Abstract
The translocon at the inner envelope membrane of chloroplasts (Tic) plays a central role in plastid biogenesis by coordinating the sorting of nucleus-encoded preproteins across the inner membrane and coordinating the interactions of preproteins with the processing and folding machineries of the stroma. Despite these activities, the precise roles of known Tic proteins in translocation, sorting, and preprotein maturation have not been defined. In this report, we examine the in vivo function of a major Tic component, Tic110. We demonstrate that Arabidopsis thaliana Tic110 (atTic110) is essential for plastid biogenesis and plant viability. The downregulation of atTic110 expression results in the reduced accumulation of a wide variety of plastid proteins. The expression of dominant negative mutants of atTic110 disrupts assembly of Tic complexes and the translocation of preproteins across the inner envelope membrane. Together, these data suggest that Tic110 plays a general role in the import of nuclear-encoded preproteins as a common component of Tic complexes.
INTRODUCTION
Plastids are essential organelles in plants that rely on the selective import of ∼2500 different nucleus-encoded proteins (Keegstra and Cline, 1999). The majority of these proteins are synthesized as preproteins carrying an N-terminal transit peptide. Translocon complexes at the outer (Toc complex) and inner (Tic complex) plastid envelope membranes control recognition of transit peptides and the translocation of preproteins across the two envelope membranes (Schnell and Hebert, 2003; Soll and Schleiff, 2004). The Toc and Tic complexes physically associate during import to allow simultaneous transport of preproteins across both envelope membranes, thereby providing a direct link between the cytoplasm and the plastid interior.
Preprotein targeting to plastids is mediated by a set of distinct Toc complexes that mediate the import of different subsets of preproteins across the outer envelope membrane (e.g., photosynthetic proteins required for chloroplast biogenesis) (Ivanova et al., 2004; Kubis et al., 2004; Smith et al., 2004). Upon insertion across the outer membrane, preproteins immediately engage Tic complexes. Tic complexes are implicated in multiple import processes, including translocation of preproteins into the stroma, sorting of integral membrane proteins into the inner membrane, sorting of some membrane proteins to the outer membrane, and coordinating the processing and folding of newly imported proteins in the stroma (Keegstra and Cline, 1999). Several proteins have been identified as possible components of the Tic complexes. Covalent cross-linking studies with isolated chloroplasts demonstrate direct roles for Tic110, Tic40, Tic22, and Tic20 in preprotein import (Kouranov et al., 1998; Chou et al., 2003; Inaba et al., 2003). Tic62, Tic55, and Tic32 are redox proteins that are proposed to play a regulatory role in protein import (Caliebe et al., 1997; Kuchler et al., 2002; Hormann et al., 2004).
Among the Tic components, Tic110 has been proposed to play a key role by binding preproteins during inner membrane translocation and serving as a scaffold for the recruitment of stromal chaperones to import sites (Kessler and Blobel, 1996; Akita et al., 1997; Nielsen et al., 1997). Tic110 is an integral membrane protein containing a hydrophilic C terminus (∼98 kD) that extends into the plastid stroma and a short N-terminal membrane anchor (∼9 kD) containing two membrane-spanning helices (Jackson et al., 1998). The large stromal domain contains a transit peptide binding site adjacent to its membrane anchor segments (Inaba et al., 2003). This site is proposed to form the initial binding site for preproteins as they emerge from the Tic channel, thereby preventing them from slipping back into the intermembrane space. The stromal domain also specifically associates with the stromal chaperones, Hsp93 (ClpC) and Cpn60, suggesting that it may function to couple preprotein import with subsequent events in protein folding (Kessler and Blobel, 1996; Akita et al., 1997; Nielsen et al., 1997). Cycles of Hsp93 binding to preproteins in the stroma also has been proposed to drive preprotein transport across the membrane, implicating Tic110 as part of the translocation motor of the Tic complex (Akita et al., 1997; Nielsen et al., 1997).
The in vitro analysis of Tic110 implies a central role for the protein in the assembly and function of the Tic complex. Here, we investigate the activities of the Arabidopsis thaliana ortholog of Tic110, atTic110, by genetic and biochemical analyses to further explore the physiological roles of Tic110 and substantiate its activities in vivo. We demonstrate that atTic110 is an essential component of protein import into multiple plastid types and substantiate its central role in Tic complex function.
RESULTS
AtTic110 Is a Component Common to All Import Complexes
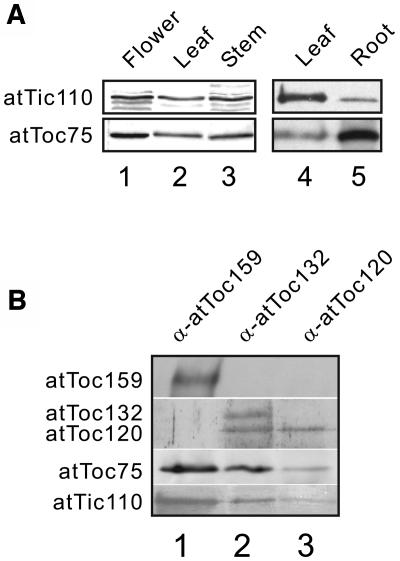
As a first step in our analysis, we examined the expression profile of atTic110 and investigated its interactions with other import components for clues to a general or specialized role in protein import. We examined the tissue distribution of atTic110 in Arabidopsis by immunoblotting extracts of various tissues using anti-atTic110 serum (Figure 1A). AtTic110 was present in flowers, leaves, stems, and roots (Figure 1A, top panel), although its levels in roots were significantly lower compared with other tissues. As a control, we immunoblotted the same protein extracts with an anti-atToc75 serum. AtToc75 is expressed in a wide variety of tissues, consistent with its roles as a component of multiple import pathways into and across the envelope (Ivanova et al., 2004; Tu et al., 2004). The expression of atToc75 exhibited a similar distribution to atTic110, although the relative levels of the two proteins varied in some tissues (Figure 1A, compare leaf and root samples). These data demonstrate that atTic110 is present in green and nongreen tissues, indicating that it functions in various plastid types and is not restricted to chloroplasts.
Figure 1.
Expression Profile and Association of atTic110 with Toc Complexes.
(A) Total protein extracts from the indicated tissues were immunoblotted with anti-atTic110 and anti- atToc75 serum.
(B) Detergent-soluble total chloroplast membranes were subjected to sequential immunoaffinity chromatography on anti-atToc159 Sepharose (α-atToc159), anti-atToc132 Sepharose (α-atToc132), and anti-atToc120 Sepharose (α-atToc120). The eluates were resolved by SDS-PAGE and immunoblotted with antisera to the proteins indicated at the left.
Recent genetic and biochemical analysis of Toc components indicate that atToc159 and its related family members, atToc132 and atToc120, assemble to form structurally and functionally distinct Toc complexes (Ivanova et al., 2004; Smith et al., 2004). We tested whether atTic110 associates with all or a subset of Toc complexes by coimmunoprecipitation using anti-atToc120, -atToc132, or -atToc159 IgG-Sepharose. As shown in Figure 1B, atTic110 is detected in association with all three Toc complexes. On the basis of these data, we conclude that atTic110 is a common component of Tic complexes that associate with multiple Toc complexes to form functional import sites.
atTic110 Is an Essential Component for Plastid Development in Arabidopsis
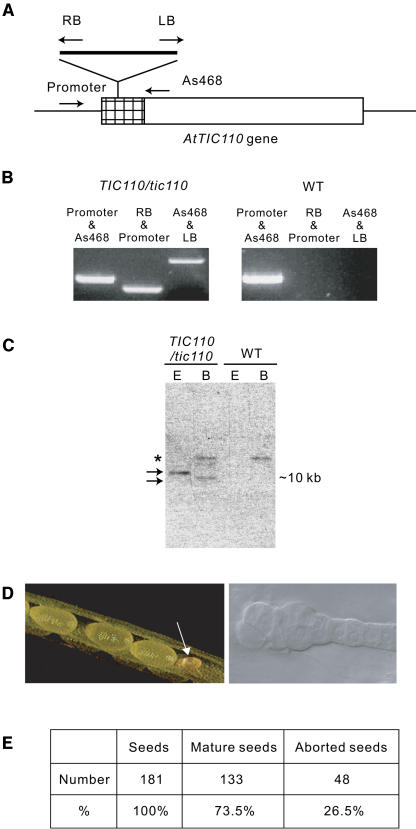
The varied activities assigned to Tic110, including preprotein and chaperone binding, as well as its presence in multiple import complexes suggest that it plays a central role in the preprotein import apparatus. To examine whether atTic110 has a detectable physiological function, we identified Arabidopsis plants from the Salk Institute Genomic Analysis Laboratory collection (Alonso et al., 2003) carrying a T-DNA insertion (SALK_094678) that disrupts the atTIC110 locus. PCR analysis of the T-DNA/atTIC110 junction established that the insertion disrupted the gene at 40 bp downstream of the start ATG start codon (Figures 2A and 2B). We refer to this line as attic110-1. DNA gel blot analysis gave a single T-DNA hybridizing band in genomic DNA isolated from heterozygous attic110-1 plants that had been digested with two different restriction enzymes, demonstrating that attic110-1 had a single T-DNA insertion in its genome (Figure 2C).
