Abstract
NONEXPRESSOR OF PR GENES1 (NPR1) is a key regulator of the plant defense response known as systemic acquired resistance. Accumulation of the signal molecule salicylic acid (SA) leads to a change in intracellular redox potential, enabling NPR1 to enter the nucleus and interact with TGACG sequence–specific binding protein (TGA) transcription factors, which in turn bind to SA-responsive elements in the promoters of defense genes. Here, we show that two NPR1-like genes, BLADE-ON-PETIOLE1 (BOP1) and BOP2, function redundantly to control growth asymmetry, an important aspect of patterning in leaves and flowers. Phenotypes in the double mutant include leafy petioles, loss of floral organ abscission, and asymmetric flowers subtended by a bract. We demonstrate that BOP2 is localized to both the nucleus and the cytoplasm, but unlike NPR1, it is highly expressed in young floral meristems and in yeast interacts preferentially with the TGA transcription factor encoded by PERIANTHIA (PAN). In support of a biological relevance for this interaction, we show that bop1 bop2 and pan mutants share a pentamerous arrangement of first whorl floral organs, a patterning defect that is retained in bop1 bop2 pan triple mutants. Our data provide evidence that BOP proteins control patterning via direct interactions with TGA transcription factors and demonstrate that a signaling mechanism similar to that formally associated with plant defense is likely used for the control of developmental patterning.
INTRODUCTION
The shoot apical meristem comprises a group of undifferentiated cells that give rise to all aerial tissues of a plant. Lateral organs (e.g., leaves) and shoot structures (e.g., flowers) arise on the flanks of the meristem. Initially, these structures appear as undifferentiated primordia, but they rapidly attain the distinct morphological features associated with specific organ and shoot types. A crucial component of patterning is the development of asymmetry. This process has three distinct phases. First, an axis must be specified, often achieved by long-range signals or unequal partitioning of a determinant within a cell before division. Second, specification of different identities occurs in domains along the axis, often the result of activation of different transcription factors that interact to reinforce distinctions between domains. Finally, the pattern is elaborated, a process that is often coupled to growth (reviewed in Hudson, 2000).
During vegetative development, leaf primordia are first initiated at specific sites on the periphery of the meristem according to a phyllotaxic pattern. Regional patterning then occurs along abaxial-adaxial, proximal-distal, and central-lateral axes to cause cells to adopt distinct developmental identities. Finally, organ growth occurs through regulated cell division and cell expansion, causing a leaf to acquire its final size and shape. Establishment of the abaxial-adaxial axis is of primary importance in leaf patterning because loss of either abaxial or adaxial identity affects blade outgrowth and symmetry, thus altering the patterning of the other two axes. This interrelationship was first revealed through analysis of extreme phenotypes of phantastica mutants in Antirrhinum majus. These mutants possess radially symmetrical leaves in which adaxial leaf tissues are absent or misexpressed (Waites and Hudson, 1995; Waites et al., 1998). Radially symmetrical leaves also result when abaxial leaf tissues are missing or misexpressed, as in phabulosa-1d mutants of Arabidopsis thaliana (McConnell and Barton, 1998). These and other observations indicate that juxtaposition of expression domains of abaxial and adaxial genes is required for the alteration of growth direction that results in blade formation in wild-type leaves (Waites and Hudson, 1995; reviewed in Bowman et al., 2002).
Although patterning of the abaxial-adaxial axis in leaves is well studied, comparatively little is known about the mechanisms involved in patterning the proximal-distal axis of leaves. Leaves are bilaterally symmetrical along this axis and are divided into two domains, consisting of a distal region, where the blade develops, and a proximal region, which is the petiole. The formation of this axis appears to correlate with the polar transport of auxin to the growing tip of young leaf primordia (discussed in Benková et al., 2003). Several genes that perturb proximal-distal leaf patterning have been identified by virtue of their ability to enhance blade development in the petiole region of leaves when mutated. These include ASYMMETRIC LEAVES1 (AS1), which encodes a MYB transcription factor (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Sun et al., 2002), AS2, which encodes a LOB domain/Leu zipper transcription factor (Iwakawa et al., 2002; Lin et al., 2003), and BLADE-ON-PETIOLE1 (BOP1), recently shown to encode a NONEXPRESSOR OF PR GENES1 (NPR1)-like transcription factor (Ha et al., 2003, 2004). AS1 and AS2 function in the same genetic pathway and bind to each other in yeast, suggesting that they are members of a complex (Byrne et al., 2002; Iwakawa et al., 2002; Lin et al., 2003; Xu et al., 2003). Analysis of double mutants shows that BOP1 has a synergistic genetic relationship with AS1 and AS2 (Ha et al., 2003), indicating that AS1/AS2 and BOP1 might function in separate genetic pathways controlling proximal-distal patterning. A common feature of each mutant, however, is the misexpression of meristem-promoting class 1 knotted1-like homeobox (knox) genes in leaf tissue, indicating that exclusion of knox gene expression and/or repression of meristematic activity might be an important determinant of proximal-distal patterning (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Ha et al., 2003).
In flowers, bilateral symmetry occurs along the plane of the abaxial-adaxial axis. However, morphological asymmetry along this axis is minor in Arabidopsis, and its flowers are considered to have radial symmetry. Nevertheless, the abaxial sepal is always larger, arising before the adaxial and lateral sepals, and grows to overlie the adaxial sepal during development, indicating that there are spatial differences along this axis (Kunst et al., 1989; Smyth et al., 1990). How this asymmetry is controlled remains unclear. It has long been understood that flowers occur in two basic designs: those with radial symmetry (two or more planes of symmetry) and those with bilateral symmetry (one plane of symmetry along the abaxial-adaxial axis). Evolutionary analyses indicate that bilateral forms of flowers are likely derived from radial forms and that bilateral symmetry is a feature that has arisen independently in many lineages, because of its association with the need to influence the behavior of pollinators (reviewed in Endress, 2001; Smyth, 2005). In species with strong bilateral symmetry, mutants with radial patterning sometimes arise, revealing that the switch between radial and bilateral symmetry is genetically controlled. The cloning of two related genes in the bilateral model species A. majus has revealed that CYCLOIDIA (CYC) and DICHOTOMA (DICH), which encode TB1, CYC, PCF basic-helix-loop-helix (TCP) domain transcription factors, act to suppress growth in the upper (adaxial) part of developing flowers and create polarity along the abaxial-adaxial axis of flowers (Luo et al., 1995, 1999). Another highly conserved feature of each floral species is the arrangement and number of each type of floral organ. Most eudicots are either tetramerous or pentamerous in form (reviewed in Endress, 2001; Smyth, 2005). Arabidopsis flowers are tetramerous in form (with four sepals, four petals, and six stamens), but mutation of the basic domain/Leu zipper (bZIP) transcription factor encoded by PERIANTHIA (PAN) is sufficient to cause most flowers to become pentamerous (with five sepals, petals, and stamens). How spatial information is generated to set up this pattern and how pan mutations perturb this process are unclear (Chuang et al., 1999).
Here, we describe an important role for two NPR1-like signaling proteins in controlling growth asymmetry, an important aspect of pattern formation during morphogenesis. The NPR1 signaling protein is a positive regulator of the plant defense response known as systemic acquired resistance (SAR), and its mechanism of action is well characterized. Upon pathogen attack, accumulation of the signal molecule salicylic acid (SA) causes a change in the redox potential of cells, leading to the conversion of NPR1 into an active monomer that enters the nucleus and interacts with members of the TGACG sequence–specific binding protein (TGA) family of transcription factors. This interaction, in turn, stimulates the binding of TGA factors to SA-responsive elements in the promoters of pathogenesis-related genes, launching the onset of SAR (Eckardt, 2003; reviewed in Dong, 2004).
The Arabidopsis genome contains five NPR1-like genes, including BOP1. BOP1 has a close homolog that we have designated BOP2. We show here that simultaneous disruption of both BOP1 and BOP2 causes numerous developmental defects, including loss of floral organ abscission and asymmetric changes in growth, that have striking effects on the morphological symmetry of both leaves and flowers. We also provide several lines of evidence that indicate similarities in the way that BOP and NPR1 proteins function. Together, these data suggest that a regulatory mechanism previously associated only with the area of plant–pathogen interactions is used in plant morphogenesis and development.
