Abstract
Interactions between proteins are essential for their functioning and the biological processes they control. The elucidation of interaction maps based on yeast studies is a first step toward the understanding of molecular networks and provides a framework of proteins that possess the capacity and specificity to interact. Here, we present a comprehensive plant protein–protein interactome map of nearly all members of the Arabidopsis thaliana MADS box transcription factor family. A matrix-based yeast two-hybrid screen of >100 members of this family revealed a collection of specific heterodimers and a few homodimers. Clustering of proteins with similar interaction patterns pinpoints proteins involved in the same developmental program and provides valuable information about the participation of uncharacterized proteins in these programs. Furthermore, a model is proposed that integrates the floral induction and floral organ formation networks based on the interactions between the proteins involved. Heterodimers between flower induction and floral organ identity proteins were observed, which point to (auto)regulatory mechanisms that prevent the activity of flower induction proteins in the flower.
INTRODUCTION
Biological processes are executed by proteins that, to a large extent, depend on interactions with other proteins for their activity. These interactions are specific, even among members of a particular protein family that contain similar interaction domains, and are often maintained during evolution. Studying these specific interactions reveals networks of molecules that may lead to potential functional linkages and molecular explanations of biological processes in an organism. These networks are complex, highly dynamic in place and time, and far from understood. The elucidation of interaction maps based on in vitro or yeast studies is a first step toward the understanding of molecular networks and provides a framework of proteins that possess the capacity and specificity to interact.
Many recent reports have presented large-scale interaction network maps from Saccharomyces cerevisiae (Uetz et al., 2000; Ito et al., 2001), Caenorhabditis elegans (Walhout et al., 2000), Drosophila melanogaster (Giot et al., 2003), Mus musculus (Suzuki et al., 2001), and humans (Lehner and Fraser, 2004) using yeast two-hybrid assays or affinity purification followed by mass spectrometry (Link et al., 1999; Gavin et al., 2002; Ho et al., 2002). Surprisingly, comparable data sets from yeast, for example, revealed hardly any overlap in interactions, suggesting that each approach provides a subset of the interactome (von Mering et al., 2002; Bader et al., 2004). Furthermore, these reports demonstrated that two-hybrid data are reliable when several validation criteria are used. Information about interactions of orthologous proteins in other species is informative and may help in validating the interaction data. The conservation of these so-called interologs has been revealed between yeast and bacteria (Kelley et al., 2003) but also between different plant species (Favaro et al., 2002). Previously, we have demonstrated that many interactions between MADS domain proteins are conserved among Arabidopsis thaliana, rice (Oryza sativa), petunia (Petunia hybrida), and Antirrhinum majus (Immink and Angenent, 2002). Another criterion for the validation of the interaction data is the colocalization of the interacting proteins in a particular cell. Several studies reported the coevolution of expression of interacting proteins and their ability to physically interact (Ge et al., 2001; Immink et al., 2002; Fraser et al., 2004). This provides a tool to validate interaction data but can also be useful to predict novel protein–protein interactions. Furthermore, other functional genomic or genetic data, such as mutants, may provide additional evidence for the in vivo existence of a particular interaction.
By zooming in on a particular group of proteins that is known from previous studies to be enriched for interactions, insight into individual pathways can be obtained. Transcription factors are an interesting class of proteins in this respect. Dimerization of transcription factor proteins increases the selectivity of protein–DNA interactions and creates a large number of diverse DNA binding complexes from a relatively small number of proteins. The gene family encoding MADS domain transcription factors in plants encompasses a relatively large family with 107 members in the Arabidopsis genome (Pařenicová et al., 2003). They are further subdivided into two groups: the class II MADS box proteins, comprising the MIKC and Mδ types, and the class I proteins that are further subdivided into the Mα, Mβ, and Mγ types (Alvarez-Buylla et al., 2000a; Pařenicová et al., 2003). A wealth of genetic and functional information is available from the MIKC group, whereas the type I subfamily with ∼60 members represents a virtually unknown group of transcription factors. Many MIKC proteins are active in a combinatorial manner to specify the identity of organs (Coen and Meyerowitz, 1991). Recent genetic and yeast two- and three-hybrid studies revealed that these MADS box proteins are able to form multimeric complexes (Honma and Goto, 2001) and as hypothesized in the quartet model as tetrameric complexes (Theissen and Saedler, 2001). These higher-order complexes are supposed to be composed of two dimers that interact at the C termini (Egea-Cortines et al., 1999). Nevertheless, information about MADS protein interactions is limited for Arabidopsis and lacks any data on the type I proteins. Besides Arabidopsis, MADS dimerization patterns have been reported for several species, including petunia, rice, Chrysanthemum Dendrathema grandiflorum, and Antirrhinum (Davies et al., 1996; Egea-Cortines et al., 1999; Favaro et al., 2002; Immink et al., 2003; Shchennikova et al., 2004), which provided data for comparative studies and revealed interactions between orthologous proteins.
