Abstract
Methyl-CpG binding domain (MBD) proteins in Arabidopsis thaliana bind in vitro methylated CpG sites. Here, we aimed to characterize the binding properties of AtMBDs to chromatin in Arabidopsis nuclei. By expressing in wild-type cells AtMBDs fused to green fluorescent protein (GFP), we showed that AtMBD7 was evenly distributed at all chromocenters, whereas AtMBD5 and 6 showed preference for two perinucleolar chromocenters adjacent to nucleolar organizing regions. AtMBD2, previously shown to be incapable of binding in vitro–methylated CpG, was dispersed within the nucleus, excluding chromocenters and the nucleolus. Recruitment of AtMBD5, 6, and 7 to chromocenters was disrupted in ddm1 and met1 mutant cells, where a significant reduction in cytosine methylation occurs. In these mutant cells, however, AtMBD2 accumulated at chromocenters. No effect on localization was observed in the chromomethylase3 mutant showing reduced CpNpG methylation or in kyp-2 displaying a reduction in Lys 9 histone H3 methylation. Transient expression of DDM1 fused to GFP showed that DDM1 shares common sites with AtMBD proteins. Glutathione S-transferase pull-down assays demonstrated that AtMBDs bind DDM1; the MBD motif was sufficient for this interaction. Our results suggest that the subnuclear localization of AtMBD is not solely dependent on CpG methylation; DDM1 may facilitate localization of AtMBDs at specific nuclear domains.
INTRODUCTION
DNA methylation is a pivotal epigenetic mark regulating genome organization and function both in plants and animals. In mammalian cells, CpG-methylated sites are targeted by a group of proteins containing the methyl-CpG binding domain (MBD), leading to chromatin compaction and gene silencing. The biological significance of MBD proteins is demonstrated in Rett syndrome, a childhood neurodevelopmental disorder caused by mutations in the gene encoding the MBD transcriptional repressor MeCP2 (Amir et al., 1999; Wan et al., 1999).
The importance of cytosine methylation for plant development was first demonstrated by treatment of plants with the hypomethylating agent 5-azacytidine and later by various genetic manipulations (Richards, 1997, and references therein). Phenotypic perturbations were reported in plants displaying a reduction in total genomic cytosine methylation, such as met1 mutants, where the DNA methyltransferase MET1 is misregulated (Finnegan et al., 1996; Ronemus et al., 1996; Kankel et al., 2003; Saze et al., 2003), or in mutants for the gene encoding the SWI2/SNF2 nucleosomal remodeling factor DDM1; in the ddm1-2 mutant, developmental abnormalities were progressively acquired during generations (Kakutani et al., 1996; Jeddeloh et al., 1999). On the other hand, plants carrying mutations in the gene coding for CHROMOMETHYLASE3 (CMT3), an enzyme required for maintenance of CpNpG methylation, displayed a wild-type phenotype (Lindroth et al., 2001).
Linkage between DNA methylation and histone methylation was demonstrated in Neurospora crassa and Arabidopsis thaliana. In N. crassa, the histone methyltransferase DIM5 is required for DNA methylation (Tamaru and Selker, 2001), whereas in Arabidopsis, the histone methyltransferase Kryptonite/SUVH4 controls CpNpG and CpNpN though not CpG methylation (Jackson et al., 2002; Malagnac et al., 2002). The interplay between DNA methylation and histone methylation in Arabidopsis is not clear. In ddm1 and met1 mutants, a reduction in H3K9 methylation was noted at certain heterochromatic regions while total H3K9 methylation remained unaffected, raising the question whether CpG methylation guides H3K9 methylation (Soppe et al., 2002; Tariq et al., 2003) or vice versa (Gendrel et al., 2002).
Recent data implicated mammalian MBD proteins in linking DNA methylation with histone methylation; MeCP2 was found to interact with histone methyltransferase and induce H3K9 methylation (Fuks et al., 2003), whereas MBD1 was found to interact with the Suv39h1-HP1 heterochromatic complex and induce DNA methylation-based transcriptional repression (Fujita et al., 2003). The finding that MBD proteins associate with histone deacetylases both in plants and animals (Hendrich and Tweedie, 2003; Zemach and Grafi, 2003) suggests that MBDs may induce heterochromatin formation by coordinating the activities of histone deacetylases and histone methyltransferases.
Several reports have characterized the MBD group of proteins in Arabidopsis and showed their capability to bind methylated CpG sites (Berg et al., 2003; Ito et al., 2003; Scebba et al., 2003; Zemach and Grafi, 2003). Although these reports demonstrated some differences among AtMBDs with respect to CpG binding activity, it becomes clear that the Arabidopsis MBD protein family is composed of at least two groups: one binds methylated CpG sites and the other does not. Binding to DNA independently of methylation was demonstrated for AtMBD11 (Scebba et al., 2003), whose downregulation by RNA interference induced developmental abnormalities (Berg et al., 2003). Using AtMBD fused to green fluorescent protein (GFP), Scebba et al. (2003) demonstrated that the heterochromatic distribution of AtMBD5 and AtMBD6 was affected by treatment with 5-azacytidine, thus confirming the importance of cytosine methylation for their subnuclear distribution.
Here, we employed genetic and biochemical approaches to study in planta the association of AtMBD proteins with methylated CpG sites and the molecular mechanism underlying their subnuclear distribution. To this end, AtMBDs were fused to GFP and either transiently expressed in Arabidopsis cells or stably transformed into Arabidopsis plants. Our results demonstrated differential subnuclear localization of AtMBDs in Arabidopsis cells, both heterochromatic and euchromatic; localization at specific nuclear domains may be facilitated by additional factors, such as the chromatin remodeling factor DDM1.
