Abstract
Although several plant microRNAs (miRNAs) have been shown to play a role in plant development, no phenotype has yet been associated with a reduction or loss of expression of any plant miRNA. Arabidopsis thaliana miR164 was predicted to target five NAM/ATAF/CUC (NAC) domain–encoding mRNAs, including NAC1, which transduces auxin signals for lateral root emergence. Here, we show that miR164 guides the cleavage of endogenous and transgenic NAC1 mRNA, producing 3′-specific fragments. Cleavage was blocked by NAC1 mutations that disrupt base pairing with miR164. Compared with wild-type plants, Arabidopsis mir164a and mir164b mutant plants expressed less miR164 and more NAC1 mRNA and produced more lateral roots. These mutant phenotypes can be complemented by expression of the appropriate MIR164a and MIR164b genomic sequences. By contrast, inducible expression of miR164 in wild-type plants led to decreased NAC1 mRNA levels and reduced lateral root emergence. Auxin induction of miR164 was mirrored by an increase in the NAC1 mRNA 3′ fragment, which was not observed in the auxin-insensitive mutants auxin resistant1 (axr1-12), axr2-1, and transport inhibitor response1. Moreover, the cleavage-resistant form of NAC1 mRNA was unaffected by auxin treatment. Our results indicate that auxin induction of miR164 provides a homeostatic mechanism to clear NAC1 mRNA to downregulate auxin signals.
INTRODUCTION
First discovered in Caenorhabditis elegans, microRNAs (miRNAs) were found recently to be expressed in several eukaryotes as well (Carrington and Ambros, 2003; Ambros, 2004). The initial cloning experiments of Reinhart et al. (2002) recovered 16 miRNAs from Arabidopsis thaliana, some of which were encoded by more than one genomic locus. These miRNAs were predicted to target mRNAs encoding transcription factors involved in plant development (Rhoades et al., 2002), and many such candidate mRNAs have been experimentally verified to be bona fide targets (Llave et al., 2002; Bartel, 2004). Subsequent computational analyses by three independent groups increased the number of potential Arabidopsis miRNAs to more than 100 (Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Wang et al., 2004). The increasing biological importance of this group of small RNAs is illustrated by the expanded list of target mRNAs, including those encoding proteins implicated in proteolysis, signaling, metabolism, ion transport, etc. These results suggest that besides controlling transcription factor mRNA abundance, miRNAs may also regulate a large number of other cellular processes (e.g., signaling pathways). In addition, direct sequencing of small RNAs from a stress-induced library has revealed several new miRNA families (Sunkar and Zhu, 2004), suggesting a role of these small RNAs in abiotic stress responses. Expression of miR393 and miR159 has been shown to be regulated by abscisic acid (Sunkar and Zhu, 2004) and gibberellin (Achard et al., 2004), respectively.
To date, the functions of plant miRNAs have been deduced from overexpression of precursor sequences encoding miRNAs and/or expression of target mRNAs that have been rendered cleavage-resistant by mutations (Bartel and Bartel, 2003; Bartel, 2004; Dugas and Bartel, 2004). Overexpression of miRNA precursors in transgenic plants leads to increased miRNA levels and decreased target mRNA levels; as expected, such transgenic plants phenocopy mutants with deficiencies of the target mRNA. Conversely, expression/overexpression of miRNA-resistant mutant mRNAs stabilizes the message and causes transgenic plants to display a phenotype similar to that obtained with target wild-type mRNA overexpression.
Further clarification of the biological role of miRNAs in plants will be aided by analysis of loss-of-function mutants that are directly affected in miRNA genes. Unfortunately, no complete loss-of-function mutant in any plant miRNA has yet been described. A recently characterized Arabidopsis T-DNA insertion mutant in the MIR164b locus (mir164b-1) produced 15-fold less miR164 than did the wild type, but no aerial phenotype was associated with this deficiency (Mallory et al., 2004a). This finding could be attributed to the fact that miR164 is encoded by at least three genomic loci in Arabidopsis (Reinhart et al., 2002). It should be pointed out that several Arabidopsis miRNAs are encoded by more than one genomic locus, thus rendering recovery of complete loss-of-function mutants difficult.
Predicted targets of miR164 include mRNAs encoding five members of the NAM/ATAF/CUC (NAC) domain transcription factor family (Rhoades et al., 2002). Of these, NAC1 is involved in transmitting auxin signals for lateral root development (Xie et al., 2000, 2002) and CUP-SHAPED COTYLEDON1 (CUC1)/CUC2 are implicated in meristem development and separation of aerial organs (Aida et al., 1997), whereas the functions of At5g07680 and At5g61430 have not yet been defined. Recent work by two independent groups has provided evidence for miRNA-mediated regulation of CUC1 (Mallory et al., 2004a) and CUC2 (Laufs et al., 2004) mRNA levels in vivo. Transgenic plants overexpressing miR164 display altered floral organ numbers and fused vegetative and floral organs. By contrast, transgenic plants expressing miRNA164-resistant CUC1 (Mallory et al., 2004a) or CUC2 (Laufs et al., 2004) are defective in leaf development, and in the case of CUC2 plants, they possess an enlarged boundary between sepals (Laufs et al., 2004). Whereas NAC1 mRNA was cleaved at the expected site in wild-type plants (Mallory et al., 2004a), no changes in NAC1 mRNA levels were detected in transgenic plants overexpressing miR164 (Laufs et al., 2004). The root phenotype of transgenic plants overexpressing miR164 was not investigated in these studies (Laufs et al., 2004; Mallory et al., 2004a).
We previously found that the transcription regulator NAC1 acts downstream of TRANSPORT INHIBITOR RESPONSE1 (TIR1) to transmit auxin signals promoting lateral root emergence (Xie et al., 2000) and that there is a positive correlation between NAC1 mRNA levels and lateral root numbers. NAC1 was subsequently found to be an unstable protein, with its stability being regulated by the RING motif E3 ligase, SINAT5 (for Arabidopsis homolog of Drosophila protein SINA) (Xie et al., 2002). Because it is an unstable protein, we asked whether its mRNA stability is also regulated. Here, we confirm that NAC1 mRNA is a target of miR164 in vivo (Rhoades et al., 2002; Mallory et al., 2004a). Analysis of transgenic plants and four mir164 mutant alleles reveals that modest changes in miR164 levels can result in substantial changes in NAC1 mRNA levels, and the transgenic plants and mir164 mutants have the expected lateral root phenotypes correlating with NAC1 mRNA abundance. Finally, we found that miR164 expression is late-auxin-responsive and that a major role of this miRNA is to clear NAC1 mRNA to attenuate auxin signaling.
