Abstract
Mitochondrial biogenesis and function require the regulated and coordinated expression of nuclear and mitochondrial genomes throughout plant development and in response to cellular and environmental signals. To investigate the levels at which the expression of nuclear and mitochondrially encoded proteins is coordinated, we established an Arabidopsis thaliana cell culture system to modulate mitochondrial biogenesis in response to sugar starvation and refeeding. Sucrose deprivation led to structural changes in mitochondria, a decrease in mitochondrial volume, and a reduction in the rate of cellular respiration. All these changes could be reversed by the readdition of sucrose. Analysis of the relative mRNA transcript abundance of genes encoding nuclear and mitochondrially encoded proteins revealed that there was no coordination of expression of the two genomes at the transcript level. An analysis of changes in abundance and assembly of nuclear-encoded and mitochondrially encoded subunits of complexes I to V of the mitochondrial inner membrane in organello protein synthesis and competence for protein import by isolated mitochondria suggested that coordination occurs at the level of protein-complex assembly. These results further suggest that expression of the mitochondrial genome is insensitive to the stress imposed by sugar starvation and that mitochondrial biogenesis is regulated by changes in nuclear gene expression and coordinated at the posttranslational level.
INTRODUCTION
The plant organelles mitochondria and chloroplasts have a bacterial origin and their own unique genomes. However, during evolution, the vast majority of the original endosymbiont genes have been transferred to the nucleus, necessitating the targeting back to the organelle of proteins synthesized in the cytosol. The organellar genomes now encode a limited number of proteins that are mainly components of the electron transport chain complexes and the transcriptional and translational machinery necessary to express these genes. Therefore, there is a requirement for coordinated expression of organellar and nuclear genomes to ensure correct assembly of complexes that contain proteins encoded in each genome. However, we are still largely ignorant of the mechanisms that lead to the coordination of expression of nuclear and organellar genomes in individual cells and tissues. This coordinated expression is essential to organelle biogenesis and to ensure that the metabolic activities of mitochondria and chloroplasts are appropriate to the changing requirements of the plant throughout its life cycle and in response to environmental stimuli.
Among the estimated 1000 or more proteins that constitute the functional plant mitochondrion (Millar et al., 2004), only 33 proteins, three rRNAs, and 20 tRNAs are encoded in the Arabidopsis thaliana mitochondrial genome (Unseld et al., 1997; Duchêne and Maréchal-Drouard, 2001). The remaining proteins and tRNAs are encoded in the nuclear genome and imported into the mitochondrion from the cytosol (Maréchal-Drouard et al., 1995; Braun and Schmitz, 1999). Therefore, precise communication and regulatory systems between nucleus and mitochondria must exist to ensure coordinated expression of both genomes. This is particularly important in the biogenesis of the multisubunit inner mitochondrial membrane complexes responsible for electron transport and ATP synthesis and for the mitochondrial ribosome, which is composed of proteins encoded in both mitochondrial and nuclear genomes.
Although our understanding of the signaling pathways between the nucleus and chloroplast has improved significantly in recent years (Leon et al., 1998; Rodermel, 2001), comparable studies on plant mitochondria have mainly focused on understanding the nuclear control of mitochondrial biogenesis (e.g., Zabaleta et al., 1998; Mackenzie and McIntosh, 1999; Edqvist and Bergman, 2002). Studies in Arabidopsis by Brennicke and coworkers have shown that regulation of mitochondrial gene expression involves complex transcriptional and posttranscriptional processes, including 5′ and 3′ RNA processing, intron splicing, RNA editing, and RNA stability (Binder and Brennicke, 2003). A comparative study of the transcriptional activities and steady state levels of the various mRNAs encoded in the Arabidopsis mitochondrial genome revealed little correlation between relative promoter activities and transcript abundance, suggesting extensive posttranscriptional regulation (Giegé et al., 2000).
A limited amount of published data supports the view that the expression of nuclear and mitochondrial genes encoding mitochondrial proteins is coordinated. However, a basic mechanistic framework for this coordination has not been established (Mackenzie and McIntosh, 1999). We have previously studied nuclear and mitochondrial gene expression during mitochondrial biogenesis in the developing wheat (Triticum aestivum) leaf (Topping and Leaver, 1990), during anther development in sunflower (Helianthus annuus) (Moneger et al., 1994; Smart et al., 1994), and during germination of maize (Zea mays) embryos (Logan et al., 2001). In each of these developmental systems, we demonstrated coordination of expression of both genomes and examples of both posttranscriptional and posttranslational regulation of gene expression. In our studies on the nuclear restoration of cytoplasmic male sterility in sunflower, we demonstrated that nuclear genes can regulate the stability of specific mitochondrial transcripts at the posttranscriptional level (Gagliardi and Leaver, 1999).
To further investigate the coordination and level of regulation of nuclear and mitochondrial genome expression during mitochondrial turnover and biogenesis, we exploited an Arabidopsis cell culture system. The selection of a cell culture system suitable for these studies was based on the earlier observations of Journet et al. (1986), who demonstrated that starvation of sycamore (Acer pseudoplatanus) cell cultures by withdrawal of sucrose from the medium led to a progressive decrease in O2 consumption (normal and uncoupled) by the cells, which they attributed to a progressive diminution of the number of mitochondria per cell. Addition of sucrose back to the medium led to a recovery of O2 uptake to near the level of normal cells within 24 h and restoration of growth. In this work, we exploited the postgenomic resources available for Arabidopsis by establishing a similar cell culture system to investigate the coordination and underlying levels of regulation of gene expression during modulation of mitochondrial biogenesis.
First, we used DNA microarrays to compare changes in transcripts of the entire mitochondrial genome and representative nuclear genes encoding mitochondrial proteins. Second, we used a proteomic approach to investigate changes in mitochondrial proteins in assembled respiratory complexes. We also investigated the competence of mitochondria to import a nuclear-encoded mitochondrial protein and in organello mitochondrial protein synthesis. A comparison of the transcript and protein changes revealed that coordination of expression of mitochondrial and nuclear genomes during modulation of sugar supply occurs at the protein level, possibly at the level of complex assembly.
RESULTS
Establishment of an Arabidopsis Cell Suspension System to Modulate Mitochondrial Biogenesis
Journet et al. (1986) demonstrated that deprivation of sucrose in sycamore cell cultures resulted in a progressive decrease in endogenous sucrose and starch that after a period of ∼30 h was associated with a progressive decline in cellular respiration. They further showed that after a long period of sucrose starvation the progressive decrease in the uncoupled rate of O2 consumption was attributable to a progressive diminution in the number of mitochondria per cell. Addition of sucrose to cells starved for 60 h resulted in a recovery in the rate of respiration and synthesis of new mitochondria.
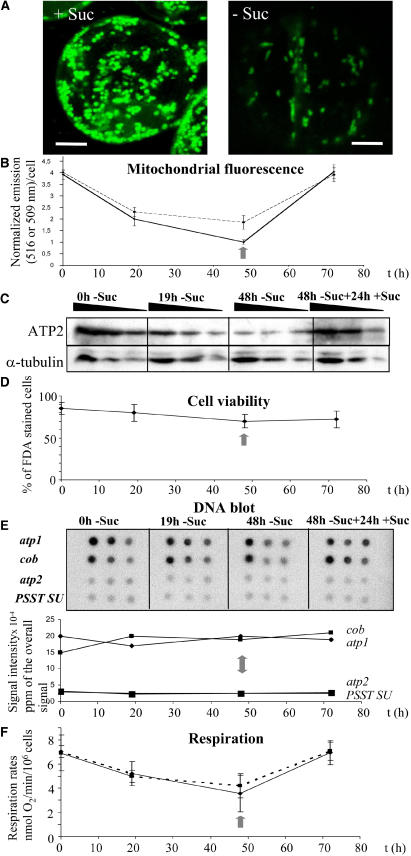
In this study, mitochondrial biogenesis was modulated in an Arabidopsis cell suspension culture (May and Leaver, 1993) by removal and subsequent readdition of sucrose. An Arabidopsis culture was grown for 7 d, divided into two, and subcultured into normal (control) medium (3% [w/v] sucrose) or sucrose-free medium, and samples were taken after a further 19 or 48 h. At 48 h, the cells from the sucrose-free culture were recovered and transferred into a culture medium containing 3% (w/v) sucrose and a further sample taken 24 h later (at 72 h). To visualize the changes in mitochondrial morphology induced by starvation, cells harvested from control and starved cultures after 19 h were stained with Mito Tracker Green FM (Molecular Probes, Eugene, OR), a dye specific for mitochondria and unaffected by membrane potential (Karbowski et al., 2001), and visualized by fluorescence confocal microscopy (Figure 1A). Mitochondria from control cells typically displayed a rounded shape, whereas those in starved cells consistently showed an elongated stick shape, suggesting that mitochondria undergo some structural changes in response to starvation. To obtain some quantitation of this change, Mito Tracker Green fluorescence was measured in starved cells at 0-, 19-, and 48-h time points. Cells were transferred back into a sucrose medium at 48 h and a further sample taken 24 h later at 72 h (Figure 1B). The fluorescence per cell decreased by ∼49% after 19 h and a further 25% after 48 h, suggesting a decrease in mitochondrial number and/or volume per cell as a result of starvation. When sucrose was restored to the medium, fluorescence per cell was restored to initial levels within 24 h.
Figure 1.
Modulation of Mitochondrial Biogenesis during Sugar Starvation of Arabidopsis Cell Cultures.
Samples were taken at 0, 19, and 48 h after transfer to a minus sucrose medium and at 24 h after transfer of 48 h starved cells (vertical gray arrow) into a plus sucrose medium (72 h).
(A) Control (+ Suc) and sucrose-starved (− Suc) cell samples were stained at 19 h with Mito Tracker Green FM and visualized by confocal microscopy. Bars = 5 μm.
