Abstract
Background
It has recently been determined that N6-methyladenosine (m6A) RNA methylation regulators have prominent effects on several cancers. However, the potential role of m6A modification in lung squamous cell carcinoma (LUSC) remains unclear.
Methods
We evaluated the modification pattern of m6A and studied the biological function of m6A regulators in LUSC. Then, we constructed the m6Ascore to predict the prognosis of LUSC and analyzed the relationship between the m6Ascore and tumor mutation burden, immune cell infiltration, and immunotherapy.
Result
In the unsupervised consensus cluster analysis, three different m6Aclusters were identified, which correspond to an immune activation state, a moderate immune activation state, and an immune tolerance state. Forty-two genes related to the m6A phenotype were used to construct the m6Ascore; subsequently, multiple validations of the m6Ascore were carried out to determine the relationship between the score and immune cell infiltration and response to CTLA-4/PD-1 inhibitor treatment. Further analysis revealed that the m6Ascore could effectively predict the prognosis of LUSC and that the m6A phenotype-related genes, FAM162A and LOM4, might be potential biomarkers.
Conclusion
These findings highlight the potential role of m6A modification in the prognosis, TME, and immunotherapy of LUSC and have profound implications for developing more effective personalized treatment strategies for LUSC.
Keywords: Lung squamous cell carcinoma, m6A, Prognosis, Tumor microenvironment, Immunotherapy
1. Introduction
Lung squamous cell carcinoma (LUSC) is a malignant epithelial tumor derived from the bronchial epithelium. As the second most common subtype of lung cancer, LUSC accounts for approximately 20% of primary lung tumors in the United States. Its incidence is mainly related to smoking, raw biofuels, chronic obstructive pulmonary disease, and other lung diseases [1]. The tumor microenvironment (TME) is composed of tumor cells, microvessels, stromal cells, and infiltrating cells. It has a variety of complex regulatory functions in LUSC and has a significant impact on the prognosis of patients [[2], [3], [4]]. Particularly, targeted immunotherapy, especially the application of immune checkpoint inhibitors (ICI) such as CTLA-4 and PD-1, has a positive effect on LUSC patients. However, due to the individual heterogeneity of LUSC patients, the effect of immunotherapy is quite different [5]. Therefore, an in-depth understanding of the molecular mechanisms and TME of LUSC is essential for developing more effective clinical diagnosis and immunotherapy.
N6-methyladenosine (M6A) is one of the most common RNA modifications in eukaryotes and is mainly found in messenger RNA (mRNA). Three types of regulators are involved in m6A methylation, namely, writers, erasers, and readers [6]. Writers are methyltransferases that promote the methylation of m6A in RNA, and the common writers are METTL3, METTL14, WTAP, KIAA1429, ZC3H13, RBM15, RBM15B, and CBLL1. Erasers are demethylases that can remove the methylation of m6A in RNA, and FTO and ALKBH5 are erasers. Readers, including IGF2BPs, YTHDCs, and YTHDFs, can bind to the methylation site of m6A in RNA and recognize m6A modifications. M6A modification is involved in circadian rhythm regulation [7], gene expression [8], lipid metabolism [9], the immune response [10], and tumorigenesis [11].
Recent reports have shown that m6A facilitates the occurrence and development of tumors [[12], [13], [14]]. For example, the upregulation of YTHDF2 expression is significantly related to the proliferation of hepatocellular carcinoma cells [15]. FTO exerts its inhibitory effect on acute myeloid leukemia by inhibiting the expression of key transcripts (ASB2, RARA, MYC, and CEBPA) [16,17]. The METTL14 mutation leads to the activation of the AKT pathway and enhances the proliferation of endometrial cancer cells [18]. Moreover, the role of m6A modification in the TME and immune response cannot be ignored [19,20]. Silencing FTO can decrease the mRNA stability of STAT1 and PPAR-γ, thus hindering the activation of macrophages [21]. METTL3-mediated m6A methylation can promote dendritic cell maturation and CD-based T cell activation [22]. Silencing YTHDF1 can reduce the translation of lysosomal cathepsin in dendritic cells, thereby enhancing the expression of tumor antigens by dendritic cells and increasing the infiltration of CD8+ T cells [23].
A recent research by Gu and his colleagues reported that the m6A-related genes WTAP, YTHDC1, and YTHDF1 independently predicted the prognosis of LUSC, and the prognostic performance was better than traditional clinical features [24]. However, the role of multiple m6A regulators in the TME and immune regulation in LUSC is largely unknown. In this study, we integrated the transcriptome data of 739 LUSC patient samples and systematically evaluated the multi-omics functions of 23 m6A regulators in LUSC. We studied copy number variation (CNV) and difference expression genes (DEGs), conducted a survival analysis based on m6A regulators in LUSC patients, constructed an interaction network of m6A regulators, and analyzed the biological function of m6A regulators and the characteristics of TME immune cells. Then, we established a m6Ascore to quantify the m6A modification pattern according to the m6A phenotype-related DEGs and verified it from the perspectives of tumor mutation burden (TMB), survival time, clinical survival status, immune cell infiltration, and response to ICI therapy.
2. Material and methods
2.1. Data collection and processing of LUSC
The primary process of this study is shown in Supplementary Fig. 1(a). Two independent patient datasets (TCGA-LUSC and GSE157010) comprising gene expression data and clinical data were obtained from The Cancer Genome Atlas (TCGA) and the Gene-Expression Omnibus (GEO) databases, respectively, and we excluded patients who lacked survival information for further study. The GEOquery R package was utilized to download the expression matrix and clinical data of the GSE157010 dataset, and the annotation file was used to convert probes into gene symbols. The limma R package was used to convert the units of RNA sequencing data in TCGA-LUSC from fragments per kilobase per million (FPKM) to transcripts per kilobase million (TPM) to be consistent with the RNA sequencing data of GEO157010 and facilitate a comparative analysis of samples. The CNV data for the TCGA-LUSC dataset were downloaded from the Xena Public database (https://xenabrowser.net). The immunotherapy score file of the TCGA-LUSC cohort was obtained from the public database The Cancer Immune Atlas (https://www.tcia.at/home). Twenty-three m6A regulators were extracted from previous studies to perform a mutation analysis in this study [[25], [26], [27], [28], [29], [30]], including 8 writers (METTL3, METTL14, WTAP, KIAA1429, ZC3H13, RBM15, RBM15B, and CBLL1), 13 readers (YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, HNRNPC, LRPPRC, HNRNPA2B1, IGFBP1, IGFBP2, IGFBP3, FMR1, and ELVAL1) and 2 erasers (FTO and ALKBH5). Among the 23 m6A regulators, KIAA1429 and ELVAL1 lack transcriptome and CNV data for the LUSC samples; thus, the remaining 21 m6A regulators were used in all subsequent analyses.
2.2. Unsupervised consensus clustering for 23 M6A regulators
We used unsupervised consensus clustering analysis to divide the LUSC patient samples into m6Aclusters according to the expression matrix of 23 m6A regulators. A consensus clustering algorithm was employed to calculate the clustering number and stability [31]. The operation used the ConsensuClusterPlus package in R, with parameters set to resample 80% of any sample (pitem = 0.8) and resample all proteins (pfeature = 1), repeated 50 times to ensure the stability of the clustering [32].
2.3. Gene Set Variation Analysis (GSVA) and estimation of immune cell infiltration in the TME
We performed GSVA analysis on the m6Aclusters to assess the enrichment differences in the pathways and biological process activities in the different subgroups and drew heatmaps for the visualization of the results. We downloaded the Kyoto Encyclopedia of Genes and Genomes (KEGG) gene set from the Molecular Signatures Database (MSIGDB) database (https://www.gsea-msigdb.org/) and then set up the parameters MinGSsize = 10, MaxGSsize = 500, and adj. p value < 0.05. The R packages used for this step were limma, GSEABase, GSVA, and pheatmap. Next, we quantified the relative abundance of various immune cells in the TME of LUSC samples by single-sample gene-set enrichment (ssGSEA) analysis. We used the gene set of 23 tumor immune infiltrating cells obtained by Charoentong et al. [33], including activated dendritic cells, activated CD4+ T cells, macrophages, neutrophils, and natural killer cells. The R packages used for this analysis were limma, GSEABase, GSVA, and ggpubr.
2.4. Identification of GeneClusters and functional analysis
We identified the genes associated with 23 m6A regulators, divided 736 LUSC patients into three different m6A geneClusters, and set the display threshold of the DEGs to adj. p value < 0.001. The empirical Bayesian approach using the R package limma (Http://www.bioconductor.org/packages/release/bioc/html/limma.html) was used to estimate the DEGs. We carried out gene ontology (GO) and KEGG enrichment analysis of the DEGs by gene annotation enrichment analysis (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) using the clusterProfiler package in R. The filter conditions of the GO terms and KEGG pathways were defined as p < 0.05, and bar charts were generated to display the results.