RESULTS
NAC1 mRNA Is Cleaved in Vivo
NAC1 is a transcription activator in the auxin signaling pathway for Arabidopsis lateral root development (Xie et al., 2000, 2002). NAC1 mRNA accumulates mainly in roots (Figure 1). Besides the full-length NAC1 mRNA, we consistently detected a shorter band (700 nucleotides) (Figure 1), which may be a 3′ cleavage product of NAC1 mRNA as it hybridized to a NAC1 3′-specific probe. Using 5′ rapid amplification of cDNA ends (RACE), we amplified cDNA corresponding to this mRNA fragment. Sequence analysis of 10 independent cDNA clones produced identical results and placed the 5′ end of the cleaved fragment at nucleotide 764 of the NAC1 mRNA. This nucleotide position is located in the middle of the miR164/NAC1 mRNA complementary region (Rhoades et al., 2002; Mallory et al., 2004a) (Figure 2A).
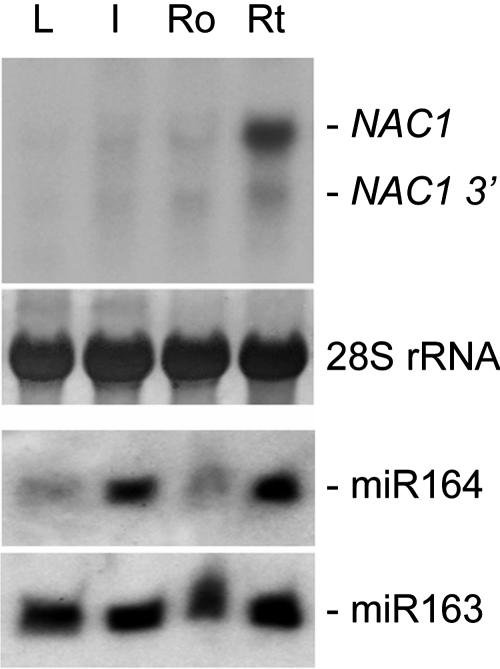
Figure 1.
Expression Patterns of NAC1 and miR164 in Wild-Type Arabidopsis RNA.
Gel blot analyses of NAC1 mRNA and miR164 from wild-type (Col) leaves (L), inflorescences (I), 2-week-old seedling rosette leaves (Ro), and roots (Rt). A gel blot containing total RNA (10 μg per lane) was hybridized with a NAC1 3′-specific fragment (nucleotides 682 to 1063; top gel). Intact NAC1 mRNA and the NAC1 3′ fragment resulting from miR164-guided cleavage are indicated. 28S rRNA visualized by methylene blue staining was used as a loading control (second gel). A gel blot containing low-molecular-weight RNA (30 μg per lane) was hybridized with an end-labeled DNA oligonucleotide complementary to miR164 (third gel). The membrane was stripped and reprobed with an end-labeled DNA oligonucleotide complementary to miR163 for a loading control (bottom gel).
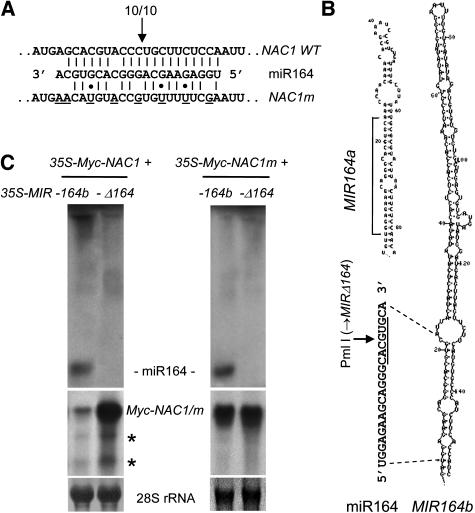
Figure 2.
Determination of NAC1 mRNA Cleavage by miR164 Using a Transient Expression System.
(A) Alignment of miR164 with NAC1 (wild type) and NAC1 mutant (NAC1m) mRNA. The NAC1/miR164 pair contains two mismatches in base pairing, whereas there are eight mismatches in the NAC1 mutant (NAC1m/miR164). The mismatched nucleotides are underlined, and the predicted G-U base pairing is shown with dots. The arrow indicates the cleavage site between nucleotides 763 and 764 of the NAC1 mRNA. Ten clones were sequenced, and an identical 5′ end was obtained in all clones.
(B) miR164 sequence and the predicted structure of the MIR164a and MIR164b synthetic precursors. A deletion mutant, MIRΔ164b (contains only 5 nucleotides of miR164), was created by digestion of the precursor MIR164b sequence with PmlI to remove 16 nucleotides of the miR164 sequence.
(C) Effect of miR164 on Myc-NAC1 (left) and Myc-NAC1m (right) transcripts in coinfiltration assays. Ten micrograms of low-molecular-weight RNAs (top gels) or 5 μg of total RNA (middle gels) from N. benthamiana leaf samples collected at 48 h after infiltration was loaded and hybridized with the appropriate probes as described for Figure 1. 28S rRNA was used as a loading control (bottom gels). The constructs used for coinfiltration are indicated above each lane. Myc-NAC1 and Myc-NAC1m mRNA are indicated. Asterisks indicate additional bands presumably derived from cleavage of Myc-NAC1 transcripts by N. benthamiana endogenous miRNA.
Plant miRNAs have recently been proposed to guide mRNAs for cleavage (Llave et al., 2002; Kasschau et al., 2003; Palatnik et al., 2003), and mRNAs for five NAC domain proteins, including NAC1, were predicted to be targets of miR164 (Rhoades et al., 2002; Laufs et al., 2004; Mallory et al., 2004a). RNA gel blot analysis showed that in Arabidopsis, miR164 expression levels were higher in roots and inflorescences compared with other organs (Figure 1) (Mallory et al., 2004a), whereas miR163 (Reinhart et al., 2002) accumulated to a similar level in all organs. The higher miR164 expression in roots suggests that this miRNA may target NAC1 mRNA in vivo.
