Abstract
SAC (for suppressor of actin) domain proteins in yeast and animals have been shown to modulate the levels of phosphoinositides, thereby regulating several cellular activities such as signal transduction, actin cytoskeleton organization, and vesicle trafficking. Nine genes encoding SAC domain–containing proteins are present in the Arabidopsis thaliana genome, but their roles in plant cellular functions and plant growth and development have not been characterized. In this report, we demonstrate the essential roles of one of the Arabidopsis SAC domain proteins, AtSAC1, in plant cellular functions. Mutation of the AtSAC1 gene in the fragile fiber7 (fra7) mutant caused a dramatic decrease in the wall thickness of fiber cells and vessel elements, thus resulting in a weak stem phenotype. The fra7 mutation also led to reduced length and aberrant shapes in fiber cells, pith cells, and trichomes and to an alteration in overall plant architecture. The AtSAC1 gene was found to be expressed in all tissues in elongating organs; however, it showed predominant expression in vascular tissues and fibers in nonelongating parts of stems. In vitro activity assay demonstrated that AtSAC1 exhibited phosphatase activity toward phosphatidylinositol 3,5-biphosphate. Subcellular localization studies showed that AtSAC1 was colocalized with a Golgi marker. Truncation of the C terminus by the fra7 mutation resulted in its localization in the cytoplasm but had no effect on phosphatase activity. Furthermore, examination of the cytoskeleton organization revealed that the fra7 mutation caused the formation of aberrant actin cables in elongating cells but had no effect on the organization of cortical microtubules. Together, these results provide genetic evidence that AtSAC1, a SAC domain phosphoinositide phosphatase, is required for normal cell morphogenesis, cell wall synthesis, and actin organization.
INTRODUCTION
Phosphoinositides have traditionally been known to be important in the generation of the second messengers inositol 1,4,5,-triphosphate and diacylglycerol. Recently, it was demonstrated that in yeast and animals, phosphoinositides themselves are regulators of a wide variety of cellular processes, such as signal transduction, actin cytoskeleton organization, vesicle trafficking, and activation of proteins such as phosphoinositide-dependent kinase 1 and phospholipase D (Martin, 1998; Takenawa and Itoh, 2001). In plant cells, all phosphoinositide forms except phosphatidylinositol 3,4,5-triphosphate [PtdIns(3,4,5)P3] have been identified, and they have been suggested to play important roles in vesicle trafficking (Matsuoka et al., 1995; Kim et al., 2001), pollen tube growth (Kost et al., 1999), and stress and hormone responses (Mikami et al., 1998; Meijer et al., 1999, 2001; Pical et al., 1999; DeWald et al., 2001). PtdIns(4,5)P2 has been shown to bind profilin (Kovar et al., 2001) and to regulate the activities of phospholipase Dδ (Qin et al., 2002) and an ATPase (Memon et al., 1989) in plants.
The synthesis and turnover of phosphoinositides are regulated by lipid kinases, lipid phosphatases, and phospholipases. Several plant kinases and phospholipase Cs involved in the metabolism of phosphoinositides have been analyzed at the molecular and genomic levels (Stevenson et al., 2000; Müeller-Röeber and Pical, 2002; Meijer and Munnik, 2003). However, much less is known about the biochemical activities and biological functions of phosphatases involved in the metabolism of phosphoinositides in plants. Recently, several plant inositol polyphosphate 5-phosphatases were demonstrated to hydrolyze phosphate from phosphoinositides (Ercetin and Gillaspy, 2004; Zhong and Ye, 2004; Zhong et al., 2004), and one of them, Fragile Fiber3 (FRA3), plays important roles in secondary wall synthesis and actin organization in fiber cells (Zhong et al., 2004).
Phosphoinositide phosphatases are classified into three main groups (i.e., 3-, 4-, or 5-phosphatases) based on the position of the phosphate on the inositol head group that they hydrolyze (Takenawa and Itoh, 2001). Recently, a novel group of phosphatases called SAC (for suppressor of actin) domain phosphatases have been demonstrated to hydrolyze phosphates on multiple positions of the inositol head group of phosphoinositides (Hughes et al., 2000a). The SAC domains of yeast Sac1p, yeast synaptojanin homologs inositol polyphosphate 5-phosphatase52p (Inp52p) and Inp53p, human synaptojanins, and rat rSac1 have been shown to possess phosphatase activities toward PtdIns(3)P, PtdIns(4)P, and PtdIns(3,5)P2 in vitro (Guo et al., 1999; Hughes et al., 2000b; Nemoto et al., 2000). By contrast, another yeast SAC domain protein, pheromone-regulated or induced gene4 (Fig4), exhibits PtdIns(3,5)P2-specific phosphatase activity (Rudge et al., 2004), and the human hSac2 possesses 5-phosphatase activity toward PtdIns(4,5)P2 and PtdIns(3,4,5)P3 (Minagawa et al., 2001). The SAC domain proteins in yeast and animals have been grouped into two classes. One class is represented by yeast Sac1p, which contains a SAC domain linked to a C-terminal region without any recognizable domains, except that some members in this class harbor two transmembrane helices. The other class includes human and yeast synaptojanins, in which the N-terminal-localized SAC domain is linked to a C-terminal type II 5-phosphatase domain (Hughes et al., 2000a).
Figure 4.
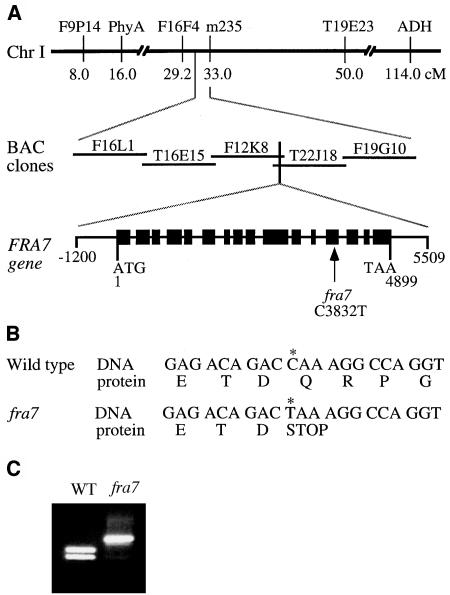
Map-Based Cloning of the FRA7 Gene and Nature of the fra7 Mutation.
(A) Map-based cloning of FRA7. The fra7 locus was mapped to a 63-kb region covered by BAC clones F12K8 and T22J18 on chromosome 1. The FRA7 gene is composed of 16 exons and 15 introns. The fra7 mutation causes a C-to-T transition in the 13th exon. Black boxes indicate exons, and lines between exons indicate introns in the FRA7 gene diagram. Chr, chromosome; cM, centimorgan.
(B) Nucleotide and amino acid sequences around the fra7 mutation site. The fra7 mutation changes a wild-type codon that encodes Gln into a stop codon (asterisk).
(C) Elimination of a XcmI site in the fra7 mutant gene. The single nucleotide mutation in fra7 occurs at a XcmI restriction endonuclease cleavage site. This is revealed by digestion of the PCR-amplified DNA fragments with XcmI, which shows that the XcmI site is missing in the fra7 mutant DNA compared with the wild type (WT).
The SAC domains from both classes of proteins are ∼400 amino acids in length and contain seven conserved motifs. In particular, the sequence RXNCXDCLDRTN within the sixth motif is highly conserved among all known SAC domain–containing proteins, and it has been proposed to be the catalytic core of SAC domain phosphatases (Hughes et al., 2000a). The importance of this core sequence in the activity of the SAC domains is demonstrated in Sac1p and Inp51p. The first conserved Asp residue was mutated in the yeast sac1-8 and sac1-22 mutants, and these mutations have been shown to abolish the phosphatase activity of Sac1p (Kearns et al., 1997). The SAC domain of Inp51p has alterations in three conserved residues of the core sequence, and concomitantly, its SAC domain does not possess any phosphatase activities (Guo et al., 1999).
Sac1p was originally identified as a suppressor of the defects associated with certain actin mutant alleles (Novick et al., 1989) and a suppressor of the defects caused by mutations of the Sec14 (an allele causing a block in the secretory pathway [sec] with the Golgi apparatus) phosphatidylinositol/phosphatidylcholine transfer protein (Cleves et al., 1989). Subsequent studies have shown that Sac1p is involved mainly in the regulation of the PtdIns(4)P pool in vivo and that this regulation is essential for many cellular activities in yeast, such as actin cytoskeleton organization, Golgi function, maintenance of vacuole morphology, and regulation of lipid storage (Foti et al., 2001). Mutational analysis of Fig4 indicates its roles in actin organization and cell morphogenesis during mating (Erdman et al., 1998) and in the regulation of vacuole morphology (Rudge et al., 2004). Recently, it was reported that mutation of Drosophila SAC1 caused a change in cell shape and an alteration in Jun N-terminal kinase signaling, accompanied by an embryo-lethal phenotype (Wei et al., 2003). The cellular functions of other animal SAC domain proteins have yet to be determined (Nemoto et al., 2000; Minagawa et al., 2001).
A recent genomic analysis revealed the presence of nine genes encoding SAC domain–containing proteins in Arabidopsis thaliana and several SAC homologs in other plant species (Zhong and Ye, 2003). Three Arabidopsis SAC proteins have been demonstrated to be able to rescue the cold-sensitive and inositol auxotroph yeast sac1-null mutant strain, and like yeast Sac1p, they are localized in the endoplasmic reticulum (ER) (Despres et al., 2003). However, the roles of SAC domain proteins in plant cellular functions and plant growth and development remain unknown.
We have been using interfascicular fiber cells in Arabidopsis inflorescence stems to study the molecular mechanisms underlying cell elongation and cell wall synthesis (Zhong et al., 2001). Among several defective fiber mutants identified, we found that the fra7 mutation caused a decrease in cell wall synthesis, a reduction in cell elongation, and an alteration in the normal organization of the actin cytoskeleton. In addition, fra7 exhibited a global change in plant architecture. The gene responsible for the fra7 mutation was cloned and shown to be AtSAC1, which encodes a SAC domain protein. Recombinant AtSAC1 protein was demonstrated to exhibit phosphatase activity toward PtdIns(3,5)P2 in vitro. Truncation of the C terminus of AtSAC1 by the fra7 mutation was found to cause an alteration in its subcellular localization but no effect on phosphatase activity. These results demonstrate that AtSAC1 plays important roles in diverse plant cellular functions.
RESULTS
The fra7 Mutant Is Defective in the Wall Thickening and Elongation of Fiber Cells
Arabidopsis inflorescence stems develop interfascicular fibers that provide the main mechanical strength to the mature stems (Zhong et al., 1997). We screened for fiber mutants based on the property of the mechanical strength of the stems. The fra7 mutant thus isolated exhibited a dramatic reduction in the breaking strength of stems (Figure 1A). The force required to break the basal mature stems was four times lower in fra7 compared with the wild type.