2.5. Construction of the M6Ascore in patients with LUSC
To evaluate the modification pattern of m6A in LUSC, we established an m6A gene signature, namely, the m6Ascore. The procedure for establishing the m6Ascore was performed as follows:
First, we normalized the DEGs in the three m6Aclusters to obtain the overlapping genes and visualized the results with a Venn diagram. Then, unsupervised consensus clustering was used to analyze the overlapping DEGs, and the number and stability of geneClusters were analyzed by a consensus clustering algorithm. Then, we carried out univariate Cox regression analysis of the above genes through the survival package in R to identify the DEGs associated with the prognosis of LUSC patients for the principal component analysis (PCA) and used principal components 1 and 2 as the signature scores. The advantage of this algorithm is that the score is concentrated on the largest related geneCluster, and the weight of undetected genes in other gene sets is reduced. The method used to define the m6Ascore is similar to that used for Gillette's gait index (GGI) [34,35], and the specific formula is as follows:
| M6Ascore = ∑(PC1i + PC2i) |
Where i represents the genes related to the m6A phenotype.
2.6. Verification of the M6Ascore
To evaluate the correlation between the m6Ascore and the survival of LUSC patients, we used the survminer R package to calculate the cut-off value of each dataset, repeatedly determined all potential cut-off points, calculated the maximum rank statistics, and divided all patients into a high m6Ascore group and a low m6Ascore group according to the median cut-off value. Then, a log-rank test was used to determine the difference in Kaplan-Meier survival curves between the two groups. We conducted the following studies on the m6Ascore to verify its reliability: the correlation between the m6Ascore and TMB was tested by the Wilcoxon signed-rank test, the correlation between the m6Ascore and clinical data was analyzed by the Wilcoxon signed-rank test, and Pearson's chi-square test, and the association between the m6Ascore and response to immunotherapy was analyzed by Spearman's test and the Wilcoxon signed-rank test.
2.7. Statistical analysis
We used the R package limma for differential gene expression analysis, Spearman's test, and distance correlation analysis to analyze the correlation between the m6Ascore and TME immune cell infiltration. The Wilcoxon signed-rank test was used to analyze the differences between the two groups. The Kruskal-Wallis test and one-way ANOVA were used to compare the differences among three or more groups [36]. We adopted the Pearson chi-square test to analyze the differences between groups and univariate Cox analysis to calculate the risk ratio of the m6A regulator-related genes and m6A phenotype-related genes. We used the waterfall function in the R package maftools to visualize the gene mutation analysis of the tumor patients in the TCGA-LUSC cohort. The specific chromosomal location of the CNVs identified in the m6A regulators was plotted through the R package RCircos [37]. The alluvial diagram was generated using the ggalluvial package in R to describe the grouping and survival status of LUSC patients by the m6Aclusters, geneClusters, and m6Ascores. All the data were analyzed by R software (version 4.0.4).
3. Results
3.1. Gene variation in M6A regulators in LUSC patient samples
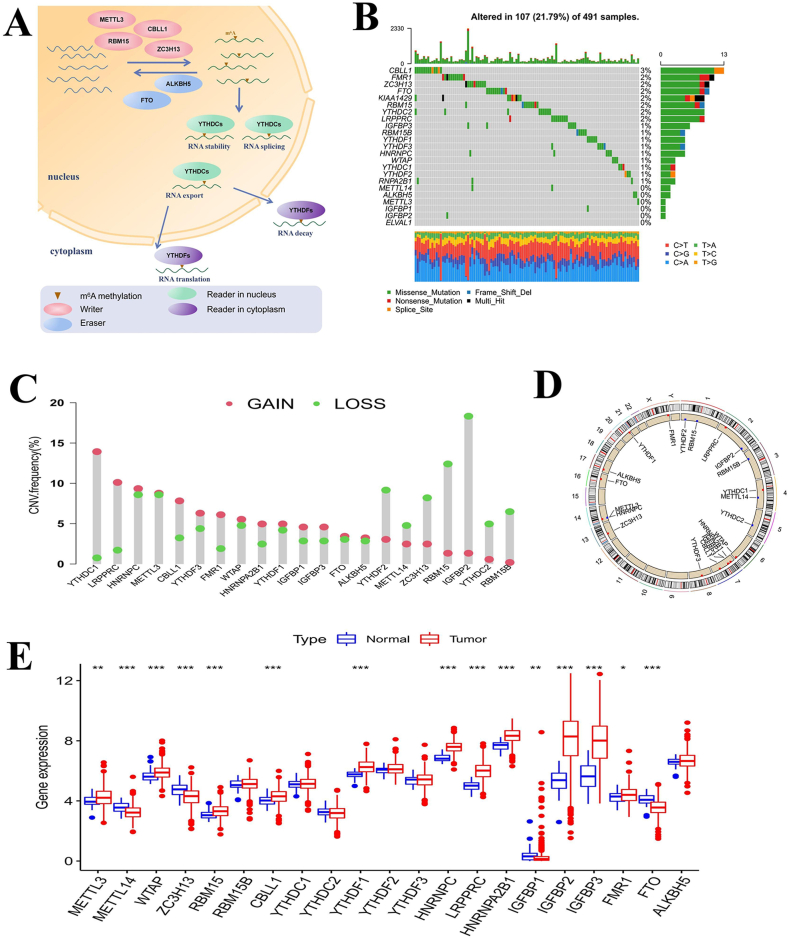
Our study included 23 m6A regulators that mediate the reversible process and biological function of mRNA methylation in cells, as shown in Fig. 1(a). We first summarized the incidence of somatic mutations and CNVs in the m6A regulators in LUSC patient samples. Of the 491 samples in the TCGA-LUSC cohort, 107 samples had mutations in 23 m6A regulators with a frequency of 21.79%. Among these regulators, the mutation rate of CBLL1 was the highest, with a mutation frequency of 3%, followed by FMR1, whereas METTL14, ALKBH5, METTL3, IGFBP1, IGFBP2, and ELVAL1 were almost unmutated in the LUSC samples (Fig. 1(b)). In the CNV study of LUSC patient samples, we found that all 21 m6A regulators with available copy number data had extensive CNVs; most regulators had copy number amplification, and only a few had copy number loss, such as YTHDF2, METTL14, ZC3H13, RBM15, IGFBP2, YTHDC2, and RBM15B (Fig. 1(c)). We also used a loop graph to show the chromosomal location of the copy number alteration in the m6A regulators (Fig. 1(d)). Then, we compared the expression levels of m6A regulators in normal and tumor samples. The results showed that the expression levels of most m6A regulators were significantly different between normal tissues and tumor tissues from LUSC patients, and the abnormal expression of m6A regulators was probably related to alterations in copy number. For example, CNV-amplified m6A regulators LRPPRC, HNRNPC, and CBLL1 showed high expression in tumor tissues, while CNV-deleted m6A regulators METTL14 and ZC3H13 showed low expression in tumor tissues (Fig. 1(e)). The above results indicated that m6A regulator gene variation is common in LUSC samples, and the expression of these regulators is heterogeneous among normal samples and LUSC samples, suggesting that abnormal m6A modification plays a vital role in the occurrence and development of LUSC.
Fig. 1.
Mature mRNA needs N6-methyladenosine (m6A) methylation to transfer from the nucleus to the cytoplasm.RNA methylation process and mutation characteristics of m6A regulators in lung squamous cell carcinoma (LUSC). A Three different types of m6A regulators (readers, erasers and writers) cooperate to mediate the dynamic and reversible RNA methylation process. The ellipses represent different kinds of m6A regulators. B The mutation characteristics of 23 m6A regulators in the TCGA-LUSC cohort. The left side of the plot shows the names of the m6A regulators, the color change in the middle of the waterfall plot indicates the mutation of the corresponding regulator, and the right side of the plot shows the mutation frequency of the regulators. The different colors in the middle of the waterfall plot indicate that the patient has the corresponding somatic mutation type. The bottom heatmap shows changes in single nucleotides in 491 samples. C The copy number variation (CNV) features of the m6A regulators of the TCGA-LUSC cohort. D The chromosomal location of the m6A regulator CNVs. Red indicates copy number gain, and blue indicates copy number loss. E The difference in the expression of m6A regulators between tumor tissues and normal tissues. The upper and lower ends of the box represent the quartile, the horizontal line in the box represents the median, the point outside the box represents the abnormal value, the upper * p < 0.05, * *p < 0.01, * * *p < 0.001.
