Abstract
The interaction of the Fip1 subunit of polyadenylation factor I with the Saccharomyces cerevisiae poly(A) polymerase (PAP) was assayed in vivo by two-hybrid analysis and was found to involve two separate regions on PAP, located at opposite ends of the protein sequence. In vitro, Fip1 blocks access of the RNA primer to an RNA binding site (RBS) that overlaps the Fip1 carboxy-terminal interaction region and, in doing so, shifts PAP to a distributive mode of action. Partial truncation of this RBS has the same effect, indicating that this site is required for processivity. A comparison of the utilization of ribo- and deoxyribonucleotides as substrates indicates the existence on PAP of a second RBS which recognizes the last three nucleotides at the 3′ end of the primer. This site discriminates against deoxyribonucleotides at the 3′ end, and interactions at this site are not affected by Fip1. Further analysis revealed that the specificity of PAP for adenosine is not simply a function of the ATP binding site but also reflects interactions with bases at the 3′ end of the primer and at another contact site 14 nucleotides upstream of the 3′ end. These results suggest that the unique specificity of PAP for ribose and base, and thus the extent and type of activity with different substrates, depends on interactions at multiple nucleotide binding sites.
Cleavage and polyadenylation of the 3′ end of eukaryotic precursor mRNA is a modification essential for proper mRNA utilization. The primary function of poly(A) polymerase (PAP) in this processing reaction is to add poly(A) tails to the cleaved precursor (for a review, see reference 34). With the help of specificity factors, PAP is directed to the appropriate substrate, exhibits increased processivity, and terminates poly(A) synthesis at the correct tail length. The factors conferring these activities to PAP are cleavage/polyadenylation specificity factor (CPSF) and poly(A) binding protein II (PAB II) in mammalian cells (34) and cleavage/polyadenylation factor I (CF I), polyadenylation factor I (PF I), and Pab1 in the yeast Saccharomyces cerevisiae (1, 4, 16, 23, 25). Direct contacts between the mammalian PAP and the p160 subunit of CPSF (24) and between yeast PAP and the Fip1 subunit of PF I (26) have been demonstrated. While specificity factors are the primary modulators of PAP activity, other regulatory mechanisms have also been documented. For example, the activity of mammalian PAP is inhibited by direct interaction with the U1A protein, as a feedback mechanism controlling the polyadenylation of mRNAs encoding U1A (9) or with the U1 70K protein as part of the U1 snRNP (8). Moreover, PAP can in turn modulate the activity of cleavage factors. The mammalian enzyme stimulates the cleavage step of the reaction (34), and a temperature-sensitive mutation in the yeast PAP can affect the choice of cleavage site in vivo (20). The biochemical mechanisms underlying these regulatory events are not understood.
While PAP is most appropriately considered a catalytic subunit of the cleavage-polyadenylation machinery, many of its enzymatic properties can be studied independently of its association with these factors. PAP does not require a nucleic acid template, a property shared with terminal deoxynucleotidyltransferase and the CCA-adding tRNA nucleotidyltransferases. Sequence comparisons (11, 21) suggest that PAP has an organization of motifs similar to these and other enzymes in the nucleotidyltransferase superfamily. Further experimental work on bovine PAP showed that three conserved aspartates within the proposed catalytic domain are necessary for catalysis (21). By analogy with better-characterized members of the nucleotidyltransferase family, the catalytic site in PAP is thought to position and activate the 3′ OH of the RNA primer to attack the α,β phosphate bond of ATP. The observation that poly(A) synthesis proceeds by a nucleophilic substitution (an SN2 in-line mechanism) without a covalent intermediate (35) is consistent with such a model.
The locations on PAP for binding of ATP and the 3′end of RNA are not known. Essential carboxyl-terminal RNA binding sites (C-RBS) in yeast and bovine PAPs have been identified (21, 36). The C-RBS of the yeast PAP does not interact with the 3′ end of the RNA (36), and its role in enzyme function has not been defined. It is thought to interact in part with the phosphate backbone of polynucleotides (36). The proposed interactions at the catalytic site of PAP and the C-RBS do not account for the preference of PAP for RNA as a primer and adenosine-containing ribonucleotides as the nucleoside triphosphate (NTP) substrate, and it is clear that our understanding of the mechanism of action of PAP, alone and when complexed with specificity factors, is not complete.
Previous reports have made conclusions regarding the specificity of PAP based on assays measuring the incorporation of radioactivity into acid-precipitable products. By examining the products of such reactions by gel electrophoresis, we have been able to more clearly dissect the properties of PAP which contribute to its substrate specificity and to determine the consequences of various protein-protein interactions on PAP’s activity. We have found that the overlap of the C-RBS with a PAP-Fip1 interaction site allows the Fip1 subunit of PF I (26) to regulate the processivity of PAP. A second site on PAP, which is not affected by Fip1, recognizes the base and ribose natures of the last three nucleotides of the primer, and a third site contacts the RNA approximately 14 nucleotides upstream of the 3′ end. Based on these results, we discuss a model for the functional organization of the S. cerevisiae poly(A) polymerase.
MATERIALS AND METHODS
Nucleic acids.
Radioactive deoxynucleoside triphosphates (dNTPs) and NTPs were from ICN and DuPont NEN. ATP, UTP, CTP, GTP, and 2′-dATP were from Promega, and 3′-dATP was obtained from Sigma. Oligo(A)2, oligo(A)3, oligo(A)4, oligo(A)12, and oligo(A)26 were from Oligos Etc. A 24-nucleotide single-stranded DNA (ATGAGCTCAGCAGCAGAAAATAAA) was used as a DNA primer. For UV cross-linking studies, randomly labeled GAL7-9 RNA was used as described previously (36).
For two-hybrid screening and assays, the S. cerevisiae PAP1 coding sequence was fused in frame to the Gal4 binding domain (GBD) in two orientations. For the first orientation, the GBD was fused to the carboxyl terminus of PAP. Using the 5′ primer GAAGATCTATGAAGCTACTGTCTTCT and the 3′ primer GAAGATCTCGATACAGTCAACTGTCT, the GBD was amplified by PCR, digested with BglII, and inserted into the BglII site of a HindIII-HindIII genomic fragment of PAP1 (18). The construct was then inserted into the HindIII site of Yeplac112 (7), yielding the plasmid Yeplac112/wtPAP-GBD. For construction of Δ14PAP-GBD, the SacI-BsmI fragment of wtPAP-GBD was replaced with the corresponding fragment from plasmid pJΔ14PAP (36). For the second orientation, the GBD was fused to the amino terminus of Pap1. In this case, the SacI-XcmI fragment of pGAD-F327 (26) was replaced with a SacI-AseI (blunted) fragment of PAP1. GBD-Δ6PAP, GBD-Δ8PAP, GBD-Δ9PAP, and GBD-Δ10PAP were constructed by truncation or replacement of the wild-type PAP coding sequence with sequences as described for the corresponding pJΔ6PAP, pJΔ8PAP, pJΔ9PAP, and pJΔ10PAP expression plasmids (36). pGAD-FIP(20–327) was obtained by two-hybrid screening (3) of the library prepared by James et al. (12), using wtPAP-GBD as a bait and Δ14PAP-GBD as a control. This fusion lacks the first 19 of the 327 amino acids of Fip1.
