281–291. [Google Scholar]
- Guilfoyle, T.J., Ulmasov, T., and Hagen, G. (1998). The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell. Mol. Life Sci. 54, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. [PubMed] [Google Scholar]
- Han, M.H., Goud, S., Song, L., and Fedoroff, N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101, 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie, L., and Estelle, M. (1995). The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 7, 211–220. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., and Zamore, P.D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., and Bartel, D.P. (2004). Computational identification of plant miRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799. [DOI] [PubMed] [Google Scholar]
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, B.A., and Timmermans, M.C. (2004). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88. [DOI] [PubMed] [Google Scholar]
- Jürgens, G. (2003). Growing up green: Cellular basis of plant development. Mech. Dev. 120, 1395–1406. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2002). Ubiquitination and auxin signaling: A degrading story. Plant Cell 14 (suppl.), S81–S95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Burge, C.B., and Bartel, D.P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., and Johnson, J.M. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773. [DOI] [PubMed] [Google Scholar]
- Liscum, E., and Reed, J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. [PubMed] [Google Scholar]
- Ljung, K., Hull, A.K., Kowalczyk, M., Marchant, A., Celenza, J., Cohen, J.D., and Sandberg, G. (2002). Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 49, 249–272. [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., and Carrington, J.C. (2000). Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97, 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Dugas, D.V., Bartel, D.B., and Bartel, B. (2004. a). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Ely, L., Smith, T.H., Marathe, R., Anandalakshmi, R., Fagard, M., Vaucheret, H., Pruss, G., Bowman, L., and Vance, V.B. (2001). HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Reinhart, B.J., Bartel, D.P., Vance, V.B., and Bowman, L.H. (2002). A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA 99, 15228–15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Reinhart, B.J., Jones-Rhoades, M.W., Tang, G., Zamore, P.D., Barton, M.K., and Bartel, D.P. (2004. b). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23, 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., and Vaucheret, H. (2004). MicroRNAs: Something important between the genes. Curr. Opin. Plant Biol. 7, 120–125. [DOI] [PubMed] [Google Scholar]
- McHale, N.A., and Koning, R.E. (2004). MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell 16, 1730–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyermans, H., et al. (2000). Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-Coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 275, 36899–36909. [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus, M., Zolman, B.K., and Bartel, B. (2003). IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 15, 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, M., Yabe, N., Ichikawa, T., Yamamoto, Y.Y., Yoshizumi, T., Hasunuma, K., and Matsui, M. (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25, 213–221. [DOI] [PubMed] [Google Scholar]
- Okushima, Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, P.H., and Ambros, V. (1999). The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216, 671–680. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Parizotto, E.A., Dunoyer, P., Rahm, N., Himber, C., and Voinnet, O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18, 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18, 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington, D.L., Vision, T.J., Guilfoyle, T.J., and Reed, J.W. (2004). Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135, 1738–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M., Reinhart, B., Lim, L., Burge, C., Bartel, B., and Bartel, D. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Rogg, L.E., and Bartel, B. (2001). Auxin signaling: Derepression through regulated proteolysis. Dev. Cell 1, 595–604. [DOI] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Stasinopoulos, T.C., and Hangarter, R.P. (1990). Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93, 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M.T., Maldonado, M.C., and Suza, W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Tiryaki, I., and Rowe, M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., and Zhu, J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup, R., Parry, G., Graham, N., Allen, T., and Bennett, M. (2002). Auxin cross-talk: Integration of signalling pathways to control plant development. Plant Mol. Biol. 49, 411–426. [DOI] [PubMed] [Google Scholar]
- Takase, T., Nakazawa, M., Ishikawa, A., Kawashima, M., Ichikawa, T., Takahashi, N., Shimada, H., Manabe, K., and Matsui, M. (2004). ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 37, 471–483. [DOI] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, C.E., Muto, H., Higuchi, K., Matamura, T., Tatematsu, K., Koshiba, T., and Yamamoto, K.T. (2004). Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 40, 333–343. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. a). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. b). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelen, F.A., Molthoff, J.W., Conner, A.J., Nap, J.P., Pereira, A., and Stiekema, W.J. (1995). pBINPLUS: An improved plant transformation vector based on pBIN19. Transgenic Res. 4, 288–290. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crété, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, F., Gasciolli, V., Crété, P., and Vaucheret, H. (2004. a). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14, 346–351. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., and Crete, P. (2004. b). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16, 69–79. [DOI] [PubMed] [Google Scholar]
- Wang, J.F., Zhou, H., Chen, Y.Q., Luo, Q.J., and Qu, L.H. (2004. a). Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Res. 32, 1688–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.J., Reyes, J.L., Chua, N.H., and Gaasterland, T. (2004. b). Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 5, R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. [DOI] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Colgan, D., and Chua, N.-H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Guo, H.-S., Dallman, G., Fang, S., Weissman, A.M., and Chua, N.-H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Kasschau, K.D., and Carrington, J.C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA. Curr. Biol. 13, 784–789. [DOI] [PubMed] [Google Scholar]
- Yekta, S., Shih, I.H., and Bartel, D.P. (2004). MicroRNA-directed cleavage of HOXB8 mRNA. Science 304, 594–596. [DOI] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (2004). amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 45, 369–385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.