Figure 2.
Identification of an atTIC110 T-DNA Insertion Line.
(A) Diagram of T-DNA insertion in the atTIC110 gene (Salk_094678). The box represents the transcribed region of the atTIC110 gene. The hatched box represents the coding region of the atTic110 transit peptide. The arrows indicate the positions of the PCR primers used for screening. LB, left border; RB, right border.
(B) Confirmation of the T-DNA insertion by PCR amplification. Genomic DNA from heterozygous attic110-1 plants (TIC110/tic110) or wild-type plants was amplified using the primers indicated in (A).
(C) DNA gel blot analysis of the attic110-1 line. Genomic DNA (10 μg) from heterozygous attic110-1 plants or wild-type plants was digested with either EcoRI (E) or BamHI (B) and hybridized to a DNA probe corresponding to the kanamycin-resistant gene of the T-DNA. Arrows indicate specific DNA fragments obtained from attic110-1 DNA, and an asterisk indicates a nonspecific signal present in both mutant and wild-type DNA.
(D) Examination of siliques of self-crossed heterozygous attic110-1 plants. Light microscopy image of a section of a silique from self-crossed plants (left panel). The arrow indicates the position of a representative aborted seed. A representative aborted embryo viewed under a light microscope with Nomarski optics (right panel).
(E) Quantitative analysis of the numbers of mature and aborted seeds from self-crossed heterozygous attic110-1 plants.
PCR analyses of the atTIC110 locus in the progeny of self-crossed attic110-1 plants failed to identify homozygous knockout plants. Furthermore, segregation of the T-DNA–linked kanamycin resistance gene gave a ratio of 62.3% resistance to 37.7% sensitive, a value close to the expected ratio of 67% resistant and 33% sensitive plants if the knockout of atTIC110 was fatal and prevented the production of viable seed. Thus, with a very high probability, the knockout of atTIC110 is not viable. Visual analysis of the siliques of self-crossed heterozygous attic110-1 plants (Figure 2D, left panel) indicated that approximately one-quarter (26.5%) of the seeds failed to develop (Figure 2E). Examination of these embryos by microscopy confirmed that they aborted very early in development before the globular stage of embryogenesis (Figure 2D, right panel). Subsequent to the analysis of the attic110-1 line, we identified a second T-DNA insertion line. The segregation and phenotypic properties of the two lines were indistinguishable (data not shown), confirming that atTIC110 null mutants are lethal. We conclude that disruption of atTIC110 leads to seed abortion during an early state of embryogenesis, confirming that atTic110 is an essential gene in Arabidopsis. Similar results were recently reported by the Jarvis group (Kovacheva et al., 2005) with independent atTIC110 T-DNA insertion lines. This group also provided evidence that heterozygous atTIC110/attic110 plants accumulated lower amounts of a wide array of plastid proteins and exhibited a noticeable pale phenotype compared with wild-type plants.
Downregulation of atTIC110 Expression Results in Defects in Plastid Biogenesis
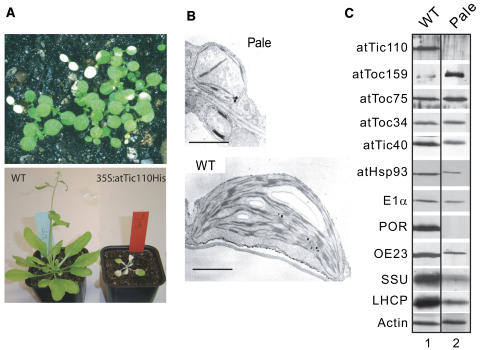
The phenotype of attic110-1 is consistent with a role for atTic110 in plastid biogenesis, but the lethality of the mutation prevented us from establishing that the defect was specific for protein import. To further analyze the role of atTic110 in vivo, we took advantage of Arabidopsis plants transformed with the atTic110 cDNA fused to a hexahistidine tag (atTic110His) under the control of the 35S promoter of Cauliflower mosaic virus. Although most transformed plants appeared normal, serious pale phenotypes arose at a relatively high frequency. Some seedlings exhibited uniform albino phenotypes upon germination and eventually arrested in growth and died (Figure 3A, top panel), whereas other plants developed pale sectors at later ages (Figure 3A, bottom panel).
Figure 3.
Analysis of Arabidopsis Plants Exhibiting atTIC110 Silencing.
(A) Phenotype of Arabidopsis carrying the 35S:atTIC110His transgene. The top panel contains a population of 5-d-old 35S:atTIC110His transgenic seedlings. The uniformly pale seedlings arrest and die at this stage of development. The bottom panel contains an adult (30-d-old) wild type and a 35S:atTIC110His transgenic line with atTIC110 silencing restricted to young tissues.
(B) Transmission electron micrographs of the wild type (bottom panel) and a pale Arabidopsis leaf exhibiting atTIC110 silencing (top panel).
(C) Immunoblot analysis of total protein extracts from wild-type plants (lane 1) and 35S:atTIC110His-induced atTIC110 silenced plants exhibiting a pale phenotype (lane 2). The immunoblots were probed with antisera corresponding to the proteins indicated at the left of the figure. The membrane also was probed with anti-actin IgG as a loading control.
We anticipated that silencing of the atTIC110 gene caused the pale/albino phenotype. To confirm this possibility, we immunoblotted proteins from wild-type plants and atTic110His transformed plants exhibiting normal and pale phenotypes with anti-atTic110 serum. In the pale plants, atTic110 protein was at undetectable levels (Figure 3C, lane 2), indicating that the pale phenotype resulted from the reduction in atTic110 expression. Examination of chloroplast ultrastructure in the pale sectors of silenced plants by transmission electron microscopy indicated a strongly reduced content of thylakoid membranes and starch granules compared with wild-type plants (Figure 3B). We conclude that the plastids in the silenced plants fail to develop into normal chloroplasts.
The atTic110 silenced plants showed significant reductions in representative nonphotosynthetic proteins (Hsp93 and pyruvate dehydrogenase E1α subunit) as well as photosynthetic proteins (protochlorophyllide oxidoreductase, 23-kD oxygen evolving complex protein, ribulose-1,5-bisphosphate carboxylase/oxygenase [Rubisco] small subunit protein, and light harvesting complex protein) (Figure 3C; see Supplemental Figure 1 online). All of these proteins are targeted to plastids via transit peptides. By contrast, two outer envelope membrane proteins that are targeted to plastids independent of transit peptides, atToc159 and atToc34, exhibit increased or stable accumulation in the silenced plants, respectively (Figure 3C). The general defect in the accumulation of a wide variety of proteins targeted to the interior compartments of plastids via transit peptides is in agreement with the embryo lethal phenotype of attic110-1 and supports a general role for Tic110 in import across the envelope in both green and nongreen plastids (Kovacheva et al., 2005).