RESULTS
NPR1 Is the Founding Member of a Small Gene Family in Arabidopsis
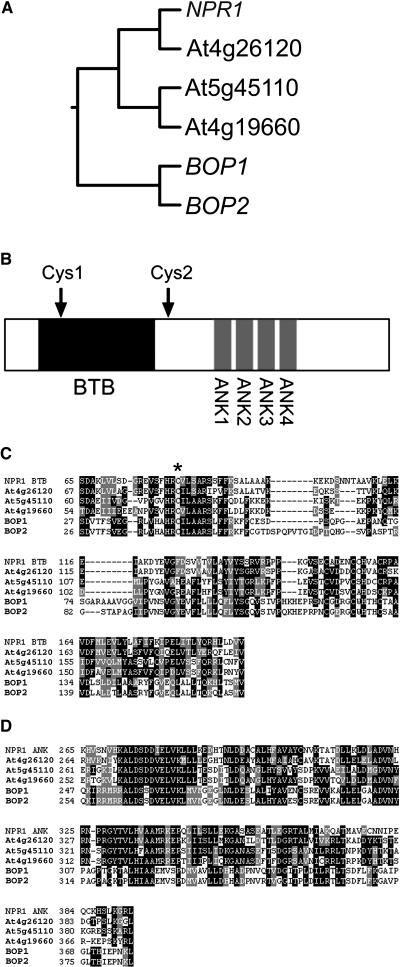
Phylogenetic analysis shows that members of the NPR1 family of proteins are closely related but form three distinct pairs (Figure 1A). This branching pattern suggests that there may be functional redundancy between members in each pair. Two protein interaction domains are conserved in all members of the NPR1 protein family (Figure 1B). The first motif is a BTB/POZ (for Broad-Complex, Tramtrack, and Bric-a-Brac/POX virus and Zinc finger) domain (Figure 1C), which in animals mediates dimerization and has been shown to interact with Cullin-3 proteins (reviewed in Collins et al., 2001; Pintard et al., 2004; van den Heuvel, 2004). Biochemical and genetic analyses indicate that BTB/POZ-containing proteins provide substrate specificity for Cullin-3–based E3 ubiquitin ligases and thus target proteins for degradation via the proteosome (reviewed in Pintard et al., 2004; van den Heuvel, 2004). Although such a role for NPR1 has not been established, a mutation in this region (npr1-2 allele, C150Y) results in the loss of NPR1 function (Cao et al., 1997). The second conserved domain is a series of four ankyrin repeats (Figure 1D), which in NPR1 have been shown to interact with members of the TGA family of transcription factors (Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000). Finally, several Cys residues that control the oligomerization state and nuclear localization of NPR1 are conserved in members of the NPR1 protein family. Nuclear localization of NPR1 is regulated by the intercellular redox potential of cells (Mou et al., 2003). In noninduced cells, intermolecular disulfide bonds hold NPR1 polypeptides together as a complex in the cytoplasm. During SAR, a biphasic change in redox potential occurs, resulting in the reduction of NPR1 to an active monomer that accumulates in the nucleus. Two Cys residues (Cys-82 and Cys-261) crucial for this form of regulation (Mou et al., 2003) are conserved in each of the NPR1 homologs. One of these Cys residues is in the BTB/POZ domain and is marked by an asterisk in Figure 1C. The other occurs in a conserved region just downstream of the BTB/POZ domain (data not shown). With respect to their positions in the phylogenetic tree, the genes that are the most distantly related to NPR1 are At3g57130 and At2g41370. They have shorter C termini than does NPR1 and lack a recognizable nuclear localization signal (data not shown). Based on their overall homology with NPR1 and their clustering in the phylogenetic tree, we reasoned that these two genes might have a redundant or shared function in an NPR1-like signaling pathway. As we were preparing to submit this article describing our analysis of these two genes, Ha et al. (2004) published an article identifying At3g57130 as BOP1. Therefore, we designate At2g41370 as BOP2.
Figure 1.
Phylogenetic Tree and Structural Motifs for the NPR1 Protein Family in Arabidopsis.
(A) Phylogenetic tree for the NPR1 protein family. Arabidopsis Genome Initiative numbers are as follows: NPR1, At1g64280; BOP1, At3g57130; and BOP2, At2g41370.
(B) Conserved structural motifs in NPR1-like proteins. BTB/POZ and ankyrin motifs are represented by boxes marked BTB and ANK, respectively. Cys1 and Cys2 indicate conserved Cys residues that mediate redox control of NPR1 oligomerization (Cys-82 and Cys-262, respectively) (Mou et al., 2003).
(C) Alignment of the predicted BTB/POZ domains for NPR1-like proteins. The asterisk indicates Cys-82 in NPR1, which mediates redox control.
(D) Alignment of the predicted ankyrin repeats for NPR1-like proteins.
BOP1 and BOP2 Function Redundantly
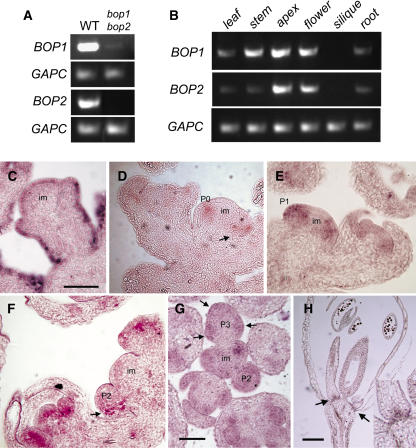
To identify a function for BOP1 and BOP2, we first obtained T-DNA insertion mutants from the Salk sequence-indexed library of insertion mutations in the Arabidopsis genome (Alonso et al., 2003). The insertion points for these T-DNAs were located at nucleotide −437 within the putative promoter region of BOP1 and at nucleotide −154 within the 5′ untranslated region of BOP2 (numbers relative to the ATG start site for each gene). In lines that were homozygous for T-DNA insertions at these loci, RT-PCR analysis indicated that few or no BOP2 transcripts were present, but the expression of BOP1 was not abolished completely (Figure 2A). As monitored by RT-PCR, transcripts for both genes were detected in leaves, stems, apices, flowers, and roots but not in siliques (Figure 2B).
Figure 2.
Expression of BOP1 and BOP2 in the Wild Type and bop1 bop2 Mutants.
(A) Analysis of BOP1 and BOP2 expression in wild-type (WT) and bop1 bop2 seedlings as monitored by RT-PCR. The gene amplified in each set of reactions is indicated at left of each gel. BOP1, 40 cycles; BOP2, 30 cycles; glyceraldehyde-3-phosphate dehydrogenase (GAPC) control, 30 cycles. A small amount of BOP1 transcript was detected in bop1 bop2 double mutants.
(B) RT-PCR analysis of tissue-specific expression of BOP1 and BOP2 in wild-type plants. The indicated tissues (top) were harvested from 6-week-old soil-grown plants, and the gene amplified in each set of reactions is indicated at left of each gel. RT-PCR conditions and cycle numbers were as in (A). (C) to (H) In situ localization of CER6 (control) or BOP2 mRNA in the inflorescence apex and flowers of wild-type plants. Hybridization is indicated by the presence of a purple precipitate.
(C) Central section of an inflorescence meristem (im) and young floral buds hybridized with an L1-specific CER6 control probe.
(D) to (F) Central longitudinal sections showing inflorescence meristems and young flower buds hybridized with a BOP2 probe. BOP2 mRNA is first detected in the floral anlagen (P0) on the flanks of the inflorescence meristem. Arrows in (D) and (F) indicate a strong band of expression at the base of stage 1 (P1) and stage 2 (P2) floral primordia in the abaxial region of these primordia corresponding to the cryptic bract.
(G) Cross section of an inflorescence meristem and young floral buds hybridized with a BOP2 probe. Arrows indicate expression in the developing sepal primordia of a stage 3 floral primordium (P3).
(H) Cross section of a mature flower showing strong bands of expression at the bases of anthers and petals (arrows). The inset shows detail at the base of the anther.
Bars = 100 μm, except 50 μm in (H).
Plants that were homozygous for single T-DNA insertions had no obvious mutant phenotype (data not shown). To test whether these genes function redundantly, we constructed plant lines that were homozygous for both mutations by crossing. Among 72 F2 progeny, 4 were genotyped to be bop1 bop2 double homozygotes. These four plants and no others had phenotypes that affected multiple aspects of development (described below). In addition, introduction of a construct harboring a fragment of genomic DNA containing the BOP2 coding sequence plus 2.2 kb of 5′ flanking sequence into bop1 bop2 double mutants by Agrobacterium tumefaciens–mediated transformation partially complemented phenotype conferred by bop1 bop2 (Figures 3F and 3G). Together, these results confirmed that the phenotype conferred by bop1 bop2 was attributable to the loss of BOP activity.
Figure 3.
Leaf and Inflorescence Phenotypes for Wild-Type, bop1, bop2, and bop1 bop2 Plants and Complementation of bop1 bop2 with BOP2.
(A) and (B) Twenty-eight-day-old plants are shown for the wild type (A) and bop1 bop2 (B). Bar = 2 cm for (A) and (B).
(C) Leaf series from 3-week-old wild-type (top row) and bop1 bop2 (bottom row) plants.
(D) Cauline leaves from wild-type (left) and bop1 bop2 (middle and right) plants.
