Summary
Alzheimer’s disease (AD) is the most common neurodegenerative disease often associated with olfactory dysfunction. Aβ is a typical AD hall marker, but Aβ-induced molecular alterations in olfactory memory remain unclear. In this study, we used a 5xFAD mouse model to investigate Aβ-induced olfactory changes. Results showed that 4-month-old 5xFAD have olfactory memory impairment accompanied by piriform cortex neuron activity decline and no sound or working memory impairment. In addition, synapse and glia functional alteration is consistent across different ages at the proteomic level. Microglia and astrocyte specific proteins showed strong interactions in the conserved co-expression network module. Moreover, this interaction declines only in mild cognitive impairment patients in human postmortem brain proteomic data. This suggests that astrocytes-microglia interaction may play a leading role in the early stage of Aβ-induced olfactory memory impairment, and the decreasing of their synergy may accelerate the neurodegeneration.
Subject areas: Classification Description, Health sciences, Disease, Biological sciences
Graphical abstract

Highlights
-
•
Aβ induces olfactory memory impairment by piriform cortex neurons in 5xFAD
-
•
Aβ accumulation mainly affects glial cells and synapse abnormal in 5xFAD
-
•
Astrocyte-microglia interaction declines appear in 4-month-old 5xFAD and MCI patient
Classification Description: Health sciences; Disease; Biological sciences
Introduction
Alzheimer’s disease (AD) is one of the most common neurodegenerative diseases that severely affects the quality of life of the elderly, placing a significant burden on individuals, families, societies, and the global economy. The pathological symptoms, such as Aβ plaques, appeared before the clinical symptoms in AD.1 But the relationship between pathological changes and clinical features is still unknown. Thus, it is important to explore the molecular alterations in Aβ induced memory impairment.1,2
APP and PS1 mutations are the most potential cause in most cases of familial AD (FAD) based on genetic analysis, and it is essential to elucidating Aβ-induced cognitive dysfunction in AD. The Aβ burden differs in various brain regions and can spread to unaffected brain regions, leading to neurodegeneration.3 And cognitive function can also be divided into multiple aspects: attention, memory, perception, and so on. Therefore, confirming a brain region that is related to one of the earliest cognitive impairments and suffers Aβ burden at the same time shows great importance in AD.
Olfactory dysfunction has been reported as a common clinical symptom in early AD before other cognitive impairments.4,5,6,7,8 In patients with mild cognitive impairment (MCI) or AD, olfaction scores highly correlate with brain Aβ levels. Specific brain regions, such as the olfactory bulb and the piriform cortex (PCx), execute olfactory functions. The PCx is related to the olfactory and memory.9,10,11,12 In human, different odorants have been reported to evoke different ensemble activity patterns in the PCx, which could discriminate between odor categories.13,14,15 In addition, several studies have also shown that the PCx may be impaired in the AD olfactory system.16,17,18 In clinic, patients at the early onset of AD usually show perceived deficits in olfaction prior to significant memory loss, which may be associated with pathological lesions in the PCx.16,17,18 Therefore, exploring the molecular changes in the PCx may provide clues to investigate the mechanism of AD pathogenesis involved with olfactory function.
In this study, we first confirmed that Aβ induced olfactory memory dysfunction, which starts at 4 months old, is earlier than sound and working memory dysfunction in the 5xFAD mouse model. Abnormal PCx neural activity appears in the olfactory-associated memory recall process, accompanied by Aβ accumulation. Then, a consensus protein co-expression network from 4 months old to later, which related to astrocytes and microglia, was identified by 5xFAD mice PCx proteomics analysis. The astrocyte-microglia interaction in the network showed significant declines only in MCI n multiple human datasets. It implies that glial cells interaction may play a pivotal role in the early progress of AD.
Result
Olfactory memory impairment accompanies by abnormal piriform cortex neuronal activity in 4-month-old 5xFAD mice
To determine if Aβ can induce olfactory dysfunction in the 5xFAD model, which was reported to be observed in the early stages of patients with AD, we designed three behavioral paradigms to detect different memory impairments in 4 months old. In the odor-associated fear memory test (Figure 1A), mice were delivered foot shocks (3 times) paired with an odor cue in context A during the encoding stage (day 1), and there was no significant difference in the freezing ratio between WT and 5xFAD (Figure 1B). However, during the recall stage (day 2), mice were exposed to one time of the same odor cue at day 1 in the context B, and showed a significantly lower freezing growth score in the 5xFAD group compared with WT (delta score = −0.94, p = 0.029, Figure 1C). It indicated that Aβ can induce olfactory memory retrieval impairment in 4-month-old mice. In contrast, both the encoding and recall stages of the sound-associated fear memory test showed no significant difference between 5xFAD and WT mice (Figures 1D–1F), indicating no auditory memory impairment in 4-month-old 5xFAD mice. These results showed that Aβ can induce olfactory memory loss but no auditory memory impairments in a 4-month-old 5xFAD mouse model.
Figure 1.
4-month-old 5xFAD mice exhibit olfactory memory retrieval impairment
(A) Behavioral design for olfactory memory.
(B and C) Behavioral performances on day 1 and day 2 (WT, n = 10; 5xFAD, n = 10).
(D) Behavioral design for auditory memory.
(E and F) Behavioral performances on day 1 and day 2 (WT, n = 8; 5xFAD, n = 8).
(G) Up, schematic of virus injection; down, Behavioral design of olfactory memory; insert, the representative confocal images of Gcamp6s expression in the PCx. Scale bar: 200μm.
(H and I) main, the mean and SEM of the corresponding Ca2+ signal of 4-month-old mice PCx neurons during day 1 and day 2; insert, quantification of the average amplitude during 2-4s (colored area) after odor stimulation on day1 and day2 between WT and 5xFAD mice (WT, n = 6; 5xFAD, n = 6, day 1 had 3 trial for each mouse, thus have 18 dots each group).
(J and K) Quantification of Aβ plaque numbers between 4-month-old WT and 5xFAD mice in PCx (WT, n = 3; 5xFAD, n = 3, 3 slices each mouse, thus 9 dots each group). Scale bar: 50μm. The numbers counting was done in the 150 μm × 150 μm area.
(L) Behavioral design for working memory.
(M) Behavioral performance (WT, n = 6; 5xFAD, n = 6).All statistical analyzes are used by t-test, ns, not significant; ∗p < 0.05, ∗∗∗p < 0.001. Data are presented as mean ± SEM.
Since PCx is a crucial region related to olfactory and memory, PCx neuronal activity was further recorded using fiber photometry in the odor fear memory test, and the fluorescence imaging showed that the virus carrying Gcamp6s was expressed in PCx neurons (Figure 1G). During the encoding stage, there was no change in PCx neuronal activity (Figure 1H, data among 2-4s was used in statistical analysis). However, during the recall stage, there was a significant decline in PCx neuronal activity (0.21-fold, p < 0.001, Figure 2I, data among 2-4s was used in statistical analysis). The decline in PCx neuronal activity was also accompanied by Aβ deposit accumulation, as shown by the immunohistochemistry (p < 0.001, Figures 1J–1K). These results indicated that abnormal PCx neuronal activity in the recall stage is involved in Aβ-induced olfactory memory impairment.
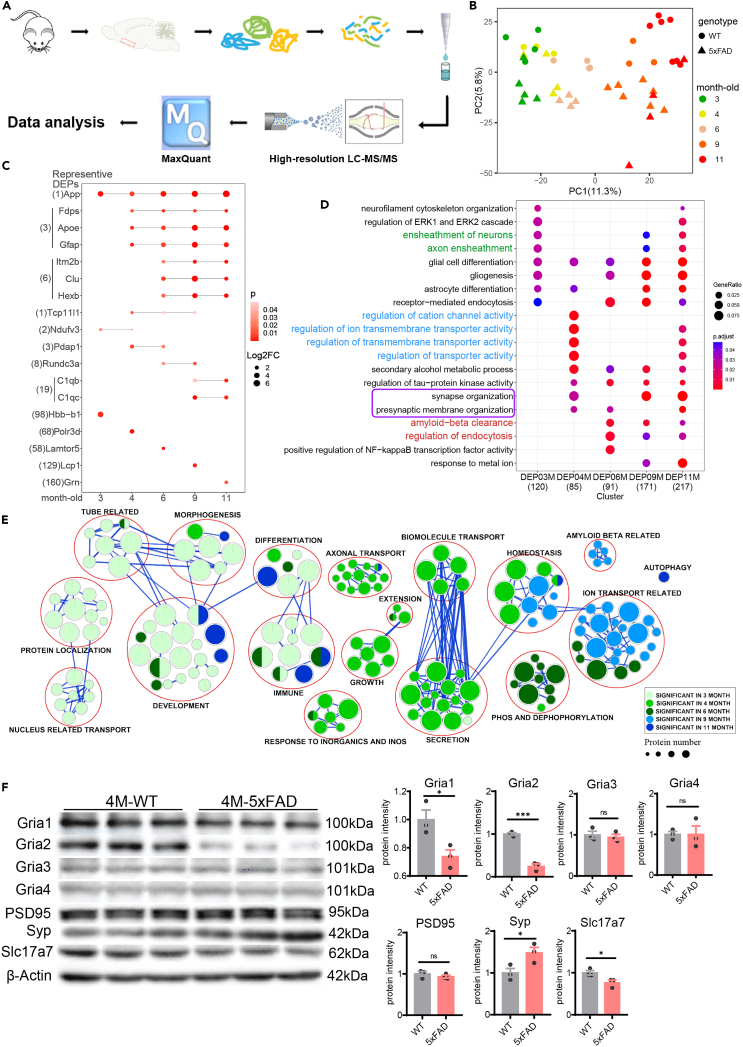
Figure 2.