Expression of atTic110 Deletion Mutants Disrupts the Function of Protein Import Complexes
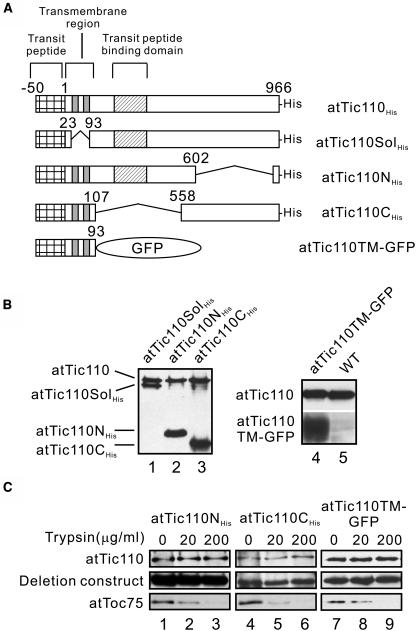
Although the reduction of atTic110 expression is consistent with an essential role in plastid biogenesis, these analyses did not address the specific functions of atTic110 in the import process. In an attempt to examine the molecular basis of atTic110 activity, we generated several atTic110 deletion constructs and expressed these proteins in Arabidopsis. In addition to atTic110His, we generated four constructs, designated as atTic110SolHis, atTic110NHis, atTic110CHis, and atTic110TM–green fluorescent protein (GFP) (Figure 4A). As shown in Figure 4B, all of the deletion constructs were successfully expressed in Arabidopsis at levels exceeding authentic atTic110. To examine the proper localization of the constructs, we tested their presence and protease sensitivity in isolated chloroplasts. AtTic110SolHis has previously been shown to localize to the plastid stroma and behave as a soluble protein in Arabidopsis (Inaba et al., 2003). atTic110NHis, atTic110CHis, and atTic110TM-GFPHis all fractionated with isolated chloroplasts, indicating proper targeting of the constructs to the organelle (Figure 4C, lanes 1, 4, and 7). The topology of these proteins in chloroplasts was further confirmed by trypsin treatment of isolated intact chloroplasts. Trypsin can permeate the outer membrane but not the inner membrane of intact chloroplasts (Cline et al., 1984). All three constructs, as well as native atTic110, were resistant to trypsin treatment, whereas atToc75, an outer membrane component, was digested (Figure 4C). These data demonstrate that all of the deletion constructs were properly targeted to chloroplasts.
Figure 4.
Construction and Expression of atTic110 Deletion Mutants in Arabidopsis.
(A) Schematic diagram of the atTic110 constructs used in this study. The known structural and functional regions of atTic110 are indicated. The numbers refer to the amino acid position, with 1 indicating the N-terminal residue of mature atTic110.
(B) Immunoblots of total protein extracts from transgenic Arabidopsis overexpressing the truncated proteins indicated at the top of the figures. The immunoblots were probed with anti-atTic110 (both panels) or anti-GFP (right panel) sera.
(C) Trypsin sensitivity of atTic110 and the atTic110 deletion constructs in isolated intact chloroplasts. Isolated chloroplasts were treated with the indicated concentrations of trypsin on ice for 30 min. Intact chloroplasts were reisolated, resolved by SDS-PAGE, and probed with anti-atTic110, anti-GFP, and anti-atToc75.
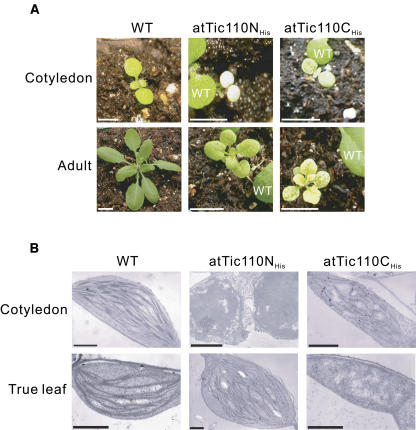
Plants stably expressing atTic110His, atTic110SolHis, and atTic110TM-GFP exhibited no apparent phenotypes (data not shown), indicating that these constructs do not interfere with the function of endogenous atTic110. By contrast, plants overexpressing either atTic110NHis or atTic110CHis showed significant phenotypes. Seedlings of both lines were markedly pale (Figure 5A). Although the atTic110NHis plants showed a uniformly pale phenotype, plants overexpressing atTic110CHis showed variegated cotyledons (Figure 5A, top panels). The expression of atTic110NHis or atTic110CHis resulted in a significant growth defect, reducing the size of adult plants >50% on soil (Figure 5A). The growth defects and visible phenotypes were not reversed by growing plants on agar plates supplemented with sucrose, indicating that the effects of atTic110NHis or atTic110CHis are not limited to photosynthetic functions.
Figure 5.
Phenotype of Plants Overexpressing atTic110NHis and atTic110CHis.
(A) Visual phenotype of wild-type plants and plants transformed with atTic110NHis and atTic110CHis in their cotyledon (top panels) and adult (bottom panels) stages. Portions of wild-type plants grown adjacent to the transgenic lines are visible in the panels of atTic110NHis and atTic110CHis plants. Bars = 0.3 cm in the top panels and 1.4 cm in the bottom panels.
(B) Ultrastructure of plastids from atTic110NHis and atTic110CHis plants. Transmission electron micrographs of atTic110NHis and atTic110CHis plants were derived from cotyledons and true leaves. Bar = 1 μm.
Chloroplasts in the atTic110NHis and atTic110CHis plants exhibited abnormal ultrastructure when examined by transmission electron microscopy. The cotyledons from atTic110NHis plants contain undeveloped plastids similar to proplastids (Figure 5B, cotyledon). In their true leaves, thylakoid membranes are present, but the number of granal stacks is significantly reduced (Figure 5B, true leaf). The plastids in atTic110CHis plants consistently showed underdeveloped granal stacks in sections from cotyledons and true leaves (Figure 5B). The visible phenotypes and disruption of plastid morphology suggest that overexpression of atTic110NHis and atTic110CHis exert dominant negative effects on the function of endogenous atTic110, resulting in the disruption of normal plastid development.
The atTic110NHis and atTic110CHis Plants Are Defective in Protein Import into Chloroplasts
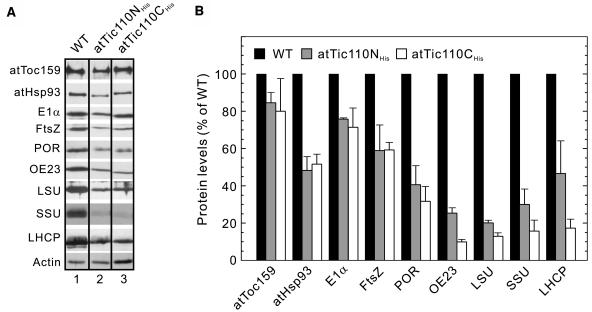
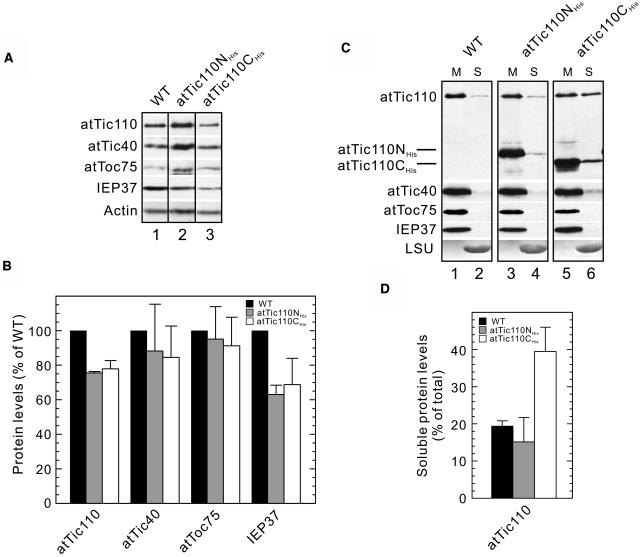
To investigate the effects of atTic110NHis and atTic110CHis expression in more detail, we examined the levels of a variety of plastid proteins in these plants. First, we examined the accumulation of representative stromal proteins that are constitutively expressed in plastids or those that represent major components of the photosynthetic machinery specific to chloroplasts. Proteins such as Rubisco small subunit (SSU) and the pyruvate dehydrogenase E1α subunit (PDH-E1α) have been shown to use distinct Toc complexes for import across the outer envelope membrane (Smith et al., 2004). Remarkably, the levels of all stromal proteins examined were significantly decreased in both atTic110NHis and atTic110CHis expressing lines, with the photosynthetic proteins exhibiting the most dramatic decrease (Figures 6A and 6B; see Supplemental Figure 2A online). The levels of atToc159 were not significantly affected in either line, consistent with the fact that its targeting does not involve a transit peptide (Figures 6A and 6B). These data are consistent with the reduced accumulation of a broad range of stromal proteins in atTIC110 silenced plants and the association of Tic110 with multiple Toc pathways. We conclude that atTic110 plays a general role in transit peptide mediated targeting to the stroma.