3.2. M6A regulator-mediated methylation modification patterns
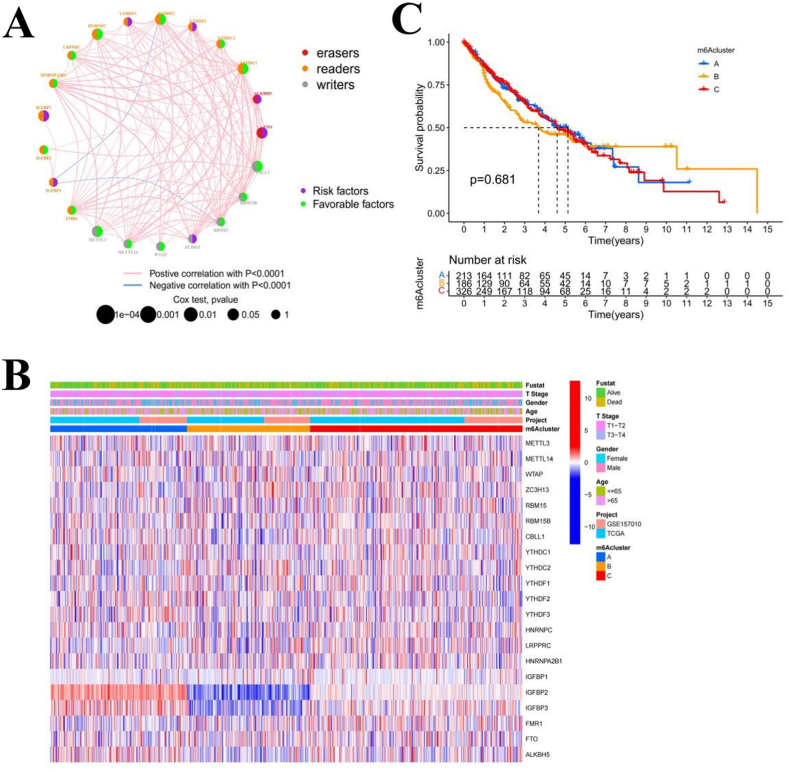
We merged two separate datasets (TCGA-LUSC and GSE157010) with survival and clinical data available into a single meta-cohort (Supplementary Table 1). A univariate Cox regression analysis showed that IGFBP1, METTL3, CBLL1, FTO, YTHDC1, YTHDC2, and HNRNPC could be used as independent prognostic factors for patients with LUSC (Supplementary Table 2). We also constructed an interaction network of the m6A regulators and analyzed the correlation among all of the m6A regulators and their prognostic effects on LUSC patients (Fig. 2(a)). We found that the positive correlations between the regulators were significantly greater than the negative correlations. Moreover, the high expression of two erasers (FTO and ALKBH5) is a risk factor for the prognosis of LUSC patients. Due to the high mutation frequency of CBLL1, FMR1, KIAA1429, and YTHDC2 in LUSC, we analyzed the expression differences between the mutated and wild-type variants of these regulators and found that differences in the expression levels of mutated and wild-type m6A regulators were related to the expression levels of different readers (Supplementary Figs. 1(b–f)). We also investigated the survival outcomes of patients with high and low expression of m6A regulators and found that different CBLL1, FMR1, FTO, HNRNPA2B1, HNRNPC, IGFBP1, IGFBP2, LRPPRC, METTL3, RBM15, WTAP, YTHDC1, and YTHDF2 expression levels were associated with significant differences in survival time. The survival time of patients with low expression of FTO and IGFBP1 was longer, while patients with low expression of other regulators had a shorter survival time (Supplementary Figs. 2(a–m)). Our results suggested that the interaction of multiple m6A regulators may have prominent effects on the formation and development of LUSC.
Fig. 2.
The interaction characteristics and grouping of m6A regulators. A The interaction network of m6A regulators. B The difference in m6A regulator expression among the m6Aclusters is depicted as a heatmap. IGFBP2 and IGFBP3 expression levels were significantly upregulated in m6Acluster A and downregulated in m6Acluster B. C There were no significant differences in patient survival times among the m6Aclusters.
To analyze the heterogeneity of m6A regulators in LUSC, we used the unsupervised consensus clustering method in the R package ConsensusClusterPlus to divide LUSC patient samples into three different m6Aclusters, m6Acluster A (n = 217), m6Acluster B (n = 191), and m6Acluster C (n = 328). The difference in the expression of m6A regulators among the three m6Aclusters is shown by the heatmap (Fig. 2(b)). Then, we analyzed the survival outcome differences of patients in each of the different m6Aclusters by Kaplan-Meier analysis, and the results showed that there were no significant differences in the survival times of patients in different m6Aclusters (Fig. 2(c)).
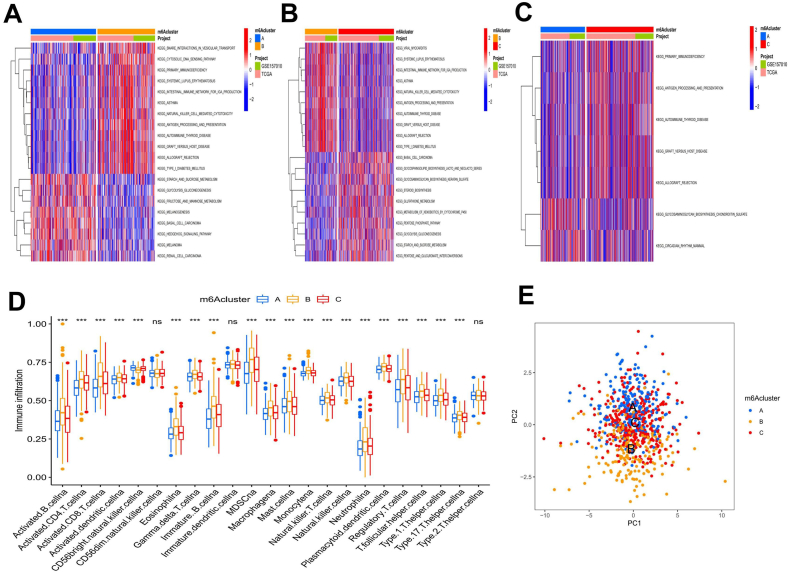
3.2.1. GSVA of the M6A modification pattern and characteristics of TME immune cell infiltration
The biological behaviors of the different m6Aclusters were analyzed by GSVA (Fig. 3(a–c)). We found that m6Acluster A was enriched in carcinogenic activation pathways, such as melanogenesis, basal cell carcinoma, renal cell carcinoma, and the hedgehog signaling pathway. M6Acluster B was enriched in various immune activation-related pathways, such as systemic lupus erythematosus, the intestinal immune network for IgA production, natural killer cell-mediated cytotoxicity, antigen processing and presentation, and autoimmune thyroid disease. The m6Acluster C was enriched in antigen processing and presentation, autoimmune thyroid disease, and several carbohydrate metabolic pathways. Then, we analyzed the characteristics of TME immune cell infiltration in the different m6Aclusters and generated a box diagram to show the results (Fig. 3(d)). Except for extensive CD56-bright natural killer cell enrichment in m6Acluster A, all the other immune infiltrating cells showed the highest enrichment in m6Acluster B and the lowest enrichment in m6Acluster A. The PCA of the three different m6A modification patterns also showed prominent differences in the transcriptome (Fig. 3(e)). According to our results, we can infer that the high expression of the immune activation pathway in m6Acluster B may lead to the infiltration of a large number of immune cells. Additionally, the disadvantage of the first 5-year survival of m6Acluster B may result from the immune activation of this cluster.
Fig. 3.
Gene Set Variation Analysis (GSVA) and single-sample gene-set enrichment (ssGSEA) of m6Aclusters. A-C Heatmaps showing the differences in pathway activation among m6Aclusters. D The differences in tumor microenvironment (TME) immune cell infiltration with different m6A modification patterns are shown by the box plot. E The scatter plot shows the differences in transcriptional profiles among m6Aclusters.
