Abstract
Cellular levels of the rapidly degraded c-myc protein play an important role in determining the proliferation status of cells. Increased levels of c-myc are frequently associated with rapidly proliferating tumor cells. We show here that myc boxes I and II, found in the N termini of all members of the myc protein family, function to direct the degradation of the c-myc protein. Both myc boxes I and II contain sufficient information to independently direct the degradation of otherwise stably expressed proteins to which they are fused. At least part of the myc box-directed degradation occurs via the proteasome. The mechanism of myc box-directed degradation appears to be conserved between yeast and mammalian cells. Our results suggest that the myc boxes may play an important role in regulating the level and activity of the c-myc protein.
The c-myc protein is a short-lived, nuclear phosphoprotein that has a role as a regulator of several biological processes, including cell proliferation and apoptosis. Elevation in the levels of c-myc is a widespread phenomenon in a large variety of tumors from a range of species. Studies of c-myc proteins in tumor cells led to the proposal that they are involved in the transcriptional control of genes required for cellular replication (5, 30). The c-myc protein has motifs that are characteristic of transcription factors, the leucine zipper and basic helix-loop-helix (bHLHZip) dimerization and DNA binding domains (20, 21, 48, 58). To become an active transactivator, c-myc dimerizes with another bHLHZip protein, max (3, 4, 12). Blackwell et al. (11) showed that c-myc recognizes the DNA sequence CACGTG, a motif which is present in various target promoters. The genes encoding α-prothomyosin (22), PAI-1 (57), ornithine decarboxylase (74), ECA39 (9), eIF-4E (37), Cdc25 (23), rcl (45), and MrDb (26) have been demonstrated to be activated by c-myc. In addition, c-myc is able to repress transcription from the adenovirus major late promoter (46) and from the promoters for c-EBPα (16), albumin (25), cyclin D (54), and gadd45 (50).
It has previously been shown that c-myc can function as a transactivator in Saccharomyces cerevisiae (3). Expression of c-myc and its dimerization partner max led to activation of a lacZ reporter gene with a CACGTG site in the promoter. In addition, Amati et al. (3) and Lech et al. (43) demonstrated that the N-terminal domain of c-myc functions as a transactivator in yeast when fused to a heterologous DNA binding domain (DBD) from serum response factor or LexA. In mammalian cells, a c-myc–GAL4 DBD chimera has been used to define an N-terminal transactivation domain of 143 amino acids (38), and the same region is a functional transactivator in yeast (51).
Identification and characterization of the N-myc, s-myc, L-myc, and B-myc proteins showed that there is a family of myc proteins that have highly homologous regions (6, 39, 44, 68, 70). Two of these regions lie within the transactivation domain and have been termed myc homology box I (MBI) and myc homology box II (MBII). MBI and MBII in human c-myc are located between amino acids 45 and 65 and amino acids 128 to 144, respectively. Both MBI and MBII are 100% homologous in human and chicken c-myc and 77 to 95% homologous when c-myc and N-myc are compared (64). MBII is important for the transforming activity of c-myc, and it has been implicated in the transcriptional repression function of the c-myc protein (46, 53). In addition, Brough et al. (15) showed that MBII forms part of the binding site for an as yet uncharacterized nuclear factor and proposed that MBII is important for specific DNA binding. The role of MBI is less clear. It contains the Thr-58 and Ser-62 phosphorylation sites, which have been reported to play a role in transactivation and transformation, but different conclusions have been drawn on the basis of results in different experimental systems (28, 32, 59). MBI is also present in protein segments that have been shown to bind to p107 (27), TBP (31, 51), and α-tubulin (2). Deletion of MBI had a deleterious effect on transforming ability (64), but a role in transactivation and/or repression has not been tested. Sequence analysis of the c-myc proteins found in cell lines derived from Burkitt’s lymphoma patients showed that mutation hot spots were located in MBI at positions 52 to 63 (1, 10). The mutated residues included the two phosphorylation sites in MBI (Thr-58 and Ser-62).
We have been investigating the N-terminal transactivation domain of c-myc by using a yeast-based assay system. A series of deletions within the transactivation domain were made, and during their analysis we noticed a significant difference in the levels of the various proteins on Western blots. In this report, we show that MBI and MBII destabilize proteins which contain them in both yeast and mammalian cells and that they target proteins for degradation via a mechanism involving the proteasome.
MATERIALS AND METHODS
Yeast strains and media.
The S. cerevisiae strain used for the investigation of the c-myc–Pho4 fusion protein was YS33 (MATa his3-11,15 leu2-3,112 ura3-Δ5 Δpho80::HIS3 Δpho4::ura3-Δ5 Canr) (71). Pho80 is a high-phosphate-dependent repressor of Pho4; by deleting it, the requirement for low-phosphate conditions for Pho4 activity is avoided. The full length c-myc and max genes were expressed in W303-1A (MATa ade2-1o can1-100o his3-11,15 leu2-3,112 trp1-1a ura3-1) and in BJ2168 (MATa ura3-52 leu2 trp1 prb1,1122 pep4-3 prc1-407 gal2-1; Yeast Genetic Stock Center). The proteasome mutants were obtained from Wolfgang Hilt (Stuttgart, Germany), YHI29/W is the wild type (MATa ura3 leu2-3,112 his3-11,5), and YHI29/14 is defective in protein degradation (MATα ura3 leu2-3,112 his3-11,5 pre1-1 pre4-1). Yeast transformations were performed by the method of Ito et al. (35). The selective yeast medium was either SD-LUW (0.67% [wt/vol] yeast nitrogen base without amino acids [Bio 101], 2% [wt/vol] glucose [Sigma], 0.2% [wt/vol] Drop Out mix [62] without tryptophan, uracil, and leucine) or SDGAL-LUW (same as SD-LUW but with glucose replaced by 2% [wt/vol] galactose [Sigma] and 2% [wt/vol] raffinose [Sigma]). For the gal2-1 strain, BJ2168, the galactose concentration was increased to 8% (wt/vol).
Plasmids and cloning.
For general cloning, E. coli XL-1blue was used (Stratagene). Transformation was carried out by electroporation (GenePulser; Bio-Rad) as specified by the manufacturer. Plasmid pP472S (2μm URA3 Ampr) contains the 1.1-kb AvaI PHO4 fragment (Pho4DBD) and the PHO4 promoter and is a mutated form of YEpΔ2 (71). The original SacI site was deleted, and a new SacI site was added upstream of the sequence encoding the Pho4 DBD and downstream of the PHO4 promoter. All c-myc N-terminal mutants were designed to have SacI ends and could be cloned in frame with the PHO4 sequences. Plasmid YCpΔ2 (ARS CEN URA3 Ampr) is a low-copy-number version of YEpΔ2 (71). The SacI site change has not been made in this plasmid, but the myc mutants could be excised from pP472S with EcoRI and BamHI and cloned into YCpΔ2 to be in frame with the PHO4 sequences there. Expression of the full-length c-myc and max genes was controlled by the GAL1/10 promoter in plasmids pSDmyc (ARS CEN TRP1 Ampr) (3) and pRSmax9 (ARS CEN LEU2 Ampr) (19), respectively. All four expression plasmids have f1+ origins for production of single-stranded template DNA. Plasmid pKVlac (2μm LEU2d Ampr) contains the lacZ gene cloned downstream of a GAL/PGK promoter in pKV50 (Delta Biotechnology Ltd., Nottingham, United Kingdom). The mammalian expression plasmid pCMVτ1cGRDBD (Ampr) contains sequences encoding τ1core (τ1c) of the glucocorticoid receptor (GR) fused to the DBD sequences from the GR. pCMVτ1cMBGRDBD (Ampr) is the same as pCMVτ1cGRDBD but with sequences encoding the two myc boxes inserted on either side of and in frame with the τ1core sequences (τ1cMB). The mammalian vector pcDNA3.1V5-HISA (Ampr; Invitrogen) was used for expression of full-length myc genes with (pcDNA3myc) and without (pcDNA3mycΔ) the myc boxes. Expression was controlled by the cytomegalovirus promoter. The proteins produced have the V5 antigen and His tag at the C terminus.