Figure 6.
Accumulation of Plastid Proteins in atTic110NHis and atTic110CHis Plants.
(A) Immunoblot analysis of total protein extracts from wild-type, atTic110NHis, and atTic110CHis plants using antisera corresponding to the proteins indicated at the left. LSU, large subunit of Rubisco.
(B) Semiquantitative analysis of plastid proteins. Serial dilutions of total protein extracts from wild-type, atTic110NHis, and atTic110CHis plants were probed with the indicated antibodies (see Supplemental Figure 2A online). X-ray films were scanned and analyzed by Image Quant software (Molecular Dynamics, Sunnyvale, CA). The signal intensities falling within the linear range of chemilumenescence detection were normalized to the actin blots. Each graph represents the mean of triplicate experiments, with bars indicating standard error.
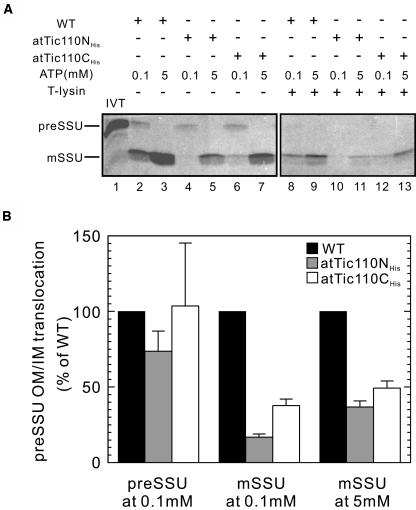
To test whether the phenotypes of plants expressing atTic110NHis and atTic110CHis result from defects in protein import, we investigated the import efficiencies of plastids isolated from the overexpressing plants. We focused on the import of the precursor to Rubisco SSU (preSSU), a substrate with a high import efficiency in wild-type plants because of the limitation in the yields of plastids from the dominant negative plants. To discriminate between effects on import at the outer and inner membranes, we incubated the plastids with 0.1 mM ATP to promote maximal insertion of preSSU across the outer membrane or 5 mM ATP to promote complete translocation across both outer and inner membranes (Fitzpatrick and Keegstra, 2001). In the presence of 0.1 mM ATP, the insertion of preSSU across the outer membrane in atTic110NHis or atTic110CHis chloroplasts was virtually unaffected (Figures 7A and 7B). On the basis of these results, we conclude that atTic110NHis and atTic110CHis do not significantly affect the activity of the Toc complex.
Figure 7.
Import of preSSU into Isolated Chloroplasts from atTic110NHis and atTic110CHis Plants.
(A) In vitro–translated (IVT) Arabidopsis [35S]preSSU was incubated with chloroplasts from atTic110NHis and atTic110CHis plants for 30 min in the dark in the presence of the indicated concentrations of ATP. One-half of each reaction was treated with thermolysin (right panel) or buffer (left panel) on ice for 30 min. The chloroplasts were analyzed directly by SDS-PAGE and phosphor imaging.
(B) Quantitative analysis of the protein import assays in (A). The amount of outer membrane insertion (preSSU at 0.1 mM) or imported preproteins (mSSU at 0.1 or 5 mM) in wild-type chloroplasts was set as 100%. Each graph represents the mean of triplicate experiments, with bars indicating standard error. IM, inner membrane; OM, outer membrane.
In the presence of 5 mM ATP, atTic110NHis and atTic110CHis chloroplasts exhibited a dramatic decrease in import capacity. The atTic110NHis and atTic110CHis chloroplasts imported <50% of preSSU compared with the wild-type control (Figures 7A and 7B). The import defect also is apparent in the low levels of import observed in the presence of 0.1 mM ATP (Figure 7A). The import of preSSU under these conditions was 15 and 35% of the levels of the wild type in atTic110NHis and atTic110CHis, respectively (Figure 7B). These data confirm that atTic110NHis and atTic110CHis disrupt translocation at the inner envelope membrane, leading to defects in plastid biogenesis and plant growth.
atTic110NHis and atTic110CHis Affect the Assembly of Import Complexes
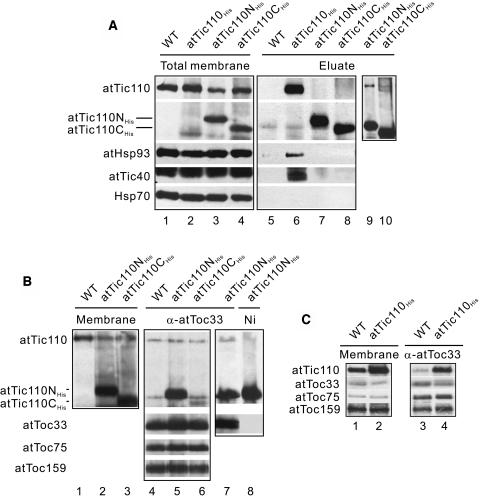
The dominant negative effects of atTic110NHis and atTic110CHis suggest that these proteins are partially functional and therefore inhibit the activity of the Tic complex by competing with authentic atTic110 for molecular interactions that are essential for import. Two such interactions that previously have been confirmed are the association of Tic110 with Hsp93 and Tic40 (Akita et al., 1997; Nielsen et al., 1997; Chou et al., 2003). To examine these associations, isolated chloroplasts from the atTic110NHis and atTic110CHis lines were treated with the homobifunctional, cleavable cross-linker dithio-bis succinimidyl propionate (DSP) to covalently stabilize the molecular complexes (Akita et al., 1997; Chou et al., 2003). The total membrane fractions from these reactions were dissolved with detergent, and the atTic110NHis and atTic110CHis complexes were purified by nickel-nitrilotriacetic acid agarose (Ni-NTA) chromatography. We used chloroplasts from wild-type plants and chloroplasts accumulating atTic110His as negative and positive controls for the chromatography, respectively.
Figure 8A demonstrates that all three hexahistidine-tagged constructs are efficiently precipitated by Ni-NTA. The precipitation is selective for the hexahistidine-tagged proteins because authentic atTic110 from wild-type extracts does not bind to the matrix (Figure 8A, lane 5). As expected, Hsp93 and atTic40 copurify with atTic110His (Figure 8A, lane 6). In contrast with atTic110His, Hsp93 and atTic40 are not detected in atTic110NHis or atTic110CHis eluates (Figure 8A, lanes 7 and 8), indicating that both constructs lack the determinants necessary to stably associate with these two proteins. The stromal Hsp70, a protein not associated with Tic complexes (Akita et al., 1997; Chou et al., 2003), is not detected in any of the Ni-NTA eluates, indicating that the cross-linking and precipitations are specific (Figure 8A).
Figure 8.
Association of Truncated atTic110NHis and atTic110CHis with Other Tic and Toc Components.
(A) Association of atTic110 deletion constructs with Tic components. Detergent-soluble total chloroplast membranes from wild-type, atTic110His, atTic110NHis, and atTic110CHis expressing plants were subjected to Ni-NTA chromatography. The Ni-NTA eluates (eluate) were resolved by SDS-PAGE under reducing conditions and immunoblotted with antisera corresponding to the proteins indicated at the left. Lanes 1 to 4 contain 5% of the total membrane fractions (total membrane) used for chromatography. Lanes 9 and 10 are duplicates of lanes 7 and 8 containing threefold more protein.