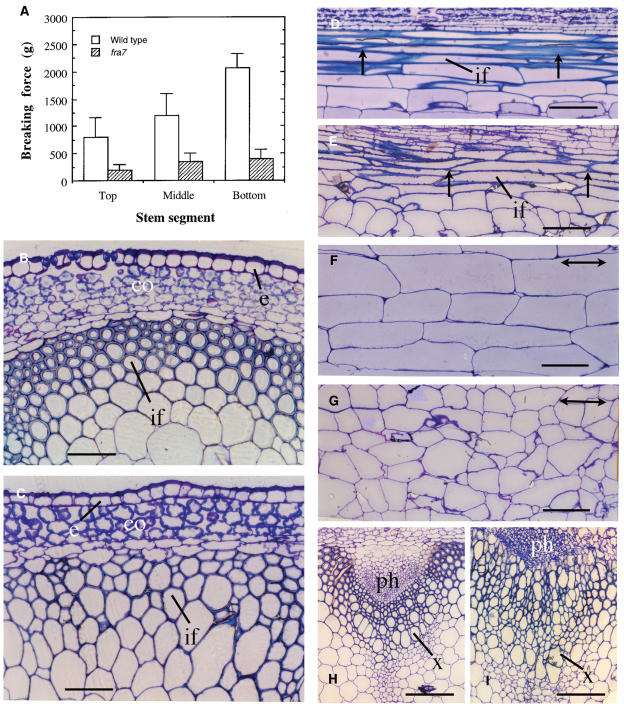
Figure 1.
Effects of the fra7 Mutation on Stem Strength and Cell Morphology.
The main inflorescence stems of 8-week-old plants were used for breaking force measurements. The bottom internodes of main inflorescence stems of 8-week-old plants were used for examination of cell morphology.
(A) Quantitative measurement of breaking force showing that the force needed to break stems apart was three to four times lower in fra7 than in the wild type. Data are means ± se of 20 plants.
(B) and (C) Cross sections of interfascicular regions of stems showing fra7 fiber cells with irregular shapes and thin walls (C) compared with the wild type (B).
(D) and (E) Longitudinal sections of interfascicular regions of stems showing fra7 fiber cells with irregular shapes and shorter length (E) compared with the wild type (D). The arrows mark the two ends of a fiber cell.
(F) and (G) Longitudinal sections of stems showing short and misshapen fra7 pith cells (G) compared with the wild type (F). Double-headed arrows represent the elongation axis of the stems.
(H) and (I) Cross sections of vascular bundles in stems showing fra7 vessel elements with irregular shapes (I) compared with the wild type (H).
co, cortex; e, epidermis; if, interfascicular fiber; ph, phloem; x, xylem. Bars = 65 μm in (B) and (C), 126 μm in (D) and (E), 85 μm in (F) and (G), and 120 μm in (H) and (I).
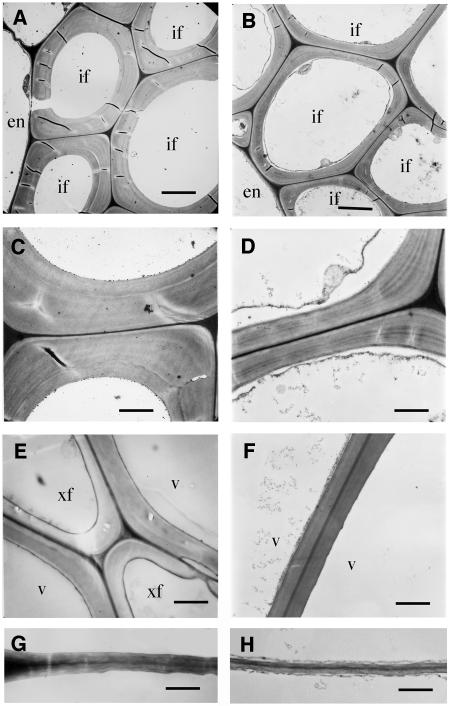
To investigate what defects in the interfascicular fibers caused the reduced stem strength, we examined the anatomical features of fiber cells. Cross sections showed that although fra7 developed interfascicular fibers as in the wild type, they apparently had an altered morphology and a reduction in wall thickness (Figures 1B and 1C). The mutant fiber cells appeared to be larger in diameter and less regular in shape compared with those of the wild type. Longitudinal sections showed that the mutant fiber cells were misshapen and much shorter compared with wild-type cells (Figures 1D and 1E). Examination of fiber wall thickness by transmission electron microscopy revealed that the wall thickness of fra7 fiber cells was reduced to 51% of that of the wild type (Figures 2A to 2D, Table 1). These results demonstrated that fra7 caused a defect in cell elongation and secondary wall synthesis in interfascicular fiber cells that likely resulted in the reduced stem strength.
Figure 2.
The fra7 Mutation Reduces the Wall Thickness of Fibers, Vessels, and Pith Cells.
The bottom internodes of main inflorescence stems of 8-week-old plants were used for transmission electron microscopy of cell walls.
(A) and (B) Interfascicular fiber cells showing thin walls in fra7 (B) compared with the wild type (A).
(C) and (D) High magnification of fiber walls showing thin secondary walls in fra7 (D) compared with the wild type (C).
(E) and (F) Vessels walls are thinner in fra7 (F) than in the wild type (E).
(G) and (H) Pith cell walls are thinner in fra7 (H) than in the wild type (G).
en, endodermis; if, interfascicular fiber; v, vessel; xf, xylary fiber. Bars = 5.2 μm in (A) and (B), 1.8 μm in (C) and (D), 2.3 μm in (E) and (F), and 1.5 μm in (G) and (H).
Table 1.
Wall Thickness of Fibers, Vessels, and Pith Cells in the Stems of Wild-Type and fra7 Mutant Plants
| Sample | Fiber Cells | Vessels | Pith Cells |
|---|---|---|---|
| Wild type | 2.53 ± 0.34 | 1.14 ± 0.10 | 0.53 ± 0.05 |
| fra7 | 1.28 ± 0.14 | 0.80 ± 0.07 | 0.34 ± 0.03 |
Wall thickness was measured from transmission electron micrographs of fibers, vessels, and pith cells. Data are means (μm) ± se from 20 cells.
The observation that the fra7 mutation affected the wall thickness of fiber cells prompted us to analyze the cell wall composition in the mutant. Analysis of cell walls from inflorescence stems showed that crystalline cellulose in the mutant was reduced to 87% of that in the wild type (Table 2). Cell wall sugar composition analysis revealed a slight decrease in the amount of glucose and xylose in the mutant compared with the wild type (Table 2). Because the reduction in fiber wall thickness was much greater than that in cellulose and glucose content, these results indicate that fra7 most likely causes a reduction in overall cell wall synthesis rather than specifically affects cellulose synthesis.
Table 2.
Cell Wall Composition of the Stems of Wild-Type and fra7 Mutant Plants
| Cell Wall Sugar Composition
|
|||||||
|---|---|---|---|---|---|---|---|
| Sample | Cellulose | Glucose | Xylose | Mannose | Galactose | Arabinose | Rhamnose |
| Wild type | 295 ± 21 | 354 ± 26 | 139 ± 13 | 13.6 ± 0.2 | 11.1 ± 2.5 | 11.5 ± 1.9 | 1.6 ± 0.9 |
| fra7 | 256 ± 14 | 293 ± 21 | 120 ± 7 | 11.8 ± 2.7 | 13.5 ± 1.4 | 11.2 ± 0.6 | 1.6 ± 0.1 |
Cell wall residues used for composition analysis were prepared from mature stems of 10-week-old plants. Data are means (mg/g dry cell wall) ± se of three independent assays.
Effects of fra7 on the Morphology of Nonfiber Cells
In addition to the abnormal fiber cells, we found that fra7 affected the morphology of other cell types in stems. The most noticeable alteration was seen in longitudinal sections of pith cells. Although wild-type pith cells were rectangular and arranged in longitudinal files along the elongation axis of stems (Figure 1F), the majority of pith cells in fra7 had irregular shapes and, consequently, did not exhibit regular cell files (Figure 1G). In addition, the shapes of vessel elements in the xylem bundles were also altered in the mutant (Figures 1H and 1I). Transmission electron microscopy showed that compared with the wild type, the wall thickness of fra7 vessel elements and pith cells was decreased by 30 and 36%, respectively (Figures 2E to 2H, Table 1). These results demonstrated that the fra7 mutation caused defects in cellular morphogenesis and cell wall synthesis in both fiber cells and nonfiber cells.
The fra7 Mutation Affects Overall Plant Growth and Architecture
To determine whether the fra7 mutation affected overall plant growth, we examined the morphology of plants at different developmental stages (Figure 3). Shorter roots and hypocotyls were seen in 4-d-old light-grown and dark-grown fra7 seedlings, respectively, compared with the wild type (Figures 3A and 3B, Table 3). The height of fra7 inflorescence stems was also reduced (Figure 3C, Table 3), apparently as a result of the reduction in cell length, as seen in pith cells (Figure 1G). Overall plant morphology was altered dramatically by the fra7 mutation. Both the main inflorescence stems and cauline branches were crooked instead of the relatively straight stature seen in the wild type (Figures 3C to 3E). The angles of cauline branches, cauline leaves, and siliques relative to the stem axis were widened in the mutant compared with the wild type. Quantitative measurement showed that the average angle of cauline branches was increased by 48% in the mutant (Table 3).
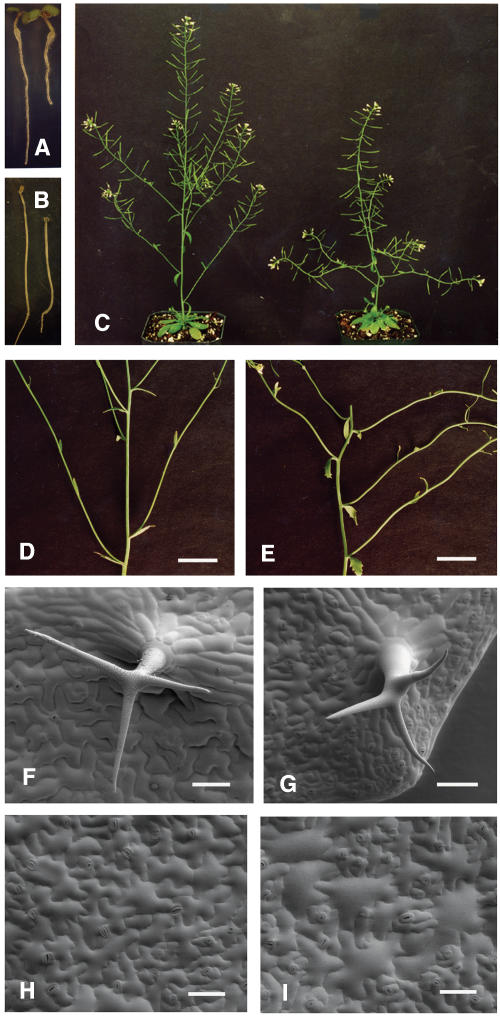
Figure 3.
Alteration of Plant Morphology by the fra7 Mutation.
(A) Four-day-old light-grown seedlings showing shorter root in fra7 (right) compared with the wild type (left).
(B) Four-day-old dark-grown seedlings showing shorter hypocotyl in fra7 (right) compared with the wild type (left).
(C) Eight-week-old plants showing shorter and wavy stems in fra7 (right) compared with the wild type (left).
