Abstract
Adeno-associated virus encodes four nonstructural proteins, which are known as Rep78, Rep68, Rep52, and Rep40. Expression of these nonstructural proteins affects cell growth and gene expression through processes that have not yet been characterized. Using a yeast two-hybrid screen, we have demonstrated that a stable interaction occurs between the viral proteins Rep78 and Rep52 and the putative protein kinase PrKX, which is encoded on the X chromosome. The stability and specificity of the Rep-PrKX interaction were confirmed by coimmunoprecipitation of complexes assembled in vitro and in vivo. Overexpressed PrKX, which was purified from cos cells, was shown to phosphorylate a synthetic protein kinase A (PKA) substrate. However, this activity was dramatically inhibited by stoichiometric amounts of Rep52 and weakly inhibited with Rep68, which lacks the carboxy-terminal sequence contained in Rep52. Similarly, a stable interaction was observed with Rep78, which also contains the carboxy-terminal sequence of Rep52. A stable interaction and inhibition were also observed between Rep52 and the catalytic subunit of PKA. By using surface plasmon resonance and kinetic studies, Kis of approximately 300 and 167 nM were calculated for Rep52 with PKA and with PrKX, respectively. Thus, Rep52 but not Rep68 can significantly inhibit the trans- and autophosphorylation activities of these kinases. The biological effects of Rep78-specific inhibition of PKA-responsive genes are illustrated by the reduction of steady-state levels of cyclic AMP-responsive-element-binding protein and cyclin A protein.
Infection with the human dependovirus adeno-associated virus type 2 (AAV) has been reported to inhibit cell proliferation rates (52) and to induce growth arrest (14), cell death (38), and tumor suppression (15–17). Some of these effects have been attributed to the viral nonstructural Rep proteins, which are central to the regulation of AAV DNA replication and transcription (2, 18, 36, 44, 46). Two of the Rep proteins (Rep68 and Rep78) have been associated with the preferential integration of AAV genomes into a region of the long or q arm of human chromosome 19 (27–29, 48). Rep68 and Rep78 are transcribed from the promoter at map unit 5 (p5) and are produced from spliced and unspliced mRNAs, respectively (see Fig. 1). In vitro assays have demonstrated helicase activities as well as site- and strand-specific nicking activities for these proteins (22, 41, 42, 50). DNA binding and nicking activities have been shown to require the binding of the Rep proteins to tandem repeats of a GAGY motif present in the inverted terminal repeat (ITR) structures at the ends of the viral genome (5). Two additional Rep proteins are produced from an internal promoter in the Rep open reading frame (ORF) at map unit 19 and are referred to by their apparent molecular masses, i.e., Rep52 and Rep40. While these two proteins lack the DNA binding ability of Rep68 and Rep78, they have recently been shown to possess ATP-dependent helicase activity (39).
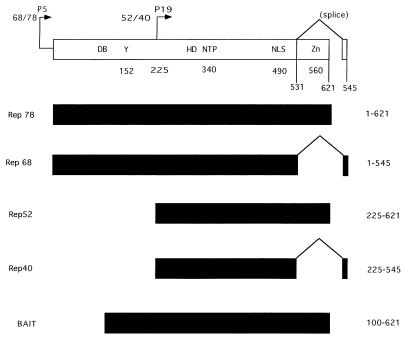
FIG. 1.
Rep functional domains. Within the Rep gene, two promoters, which are located at map units 5 and 19, direct the expression of four proteins, Rep78 and Rep68 and Rep52 and Rep40, respectively. Two proteins are produced from each promoter, one of which is spliced (Rep68 and Rep40) and one of which is unspliced (Rep78 and Rep52). Within the Rep ORF, several functional domains have been identified and are indicated at the top. These include the DNA DB, terminal resolution site (Y), NTP binding pocket (NTP), helicase domain (HD), nuclear localization domain (NLS), zinc finger (Zn), and splice sites, with their relative amino acid positions indicated below. The four Rep proteins as well as the portion of Rep used as bait in the yeast two-hybrid screen are indicated.
In addition to their role in the life cycle of AAV, Rep expression results in a characteristic phenotype. Overexpression of Rep68 and Rep78 has been shown to either negatively or positively affect transcription of heterologous promoters, to inhibit cellular transformation by papillomavirus or by adenovirus E1a plus an activated ras oncogene (15–17, 19, 24, 30, 31, 51), and to inhibit progression through the cell cycle (14, 52). Recent data have demonstrated that infection with AAV can affect cell growth by inhibiting phosphorylation of the pRB protein family as well as by stimulating expression of the regulatory protein of the cyclin-dependent protein kinase p21WAF1 (14). While some of these activities may be mediated by the direct interaction of Rep with DNA, other effects may result from interactions with cellular regulatory proteins. Furthermore, the AAV p19-promoted protein Rep52, which does not bind the ITR, has also been shown to inhibit expression of heterologous promoters. This observation suggests that Rep52 may affect cellular gene expression through interactions with cellular proteins (19, 31).
One technique used to examine protein-protein interaction is the yeast two-hybrid system. The results presented here demonstrate a strong interaction of Rep52 and Rep78 but not Rep68Δ with a protein kinase A (PKA)-type kinase, which is referred to as PKX1 (or PrKX). PrKX is located on the human X chromosome at Xp22.3. This novel protein kinase has widespread expression, with the highest levels of expression in fetal and adult brain, kidney, and lung tissue as detected by Northern blot analysis (25).
Based on surface plasmon resonance (SPR) and enzyme kinetics, our data show that Rep52 associates with PrKX and inhibits kinase activity. The catalytic subunit of PKA was similarly inhibited by Rep52. While the physiological role of PrKX remains to be elucidated, Rep-mediated inhibition of PKA alone or both PKA and PrKX may explain some of the effects of Rep expression on cell growth and cell cycle progression.
MATERIALS AND METHODS
Plasmids and yeast transformation.
The Gal4 DNA binding domain (BD)-Rep fusion expression plasmid (bait plasmid) was constructed by ligating the NcoI/XhoI fragment of the AAV Rep ORF into the NdeI/SalI sites of the pAS1-CYH2 plasmid (kindly provided by Stephen Elledge) via an NdeI/NcoI linker. The random-primed HeLa cell cDNA library was obtained from Clonetech Laboratories. Yeast (Saccharomyces cerevisiae Y1090) were transformed with the Yeastmaker kit (Clonetech Laboratories) according to the manufacturer’s instructions. The plasmid pGem Rep52 (pSR378) contains AAV nucleotides 989 to 2187 (containing the Rep52 ORF) in plasmid pGEM-3Z (Promega). The maltose-binding protein (MBP)-Rep fusion proteins MBP-Rep52, MBP-Rep68, and MBP-LacZ were purified as previously described (3). The cDNA for PrKX was isolated from a random-primed HeLa cell cDNA library. Several independent clones were isolated and sequenced. The longest clone contained bases 450 to 1880 of the cDNA originally reported (25) (GenBank accession no. X85545). This cDNA has a valine-to-alanine substitution at position 42, compared to the original isolate. A full-length clone was constructed (SVPrKX) by using synthetic oligonucleotides which incorporated an amino-terminal poly-His tag (MGSSHHHHHHSSG) for affinity purification. The final construct was produced by ligation into the StuI and HindIII sites of pSV-Sport (BRL). This plasmid was introduced into cos cells by electroporation as previously described (5). A glutathione S-transferase (GST)-PrKX fusion protein was produced with the ESP yeast expression system in Schizosaccharomyces pombe (Stratagene). An SmaI/HindIII fragment was isolated from SVPrKX and cloned into the StuI/HindIII sites of the HTb shuttle vector (BRL); a BamHI/HindIII fragment was then isolated, and the HindIII site was filled in with the Escherichia coli DNA polymerase Klenow fragment. This fragment was then cloned into the BamHI/SmaI sites of ESP. Recombinant protein was produced following the manufacturer’s instructions (Stratagene).
