Abstract
The centromere of the maize (Zea mays) B chromosome contains several megabases of a B-specific repeat (ZmBs), a 156-bp satellite repeat (CentC), and centromere-specific retrotransposons (CRM elements). Here, we demonstrate that only a small fraction of the ZmBs repeats interacts with CENH3, the histone H3 variant specific to centromeres. CentC, which marks the CENH3-associated chromatin in maize A centromeres, is restricted to an ∼700-kb domain within the larger context of the ZmBs repeats. The breakpoints of five B centromere misdivision derivatives are mapped within this domain. In addition, the fraction of this domain remaining after misdivision correlates well with the quantity of CENH3 on the centromere. Thus, the functional boundaries of the B centromere are mapped to a relatively small CentC- and CRM-rich region that is embedded within multimegabase arrays of the ZmBs repeat. Our results demonstrate that the amount of CENH3 at the B centromere can be varied, but with decreasing amounts, the function of the centromere becomes impaired.
INTRODUCTION
The chromosomes of most higher eukaryotes contain a single centromere, the region that serves as the attachment site for spindle fibers and sister chromatid cohesion during cell division. The proteins that are involved in centromere function are highly conserved among all eukaryotes. Despite the similarities of form and function, the primary DNA sequence that underlies centromeres has no discernable conservation among various model organisms, underscoring the present lack of understanding of factors that determine centromere identity.
Although there is no obvious relationship among sequences found at centromeres, there are commonalities in organization. In most complex eukaryotes studied so far, including Drosophila melanogaster, humans, mice, maize (Zea mays), rice (Oryza sativa), and Arabidopsis thaliana, the centromeres are embedded within long tracks of highly repetitive DNA sequences consisting primarily of satellite repeats and transposons (Henikoff et al., 2001; Jiang et al., 2003). Human centromeres are the best studied among multicellular eukaryotes and appear to be a representative model. The most abundant DNA sequence in human centromeres is the ∼171-bp α-satellite repeat (Willard, 1998). The amount of α-satellite DNA varies from ∼250 kb to >4 Mb in different centromeres (Wevrick and Willard, 1989; Oakey and Tyler-Smith, 1990). Human artificial chromosomes were successfully assembled using either synthetic or cloned α-satellite DNA (Harrington et al., 1997; Ikeno et al., 1998), suggesting that long stretches of α-satellite DNA could act as a functional human centromere. Structural and functional analyses of DNA in the X chromosome centromere revealed that the α-satellite repeats are more diverged in the flanks of the centromere and become increasingly homogenized toward a functional core (Schueler et al., 2001). These data suggest that the centromeres evolve by selecting new repeats at their functional core, pushing older repeats to the flanking regions (Henikoff, 2002).
In contrast with the lack of conservation of centromeric DNA, several proteins specific to the centromere/kinetochore complex are highly conserved (Henikoff et al., 2001; Sullivan et al., 2001; Houben and Schubert, 2003). A centromere-specific histone H3-like protein, referred to as CENP-A or CENH3, has been found to underlie the kinetochore (Henikoff et al., 2001; Sullivan et al., 2001). CENH3 has been found in all model eukaryotes, including several plant species (Talbert et al., 2002; Zhong et al., 2002; Nagaki et al., 2004). There are numerous lines of evidence that CENH3 plays the key role in the establishment and function of kinetochores in various organisms (Henikoff et al., 2001; Sullivan et al., 2001). In maize, CENH3-associated chromatin is exclusively associated with the centromeric satellite and the CRM retroelements from the centromeric retrotransposon (CR) family in grasses (Zhong et al., 2002; Jin et al., 2004; Topp et al., 2004).
Because the stretches of repetitive DNA in centromeres are highly homogeneous and are often megabases in length, they are left as gaps in most sequencing projects of complex eukaryotic genomes (Henikoff, 2002). Currently, nearly complete sequences are available only from two native centromeres of rice (Nagaki et al., 2004; Wu et al., 2004; Zhang et al., 2004) and human neocentromeres (Saffery et al., 2003), which contain minimal or no satellite repeats. Therefore, methods other than sequencing must be used to study the relationship between the centromeric protein structure and the underlying DNA sequences. The centromere of the maize B chromosome provides an excellent model to study centromeres that contain an excess amount of satellite repeats. This supernumerary chromosome is present in some but not all maize varieties and is maintained by an accumulation mechanism involving nondisjunction at the second pollen mitosis followed by preferential fertilization of the egg by the B-containing sperm in the process of double fertilization (Roman, 1947). The maize B centromere region contains a repetitive element ZmBs that is not present on the A chromosomes (Alfenito and Birchler, 1993). The specificity of the ZmBs repeat to the B chromosome allows molecular and cytological studies of a single centromere among others in the genome (Kaszas and Birchler, 1996, 1998). In addition, B centromere misdivision derivatives can be readily developed from specific cytogenetic stocks, and the transmission of these derivatives can be easily tracked (Kaszas and Birchler, 1998). These materials provide a unique system to study a centromere deletion series to establish the molecular correlates of centromere function.
In this study, we report the fine structure and DNA composition of the centromeres from the normal B chromosome and a set of B centromere misdivision derivatives. We describe an ∼700-kb domain that consists of all three previously described repeats (ZmBs repeat, CentC, and CRM). Five misdivision breakpoints are mapped within this domain and the amount of the B centromere–bound CENH3 is correlated with the amount of sequence from this domain. These results provide support for the view that CentC/CRM is present at all sites in the maize genome that specify a centromere (Ananiev et al., 1998; Jin et al., 2004). They also demonstrate that the amount of CENH3 can be altered and correlates with centromere function. These data demonstrate the minimum requirements for a representative centromere in a multicellular eukaryote.