Here, we report a comprehensive plant interactome map of nearly all members of the Arabidopsis MADS box family. It reveals interactions between type I, type II, and between the two types of proteins. Combined with phylogenetic analysis, it sheds light on evolutionary aspects of this protein family. Clustering of proteins based on their interaction pattern pinpoints proteins involved in the same developmental program and provides evidence for the participation of uncharacterized proteins in these programs. Finally, we propose a model that integrates the network of floral induction proteins with the network of floral organ identity proteins, and we predict feedback loops between the two subnetworks.
RESULTS
Comprehensive Analysis of MADS Box Transcription Factor Dimerization
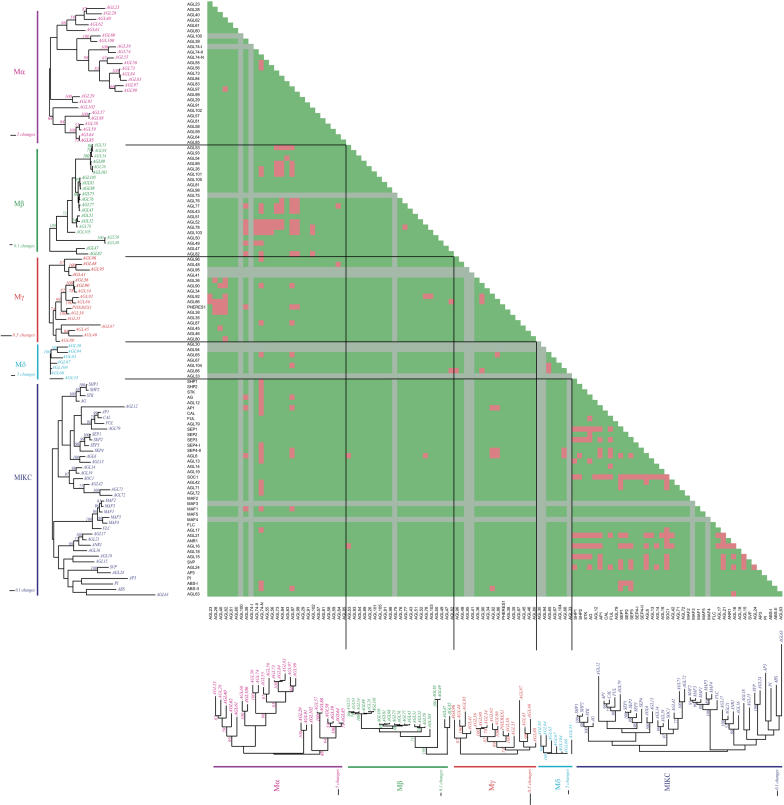
Several studies with various plant species have revealed that MADS domain transcription factors form specific homodimers and heterodimers. In general, individual screens of cDNA expression libraries with the yeast two-hybrid GAL4 system have been used for this purpose. These assays are laborious, they result in the identification of a relatively high number of false positives, and they are often limited because of autoactivation of yeast reporters by the presence of an intrinsic activation domain in the bait protein. Therefore, in this study, a matrix-based yeast two-hybrid approach has been followed to identify specific dimerization among the members of the Arabidopsis MADS domain transcription factor family. The complete data set with all the scores is presented in Supplemental Table 1 online, and the interactions are summarized in a matrix in Figure 1 and in Supplemental Table 2 online.
Figure 1.
The Arabidopsis MADS Box Transcription Factor Interaction Matrix.
The MADS box transcription factors are arranged according to their phylogenetic relationship as has been reported by Pařenicová et al. (2003). The phylogenetic trees are indicated on the x and y axis with the different groups indicated (Mα, Mβ, Mγ, Mδ, and MIKC). Protein–protein interactions are represented by red blocks, no interactions by green blocks, and interactions that could not be tested by gray blocks.
Remarkably, the MIKC proteins that contain the K-box, a domain specific for type II plant MADS box proteins that is presumed to fold into an amphipathic α-helical structure (Riechmann and Meyerowitz, 1997; Alvarez-Buylla et al., 2000a), interact preferably with other type II proteins and hardly form dimers with the type I MADS box proteins. However, there are some exceptions. In particular, there is a preference for interactions with type I proteins from the Mα subclade. Among the type I proteins, most heterodimers are found between members of different subclades. Interactions among Mα proteins are rare, but they dimerize preferentially with many proteins of the Mβ and Mγ clades. Similarly, only a few interactions among members of the Mβ and Mγ clades were observed, and Mβ-Mγ heterodimers are rare. This suggests that the participation of a Mα protein is a prerequisite for a stable dimer consisting of only type I proteins. Although many interactions were observed, a relatively large number of MADS domain proteins appeared to have no interactions at all. Possibly these proteins interact only with non-MADS box proteins, or alternatively, particular interactions are not formed in a yeast two-hybrid assay. For example, the interaction between the B-type proteins APETALA3 (AP3) and PISTILLATA (PI) was not found in this screen. Previously, these proteins appeared to interact exclusively in a higher-order complex, with either SEPALLATA3 (SEP3) or AP1 (Honma and Goto, 2001), suggesting that the additional factors stabilize the AP3-PI dimer. This requirement for stabilizing factors to maintain specific dimers could be more general. Homodimerization is another form of MADS domain transcription factor interaction that is difficult to detect by yeast two-hybrid analysis (Immink et al., 2002), and hence, many homodimers have probably been missed in this screening.