The alteration of PCx proteome between WT vs. 5xFAD in different stages
(A) Workflow summarizing the process of mass-spectrum based proteomics.
(B) PCA of proteome data showed main difference in age (3-month-old: WT = 4, 5xFAD = 5; 4-month-old: WT = 4, 5xFAD = 4; 6-month-old: WT = 4, 5xFAD = 5; 9-month-old: WT = 3, 5xFAD = 7; 11-month-old: WT = 7, 5xFAD = 4).
(C) Diagram of DEPs in different month-old mice, degree of red color indicated their p values, nodes size indicates their log2FC, bar in nodes indicate their overlapping status among different month-old.
(D) GO pathway enrichment of DEPs of different month-old mice. The key pathways were colored in green, blue, red fonts which show specific enrichment in different month-old and purple box showed synapse related functions.
(E) GSEA enrichment of DEPs in different month-old mice. Results from left to right were sorted in the order of 3-month to 11-month.
(F) Classic synaptic proteins expression showed synapse impairment in 4-month-old 5xFAD. Left, Western blot images; right, quantification of western blots, t-test, ns, not significant; ∗p < 0.05, ∗∗∗p < 0.001. Data are presented as mean ± SEM.
To further investigate the different memory declines in 5xFAD mice, we assessed one more behavior test, the T-maze test, in 4-month-old mice, olfactory and auditory memory test in 6-month-old mice, olfactory memory and novel odor recognition test in 3-month-old mice. The T-maze test in 4-month-old mice showed no difference in working memory between 5xFAD and WT mice (Figures 1L and 1M). Olfactory and auditory memory tests in 6-month-old mice showed that olfactory and auditory fear memory were both impaired in 6-month-old 5xFAD mice (Figures S1A–S1D). In 3-month-old mice, olfactory fear memory showed no decline (Figures S2A and S2B) but olfactory recognition function is decline by novel odor recognition test (Figures S2C and S2D, delta index = −0.24, p = 0.029). These results imply that Aβ damage olfactory related function earlier than auditory and working memory.
Therefore, investigating the molecular alterations in PCx at different ages, especially in 4-month-old mice, may help elucidate the molecular mechanism of Aβ induced memory progress.
Proteomics revealed various function changes of piriform cortex in different ages of 5xFAD mice
To discover the Aβ accumulated effect in PCx, age- and sex-matched WT and 5xFAD mice were chosen to conduct PCx proteomics experiments across 5 ages (3-month-old: WT = 4, 5xFAD = 5; 4-month-old: WT = 4, 5xFAD = 4; 6-month-old: WT = 4, 5xFAD = 5; 9-month-old: WT = 3, 5xFAD = 7; 11-month-old: WT = 7, 5xFAD = 4). The PCx was isolated based on a mouse brain atlas, and a total of 47 samples from 5 different ages were subjected to LC-MS/MS for proteomic analysis, as shown in Figure 2A. Outlier samples were removed, and proteins with high missing rates were excluded, resulting in the quantification and analysis of 3913 out of 5359 proteins and 45 out of 47 samples (Tables S1 and S2).
Principal component analysis (PCA) indicated that proteomic patterns mainly differed among ages in the PC1 dimension (younger group prefers to cluster in the left, older in the right), differed between genotypes in the PC2 dimension (WT prefers to cluster in the upper area, 5xFAD in the lower area) (Figure 2B). Then, the differentially expressed proteins (DEPs) between WT and 5xFAD in these 5 ages were calculated based on FDR <0.05 and absolute log2FC > 1.2. The numbers of DEPs increased with age, except for the 3-month-old stage (3-month-old = 120; 4-month-old = 85; 6-month-old = 91; 9-month-old = 171; 11-month-old = 217, Table S3). Interestingly, the DEPs in these ages had few overlapping proteins (Chi-Squared Test, P3vs.4 = 0.74, P4vs.6 = 0.90, P6vs.9 = 0.15, P9vs.11 = 0.013). And the overlapping DEPs number between 3 and 4, 4–6, 6–9, 9–11 increased with age (2, 3, 8, 19 respectively, Figure 2C). Only one protein (APP) was shared among all ages (Figure 2C). The protein expression changes between WT and 5xFAD in the proteomics levels also showed a lower conservation property in each age, except for the relatively high conservation property seen in 6 and 9 months old (Figure S3). These results suggest that the PCx proteomics alteration compared to WT vs. 5xFAD have distinct features in each age, but showed relatively high similarity among 6/9/11-month-old.
GO enrichment analysis of DEPs further confirmed the observed changes in protein functions at different stages (Figure 2D). The 3-month-old DEPs were found to be enriched in ensheathment-related functions (Figure 2D, colored in green), while the 4-month-old and 6/9/11-month-old DEPs were enriched in transporter activity- and Aβ clearance-related functions (Figure 2D, colored in blue and red), respectively. This finding suggests that there is a shift in the protein functions involved in the progression of olfactory memory impairment. Moreover, the 4-month-old stage is a critical turning point in the dysfunction process, as the enriched pathways are strongly different from those observed in the 3-month-old stage and start to be involved in synapse-related pathways (Figure 2D, boxed in purple). This finding provides supportive evidence for the behavioral changes observed in olfactory memory dysfunction starting in the 4-month-old 5xFAD mice. In addition, gene set enrichment analysis (GSEA) provided an overall view of functional changes at the proteomic level in different ages (Figure 2E). The significant functional changes observed in the 3-month-old mainly involved morphogenetic, tube-related, and development-related pathways, while those observed in the 4-month-old were related to axonal transport, secretion, and extension, which may all potentially contribute to the pathological changes observed in early AD. The significant function terms of the 6, 9, and 11-month-olds were not as prominent in the GSEA results, but were mainly related to phos/dephosphorylation, Aβ-related, ion transport, and autophagy-related functions. These results suggest that the altered proteins and functions are dynamic and shift along with Aβ accumulation, especially showing a turning point at the 4-month-old stage and starting to be more involved in AD progress.
Since the results of different analyses all showed several synapse-related functions and pathways, especially in 4-month-olds (Figure 2D purple box; Figure 2E), Western blot (WB) was performed. Classic synaptic biomarkers were used to investigated the changes in the synapse. The result of WB confirmed Aβ can induced synapse impairment in 4-month-old 5xFAD, with the up regulation of pre-synaptic protein synaptophysin (Syp, p = 0.047), and down regulation of glutamate receptors glutamate receptor 1, 2 (Gria1, p = 0.030, Gria2, p = 3.0e-4) and vesicular glutamate transporter1 (Slc17a7, p = 0.043) (Figures 2F and S7A).
Differentially expressed proteins in different stages shared the conserved network connection associated with Alzheimer’s disease and glial cells
The significant variation in protein changes between WT and 5xFAD in different stages begs the question of how these changes are reflected in the co-expression network. To address this, PCx proteomics data from all stages (45 samples x 3913 proteins) were used to construct a co-expression network using weighted correlation network analysis (WGCNA) (Figure 3A). The analysis generated 14 co-expression modules (M1-M14). We used module eigenvalue (ME) to calculate the correlation of the module and different traits (group, month-old, sex and genotype (WT or 5xFAD)). The results showed that one module was negatively correlated to group (M6), eight modules were correlated to month-old (M1, M4, M5, M7 were negatively correlated, M3, M6, M8, and M13 were positively correlated), five modules were correlated to genotype (M3, M4 were negatively correlated, M2, M6, and M13 were positively correlated), and four modules were correlated to both month-old and genotype (M3, M4, M6, and M13) (Figures 3B and S4).
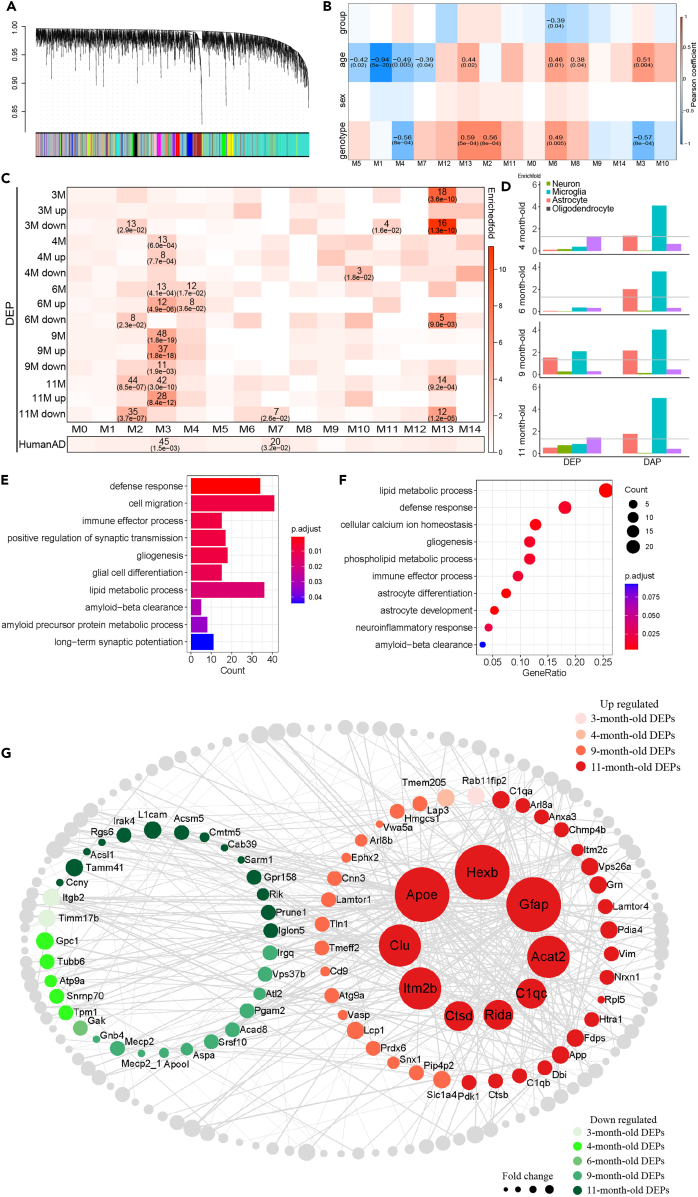
Figure 3.