Rep68 and Rep78 expression plasmids were created by cloning the Rep68 and Rep78 ORFs into the expression plasmid Cb6 (37). Rep68 and Rep78 ORFs were generated by PCR amplification with Pfu DNA polymerase (Stratagene), with BglII restriction sites added to the coding primer and XhoI sites added to the noncoding primer. The sequence was confirmed by dye terminator sequencing reactions on an automated sequencer (ABI 373a).
Purification of PrKX from cos cells.
cos cells (2 × 108) were electroporated with pSVPrKX as previously described (4). The medium was changed 24 h postelectroporation, and the cells were scraped from the tissue culture plates 12 to 24 h later (36 to 48 h postelectroporation). The detached cells were rinsed once with cold phosphate-buffered saline (PBS), and the cells were pelleted and resuspended in 10 ml of 1× binding buffer (BB) (20 mM Tris-HCl [pH 7.9], 500 M NaCl containing 5 mM imidazole) (Novagen). The cells were lysed by the addition of 10 ml of 1× binding buffer containing 2% Nonidet P-40 (NP-40) and incubated on ice for 5 min. Following sonication (2× 10-s pulses), the lysate was centrifuged for 5 min at 12,000 × g, and the supernatant was removed. The pellet was resuspended in 10 ml of BB containing 1% NP-40 and 5 mM imidazole, sonicated, and centrifuged for 20 min at 12,000 × g. Finally, the supernatant was applied to a 0.5-ml column of charged nickel resin (Novagen) and washed sequentially with (i) 40 ml of BB with 1% NP-40, (ii) 10 ml of 25 mM imidazole BB–1% NP-40, (iii) 10 ml of 25 mM imidazole BB containing 1 mM fresh cyclic AMP (cAMP) and 1% NP-40, (iv) 20 ml of 25 mM imidazole BB, (v) 20 ml of 40 mM imidazole BB, and (vi) 10 ml of 60 mM imidazole BB buffer with 1% NP-40. The remaining adsorbed protein was eluted with 150 mM imidazole wash buffer. The peak protein fractions, as determined by Coomassie staining of sodium dodecyl sulfate (SDS)-polyacrylamide gels, were pooled and dialyzed against 0.25% NP-40–20% glycerol–1 mM dithiothreitol (DTT)–20 mM Tris-HCl (pH 8) or 20 mM 3-(N-morpholino)propanesulfonic acid–150 mM NaCl–1 mM DTT–0.005% surfactant P20 (pH 7.0; Biacore buffer B).
Coimmunoprecipitation.
35S-labeled proteins were produced with a coupled transcription-translation reaction (TNT system; Promega Corp.) following the manufacturer’s instructions. Glutathione-Sepharose beads (Pharmacia) were washed three times in GST-BB (PBS [pH 7.2], 1 mg of bovine serum albumin [BSA] per ml, 0.5% NP-40). Radiolabeled proteins were first diluted in GST-BB and centrifuged at 10,000 × g for 2 min. The supernatant was transferred to a fresh tube and recentrifuged with 60 μl of a 50% slurry (vol/vol) of glutathione beads to remove proteins which bound to the glutathione beads alone. The supernatant was then divided into two tubes, and one tube was supplemented with 1 μg of purified yeast GST-PrKX and allowed to incubate for 1 h at 8°C. Then, glutathione beads were added to all samples, and the samples were incubated for an additional 30 min followed by three washes with GST-BB and three washes with GST-BB without BSA. The pellet containing the beads was then resuspended in SDS sample buffer and fractionated by electrophoresis on a Tris-glycine polyacrylamide gel. The bacterial expression plasmid for PKA was kindly provided by Susan S. Taylor (University of California, San Diego).
In vivo coimmunoprecipitations were done by electroporating cos cells with the indicated plasmids as described above. At 36 h postelectroporation, the plates were rinsed with cold PBS and lysed by adding 3 ml of lysis buffer (PBS [pH 7.4], 0.5% NP-40, complete protease inhibitors [Boehringer Mannheim]) and rocking in the cold for 30 min. The plates were then scraped and rinsed with 1 ml of lysis buffer followed by centrifugation for 15 min. The supernatant was then transferred to a fresh tube, and the pellet was resuspended in lysis buffer supplemented with an additional 0.5% NP-40 and briefly sonicated. Following microcentrifugation, the supernatant was removed and pooled with the supernatant from the previous centrifugation. The lysates were then cleared by adding 1 μg of normal rabbit immunoglobulin G (Santa Cruz Biotechnology) and protein A beads followed by centrifugation. This step was repeated with protein A beads alone. Approximately 1 ml of the lysate was recentrifuged in an Eppendorf tube, and 3 μg of anti-His antibody (Santa Cruz Biotechnology) was incubated with the lysate for 1 h on a rotating platform. A 20-μl aliquot of protein A beads (Santa Cruz Biotechnology) was then added, and the incubation was continued for an additional 5 h. Following this incubation, the beads were pelleted and washed three times with lysis buffer and then resuspended in SDS sample buffer. Western blots were prepared by electrophoresis of the precipitated protein samples on SDS-10% polyacrylamide gels, followed by transfer to nitrocellulose membranes and probing with an anti-Rep antibody (303.9; American Research Products).
SPR.
Binding experiments between immobilized PrKX or PKA and Rep52 and Rep68 were performed on a BIAcore 2000 system (BIAcore AB) in 20 mM morpholinepropanesulfonic acid (MOPS)–150 mM KCl–1 mM DTT–0.005% surfactant P20 (BIAcore AB no. BR1000-54) (pH 7.0; buffer B). A GST-specific sensor surface was created by immobilizing an αGST antibody (BIAcore GST capture kit) on carboxymethyl (CM) 5 sensor chips (research grade). A total of 6,000 response units (RU) of the antibody was coupled with a standard amine coupling reaction via N-hydroxysuccinimide/N-ethyl-N7-(3-diethylaminopropyl)carbodiimide (NHS/EDC) according to the instructions of the manufacturer (BIAcore). A total of 1,000 RU corresponds to 1 ng of protein per mm2 bound to the CM surface. Both PrKX overexpressed and purified from S. pombe and the catalytic subunit of PKA overexpressed and purified from E. coli were immobilized as GST-fusion proteins. A 5-μg/ml solution of each protein was injected over the αGST surface with a flow rate of 5 μl/min, resulting in 480 to 600 RU bound to the GST antibody. With a baseline drift of 2 to 3 RU/min, this surface was stable enough for binding of Rep with a flow rate of 30 μl/min. After each binding event, the surface was regenerated with 10 mM glycine (pH 2.2), removing the PrKX-Rep or PKA-Rep complex from the antibody surface, followed by a novel coupling of GST-PrKX or GST-PKA. To obtain association and dissociation rate constants and to calculate apparent Kds, the interactions between GST-PrKX or GST-PKA and the Rep proteins were measured by injecting Rep at a concentration ranging from 150 to 1,400 nM in buffer B. In the association phase, the interactions were monitored for 4 min. The dissociation phase was then monitored for 6 min. To correct for unspecific binding, blank runs were performed on a surface with the same amount of antibody immobilized and this value was subtracted. To prevent nonspecific binding to the CM surface, runs were performed in the presence of 1 mg of CM dextran (Fluka) per ml. Kinetic constants were calculated by global fit analysis employing BIAevaluation 3.0 software. A single set of rate constants was used to fit association and dissociation rate constants simultaneously at different concentrations.