NAC1 mRNA Cleavage Is Directed by miR164
To obtain direct evidence that miR164 can mediate the cleavage of NAC1 mRNA, we used a transient expression system involving Agrobacterium tumefaciens infiltration of Nicotiana benthamiana leaves (English et al., 1997). miR164 is presumably encoded by three genomic loci, MIR164a, MIR164b, and MIR164c (Reinhart et al., 2002; Dugas and Bartel, 2004; http://www.sanger.ac.uk/cgi-bin/Rfam/mirna/browse.pl). It has been shown that transcription of genomic sequences (∼2 kb) containing the miRNA foldback region by a 35S promoter can produce the mature miRNA (Llave et al., 2002; Aukerman and Sakai, 2003; Chen, 2004; Parizotto et al., 2004). In our experiments, we used a 153-bp sequence derived from MIR164b as a synthetic precursor of miR164 (Figure 2B). As a negative control, we deleted from the synthetic precursor sequences encoding 16 nucleotides of miR164 and designated the deletion mutant 35S-MIRΔ164 (Figure 2B). Figure 2C shows that the synthetic precursor 35S-MIR164b, but not the deletion mutant 35S-MIRΔ164, could produce miR164 in N. benthamiana. The levels of NAC1 (Myc-NAC1) mRNA (Xie et al., 2002) were decreased significantly when coexpressed with 35S-MIR164b but not 35S-MIRΔ164 (Figure 2C). This result provides direct evidence that miR164 produced from 35S-MIR164b triggers Myc-NAC1 mRNA cleavage. As a negative control, we introduced eight nucleotide mismatches into the miR164/NAC1 mRNA complementary region of NAC1 mRNA sequence without changing the encoded amino acids, and the mutant was designated NAC1m (Figure 2A). In contrast with Myc-NAC1 mRNA, no cleavage was seen with Myc-NAC1m mRNA (Figure 2C), confirming the importance of the complementary region in miR164-mediated cleavage.
Transgenic Plants Expressing a Cleavage-Resistant Form of NAC1 mRNA
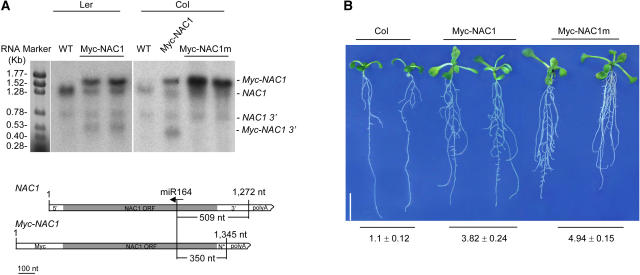
We generated Arabidopsis transgenic plants expressing 35S-Myc-NAC1 and 35S-Myc-NAC1m. Figure 3A shows that a 3′ cleavage fragment of Myc-NAC1 (500 nucleotides) was present in 35S-Myc-NAC1 transgenic plants but was not detected in 35S-Myc-NAC1m transgenic plants. This observation indicates that the Myc-NAC1m transcript is cleavage-resistant and not a target of endogenous miR164, consistent with the transient coexpression results obtained from N. benthamiana (Figure 2C). In 7-d-old seedlings, no lateral root initials were apparent in the wild type, whereas short lateral roots and three to four lateral root initials were found in some 35S-Myc-NAC1 transgenic lines (data not shown). Most of the 35S-Myc-NAC1m transgenic seedlings produced five to six lateral root initials and more short lateral roots compared with 35S-Myc-NAC1 lines (data not shown). At 12 d after germination (Figure 3B), the average number of lateral roots per centimeter of primary root for each line (n = 20) was 1.1 ± 0.12, 3.8 ± 0.24, and 4.9 ± 0.15 for the wild type (Columbia [Col]), 35S-Myc-NAC1, and 35S-Myc-NAC1m, respectively. Student's t test showed that the unit lateral root numbers differed significantly between 35S-Myc-NAC1 and 35S-Myc-NAC1m lines (P < 0.001). Together with the observation that Myc-NAC1m was resistant to miR164-mediated cleavage, our results provide evidence that the regulation of NAC1 mRNA levels by miR164 is an important control point for lateral root initiation. In most seedlings of transgenic lines carrying either 35S-Myc-NAC1 or 35S-Myc-NAC1m, two lateral roots emerged at the hypocotyl/root junction and secondary lateral roots developed at a very early stage (data not shown).
Figure 3.
RNA Analysis and Phenotypes of 35S-NAC1 Transgenic Plants.
(A) Myc-NAC1 but not Myc-NAC1m transcripts were cleaved by endogenous miR164 (top). RNAs were extracted from roots of 2-week-old seedlings of the wild type (Ler), 35S-Myc-NAC1 (Ler), the wild type (Col), 35S-Myc-NAC1 (Col), and 35S-Myc-NAC1m (Col). Each lane contained 5 μg of RNA, and blots were hybridized to a NAC1 gene-specific probe as described for Figure 1. Full-length endogenous NAC1 and Myc-NAC1 mRNA and their respective 3′ fragments resulting from miR164-guided cleavage are indicated. At bottom is a scheme of endogenous NAC1 and Myc-NAC1m transcript constructs and the positions complementary to miR164.
(B) Effects of overexpression of 35S-Myc-NAC1 and 35S-Myc-NAC1m on lateral root development. Wild-type (Col) and transgenic (Col) seedlings expressing 35S-Myc-NAC1 or 35S-Myc-NAC1m transgene were grown on MS medium with 2% sucrose. Seedlings were photographed 12 d after germination. Lateral root initials were identified by staining whole seedlings with a mixture of toluidine blue and basic fuchsin and counted with a binocular microscope. Numbers of lateral roots per centimeter of primary root of each seedling (average ± se) for wild-type Col, 35S-Myc-NAC1 (12 independent lines), and 35S-Myc-NAC1m (18 independent lines) are presented at bottom (n = 20). Bar = 1 cm.
Overexpression of miR164
To further confirm the regulation of NAC1 mRNA by miR164, we introduced the synthetic precursor MIR164b into the 17-β-estradiol–inducible system, pX7 (Guo et al., 2003). The resulting construct, pX7-MIR164 (pX7-164), was used to retransform Arabidopsis transgenic plants (Landsberg erecta [Ler]) expressing either 35S-Myc-NAC1 or 35S-Myc (vector control) (Xie et al., 2002). Figure 4A shows that the inducer increased miR164 levels by 2- to 10-fold (top panels, lanes 2, 4, and 6), indicating appropriate maturation of the synthetic precursor MIR164b transcript in Arabidopsis. Transgenic Myc-NAC1 and endogenous NAC1 mRNA levels were significantly reduced in inducer-treated roots (Figure 4A, middle panel, lanes 2, 4, and 6) compared with untreated controls (lanes 1, 3, and 5), although there was no detectable increase in the levels of Myc-NAC1 mRNA 3′ cleavage product. Changes in the NAC1 mRNA levels were accompanied by the expected changes in lateral root initials (Figure 4B). Some double transgenic plant lines displayed shoot developmental defects upon inducer treatment (data not shown), presumably resulting from miR164-mediated cleavage of other NAC domain mRNAs (e.g., CUC1/CUC2 mRNAs) (Laufs et al., 2004; Mallory et al., 2004a).