Subsequently, the proteins were clustered based on the obtained interaction patterns, which allows the identification of proteins with similar interactions and groups of proteins that are highly connected (Figure 2). This analysis gives clues about the involvement of proteins in certain developmental programs. It reveals groups of proteins with common known functions, but more informatively, also shows clusters containing uncharacterized proteins, for which a function can now be predicted, based on their presence in a particular interaction cluster.
Figure 2.
Interactome Map of the Arabidopsis MADS Box Transcription Factor Family.
Proteins are organized based on hierarchical clustering of their protein–protein interaction patterns. Proteins that do not interact in the screen are omitted from this figure. Protein–protein interactions are indicated with red blocks and no interactions with green blocks. Presence of clustered proteins with a putative similar function is indicated with a colored bar on the left and bottom side of the figure: red for embryo, green for root, blue for flowering, and yellow for floral organs.
Data Validation of Yeast Two-Hybrid Experiments
To obtain more insight into the reliability of the data, a comparison was made between our interaction data and Arabidopsis MADS domain protein interactions described in the literature. In contrast with the wealth of genetic data, virtually nothing is known about molecular interactions among members of the Arabidopsis MADS box transcription factor family. In Supplemental Table 3 online, an overview of the published interactions is given. Of 16 previously reported interactions, nine were also found in our study. The majority of the remaining seven interactions were only identified between truncated forms of the proteins, which provides a possible explanation why we did not detect them in our study with full-length proteins.
We also used information on interactions between orthologous MADS domain proteins from other species. MADS factors are key regulators of plant development, and many of their important roles as developmental selector genes are conserved among various plant species, although it has also been suggested that diversification of MADS activity after gene duplication may contribute to floral diversity (reviewed in Ferrario et al., 2004a). In line with the evolutionary conservation of MADS box transcription factor functions, the interaction patterns for specific MADS box proteins with identical functions, but from different species, have proven to be conserved (Favaro et al., 2002; Immink and Angenent, 2002). To validate the data presented here, the literature was screened for putative interologs of the various Arabidopsis MADS box protein combinations. This analysis could be performed for the type II proteins only because no interaction with type I proteins have yet been reported for any plant species. Figure 3 shows the subset of Arabidopsis MADS box protein–protein interactions for which at least one homologous interaction has been found.
Figure 3.
Subset of Arabidopsis MADS Box Transcription Factor Interactions Confirmed by Interologs.
Proteins are indicated by ovals and protein interactions with lines. The color of the line corresponds with the species for which orthologous interactions have been reported (Arabidopsis [At], purple; petunia [Ph], orange; A. majus [Am], black; rice [Os], green; Gerbera hybrida [Gh], red; tobacco [Nt], blue; tomato [Le], gray; maize [Zm], yellow; lily [Ll], cyan; chrysanthemum [Cd], pink). The AP3-PI heterodimer is included in the figure because many interologs have been reported, although the full-length Arabidopsis proteins do not interact detectably in yeast. The references used to create this subset of conserved interactions are presented online (see Supplemental Table 4 online).
An interaction between proteins observed in a yeast two-hybrid assay can only be biologically relevant when they are present in the same cell and at the same moment. Hence, coexpression of the corresponding genes can be used for the validation of protein interaction data, even though the correlation of RNA and protein levels varies for different genes (Gygi et al., 1999; Beyer et al., 2004). We used the developmental data set of the AtGenExpress project (Schmid et al., 2005) (see Methods) to investigate whether there is a correlation between gene expression and protein interaction. In general, genes with similar functions, such as the ABC homeotic genes and the SEP genes (Pelaz et al., 2000) or the redundantly acting SHATTERPROOF1 (SHP1), SHP2, and SEEDSTICK (Pinyopich et al., 2003), genes clustered together (see Supplemental Figure 1 online). We asked more specifically how often genes are coexpressed in at least one sample using an absolute criterion for expression. This comparison revealed that almost 100% of the interacting proteins have an overlap in expression pattern of the corresponding genes, which is a prerequisite for a possible in planta interaction and relevance in Arabidopsis tissues (Figure 4).
Figure 4.
Comparison of Expression Patterns of Genes That Encode Interacting Proteins.
The data from the AtGenExpress expression atlas are represented such that expression of each gene is normalized across the entire data set. The most important groups of tissues are indicated at the top and in detail numbered in the bottom (list of all tissues is presented in Supplemental Table 5 online). Blue indicates underexpression and red overexpression relative to the mean, with yellow expression levels that are close to the average for the corresponding gene.