(E) Inflorescences from wild-type (left) and bop1 bop2 (right) plants. Arrows indicate lack of floral organ abscission. The inset shows wild-type (left) and bop1 bop2 (right) siliques.
(F) Leaf series showing representative rosette leaves from a wild-type plant (left), a bop1 bop2 plant (middle), and a bop1 bop2 plant complemented by a fragment of genomic DNA containing the BOP2 gene (right).
(G) Inflorescence of a bop1 bop2 plant partially complemented for floral organ abscission by a fragment of genomic DNA containing the BOP2 gene.
To check the disease resistance status of bop1 bop2 mutants, leaves of 4-week-old plants were infiltrated with the virulent bacteria Pseudomonas syringae maculicola ES4326 at a dose of OD600 = 0.0001. The bacteria grew to a similar titer in bop1 bop2 mutants as in wild-type control plants, indicating that bop1 bop2 plants were neither more susceptible nor more resistant than were wild-type plants to challenge with the pathogen (data not shown). Thus, we concluded that these two NPR1 homologs are probably not involved in disease resistance signaling.
BOP1 and BOP2 Control Leaf Patterning and Floral Organ Abscission
The earliest notable defect in the bop1 bop2 double mutant was in leaf morphology. Wild-type Arabidopsis rosette leaves are divided into distinct proximal (petiole) and distal (blade) zones. In contrast with wild-type leaves, leaf growth in bop1 bop2 was indeterminate along the proximal-distal leaf axis, resulting in abnormally long leaves (cf. Figures 3A and 3B). In addition, blade development was derepressed along the petiole, resulting in the loss of distinct proximal and distal zones in the leaf. Leaflets were initiated along the petiole repeatedly throughout development (Figure 3C). This one aspect of the leaf phenotype conferred by bop1 bop2 is similar to that described previously for bop1-1 single mutants (Ha et al., 2003, 2004) (see Discussion). The inflorescences of bop1 bop2 double mutants contain several notable defects as well. First, cauline leaves in the double mutant were serrated at their bases, as opposed to smooth in the wild type (Figure 3D); second, floral organs failed to abscise (Figure 3E). Abscission normally occurs after pollination and is a form of developmental programmed cell death that is promoted by ethylene and inhibited by auxin (reviewed in Bleecker and Patterson, 1997; Roberts et al., 2002). It is not yet known whether the abscission defect in bop1 bop2 reflects a structural defect in the abscission zone or an altered response to the hormonal regulation of this process.
BOP1 and BOP2 Control Floral Patterning
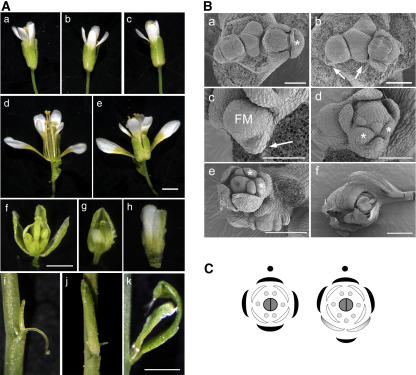
The most dramatic aspect of the phenotype conferred by bop1 bop2 was observed in flowers, and it is here that we focus most of our analysis. Wild-type Arabidopsis flowers develop in the absence of a subtending bract and possess a high degree of radial symmetry. Four types of floral organs are formed on the flanks of the floral meristem, and these are arranged in concentric whorls composed of four sepals in the first (outer) whorl, four petals in the second whorl, six stamens in the third whorl, and two carpels in the fourth whorl, as diagrammed in Figure 4C (left). In contrast with the wild type, bop1 bop2 flowers were often subtended by a small floral bract that curled around the pedicel toward the adaxial side of the flower (Figures 4A [compare panels a to c with d and e] and 4B, panel f). Development of this bract was sometimes incomplete (Figure 4A, panel i), and sepals or filaments often were produced in the axil of this bract, causing it to appear inflorescence-like (Figure 4A, panel k). For reasons that are unclear, the floral bract was not always located at the base of the pedicel but at variable locations on the floral pedicel.
Figure 4.
Comparison of Wild-Type and bop1 bop2 Floral Phenotypes.
(A) Morphology of mature flowers. (a) Wild type. (b) bop1. (c) bop2. (d) bop1 bop2 abaxial side. (e) Adaxial side. (f) Abaxial view of a flower with abaxial first whorl organs removed. (g) Abaxial view of a young flower showing retarded growth of abaxial organs in the first whorl. (h) Example of fused abaxial first whorl organs with sepal-petal characteristics. Bars = 1 mm. (i) to (k) Examples of bracts and bract-like structures that subtend flowers. Bar = 0.5 mm.
(B) Scanning electron micrographs of wild-type and bop1 bop2 inflorescence apices depicting inflorescence meristems, floral meristems, and flowers. (a) Wild-type inflorescence apex. The asterisk marks the abaxial sepal. (b) bop1 bop2 inflorescence apex. Arrows indicate floral bracts. (c) Stage 2 floral primordia with subtending bract (arrow). FM, floral meristem. (d) Stage 5 flower with subtending bract and five evenly spaced organs in the sepal whorl such that the adaxial sepal is in the same position as in the wild type. An extra stamen primordia occurs between the two abaxial sepal whorl organs (asterisks). (e) Stage 7 flower showing retarded development of sepals. Smaller abaxial first whorl organs are marked by asterisks. (f) Example of a flower with abaxial first whorl organs that are half-sepal (bumpy cells) and half-petal (smooth inner cells). Bars = 50 μm except 100 μm in (e) and 500 μm in (f).
(C) Floral diagrams for wild-type and bop1 bop2 flowers. The adaxial side is marked with a closed circle and indicates the position of the stem. The wild-type flower shows radial symmetry (left), and the bop1 bop2 flower shows bilateral symmetry (right).
In bop1 bop2 flowers, there was also an altered pattern of floral organ formation. Mature flowers had an asymmetric appearance and typically possessed five organs in the sepal whorl, four organs in the petal whorl, six or seven organs in the stamen whorl, and two organs in the carpel whorl, as diagrammed in Figure 4C (right; see also Table 1). The extra floral organs in bop1 bop2 mutants were always formed on the abaxial side of the flowers. In the sepal whorl, the adaxial sepal was always positioned as in the wild type, with the other organs placed equidistantly in the whorl. The two organs straddling the abaxial position in the sepal whorl grew more slowly compared with sepals in the lateral and adaxial positions and were usually petaloid in appearance. Sometimes the abaxial organs fused together (Figure 4A, panel h). The first whorl abaxial organs also had a unique growth trajectory compared with the other organs in the whorl, growing outward like wings toward the adaxial side of the flower (Figure 4A, panels d and e). Thus, mutation of the BOP genes altered growth specifically on the abaxial side of the flower, causing the loss of radial symmetry. Such bilateral symmetry is not seen in flowers of the Brassicaciae family but is common in many species of flowering plants, including the model plant species A. majus (reviewed in Endress, 2001).
Table 1.
Average Number of Floral Organ Types in Wild-Type and Mutant Flowers
| Genotype | Sepal Whorl | Petal Whorl | Stamens | Carpels | Subtending Sepal, Bract, or Filament | n |
|---|---|---|---|---|---|---|
| Columbia-2 (wild type) | 4.0 ± 0.0 | 4.0 ± 0.0 | 5.9 ± 0.4 | 2.0 ± 0.0 | 0.0 ± 0.0 | 21 |
| bop1-3 | 4.0 ± 0.0 | 4.0 ± 0.0 | 5.9 ± 0.3 | 2.0 ± 0.0 | 0.0 ± 0.0 | 28 |
| bop2-1 | 4.0 ± 0.0 | 4.0 ± 0.0 | 5.9 ± 0.3 | 2.0 ± 0.0 | 0.0 ± 0.0 | 51 |
| bop1-3 bop2-1 | 4.9 ± 0.3 | 4.0 ± 0.2 | 6.4 ± 0.6 | 2.0 ± 0.0 | 0.6 ± 0.5 | 94 |
| pan-3 (Landsberg erecta) | 4.6 ± 0.5 | 4.4 ± 0.6 | 5.3 ± 0.8 | 2.0 ± 0.0 | n.d. | 33 |
| pan-1 (Wassilewskija) | 4.9 ± 0.6 | 4.5 ± 0.5 | 5.3 ± 0.8 | 2.0 ± 0.0 | n.d. | 50 |
| pan-3 (Columbia) | 4.6 ± 0.5 | 4.3 ± 0.5 | 4.5 ± 0.5 | 2.0 ± 0.0 | n.d. | 39 |
| bop1-3 bop2-1 pan-3 | 4.9 ± 0.5 | 4.1 ± 0.4 | 4.4 ± 1.2 | 2.0 ± 0.0 | n.d. | 59 |
Bracts, filaments, or outgrowths on the floral pedicel were scored as subtending bracts. Values are averages ± sd of the number of floral organs per flower. n.d., not determined.
