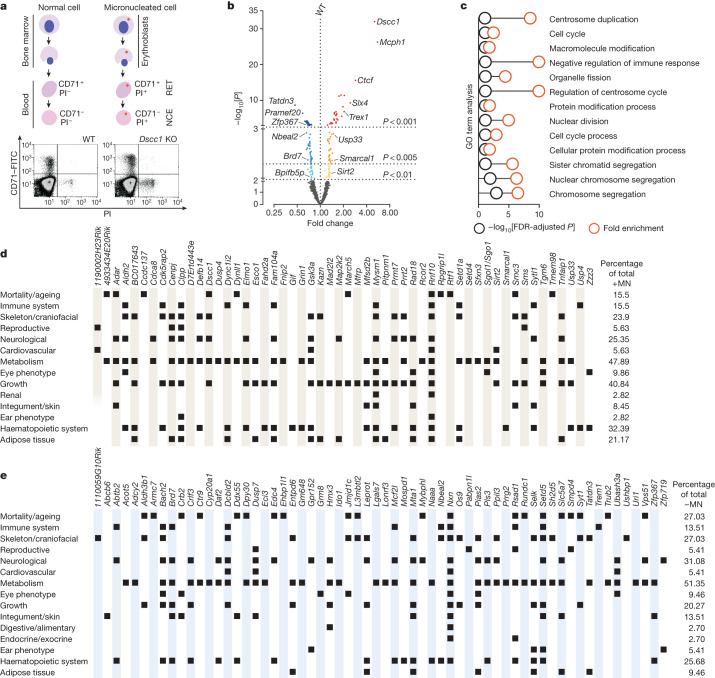
Fig. 1. An in vivo screen for genetic regulators of MN formation.
a, Schematic of the in vivo micronucleus assay. Full protocol details have been described previously20. Data for a Dscc1-KO mouse and a WT littermate control are shown. CD71, transferrin receptor; NCE, normochromatic erythrocyte; PI, propidium iodide; RET, reticulocyte. b, The MN screen results indicating mutants that, compared with the WT, have lower (−MN; left) or higher (+MN; right) MN formation and accumulation. Three statistical tiers are indicated on the basis of P-value cut-offs and false-discovery rates (FDR): tier 1 (P < 0.001; FDR < 0.017; +MN, red dots; −MN, dark blue dots); tier 2 (P < 0.005; FDR < 0.046; +MN, orange dots; −MN, blue dots); and tier 3 (P < 0.01; FDR < 0.068; +MN, yellow dots; −MN, light blue dots). The effect of genotype on the percentage of MN was assessed using a mixed linear effect beta regression model in R with baseline WT mice (n = 285) together with mice of each genotype. A total of n = 6,210 mice were analysed. Multiple testing was managed by adjusting the P values to control the FDR (Methods). The full dataset and statistics are provided in Supplementary Table 1. c, Pathway analysis for +MN screen hits, aligning them with biological processes. GO, Gene Ontology. d,e, Statistically significant phenotypes of mouse lines with increased (+MN; d) or decreased (−MN; e) MN59. Out of 71 +MN mutant lines, 54 had additional phenotypes; out of 74 −MN mutant lines, 62 had additional phenotypes. The squares indicate the related organ system affected. The percentage representation of phenotypes within the +MN and −MN genes is shown on the right. The full dataset and statistical methods are available through the International Mouse Phenotyping Consortium (IMPC) (www.mousephenotype.org). The individual mouse was considered to be the experimental unit in these studies. The data presented are a snapshot from September 2023 (Methods). Tabular data are also available at GitHub (https://github.com/team113sanger/Large-scale-analysis-of-genes-that-regulate-micronucleus-formation/tree/main/Mouse_Phenotyping_Data).