Oligonucleotides for PCR and deletion mutagenesis.
The SacI PCR-generated N- and C-terminal deletion mutations were produced in standard PCRs with VENT polymerase (New England Biolabs). A full-length human c-myc clone was used as a template, and the primers were as follows (the name indicates whether it is an N- or C-terminal primer and the first amino acid encoded), with the SacI sites highlighted: N1, 5′GCGATAGAGCTCGATGCCCCTCAACGTTAGCTTCAC3′; N41, 5′CGCAGCGAGCTCTCAGCCCCCGGCGCCCAGCGA3′; N66, 5′CCGGGGGAGCTCTCGCTCCGGGCTCTGCTCGCCCT3′; N94, 5′CGCAGCGAGCTCGTCCACGGCCGACCAGCTGGAG3′; C41, 5′CGCGGCGAGCTCTGCAGCTCGCTCTGCTGCTGC3′; C127, 5′CGCGGCGAGCTCCGGTTTTTGATGAAGGTCTCGTCGTC3′; and C149, 5′CGAGACGAGCTCCGCAGCTTCTCTGAGACGAGCTTG3′. Following purification and SacI digestion, the PCR products were cloned into pP472S. All the PCR products were sequenced (Sequenase, United States Biochemicals) to check for errors.
Internal deletions of regions 41 to 66, 66 to 127, and 127 to 149 (full-length myc only) were made by using single oligonucleotides spanning the deletion site. Mutagenesis was carried out as described by Kunkel et al. (41), with the conditions for annealing as follows: single-stranded template DNA and the appropriate oligonucleotide were heated to 75°C for 5 min and then transferred immediately to 37°C for 30 min. The extension reaction was allowed to proceed at 37°C for 90 to 120 min. The oligonucleotides were designed with two 15-bp “clamps” homologous to regions 5′ and 3′ of the deletion site, and a BglII site was inserted at the point of deletion to act as a diagnostic restriction site. The oligonucleotides had the following sequences (the name indicates the region deleted), with the BglII site highlighted: 41–66, 5′GCAGAGCGAGCTGAGATCTTCCGGGCTCTGCT3′; 66–127, 5′CCCTAGCCGCAGATCTAACATCATCATCCAGG3′; and 127–149, 5′CGAGACCTTCATCAAAAGATCTGCTCCTACCAGG3′. All the oligonucleotides were synthesized by CyberGene AB (Sweden). Mutant clones were isolated by PCR screening for reduced insert size and by the presence of the BglII site. Positive clones were sequenced to check the deletion site.
The τ1cMB construct was produced by cloning a PCR-amplified τ1core sequence between the two myc boxes in the construct Δ1–41/66–127P. There is a BglII site between the myc boxes, and the τ1core PCR primers were designed with BglII and SacI sites at the 5′ and 3′ ends. The τ1cMBI and τ1cMBII constructs were produced from τ1cMB by using appropriate pairs of primers (τ1N and C149 or N41 and τ1C).
The τ1core primers had the following sequences, with SacI and BglII sites highlighted: τ1N, 5′CGAGACGAGCTCGAGATCTGACCAAAGCACCTTTGACATTTTGC3′; and τ1C, 5′CGCGGCGAGCTCCGAGATCTGTCCTCATTCGAGTTTCCTTCC3′. The full-length myc genes for mammalian expression were amplified from the yeast pSD plasmids by using Pfu polymerase (Stratagene) under standard PCR conditions. The PCR primers were designed with XhoI or HindIII restriction sites to allow cloning into pcDNA3.1V5-HISA. The primers had the following sequences (restriction sites highlighted): FN1, 5′CGCAGCAAGCTTCGATGCCCCTCAACGTTAGCTTCACC3′; and F439C, 5′CGAGACCTCGAGCGCACAAGAGTTCCGTAGCTGTTC3′.
SDS-PAGE and Western blotting.
Yeast extracts were prepared by using the “rapid” protein extraction procedure described by Horvath and Riezman (34). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with 12.5% (myc-Pho4) or 7.5% (full length c-myc) polyacrylamide resolving gels (102 by 72 by 0.75 mm). Proteins were transferred electrophoretically to Hybond C-super membrane (Amersham). The membranes were blocked in 5% milk, and proteins were detected with primary antibody, secondary anti-rabbit or anti-mouse immunoglobulin G (IgG)-horseradish peroxidase (HRP) conjugates and chemiluminescence with an enhanced chemiluminescence cocktail (68 mM p-coumaric acid in dimethyl sulfoxide, 1.25 mM luminol in Tris [pH 8.5], 0.009% H2O2). Primary antibodies were raised against Pho4 DBD in rabbits (antibodies obtained from Colin Gooding) or against a C-terminal c-myc-specific epitope (408–438, 9E10) in mice (Genosys Biotechnologies Inc.).
RNA extraction and Northern blotting.
Total RNA was extracted from yeast transformant cells grown to mid-log phase under conditions where c-myc expression was induced. RNA was prepared as described by Schmitt et al. (66), run on denaturing morpholinepropanesulfonic acid (MOPS) gels, and blotted onto a Hybond-N nylon membrane (Amersham). Prehybridization and hybridization were carried out at 65°C in 7% (wt/vol) SDS–0.5 M Na2HPO4 (pH 7.2)–1 mM EDTA for 20 h. The membrane was washed four times at 65°C in 5% (wt/vol) SDS–40 mM NaH2PO4 (pH 7.2)–1 mM EDTA (17). The probes for hybridization were [α-32P]dCTP-labelled BamHI-XhoI Pho4 DBD and HindIII-BamHI actin fragments. The relative amounts of each mRNA detected were calculated after phosphorimager analysis of the hybridization intensity (Fujix BAS 2000). The actin levels were used as a loading control, and the levels of the myc-Pho4 mRNAs were adjusted to correct for loading differences. The mRNA level measured for the 1–149P construct was given an arbitrary value of 1, and the levels of the other mRNAs were compared to this.
Estimation of protein degradation rates.
Cultures of W303-1A or BJ2168 carrying pRSmax9 and either pSDmyc or pSDmycΔMBI+II plasmids were grown in selective galactose medium at 30°C until the cells reached a steady growth rate. Glucose was then added to a final concentration of 5%, and the incubation was continued. Samples were removed at measured time intervals, and total protein extracts and Western blotting were performed as described above. The optical density at 600 nm (OD600) of all samples was measured and the volume of culture was adjusted to ensure that equal amounts of cells were used in each extract. The intensity of each band on the Western blot was determined with GelPro image analysis software (Media Cybernetics). Half-time values for each protein were calculated from the measured, integrated optical density of each band, expressed as a percentage (%IOD) plotted against time or the log %IOD plotted against time.