RESULTS
AtMBDs Exhibit Differential Subnuclear Localization in Arabidopsis
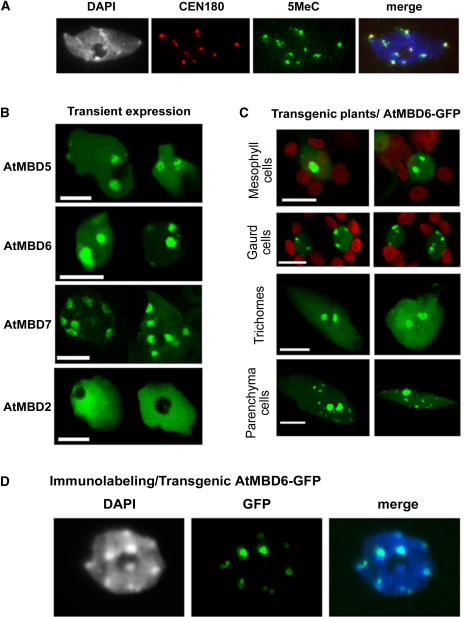
We previously have shown that AtMBD5, AtMBD6, and AtMBD7 bind in vitro methylated CpG sites, whereas AtMBD1, AtMBD2, and AtMBD4 do not (Zemach and Grafi, 2003). To study the interaction of AtMBDs with methylated DNA in vivo, AtMBDs were tagged at their C termini with GFP and placed downstream from the 35S promoter. The GFP fusion constructs were transformed into Arabidopsis leaf protoplasts by the polyethylene glycol methodology. In Arabidopsis, methylated DNA is found mainly at heterochromatic chromocenters (Figure 1A; see also Fransz et al., 2002; Soppe et al., 2002). In transient expression assays, AtMBD7-GFP was evenly distributed at all chromocenters (Figure 1B), whereas AtMBD5 and 6 showed preference for two large domains adjacent to the nucleolus (Figure 1B; see Supplemental Figure 1A online). By contrast, AtMBD2, previously shown incapable of binding in vitro methylated CpG sites (Zemach and Grafi, 2003), was dispersed within the nucleus but excluded from chromocenters and the nucleolus (Figure 1B; see Supplemental Figure 1B online).
Figure 1.
AtMBD5, 6, and 7 Are Localized to Highly Methylated Chromocenters.
(A) Immunolabeling/FISH assay showing that 5-methylcytosine signals are associated with CEN180 at chromocenters in wild-type Arabidopsis.
(B) Transient expression showing different patterns of subnuclear localization of AtMBD-GFP proteins. Arabidopsis protoplasts were polyethylene glycol–transformed with the indicated AtMBD-GFP constructs and inspected under a confocal microscope. Note the preference of AtMBD5 and 6 for two perinucleolar chromocenters. Bars = 5 μm.
(C) Subnuclear localization of AtMBD6-GFP in various types of leaf cells in transgenic plants. Bars = 5 μm.
(D) Immunolabeling assay showing the localization of AtMBD6-GFP at the intensely DAPI-stained chromocenters. Fixed nuclei from Arabidopsis plants expressing AtMBD6-GFP were immunolabeled with anti-GFP and inspected under a fluorescence microscope. Note the intense GFP signal at perinucleolar chromocenters and the lower signals at other chromocenters.
In stably transformed plants, AtMBD5 and 6 generally followed the subnuclear localization pattern found in transient expression, except for guard cells, where the distribution was at multiple subnuclear domains (Figure 1C). Immunolabeling assays using anti-GFP on nuclei prepared from transgenic plants expressing AtMBD6-GFP showed colocalization with the intensely 4′,6-diamidino-2-phenylindole (DAPI)-stained chromocenters (Figure 1D). Together, these results demonstrate the association of AtMBDs (5, 6, and 7) with methylated CpG-rich chromatin in planta and suggest the involvement of additional factors in determining their preference for specific nuclear domains.
AtMBD6 Is Tightly Bound to Chromatin and Is Associated with Centromeric Repeats and the 18S rDNA
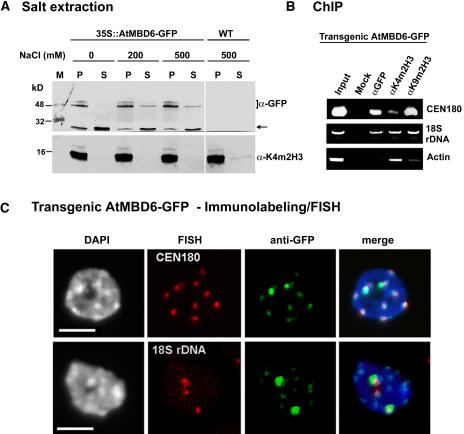
We estimated the strength of AtMBD6-GFP binding with chromatin by salt extraction of nuclei prepared from transgenic plants expressing AtMBD6-GFP. Nuclei were incubated in the presence of increasing concentrations of NaCl after which the soluble and the insoluble pellet fractions were resolved by SDS-PAGE. Immunoblotting analysis using αGFP showed that AtMBD6-GFP was composed of two fractions: a major fraction (∼85%), which was tightly associated with chromatin and could not be released even under 500 mM NaCl, and a minor fraction (∼15%), which was loosely associated with chromatin and was released to the soluble fraction with as low as 200 mM NaCl (Figure 2A). As expected, a control histone H3 was strongly associated with chromatin as revealed with anti-K4–methylated histone H3 (α-K4m2H3). We also analyzed whether the dispersal of AtMBD2-GFP within the nucleus reflects the presence of AtMBD2 in the soluble nuclear fraction. Nuclei prepared from transgenic Arabidopsis expressing AtMBD2-GFP were subjected to salt extraction. Results showed that AtMBD2-GFP is not found in the soluble fraction but rather strongly bound to chromatin (see Supplemental Figure 2 online).
Figure 2.
AtMBD6-GFP Is Tightly Bound to Chromatin and Is Associated with Centromeric Repeats and the 18S rDNA.
(A) Nuclei prepared from leaves of transgenic Arabidopsis expressing AtMBD6-GFP or of wild-type (ecotype Wassilewskija) plants were extracted with increasing concentrations of NaCl. Soluble (S) and insoluble pellet (P) fractions were analyzed for the presence of AtMBD6-GFP or histone H3 methylated at Lys 4 by immunoblotting using anti-GFP (αGFP) and anti-dimethylated K4 histone H3 (αK4m2H3), respectively. M indicates molecular weight markers. Note that AtMBD6-GFP appears as multiple protein bands that are absent from wild-type plants. Arrow indicates a breakdown product of the AtMBD6-GFP, which is loosely bound to chromatin.
(B) ChIP assay demonstrating the association of AtMBD6-GFP with CEN180 but not with actin-encoding sequence. ChIP was performed on nuclei prepared from AtMBD6-GFP–expressing plants. Chromatin was immunoprecipitated using αGFP, αK4m2H3, or antidimethylated K9 histone H3 (αK9m2H3). DNA was extracted and subjected to PCR using sets of primers to amplify the centromeric 180-bp repeats (CEN180), the 18S rDNA, or actin-encoding sequence as a control.