(B) Fluorescence emission of cells at 516 nm for Mito Green FM (solid line) and at 509 nm for GFP (dashed line), divided by the emission of a blank and normalized for the number of cells.
(C) Protein gel blot analysis of mitochondrial ATP2 and cytosolic α-tubulin of 20, 10, and 5 μg of total cellular protein isolated at the four time points.
(D) Cell viability estimated by counting the percentage of fluorescent cells after FDA staining.
(E) Changes in nuclear and mitochondrial genome copy number. Labeled total DNA extracted from cells isolated at the four time points was hybridized on nylon filters spotted with 500, 150, and 50 ng of atp1, cob, atp2, and complex I PSST subunit-specific PCR products. The average signal, quantified with a phosphor imager, was plotted against time.
(F) Respiration rate of an equal number of cells at each time point measured either in the growth medium in which they were cultured (solid line) or after transfer into fresh sucrose-containing medium 1 h before measurement (dashed line). Values displayed are the average of three independent experiments, with standard deviation as error bars.
The effect of starvation on mitochondrial number/volume was confirmed by repeating the experimental protocol with a transgenic Arabidopsis cell culture derived, as described by May and Leaver (1993), from transgenic plants expressing a fusion of green fluorescence protein (GFP) with the mitochondrial targeting sequence of ATP synthase subunit 2 (ATP2) (Logan and Leaver, 2000). GFP fluorescence per cell decreased by ∼48% after 48 h starvation, paralleling the result obtained with Mito Tracker Green, and increased to initial 0 h levels at 72 h after readdition of sucrose at 48 h (Figure 1B). These observations confirm that the amount of mitochondria (number and/or volume of mitochondria/cell) can be modulated by changes in the external concentration of sucrose.
The effect of starvation on the relative abundance of mitochondrial protein was further investigated by protein blot analysis (Figure 1C). Whole-cell protein samples were taken at 0, 19, and 48 h after removal of sucrose and 24 h after the readdition of sucrose, fractionated by SDS-PAGE, blotted onto a membrane, and probed with antibodies against the mitochondrial protein ATP2 and the cytosolic protein α-tubulin. Results showed that the abundance of ATP2 decreased during starvation and increased back to the initial level after refeeding of sucrose, whereas α-tubulin abundance remained unchanged during these treatments. This suggested that the mitochondrial protein mass per cell decreased during starvation and was restored to initial levels after readdition of sucrose.
To ensure that the decrease in fluorescence in the cultures was not due to an increase in the proportion of dead cells, cell viability was measured at each time point by staining with fluorescein diacetate (FDA), which only stains living cells. There was no significant change in FDA staining during starvation, suggesting that cell viability is unaffected by the treatment (Figure 1D). Although the proportion of living cells decreased slightly after starvation, the observed changes did not correlate with the far more significant changes in Mito Tracker Green and GFP fluorescence, suggesting that the latter was indeed a true reflection of the change in mitochondria per cell.
To follow changes in the ratio of nuclear to mitochondrial genome copy number during the course of the experimental protocol, total DNA was extracted from cultures at 0, 19, and 48 h after removal of sucrose and 24 h after the readdition of sucrose. Total DNA samples were radiolabeled and hybridized to filters spotted with cDNAs of the mitochondrially encoded atp1 and cytochrome b (cob) genes, the nuclear-encoded mitochondrial subunit atp2, and the PSST subunit of complex I (Figure 1E). Quantification of the hybridization signal revealed no major change in the relative amount of nuclear and mitochondrial DNA during the course of the experimental treatment, suggesting that the mitochondrial genome copy number relative to the nuclear genome copy number per cell remains the same during starvation and subsequent readdition of sucrose to the medium.
The rate of respiration of cell cultures deprived of sucrose decreased by ∼26% after 19 h and ∼49% after 48 h but recovered to the initial 0 h rate within 24 h after the readdition of sucrose (Figure 1F). It is possible that the observed decrease in respiration rate is due to a reduction in the available respiratory substrates in the cell. Therefore, as a control, cells were harvested 1 h before the selected time points and resuspended in a medium containing 3% (w/v) sucrose, and respiration was measured after a further hour (Figure 1F). Essentially the same changes in respiration rates were observed as in the previous experiments, confirming that the changes in respiratory activity together with the diminution of mitochondrial protein mass (Figure 1C) reflected a change in mitochondrial number and/or volume per cell. These changes are in agreement with the observations of Journet et al. (1986) on the effect of sucrose deprivation on mitochondrial number/cell in cultured sycamore cells.
Changes in mRNA Transcript Abundance in Response to Sugar Starvation
To follow changes in expression profiles (transcript abundance) of genes involved in mitochondrial biogenesis and other aspects of cellular metabolism during the experimental modulation induced in cell cultures after sucrose starvation and refeeding, we constructed a customized DNA microarray containing 119 gene-specific probes. The microarray contained the entire gene content of the Arabidopsis mitochondrial genome (Unseld et al., 1997) and the genes of nuclear-encoded mitochondrial proteins representative of subunits of the major respiratory complexes of the inner mitochondrial membrane (complexes I, II, III, IV, and V) and the mitochondrial ribosome. Gene probes encoding representative proteins of the transcription and translation apparatus in different compartments of the plant cell, as well as proteins involved in carbon metabolism, lipid synthesis, amino acid synthesis, pyruvate metabolism, starch synthesis, response to oxidative stress, proteases, and chloroplast metabolism, were also included on the microarray (Table 1). Each gene was present as three replicate spots on the microarray.
Table 1.
Changes in Transcript Abundance during Sugar Starvationa
| Function/Transcript | AGI | S1 | S2 | RS |
|---|---|---|---|---|
| Complex I | ||||
| nad1 | AtMg00516 | 0.1 | 0.6 | −3.4 |
| nad2 | AtMg00285 | 0.6 | 0.6 | −2.4 |
| nad3 | AtMg00990 | 0.3 | −0.3 | 0.3 |
| nad4 | AtMg00580 | 0.6 | −0.3 | −1.8 |
| nad4L | AtMg00650 | 0.5 | 0.0 | −3.5 |
| nad5 | AtMg00513 | 0.2 | 0.1 | −1.2 |
| nad6 | AtMg00270 | 0.6 | 0.5 | −4.0 |
| nad7 | AtMg00510 | 0.1 | 1.2 | −2.7 |
| nad9 | AtMg00070 | 0.3 | 0.5 | −3.1 |
| B14 SU | At3g12260 | −0.4 | −0.9 | 0.2 |
| 40-kD SU | At2g20360 | 0.1 | −1.1 | 0.8 |
| 76-kD Fe-S | At5g37510 | −0.9 | −0.1 | 0.0 |
| PSST SU | At5g11770 | −0.3 | −0.8 | −0.9 |
| Complex II | ||||
| Ψsdh4 | AtMg01420 | 0.6 | 1.3 | −1.3 |
| sdh2 Fe-S | At5g40650 | 0.2 | 1.7 | 0.1 |
| Complex III | ||||
| cob | AtMg00220 | 0.4 | 0.0 | −2.1 |
| Cytochrome c1 | At5g40810 | 1.5 | −0.7 | 2.1 |
| 11-kD SU | At1g15120 | −1.5 | −1.5 | 1.4 |
| 14-kD SU | At5g25450 | −0.9 | −0.6 | 0.2 |
| MPP α | At1g51980 | −1.2 | −0.6 | 1.6 |
| MPP β | At3g02090 | −1.3 | −1.1 | −0.5 |
| Complex IV | ||||
| cox1 | AtMg01360 | 0.6 | 0.9 | −2.1 |
| cox2 | AtMg00160 | 0.4 | 0.9 | 2.1 |
| cox3 | AtMg00730 | 0.3 | 0.8 | 1.4 |
| coxVc | At2g47380 | −0.2 | −0.7 | 0.5 |
| coxVIb | At4g28060 | −0.7 | 0.9 | −0.1 |
| coxVb | At1g80230 | −0.6 | 0.4 | −1.1 |
| Complex V | ||||
| atp1 | AtMg01190 | −0.1 | 0.6 | −1.9 |
| atp6 | AtMg01170 | 0.3 | 0.8 | −1.6 |
| atp8 | AtMg00480 | 0.3 | 0.2 | −2.9 |
| atp9 | AtMg01080 | 0.3 | −0.2 | −2.4 |
| atp2 | At5g08670 | −1.1 | 0.3 | −0.3 |
| atp2 | At5g08690 | −1.3 | −0.6 | 0.6 |
| atpδ′ | At5g47030 | −0.5 | −1.1 | 1.1 |
| atpγ | At2g33040 | 0.3 | −2.4 | 2.4 |