Transient transfections and proteolysis inhibition.
COS7 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and antibiotics. The cells were transfected with 2.5 μg of expression plasmid and 30 μg of the liposomal transfection reagent DOTAP (Boehringer Mannheim) in 60-mm plates for 14 h and then incubated in fresh medium for a further 30 h. For protease inhibition, the inhibitor carboxybenzyl-leucyl-leucyl-leucine vinyl sulfane (Z-L3 VS [13]), leupeptin (Sigma), or pepstatin (Sigma) was added to the plates, at a final concentration of 10 μM, 4 h before the cell extracts were made. Equal amounts of total protein from whole-cell extracts were analyzed by SDS-PAGE on 20% polyacrylamide gels. Western blotting was performed as described above, and the τ1c and τ1cMB proteins were detected with a monoclonal antibody to the GR-DBD (Ann-Charlotte Wikström and Marika Rönnholm, Karolinska Institute). The pcDNA3myc and pcDNA3mycΔ constructs were transfected into COS7 cells with FuGENE transfection reagent (Boehringer Mannheim) as specified by the manufacturer. The cells were incubated with the FuGENE-DNA mix for 36 h before extracts were made. Proteins were detected on Western blots with an antibody to the V5 epitope (Invitrogen).
RESULTS
Role of myc boxes in determining protein levels.
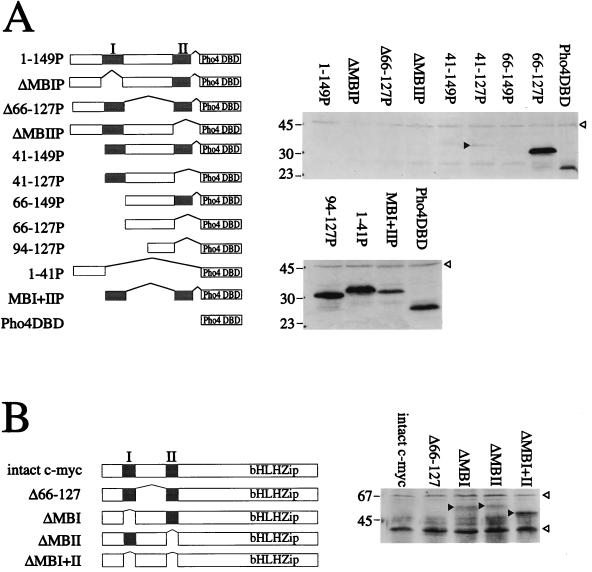
During analysis of the N-terminal transactivation domain of c-myc with a yeast system, various deletions were made in the context of both the intact c-myc protein and a c-myc–Pho4 fusion protein, which consists of the N terminal of c-myc fused to the DBD of the yeast Pho4 protein. Western blot analysis of the intact proteins and deletion mutants showed that there was a dramatic variation in the levels of protein produced (Fig. 1). This variation did not reflect differential extraction of the proteins from yeast since similar results were obtained when proteins were extracted in loading buffer from physically disrupted cells (results not shown). Analysis of the various myc-Pho4 constructs (Fig. 1A) showed that low protein levels were correlated with the presence of one or both of the conserved myc boxes. We concluded that the absence of both myc boxes led to an increase in the amount of myc-Pho4 protein present in the cell extracts. The only exception to this pattern is a construct in which the two myc boxes are juxtaposed (MBI+II); under these circumstances, the protein is only slightly destabilized (see Discussion). When the protein was not detectable, we could confirm that it was being produced, since the proteins were able to induce a lacZ reporter gene fused to a Pho4-regulated promoter (data not shown). Using a construct expressing the full-length c-myc protein, we obtained similar results; that is, the full-length c-myc protein was not detectable. The amount of protein detected increased on removal of either myc box, and when both were removed, the levels increased further (Fig. 1B). Taken together, these results suggest that each of the myc boxes contributes independently to the low level of the c-myc protein observed. Since c-myc is generally found as a dimer with max, we included a max expression plasmid in all the yeast strains expressing full-length c-myc and its derivatives. The levels of c-myc proteins were unaffected by the presence or absence of max.
FIG. 1.
Western blot analysis of the myc-Pho4 and full-length c-myc constructs in yeast cells. (A) Schematic representation of the myc-Pho4 fusions (left). The myc boxes are shaded. Western blots showing the levels of myc-Pho4 fusion proteins are shown on the right. Whole-cell extracts from YS33 cells expressing each of the myc-Pho4 fusions were made by boiling 1.5 OD600 units of cells in 100 μl of 1× loading buffer (34). A 30-μl volume of each supernatant was subjected to SDS-PAGE (12.5% polyacrylamide). After electroblotting to Hybond C-super, the proteins were detected with anti-Pho4 DBD antibodies (1:6,000 in 1% milk–phosphate-buffered saline–Tween) and secondary donkey anti-rabbit IgG-HRP conjugate (1:4,000 in phosphate-buffered saline–Tween). Interaction with the antibodies was visualized by chemiluminescence. The positions of the molecular mass markers are indicated in kilodaltons. The solid arrowhead indicates the 41-127P protein, which is barely visible on the blot; the open arrowhead indicates a nonspecific band detected by the antibody. (B) Schematic representation of the full-length c-myc protein deletions (left). The myc boxes are shaded. The c-myc DBD is indicated (bHLHZip). Western blots of intact c-myc and deletion derivatives of c-myc are shown on the right. Whole-cell extracts were made as described above, from W303-1A cells carrying the c-myc constructs and a max expression plasmid. A 30-μl volume of each extract was subjected to SDS-PAGE (7.5% polyacrylamide). Proteins were detected with an anti-myc C-terminal antibody (9E10; 1:500 in 1% milk–phosphate-buffered saline–Tween), secondary sheep anti-mouse IgG-HRP conjugate (1:4,000 in phosphate-buffered saline–Tween), and chemiluminescence. Molecular mass standards are indicated in kilodaltons. Solid arrowheads indicate the c-myc proteins; open arrowheads indicate nonspecific bands.
Changes in mRNA levels do not account for the protein destabilization effects of the myc boxes.
Previous studies have shown that expression of c-myc is regulated at the mRNA level (42, 65, 76). Interestingly, exon 2, which encodes the myc boxes, has been reported to contain sequences which can affect the amounts of c-myc mRNA (56, 75). To determine whether the effect on protein levels was due to an effect of the myc boxes at the mRNA level, we measured the transcript levels of the different constructs. Total RNA was extracted from YS33 yeast cells expressing each of the myc-Pho4 constructs. Northern blots of the RNA were incubated with a probe to Pho4 DBD, which could detect all the myc-Pho4 mRNAs regardless of the c-myc sequences present. The mRNA levels were quantitated and expressed relative to an internal actin control (Fig. 2). Although the mRNA levels for the various constructs vary considerably, the observed variation does not account for the variations at the protein level, which we attribute to the myc boxes. Similar conclusions were drawn from results obtained with Northern blots, in which transcripts representing the full-length c-myc proteins, both intact and with the myc box deleted, were studied (data not shown).