Kinase assay.
Kinase reactions were typically carried out under conditions that consisted of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 200 μM ATP supplemented with [γ-32P]ATP to a final specific activity of 200 μCi/μmol, with incubation at 30°C. Peptide substrates were purchased from New England Biolabs and used at a final concentration of 200 μM. For kinetic determinations, 25-μl reaction mixtures were started by the addition of 100 ng (2.6 pmol) of purified PrKX, and 3-μl samples were removed at 1-min intervals (starting at 30 s) for 5 min. Samples were spotted onto 25-cm P81 filters (Whatman) and then washed four times for 3 min with 75 mM phosphoric acid. The washed filters were then dried under a heat lamp, and the bound radioactivity was measured by liquid scintillation counting. The kinase activity of PKA was measured in a similar manner, except that 10 nM (0.25 pmol) purified PKA was used.
Western blots.
Whole-cell lysates were prepared from electroporated cos cells at 36 h postelectroporation. The cells were washed in PBS and lysed in 1× SDS sample buffer, and the DNA was sheared with a Qiashredder column (Qiagen). Equal loading of protein was determined by Coomassie staining of polyacrylamide gels with serial dilutions of the samples. Western blots were prepared by electrophoresis of equivalent amounts of the protein samples on 10% Tris-glycine gels (Novex) and transferred to a polyvinylidene difluoride membrane. The following antibodies were used according to the suppliers’ instructions: Rep (303.9; American Research Products), cAMP-responsive-element-binding protein (CREB; Calbiochem), cyclin A (H-432; Santa Cruz Biotechnology), and actin (C11; Santa Cruz Biotechnology).
RESULTS
In order to identify cellular proteins which interact with the Rep proteins of AAV, a yeast two-hybrid screen was performed. Initial constructs fusing the Rep78 ORF with the Gal4 DNA BD vector (12) appeared to be unstable in yeast and also autoactivated expression of His and LacZ reporter genes. Therefore, fragments of the Rep ORF were cloned into the BD vector and tested for autoactivation. One Rep fragment (amino acids 100 to 621) did not autoactivate expression of the marker genes and was subsequently used to screen a random-primed HeLa cell cDNA library. This fragment is referred to as the bait (Fig. 1). Transformants (>107) were screened, and 50 were scored as His and LacZ positive. Of these 50 clones, 4 were represented more than once, with 1 clone isolated eight times. A search of GenBank identified the clone as coding for the protein kinase X 1 (PrKX) (25) (GenBank accession no. X85545). Multiple cDNAs were sequenced and found to differ at amino acid 42 from the original PrKX cDNA (Val-to-Ala substitution). While there were no available functional data for the gene product, previous sequence homology analysis indicated that it had strong homology to PKA-type kinases isolated from invertebrates (25).
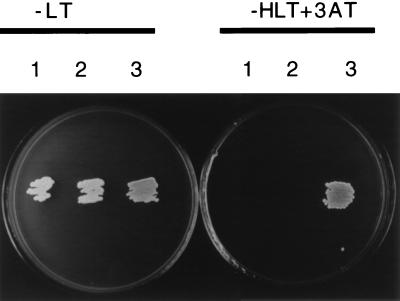
The specificity of the interaction between Rep and PrKX was confirmed with the yeast two-hybrid system. Plasmids encoding the activation domain-PrKX fusion (AD-PrKX) or BD-Rep fusion were transformed into yeast that contained either empty Gal4 AD, empty BD vectors, or both fusion proteins. While growth was detected on plates lacking leucine and tryptophan (−LT) (Fig. 2, plate 1, colonies 1, 2, and 3), growth was visible on selective media only if both fusion plasmids were present (Fig. 2, plate 2, colony 3), confirming that activation of the reporter genes resulted from at least transient interaction between the Rep and PrKX moieties.
FIG. 2.
Yeast two-hybrid screen. Yeast strain Y1090 was transformed with either the BD-Rep fusion plus empty AD plasmids (colony 1), AD-PrKX fusion plus empty BD plasmids (colony 2), or BD-Rep plus AD-PrKX (colony 3) and then streaked onto a plate lacking leucine and tryptophan (−LT) (left plate) or a selective plate lacking histidine, tryptophan, and leucine (−HTL) supplemented with 3-aminotriazole (3AT) (right plate). While all clones grew under nonselective conditions (left plate), only yeast which had been transformed with both the Rep and PrKX fusion grew under selective conditions (right plate, colony 3).
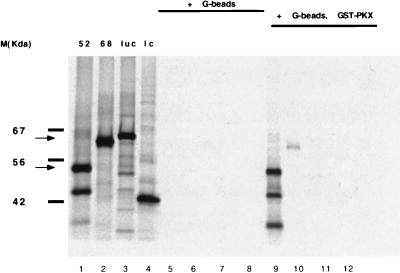
To corroborate the results of the yeast genetic screen, in vitro coimmunoprecipitation experiments were performed. Rep52, Rep68, luciferase, or a randomly selected library clone were transcribed and translated with [35S]methionine in rabbit reticulocyte lysates (Promega) (Fig. 3, lanes 1 to 4). The three bands observed in the in vitro translation of Rep52 result from internal initiation of translation, and all contain a common carboxy terminus (data not shown). No precipitation of the four proteins translated in reticulocyte lysate supplemented with glutathione-Sepharose beads alone was observed (Fig. 3, lanes 5 to 8). However, all three Rep52-derived polypeptides were precipitated with glutathione beads in the presence of GST-PrKX fusion protein produced in yeast (Fig. 3, lane 9). A weak interaction between PrKX and Rep68Δ may account for the relatively small amount of Rep68Δ recovered (Fig. 3, lane 10). No 35S-labeled protein was precipitated with GST-PrKX and luciferase or the random library clone (Fig. 3, lanes 11 and 12).
FIG. 3.
In vitro coimmunoprecipitation. Rabbit reticulocyte lysates programmed with Rep52 (lanes 1, 5, and 9) Rep68Δ (lanes 2, 6, and 10) luciferase (lanes 3, 7, and 11), or a random library clone (1c) (lanes 4, 8, and 12) were incubated in the presence of glutathione beads alone (lanes 5 to 8) or were supplemented with a GST-PrKX fusion protein (lanes 9 to 12). The bound protein was then precipitated and separated on a polyacrylamide gel. As a control, total protein in the programmed reticulocyte lysates was also analyzed (lanes 1 to 4). The products of the in vitro translations of Rep68 and Rep52 are indicated by the upper and lower arrows, respectively, on the left.
SPR was used to quantify the interaction between Rep and PrKX with immobilized GST-PrKX. The association and dissociation of MBPRep68Δ and MBPRep52 were monitored at a buffer flow rate of 30 μl/min. Both proteins were shown to be greater that 90% pure according to Coomassie-stained SDS-polyacrylamide gels. Both Rep proteins showed some unspecific binding which could be subtracted by using a control GST surface. A binding constant was calculated from the respective rate constants and protein concentrations. The interaction between Rep68Δ and PrKX had a Kd approximately sixfold higher than that of the interaction between Rep52 and PrKX (Table 1). This difference was mainly due to the combined effect of a threefold lower association rate constant and a twofold lower dissociation rate constant. The difference in relative binding affinities measured by SPR agrees with the immunoprecipitation data, which show a weaker interaction of PrKX with Rep68Δ compared to that with Rep52.
TABLE 1.