| 6-kD SU | At3g46430 | −0.9 | 0.0 | 0.0 |
| Mito ribosome | ||||
| rrn26 | NA | 0.2 | 0.2 | −3.1 |
| rrn18 | NA | 0.2 | 0.2 | −3.1 |
| rrn5 | NA | 0.0 | 0.7 | −1.7 |
| rpl2 | AtMg00560 | 0.8 | 0.4 | −3.2 |
| rpl5 | AtMg00210 | 0.5 | 0.2 | −0.1 |
| rpl16 | AtMg00080 | 0.1 | 0.1 | −3.0 |
| rps3 | AtMg00090 | 0.1 | 0.3 | −3.3 |
| rps4 | AtMg00290 | 0.3 | 1.5 | 1.4 |
| rps7 | AtMg01270 | −0.5 | 0.3 | −1.0 |
| rps12 | AtMg00980 | 0.5 | 1.7 | −0.2 |
| rps24 | At5g28060 | −0.2 | −0.8 | 0.2 |
| rps14 | At2g34520 | 0.0 | −1.1 | 0.0 |
| Cytochrome c maturation | ||||
| ccmFN2 | AtMg00960 | 0.4 | 0.4 | −3.0 |
| ccmB | AtMg00110 | 0.1 | −0.2 | −3.5 |
| ccmC | AtMg00900 | 0.6 | 0.4 | −2.0 |
| ccmFN1 | AtMg00830 | 0.6 | 1.0 | −2.9 |
| ccmFC | AtMg00180 | −0.2 | 0.1 | −2.8 |
| Other | ||||
| orf240a | AtMg00690 | 0.4 | 0.3 | −3.4 |
| orf262 | AtMg01090 | 0.0 | −0.2 | −0.2 |
| orf291 | AtMg01280 | 0.8 | 0.2 | −0.3 |
| orf294 | AtMg01200 | −0.7 | 0.3 | −2.6 |
| orf25 | AtMg00640 | 0.5 | −0.9 | −2.6 |
| matR | AtMg00520 | −0.1 | 0.0 | −2.0 |
| mtt2 | AtMg00570 | 0.3 | 0.2 | 0.1 |
| Transcription apparatus | ||||
| RNA pol III 160 kD | At5g60040 | −0.9 | −1.9 | −0.3 |
| Chloro RNA pol (PEPα chain) | AtCg00740 | 0.0 | −1.3 | −0.9 |
| Mito RNA pol | At1g68990 | 0.2 | −0.3 | −0.5 |
| RNA pol II 13.3-kD SU | At3g52090 | −0.4 | −0.5 | 1.1 |
| RNA pol II elongation factor | At1g75950 | 0.5 | −4.7 | −1.7 |
| Transcription factor tga1 | At5g65210 | 1.0 | 4.8 | −0.7 |
| Transcription factor SF3 | At1g10200 | 0.2 | −0.2 | 0.0 |
| Transcription factor BTF3 | At1g17880 | −2.2 | −0.9 | 0.7 |
| Heat shock transcription factor (HSTF8) | At3g51910 | 0.7 | 0.6 | −0.3 |
| Translation apparatus | ||||
| Glu tRNA synthetase | At3g62120 | −1.5 | 0.2 | 0.6 |
| Arg tRNA synthetase | At4g26300 | −0.7 | 0.5 | 0.7 |
| Gly tRNA synthetase | At1g29880 | −0.9 | 0.4 | 1.0 |
| Mito elongation factor G | At1g45332 | −0.8 | 1.0 | −2.3 |
| Elongation factor eEF-1 | At1g07930 | −2.9 | 0.0 | 0.2 |
| Elongation factor Tu | At4g02930 | −1.1 | 1.5 | 0.1 |
| Carbon metabolism | ||||
| Citrate synthase | At2g44350 | −0.9 | 0.0 | 0.4 |
| Aconitase | At4g35830 | 0.0 | 0.6 | 0.2 |
| Aldolase c1 | At3g52930 | −1.8 | −1.3 | 1.3 |
| Enolase | At2g36530 | −1.3 | −1.9 | 1.5 |
| Hexokinase 1 | At4g29130 | −0.1 | −0.2 | 0.3 |
| Mito malate dehydrogenase | At3g15020 | −0.6 | 0.3 | 1.0 |
| Cytochrome malate dehydrogenase | At1g04410 | −0.4 | 0.0 | −0.1 |
| Lipid synthesis | ||||
| Phospholipase D | At4g11850 | 0.1 | 0.1 | −0.2 |
| Phosphinositide-spec phospholipase C | At3g47220 | 0.7 | 0.3 | 0.6 |
| Acyl-desaturase | At2g43710 | −1.1 | 0.2 | 0.1 |
| Acetyl-coA carboxylase | At1g36160 | 0.0 | 0.0 | 0.0 |
| Amino acid synthesis | ||||
| Glu dehydrogenase | At5g18170 | 1.1 | −1.3 | −2.0 |
| Asp aminotransferase Asp2 | At5g19550 | −0.2 | −3.3 | 0.2 |
| Pyruvate metabolism | ||||
| Pyruvate dehydrogenase E1, β SU mito | At5g50850 | −0.6 | −0.1 | 0.1 |
| Pyruvate dehydrogenase E1, α SU mito | At1g59900 | −1.1 | 1.5 | 0.1 |
| NADP-dep malic enzyme | At5g11670 | 0.0 | 0.2 | 0.7 |
| Cytosol pyruvate kinase | At2g36580 | −0.7 | −0.9 | 0.3 |
| Plastid pyruvate kinase | At3g22960 | −1.1 | −0.3 | 0.7 |
| Starch synthesis | ||||
| ADP-glucose pyrophosphorylase | At5g48300 | −0.6 | 0.1 | 0.6 |
| Oxidative stress | ||||
| Glutathione S-transferase ERD11 | At1g02920 | 0.9 | −0.4 | −0.1 |
| Glutathione S-transferase PM24 | At2g02930 | 1.4 | 0.9 | −0.1 |
| Glutathione peroxidase, cytosolic | At2g31570 | 2.1 | 2.0 | −1.8 |
| Glutathione peroxidase, chloroplast | At2g25080 | 1.0 | −0.1 | −0.1 |
| l-ascorbate peroxidase, cytosolic | At4g09010 | 0.3 | 0.5 | −0.1 |
| l-ascorbate peroxidase, chloroplast | At1g07890 | −1.0 | 1.0 | 0.3 |
| Aldose reductase, AR (aldehyde reductase) | At2g37790 | −0.4 | 0.0 | −0.2 |
| Mn superoxide dismutase | At3g10920 | −0.6 | 0.0 | 0.3 |
| Proteases | ||||
| LON protease (Ser prot, ATP dep) | At5g26860 | −1.7 | 0.0 | 0.3 |
| Subtilisin-like protease (Ser) | At2g05920 | −2.0 | 0.5 | 0.7 |
| Cys protease (drought inducible) | At4g11320 | 1.1 | −2.0 | 0.0 |
| 26S protease AAA ATPase SU (RPT3) | At5g58290 | −0.4 | 2.5 | 0.0 |
| CLPP-like protease (nClpP2) | At5g23140 | 0.2 | −2.0 | −0.1 |
| Thiol (Cys) protease rd21A | At5g43060 | 2.2 | −0.2 | −2.0 |
| ATP dep CLP protease (ClpP1) | At1g02560 | 0.0 | 0.0 | −0.3 |
| Chloroplast metabolism | ||||
| ATPase δ | At4g09650 | 1.6 | −0.4 | 0.1 |
| ATPase ɛ | AtCg00470 | 0.5 | −0.6 | −1.3 |
| Rubisco activase | At2g39730 | 0.8 | 0.0 | −0.1 |
| Rubisco BPβ SU | At1g26230 | −0.3 | −1.1 | −0.6 |
Individual genes represented on the microarray are listed and classified according to functions. Gene identifiers (Arabidopsis Genome Initiative [AGI]) are given for all the transcripts. The values displayed in the table are the triplicate average log ratio for each independent experiment. S1 compares 19 h +sucrose (Cy5) with 19 h −sucrose (Cy3); S2 compares 19 h −sucrose (Cy5) with 48 h −sucrose (Cy3); RS compares 48 h −sucrose (Cy3) with 72 h (48 h −sucrose + 24 h +sucrose (Cy5). NA, not applicable; SU, subunit; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
Total RNA was isolated from control cells cultured for 19 h in the presence of sucrose and from sucrose-starved cells after 19, 48, and 24 h after the readdition of sucrose to cells starved for 48 h. Pairwise combinations of cDNAs, synthesized from the RNA extracted from different time points and treatments, labeled with either Cy5 or Cy3-dCTP (Table 1) were hybridized to the microarrays. S1 experiments compare 19 h +sucrose with 19 h −sucrose samples; S2 experiments compare 19 h −sucrose with 48 h −sucrose samples; RS experiments compare 48 h −sucrose with 24 h +sucrose after 48 h −sucrose (Table 1). Data were acquired with an Affymetrix A428 scanner (Santa Clara, CA). The Cy5 and Cy3 signal intensities were normalized against the overall signal intensity acquired in the Cy3 channel. The normalized intensities were averaged for the three technical replicate spots on the array, and log ratios (log base 2) were calculated. For each experiment, three biological replicates were performed (i.e., pairs of RNA samples were extracted during three independent experiments). In one experiment, a reverse labeling reaction was done in which the Cy5 and Cy3 labels were swapped. A Welch two-sample t test was applied to show that the Cy5 and Cy3 intensities were significantly above background (see Supplemental Table 1 online). The reproducibility of the biological replicates was assessed by calculating correlation coefficients (Table 2). The average values were 0.83 for S1, 0.66 for S2, and 0.92 for RS, well within the range of significance (P < 0.05). To compare the overall expression variation between the different microarray experiments, box plots were made, and the median absolute deviations (MADs) were calculated (see Supplemental Figure 1 online). Expression levels between the biological replicates in S1, S2, and RS experiments were found to be homogeneous, which, together with the high correlation coefficients between the genes, indicated a high reproducibility of the data. Nevertheless, the MAD values were more variable for S1 than for the other experiments (see Supplemental Figure 1 online). For this reason, a between-array normalization based on the quantile method was performed for each experiment (S1, S2, and RS). Altogether, when comparing the MAD values, it was observed that the average MAD value for S1, S2, and RS was 57 and 41% higher in RS than in S1 and S2, respectively, suggesting that the overall expression variation was higher after sucrose was added back. A differential expression analysis was performed using the Limma software package to determine which genes were significantly differentially expressed across treatments (e.g., between 19 h +sucrose and 19 h −sucrose in S1). An observation of results taking a false discovery rate of 5% shows a total of 76 (61%), 74 (59%), and 67 (54%) genes significantly upregulated or downregulated for S1, S2, and RS, respectively (see Supplemental Table 1 online for a detailed list).
Table 2.