Proteome in different stages shared the conserved co-expression network
(A) Dendro-tree plot of WGCNA. All protein were used to build up WGCNA module.
(B) Heatmap of module-trait relationship. Diagnose, age, sex and disease were all included to see their pearson correlation with different module from WGCNA (Module eigenvalue (ME) were used to calculate pearson coefficient, number in the upper cell block is Pearson coefficient and in lower cell block is p value).
(C) Heatmap of DEPs and Human postmortem brain data enrichment in WGCNA module. Enriched number and p value were showed in the matched cell block (Enriched fold in upper cell block and p value in lower cell block).
(D) Histogram of DEPs and DAPs enrichment in different type of cell. Enrichfold was taken –log10 and gray line indicate p < 0.05.
(E) KEGG pathway enrichment of proteins in M3.
(F) KEGG pathway enrichment of DAPs core (overlapping DAPs in M3).
(G) Network of all DEPs and DAPs. Size of the nodes indicate their fold change. Green nodes indicate they are down-regulated, red nodes indicate they are up-regulated, both color shade along with age. The size of hub protein nodes are 5 times enlarged and are in the middle of up-regulated proteins.
The DEPs at different ages were then subjected to an enrichment analysis with the co-expression modules. Surprisingly, almost all DEPs were enriched in M3, with the exception of DEPs in 3-month-old (DEPs 3M), and the extent of enrichment increased with age (4-month-old: enrichfold = 2.54, p = 6.0e-4; 6-month-old: enrichfold = 2.47, p = 4.1e-4; 9-month-old: enrichfold = 4.84, p = 1.8e-19; 11-month-old: enrichfold = 3.36, p = 3.0e-10; Figure 3C). This finding indicated that DEPs across different ages shared a conserved protein network dysfunction that began with 4-month-old mice, despite high DEP heterogeneity. Furthermore, the module enrichment analysis of AD DEPs in human postmortem prefrontal cortex (ROSMAP dataset)19 also enriched in M3 (enrichfold = 1.55, p = 1.5e-3, Figure 3C) indicated that M3 is highly correlation with AD. Summarizing the findings with the previous behavioral results suggests that PCx proteome changes accompany with olfactory memory impairment are associated with AD progress. Additionally, KEGG enrichment revealed that proteins in M3 participate in immune-, synaptic-, glia- and even Aβ-related pathways (Figure 3E), all of which are the main pathological impairments of AD.20,21,22
In addition, we utilized the DDPNA method to obtain DEPs-associated proteins (DAPs) in order to gain a better understanding of their underlying mechanisms and relations. DAPs are sets of proteins that are significantly correlated with DEPs in M3 and are considered to be involved in a common function. DAPs from various stages were combined and constructed through a co-expression protein network based on the Pearson’s correlation coefficient (Figure 3G), where the up-regulated and down-regulated DAPs are presented in the middle circle. The network showed highly connected and a total of nine hub genes were identified, all of which were up-regulated. Most of these hub genes, such as Apoe, Clu, Itm2b, Ctsd, and C1qc, have been reported to be associated with AD.23,24,25,26,27 Many of these genes are primarily expressed in microglia and astrocytes, such as Hexb, Apoe, and C1qc in microglia and Gfap in astrocytes. This indicates that these DEPs or DAPs may have a cell preference.
To answer wheather DEPs and DAPs present cell preferences, we performed a cell-type enrichment analysis based on the cell-type specific protein markers list, which was obtained from the supplementary file in Seyfried’s work.28 All DAPs in M3 derived from 4-month-old to 11-month-old DEPs (labeled as DAP4, DAP6, DAP9, and DAP11) were enriched in both astrocyte and microglia, while only DEPs in 9-month-old were enriched in both astrocyte and microglia (Figure 3D). The KEGG pathway analysis of the core DAPs (97 proteins), overlapping DAPs among 4/6/9/11-month-old in M3, is enriched in lipid metabolic processes, neuroinflammatory responses, glial functions, and Aβ clearance function which is closely approaching significant enrichment (FDR = 0.093, p = 3.23e-3) (Figure 3F). In conclusion, DAPs in M3 showed strong cell preference with astrocytes and microglia, which indicates that those two cell types may play essential roles in Aβ induced PCx molecular change.
To validate the pathology-related changes in microglia and astrocyte in the PCx, WB and immunohistochemistry were conducted. The results of the WB analysis showed significant increase in both Iba1 (a biomarker of microglia) and Gfap (a biomarker of astrocyte) in 4-month-old 5xFAD mice, and also Gfap in 3-month-old mice (Iba1, 3-month-old: p = 0.081; 4-month-old: p = 3.5e-3; Gfap, 3-month-old: p = 9.9e-3; 4-month-old: p = 8.9e-3, Figures 4D–4F; Figure S7B). The immunohistochemistry results (Iba1 3-month-old: p = 0.66 4-month-old: 1.24-fold, p = 0.024; Gfap 3-month-old: 1.47-fold, p = 0.047; 4-month-old: 2.06-fold, p < 0.001; Figures 4A–4C; Figure S7B) were consistent with the WB results. Interestingly, although the number of astrocytes increased in 3-month-old 5xFAD mice, only in 4-month-old 5xFAD mice were microglia and astrocyte observed to be closely gathering around Aβ deposits. These findings confirm our previous results and also emphasize the microglia and astrocyte functional changes and their potential role in Aβ-induced olfactory memory impairment at the molecular level.
Figure 4.
Immunohistochemistry and WB of astrocyte and microglia in the PCx
(A) Immunohistochemistry images of Aβ, Iba1, and Gfap in the PCx from WT and 5xFAD mice (3 and 4-month-old). Scale bar, 50μm.
(B and C) Quantification of Gfap+ (b) and Iba1+ (c) cell numbers in the immunohistochemistry images (3 mice each group, 3 slides each mouse, counting in the same area). White boxed area is for zoom in.
(D) Western blot images of β-actin, Iba1, and Gfap in the PCx from WT and 5xFAD mice (3 and 4-month-old). The calculation was done in the 640 μm × 640 μm area.
(E and F) Quantification of Gfap Iba1 in the western blots shown in (d). Values are mean ± SEM (3 and 4-month-old, n = 3 mice/genotype). All values are mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Multiple t test.
Interaction between microglia and astrocyte significantly decreased in mild cognitive impairment
Since 4-month-old is a turning point of Aβ-induced olfactory memory impairment and PCx molecular change, a 4-month-old DAP co-expression network was performed and showed plenty of significant interaction astrocyte specific proteins (ASPs) and microglia specific proteins (MSPs) in expression correlation and STRING protein-protein interaction levels (Figure S5A; Figure 5A; Table S4). The heatmap (Figure 5A) showed that almost all ASPs and MSPs in M3 DAPs were upregulated in 5xFAD vs. WT starting from 4-month-old. WB provide additional support to significant changes in 4-month-old mice with up regulation in Anxa3 (p = 0.010) and Vim (p = 1.1e-3) and down regulation in Lcp1 (p = 0.020) (Figure 5B). Further GO enrichment analysis of 4-month-old DAP implies that these ASPs and MSPs may affect cognitive function by regulating synaptic function through Golgi vesicle transport and synapse structure (Figure 5C). Moreover, The ASPs have the most interaction with MSPs (p = 8.0e-3, 1000 times permutation test, Figure S5B). The STRING network analysis between astrocyte and microglia specific proteins also showed the highest connectivity compared to randomly picked protein interactions in DAP4 (p < 0.001, 1000 times permutation test, Figure S5C). These astrocyte-microglia interactions predominantly contribute to the early stage of 5xFAD mice olfactory dysfunction.
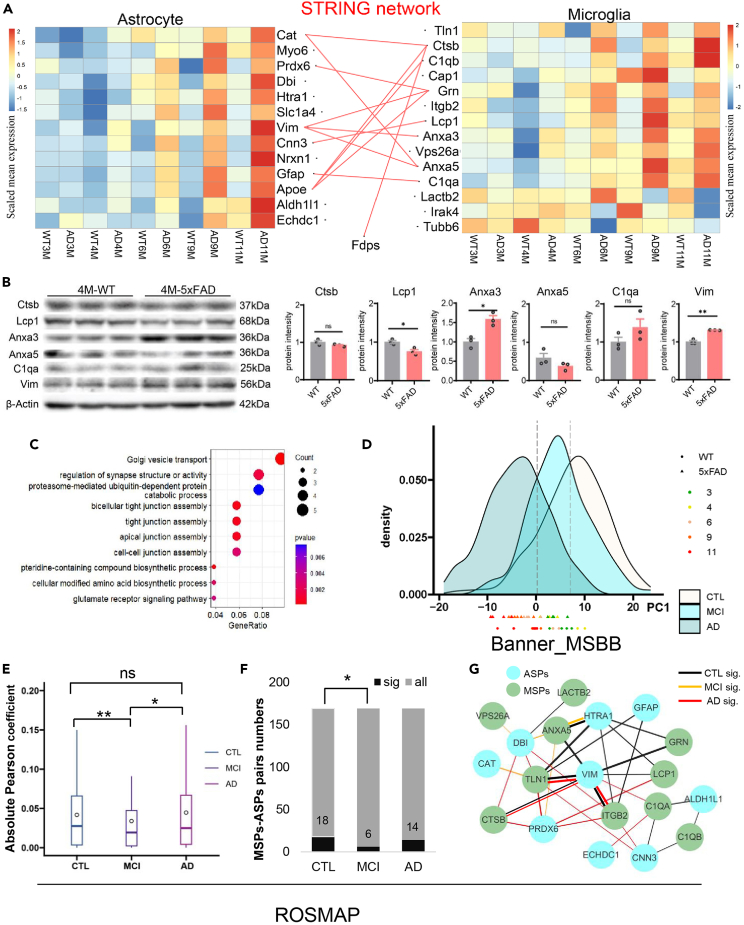
Figure 5.