We next asked whether the expression patterns of interacting pairs were on average more similar than those of noninteracting pairs. Although the average Pearson correlation of expression levels of interacting genes was only slightly higher than of noninteracting genes, the distribution of noninteracting and interacting genes was significantly different. Specifically, the interacting pairs included a larger group of genes with more similar expression patterns (see Supplemental Figure 2 online), although there was also an excess of genes with contrasting expression patterns. A prominent case in this latter group was SHORT VEGETATIVE PHASE (SVP), a floral repressor (Hartmann et al., 2000) whose expression pattern is negatively correlated with those of SEP1, SEP3, and AP1, all of which play positive roles in flowering (Ferrario et al., 2004a).
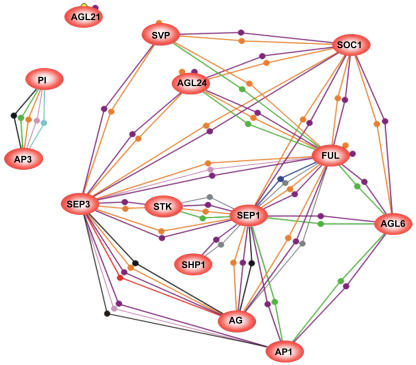
The Flower Induction and Flower Organ Formation Subnetworks
The regulation of flowering time is a complex process in which many environmental and internal signals are integrated, finally giving rise to a switch from vegetative to generative development at the appropriate time. MADS box transcription factors have shown to play pivotal roles in the flowering program and occupy many important positions in the hierarchical network (summarized and reviewed in Blazquez, 2000; Simpson and Dean, 2002). Based on the interaction data obtained in this study, we tried to unravel two subnetworks composed of interactions between known MADS box proteins involved in flower induction and flower organ formation (Figure 5). The proteins AP1 and FRUITFULL (FUL) are present in both subnetworks, which would refer to their early and late function in flowering (Mandel et al., 1992; Ferrandiz et al., 2000). However, the most striking observation is that many of the floral organ identity proteins, such as AGAMOUS (AG), SEP1/2/3, and SHP1/2 proteins, interact not only with positive regulators of flowering, such as SUPPRESSOR OF CONSTANS 1 (SOC1) and AGAMOUS-LIKE 24 (AGL24), but also with a negative regulator, SVP, implying that there is both positive and negative crosstalk between the two pathways via protein interactions, as pointed out above.
Figure 5.
Representation of the Flower Induction and Flower Formation Networks.
Proteins are indicated by ovals (red for the flower induction, blue for the flower formation network, green for the hubs), and interactions are represented by lines. The proteins SOC1 and AGL24 form a homodimer, which is indicated with a small dot next to the oval of the protein.
DISCUSSION
Interactions between proteins are essential for their activity and serve as the building blocks for the molecular networks that control biological processes in organisms. Here, we report a large protein–protein interaction study performed in plants, resulting in a near-complete interactome of Arabidopsis MADS domain transcription factors. Although derived from a heterologous system, these interaction patterns give valuable clues about the involvement of the MADS factors in certain processes. Some of the unexpected interactions, such as those between regulators of flowering time and floral pattern, may indicate the existence of hitherto unsuspected regulatory mechanisms. Duplication of MADS box genes appears to be a common phenomenon, not only giving rise to functionally redundant genes, but also allowing diversification of developmental processes through changes in expression pattern or protein functions (reviewed in Smyth, 2000; Ferrario et al., 2004a). Completely redundant proteins are expected to have identical interaction patterns, and proteins playing a role in the same process are likely to have shared interaction partners. As expected, redundant proteins such as SEP1 and SEP3 (Pelaz et al., 2000) cluster together in the interaction matrix, as do members of the AG clade, which have partially overlapping functions (Favaro et al., 2003; Pinyopich et al., 2003). Similarly, proteins that may play a role in root development are grouped (AGL19, AGL42, AGL12, ANR1, and AGL17) (Rounsley et al., 1995; Zhang and Forde, 1998; Alvarez-Buylla et al., 2000b; Burgeff et al., 2002), as are proteins known to be involved in the timing of flowering (e.g., SVP, AGL24, and FUL) (Ferrandiz et al., 2000; Hartmann et al., 2000; Yu et al., 2002; Michaels et al., 2003). The AGL6 protein has an interaction pattern closely resembling the AP1 interactions, suggesting that this protein plays a role in the flowering program as well. This hypothesis is strengthened by the fact that overexpression of OMADS1 from orchid (Oncidium Gower Ramsey), a gene closest in sequence to AGL6, resulted in extremely early flowering in Arabidopsis and loss of inflorescence indeterminacy (Hsu et al., 2003). Another protein for which the function recently has been elucidated by mutant analysis is AGL3 and based on its determined function has been renamed SEP4 (Ditta et al., 2004). Besides its function in floral organ formation, this protein appears to play a role in determining the floral meristem identity, redundantly with AP1 and CAULIFLOWER (CAL). Remarkably, SEP4 and CAL cluster together based on their interaction patterns, which also points to their redundant function.