FIG. 2.
Northern blot analysis of myc-Pho4 mRNA expression in yeast cells. Total RNA was extracted from YS33 cells expressing each of the myc-Pho4 fusions. The RNA was separated on a 1.5% denaturing gel and blotted onto Hybond N. The mRNA transcripts were detected with a α-32P-labelled probe to Pho4 DBD or actin. The intensity of hybridization was calculated by phosphorimager analysis, and the relative amounts of each myc-Pho4 mRNA, normalized for actin levels, are shown below each lane. ∗, constructs which have at least one myc box present.
The myc boxes cause a decrease in the stability of the c-myc protein.
Since mRNA stability did not account for the variation in the level of myc box-specific protein, we decided to examine the rates of protein degradation. If some proteins were more stable than others, this could account for the difference in steady-state levels seen on Western blots. To allow the detection of all c-myc derivatives, we expressed the wild-type and mutant c-myc proteins in a protease-deficient yeast strain (BJ2168). BJ2168 is deficient for PEP4, PRB1, and PRC1, key components of the yeast vacuolar protease system (reviewed in reference 36). The full length c-myc expression vectors (pSDmyc and pSDmycΔMBI+II) were cotransformed with a max expression plasmid (pRSmax9), and whole-cell extracts were analyzed by Western blotting. In this strain, the previously nondetectable constructs were visible, although still at considerably lower levels than the other constructs.
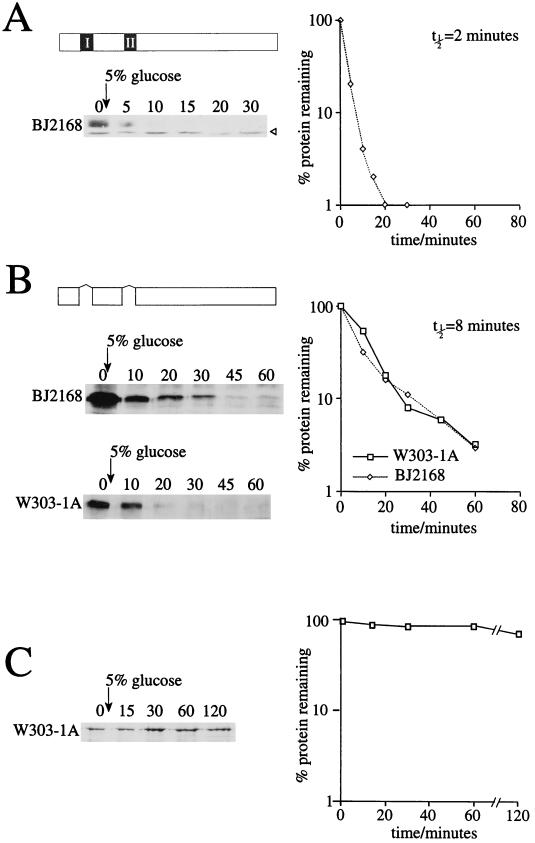
The cells expressing intact c-myc and the derivative with the myc boxes deleted were grown under inducing (galactose) conditions to allow production of the c-myc proteins. Glucose was then added to 5%, which inhibits galactose-induced expression. Samples were removed from the cultures at various times thereafter, and cell extracts were analyzed by Western blotting (Fig. 3). The intensity of the c-myc band on the blot was calculated for each time point, and the half-time for its disappearance was determined. The half-time of intact c-myc was calculated to be 2 min (Fig. 3A). This was increased fourfold to 8 min for the construct lacking the two myc boxes (Fig. 3B, upper panel). Interestingly, the same half-time of 8 min was measured in a wild-type yeast strain (lower panel); therefore, the vacuolar proteases (mutated in BJ2168) do not appear to contribute to in vivo degradation of the construct with the myc boxes deleted. However, it is possible that they contribute directly to the degradation of intact c-myc and that a half-time of 2 min is an overestimate of its stability in yeast. To ensure that the decrease in protein levels was not an artifact associated with the experimental procedure, we analyzed the half-time of β-galactosidase (expressed from a GAL-lacZ construct). β-Galactosidase is a very stable protein and exhibited only a slight change in levels during the assay (Fig. 3C). Furthermore, the same half-times for the c-myc proteins are observed if the glucose addition step is replaced by transfer to medium containing raffinose as a carbon source (data not shown).
FIG. 3.
Half-time determinations for c-myc proteins in yeast cells. W303-1A or BJ2168 cells expressing the protein of interest were grown to mid-log phase in galactose selective medium. Glucose was added to 5% to repress expression of the c-myc derivatives, and 10-ml aliquots were removed from the cultures at various times thereafter. Cell extracts were made as described for Fig. 1B. The volumes of cells for each extract were adjusted if necessary after the OD600 was determined. A 30-μl volume of each sample was subjected to SDS-PAGE (7.5% polyacrylamide), and Western blotting was performed as described for Fig. 1B, except where indicated. The intensity of each band was determined with GelPro image analysis software. The amount of protein present at time zero was set to 100%, and the percentages remaining were plotted on a logarithmic scale against time to calculate the half-time. (A) Half-time determination for intact full-length c-myc. The half-time was measured for two different BJ2168 transformants. A representative Western blot is shown, and the values used to calculate the half-time are the means from two experiments. The c-myc protein is undetectable in W303-1A cells, so the half-time could not be measured in this strain. The open arrow indicates a nonspecific band detected by the antibody. (B) Half-time determinations for full-length c-myc lacking the two myc boxes. The half-time was measured in duplicate for transformants of both W303-1A and BJ2168. Representative Western blots are shown, and the values used to calculate the half-time are the means from two experiments. (C) Half-time determination of β-galactosidase. The primary antibody used was against β-galactosidase (1:500 in 1% milk–phosphate-buffered saline–Tween), and the secondary antibody was sheep anti-mouse IgG-HRP conjugate. The measured values for the intensity of the β-galactosidase bands were corrected with an internal loading control (results not shown).
The myc boxes can destabilize other proteins.
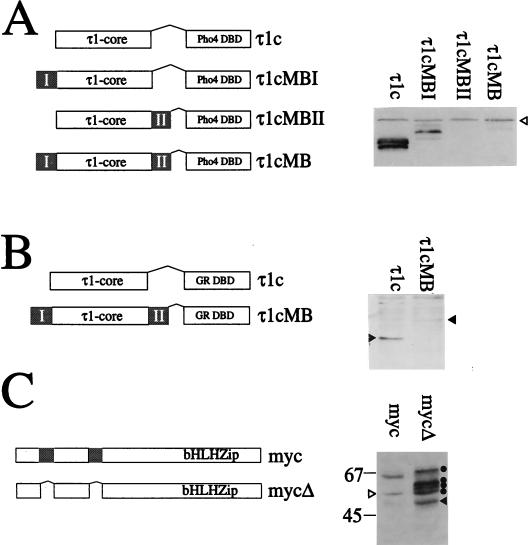
Rapidly degraded proteins have sometimes been shown to have a “destruction box” or sequence that targets the protein for degradation. In some cases, sequences containing the destruction box are sufficient to destabilize any protein to which they are attached, as was shown for cyclin B (24) and the MATα2 repressor (33). To determine whether the myc boxes contained all the necessary sequence information to target proteins for degradation, we constructed a hybrid gene in which sequences encoding the glucocorticoid receptor transactivation domain (GR τ1c) were cloned in frame with either one or both of the myc boxes, upstream of the Pho4 DBD sequences (Fig. 4A). The τ1c-Pho4 protein is usually produced in large amounts in yeast, indicating that it is not as rapidly degraded as c-myc. Addition of one or both of the myc boxes to this construct resulted in a striking decrease in protein levels (Fig. 4A). The effect of MBI alone on the levels of the protein was less dramatic than that of MBII alone. As described above, the presence of the less stable proteins in yeast cells could be confirmed by using a lacZ reporter gene to demonstrate that the τ1cMB constructs were being produced and were still able to function as transactivators of gene expression (data not shown).