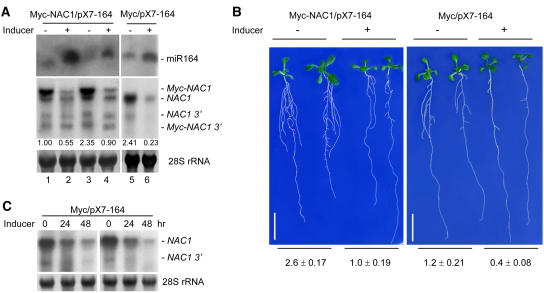
Figure 4.
RNA Analysis and Phenotypes of Transgenic Plants with Inducible Expression of miR164.
(A) Induction of miR164 and effect on target NAC1 mRNA accumulation. Seedlings containing either 35S-Myc-NAC/pX7-164 or 35S-Myc/pX7-164 were germinated on selective medium for 6 d before being transferred to MS medium or MS + 17-β-estradiol (2 μM). After 10 d, root samples were collected for analysis. Ten micrograms of low-molecular-weight RNAs (top gel) or 20 μg of total RNA (middle gel) was loaded and hybridized with the appropriate probes as described for Figure 1. 28S rRNA was used as a loading control (bottom gel). miR164, full-length Myc-NAC1, and NAC1 mRNAs and their derived 3′ fragments are indicated. Plus and minus signs indicate with or without inducer treatment, respectively. Numbers below the middle gel indicate relative endogenous NAC1 mRNA levels.
(B) Effect of inducible expression of miR164 on lateral root development. Representative plants from transgenic lines in (A) were photographed 10 d after transfer to medium with (+) or without (−) inducer. Numbers of lateral roots per centimeter of primary root (average ± se) were determined as described for Figure 3B. Bars = 1 cm.
(C) Effect of a shorter time course of induction on endogenous NAC1 mRNA levels. Two independent lines of 35S-Myc/pX7-164 seedlings were germinated on selective medium for 9 d before being transferred to MS + 17-β-estradiol (2 μM). Root samples were collected 24 and 48 h after treatment. Each lane contained 10 μg of total RNA (top gel). Blots were hybridized with NAC1 3′-specific probes as described for Figure 1. 28S rRNA was used as a loading control (bottom gel).
Because NAC1 mRNA is expressed in lateral root initials, it could be argued that the reduction of NAC1 mRNA levels in the induced pX7-164 plants was attributable to the fewer number of lateral roots in the inducer-treated transgenic plants and not to the direct action of miR164. To investigate this possibility, we performed a shorter time course of induction with two independent lines of 35S-Myc/pX7-164 plants. Figure 4C shows that 24 h of inducer treatment reduced NAC1 mRNA levels by fourfold to fivefold, and during this period there was no decrease in lateral root initials/numbers. This result provides evidence that miR164-directed changes in NAC1 mRNA levels precede changes in lateral root development.
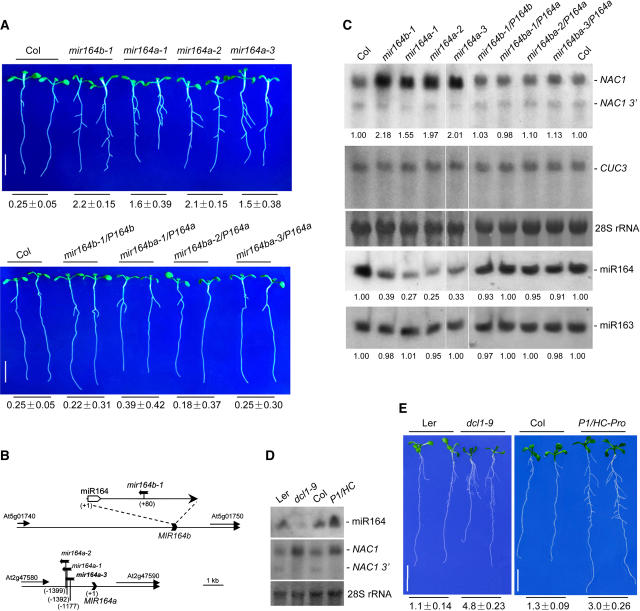
Phenotypes of T-DNA Insertion Mutants in MIR164a and MIR164b
To investigate possible loss-of-function phenotypes, we obtained three T-DNA insertion mutants in MIR164a and one in MIR164b from the SALK collection of T-DNA insertion mutants. At 7 d after germination, seedlings of all four insertion mutants (mir164a-1, mir164a-2, mir164a-3, and mir164b-1) but not wild-type (Col) seedlings developed lateral roots (Figure 5A, top). Some mutant seedlings developed two lateral roots at the hypocotyl/root junction and produced secondary lateral roots at an early stage, similar to Myc-NAC1 or Myc-NAC1m transgenic plants (data not shown). At 10 d after germination, mutant seedlings produced ∼10- fold more lateral roots per unit of primary root than wild-type controls. Figure 5B shows the T-DNA insertion sites of these mutants, which have been verified by PCR and sequence analysis. In mir164b-1, the T-DNA was inserted in the loop region (+80) of the putative hairpin structure (Figures 2B and 5B) (Mallory et al., 2004a). This mutant is likely to be a miR164b null allele. For the three mutant alleles in the MIR164a locus, the T-DNA insertion sites are ∼1.2 to 1.4 kb 5′ of miR164a (Figure 5B). RNA gel blot analysis showed that miR164 expression was reduced two to three times not only in mir164b-1 but also in mir164a-1, mir164a-2, and mir164a-3 (Figure 5C). In all cases, miR163 expression was not affected (Figure 5C). These results suggest that miR164 expression was affected in the mir164a-1, mir164a-2, and mir164a-3 insertion mutants. The specific reduction of miR164 levels was accompanied by an increased NAC1 mRNA level, but levels of CUC3 mRNA, which is not targeted by miRNA164, remained unaltered (Figure 5C).
Figure 5.
RNA Gel Blot Analysis and Root Phenotypes of mir164a and mir164b Mutants, dcl1-9 Mutant, and P1/HC-Pro Transgenic Plants.