Interaction patterns may also provide clues about the role of the interacting proteins in a certain pathway, even when the majority of the proteins in the interaction cluster are unknown. An example is provided by the type I proteins, for which virtually no functional information is available. An exception is PHERES1 (PHE1; Köhler et al., 2003), a target of the polycomb protein MEDEA that is involved in seed development (Grossniklaus et al., 1998). PHE1 interacts with AGL28, AGL40, and AGL62, which are all coexpressed with PHE1 in the embryo and cluster together according to their interacting patterns. This clearly points to their involvement in the same developmental process.
Protein interactions that are clustered based on similar interaction patterns can serve as backbones for more complex molecular networks responsible for a particular function or pathway. We have focused on two subnetworks, one for the flower induction and one for the flower organ formation pathway, which appear to be highly interconnected. Highly connected proteins can function as hubs to interconnect pathways that are either spatially or temporally separated. The proteins AP1 and FUL could serve as hubs between the flower induction pathway comprising interacting proteins such as SVP, SOC1, and AGL24, and the floral organ identity proteins. Both AP1 and FUL have a dual function in floral meristem identity (early function) and floral organ determination (late function) (Mandel et al., 1992; Ferrandiz et al., 2000), which is in line with the fact that dimers are formed with both the flowering proteins and the floral homeotic proteins.
Surprisingly, SVP, SOC1, and AGL24 also interact directly with the floral organ identity proteins. Both groups of genes share similar expression at the shoot apex, although the overlap on the cellular level is relatively limited. One possibility is that there is mutual negative feedback regulation, which would sharpen contrasting expression patterns (Heck et al., 1997; McKay and Cidlowski, 1998). In such a scenario, the corresponding dimers would repress expression of both SVP and AP1/SEP1/SEP3, thus ensuring that overlap in expression pattern is minimized. An even more intriguing possibility is that there is overlap in expression pattern precisely at the moment when the shoot apical meristem is transformed into a generative meristem. Then, these dimers could serve not only as repressors of the early flowering genes, but also as activators of the floral organ identity genes, further sharpening the transition to flowering.
Positive autoregulatory feedback loops have been reported for the class B homeotic genes in Arabidopsis (Goto and Meyerowitz, 1994; Samach et al., 1997) and Antirrhinum (Schwarz-Sommer et al., 1992; Tröbner et al., 1992) and more recently for AG in Arabidopsis (Gómez-Mena et al., 2005). A prerequisite for the negative autoregulatory feedback loop theory presented here is that the potentially repressed genes contain the motif for MADS box protein binding, the so-called CArG-box (Shore and Sharrocks, 1995). All three genes, SVP, SOC1, and AGL24, contain a perfect CArG-box [CC(A/T)6GG] in their putative regulatory sequences, which is, for example, lacking in the CAL gene for which the gene product did not reveal interactions with the floral identity proteins.
Further analyses are required to provide evidence for these negative feedback loops, which could facilitate the major switches in meristem identity. However, first indications for this theory are already available from genetic data. The SVP and AGL24 proteins, which are very close in sequence and have similar interaction patterns, have an opposite effect on flowering time (Hartmann et al., 2000; Yu et al., 2002; Michaels et al., 2003). This suggests that SVP and AGL24 are acting at the molecular level as floral repressor and inducer, respectively, by dimerization with the same partners. In recent studies, constitutive expression of either SVP or AGL24 resulted, as expected, in late and early flowering, respectively (Masiero et al., 2004; Yu et al., 2004). However, in contradiction with the opposite flowering time phenotypes, these transgenic Arabidopsis plants revealed similar alterations in the flower. The flowers have features of ap1 mutant flowers and often contain greenish sepaloid petals and showed indeterminacy. These observations are in accordance with the proposed model that the flowering time proteins are normally switched off in the flower by negative feedback mechanisms, which are controlled by heterodimers containing both flowering time and floral organ identity proteins. In case of ectopic expression using the strong constitutive 35S promoter of Cauliflower mosaic virus, the negative feedback loops are overruled, giving rise to floral mutations. The altered floral phenotypes from 35S:SVP and 35S:AGL24 plants can be explained by our observed protein interactions. Both SVP and AGL24 form interactions with the floral organ identity proteins, such as AP1, AG, and SEP3. In the overexpressers, these protein complexes may act in a dominant-negative manner on the floral organ identity proteins. Similar floral defects were obtained upon overexpression of SOC1, which functions as an accelerator of flowering (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000). Detailed analyses with the petunia UNSHAVEN protein, the putative functional homolog of SOC1, has also shown that in this case the floral phenotype is obtained by a dominant-negative effect on the floral organ identity proteins (Ferrario et al., 2004b). In summary, all results from mutant and overexpression analyses and the interaction data presented here for SVP, AGL24, and SOC1 give strong indications for the proposed negative feedback loop model.