FIG. 4.
The myc boxes are signals for proteolysis. (A) Constructs used to investigate the effect of the myc boxes on protein levels in yeast cells (left). The τ1core (τ1c) of the human glucocorticoid receptor was fused to one or both of the myc boxes (indicated as shaded regions) and the Pho4 DBD. Whole-cell extracts from yeast strain YS33 expressing either the τ1c or the various τ1cMB constructs were analyzed by Western blotting (right) as described for Fig. 1A. The open arrowhead indicates a nonspecific band recognized by the antibody. (B) Schematic representation of the mammalian expression constructs used to investigate the destabilizing role of the myc boxes in COS7 cells (left). The myc boxes are shaded. COS7 cells were transiently transfected with the expression plasmids for τ1c and τ1cMB. A 30-μg portion of total protein from whole-cell extracts was subjected to SDS-PAGE (20% polyacrylamide) and blotted onto Hybond C-super membrane. The τ1c and τ1cMB proteins were detected with anti GR-DBD antibody (1:3,333 in 1% milk–phosphate-buffered saline–Tween) and secondary sheep anti-mouse IgG-HRP conjugate (1:2,000 in phosphate-buffered saline–Tween). Interaction with the antibodies was visualized by chemiluminescence (right). Solid arrowheads indicate the positions of the τ1c and τ1cMB proteins. (C) Western blot analysis of the levels of full-length c-myc proteins in mammalian cells. A schematic representation of the mammalian expression constructs used is shown on the left. The myc boxes are shaded. COS7 cells were transiently transfected with 0.1 μg of the expression plasmid pcDNA3myc or pcDNA3mycΔ. A 50-μg portion of total protein from whole-cell extracts subjected to SDS-PAGE (7.5% polyacrylamide) and blotted onto Hybond C-super membrane. The c-myc proteins were detected with anti-V5 antibody (1:10,000 in 1% milk–phosphate-buffered saline–Tween) and secondary sheep anti-mouse IgG-HRP conjugate (1:2,000 in phosphate-buffered saline–Tween). Interaction with the antibodies was visualized by chemiluminescence. The mycΔ protein is indicated by the solid arrowhead; the derivatives are indicated by solid circles. A nonspecific band is indicated by an open arrowhead. Standard molecular mass markers are indicated in kilodaltons.
To determine whether the myc boxes were also able to destabilize the τ1c protein in mammalian cells, plasmids expressing similar constructs (Fig. 4B) were transiently transfected into COS7 cells. In these experiments, we have repeatedly observed that the protein containing the myc boxes is present at much lower levels than is the τ1c protein alone (Fig. 4B). We conclude that the myc boxes can target proteins for degradation in both yeast and mammalian cells.
The myc boxes destabilize c-myc in mammalian cells.
Having demonstrated that the myc boxes can function as degradation signals in mammalian cells, we investigated the effect of deleting them from a full-length c-myc protein produced in COS7 cells (Fig. 4C). We observed that the intact protein (myc) was present at a lower level than that of the protein lacking the myc boxes (mycΔ). In addition, we observed four extra bands of higher molecular weight in the mycΔ track. These were purified by using the His tag fused to this construct and shown to cross-react with a myc antibody (9E10), confirming that they are derived from the c-mycΔ protein (result not shown). No higher-molecular-weight forms were detected for the intact protein. We propose that these higher-molecular-weight derivatives might be intermediates in the degradation pathway which are visible in the absence of the myc boxes because the rate of degradation has been decreased significantly, allowing the intermediates to accumulate in the cell.
The myc boxes target protein degradation via the proteasome.
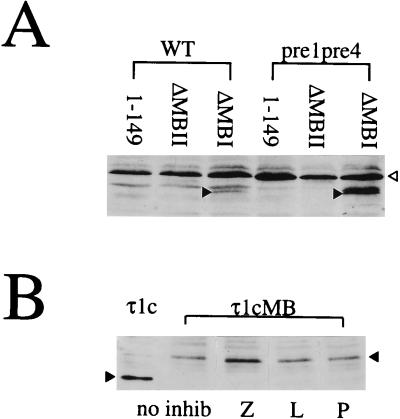
The 26S proteasome is a major proteolysis system for short-lived proteins and is highly conserved between yeast and mammalian cells (reviewed in reference 55). There are yeast strains available in which one or more of the proteasome subunits have been mutated. In these mutants, the proteasome is still partially active so that the cells can survive, but in many cases it is possible to detect a decreased turnover rate of proteins. We looked at the protein levels of the myc-Pho4 constructs 1–149P, ΔMBIP, and ΔMBIIP in a strain which produces mutant forms of the catalytic subunits, Pre1 and Pre4. Figure 5A shows that there is a dramatic increase in the level of ΔMBIP in the mutant strain, strongly suggesting that MBII targets proteins for degradation via the proteasome in yeast. There was no effect on the MBI-targeted degradation in this strain.
FIG. 5.
Role of the proteasome in myc box-directed degradation. (A) Whole-cell extracts from yeast strains YHI29/W (WT) and YHI29/14 (pre1, pre4) expressing the myc-Pho4 fusion proteins 1–149P, ΔMBIP, and ΔMBIIP were analyzed by Western blotting as described for Fig. 1A. The open arrowhead indicates a nonspecific band recognized by the antibody; the solid arrowheads indicate the ΔMBIP protein band. (B) COS7 cells were transiently transfected with the expression plasmid for τ1cMB. After 42 h of incubation, 10 μM inhibitor (Z-L3 VS [Z], leupeptin [L], or pepstatin [P]) was added. Incubation was continued for a further 4 h. A 30-μg portion of total protein from whole-cell extracts of treated (lanes Z, L, and P) and untreated (lanes no inhib) cells were subjected to SDS-PAGE (20% polyacrylamide) and blotted onto Hybond C-super membrane. The τ1cMB protein was detected with anti GR-DBD antibody (1:3,333 in 1% milk–phosphate-buffered saline–Tween) and secondary sheep anti-mouse IgG-HRP conjugate (1:2,000 in phosphate-buffered saline–Tween). Interaction with the antibodies was visualized by chemiluminescence. An extract from untreated cells producing the τ1c protein was included as a quantification control. Solid arrowheads indicate the positions of the proteins.
To test whether the τ1cMB fusion protein is degraded by the proteasome in mammalian cells, we investigated the effects of various different diffusible proteolysis inhibitors on the level of the τ1cMB protein (Fig. 4B) expressed in COS7 cells. Figure 5B shows that the Z-L3VS inhibitor, which is a specific and effective inhibitor of the proteasome (13), stabilizes the myc box-containing protein, whereas leupeptin and pepstatin, which inhibit thiol and acid proteases, respectively, have no stabilizing effect. Taken together, our data support a model in which the myc boxes target protein degradation via the proteasome in both yeast and mammalian cells.