Two-hybrid assay.
S. cerevisiae EGY40 (ura3-1 his3-11 trp1-1 leu2-3,112), a gift from R. Brent, was used for two-hybrid screening and analysis of PAP-Fip1 interactions in vivo. The interactions of pGAD-FIP(20–327) with wtPAP-GBD, Δ14PAP-GBD, GBD-wtPAP, and GBD-Δ6PAP, -Δ8PAP, -Δ9PAP, and -Δ10PAP were tested by two-hybrid analysis (3), using the reporter plasmid pBS-GAL (a gift from E. Androphy, Tufts University). Interactions were quantitated by β-galactosidase assays using permeabilized cells (13).
Recombinant proteins.
Wild-type PAP, Δ14PAP, and Δ9PAP were expressed and purified as described previously (36). The Fip1-His6 fusion was expressed from the pFL11 plasmid, a gift of W. Keller (26), using the expression conditions and crude extract preparation described for PAP (36), except that the composition of the lysis buffer was 10 mM Tris HCl (pH 8), 500 mM NaCl, 10% glycerol, 20 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 0.6 μM leupeptin, 2 μM pepstatin A, and 0.5% Nonidet P-40. This His6-tagged Fip1 was then purified by loading the supernatant onto a nickel-charged 2.5-ml iminodiacetic acid column (Sigma) which had been equilibrated with starting buffer (lysis buffer without Nonidet P-40). Bound protein was eluted by applying consecutive step elutions of five column volumes of starting buffer containing 5, 20, 60, 125, 250, and 400 mM imidazole. Fractions containing Fip1 (60 mM imidazole) were pooled, dialyzed twice, for 3 h each, against 1 liter of 10 mM Tris-HCl (pH 8)–100 mM KCl–10% glycerol–1 mM phenylmethylsulfonyl fluoride–0.5 mM dithiothreitol, and loaded onto an equilibrated 15-ml DEAE-Sephacel column (Pharmacia). The column was developed with a 100 to 500 mM KCl gradient, and fractions containing Fip1 (150 mM KCl) were combined. Attempts to purify Fip1 further by means of a 1-ml HiTrap heparin column (Pharmacia) and a 1-ml Mono S column (Pharmacia) were unsuccessful because Fip1 and contaminating proteins did not bind to these matrices. The Fip1-containing flowthrough from the Mono S column was applied to a 1-ml Mono Q column (Pharmacia) equilibrated with a buffer containing 50 mM HEPES-KOH (pH 7.5), 100 mM KCl, 10% glycerol, 0.25 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 0.5 mM dithiothreitol and eluted with a 100 to 500 mM KCl gradient. Fip1 eluted at a KCl concentration of approximately 200 mM. The S. cerevisiae poly(A) binding protein (Pab1) was a kind gift of Allan Jacobson.
PAP assays.
Polymerization reactions were carried out as described previously (36) in 10 μl of a solution containing 20 mM HEPES-KOH (pH 7.5), 10% glycerol, 1 mM MnCl2, 50 mM KCl, 0.25 mM EDTA, 0.5 mM dithiothreitol, 0.5 mg of bovine serum albumin per ml, 250 μM NTP, 1 μCi of [32P]NTP, 1 μM primer, and 20 to 250 ng of PAP (32 to 400 nM) at 30°C for the various periods of time. The various substrates, proteins, and PAP enzymes used in the different assays are indicated in the figure legends. Reaction products were analyzed either by Cherenkov counting of acid-precipitable counts as described previously (36) or by fractionation on 18% polyacrylamide–8.3 M urea gels and visualization by autoradiography, using a PhosphorImager (Molecular Dynamics). Cross-linking of PAP to RNA with UV light and analysis of products were performed as described previously (36), using 200 ng of PAP per 10-μl reaction volume.
RESULTS
Two PAP regions mediate interaction with Fip1.
In eukaryotic cells, PAP functions as a complex with other proteins to specifically polyadenylate cleaved mRNA precursor. This type of reaction, when recreated in vitro with purified and/or partially purified factors, is referred to as the specific activity of PAP. The ability of PAP to extend any RNA primer in the absence of other proteins is known as its nonspecific activity. In our previous analysis of PAP truncations (36), we discovered that the N-terminal 18 amino acids of yeast PAP are essential for its specific activity in vitro and in vivo yet are entirely dispensable for nonspecific poly(A) addition. We have called this 18-amino-acid region specificity domain 1 (SpD1) (see Fig. 1A), reflecting its probable role in interactions with other components of the polyadenylation machinery. Preker et al. (26) have shown that Fip1, a component of PF I, interacts directly and specifically with yeast PAP. However, the Fip1 binding sites on PAP and the consequences of its interaction for PAP activity have not been studied.
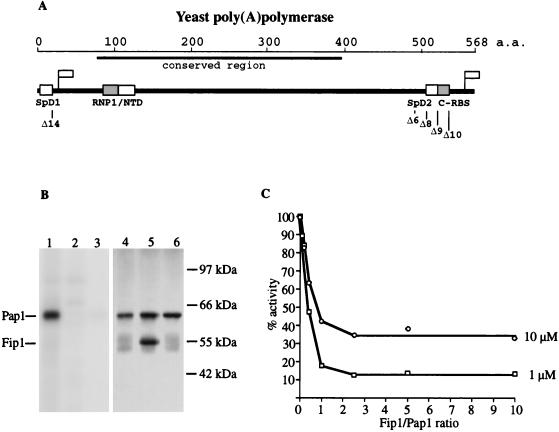
FIG. 1.
(A) Schematic diagram of yeast poly(A) polymerase showing functional domains and homologies discussed in the text. RNP1, homology to ribonucleoprotein motif 1 of the RRM-type RNA binding sites (27); NTD, nucleotidyltransferase domain. Flags indicate the positions of epitopes recognized by the monoclonal antibodies specific for the amino or carboxyl terminus of yeast PAP. a.a., amino acids. (B) UV-induced cross-linking of radioactive RNA to PAP. Samples were separated on an SDS–10% polyacrylamide gel. The same gel was scanned with a PhosphorImager to detect radioactive protein (lanes 1 to 3) and stained with silver to visualize the protein bands (lanes 4 to 6). Lanes 1 and 4, PAP alone; lanes 2 and 5, PAP plus Fip1 at a PAP/Fip1 molar ratio of 1:2; lanes 3 and 6, PAP plus 14 μg of poly(A). (C) Fip1 inhibits poly(A) addition as measured by the incorporation of acid-precipitable counts into oligo(A) primer. Increasing amounts of Fip1 were added to standard poly(A) addition reaction mixtures containing either 1 or 10 μM oligo(A)12 and 20 ng of PAP, and the mixtures were incubated for 15 min at 30°C.