RESULTS
DNA Repeats and Their Organization in the Intact Maize B Centromere
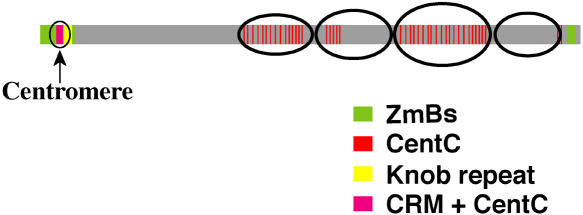
It was previously demonstrated that the centromeres of maize A chromosomes contain a 156-bp centromere-specific satellite repeat CentC (Ananiev et al., 1998) and retrotransposons from the CR family, including CRM1, CRM2 (full-size CR elements), and CentA (a truncated CR element) (Ananiev et al., 1998; Zhong et al., 2002; Nagaki et al., 2003b; Jin et al., 2004; Lamb et al., 2005). In addition to the elements found in A centromeres, the centromere of the maize B chromosome contains a B-specific repeat ZmBs (Alfenito and Birchler, 1993; Kaszas and Birchler, 1996). High-resolution fluorescence in situ hybridization (FISH) mapping on pachytene B chromosomes revealed that the B chromosome contains two major hybridization sites of the ZmBs repeat: one at the centromeric region and another at the subtelomeric region on the long arm (Lamb et al., 2005) (Figure 1). Major CRM signals were only detected in the centromeric regions, although minor CRM signals were also observed outside of the centromere (Lamb et al., 2005). The CentC signals, which are specific to the centromeres of the A chromosomes, were dispersed throughout the B chromosome (Lamb et al., 2005) (Figure 1).
Figure 1.
Locations of the CentC, CRM, ZmBs, and Knob Repeats on the Maize Pachytene B Chromosome.
Minor sites of these repeats on the B chromosome are not shown. The four distinctive heterochromatin blocks on the long arm are illustrated. The diagram is based on Lamb et al. (2005).
We used a fiber-FISH approach to analyze the organization of CentC, CRM, and the ZmBs repeat within the B centromere. We found that the longest array of the ZmBs repeat was ∼1150 μm (∼3.7 Mb) in length. Because it is technically difficult to obtain DNA fibers >2 Mb, we estimate that the ZmBs array in the centromeric region is at least 3.7 Mb and is potentially longer. The fiber-FISH–based estimate excludes the ZmBs arrays that are near the centromere but separated from the major cluster as well as those in the distal long arm tip (Figure 1).
Two of the longest fiber-FISH signals are shown in Figure 2. Figure 2A shows signals for ZmBs (green) and CentC (red), and Figure 2C shows signals for ZmBs (green) and CRM (red). The CentC signals were restricted to an ∼700-kb domain that is also rich in CRM sequences. The CentC and CRM sequences in this domain are intermingled (Figures 2B, 2D, 2E, and 2F), similar to the organization of these two sequences in the A centromeres (Jin et al., 2004). Unlike A centromeres, however, the intermingled CentC/CRM sequences are disrupted by five ZmBs arrays (Figures 2E and 2F). This ∼700-kb CentC/CRM/ZmBs repeat domain is flanked on either side by long ZmBs arrays that contain short CRM signals (Figure 2D). The ZmBs array flanking on one side (arbitrarily called “left”) is longer than the array on the “right” side, with lengths measuring ∼2.2 and 0.8 Mb, respectively (Figures 2B and 2D). The fiber-FISH signal patterns in the flanking domains are remarkably uniform, suggesting a high concentration of the ZmBs repeats in these regions. A diagram of the fiber-FISH–based pattern map of the CentC-rich domain is depicted in Figure 2F.
Figure 2.
Organization of CentC, CRM, and the ZmBs Repeat in the Centromere of Maize B Chromosomes.
(A) A fiber-FISH signal derived the ZmBs repeat (green) and CentC (red). Arrows mark the border of the CentC-enriched domain.
(B) A diagram based on the fiber-FISH signal pattern in (A). Unambiguous signals from CentC were only observed between the two arrows.
(C) A fiber-FISH signal derived the ZmBs repeat (green) and CRM (red). Arrows mark the border of the CentC-enriched domain.
(D) A diagram based on the fiber-FISH signal pattern in (C). Additional CRM signals located outside of the CentC-enriched domain are marked by arrowheads.
(E) An expanded image of the domain in (A). The five ZmBs repeat arrays are marked.
(F) An expanded image of the domain in (C). The five ZmBs repeat arrays are marked.
(G) A diagram depicting the distribution patterns of the major blocks of CentC, CRM, and ZmBs. Note: the first ZmBs repeat array contains a distinctive CRM insertion in the middle.
Bars = 100 μm in (A) and (C) and 20 μm in (E) and (F).
Mapping the Breakpoints of Misdivided B Centromeres
Early cytogenetic analysis of univalent chromosomes in meiosis demonstrated that the centromere is a compound structure consisting of multiple functional units (Darlington, 1939; Sears, 1952). A misdivision of a centromere will result in two functional derivatives. Therefore, analysis of misdivision breakpoints is a powerful approach to dissect the functional domains of centromeres. Numerous B centromere misdivision derivatives were previously isolated from progenies of a TB-9Sb translocation chromosome (Carlson, 1970; Carlson and Chou, 1981; Kaszas and Birchler, 1996, 1998). A set of seven B misdivision derivatives (Figure 3) was selected for fine mapping of the B centromeric regions. The centromeres of these selected misdivision derivatives have significantly different sizes based on our previous pulsed field gel electrophoresis (PFGE) mapping data (Kaszas and Birchler, 1998).
Figure 3.
The Pedigree and Structure of the B Centromere Misdivision Derivatives.
(A) The pedigree of B centromere misdivision derivatives used in this study.
(B) Graphical representation of the centromere size of B chromosome misdivision derivatives based on Kaszas and Birchler (1998). The closed boxes represent the centromeric region.