DISCUSSION
We have shown that MBI and MBII, which are highly conserved in all myc protein family members (Fig. 6), are signals for rapid turnover of the c-myc protein. In addition, we have demonstrated that the myc boxes contain sufficient sequence information to reduce the levels of heterologous proteins to which they are attached in both yeast and mammalian cells. At least part of myc box-directed degradation is targeted via the proteasome in both yeast and mammalian cells (Fig. 5). The high conservation of the myc box sequences throughout the myc family proteins suggests that the proteolysis-signalling function may also be present in other myc family members.
FIG. 6.
Alignment of the N-terminal sequences from different members of the myc protein family. The alignment was produced with DNASTAR/Lasergene software. Residues identical to the consensus are shaded. The positions of MBI and MBII, described by Sarid et al. (64), are indicated.
Previously, it has been proposed that the stability of the c-myc mRNA is important for regulating levels of c-myc protein (reviewed in references 42 and 65). Many different stability-determining sequences have been identified in both the untranslated and the coding regions of the c-myc transcript. In addition, the c-myc protein has a rapid turnover, with a half time of 30 min in mammalian cells (29, 60), so that factors influencing the stability of the c-myc protein could also be involved in regulating the cellular levels. We measured a half time of 2 min for the c-myc protein in yeast. The shorter half time observed in yeast may be correlated with the shorter duration of the yeast cell cycle. When the two myc boxes were deleted, the half-time was increased fourfold; therefore, the myc boxes appear to be important in determining the stability of c-myc in yeast. The very high degree of myc box homology across a range of myc proteins, as well as the conservation across species (64), suggests that the myc boxes may play an important role in determining the protein levels of all the myc family members. Sequence analysis of the c-myc proteins found in Burkitt’s lymphoma revealed the existence of mutation hot spots that colocalize with one of the myc boxes (1, 10). The c-myc protein in Burkitt’s lymphoma is characterized by deregulated expression caused by a translocation to the IgG locus (49). Mutations within the myc boxes may also override normal cellular control mechanisms and lead to an increase in cellular levels of c-myc. The binding of c-myc to p107 has been proposed to be an important step in cell cycle control (8). This correlates with the observation that c-myc stimulates the G1-to-S transition and p107 causes G1 arrest. Beijersbergen et al. (8) showed that increasing the cellular levels of c-myc could partly reverse the p107 effect and that this might be caused by direct binding of c-myc to p107. Thus, increased levels of c-myc due to an increase in stability of the protein could lead to a reversal of p107 arrest and cause cellular proliferation.
Attempts have been made to delineate the function of the myc boxes. MBII is essential for the transcriptional repression activity of c-myc (46), and both myc boxes are required for transformation of cells (69). Deletion of the myc boxes does not affect transactivation activity (22a, 46). Several proteins have been shown to bind to N-terminal sequences containing the myc boxes (TATA binding protein [31], p107 [27], and α-tubulin [2]), but in only one case has a myc box been shown to be specifically involved in protein binding (15). This study reported that MBII is specifically involved in binding to a nuclear factor that may be involved in target sequence recognition. It is conceivable that the myc box-targeted protein degradation could provide a universal mechanism to account for the previously reported functions of the myc boxes, but it is equally possible that they function by several distinct mechanisms.
To investigate whether the myc boxes alone were sufficient to cause a decrease in heterologous protein levels, we coupled one or both of the myc boxes to the τ1core of the glucocorticoid receptor. The 58-amino-acid τ1core is similar in size to the 62-amino-acid region which normally separates the myc boxes. In these constructs, the myc boxes caused a significant decrease in protein levels. These results show that the myc boxes alone contain sufficient sequence information to lead to the degradation of heterologous proteins. This property has also been reported for other, short “destabilizing” sequences found in c-jun (72), cyclin B (24), and MATα2 (33). The τ1cMB constructs show that there is a clear difference in the relative effectiveness of the two myc boxes. In these constructs, MBII appears to cause a much greater decrease in protein levels than does MBI. It is interesting that when the myc boxes are deleted in the full-length c-myc protein, there is no significant difference between the levels of the mutant proteins, suggesting that they contribute similarly in this context. It may be that although deletion of MBI is sufficient to disrupt its destabilizing effect, the MBI sequence alone is not sufficient for efficient protein destabilization. Our data suggest that there may be other sequences, located N terminal of MBI, which are required for its efficient function (compare ΔMBIIP and 41–127P in Fig. 1A). MBII, in contrast, is an efficient and effective destabilization motif and apparently contains all the necessary sequence information. The c-mycS protein is a result of initiation of transcription from an AUG codon downstream from the normal start sites (66), and thus the protein product is a shortened form of c-myc lacking nearly all of the N-terminal transactivation region. The c-mycS protein starts at the equivalent of amino acid 100 in the human c-myc protein, just upstream of MBII sequences. This protein retains a half-life of 30 min in mammalian cells (67), and thus, at least under these conditions, MBI was not required to cause the rapid turnover of the protein. Our results clearly show that MBI and MBII represent destabilization sequences that can work independently. However, one construct in which MBI and MBII are juxtaposed (MBI+IIP; Fig. 1A) is relatively poorly degraded; therefore, it could be that their activity is partly dependent on context, but we have not investigated this further.
Our evidence suggests that myc box-containing proteins are degraded by the proteasome in both yeast and mammalian cells. We have shown that MBII-directed proteolysis can be inhibited in the presence of mutations affecting some of the catalytic subunits of the yeast proteasome (Pre1 and Pre4; Fig. 5A). MBI-directed proteolysis appears to be unaffected in these mutants, suggesting either that MBI-targeted degradation is dependent on other proteasome subunits or that the proteasome is not involved. Results from mammalian cell studies show that inhibition of the proteasome quantitatively overcomes the effect of both myc boxes. This suggests that the proteasome also plays a role in MBI-directed degradation, but this remains to be shown directly.
Many short-lived proteins which are degraded by the 26S proteasome are ubiquitin tagged prior to degradation. There are no specific sequences that mark a protein as a substrate for degradation by the 26S proteasome. However, it has been reported that many ubiquitinated proteins have a destabilizing N-terminal amino acid, as defined by the N-end rule (7). In addition, sequences rich in proline, glutamic acid, serine, and threonine, the so-called PEST sequences, can function as degradation signals (61). The ubiquitin tag is always conjugated to a lysine residue on the target protein, but the positioning of the lysine residue does not appear to be of great importance (40). The myc boxes do not contain any PEST sequences, and none of the constructs tested have a destabilizing N-terminal amino acid. Each myc box does contain lysine residues, and this amino acid is not found anywhere else in the N terminus. High-molecular-weight forms of c-myc, which would normally be associated with ubiquitination, have not been detected (47), but using an in vitro assay system, Ciechanover et al. (18) showed that c-myc and other oncoproteins could be rapidly degraded via the ubiquitin pathway in vitro. The higher-molecular-weight forms present in the absence of the myc boxes in Fig. 4C may be a result of ubiquitin conjugation, but we have been unable to confirm this. Thus, to date, we do not have direct evidence for or against ubiquitination of the c-myc protein in vivo. However, the human N-myc protein has recently been shown to be ubiquitinated in vivo before undergoing degradation by the 26S proteasome (14). At least one protein, ornithine decarboxylase, has been shown to be degraded by the proteasome without being modified by the ubiquitin pathway (52, 63), so it remains a possibility that c-myc is also degraded in this way. Since the myc box-directed proteolysis is conserved between yeast and mammalian cells, it may be possible to use the powerful genetic approaches available for yeast to identify the components involved in the myc box-targeting and proteolysis systems which regulate c-myc levels. The prospects for such an approach are promising, since investigation of proteolysis events that regulate the cell division cycle has shown that the mechanisms involved are highly conserved in yeast, Xenopus, and humans (73).