(A) Root phenotypes of wild-type Col, mir164a, and mir164b mutants (top) and of complemented lines (bottom). Seedlings were photographed 10 d after germination on MS medium with 2% sucrose. Bars = 1 cm.
(B) T-DNA insertion sites of mir164b-1, mir164a-1, mir164a-2, and mir164a-3 relative to the annotated genes.
(C) RNA analysis of mir164a and mir164b mutants and complemented lines. Ten micrograms of total RNA (top gel) or low-molecular-weight RNAs (fourth gel) from roots of 10-d-old seedlings was loaded and hybridized with the appropriate probes as described for Figure 1. The filters were then stripped and rehybridized with a CUC3 3′ gene-specific sequence (nucleotides 1273 to 1863; second gel) or with a 5′ end-labeled DNA oligonucleotide complementary to miR163 (fifth gel) to serve as loading controls. 28S rRNA was used as a total RNA loading control (third gel). For each plant line, the relative accumulation levels of full-length NAC1 mRNA, miR164, and miR163 are indicated below the lanes. Levels in wild-type Col were arbitrarily designated as 1.00.
(D) and (E) RNA analysis and root phenotypes of the dcl1-9 mutant and P1/HC-Pro transgenic plants. dcl1-9 (Ler) and P1/HC-Pro (Col) transgenic seedlings and their respective wild-type controls were grown on MS medium with 2% sucrose for 2 weeks.
(D) Root RNA was analyzed as described for Figure 1 using 10 μg of low-molecular-weight RNAs (top gel) or 3 μg of total RNA (middle gel). 28S rRNAs bands were used as loading controls (bottom).
(E) Representative plants were photographed. dcl1-9 and P1/HC-Pro seedlings were identified by their developmental phenotypes. Bars = 1 cm.
Numbers below (A) and (E) indicate the number of lateral roots per centimeter of primary root (average ± se) as described for Figure 3B (n = 20).
We introduced into the four T-DNA insertion mutants ∼2.2 kb of the appropriate genomic sequences, including the miR164 coding region (Figure 2B). Expression of miR164 and NAC1 was restored to wild-type levels, indicating complementation of the deficiency (Figure 5C). Phenotype analysis confirmed restoration of the wild-type root phenotype (Figure 5A, bottom).
Phenotypes of Plants with Disrupted miRNA Metabolism
The Arabidopsis mutant dicer-like1 (dcl1-9) displays reduced miRNA accumulation (Park et al., 2002) because of a defect in miRNA biogenesis. Increased miRNA accumulation was seen in transgenic plants expressing P1/HC-Pro, a viral suppressor, but these plants phenocopy the miRNA-deficient dcl1-9 because P1/HC-Pro interferes with miRNA functions (Kasschau et al., 2003). Figure 5D shows that miR164 abundance was reduced in dcl1-9 but enhanced in P1/HC-Pro–overexpressing plants compared with their respective wild-type controls. NAC1 mRNA accumulation, however, increased in both dcl1-9 and P1/HC-Pro plants relative to either wild-type control. Consistent with the higher accumulation level of NAC1 mRNA, more lateral roots were observed in both dcl1-9 and P1/HC-Pro plants (Figure 5E).
miR164 Clears NAC1 mRNA during Auxin Response
As an early auxin-responsive gene, an increase in NAC1 mRNA was evident within 30 min of 2 μM 1-naphthalene acetic acid (NAA) treatment and reached a maximum level after 2 h (Xie et al., 2000, 2002). We found that under this condition, there was no change in miR164 levels (data not shown). However, at 10 μM NAA, we consistently detected an increase of miR164, but not of miR163, by ∼1.5-fold at 6 to 8 h after treatment (Figure 6A). Coincident with the miR164 increase at 6 h, there was a reduction in NAC1 mRNA levels and a concomitant increase in the levels of NAC1 mRNA 3′ fragment (Figure 6A). This observation suggests that the reduction of NAC1 mRNA levels after prolonged auxin treatment may result from miR164-mediated cleavage. The NAC1 mRNA 3′ fragment was unstable, as its intensity had decreased at 14 h after application of NAA.
Figure 6.
RNA Analysis of Wild-Type (Col) Plants, 35S-Myc-NAC1m Transgenic Plants, and axr1-12, axr2-1, and tir1-1 Plants.
Ten-day-old seedlings were treated with NAA (10 μM), and root samples were collected at the times indicated at top. Each lane contained 10 μg of total RNA (top two gels) or low-molecular-weight RNAs (bottom two gels). Blots were hybridized with the appropriate probes as described for Figure 1. Stripped low-molecular-weight RNA filter was rehybridized with a miR163 probe (bottom gels). 28S rRNA was used as a loading control (second gel). Numbers indicate relative levels of NAC1 mRNA (top gel), miR164 (third gel), and miR163 (bottom gel). Relative mRNA levels (average ± se) are 1.89 ± 0.03 (2 h), 1.86 ± 0.03 (4 h), 0.11 ± 0.02 (6 h), and 0.12 ± 0.02 (8 h) for NAC1 and 1.03 ± 0.05 (2 h), 1.15 ± 0.05 (4 h), 1.57 ± 0.05 (6 h), and 1.57 ± 0.10 (8 h) for miR164. Positions of full-length endogenous NAC1, transgenic Myc-NAC1 mRNA, their respective 3′ fragments, and miR164 and miR163 are shown.
The specificity and reproducibility of this auxin-induced cleavage were further confirmed by analysis of transgenic plants expressing 35S-Myc-NAC1m. Figure 6A shows that the transgenic Myc-NAC1m mRNA was resistant to auxin-induced cleavage and the endogenous NAC1 mRNA remained susceptible. As additional controls, we also investigated miR164 and NAC1 mRNA levels in three Arabidopsis auxin-insensitive mutants, auxin resistant1 (axr1-12) (del Pozo and Estelle, 1999; del Pozo et al., 2002), axr2-1 (Timpte et al., 1994), and tir1-1 (Gray et al., 2001), and found no changes in either miR164 levels or NAC1 mRNA levels with auxin treatment (Figure 6B). AXR1 and TIR1 are both implicated in the auxin-induced rapid degradation of indoleacetic acid repressors (Gray et al., 2001), whereas axr2-1 expresses a dominant mutant of Indoleacetic Acid7 (Timpte et al., 1994). Our results confirm that the increase of miR164 levels and of miR164-mediated NAC1 mRNA cleavage is, in part, auxin-dependent. The slower induction kinetics of miR164 suggests that it mediates the clearance of NAC1 mRNA, which is induced faster.