Fiber-FISH was used to analyze the organization of CentC, CRM, and the ZmBs repeat in the misdivision derivatives. Four lines, Telo4-11(-), Ring4-8(-), Telo4-4(-), and Ring4-12(-), were derived from the same progenitor, Iso3(-) (Figure 3A). The sizes of the fiber-FISH signals from Telo4-11(-) and Ring4-8(-) were 825 ± 55 kb and 440 ± 42 kb, respectively (Figures 4A to 4D), in reasonable agreement with the 490- and 500-kb estimates made previously from PFGE (Kaszas and Birchler, 1998) (Table 1). Both Telo4-11(-) and Ring4-8(-) contain blocks of CentC/CRM repeat arrays that align with the patterns obtained from the normal B centromere (Figures 4A to 4D and 5). Telo4-11(-) contains the first four of the five short ZmBs arrays within the CentC-rich domain, whereas Ring4-8(-) contains only the first ZmBs cluster (Figures 4A to 4D and 5).
Figure 4.
Distribution Patterns of CentC, CRM, and the ZmBs Repeat in the Centromeres of B Centromere Misdivision Derivatives.
The ZmBs repeat arrays derived from the CentC-enriched domain are marked. Bars = 30 μm in (E), 25 μm in (A) and (B), 20 μm in (G), 10 μm in (C), (D), (H), and (I), and 5 μm in (F).
(A) A fiber-FISH signal of ZmBs (green) and CRM (red) from the centromere of Telo4-11(-).
(B) A fiber-FISH signal of ZmBs (green) and CentC (red) from the centromere of Telo4-11(-).
(C) A fiber-FISH signal of ZmBs (green) and CentC (red) from the centromere of Ring4-8(-).
(D) A fiber-FISH signal of ZmBs (green) and CRM (red) from the centromere of Ring4-8(-).
(E) A fiber-FISH signal of ZmBs (green) and CRM (red) from the centromere of Telo4-4(-).
(F) A fiber-FISH signal of ZmBs (green) and CRM (red) from the centromere of Ring4-12(-). Note: the ZmBs array is disrupted by a CRM insertion (arrowhead), which is characteristic of the first ZmBs array in the domain.
(G) A fiber-FISH signal from ZmBs (green) and a mixed probe of CRM and CentC (red) from the centromere of Telo2-2(-) (fragment 1).
(H) A fiber-FISH signal of ZmBs (green) from the centromere of Telo2-2(-) (fragment 2). No CRM and CentC signals were detected within this fragment.
(I) A fiber-FISH signal of ZmBs (green) from the centromere of Telo3-3(-). No CRM and CentC signals were detected within this fragment.
Table 1.
Distribution of CentC, CRM, and the ZmBs Repeat among Normal and Misdivision Derivatives of Maize B Centromeres
| Lines | n | Full Size (μm) | Full Size (kb)a | ZmBs Repeats (kb) | ZmBs Repeats (kb)b | Presence of CentC | Presence of CRM | Transmissionc |
|---|---|---|---|---|---|---|---|---|
| Intact Bd | 10 | >1150 | >3690 | >3000 | >9000 | Yes | Yes | – |
| Core domain of B | 10 | 220 ± 14 | 705 ± 45 | ∼245 | NDe | Yes | Yes | – |
| Ring4-8(-) | 17 | 137 ± 13 | 440 ± 42 | ∼220 | ∼500 | Yes | Yes | ND |
| Telo4-11(-) | 21 | 257 ± 17 | 825 ± 55 | ∼500 | ∼2360 | Yes | Yes | 42% |
| Telo4-4(-) | 12 | 228 ± 19 | 732 ± 62 | ∼500 | ∼490 | Yes | Yes | 10% |
| Ring4-12(-) | 12 | 29 ± 3 | 93 ± 10 | ∼60 | ND | No | Yes | ND |
| Telo2-2(-) 1 | 8 | 225 ± 15 | 722 ± 48 | ∼330 | ∼2150 | Yes | Yes | 43% |
| Telo2-2(-) 2 | 7 | 95 ± 14 | 305 ± 45 | ∼200 | ND | No | No | – |
| Telo3-3(-) | 14 | 87 ± 6 | 280 ± 19 | ∼185 | ∼280 | No | No | 13% |
The length of the fiber-FISH signals (μm) was converted to kilobases using a 3.21 kb/μm conversion rate (Cheng et al., 2002).
The sum of fragments containing the ZmBs repeat arrays was estimated by PFGE (Kaszas and Birchler, 1998).
Data from Kaszas and Birchler (1998).
We used the longest fiber-FISH signals to represent the minimum size.
ND, not determined.
Figure 5.
Diagrams of DNA Organization of Five B Centromere Misdivision Derivatives.
All five lines, Telo4-11(-), Telo4-4(-), Ring4-8(-), Ring4-12(-), and Telo2-2(-), contain a DNA fragment derived from the CentC-enriched domain of the original B centromere. The misdivision breakpoints of these five derivatives are all located in the middle of this domain. Vertical arrows point to the borders of the ∼700-kb CentC-enriched domain.
The size and pattern of the fiber-FISH signals derived from Telo4-4(-) is almost identical to those from Telo4-11(-) (Figures 4E and 5). PFGE analysis, however, suggested that Telo4-4(-) contains >2 Mb of the ZmBs repeat (Kaszas and Birchler, 1998) (Table 1). In fiber-FISH analysis, we only collected and analyzed the signals associated with CentC and/or with a significant amount of CRM. Additional fiber-FISH signals consisting of solely ZmBs repeats cannot be reliably identified because these signals may be derived from broken DNA fibers. PFGE analysis, by contrast, identifies all DNA fragments associated with the ZmBs repeat. In addition, analysis of progenies from a single individual containing Telo4-4(-) revealed considerable differences in the amount of the ZmBs repeat (J. Lamb and J.A. Birchler, unpublished data), suggesting that this line may not be stable. Taken together, differences in what the two techniques detect and possibly an instability of Telo4-4(-) might explain the discrepancy of the amount of the ZmBs repeats estimated by fiber-FISH and PFGE.