Association and dissociation ratesa
| Complex | Result by BIAcore
|
Result by kinetic studies
|
|||
|---|---|---|---|---|---|
| Ka (M−1s−1) | Kd (s−1) | KD (nM) | IC50 (nM) | Ki (nM) | |
| PrKX–MBP-Rep52 | 2.4 × 104 ± 3 × 102 | 9 × 10−3 ± 8 × 10−5 | 385 | 210 | 90 |
| PrKX–MBP-Rep68Δ | 8.2 × 103 ± 2 × 102 | 1.8 × 10−2 ± 2 × 10−3 | 2,100 | ND | ND |
| PrKX–MBP-Rep78 | 1.5 × 104 ± 2 × 102 | 1 × 10−2 ± 1.2 × 10−4 | 680 | ND | ND |
| PKA–MBP-Rep52 | 6.5 × 103 ± 0.98 × 102 | 1.95 × 10−3 ± 2 × 10−5 | 320 | 340 | 330 ± 189 |
| PKA–MBP-Rep68Δ | NSBb | NSBb | NDc | NDc | NDc |
Association rate constants (Ka), dissociation rates (Kd), and binding constants (KD) were determined by SPR. IC50s were determined by in vitro kinase assays as described in Materials and Methods. Ki, IC50/(1 + [ligand/Kd]). Standard errors are given as standard deviations. ND, not determined.
NSB, no specific binding detected.
Because no specific binding was detected, value could not be determined.
Rep68Δ contains an amino-terminal sequence absent in Rep52 and lacks the carboxy-terminal sequence of Rep52, which is spliced out (Fig. 1). To distinguish whether the carboxy terminus is important in the interaction with PrKX, the association and dissociation rates of MBPRep78, which contains the same carboxy terminus as MBPRep52, and the amino terminus of MBPRep68Δ were measured by SPR. Values similar to those obtained with MBPRep52 were obtained (Table 1), confirming the importance of the carboxy terminus of Rep in the interaction with PrKX.
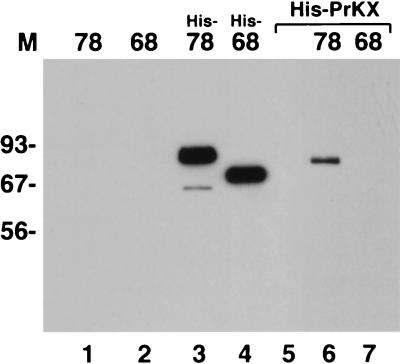
The stability of the Rep-PrKX complex under more physiological conditions was examined by in vivo coimmunoprecipitation experiments. cos cells were transfected with a His-tagged PrKX expression plasmid and either Rep68 or Rep78 expression plasmid. Whole-cell lysates were prepared from the transfected cells immunoprecipitated with a His-tag-specific antibody. Proteins which coimmunoprecipitate with the His-PrKX were then separated on polyacrylamide gels and probed for Rep by Western blotting with a Rep-specific antibody (Fig. 4). As a control, cos cells were also transfected with His-tagged Rep78 or His-tagged Rep68 alone and precipitated with the His-tag-specific antibody as described above. Western blots of immunoprecipitated, His-tagged, Rep78- and Rep68-transfected cell lysates showed bands of the appropriate sizes for His-Rep68 or His-Rep78 (Fig. 4, lanes 3 and 4). No Rep protein was detected in the immunoprecipitated lysate prepared from cells transfected with non-His-tagged Rep78 or Rep68 or cells transfected with His-tagged PrKX alone (Fig. 4, lanes 1, 2, and 5, respectively). However, a Rep78-specific band was detected in lysates cotransfected with His-PrKX and Rep78, but no Rep band was detected when His-PrKX was cotransfected with Rep68 (Fig. 4, lanes 6 and 7). The slight difference in the mobilities of the Rep78 band in lanes 3 and 6 is the result of the His tag on the Rep78 protein in the positive control in lane 3. Thus, the stable interaction observed in vitro between Rep78 and PrKX can also be detected in vivo.
FIG. 4.
In vivo coimmunoprecipitation. Whole-cell lysates were prepared from transfected cells, and the proteins complexed with PrKX were precipitated with an antibody to the poly-His tag fused to the amino terminus of PrKX. cos cells were transfected with a His-tagged PrKX expression plasmid only (lane 5) or were cotransfected with either Rep78 or Rep68 (lanes 6 and 7). As a control, cos cells were also transfected with either His-tagged Rep78 or Rep68 or untagged Rep78 or Rep68 and also precipitated (lanes 3, 4, 1, and 2, respectively). The complexes were then separated on polyacrylamide gels and probed for Rep by Western blotting with a Rep-specific antibody. Rep78 but not Rep68 was precipitated only when transfected with His-tagged PrKX (lanes 6 and 7, respectively).
Comparison of the PrKX sequence with the crystal structure of the PKA catalytic subunit indicates that the catalytic site is conserved (26). An alignment of the amino acid sequence of PKA and PrKX shows conservation of residues important within the serine-threonine protein kinase family (Fig. 5). Furthermore, the residues important for the interaction with a synthetic substrate for the PKA-type kinases, the heptapeptide Kemptide (LRRASLG), are conserved. The target serine in Kemptide is referred to as the P site (43). The residues important for recognition by the PKA catalytic subunit are located at position P(−)3, which interacts with PKA residues D328 and E127; position P(−)2, which interacts with PKA residues E170 and E230; and the PKA residues L198, P202, and L205, forming a hydrophobic pocket for the P(+)1 residue. All of these residues are conserved between PKA and PrKX, except for one conservative exchange (D to E) at amino acids 328 and 333 in PKA and PrKX, respectively (Fig. 5). The conservation of these residues suggests that PrKX may function as a protein kinase.
FIG. 5.
Comparison of PKA and PrKX; sequence alignment between the human catalytic subunit of PKA alpha (Swiss-Prot accession no. P17612) and PrKX (human PKX1 Swiss-Prot accession no. P51817) with the program CLUSTAL (pcgene). Asterisks, identical amino acids; ellipses, conserved amino acids; boldface, amino acids conserved within protein kinases; underlining, functional residues identified within PKA as previously identified (11a, 43a).
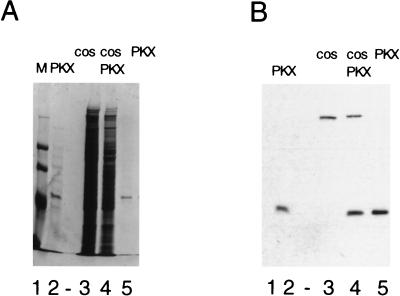
While a high level of expression of PrKX was achieved in bacteria, the majority of the protein was insoluble and inactive (data not shown). Activity was not restored by either a urea denaturation-renaturation procedure or expression as a fusion protein with the MBP. Expression of His-tagged PrKX in transformed African green monkey kidney cells (cos) and subsequent purification by nickel affinity chromatography yielded a soluble and catalytically active protein of approximately 42 kDa. The eluted PrKX fractions were estimated by Coomassie staining to be greater than 90% free of contaminating proteins (Fig. 6A, lane 5). Western blot analysis with an antibody directed at the His tag further confirmed the identity of this purified protein, as well as its absence in control cells electroporated with pUC19 (Fig. 6B). The highest level of kinase activity, as measured in a Kemptide phosphorylation assay, was observed in the peak protein elution fractions. In contrast, no phosphorylation activity of a casein kinase II substrate at a similar concentration was observed in these fractions, and Western blot analysis of the affinity-purified PrKX with antibodies against PKAα was negative (data not shown). A control lysate from cos cells transfected with pUC19, which was passed over the nickel affinity column and eluted in the same manner as that for the PrKX-transfected cos cells, showed no phosphorylation of Kemptide over background levels (data not shown).