Interaction of astrocyte and microglia alter in the early stage of AD
(A) STRING network of astrocyte and microglia specific proteins in 4-month-old DAPs combined with a heatmap of their relative abundance.
(B) WB of astrocyte and microglia specific proteins in the 4-month-old mice.
(C) GO biological process enrichment of 4-month-old DAPs.
(D) PCA analysis with combined human (Banner_MSBB dataset) and mouse proteomic data.
(E and F) ROSMAP data analysis (CTL = 117, MCI = 155, AD = 94). e, Absolute Pearson coefficient of ASPs-MSPs interaction showed in a significant decreased in MCI from the ROSMAP cohort, Wilcox test. f, Significant interaction numbers between ASPs and MSPs only decreased in MCI from the ROSMAP cohort, chi-square test.
(G) The disease status specific interaction between ASPs and MSPs from the ROSMAP cohort. ∗, p < 0.05, ∗∗p < 0.01.
Aβ deposit is accumulated in 5xFAD over time and has also increased among healthy control (CTL), MCI, and AD in human, PCA analysis with combined human (Banner_MSBB dataset) and mouse proteomic data was processed. Result showed that 3/4-month-old 5xFAD distributed in MCI stage and 6/9/11-month-old 5xFAD distributed in AD stage in PC1 level (Figure 5D). It suggested that 4-month-old 5xFAD mice may reflect the Aβ effect in MCI, which has the potential to develop into AD.
Next, the ROSMAP proteomic dataset was analyzed to test the consistency with 5xFAD mice results. The 13 ASPs and 13 MSPs of DAP4 (IRAK4 is missing in the dataset) were picked and separately calculated the Pearson’s correlation between ASPs and MSPs based on different disease status. Interestedly, MCI has the lowest absolute Pearson coefficient (Figure 5E, Wilcoxon test, p = 0.014, CTL vs. MCI; p = 0.036, CTL vs. AD) and the lowest significant interaction pairs (Figure 5F, p = 0.011, chi square test, CTL vs. MCI). There were more ASPs-MSPs interactions in the CTL and AD groups than in the MCI group. Most of the ASPs-MSPs interactions in the CTL disappeared in MCI, retaining only ANXA5-HTRA1 protein interaction. But more ASPs-MSPs interactions in the CTL remained in the AD than in the MCI, for example, TLN1-VIM, CTSB-VIM, ITGB2-VIM pairs. At the same time, AD appear some additional interaction (Figure 5G). Another human postmortem dataset (Banner_MSBB dataset) also showed the similar results. MCI have lowest absolute Pearson’s coefficient in the Banner_MSBB dataset (Figure S6A, Wilcoxon test, p = 0.041, CTL vs. MCI; p = 7.2e-4, CTL vs. AD), and MCI have lowest significant interaction (Figures S6B and S6C). Notably, TLN1-VIM pair in these two human datasets are both significant in CTL and AD, DBI-LACTB2 pair in CTL, VPS26A-DBI pair in MCI. Above all, these results shown that the decline of the astrocytes-microglial interaction is the characteristics in the MCI stage. These protein-protein interactions may be important in Aβ involved AD pathogenesis, but not only the protein expression change.
To further investigate the interaction between astrocyte and microglia, in vitro cell cultured experiment was performed. We utilized astrocyte induced medium (AIM) with different treatments and regular medium (RM) to culture microglia. Then, WB was performed to see the changes in certain proteins which showed interaction in previous results (Figure 6; Figure S8). In WB results, astrocyte cannot induce MSPs change (Figure 6C, AIM vs. RM), but Aβ induced astrocyte can alter microglia Anxa3, C1qa, Ctsb and Lcp1 expression (Figure 6C, both significant difference between Aβ∗ + AIM vs. DMSO+AIM, Anxa3 P = 5e-3, C1qa P = 1e-3, Ctsb p = 0.048, Lcp1 p = 0.02; Anxa3 and C1qa significant between Aβ∗ + AIM vs. DMSO + RM, Anxa3 p = 0.02, C1qa p = 1.9e-3). Interestedly, only C1qa have change directly by Aβ induced microglia without astrocyte (Figure 6C, Aβ + RM vs. DMSO+ RM, p = 3.3e-3), but Aβ + Aβ induced AIM have no synergistic effect (Figure 6C, no significant between Aβ∗ + AIM vs. Aβ+AIM). They showed that Anxa3, C1qa, Ctsb and Lcp1 protein change in microglia need astrocyte which activated by Aβ. Additionally, Lcp1 and C1qa also showed more and stronger connection signal in AD, while Ctsb showed more connection signal in MCI (Figure 5G; Figure S6C). These results confirm our previous finding that Aβ can induce astrocyte and microglia interaction.
Figure 6.
Interaction between astrocyte and microglia
(A) Workflow of astrocyte and microglia in vitro experiment. Microglia were cultured with astrocyte medium and regular medium separately. ∗ Replacing medium with regular DMEM+10%FBS after 24h of Aβ stimulate, and collected medium after cultured with regular medium for 24h.
(B) Immunoblots images of β-actin, Anxa3, Anxa5, C1qA, Lcp1, and Ctsb in the PCx from cultured microglia.
(C) Quantification of Anxa3, Anxa5, C1qA, Lcp1, and Ctsb in the immunoblots shown in (b). Values are mean ± SEM (4-month-old, n = 3 mice/genotype). One-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
AD is the common neurodegenerative disease and Aβ induced molecular feature in MCI is not well understood. Based on the National Institute on Aging and Alzheimer’s Association created separate diagnostic recommendations,29 MCI is the previous stage of dementia in AD. Identifying and characterizing the molecular mechanism of MCI may contribute to understanding early AD pathology and the treatment of AD. In our results, we found that olfactory memory impairment starts from 4-month-old and earlier than sound memory impairment and working memory impairment, which is consistent with previous reports that early olfactory dysfunction may be a unique symptom in MCI or early AD.2,4,5,6,7,8,30,31 Combined with the proteome PCA result suggested that 4-month-old 5xFAD mice might be as the MCI mouse model. The proteomics analysis of the PCx region, a key brain region associated with Aβ plaque and olfactory memory, in 5xFAD mice was performed to identified the core molecular function alteration in the MCI stage. The DEPs in 4-month-old enriched in transporter activity specificity and start to have alteration in microglia and astrocyte related functions.
Glia cells play a pivotal role in AD pathogenesis. Microglia is considered to most important in AD progress. It can establish an intimate contact with Aβ plaques and became reactive. AD-associated microglia were identified in postmortem human brains and AD mouse models and were involved in Aβ spreading and Aβ plaque growth.32 APOE is a key risk factor in AD and is mainly expressed in astrocyte, which produced along with Aβ.33 It is upstream of Aβ plaque formation and part of an essential feedforward loop between astroglia and microglia which also plays a hub role in our data. Combined with mouse proteomics and human postmortem brain proteomics, the interaction of several proteins enriched in microglia and astrocyte showed a significant change only in the MCI stage. In the network, the hub proteins are Vim, Gfap, Apoe, and Fdps. Except Fdps, other hub proteins are all ASPs. It indicated that astrocyte may play a leading role in MCI stage. Moreover, Apoe interact with microglia specific proteins (MSPs: Αnxa3/5, C1qa/b, Ctsb and and so forth), oligodendrocyte specific proteins (OSPs: Cntn1, Enpp6), neuron specific protein (NSP: L1cam) and almost one-third of non-specific cell type proteins (28/69). Gfap also interact with MSPs (C1qa/b, Anxa5, Tln1, and Lcp1), OSPs (Gsn, Tpm1, Rab39b, Gpr158, and Unc80) and another one-third of non-specific cell type proteins (27/69), but not with neuron. While Vim only interacts with MSPs (Lactb2 and Anxa5) (Figure S5A). This implies astrocyte may subtly bridge the interaction among microglia, oligodendrocyte and neuron. Combined the co-expression network and STRING network, Apoe, the strongest risk factor for AD, showed interactions with Ctsb and C1q. Ctsb, a powerful lysosomal protease, has been reported to improve behavioral deficits in AD when the gene is knocked out.34 C1q also reported can binding to Aβ.35 The APOE-C1QB pair also validated in human dataset (Figure S6C). In microglia, C1q a/b and Anxa3/5 were highly interacting with all hub genes of astrocyte. ANXA3/5-VIM pairs also validated in human dataset (Figure 6E; Figure S6C). This suggests that these proteins may act synergistically to accelerate memory impairment and the decreased astrocyte-microglia interaction reflects the early characteristics of AD.
The hub proteins Vim and Gfap are both involved in reactive astrocytes formation in AD mice and patients.36 Apoe can bind to Aβ and involved in neuroinflammation. These ASPs interacted with several MSPs which also highly associated with AD. Anxa5 enriched in Aβ plaque.37 Ctsb involved in Aβ clearance.25 Elevating C1q expression in microglia is important in harmful A1 astrocyte activation, and also bind to Aβ plaque to release an “eat-me” signal to recruit microglia to clear it.38 These proteins are both associated with Aβ or astrocyte activation. Vim and Gfap are both highly expressed in reactive astrocytes. All of these MSPs and ASPs in 4-month-old mice and patients with MCI are crucial in AD impairment and numerous studies have confirmed that astrocytes and microglia interaction are associated with AD characteristic,39,40,41 but activated microglia were insufficient to kill neurons alone.38 Overall, these findings suggest that astrocytes-microglia interaction not only astrocyte and microglia individually may play a leading role in the early stage of AD and the decreasing of astrocytes-microglia interaction may accelerate the neurodegeneration (Figure 7).