The results presented here provide a glimpse of the complex interaction network for the Arabidopsis MADS domain transcription factor family. The current available protein interaction map still represents a largely static view of the cellular processes regulated by the interactome. Technologies such as fluorescence resonance energy transfer (Immink et al., 2002) and bimolecular fluorescence complementation (Bracha-Drori et al., 2004; Walter et al., 2004) are powerful tools for in vivo studies aimed at analyzing the dynamics of changing protein interactions. Unraveling the dynamic spatial and temporal changes in binary and macromolecular assemblies and the de novo complex assembly in response to varying external stimuli will provide a detailed understanding of biological systems.
METHODS
Cloning of the Full-Length MADS Box Transcription Factors
A detailed description of the amplification of the open reading frames and subsequent cloning in yeast two-hybrid vectors is given in Supplemental Text 1 online. In summary, 102 open reading frames were cloned: 99 in the bait vector and 102 in the prey vector.
Yeast Two-Hybrid Analysis
The bait vectors were transformed into yeast strain PJ69-4a (MATa; James et al., 1996) and all prey vectors into strain PJ69-4α (MATα; James et al., 1996) and selected on SD plates lacking Leu and Trp, respectively. Subsequently, overnight cultures were grown (30°C, 300 rpm) from single colonies of each transformant in selective SD medium and systematically mated with each other by spotting 5-μL droplets of the liquid cultures on top of each other on SD complete plates (Nunc Omnitray; VWR International, Amsterdam, The Netherlands) containing all the essential amino acids. The spotting was performed in a systematic manner in a grid of 96 spots/plate by a pipetting robot (Genesis RSP150 workstation; Tecan, Maennedorf, Switzerland). In addition, some negative control combinations were spotted, for which water was used instead of either a bait or prey culture. Subsequently, the plates were incubated at 30°C for 16 h, and afterwards the yeast was transferred to SD plates lacking both Leu and Trp with disposable 96-pin replicators (Nunc-TSP; VWR International) to select for diploid yeast containing both plasmids. After 2 d of growth at 30°C, the yeast was transferred to two different selection plates containing SD medium lacking Leu, Trp, and Ade and SD lacking Leu, Trp, and His, supplemented with 5 mM 3-amino-1,2,4-triazole. These plates were incubated at 20°C and scored for growth of yeast and hence protein–protein interaction events after 5 d.
The screening was performed in duplicate, yielding in theory eight data points for each combination, four times with protein A as bait and B as prey (two scores from the Ade selection and two scores from the His selection) and four times reciprocally, with protein B as bait and A as prey. In case of autoactivation for one of the two proteins, just four data points were obtained for the specific combination. The mating efficiency appeared to be 100%, and where water was used for mating, either instead of a bait culture or instead of a prey culture, no growth was obtained on medium selecting for the presence of the two plasmids or on the media selecting for interactions (see Supplemental Figure 3 online). This shows that no cross-contamination occurred as a result of the procedure that followed. A combination was scored as a true interaction when it resulted in growth for at least one of the two selection markers in both screenings, but almost all positively scored combinations grew on both selection media.
Data Analysis
All protein–protein interaction data were transferred to Microsoft Excel sheets (Redmond, WA), and for easier data analyses, the interaction data was made reciprocal. One data matrix was made with all MADS box proteins, Matrix1, and one matrix with MADS box proteins that had at least one protein–protein interaction, Matrix2. Both matrixes were subjected to GeneMaths software (Applied Maths BVBA, Sint-Martens-Latem, Belgium) for further data analyses. Matrix1 was organized based on the phylogenetic distribution of all Arabidopsis thaliana MADS box proteins according to Pařenicová et al. (2003). Cluster analysis was performed on Matrix2 with Pearson correlation coefficient and UPGMA algorithm on both the rows and columns. In both cases, the data are represented in one direction (not reciprocal).
Coexpression Analysis
The developmental set of the AtGenExpress expression atlas (ftp://ftp.arabidopsis.org/home/tair/Microarrays/Datasets/AtGenExpress/; http://weigelworld.org/resources/microarray/AtGenExpress) (Schmid et al., 2005) was analyzed for expression of MADS box genes. Expression estimates were obtained using gcRMA (http://bioconductor.org), a modification of the robust multiarray analysis algorithm (Irizarry et al., 2003). A threshold of log2 ≥ 3 was applied to identify overlap in tissues with expression of genes. Approximately 75% of the Arabidopsis MADS family is represented on the Affymetrix GeneChip ATH1 (Santa Clara, CA).
Supplementary Material
Acknowledgments
We thank H. Jonker for technical assistance with the pipetting robot and J. Busscher for contributing to cloning procedures. The project is in part sponsored by the Netherlands Proteomics Centre, the EU-Regulatory Gene Initiative in Arabidopsis project (QLG2-1999-00876), the Centre for BioSystems Genomics, Fondo per gli Investimenti della Ricerca di Base 2002, and Ministero dell'Universita e della Ricerca Scientifica e Technologica 2003. We acknowledge the use of microarray data from the AtGenExpress project, coordinated by L. Nover, T. Altmann, and D.W. and supported by funds from the Deutsche Forschungsgemeinschaft and the Max Planck Society. The developmental portion of AtGenExpress was produced by M. Schmid and J. Lohmann (Max Planck Institute, Tübingen, Germany).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gerco C. Angenent (gerco.angenent@wur.nl).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031831.