FIG. 6.
Purification of PrKX. cos cells were transfected with pSVPrKX, and His-tagged PrKX was isolated as described in Materials and Methods. Bacterially expressed His-PrKX (lane 2) and total protein from either pUC19 (lane 3)- or SVPrKX (lane 4)-electroporated cos cells was separated by SDS-polyacrylamide gel electrophoresis and compared with nickel affinity-purified cos His-PrKX (lanes 5). Duplicate gels were either stained with Coomassie brilliant blue (A) or Western blotted and probed with an antibody directed against the His tag (B). The eluted 42-kDa protein was found to be greater than 90% pure and was found only in the SVPrKX-electroporated cells when probed with the anti-His tag antibody.
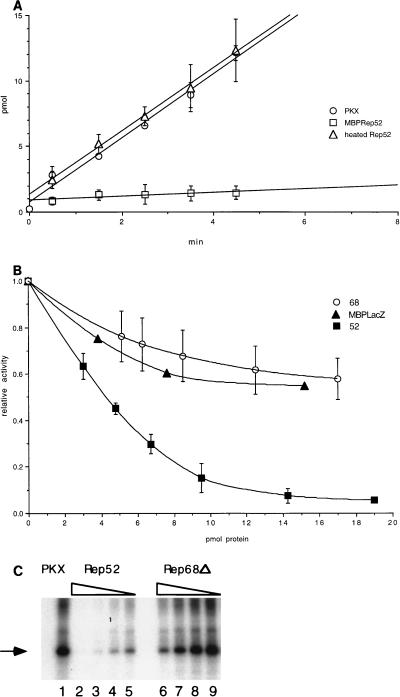
Kinetic analysis of PrKX phosphorylation of Kemptide indicates a rate of incorporation of 0.2 U/mg of protein (1 U = 1 μmol of 32P incorporation/min). Addition of 19 pmol of MBPRep52 dramatically inhibited PrKX (2.6 pmol) kinase activity (Fig. 7A). However, MBPRep52 heated for 10 min at 65°C prior to addition resulted in a lack of inhibition (Fig. 7A), indicating that the native protein was responsible for the inhibition. This inhibition appeared to be specific, since no inhibition of CKII could be detected in a similar assay (data not shown). Titration of MBPRep52 showed a strong interaction between MBPRep52 and PrKX (Fig. 7B), with a 50% inhibition concentration (IC50) of 200 nM. At the concentrations tested, only weak inhibition of PrKX kinase activity was observed with MBP-Rep68Δ.
FIG. 7.
Inhibition of PrKX trans and autophosphorylation activity. (A) Incubation of purified cos His-PrKX with Kemptide and γ-32P-labeled ATP showed transfer of 32P to the Kemptide substrate. Addition of a 7-fold molar excess of MBP-Rep52 inhibited greater than 90% of this kinase activity. Rep52 heated to 65°C for 10 min prior to addition did not inhibit kinase activity. (B) The kinetics of inhibition of PrKX by MBP-Rep68Δ and MBP-LacZ were compared with that for MBP-Rep52. Increasing amounts of each protein were added to the kinase reaction mixture containing 2.6 pmol of His-PrKX protein each. The resulting kinase activity was then compared with that of a control reaction mixture containing only PrKX, and percent inhibition was determined. Similar levels of PrKX activity were measured in the presence of either MBP-Rep68Δ or MBP-LacZ. (C) Inhibition of autophosphorylation of PrKX, which was measured by incubation of 100 nM PrKX in the presence of [γ-32P]ATP, results in phosphorylation of the 42-kDa His-PrKX fusion protein, as indicated by the arrow (lane 1). This activity is inhibited upon addition of fourfold molar excess of MBP-Rep52 (lane 2). Twofold serial dilution of MBP-Rep52 (200, 100, and 50 nM) eventually restored some activity (lanes 3, 4, and 5). A similar titration of MBP-Rep68Δ showed only slight inhibition at the highest concentrations tested (lanes 6 to 9).
The data presented above demonstrate that Rep52 is able to inhibit PrKX phosphorylation of the synthetic substrate Kemptide. The effect of Rep52 on PrKX autophosphorylation activity was determined by incubation of PrKX with [γ-32P]ATP and MgCl2 alone or in the presence of either MBP-Rep52 or MBP-Rep68Δ, followed by separation on a polyacrylamide gel. Incubation of PrKX alone showed autophosphorylation, which has been reported for other PKA-type kinases (1) (Fig. 7C, lane 1). However, addition of an equimolar amount of MBPRep52 inhibited greater than 90% of the autophosphorylation activity (Fig. 7C, lane 4). Autophosphorylation was detected when the ratio of Rep52 to PrKX was decreased. In contrast, only minor inhibition of autophosphorylation was detected with MBPRep68Δ at the same concentrations (Fig. 7C, lane 8).
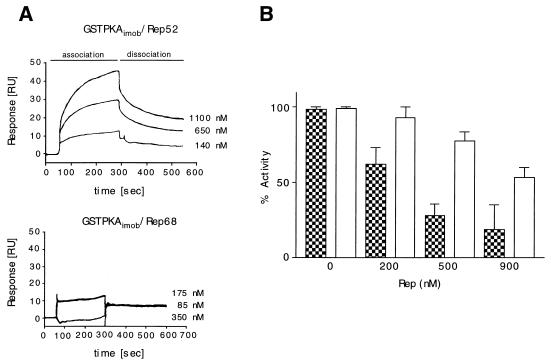
Previous research has demonstrated that the physiological inhibitors of PKA exert their influence by binding as pseudosubstrates within the catalytic core domain. This core catalytic region is conserved between PrKX and PKA. The interaction between MBPRep52 or MBPRep68Δ with PKA was measured by SPR and was found to be similar to that of MBPRep52 or MBPRep68Δ with PrKX, i.e., MBPRep52 bound to PKA with higher affinity than MBP-Rep68Δ (Fig. 8A). GST-PKA was immobilized via an anti-GST antibody, and the association and dissociation rates of MBPRep68Δ and MBPRep52 were monitored at a buffer flow rate of 30 μl/min. MBPRep52 again had a Kd of approximately 320 nM (Table 1). MBPRep68Δ showed only unspecific interaction with PKA, and a Kd value could not be determined at the concentrations used. As in the case of PrKX, the addition of MBPRep52 to a phosphorylation assay mixture containing PKA resulted in inhibition of the PKA kinase activity (Fig. 8B). Only minimal inhibition was observed when MBPRep68Δ was added to the reaction mixture. Kis of approximately 190 and 330 nM (±189 nM) could be calculated for Rep52 with either PrKX or PKA, respectively.
FIG. 8.
Interaction of MBP-Rep52 and MBP-Rep68 with immobilized GST-PKA and inhibition. (A) A total of 500 to 600 RU of GST-PrKX was immobilized via an anti-GST antibody to a CM5 chip. MBP-Rep52 (upper panel) and MBP-Rep68 (lower panel) at the concentrations indicated were injected for 240 s (association phase). After the injection was stopped, the dissociation rate was monitored for 300 s (dissociation phase). (B) GST-PKA (10 nM) was inhibited with Rep52 (checkered bars) and Rep68 (open bars) by using the concentrations indicated. Mean values with standard deviations of several different experiments are shown. The inhibitions were determined by the radioactive kinase assay in duplicate. Three time points for each Rep concentration were assayed.