To determine domains in PAP necessary for its interaction with Fip1, we employed yeast two-hybrid analysis (3). The GBD was fused in frame with the PAP coding sequence at a site 20 amino acids up from the carboxyl end of PAP. We had previously shown that the last 20 amino acids of PAP are not necessary for any of its activities (36). This construct (wtPAP-GBD) was tested for interaction with an amino-terminal fusion of the Gal4 activation domain (GAD) to Fip1 sequence GAD-FIP(20–327), using a Gal4-dependent β-galactosidase reporter gene. The wtPAP-GBD fusion yielded a high level of β-galactosidase activity when paired with GAD-FIP(20–327) (Table 1). With a construct lacking the amino-terminal specificity domain (Δ14PAP-GBD) (Table 1), β-galactosidase activity was reduced over 50-fold, implicating this region in Fip1 interaction.
TABLE 1.
Interaction of different PAP constructs with FIP(20–327) as determined by β-galactosidase activity via two-hybrid analysis
| PAP construct | No. of amino acids deleted | β-galactosidase activity (U)a |
|---|---|---|
| C-terminal truncations | ||
| GBD-wtPAP | 0 | 107 |
| GBD-Δ10PAP | 31 | 149 |
| GBD-Δ9PAP | 43 | 194 |
| GBD-Δ8PAP | 55 | 1.9 |
| GBD-Δ6PAP | 67 | 1.1 |
| N-terminal truncations | ||
| wtPAP-GBD | 0 | 93 |
| Δ14PAP-GBD | 18 | 1.5 |
Expressed as average units per minute per number of cells from two independent assays.
A construct in which the GBD was fused in frame to the amino terminus of the PAP (GBD-wtPAP) was also able to strongly activate the β-galactosidase reporter gene (Table 1). Interestingly, only the wtPAP-GBD fusion, and not GBD-wtPAP, could rescue a lethal disruption of the chromosomal PAP1 gene (data not shown). Since we have previously shown that the SpD1 domain at the amino terminus of PAP is essential for cell viability (36), the failure of GBD-wtPAP to rescue a PAP1 knockout is likely to be the result of a steric constraint imposed by the GBD, rendering SpD1 inaccessible to interaction with specificity factors. Nevertheless, our two-hybrid results, as well as data from the original screen which identified Fip1 (26), showed that GBD-wtPAP is still capable of Fip1 interaction (Table 1) and indicated the presence of an interaction domain separate from SpD1. To map this second domain, we performed additional tests, using truncations of PAP from the C terminus of the GBD-PAP fusion, and found that a deletion of 55 amino acids (GBD-Δ8PAP) led to a 50-fold drop in β-galactosidase activity (Table 1). Interestingly, truncations of 31 and 43 amino acids from the carboxyl end of PAP (GBD-Δ10PAP and GBD-Δ9PAP, respectively) led to an increase in β-galactosidase activity in comparison to that of the wild-type PAP construct (Table 1). This activation implies that the carboxyl Fip1 interaction domain initially becomes more accessible in these constructs before being affected by the truncation of amino acids 44 through 55. The presence of the GBD at the carboxyl terminus of PAP in the wtPAP-GBD fusion may exert an even greater inhibitory effect, explaining the negative result with Δ14PAP-GBD. We have designated the interaction site defined by the Δ8 deletion as SpD2 (Fig. 1A). SpD2 has no homology to SpD1, implying that the two PAP regions interact with Fip1 in different ways. Because the two-hybrid analysis is conducted in vivo, either interaction with Fip1 could be direct or mediated through other cellular factors.
We had previously demonstrated that the region removed in the Δ8 and Δ9 mutants is important for interaction with the RNA primer (36). This study showed that the majority of PAP-RNA cross-links introduced by UV light occur at this C-RBS (Fig. 1A). Since our two-hybrid results indicate that the second Fip1 interaction region partly overlaps the C-RBS, we decided to test whether purified recombinant Fip1 could affect the ability of purified PAP to cross-link to RNA. PAP was incubated with uniformly labeled RNA in the presence or absence of Fip1 and exposed to UV light. The samples were then treated with RNase and resolved on an SDS-polyacrylamide gel, and PAP was detected by silver staining and autoradiography, as described by Zhelkovsky et al. (36). Cross-linking of RNA to PAP is eliminated by the presence of a twofold molar excess of Fip1 (Fig. 1B; compare lanes 1 and 2), suggesting that Fip1 is blocking the interaction of C-RBS with RNA. A similar decrease in the amount of PAP cross-linked to radioactive RNA is found when an excess of unlabeled poly(A) is included in the reaction (Fig. 1B, lane 3).
The processivity of PAP depends on interactions at the C-RBS.
We also examined the effect of Fip1 on the nonspecific activity of PAP. The addition of increasing amounts of recombinant Fip1 progressively inhibited the activity of PAP, as determined by incorporation of [α-32P]AMP into oligo(A)12 primer (Fig. 1C). The inhibition was close to maximal at an Fip1/PAP molar ratio of 1:1, and further inhibition was not observed at ratios beyond 2.5:1. Raising the concentration of RNA primer 10-fold partially rescued the PAP activity, indicating that the mechanism of Fip1 inhibition is competitive in nature. Further kinetic analysis indicated that at a Fip1/PAP molar ratio of 2:1 the Km for RNA increased to 10 μM, which is 20-fold higher than that for PAP alone. These observations are consistent with the Fip1-mediated inhibition of UV cross-linking but are surprising given the report of Preker et al. (25) that nonspecific activity of PAP is stimulated by association with the Fip1-containing PF I factor.
To gain additional insight into the mechanism of Fip1-induced inhibition, we examined the RNA products of poly(A) addition reactions carried out in the presence of a twofold molar excess of Fip1. This analysis showed a striking difference in the products formed by PAP in the reactions containing Fip1 compared to those containing only PAP. In the absence of Fip1, PAP rapidly and processively elongated the primer after an initial oligoadenylation (Fig. 2A). In the presence of Fip1, the poly(A) tails were substantially shorter. Moreover, the enzyme functioned in a more distributive mode, as evidenced by the gradual increase in length of these short products over time. Δ14PAP, which lacks SpD1, displayed the same distribution of products as wild-type PAP, both in the absence and presence of Fip1 (Fig. 2B). Examination of the reaction products generated by Δ9PAP, which lacks part of the C-RBS, revealed a product distribution similar to that of wild-type PAP in the presence of Fip1 (Fig. 2C). Addition of Fip1 to reactions containing Δ9PAP further slowed polymerization of the poly(A) tails, consistent with the presence of SpD2 on this mutant enzyme.