The final Iso3(-) derivative characterized was Ring4-12(-), which contains only 93 ± 10 kb of CRM and the ZmBs repeat and no detectable CentC. This short fragment also aligns with a small part of the CentC-rich domain that includes the first of the five short ZmBs arrays. The first ZmBs array in this region can be identified based on the presence of a distinctive CRM insertion (Figures 4F and 5).
Line Telo3-3(-) and its parental line Telo2-2(-) have a different lineage from line Iso3(-) (Figure 3A). Two DNA fragments containing the ZmBs repeat were detected by fiber-FISH in Telo2-2(-). The first fragment is ∼800 kb long and consists of all three elements: CentC, CRM, and ZmBs. The organization of the three repeats in Telo2-2(-) is nearly identical to that of Telo4-11(-) and Telo4-4(-) (Figures 4G and 5). The second fragment is ∼240 kb in size and consists exclusively of the ZmBs repeat (Figure 4H). We were not able to detect CentC or CRM signals within this fragment using fiber-FISH and could not align it to the CentC-rich domain. Furthermore, we could not find any unambiguous connection between these two centromeric DNA fragments, suggesting that the two fragments are separated by several hundred kilobases (Jackson et al., 1998).
Pachytene FISH analysis of Telo2-2(-) revealed a larger ZmBs repeat signal, which colocalizes with CentC and CRM, and a separate, nearby but smaller ZmBs repeat signal, which does not colocalize with other centromeric elements (Figure 6). This smaller FISH site is also present in the normal B chromosome (Lamb et al., 2005). Thus, the ∼240-kb ZmBs array is likely to coincide with this centromere distal ZmBs site. A single DNA fragment containing ZmBs was found in Telo3-3(-), which appeared identical to the ∼240-kb ZmBs fragment in Telo2-2(-) (Figure 4I). Both fiber-FISH and pachytene FISH did not detect CentC and CRM signals in Telo3-3(-). Interestingly, Telo3-3(-) is the most unstable B centromere derivative recovered to date (Kaszas and Birchler, 1998). Telo3-3(-) is frequently lost both mitotically and meiotically and routinely shows developmental loss of genetic markers present on the chromosome (Kaszas and Birchler, 1998). It is also unstable structurally, being found alternately in our stocks as a telocentric or isochromosome.
Figure 6.
Pachytene Localization of the ZmBs and CENH3.
(A) Pachytene spreads from Telo2-2(-), Telo4-4(-), and Telo3-3(-) (as an isochromosome in this line) were immunolabeled with CENH3 (in green), hybridized with ZmBs (in red), and counterstained with 4′,6-diamidino-2-phenylindole.
(B) Only the ZmBs hybridization signal is shown. The arrow indicates a minor hybridization signal distinct from the main ZmBs hybridization site. The arrowhead indicates a cluster of ZmBs on the distal portion of the reciprocal A-B translocation chromosome in this line.
(C) Only the CENH3 signal is shown. Arrows point to the CENH3 signals associated with the misdivided B centromeres. Complete or partial colocalization of ZmBs and CENH3 is always observed for Telo2-2(-) and Telo4-4(-), whereas Telo3-3(-) shows no colocalization in some spreads.
(D) A Telo2-2(-) pachytene chromosome labeled with ZmBs (in red) and CentC (in green). Arrow indicates a minor hybridization signal distinct from the main ZmBs hybridization site. Arrowhead indicates the location of CentC hybridization to the B chromosome heterochromatin of the B-A translocation.
The ZmBs Repeat Is Associated with Maize CENH3
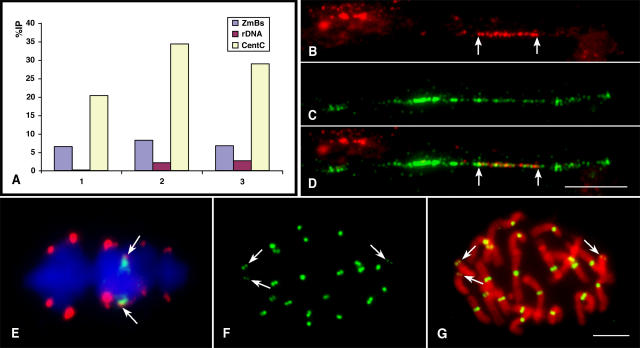
Transmission studies of misdivision derivatives suggest that the ZmBs repeat is present in the functional region of the B centromere (Kaszas and Birchler, 1998). We conducted chromatin immunoprecipitation (ChIP) experiments using an antibody against maize CENH3. Data from three independent experiments (Figure 7A) showed that, on average, the percentage of immunoprecipitation (%IP) of the ZmBs repeat was 7.1% (se = ±0.9%, n = 3), whereas only 1.6% (se = ±1.4%) was detected with the rDNA control, a statistically significant difference (Student's t test, P = 0.0022). By contrast, the average %IP for the CentC repeat was 27.8% in the same experiments, similar to our previous results (Zhong et al., 2002). The association of the ZmBs repeat with CENH3 was also visualized by sequential detection of CENH3 and the ZmBs repeat on stretched B centromeres. On stretched chromatin fibers, the CENH3 domain appeared to colocalize with the second quarter of the ZmBs repeat array (Figures 7B to 7D), a similar position as the CentC-rich domain in the B centromere from fiber-FISH analysis (Figures 2A and 2C). On labeled pachytene chromosomes, the location of CENH3 appeared to be near the middle of the major ZmBs array and colocalized with the CentC-containing domain (Lamb et al., 2005). At metaphase I and anaphase I of meiosis, FISH signals derived from CentC were consistently located at the most poleward position on B chromosome bivalents as well as bivalents of Telo2-2(-) (Figure 7E). Taken together, these results provide further support that the CentC-enriched domain within the ZmBs array is the region associated with kinetochore formation.
Figure 7.
ChIP Analysis and Cytological Mapping of CENH3 on Stretched Chromatin Fibers and Meiotic and Mitotic Chromosomes.
(A) ChIP analysis of CentC and the ZmBs repeat in maize line B73+3B. The ZmBs repeat was coimmunoprecipitated with the anti-CENH3 antibody in all three experiments. The %IP of ZmBs repeat was significantly different from that of the negative control rDNA.