ACKNOWLEDGMENTS
We thank W. Hörz (Ludwig-Maximilians Universität, Munich, Germany), C. Goding (Marie Curie Research Institute, United Kingdom), T. Almlöf (Karolinska Institute), and P. Ljungdahl (Ludwig Institute for Cancer Research, Stockholm, Sweden) for plasmids; W. Hilt (Universität Stuttgart, Stuttgart, Germany) and W. Hörz for yeast strains; A.-C. Wikström (Karolinska Institute) and C. Gooding for antibodies; and G. Mason (Bristol University, Bristol, United Kingdom) for the inhibitor Z-L3VS. We thank S. Hermann for critically reading the manuscript and members of the Steroid Receptor Group and the Yeast Molecular Genetics Group for useful discussions and technical advice.
This work was supported by a grant from the Swedish Cancer Fund (3831-B97-02YBB).
REFERENCES
- 1.Albert T, Urlbauer B, Kohlhuber F, Hammersen B, Eick D. Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt’s lymphoma cell lines. Oncogene. 1994;9:759–763. [PubMed] [Google Scholar]
- 2.Alexandrova N, Niklinski J, Bliskovsky V, Otterson G A, Blake M, Kaye F J, Zajac-Kaye M. The N-terminal domain of c-Myc associates with alpha-tubulin and microtubules in vivo and in vitro. Mol Cell Biol. 1995;15:5188–5195. doi: 10.1128/mcb.15.9.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amati B, Dalton S, Brooks M W, Littlewood T D, Evan G I, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- 4.Amin C, Wagner A J, Hay N. Sequence-specific transcriptional activation by Myc and repression by Max. Mol Cell Biol. 1993;13:383–390. doi: 10.1128/mcb.13.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ariga H, Imamura Y, Iguchi-Ariga S M. DNA replication origin and transcriptional enhancer in c-myc gene share the c-myc protein binding sequences. EMBO J. 1989;8:4273–4279. doi: 10.1002/j.1460-2075.1989.tb08613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asker C E, Magnusson K P, Piccoli S P, Andersson K, Klein G, Cole M D, Wiman K G. Mouse and rat B-myc share amino acid sequence homology with the c-myc transcriptional activator domain and contain a B-myc specific carboxy terminal region. Oncogene. 1995;11:1963–1969. [PubMed] [Google Scholar]
- 7.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 8.Beijersbergen R L, Hijmans E M, Zhu L, Bernards R. Interaction of c-Myc with the pRb-related protein p107 results in inhibition of c-Myc-mediated transactivation. EMBO J. 1994;13:4080–4086. doi: 10.1002/j.1460-2075.1994.tb06725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benvenisty N, Leder A, Kuo A, Leder P. An embryonically expressed gene is a target for c-Myc regulation via the c-Myc-binding sequence. Genes Dev. 1992;6:2513–2523. doi: 10.1101/gad.6.12b.2513. [DOI] [PubMed] [Google Scholar]
- 10.Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 11.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 12.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 13.Bogyo M, McMaster J S, Gaczynska M, Tortorella D, Goldberg A L, Ploegh H. Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog Hslv by a new class of inhibitors. Proc Natl Acad Sci USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonvini P, Nguyen P, Trepel J, Neckers L. In vivo degradation of N-myc in neuroblastoma cells is mediated by the 26S proteasome. Oncogene. 1998;16:1131–1140. doi: 10.1038/sj.onc.1201625. [DOI] [PubMed] [Google Scholar]
- 15.Brough D E, Hofmann T J, Ellwood K B, Townley R A, Cole M D. An essential domain of the c-myc protein interacts with a nuclear factor that is also required for E1A-mediated transformation. Mol Cell Biol. 1995;15:1536–1544. doi: 10.1128/mcb.15.3.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christy R J, Kaestner K H, Geiman D E, Lane M D. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci USA. 1991;88:2593–2597. doi: 10.1073/pnas.88.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciechanover A, DiGiuseppe J A, Bercovich B, Orian A, Richter J D, Schwartz A L, Brodeur G M. Degradation of nuclear oncoproteins by the ubiquitin system in vitro. Proc Natl Acad Sci USA. 1991;88:139–143. doi: 10.1073/pnas.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crouch D H, Fisher F, Clark W, Jayaraman P S, Goding C R, Gillespie D A. Gene-regulatory properties of Myc helix-loop-helix/leucine zipper mutants: Max-dependent DNA binding and transcriptional activation in yeast correlates with transforming capacity. Oncogene. 1993;8:1849–1855. [PubMed] [Google Scholar]
- 20.Dang C V, McGuire M, Buckmire M, Lee W M. Involvement of the ‘leucine zipper’ region in the oligomerization and transforming activity of human c-myc protein. Nature. 1989;337:664–666. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- 21.Dang C V, van Dam H, Buckmire M, Lee W M. DNA-binding domain of human c-Myc produced in Escherichia coli. Mol Cell Biol. 1989;9:2477–2486. doi: 10.1128/mcb.9.6.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eilers M, Schirm S, Bishop J M. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Flinn, E. M. Unpublished data.