DNA Microarray Biological Replicates Are Highly Reproduciblea
| Replicate | S1 I | S1 IIIb | S2 I | S2 III | RS I | RS III |
|---|---|---|---|---|---|---|
| S1 II | 0.866 | 0.926 | – | – | – | – |
| S1 III | 0.805 | 1.000 | – | – | – | – |
| S2 II | – | – | 0.701 | 0.559 | – | – |
| S2 III | – | – | 0.732 | 1 | – | – |
| RS II | – | – | – | – | 0.956 | 0.893 |
| RS III | – | – | – | – | 0.906 | 1.000 |
Correlation coefficients for microarray biological replicates I, II, and III were calculated before between-array normalization.
Cy5 and Cy3 dyes were reversed in experiment III.
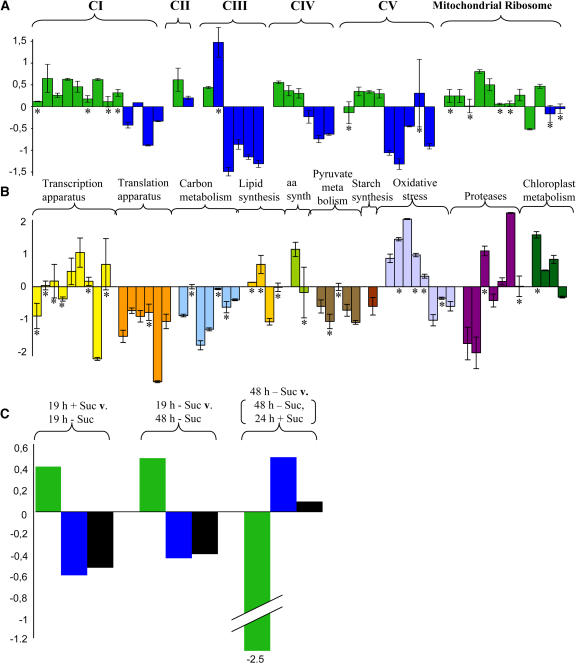
A comparison of changes in transcript abundance of mitochondrial and nuclear-encoded protein subunits of the major respiratory complexes of the inner mitochondrial membrane and representative components of the mitochondrial ribosome in cultures grown plus and minus sucrose for 19 h revealed an increase in transcript abundance of nearly all of the mitochondrially encoded proteins (green bars in Figure 2A, column S1 in Table 1) and a decrease in transcript abundance of nearly all of the nuclear-encoded proteins present in these complexes (blue bars in Figure 2A, Table 1). The steady state levels of transcripts derived from genes whose protein products are involved in other aspects of cellular metabolism revealed several consistent changes (Figure 2B, Table 1). In response to 19 h starvation, there was a significant decrease in transcript abundance of genes involved in mitochondrial translation and carbon and pyruvate metabolism, which is consistent with an overall slowdown of cellular metabolism during starvation. These results are also consistent with the downregulation of genes involved in translation and metabolism during sucrose starvation observed by Contento et al. (2004).
Figure 2.
The Effect of Sugar Starvation on Changes in Transcript Abundance of All Mitochondrial Genes, Nuclear Genes Encoding Major Subunits of the Major Respiratory Enzyme Complexes, and a Selection of Nuclear Genes Encoding Representative Proteins Involved in Gene Expression and Other Areas of Cellular Metabolism.
Changes in transcript abundance of the individual genes are expressed as base 2 log ratios, in the order listed in Table 1, in cultures grown for 19 h plus and minus sucrose.
(A) Mitochondrial genes encoding components of the respiratory chain complexes (I to V) and the mitochondrial ribosome are shown in green and the nuclear-encoded components in blue.
(B) Genes encoding proteins involved in other aspects of cellular metabolism and color coded according to functional families. Genes marked with an asterisk were not significantly regulated between 19 h plus and 19 h minus sucrose.
(C) Changes in the average transcript abundance (average log ratio of the genes significantly regulated) for mitochondrially encoded proteins (green), nuclear-encoded proteins present in respiratory enzyme complexes with mitochondrial gene products (blue), and all of the nuclear-encoded gene products represented on the microarray (black), corresponding to the sampling times described in Figure 1. Error bars represent standard deviations for three independent experiments.
A comparison of the averages of only the statistically significant changes in transcript abundance (only significantly regulated genes) of all of the mitochondrially encoded gene products with those of the nuclear-encoded proteins in the same complexes revealed that after 19 and 48 h of starvation the abundance of the mitochondrially encoded gene transcripts increased (Figure 2C, green bars), whereas the nuclear-encoded gene transcripts decreased significantly (Figure 2C, blue bars). The average of the statistically significant changes in transcript abundance of all of the nuclear-encoded genes on the array also revealed a marked decrease after starvation for 19 or 48 h (Figure 2C, black bars). This difference in transcript abundance between mitochondrially and nuclear-encoded gene products was reversed after readdition of sucrose to the 48 h starved cultures with a marked decrease in the abundance of mitochondrially encoded transcripts and an increase in nuclear-encoded transcripts of proteins in the same complex (Figure 2C).
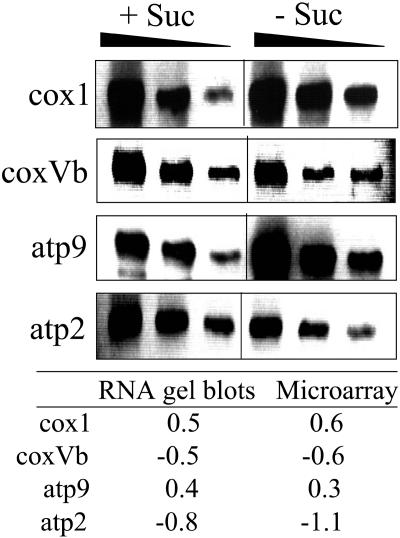
To validate the microarray data, RNA gel blot experiments were performed with a representative selection of nuclear-encoded and mitochondrially encoded genes (Figure 3). Total RNA was isolated from cells cultured for 19 h in the presence or absence of sucrose. Samples of 10, 5, and 2.5 μg of RNA were separated on agarose denaturing gels, blotted on nylon membranes, and hybridized with gene-specific probes. The quantified signals were used to calculate log ratios. These were compared with the ones obtained with the microarray experiments, revealing similar results (i.e., a similar change in transcript abundance in response to starvation, thus confirming the microarray data) (Figure 3).
Figure 3.
Validation of Microarray Data.
RNA gel blots with 10, 5, and 2.5 μg of total RNA extracted from cells grown for 19 h plus and minus sucrose were hybridized with specific labeled PCR products representing the mitochondrially encoded cytochrome oxidase subunit 1 (cox1) and atp9 and the nuclear-encoded cytochrome oxidase subunit Vb (coxVb) and atp2 genes. Signals were quantified with a phosphor imager and averaged for the three amounts of RNA used. A base 2 log ratio of the +S and –S signal intensities was calculated and compared in the table with the results obtained for the microarray experiments.
Changes in Mitochondrial Proteins in Response to Sugar Starvation
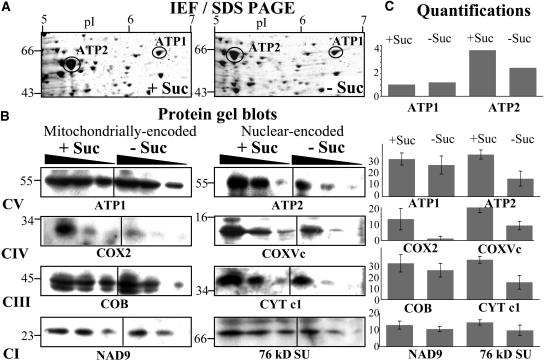
To further investigate the effect of sucrose starvation on the turnover of mitochondria in Arabidopsis cell cultures, we performed an analysis of the total mitochondrial proteome and analyzed changes in the amounts of the mitochondrially encoded F1 α-subunit ATP1 and the nuclear-encoded F1 β-subunit ATP2 of the mitochondrial F1-F0 ATPase (complex V) by two-dimensional gel electrophoresis (isoelectric focusing, SDS-PAGE). We had previously identified these two subunits of the mitochondrial ATPase on this type of gel (Millar et al., 2001), ATP1 being the only mitochondrially encoded protein identified. A comparison of the relative abundance of these proteins among total mitochondrial proteins extracted from 19 h, control, and minus sucrose cultures revealed that abundance of the nuclear-encoded ATP2 protein consistently decreased by ∼40% after 19 h starvation, whereas the abundance of the mitochondrial-encoded ATP1 protein remained unchanged (Figures 4A and 4C).
Figure 4.
The Effect of Sucrose Starvation on the Amount of Mitochondrially and Nuclear-Encoded Proteins from Respiratory Complexes I to V (CI to CV) in Mitochondria from Arabidopsis Cell Cultures.
(A) Mitochondria were isolated from cultures grown for 19 h plus (+ Suc) or minus (− Suc) sucrose and total protein analyzed by two-dimensional isoelectric focusing (IEF), SDS-PAGE. ATP1 and ATP2 were localized on these gels as described previously (Millar et al., 2001).
(B) Protein gel blots of equal amounts (20, 10, and 5 μg) of total mitochondrial protein isolated from cultures grown for 19 h plus (+ Suc) or minus (− Suc) sucrose and probed with antibodies against ATP1, ATP2, COX2, COXVc, COB, CYTc1, NAD9, and the 76-kD subunit of complex I.
(C) Quantitation of the proteins on the two-dimensional gels and protein gel blots. The intensities of the stained spots were analyzed with the Z3 proteome analysis software (Compugen, Tel-Aviv, Israel), averaged for the three protein loadings, and expressed as histograms of ppm of the overall intensity. The values obtained were averaged for three independent experiments, with error bars representing the standard deviation.