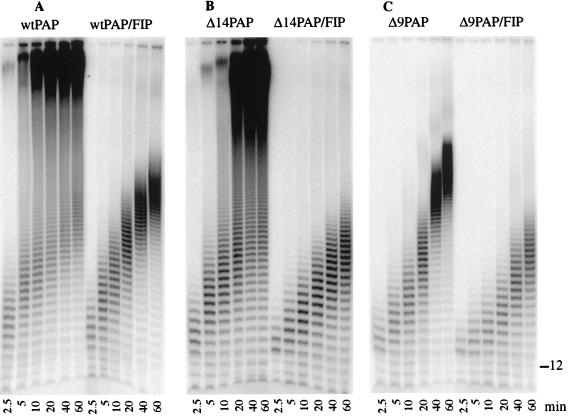
FIG. 2.
Kinetic analysis of the effect of Fip1 on PAP activity. Twenty-five nanograms of wild-type (wt) or mutant PAP was incubated under standard conditions with oligo(A)12 primer, ATP, and [α-32P]ATP. For these reactions, the molar ratio of oligo(A)12 primer to PAP was 30:1. Samples were removed from the reaction mixture after durations of 2.5, 5, 10, 20, 40, and 60 min. Products were resolved on an 18% acrylamide–8.3 M urea gel and visualized by PhosphorImager scanning. (A) Wild-type PAP ± 28 ng of Fip1 (1:2 molar ratio). (B) Δ14PAP ± 28 ng of Fip1. (C) Δ9PAP ± 28 ng of Fip1. The position of the unmodified oligo(A)12 primer is indicated on the right.
These results indicate that the inhibitory effect of Fip1 in vitro is propagated through the C-terminal Fip1 interaction domain (SpD2) rather than the N-terminal site (SpD1). We have previously shown that the Δ9PAP deletion infringes on the C-RBS, resulting in a 50-fold higher Km for RNA and a decreased ability to cross-link to the RNA substrate (36). Our results presented here implicate the C-RBS in enzyme processivity and suggest that one consequence of the interaction of Fip1 with SpD2 may be to limit the interaction of Pap1 with the primer, resulting in a shift to a distributive mode of action.
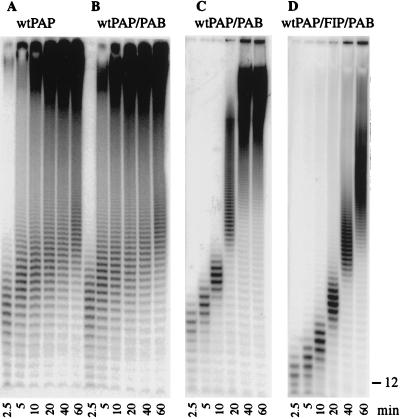
The S. cerevisiae poly(A) binding protein, Pab1 (29), is another protein known to affect the activity of yeast PAP. It is responsible for limiting the length of the poly(A) tails in the specific polyadenylation reaction (1, 16, 23), and it inhibits nonspecific poly(A) addition (19), as measured by incorporation of radioactivity into acid-precipitable counts. When Pab1 was added to poly(A) addition reactions at a PAP/Pab1 ratio of 1:2, there was no effect on polymerization (Fig. 3A and B). In the absence of Pab1 or at a low concentration of Pab1, very long poly(A) species rapidly appeared as products of the reaction (Fig. 3A and B). In contrast, when the amount of Pab1 was increased 20-fold to a molar concentration 1.5 times that of the RNA primer, PAP acted in a distributive mode, elongating the primer slowly and uniformly, without a jump from short to very long products (Fig. 3C, as seen up to the 10-min time point). However, processive poly(A) addition resumed as the amount of de novo-synthesized poly(A) increased and Pab1 became limiting (Fig. 3C, 20-min time point). The inclusion of Fip1 as well as Pab1 in the reaction further slowed the enzyme (Fig. 3D), as expected from the analysis of Fip1-PAP interactions described above.
FIG. 3.
The effect of Pab1 on PAP activity. The assays were performed as described in the legend to Fig. 2. (A) Control reaction with wild-type (wt) PAP only. (B) Wild-type PAP ± 48 ng of Pab1 (1:2 molar ratio). (C) Wild-type PAP ± 960 ng of Pab1 (Pab1/RNA ratio of 1.5:1). (D) Wild-type PAP ± 28 ng of Fip1 and 960 ng of Pab1. The position of the unmodified oligo(A)12 primer is indicated on the right.
Fip1 does not inhibit the initial steps of poly(A) addition.
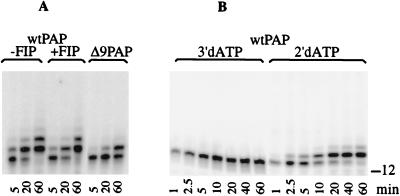
2′-dATP can be utilized by PAP and has been shown to be an effective and economical means of 3′-end labeling RNA (15, 22). Because the primer is extended by only a few 2′-dAMPs, incorporation of this nucleotide into the ends of the oligo(A)12 primer can also serve as an assay to examine factors affecting the initial steps of the poly(A) addition reaction. We tested whether the addition of Fip1 at a concentration that inhibits the processivity of PAP in nonspecific poly(A) addition had any effect on this reaction. Figure 4A shows that the efficiency of 2′-dAMP addition is the same regardless of the presence of Fip1. Furthermore, Δ9PAP, with a partial truncation of the C-RBS, also works well in the addition of 2′-dAMP (Fig. 4A). This result demonstrates that the catalytic function of PAP is not altered when it is complexed with Fip1 and suggests that the PAP-RNA interactions at the C-RBS are separate from interactions involved in positioning the 3′ end of the primer in the active site.
FIG. 4.
Utilization of dATP substrates by PAP. Reaction mixtures contained unlabeled oligo(A)12 primer and the indicated PAPs and radioactive dATP substrates. Reaction conditions were as described in the legend to Fig. 2. The products were separated on an 18% polyacrylamide–8.3 M urea gel and visualized by PhosphorImager scanning. (A) Time course of dAMP incorporation, using radioactive 2′-dATP, oligo(A)12 primer, and either wild-type (wt) PAP, wild-type PAP plus 28 ng of Fip1, or Δ9PAP. (B) Time course of dAMP incorporation, using wild-type PAP and radioactive 3′-dATP or 2′-dATP. The position of the unmodified oligo(A)12 primer is indicated on the right.
A second RBS on PAP recognizes the last three nucleotides of the primer.