References
- Alvarez-Buylla, E.R., Liljegren, S.J., Pelaz, S., Gold, S.E., Burgeff, C., Ditta, G.S., Vergara-Silva, F., and Yanofsky, M.F. (2000. b). MADS-box gene evolution beyond flowers: Expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 24, 457–466. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla, E.R., Pelaz, S., Liljegren, S.J., Gold, S.E., Burgeff, C., Ditta, G.S., de Pouplana, L.R., Martinez-Castilla, L., and Yanofsky, M.F. (2000. a). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97, 5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader, J.S., Chaudhuri, A., Rothberg, J.M., and Chant, J. (2004). Gaining confidence in high-throughput protein interaction networks. Nat. Biotechnol. 22, 78–85. [DOI] [PubMed] [Google Scholar]
- Beyer, A., Hollunder, J., Nasheuer, H.-P., and Wilhelm, T. (2004). Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol. Cell. Proteomics 3, 1083–1092. [DOI] [PubMed] [Google Scholar]
- Blazquez, M.A. (2000). Flower development pathways. J. Cell Sci. 113, 3547–3548. [DOI] [PubMed] [Google Scholar]
- Borner, R., Kampmann, G., Chandler, J., Gleissner, R., Wisman, E., Apel, K., and Melzer, S. (2000). A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 24, 591–599. [DOI] [PubMed] [Google Scholar]
- Bracha-Drori, K., Shichrur, K., Katz, A., Oliva, M., Angelovici, R., Yalovsky, S., and Ohad, N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40, 419–427. [DOI] [PubMed] [Google Scholar]
- Burgeff, C., Liljegren, S.J., Tapia-López, R., Yanofsky, M.F., and Alvarez-Buylla, E.R. (2002). MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 214, 365–372. [DOI] [PubMed] [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Davies, B., Egea-Cortines, M., de Andrade Silva, E., Saedler, H., and Sommer, H. (1996). Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 15, 4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., and Yanofsky, M.F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18, 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro, R., Immink, R.G.H., Ferioli, V., Bernasconi, B., Byzova, M., Angenent, G.C., Kater, M., and Colombo, L. (2002). Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol. Genet. Genomics 268, 152–159. [DOI] [PubMed] [Google Scholar]
- Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., Yanofsky, M.F., Kater, M.M., and Colombo, L. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15, 2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz, C., Gu, Q., Martienssen, R., and Yanofsky, M.F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127, 725–734. [DOI] [PubMed] [Google Scholar]
- Ferrario, S., Busscher, J., Franken, J., Gerats, T., Vandenbussche, M., Angenent, G.C., and Immink, R.G.H. (2004. b). Ectopic expression of the petunia MADS box gene UNSHAVEN accelerates flowering and confers leaf-like characteristics to floral organs in a dominant-negative manner. Plant Cell 16, 1490–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario, S., Immink, R.G., and Angenent, G.C. (2004. a). Conservation and diversity in flower land. Curr. Opin. Plant Biol. 7, 84–91. [DOI] [PubMed] [Google Scholar]
- Fraser, H.B., Hirsh, A.E., Wall, D.P., and Eisen, M.B. (2004). Coevolution of gene expression among interacting proteins. Proc. Natl. Acad. Sci. USA 101, 9033–9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin, A.-C., et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Ge, H., Liu, Z., Church, G.M., and Vidal, M. (2001). Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 29, 482–486. [DOI] [PubMed] [Google Scholar]
- Giot, L., et al. (2003). A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736. [DOI] [PubMed] [Google Scholar]
- Gómez-Mena, C., de Folter, S., Costa, M.M.R., Angenent, G.C., and Sablowski, R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132, 429–438. [DOI] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Gygi, S.P., Rochon, Y., Franza, B.R., and Aebersold, R. (1999). Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, U., Hohmann, S., Nettesheim, K., Wisman, E., Saedler, H., and Huijser, P. (2000). Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 21, 351–360. [DOI] [PubMed] [Google Scholar]
- Heck, S., Bender, K., Kullmann, M., Gottlicher, M., Herrlich, P., and Cato, A.C.B. (1997). I kappaB alpha-independent downregulation of NF-kappaB activity by glucocorticoid receptor. EMBO J. 16, 4698–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. [DOI] [PubMed] [Google Scholar]
- Hsu, H.-F., Huang, C.-H., Chou, L.-T., and Yang, C.-H. (2003). Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol. 44, 783–794. [DOI] [PubMed] [Google Scholar]
- Immink, R.G.H., and Angenent, G.C. (2002). Transcription factors do it together: The hows and whys of studying protein-protein interactions. Trends Plant Sci. 7, 531–534. [DOI] [PubMed] [Google Scholar]
- Immink, R.G.H., Ferrario, S., Busscher Lange, J., Kooiker, M., Busscher, M., and Angenent, G.C. (2003). Analysis of the petunia MADS-box transcription factor family. Mol. Genet. Genomics 268, 598–606. [DOI] [PubMed] [Google Scholar]