(C) Immunolabeling/FISH assay showing that AtMBD6-GFP is associated with CEN180 and has preference for perinucleolar chromocenters adjacent to the 18S rRNA genes. DAPI was used as a counterstain. Bars = 5 μm.
To verify the association of AtMBDs with chromocenters, we employed chromatin immunoprecipitation (ChIP) assay on nuclei prepared from transgenic Arabidopsis expressing AtMBD6-GFP. The antibodies used recognize GFP (αGFP), dimethylated H3K4 (αK4m2H3), a known histone modification associated with transcriptionally active chromatin (Fischle et al., 2003; Lachner et al., 2003), as well as dimethylated H3K9 (αK9m2H3), a known histone modification associated with chromatin compaction and gene silencing (Jenuwein and Allis, 2001; Zhang and Reinberg, 2001). Precipitated DNAs were subjected to PCR to amplify a set of DNA sequences, including centromeric 180-bp repeats (CEN180), the 18S rDNA, as well as actin-encoding sequence. Results showed that anti-GFP precipitated CEN180 and 18S rDNA but not the actin-encoding sequence (Figure 2B). The association of the 18S rDNA with both K4- and K9-dimethylated histone H3 reflects the diverse chromatin configurations of rRNA gene clusters, being either transcriptionally active or inactive. As expected, the actin-encoding sequence was precipitated mainly with αK4m2H3, slightly with αK9m2H3, but not with αGFP (Figure 2B), confirming the association of AtMBD6 with the transcriptionally inactive chromatin.
The two large perinucleolar domains to which AtMBD5 and 6 bind are likely to define chromocenters of the acrocentric chromosomes 2 and/or 4, which together with the nucleolar organizing region form a large domain of heterochromatin. To assess this possibility, fixed nuclei from transgenic Arabidopsis expressing AtMBD6-GFP were first immunolabeled with anti-GFP followed by fluorescence in situ hybridization (FISH) with tetramethylrhodamine-5-dUTP–labeled CEN180 or 18S rDNA. Results showed that AtMBD6-GFP colocalized with the intensely DAPI-stained chromocenters and was associated with CEN180, displaying preference for perinucleolar chromocenters adjacent to the 18S rDNA (Figure 2C). Thus, AtMBD5 and 6 preferentially bind chromocenters of chromosome 2 and/or 4, where they might regulate chromatin compaction and silencing of rRNA genes.
Mutations in DDM1 and MET1, but Not in CMT3, Disrupt the Localization of AtMBD Proteins
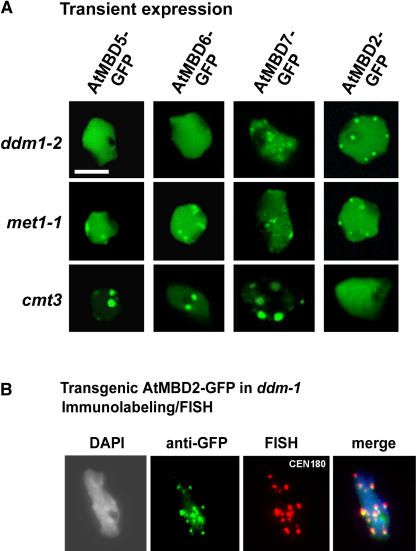
To confirm the preference of AtMBD5, 6, and 7 for methylated CpG sites, we examined the distribution of AtMBD-GFPs in three Arabidopsis DNA methylation mutants: ddm1-2, a mutation in the DDM1 gene encoding the SWI2/SNF2 chromatin remodeling factor; met1-1, a mutation in MET1 DNA methyltransferase gene—both of which show reduction in CpG methylation (Kakutani et al., 1996; Kankel et al., 2003); cmt3, a mutation in the CMT3 gene that reduces CpNpG and CpNpN methylation (Bartee et al., 2001; Lindroth et al., 2001). Reduced DNA methylation in ddm1-2 and met1-1 was demonstrated by the sensitivity of the centromeric 180-bp repeats as well as the 18S rDNA to digestion by the methylation-sensitive HpaII enzyme (data not shown). In ddm1-2 and met1-1 cells, the subnuclear localization of AtMBD5, 6, and 7 was disrupted (Figure 3A). A greater effect was observed in ddm1-2 cells where AtMBD5 and 6 were evenly dispersed within the nucleus. In met1-1 (Figure 3A), as well as in met1-3 cells, where CpG methylation is thought to be almost completely lacking (Tariq et al., 2003), AtMBD5 and 6 showed patchy distribution with a certain fraction of these proteins still associated with chromocenters (see Supplemental Figure 3A online). To quantify these differences, we compared the magnitude of fluctuation in fluorescence intensity by determining the coefficient of variation for each population of nuclei (shown in Supplemental Figure 3A online) using the NIH Image program (Htun et al., 1999; see Methods). In wild-type cells, the coefficient of variation (see Supplemental Figure 3B online) was the highest (0.772 ± 0.18), indicating very high fluctuation in fluorescence intensity of AtMBD6-GFP within the nucleus. The coefficient of variation for met1-1 (0.239 ± 0.06) and met1-3 (0.225 ± 0.062) was significantly higher (approximately twofold) than that of ddm1-2 (0.114 ± 0.029), thus confirming higher fluctuation in fluorescence intensity for AtMBD6-GFP in met1 mutants compared with ddm1-2. In the cmt3 mutant, subnuclear localization of AtMBD-GFP proteins was similar to that found in wild-type plants (Figure 3A). These results verified the importance of CpG methylation in controlling AtMBD5, 6, and 7 subnuclear localization. Interestingly, in ddm1-2 and met1-1 mutant cells, AtMBD2-GFP was accumulated at chromocenters as confirmed by immunolabeling/FISH assay on nuclei derived from transgenic ddm1-2 expressing AtMBD2-GFP (Figures 3A and 3B). This redistribution, however, cannot be accounted for by direct binding of AtMBD2 to unmethylated CpG sites inasmuch as glutathione S-transferase (GST)-AtMBD2 failed to form complexes with unmethylated sites in electrophoretic mobility shift assays (data not shown).
Figure 3.
Localization of AtMBD-GFPs to Chromocenters Requires CpG Methylation.