(B) A stretched B centromere is stained by the maize anti-CENH3 antibody. The arrows mark the borders of the CENH3 domain.
(C) FISH mapping on the same stretched B centromere using the ZmBs repeat.
(D) A merged image of the immunostaining and FISH signals. Bar = 10 μm.
(E) FISH mapping of CentC (red) and the ZmBs repeat (green) on metaphase I chromosomes of maize B73 line containing a pair of B chromosomes. The Cent repeats (arrows) are located at the most poleward position (ahead of the ZmBs repeats) on the B chromosome bivalent.
(F) Immunostaining signals from the anti-CENH3 antibody on the somatic metaphase chromosomes of maize line B73+3B. Arrows point to the signals on the B chromosomes, which are weaker than those on the A centromeres.
(G) A merged image of the immunostaining signals with chromosomes, which are stained with 4′,6-diamidino-2-phenylindole and are pseudocolored in red. Bar = 5 μm.
Relationship between the Amount of CENH3 and the Size of Misdivided B Centromeres
Immunostaining analysis revealed that CENH3 signals are only located at the B centromeres and not at any of the B chromosomal regions containing CentC or the ZmBs repeat (Figures 7F and 7G). Notably, the CENH3 signals in the B centromeres were often weaker than those in the A centromeres (Figure 7F). To investigate the relationship between the amount of centromeric DNA and CENH3, we analyzed several misdivision lines using a combination of FISH and immunolocalization procedures. The CENH3 immunostaining signals derived from the misdivided B centromeres can be unambiguously identified based on their colocalization with the ZmBs repeat (Figure 8). To analyze the results quantitatively, the immunostaining signals were divided into five classes: I, the intensity of the immunostaining signal was similar to those in A centromeres; II, signals weaker than those in A centromeres; III, signals significantly weaker than those in A centromeres; IV, the signal could not be observed without contrast adjustment by computer software; V, signal could not be observed even after contrast adjustment.
Figure 8.
Interphase Detection of CENH3 and the ZmBs Repeat in a Maize B73 Line with a Single B Chromosome, Telo4-11(-), Telo4-4(-), Ring4-8(-), and Telo3-3(-).
(A) Maize B73 line with a single B chromosome.
(B) Telo4-11(-).
(C) Telo4-4(-).
(D) Ring4-8(-).
(E) Telo3-3(-).
The signals associated with normal or the misdivided centromeres are indicated by arrows. Note: the ZmBs site in (E) is not associated with any CENH3 staining.
We scored 14% class I, 68% class II, 18% class III, but no class IV and V signals from the intact B centromere (Table 2). Telo4-11(-), which contains ∼545 kb of the ∼700-kb CentC-rich domain, showed a similar amount of CENH3 as intact B centromeres (Table 2). Telo4-4(-), however, with slightly less of this domain (495 kb), contained no class I signals and an increase in the number of class IV and class V signals (Table 2). Ring 4-8(-), with only ∼240 kb from the CentC-rich domain, recruited much less CENH3 than the other derivatives. The vast majority of the Ring 4-8(-) signals fell into the class IV and V categories. Finally, Telo3-3(-), which has undetectable amounts of CentC and CRM in the centromeric region, recruited so little CENH3 that it was only weakly detected in 12% of the cells (class IV) and undetectable (class V) in the remaining cells (Table 2). The very low association of CENH3 with Telo3-3(-) explains the highly unstable nature of this chromosome as noted above.
Table 2.
Signal Intensities of CENH3 Immunostaining in Normal and Misdivision Derivatives of the Maize B Chromosome
| Lines | Size (kb) | Core Domain (kb) | n | Ia | IIb | IIIc | IVd | Ve |
|---|---|---|---|---|---|---|---|---|
| Intact B | >3690 | ∼705 | 89 | 13 (14.6%) | 59 (66.3%) | 17 (19.1%) | 0 (0.0%) | 0 (0.0%) |
| Telo4-11(-) | 825 | ∼545 | 98 | 17 (17.3%) | 65 (66.3%) | 15 (15.3%) | 1 (1.0%) | 0 (0.0%) |
| Telo4-4(-) | 732 | ∼495 | 87 | 0 (0.0%) | 43 (49.4%) | 36 (41.4%) | 6 (6.9%) | 5 (5.7%) |
| Ring4-8(-) | 440 | ∼240 | 65 | 0 (0.0%) | 0 (0.0%) | 2 (3.1%) | 26 (40.0%) | 37 (56.9%) |
| Telo3-3(-) | 280 | 0 | 34 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 4 (11.8%) | 30 (88.2%) |
The intensity of CENH3 signals on the B centromere is similar to those of A centromeres.
The intensity of CENH3 signals on the B centromere is weaker than those of A centromeres.
The intensity of CENH3 signals on the B centromere is significantly weaker than those of A centromeres.
The signal can not be observed without contrast adjustment using computer software.
The signal can not be observed even with contrast adjustment.
DISCUSSION
Dissection of the Functional Domain of Maize B Chromosome Centromere
Centromeres in complex eukaryotes often contain arrays of a single class of repetitive DNA elements up to several megabases. However, generally only a portion of such long arrays is associated with CENH3. The repetitive DNA outside of the CENH3-associated domain presumably plays a role in other centromeric functions, such as sister chromatid cohesion and chromosomal condensation (Grewal and Moazed, 2003). For example, the centromere of the human X chromosome contains ∼3 Mb of the α-satellite but only a portion of this α-satellite array is associated with centromeric proteins (Schueler et al., 2001; Spence et al., 2002). Association of CENH3 with a subdomain of the centromeric satellite DNA has also been demonstrated in maize (Jin et al., 2004) and Arabidopsis (Shibata and Murata, 2004).