3.2.2. Functional analysis of M6A phenotype-related genes and construction of GeneClusters
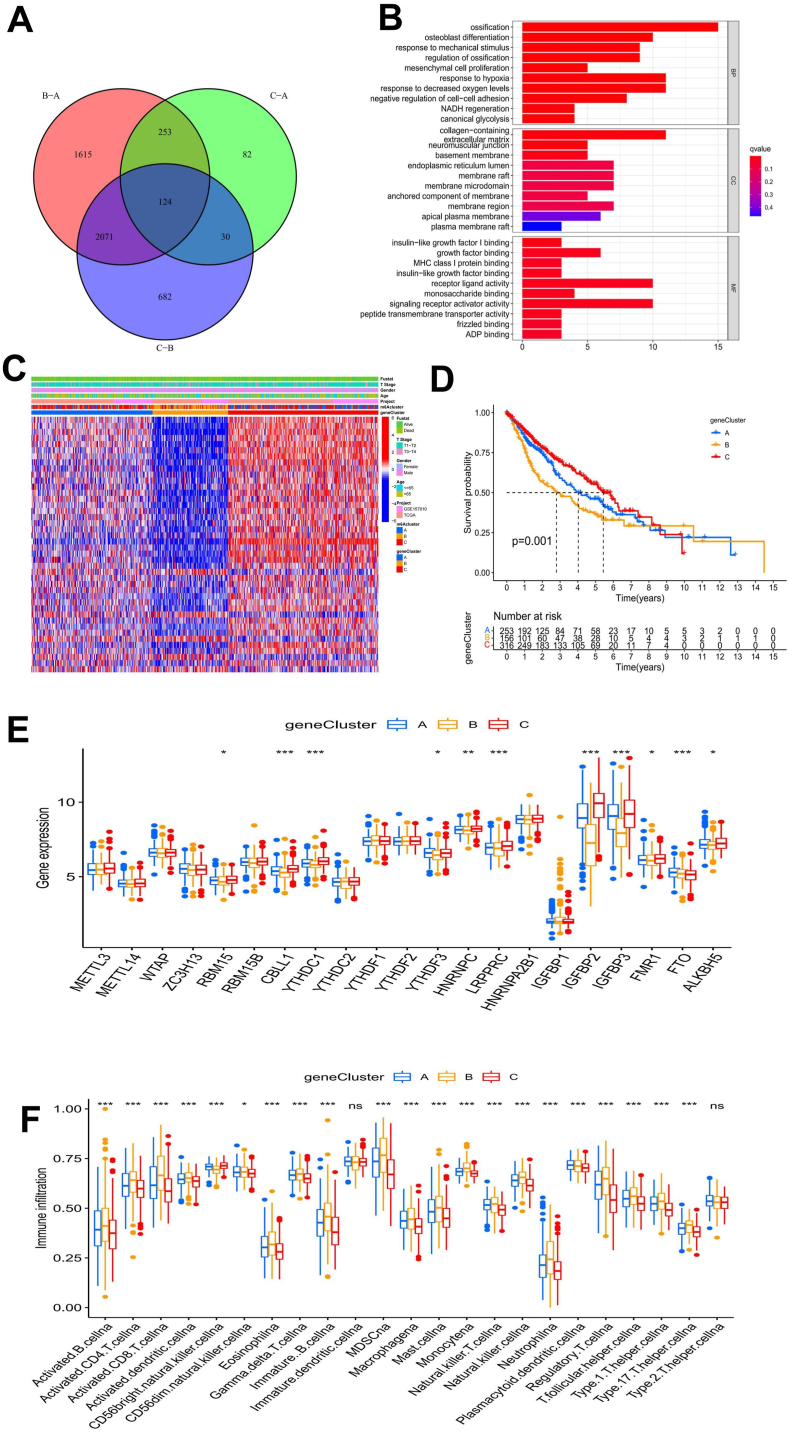
The above results demonstrated the critical role of m6A modification in the prognosis and TME of LUSC patients. To further analyze the biological behavior of the m6A modification pattern in LUSC, we screened 124 m6A phenotype-related DEGs (Fig. 4(a) and Supplementary Table 3). Furthermore, we carried out GO and KEGG enrichment analyses of the DEGs. Surprisingly, the DEGs were associated with cell division, transmembrane transport, and receptor-ligand activity (Fig. 4(b)). KEGG enrichment analysis showed that these DEGs were associated with extracellular matrix (ECM) receptors, antigen processing, the Wnt signaling pathway, and the HIF-1 signaling pathway (Supplementary Fig. 3(a)). These results confirmed that m6A modification has a prominent effect on the immune regulation of the TME.
Fig. 4.
Functional analysis of phenotype-related difference expression genes (DEGs) of m6A and geneClusters. A The Venn diagram depicts 124 m6A phenotype-related DEGs. B The results of gene ontology (GO) enrichment analysis of m6A phenotype-related DEGs are shown by a histogram, and the color represents the significant degree of enrichment. The abscissa indicates the number of DEGs enriched. C The heatmap shows the m6A-related DEGs among different geneClusters. D There was a significant difference in the survival outcomes among the geneClusters. E The box plot was used to show the difference in m6A phenotype-related DEGs among the geneClusters. F The characteristics of TME infiltrating cells among different geneClusters are shown in a box plot.
To further verify the mechanism of m6A modification in LUSC, we carried out an unsupervised consensus clustering analysis of these 124 DEGs. The LUSC samples were divided into three gene subgroups, named geneCluster A (n = 253), geneCluster B (n = 160), and geneCluster C (n = 323) (Supplementary Figs. 3(b–e)). In addition, univariate Cox regression analysis was carried out on the DEGs, and 42 related genes with independent prognostic values were obtained for follow-up analysis (Supplementary Table 4). The results showed that the three geneClusters had different m6A modification patterns in LUSC. DEGs in geneCluster C were upregulated, while DEGs in geneCluster B were downregulated. The number of stage T3-T4 patients and dead patients in geneCluster B was remarkably greater than that in other geneClusters (Fig. 4(c)). Kaplan-Meier survival analysis revealed a survival advantage in geneCluster C > geneCluster A > geneCluster B, and the difference was statistically significant (Fig. 4(d)). Through the analysis of the differences in the expression of m6A-related genes in each of the three geneClusters, it was also revealed that the expression of m6A-related genes in geneCluster B was downregulated (Fig. 4(e)). Our results suggested that the upregulation of m6A phenotype-related DEGs might indicate a better prognosis in patients with LUSC. We also conducted GSVA on the geneClusters and found that the enrichment of the pathways in geneCluster B was almost the same as that in m6Acluster B, while geneCluster A was more similar to m6Acluster C and geneCluster A was more consistent with m6Acluster C (Supplementary Figs. 3(f–h)). The characteristics of TME immune cell infiltration in the different geneClusters are shown in Fig. 4(f), with geneCluster B > geneCluster A > geneCluster C in most infiltrating immune cell subsets. The patients in geneCluster B had the highest degree of immune activation and the largest level of immune cell infiltration, and the survival prognosis was poor.
3.2.3. Construction of the M6Ascore for LUSC patients
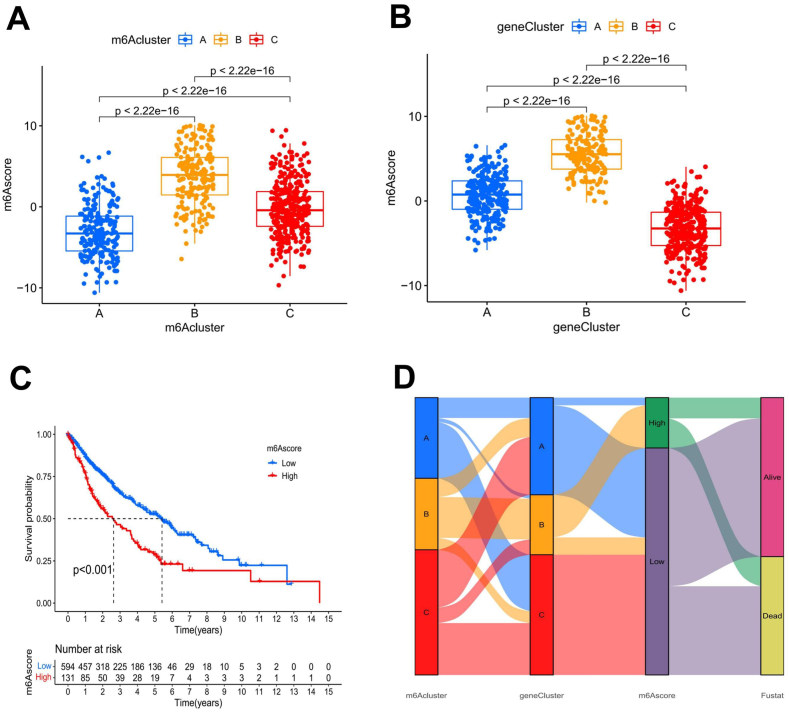
Due to the complexity and heterogeneity of m6A methylation patterns in tumor patients, we constructed the m6Ascore for LUSC patients based on the 42 m6A-related DEGs obtained above, which improved the prediction reliability of the m6A methylation pattern in individual LUSC patients. Through the Kruskal-Wallis test, we found a significant difference in the m6Ascore between patients in different m6Aclusters and different geneClusters. The results revealed that among the m6Aclusters, the median m6Ascore of m6Acluster B patients was the highest, and the median m6Ascore of m6Acluster A patients was the lowest (Fig. 5(a)). The median m6Ascore of geneCluster B patients was the highest, while the m6Ascore of geneCluster C patients was the lowest among the three geneClusters (Fig. 5(b)). Next, to study the prognostic value of the m6A modification pattern in LUSC patients, we divided LUSC patients into high and low m6Ascore groups, with 131 patients in the high m6Ascore group and 607 patients in the low m6Ascore group (Supplementary Table 5). Kaplan-Meier survival analysis was carried out for the high and low m6Ascore groups, and the results showed that the patients in the low m6Ascore group had an obvious survival advantage (p < 0.001; Fig. 5(c)). We also plotted an alluvial diagram to show the differences in the attributes of the LUSC patients in each group (Fig. 5(d)).