(B) Association of atTic110 deletion constructs with Toc-Tic supercomplexes. Detergent-soluble total chloroplast membranes from wild-type, atTic110His, atTic110NHis, and atTic110CHis expressing plants were subjected to immunoaffinity chromatography on anti-atToc33 Sepharose (α-atToc33). The eluates (lanes 4 to 7) were resolved by SDS-PAGE and probed with antibodies corresponding to the proteins indicated at the left of the panels. Lanes 1 to 3 contain 5% of the total membrane fractions (membrane) used for chromatography. The unbound fraction from the atTic110NHis chloroplast extracts that had been immunodepleted on α-atToc33 was subjected to subsequent chromatography on Ni-NTA matrix to precipitate residual atTic110NHis (lane 8).
(C) Association of overexpressed atTic110His with Toc-Tic supercomplexes. Detergent extracts of total chloroplast membranes from wild-type plants and plants overexpressing atTic110His were subjected to immunoaffinity chromatography on anti-atToc33 Sepharose (α-atToc33). The eluates (lanes 3 and 4) were resolved by SDS-PAGE and probed with antibodies corresponding to the proteins indicated at the left. Lanes 1 and 2 contain 5% of the total membrane fractions (membrane) used for chromatography.
Overloading of the immunoblots of the atTic110NHis and atTic110CHis eluates reveals the presence of a significant amount of authentic atTic110 that coprecipitates with atTic110NHis (Figure 8A, lane 9). The presence of both hexahistidine-tagged and untagged forms of atTic110 in the precipitate suggests that multiple copies of Tic110 are present in Tic complexes and could account for part of the dominant negative effect of this construct. The ability of atTic110NHis to mimic some critical interactions, such as direct or indirect self-association, but lack the determinants for other interactions, such as binding to atTic40 and Hsp93, could generate disabled Tic complexes that disrupt translocon function.
In addition to its interactions with other Tic components, previous studies in pea (Pisum sativum) have shown that a fraction of Tic110 (∼5%) associates with Toc components to form supercomplexes (Kouranov et al., 1998). Preproteins trapped in the process of import are enriched in supercomplexes, indicating that they correspond to functional translocons at the envelope (Akita et al., 1997; Nielsen et al., 1997; Kouranov et al., 1998). To examine the ability of atTic110NHis or atTic110CHis to associate with Toc components, we quantitatively immunoprecipitated detergent-solubilized chloroplast membranes from wild-type, atTic110NHis, or atTic110CHis plants with IgG-Sepharose containing antibodies against atToc33, the Arabidopsis Toc34 ortholog. Figure 8B indicates that the three core Toc components, atToc33, atToc75, and atToc159, are efficiently immunoprecipitated by this method (Kouranov et al., 1998). As previously demonstrated with pea chloroplasts, a fraction of authentic atTic110 coprecipitates with the Toc components (Figure 8B, lane 4).
atTic110NHis also efficiently coimmunoprecipitates with atToc33 (Figure 8B, lane 5), indicating that this mutant possesses the determinants required to associate with Toc components. The amount of atTic110NHis coimmunoprecipitating with atToc33 is substantially higher than authentic atTic110. Previous studies proposed that supercomplex formation is dynamic (Akita et al., 1997; Nielsen et al., 1997; Kouranov et al., 1998). Therefore, the increased amounts of atTic110NHis in supercomplexes could result from its relatively high levels of expression or a propensity to get trapped in supercomplexes. The ratio of atTic110NHis to authentic atTic110 in supercomplexes (7.1:1) is similar to the ratio of the two proteins in the total membrane fraction (8.25:1) (Figure 8B, compare lanes 2 and 5), consistent with the idea that assembly of Tic110 into supercomplexes is proportional to its expression level. To confirm this possibility, we examined the relative amounts of authentic atTic110 that coimmunoprecipitate with atToc33 in the line overexpressing atTic110His. As with atTic110NHis, the amount of atTic110His in supercomplexes increases significantly (Figure 8C, compare lanes 3 and 4) and appears to be proportional to its relatively high levels of expression (Figure 8C, compare lanes 1 and 2 with 3 and 4). These observations are consistent with the proposal that the assembly of supercomplexes is dynamic and indicate that the distribution of Tic110 represents an equilibrium situation between supercomplexes and a state that is not associated with Toc components.
The ability of atTic110NHis to assemble with Toc components (Figure 8B) and its ability to associate with authentic atTic110 (Figure 8A) raises the possibility that the association of atTic110NHis with authentic atTic110 (Figure 8A, lane 9) might be mediated by supercomplex formation. Indeed, atTic110 is not detected in atTic110NHis eluates from Ni-NTA chromatography of chloroplast membranes that previously had been quantitatively depleted of supercomplexes by anti-atToc33 Sepharose chromatography (Figure 8B, compare lanes 7 and 8). On the basis of these results, we conclude that the association appears to be exclusive to supercomplexes. On the basis of the data in Figure 8, we conclude that additional dominant negative effects of atTic110NHis derive from its ability to associate with Toc complexes and thereby generate inactive or partially active import sites.
In contrast with atTic110NHis, low levels of atTic110CHis are detected in the anti-atToc33 immunoprecipitates (Figure 8B, lane 6). The ratio of atTic110CHis to authentic atTic110 in supercomplexes (3:1) is significantly lower than that observed in total membrane fractions (10.3:1) (Figure 8B, compare lanes 3 and 6). It appears that atTic110CHis is less efficient than native atTic110 at assembling into supercomplexes. Nonetheless, the 3:1 ratio of atTic110CHis to atTic110 is likely to be sufficient to interfere with the function of supercomplexes in this line.
atTic110CHis Interferes with the Proper Sorting of Authentic atTic110 into the Membrane
The results in Figures 6 and 7 indicate that the dominant negative constructs disrupt the import of stromal proteins across the inner envelope membrane. In addition to these soluble proteins, we wished to examine the effects of the deletion mutants on the import and insertion of integral inner envelope membrane proteins. To test these possibilities, we examined the accumulation and membrane association of several envelope membrane proteins in the dominant negative lines.
The accumulation of atToc75 in atTic110CHis or atTic110NHis plants was not significantly different from that in wild-type plants (Figures 9A and 9B; see Supplemental Figure 2B online). This is consistent with results from the atTIC110 silenced plants (Figure 3). Although we cannot rule out the possibility that the rate of insertion of atToc75 is affected in these lines, the data suggest that the full activity of atTic110 is not required for the targeting and insertion of atToc75. As representative integral inner membrane proteins, we examined the accumulation of atTic40, IEP37, and atTic110 itself in the atTic110NHis and atTic110CHis expressing lines. Although we did not detect a significant effect on atTic40 accumulation, the accumulation of IEP37 and atTic110 was moderately reduced by ∼32 and ∼25% in the two lines, respectively (Figures 9A and 9B; see Supplemental Figure 2B online).
Figure 9.
Accumulation and Distribution of Chloroplast Envelope Membrane Proteins in atTic110NHis and atTic110CHis Transgenic Plants.
(A) Accumulation of envelope membrane proteins in wild-type, atTic110NHis, and atTic110CHis plants using immunoblotting as described in Figure 6.
(B) Semiquantitative analysis of the samples shown in (A) was performed as described in the legend to Figure 6. The serial dilutions of protein extracts used for quantitation are shown in Supplemental Figure 2B online. Each graph represents the mean of triplicate experiments, with bars indicating standard error.
(C) Distribution of envelope membrane proteins in wild-type and dominant negative plants. Isolated chloroplasts from wild-type, atTic110NHis, and atTic110CHis plants were separated into membrane (M) or soluble (S) fractions by extraction with 0.1 M Na2CO3, pH 11.5. The samples were resolved by SDS-PAGE and immunoblotted with antibodies corresponding to the proteins indicated at the left.
(D) Quantitative analysis of the levels of authentic atTic110 in the soluble fractions from wild-type, atTic110NHis, and atTic110CHis plants. Each graph represents the mean of triplicate experiments, with bars indicating standard error.