Previous studies demonstrated that the maize B chromosome contains up to 9 Mb of the ZmBs repeat (Kaszas and Birchler, 1996). This ZmBs repeat was considered to be represented in the functional region of the B centromere because all of the B centromere misdivision derivatives retained a portion of the ZmBs repeat and rearranged the restriction pattern. There is a strong correlation between the size of the retained ZmBs repeat array and meiotic transmission (Kaszas and Birchler, 1998). However, it was not known if the ZmBs repeat was the sole DNA element located within the functional B centromere. It was also found that some misdivided B centromeres retained up to 2.5 Mb of the ZmBs repeat but still showed poor meiotic transmission (Kaszas and Birchler, 1998).
We provide several lines of evidence that an ∼700-kb CentC-rich domain represents the primary CENH3 binding and functional kinetochore of the B centromere. (1) The breakpoints of all of the misdivision derivatives analyzed, except for Telo3-3(-), could be recognized within the ∼700-kb domain (Figure 5). (2) The relative position of the CENH3 binding domain on stretched B centromeres (Figures 7B to 7D) is similar to the position of the ∼700-kb domain on mapped DNA fibers (Figures 2A and 2C). The leading cytological location of the CentC repeats on the B chromosome bivalent at metaphase I also suggests that they are bound within the kinetochore (Figure 7E). (3) The amount of CENH3 in misdivided B centromeres is correlated with the size of the DNA fragment derived from the ∼700-kb domain (Table 2). Because the ZmBs repeat is present within this CentC-rich domain (Figure 5), this interpretation is also consistent with prior findings (Kaszas and Birchler, 1996, 1998).
An Estimate of the Normative Size of a Functional Plant Centromere
The size of a functional centromere has been estimated in different species using different approaches. Deletion mapping revealed that the minimum size of a human minichromosome is ∼100 kb (Yang et al., 2000). Similarly, deletion mapping placed the minimum centromere of a Drosophila minichromosome within a 420-kb region (Murphy and Karpen, 1995; Sun et al., 1997). The CENP-A binding domains of several human neocentromeres have been determined using ChIP analysis. The chromatin domain associated with CENP-A in these neocentromeres ranges from ∼130 to 460 kb (Lo et al., 2001a, 2001b; Alonso et al., 2003).
In plants, the centromere of rice chromosome 8 (CEN8) contains an ∼750-kb region associated with rice CENH3 (Nagaki et al., 2004). Here, we demonstrate that the CENH3 binding region in the maize B centromere is ∼700-kb, close in size to the CENH3 binding domain in rice CEN8. We have also previously shown that the centromeres of five of seven maize A chromosomes analyzed contain 300 to 700 kb of intermingled CentC/CRM sequences that interact with CENH3 (Jin et al., 2004). Our deletion analysis supports the view that these domains correspond to functional centromeric regions. Thus, the functional centromeres of maize A and B chromosomes have a similar size and contain ∼300 to 700 kb DNA in the CENH3-associated chromatin.
The B Centromere Is Defined by Both Genetic and Epigenetic Mechanisms
Extensive research in several model eukaryotes has revealed that centromere formation can be controlled by both genetic and epigenetic mechanisms. In humans, fully functional artificial centromeres can be assembled from the α-satellite alone (Harrington et al., 1997; Ikeno et al., 1998), and minor manipulations of the sequence alter this capacity (Ohzeki et al., 2002). However, many human neocentromeres contain no detectable α-satellite DNA, yet associate with the same proteins as normal human centromeres (du Sart et al., 1997; Saffery et al., 2000) and are stable in both mitosis and meiosis (Tyler-Smith et al., 1999; Amor et al., 2004).
The ∼700-kb domain in the B centromere contains arrays of three repeats, the ZmBs repeat, CentC, and CRM. These three repeats, however, are not specific to the B centromere and show multiple locations throughout the B chromosome (Lamb et al., 2005). Nevertheless, CENH3 is only associated with the CentC-rich domain in the B centromere. Therefore, the repetitive DNA within this domain is possibly marked epigenetically for CENH3 recognition. The CentC repeat is highly specific to the centromeres of maize A chromosomes (Ananiev et al., 1998). It is interesting to note that although the CentC repeat is distributed throughout the B chromosomes, the CentC sequence within the B centromere is restricted to the ∼700-kb domain. We previously also demonstrated that maize CENH3 is always associated with intermingled CentC/CRM sequences in A centromeres, although not all of the CentC/CRM sequences are associated with CENH3 (Jin et al., 2004). The CentC/CRM sequences in the ∼700-kb region of the B centromere appears to serve a similar role in CENH3 recognition. It has recently been demonstrated in a human neocentromere that the CENH3-associated chromatin is divided into seven blocks by H3-associated chromatin within a 330-kb region (Chueh et al., 2005).
Based on these data, the CentC-containing region within the B centromere appears to be critical for association with CENH3 because removal of CentC within the ∼700-kb domain diminishes CENH3 staining. However, when CentC is reduced below detection, as on Telo3-3(-), some CENH3 remains. The small amount of CENH3 remaining on the Telo3-3(-) derivative may be binding to a small amount of epigenetically marked ZmBs repeat derived from the ∼700-kb domain. Alternatively, it is possible that sequences adjacent to the CentC-enriched domain are recruited to the centromere after misdivision, in an analogous fashion to neocentromere formation in Drosophila (Platero et al., 1999; Maggert and Karpen, 2001).
Taken together, the results suggest that the functional centromere of the B chromosome is a small CentC- and CRM-rich domain that is embedded within a much larger array of the ZmBs repeat. The amount of CENH3 found at the B centromere is correlated with the size of this domain and the stability of the chromosome. These findings bolster the view that CentC and CRM are key elements of maize centromeres (Zhong et al., 2002; Jin et al., 2004) and provide genetic support for the idea that CENH3 is necessary for proper kinetochore formation.