We further investigated these floral defects by examining the inflorescence apices of wild-type and bop1 bop2 flowers using scanning electron microscopy. Wild-type and bop1 bop2 mutant plants produced a similar number of stage 1 floral primordia of similar size on the flanks of the inflorescence meristem (Figure 4B, compare panels a and b). A small buttress corresponding to the floral bract was visible on the flanks of stage 1 and stage 2 floral primordia in the bop1 bop2 double mutant but not in the wild type (Figure 4B, panels b and c, arrows). We next compared the development of sepal primordia in wild-type and bop1 bop2 inflorescence meristems. Sepal primordia are first visible in the wild type as four ridges that appear high on the flanks of the floral meristem; their formation marks the beginning of stage 3 of floral development (Kunst et al., 1989; Smyth et al., 1990). Although Arabidopsis flowers are described as radially symmetrical, the abaxial sepal arises first, is slightly larger than the others, and grows to overlie the lateral and adaxial sepals (Kunst et al., 1989; Smyth et al., 1990) (Figure 4B, panel a, asterisk). In the wild type, the sepals grow rapidly to enclose the interior of the floral meristem such that development of the stamens, petals, and carpels is obscured. In the floral meristem of bop1 bop2, the appearance of the sepal primordia was slightly delayed relative to the plastochron compared with the wild-type. In addition, two first whorl primordia were formed on the abaxial side of the flower, slightly to the interior of where a wild-type abaxial sepal primordium would form, resulting in an apparent whorl of five evenly spaced organs (Figure 4B, panel d, asterisks). The two abaxial primordia in the first whorl were smaller and developed more slowly than the primordia in the adaxial and lateral positions (Figure 4B, panel e). However, in the double mutant, the growth of all first whorl organs was retarded compared with that in the wild type, allowing us to see the stamen and carpel primordia without having to dissect the sepals away. At later stages in floral development, the two abaxial organs in the first whorl of bop1 bop2 flowers differentiated as petals or sepal-petals. We speculate that ectopic development of the bract and/or changes in growth pattern on the abaxial side of the floral meristem may cause displacement of the abaxial organs in the sepal whorl, causing them to be lodged partially or entirely within the anlagen specifying petal identity. Evidence of this comes from the observation that some of these abaxial organs develop as half-sepal and half-petal (Figures 4A, panels g and h, and 4B, panel f).
BOP2 Is Expressed in Young Floral Primordia and Encodes a Protein Localized to Both the Cytoplasm and the Nucleus
The developmental pattern of BOP2 mRNA localization was determined in flowers by in situ hybridization. This analysis showed that BOP2 transcript is not expressed above background in the central part of the inflorescence meristem but is expressed strongly in floral anlagen situated on the flanks of the inflorescence meristem (Figure 2D, P0). Expression persisted throughout stage 1 and stage 2 floral primordia, with the strongest expression often seen near the base of these structures in the region that gives rise to the floral bract (Figures 2D to 2G, see arrows in 2D and 2F). At stage 3 of flower development, expression of BOP2 subsided from the central part of the floral meristem but was detected throughout very young organ primordia in all four whorls as these structures developed (Figures 2E to 2G and data not shown). As flowers matured, a strong band of BOP2 transcript was detected at the base of floral organs in the region corresponding to the abscission zone (e.g., as seen for the mature flower shown in Figure 2H). All features of the BOP2 expression pattern were distinct from that of CER6, a control probe (Figure 2C).
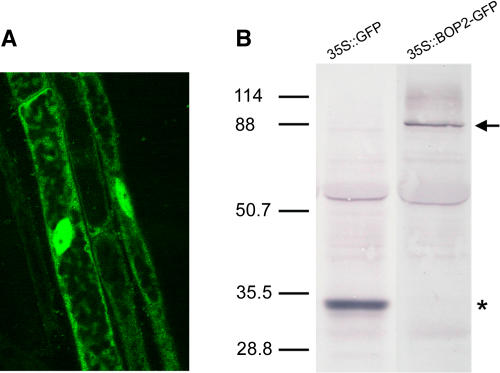
To determine the subcellular localization of the BOP2 protein, a 35S:BOP2–green fluorescent protein (GFP) transgene was introduced into bop1 bop2 plants by Agrobacterium-mediated transformation. Partial complementation of the phenotype conferred by bop1 bop2 was obtained for many of the resulting transgenic plant lines, indicating that the fusion protein had biological activity (data not shown). Confocal laser scanning of a seedling at 4 d after germination shows that in root cells, the BOP2-GFP fusion protein was localized to both the cytoplasm and the nucleus (Figure 5A). We then used protein gel blotting to correlate this GFP fluorescence pattern with the presence of full-length BOP2-GFP in extracts derived from complemented transgenic plant lines. No band corresponding to free GFP was observed in these extracts (Figure 5B). Therefore, we concluded that BOP2-GFP is targeted to the nucleus despite lacking a canonical nuclear localization sequence, because the predicted size of BOP2-GFP (∼88 kD) well exceeds the estimated size exclusion limit (∼60 kD) for passive diffusion of proteins through nuclear pores (Haasen et al., 1999). A similar pattern of subcellular localization was observed for NPR1-GFP transgenic seedlings in uninduced guard cells. However, when such seedlings were grown in the presence of a signal molecule (SA or 2,6-dichloroisonicotinic acid), strong NPR1-GFP florescence was detected exclusively in the nuclei of guard cells, indicating that repartitioning had occurred (Kinkema et al., 2000).
Figure 5.
Subcellular Localization of BOP2-GFP Protein.
(A) Confocal image of GFP fluorescence in root cells of a 35S:BOP2-GFP transgenic seedling.
(B) Protein gel blot of whole cell extracts from leaves of 35S:GFP and 35S:BOP2-GFP transgenic plants. Blots were probed with an anti-GFP antibody and deliberately overexposed to reveal the presence of minor bands. No major degradation bands in the form of free GFP were seen. An arrow denotes the position of a band corresponding to the full-length BOP2-GFP fusion protein. An asterisk denotes the position of a band corresponding to free GFP.
We concluded that BOP2 localized to the nucleus and the cytoplasm and has a discrete pattern of gene expression that correlates with phenotypic defects observed in bop1 bop2 flowers. Transcripts are excluded from the inflorescence meristem but are detected in floral anlagen, throughout young floral meristems, in young floral organ primordia, and at the base of mature floral organs at the predicted location of the abscission zone.
Biochemical and Genetic Interactions between BOP Proteins and the TGA Transcription Factor Encoded by PAN
Central to the accepted model for NPR1 signaling is its ability to modulate the activity of specific members of the TGA family of bZIP transcription factors. BOP proteins contain four conserved ankyrin motifs that in NPR1 mediate interactions with TGA transcription factors (Zhang et al., 1999; Després et al., 2000, 2003; Zhou et al., 2000). Moreover, disruption of TGA2, TGA5, and TGA6 together leads to the loss of SAR, a phenocopy of the npr1 mutant phenotype, indicating that NPR1 controls defense responses primarily via posttranscriptional regulation of TGA transcription factor activity (Zhang et al., 2003).
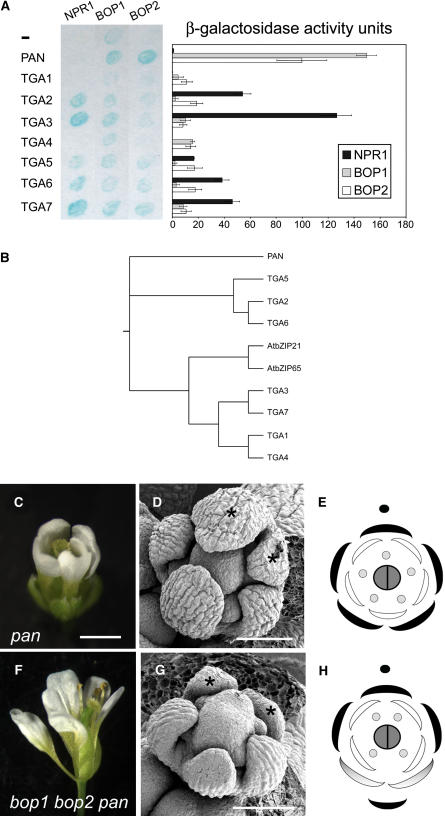
Therefore, we sought to test whether BOP proteins can interact directly with members of the TGA class of transcription factors and to ascertain whether any of these interactions are biologically significant. To test for a direct interaction of BOP proteins with specific TGA transcription factors, we used the yeast two-hybrid system to evaluate in a pair-wise manner the ability of NPR1 (a control) and BOP proteins to interact with eight members of the Arabidopsis TGA transcription factor family (Figure 6B shows a phylogenetic tree illustrating the relationship between the 10 members in this gene family). For each pair-wise test, NPR1 or BOP fused to the DNA binding domain of GAL4 served as the bait protein, whereas a TGA transcription factor fused to the transcriptional activation domain of GAL4 served as the prey protein. Each bait and prey combination was coexpressed in yeast, and an X-Gal filter assay was first used to monitor for interactions visually (Figure 6A, left; the blue precipitate indicates reporter gene activity). We then quantified the relative strength of these interactions by measuring β-galactosidase activity for each strain (Figure 6A, right). As expected, NPR1 failed to interact with TGA1 and TGA4 and showed preference for interactions with TGA2, TGA3, TGA6, and TGA7 (Zhang et al., 1999; Després et al., 2000). BOP1 and BOP2 also interacted with several different TGA transcription factors in yeast, but reporter gene activity was greatest (5- to 10-fold) when BOP proteins were paired with the TGA transcription factor encoded by PAN. BOP and PAN proteins also interact in the reciprocal orientation, when PAN is fused to the GAL4 DNA binding domain and BOP is fused to the GAL4 activation domain (data not shown). By contrast, NPR1 failed to interact with PAN. We concluded that BOP proteins show a different specificity for TGA transcription factor interactions in yeast than does NPR1 and that the strong interaction between BOP proteins and PAN may be of biological relevance.