To gain a better understanding of the mechanism of action of PAP, we further analyzed the kinetics of incorporation of dAMP into the RNA primer. 3′-dATP is a substrate for PAP but terminates polyadenylation because of the lack of an acceptor hydroxyl group (17). The chain-terminating ability of 3′-dATP is evident from the rapid appearance and accumulation of a single band, representing the incorporation of radioactive 3′-dAMP into the oligo(A)12 primer (Fig. 4B). From this time course, it is clear that 3′-dATP is an efficient substrate for PAP, and after 10 min of incubation, there was no further increase in incorporation of label into the end of the primer, indicating that all of the primer had been modified. When PAP is presented with radioactive 2′-dATP instead of 3′-dATP (Fig. 4B), the initial rate of incorporation of 2′-dAMP is similar to that of 3′-dATP. However, unlike 3′-dATP, incorporation of the first 2′-dAMP into the RNA primer does not block the reaction. The resulting hybrid primer can accept a second dAMP residue, but at a rate much lower than that for the incorporation of the initial 2′-dAMP. The accumulation of product with a third 2′-dAMP is even slower, and a very slow distributive elongation beyond this point is observed only with a 10-fold-higher concentration of PAP (see Fig. 7B). These results indicate that PAP uses 2′-dATP and 3′-dATP as its substrates equally well. However, PAP is highly sensitive to the lack of 2′ OH groups at the end of the RNA primer, and even though such a primer is a catalytically competent substrate, PAP does not extend it very efficiently.
FIG. 7.
Utilization of various NTPs as substrates. (A and D) Reactions conducted with the indicated radioactive NTPs, 30 nM PAP, and oligo(A)12 primer. (B and C) Reaction mixtures contained 300 nM PAP and either oligo(A)12 (B) or oligo(A)26 (C). The products were separated on an 18% polyacrylamide–8.3 M urea gel and visualized by PhosphorImager scanning. The positions of the unmodified oligo(A)12 and oligo(A)26 primers are indicated on the right.
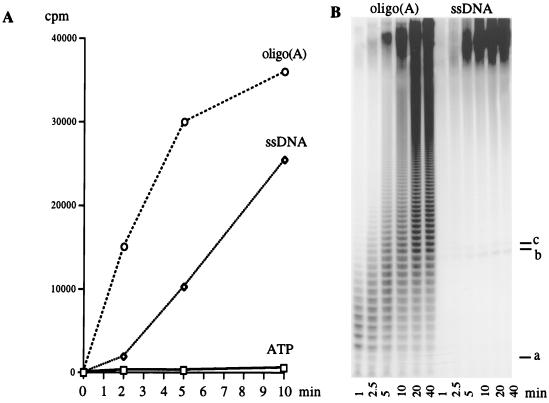
Since we had previously shown that single-stranded DNA could effectively compete for binding at the C-RBS of PAP (36), we decided to examine the kinetics of poly(A) addition with a 24-base DNA oligonucleotide as a primer. The last three bases of the DNA primer were deoxyadenylates (see Materials and Methods). With an oligo(A)12 riboprimer, incorporation of radioactivity into acid-precipitable material was evident at the earliest time point. In contrast, with DNA as the primer, a lag was observed in the initial phase of the reaction (Fig. 5A). Analysis of the reaction products by denaturing acrylamide gel electrophoresis provided an explanation for this lag. With an RNA primer, short heterogeneous poly(A) products are rapidly converted into very long poly(A) molecules (Fig. 5B). In contrast, with a DNA primer, the only detectable products are very long poly(A) species and a low level of primer which has been elongated by one or two AMPs. The accumulation of these short products (b and c in Fig. 5B) suggests that the addition of the first three AMP residues to the DNA primer is rate limiting. The marked increase in processivity following this short extension indicated that the ribonucleotide-extended DNA is highly preferred as a substrate, even with an excess of unextended DNA. These results, presented in Fig. 4 and 5, demonstrate that much of the specificity of PAP for ribose-containing substrates comes from interactions at the 3′-end RBS. Once a few dAMPs are added to an RNA primer, PAP treats the primer as DNA and does not extend it further. In contrast, once a few AMP residues are added to a DNA primer, it becomes as efficient a substrate as RNA.
FIG. 5.
Kinetic analysis of polyadenylation reactions using oligo(A)12 or single-strand (ss) DNA primers. Standard reactions, using the conditions described in the legend to Fig. 2, were conducted for the indicated periods of time, stopped by acid precipitation, and quantitated by scintillation counting (A) or directly loaded onto an 18% polyacrylamide–8.3 M urea gel and visualized by autoradiography (B). a, position of oligo(A)12 primer; b and c, positions of the first and second intermediate products in the ssDNA priming reaction, respectively. A control reaction, containing ATP without a nucleic acid primer, is included in panel A.
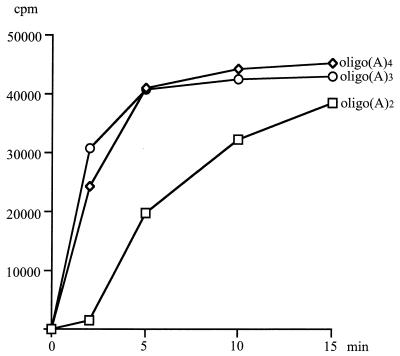
One implication of these findings is that very small RNAs could serve as primers. Early studies of bovine PAP showed that trinucleotides are sufficient for priming (14, 32). This is also the case for yeast PAP (Fig. 6). Oligo(A) trimers and tetramers worked very efficiently as primers, while a dimer exhibited a lag before the rate of polymerization reached that seen with the longer primers. Therefore, a trinucleotide is the minimum primer length needed for proper positioning at the 3′ RBS. In summary, these results show that a site on PAP specifically recognizes the ribose nature of the polynucleotide primer through interactions with the three 3′-terminal nucleotides.
FIG. 6.
Kinetic analysis of polyadenylation, using oligo(A)2, oligo(A)3, or oligo(A)4 as the primer. Reactions were conducted as described in Materials and Methods, using 50 ng of PAP for the indicated periods of time, and then stopped by acid precipitation and quantitated by scintillation counting.
Nonspecific nucleotidyl transfer reveals another level of substrate specificity and points to the presence of an additional RBS.
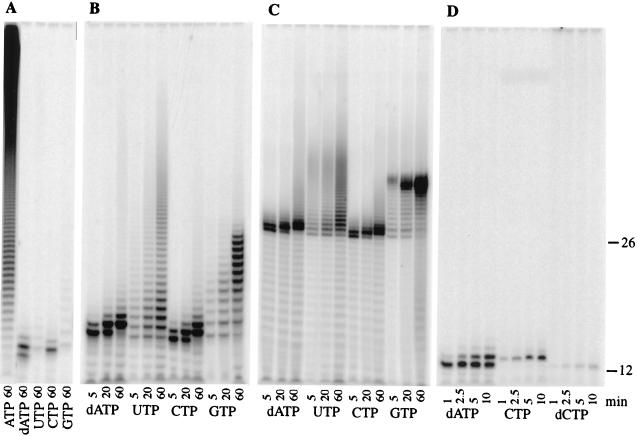
It has been reported that eukaryotic PAPs have a high specificity for ATP as a substrate and little affinity for other NTPs (10, 32). This conclusion, based on assays using incorporation of acid-precipitable counts into primer, has led to the general assumption that the preference for adenosine is a function of the ATP binding site of the enzyme. However, analysis of the products derived from reactions using other NTPs as substrates suggests that this interpretation may not give a complete picture of how PAP selects its substrates. We performed polyadenylation reactions using [α-32P]-labeled NTPs under our standard conditions of 30 nM PAP, 0.5 mM NTP, and 1 μM oligo(A)12 primer and analyzed the products by gel electrophoresis (Fig. 7A). As expected, no phase of the reaction is limited with ATP, and primer is rapidly polyadenylated. In agreement with previously reported decreases in incorporation of radioactivity with other NTPs, the amounts of radioactive product in reactions containing GTP, UTP, CTP, or 2’-dATP were much smaller than that found for ATP. However, the distribution of products varied depending on the NTP used.