These results were confirmed by protein gel blot analysis and extended to proteins of the respiratory complexes I, III, and IV. Duplicate mitochondrial protein samples were fractionated by one-dimensional SDS-PAGE, blotted onto membranes, and probed with antibodies specific for ATP1 and ATP2, for the mitochondrially encoded cytochrome oxidase subunit 2 (COX2), COB, and NADH dehydrogenase subunit 9 (NAD9), for the nuclear-encoded COXVc and cytochrome c1 (CYTC1), and the 76-kD subunit (SU) of complex I (Figures 4B and 4C). A consideration of the average of three independent experiments indicated that the amount of the nuclear-encoded proteins was reduced by 58 (ATP2), 45 (COXVC), 52 (CYTC1), and 36% (76 kD SU) in mitochondria from starved cells, whereas the mitochondrially encoded protein levels were reduced by 10 (ATP1), 78 (COX2), 12 (COB), and 17% (NAD9) during starvation. With the exception of COX2, this result suggested that the mitochondrially encoded protein levels were reduced to a lesser degree (i.e., less affected by starvation compared with their nuclear-encoded counterparts) (Figures 4B and 4C).
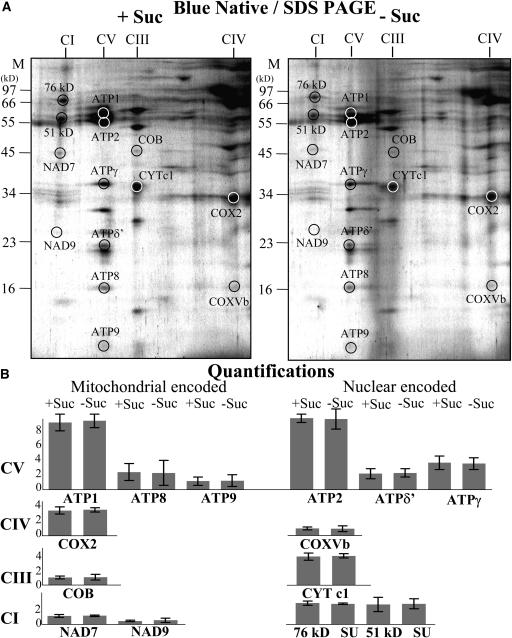
The Relative Stoichiometry and Protein Subunit Composition of Major Enzyme Complexes of the Inner Mitochondrial Membrane Are Unaffected by Starvation
The effect of starvation on the protein subunit composition of representative respiratory enzyme complexes of the inner mitochondrial membrane was investigated by two-dimensional Blue-Native SDS gel electrophoresis (Schagger, 2001). Mitochondria were isolated from 19 h control and sucrose-starved cell cultures. Assembled enzyme complexes were fractionated in the first dimension in their native form according to size on nondenaturing gels, and then subunits of individual complexes were resolved in the second dimension on SDS-denaturing gels (Figure 5). We have previously described the fractionation, identification, and subunit composition of the major enzyme complexes of the inner mitochondrial membrane in Arabidopsis (Giegé et al., 2003; Sabar et al., 2003). We consistently observed that the relative proportion of protein subunits in the major respiratory complexes (i.e., complexes I, III, IV, and V) was virtually identical in mitochondria from control and starved cells (Figure 5A). To further confirm this, the abundance of seven mitochondrially encoded and seven nuclear-encoded protein subunits of complexes I, III, IV, and V identified according to Jänsch et al. (1996), Brumme et al. (1998), Giegé et al. (2003), Heazlewood et al. (2003a) (2003b), and Sabar et al. (2003) was quantified for three replicate gels (Figure 5B) and shown to be equally abundant in mitochondria from control and starved cells. It is not possible to assess the amount of each enzyme complex relative to total mitochondrial protein because the complexes dominate Blue-Native gels; therefore, changes in other mitochondrial proteins are not accounted for in relative abundance estimations of individual proteins on the gel.
Figure 5.
The Relative Stoichiometry and Protein Subunit Composition of Major Enzyme Complexes of the Inner Mitochondrial Membrane Are Unaffected by Starvation.
Mitochondria were isolated from cultures grown for 19 h plus (+ Suc) or minus (− Suc) sucrose and fractionated by Blue-Native gels in the first dimension and SDS-denaturing gels in the second dimension to resolve proteins from respiratory complexes I to V (A). The spot intensities of seven mitochondrially encoded and seven nuclear-encoded proteins from complexes I to V were quantified with the Z3 proteome analysis software (Compugen), expressed as ppm of the overall intensity, and averaged for three replicate gels. Error bars show the standard deviation between the gels (B).
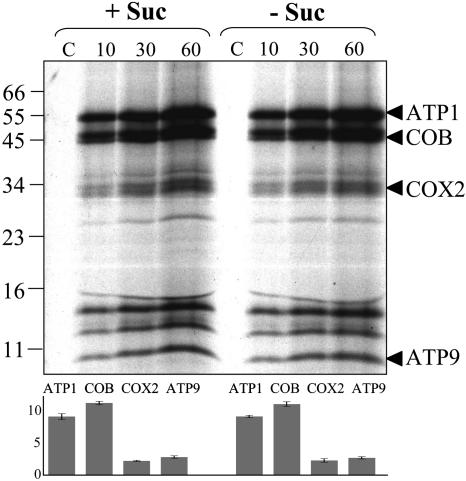
In Organello Translation by Mitochondria Isolated from Control and Starved Cultures Is Not Affected by Sugar Starvation
To investigate whether the capacity for in organello translation by mitochondria was affected by starvation of cell cultures, mitochondria were isolated from 19 h control and starved cells and labeled with 35S-Met. Incorporation of 35S-Met was estimated as previously described, and labeled mitochondrial proteins were visualized by SDS gel electrophoresis followed by autoradiography of the dried gels (Leaver et al., 1983). The incorporation of 35S-Met into trichloroacetic acid–precipitable mitochondrial protein was ∼7000 cpm/μg protein by mitochondria from both control and starved cells after 60 min of incorporation, and the pattern of labeled mitochondrial proteins was indistinguishable (Figure 6). Four proteins were identified according to their calculated molecular weight (Unseld et al., 1997), and the amount of 35S-Met incorporated into each was quantified at three different time points during the incubation (data not shown). The abundance of the four proteins was found to be identical for mitochondria isolated from both control and starved cells. To demonstrate the absence of bacterial contamination, control experiments were performed where sodium acetate was used as an energy source. Sodium acetate is metabolized by bacteria only and cannot be used as a substrate by mitochondria (Leaver et al., 1983). No proteins were synthesized in the control experiments (Figure 6, lane C).
Figure 6.
There Is No Difference in the Capacity for in Organello Protein Synthesis by Mitochondria Isolated from Control and Sucrose-Starved Arabidopsis Cell Cultures.
Autoradiogram of proteins synthesized in organello for 10, 30, and 60 min by mitochondria isolated from 19 h plus (+ Suc) or minus (− Suc) sucrose cultures and fractionated on 12% (w/v) PAGE. Lanes marked with a C are bacterial contamination controls where sodium acetate was used as an energy source. Proteins are identified according to their calculated molecular weight. The histogram shows the quantification with the Z3 proteome analysis software (Compugen) of the four identified proteins, expressed as ppm of the overall intensity, in plus sucrose and minus sucrose experiments. The average for the three different time points averaged for three independent experiments is represented, with standard deviations as error bars.
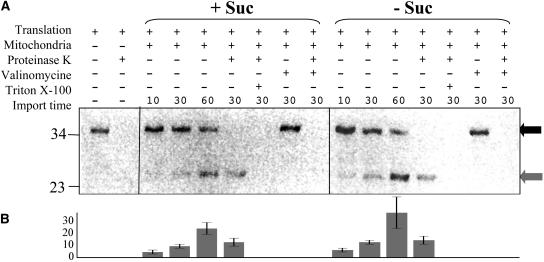
Import of Proteins by Mitochondria Isolated from Control and Starved Cultures Is Not Affected by Sugar Starvation
Another function that could be affected by starvation is the import of nuclear-encoded, cytosolically synthesized proteins in mitochondria. Indeed, the import process could potentially be involved in the regulation of mitochondrial biogenesis by controlling the capacity for import of individual nuclear-encoded proteins into the mitochondrion. To conduct a preliminary investigation as to whether starvation affected the competence of mitochondria to import protein, the mitochondrial targeting sequence of ATP2 fused to GFP (Logan and Leaver, 2000) was radiolabeled in an in vitro protein synthesizing system and imported into equal amounts of purified mitochondria from 19 h control and starved cells (Figure 7). The ATP2 targeting sequence/GFP fusion protein has previously been shown to be efficiently targeted to Arabidopsis mitochondria (Logan and Leaver, 2000). Import was monitored over the course of three different incubation periods (10, 30, and 60 min). Two radiolabeled proteins were detected; the 36-kD in vitro–synthesized protein precursor decreased in abundance during the import reaction, whereas the 27-kD mature-processed GFP was found to accumulate during import. Proteinase K treatment of the reisolated mitochondria removed the residual 36-kD precursor protein but not the imported 27-kD protein, which was protected from protease digestion. Control experiments with valinomycin showed that import depended on membrane potential. In other control experiments Triton X-100 was used to rupture the mitochondrial membrane, thus exposing the imported protein to protease digestion. The intensities of the bands corresponding to imported proteins were quantified with a phosphor imager as ppm of the overall intensity for three independent experiments, revealing that the abundance of imported proteins did not decrease in mitochondria isolated from starved cells compared with control cells (Figure 7B). In fact, the intensity of the protein imported at 60 min by mitochondria from starved cells was found to increase, on average, by 32%. This suggests that the capacity to import proteins from the cytosol is not affected by starvation. Given the dependence of protein import upon membrane potential, this also indicates that mitochondrial membrane potential is not dramatically reduced during starvation.
Figure 7.
The Capacity for in Vitro Mitochondrial Protein Import Is Not Impaired in Mitochondria Isolated from Sucrose-Starved Arabidopsis Cell Cultures.