Fig. 5.
Construction of the m6Ascore and the correlation among m6Ascores, m6Aclusters, and geneClusters. A and B Box plots showed significant differences among m6Aclusters A, geneClusters B, and m6Ascores. C Kaplan-Meier curves were used to analyze the survival advantage among the groups of patients with LUSC. Patients with low m6Ascores had a significant survival advantage. D The alluvial diagram depicts the changes in LUSC patient characteristics based on m6Acluster, geneCluster, m6Ascore, and survival status.
3.2.4. The correlation between the M6Ascore and tumor somatic mutations
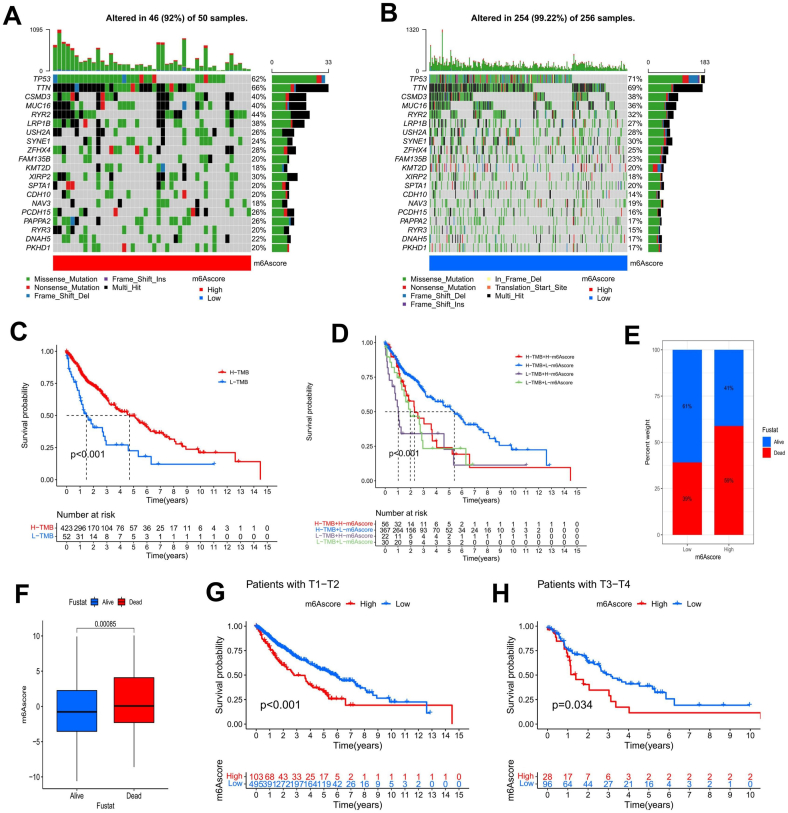
We used the maftool package in R to analyze the differences in somatic mutations in TCGA-LUSC samples stratified into different m6Ascore groups (Fig. 6(a and b)). The tumor somatic mutation rate of the high m6Ascore group was 92%, while that of the low m6Ascore group was 99.22%; the top 20 mutated genes were the same in both groups. Although we could not confirm that there was an apparent correlation between the m6Ascore and TMB, we analyzed the survival outcomes of the high and low TMB groups by Kaplan-Meier analysis and found that the high TMB group had a significant survival advantage (Fig. 6(c)). The survival analysis conducted on the TMB combined with m6Ascore suggested that the survival advantage of the low m6Ascore + high TMB group was the best, and the survival time of the high m6Ascore + low TMB group was the shortest (Fig. 6(d)). The survival advantage of patients with high TMB may be related to their high sensitivity to clinical immunotherapy.
Fig. 6.
The m6Ascore is associated with somatic mutations and clinical features. A, B Waterfall plots showing the results of tumor somatic mutation analysis in 50 patients with low m6Ascores A and 256 patients with high m6Ascores B in the TCGA-LUSC cohort. C Kaplan-Meier curves indicated that the survival of LUSC patients in the high TMB group was significantly better than that in the low TMB group. D The survival analysis results of LUSC patients combining TMB and the m6Ascore suggested that patients with a high TMB + a low m6Ascore had a significant survival advantage. E There were remarkable differences in survival and death rates between m6Ascore groups. F The comparison of the m6Ascores between surviving and nonsurviving LUSC patients showed that the difference was significant. G, H The Kaplan-Meier curves showed the difference in survival rates between the high and low m6Ascore groups with different T stages.
3.2.5. Correlation of the M6Ascore with clinical survival status and immunity
When we investigated the clinical survival status of LUSC patients in the high and low m6Ascore groups, we found that 59% of patients in the high m6Ascore group had died, while only 39% in the low m6Ascore group had died (Fig. 6(e)). There were striking differences in the m6Ascores associated with different survival states among the LUSC patients (Fig. 6(f)). We also analyzed the survival status of patients with different T stages in different m6Ascore groups and found that patients in the low m6Ascore group had significant survival advantages at both stages T1-T2 and T3-T4 (Fig. 6(g, h)).
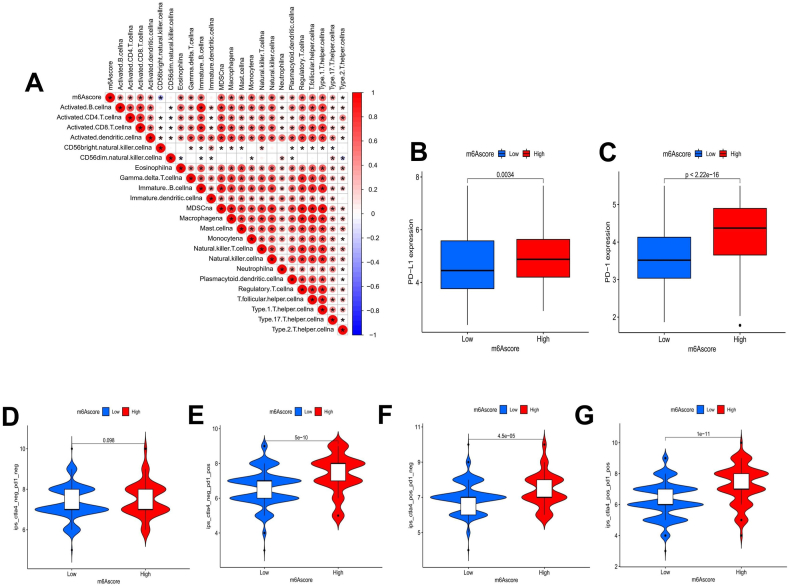
As a recent study revealed a prominent effect of ICI on cancer, we verified the correlation between the m6Ascore and immunity. We analyzed the correlation between the m6Ascore and TME immune cell infiltration and found that the m6Ascore was significantly correlated with 22 immune cell types with the exception of CD56-dim natural killer cells and immature dendritic cells. Except for a negative correlation with CD56-bright natural killer cells, the m6Ascore was positively correlated with the levels of all of the immune cells (Fig. 7(a)). We also selected a representative PD-L1/PD-1 immune checkpoint to investigate the response of LUSC patients with high and low m6Ascores to immunosuppressant therapy. The results showed that the expression of PD-L1 and PD-1 was significantly higher in the high m6Ascore group (Fig. 7(b and c)), suggesting that LUSC patients in the high m6Ascore group may have a better response to PD-L1/PD-1 blockade therapy. Then, we investigated the CTLA-4 and PD-1 blockade treatment scores of the TCGA-LUSC cohort. Whether CTLA-4 inhibitors or PD-1 inhibitors were used alone or in combination, the immunotherapy score of patients in the high m6Ascore group indicated a prominent advantage (Fig. 7(d–g)). Our results suggest that although the prognosis of LUSC patients in the high m6Ascore group is poor, this group has better ICI therapy advantages. ICI therapy may prolong the survival time of patients in the high m6Ascore group.
Fig. 7.
Correlation of the m6Ascore with immune cell infiltration and ICI therapy. A The correlation between the m6Ascore and TME-infiltrating cells was analyzed by detailed Spearman's correlation. * indicates that there was a correlation between the two endpoints. B, C The Wilcoxon signed-rank test was utilized to analyze the gene expression levels of PD-L1 B and PD-1 C in different m6Ascore groups. D-G Violin plots depicting the difference in CTLA-4/PD-1 blocking treatment scores between the high and low m6Ascore groups.