(D) and (E) Inflorescence stem segments showing wavy stems and wider angles between the main stem and cauline branches in fra7 (E) compared with the wild type (D).
(F) and (G) Scanning electron micrographs of trichomes showing thicker stalk and shorter, wavy branches in fra7 (G) compared with the wild type (F).
(H) and (I) Scanning electron micrographs of leaf epidermis showing some swollen epidermal cells in fra7 (I) compared with the wild type (H).
Bars = 9 mm in (D) and (E), 84 μm in (F) and (G), and 63 μm in (H) and (I).
Table 3.
Length of Cells and Organs of Wild-Type and fra7 Mutant Plants
| Sample | Wild Type | fra7 |
|---|---|---|
| Pith cell lengtha (μm) | 184 ± 56 | 62 ± 20 |
| Interfascicular fiber cell lengtha (μm) | 422 ± 125 | 232 ± 79 |
| Inflorescence stem lengthb (cm) | 29.5 ± 3.6 | 17.8 ± 4.2 |
| Cauline branch anglec (°) | 35.4 ± 5.6 | 52.6 ± 11.5 |
| Root lengthd (mm) | 10.4 ± 1.5 | 7.1 ± 1.2 |
| Hypocotyl lengthe (mm) | 15.4 ± 1.5 | 11.3 ± 1.1 |
Data are means ± se from 20 to 25 samples.
Pith and fiber cells were from basal parts of inflorescence stems of 8-week-old plants.
Eight-week-old plants were measured for stem height.
Angles between cauline branches and main inflorescence stems were measured.
Roots of 4-d-old light-grown seedlings were used for measurement.
Hypocotyls of 4-d-old dark-grown seedlings were measured.
Furthermore, we found that the development of trichomes was affected by the fra7 mutation. Trichomes in the wild type typically had a short stalk and three long, pointy branches (Figure 3F). Although the number of branches was not altered in the trichomes of fra7, the fra7 trichomes appeared to have a much thicker stalk and shorter, crooked branches (Figure 3G). Examination of leaf epidermal cells revealed that some ordinary epidermal cells in the fra7 mutant were swollen and less sinuous in shape compared with those in the wild type (Figures 3H and 3I). Together, these results demonstrated that fra7 caused morphological alterations at both the cellular and organ levels.
Map-Based Cloning of FRA7
To investigate the molecular mechanisms responsible for the cellular defects in fra7, we undertook the cloning of the FRA7 gene. Because fra7 was isolated from an ethyl methanesulfonate–mutagenized population of Arabidopsis (ecotype Columbia), a map-based cloning approach was used. The mutant was crossed with the wild-type ecotype Landsberg erecta, and homozygous fra7 plants were selected from the F2 plants and used for mapping with the codominant amplified polymorphic sequence (CAPS) markers (Konieczny and Ausubel, 1993). The fra7 locus was found to be closely linked to the F9P14 marker on chromosome 1 (Figure 4A). Further mapping with adjacent markers indicated that the fra7 locus resided between markers F16F4 and m235. Based on the sequence information of overlapping BAC clones between these two markers, we designed additional CAPS markers and used them to gradually narrow the fra7 locus to a 63-kb region covered by BAC clones F12K8 and T22J18 (Figure 4A).
According to the gene annotations of chromosome 1 from the Arabidopsis genome database, the 63-kb region where the fra7 locus resides encompasses 11 putative genes. To determine which of these genes carried the fra7 mutation, we sequenced all 11 genes from fra7. By comparing the gene sequences from the mutant with those from the wild type, we found a point mutation (C-to-T) in one of the genes, F12K8.3. The F12K8.3 gene is also named T22J18.20 because it is located in the overlapping region of BAC clones F12K8 and T22J18. The C-to-T mutation in F12K8.3 was further revealed by loss of the XcmI site in fra7 (Figure 4C).
To confirm that the C-to-T mutation in F12K8.3 was responsible for the phenotypes conferred by fra7, we introduced the wild-type F12K8.3 gene into fra7 by Agrobacterium tumefaciens–mediated transformation. Expression of the wild-type F12K8.3 gene in fra7 plants completely rescued the phenotypes conferred by fra7, including the stem mechanical strength, the length and wall thickness of fiber cells, the shape of pith cells, and the whole plant morphology (data not shown). These results unequivocally demonstrated that the C-to-T mutation in F12K8.3 resulted in the phenotypes conferred by fra7; therefore, F12K8.3 represents the FRA7 gene.
Nature of the fra7 Mutation
The FRA7 gene consists of 4899 bp from the start codon to the stop codon. It is organized into 16 exons and 15 introns (Figure 4A). The fra7 mutation occurs in the 13th exon. Comparison of the wild-type and mutant cDNAs and their deduced amino acid sequences revealed that the fra7 mutation changed a Gln codon CAA into a stop codon TAA (Figure 4B), which results in a truncated protein with a deletion of 199 amino acid residues. The deduced FRA7 protein consists of 912 amino acids with a predicted molecular mass of 102,812 D and a predicted pI of 6.2.
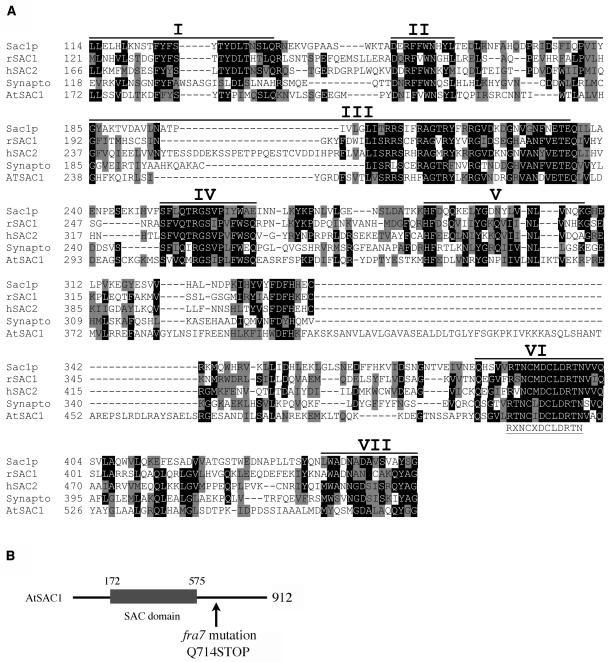
A BLAST search of the GenBank database revealed that FRA7 contains a domain that shares high sequence similarity with the SAC domains of a group of proteins found in yeast and animals. The SAC domains in yeast and animal proteins are ∼400 amino acids in length and contain seven conserved motifs (Hughes et al., 2000a). The putative SAC domain in FRA7 is also ∼400 amino acids in length and shares 26% identity and 55% similarity with the yeast and animal SAC domains (Figure 5A). The FRA7 SAC domain retained all seven conserved motifs. In particular, the proposed catalytic core sequence RXNCXDCLDRTN, which is located in the sixth motif, is completely conserved in the putative FRA7 SAC domain (Figure 5A). The Arabidopsis genome has been shown to contain nine genes encoding SAC domain proteins AtSAC1 to AtSAC9 (Zhong and Ye, 2003), and FRA7 represents AtSAC1. Therefore, the name AtSAC1 will be used hereafter. The fra7 nonsense mutation occurred in the C-terminal region outside the SAC domain (Figure 5B), indicating that the C-terminal region of AtSAC1 is essential for its cellular functions.
Figure 5.
Amino Acid Sequence Alignment of the SAC Domains of AtSAC1 and Several SAC Domain Proteins from Yeast and Animals.
(A) The SAC domain sequences from yeast Sac1 (Sac1p), rat SAC1 (rSAC1), human SAC2 (hSAC2), human synaptojanin I (Synapto), and Arabidopsis SAC1 (AtSAC1) were aligned using the CLUSTAL W 1.8 program. The numbers at left of the individual sequences indicate the positions of amino acid residues in the corresponding proteins. Gaps (marked with dashes) were introduced to maximize the sequence alignment. Identical and similar amino acid residues are shaded with black and gray, respectively. The seven conserved motifs defined for the SAC domains of yeast and animal proteins (Hughes et al., 2000a) are marked with solid lines above the sequences. The putative catalytic core sequence of the SAC domain phosphatase is located in motif VI, and its consensus is shown below the sequence.
(B) Diagram of the AtSAC1 protein showing the location of the SAC domain and the site of the nonsense fra7 mutation.
Expression of the AtSAC1 Gene Is Developmentally Regulated
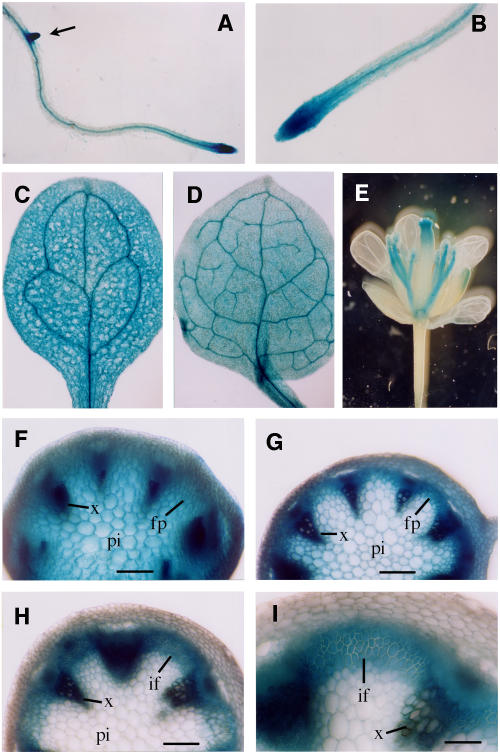
To investigate the expression pattern of AtSAC1, we used the β-glucuronidase (GUS) reporter gene to examine its expression in different tissues. A 1.2-kb 5′ upstream sequence and the entire exon and intron region of AtSAC1 were ligated in frame with the GUS reporter gene in the binary vector pBI101. The AtSAC1-GUS construct was introduced into both wild-type and fra7 plants. Expression of the AtSAC1-GUS construct in fra7 plants was found to completely rescue the phenotypes conferred by fra7 (data not shown), indicating that the AtSAC1 sequence used in the construct contains all elements responsible for the expression of the endogenous AtSAC1 gene. Analysis of GUS activity in transgenic plants revealed that GUS staining was highly present in the root apex and gradually became concentrated in vascular tissues in the mature region of the root (Figures 6A and 6B). In young cotyledons (Figure 6C) and leaves (Figure 6D), GUS staining was seen in all tissues, with more prominent staining in vascular strands. In flowers, the staining was seen mainly in filaments and in the apical region of the style (Figure 6E).
Figure 6.
Expression Pattern of AtSAC1 Revealed by the GUS Reporter Gene.
The AtSAC1 gene, including a 1.2-kb 5′ upstream sequence and the entire exon and intron sequence, was ligated in frame with the GUS reporter gene, and the construct was introduced into Arabidopsis plants. Various organs of the transgenic plants were examined for GUS activity, which is shown as blue coloring.
(A) Primary root of a 3-d-old seedling showing strong GUS staining at the apex and an emerging lateral root (arrow).
(B) High magnification of (A) showing intense GUS staining at the root apex and more concentrated staining in the vascular cylinder in the mature region.