The modest effects on the accumulation of IEP37 and atTic110 in the dominant negative lines suggested that the import of these proteins was not severely affected by atTic110NHis and atTic110CHis. Next, we examined the membrane association of the inner membrane markers using alkaline extraction as a measure of membrane integration to test whether they were properly inserted into the inner membrane in the atTic110NHis and atTic110CHis lines. The Rubisco large subunit was used as a control for a soluble protein. As expected, authentic atTic110, atTic40, atToc75, and IEP37 partitioned predominantly with the membrane fraction after alkaline extraction of whole chloroplasts from wild-type plants (Figure 9C, lanes 1 and 2). A similar pattern of distribution was observed in atTic110NHis expressing plants (Figure 9C, lanes 3 and 4). By contrast, the amounts of atTic110CHis and authentic atTic110 in the soluble fraction of the atTic110CHis plants increased significantly (Figure 9C, lanes 5 and 6). In the case of authentic atTic110, we found that ∼40% of the total protein was consistently observed in the soluble fraction compared with <20% in wild-type and atTic110NHis plants (Figure 9D). Approximately 20% of atTic110CHis itself was observed in the soluble fraction (Figure 9C, lane 6). We also observed a slight increase in the levels of atTic40 in the soluble fraction in this line (Figure 9C, compare lanes 2, 4, and 6), but the increase was too small to provide reliable quantitation. We did not observe measurable amounts of soluble IEP37 in the atTic110CHis plants.
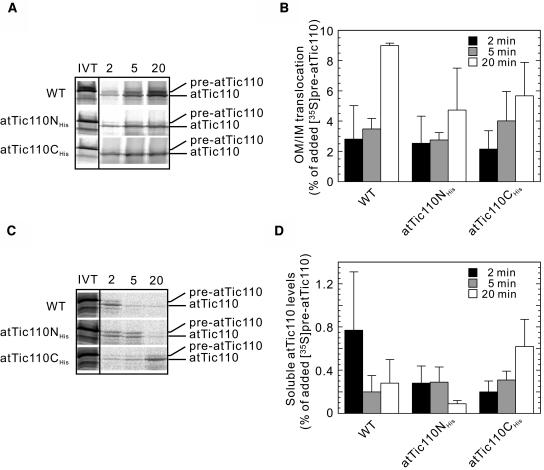
The displacement of atTic110 to the soluble fraction in atTic110CHis plants suggested that the proper insertion of this component was disrupted in this line. To investigate this in more detail, we examined the rate of inner membrane insertion using an in vitro import assay with in vitro–translated [35S] pre-atTic110 to determine whether or not the rate of insertion was reduced in atTic110CHis mutant chloroplasts. Although both mutants showed a time-dependent accumulation of imported atTic110, the overall levels of atTic110 import were significantly lower than those observed in wild-type chloroplasts (Figures 10A and 10B), consistent with the data in Figure 7. We next measured the fraction of imported [35S] atTic110 that remained soluble during the course of the import assay. The highest levels of the soluble form of imported atTic110 were observed at 2 min and decreased in a time-dependent manner in wild-type chloroplasts (Figures 10C and 10D). A similar pattern was observed in atTic110NHis chloroplasts (Figures 10C and 10D). By contrast, the amount of soluble atTic110 in atTic110CHis chloroplasts continued to accumulate during the import assay and reached a maximum at 20 min (Figures 10C and 10D). These data suggest that the overexpression of atTic110CHis interferes with the proper sorting or insertion of authentic atTic110 to the inner membrane, resulting in its displacement to the stromal fraction. The mislocalization of authentic atTic110 likely makes additional contributions to the dominant negative effects of atTic110CHis.
Figure 10.
Import of pre-atTic110 into Isolated Chloroplasts from atTic110NHis and atTic110CHis Plants.
(A) In vitro–translated (IVT) Arabidopsis [35S]pre-atTic110 was incubated with chloroplasts from wild-type, atTic110NHis, and atTic110CHis plants in the presence of 5 mM ATP. Equivalent samples of each reaction were removed at the indicated time points, and the chloroplasts were reisolated through 40% Percoll. The chloroplasts were analyzed directly by SDS-PAGE and phosphor imaging. The positions of pre-atTic110 and atTic110 are indicated at the right.
(B) Quantitative analysis of the protein import assays in (A). The amount of imported mature [35S]atTic110 was calculated as a percentage of the total [35S]pre-atTic110 in vitro translation product added to each reaction. Each graph represents the mean of three experiments, with bars indicating standard error. IM, inner membrane; OM, outer membrane.
(C) Accumulation of soluble [35S]atTic110 during the time course of import. A portion of each time point sample in the import reactions in (A) was extracted with alkaline buffer, and the soluble fraction was recovered by trichloroacetic acid precipitation. Samples were analyzed by SDS-PAGE and phosphor imaging.
(D) Quantitative analysis of the soluble [35S]atTic110 in (C). The amount of soluble mature [35S]atTic110 was calculated as a percentage of the total [35S]pre-atTic110 in vitro translation product added to the original import reaction. Each graph represents the mean of three experiments, with bars indicating standard error.
DISCUSSION
The in vitro analysis of Tic110 has revealed potential roles in several key elements of protein import into plastids, including Tic complex assembly, preprotein binding, and the recruitment of molecular chaperones to import sites. In this article, we examined the physiological significance of these activities by assessing the role of atTic110 in vivo. Using several reverse genetic approaches, we have demonstrated that atTic110 is essential for plastid biogenesis in Arabidopsis. Two independent T-DNA insertion mutants lacking atTic110 expression exhibit an embryo lethal phenotype (Figure 2). In addition, reduction in atTic110 expression by gene silencing resulted in significant defects in plant growth and the development of chloroplasts as a result of the reduced accumulation of a broad range of nuclear-encoded plastid proteins (Figure 3). The expression of deletion constructs of atTic110 in transgenic plants generated dominant negative effects with specific defects in the structure of the Tic complex (Figure 8) and consequent inhibition in preprotein translocation across the inner membrane (Figure 7). These data substantiate the central role of Tic110 in Tic complex function as an essential component of the protein import apparatus.
The combined analyses of the attic110-1 null mutant and atTIC110 silenced plants suggest that atTic110 activity is required for the import of a variety of preproteins at multiple stages in plant development. The embryo lethal phenotype of attic110-1 indicates that atTic110 function is necessary for plastid development at an early stage during embryogenesis. Kovacheva et al. (2005) performed a more detailed study of atTIC110 T-DNA knockout plants. They demonstrated that embryos homozygous for the atTIC110 knockout exhibit defects before the globular stage of development, well before embryo greening, consistent with a critical role for Tic110 at all stages of plastid biogenesis. Furthermore, seedlings that exhibit atTIC110 silencing arrest in growth and eventually die (Figure 3). These observations indicate that atTic110 function is required throughout the life cycle of the plant and is consistent with the fact that the accumulation of a wide variety of plastid proteins is affected in the silenced plants. As such, Tic110 appears to be a common, essential component of the import machinery of most, if not all, plastids.
The general role of atTic110 in import is further supported by its association with components of multiple, distinct Toc complexes (Figure 1). Recent studies have demonstrated that members of the Toc GTPase families represent independent Toc complexes with unique specificities for different classes of nucleus-encoded preproteins. In particular, atToc159 and atToc132/120 form structurally and functionally distinct Toc complexes by differentially associating with atToc33 and atToc34, respectively (Ivanova et al., 2004). Genetic analysis of the GTPases supports specific roles for these receptors in plastid development (Ivanova et al., 2004; Kubis et al., 2004). Our data demonstrate that atTic110 assembles into Toc-Tic supercomplexes with the atToc159, atToc132, and atToc120 GTPases. These data suggest that different Toc complexes might converge at a common Tic machinery containing atTic110. This scenario would account for the observations that a wide variety of plastid preproteins compete with one another for import into isolated chloroplasts (Row and Gray, 2001). Alternatively, Tic110 could be a common component of distinct Tic complexes that associate individually with different Toc complexes. This model is analogous to that proposed for the Toc channel component, Toc75, which is detected in all known Toc complexes (Ivanova et al., 2004). In either case, Tic110 appears to participate in the import of most, if not all, transit peptide mediated import pathways.