4. Discussion
Compared to lung adenocarcinoma, treatment strategies for LUSC are limited [38]. Because LUSC has a higher recurrence and metastasis rate, the prognosis is worse [39]. Therefore, exploring new biomarkers and accurate prognostic models for LUSC patients is necessary. M6A modification refers to the dynamic, reversible biological process of RNA regulation by m6A regulators. It can be activated by methyltransferase in the nucleus or removed by demethylase [6] and has the important function of regulating gene expression in malignant tumors and the innate immune system. However, to date, most studies on the mechanism of m6A modification in tumors have focused on only a single m6A regulator. In contrast, the mechanisms of the occurrence and development of tumors are very complex and involve the joint action of multiple m6A regulators. Therefore, a comprehensive understanding of the molecular and biological characteristics of multiple m6A regulators will improve our understanding of the integration of m6A modification patterns in LUSC, identify new potential prognostic biomarkers, and more effectively guide the application of clinical immunotherapy.
Previous studies have revealed that mutations in m6A regulators cause colorectal cancer susceptibility [40]. Changes in the expression of as little as 3% of the m6A regulator genes shorten the overall survival (OS) time of acute myelocytic leukemia patients [41]. Our study revealed that the gene mutation rate of m6A regulators in LUSC is 0%–3%, and differences in the gene expression levels of 13 m6A regulators (CBLL1, FMR1, FTO, HNRNPA2B1, HNRNPC, IGFBP1, IGFBP2, LRPPRC, METTL3, RBM15, WTAP, YTHDC1, and YTHDF2) have a substantial impact on the prognosis of LUSC. CNV is a variation in DNA fragments longer than 1 kb and is one of the important pathogenic factors in human diseases [42]. Our study also indicated that CNV occurred in all 21 m6A regulators, and in most m6A regulators, such as IGFBP2, LRPPRC, and HNRNPC, the expression level of the regulators was positively correlated with CNV frequency. As the CNV genome coverage may be only 5%–10%, the changes in the number of copies of YTHDF2, RMB15B, and YTHDC1 do not cause significant differences in gene expression. These findings still suggest that the roles played by CNVs of m6A regulators in the occurrence and development of LUSC must be considered.
Because the function of m6A regulators is complex and diverse, and their interactions affect several biological processes, such as mRNA splicing, translation, and expression, it is more valuable to analyze the effect of multiple m6A regulators on LUSC than to study a single m6A regulator. In the m6A regulator interaction network, all of the m6A regulators analyzed in our study had correlations with LUSC, and positive correlations outnumbered negative correlations by a large margin. FTO, CBLL1, METT3, YTHDF2, and IGFBP1 were identified in the interaction network as prognostic factors for LUSC. Li et al. [43] found that some miRNAs and genes may play different roles in 11 types of tumors, including LUSC, when FTO is knocked out or if FTO activity is low. Experimental studies have shown that FTO can promote the proliferation of LUSC cells and reduce tumor cell apoptosis by enhancing the expression of MZF1 [44]. CBLL1 can promote the proliferation of non-small cell lung cancer (NSCLC) cells by enhancing the expression of MMP2 and MMP9 [45]. Moreover, METTL3 downregulates the expression of the effector factor Cyclin D1 through the AKT signaling pathway to promote the proliferation of ovarian cancer cells [46]. METTL3 can also methylate pri-miR-1246 and downregulate the expression of the tumor suppressor gene SPRED2 to promote the metastasis of colorectal cancer [47]. YTHDF2 can regulate Akt phosphorylation and induce the proliferation and migration of prostate cancer cells by inhibiting the mRNA and protein expression of the tumor suppressor genes LHPP and NKX3-1 [48]. Interestingly, YTHDF2 destroys the stability of EGFR mRNA in hepatocellular carcinoma cells and inhibits hepatocellular carcinoma by binding to the m6A modification site EGFR-3′-UTR [49]. IGFBP1 can regulate the insulin sensitivity of leukemia, inhibit insulin secretion, and hijack patient glucose, thus accelerating the progression of leukemia [50]. Surprisingly, IGFBP1 is highly expressed in colorectal cancer, and has the dual effect of inhibiting tumor cell proliferation and promoting liver metastasis [51].
In our study, the LUSC patient samples were divided into three different m6Aclusters. m6Acluster B was enriched in systemic lupus erythematosus, the intestinal immune network for IgA production, natural killer cell-mediated cytotoxicity, antigen processing and presentation, and autoimmune thyroid disease pathways, and the number of TME immune infiltrating cells was highest in this cluster, corresponding to a state of strong immune activation. Meanwhile, m6Acluster C was enriched in antigen processing and presentation, autoimmune thyroid disease, and primary immunodeficiency pathways, and the number of immune infiltrating cells was moderate, corresponding to moderate immune activation. However, m6Acluster A was enriched in the melanoma, basal cell carcinoma, and renal cell carcinoma pathways, and the number of infiltrating immune cells was the lowest in this group, corresponding to the immunosuppressive phenotype and carcinogenic status. Accordingly, it was inferred that the high expression of the immune activation pathway in m6Acluster B might lead to the infiltration of a large number of immune cells, which was consistent with previous findings that immune activators induced the proliferation, activation, and cytotoxicity of dendritic cells, CD8+ T cells, and other immune cells in TME [52,53]. Consistent with the findings of previous studies [[54], [55], [56], [57]], we also found that the m6A phenotype-related genes were significantly related to ECM-receptor interaction, receptor-ligand activity, basal cell carcinoma, gastric cancer, antigen processing and presentation, primary immunodeficiency and other functions of cancer, m6A modification, and immune regulation. Hence, m6A modification patterns play an essential role in the TME and in immune regulation in LUSC [58,59]. These results also provide clues to further explore the potential mechanisms of m6A regulators in the TME.
Given the apparent individual heterogeneity in m6A modifications in tumor patients, it is essential to quantify the m6A modifications. This m6Ascore has been confirmed in m6A-related gastric cancer studies [60]. We established the m6Ascore to predict the prognosis of patients with LUSC, which avoided errors resulting from patient tumor heterogeneity. In our signature, there were 42 m6A phenotype-related DEGs, which were independent and effective prognostic factors for LUSC patients, among which ZNF703, FAM162A, LOM4, and APTR were significantly differentially expressed (p < 0.001). The overexpression of ZNF703 is associated with many types of cancer, and its expression is significantly upregulated in NSCLC and medullary thyroid carcinoma, which may be related to the activation of the Akt/mTOR pathway [61,62]. The overexpression of ZNF703 in oral squamous cell carcinoma activates PI3K/Akt/GSK-3β signaling, promoting the proliferation and migration of oral squamous cell carcinoma [63]. APTR regulates lncRNA APTR, trans-acting on the effector of tumor suppressor p53, CDKN1A/p21, and lncRNA APTR also recruits polycomb inhibitory complex 2 (PRC2) to inhibit the p21 promoter, thus promoting the proliferation and invasion of glioblastoma cells [64,65]. The role of FAM162A and LOM4 in cancer is less well understood. Nevertheless, these genes may be potential prognostic biomarkers for LUSC patients.
Our data also exhibited the correlation of m6Ascore with TMB, clinical survival status, immune cell infiltration, and response to ICI therapy, which verified the prognostic value of the m6Ascore in LUSC. In the survival analysis of the high and low TMB groups, the survival status of LUSC patients with high TMB showed obvious superiority. This result is perhaps owing to the fact that tumor somatic cell mutation increases the immunogenicity of somatic cells [66] and improves the sensitivity of tumors to ICI [67]. Previous studies have also found that high TMB continuous selection has become a biomarker of ICI [68,69]. The immune system plays a role in destroying and monitoring the cells of cancer patients. Unfortunately, tumor cells can use immune checkpoints to escape the immune response. Thus, the therapeutic effects of ICI therapy for cancer cannot be ignored. A number of studies have demonstrated that ICI targeting PD-L1 and PD-1 can prolong the OS rate of patients with hepatocellular carcinoma, colon cancer, melanoma, NSCLC, and other cancers [[70], [71], [72], [73]]. Our data indicated that the treatment effect of CTLA-4/PD-1 inhibitors was significantly better in patients with a high m6Ascore. This may be because most patients in the high m6Ascore group were in m6Acluster B and geneCluster B, both associated with strong immune activation. Through our m6Ascore, LUSC patients more sensitive to CTLA-4/PD-1 inhibitors can be screened. Therefore, the m6Ascore represents a new contribution to developing individualized immunotherapy for tumor patients.
5. Conclusion
In recent years, the study of prognostic signatures related to m6A modification has become a hot spot. Zhou et al. established a signature of seven m6A phenotype-related DEGs, which can predict the prognosis and the expression of immune checkpoint-related molecules in patients with lung adenocarcinoma [74]. In addition, the expression level of SNRPC of the m6A methylation regulator proposed by Cai et al. may be a potential indicator of prognosis and anti-CTLA-4/PD-1 immunotherapy for hepatocellular carcinoma [75]. We established a prognostic signature of LUSC based on 42 m6A phenotype-related DEGs, which showed application value in TME and anti-CTLA-4/PD-1 treatment.