METHODS
Materials
A maize (Zea mays) line B73 containing multiple B chromosomes was used for cytological analysis of the intact B chromosomes. B centromere misdivision derivatives were produced from the A-B translocation line TB-9Sb (Kaszas and Birchler, 1996, 1998) (Figure 3A). These derivatives were classified by their chromosome type (telocentric chromosome, isochromosome, or ring chromosome) and presence (+) or absence (−) of a knob adjacent to the centromeric region (Kaszas and Birchler, 1996). The centromeric DNA probes, including CentC, CRM, and the ZmBs repeat, were described previously (Alfenito and Birchler, 1993; Nagaki et al., 2003b).
FISH and Fiber-FISH
The FISH and fiber-FISH were performed according to published protocols (Jiang et al., 1995; Jackson et al., 1998). DNA probes were labeled with biotin-dUTP or digoxigenin-dUTP (Roche, Indianapolis, IN). Chromosomes were counterstained by 4′,6-diamidino-2-phenylindole in an antifade solution Vectashield (Vector Laboratories, Burlingame, CA). Images were captured digitally using a Sensys CCD camera (Roper Scientific, Tucson, AZ) attached to an Olympus BX60 epifluorescence microscope (Tokyo, Japan). The camera control and imaging analysis were performed using IPLab Spectrum v3.1 software (Signal Analytics, Vienna, VA).
Sequential Detection of CENH3 and the ZmBs Repeat on Chromosomes and Stretched Centromeres
Preparation of somatic chromosomes and stretched centromeres for immunostaining was performed according to Jin et al. (2004) with only minor modifications. Maize nuclei were isolated from young maize kernels rather than callus. The preparations were fixed in 4% paraformaldehyde in PBS for 10 min. Approximately 100 μL rabbit anti-CENH3 antibody (1:200 in TNB buffer [0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.5% blocking reagent]) was added to the slides. After incubation in a humid chamber at 37°C for 3 h, the slides were washed three times in PBS before adding 100 μL of rhodamine anti-rabbit secondary antibody (1:50 in TNB buffer). The slides were incubated and washed again and fixed in 4% paraformaldehyde. The slides were then probed with the ZmBs repeat. Chromosomes and interphase nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Labeling and detection of DNA repeats and CENH3 on maize pachytene chromosomes were described previously (Lamb et al., 2005).
ChIP
ChIP was performed as described previously (Nagaki et al., 2003a). The nuclei were isolated from leaf tissue and digested with micrococcal nuclease. The resultant nucleosomes were incubated with the maize anti-CENH3 antibody (Zhong et al., 2002). The immune complexes were precipitated and separated into unbound (Sup, for supernant) and bound (Pel, for pellet) fractions. Equal amounts of the Sup and Pel fractions were blotted on membranes and hybridized with 32P-labeled ZmBs repeat. The hybridization was quantified using a phosphor imager. Mock experiments using preimmunized rabbit serum served as nonspecific binding control for each ChIP assay. The %IP, defined as pel/(pel + sup) of the mock experiments, was subtracted in each case from %IP of the anti-CENH3 treatments. We used 18S-26S rRNA genes (rDNA) and CentC as negative and positive controls, respectively.
Acknowledgments
This research was supported by National Science Foundation Grant 9975827 to R.K.D., J.A.B., and J.J. J.L. was supported by a University of Missouri Life Sciences Graduate Fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: James A. Birchler (birchlerj@missouri.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.030643.
References
- Alfenito, M.R., and Birchler, J.A. (1993). Molecular characterization of a maize B chromosome centric sequence. Genetics 135, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, A., Mahmood, R., Li, S., Cheung, F., Yoda, K., and Warburton, P.E. (2003). Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 12, 2711–2721. [DOI] [PubMed] [Google Scholar]
- Amor, D.J., Bentley, K., Ryan, J., Perry, J., Wong, L., Slater, H., and Choo, K.H.A. (2004). Human centromere repositioning “in progress”. Proc. Natl. Acad. Sci. USA 101, 6542–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95, 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, W.R. (1970). Nondisjunction and isochromosome formation in the B chromosome of maize. Chromosoma 30, 356–365. [Google Scholar]
- Carlson, W.R., and Chou, T.-S. (1981). B chromosome nondisjunction in corn: Control by factors near the centromere. Genetics 97, 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z.K., Buell, C.R., Wing, R.A., and Jiang, J. (2002). Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes and extended DNA fibers. Chromosome Res. 10, 379–387. [DOI] [PubMed] [Google Scholar]
- Chueh, A.C., Wong, L.H., Wong, N., and Choo, K.H.A. (2005). Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum. Mol. Genet. 14, 85–93. [DOI] [PubMed] [Google Scholar]
- Darlington, C.D. (1939). Misdivision and the genetics of the centromere. J. Genet. 37, 341–364. [Google Scholar]
- du Sart, D., Cancilla, M.R., Earle, E., Mao, J.I., Saffery, R., Tainton, K.M., Kalitsis, P., Martyn, J., Barry, A.E., and Choo, K.H.A. (1997). A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat. Genet. 16, 144–153. [DOI] [PubMed] [Google Scholar]
- Grewal, S.I.S., and Moazed, D. (2003). Heterochromatin and epigenetic control of gene expression. Science 301, 798–802. [DOI] [PubMed] [Google Scholar]
- Harrington, J.J., Bokkelen, G.V., Mays, R.W., Gustashaw, K., and Willard, H.F. (1997). Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 15, 345–355. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. (2002). Near the edge of a chromosome's ‘black hole’. Trends Genet. 18, 165–167. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Houben, A., and Schubert, I. (2003). DNA and proteins of plant centromeres. Curr. Opin. Plant Biol. 6, 554–560. [DOI] [PubMed] [Google Scholar]
- Ikeno, M., Grimes, B., Okazaki, T., Nakano, M., Saitoh, K., Hoshino, H., McGill, N.I., Cooke, H., and Masumoto, H. (1998). Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 16, 431–439. [DOI] [PubMed] [Google Scholar]
- Jackson, S.A., Wang, M.L., Goodman, H.M., and Jiang, J. (1998). Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41, 566–572. [PubMed] [Google Scholar]