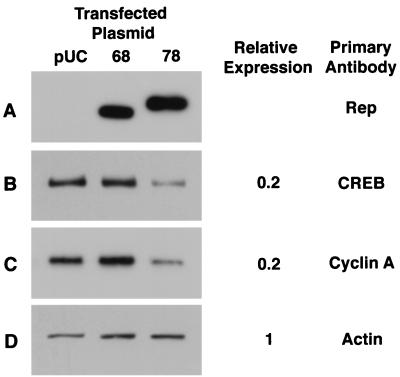
The genetic and biochemical data demonstrate a stable interaction between Rep52 or Rep78 and PrKX and only a weak interaction with Rep68, which lacks the carboxy-terminal sequence of Rep52 and Rep78. In order to examine the inhibitory effects of Rep52 and Rep78 on the expression of genes which are responsive to PKA phosphorylation activity, cos cells were transfected with either Rep78 or Rep68 expression plasmids or pUC19 and whole-cell lysates were prepared. These lysates were analyzed by Western blotting for changes in the expression of cAMP-responsive genes. Western blots probed with Rep-specific antibodies demonstrated expression of Rep68 and Rep78 (Fig. 9A).
FIG. 9.
Effect of Rep68 and Rep78 on cellular protein expression. Whole-cell lysate was prepared from cos cells transfected with either Rep68 or Rep78 expression vector or pUC as a control. Equal loading of protein was measured by Coomassie staining of duplicate gels. Western blots were probed with antibodies to Rep, CREB, or cyclin A (A, B, and C). As an additional control, the blots were also probed with actin (D). Horseradish peroxidase-conjugated secondary antibodies were used and detected by chemiluminescence. Changes in expression were determined by laser densitometry of the films and are reported relative to the level of expression observed in Rep68 or pUC.
The CREB protein is a transcription factor which requires phosphorylation by PKA for activation. Furthermore, it has been demonstrated that expression of this gene is also stimulated by cAMP via cAMP-responsive elements (CRE) in its promoter (34). Western blots probed with a CREB-specific antibody show a 40 to 80% inhibition of expression in Rep78-transfected cells compared to Rep68- or pUC-transfected cells (Fig. 9B). Expression of cyclin A is also induced by cAMP via PKA activation (7). Western blots probed with a cyclin A-specific antibody also showed a 40 to 80% inhibition in expression in Rep78-transfected cell lysates compared to Rep68 or pUC-transfected cells (Fig. 9C). In contrast to CREB and cyclin A expression, the level of actin was unchanged in Rep78-transfected cells (Fig. 9D).
DISCUSSION
Yeast two-hybrid screens have been used to identify a number of kinase-inhibitor interactions such as p21WAF1 and the G1 cyclin-dependent kinase Cdk2 (12). The interaction of Rep52 with PrKX was identified with a yeast two-hybrid screen and confirmed biochemically by coimmunoprecipitations and SPR. The difference in the binding behavior of Rep78-Rep52 and PrKX versus Rep68 and PrKX indicates that an interaction domain may be located in the carboxy terminus that is present in the Rep78 as well as in the Rep52 protein. These data would imply a functional role for the carboxy terminus of Rep78-Rep52, which is spliced out in the Rep68 and Rep40 proteins. To date, this region has not been associated with any functional significance, although it contains a zinc finger motif and a PVS motif similar to those of the adenovirus protein E1a (47). The absence of these residues in Rep68-Rep40 further suggests distinct functions for these proteins compared to Rep78-Rep52. Furthermore, the only activity attributed to the p19-derived polypeptides, Rep52 and Rep40, is that of a DNA helicase (40). The interaction of Rep52 with PrKX demonstrates an additional role for this protein.
Based on sequence homology to known protein kinases, PrKX is a PKA-type kinase. This novel protein kinase encoded on the human X chromosome is expressed in a variety of tissues, with the highest level of expression in fetal and adult brain, kidney, and lung tissue (25). PrKX expressed and purified from cos cells showed kinase activity with a synthetic peptide substrate for PKA, Kemptide, with a specific activity of 0.2 U/mg. The specific activity of PrKX is lower than that of PKA (5 U/mg). Addition of a sevenfold molar excess of Rep52 inhibited 90% of the PrKX activity. Gel filtration chromatography (with a prepacked Superdex 200 3.2/30 column) indicates that 50% of the protein in the MBPRep52 preparation elutes in the void volume of the column (data not shown). This suggests that a significant portion of MBPRep52 may form a high-molecular-weight aggregate which cannot interact with PKA or PrKX. Thus, the estimate of the molar ratio of MBPRep52 to PrKX required for inhibition in these studies may be conservative.
Kinetic analysis of the inhibitory activity of MBPRep52 on PrKX phosphotransferase activity shows an IC50 of 200 nM. SPR determined a Kd of 375 nM, which is in agreement with the IC50. In contrast, only weak interaction with another AAV2-derived protein, Rep68Δ, was observed with minimal inhibition of PrKX kinase activity. Similar results concerning the inhibitory effects of Rep52 versus Rep68 were observed for the autophosphorylation of PrKX.
PKA is also inhibited by Rep52 with a Kd close to the value of PrKX (320 nM), suggesting a similar interaction mechanism. However, the exact mechanism of the Rep-mediated inhibition of PrKX and PKA is unclear. Known inhibitors of PKA, such as the heat-stable protein kinase inhibitor PKI and the type I regulatory subunit (RI), contain pseudosubstrate sites which inhibit PKA activity. However, the type II regulatory subunit (RII) contains a phosphorylation site but still inhibits with subnanomolar binding constants (23). Sequence analysis of the carboxy terminus of Rep identified one region which has homology to the pseudosubstrate consensus sequence (amino acids 526 to 545). However, initial experiments with a peptide of this region failed to inhibit PrKX activity (data not shown). Furthermore, two additional PKA phosphorylation sites were identified at amino acids 71 and 329 in the Rep ORF. While the carboxy terminus appears to be important for inhibition, it is not clear if it is sufficient or requires additional interaction sites present in the region common to all four Rep proteins.
The cAMP signal transduction pathway and the cAMP-dependent PKA have been studied extensively and have been shown to play a central role in cell growth and development (6, 35). While PrKX bears homology to the catalytic core region of PKA (26), the role of this kinase in the cell remains unclear; however, it is capable of activating PKA-sensitive pathways in vivo (unpublished observations). The activity of PKA is regulated by the regulatory subunits RI and RII. In the absence of cAMP, these subunits bind to the catalytic subunit with subnanomolar affinity and inhibit kinase activity (13). However, in the presence of cAMP, the inactive holoenzyme complex dissociates and the free catalytic subunit is then able to enter the nucleus. Nuclear targets include the CREB/ATF family of transcription factors, which induce expression of genes containing CRE, thus affecting transcriptional activity. Since the Rep proteins of AAV are predominantly found in the nucleus (21), inhibition of this signal transduction pathway could explain some of the global effects of Rep on gene expression, namely, the inhibition of gene expression from promoters which do not have identified Rep binding sites and the inhibition of gene expression by the P19 Rep proteins which do not bind DNA.
In addition to the regulatory subunits, PKI has been shown to be a potent inhibitor of PKA (45). Microinjection of labeled PKI indicates a nuclear localization, while injections of the catalytic subunit indicate free diffusion between the cytoplasm and the nucleus (9). In contrast, the regulatory subunits are unable to cross the nuclear membrane and further serve to anchor the C subunit in the cytoplasm or via specific A-kinase anchoring proteins (AKAPs) to specific organelles (10). Furthermore, PKI has been reported to actively transport PKA from the nucleus to the cytoplasm (8).