(C) and (D) Young cotyledon (C) and leaf (D) showing GUS staining in all tissues with more prominent staining in vascular strands.
(E) Flower showing GUS staining in filaments and the apical region of the style.
(F) Section from a young elongating internode showing GUS staining in all tissues.
(G) Section from an internode near the end of elongation showing intense GUS staining in vascular bundles and interfascicular regions.
(H) Section of a nonelongating internode showing GUS staining predominantly in vascular bundles and interfascicular fibers.
(I) High magnification of (H) showing fiber cells with thick secondary walls and intense GUS staining.
fp, fiber precursor; if, interfascicular fiber; pi, pith; x, xylem. Bars = 180 μm in (F) to (H) and 360 μm in (I).
Examination of the GUS activity in inflorescence stems showed that although staining was present in all tissues in rapidly elongating internodes (Figure 6F), it was more concentrated in interfascicular fiber cells and vascular bundles in the internodes that were near the end of elongation (Figure 6G). In nonelongating internodes in which fiber cells were undergoing secondary wall thickening (Ye et al., 2002), GUS staining was found only in fiber cells and vascular bundles (Figures 6H and 6I). These results indicate that the expression of AtSAC1 is developmentally regulated. Its ubiquitous expression in elongating cells and continuous expression in developing fiber cells are consistent with the phenotypes conferred by fra7 of defective cell elongation and secondary wall thickening.
AtSAC1 Exhibits Phosphatase Activity toward PtdIns(3,5)P2
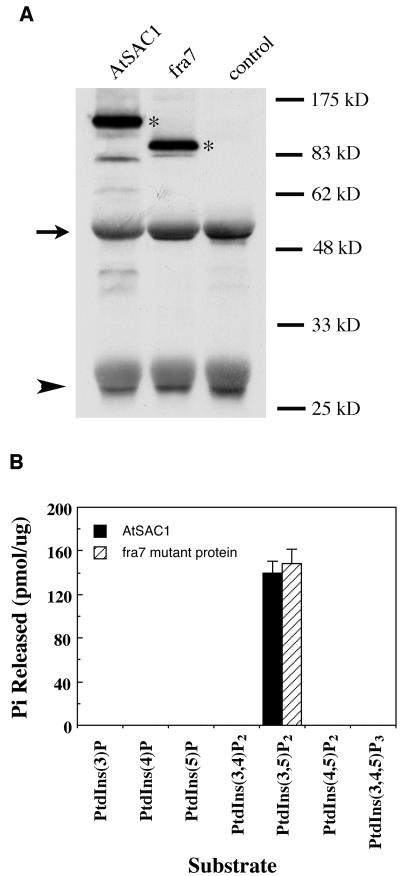
To investigate whether AtSAC1 possesses phosphoinositide phosphatase activity, we expressed a V5 epitope–tagged recombinant AtSAC1 protein in yeast for the activity assay. The recombinant protein was purified by immunoprecipitation with a monoclonal antibody against the V5 epitope (Figure 7A) and assayed for its phosphatase activity toward various phospholipids. AtSAC1 was found to hydrolyze phosphate from PtdIns(3,5)P2 but not from other phospholipids (Figure 7B). AtSAC1 did not exhibit any phosphatase activity toward the water-soluble inositol polyphosphates Ins(1,4,5)P3 and Ins(1,3,4,5)P4 (data not shown), in agreement with yeast and animal SAC domain phosphatases that only hydrolyze phosphate from phospholipids (Hughes et al., 2000a). Because the fra7 mutation caused a truncation of the C terminus beyond the SAC domain, we tested whether the fra7 mutant protein still retains phosphatase activity. It was found that recombinant fra7 mutant protein exhibited phosphatase activity toward PtdIns(3,5)P2 (Figure 7B) identical to that of the full-length AtSAC1. These results demonstrate that AtSAC1 is a PtdIns(3,5)P2 phosphatase and that truncation of the C terminus by fra7 does not affect its activity.
Figure 7.
AtSAC1 Exhibits Phosphatase Activity toward PtdIns(3,5)P2.
The V5-tagged wild-type AtSAC1 and fra7 mutant proteins were expressed in yeast, purified by immunoprecipitation, and used for assay of their phosphatase activities.
(A) Detection of recombinant AtSAC1 and fra7 expressed in yeast. Immunoprecipitated proteins were detected with a monoclonal antibody against the V5 epitope. Note that in additional to the recombinant AtSAC1 (107 kD) and fra7 (84 kD) proteins (asterisks), the heavy chain (arrow) and light chain (arrowhead) of the antibody used for immunoprecipitation were also revealed. Immunoprecipitates from yeast cells harboring the expression vector alone were used as a control.
(B) Phosphatase activity of AtSAC1 and fra7 mutant protein toward various phosphoinositides. Various phosphoinositides were incubated with the fusion protein immunoprecipitates, and the amount of free phosphate released was measured by the malachite green method. Both wild-type AtSAC1 and fra7 mutant proteins exhibit phosphatase activity toward PtdIns(3,5)P2 but not any other phosphoinositides. The immunoprecipitates from yeast cells harboring the expression vector alone were used as a control in the assay, and no activity was detectable. Data are means ± se of two assays.
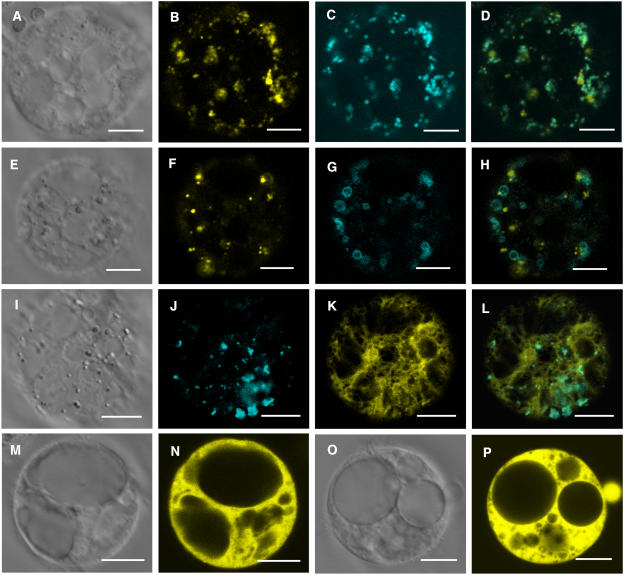
Subcellular Localization of the AtSAC1 Protein
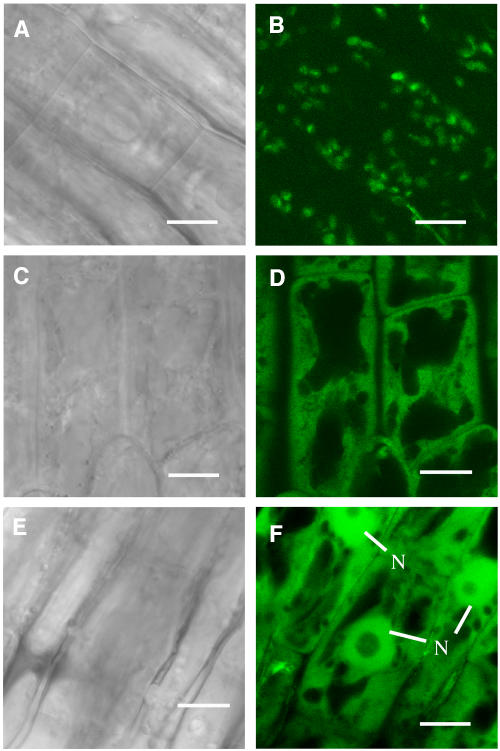
Because truncation of the AtSAC1 C terminus by the fra7 mutation did not affect its phosphatase activity, we next investigated whether there were any differences in the subcellular localization of AtSAC1 and the fra7 mutant protein. AtSAC1 and fra7 were expressed as green fluorescent protein (GFP) fusion proteins in transgenic Arabidopsis plants, and the GFP signals in root epidermal cells were visualized with a confocal microscope. It was found that although AtSAC1-GFP displayed a punctate pattern (Figures 8A and 8B), fra7-GFP was distributed throughout the cytoplasm (Figures 8C and 8D). These results indicate that AtSAC1-GFP is associated with certain organelles and that truncation of the AtSAC1 C terminus by fra7 alters its subcellular localization. The GFP control protein was seen in both cytoplasm and nucleus (Figures 8E and 8F), consistent with the subcellular localization pattern of the GFP control protein reported previously (Li et al., 2001; Zhong et al., 2002).
Figure 8.
Subcellular Localization of GFP-Tagged AtSAC1 and fra7 Mutant Protein in Arabidopsis Root Cells.
GFP-tagged AtSAC1 and fra7 mutant proteins were expressed in Arabidopsis plants, and their subcellular locations were examined with a laser confocal microscope.
(A) and (B) Differential interference contrast (DIC) image (A) of root epidermal cells of 3-d-old Arabidopsis seedlings expressing AtSAC1-GFP and the corresponding AtSAC1-GFP signals (B). Note that the GFP signals show a punctate pattern in the cells.
(C) and (D) DIC image (C) of root epidermal cells of 3-d-old seedlings expressing fra7-GFP and the corresponding fra7-GFP signals (D). The GFP signals are distributed throughout the cytoplasm.
(E) and (F) DIC image (E) of root epidermal cells of 3-d-old seedlings expressing GFP alone and the corresponding GFP signals (F). The GFP signals are present throughout the cytoplasm and the nucleus.
N, nucleus. Bars = 8 μm.
To ascertain the exact subcellular location of AtSAC1, we used carrot (Daucus carota) protoplasts to study the colocalization of AtSAC1 with various organelle markers. Like AtSAC1-GFP in Arabidopsis root cells, AtSAC1–enhanced yellow fluorescent protein (EYFP) and AtSAC1–enhanced cyan fluorescent protein (ECFP) displayed punctate localization patterns in carrot protoplasts (Figure 9). It was found that the punctate pattern of AtSAC1-EYFP was almost identical to the localization pattern of the Golgi marker Arabidopsis proton-translocating pyrophosphatase2 (AVP2) (Figures 9A to 9D) (Mitsuda et al., 2001) but showed obvious differences from those of the prevacuolar membrane marker Ras-related small GTP binding protein1 (Rha1) (Figures 9E to 9H) (Lee et al., 2004) and an ER marker (Figures 9I to 9L). These results suggest that AtSAC1 is associated with Golgi in the cells.
Figure 9.
Subcellular Localization of EYFP- or ECFP-Tagged AtSAC1 and fra7 Mutant Protein in Carrot Protoplasts.
EYFP- or ECFP-tagged AtSAC1 and fra7 were expressed in carrot protoplasts, and their subcellular locations were examined with a laser confocal microscope.
(A) to (D) DIC image (A) of a carrot cell expressing AtSAC1-EYFP and the Golgi marker AVP2-ECFP and the corresponding AtSAC1-EYFP signals (B), AVP2-ECFP signals (C), and a merged image (D). It is evident that the AtSAC1-EYFP and AVP2-ECFP signals are overlapped.