(A) Subnuclear localization of AtMBD-GFPs is disrupted in ddm1-2 and met1-1 but not in the cmt3 mutant. The indicated AtMBD-GFPs were transiently expressed in ddm1-2, met1-1, and cmt3 mutant cells. Note the dispersion of AtMBD5 and 6 throughout the nucleus in ddm1-2 and to a lesser extent in met1-1. Also note that AtMBD2 accumulates at chromocenters in ddm1-2 and met1-1 but not in cmt3 cells. Bar = 5 μm.
(B) AtMBD2 is associated with CEN180 in the ddm1-2 mutant. Immunolabeling (anti-GFP) followed by FISH (CEN180) was performed on nuclei derived from transgenic ddm1-2 expressing AtMBD2-GFP.
DDM1 Colocalizes and Interacts with AtMBD Proteins
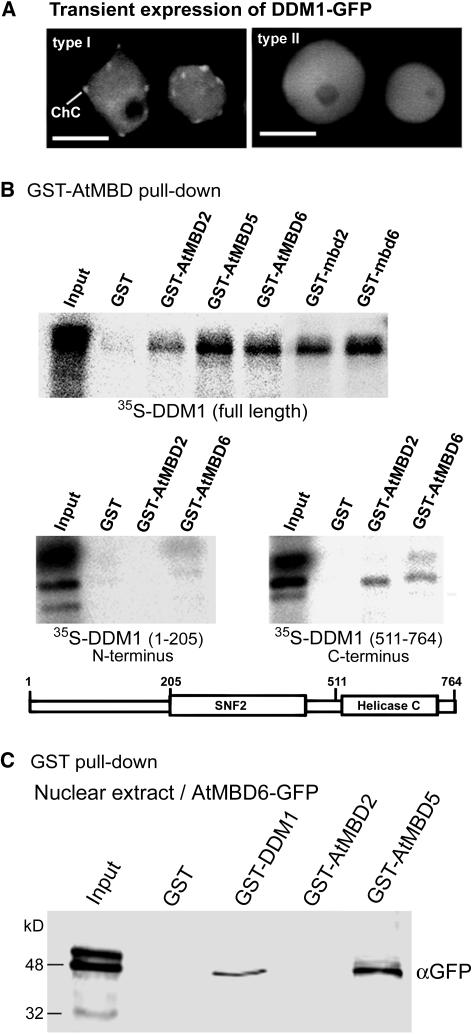
Our results demonstrated a role for CpG methylation in the subnuclear localization of AtMBDs. The difference, however, between ddm1-2 and met1-1 concerning the localization of AtMBD5 and 6 prompted us to investigate possible association between DDM1 and AtMBD proteins. To this end, we first tested whether DDM1 and AtMBDs occupy common nuclear domains. DDM1 cDNA was subcloned downstream from the 35S promoter and upstream from GFP to generate DDM1-GFP fusion protein. This construct was transformed into Arabidopsis leaf protoplasts and inspected under a confocal microscope 24 h after transformation. Results showed two types of localization for DDM1-GFP in Arabidopsis leaf nuclei (Figure 4A): DDM1-GFP was either localized at chromocenters, similarly to AtMBD5, 6, and 7 (type I), or evenly dispersed throughout the nucleus, similarly to the distribution pattern of AtMBD2 in wild-type cells (type II). Thus, AtMBDs and DDM1 occupy common nuclear domains.
Figure 4.
AtMBDs Colocalize and Interact in Vitro with DDM1.
(A) DDM1 fused to GFP displays two types of subnuclear localization in Arabidopsis; type I, showing localization to chromocenters; type II, showing dispersion throughout the nucleus.
(B) GST-AtMBD proteins bind DDM1. GST pull-down assay was performed with the indicated AtMBD proteins fused to GST using in vitro–translated, 35S-labeled, full-length DDM1, N-terminal DDM1/1-205, or C-terminal DDM1/511-764. GST alone was used as a negative control. A schematic representation of the DDM1 protein and its unique domains is shown. Input indicates 15% of the input 35S-labeled proteins.
(C) GST-DDM1 precipitates AtMBD6-GFP from nuclear extract. GST alone, GST-DDM1, GST-AtMBD2, and GST-AtMBD5 were mixed with nuclear extract derived from transgenic Arabidopsis expressing AtMBD6-GFP. Precipitated proteins were resolved by SDS-PAGE and immunoblotted using anti-GFP to detect AtMBD6-GFP. Input lane indicates 10% of the input proteins.
We next investigated physical interaction between AtMBDs and the DDM1 protein using the GST pull-down assay. Glutathione sepharose containing GST alone or GST fused with various AtMBDs were incubated with in vitro–translated 35S-labeled DDM1 full-length protein and after extensive washing samples were resolved on SDS-PAGE and exposed to a phosphor imager. Results showed that all GST-AtMBD proteins, but not GST alone, bound to the full-length DDM1 protein (Figure 4B). The MBD motif alone, either of AtMBD6 (GST-mbd6) or of AtMBD2 (GST-mbd2), was sufficient for this interaction (Figure 4B). The deletion/frameshift caused by the ddm1-2 allele occurs at amino acid 524, leading to premature translation termination upstream from the predicted helicase C-terminal domain (Jeddeloh et al., 1999). We therefore analyzed possible interaction between AtMBDs and the C-terminal region of DDM1 (DDM1/511-764) using the N-terminal region of DDM1 (DDM1/1-205) as a control. Both AtMBD2 and AtMBD6 were capable of binding to the C-terminal region of DDM1, though at a reduced affinity compared with the full-length DDM1. No significant interaction of AtMBDs with the DDM1 N-terminal region (DDM1/1-205) could be detected (Figure 4B).
To confirm this interaction, we performed reciprocal assays and analyzed the capability of the full-length DDM1 fused to GST (GST-DDM1) to precipitate the AtMBD6 from nuclear extract derived from transgenic plants expressing AtMBD6-GFP. To this end, GST alone, GST-DDM1, as well as GST-AtMBD2 and GST-AtMBD5 immobilized onto glutathione sepharose were mixed with nuclear extract, and after incubation (4°C, 12 h) and extensive washing precipitated proteins were resolved on SDS-PAGE and immunoblotted using anti-GFP. Results showed that GST-DDM1 and GST-AtMBD5, but not GST alone or GST-AtMBD2, were capable of precipitating AtMBD6-GFP from nuclear extract. These results further support physical interaction between AtMBD6 and the chromatin remodeling factor DDM1.