Figure 6.
Analysis of Genetic and Biochemical Interactions between BOP and TGA Transcription Factors.
(A) Yeast two-hybrid interactions between NPR1, BOP1 and BOP2, and TGA transcription factors. Left, X-Gal filter-lift assay. Bait proteins are indicated at top and prey proteins are indicated at left. Accumulation of blue precipitate indicates β-galactosidase activity. Right, quantitative determination of reporter gene expression for each corresponding interaction shown at left, presented in chart format as β-galactosidase activity units. All data shown were corrected for self-activation of reporter gene expression by bait protein.
(B) Phylogenetic tree for the Arabidopsis TGA transcription factor family (Jakoby et al., 2002). Arabidopsis Genome Initiative numbers are as follows: PAN, At1g68640; TGA5, At5g06960; TGA2, At5g06950; TGA6, At3g12250; AtbZIP21, At1g08320; AtbZIP65, At5g06839; TGA3, At1g22070; TGA7, At1g77920; TGA1, At5g65210; TGA4, At5g10030.
(C) to (H) Comparison of pan-3 and bop1 bop2 pan-3 mutant floral phenotypes.
(C) Typical pan-3 mutant flower with pentamerous arrangement of floral organs: five sepals, five petals, and five stamens (abaxial view).
(D) Scanning electron micrograph of a stage 5 pan-3 flower with pentamerous arrangement of sepals. Abaxial sepals are marked with asterisks.
(E) Floral diagram of an average pan-3 flower. The adaxial side is marked with a closed circle.
(F) Typical bop1 bop2 pan-3 flower with pentamerous arrangement of first whorl organs (lateral view).
(G) Scanning electron micrograph of a stage 5 bop1 bop2 pan-3 flower that retains a pentamerous arrangement of first whorl organs. Abaxial organs are marked with asterisks.
(H) Floral diagram of an average bop1 bop2 pan-3 triple mutant flower. The adaxial side is marked with a closed circle.
Bars = 1 mm for (C) and (F) and 100 μm for (D) and (G).
To examine the biological relevance of the interactions between BOP and PAN, we first compared the mutant phenotypes for bop1 bop2 and pan. To date, PAN is the only TGA transcription factor in Arabidopsis with a known developmental function (Jakoby et al., 2002), and its mutant phenotype is restricted to the flower, despite a more widespread expression pattern (Chuang et al., 1999). The fact that pan mutants have a phenotype correlates well with the unique unpaired grouping of PAN in the phylogenetic tree for Arabidopsis TGA transcription factors (Figure 6B). As observed for bop1 bop2, the arrangement of first whorl organs in pan mutants is typically pentamerous (Table 1, cf. Figures 4B, panel d, and 4C, right, with 6D and 6E) (Running and Meyerowitz, 1996). In the majority of cases, an extra organ forms in the sepal whorl on the abaxial side of the flower, with an additional extra organ occurring in the petal whorl also on the abaxial side, as diagrammed in Figure 6E (Running and Meyerowitz, 1996). We found this change in patterning that occurs on the abaxial side of pan floral meristems strikingly similar to the change that occurs in bop1 bop2 mutants.
Given the proven biological relevance of NPR1–TGA interactions, together with the result that BOP and PAN proteins interact directly with apparent specificity in yeast, we concluded that similar phenotypic defects in floral patterning seen for these two mutants most likely occur because BOP and PAN have a shared function in perianth organ patterning. Consistent with this notion, BOP and PAN have an overlapping pattern of expression in developing flowers and are coexpressed throughout stage 1 and stage 2 floral primordia, in young floral organ primordia, and at the base of petals (Figure 2; data not shown) (Chuang et al., 1999; Ha et al., 2004). Therefore, we see the phenotypic differences between these two mutants as an indication that BOP and PAN have additional independent functions in plant development and/or that PAN has functions that are redundant with those of other TGA transcription factors.
To further examine the genetic relationship between BOP and PAN and to test the hypothesis that these genes function in the same genetic pathway, we constructed a triple mutant that was defective in both bop and pan activities. For this, we crossed the bop1 bop2 double mutant with the pan-3 mutant and used PCR genotyping to identify bop1 bop2 pan-3 triple mutants in the resulting F2 population. The vegetative leaf phenotype and floral organ abscission phenotype of these triple mutants was identical to that of the bop1 bop2 parent. We then examined the floral phenotype of these triple mutant flowers by scoring the number of floral organs in each whorl and by examining the floral organ patterning in young floral buds using scanning electron microscopy (Table 1, Figures 6F and 6G). Similar to both bop1 bop2 and pan parents, triple mutant flowers had an average of five organs in the first whorl. Similar to the bop1 bop2 parent, a small bract or filament subtended many flowers, the abaxial organs in the sepal whorl often developed into petaloid wings, and there were on average four organs in the petal whorl. Similar to the pan-3 parent (as monitored after backcrossing to the Columbia background), bop1 bop2 pan-3 flowers had an average of four to five stamens and the occasional flower with unfused carpels. The overall phenotype of the triple mutant could best be described as a fusion between the phenotypes conferred by bop1 bop2 and pan individually, because we did not observe an additive or synergistic increase in patterning defects or in the number of perianth organs. Together, these data support the hypothesis that BOP and PAN function in the same genetic pathway and have a joint role in abaxial patterning of the floral meristem; they also provide evidence that, in accord with an NPR1-like signaling activity, BOP proteins form biologically relevant interactions with TGA-class bZIP transcription factors.
DISCUSSION
Our study has revealed a redundant role for the NPR1-like signaling proteins BOP1 and BOP2 in controlling leaf and floral patterning and in promoting floral organ abscission. Several lines of evidence suggest that BOP1 and BOP2 function using an NPR1-like mechanism. First, protein motifs (BTB/POZ domain and ankyrin repeats) shown to be important for NPR1 function are conserved in BOP proteins, along with Cys residues shown to be important for the redox control of NPR1 activity. Second, BOP2 is localized to both the nucleus and the cytoplasm, as shown previously for NPR1 (Després et al., 2000; Kinkema et al., 2000). Third, BOP proteins interact directly with TGA transcription factors in yeast and show specificity for PAN. This specificity appears to have biological significance because bop1 bop2 and pan mutants have an overlapping defect in perianth patterning and because this patterning defect does not change in an additive or synergistic manner in bop1 bop2 pan triple mutants, consistent with these two proteins functioning in the same genetic pathway. Collectively, these results provide evidence that BOP1 and BOP2 function in a manner similar to that of NPR1, and we speculate that in response to an appropriate signal, BOP proteins may accumulate in the nucleus and/or become activated so that they may complex with TGA transcription factors such as PAN to regulate the transcription of target genes involved in the control of leaf and floral patterning and floral organ abscission.
BOP and Proximal-Distal Leaf Patterning
The mechanisms involved in establishing pattern formation along the proximal-distal leaf axis are still not well defined. Leaves are bilaterally symmetric along this axis and are divided into two distinct domains consisting of a distal blade region and a proximal petiole region. Differentiation in leaves tends to occur basipetally, with distal blade forming first, followed by proximal petiole. This pattern has an inverse relationship to cell division rates, which are highest in the proximal region of leaves, as monitored through genetic clonal analysis (Poethig and Sussex, 1985) and through observation of a cyclin AT:β-glucuronidase cell cycle reporter gene (Donnelly et al., 1999).