(E) to (H) DIC image (E) of a carrot cell expressing AtSAC1-EYFP and the prevacuolar membrane marker Rha1-ECFP and the corresponding AtSAC1-EYFP signals (F), Rha1-ECFP signals (G), and a merged image (H).
(I) to (L) DIC image (I) of a carrot cell expressing AtSAC1-ECFP and the ER marker EYFP-ER and the corresponding AtSAC1-ECFP signals (J), EYFP-ER signals (K), and a merged image (L).
(M) and (N) DIC image (M) of a carrot cell expressing fra7-EYFP and the corresponding fra7-EYFP signals (N). Note that the signals are present throughout the cytoplasm.
(O) and (P) DIC image (O) of a carrot cell expressing EYFP alone and the corresponding EYFP signals (P) that show cytoplasmic localization.
Bars = 22 μm in (A) to (H), 26 μm in (I) to (L), and 40 μm in (M) to (P).
As in the Arabidopsis root cells, fra7-EYFP was distributed throughout the cytoplasm in carrot protoplasts (Figures 9M and 9N), a localization pattern similar to that of the cytoplasm-localized EYFP control protein (Figures 9O and 9P). These observations indicate that the C terminus of AtSAC1 is essential for its proper subcellular localization in the cells. It is interesting that the C terminus of yeast Fig4, another SAC domain PtdIns(3,5)P2 phosphatase, is also required for its proper subcellular localization (Rudge et al., 2004).
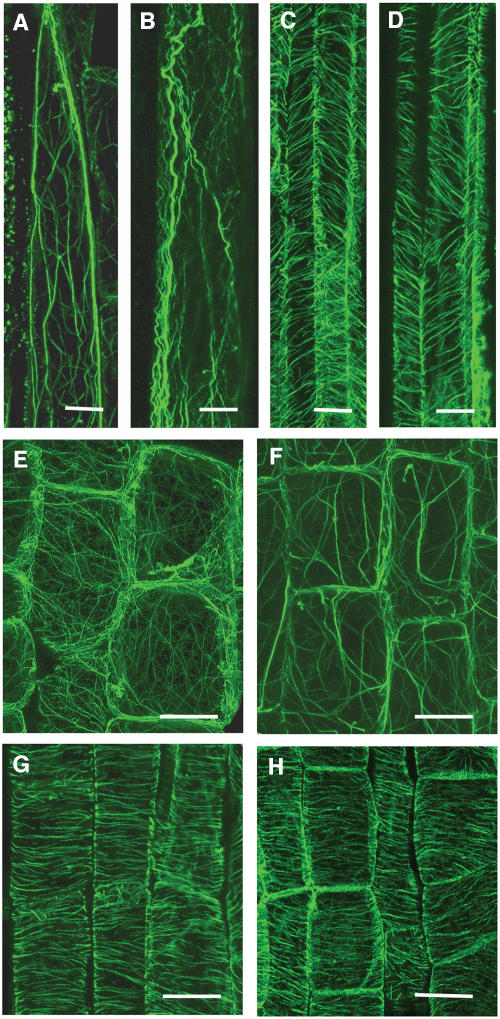
Organization of Actin and Microtubule Cytoskeletons in fra7 Cells
It has been demonstrated that yeast Sac1p influences the organization of the actin cytoskeleton in yeast cells (Foti et al., 2001). To investigate whether AtSAC1 was involved in cytoskeletal regulation, we examined the organization of filamentous actin (F-actin) and cortical microtubules in interfascicular fiber cells and pith cells in the stems (Figure 10). Immunocytochemical study showed that in the elongating fiber cells of the wild type, the F-actin cables formed a fine network (Figure 10A). By contrast, in the elongating fiber cells of fra7, the F-actin cables appeared to lose their fine network organization (Figure 10B). A similar alteration in F-actin organization was also seen in the elongating pith cells of fra7 stems (Figures 10E and 10F). Examination of the organization of cortical microtubules in elongating fibers and pith cells did not show any apparent differences between fra7 and the wild type (Figures 10C, 10D, 10G, and 10H). These results demonstrated that the fra7 mutation affected the organization of the actin cytoskeleton but had no effect on cortical microtubules.
Figure 10.
The fra7 Mutation Causes an Alteration in Actin Organization in Elongating Fibers and Pith Cells.
Longitudinal sections of elongating stems were immunodetected for microtubules and actin filaments with monoclonal antibodies against actin or α-tubulin and fluorescein isothiocyanate–conjugated secondary antibodies. Fluorescence-labeled actin filaments and cortical microtubules were viewed with a confocal laser microscope.
(A) and (B) Elongating fiber cells showing fine actin filaments in the wild type (A) and thick actin cables in fra7 (B).
(C) and (D) Elongating fiber cells showing cortical microtubules aligned in parallel in both the wild type (C) and fra7 (D).
(E) and (F) Elongating pith cells showing fine actin filaments in the wild type (E) and thick actin cables in fra7 (F).
(G) and (H) Elongating pith cells showing cortical microtubules aligned transversely in both the wild type (G) and fra7 (H).
Bars = 6 μm in (A) and (B), 15 μm in (C) to (F), and 20 μm in (G) and (H).
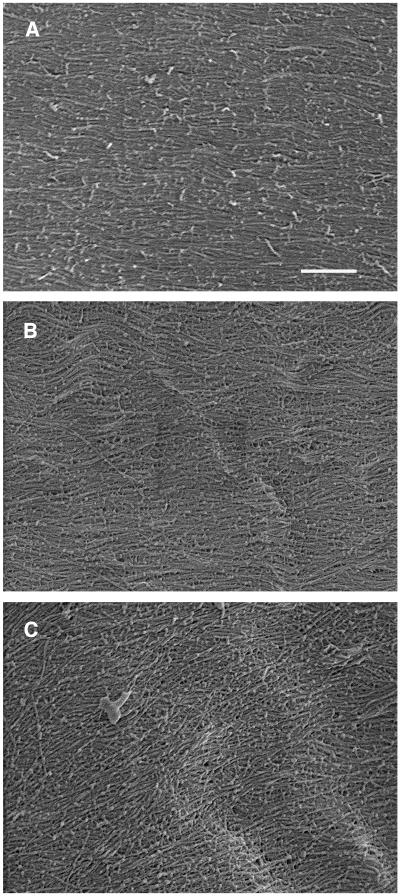
Orientation of Cellulose Microfibrils in the Walls of fra7 Pith Cells
It has been shown that F-actin influences cellulose microfibril deposition in brown algal cells (Katsaros et al., 2002). To investigate whether the cellulose microfibril deposition was altered in the fra7 mutant, we examined the cellulose microfibril orientation in pith cells using field emission scanning electron microscopy. In the innermost layer of walls of wild-type elongating cells, the cellulose microfibrils ran in parallel and were oriented transversely along the elongation axis (Figure 11A). This is the typical orientation pattern of cellulose microfibrils that has been reported in elongating cells of several Arabidopsis organs (Sugimoto et al., 2000; Burk and Ye, 2002). The cellulose microfibrils in the walls of elongating fra7 pith cells were generally oriented transversely along the long axis (Figure 11B). It was noticed that occasionally, in the walls of some misshapen cells, the cellulose microfibrils were deposited with slight deviations from the transverse orientation (Figures 11C), but no random orientation was observed. These results indicate that the fra7 mutation had no apparent effect on cellulose microfibril deposition.
Figure 11.
Visualization of Cellulose Microfibrils in the Innermost Layer of Pith Cell Walls.
Elongating stems were sectioned longitudinally through pith cells, and microfibrils in the innermost layer of pith cell walls were visualized using a field emission scanning electron microscope. The vertical direction of the images corresponds to the elongating axis of pith cells.
(A) Cellulose microfibrils in a wild-type cell showing their parallel and transverse alignment.
(B) and (C) Cellulose microfibrils in fra7 cells showing their parallel and transverse alignment (B) and, occasionally, parallel alignment with a small deviation (C).
Bar in (A) = 323 nm for (A) to (C).
DISCUSSION
SAC domain proteins were first identified in yeast and subsequently in animals, and their essential roles in diverse cellular activities have been ascertained in yeast. However, little is known about the cellular functions of SAC domain–containing proteins in plants. A recent genome analysis revealed that the Arabidopsis genome contains nine genes encoding SAC domain–containing proteins (Zhong and Ye, 2003). Three of these SAC domain proteins contain transmembrane helices and have been shown to be localized in the ER (Despres et al., 2003). Our study provides genetic evidence that AtSAC1, a plant SAC domain protein, plays important roles in cell morphogenesis, cell wall synthesis, and actin cytoskeleton organization.
AtSAC1 Is a PtdIns(3,5)P2 Phosphatase Colocalized with a Golgi Marker
Like the SAC domains of yeast and animal proteins, the SAC domain of AtSAC1 contains all of the seven conserved motifs thought to be important for SAC phosphatase activities. In particular, the putative catalytic core sequence located in the sixth motif is completely conserved in AtSAC1. Biochemical analysis revealed that AtSAC1 possesses phosphatase activity toward PtdIns(3,5)P2, demonstrating that AtSAC1 is indeed a phosphoinositide phosphatase. It is apparent that AtSAC1 exhibits the same biochemical activity as yeast Fig4, a SAC domain protein that hydrolyzes only PtdIns(3,5)P2 (Rudge et al., 2004).
It is intriguing that AtSAC1 is colocalized with a Golgi marker and that truncation of its C terminus results in its localization in the cytoplasm. This finding indicates that the C terminus is required for the proper targeting of AtSAC1 to the Golgi. This scenario is similar to that of Fig4, of which the C terminus is essential for proper subcellular targeting. Full-length Fig4 has been shown to be localized at the vacuole membrane, and deletion of its C terminus leads to cytoplasmic localization (Rudge et al., 2004). Fig4 does not contain any transmembrane helices or subcellular targeting signal sequences, and its vacuolar targeting has been demonstrated to be mediated by binding to a vacuole membrane–localized protein, integral vacuolar membrane protein14, through its C terminus (Rudge et al., 2004). Because AtSAC1 also lacks transmembrane helices or subcellular targeting signal sequences, it is reasonable to propose that the C terminus of AtSAC1 is most likely involved in interacting with another protein that mediates the targeting of AtSAC1 to the Golgi. The molecular mechanisms underlying the subcellular targeting of AtSAC1 remain to be investigated.
Although the fra7 mutant protein lacks the majority of the C terminus, it retains the intact SAC domain and possesses phosphatase activity. This fact indicates that the phenotypes conferred by fra7 are not caused by the loss of its phosphatase activity. Because fra7 caused mislocation of the AtSAC1 protein, it is very likely that the phenotypes conferred by fra7 are the result of improper localization of the protein. Mistargeting of the fra7 mutant protein might lead to a disruption of the tight regulation of the local phosphoinositide pool at the Golgi or might alter the phosphoinositide pools at membranes of other subcellular organelles, thus affecting their normal cellular functions. The functional importance of proper subcellular targeting of SAC domain phosphatases has also been reported for yeast Sac1p and Fig4. Mistargeting of Sac1p caused by deletion of its C terminus has been shown to result in altered phosphoinositide levels and defective growth phenotypes (Foti et al., 2001). Similarly, mislocation of Fig4 has been proposed to cause misregulation of the vacuolar pool of PtdIns(3,5)P2 (Rudge et al., 2004). To understand exactly how fra7 causes the mutant phenotypes, it will be important to analyze the overall profile of phosphoinositides and test the subcellular concentration of phosphoinositides in the fra7 mutant.