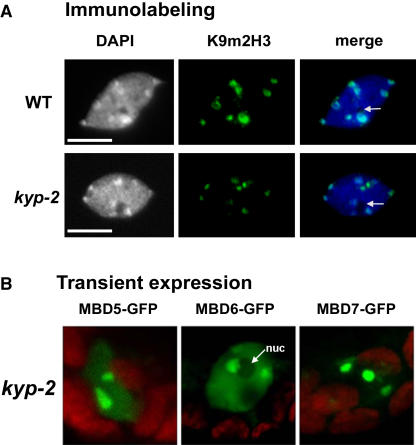
Association of AtMBDs with Chromocenters Is Not Altered in the kyp-2 Mutant
In ddm1-2 and met1 mutants, H3K9 dimethylation at chromocenters is reduced (Soppe et al., 2002; Tariq et al., 2003). To assess the involvement of H3K9 dimethylation in recruiting AtMBDs to chromocenters, we transiently expressed AtMBD-GFP constructs in kyp-2 cells where H3K9 dimethylation at chromocenters is significantly reduced (Figure 5A; see also Jasencakova et al., 2003; Jackson et al., 2004), whereas CpG methylation at centromeres is unaffected (Jackson et al., 2002; Malagnac et al., 2002). Results showed that the distribution of AtMBD-GFP proteins in kyp-2 was similar to that of wild-type plants (Figure 5B), suggesting that H3K9 dimethylation is dispensable for the localization of AtMBDs at chromocenters.
Figure 5.
Reduced Methylation of Histone H3 at Lys 9 Does Not Affect AtMBDs Localization at Chromocenters.
(A) Immunolabeling using anti-Lys 9 dimethylated histone H3 (K9m2H3) showing intense labeling at chromocenters in the wild-type nucleus and reduced labeling in the kyp-2 nucleus. Arrows indicate nucleoli. Bars = 5 μm.
(B) Localization of AtMBD-GFPs in kyp-2 cells is indistinguishable from that of wild-type cells. nuc, nucleolus.
DISCUSSION
Results presented here demonstrated the preference of AtMBD5, 6, and 7 for binding CpG-methylated sites in planta. First, these AtMBDs were localized at the highly CpG-methylated chromocenters, and second, their localization was disrupted in the DNA methylation mutants ddm1-2 and met1-1 (both display a significant reduction in CpG methylation). Indeed, treatment of plant cells with the hypomethylating agent 5-azacytidine modified the heterochromatic distribution of AtMBD5 and AtMBD6 (Scebba et al., 2003). Also, in mammalian cells, MBD proteins are localized mainly to nuclear foci enriched in methyl CpG; in mouse cells deficient in CpG methylation, most MBDs are dispersed within the nucleus (Hendrich and Bird, 1998). In this work, we showed that AtMBD proteins capable of binding methylated CpG sites displayed differential subnuclear localization in Arabidopsis cells. Whereas AtMBD5 and 6 preferentially bound to perinucleolar chromocenters adjacent to the rRNA gene clusters, AtMBD7, a unique member of the AtMBD family containing three putative MBD motifs (Berg et al., 2003), displayed even distribution at all chromocenters. These localization patterns imply that CpG methylation, while essential, may not be sufficient to determine the subnuclear localization of AtMBD proteins; other factors may be involved in determining the localization of AtMBDs at specific nuclear domains. The findings that DDM1 and AtMBDs share common nuclear sites and both proteins interact with each other suggest that DDM1 may play an active role in determining AtMBDs subnuclear localization. Consistent with our finding, Harikrishnan et al. (2005) demonstrated an interaction between the chromatin remodeling factor Brahma and the human MBD protein MeCP2 and raised the hypothesis that chromatin remodeling activity at methylated sites is required for the assembly of MeCP2 to establish heterochromatin.
Notably, similarly to DDM1, the Lsh gene, a mouse homolog of DDM1, is required for genome-wide methylation (Dennis et al., 2001). In Lsh-deficient tissues as well as in ddm1 mutant plants, the activity of DNA methyltransferases is not altered significantly compared with wild-type tissues (Kakutani et al., 1995; Dennis et al., 2001), suggesting that Lsh and DDM1 do not act in enhancing the DNA methylation machinery. Instead, DDM1 and Lsh have been suggested to function as chromatin remodeling factors regulating the accessibility of chromatin to DNA methyltransferases (Jeddeloh et al., 1999; Dennis et al., 2001). However, no effect on localization of the DNA methyltransferase 1 (Dnmt1) was observed in Lsh−/− cells, suggesting that Lsh is not essential for proper subnuclear localization of Dnmt1 (Yan et al., 2003). An alternative explanation for DDM1 function emerged from the finding that in N. crassa and Arabidopsis DNA methylation is tightly associated with histone H3K9 methylation (Tamaru and Selker, 2001; Jackson et al., 2002; Malagnac et al., 2002; Tamaru et al., 2003). In the ddm1 mutant, a shift from H3K9 methylation toward H3K4 methylation was noted in the heterochromatic knob of chromosome 4, raising the hypothesis that the primary effect of DDM1 is to facilitate H3K9 methylation, which in turn induces cytosine methylation (Gendrel et al., 2002). In the Arabidopsis kyp-2 mutant, however, reduction in H3K9 methylation at chromocenters had no effect on CpG methylation (Jackson et al., 2002), indicating that in Arabidopsis, CpG methylation is not necessarily dependent on H3K9 methylation (Richards, 2002). Our results point to the possibility that the effect of DDM1 on DNA methylation is mediated, at least partly, by MBD proteins. Accordingly, the ATP-dependent chromatin remodeling activity of DDM1 (Brzeski and Jerzmanowski, 2003) is required for making CpG-methylated sites accessible for binding AtMBD proteins. Alternatively, DDM1 may function as an assembly platform guiding AtMBDs to their binding sites, thus protecting CpG-methylated sites from demethylating activities. It remains to be determined whether such a relationship also exists between mammalian MBDs and the chromatin remodeling factor Lsh.
Transient expression of AtMBD2-GFP in wild-type Arabidopsis cells showed a nearly uniform distribution within the nucleus, excluding the heterochromatic, CpG-methylated chromocenters and the nucleolus. However, in ddm1-2 and met1-1 cells, AtMBD2 was found at chromocenters, suggesting that reduction in cytosine methylation plays a major role in the accumulation of AtMBD2 at these sites. Presently, we do not know what brings AtMBD2 to chromocenters in these mutants. Because AtMBD2 did not bind in vitro–unmethylated DNA, it is likely that its association with chromocenters is mediated through interaction with an as yet unknown factor(s).