DISCUSSION
miR164 Mediates NAC1 mRNA Cleavage in Vivo
Plant miRNAs have been implicated in the control of leaf morphogenesis (Palatnik et al., 2003), leaf polarity (Kidner and Martienssen, 2004; Juarez et al., 2004; Mallory et al., 2004b), flowering time (Aukerman and Sakai, 2003; Achard et al., 2004; Chen, 2004), and flower development (Laufs et al., 2004; Mallory et al., 2004a). In contrast with animal miRNAs, most plant miRNAs have been shown to mediate cleavage of their target mRNAs (Dugas and Bartel, 2004), whereas only miRNA172 has been implicated in translational control (Aukerman and Sakai, 2003; Chen, 2004). Using coexpression in N. benthamiana, we show here that miR164 directs NAC1 mRNA cleavage in vivo at a position complementary to the 10th nucleotide from the 5′ end of miR164 and that disruption of the base-pairing region compromises cleavage. These results confirm and extend previous findings (Rhoades et al., 2002; Mallory et al., 2004a). It is possible that NAC1 mRNA cleavage might be developmentally regulated in Arabidopsis, because no cleavage was detected in flowers of transgenic plants overexpressing miR164 (Laufs et al., 2004).
miR164 Negatively Regulates Lateral Root Development
By manipulating miR164 levels or expressing the cleavage-resistant NAC1m mRNA in vivo, we have provided several lines of evidence that this miRNA functions as a negative regulator of auxin-mediated lateral root development by controlling NAC1 mRNA levels. (1) Plants overexpressing NAC1m produce more lateral roots compared with NAC1-overexpressing plants. (2) Mutant plants (dcl1-9) defective in miRNA biogenesis possess higher NAC1 mRNA levels and develop more lateral roots. (3) Conditional overexpression of miR164 reduces NAC1 mRNA levels as well as lateral root numbers. (4) Mutants partially affected in miR164 production accumulate higher levels of NAC1 mRNA and produce more lateral roots. In all of these instances, there is a strict inverse correlation between changes in miR164 levels and changes in both NAC1 mRNA levels and lateral root initials. An exception is seen in P1/HC-Pro transgenic plants, which accumulate miR164 but in which cleavage of NAC1 mRNA is blocked by the viral suppressor. Therefore, it is not surprising that these plants maintain high NAC1 mRNA levels and develop more lateral roots.
We have characterized four T-DNA insertion mutants of Arabidopsis that accumulate reduced miR164 levels. All of these mutants have a single T-DNA insertion in either MIR164a (three alleles) or MIR164b (one allele), and the phenotype segregates with the T-DNA. In addition to these two loci, Arabidopsis contains a third locus, MIR164c, which can potentially produce miR164 (Dugas and Bartel, 2004). However, their relative contributions to miR164 expression are not clear. Nonetheless, our results show that both MIR164a and MIR164b contribute to miR164 expression in roots, as mutations in these loci reduce miR164 levels but do not eliminate expression totally. Based on the site of T-DNA disruption (Figure 5B) (Mallory et al., 2004a), mir164b-1 is likely to be a mir164b null allele. The T-DNA insertion sites of the three mir164a mutant alleles are located ∼1.2 kb upstream of the first nucleotide of miR164 (Figure 5B). Because of their reduced miR164 expression, we propose that transcription of this locus is affected in all three mutants. Notwithstanding the functional redundancy, a partial decrease (twofold to threefold) of miR164 level is sufficient to increase NAC1 mRNA expression levels by approximately twofold, indicating a fine balance between the regulator and the regulated mRNA.
Recent results have shown that in both animals and plants, miRNAs are transcribed from Pol II promoters (Kurihara and Watanabe, 2004; Parizotto et al., 2004). In our complementation experiments, we used genomic fragments with ∼1.9- to 2.25-kb upstream sequences. Because rescue of molecular and morphological phenotypes was seen in the complemented lines, we conclude that these genomic fragments contain the requisite signals for miR164a and miR164b expression.
Auxin Regulation of miR164
Several plant miRNAs regulate or are predicted to regulate mRNAs involved in various phytohormone signaling pathways (Rhoades et al., 2002; Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Wang et al., 2004). To date, however, only miR159 has been shown to respond to gibberellins (Achard et al., 2004) and miR393 to abscisic acid (Sunkar and Zhu, 2004). Here, we show that miR164 levels in Arabidopsis roots can be increased by another phytohormone, auxin, through a signaling pathway dependent on AXR1, AXR2, and TIR1. These signaling components are known to be required for auxin-mediated lateral root development (Timpte et al., 1994; del Pozo and Estelle, 1999; Gray et al., 2001; del Pozo et al., 2002). Time-course studies showed that the miR164 increase, which occurs late in auxin treatment (after 6 h), is rather modest, being only 1.5-fold. Nevertheless, there is a clear and concomitant effect on NAC1 mRNA, whereas cleavage-resistant NAC1m mRNA is unaffected (Figure 6A). Consistent with the results obtained with the miR164 mutants, these results show that cleavage of NAC1 mRNA requires only a modest increase in its regulator. NAC1 has been shown to be expressed early (1 to 2 h) in pericycle cells during auxin-induced lateral root initiation (Xie et al., 2000, 2002). The modest auxin-induced increase of miR164 might result from restricted expression in these specialized cells but at a later developmental time. This issue remains to be resolved by future in situ hybridization experiments.
NAC1 is a transcriptional activator for auxin-induced lateral root initiation (Xie et al., 2000) that has been shown to be unstable (Xie et al., 2002). The NAC1 instability is attributable to its polyubiquitination by the SINAT5 E3 ligase, which targets it for degradation by 26S proteasomes (Xie et al., 2002). Our work here extends the signal desensitization to NAC1 mRNA levels, which, together with targeted NAC1 degradation, ensures a more rapid signal suppression. The induction of miR164, at least in part, by auxin suggests an autoregulatory loop by which the miRNA mediates the clearance of NAC1 mRNA to attenuate and terminate auxin signaling. A corollary of this observation is that we expect many mRNA targets of miRNAs to encode unstable proteins such as NAC1, because unstable mRNAs are less likely to encode stable proteins.
Among the five NAC domain mRNAs targeted by miR164, CUC1 and CUC2 are involved in cotyledon separation and meristem formation (Aida et al., 1997; Furutani et al., 2004). Expression of these two genes is repressed by auxin, and a localized increase in auxin concentration has been shown to phenocopy cuc1 cuc2 double mutant plants (Furutani et al., 2004). Transgenic plants overexpressing miR164 also have a similar phenotype (Laufs et al., 2004; Mallory et al., 2004a). It is possible that the CUC1/CUC2 repression is brought about, at least in part, by an auxin-induced increase in miR164 expression, as shown here. Future work should be directed toward understanding the mechanism of miRNA164 regulation by phytohormones.