The leafy petiole defect found in bop1 bop2 mutants is similar to that reported for the bop1-1 single mutant. This mutation causes leaflet-like structures to form on leaf petioles and the proximal parts of leaf blades but does not affect growth determinacy along the proximal-distal leaf axis (Ha et al., 2003). In our analysis of plants homozygous for single alleles of bop1 or bop2, we failed to detect a leaf phenotype. Although bop1-3 is not a complete loss-of-function allele (we detected a small amount of transcript), the lack of a prominent leaf defect is also seen for bop1-4, which is a transcript null mutation (Ha et al., 2004). Such phenotypic differences between alleles may be the result of sensitivity to growth conditions or different genetic backgrounds (bop1-1 is a Landsberg erecta allele, whereas the other alleles are in the Columbia background), but another possibility is that bop1-1 is an atypical allele. Ha et al. (2004) point out that bop1-1 has a weak semidominant phenotype, is expressed at slightly higher levels than is wild-type transcript, and contains an unusual mutation in which the stop codon is altered, adding an additional four amino acids onto the C terminus of the protein. These details suggest that the protein encoded by bop1-1 may exert a weak dominant negative effect by interfering with the function of an interacting partner, such as BOP2. BOP1 and BOP2 can form self-dimers and heterodimers in yeast (S.R. Hepworth and G.W. Haughn, unpublished results) and have the same pattern of gene expression in flowers (Ha et al., 2004; this study), an indication that these two proteins may form a complex together and function interchangeably in some cases. These data are also consistent with redundant functions for these two genes. Nevertheless, the fact that bop1-1 primarily affects the petiole suggests that BOP1 may make a greater contribution to the control of leaf patterning than does BOP2 and that there may be some specialization of function for these two proteins.
Leafy petioles are also caused by the ectopic expression of LEAFY PETIOLE (LEP), which encodes an EREBP/AP2-type transcription factor (van der Graaff et al., 2000), and JAGGED (JAG), which encodes a C2H2 zinc finger transcription factor (Dinneny et al., 2004; Ohno et al., 2004). One possibility is that BOP activity regulates the spatial expression of leaf-promoting transcription factors such as JAG or LEP in the proximal parts of leaves and in floral bracts. Consistent with this notion, we detected upregulation of JAG transcript in bop1 bop2 leaves (S.R. Hepworth and G.W. Haughn, unpublished results). Plants misexpressing JAG also develop floral bracts (Dinneny et al., 2004; Ohno et al., 2004), another aspect of the bop1 bop2 mutant phenotype. Previous analyses of bop1-1 plants have shown that knox genes are also misexpressed in the leaves, predominantly in the petiole region (Ha et al., 2003). Thus, an additional function of BOP may be to exclude knox gene expression from leaf tissues and maintain a developmentally determinate state in cells. knox gene misexpression is often correlated with changes in proximal-distal leaf patterning (reviewed in Byrne et al., 2001; Hake et al., 2004).
BOP and Floral Symmetry
Wild-type Arabidopsis flowers are nearly radially symmetrical but show some asymmetry along the abaxial-adaxial axis. Mutation of the BOP genes appears to exaggerate this asymmetry, as indicated by the formation of an ectopic floral bract, extra abaxial floral organs, and the transformation of the abaxial sepal whorl organs into petaloid structures that grow outward like wings toward the adaxial side of the flower. Information on how asymmetries develop along the abaxial-adaxial axis is best understood in the model species A. majus, which possesses flowers with strong bilateral asymmetry. In this species, flowers are subtended by floral bracts and have a pentamerous arrangement of sepals, petals, and stamens, similar to that observed in bop1 bop2 pan triple mutants; additionally, the abaxial petals of each flower are of a different shape than the adaxial petals. These asymmetries depend on the actions of CYC and DICH, which are members of the TCP family of DNA binding proteins. These transcription factors are thought to respond to an abaxial-adaxial prepattern in floral meristems (Luo et al., 1995, 1999). In Arabidopsis flowers, although morphological asymmetries along the abaxial-adaxial axis are slight, there is retained adaxial-specific expression of TCP1 (an Arabidopsis ortholog of CYC) in floral meristems (Cubas et al., 2001). The abaxial-specific floral defects that we see in bop1 bop2 mutants are further evidence of asymmetries along this axis, and it is possible that BOP activity limits abaxial growth as part of a mechanism to promote radial symmetry in Arabidopsis flowers.
We find it intriguing that bop1 bop2 mutations both promote bilateral symmetry and cause flowers to switch to a pan-like pentamerous arrangement of sepal whorl organs. Interaction of BOP with PAN suggests that BOP may modulate PAN activity, possibly to control growth on the abaxial side of the floral meristem. However, no measurable increase in floral meristem size is detected in bop1 bop2 mutants (this study) or in pan mutants (Running and Meyerowitz, 1996). Double mutant analyses, however, have revealed that PAN functions redundantly with ULTRAPETALA and ETTIN/AUXIN RESPONSE FACTOR3 to control the size of the floral meristem and the initiation of floral organs, respectively (Sessions et al., 1997; Fletcher, 2001). Thus, a clearer picture of the role of BOP in controlling the size of the floral meristem and/or the initiation of floral organs may be obtained from further genetic analyses.
BOP and Floral Organ Abscission
In addition to its role in leaf and floral patterning, BOP activity is required for floral organ abscission. Abscission is a developmentally controlled process commencing soon after pollination in which floral organs senesce and are shed through separation at a specialized zone of cells located at the base of stamens and perianth organs. This late role for BOP activity in floral development correlates with the expression of BOP1 and BOP2 at the base of floral organs in mature flowers (Ha et al., 2004; this study). It is possible that this defect represents a structural defect within the abscission zone, but alternatively, it may represent failure to respond to hormonal signals controlling this process. The predominant view is that auxin delays abscission and ethylene promotes abscission. The roles of other plant hormones (e.g., abscisic acid, gibberellic acid, and cytokinins) involved in this process have been postulated primarily to involve altering the levels of or sensitivity to auxin and ethylene in abscising tissues (Sexton and Roberts, 1982). Although a link between growth asymmetry and floral organ abscission is not immediately obvious, it is possible that these two processes are controlled by a common signal to which BOP responds.
Spatial Regulation of BOP Activity
A unique feature of the phenotype conferred by bop1 bop2 is that ectopic growth and development is restricted to specific domains along the axes of bilateral symmetry within leaves and flowers. In leaves, excess growth occurs along the proximal-distal axis and ectopic blade development occurs in the proximal region. In flowers, ectopic bract development and floral defects are centered on the abaxial side. The asymmetric nature of these defects suggests that BOP activity links spatial information with pattern elaboration. In part, this is accomplished through the spatial regulation of BOP expression. BOP1 and/or BOP2 are expressed in the proximal part of young leaves, in young floral meristems, and at the base of mature floral organs (Ha et al., 2004; this study). Each of these locations closely correlates with the phenotypes in bop1 bop2 mutants affecting leaf petioles, floral patterning, and floral organ abscission. However, although BOP2 transcript is present throughout stage 1 and stage 2 floral primordia, defects are centered on the abaxial side of the flower, raising the possibility that BOP activity is also controlled at the posttranscriptional level by a developmental signal.
Characterization of the NPR1 protein has shown that its activity is redox-sensitive and that the signal molecule SA controls activity indirectly by causing changes in the intracellular redox potential of cells (reviewed in Eckardt, 2003; Dong, 2004). In the absence of signal, intermolecular disulfide bonds hold NPR1 proteins together in an inactive complex. The signal molecule SA causes a rapid oxidative burst, with the cellular redox state recovering to a more reduced state, so that the intermolecular disulfide bonds holding NPR1 proteins together are reduced and the majority of NPR1 is present in an active monomeric form that accumulates in the nucleus (Mou et al., 2003). TGA1 also contains redox-sensitive Cys residues that modulate its ability to interact with NPR1 (Després et al., 2003). Because the Cys residues required for the redox control of NPR1 are conserved in the BOP proteins, it is possible that BOP activity is controlled by a signal that alters the redox state of the cell, but this remains to be tested. Although crucial for the activation of plant defenses, SA does not appear to play a major role in growth or development. One report suggests that SA may regulate the balance between cell growth and cell death, but the significance of this finding with respect to development is unclear (Vanacker et al., 2001). Another possibility is that signals other than SA are perceived in the cell as a change in redox potential. Consistent with this notion, a role for reactive oxygen species as a signal for the hormonal regulation of various aspects of plant development has been proposed in recent studies (Sagi et al., 2004, and references therein). Also, TGA binding elements (also known as as1 or ocs elements) appear to mediate activation by hydrogen peroxide and by plant phytohormones such as auxin, SA, and jasmonic acid (Xiang et al., 1996, and references therein). Auxin is particularly attractive as a signal molecule for the control of BOP activity because of its roles in regulating cell growth and cell expansion, patterning of lateral organs, and floral organ abscission, combined with its polarized transport throughout the plant (Rogg and Bartel, 2001; Friml, 2003; reviewed in Roberts et al., 2002). Nevertheless, other signals cannot be excluded, and further experiments are needed to resolve this issue.