- Jiang, J., Birchler, J.B., Parrott, W.A., and Dawe, R.K. (2003). A molecular view of plant centromeres. Trends Plant Sci. 8, 570–575. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Gill, B.S., Wang, G.L., Ronald, P.C., and Ward, D.C. (1995). Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 92, 4487–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W.W., Melo, J.R., Nagaki, K., Talbert, P.B., Henikoff, S., Dawe, R.K., and Jiang, J. (2004). Maize centromeres: Organization and functional adaptation in the genetic background of oat. Plant Cell 16, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaszas, E., and Birchler, J.A. (1996). Misdivision analysis of centromere structure in maize. EMBO J. 15, 5246–5255. [PMC free article] [PubMed] [Google Scholar]
- Kaszas, E., and Birchler, J.A. (1998). Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics 150, 1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J.C., Kato, A., and Birchler, J.A. (2005). Sequences associated A chromosome centromeres are present throughout the maize B chromosome. Chromosoma 113, 337–349. [DOI] [PubMed] [Google Scholar]
- Lo, A.W., Craig, J.M., Saffery, R., Kalitsis, P., Irvine, D.V., Earle, E., Magliano, D.J., and Choo, K.H. (2001. a). A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 20, 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, A.W.I., Magliano, D.J., Sibson, M.C., Kalitsis, P., Craig, J.M., and Choo, K.H.A. (2001. b). A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 11, 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert, K.A., and Karpen, G.H. (2001). The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics 158, 1615–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, T.D., and Karpen, G.H. (1995). Localization of centromere function in a Drosophila minichromosome. Cell 82, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z.K., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36, 138–145. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Song, J., Stupar, S.M., Parokonny, A.S., Yuan, Q., Ouyang, S., Liu, J., Hsiao, J., Jones, K.M., Dawe, R.K., Buell, C.R., and Jiang, J. (2003. b). Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics 163, 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Talbert, P.B., Zhong, C.X., Dawe, R.K., Henikoff, S., and Jiang, J.M. (2003. a). Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163, 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakey, R., and Tyler-Smith, C. (1990). Y chromosome DNA haplotyping suggests that most European and Asian men are descended from one of two males. Genomics 7, 325–330. [DOI] [PubMed] [Google Scholar]
- Ohzeki, J., Nakano, M., Okada, T., and Masumoto, H. (2002). CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 159, 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero, J.S., Ahmad, K., and Henikoff, S. (1999). A distal heterochromatic block displays centromeric activity when detached from a natural centromere. Mol. Cell 4, 995–1004. [DOI] [PubMed] [Google Scholar]
- Roman, H. (1947). Mitotic non-disjunction in the case of interchanges involving the B-type chromosome in maize. Genetics 32, 391–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffery, R., Irvine, D.V., Griffiths, B., Kalitsis, P., Wordeman, L., and Choo, K.H.A. (2000). Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum. Mol. Genet. 9, 175–185. [DOI] [PubMed] [Google Scholar]
- Saffery, R., Sumer, H., Hassan, S., Wong, L.H., Craig, J.M., Todokoro, K., Anderson, M., Stafford, A., and Choo, K.H.A. (2003). Transcription within a functional human centromere. Mol. Cell 12, 509–516. [DOI] [PubMed] [Google Scholar]
- Schueler, M.G., Higgins, A.W., Rudd, M.K., Gustashaw, K., and Willard, H.F. (2001). Genomic and genetic definition of a functional human centromere. Science 294, 109–115. [DOI] [PubMed] [Google Scholar]
- Sears, E.R. (1952). Misdivision of univalents in common wheat. Chromosoma 4, 535–550. [DOI] [PubMed] [Google Scholar]
- Shibata, F., and Murata, M. (2004). Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana. J. Cell Sci. 117, 2963–2970. [DOI] [PubMed] [Google Scholar]
- Spence, J.M., Critcher, R., Ebersole, T.A., Valdivia, M.M., Earnshaw, W.C., Fukagawa, T., and Farr, C.J. (2002). Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha satellite array. EMBO J. 21, 5269–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B.A., Blower, M.D., and Karpen, G.H. (2001). Determining centromere identity: Cyclical stories and forking paths. Nat. Rev. Genet. 2, 584–596. [DOI] [PubMed] [Google Scholar]
- Sun, X.P., Wahlstrom, J., and Karpen, G. (1997). Molecular structure of a functional Drosophila centromere. Cell 91, 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P.B., Masuelli, R., Tyagi, A.P., Comai, L., and Henikoff, S. (2002). Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14, 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp, C.N., Zhong, C.X., and Dawe, R.K. (2004). Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA 101, 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith, C., Gimelli, G., Giglio, S., Floridia, C., Pandya, A., Terzoli, G., Warburton, P.E., Earnshaw, W.C., and Zuffardi, O. (1999). Transmission of a fully functional human neocentromere through three generations. Am. J. Hum. Genet. 64, 1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevrick, R., and Willard, H.F. (1989). Long-range organization of tandem arrays of a satellite DNA at the centromeres of human chromosomes: High frequency array-length polymorphism and meiotic stability. Proc. Natl. Acad. Sci. USA 86, 9394–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard, H.F. (1998). Centromeres: The missing link in the development of human artificial chromosomes. Curr. Opin. Genet. Dev. 8, 219–225. [DOI] [PubMed] [Google Scholar]
- Wu, J.Z., et al. (2004). Composition and structure of the centromeric region of rice chromosome 8. Plant Cell 16, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J.W., Pendon, C., Yang, J., Haywood, N., Chand, A., and Brown, W.R. (2000). Human mini-chromosomes with minimal centromeres. Hum. Mol. Genet. 9, 1891–1902. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Huang, Y.C., Zhang, L., Li, Y., Lu, T.T., Lu, Y.Q., Feng, Q., Zhao, Q., Cheng, Z.K., Xue, Y.B., Wing, R.A., and Han, B. (2004). Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 32, 2023–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C.X., Marshall, J.B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J.A., Jiang, J.M., and Dawe, R.K. (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14, 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]