Compared with other studies [58,59,[76], [77], [78], [79]], our study still has many shortcomings, such as the delayed data update in the TCGA-LUSC and GSE157010 datasets we adopted, the inconsistent sample sequencing techniques and quality control methods in the two datasets, and the vast majority of patients in the samples were from North America, which could not represent patients from other regions. We also lack external experimental and clinical validation to confirm the reliability of the built m6Ascore. However, by combining multi-center LUSC sample data, converting the annotation format of the gene files, and verifying the prediction reliability of the m6Ascore from multiple perspectives, we minimized the error to the greatest extent and more comprehensively combined clinical, immune and tumor somatic mutation analysis. In conclusion, by studying the molecular biological processes, gene mutations, and characteristics of TME immune cell infiltration related to m6A modifications in LUSC, we determined that m6A modification abnormalities are closely related to the immune phenotype and play a key role in LUSC. Then, we constructed the m6Ascore to predict the prognosis of LUSC patients and verified the predictive ability of the m6Ascore by analyzing immune cell infiltration, TMB, clinical survival status, and response to CTLA-4/PD-1 inhibitor therapy. These findings indicate that the m6Ascore is reliable for predicting the prognosis of LUSC patients. Because the patients in different m6Ascore groups had different sensitivities to CTLA-4/PD-1 inhibitor therapy, the m6Ascore combined with CTLA-4/PD-1 immunotherapy score offers a new basis for personalized diagnosis and the determination of the potential immune-targeted treatment approaches for LUSC patients. Notably, it was also found for the first time that m6A phenotype-related genes FAM162A and LOM4 might be potential biomarkers of LUSC, which has profound implications for the identification of new diagnostic markers and the development of new anticancer drugs for LUSC. Nevertheless, further experiments are needed to verify the signature established in our study and unravel the molecular mechanisms of m6A RNA modification in LUSC and other types of malignant tumors. Subsequent cellular experiments are required to investigate the relationship between the m6A regulator and the phenotype genes FAM162A and LOM4, together with animal and clinical studies to confirm FAM162A and LOM4 expression and biological functions.
Funding statement
This work was supported by a grant from The National Natural Science Foundation of China (No. 82074327).
Ethics approval
The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) database are publicly available for research, thus ethical approval is not required.
Data availability statement
The datasets analyzed in this study are available from the corresponding authors on request.
CRediT authorship contribution statement
Pei Li: Writing – original draft, Formal analysis, Data curation. Peiyu Xiong: Writing – review & editing, Writing – original draft, Formal analysis. Xinyun Li: Formal analysis, Data curation. Xiaobo Zhang: Formal analysis. Xu Chen: Visualization. Wei Zhang: Visualization. Bo Jia: Writing – review & editing. Yu Lai: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are thankful to all patients who participated in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e26851.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Barta J.A., Powell C.A., Wisnivesky J.P. Global epidemiology of lung cancer. Annals of Global Health. 2019;85:1–16. doi: 10.5334/aogh.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen S., Giannakou A., Wyman S., et al. Cancer-associated fibroblasts suppress SOX2-induced dysplasia in a lung squamous cancer coculture. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E11671–E11680. doi: 10.1073/pnas.1803718115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao J., Wang H., Song L., et al. Infiltration of CD8+ FOXP3+ T cells, CD8+ T cells, and FOXP3+ T cells in non-small cell lung cancer microenvironment. Int. J. Clin. Exp. Pathol. 2020;13:880–888. [PMC free article] [PubMed] [Google Scholar]
- 4.Li L., Liu Y.D., Zhan Y.T., et al. High levels of CCL2 or CCL4 in the tumor microenvironment predict unfavorable survival in lung adenocarcinoma. Thoracic Cancer. 2018;9:775–784. doi: 10.1111/1759-7714.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan H., Liu J., Zhang J. The current landscape of immune checkpoint blockade in metastatic lung squamous cell carcinoma. Molecules. 2021;26:1392. doi: 10.3390/molecules26051392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y., Hsu P.J., Chen Y.-S., Yang Y.-G. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fustin J.-M., Doi M., Yamaguchi Y. RNA-Methylation-Dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 8.Patil D.P., Chen C.-K., Pickering B.F., et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu R., Liu Y., Yao Y., et al. FTO regulates adipogenesis by controlling cell cycle progression via m6A-YTHDF2 dependent mechanism. BBA - Molecular and Cell Biology of Lipids. 2018;1863:1323–1330. doi: 10.1016/j.bbalip.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Liu J., Zhang X., Chen K., et al. CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-Mediated glycolysis. Immunity. 2019;50:600. doi: 10.1016/j.immuni.2019.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Zhang S., Zhao B.S., Zhou A., et al. A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. (e596) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui Q., Shi H., Ye P., et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le D.T., Durham J.N., Smith K.N., et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y., Hu H., Wang Y., et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell. Physiol. Biochem. 2018;48:838–846. doi: 10.1159/000491915. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z., Li J., Feng G., et al. MicroRNA-145 Modulates N6-methyladenosine levels by targeting the 3'-untranslated mRNA Region of the N6-Methyladenosine Binding YTH domain family 2 protein. J. Biol. Chem. 2017;292:3614–3623. doi: 10.1074/jbc.M116.749689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., Weng H., Su R., et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su R., Dong L., Li C., et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. 2018;172:90. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., Eckert M.A., Harada B.T., et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H.-B., Tong J., Zhu S., et al. M6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang B., Chunxing Y., Runliu W., et al. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front. Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu X., Zhang Y., Li D., et al. N6-methyladenosine demethylase FTO promotes M1 and M2 macrophage activation. Cell. Signal. 2020;69 doi: 10.1016/j.cellsig.2020.109553. [DOI] [PubMed] [Google Scholar]
- 22.Wang H., Hu X., Huang M., et al. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat. Commun. 2019;10:1898. doi: 10.1038/s41467-019-09903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song P., Zhang J.C., Shang C.C., Zhang L. Curative effect assessment of immunotherapy for non‐small cell lung cancer: the "blind area" of Immune Response Evaluation Criteria in Solid Tumors (iRECIST) Thoracic Cancer. 2019;10:587–592. doi: 10.1111/1759-7714.13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu C., Shi X., Qiu W., et al. Comprehensive analysis of the prognostic role and mutational characteristics of m6A-related genes in lung squamous cell carcinoma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.661792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arguello A.E., DeLiberto A.N., Kleiner R.E. RNA chemical proteomics reveals the N-6-Methyladenosine (m(6)A)-regulated protein-RNA interactome. J. Am. Chem. Soc. 2017;139:17249–17252. doi: 10.1021/jacs.7b09213. [DOI] [PubMed] [Google Scholar]
- 26.Chen X.-Y., Zhang J., Zhu J.-S. The role of m6A RNA methylation in human cancer. Molecular Cancer. 2019;vol. 18 doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frankiw L., Baltimore D., Li G. Alternative mRNA splicing in cancer immunotherapy. Immunology. 2019;19:675–687. doi: 10.1038/s41577-019-0195-7. [DOI] [PubMed] [Google Scholar]
- 28.Liu S., Li Q., Chen K., et al. The emerging molecular mechanism of m6A modulators in tumorigenesis and cancer progression. Biomed. Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110098. [DOI] [PubMed] [Google Scholar]
- 29.Niu Y., Zhao X., Wu Y.-S., et al. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Dev. Reprod. Biol. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Z., Lv J., Yu H., et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol. Cancer. 2020;19:1–20. doi: 10.1186/s12943-020-01216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartigan J.A., Wong M.A. A K-means clustering algorithm: algorithm AS 136. Applied Statistics. 1979;28:100–108. [Google Scholar]
- 32.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charoentong P., Finotello F., Angelova M., et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 34.Sotiriou C., Wirapati P., Loi S., et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 35.Zeng D.Q., Li M.Y., Zhou R., et al. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol. Res. 2019;7:737–750. doi: 10.1158/2326-6066.CIR-18-0436. [DOI] [PubMed] [Google Scholar]