(A) In vitro–synthesized, 35S-labeled, fusion protein between the nucleotide sequence of ATP2 and GFP (translation) was incubated with mitochondria isolated from 19 h plus (+ Suc) or minus (− Suc) sucrose cultures for 10, 30, and 60 min. Experiments were also performed in the presence (+) or absence (−) of Proteinase K (10 units/mg proteins for 30 min), Valinomycin (50 pM), and Triton X-100 (0.1%) reisolated, fractionated on 15% (w/v) PAGE, dried, and autoradiographed. The 36-kD in vitro–synthesized precursor protein is indicated by a black arrow, the 27-kD imported protein product by a gray arrow.
(B) Quantitation with the Z3 proteome analysis software (Compugen) of the 27-kD imported protein, expressed as ppm of the overall intensity. The histogram shows the average for three independent experiments, with standard deviation as error bars.
DISCUSSION
We describe the establishment of an Arabidopsis cell culture system to modulate mitochondrial biogenesis in response to sugar starvation and refeeding to investigate the level at which the expression of nuclear and mitochondrial genes is coordinated. This coordination is essential for mitochondrial biogenesis in general and for the assembly of multisubunit complexes of the respiratory chain and mitochondrial ribosome, which are composed of both mitochondrial and nuclear-encoded subunits in particular. The stress induced by sucrose deprivation in the cultures led to structural changes in mitochondria, a decrease in mitochondrial volume, and a reduction in the rate of cellular respiration. All these changes could be reversed by the readdition of sucrose (Figure 1). During the course of these experiments, the mitochondrial genome copy number relative to nuclear genome copy number per cell remained constant, suggesting that the pathway regulating relative genome copy number is distinct from those controlling mitochondrial volume and structure.
We first performed a customized cDNA microarray analysis to better understand how and at which level the coordination of nuclear and mitochondrial genome expression was achieved during these changes in mitochondrial structure and activity. This clearly showed that the expression of the mitochondrial and nuclear genomes in response to starvation is not coordinated at the level of transcript abundance (Figures 2 and 3). The transcripts for the nuclear-encoded proteins of the respiratory enzyme complexes decreased in relative abundance during sugar starvation, whereas the mitochondrially encoded protein transcripts increased. This divergent trend in transcript abundance was reversed on readdition of sucrose to the cultures (Figure 2).
If we assume that nuclear and mitochondrial gene expression levels were coordinated before sucrose deprivation, this suggests that the abundance of nuclear transcripts could become the limiting factor for the synthesis and assembly of functional mitochondrial complexes, at least in response to sugar starvation. The observed increase in mitochondrial transcript abundance need not be due to an increase in gene expression in the mitochondria. Indeed, if there was an overall decrease of nuclear transcript abundance after starvation, as suggested by the data, then mitochondrial RNA as a proportion of total cellular RNA would increase.
The microarray data is a measure of steady state transcript levels and consequently reflects not only changes in transcription itself, but also other posttranscriptional regulatory mechanisms, including RNA processing and turnover. The apparent lack of coordination of gene expression between nuclear and mitochondrial genomes observed in this analysis suggests that translational and posttranslational regulatory processes may play an important role in the coordination of genome expression during mitochondrial biogenesis.
To investigate these possibilities further, we performed preliminary experiments to measure changes in abundance and assembly of nuclear-encoded and mitochondrially encoded subunits of specific enzyme complexes of the mitochondrial inner membrane and to assess in organello protein synthesis by isolated mitochondria and competence for mitochondrial protein import.
Protein gel blot analysis of total mitochondrial protein from 19 h control and sucrose-starved cell cultures revealed that the level of the nuclear-encoded subunits of respiratory complexes were consistently reduced, whereas the level of mitochondrially encoded subunits of the same complexes (with the exception of COX2) decreased to a lesser extent in response to starvation (Figure 4). However, the relative stoichiometry and protein subunit composition of complexes I, III, IV, and V of the mitochondrial inner membrane are unaffected by starvation in 19 h cultures (Figure 5).
These results support the hypothesis that the availability of nuclear-encoded subunits of respiratory enzyme complexes could be a limiting factor in the regulation of mitochondrial biogenesis in response to sugar starvation. The decrease in the levels of mitochondrially encoded protein was not due to a loss in the capacity for de novo protein synthesis by mitochondria because this capacity was unchanged in mitochondria from starved cultures. This was investigated by studying the capacity for in organello protein synthesis by mitochondria isolated from 19 h control and sucrose-starved cell cultures. No significant differences were detected between mitochondria from control and starved cells, in either the protein synthetic activity or translation products (Figure 6).
We also investigated whether sugar starvation affected the ability of mitochondria to import nuclear-encoded, cytosolically synthesized proteins and could thus play a regulatory role in the assembly of mitochondrial enzyme complexes. Mitochondria isolated from 19 h control and sucrose-starved cultures showed no reduction in the capacity to import the nuclear-encoded mitochondrial targeting sequence of ATP2 fused to GFP (Figure 7).
Taken together, these results suggest that the coordination of gene expression between mitochondria and nucleus during modulation of mitochondrial biogenesis is posttranslational and at the level of protein complex assembly. Thus, it would be expected that the mitochondrially encoded proteins that are synthesized in excess and not assembled into complexes would be degraded by specific protease activities, such as the one described by Sarria et al. (1998). This does not seem to be happening to the same extent in the case of ATP1, COB, and NAD9 (i.e., their abundance does not decrease as much as the abundance of the nuclear-encoded subunits of the same complexes), but results from other experimental plant systems (Lu et al., 1996) and yeast (reviewed in Rep and Grivell, 1996) suggest that rapid degradation of nonfunctional or unassembled mitochondrially encoded proteins seems to be the norm in mitochondria.
In summary, we have described an experimental cell culture system in which mitochondrial biogenesis was affected after sugar deprivation. This system has been exploited to investigate the level at which the expression of nuclear and mitochondrial genes is coordinated. Sugar deprivation results in structural changes and a reduction in mitochondrial volume. The observed reduction of mitochondrial respiration after sugar deprivation could be explained by a decrease in assembled mitochondrial respiratory complexes. Nevertheless, it cannot be excluded that the effect observed could be due to the disassembly of respiratory supercomplexes (Eubel et al., 2003) without disassembly of the individual respiratory complexes and turnover of subunits. This is parallelled by decreases in the transcript abundance of nuclear-encoded components of mitochondrial respiratory complexes and ribosomes but not in the transcript abundance of mitochondrially encoded components, which generally increase. The conclusion that can be drawn from this observation is that the expression of mitochondrial and nuclear genes is not coordinated at the transcript level. Further investigations of posttranscriptional events suggested that coordination is in fact posttranslational and occurs at the level of protein-complex assembly. These results can be formulated into a model that suggests a mechanism of coordinated expression of mitochondrial and nuclear genes during changes in mitochondrial biogenesis caused by sugar deprivation. The sequence of events is as follows. Deprivation of sugar leads to changes in nuclear gene expression (probably because of the activity of specific transcription factors; see, e.g., Chen et al., 2002; Vrebalov et al., 2002). This results in reduced levels of nuclear transcripts encoding a wide range of cellular proteins, including mitochondrial proteins of the respiratory enzyme complexes and mitochondrial ribosomes (notable exceptions being genes encoding some proteins involved in oxidative stress and proteolytic activity). However, transcripts of the mitochondrially encoded proteins, which are components of the same complexes, do not decrease, and the capacity for mitochondrial protein synthesis is unaffected, leading to an excess of unassembled mitochondrially encoded subunits. Despite this, the stoichiometry of the complexes in the inner mitochondrial membrane is maintained, confirming that the excess mitochondrially encoded subunits are not assembled.
This suggests, at least in this case, that cellular stress is not perceived by the mitochondrial genome. There is ample evidence that the semiautonomous mitochondrial genetic system regulates its own activity (e.g., in transcription and RNA degradation) (Giegé and Brennicke, 2001) and could possibly also have a self-regulation system similar to the control of epistasy of synthesis system described for plastids (Choquet et al., 1998). However, the work described here suggests that this regulatory system is not sensitive to cellular starvation. This is not to say that the mitochondrion as a whole does not perceive this stress. Indeed, retrograde signals between the mitochondrion and the nucleus in response to stress situations are well established (Millar et al., 2004). It is just that there is no genetic response within the mitochondrion. Thus, in response to sugar deprivation, it is changes in nuclear-encoded gene expression that modulate mitochondrial biogenesis, with these changes being regulated at the level of assembly of protein complexes within the mitochondrion.
METHODS
Arabidopsis Cell Cultures
Arabidopsis thaliana var Landsberg erecta suspension cultures (May and Leaver, 1993) were maintained in an MS basal medium containing 0.5 mg/L naphthalene acetic acid, 0.05 mg/L kinetin, and 3% (w/v) sucrose, pH 5.75. One-hundred-milliliter cell cultures were maintained in 250-mL conical flasks in the dark at 22°C on a rotary shaker at 150 rpm. Every 7 d, 10 mL of the culture was subcultured into 100 mL of fresh media. Seven-day-old cultures were starved by sterile filtration of the cells and resuspension in fresh media without sucrose. Cells that had been starved for 48 h were recovered by sterile filtration and resuspended in fresh media containing 3% (w/v) sucrose. Cell viability was estimated by taking an equivalent number of cells (in 50 μL of cell suspension) and staining with FDA 1:1 (v/v) for 15 min at 22°C. The proportion of living fluorescent cells was counted using a fluorescence microscope.
Mito Tracker Green FM Staining and Fluorescence Measurements
Cell samples were sedimented at 1000g for 2 min at room temperature and resuspended in 200 μL of culture medium containing 75 nM Mito Tracker Green FM (Molecular Probes). Cells were stained for 60 min under normal cell growth conditions. Fluorescence was quantified with a Kontron SFM 25 fluorimeter (Munich, Germany) set to measure excitation at 490 nm and emission at 516 nm.
GFP Fluorescence Measurements
A stably transformed Arabidopsis cell suspension with GFP fused to the mitochondrial protein ATP2 N-terminal sequence downstream of the 35S promoter of Cauliflower mosaic virus (Logan and Leaver, 2000) was used. Fluorescence was quantified with a Kontron SFM 25 fluorimeter set to measure excitation at 395 nm and emission at 509 nm.