This variation was even more obvious if the assays were performed with oligo(A)12 or oligo(A)26 primer and a 10-fold higher concentration of PAP (300 nM). For example, the pattern seen with CTP was surprisingly similar to that seen with 2′-dATP, regardless of the primer used (Fig. 7B and C). Incorporation of the first cytidylate residue was rapid, but incorporation stalled after the addition of three nucleotides, indicating that a primer with a cytidylate residue at the 3′ end is not a favored substrate for PAP. Additional experiments showed that incorporation of the first CMP into the primer was more efficient than that of 2′-dCMP and that incorporation of the first 2′-dCMP was much less efficient than that of 2′-dAMP (Fig. 7D). These results show that discrimination at the 3′-end binding site is not based exclusively on the ribose nature of the last nucleotides of the primer but can, as in the case of CMP incorporation, include the base. Furthermore, they demonstrate that the ATP binding site, while being primarily involved in base discrimination, also contributes to the distinction between NTP and dNTP substrates.
Previous studies showed that at a high concentration of enzyme, yeast and vaccinia virus PAPs could incorporate GMP into the ends of a primer but that such incorporation abruptly stalled after addition of 14 nucleotides (22, 31). We reproduced this assay using an oligo(A)12 or oligo(A)26 primer. With an oligo(A)12 primer and GTP as substrates, products with 14 or fewer guanidylate residues accumulated (Fig. 7B). The elongation of the 26-mer was a more efficient reaction but also halted after the addition of 14 nucleotides (Fig. 7C). In contrast, the incorporation of uridylate continued without restriction to longer lengths on both primers, again at a higher efficiency with the 26-mer (Fig. 7B and C). These results imply the existence of base-specific RNA contacts with PAP which are different from those at the 3′-end binding site. This interaction, by discriminating against poly(G), provides an additional level of base-mediated, primer-specific recognition. Poly(G) addition by PAP onto the oligo(A)26 primer was less processive and less efficient in the presence of Fip1 but still halted after the addition of 14 GMPs (data not shown), indicating that the interaction between this site on PAP and the RNA is not obstructed by Fip1 binding. Similar data were obtained with the Δ9PAP mutant, suggesting that this part of the C-RBS is not involved in the poly(G) interaction (data not shown).
DISCUSSION
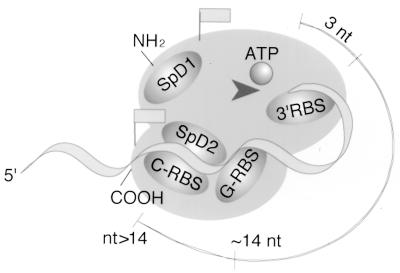
Based on the data described in this report, we propose a model for the functional organization of S. cerevisiae PAP that is shown schematically in Fig. 8. We have found that PAP operates in different modes, depending on the length and composition of the primer and the presence of other proteins or mutations in PAP which limit the access of the enzyme to its RNA substrate. These modes, discussed below, suggest that PAP is roughly organized into two functionally distinct domains: a catalytic pocket consisting of the N-terminal part of the molecule and a primer grip region found in the carboxyl half. These domains represent at least two points of contact of PAP with the RNA substrate, one with the 3′ end of the primer, at a site on PAP presumably near the nucleotidyltransferase catalytic triad, and the second with the body of the primer in the carboxyl part of PAP.
FIG. 8.
Model of a two-domain structure for the S. cerevisiae PAP. SpD1 and SpD2 indicate Fip1 interaction sites. Flags mark epitopes recognized by the two PAP-specific antibodies. The arrowhead indicates the nucleotidyltransferase catalytic site. The 3′ RBS recognizes the last 3 nucleotides (nt) of the primer. The G-RBS interacts with the primer at a site approximately 14 nucleotides from the 3′ end. The C-RBS, which probably recognizes the body of the primer nonspecifically at a point beyond 14 nucleotides, overlaps with SpD2.
The catalytic domain of PAP is a region defined by the nucleotidyltransferase motif and sites which interact with the incoming NTP and the very-3′ end of the nucleic acid primer. Structural analysis of enzymes in the nucleotidyltransferase family (30) predicts that the carboxylate triad in PAP participates in metal-coordinated binding of the phosphate groups of ATP, as well as in positioning the nucleophilic 3′ OH of the primer terminus to facilitate the nucleotidyltransferase reaction. Regions in PAP which interact with the base and/or sugar group of ATP and with the nucleotides at the 3′ end of the primer have not been identified. However, properties of these sites which contribute to the specificity of PAP can be deduced from the experimental data collected here and in other published studies.
We have previously shown that ATP analogs with a 2- or 8-azido group, commonly used to map ATP binding sites by cross-linking, did not work for this purpose with yeast PAP (reference 36 and unpublished data), suggesting that bulky substitutions on the base of the incoming NTP are not tolerated. The fact that all nonadenosine NTPs are transferred poorly to a primer (10, 32) suggests that the primary discrimination of NTP on the base level occurs at the ATP binding site. However, the fact that some incorporation of other nucleotides can be detected, especially at a high enzyme concentration (10, 22), suggests that PAP is not absolutely specific for adenosine as the base of the incoming NTP. In this regard, one of the original studies of the yeast PAP found cytidylate residues in the poly(A) tracts of RNA made in vivo (10). Furthermore, our comparison of CMP and dCMP incorporation indicates that the 2′ hydroxyl group of ribose, while not essential, does stabilize the interaction at the ATP binding site.
Our results show that interactions at a 3′-end binding site (3′RBS, Fig. 8) also contribute to the ribose and base specificity of PAP. An oligo(A) trimer or a DNA primer that has been extended by three AMPs can efficiently prime poly(A) addition. These observations suggest that a minimum of three ribonucleotides at the 3′ end are required to accurately position the 3′ OH of the primer and form a productive complex in the active site of the enzyme (Fig. 8). Since DNA can serve as a primer, albeit a poor one, the 2′ hydroxyl group of ribose must not be directly involved in catalysis and more likely stabilizes the primer binding. The fact that 2′-dAMP addition renders an RNA primer a poor substrate supports this model. Interestingly, terminal deoxynucleotidyltransferase, the closest analog of PAP in terms of biochemical activity, can incorporate ribonucleotides into an oligodeoxynucleotide primer, but incorporation also stalls after the addition of two nucleotides (28). This modified primer can then be used for deoxynucleotidyl elongation, in a reaction which also displays a lag ressembling that seen for PAP-catalyzed addition of AMP to DNA.