Figure 7.
Illustration of potential mechanism among Aβ, astrocytes, and microglia in initial memory impairment stage
Limitations of the study
Although we found Aβ induced ASPs-MSPs interactions to have pivotal roles in early AD. However, some ASPs-MSPs interactions disappear in MCI and another ASPs-MSPs interactions appear in AD need more research to verified and discovered the potential mechanism. And how Aβ, astrocytes and microglia joint effect involved in memory loss still need more experiments to elucidate.
What’s more, inconsistent in different stages proteome level implies that the observation in the later stage of AD may hard to elucidate the pathogenesis of AD. Here, we confirmed that the interaction of astrocytes and microglia decreased is important in the MCI stage of AD progress and give a new insight that the synergy effect of astrocyte and microglia decreased might be more harmful to initial cognitive impairment in AD progress. Finding a drug that can synergy the astrocyte and microglia interaction in the MCI stage may be useful in AD treatment research.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Iba1 | FUJIFILM Wako Shibayagi | Cat# 019–19741, RRID: AB_839504 |
| Anti-GFAP | Abcam | Cat# ab53554, RRID: AB_880202 |
| Anti-6E10 | BioLegend | Cat# 803014, RRID: AB_2565329 |
| Anti-Synaptophysin | Abcam | Cat# ab14692, RRID: AB_301417 |
| Anti-Gria1 | Proteintech | Cat# 67642-1-Ig, RRID: AB_2882842 |
| Anti-Gria2 | Proteintech | Cat# 11994-1-AP, RRID: AB_2113725 |
| Anti-Gria3 | Proteintech | Cat# 29588-1-AP |
| Anti-Gria4 | Proteintech | Cat# 23350-1-AP, RRID: AB_2879262 |
| Aniti-PSD95 | Proteintech | Cat# 20665-1-AP, RRID: AB_2687961) |
| Anti- VGlut1 | Cell Signaling Technology | Cat# 12331, RRID: AB_2797887 |
| Anti-Vim | Immunoway | Cat# YT4880 |
| Aniti-C1qa | Immunoway | Cat# YN0612 |
| Anti-LCP1 | Immunoway | Cat# YN0002 |
| Anti-Anxa3 | Immunoway | Cat# YT0237 |
| Anti-Anxa3 | Abcam | Cat# ab14196, RRID: AB_300979 |
| Anti- Ctsb | R&D | Cat# AF965 |
| Anti-β-Actin | Sigma-Aldrich | Cat# A5441, RRID: AB_476744 |
| Bacterial and virus strains | ||
| AAV9-CamkII-Gcamp6s | Taitool, Shanghai | Cat# S0290-9 |
| Critical commercial assays | ||
| MonoSpin Reversed Phase Columns (C18, C18 FF, Ph) | GL sciences | Cat# 5010-21700 |
| BCA Protein Assay Kit | Solarbio | Cat# PC0020 |
| Deposited data | ||
| Raw Mass Spectrometry Data Files | This paper | ProteomeXchange,PXD040802 |
| ROSMAP TMT-labeled proteomics data | Synapse website | https://www.synapse.org/#!Synapse:syn17015098 |
| Banner label-free proteomics data | Synapse website | https://www.synapse.org/#!Synapse:syn20818199 |
| MSBB label-free proteomics data | Synapse website | https://www.synapse.org/#!Synapse:syn20801227 |
| Experimental models: Cell lines | ||
| MG6 | Cellosaurus | RRID: CVCL_8732 |
| U87 | ATCC | Cat# HTB-14 |
| Experimental models: Animal | ||
| Mouse: 5xFAD Tg6799 | The Jackson Laboratory | Cat# 34848-JAX |
| Mouse: C57BL/6J | The Jackson Laboratory | Cat# 000664 |
| Software and algorithms | ||
| R | R Foundation for Statistical Computing | https://www.r-project.org/ |
| WGCNA | Langfelder and Horvath, 200842 | https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/ |
| MATLAB | The MathWorks | https://au.mathworks.com/products/matlab.html |
| Maxquant | Cox and Mann, 2008; Version 1.5.0.38 | https://www.maxquant.org/ |
| Cytoscape | Cytoscape Consortium, version 3.10.0. | https://cytoscape.org/ |
| Anymaze | Stoelting | version 6.0 |
Resource availability
Lead contact
Lead contact Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hong Qing (hqing@bit.edu.cn).
Materials availability
This study did not generate new reagents.
Data and code availability
-
•
The original mass spectra and the protein sequence databases have been deposited at ProteomeXchange (PXD040802) (https://www.proteomexchange.org/) and are publicly accessible at https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD040802.
-
•
Human postmortem proteomic data (ROSMAP: syn17015098, MSBB: syn20801227and Banner: syn20818199) were not generate by this work and can be obtained from synapse website upon request. Customized code was uploaded to github (https://github.com/liukf10/5xFAD-PCx-proteomic-data-analysis).
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and study participant details
Animals
All surgical and experimental procedures were approved by the Institutional Animal Care and Use Committee of the Beijing Institute of Technology, Beijing, China. The following animals were used in this study: adult (3, 4, 6, 9, 11-month-old) 5xFAD and wild-type (WT) mice (C57/BL6). WT and 5xFAD mice of C57/BL6 background were kept and bred in the Animal room of Beijing Institute of Technology. Mice were group-housed in a 12/12 h light/dark cycle (2–5 animals per cage) at a consistent ambient temperature (23 ± 1°C) and humidity (50 ± 5%), and all experiments were performed during the light cycle. Food and water were accessed ad libitum. Littermates were randomly assigned to each condition by the experimenter. WT and 5xFAD were bred together, and the survived offspring were both used for the experiment. The littermates were identified by genotyping and separated into two groups. Sex information can be found in the Table S1.
Cell lines
U87 (Astrocyte related cell line) and MG6 (microglia related cell line) were cultured in vitro, both using DMEM+10%FBS medium. The cells were used after passage and maintained at 37°C in a 5% CO2 95% air atmosphere for a week.
To simulate Aβ related environment, Aβ1-42 were added to partial dishes of U87 and MG6 at the concentration of 4μM according to the design.
More detailed design was described in method details (astrocyte and microglia interaction in vitro)
Method details
Odor or sound associated fear memory test
10 WT and 10 5xFAD were used for odor associated fear memory test, 8 WT and 8 5xFAD were used for sound associated fear memory test. The sample size was used based on experience and previous experiment.43 Sex effect was not included in this work. Experiment was performed blindly, with only mice number can be seen but not genotype. All mice were used for statistical analysis.
All mice were handled for 1h per day for two weeks prior to the behavioral tests. During the handling session, mice were habituated by petting the mouse gently. For fiber photometry recording experiment, mice were connecting to the optical fiber in the same time. The behavioral session was conducted in a fear conditioning box (46001, UGO BASILE S.r.l, Italy).
The freezing was defined as a complete absence of movement, except for respiration. Scoring of the duration of the freezing response was started after 1 s of sustained freezing behavior. The freezing was quantified by Anymaze (version 6.0, Stoelting) software based on a threshold of change in video image pixels.
At the encoding stage (day 1), after 5 min habituation in context A, mice received olfactory cue or auditory cue with electric foot shocks 3 times. The cues have 15 s duration (olfactory: limonene or sound: 70dB 10kHz pure tone). Electric foot shocks only appear in the last 2 s of cues duration. The interval time between each trial is 60 s. The freezing time on 60 s after receiving cue was used to calculated the freezing ratio.
The freezing ratio = odor or auditory cue freezing time (s)/60 s.
At the recall stage (day 2), after 10 min habituation, the mice were received same cue on the day1 in different context B. The freezing time on 10 min habituation and the freezing time on 60 s after receiving cue was used to calculate freezing growth score:
The freezing growth score = (odor or auditory cue freezing time - habituation freezing time)/(odor or auditory cue freezing time + habituation freezing time).
The novel odor recognition test
9 WT and 14 5xFAD were used for novel odor recognition test. The sample size was used based on experience and previous experiment.43 Sex effect was not included in this work. Experiment was performed blindly, with only mice number can be seen but not genotype. All mice were used for statistical analysis.
A common program for novel odor recognition involves two stages: the training stage and the testing stage. During the training stage, the limonene-scented dish is placed in an open field with two identical dishes and mice can freely explore those dishes over a period of time. After 90 min, mice are sent back to the same open field for the testing phase, to which the familiar limonene-scented dish and a new pine-scented dish are placed during the testing stage. Mice exploration time near the odor dish was recorded. Novel odor recognition index calculation formula:
Novel odor recognition index = new odor exploration time - old odor exploration time/(new odor exploration time + old odor exploration time) ×100%.
T-maze test
6 WT and 6 5xFAD were used for working memory T-maze test. The sample size was used based on experience and previous experiment.43 Sex effect was not included in this work. Experiment was performed blindly, with only mice number can be seen but not genotype. All mice were used for statistical analysis.
T-maze was performed to test working memory by manual operation. Mice were gradually food restricted until they reached 85% body weight. Mice were habituated to the T-maze over two days, during which mice were placed into the T-maze for 10 min 50 μL of 50% sweet-milk reward was placed in the reward tube at the end of choice arms. Once experiments began, mice were given ten trials per day. Each trial was consisted of the sample run, delay and choice run.