Total DNA Extraction and DNA Gel Blot Hybridization
Cells were frozen and ground in liquid nitrogen, then transferred to extraction medium containing 500 mM NaCl, 50 mM Tris-HCl, pH 8.0, 50 mM EDTA, 1% (v/v) β-mercaptoethanol, and 400 mg/mL RNase I. After mixing, 5% (w/v) polyvinylpyrrolidone-40 and 1% (w/v) SDS were added. The solution was incubated for 15 min at 65°C, 0.45 M potassium acetate was added, and the solution was further incubated for 30 min on ice and centrifuged for 10 min at 21,000g at 4°C. The supernatant was extracted with phenol-chloroform, precipitated twice with isopropanol, and washed in 70% (v/v) ethanol. The DNA pellet was finally resuspended in water. Total DNA extracted from cells at the four treatment time points was labeled and hybridized on nylon filters spotted with 500, 150, and 50 ng of atp1, cob, atp2, and complex I PSST subunit-specific PCR products. The average signal, quantified with a phosphor imager, was plotted against time.
Total cell DNA was radiolabeled in the presence of 50 μCi of α dCTP 3000 Ci/mmol with random primers and Klenow enzyme according to the manufacturer's instruction (Invitrogen, Carlsbad, CA). DNA gel blot hybridization was performed following standard molecular biology techniques (Sambrook et al., 1989).
Mitochondrial Extraction, Purification, and Measurement of Respiration Rates
Cells were harvested by filtration through Miracloth (Calbiochem, San Diego, CA) and mitochondria extracted and purified by Percoll gradient centrifugation as described previously (Giegé et al., 2003). Oxygen consumption of control or sucrose-starved cells was measured in a Clark-type oxygen electrode in 1 mL volume.
RNA Isolation and Fluorescent cDNA Synthesis
Total RNA was isolated from cell cultures using the TRIzol method as described on the Arabidopsis Functional Genomics Consortium Web site (http://www.arabidopsis.org/info/2010_projects/comp_proj/AFGC/). Fifty micrograms of total RNA were primed with 3 μg of random hexamers, and fluorescent cDNA was synthesized in the presence of 25 μM of Cy5 or Cy3-dCTP (Amersham, Little Chalfont, UK) with Superscript II reverse transcriptase according to the manufacturer's instructions (Invitrogen).
Microarray Slide Printing, Processing, and Hybridization
One hundred and nineteen unique features were spotted as triplicates on the microarray. Among these, 40 were mitochondrially encoded genes and 79 were nuclear-encoded genes. Of the latter, 20 were nuclear-encoded genes for proteins found, along with mitochondrially encoded proteins, in the respiratory enzyme complexes of mitochondria. The feature representing atpase γ was spotted seven times at different places on the array. This made an array with 375 spots. Mitochondrially encoded gene-specific probes were amplified from custom primers (Giegé and Brennicke, 1999; Giegé et al., 2000). Gene probes for nuclear-encoded proteins were amplified from EST clones available from The Arabidopsis Information Resource, and the identity was confirmed by sequence analysis. Sequences of the custom primers and accession numbers of the EST clones used in this analysis are detailed in Supplemental Table 2 online. Gene-specific DNA probes were resuspended in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), DMSO (50% v/v; at a concentration of 200 ng/μL) and printed on CMT GAPS II slides (Corning, Corning, NY) with an Affymetrix A417 arrayer using a pin-ring printing device. Printed slides were cross-linked at 200 mJ, baked at 80°C for 2 h, rehydrated for 1 min at 95°C in water, and snap dried on a 100°C heating block. Slides were then immersed and shaken for 15 min at room temperature in an n-methyl-pyrrolidinone solution containing 1.6% (w/v) succinic anhydride and 20 mM sodium borate, pH 8.0. Slides were then immersed for 2 min at 95°C in water, 1 min at room temperature in 95% (v/v) ethanol, air dried, and stored in a dry, sealed environment.
DNA microarrays were prehybridized for 45 min at 42°C in a solution containing 0.1% (w/v) SDS, 25% (v/v) formamide, 5× SSC, and 10 mg/mL BSA, rinsed with water, and dried. Cy5- and Cy3-labeled cDNA pellets were resuspended in 10 μL of hybridization mix containing 0.1% (w/v) SDS, 25% (v/v) formamide, and 5× SSC. The two samples were mixed and centrifuged at 21,000g for 3 min. Supernatants were transferred to a clean tube, 1 μL of Liquid block (Amersham) was added, and samples were denatured for 5 min at 95°C and deposited on the microarray. Slides were covered with Hybri-slips (Sigma-Aldrich, St. Louis, MO) and hybridized overnight at 42°C in a sealed hybridization chamber. The slides were washed for 5 min at 42°C in 2× SSC and 0.1% (w/v) SDS, for 10 min at room temperature in 0.1× SSC and 0.1% (w/v) SDS, and four times for 1 min at room temperature in 0.1× SSC. Finally, the slides were rinsed with water and ethanol and dried.
Microarray Scanning, Data Analysis, and Statistical Analysis
Microarrays were scanned at 635 and 532 nm (Cy5 and Cy3 absorbance) with an Affymetrix A428 array scanner and acquisition software according to the manufacturer's instructions. After scanning, images were analyzed with Affymetrix Jaguar 1.0 software. The Cy5 and Cy3 intensities for each spot were extracted and normalized with the overall Cy3 intensity (with the “all spots normalization” command of Jaguar 1.0) (Nunes et al., 2003). The technical triplicates were then averaged to obtain a single value for each feature on each slide. A Welsh t test was applied to show that the intensities were significantly (P < 0.05) above background. For further analysis, the raw data obtained this way were collected and imported into the statistics software R (http://www.r-project.org/) and analyzed as well with the Limma package (Smyth, 2004) from Bioconductor (Gentleman et al., 2004). The log ratios (log base 2) were calculated for each slide with the raw intensities without previous substraction of the background. The values obtained were transformed for the reverse labeling experiments for easier comparison of the results between the slides. At this point, no filtering of the intensity values or ratio was performed. All the genes were considered for further analysis. The overall expression difference between slides was visualized with box plots (see Supplemental Figure 1 online) and by calculating MAD. A between-array normalization for a given treatment was performed with the quantile method (Yang and Thorne, 2003) to homogenize the data (see Supplemental Figure 1 online). Correlation coefficients between slides in each treatment were calculated to show the reproducibility of the experiments. Genes with statistically significant expression variations across time points in S1, S2, and RS were identified with Limma by performing a linear modeling and an empirical Bayes analysis on log ratios. The raw P values were adjusted by the multiple testing correction of Benjamini and Hochberg (Benjamini and Hochberg, 1995), which controls the false discovery rate. We considered genes with adjusted P values < 0.05 (i.e, taking a false discovery rate of 5% as significant for further analysis) (see Supplemental Table 1 online).
RNA Gel Blot Analysis
RNA samples extracted as described above were separated on agarose denaturing gels, blotted on Hybond-NX membranes (Amersham), and hybridized using standard methods (Sambrook et al., 1989). Specific PCR products used to probe the blots were 32P-labeled with a DECA-Prime II kit (Ambion, Austin, TX) following the manufacturer's instructions.
Two-Dimensional Electrophoresis
Samples of 800 μg of mitochondrial protein were acetone precipitated and separated by isoelectric focusing in the first dimension and SDS-PAGE in the second and stained with colloidal Coomassie Brilliant Blue G 250 as described previously (Millar et al., 2001).
Protein Gel Blot Analysis
Equal amounts of mitochondrial or total protein (20, 10, and 5 μg) were fractionated by SDS-PAGE and transferred to Protran BA83 cellulose nitrate membranes (Schleicher and Schuell, Keene, NH). Proteins were visualized by brief staining with Ponceau S. Membranes were blocked with 5% (w/v) BSA and incubated overnight with mouse monoclonal antibodies at a dilution of 1:500 for maize (Zea mays) ATP1 (obtained from T.E. Elthon, Lincoln, NE), 1:1000 for maize ATP2 (obtained from T.E. Elthon), and 1:2000 for human α-tubulin (Amersham), with rabbit polyclonal antibodies at a dilution of 1:5000 for sweet potato (Ipomoea batatas) COX2 (otained from T. Asahi, Nagoya, Japan), 1:5000 for sweet potato COXVc (otained from T. Asahi), 1:20000 for yeast COB (obtained from G. Schatz, Basel, Switzerland), 1:50,000 for yeast CYTC1 (obtained from G. Schatz), 1:100,000 for wheat (Triticum aestivum) NAD9 (Lamattina et al., 1993), and 1:10,000 for beef 76-kD subunit of complex I (obtained from J.E. Walker, Cambridge, UK). Sheep anti-mouse and goat anti-rabbit antibodies conjugated with horseradish peroxidase (Amersham) were used as secondary antibodies and visualized with enhanced chemiluminescent reagents (Pierce, Rockford, IL).
Solubilization of Mitochondrial Complexes and Separation on Two-Dimensional Blue-Native Gels
Five hundred micrograms of purified mitochondria were solubilized in 1% (w/v) n-dodecyl maltoside in 40 μL, and mitochondrial enzyme complexes were resolved by Blue-Native gel PAGE in the first dimension followed by SDS-PAGE to separate the component subunits in the second dimension as described previously (Giegé et al., 2003).
In Organello Protein Synthesis
Three hundred micrograms of mitochondrial proteins were resuspended in a solution containing 5 mM KH2PO4, pH 7.0, 300 mM mannitol, 60 mM KCl, 50 mM Hepes, 10 mM MgCl2, 10 mM Na-malate, 10 mM Na-pyruvate, 2 mM GTP, 2 mM DTT, 4 mM ADP, 0.1% (w/v) BSA, 25 μM of an unlabeled 19–amino acid solution, and 30 μCi (>1000 Ci/mmol 35S-Met). Reactions were performed in 100 μL for 10, 30, and 60 min at 25°C on an orbital shaker and incorporations stopped by addition of 1 mL mitochondria wash buffer containing 10 mM Met. In control experiments, 25 mM Na-acetate was used instead of Na-malate and Na-pyruvate. Radiolabeled proteins were separated by SDS-PAGE and visualized by autoradiography as previously described (Leaver et al., 1983).