Based on our data, the 3′ RBS of PAP recognizes the last three nucleotides of the primer and strongly discriminates against deoxyribonucleotidyl residues at this position. Our results also imply a role for the 3′ RBS in base recognition. The incorporation of a single CMP onto a primer is quite rapid, but the resulting product is a poor substrate for further elongation. Similarly, a lag phase in the utilization of poly(U) primer by the bovine PAP (33) and of poly(C) primer by the yeast PAP (19) has been interpreted to mean that the addition of a few adenosines to the ends of these substrates converts them to more-efficient substrates. It is also interesting to note that the penultimate nucleotide in cleaved mRNA precursor is often cytidine, followed by adenosine (34). This configuration is probably unfavorable for PAP and may provide time for the processing machinery to reorganize from a cleavage to a polyadenylation complex.
An additional RBS on PAP may also contribute to the specificity of the enzyme. The existence of this site is indicated by the surprising observation that the polymerization of oligo(G) tracts into primer abruptly stops after 14 nucleotides have been added (references 22 and 31 and results reported herein). At this point, the PAP does not switch to a slow distributive elongation mode like that seen with 2′-dAMP or CMP incorporation. Instead, a Gaussian distribution of products occurs, with an average of 12 GMP residues accumulating. One possible explanation for this phenomenon is the existence on PAP of an RBS which interacts with the primer at a point approximately 14 nucleotides from the 3′ end. This site, which was termed guanylyl-RBS (G-RBS, Fig. 8) for the discrimination against poly(G), is base specific, allowing PAP to elongate a primer containing poly(A) and poly(U), but not poly(G), beyond 14 nucleotides. The effect of poly(C) on this site has not been tested. In support of this interpretation, unpublished work referenced by Martin and Keller (22) suggests that the yeast PAP binds very poorly to primer with a G14 tail in a mobility shift experiment. The location of this site is not known, but our data suggest that it is different from the 3′ RBS and the portion of the C-RBS defined by the Δ9 deletion of yeast PAP. A previous report (10) has pointed out that GTP, unlike CTP and UTP, is a noncompetitive inhibitor of yeast PAP. Based on this fact and the unusual distribution of reaction products formed with GTP, we speculate that the G-RBS may be involved in primer translocation, but additional research will be necessary to define its function. It is interesting that the vaccinia virus PAP, which exhibits very little sequence homology to the eukaryotic PAPs, also stalls after addition of 14 guanidylate residues (31). This common property strengthens the hypothesis that the eukaryotic and vaccinia virus PAPs share a similar structure-function organization.
Even though PAP is dedicated to the synthesis of poly(A) on precursor mRNA, its ATP binding site is not absolutely specific. However, PAP clearly contains motifs which place it in the nucleotidyltransferase superfamily, a group which includes mostly DNA polymerases. If PAP shares with these enzymes a common ancestor which lacks NTP specificity, it has, perhaps through evolution, been adjusted at several levels to create its specificity for adenosine and ribose, in this case at the ATP binding site, the 3′ RBS, and the G-RBS. The interactions at these sites, together with the higher concentration of ATP in the cell relative to the other NTPs, ensures that the tails added to mRNA are poly(A).
Another RBS (C-RBS, Fig. 8) is located at the carboxyl terminus of yeast PAP (36), and a similar site was found in bovine PAP (21). PAP can successfully elongate a single-stranded DNA primer after the rate-limiting addition of three AMP residues. Moreover, single-stranded DNA competes efficiently for binding at the C-RBS (36). Based on these findings, we conclude that the C-RBS does not discriminate the sugar nature of the primer. It is not involved in 3′-end recognition, since radioactivity cannot be transferred to this site during UV cross-linking experiments if the RNA is 3′-end labeled (36). It is not known whether this site exhibits any preference for the base composition of the RNA substrate. However, as discussed below, we have found that the C-RBS, by gripping the body of the primer, confers processivity to yeast PAP.
We have shown that poor interactions at the 3′ RBS cause a very slow elongation of unfavorable substrates such as oligo(A) dimer, single-stranded DNA, or primer with cytidylates at the 3′ end. Our results indicate that during poly(A) addition without such limitations, the processivity of yeast PAP depends on the ability of the RNA molecule to interact simultaneously with the 3′ RBS and the C-RBS (Fig. 8). Disruption of the C-RBS, as seen with Δ9PAP, keeps the enzyme in a distributive mode. This C-RBS does not seem to participate in the block to elongation caused by poly(G)14. Based on this fact and the observation that oligo(A)26 is a better substrate for PAP than is oligo(A)12, it is likely that the C-RBS binds RNA most effectively at some point beyond 14 nucleotides from the 3′ end (Fig. 8).
It is also intriguing that interactions of other proteins with PAP or with the RNA substrate can shift PAP from a processive to a distributive mode of action. Our data shown that Fip1 has this property and probably acts through a direct interaction at SpD2 to limit access of the RNA to the C-RBS (Fig. 8). Preker et al. (26) have proposed that Fip1 recruits PAP to the rest of the polyadenylation machinery, thereby directing its activity to the appropriate substrate, i.e., the cleaved ends of the mRNA precursor. PF I-associated PAP shows an increased processivity in the nonspecific poly(A) addition reaction in comparison to PAP alone (25), suggesting that other components of PF I must be able to restore the interaction with RNA occluded by Fip1. Moreover, the Δ9PAP mutant, with a truncated C-RBS, can rescue a yeast strain with a chromosomal disruption of the PAP1 gene and is active in specific polyadenylation assays in vitro, giving tails of the correct length (36). This further supports the idea that other factors can overcome a block in processivity. PF I subunits with such a property could include Yth1, which interacts with Fip1 and nonspecifically with RNA (2), or Pfs1, which has a zinc knuckle motif predicted to bind RNA (25). The interaction of Fip1 with Rna14 (26) provides additional bridges to the RNA substrate, via the Rna15 subunit of CF IA as well as Hrp1 (CF IB), which directly contacts the UA-rich efficiency element (15).
An unanswered question concerns the function of the SpD1 domain of yeast PAP. This region is clearly needed for PAP’s participation in specific polyadenylation but is not necessary for Fip1’s effect on PAP processivity. Further experiments will be needed to determine the consequences of the SpD1-mediated interaction of PAP with Fip1. Since it is possible that this interaction is mediated through other factors, it may also be involved in restoring PAP processivity. In summary, interaction of PAP with the Fip1 subunit of PF I may have several functions in controlling the activity of PAP. It could help prevent PAP from engaging inappropriate substrates. If the PAP-Fip1 interaction is maintained throughout the lengthening of the poly(A) tail to 50 to 90 nucleotides, disenabling the intrinsic processivity of PAP may make it easier to regulate PAP activity and facilitate the termination of poly(A) addition provoked by Pab1 (1, 16, 23).