The capability of GST-AtMBD5 to precipitate AtMBD6-GFP from nuclear extract suggests either that these proteins interact physically with each other or they are present in the same protein complex. GST pull-down assay using in vitro–translated, 35S-Met–labeled AtMBD6 showed that neither GST-AtMBD5 nor GST-AtMBD6 binds 35S-labeled AtMBD6 protein (data not shown). Taken together, our results imply that the plant MBD protein complex possesses the chromatin remodeling factor DDM1 and at least two AtMBD molecules. This is similar to mammalian cells where the main multiprotein repressory complex MeCP1 contains the SNF2 chromatin remodeling factor Mi2, MBD2, MBD3, and histone deacetylases as major components (Wade et al., 1999; Feng and Zhang, 2001).
METHODS
Plant Materials and Protoplast Transformation
Seeds of Arabidopsis thaliana mutants for ddm1-2 and met1-1 were kindly provided by E. Richards and seeds of met1-3 from J. Paszkowski. These mutants were confirmed by the sensitivity of the centromeric 180-bp repeats as well as the 18S rDNA to digestion by the methylation-sensitive HpaII enzyme. The ddm1-2 mutant was further verified by RT-PCR as described (Jeddeloh et al., 1999) and met1-3 by PCR as described (Saze et al., 2003). Wild-type and Arabidopsis mutants (all in the background of Columbia ecotype) were grown under short-day conditions at 20°C. At 4 to 6 weeks after germination, rosette leaves were collected for the isolation and transformation of protoplasts essentially as described (Sheen, 2002). The GFP signal was detected 24 h after transfection using a laser confocal microscope (Olympus IX70; Hamburg, Germany). Images were obtained using an excitation wavelength of 488 nm, and images for GFP and chlorophyll signals were collected through 505 to 525- and 630-nm filters, respectively.
Immunolabeling and FISH
Nuclei were isolated from leaves as previously described (Fass et al., 2002) and fixed in 4% paraformaldehyde dissolved in PBS for 15 min at room temperature followed by washing twice with PBS. Nuclei were placed on slides, air dried, permeabilized in cold acetone (100%) for 7 min in −20°C, and washed twice with PBS. Slides were blocked in 2% BSA in PBS for 2 h at room temperature followed by overnight incubation at 4°C with 100 μL of primary antibody mixture containing 2% BSA and 2 μg of antidimethylated Lys 9 histone H3 (Upstate Biotechnology, Lake Placid, NY) or anti-GFP (Roche, Indianapolis, IN). Slides were washed three times, 5 min each, in PBS, followed by incubation at room temperature for 2 h with the appropriate secondary antibody tagged with fluorescein (Sigma-Aldrich, St. Louis, MO). For FISH assays, slides washed as above were fixed with 1% paraformaldehyde for 5 min, denatured with formamide, and probed with the 180-bp repeats (CEN180) and the 18S rDNA, both labeled with tetramethylrhodamine-5-dUTP (Roche) as described (Avivi et al., 2004). After hybridization, slides were washed, stained with 10 μg/mL of DAPI, washed twice, and mounted in Vectashield (Vector Laboratories, Burlingame, CA). Hybridization signals were visualized by a fluorescence microscope (Olympus) equipped with a CCD camera (Imago; TILL Photonics, Hamburg, Germany) using Olympus filters U-MNU, U-MWIBA2, and U-MNG to detect DAPI, fluorescein, and rhodamine, respectively. Images were pseudocolored and merged using TILL Vision version 3.3 software (TILL Photonics). All images were processed using Adobe Photoshop software (Mountain View, CA).
Immunolabeling with polyclonal anti-5-methyl cytosine (Megabase Research Products, Lincoln, NE) was performed on nuclei fixed with ethanol/acetic acid (3:1) essentially as described above except that before blocking with BSA, nuclei were processed to DNA denaturation as previously described for FISH (Avivi et al., 2004).
Construction of AtMBD-GFP Plasmids and Generation of Transgenic Plants
Fusion of AtMBD to GFP was performed by subcloning each of the indicated AtMBD cDNA into pUC19-35S-GFP (a gift from A. Levitan and A. Danon), downstream from the 35S promoter and in frame with the GFP. Each AtMBD cDNA was amplified by PCR to eliminate the stop codon using the following primers: AtMBD2-S, 5′-GAGAGGATCCATGCCTTCAATGCAGAAGTATGAA-3′; AtMBD2-AS, 5′-TCTCCCCGGGTCTATCAGCAAGTTCGTCGTTGG-3′; AtMBD5-S, 5′-TGATATCAGATCTATGTCGAACGGCACGGATCAG-3′; AtMBD5-AS, 5′-TCTCCCCGGGGAACATCGTTTTTCCAGCGTT-3′; AtMBD6-S, 5′-GAGATCTAGAATGTCAGATTCTGTGGCCGGC-3′; AtMBD6-AS, 5′-TCTCCCCGGGAGCCGACACTTTACTAGGG-3′; AtMBD7-S, 5′-GAGAAGATCTAGAATGCAGACGAGATCCTCTTCCTCTCC-3′; AtMBD7-AS, 5′-GAGACCCGGGAATTCTTAAGAGCGGTCTTCGATCAGTG-3′.
The various AtMBD PCR products were digested either with BamHI and SmaI or with BglII and SmaI and subcloned into BglII-SmaI sites of pUC19-35S-GFP. The integrity of each construct was verified by sequencing. These constructs were used for protoplast transformation experiments using polyethylene glycol essentially as described (Sheen, 2002). To generate transgenic Arabidopsis plants expressing AtMBD fused to GFP, the 35S-AtMBD-GFP fragment was excised out using EcoRI and subcloned into the same site of the binary vector pPZP-111 to generate pPZP-35S-AtMBD-GFP followed by transformation into Agrobacterium tumefaciens and Arabidopsis plants (ecotypes Colombia and Wassilewskija as well as the ddm1-2 mutant).