- 23.Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 24.Glotzer M, Murray A W, Kirschner M W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 25.Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 26.Grandori C, Mac J, Siebelt F, Ayer D E, Eisenman R N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 27.Gu W, Bhatia K, Magrath I T, Dang C V, Dalla-Favera R. Binding and suppression of the Myc transcriptional activation domain by p107. Science. 1994;264:251–254. doi: 10.1126/science.8146655. [DOI] [PubMed] [Google Scholar]
- 28.Gupta S, Seth A, Davis R J. Transactivation of gene expression by Myc is inhibited by mutation at the phosphorylation sites Thr-58 and Ser-62. Proc Natl Acad Sci USA. 1993;90:3216–3220. doi: 10.1073/pnas.90.8.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hann S R, Eisenman R N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hann S R, King M W, Bentley D L, Anderson C W, Eisenman R N. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt’s lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 31.Hateboer G, Timmers H T, Rustgi A K, Billaud M, van’t Veer L J, Bernards R. TATA-binding protein and the retinoblastoma gene product bind to overlapping epitopes on c-Myc and adenovirus E1A protein. Proc Natl Acad Sci USA. 1993;90:8489–8493. doi: 10.1073/pnas.90.18.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henriksson M, Bakardjiev A, Klein G, Luscher B. Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene. 1993;8:3199–3209. [PubMed] [Google Scholar]
- 33.Hochstrasser M, Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell. 1990;61:697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- 34.Horvath A, Riezman H. Rapid protein extraction from Saccharomyces cerevisiae. Yeast. 1994;10:1305–1310. doi: 10.1002/yea.320101007. [DOI] [PubMed] [Google Scholar]
- 35.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones E W. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J Biol Chem. 1991;266:7963–7966. [PubMed] [Google Scholar]
- 37.Jones R M, Branda J, Johnston K A, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt E V. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato G J, Barrett J, Villa-Garcia M, Dang C V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990;10:5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohl N E, Legouy E, DePinho R A, Nisen P D, Smith R K, Gee C E, Alt F W. Human N-myc is closely related in organization and nucleotide sequence to c-myc. Nature. 1986;319:73–77. doi: 10.1038/319073a0. [DOI] [PubMed] [Google Scholar]
- 40.Kornitzer D, Raboy B, Kulka R G, Fink G R. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 42.Laird-Offringa I A. What determines the instability of c-myc proto-oncogene mRNA? Bioessays. 1992;14:119–124. doi: 10.1002/bies.950140209. [DOI] [PubMed] [Google Scholar]
- 43.Lech K, Anderson K, Brent R. DNA-bound Fos proteins activate transcription in yeast. Cell. 1988;52:179–184. doi: 10.1016/0092-8674(88)90506-5. [DOI] [PubMed] [Google Scholar]
- 44.Legouy E, DePinho R, Zimmerman K, Collum R, Yancopoulos G, Mitsock L, Kriz R, Alt F W. Structure and expression of the murine L-myc gene. EMBO J. 1987;6:3359–3366. doi: 10.1002/j.1460-2075.1987.tb02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis B C, Shim H, Li Q, Wu C S, Lee L A, Maity A, Dang C V. Identification of putative c-myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol Cell Biol. 1997;17:4967–4978. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L H, Nerlov C, Prendergast G, MacGregor D, Ziff E B. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 1994;13:4070–4079. doi: 10.1002/j.1460-2075.1994.tb06724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luscher B, Eisenman R N. c-myc and c-myb protein degradation: effect of metabolic inhibitors and heat shock. Mol Cell Biol. 1988;8:2504–2512. doi: 10.1128/mcb.8.6.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luscher B, Eisenman R N. New light on Myc and Myb. I. Myc. Genes Dev. 1990;4:2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- 49.Magrath I. The pathogenesis of Burkitt’s lymphoma. Adv Cancer Res. 1990;55:133–270. doi: 10.1016/s0065-230x(08)60470-4. [DOI] [PubMed] [Google Scholar]
- 50.Marhin W W, Chen S, Facchini L M, Fornace A J, Jr, Penn L Z. Myc represses the growth arrest gene gadd45. Oncogene. 1997;14:2825–2834. doi: 10.1038/sj.onc.1201138. [DOI] [PubMed] [Google Scholar]
- 51.McEwan I J, Dahlman-Wright K, Ford J, Wright A P. Functional interaction of the c-Myc transactivation domain with the TATA binding protein: evidence for an induced fit model of transactivation domain folding. Biochemistry. 1996;35:9584–9593. doi: 10.1021/bi960793v. [DOI] [PubMed] [Google Scholar]
- 52.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 53.Penn L J Z, Brooks M W, Laufer E M, Littlewood T D, Morgenstern J P, Evan G I, Lee W M F, Land H. Domains of human c-myc protein required for autosuppression and cooperation with ras oncogenes are overlapping. Mol Cell Biol. 1990;10:4961–4966. doi: 10.1128/mcb.10.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Philipp A, Schneider A, Vasrik I, Finke K, Xiong Y, Beach D, Alitalo K, Eilers M. Repression of cyclin D1: a novel function of MYC. Mol Cell Biol. 1994;14:4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pickart C M. Targeting of substrates to the 268 proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 56.Pistoi S, Roland J, Babinet C, Morello D. Exon 2-mediated c-myc mRNA decay in vivo is independent of its translation. Mol Cell Biol. 1996;16:5107–5116. doi: 10.1128/mcb.16.9.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prendergast G C, Diamond L E, Dahl D, Cole M D. The c-myc-regulated gene mrl encodes plasminogen activator inhibitor 1. Mol Cell Biol. 1990;10:1265–1269. doi: 10.1128/mcb.10.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prendergast G C, Ziff E B. A new bind for Myc. Trends Genet. 1992;8:91–96. doi: 10.1016/0168-9525(92)90196-b. [DOI] [PubMed] [Google Scholar]
- 59.Pulverer B J, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett J R. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 60.Ramsay G, Evan G I, Bishop J M. The protein encoded by the human proto-oncogene c-myc. Proc Natl Acad Sci USA. 1984;81:7742–7746. doi: 10.1073/pnas.81.24.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 62.Rose M D, Winston F, Heiter P. Yeast genetics: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 63.Rosenberg-Hasson Y, Bercovich Z, Ciechanover A, Kahana C. Degradation of ornithine decarboxylase in mammalian cells is ATP dependent but ubiquitin independent. Eur J Biochem. 1989;185:469–474. doi: 10.1111/j.1432-1033.1989.tb15138.x. [DOI] [PubMed] [Google Scholar]
- 64.Sarid J, Halazonetis T D, Murphy W, Leder P. Evolutionarily conserved regions of the human c-myc protein can be uncoupled from transforming activity. Proc Natl Acad Sci USA. 1987;84:170–173. doi: 10.1073/pnas.84.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schiavi S C, Belasco J G, Greenberg M E. Regulation of proto-oncogene mRNA stability. Biochim Biophys Acta. 1992;1114:95–106. doi: 10.1016/0304-419x(92)90009-n. [DOI] [PubMed] [Google Scholar]
- 66.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spotts G D, Patel S V, Xiao Q, Hann S R. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol Cell Biol. 1997;17:1459–1468. doi: 10.1128/mcb.17.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stanton L W, Schwab M, Bishop J M. Nucleotide sequence of the human N-myc gene. Proc Natl Acad Sci USA. 1986;83:1772–1776. doi: 10.1073/pnas.83.6.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stone J, de Lange T, Ramsay G, Jakobovits E, Bishop J M, Varmus H, Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987;7:1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugiyama A, Kume A, Nemoto K, Lee S Y, Asami Y, Nemoto F, Nishimura S, Kuchino Y. Isolation and characterization of s-myc, a member of the rat myc gene family. Proc Natl Acad Sci USA. 1989;86:9144–9148. doi: 10.1073/pnas.86.23.9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Svaren J, Schmitz J, Horz W. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Treier M, Staszewski L M, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 73.Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 74.Wagner A J, Meyers C, Laimins L A, Hay N. c-Myc induces the expression and activity of ornithine decarboxylase. Cell Growth Differ. 1993;4:879–883. [PubMed] [Google Scholar]
- 75.Yeilding N M, Lee W M. Coding elements in exons 2 and 3 target c-myc mRNA downregulation during myogenic differentiation. Mol Cell Biol. 1997;17:2698–2707. doi: 10.1128/mcb.17.5.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yeilding N M, Rehman M T, Lee W M. Identification of sequences in c-myc mRNA that regulate its steady-state levels. Mol Cell Biol. 1996;16:3511–3522. doi: 10.1128/mcb.16.7.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]