Pab1 is required in the specific poly(A) addition reaction to limit the extension of the poly(A) tails to the normal length of 50 to 100 nucleotides (1, 16, 23). Pab1 also inhibits the nonspecific activity of yeast PAP (19). In this report, we have shown that the reason for this inhibition is a decrease in the processivity of PAP. The mechanism of Pab1 inhibition is most likely different from but related to that underlying the Fip1 inhibition. While Fip1 will block PAP from contacting the RNA through its C-RBS, the formation of Pab1-RNA complex leaves only the 3′ end accessible, thereby preventing the RNA from interacting with the C-RBS. To have this effect, Pab1 would not need to interact directly with PAP. However, a combination of Fip1 and Pab1 was not sufficient to terminate the addition of poly(A) to oligo(A) primer; other components of the polyadenylation machinery must be needed for full termination.
Similar regulatory mechanisms may be operating with the mammalian PAP. The mammalian holoenzyme (PAP plus CPSF and PAB II) synthesizes the poly(A) tail in a bimodal fashion, first slowly extending the RNA by about 10 to 15 nucleotides to create a PAB II binding site and then rapidly and processively elongating it to 250 nucleotides (34). p160 of CPSF has been shown to inhibit specific and nonspecific polyadenylation (24). It will be interesting to see whether interactions with CPSF subunits or PAB II modulate the binding of RNA to the RBS found in the carboxyl half of mammalian PAP (21), in a manner analogous to what we have found for yeast PAP. The region of mammalian PAP containing this site becomes hyperphosphorylated during the M phase of the cell cycle, resulting in inactivation of the enzyme (5, 6). The increased negative charge may also prevent RNA interaction at this site on PAP. Further research should allow us to draw additional correlations between the structural organization of the PAP family and mechanisms which define and regulate its enzymatic properties.
ACKNOWLEDGMENTS
We thank Debu Raychaudhuri and David Lazinsky for helpful discussions and review of the manuscript.
This work was supported by grant RPG-95-048-03-NP from the American Cancer Society to C.M.
REFERENCES
- 1.Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barabino S, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 3.Bartel P, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Moore C. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol Cell Biol. 1992;12:3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colgan D, Murthy K, Prives C, Manley J. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- 6.Colgan D, Murthy K, Zhao W, Prives C, Manley J. Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. EMBO J. 1998;17:1053–1062. doi: 10.1093/emboj/17.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gietz R, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 8.Gunderson S, Polycarpou-Schwarz M, Mattaj I. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U170K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 9.Gunderson S, Vagner S, Polycarpou-Schwarz M, Mattaj I. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 1997;11:761–773. doi: 10.1101/gad.11.6.761. [DOI] [PubMed] [Google Scholar]
- 10.Haff L, Keller E. The polyadenylate polymerases from yeast. J Biol Chem. 1975;250:1838–1846. [PubMed] [Google Scholar]
- 11.Holm L, Sander C. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 12.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser C, Michaelis S, Mitchel A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 14.Keshgegian A A, Meltzer S M, Furth J J. Poly(A) polymerase of bovine lymphosarcoma. Cancer Res. 1975;35:1141–1146. [PubMed] [Google Scholar]
- 15.Kessler M, Henry M, Gross S, Shen E, Zhao J, Silver P, Moore C. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessler M M, Zhao J, Moore C L. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. J Biol Chem. 1996;271:27167–27175. doi: 10.1074/jbc.271.43.27167. [DOI] [PubMed] [Google Scholar]
- 17.Lingner J, Keller W. 3′-end labeling of RNA with recombinant yeast poly(A) polymerase. Nucleic Acids Res. 1993;21:2917–2920. doi: 10.1093/nar/21.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lingner J, Kellermann J, Keller W. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae. Nature. 1991;354:496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- 19.Lingner J, Radtke I, Wahle E, Keller W. Purification and characterization of poly(A) polymerase from Saccharomyces cerevisiae. J Biol Chem. 1991;266:8741–8746. [PubMed] [Google Scholar]
- 20.Mandart E, Parker R. Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol Cell Biol. 1995;15:6979–6986. doi: 10.1128/mcb.15.12.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and a catalytic domain, homologous to the family χ polymerases and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 22.Martin G, Keller W. Tailing and 3′-end labeling of RNA with yeast poly(A) polymerase and various nucleotides. RNA. 1998;4:226–230. [PMC free article] [PubMed] [Google Scholar]
- 23.Minvielle-Sebastia L, Preker P, Wiederkehr T, Strahm Y, Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc Natl Acad Sci USA. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murthy K G K, Manley J L. The 160 kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′ end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 25.Preker P, Ohnacker M, Minvielle-Sebastia L, Keller W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preker P J, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 27.Raabe T, Murthy K G K, Manley J L. Poly(A) polymerase contains multiple functional domains. Mol Cell Biol. 1994;14:2946–2957. doi: 10.1128/mcb.14.5.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roychoudhury R, Kossel H. Enzymic synthesis of ribonucleotide terminated oligodeoxynucleotides and their use as primers for the enzymic synthesis of polydeoxynucleotides. Eur J Biochem. 1971;22:310–320. doi: 10.1111/j.1432-1033.1971.tb01546.x. [DOI] [PubMed] [Google Scholar]
- 29.Sachs A, Bond M, Kornberg R. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 30.Sousa R. Structural and mechanistic relationships between nucleic acid polymerases. Trends Biochem Sci. 1996;21:186–191. [PubMed] [Google Scholar]
- 31.Thomson J, Gershon P. Use of vaccinia virus poly(A) polymerase for RNA 3′-end labeling with a chain-terminating nucleotide or a short 3′ homopolymer tract. BioTechniques. 1995;19:416–425. [PubMed] [Google Scholar]
- 32.Tsiapalis C M, Dorson J W, De Sante D M, Bollum F J. Terminal riboadenylate transferase: a polyadenylate polymerase from calf thymus gland. Biochem Biophys Res Commun. 1973;50:737–743. doi: 10.1016/0006-291x(73)91306-5. [DOI] [PubMed] [Google Scholar]
- 33.Wahle E. Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J Biol Chem. 1991;266:3131–3139. [PubMed] [Google Scholar]
- 34.Wahle E, Kuhn U. The mechanism of 3′ cleavage and polyadenylation of eukaryotic pre-mRNA. Prog Nucleic Acid Res Mol Biol. 1997;57:41–70. doi: 10.1016/s0079-6603(08)60277-9. [DOI] [PubMed] [Google Scholar]
- 35.Wittmann T, Wahle E. Purification and characterization of full-length mammalian poly(A) polymerase. Biochim Biophys Acta. 1997;1350:293–305. doi: 10.1016/s0167-4781(96)00164-9. [DOI] [PubMed] [Google Scholar]
- 36.Zhelkovsky A M, Kessler M M, Moore C L. Structure-function relationships in the Saccharomyces cerevisiae poly(A) polymerase. J Biol Chem. 1995;270:26715–26720. doi: 10.1074/jbc.270.44.26715. [DOI] [PubMed] [Google Scholar]