Salt Extraction and ChIP
Salt extraction of nuclei was performed as described (Fass et al., 2002). ChIP was performed on nuclei prepared from Arabidopsis plants expressing AtMBD6-GFP essentially as described (Lawrence et al., 2004) using anti-GFP (Roche), anti-dimethylated K4 histone H3 (Upstate Biotechnology), as well as antidimethylated K9 histone H3 (Upstate Biotechnology). Precipitated DNAs were subjected to PCR using the following primers: CEN180-S, 5′-GAGAGGATCCCGTAAGAATTGTATCCTTGTTAG-3′; CEN180-AS, 5′-GAGAGAATTCCCTTTAAGATCCGGTTGTGG-3′; 18SrDNA-S, 5′-GCTACCTGGTTGATCCTGCCAGTAGTC-3′; 18SrDNA-AS, 5′-CGACCTTTTATCTAATAAATGCGTCCC-3′; Actin-S, 5′-GGTTTTGCTGGGGATGATGC-3′; Actin-AS, 5′-CATTGAATGTCTCAAACATGATTTGAGTC-3′.
ChIP PCR conditions were as follows: 94°C, 5 min; 30 cycles of 94°C, 30 s; 56°C, 30 s; 72°C, 45 s; 72°C, 10 min. PCR products were resolved on 1.2% agarose gel and stained with ethidium bromide.
DNA Extraction, DNA Gel Blot Analysis, and Electrophoretic Mobility Shift Assay
DNA was extracted from Arabidopsis leaves by modification of the C-elyltrimethyl ammonium bromide method (Wagner et al., 1987). To determine the DNA methylation status at chromocenters, genomic DNA was digested with the methylation-sensitive enzymes HpaII and MspI, and samples were run on 1% agarose gel, blotted onto nylon membrane, and hybridized with the 180-bp repeats labeled with [α32P]dCTP using the Nick Translation kit (Roche) according to the manufacturer's protocol.
Electrophoretic mobility shift assay was performed as previously described using GST alone, GST-AtMBD2, and GST-AtMBD5 and 32P-labeled umCG or 1mCG double-stranded oligonucleotides as described (Zemach and Grafi, 2003).
Construction of DDM1 Plasmids and GST Pull-Down Assay
DDM1 and its truncated forms were constructed in pBluescript KS (Stratagene, La Jolla, CA) by PCR using the full length of DDM1 (kindly provided by Kazusa DNA Research Institute, Chiba, Japan) as a template and the following primers: D(1-205)-S, 5′-CACAGGATCCCCTTCGATGGTTAGTCTTCGCTCC-3′; D(1-205)-AS, 5′-CTCCCCGGGTCAATAAGACTTTAACTGTCC-3′; D(511-764)-S, 5′-GAGAGGATCCATGTATCTCTACCCTCCTGTTG-3′; D(511-764)-AS, 5′-TGTGGAATTCCTAACTGTTCAGGGAAGACAGC-3′.
The PCR products were digested either with BamHI and SmaI or BamHI and EcoRI and subcloned into the same sites of either pGEX2T or pBluescript KS+ downstream from the T7 promoter to generate pBs-DDM1(FL), pBs-DDM1(1-205), and pBs-DDM1(511-764). These plasmids were subjected to in vitro transcription–coupled translation in the presence of 35S-Met using a Promega kit (Madison, WI) and according to the supplied protocol. Labeled proteins were subjected to GST pull-down assay using GST-AtMBDs (Zemach and Grafi, 2003) or GST alone (as a negative control) essentially as described (Grafi et al., 1996). GST-MBD2 was reconstructed using a cDNA library (kindly provided by the ABRC) as a template and 5′-GAGAGGATCCATGAGTATGTCGCAGTCTCGAGC-3′ as sense primer and 5′-GAGAGAATTCTTATCTATCAGCAAGTTCGTCG-3′ as antisense primer. The PCR product was digested with BamHI and EcoRI and subcloned into the same sites of pGEX-2T.
Image Analysis
Image analysis was performed using the NIH Image program version 1.61 (http://rsb.info.nih.gov/nih-image). Briefly, to obtain the coefficient of variation for fluorescence intensity, the sd of the pixel values for each nucleus was divided by the mean pixel value. The mean of the coefficient of variation for a given population and the sd were determined for each set of nuclei transformed with AtMBD6-GFP (shown in Supplemental Figure 3A online). Statistical significance was determined using TTEST.
Supplementary Material
Acknowledgments
We thank E. Richards for providing seeds of Arabidopsis mutants ddm1-2 and met1-1, J. Paszkowski for providing met1-3, A. Levitan and A. Danon for providing the GFP construct (pUC19-35S-GFP), and anonymous reviewers for their helpful comments. We also thank the Kazusa DNA Research Institute (Chiba, Japan) for providing DDM1 cDNA clone, and the ABRC for providing Arabidopsis cDNA libraries. This research was supported by the Israel Science Foundation (Grant 385/02-1 to G.G.), by the Ministry of Science of Israel (Eshkol Fellowship to A.Z.), and by the Raymond Burton Fund for Plant Genomic Research.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Assaf Zemach (assaf.zemach@weizmann.ac.il) and Gideon Grafi (gideon.grafi@weizmann.ac.il).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031567.