METHODS
Plasmid Construction
Genomic sequence (153 bp) containing the MIR164b foldback was used as a synthetic precursor sequence (Figure 2B). The sequence was amplified from Arabidopsis thaliana genomic DNA by PCR using two primers, 164b5′ (5′-GATGGAGAAGCAGGGCACGTGCATTAC-3′) and 164b3′ (5′-GATGGTGAAGATGGGCACATGAAGAAC-3′), and cloned into pGEM-T Easy Vector (Promega, Madison, WI), giving pT-MIR164b, which was verified by sequencing. A NcoI/Klenow-SacI fragment from pT-MIR164b containing the MIR164b sequence was then cloned into the SmaI and SacI sites in pCAMBIA-1300 (AF234296) between the 35S promoter and the NOS 3′ poly(A) addition signal. The resulting construct was named 35S-MIR164b. The deletion mutant 35S-MIRΔ164 was created by digesting 35S-MIR164b with BamHI/Klenow+PmlI followed by self-ligation to remove sequences encoding 16 of the 21 nucleotides of the miR164 sequence (Figure 2B). A SphI/Klenow-SpeI fragment from pT-MIR164b containing the MIR164b sequence was also cloned into pX7-GFP (Guo et al., 2003) digested with XhoI/Klenow+SpeI to give pX7-164. For complementation of the three miR164a mutants, we used the construct p164a-164a-NOS, which contains a 5′ upstream fragment (−1911 to + 1, with + 1 being the first nucleotide of miR164) of MIR164a, the synthetic MIR164a precursor sequence, and the NOS poly(A) addition sequences. The synthetic MIR164a precursor contains 84 bp, which includes the foldback region of miR164 (Figure 2B). For complementation of the mir164b-1 mutant, we used the construct p164b-164b-NOS, which contains a 5′ upstream fragment of MIR164b (−2255 to +1, with +1 being the first nucleotide of miR164b), the synthetic MIR164b precursor (Figure 2B), and the NOS poly(A) addition sequences.
Site-directed mutagenesis of the NAC1 cDNA in the nucleotide sequence of the miR164 complementary site was performed with two primers in opposite orientations: NAC1m5′ (5′-CATCATCAATGAACATGTACCGTGTTTTTCGAATTTGTCACAGAACCAAACC-3′) and NAC1m3′ (5′-CTGTGACAAATTCGAAAAACACGGTACATGTTCATTGATGATGTAGTGATG-3′). NAC1 cDNA intermediate plasmid pBS-NAC1 (Xie et al., 2000) was used as template, using the QuickChange Multi Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). The resulting DNA was digested with DpnI to eliminate the methylated, nonmutated original template DNA before transformation of Escherichia coli DH5α. Correct mutagenesis was verified by sequencing. A HincII-XhoI (586 bp) fragment from pBS-NAC1m, which contains the mutations, was cloned into 35S-Myc-NAC1 (Xie et al., 2000) digested with SpeI/Klenow+XhoI to replace the corresponding original wild-type sequence. The resulting clone was designated 35S-Myc-NAC1m.
Plant Materials and Transformation
Arabidopsis (Ler) plants expressing either 35S-Myc-NAC1 or 35S-Myc (vector control) (Xie et al., 2002) were used for retransformation with pX7-MIR164 (Guo et al., 2003). Wild-type Arabidopsis (Col) was used for 35S-Myc-NAC1 and 35S-Myc-NAC1m transformation. Transgenic plants were generated by the floral dip method (Clough and Bent, 1998). Double transgenic plants (Myc-NAC1/pX7-164 and Myc/pX7-164) were selected on MS medium (Sigma-Aldrich, St. Louis, MO) containing 10 μg/mL Basta and 20 μg/mL hygromycin B. For chemical-induced expression of miRNA, 6-d-old T2 seedlings germinated on selective medium were transferred to inductive medium containing 2 μM 17-β-estradiol. Myc-NAC1 and Myc-NAC1m transgenic Arabidopsis (Col) plants were selected on MS medium containing 10 μg/mL Basta. Twelve Myc-NAC1 and 18 Myc-NAC1m T2 single-copy lines were selected for the population study of lateral root phenotype.
The T-DNA insertion mutants mir164b-1 (SALK_136105), mir164a-1 (SALK_061177), mir164a-2 (SALK_068270), and mir164a-3 (SALK_047619) were obtained from the ABRC. The SALK_136105 line obtained from the ABRC was homozygous with respect to kanamycin resistance, and the single-copy T-DNA was inserted into the loop of the predicted hairpin structure (Mallory et al., 2004a) (Figure 2B). Lines from SALK_061177, SALK_068270, and SALK-047619 showed a 3:1 segregation of kanamycin resistance, indicating a single T-DNA insertion in each. The T-DNA insertion positions of all mir164 mutants were confirmed by PCR using primers LBa1 (SALK T-DNA primer, 5′-TGGTTCACGTAGTGGGCCATCG-3′) and P164b (5′-GGGTAGCATGTTCATGGGTCAGCATGCTGTCTCC-3′) for mir164b-1 and LBa1 and P164a (5′-GCCTCTTCTTCCATATTTGATCAGATAACTGAAGACC-3′) for mir164a-1, mir164a-2, and mir164a-3. Homozygous mutant lines were selected for RNA and phenotypic analysis.
The mutants dcl1-9 (Park et al., 2002), axr1-12 (del Pozo and Estelle, 1999), axr2-1 (Timpte et al., 1994), and tir1-1 (Gray et al., 2001) and P1/HC-Pro transgenic plants (Kasschau et al., 2003) have been described.
Plant Growth Conditions, Auxin Treatment, and Observation of Root Phenotypes
Arabidopsis seeds were surface sterilized with 20% bleach and 0.01% Triton X-100 and washed three times with sterile water. Sterilized seeds were suspended in 0.15% agarose and plated on MS medium. Plates were vernalized in darkness for 2 d at 4°C and then transferred to a tissue culture room at 22°C under a 16-h-light/8-h-dark photoperiod. For observation of root phenotypes, seedlings were grown on vertical MS agar medium containing 2% sucrose. Lateral root initials were identified by staining whole seedlings with a mixture of toluidine blue and basic fuchsin and counted with a binocular microscope. For auxin treatment, seedlings were grown vertically on MS agar medium for 12 d, then incubated with 10 μM NAA. Root samples were collected at various times for RNA analysis.