On the sample run, the mouse was forced to enter the left or right arm to consume the sweet-milk reward, while the other arm was blocked by a door. Then the mouse came back in the start position. On the choice run, the blocked door was removed and the mouse was allowed to choose one of the goal arms. The time interval between the sample and choice run was 10s. If the mouse visited the non-sample arm, it was allowed to acquire the reward. If the animal visited sample arm, it could not receive the reward. If the mouse was consumed the reward, this was scored as a correct trial. The time interval from one trial to the next was about 1 min. After rewards, the mice were allowed to return to the start point by themselves; however, if the mice could not return to the start point by themselves 15 s after feeding, they will be forced to return to the start point. Mice were trained for 10 trials per day during 7 days.
Brain stereotactic surgery for fiber photometry recording
The animals were deeply anesthetized and placed in a stereotactic frame (RWD, Shenzhen, China). Ophthalmic ointment was applied to prevent dehydration. 300 nL viruses of AAV9-CamkII-Gcamp6s (titer, 3.1 × 1012gc/ml, Taitool, Shanghai, China) were injected by a 10 μL Hamilton microsyringe at a constant speed with a microsyringe pump (UMP3; WPI, Sarasota, FL, USA) and controller (Micro4; WPI, Sarasota, FL, USA). The viruses were injected into PCx region (anteroposterior (AP): −1.0 mm; mediolateral (ML): ±4.5 mm; dorsoventral (DV): −4.0 mm). The needle was kept still for 10 min to allow the diffusion of the virus once the injection was completed and then was withdrawn slowly and completely. We also detected the optic fiber transmissivity by measuring the light intensity as the laser launch, tip of the coupled fiber (diameter, 200 μm; Ferrule O.D, 1.25 mm; N. A., 0.37; length, 2.0 or 4.0 mm; OriginOpto Inc., China), and the fiber ferrule implant by optical power meter (PM121D, Thorlab, USA). Fiber ferules were selected based on optical transmissivity (>80%).
We measured the intensity of coupled fibers before that were connected to the implanted fiber ferrule and to each animal every day. The intensity was adjusted to a certain power. Also, the fiber ferrule was placed 50–100 μm above the viral injection site. The optical fiber was secured to the skull using jeweler′s screws and dental cement.
Fiber photometry recording
6 WT and 6 5xFAD were used for fiber photometry recording. The sample size was used based on experience and previous experiment.43 Sex effect was not included in this work. Experiment was performed blindly, with only mice number can be seen but not genotype. All mice were used for statistical analysis.
The fiber photometry recording was performed in an apparatus obtained from the Thinker Tech Nanjing Biotech Limited Co., Nanjing, China. The signal was digitized and collected by ThorCam-DAQ software. The calcium signal was recorded after three weeks of GcamP6s virus expression. The timing of behavioral variables was recorded by the same system. The behavioral processes and calcium signal were synchronized and analyzed by the custom-written MATLAB software (The MathWorks, Inc., USA).
GcamP protein is composed of cpGFP protein, CaM (calmodulin) and M13 peptide, and when calcium ions are present, CaM and M13 bind, which jointly cause the conformation of cpGFP protein and enhance the fluorescence signal. It is used in experiments to indicate Ca signaling activity in cells. GcamP6s is a new generation of GcamP proteins. GcamP6s is an ultra-sensitive fluorescent protein that can specifically and efficiently reflect changes in intracellular free Ca2+ concentrations.
For calculating GcamP6s signal, the relative fluorescence changes of ΔF/F were calculated as Ca2+ signal and the fluorescence responses of ACh signal as follows: ΔF/F= (Fraw- Fbaseline)/Fbaseline.
Immunohistochemistry
Mice were deeply anesthetized and transcardially perfused with 0.9% saline followed by 4% paraformaldehyde (PFA) in PBS. Brains were extracted, removed and kept in 4% PFA for at least 24 h. Next, brains were transferred to 30% sucrose dissolved in PBS until they sank to the bottom of the container and were then sliced into 30 μm coronal sections using a freezing microtome (Leica, CM3050 S, Germany). The sections were stored at −20°C in PBS containing 30% glycerol (v per v), 30% ethylene glycol (v per v) until they were processed. For immunofluorescence staining, free-floating sections were washed with PBS three times (5 min each) and incubated with blocking buffer that contains 10% donkey serum dissolved in 0.3% PBST (0.3% Triton X-100 in PBS) for 2 h. Sections were then incubated with primary antibodies diluted in blocking buffer (5% donkey serum) overnight at 4°C. After incubation, the sections were washed three times (5 min each) with PBST and incubated with a fluorescent dye-conjugated secondary antibody (1:500, Abcam, UK) for 2h at room temperature. Following three washes (5 min each time) with PBS, sections were mounted under coverslips. Primary antibodies used were: anti-Iba1 (1:1000, rabbit, 019–19741, Wako), anti-GFAP (1:1000, rabbit, goat, ab53554, Abcam), anti-6E10 (1:1000, mouse, 803014, biolegends), anti- Synaptophysin (1:1000, rabbit, ab14692, Abcam), anti-Gria1 (1:20000, mouse, 67642-1-IG, proteintech), anti-Gria2 (1:500, rabbit, 11994-1-IG, proteintech), anti-Gria3 (1:1000, rabbit, 29588-1-IG, proteintech), anti-Gria4 (1:500, rabbit, 23350-1-IG, proteintech), anti-PSD95 (1:2000, rabbit, 20665-1-IG, proteintech), anti-Vglut1 (1:1000, rabbit, 12331, Cell signaling), anti-Vim (1:1000, rabbit, YT4880, immunoway), anti-C1qa (1:1000, rabbit, YN0612, immunoway), anti-Lcp1 (1:1000, rabbit, YN0002, immunoway), anti-Anxa3 (1:1000, rabbit, YT0237, immunoway), anti-Anxa5 (1:1000, rabbit, ab14196, Abcam), anti- Ctsb (1:1000, goat, AF965, RD), anti-β-actin (1:5000, mouse, A5441, sigma), Confocal fluorescence images were acquired using a Nikon A1 confocal laser scanning microscope with ×20 objectives for imaging stained or autofluorescent neurons. Images were analyzed by NIS-Elements AR and ImageJ software.
WB normalization: First, the gray value of each sample is divided by the corresponding gray value of β-actin. Then, average of control was used to divide every sample.
Protein extraction
The frozen samples of piriform cortex were homogenized in lysis buffer (40Mm Tris-HCl pH7.4, 10mM DTT, 8M Urea) with 1x Proteinase inhibitor and 1x PhosSTOP (Roche). The homogenized samples were then sonicated and centrifuged to fully extract proteins (4°C, 12000 rpm × 20 min). The supernatant was transfer to new tube and the protein concentration was measured using BCA kit (Solarbio) according to protocol.
Western blot
The samples in equal 30ug amount of protein were loaded on 10% SDS-polyacrylamide gels and then transferred to 0.22μm PVDF membranes. The blots were probed with the following antibodies: anti-Iba1 (1:500, rabbit, 019–19741, Wako), anti-GFAP (1:500, rabbit, goat, ab53554, Abcam). Primary antibodies were incubated overnight at 4°C and followed by incubation with second antibodies for 1h at room temperature before washing by TBST for 3 times. The HRP-labeled antibodies were detected by an ECL kit with image analyzer.
Proteome preparation for MS
360ug protein of each sample was obtained and diluted into 100uL using lysis buffer, according to their concentration. The samples were treated with IAA (50mM, 10uL) for 30min in dark after 60min degeneration of protein in 37°C. Then the sample were diluted to 800uL by using 50mM NH4HCO3 and 160uL (72ug protein) were transfer to new tube to be digested into peptides by 1.5ug trypsin (Promega) for 18-20h. After digestion, desalination was performed using Mono Spin C18 kit (GL Science Inc) followed its instruction. Peptide concentration of samples was estimated by BCA kit (Solarbio).
Label-free LC-MS analysis and protein identification
The analysis of PCx tissue was performed by an Ultimate 3000 nano LC system coupled to Q Exactive HFx mass spectrometer (Thermo Fisher Scientific). The C18 separation column is Acclaim PepMapTM RSLC 75um×25cm, Nano Viper (Thermo Fisher Scientific). Samples were eluted with solvent A (100% LC-MS grade water and 0.1% LC-MS grade formic acid) and solvent B (20% LC-MS grade water, 80% LC-MS grade acetonitrile and 0.08% LC-MS grade formic acid). Elution process is as follows: 95% solvent A and 5% solvent B for 4min, 92% solvent A and 8% solvent B for 120min, 68% solvent A and 32% solvent B for 5min, 1% solvent A and 99% solvent B for 6min, 95% solvent A and 5% solvent B for 5min. After 140min eluting process, raw data were acquired with Xcalibur software. Then, the raw data of LC-MS were further analyzed using Maxquant software and matching with Uniprot mus musculus database (Swiss-Prot + TrEMBL, downloaded at Feb,2021). 5359 proteins were identified in total. The raw proteomic data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository44,45 with the dataset identifier PXD040802.
Astrocyte and microglia interaction in vitro
U87 (Astrocyte related cell line) and MG6 (microglia related cell line) were cultured in vitro, both using DMEM+10%FBS medium. 7 group were designed to investigate how can astrocyte influence microglia protein expression. 7 group include AIM+microglia, DMSO+AIM+microglia, Aβ1-42+AIM+microglia, Aβ1-42∗+AIM+microglia,RM + microglia,DMSO+RM+microglia and Aβ1-42+RM+microglia. Aβ1-42 was pre-processed according to established protocol.46 Briefly, Aβ1-42 was dissolved in hexafluoro-2-propanol and dried by evaporation. Then, Aβ1-42 was dissolved in DMSO to 5 mM, which was then added to ice-cold F12 medium to 100 μM at 4°C for 24h. After centrifuged at 14 000g for 10min, Aβ1-42 was ready for use. Astrocytes were cultured with regular medium with DMSO and Aβ1-42 separately for 24h, then the medium were collected to culture microglia. One of the astrocytes were continued to culture for 24h after removing the medium with Aβ1-42. After that, this medium of astrocyte was also collected to cultured microglia. Half of the microglia were cultured with regular medium, medium with DMSO and Aβ1-42, another half were cultured with astrocyte medium collected previously. All microglia were collected after 24h culturing. Then, microglia were lysed and applied for western blot to investigate related protein changes found in previous analysis.