In an attempt to dissect the functions of the structural domains of atTic110, we overexpressed a series of deletion mutants in Arabidopsis. Expression of the transmembrane region of atTic110 as a fusion protein with GFP or the entire stromal domain as a soluble stromal protein had no detectable effects on plant growth or development. Therefore, it appears that the membrane anchor region and the stromal domain on their own do not interfere with critical functions of atTic110 in vivo. By contrast, the expression of deletion constructs lacking portions of the N-terminal (atTic110CHis) and C-terminal (atTic110NHis) regions of the stromal domain of atTic110 disrupt plastid and plant development (Figure 4). The import-associated stromal chaperone, Hsp93, did not coprecipitate with either construct, indicating that the structural determinants for chaperone binding rely on both regions of the stromal domain. Likewise, atTic40 associates with native atTic110 but not atTic110NHis or atTic110CHis. atTic40 is an integral membrane Tic component implicated in binding chaperones in cooperation with Tic110 (Chou et al., 2003). These data provide in vivo evidence of a direct role for Tic110 in mediating the association of molecular chaperones with the import machinery. This is consistent with the hypothesis that Tic110 provides an essential function as a scaffold that coordinates the interactions between the translocating preprotein and the stromal chaperone machinery, thereby coupling the preprotein to the translocation motor and protein folding machinery (Kessler and Blobel, 1996; Akita et al., 1997; Nielsen et al., 1997; Inaba et al., 2003). Recent studies using attic110, attic40, and hsp93-V double mutants support the in vivo interactions among these components (Kovacheva et al., 2005). Interestingly, mutants with reduced levels of atTic110 or Hsp93 partially suppressed the pale phenotype of attic40 null plants. Kovacheva et al. (2005) suggested that the reduction in atTic110 and Hsp93 might partially compensate for a stoichiometric imbalance imposed by the absence of atTic40. The high rate of gene silencing observed with the atTic110His overexpressing lines (Figure 3) and severe phenotypes generated by the atTic110NHis and atTic110CHis constructs (Figure 5) underscore the necessity to maintain a stoichiometric balance among the components that is necessary for the proper function of import complexes.
Although the interactions between Toc and Tic translocons are essential for the assembly of import sites, the mechanisms of the formation and regulation of supercomplexes have not been defined. The analysis of atTic110NHis provides evidence that amino acids 93 to 602 of the stromal domain of atTic110 are sufficient for assembly into supercomplexes. This region of atTic110 has previously been shown to possess a transit peptide binding site (Inaba et al., 2003) providing a possible mechanism for the efficient recruitment into supercomplexes. In this model, Tic110 would bind to preproteins on the trans side of the inner membrane as they emerge from the Tic channel, thereby delivering its associated stromal chaperones (e.g., Hsp93) to import sites. AtTic110NHis would retain the ability to bind preproteins and form supercomplexes, but its inability to assemble additional Tic components would prevent the formation of a fully functional translocon, resulting in its dominant negative effects. This interpretation is supported by the observation that isolated atTic110NHis chloroplasts form early import intermediates spanning the outer membrane but are defective in translocation at the inner membrane (Figure 7). The assembly of supercomplexes appears to involve multiple atTic110 molecules (Figure 8). It remains to be determined whether or not the association of atTic110 oligomers in supercomplexes is direct or mediated by an indirect association with other Tic components. Nonetheless, the presence of distinct supercomplex and free atTic110 fractions is consistent with models proposing a dynamic assembly of Tic complexes. The assembly of supercomplexes in response to membrane translocation could be important to prevent the accumulation of inactive translocons that could compromise the membrane permeability barrier of the inner membrane. Similar models of dynamic protein translocon assembly have been proposed for peroxisome and thylakoid Tat import pathways (Schnell and Hebert, 2003).
The effects of atTic110CHis on chloroplast biogenesis and protein import also can be attributed, at least in part, to its ability to disrupt the assembly of Tic complexes (Figure 8). In addition to this assembly defect, the phenotype of this mutant can also be attributed to the considerable negative impact it has on the insertion of authentic atTic110 into the inner membrane. Significant amounts of authentic atTic110 and atTic110CHis were displaced to the stroma in atTic110CHis plants (Figure 9), and in vitro import studies suggest that the rate of insertion of newly imported atTic110 into the membrane is significantly lower in atTic110CHis plants compared with wild-type and atTic110NHis plants (Figure 10). The N-terminal region of pea Tic110 has been shown previously to possess targeting information critical for inner membrane insertion (Lubeck et al., 1997), providing an explanation for the inefficient targeting of atTic110CHis to the inner membrane in this line. However, it is less clear how atTic110CHis also disrupts integration of native atTic110 to the membrane. AtTic110CHis might interact with factors that are necessary for the insertion of atTic110 into the membrane, thereby competing with authentic atTic110 for insertion. The insertion factors could be unknown proteins in the stroma or inner membrane that facilitate inner membrane insertion. Alternatively, atTic110 could require the activity of the Tic translocon for insertion into the inner membrane. In this scenario, atTic110CHis would disrupt atTic110 reinsertion by incorporating into and disabling Tic complexes in a manner similar to its effects on preprotein import. It will be of great interest to determine the precise nature of the atTic110 targeting defect in the atTic110CHis plants and examine if proper sorting of other inner envelope proteins also involves the activity of Tic110.
Taken together, these data indicate that Tic110 plays a central role in protein translocation and sorting to multiple subcompartments in plastids. These results and those of previous studies (Kessler and Blobel, 1996; Akita et al., 1997; Nielsen et al., 1997; Inaba et al., 2003) extend the functions of Tic110 to include the binding of preproteins to maintain unidirectional transport across the envelope, participation in the recruitment of stromal chaperones to participate as translocation motors and assist in folding, and the sorting of stromal and inner membrane proteins. As such, Tic110 clearly plays a key role in translocon function by coordinating the stromal events that are necessary for preprotein import. Further investigation of these and other dominant negative mutants should provide valuable insight into the molecular basis for Tic110 function in the import and sorting processes.
METHODS
DNA Constructs
The atTic110His and atTic110SolHis constructs were identical to the pre-atTic110 and pre-atTic11093-966 constructs, respectively, that were described previously (Inaba et al., 2003). The atTic110NHis and atTic110CHis constructs, corresponding to the regions indicated in Figure 4, were generated by PCR-based cloning. For atTic110TM-GFP, the transmembrane domain of atTic110 was amplified by PCR and inserted into pCL60, a pBluescript SK+–based vector containing GFP (Bauer et al., 2002). All constructs were subcloned into the NcoI and XbaI sites of the pCAMBIA3300.1 binary vector. All these constructs were introduced into Arabidopsis thaliana via Agrobacterium tumefaciens–mediated transformation by the floral dip method (Clough and Bent, 1998).
Identification of T-DNA Insertion Mutants
The T-DNA insertion line (SALK_094678) in the atTIC110 gene (GenBank accession number AY099850) was identified in silico in the T-DNA collection of the Salk Institute Genome Analysis Laboratory (Alonso et al., 2003) using primers indicated in Figure 2A. Siliques of TIC110/tic110 plants were observed using a dissecting microscope, and embryos were observed by Nomarski optics.
Arabidopsis Chloroplast Isolation, Protein Extraction, Immunoblotting, and Electron Microscopy
Chloroplasts were isolated from 15- to 20-d-old seedlings grown on 0.5× MS plates supplemented with sucrose as described previously (Smith et al., 2002). Isolated intact chloroplasts were separated into membrane and soluble fractions by lysis in 0.1 M Na2CO3, pH 11.5, followed by centrifugation at 200,000g for 20 min. The pellets (membrane fraction) were directly dissolved in SDS-PAGE sample buffer. The soluble proteins were recovered by precipitation with trichloroacetic acid and dissolved into SDS-PAGE sample buffer. Intact chloroplasts were treated with trypsin as described previously (Jackson et al., 1998).
Total protein extracts from Arabidopsis were obtained by directly homogenizing leaves in SDS-PAGE sample buffer unless specified. To avoid proteolytic degradation, the extraction buffer was supplemented with 2000-fold diluted protease inhibitor cocktail for plant cell extracts (Sigma-Aldrich, St. Louis MO). Extraction of total proteins from different organs of soil-grown plants was done as described (Rensink et al., 1998).