AtSAC1 Is Required for Normal Actin Organization
The fra7 mutation was shown to cause an alteration in actin cytoskeleton organization in fibers and pith cells. This finding suggests that the cellular functions of AtSAC1 may overlap, at least in part, with the functions of yeast Sac1p and Fig4. The yeast Sac1p was originally discovered in a screen for sac mutations (Novick et al., 1989). Mutations of both Sac1p and Fig4 were found to cause aberrant organization of the actin cytoskeleton (Erdman et al., 1998; Foti et al., 2001). Phosphoinositides are known to modulate the activities of various actin regulatory proteins, such as profilin, the capping proteins CapZ and gelsolin, actin-depolymerizing factors, and α-actinin (Takenawa and Itoh, 2001).
In plants, a growing line of evidence indicates that many of the actin regulatory proteins, such as profilin, gelsolin, and the capping protein CapZ, are functionally conserved, and the activities of profilin and gelsolin have been shown to be modulated by PtdIns(4,5)P2 (Kovar et al., 2001; Huang et al., 2003, 2004). However, it is not known whether other phosphoinositides, such as PtdIns(3,5)P2, are involved in the modulation of the activities of actin regulatory proteins. The regulation of PtdIns(3,5)P2 by Fig4 has been demonstrated to be essential for normal actin organization in yeast, although how the fig4 mutation causes actin defects remains unknown (Erdman et al., 1998; Rudge et al., 2004). PtdIns(3,5)P2 has been detected in plants, but its cellular functions have not been studied (Meijer et al., 1999; Meijer and Munnik, 2003). We propose that like its yeast counterparts Sac1p and Fig4, AtSAC1 regulates the metabolism of phosphoinositides, thereby influencing actin organization. It should be noted that AtSAC1 appears not to be colocalized with actin filaments, although the fra7 mutation causes a defect in actin organization. This is similar to the situation with yeast Sac1p and Fig4, which are localized in the ER and at the vacuolar membrane, respectively, and are known to be important for normal actin organization (Erdman et al., 1998; Foti et al., 2001). A correlation between phosphoinositide metabolism and actin organization has been demonstrated in fra3 (Zhong et al., 2004). The FRA3 gene encodes a type II inositol polyphosphate 5-phosphatase with the highest substrate affinity toward PtdIns(4,5)P2. Mutation of FRA3 causes an abnormal bundling of F-actin cables, suggesting an important role of phosphoinositide metabolism in F-actin organization in plants.
AtSAC1 Is Essential for Normal Cell Wall Biosynthesis
The dramatic reduction in cell wall thickness caused by the fra7 mutation suggests that AtSAC1 is important for cell wall biosynthesis. It is possible that the reduced cell wall synthesis in fra7 is, at least in part, a result of aberrant actin organization. F-actin is known to regulate the transport of vesicles carrying cell wall polysaccharides and enzymes involved in cell wall synthesis. The altered F-actin organization caused by the fra7 mutation likely impedes the process of vesicle trafficking, thus causing the reduced cell wall synthesis.
A correlation between actin organization and cell wall synthesis has been demonstrated in fra4 with a mutation in the Root Hair Defective3 gene (Hu et al., 2003). The fra4 mutation caused a dramatic reduction in cell wall thickness and an aggregation of F-actin cables, a phenotype similar to that seen in fra7. The importance of F-actin organization in cell wall synthesis was also substantiated by the finding that interference with F-actin organization by cytochalasin D reduces secondary wall thickening in fiber cells (Hu et al., 2003). Another example of the close correlation between actin organization and cell wall synthesis is seen in the fra3 mutant, which displays an aberrant organization of F-actin cables and a dramatic reduction in secondary wall thickness in fiber cells (Zhong et al., 2004). However, the possibility that the phosphoinositides regulated by AtSAC1 act as signaling molecules to directly influence vesicle trafficking and cell wall synthesis could not be excluded.
AtSAC1 Plays an Important Role in Cell Morphogenesis
Mutation of AtSAC1 not only reduces cell length but also alters cell shape, indicating that AtSAC1 is required for cell morphogenesis. The aberrant cell shapes seen in fra7 could be a result of the defective F-actin organization. Alterations of F-actin organization in plants by actin-disrupting drugs or genetic mutations have been shown to have dramatic effects on cell morphogenesis (Baluska et al., 2001a, 2001b; Dong et al., 2001; Ringli et al., 2002; Le et al., 2003; Li et al., 2003; Mathur et al., 2003a, 2003b). The common phenotypic defects associated with abnormal F-actin organization are reduced cell expansion and the formation of aberrant cell shapes. These defects were observed in elongating fra7 fibers and pith cells, which concomitantly exhibited altered F-actin organization. These results suggest a close correlation between aberrant cell morphology and abnormal F-actin organization. The thick F-actin cables seen in elongating fra7 cells resemble the F-actin organization observed in cells near the end of growth (Waller et al., 2002), which was thought to restrict cell expansion by attenuating membrane and vesicle trafficking. Aberrant formation of aggregated F-actin cables was also observed in expanding cells in the crooked, wurm, and distorted1 mutants, which were associated with crooked trichome phenotypes (Mathur et al., 2003a, 2003b). The aggregation of F-actin in these mutants was proposed to cause aberrant cell morphogenesis by reducing the directional transport of vesicles to the plasma membrane (Mathur et al., 2003b).
It is also possible that the phosphoinositides regulated by AtSAC1 act as signaling molecules to directly influence cell morphogenesis. It has been shown that PtdIns(4,5)P2 and PtdIns(4,5)P2-modulated proteins play roles in plant cell morphogenesis (Braun et al., 1999; Kost et al., 1999; Dong et al., 2001; McKenna et al., 2004). However, little is known about the functional roles of other phosphoinositides, such as PtdIns(3,5)P2. Further investigation of the cellular functions of AtSAC1 is expected to provide novel insights into the roles of phosphoinositides in cell morphogenesis. It is intriguing that although both fra3 (Zhong et al., 2004) and fra7 affect secondary wall synthesis and actin organization, only the fra7 mutation causes a defect in cell morphogenesis.
In addition to altered cell morphogenesis, the fra7 mutant displayed abnormal cell files in the pith of inflorescence stems. These aberrant cell files suggest an abnormal placement of cell division planes, because the linear cell files are determined during the division of pith precursor cells. The placement of cell division planes has been known to be affected by cytoskeletons (Granger and Cyr, 2000). Microtubules in the preprophase band are thought to influence the proper orientation of the cell division plane during mitosis. Mutants such as fra2/botero1 (Bichet et al., 2001; Burk et al., 2001) and tonneau2 (Traas et al., 1995) with alterations in microtubule organization have been shown to disrupt the normal linear cell files, possibly by altering the normal placement of cell division planes. The actin cytoskeleton is also an important player in cell division plane placement. Alteration of actin organization by actin-disrupting drugs or genetic mutations has been known to cause aberrant formation of cell division planes (Mineyuki and Palevitz, 1990; Gallagher and Smith, 1999; Granger and Cyr, 2000; Mathur et al., 2003b). Because the fra7 mutation alters actin organization, it is tempting to propose that the aberrant cell files seen in the fra7 pith are caused by abnormal F-actin organization.
In conclusion, we have demonstrated that AtSAC1, a plant SAC domain phosphoinositide phosphatase, plays important roles in cellular processes such as cell elongation and cell wall biosynthesis. Because these cellular processes are known to be regulated by the actin cytoskeleton and mutation of AtSAC1 disrupts normal actin organization, it is conceivable that AtSAC1 exerts its cellular functions, at least in part, by modulating the actin organization. Further investigation of the cellular functions of AtSAC1 is expected to further our understanding of the molecular mechanisms underlying plant cell morphogenesis.
METHODS
Mutant Isolation
M2 Arabidopsis thaliana (ecotype Columbia) plants generated from ethyl methanesulfonate mutagenization were grown in a greenhouse, and their inflorescence stems were screened for mutants with reduced breaking strength. Stems were divided into three equal segments, and each segment was measured for its breaking force using a digital force/length tester (model DHT4-50; Larson System, Minneapolis, MN). The breaking force was calculated as the force needed to break apart a stem segment (Zhong et al., 1997). Putative mutants with reduced stem-breaking strength were selected and backcrossed with wild-type Columbia three times before analysis.
Microscopy
Stem samples were fixed in 2% (v/v) glutaraldehyde in PEMT buffer (50 mM Pipes, 2 mM EGTA, 2 mM MgSO4, and 0.05% [v/v] Triton X-100, pH 7.2) at 4°C overnight. After being washed in phosphate buffer (50 mM, pH 7.2), samples were postfixed in 2% (v/v) OsO4 for 2 h and then dehydrated through a gradient of ethanol, cleared in propylene oxide, and embedded in Araldite/Embed 812 resin (Electron Microscopy Sciences, Fort Washington, PA). One-micrometer-thick sections were cut, stained with toluidine blue, and viewed with a light microscope. For transmission electron microscopy, 90-nm ultrathin sections were cut, mounted on Formvar-coated gold slot grids, poststained with uranyl acetate and lead citrate, and observed with a Zeiss EM 902A electron microscope (Zeiss, Jena, Germany).
For visualization of leaf epidermis, mature leaves were cryoprepared, surface-coated with gold, and observed with a LEO982 FE scanning electron microscope (Leo Electron Microscopy, Thornwood, NY).
Cell Wall Analysis
Inflorescence stems of 10-week-old plants were collected for cell wall isolation. Stems were ground into fine powder in liquid nitrogen with a mortar and pestle, homogenized with a Polytron, and extracted in 70% ethanol at 70°C. The remaining cell wall residues were dried in a vacuum oven at 60°C and used for analysis of cellulose amount and sugar composition. Crystalline cellulose was measured with the acetic-nitric anthrone reagent according to Updegraff (1969). Cell wall sugars (as alditol acetates) were determined according to the procedure described by Hoebler et al. (1989). All samples were run in triplicate.
Map-Based Cloning
The fra7 mutant (ecotype Columbia) was crossed with Arabidopsis ecotype Landsberg erecta to generate 1250 F2 mapping plants. Fine mapping of the fra7 locus was done with CAPS markers according to Konieczny and Ausubel (1993). CAPS markers were developed based on sequence information from the Cereon Arabidopsis polymorphic database (http://www.arabidopsis.org/cereon).
For complementation analysis, the wild-type AtSAC1 gene was amplified by PCR, confirmed by sequencing, and cloned into the binary vector pBI101. The construct was then introduced into fra7 by Agrobacterium tumefaciens–mediated transformation (Bechtold and Bouchez, 1994). Transgenic plants were selected on kanamycin and grown to maturity for analysis of their ability to complement the mutant phenotypes.