Quantification and statistical analysis
Human postmortem brain proteomic resources
ROSMAP TMT-labeled proteomics dataset (165 CTL,97 MCI, AD 121, syn17015098) is obtained from synapse website. The quantification matrix was removing batch effect by TAMPOR algorithm (https://github.com/edammer/TAMPOR/) and then run the preprocess flow below. Banner (73 CTL,26 MCI, AD 92, syn20818199) and MSBB (35 CTL,41 MCI, AD 178, syn20801227) datasets are label-free proteomics data which is obtained from synapse website.
Preprocess of proteomic data
For mouse proteomic data, quantified proteomic data acquired from Maxquant were checked for contaminated and reversed protein first. After removing contaminated and reversed protein, 4985/5393 proteins were remained for later evaluation. First, data were checked for sample outlier and 2 outliers out of 47 samples were removed. Then, samples with less than 80% of missing value were imputed. All processes were running in R with DDPNA package (https://cran.r-project.org/web/packages/DDPNA/index.html). 3913 out of 4985 proteins were left and log2 transformed. Unknown covariates were searched by sva packages. The unknown covariates and known covariates (including sex, age, passaged) will adjusted by linear regression. For human proteomic data, quantified matrix followed the sample outlier removal, missing value imputation and log2 transformed. And then, Banner and MSBB datasets are merged by ComBat function in sva package as Banner_MSBB dataset.
Principal component analysis
Principal component analysis (PCA), based on the relative expression of all proteins as features, was used to investigate and visualize the differences among 5xFAD model mouse, control wild-type mouse and also different age of mouse.
Differential expression analysis
Student’s t test was used to define DEPs in different age respectively. Absolute log2(FC) > 1.2 and FDR <0.05 were used to further filtering DEPs.
Weighted gene co-expression network analysis (WGCNA)
Weighted gene co-expression network analysis was carried out using R package WGCNA.42 The whole dataset was used to build up the clusters and modules at soft-threshold power of 4. All proteins were assigned to the most correlated cluster based on the Pearson correlation coefficient and building up different module, in which contain proteins with similar expression pattern. Then, DEGs was used to further investigate their role and function base on the DEGs enrichment performance in different modules. Dataset from other studies (including human and mice) were also used for deeper exploration of DEGs and WGCNA modules.
Differential expression associated proteins analysis
Proteins in one WGCNA modules were considered have strong interaction each other. Pearson correlation of each protein pairs in one module was calculated and filtered by fdr adjust. The Planar Maximally Filtered Graph algorithm was used to filter the most significant proteins linked with DEPs. It will be constructed a compact DEPs associated protein interaction network. The proteins in the network were called DAPs for further analysis. The process was running in R by DDPNA package.
Enrichment analysis
Gene ontology and KEGG pathway enrichment analysis was carried out using R package cluster Profiler.47 GSEA enrichment analysis and visualization were using GESA, Cytoscape and Enrichment Map.48 The different age proteomics change was evaluated by RRHO.49
DEPs-modules association and cell type specific enrichment analysis was also performed by hypergeometric test. Brain cell specific genes marker list was collected from supplementary files of Seyfried’s work.28 Enrich folds were showed in enrichment heatmap.
Enrich fold = (NumberDEPs i in module x/Numbermodule x)/(NumberDEPs i/Total Protein Number)
Protein-protein interaction (PPI) analysis
DAPs enriched in M3 from all stages were superimposed onto a composite PPI network, in which showing overall interaction and regulation of DAPs. Also, DAPs in M3 from 3- and 4-month-old were separately performed to observe the interaction of cell type specific proteins. Network from R were and visualized by Cytoscape. In addition, STRING network of astrocyte and microglia were obtained from STRING V1150 to validate PPI network from R. Permutation test was random 1000 times pick 13 and 14 proteins groups in network and calculated the two groups connective degrees.
The MSPs and ASPs were picked from human proteomic dataset and divided into CTL, MCI and AD three data to calculated Pearson coefficient. Due to the Pearson coefficient and p value are sensitivity with sample size. The equivalent sample size was used. 80 samples were random sampling 10 times in ROSMAP dataset and 50 samples sampling in Banner_MSBB dataset. The average Pearson coefficients were used. p values were calculated based on the average Pearson coefficient and sample size. Wilcoxon tests were used in differential analysis of absolute Pearson coefficients among the three groups. Chiq square test were used to evaluate the significant pairs number alter among the three groups. The significant protein-protein interaction was visualized by Cytoscape.
Statistical analysis
For behavior test, immunohistochemistry and WB, the unpaired two-tailed t-test was used. All data was presented as means ± standard errors of the means (SEM). All the statistical analyses were performed with GraphPad Prism 8.0. All of the statistical details of experiments can be found in the figure legends. Each experiment has been repeated at least three times in the manuscript with similar results. The statistical analysis of the experimental data was based on previous works43 and did not undergo a normal distribution test.
Acknowledgments
We thank the Analysis & Testing Center of Beijing Institute of Technology for MS instrument support. This research was supported by the Ministry of Science and Technology Key project (Grant No.2022ZD0206800), the National Natural Science Foundation of China (Grant No. 92049102), Beijing Municipal Natural Science Foundation (Grant No. 7222113). The First Hospital of Hebei Medical University Spark Program, Key Science and Technology Research Program of Hebei Provincial Health Commission Grant No. ZF2024142.The results published here are in whole or in part based on data obtained from the AD Knowledge Portal (∗∗https://adknowledgeportal.org/∗∗). These data were generated from multiple studies:MSBB dataset: postmortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank provided by Dr. Eric Schadt from Mount Sinai School of Medicine, with the proteomics provided by Dr. Levey from Emory University. Banner dataset: proteomics data provided by Dr. Levey from Emory University. A portion of these data were generated from samples collected through the Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona. The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer1s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer1s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson's Disease Consortium) and the Michael J. Fox Foundation for Parkinson's Research. ROSMAP TMT dataset: Study data were provided through the Accelerating Medicine Partnership for AD (U01AG046161 and U01AG061357) based on samples provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago. Data collection was supported through funding by NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG30146.
Author contributions
KF.Liu, A.Aierken, Y.Yan, and H.Qing conceived and designed the studies and wrote the articles; J.Hong and A.Aierken, Label-free LC-MS analysis and protein identification; KF.Liu, A.Aierken, C.Chao, and WJ.Peng proteomic data analysis; W.Kong, XY.Liu, and XY.Ying carried out astrocyte and microglia interaction test; MY.Liu and N.Parhat, carried out fiber photometry recordings and immunohistochemistry and cell counting. G.Liu, D. Yu, JJ.Ni, ZZ.Quan, SM. Ji, and J.Mao. discussed and commented on the article.
Declaration of interests
The authors declare that they have no competing interests.
Published: February 20, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109281.
Contributor Information
Yan Yan, Email: 5903974@hebmu.edu.cn.
Hong Qing, Email: hqing@bit.edu.cn.