Potted plants were grown in growth chambers at 22°C and 75% humidity under a 16-h-light/8-h-dark photoperiod.
Agrobacterium tumefaciens Infiltration in Nicotiana benthamiana
The constructs 35S-MIR164b, 35S-MIRΔ164, 35S-Myc-NAC1, and 35S-Myc-NAC1m were transformed into A. tumefaciens strain EHA105 by electroporation and selected on Luria-Bertani medium containing rifampicin at 10 μg/mL and kanamycin at 50 μg/mL (for 35S-MIR164b and 35S-MIRΔ164) or spectinomycin at 100 μg/mL (for 35S-Myc-NAC1 and 35S-Myc-NAC1m). Agrobacterial cells were infiltrated into leaves of N. benthamiana as described (English et al., 1997). For coinfiltration experiments, equal volumes of an Agrobacterium culture containing 35S-MIR164b or 35S-MIRΔ164 (OD600 = 1.75) and 35S-Myc-NAC1 or 35S-Myc-NAC1m (OD600 = 0.25) were mixed before infiltration into N. benthamiana leaves.
RNA Isolation and Analysis
Total RNA was isolated from plant tissues by LiCl precipitation (Verwoerd et al., 1989). For RNA gel blot analysis, total RNA was separated on a 1.2% agarose gel containing 6% formaldehyde and transferred to Hybond-N+ membranes. To specifically detect NAC1 transcripts, blots were hybridized with a BamHI-SalI fragment from pBS-NAC1 encoding the NAC1 C-terminal domain labeled with [α-32P]dCTP using a Ready-primed labeling kit (Amersham International, Buckinghamshire, UK). To specifically detect CUC3 transcripts, the membrane was stripped and reprobed with a PCR fragment encoding the CUC3 C-terminal domain. Low-molecular-weight RNAs were precipitated from the LiCl supernatant fraction of total RNA extraction with three volumes of ethanol. Low-molecular-weight RNAs were separated by electrophoresis on denaturing 17% polyacrylamide gels, and miRNA gel blot hybridizations were performed as described (Reinhart et al., 2002). DNA oligonucleotides complementary to miR164 or miR163 were end-labeled using T4 polynucleotide kinase (Roche Applied Science, Penzberg, Germany) and used for hybridizations. Signal intensity was measured using a PhosphorImager (Bio-Rad, Hercules, CA).
Mapping of miRNA-Guided Cleavage Site
The FirstChoice RLM-RACE kit (Ambion, Austin, TX) was used for RLM-RACE assay according to the manufacturer's instructions. Briefly, total RNAs were extracted from roots of wild-type Arabidopsis (Col) seedlings, and poly(A) mRNA was purified using an Oligotex mRNA mini kit (Qiagen, Valencia, CA). Poly(A) mRNA was directly ligated to the RLM-RACE 5′ RACE RNA Oligo adaptor (45 nucleotides) from the FirstChoice RLM-RACE kit (Ambion). The oligo(dT) (15-mer) primer was used to prime cDNA synthesis with reverse transcriptase. The resulting cDNA was used for the first round of nested PCR using the 5′ RACE Outer Primer together with a NAC1 gene-specific primer (5′-GCAATTCCAAACAGTGCTTGG-3′) complementary to nucleotides 1040 to 1060 of the NAC1 mRNA sequence. The 5′ RACE Inner Primer and the NAC1 gene-specific primer were used for the second round of nested PCR. Gel-purified PCR products were cloned into pGEM-T Easy vector (Promega) for sequencing.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AAF21437 (NAC1) and AF234296 (pCAMBIA-1300).
Acknowledgments
We thank Peter Hare and Jose Reyes for discussion, Vicki Vance for P1/HC-Pro transgenic seeds, and the ABRC for dc11-9 and the mir164 insertion mutants. N.-H.C. was supported by National Institutes of Health Grant GM-44640. Q.X. was partially supported by Chinese MST 973 Project 2003CB114304.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Nam-Hai Chua (chua@mail.rockefeller.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.030841.
References
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131, 3357–3365. [DOI] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros, V. (2004). The functions of animal microRNAs. Nature 431, 350–355. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, B., and Bartel, D.P. (2003). MicroRNAs: At the root of plant development? Plant Physiol. 132, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Bonnet, E., Wuyts, J., Rouze, P., and Van de Peer, Y. (2004). Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA 101, 11511–11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301, 336–338. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96, 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., Dharmasiri, S., Hellmann, H., Walker, L., Gray, W.M., and Estelle, M. (2002). AXR1–ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas, D.V., and Bartel, B. (2004). MicroRNA regulation of gene expression in plants. Curr. Opin. Plant Biol. 7, 512–520. [DOI] [PubMed] [Google Scholar]
- English, J.J., Davenport, G.F., Elmayan, T., Vaucheret, D., and Baulcombe, C. (1997). Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J. 12, 597–603. [Google Scholar]
- Furutani, M., Vernoux, T., Traas, J., Kato, T., Tasaka, M., and Aida, M. (2004). PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131, 5021–5030. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Guo, H.S., Fei, J.F., Xie, Q., and Chua, N.H. (2003). A chemical-regulated inducible RNAi system in plants. Plant J. 34, 383–392. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., and Bartel, D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799. [DOI] [PubMed] [Google Scholar]
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, B.A., and Timmermans, M.C. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84. [DOI] [PubMed] [Google Scholar]
- Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101, 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. [DOI] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Dugas, D.V., Bartel, D.P., and Bartel, B. (2004. a). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Reinhart, B.J., Jones-Rhoades, M.W., Tang, G., Zamore, P.D., Barton, M.K., and Bartel, D.P. (2004. b). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23, 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Parizotto, E.A., Dunoyer, P., Rahm, N., Himber, C., and Voinnet, O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18, 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., and Zhu, J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpte, C., Wilson, A.K., and Estelle, M. (1994). The axr2-1 mutation of Arabidopsis thaliana is a grain-of-function mutation that disrupts an early step in auxin response. Genetics 138, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd, T.C., Dekker, B.M., and Hoekema, A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.J., Reyes, J.L., Chua, N.-H., and Gaasterland, T. (2004). Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 5, R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Colgan, D., and Chua, N.H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Guo, H.S., Dallman, G., Fang, S., Weissman, A.M., and Chua, N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170. [DOI] [PubMed] [Google Scholar]