Gene Expression Analysis
The expression pattern of AtSAC1 was studied using the GUS reporter gene. The AtSAC1 gene, including a 1.2-kb upstream sequence and the entire exon and intron region, was amplified by PCR using high-fidelity DNA polymerase with gene-specific primers (5′-TTGCCACATTTAGGACAACTCTTC-3′ and 5′-ATTGACTTGCTTAGGCATCAGGCG-3′), confirmed by sequencing, and ligated in frame with the GUS reporter gene in the pBI101 vector. The construct was transformed into wild-type and fra7 mutant plants by the Agrobacterium-mediated transformation procedure (Bechtold and Bouchez, 1994). Transgenic plants were selected on kanamycin and used for expression analysis of the GUS reporter gene. Tissues were first immersed in 90% ice-cold acetone for 20 min and then incubated in GUS staining solution (100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, and 1 mM 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid) at 37°C. After being cleared in 70% ethanol, the tissues were observed for GUS staining with a light microscope.
Expression of Recombinant Proteins in Yeast
The full-length cDNA of wild-type AtSAC1 and the truncated fra7 mutant cDNA were amplified by PCR using high-fidelity DNA polymerase from cDNAs synthesized from wild-type or fra7 stems, respectively. The amplified cDNAs were confirmed by sequencing and ligated in frame into the yeast expression vector pYES3/CT that is tagged with the V5 epitope (GKPIPNPLLGLDST) at the C terminus (Invitrogen, Carlsbad, CA). The constructs were transformed into the yeast strain INVSc1 (Invitrogen). The expression of recombinant proteins was induced in the presence of 2% galactose for 24 h. After induction, yeast cells were broken using glass beads, and the crude protein extracts were used for immunoprecipitation of recombinant proteins by incubating with a monoclonal antibody against the V5 epitope (Invitrogen) and agarose bead–conjugated secondary antibodies. The immunoprecipitates were washed six times before use for further analysis.
The expression of recombinant proteins was confirmed by immunoblot analysis. Purified recombinant proteins were separated on a 12.5% SDS gel and transferred onto a nitrocellulose membrane. The recombinant proteins were detected by incubation with a monoclonal antibody against the V5 epitope and horseradish peroxidase–conjugated secondary antibodies. The control used was the immunoprecipitates from yeast cells harboring the expression vector pYES3/CT alone. Both recombinant wild-type AtSAC1 and mutant fra7 proteins were confirmed to be expressed in yeast cells.
Phosphatase Activity Assay
Purified recombinant protein immunoprecipitates were used for phosphoinositide phosphatase activity assay. Phospholipids (Echelon, Salt Lake City, UT) were dissolved in the reaction buffer (50 mM Tris-HCl, pH 7.0, 0.25% β-d-octylglucoside, 5 mM MgCl2, 10 mM KCl, and 1 mM phenylmethylsulfonyl fluoride) with the aid of sonication. The phosphatase activity of recombinant proteins was measured by mixing 100 μM substrates in the reaction buffer with the immunoprecipitates, and the mixture was incubated at 30°C for 50 min. The reactions were agitated every 10 min during incubation. The free phosphate released from the substrates was detected by the malachite green method (Kodama et al., 1986). The immunoprecipitates from yeast cells harboring the expression vector alone were used as a control in the assay, and no activity was detected. The amount of fusion protein used in the assay was estimated by comparing the fusion protein bands with known amounts of V5-tagged β-galactosidase on the immunoblots. All assays were run in duplicate and repeated twice, and identical results were obtained.
Localization of Fluorescent Protein–Tagged AtSAC1
The AtSAC1 gene, including a 1.2-kb upstream sequence and the entire exon and intron region, was amplified by PCR, confirmed by sequencing, and then fused in frame with the GFP cDNA (ABRC, Columbus, OH; developed by S.J. Davis and R.D. Vierstra) in the binary vector pBI101. For expression of fra7-GFP, AtSAC1 truncated at the fra7 mutation site was used. The AtSAC1-GFP and fra7-GFP constructs were introduced into Arabidopsis plants by Agrobacterium-mediated transformation. Transgenic plants were selected on kanamycin, and T2 progeny were used for GFP localization.
The GFP signals from roots of 3-d-old transgenic seedlings were viewed with a Leica TCs SP2 spectral confocal microscope (Leica Microsystems, Heidelberg, Germany). Images were saved and processed with Adobe Photoshop version 7.0 (Adobe Systems, Mountain View, CA).
To determine the exact subcellular localization of AtSAC1, the AtSAC1 cDNA was fused in frame with an EYFP or an ECFP and ligated between the Cauliflower mosaic virus 35S promoter and the nopaline synthase terminator in a high-copy vector. The expression constructs were cotransfected into carrot (Daucus carota) protoplasts with several EYFP- or ECFP-tagged markers for various organelles. These markers include the Golgi-localized AVP2 (Mitsuda et al., 2001), the prevacuolar membrane marker Rha1 (Lee et al., 2004), and an ER-localized marker containing the calrecticulin targeting sequence and the ER retention sequence (BD Biosciences, Palo Alto, CA). Three-day-old subcultured carrot cells were digested with a mixture of cell wall–digesting enzymes (Driselase; Sigma-Aldrich, St. Louis, MO) to generate protoplasts. The protoplasts (6 × 105) in 300 μL of transfection solution (154 mM NaCl, 5 mM KCl, 125 mM CaCl2, and 5 mM glucose, pH 6.0) were mixed with 20 μg of AtSAC1-EYFP or AtSAC1-ECFP and 20 μg of marker constructs (Liu et al., 1994). After the addition of 300 μL of 40% (w/v) polyethylene glycol, the mixture was incubated for 5 min with mixing and then transferred to 4 mL of cell culture media. The transfected protoplasts were incubated in darkness for 20 h before examination with a Leica TCs SP2 spectral confocal microscope. Images from single optical sections were collected and processed with Adobe Photoshop version 7.0.
Immunolocalization of Actin Filaments and Microtubules
Actin filaments and cortical microtubules in wild-type and fra7 mutant cells were immunolocalized according to Sugimoto et al. (2000). Elongating inflorescence stems of 6-week-old plants were fixed in PEMT buffer containing 1.5% (v/v) formaldehyde and 0.5% (v/v) glutaraldehyde. It has been reported that aldehyde fixation preserves actin filaments well in plant cells and that the common failures to localize actin filaments are attributable to such subsequent steps as wax embedding after fixation (Vitha et al., 2000; Wasteneys and Galway, 2003). Therefore, fixed stems were used directly for immunolocalization. It has been shown that the F-actin patterns revealed by histolocalization and GFP-tagged talin are comparable (Frank et al., 2003). Thin sections (100 μm thick) were cut longitudinally with a vibratome. Sections were incubated with monoclonal antibodies against chicken actin (ICN, Aurora, OH) or chicken α-tubulin (Sigma-Aldrich) and fluorescein isothiocyanate–conjugated secondary antibodies. The fluorescence-labeled microtubules and F-actins were viewed with a Leica TCs SP2 spectral confocal microscope. Images were saved and processed with Adobe Photoshop version 7.0.
Visualization of Cellulose Microfibrils
Pith cells from elongating inflorescence stems of 6-week-old plants were used for visualization of cellulose microfibrils in the innermost layer of cell walls using field emission scanning electron microscopy according to Sugimoto et al. (2000). Stem segments were sectioned longitudinally through pith cells and fixed in PEMT buffer containing 4% (v/v) formaldehyde. After dehydration, sections were dried in a semidry critical point drier (Tousimis, Rockville, MD) and mounted on stubs with carbon paste. Sections were coated with platinum using an Edwards 306 vacuum evaporator (Edwards High Vacuum International, Wilmington, MA) and then examined for cellulose microfibrils using a LEO 982 FE scanning electron microscope.
The GenBank accession numbers for the sequences shown in Figure 5 are AY227244 (AtSAC1), X51672 (Sac1p), NM_053798 (rSAC1), NM_014937 (hSAC2), and NM_003895 (Synapto).
Acknowledgments
We thank Beth Richardson and John Shields for their help with microscopy and the editor and reviewers for their constructive suggestions. D.H.B. was supported by a Plant Evolution Training Grant from the National Science Foundation. This work was supported by grants from the U.S. Department of Energy-Bioscience Division (DE-FG02-03ER15415) and the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (2002-35304-12709).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instruction for Authors (www.plantcell.org) is: Zheng-Hua Ye (zhye@plantbio.uga.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031377.
References
- Baluska, F., Busti, E., Dolfini, S., Gavazzi, G., and Volkmann, D. (2001. a). Lilliputian mutant of maize lacks cell elongation and shows defects in organization of actin cytoskeleton. Dev. Biol. 236, 478–491. [DOI] [PubMed] [Google Scholar]
- Baluska, F., Jasik, J., Edelmann, H.G., Salajova, T., and Volkmann, D. (2001. b). Latrunculin B-induced plant dwarfism: Plant cell elongation is F-actin-dependent. Dev. Biol. 231, 113–124. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Bouchez, D. (1994). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants, I. Potrykus and G. Spangenberg, eds (Berlin: Springer-Verlag), pp. 19–23.