Antisera to atToc159, atToc132, atToc120, atToc75, atToc33, atToc34, SSU, large subunit, and protochlorophyllide oxidoreductase were described previously (Ivanova et al., 2004). The anti-atTic110 serum was generated to amino acids 370 to 966 of atTic110 that had been expressed and purified from Escherichia coli (Inaba et al., 2003). The light-harvesting complex protein and OE23 antibodies were a generous gift of Kenneth Cline (University of Florida, Gainesville, FL). The anti-FtsZ, anti-PDH-E1α, anti-atTic40, and anti-Hsp93 sera were generous gifts of Katherine Osteryoung (Michigan State University, East Lansing, MI), Douglas Randall (University of Missouri, Columbia, MO), Hsou-min Li (Academia Sinica, Taiwan, China), and Kenneth Keegstra (Michigan State University), respectively. All immunoblots of atTic110CHis and atTic110CHis expressing plants were performed with extracts obtained from plate-grown plants.
The preparation of cotyledons and true leaves from transgenic and wild-type plants for electron microscopy was performed as described previously (Ivanova et al., 2004). The grids were observed using a Philips-Tecnai 12 transmission electron microscope (Hillsboro, OR).
In Vitro Translation and Protein Import Assay
[35S]Met-labeled Arabidopsis preSSU and pre-atTic110 were generated in a coupled transcription-translation system containing reticulocyte lysate according to the manufacturer's instructions (Promega, Madison, WI). The mixture of preSSU was depleted of free nucleotides by gel filtration as described previously (Chen and Schnell, 1997). Chloroplast import assays were performed using intact chloroplasts as described (Smith et al., 2002).
Cross-Linking of Chloroplasts
For cross-linking, isolated chloroplasts were incubated in the dark for 30 min to deplete internal ATP and reducing potential. The depleted chloroplasts were subsequently incubated with 2.5 mM DSP on ice for 15 min in the dark (Akita et al., 1997). The reaction was quenched by incubating with glycine at a final concentration of 50 mM on ice for an additional 15 min.
Affinity Chromatography
For Ni-NTA affinity chromatography under denaturing conditions, the total membranes (400 μg of chlorophyll) of DSP cross-linked chloroplasts were solubilized in 25 mM Hepes-KOH, pH 8.0, and 100 mM NaCl with 1% (w/v) Triton X-100 and protease inhibitor cocktail for 10 min on ice. The solubilized membrane was diluted to 6 M urea in buffer containing 25 mM Hepes-KOH, pH 8.0, 100 mM NaCl, and 20 mM imidazole with 1% (w/v) Triton X-100. The denatured extracts were sonicated and clarified by centrifugation at 100,000g for 10 min. The supernatant was incubated with 30 μL of Ni-NTA resin for 1 h at room temperature. The bound proteins were eluted by SDS-PAGE sample buffer containing 500 mM imidazole. To cleave the cross-linker, the samples were heated at 95°C for 5 min in the presence of 100 mM DTT before SDS-PAGE.
Immunoaffinity chromatography on anti-atToc33 IgG-Sepharose under native conditions was performed with total membranes (800 μg of chlorophyll) from DSP cross-linked chloroplasts as described previously (Kouranov et al., 1998). The eluates and total membranes were analyzed by SDS-PAGE and immunoblotting.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant MCB-0090727 to D.J.S. T.I. was a recipient of a Japan Society for the Promotion of Science postdoctoral fellowship for research abroad. F.K. was supported by Swiss National Science Foundation Grant 3100-067764 and in part by the National Center of Competence in Research “Plant Survival.”
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Felix Kessler (felix.kessler@unine.ch) and Danny J. Schnell (dschnell@biochem.umass.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.030700.
References
- Akita, M., Nielsen, E., and Keegstra, K. (1997). Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Bauer, J., Hiltbrunner, A., Weibel, P., Vidi, P.A., Alvarez-Huerta, M., Smith, M.D., Schnell, D.J., and Kessler, F. (2002). Essential role of the G-domain in targeting of the protein import receptor atToc159 to the chloroplast outer membrane. J. Cell Biol. 159, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliebe, A., Grimm, R., Kaiser, G., Lubeck, J., Soll, J., and Heins, L. (1997). The chloroplastic protein import machinery contains a Rieske-type iron-sulfur cluster and a monoculear iron-binding protein. EMBO J. 16, 7342–7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., and Schnell, D.J. (1997). Insertion of the 34-kDa chloroplast protein import component, IAP34, into the chloroplast outer membrane is dependent on its intrinsic GTP-binding capacity. J. Biol. Chem. 272, 6614–6620. [DOI] [PubMed] [Google Scholar]
- Chou, M.L., Fitzpatrick, L.M., Tu, S.L., Budziszewski, G., Potter-Lewis, S., Akita, M., Levin, J.Z., Keegstra, K., and Li, H.M. (2003). Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 22, 2970–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, K., Werner-Washburne, M., Andrews, J., and Keegstra, K. (1984). Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 74, 675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick, L.M., and Keegstra, K. (2001). A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J. 27, 59–65. [DOI] [PubMed] [Google Scholar]
- Hormann, F., Kuchler, M., Sveshnikov, D., Oppermann, U., Li, Y., and Soll, J. (2004). Tic32, an essential component in chloroplast biogenesis. J. Biol. Chem. 279, 34756–34762. [DOI] [PubMed] [Google Scholar]
- Inaba, T., Li, M., Alvarez-Huerta, M., Kessler, F., and Schnell, D.J. (2003). atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 278, 38617–38627. [DOI] [PubMed] [Google Scholar]
- Ivanova, Y., Smith, M.D., Chen, K., and Schnell, D.J. (2004). Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell 15, 3379–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D.T., Froehlich, J.E., and Keegstra, K. (1998). The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 273, 16583–16588. [DOI] [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, F., and Blobel, G. (1996). Interaction of the protein import and folding machineries in the chloroplast. Proc. Natl. Acad. Sci. USA 93, 7684–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov, A., Chen, X., Fuks, B., and Schnell, D.J. (1998). Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva, S., Bedard, J., Patel, R., Dudley, P., Twell, D., Rios, G., Koncz, C., and Jarvis, P. (2005). In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 41, 412–428. [DOI] [PubMed] [Google Scholar]
- Kubis, S., Patel, R., Combe, J., Bedard, J., Kovacheva, S., Lilley, K., Biehl, A., Leister, D., Rios, G., Koncz, C., and Jarvis, P. (2004). Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16, 2059–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler, M., Decker, S., Hormann, F., Soll, J., and Heins, L. (2002). Protein import into chloroplasts involves redox-regulated proteins. EMBO J. 21, 6136–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck, J., Heins, L., and Soll, J. (1997). A nuclear-encoded chloroplastic inner envelope membrane protein uses a soluble sorting intermediate upon import into the organelle. J. Cell Biol. 137, 1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, E., Akita, M., Davila-Aponte, J., and Keegstra, K. (1997). Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink, W.A., Pilon, M., and Weisbeek, P. (1998). Domains of a transit sequence required for in vivo import in Arabidopsis chloroplasts. Plant Physiol. 118, 691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row, P.E., and Gray, J.C. (2001). Chloroplast precursor proteins compete to form early import intermediates in isolated pea chloroplasts. J. Exp. Bot. 52, 47–56. [PubMed] [Google Scholar]
- Schnell, D.J., and Hebert, D.N. (2003). Protein translocons: Multifunctional mediators of protein translocation across membranes. Cell 112, 491–505. [DOI] [PubMed] [Google Scholar]
- Smith, M.D., Fitzpatrick, L.M., Keegstra, K., and Schnell, D.J. (2002). In vitro analysis of chloroplast protein import. In Current Protocols in Cell Biology, J.S. Bonifacino, M. Dasso, J. Lippincott-Schwartz, J.B. Harford, and K.M. Yamada, eds (New York: John Wiley & Sons), pp. 11.16.11–11.16.21. [DOI] [PubMed]
- Smith, M.D., Rounds, C.M., Wang, F., Chen, K., Afitlhile, M., and Schnell, D.J. (2004). atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J. Cell Biol. 165, 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll, J., and Schleiff, E. (2004). Protein import into chloroplasts. Nat. Rev. Mol. Cell Biol. 5, 198–208. [DOI] [PubMed] [Google Scholar]
- Tu, S.L., Chen, L.J., Smith, M.D., Su, Y.S., Schnell, D.J., and Li, H.M. (2004). Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 16, 2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.