- Immink, R.G.H., Gadella, T.W.J., Jr., Ferrario, S., Busscher, M., and Angenent, G.C. (2002). Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. USA 99, 2416–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R.A., Bolstad, B.M., Collin, F., Cope, L.M., Hobbs, B., and Speed, T.P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, B.P., Sharan, R., Karp, R.M., Sittler, T., Root, D.E., Stockwell, B.R., and Ideker, T. (2003). Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc. Natl. Acad. Sci. USA 100, 11394–11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., and Grossniklaus, U. (2003). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17, 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., Suh, S.-S., Park, E., Cho, E., Ahn, J.H., Kim, S.-G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner, B., and Fraser, A. (2004). A first-draft human protein-interaction map. Genome Biol. 5, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, A.J., Eng, J., Schieltz, D.M., Carmack, E., Mize, G.J., Morris, D.R., Garvik, B.M., and Yates, J.R. (1999). Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Masiero, S., Li, M.-A., Will, I., Hartmann, U., Saedler, H., Huijser, P., Schwarz-Sommer, Z., and Sommer, H. (2004). INCOMPOSITA: A MADS-box gene controlling prophyll development and floral meristem identity in Antirrhinum. Development 131, 5981–5990. [DOI] [PubMed] [Google Scholar]
- McKay, L.I., and Cidlowski, J.A. (1998). Cross-talk between nuclear factor-kappa B and the steroid hormone receptors: Mechanisms of mutual antagonism. Mol. Endocrinol. 12, 45–56. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., Ditta, G., Gustafson-Brown, C., Pelaz, S., Yanofsky, M., and Amasino, R.M. (2003). AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 33, 867–874. [DOI] [PubMed] [Google Scholar]
- Pařenicová, L., de Folter, S., Kieffer, M., Horner, D.S., Favalli, C., Busscher, J., Cook, H.E., Ingram, R.M., Kater, M.M., Davies, B., Angenent, G.C., and Colombo, L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15, 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Pinyopich, A., Ditta, G.S., Savidge, B., Liljegren, S.J., Baumann, E., Wisman, E., and Yanofsky, M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1997). MADS domain proteins in plant development. Biol. Chem. 378, 1079–1101. [PubMed] [Google Scholar]
- Rounsley, S.D., Ditta, G.S., and Yanofsky, M.F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7, 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Kohalmi, S.E., Motte, P., Datla, R., and Haughn, G.W. (1997). Divergence of function and regulation of class B floral organ identity genes. Plant Cell 9, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Schölkopf, B., Weigel, D., and Lohmann, J. (2005). A gene expression map of Arabidopsis development. Nat. Genet., in press. [DOI] [PubMed]
- Schwarz-Sommer, Z., Hue, I., Huijser, P., Flor, P., Hansen, R., Tetens, F., Lonnig, W., Saedler, H., and Sommer, H. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchennikova, A.V., Shulga, O.A., Immink, R., Skryabin, K.G., and Angenent, G.C. (2004). Identification and characterization of four chrysanthemum MADS-box genes, belonging to the APETALA1/FRUITFULL and SEPALLATA3 subfamilies. Plant Physiol. 134, 1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore, P., and Sharrocks, A.D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229, 1–13. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Smyth, D. (2000). A reverse trend: MADS functions revealed. Trends Plant Sci. 5, 315–317. [DOI] [PubMed] [Google Scholar]
- Suzuki, H., Fukunishi, Y., Kagawa, I., Saito, R., Oda, H., Endo, T., Kondo, S., Bono, H., Okazaki, Y., and Hayashizaki, Y. (2001). Protein-protein interaction panel using mouse full-length cDNAs. Genome Res. 11, 1758–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen, G., and Saedler, H. (2001). Plant biology: Floral quartets. Nature 409, 469–471. [DOI] [PubMed] [Google Scholar]
- Tröbner, W., Ramirez, L., Motte, P., Hue, I., Huijser, P., Lönnig, W., Saedler, H., Sommer, H., and Schwarz-Sommer, Z. (1992). GLOBOSA: A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11, 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627. [DOI] [PubMed] [Google Scholar]
- von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S.G., Fields, S., and Bork, P. (2002). Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417, 399–403. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J.M., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. (2000). Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287, 116–122. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Yu, H., Ito, T., Wellmer, F., and Meyerowitz, E.M. (2004). Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat. Genet. 36, 157–161. [DOI] [PubMed] [Google Scholar]
- Yu, H., Xu, Y., Tan, E.L., and Kumar, P.P. (2002). AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. USA 99, 16336–16341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., and Forde, B.G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.