METHODS
Plant Materials and Growth Conditions
Wild type was the Columbia-0 ecotype of Arabidopsis thaliana. All plants were grown in continuous light as described (Bellaoui et al., 2001) on GM agar plates or on prepared soil mix (Sunshine Mix 5; SunGro, Seba Beach, Alberta, Canada). The bop1-3 and bop2-1 T-DNA insertion mutants used in this study were obtained from the ABRC (Ohio State University, Columbus, OH; stock numbers SALK_012994 and SALK_075879, respectively); the pan-3 mutant (Landsberg erecta background) was a gift from Elliot Meyerowitz and the pan-1 mutant (Wassilewskija background) was a gift from Mark Running (Running and Meyerowitz, 1996). bop1 bop2 double mutants and bop1 bop2 pan-3 triple mutants were constructed by crossing, and their genotypes were confirmed by PCR-based genotyping. Floral stages were determined according to Smyth et al. (1990). For the definition of floral bract, see Dinneny et al. (2004).
Sequence Analysis
The complete protein sequences for members of the NPR1 gene family were aligned using ClustalW (Thompson et al., 1994), and phylogenetic trees (Neighbor Joining method with 1000 Bootstrap output) were produced using the Web-based Phylodendron program (http://iubio.bio.indiana.edu/soft/molbio/java/apps/trees). The Web-based SMART program was used to predict the BTB/POZ domains and to confirm the locations of ankyrin repeats for each protein (Schultz et al., 2002). The individual domains were then realigned and shaded using ClustalW and BOXSHADE.
Complementation of bop1 bop2 Double Mutants
To create plasmid pGreen0229/BOP2, a 6.1-kb EagI-PstI fragment was excised from Arabidopsis BAC F13H10 (AC005662) and cloned into the corresponding sites of the binary vector pGreen0229 (Hellens et al., 2000). This fragment contained the BOP2 gene plus ∼2.2 kb of sequence 5′ to the ATG start codon. The construct was introduced into bop1 bop2 plants by floral dipping (Clough and Bent, 1998). Basta-resistant transformants were selected on soil by treatment of seedlings with the herbicide Final ev 150 (AgrEvo, Paris, France). The Agrobacterium tumefaciens strain used was C58C1 pGV3101 pMP90 (Koncz and Schell, 1986).
Subcellular Localization of BOP2 Protein
To create the 35S:BOP2-GFP transgene, full-length BOP2 cDNA was amplified by PCR using pCR2-BOP2 as the template (see below) and with primers that incorporated EcoRI and BamHI sites at the 5′ and 3′ ends of the gene, respectively. The resulting fragment was digested with EcoRI and BamHI and cloned into corresponding sites of pBS-GFP5 (Haseloff et al., 1997). The resulting plasmid was sequenced to confirm that the fusion was in-frame and without PCR-induced mistakes. The fragment containing the BOP2-GFP fusion was then excised from pBS-GFP by digestion with EcoRI and SacI and cloned into the corresponding sites of pBI1.4T to yield pBI-BOP2-GFP (Mindrinos et al., 1994). This construct was introduced into bop1 bop2 plants by Agrobacterium-mediated transformation as described above. Transgenic plants were selected on MS medium containing 50 μg/mL kanamycin sulfate. Roots of the transgenic seedlings were examined for GFP fluorescence using confocal microscopy as described (Haseloff et al., 1997) using a Bio-Rad Radiance 2000 multiphoton microscope (Bio-Rad, Hercules, CA).
Immunoblot Analysis of GFP and GFP Fusion Proteins
Ten microliters of a crude protein extract from leaves of 35S:BOP2-GFP or 35S:GFP transgenic plants was resolved on a 10% SDS–polyacrylamide gel by electrophoresis and blotted onto a nitrocellulose membrane. The anti-GFP antibody (A6455; Molecular Probes, Eugene, OR) was used as recommended by the supplier. The blots were developed using an alkaline phosphatase detection system according to the manufacturer's instructions (Roche, Indianapolis, IN).
Scanning Electron Microscopy
Scanning electron microscopy samples were prepared as described by Modrusan et al. (1994). Flower buds and inflorescences were mounted on stubs. If necessary, the organs surrounding the inflorescence meristem were dissected away. The stubs were coated with gold-palladium in a SEMPrep2 sputter coater (Nanotech, Manchester, UK) and observed using a Hitachi S4700 scanning electron microscope (Hitachi, Tokyo, Japan) with an accelerating voltage of 0.8 or 2 kV.
In Situ Hybridization Analysis
Tissue fixation, sectioning, hybridization, signal detection, and strategy for probe synthesis were as described previously (Hooker et al., 2002). To synthesize a BOP2 antisense probe, a DNA template was amplified by PCR using pCR2-BOP2 as the template and using 5′-CTCATATGAATGAGGAGCAC-3′ and 5′-GATAATACGACTCACTATAGGGACTCATACCTTCCCTCTGA-3′ (which incorporates a binding site for T7 polymerase) as primers. CER6 probe was as described previously (Hooker et al., 2002). Sections were photographed using bright-field optics.
Photography and Light Microscopy
Photographs were taken with a digital camera (Coolpix; Nikon, Tokyo, Japan). A dissecting microscope fitted with a digital camera (SPOT Diagnostics) was used to take close-up pictures of flowers, cauline leaves, and siliques. Digital photographs and micrographs were manipulated using Photoshop 5.0 (Adobe Systems, San Jose, CA).
Construction of Yeast Two-Hybrid Plasmids
Full-length cDNAs corresponding to BOP1 and BOP2 were amplified by RT-PCR using the Expand Hi-Fidelity PCR system (Roche). The template used for these reactions was cDNA derived from total RNA isolated from 20-d-old seedlings grown on agar plates. The BOP1 and BOP2 cDNAs were subcloned into pCR2 using the TA cloning system (Invitrogen, Carlsbad, CA) to yield pCR2-BOP1 and pCR2-BOP2, respectively. Full-length cDNAs corresponding to PAN (stock No. U50929) and NPR1 (stock number U13446) were obtained from the ABRC. The sequences of all four cDNAs were verified by sequencing. To create the constructs used for the yeast two-hybrid assay, the full-length coding sequences of BOP1, BOP2, PAN, and NPR1 were amplified by PCR using pCR2-BOP1, pCR2-BOP2, pU1539, and pU13446, respectively, as the templates for Pwo polymerase (Roche). Recognition sites for restriction enzymes were incorporated at the 5′ ends of the primers used in these PCR procedures to facilitate the directional cloning of these cDNA fragments into the bait (GAL4-DB) plasmid pBI-880 or the prey (GAL4-TA) plasmid pBI-881 (Kohalmi et al., 1998). The BOP1, BOP2, and NPR1 cDNA inserts were ligated as SalI-NotI fragments into the corresponding sites of pBI-880 to create pBI-880/BOP1, pBI-880/BOP2, and pBI-880/NPR1, respectively. The PAN cDNA insert was ligated as a BamHI-NotI fragment into the corresponding sites of pBI-881 to create pBI-881/PAN. The sequences of all primers are available upon request. pBI-881 prey plasmids containing TGA1-7 were described previously (Després et al., 2000).
Two-Hybrid Assays
We used a GAL4-based yeast two-hybrid system that has been described previously (Kohalmi et al., 1998). Briefly, the appropriate bait (GAL4-DB) and prey (GAL4-TA) constructs were introduced into yeast two-hybrid strain YPB2 (Fields and Song, 1989). Transformants were selected on SD plates lacking Leu and Trp and then spread in patches onto fresh plates of the same composition. Interactions were detected visually using an X-Gal filter assay performed as described previously (Kohalmi et al., 1998). Quantitative assays for β-galactosidase were performed using the liquid culture assays with 2-nitrophenyl-β-d-galactopyranoside as the substrate as described in the Yeast Protocol Handbook (Clontech Laboratories, Palo Alto, CA).
RT-PCR
RT was performed using 1 μg of total RNA isolated from seedlings or individual tissues of wild-type or bop1 bop2 plants and Superscript II RT (Invitrogen). To amplify transcripts, PCR was performed using 1 μL of cDNA as the template with 1 unit of Taq polymerase. Primers used to amplify BOP1 were 5′-TCTGTGAATCTGATCCTTCGCAACC-3′ and 5′-CCTGATCTTCTGGTCTTCGAGGTCT-3′ or 5′-ACTGCAGCCGTCGATCTC-3′ and 5′-ATCTCTTGTGGAAGCCCGGA-3′. Primers used to amplify BOP2 were 5′-ACAGACTCACCACAACCTGTCACAG-3′ and 5′-GATCTTCTAGGTCTTGAGCCACGCT-3′. Amplification of the cytosolic glyceraldehyde-3-phosphate dehydrogenase cDNA from the same cDNA pools was performed under the same conditions and served as a control (Western et al., 2004).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY928830 (BOP2 cDNA).