- 36.Hazra A., Gogtay N. Biostatistics series module 3: comparing groups: numerical variables. Indian J. Dermatol. 2016;61:251–260. doi: 10.4103/0019-5154.182416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayakonda A., Lin D.-C., Assenov Y., et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yaru T., Xiaoyang Z., Weiwei Y., et al. Clinical outcomes of immune checkpoint blockades and the underlying immune escape mechanisms in squamous and adenocarcinoma NSCLC. %J Cancer medicine. 2020;10 doi: 10.1002/cam4.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q., Longlong L., Yufeng D., et al. MicroRNA-588 suppresses tumor cell migration and invasion by targeting GRN in lung squamous cell carcinoma. %J Molecular medicine reports. 2016;14:3021–3028. doi: 10.3892/mmr.2016.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwok C., Marshall A.D., Rasko J.E., Wong J.J. Erratum to: genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J. Hematol. Oncol. 2017;10:49. doi: 10.1186/s13045-017-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng Y., Li S., Gu D., et al. Genetic variants in m6A modification genes are associated with colorectal cancer risk. Carcinogenesis. 2020;41:8–17. doi: 10.1093/carcin/bgz165. [DOI] [PubMed] [Google Scholar]
- 42.Zarrei M., MacDonald J.R., Merico D., Scherer S.W. A copy number variation map of the human genome. Springer Nature. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 43.Li M.Y., Ma C.X., Wu H.X., et al. Prediction of target genes has-miR-22-3p and has-miR-671-5p and their relationship to survival rate of patients with cancer. Chinese Journal of Biologicals. 2018;31:591–597. [Google Scholar]
- 44.Liu J., Ren D., Du Z., et al. m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem. Biophys. Res. Commun. 2018;502:456–464. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 45.Hui L., Zhang S., Wudu M., et al. CBLL1 is highly expressed in non‐small cell lung cancer and promotes cell proliferation and invasion. Thoracic Cancer. 2019;10:1479–1488. doi: 10.1111/1759-7714.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang S., Guan H., Lin X., et al. METTL3 serves an oncogenic role in human ovarian cancer cells partially via the AKT signaling pathway. Oncol. Lett. 2020;19:3197–3204. doi: 10.3892/ol.2020.11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng W., Li J., Chen R., et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:393. doi: 10.1186/s13046-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo L.-H., Rao L., Luo L.-F., et al. Long non-coding RNA NKILA inhibited angiogenesis of breast cancer through NF-κB/IL-6 signaling pathway. Microvasc. Res. 2020;129 doi: 10.1016/j.mvr.2019.103968. [DOI] [PubMed] [Google Scholar]
- 49.Zhong L., Liao D., Zhang M., et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 50.Ye H., Adane B., Khan N., et al. Subversion of systemic glucose metabolism as a mechanism to support the growth of leukemia cells. Cancer Cell. 2018;34:659–673. doi: 10.1016/j.ccell.2018.08.016. (e656) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J.C., Ha Y.J., Tak K.H., et al. Complex behavior of ALDH1A1 and IGFBP1 in liver metastasis of colorectal cancer. Cancer Res. 2016;76:1641. doi: 10.1371/journal.pone.0155160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sadeghlar F., Vogt A., Mohr R.U., et al. Induction of cytotoxic effector cells towards cholangiocellular, pancreatic, and colorectal tumor cells by activation of the immune checkpoint CD40/CD40L on dendritic cells Cancer Immunology. Immunotherapy. 2021;70:1451–1464. doi: 10.1007/s00262-020-02746-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hedlund G., Dohlsten M., Petersson C., Kalland T. Superantigen-based tumor therapy: in vivo activation of cytotoxic T cells Cancer Immunology. Immunotherapy. 1993;36:89–93. doi: 10.1007/BF01754407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge L., Zhang N., Chen Z., et al. Level of N6-Methyladenosine in peripheral blood RNA: a novel predictive biomarker for gastric cancer. Clin. Chem. 2020;66:342–351. doi: 10.1093/clinchem/hvz004. [DOI] [PubMed] [Google Scholar]
- 55.Kim G.-W., Siddiqui A. N6-methyladenosine modification of HCV RNA genome regulates cap-independent IRES-mediated translation via YTHDC2 recognition. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2022024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogawa A., Nagiri C., Shihoya W., et al. N6-methyladenosine (m6A) is an endogenous A3 adenosine receptor ligand. Mol. Cell. 2021;81:659–674. doi: 10.1016/j.molcel.2020.12.038. (e657) [DOI] [PubMed] [Google Scholar]
- 57.Zeng Y., Huang T., Zuo W., et al. Integrated analysis of m6A mRNA methylation in rats with monocrotaline-induced pulmonary arterial hypertension. Aging. 2021;13:18238–18256. doi: 10.18632/aging.203230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chengyin W., Lina W., Guolong L., et al. Identification of a N6-methyladenosine (m6A)-Related lncRNA signature for predicting the prognosis and immune landscape of lung squamous cell carcinoma. J Frontiers in Oncology. 2021;11:763027. doi: 10.3389/fonc.2021.763027. 763027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Z., Xuhui G., Shuncong W., et al. N6-Methyladenosine (m6A)-Related lncRNAs are potential signatures for predicting prognosis and immune response in lung squamous cell carcinoma. J Journal of Oncology. 2022:5240611. doi: 10.1155/2022/5240611. 5240611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang B., Wu Q., Li B., et al. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer. 2020;19:53. doi: 10.1186/s12943-020-01170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baykara O., Dalay N., Kaynak K., Buyru N. ZNF703 overexpression may act as an oncogene in non-small cell lung cancer. Cancer Med. 2016;5:2873–2878. doi: 10.1002/cam4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang X., Liu G., Li W., et al. Silencing of zinc finger protein 703 inhibits medullary thyroid carcinoma cell proliferation in vitro and in vivo. Oncol. Lett. 2020;19:943–951. doi: 10.3892/ol.2019.11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H., Deng X.B., Zhang J.S., et al. Elevated expression of Zinc finger protein 703 promotes cell proliferation and metastasis through PI3K/AKT/GSK-3β signalling in oral squamous cell carcinoma. Cell. Physiol. Biochem: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;44:920–934. doi: 10.1159/000485360. [DOI] [PubMed] [Google Scholar]
- 64.Negishi M., Wongpalee S.P., Sarkar S., et al. A new lncRNA, APTR, associates with and represses the CDKN1A/p21 promoter by recruiting polycomb proteins. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang S.-S., Yu L. The potential effect of APTR amplification on glioma patients' prognosis. Chinese Journal of Contemporary Neurology & Neurosurgery. 2019;19:507–513. [Google Scholar]
- 66.Rizvi N., Hellmann M., Snyder A., et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yarchoan M., Hopkins A., Jaffee E.M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chumsri S., Sokol E.S., Soyano-Muller A.E., et al. Durable complete response with immune checkpoint inhibitor in breast cancer with high tumor mutational burden and APOBEC signature. J. Natl. Compr. Cancer Netw. 2020;18:517–521. doi: 10.6004/jnccn.2020.7543. [DOI] [PubMed] [Google Scholar]
- 69.Chan T.A., Yarchoan M., Jaffee E., et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann. Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.El-Khoueiry A.B., Sangro B., Yau T., et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goldberg S.B., Gettinger S.N., Mahajan A., et al. Pembrolizumab for patients with melanoma or non-small-cell lung cancer and untreated brain metastases: early analysis of a non-randomised, open-label, phase 2 trial. Lancet Oncol. 2016;17:976–983. doi: 10.1016/S1470-2045(16)30053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morse M.A., Overman M.J., Hartman L., et al. Safety of nivolumab plus low‐dose ipilimumab in previously treated microsatellite instability‐high/mismatch repair‐deficient metastatic colorectal cancer. Oncol. 2019;24:1453–1461. doi: 10.1634/theoncologist.2019-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Overman M.J., McDermott R., Leach J.L., et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou H., Zheng M., Shi M., et al. Characteristic of molecular subtypes in lung adenocarcinoma based on m6A RNA methylation modification and immune microenvironment. BMC Cancer. 2021;21:938. doi: 10.1186/s12885-021-08655-1. 938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cai J., Zhou M., Xu J. N6-methyladenosine (m6A) RNA methylation regulator SNRPC is a prognostic biomarker and is correlated with immunotherapy in hepatocellular carcinoma. World J. Surg. Oncol. 2021;19:241. doi: 10.1186/s12957-021-02354-8. 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gu C., Shi X., Qiu W., et al. Comprehensive analysis of the prognostic role and mutational characteristics of m6A-related genes in lung squamous cell carcinoma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.661792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hao J., Wang H., Song L., et al. Infiltration of CD8(+) FOXP3(+) T cells, CD8(+) T cells, and FOXP3(+) T cells in non-small cell lung cancer microenvironment. Int. J. Clin. Exp. Pathol. 2020;13:880–888. [PMC free article] [PubMed] [Google Scholar]
- 78.Huang H., Wu W., Lu Y., Pan X. The development and validation of a m6A-lncRNAs based prognostic model for overall survival in lung squamous cell carcinoma. J. Thorac. Dis. 2022;14:4055–4072. doi: 10.21037/jtd-22-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang W., Zhang Q., Xie Z., et al. N (6) -Methyladenosine-Related long non-coding RNAs are identified as a potential prognostic biomarker for lung squamous cell carcinoma and validated by real-time PCR. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.839957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed in this study are available from the corresponding authors on request.