- Bichet, A., Desnos, T., Turner, S., Grandjean, O., and Höfte, H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25, 137–148. [DOI] [PubMed] [Google Scholar]
- Braun, M., Baluska, F., von Witsch, M., and Menzel, D. (1999). Redistribution of actin, profilin and phosphatidylinositol-4,5-bisphosphate in growing and maturing root hairs. Planta 209, 435–443. [DOI] [PubMed] [Google Scholar]
- Burk, D.H., and Ye, Z.-H. (2002). Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule severing protein. Plant Cell 14, 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk, D.H., Liu, B., Zhong, R., Morrison, W.H., and Ye, Z.-H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Cleves, A.E., Novick, P.J., and Bankaitis, V.A. (1989). Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J. Cell Biol. 109, 2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres, B., Bouissonnié, F., Wu, H.-J., Gomord, V., Guilleminot, J., Grellet, F., Berger, F., Delseny, M., and Devic, M. (2003). Three SAC1-like genes show overlapping patterns of expression in Arabidopsis but are remarkably silent during embryo development. Plant J. 34, 293–306. [DOI] [PubMed] [Google Scholar]
- DeWald, D.B., Torabinejad, J., Jones, C.A., Shope, J.C., Cangelosi, A.R., Thompson, J.E., Prestwich, G.D., and Hama, H. (2001). Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-triphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol. 126, 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C.-H., Xia, G.-X., Hong, Y., Ramachandran, S., Kost, B., and Chua, N.-H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13, 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercetin, M.E., and Gillaspy, G.E. (2004). Molecular characterization of an Arabidopsis gene encoding a phospholipid-specific inositol polyphosphate 5-phosphatase. Plant Physiol. 135, 938–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman, S., Lin, L., Malczynski, M., and Snyder, M. (1998). Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140, 461–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti, M., Audhya, A., and Emr, S.D. (2001). Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol. Biol. Cell 12, 2396–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, M.J., Cartwright, H.N., and Smith, L.G. (2003). Three brick genes have distinct functions in a common pathway promoting polarized cell division and cell morphogenesis in the maize leaf epidermis. Development 130, 753–762. [DOI] [PubMed] [Google Scholar]
- Gallagher, K., and Smith, L.G. (1999). discordia mutations specifically misorient asymmetric cell divisions during development of the maize leaf epidermis. Development 126, 4623–4633. [DOI] [PubMed] [Google Scholar]
- Granger, C.L., and Cyr, R.J. (2000). Use of abnormal preprophase bands to decipher division plane determination. J. Cell Sci. 114, 599–607. [DOI] [PubMed] [Google Scholar]
- Guo, S., Stolz, L.E., Lemrow, S.M., and York, J.D. (1999). SAC1-like domains of yeast SAC1, INP52, INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274, 12990–12995. [DOI] [PubMed] [Google Scholar]
- Hoebler, C., Barry, J.L., David, A., and Delort-Laval, J. (1989). Rapid acid-hydrolysis of plant cell wall polysaccharides and simplified quantitative determination of their neutral monosaccharides by gas-liquid chromatography. J. Agric. Food Chem. 37, 360–367. [Google Scholar]
- Hu, Y., Zhong, R., Morrison, W.H., and Ye, Z.-H. (2003). The Arabidopsis RHD3 gene is required for cell wall biosynthesis and actin organization. Planta 217, 912–921. [DOI] [PubMed] [Google Scholar]
- Huang, S., Blanchoin, L., Chaudhry, F., Franklin-Tong, V.E., and Staiger, C.J. (2004). A gelsolin-like protein from Papaver rhoeas pollen (PrABP80) stimulates calcium-regulated severing and depolymerization of actin filaments. J. Biol. Chem. 279, 23364–23375. [DOI] [PubMed] [Google Scholar]
- Huang, S., Blanchoin, L., Kovar, D.R., and Staiger, C.J. (2003). Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 278, 44832–44842. [DOI] [PubMed] [Google Scholar]
- Hughes, W.E., Cooke, F.T., and Parker, P.J. (2000. a). Sac phosphatase domain proteins. Biochem. J. 350, 337–352. [PMC free article] [PubMed] [Google Scholar]
- Hughes, W.E., Woscholski, R., Cooke, F.T., Patrick, R.S., Dove, S.K., McDonald, N.Q., and Parker, P.J. (2000. b). SAC1 encodes a regulated lipid phosphoinositide phosphatase, defects in which can be suppressed by the homologous Inp52p and Inp53p phosphatases. J. Biol. Chem. 275, 801–808. [DOI] [PubMed] [Google Scholar]
- Katsaros, C.I., Karyophyllis, D.A., and Galatis, B.D. (2002). Cortical F-actin underlies cellulose microfibril patterning in brown algal cells. Phycologia 41, 178–183. [Google Scholar]
- Kearns, B.G., McGee, T.P., Mayinger, P., Gedvilaite, A., Philips, S.E., Kagiwada, S., and Bankaitis, V.A. (1997). Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature 387, 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.H., Eu, Y.-J., Yoo, C.M., Kim, Y.-W., Pih, K.T., Jin, J.B., Kim, S.J., Stenmark, H., and Hwang, I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama, T., Fukui, K., and Kometani, K. (1986). The initial phosphate burst in ATP hydrolysis by myosin and subfragment-1 as studied by a modified malachite green method for determination of inorganic phosphate. J. Biochem. (Tokyo) 99, 1465–1472. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.-H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar, D.R., Drobak, B.K., Collings, D.A., and Staiger, C.J. (2001). The characterization of ligand-specific maize (Zea mays) profilin mutants. Biochem. J. 358, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, J., El-Assal, S.E.-D., Basu, D., Saad, M.E., and Szymanski, D.B. (2003). Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr. Biol. 13, 1341–1347. [DOI] [PubMed] [Google Scholar]
- Lee, G.-J., Sohn, E.J., Lee, M.H., and Hwang, I. (2004). The Arabidopsis Rab5 homologs Rha1 and Ara7 localize to the prevacuolar compartment. Plant Cell Physiol. 45, 1211–1220. [DOI] [PubMed] [Google Scholar]
- Li, L., Tutone, A.F., Drummond, R.S.M., Gardner, R.C., and Luan, S. (2001). A novel family of magnesium transport genes in Arabidopsis. Plant Cell 13, 2761–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Blanchoin, L., Yang, Z., and Lord, E.M. (2003). The putative Arabidopsis Arp2/3 complex controls leaf cell morphogenesis. Plant Physiol. 132, 2034–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.B., Ulmasov, T., Shi, X., Hagen, G., and Guilfoyle, T.J. (1994). Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, T.F.J. (1998). Phosphoinositide lipids as signaling molecules: Common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu. Rev. Cell Dev. Biol. 14, 231–264. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kernebeck, B., and Hülskamp, M. (2003. a). Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15, 1632–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kirik, V., Kernebeck, B., Srinivas, B.P., and Hülskamp, M. (2003. b). Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130, 3137–3146. [DOI] [PubMed] [Google Scholar]
- Matsuoka, K., Bassham, D.C., Raikhel, N.V., and Nakamura, K. (1995). Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J. Cell Biol. 130, 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, S.T., Vidali, L., and Hepler, P.K. (2004). Profilin inhibits pollen tube growth through actin-binding, but not poly-l-proline-binding. Planta 218, 906–915. [DOI] [PubMed] [Google Scholar]
- Meijer, H.J., Berrie, C.P., Iurisci, C., Divecha, N., Musgrave, A., and Munnik, T. (2001). Identification of a new polyphosphoinositide in plants, phosphatidylinositol 5-monophosphate (PtdIns5P), and its accumulation upon osmotic stress. Biochem. J. 360, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, H.J., Divecha, N., van den Ende, H., Musgrave, A., and Munnik, T. (1999). Hyperosmotic stress induces rapid synthesis of phosphatidyl-d-inositol 3,5-bisphosphate in plant cells. Planta 208, 294–298. [Google Scholar]
- Meijer, H.J.G., and Munnik, T. (2003). Phospholipid-based signaling in plants. Annu. Rev. Plant Biol. 54, 265–306. [DOI] [PubMed] [Google Scholar]
- Memon, A.R., Chen, Q.Y., and Boss, W.F. (1989). Inositol phospholipids activate plasma membrane ATPase in plants. Biochem. Biophys. Res. Commun. 162, 1295–1301. [DOI] [PubMed] [Google Scholar]
- Mikami, K., Katagiri, T., Luchi, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 15, 563–568. [DOI] [PubMed] [Google Scholar]
- Minagawa, T., Ijuin, T., Mochizuki, Y., and Takenawa, T. (2001). Identification and characterization of a Sac domain-containing phosphoinositide 5-phosphatase. J. Biol. Chem. 276, 22011–22015. [DOI] [PubMed] [Google Scholar]
- Mineyuki, Y., and Palevitz, B.A. (1990). Relationship between preprophase band organization, F-actin and the division site in Allium. J. Cell Sci. 97, 283–295. [Google Scholar]
- Mitsuda, N., Enami, K., Nakata, M., Takeyasu, K., and Sato, M.H. (2001). Novel type Arabidopsis thaliana H+-PPase is localized to the Golgi apparatus. FEBS Lett. 488, 29–33. [DOI] [PubMed] [Google Scholar]
- Müeller-Röeber, B., and Pical, C. (2002). Inositol phospholipid metabolism in Arabidopsis: Characterization and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 130, 22–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto, Y., Kearns, B.G., Wenk, M.R., Chen, H., Mori, K., Alb, J.G., Camilli, P.D., and Bankaitis, V.A. (2000). Functional characterization of a mammalian Sac1 and mutants exhibiting substrate-specific defects in phosphoinositide phosphatase activity. J. Biol. Chem. 275, 34293–34305. [DOI] [PubMed] [Google Scholar]
- Novick, P., Osmond, B.C., and Botstein, D. (1989). Suppressors of yeast actin mutations. Genetics 121, 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pical, C., Westergren, T., Dove, S.K., Larsson, C., and Sommarin, M. (1999). Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4,5-bisphosphate, diacylglycerol pyrophosphate, and phosphatidylcholine in Arabidopsis thaliana cells. J. Biol. Chem. 274, 38232–38240. [DOI] [PubMed] [Google Scholar]
- Qin, C., Wang, C., and Wang, X. (2002). Kinetic analysis of Arabidopsis phospholipase Dδ. J. Biol. Chem. 277, 49685–49690. [DOI] [PubMed] [Google Scholar]
- Ringli, C., Baumberger, N., Diet, A., Frey, B., and Keller, B. (2002). ACTIN2 is essential for bulge site selection and tip growth during root hair development of Arabidopsis. Plant Physiol. 129, 1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge, S.A., Anderson, D.M., and Emr, S.D. (2004). Vacuole size control: Regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol. Biol. Cell 15, 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, J.M., Perera, I.Y., Heilmann, I., Persson, S., and Boss, W.F. (2000). Inositol signaling and plant growth. Trends Plant Sci. 5, 252–258. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa, T., and Itoh, T. (2001). Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim. Biophys. Acta 1533, 190–206. [DOI] [PubMed] [Google Scholar]
- Traas, J., Bellini, C., Nacry, P., Kronenberger, J., Bouchez, D., and Caboche, M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375, 676–677. [Google Scholar]
- Updegraff, D.M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32, 420–424. [DOI] [PubMed] [Google Scholar]
- Vitha, S., Baluska, F., Braun, M., Samaj, J., Volkmann, D., and Barlow, P.W. (2000). Comparison of cryofixation and aldehyde fixation for plant actin immunocytochemistry: Aldehydes do not destroy F-actin. Histochem. J. 32, 457–466. [DOI] [PubMed] [Google Scholar]
- Waller, F., Riemann, M., and Nick, P. (2002). A role for actin-driven secretion in auxin-induced growth. Protoplasma 219, 72–81. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O., and Galway, M.E. (2003). Remodeling the cytoskeleton for growth and form: An overview with some new views. Annu. Rev. Plant Biol. 54, 691–722. [DOI] [PubMed] [Google Scholar]
- Wei, H.-C., Sanny, J., Shu, H., Baillie, D.L., Brill, J.A., Price, J.V., and Harden, N. (2003). The Sac1 lipid phosphatase regulates cell shape change and the JNK cascade during dorsal closure in Drosophila. Curr. Biol. 13, 1882–1887. [DOI] [PubMed] [Google Scholar]
- Ye, Z.-H., Freshour, G., Hahn, M.G., Burk, D.H., and Zhong, R. (2002). Vascular development in Arabidopsis. Int. Rev. Cytol. 220, 225–256. [DOI] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (2003). The SAC domain-containing protein gene family in Arabidopsis. Plant Physiol. 132, 544–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (2004). Molecular and biochemical characterization of three WD-repeat domain-containing inositol polyphosphate 5-phosphatases in Arabidopsis thaliana. Plant Cell Physiol. 45, 1720–1728. [DOI] [PubMed] [Google Scholar]
- Zhong, R., Burk, D.H., Morrison, W.H., III, and Ye, Z.-H. (2002). A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14, 3101–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Burk, D.H., Morrison, W.H., III, and Ye, Z.-H. (2004). FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells. Plant Cell 16, 3242–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Burk, D.H., and Ye, Z.-H. (2001). Fibers: A model for studying cell differentiation, cell elongation, and cell wall biosynthesis. Plant Physiol. 126, 477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Taylor, J.J., and Ye, Z.-H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9, 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]