Acknowledgments
We thank Elliot Meyerowitz for providing the pan-3 mutant, Mark Running for providing the pan-1 mutant, Tanya Hooker for providing the CER6 in situ probe, Owen Rowland for providing tissue-specific total cDNAs, and Pierre Fobert for providing TGA-containing two-hybrid constructs. We are grateful to the Biological Sciences Microscopy and Imaging Centre at the University of British Columbia for providing microscopy support and to Hugo Zheng for confocal imaging. We also thank Owen Rowland, Ravi Kumar, and Gillian Dean for critical reading of the manuscript. S.M. was a recipient of a postgraduate scholarship B award from the Natural Science and Engineering Research Council (NSERC) of Canada. Work in G.W.H.'s laboratory is supported by an NSERC Discovery Grant. Work in X.L.'s laboratory is supported by grants from the NSERC and the Canadian Foundation for Innovation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: George W. Haughn (haughn@interchange.ubc.ca).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.030536.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Bellaoui, M., Pidkowich, M.S., Samach, A., Kushalappa, K., Kohalmi, S.E., Modrusan, Z., Crosby, W.L., and Haughn, G.W. (2001). The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13, 2455–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková, E., Michneiwicz, M., Suaer, M., Teichmann, T., Seifertová, D., Jürgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Bleecker, A.B., and Patterson, S.E. (1997). Last exit: Senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9, 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Eshed, Y., and Baum, S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18, 134–141. [DOI] [PubMed] [Google Scholar]
- Byrne, M., Timmermans, J., Kidner, C., and Martienssen, R. (2001). Development of leaf shape. Curr. Opin. Plant Biol. 4, 38–43. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965. [DOI] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Chuang, C.-F., Running, M.P., Williams, R.W., and Meyerowitz, E.M. (1999). The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 13, 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Collins, T., Stone, J.R., and Williams, A.J. (2001). All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 21, 3609–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas, P., Coen, E., and Martínez-Zapater, J.M. (2001). Ancient asymmetries in the evolution of flowers. Curr. Biol. 11, 1050–1052. [DOI] [PubMed] [Google Scholar]
- Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., and Fobert, P.R. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15, 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Dinneny, J.R., Yadegari, R., Fischer, R.L., Yanofsky, M.F., and Weigel, D. (2004). The role of JAGGED in shaping lateral organs. Development 131, 1101–1110. [DOI] [PubMed] [Google Scholar]
- Dong, X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. [DOI] [PubMed] [Google Scholar]
- Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419. [DOI] [PubMed] [Google Scholar]
- Eckardt, N. (2003). A new twist on systemic acquired resistance: Redox control of the NPR1–TGA1 interaction by salicylic acid. Plant Cell 15, 1947–1949. [DOI] [PMC free article] [Google Scholar]
- Endress, P.K. (2001). Evolution of floral symmetry. Curr. Opin. Plant Biol. 4, 86–91. [DOI] [PubMed] [Google Scholar]
- Fields, S., and Song, O. (1989). A novel genetic system to detect protein-protein interactions. Nature 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C. (2001). The ULTRAPETALA gene controls shoot and floral meristem size in Arabidopsis. Development 128, 1323–1333. [DOI] [PubMed] [Google Scholar]
- Friml, J. (2003). Auxin transport: Shaping the plant. Curr. Opin. Plant Biol. 6, 7–12. [DOI] [PubMed] [Google Scholar]
- Ha, C.M., Jun, J.H., Nam, H.G., and Fletcher, J.C. (2004). BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 45, 1361–1370. [DOI] [PubMed] [Google Scholar]
- Ha, C.M., Kim, G.-T., Kim, B.C., Jun, J.H., Soh, M.S., Ueno, Y., Mahida, Y., Tsukaya, H., and Nam, H.G. (2003). The BLADE-ON-PETIOLE1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130, 161–172. [DOI] [PubMed] [Google Scholar]
- Haasen, D., Kohler, C., Neuhaus, G., and Merkle, T. (1999). Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 20, 695–705. [DOI] [PubMed] [Google Scholar]
- Hake, S., Smith, H.M., Holtan, H., Magnani, E., Mele, G., and Ramirez, J. (2004). The role of KNOX genes in plant development. Annu. Rev. Cell Dev. Biol. 20, 125–151. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Hooker, T.S., Millar, A.A., and Kunst, L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129, 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, A. (2000). Development of symmetry in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 349–370. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478. [DOI] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vincente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohalmi, S.E., Reader, L.J.W., Samach, A., Nowak, J., Haughn, G.W., and Crosby, W.L. (1998). Identification and characterization of protein interactions using the yeast 2-hybrid system. In Plant Molecular Biology Manual M1, S.B. Gelvin and R. A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–30.
- Koncz, C., and Schell, J. (1986). The promoter of T1-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Kunst, L., Klenz, J.E., Martinez-Zapater, J., and Haughn, G.W. (1989). AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1, 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W., Shuai, B., and Springer, P. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES–domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15, 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, D., Carpenter, R., Copsey, L., Vincent, C., Clark, J., and Coen, E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99, 367–376. [DOI] [PubMed] [Google Scholar]
- Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1995). Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 121, 2935–2942. [DOI] [PubMed] [Google Scholar]
- Mindrinos, M., Katagiri, F., Yu, G.-L., and Ausubel, F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Modrusan, Z., Reiser, L., Feldmann, K.A., Fischer, R.L., and Haughn, G.W. (1994). Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. [DOI] [PubMed] [Google Scholar]
- Ohno, C.K., Reddy, G.V., Heisler, M.G.B., and Meyerowitz, E.M. (2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131, 1111–1122. [DOI] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532. [DOI] [PubMed] [Google Scholar]
- Poethig, R.S., and Sussex, I.M. (1985). The cellular parameters of leaf development in tobacco: A clonal analysis. Planta 165, 170–184. [DOI] [PubMed] [Google Scholar]
- Pintard, L., Willems, A., and Peter, M. (2004). Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 23, 1681–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, J.A., Elliot, K.A., and Gonzalez-Carranza, Z.H. (2002). Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 53, 131–158. [DOI] [PubMed] [Google Scholar]
- Rogg, L.E., and Bartel, B. (2001). Auxin signaling: Derepression through regulated proteolysis. Dev. Cell 1, 595–604. [DOI] [PubMed] [Google Scholar]
- Running, M.P., and Meyerowitz, E.M. (1996). Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122, 1261–1269. [DOI] [PubMed] [Google Scholar]
- Sagi, M., Davydov, O., Orazova, S., Yesbergenova, Z., Ophir, R., Stratmann, J.W., and Fluhr, R. (2004). Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum. Plant Cell 16, 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., Copley, R.R., Doerks, T., Ponting, C.P., and Bork, P. (2002). SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakaka, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Nemhauser, J.L., McCall, A., Roe, J.L., Feldmann, K.A., and Zambryski, P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491. [DOI] [PubMed] [Google Scholar]
- Sexton, R., and Roberts, J.A. (1982). Cell biology of abscission. Annu. Rev. Plant Physiol. 33, 133–162. [Google Scholar]
- Smyth, D.R. (2005). Morphogenesis of flowers: Our evolving view. Plant Cell 17, 330–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Zhou, Q., Zhang, W., Fu, Y., and Huang, H. (2002). ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 214, 694–702. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). Clustal W: Improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position-specific gap penalties and weight matrix choices. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker, H., Lu, H., Rate, D.N., and Greenberg, J.T. (2001). A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J. 28, 209–216. [DOI] [PubMed] [Google Scholar]
- van den Heuvel, S. (2004). Protein degradation: CUL-3 and BTB—partners in proteolysis. Curr. Biol. 14, R59–R61. [PubMed] [Google Scholar]
- van der Graaff, E., Dulk-Ras, A.D., Hooykaas, P.J.J., and Keller, B. (2000). Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development 127, 4971–4980. [DOI] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121, 2143–2154. [Google Scholar]
- Waites, R., Slevadurai, H.R.N., Oliver, I.R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93, 779–789. [DOI] [PubMed] [Google Scholar]
- Western, T.L., Young, D.S., Dean, G.H., Tan, W.L., Samuels, A.L., and Haughn, G.W. (2004). MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 134, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., Miao, Z.H., and Lam, E. (1996). Coordinated activation of as-1-type elements and a tobacco glutathione S-transferase gene by auxins, salicylic acid, methyl jasmonate and hydrogen peroxide. Plant Mol. Biol. 32, 415–426. [DOI] [PubMed] [Google Scholar]
- Xu, L., Xu, Y., Dong, A., Sun, Y., Pi, L., Xu, Y., and Huang, H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial polarity. Development 130, 4097–4107. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Tessaro, M.J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15, 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., and Klessig, D.F. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant-Microbe Interact. 13, 191–202. [DOI] [PubMed] [Google Scholar]