PrKX has the greatest similarity (62.4% identity in the catalytic core region) to the DC2 kinase of Drosophila melanogaster, which behaves as a PKA catalytic subunit in vitro (33). In contrast, PrKX has only 53.2% identity with the human C subunit of PKA. This degree of homology is much lower than the degree of similarity of the different human PKA catalytic subunit isoforms (90.5% identity), suggesting a distinct role for PrKX. Genetic analysis of Drosophila indicates that deletion of DC2 is not lethal, and animals lacking DC2 appear to develop normally. However, DC2 kinase cannot substitute efficiently for the DC0 kinase, the major Drosophila PKA catalytic subunit. While DC2 was shown to be regulated by the RI subunit and cAMP (33), studies are under way to investigate the mechanism of PrKX regulation (54).
The initial purpose of the yeast two-hybrid screen was to identify proteins which interact with Rep and thus explain some of the in vivo effects resulting from overexpression of the Rep ORF or from AAV infection. Although the mechanism of Rep-mediated inhibition of cell growth has not been established, the direct involvement of Rep in cell cycle regulatory processes may provide the basis for this phenomenon. Recently, infection of primary human fibroblasts with wild-type AAV2 has been shown to inhibit phosphorylation of the Rb family of proteins and to upregulate the kinase inhibitor p21WAF1 (14). Increased intracellular levels of cAMP have also been shown to result in either phosphorylation or dephosphorylation of Rb, depending on the cell type, as well as to affect the levels of p27KIP inhibitor. The net effect is that infection with AAV can inhibit cells from progressing through S phase.
Inhibition of PKA by PKI plays a central role in the induction of mitosis (32), and the expression and intracellular localization of PKI are cell cycle regulated (49). Furthermore, the expression of several proteins involved in cellular regulation, i.e., cyclin A (53), a cell cycle-regulated histone (H4) (11), or proliferating-cell nuclear antigen (PCNA) (20) is regulated via CRE in their promoters and is sensitive to the phosphorylation state of the CREB proteins. While the exact role of PrKX and its response to cAMP is not known, PrKX, as a PKA-type kinase, might have similar or modulatory effects on gene expression and cell growth. Thus, the observed effects of infection with AAV2 or overexpression of Rep proteins on cell growth might be explained by the inhibition of PKA and PrKX by the nonstructural AAV2 proteins Rep52 and Rep78.
ACKNOWLEDGMENTS
We thank Susan S. Taylor for providing the PKA bacterial expression plasmid and Yoon S. Cho-Chung for review of the manuscript.
This work was supported in part by the Deutsche Forschungsgemeinschaft (grant SFB394 [B4] to B.Z. and F.W.H.) and the Bennigsen-Foerder-Preis (F.W.H.).
REFERENCES
- 1.Adams J A, McGlone M L, Gibson R, Taylor S S. Phosphorylation modulates catalytic function and regulation in the cAMP-dependent protein kinase. Biochemistry. 1995;34:2447–2454. doi: 10.1021/bi00008a007. [DOI] [PubMed] [Google Scholar]
- 2.Chejanovsky N, Carter B J. Replication of a human parvovirus nonsense mutant in mammalian cells containing an inducible amber suppressor. Virology. 1989;171:239–247. doi: 10.1016/0042-6822(89)90531-x. [DOI] [PubMed] [Google Scholar]
- 3.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiorini J A, Wendtner C M, Urcelay E, Safer B, Hallek M, Kotin R M. High-efficiency transfer of the T cell co-stimulatory molecule B7-2 to lymphoid cells using high-titer recombinant adeno-associated virus vectors. Hum Gene Ther. 1995;6:1531–1541. doi: 10.1089/hum.1995.6.12-1531. [DOI] [PubMed] [Google Scholar]
- 5.Chiorini J A, Yang L, Safer B, Kotin R M. Determination of adeno-associated virus Rep68 and Rep78 binding sites by random sequence oligonucleotide selection. J Virol. 1995;69:7334–7338. doi: 10.1128/jvi.69.11.7334-7338.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho-Chung Y S, Pepe S, Clair T, Budillon A, Nesterova M. cAMP-dependent protein kinase: role in normal and malignant growth. Crit Rev Oncol-Hematol. 1995;21:33–61. doi: 10.1016/1040-8428(94)00166-9. [DOI] [PubMed] [Google Scholar]
- 7.Desdouets C, Matisic G, Molina C A, Foulkes N S, Sassone-Corsi P, Brechot C, Sobczak-Thepot J. Cell cycle regulation of cyclin A gene expression by the cyclic AMP-responsive transcription factor CREB and CREM. Mol Cell Biol. 1995;15:3301–3309. doi: 10.1128/mcb.15.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fantozzi D A, Harootunian A T, Wen W, Taylor S S, Feramisco J R, Tsien R Y, Meinkoth J L. Thermostable inhibitor of cAMP-dependent protein kinase enhances the rate of export of the kinase catalytic subunit from the nucleus. J Biol Chem. 1994;269:2676–2686. [PubMed] [Google Scholar]
- 9.Fantozzi D A, Taylor S S, Howard P W, Maurer R A, Feramisco J R, Meinkoth J L. Effect of the thermostable protein kinase inhibitor on intracellular localization of the catalytic subunit of cAMP-dependent protein kinase. J Biol Chem. 1992;267:16824–16834. [PubMed] [Google Scholar]
- 10.Faux M C, Scott J D. Molecular glue—kinase anchoring and scaffold proteins. Cell. 1996;85:9–12. doi: 10.1016/s0092-8674(00)81075-2. [DOI] [PubMed] [Google Scholar]
- 11.Guo B, Stein J L, van Wijnen A J, Stein G S. ATF1 and CREB trans-activate a cell cycle regulated histone H4 gene at a distal nuclear matrix associated promoter element. Biochemistry. 1997;36:14447–14455. doi: 10.1021/bi971781s. [DOI] [PubMed] [Google Scholar]
- 11a.Hanks S K, Hunter T. Protein kinases. 6. The eukaryotic protein kinases superfamily—kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 12.Haper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 13.Herberg F W, Taylor S S. Physiological inhibitors of the catalytic subunit of cAMP-dependent protein kinase: effect of MgATP on protein-protein interactions. Biochemistry. 1993;32:14015–14022. doi: 10.1021/bi00213a035. [DOI] [PubMed] [Google Scholar]
- 14.Hermanns J, Schulze A, Durr P, Kleinschmidt J A, Schmidt R, zur Hausen H. Infection of primary cells by adeno-associated virus type 2 results in a modulation of cell cycle-regulating proteins. J Virol. 1997;71:6020–6027. doi: 10.1128/jvi.71.8.6020-6027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermonat P L. The adeno-associated virus Rep78 gene inhibits cellular transformation induced by bovine papillomavirus. Virology. 1989;172:253–261. doi: 10.1016/0042-6822(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 16.Hermonat P L. Inhibition of H-ras expression by the adeno-associated virus Rep78 transformation suppressor gene product. Cancer Res. 1991;51:3373–3377. [PubMed] [Google Scholar]
- 17.Hermonat P L. Inhibition of bovine papillomavirus plasmid DNA replication by adeno-associated virus. Virology. 1992;189:329–333. doi: 10.1016/0042-6822(92)90710-7. [DOI] [PubMed] [Google Scholar]