References
- Amir, R.E., Van den Veyver, I.B., Wan, M., Tran, C.Q., Francke, U., and Zoghbi, H.Y. (1999). Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188. [DOI] [PubMed] [Google Scholar]
- Avivi, Y., Morad, V., Ben-Meir, H., Zhao, J., Kashkush, K., Tzfira, T., Citovsky, V., and Grafi, G. (2004). Reorganization of specific chromosomal domains and activation of silent genes in plant cells acquiring pluripotentiality. Dev. Dyn. 230, 12–22. [DOI] [PubMed] [Google Scholar]
- Bartee, L., Malagnac, F., and Bender, J. (2001). Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 15, 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, A., Meza, T.J., Mahic, M., Thorstensen, T., Kristiansen, K., and Aalen, R.B. (2003). Ten members of the Arabidopsis gene family encoding methyl-CpG-binding domain proteins are transcriptionally active and at least one, AtMBD11, is crucial for normal development. Nucleic Acids Res. 31, 5291–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeski, J., and Jerzmanowski, A. (2003). Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J. Biol. Chem. 278, 823–828. [DOI] [PubMed] [Google Scholar]
- Dennis, K., Fan, T., Geiman, T., Yan, Q., and Muegge, K. (2001). Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 15, 2940–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass, E., Shahar, S., Zhao, J., Zemach, A., Avivi, Y., and Grafi, G. (2002). Phosphorylation of histone H3 at serine 10 cannot account directly for the detachment of human heterochromatin protein 1γ from mitotic chromosomes in plant cells. J. Biol. Chem. 277, 30921–30927. [DOI] [PubMed] [Google Scholar]
- Feng, Q., and Zhang, Y. (2001). The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 15, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle, W., Wang, Y., and Allis, C.D. (2003). Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15, 172–183. [DOI] [PubMed] [Google Scholar]
- Fransz, P., De Jong, J.H., Lysak, M., Castiglione, M.R., and Schubert, I. (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99, 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, N., Watanabe, S., Ichimura, T., Tsuruzoe, S., Shinkai, Y., Tachibana, M., Chiba, T., and Nakao, M. (2003). Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J. Biol. Chem. 278, 24132–24138. [DOI] [PubMed] [Google Scholar]
- Fuks, F., Hurd, P.J., Wolf, D., Nan, X., Bird, A.P., and Kouzarides, T. (2003). The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 278, 4035–4040. [DOI] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297, 1871–1873. [DOI] [PubMed] [Google Scholar]
- Grafi, G., Burnett, R.J., Helentjaris, T., Larkins, B.A., DeCaprio, J.A., Sellers, W.R., and Kaelin, W.G., Jr. (1996). A maize cDNA encoding a member of the retinoblastoma protein family: Involvement in endoreduplication. Proc. Natl. Acad. Sci. USA 93, 8962–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikrishnan, K.N., Chow, M.Z., Baker, E.K., Pal, S., Bassal, S., Brasacchio, D., Wang, L., Craig, J.M., Jones, P.L., Sif, S., and El-Osta, A. (2005). Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 37, 254–264. [DOI] [PubMed] [Google Scholar]
- Hendrich, B., and Bird, A. (1998). Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18, 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich, B., and Tweedie, S. (2003). The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 19, 269–277. [DOI] [PubMed] [Google Scholar]
- Htun, H., Holth, L.T., Walker, D., Davie, J.R., and Hager, G.L. (1999). Direct visualization of the human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor. Mol. Biol. Cell 10, 471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, M., Koike, A., Koizumi, N., and Sano, H. (2003). Methylated DNA-binding proteins from Arabidopsis. Plant Physiol. 133, 1747–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, J.P., Johnson, L., Jasencakova, Z., Zhang, X., Perez-Burgos, L., Singh, P.B., Cheng, X., Schubert, I., Jenuwein, T., and Jacobsen, S.E. (2004). Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112, 308–315. [DOI] [PubMed] [Google Scholar]
- Jackson, J.P., Lindroth, A.M., Cao, X., and Jacobsen, S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560. [DOI] [PubMed] [Google Scholar]
- Jasencakova, Z., Soppe, W.J., Meister, A., Gernand, D., Turner, B.M., and Schubert, I. (2003). Histone modifications in Arabidopsis—High methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 33, 471–480. [DOI] [PubMed] [Google Scholar]
- Jeddeloh, J.A., Stokes, T.L., and Richards, E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kakutani, T., Jeddeloh, J.A., Flowers, S.K., Munakata, K., and Richards, E.J. (1996). Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. USA 93, 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani, T., Jeddeloh, J.A., and Richards, E.J. (1995). Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 23, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel, M.W., Ramsey, D.E., Stokes, T.L., Flowers, S.K., Haag, J.R., Jeddeloh, J.A., Riddle, N.C., Verbsky, M.L., and Richards, E.J. (2003). Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163, 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner, M., O'Sullivan, R.J., and Jenuwein, T. (2003). An epigenetic road map for histone lysine methylation. J. Cell Sci. 116, 2117–2124. [DOI] [PubMed] [Google Scholar]
- Lawrence, R.J., Earley, K., Pontes, O., Silva, M., Chen, Z.J., Neves, N., Viegas, W., and Pikaard, C.S. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13, 599–609. [DOI] [PubMed] [Google Scholar]
- Lindroth, A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S., and Jacobsen, S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Malagnac, F., Bartee, L., and Bender, J. (2002). An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 21, 6842–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E.J. (1997). DNA methylation and plant development. Trends Genet. 13, 319–323. [DOI] [PubMed] [Google Scholar]
- Richards, E.J. (2002). Chromatin methylation: Who's on first? Curr. Biol. 12, R694–R695. [DOI] [PubMed] [Google Scholar]
- Ronemus, M.J., Galbiati, M., Ticknor, C., Chen, J., and Dellaporta, S.L. (1996). Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273, 654–657. [DOI] [PubMed] [Google Scholar]
- Saze, H., Scheid, O.M., and Paszkowski, J. (2003). Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34, 65–69. [DOI] [PubMed] [Google Scholar]
- Scebba, F., Bernacchia, G., De Bastiani, M., Evangelista, M., Cantoni, R.M., Cella, R., Locci, M.T., and Pitto, L. (2003). Arabidopsis MBD proteins show different binding specificities and nuclear localization. Plant Mol. Biol. 53, 715–731. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (2002). A transient expression assay using Arabidopsis mesophyll protoplasts. http://genetics.mgh.harvard.edu/sheenweb/.
- Soppe, W.J., Jasencakova, Z., Houben, A., Kakutani, T., Meister, A., Huang, M.S., Jacobsen, S.E., Schubert, I., and Fransz, P.F. (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21, 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru, H., and Selker, E.U. (2001). A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414, 277–283. [DOI] [PubMed] [Google Scholar]
- Tamaru, H., Zhang, X., McMillen, D., Singh, P.B., Nakayama, J., Grewal, S.I., Allis, C.D., Cheng, X., and Selker, E.U. (2003). Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 34, 75–79. [DOI] [PubMed] [Google Scholar]
- Tariq, M., Saze, H., Probst, A.V., Lichota, J., Habu, Y., and Paszkowski, J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100, 8823–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, P.A., Gegonne, A., Jones, P.L., Ballestar, E., Aubry, F., and Wolffe, A.P. (1999). Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 23, 62–66. [DOI] [PubMed] [Google Scholar]
- Wagner, D.B., Furnier, G.R., Saghai-Maroof, M.A., Williams, S.M., Dancik, B.P., and Allard, R.W. (1987). Chloroplast DNA polymorphisms in lodgepole and jack pines and their hybrids. Proc. Natl. Acad. Sci. USA 84, 2097–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, M., et al. (1999). Rett syndrome and beyond: Recurrent spontaneous and familial MECP2 mutations at CpG hotspots. Am. J. Hum. Genet. 65, 1520–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Q., Cho, E., Lockett, S., and Muegge, K. (2003). Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin. Mol. Cell. Biol. 23, 8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach, A., and Grafi, G. (2003). Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J. 34, 565–572. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., and Reinberg, D. (2001). Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.