Mitochondrial Import in Vitro Assays
The mitochondrial protein ATP2 N-terminal sequence fused with mGFP5 (Logan and Leaver, 2000) was synthesized from the corresponding cDNA clone in pGEM-T vector by coupled transcription/translation in presence of 35S-Met according to the manufacturer's instructions (Promega, Fitchburg, WI). In vitro import assays were performed with freshly prepared intact mitochondria as described by Wischmann and Schuster (1995).
Supplementary Material
Acknowledgments
P.G. was funded by a European Union Marie Curie long-term fellowship and by the Centre National de la Recherche Scientifique. L.J.S. was supported by a Biotechnology and Biological Science Research Council David Phillips Research Fellowship. Grants from the Biotechnology and Biological Science Research Council to C.J.L. are gratefully acknowledged.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christopher J. Leaver (chris.leaver@plants.ox.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.030254.
References
- Binder, S., and Brennicke, A. (2003). Gene expression in plant mitochondria: Transcriptional and posttranscriptional control. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57, 289–300. [Google Scholar]
- Braun, H.P., and Schmitz, U.K. (1999). The protein-import apparatus of plant mitochondria. Planta 209, 267–274. [DOI] [PubMed] [Google Scholar]
- Brumme, S., Kruft, V., Schmitz, U.K., and Braun, H.P. (1998). New insights into the co-evolution of cytochrome c reductase and the mitochondrial processing peptidase. J. Biol. Chem. 273, 13143–13149. [DOI] [PubMed] [Google Scholar]
- Chen, W., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet, Y., Stern, D.B., Wostrikoff, K., Kuras, R., Girard-Bascou, J., and Wollman, F.A. (1998). Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 95, 4380–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento, A.L., Kim, S.J., and Bassham, D.C. (2004). Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 135, 2330–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne, A.-M., and Maréchal-Drouard, L. (2001). The chloroplast-derived trnW and trnM-e genes are not expressed in Arabidopsis mitochondria. Biochem. Biophys. Res. Commun. 285, 1213–1216. [DOI] [PubMed] [Google Scholar]
- Edqvist, J., and Bergman, P. (2002). Nuclear identity specifies transcriptional initiation in plant mitochondria. Plant Mol. Biol. 49, 4959–4968. [DOI] [PubMed] [Google Scholar]
- Eubel, H., Jansch, L., and Braun, H.P. (2003). New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol. 133, 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi, D., and Leaver, C.J. (1999). Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 18, 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman, R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé, P., and Brennicke, A. (1999). RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 96, 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé, P., and Brennicke, A. (2001). From gene to protein in higher plant mitochondria. C. R. Acad. Sci. III 324, 209–217. [DOI] [PubMed] [Google Scholar]
- Giegé, P., Hoffmann, M., Binder, S., and Brennicke, A. (2000). RNA degradation buffers asymmetries of transcription in Arabidopsis mitochondria. EMBO Rep. 1, 164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé, P., Sweetlove, L., and Leaver, C.J. (2003). Identification of mitochondrial protein complexes in Arabidopsis using two-dimensional Blue-Native polyacrylamide gel electrophoresis. Plant Mol. Biol. Rep. 21, 1–12. [Google Scholar]
- Heazlewood, J.L., Howell, K.A., and Millar, A.H. (2003. a). Mitochondrial complex I from Arabidopsis and rice: Orthologs of mammalian and fungal components coupled with plant-specific subunits. Biochim. Biophys. Acta 1604, 159–169. [DOI] [PubMed] [Google Scholar]
- Heazlewood, J.L., Whelan, J., and Millar, A.H. (2003. b). The products of the mitochondrial orf25 and orfB genes are FO components in the plant F1FO ATP synthase. FEBS Lett. 540, 201–205. [DOI] [PubMed] [Google Scholar]
- Jänsch, L., Kruft, V., Schmitz, U.K., and Braun, H.P. (1996). New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J. 9, 357–368. [DOI] [PubMed] [Google Scholar]
- Journet, E.-P., Bligny, R., and Douce, R. (1986). Biochemical changes during sucrose deprivation in higher plant cells. J. Biol. Chem. 261, 3193–3199. [PubMed] [Google Scholar]
- Karbowski, M., Spodnik, J.H., Teranishi, M., Wozniak, M., Nishizawa, Y., Usukura, J., and Wakabayashi, T. (2001). Opposite effects of microtubule-stabilizing and microtubule-destabilizing drugs on biogenesis of mitochondria in mammalian cells. J. Cell Sci. 114, 281–291. [DOI] [PubMed] [Google Scholar]
- Lamattina, L., Gonzalez, D., Gualberto, J., and Grienenberger, J.M. (1993). Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur. J. Biochem. 217, 831–838. [DOI] [PubMed] [Google Scholar]
- Leaver, C.J., Hack, E., and Forde, B.G. (1983). Protein synthesis by isolated plant mitochondria. Methods Enzymol. 43, 476–484. [DOI] [PubMed] [Google Scholar]
- Leon, P., Arroyo, A., and Mackenzie, S. (1998). Nuclear control of plastid and mitochondrial development in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 453–480. [DOI] [PubMed] [Google Scholar]
- Logan, D.C., and Leaver, C.J. (2000). Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J. Exp. Bot. 51, 865–871. [PubMed] [Google Scholar]
- Logan, D.C., Millar, A.H., Sweetlove, L.J., Hill, S.A., and Leaver, C.J. (2001). Mitochondrial biogenesis during germination in maize embryos. Plant Physiol. 125, 662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, B., Wilson, R., Phreaner, C.G., Mulligan, R.M., and Hanson, M.R. (1996). Protein polymorphism generated by differential RNA editing of a plant mitochondrial rps12 gene. Mol. Cell. Biol. 16, 1543–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, S., and McIntosh, L. (1999). Higher plant mitochondria. Plant Cell 11, 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal-Drouard, L., Small, I., Weil, J.H., and Dietrich, A. (1995). Transfer RNA import into plant mitochondria. Methods Enzymol. 260, 310–327. [DOI] [PubMed] [Google Scholar]
- May, M.J., and Leaver, C.J. (1993). Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 103, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.H., Day, A.D. and Whelan, J. (2004). Mitochondrial biogenesis and function in Arabidopsis. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0105, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Millar, A.H., Sweetlove, L.J., Giegé, P., and Leaver, C.J. (2001). Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127, 1711–1727. [PMC free article] [PubMed] [Google Scholar]
- Moneger, F., Smart, C.J., and Leaver, C.J. (1994). Nuclear restoration of cytoplasmic male sterility in sunflower is associated with the tissue-specific regulation of a novel mitochondrial gene. EMBO J. 13, 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes, L., Rosato, Y., Muto, N., Yanai, G., da Silva, V., Leite, D., Gonçalves, E., de Souza, A., Coletta-Filho, H., Machado, M., Lopes, S.A., and Costa de Oliveira, R. (2003). Microarray analyses of Xylella fastidiosa provide evidence of coordinated transcription control of laterally transferred elements. Genome Res. 13, 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep, M., and Grivell, L.A. (1996). The role of protein degradation in mitochondrial function and biogenesis. Curr. Genet. 30, 367–380. [DOI] [PubMed] [Google Scholar]
- Rodermel, S. (2001). Pathways of plastid to nucleus signalling. Trends Plant Sci. 6, 471–478. [DOI] [PubMed] [Google Scholar]
- Sabar, M., Gagliardi, D., Balk, J., and Leaver, C.J. (2003). ORFB is a subunit of F1F(O)-ATP synthase: Insight into the basis of cytoplasmic male sterility in sunflower. EMBO Rep. 4, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sarria, R., Lyznik, A., Vallejos, C.E., and Mackenzie, S.A. (1998). A cytoplasmic male sterility-associated mitochondrial peptide in common bean is posttranslationally regulated. Plant Cell 10, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger, H. (2001). Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 65, 231–244. [DOI] [PubMed] [Google Scholar]
- Smart, C.J., Moneger, F., and Leaver, C.J. (1994). Cell-specific regulation of gene expression in mitochondria during anther development in sunflower. Plant Cell 6, 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, art3. [DOI] [PubMed]
- Topping, J.F., and Leaver, C.J. (1990). Mitochondrial gene expression during wheat leaf development. Planta 182, 399–407. [DOI] [PubMed] [Google Scholar]
- Unseld, M., Marienfeld, J.R., Brandt, P., and Brennicke, A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15, 57–61. [DOI] [PubMed] [Google Scholar]
- Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., Schuch, W., and Giovannoni, J. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. [DOI] [PubMed] [Google Scholar]
- Wischmann, C., and Schuster, W. (1995). Transfer of rps10 from the mitochondrion to the nucleus in Arabidopsis thaliana: Evidence for RNA-mediated transfer and exon shuffling at the integration site. FEBS Lett. 374, 152–156. [DOI] [PubMed] [Google Scholar]
- Yang, Y.H., and Thorne, N. (2003). Normalization for two-color cDNA microarray data. In Science and Statistics: A Festschrift for Terry Speed, D. Goldstein, ed, IMS Lecture Notes, Monograph Series, Vol. 40. (Beachwood, OH: Institute of Mathematical Statistics), pp. 403–418.
- Zabaleta, E., Heiser, V., Grohmann, L., and Brennicke, A. (1998). Promoters of nuclear-encoded respiratory chain complex I genes from Arabidopsis thaliana contain a region essential for anther/pollen-specific expression. Plant J. 15, 49–59. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.