- 18.Hong G, Ward P, Berns K I. In vitro replication of adeno-associated virus DNA. Proc Natl Acad Sci USA. 1992;89:4673–4677. doi: 10.1073/pnas.89.10.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hörer M, Weger S, Butz K, Hoppe-Seyler F, Geisen C, Kleinschmidt J A. Mutational analysis of adeno-associated virus rep protein-mediated inhibition of heterologous and homologous promoters. J Virol. 1995;69:5485–5496. doi: 10.1128/jvi.69.9.5485-5496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang D, Shipman-Appasamy P M, Orten D J, Hinrichs S H, Prystowsky M B. Promoter activity of the proliferating-cell nuclear antigen gene is associated with inducible CRE-binding proteins in interleukin 2-stimulated T lymphocytes. Mol Cell Biol. 1994;14:4233–4243. doi: 10.1128/mcb.14.6.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter L A, Samulski R J. Colocalization of adeno-associated virus Rep and capsid proteins in the nuclei of infected cells. J Virol. 1992;66:317–324. doi: 10.1128/jvi.66.1.317-324.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Im D-S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 23.Kerlavage A R, Taylor S S. Covalent modification of an adenosine 3′:5′-monophosphate binding site of the regulatory subunit of cAMP-dependent protein kinase II with 8-azidoadenosine 3′:5′-monophosphate. Identification of a single modified tyrosine residue. J Biol Chem. 1980;255:8483–8488. [PubMed] [Google Scholar]
- 24.Khleif S N, Myers T, Carter B J, Trempe J P. Inhibition of cellular transformation by the adeno-associated virus rep gene. Virology. 1991;181:738–741. doi: 10.1016/0042-6822(91)90909-u. [DOI] [PubMed] [Google Scholar]
- 25.Klink A, Schiebel K, Winkelmann M, Rao E, Horsthemke B, Lüdecke H-J, Claussen U, Scherer G, Rappold G. The human protein kinase gene PKX1 on Xp22.3 displays Xp/Xp homology and is a site of chromosomal instability. Hum Mol Genet. 1995;4:869–878. doi: 10.1093/hmg/4.5.869. [DOI] [PubMed] [Google Scholar]
- 26.Knighton D R, Zheng J, Ten Eyck L F, Ashford V A, Xuong N-H, Taylor S S, Sowadski J M. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–413. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 27.Kotin R M, Linden R M, Berns K I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotin R M, Menninger J C, Ward D C, Berns K I. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 29.Kotin R M, Siniscalco M, Samulski R J, Zhu X, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labow M A, Berns K I. The adeno-associated virus rep gene inhibits replication of an adeno-associated virus/simian virus 40 hybrid genome in cos-7 cells. J Virol. 1988;62:1705–1712. doi: 10.1128/jvi.62.5.1705-1712.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labow M A, Graf L H, Jr, Berns K I. Adeno-associated virus gene expression inhibits cellular transformation by heterologous genes. Mol Cell Biol. 1987;7:1320–1325. doi: 10.1128/mcb.7.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamb N J, Cavadore J C, Labbe J C, Maurer R A, Fernandez A. Inhibition of cAMP-dependent protein kinase plays a key role in the induction of mitosis and nuclear envelope breakdown in mammalian cells. EMBO J. 1991;10:1523–1533. doi: 10.1002/j.1460-2075.1991.tb07672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meléndez A, Li W, Kalderon D. Activity, expression and function of a second Drosophila protein kinase A catalytic subunit gene. Genetics. 1995;141:1507–1520. doi: 10.1093/genetics/141.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer T E, Waeber G, Lin J, Beckmann W, Habener J F. The promoter of the gene encoding 3′,5′-cyclic adenosine monophosphate (cAMP) response elements binding protein contains cAMP response elements: evidence for positive autoregulation of gene transcription. Endocrinology. 1993;132:770–780. doi: 10.1210/endo.132.2.8381074. [DOI] [PubMed] [Google Scholar]
- 35.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 36.Ni T-H, Zhou X, McCarty D M, Zolotukhin I, Muzyczka N. In vitro replication of adeno-associated virus DNA. J Virol. 1994;68:1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patwardhan S, Gashler A, Siegel M G, Chang L C, Joseph L J, Shows T B, Le Beau M M, Sukhatme V P. EGR3, a novel member of the Egr family of genes encoding immediate-early transcription factors. Oncogene. 1991;6:917–928. [PubMed] [Google Scholar]
- 38.Smith, R. A., J. A. Chiorini, and R. M. Kotin. Unpublished observations.
- 39.Smith R H, Kotin R M. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol. 1998;72:4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith R H, Kotin R M. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol. 1998;72:4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snyder R O, Im D-S, Ni T, Xiao X, Samulski R J, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snyder R O, Im D S, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor S S, Radzioandzelm E, Hunter T. Protein kinases. How do protein kinases discriminate between serine threonine and typrosine-structural insights from the insulin receptor protein-tyrosine kinase? FASEB J. 1995;9:1255–1266. doi: 10.1096/fasebj.9.13.7557015. [DOI] [PubMed] [Google Scholar]
- 43a.Taylor S S, Radzio-Andzelm E. Cyclic AMP-dependent protein kinase. In: Woodgett J R, editor. Protein kinases. Oxford, United Kingdom: Oxford University Press; 1994. [Google Scholar]
- 44.Urcelay E, Ward P, Wiener S M, Safer B, Kotin R M. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh D A, Ashby C D, Gonzalez C, Calkins D, Fischer E H, Kerbs E G. Purification and characterization of a protein inhibitor of adenosine 3′,5′-monophosphate-dependent protein kinases. J Biol Chem. 1971;246:1977–1987. [PubMed] [Google Scholar]
- 46.Ward P, Berns K I. In vitro rescue of an integrated hybrid adeno-associated virus/simian virus 40 genome. J Mol Biol. 1991;218:791–804. doi: 10.1016/0022-2836(91)90267-a. [DOI] [PubMed] [Google Scholar]
- 47.Webster L C, Ricciardi R P. Trans-dominant mutants of E1a provide genetic evidence that the inc finger of the trans-activating domain binds a transcription factor. Mol Cell Biol. 1991;11:4287–4296. doi: 10.1128/mcb.11.9.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weitzman M D, Kyöstiö S R M, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wen W, Taylor S S, Meinkoth J L. The expression and intracellular distribution of the heat-stable protein kinase inhibitor is cell cycle regulated. J Biol Chem. 1995;270:2041–2046. doi: 10.1074/jbc.270.5.2041. [DOI] [PubMed] [Google Scholar]
- 50.Wonderling R S, Kyöstiö S R M, Owens R A. A maltose-binding protein/adeno-associated virus rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wonderling R S, Kyostio S R M, Walker S L, Owens R A. The rep68 protein of adeno-associated virus type 2 increases RNA levels from the human cytomegalovirus major immediate early promoter. Virology. 1997;236:167–176. doi: 10.1006/viro.1997.8724. [DOI] [PubMed] [Google Scholar]
- 52.Yang Q, Chen F, Trempe J P. Characterization of cell lines that inducibly express the adeno-associated virus Rep proteins. J Virol. 1994;68:4847–4856. doi: 10.1128/jvi.68.8.4847-4856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshizumi M, Wang H, Hsieh C M, Sibinga N E, Perrella M A, Lee M E. Down-regulation of the cyclin A promoter by transforming growth factor-beta 1 is associated with a reduction in phosphorylated activating transcription factor-1 and cyclic AMP-responsive element-binding protein. J Biol Chem. 1997;272:22259–22264. doi: 10.1074/jbc.272.35.22259. [DOI] [PubMed] [Google Scholar]
- 54.Zimmermann, B., J. A. Chiorini, R. M. Kotin, and F. Herberg. Unpublished data.