Supplemental information
References
- 1.Crous-Bou M., Minguillón C., Gramunt N., Molinuevo J.L. Alzheimer's disease prevention: from risk factors to early intervention. Alzheimers Res. Ther. 2017;9:71. doi: 10.1186/s13195-017-0297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy C. Olfactory and other sensory impairments in Alzheimer disease. Nat. Rev. Neurol. 2019;15:11–24. doi: 10.1038/s41582-018-0097-5. [DOI] [PubMed] [Google Scholar]
- 3.Sepulcre J., Grothe M.J., d'Oleire Uquillas F., Ortiz-Terán L., Diez I., Yang H.S., Jacobs H.I.L., Hanseeuw B.J., Li Q., El-Fakhri G., et al. Neurogenetic contributions to amyloid beta and tau spreading in the human cortex. Nat. Med. 2018;24:1910–1918. doi: 10.1038/s41591-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devanand D.P., Lee S., Manly J., Andrews H., Schupf N., Doty R.L., Stern Y., Zahodne L.B., Louis E.D., Mayeux R. Olfactory deficits predict cognitive decline and Alzheimer dementia in an urban community. Neurology. 2015;84:182–189. doi: 10.1212/wnl.0000000000001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devanand D.P., Tabert M.H., Cuasay K., Manly J.J., Schupf N., Brickman A.M., Andrews H., Brown T.R., DeCarli C., Mayeux R. Olfactory identification deficits and MCI in a multi-ethnic elderly community sample. Neurobiol. Aging. 2010;31:1593–1600. doi: 10.1016/j.neurobiolaging.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahayel S., Frasnelli J., Joubert S. The effect of Alzheimer's disease and Parkinson's disease on olfaction: a meta-analysis. Behav. Brain Res. 2012;231:60–74. doi: 10.1016/j.bbr.2012.02.047. [DOI] [PubMed] [Google Scholar]
- 7.Roalf D.R., Moberg M.J., Turetsky B.I., Brennan L., Kabadi S., Wolk D.A., Moberg P.J. A quantitative meta-analysis of olfactory dysfunction in mild cognitive impairment. J. Neurol. Neurosurg. Psychiatry. 2017;88:226–232. doi: 10.1136/jnnp-2016-314638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts R.O., Christianson T.J.H., Kremers W.K., Mielke M.M., Machulda M.M., Vassilaki M., Alhurani R.E., Geda Y.E., Knopman D.S., Petersen R.C. Association Between Olfactory Dysfunction and Amnestic Mild Cognitive Impairment and Alzheimer Disease Dementia. JAMA Neurol. 2016;73:93–101. doi: 10.1001/jamaneurol.2015.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes D.C., Hofacer R.D., Zaman A.R., Rennaker R.L., Wilson D.A. Olfactory perceptual stability and discrimination. Nat. Neurosci. 2008;11:1378–1380. doi: 10.1038/nn.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meissner-Bernard C., Dembitskaya Y., Venance L., Fleischmann A. Encoding of Odor Fear Memories in the Mouse Olfactory Cortex. Curr. Biol. 2019;29:367–380.e4. doi: 10.1016/j.cub.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Sacco T., Sacchetti B. Role of secondary sensory cortices in emotional memory storage and retrieval in rats. Science (New York, N.Y.) 2010;329:649–656. doi: 10.1126/science.1183165. [DOI] [PubMed] [Google Scholar]
- 12.Schoonover C.E., Ohashi S.N., Axel R., Fink A.J.P. Representational drift in primary olfactory cortex. Nature. 2021;594:541–546. doi: 10.1038/s41586-021-03628-7. [DOI] [PubMed] [Google Scholar]
- 13.Donoshita Y., Choi U.S., Ban H., Kida I. Assessment of olfactory information in the human brain using 7-Tesla functional magnetic resonance imaging. Neuroimage. 2021;236 doi: 10.1016/j.neuroimage.2021.118212. [DOI] [PubMed] [Google Scholar]
- 14.Lu J., Wang X., Qing Z., Li Z., Zhang W., Liu Y., Yuan L., Cheng L., Li M., Zhu B., et al. Detectability and reproducibility of the olfactory fMRI signal under the influence of magnetic susceptibility artifacts in the primary olfactory cortex. Neuroimage. 2018;178:613–621. doi: 10.1016/j.neuroimage.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Tabert M.H., Steffener J., Albers M.W., Kern D.W., Michael M., Tang H., Brown T.R., Devanand D.P. Validation and optimization of statistical approaches for modeling odorant-induced fMRI signal changes in olfactory-related brain areas. Neuroimage. 2007;34:1375–1390. doi: 10.1016/j.neuroimage.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Ebadi A., Dalboni da Rocha J.L., Nagaraju D.B., Tovar-Moll F., Bramati I., Coutinho G., Sitaram R., Rashidi P. Ensemble Classification of Alzheimer's Disease and Mild Cognitive Impairment Based on Complex Graph Measures from Diffusion Tensor Images. Front. Neurosci. 2017;11:56. doi: 10.3389/fnins.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W., Howard J.D., Gottfried J.A. Disruption of odour quality coding in piriform cortex mediates olfactory deficits in Alzheimer's disease. Brain. 2010;133:2714–2726. doi: 10.1093/brain/awq209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasavada M.M., Martinez B., Wang J., Eslinger P.J., Gill D.J., Sun X., Karunanayaka P., Yang Q.X. Central Olfactory Dysfunction in Alzheimer's Disease and Mild Cognitive Impairment: A Functional MRI Study. J. Alzheimers Dis. 2017;59:359–368. doi: 10.3233/jad-170310. [DOI] [PubMed] [Google Scholar]
- 19.Johnson E.C.B., Dammer E.B., Duong D.M., Ping L., Zhou M., Yin L., Higginbotham L.A., Guajardo A., White B., Troncoso J.C., et al. Large-scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med. 2020;26:769–780. doi: 10.1038/s41591-020-0815-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heckmann B.L., Teubner B.J.W., Tummers B., Boada-Romero E., Harris L., Yang M., Guy C.S., Zakharenko S.S., Green D.R. LC3-Associated Endocytosis Facilitates β-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer's Disease. Cell. 2019;178:536–551.e514. doi: 10.1016/j.cell.2019.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong S., Beja-Glasser V.F., Nfonoyim B.M., Frouin A., Li S., Ramakrishnan S., Merry K.M., Shi Q., Rosenthal A., Barres B.A., et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science (New York, N.Y.) 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng L., Bestard-Lorigados I., Song W. The synapse as a treatment avenue for Alzheimer's Disease. Mol. Psychiatr. 2022;27:2940–2949. doi: 10.1038/s41380-022-01565-z. [DOI] [PubMed] [Google Scholar]
- 23.Del Campo M., Teunissen C.E. Role of BRI2 in dementia. J. Alzheimers Dis. 2014;40:481–494. doi: 10.3233/jad-131364. [DOI] [PubMed] [Google Scholar]
- 24.Foster E.M., Fernandes M., Dangla-Valls A., Hublitz P., Pangalos M., Lovestone S., Ribe E.M., Buckley N.J. Glycosylated clusterin species facilitate Aβ toxicity in human neurons. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-23167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallwitz L., Schmidt L., Marques A.R.A., Tholey A., Cassidy L., Ulku I., Multhaup G., Di Spiezio A., Saftig P. Cathepsin D: Analysis of its potential role as an amyloid beta degrading protease. Neurobiol. Dis. 2022;175 doi: 10.1016/j.nbd.2022.105919. [DOI] [PubMed] [Google Scholar]
- 26.Martens Y.A., Zhao N., Liu C.C., Kanekiyo T., Yang A.J., Goate A.M., Holtzman D.M., Bu G. ApoE Cascade Hypothesis in the pathogenesis of Alzheimer's disease and related dementias. Neuron. 2022;110:1304–1317. doi: 10.1016/j.neuron.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J., Zhou H., Xiang G. Identification of Key Biomarkers and Pathways for Maintaining Cognitively Normal Brain Aging Based on Integrated Bioinformatics Analysis. Front. Aging Neurosci. 2022;14 doi: 10.3389/fnagi.2022.833402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seyfried N.T., Dammer E.B., Swarup V., Nandakumar D., Duong D.M., Yin L., Deng Q., Nguyen T., Hales C.M., Wingo T., et al. A Multi-network Approach Identifies Protein-Specific Co-expression in Asymptomatic and Symptomatic Alzheimer's Disease. Cell Syst. 2017;4:60–72.e4. doi: 10.1016/j.cels.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jack C.R., Jr., Bennett D.A., Blennow K., Carrillo M.C., Dunn B., Haeberlein S.B., Holtzman D.M., Jagust W., Jessen F., Karlawish J., et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloret A., Esteve D., Lloret M.A., Cervera-Ferri A., Lopez B., Nepomuceno M., Monllor P. Vol. 19. American Psychiatric Publishing; 2021. When Does Alzheimer's Disease Really Start? The Role of Biomarkers; pp. 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodward M.R., Amrutkar C.V., Shah H.C., Benedict R.H.B., Rajakrishnan S., Doody R.S., Yan L., Szigeti K. Validation of olfactory deficit as a biomarker of Alzheimer disease. Neurol. Clin. Pract. 2017;7:5–14. doi: 10.1212/cpj.0000000000000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.d'Errico P., Ziegler-Waldkirch S., Aires V., Hoffmann P., Mezö C., Erny D., Monasor L.S., Liebscher S., Ravi V.M., Joseph K., et al. Microglia contribute to the propagation of Aβ into unaffected brain tissue. Nat. Neurosci. 2022;25:20–25. doi: 10.1038/s41593-021-00951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tcw J., Qian L., Pipalia N.H., Chao M.J., Liang S.A., Shi Y., Jain B.R., Bertelsen S.E., Kapoor M., Marcora E., et al. Cholesterol and matrisome pathways dysregulated in astrocytes and microglia. Cell. 2022;185:2213–2233.e25. doi: 10.1016/j.cell.2022.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hook G., Reinheckel T., Ni J., Wu Z., Kindy M., Peters C., Hook V. Cathepsin B Gene Knockout Improves Behavioral Deficits and Reduces Pathology in Models of Neurologic Disorders. Pharmacol. Rev. 2022;74:600–629. doi: 10.1124/pharmrev.121.000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webster S., Glabe C., Rogers J. Multivalent binding of complement protein C1Q to the amyloid beta-peptide (A beta) promotes the nucleation phase of A beta aggregation. Biochem. Biophys. Res. Commun. 1995;217:869–875. doi: 10.1006/bbrc.1995.2852. [DOI] [PubMed] [Google Scholar]
- 36.Chen K.Z., Liu S.X., Li Y.W., He T., Zhao J., Wang T., Qiu X.X., Wu H.F. Vimentin as a potential target for diverse nervous system diseases. Neural Regen. Res. 2023;18:969–975. doi: 10.4103/1673-5374.355744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedrero-Prieto C.M., Flores-Cuadrado A., Saiz-Sánchez D., Úbeda-Bañón I., Frontiñán-Rubio J., Alcaín F.J., Mateos-Hernández L., de la Fuente J., Durán-Prado M., Villar M., et al. Human amyloid-β enriched extracts: evaluation of in vitro and in vivo internalization and molecular characterization. Alzheimer's Res. Ther. 2019;11:56. doi: 10.1186/s13195-019-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liddelow S.A., Guttenplan K.A., Clarke L.E., Bennett F.C., Bohlen C.J., Schirmer L., Bennett M.L., Münch A.E., Chung W.S., Peterson T.C., et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carter S.F., Herholz K., Rosa-Neto P., Pellerin L., Nordberg A., Zimmer E.R. Astrocyte Biomarkers in Alzheimer's Disease. Trends Mol. Med. 2019;25:77–95. doi: 10.1016/j.molmed.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Hansen D.V., Hanson J.E., Sheng M. Microglia in Alzheimer's disease. J. Cell Biol. 2018;217:459–472. doi: 10.1083/jcb.201709069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oksanen M., Petersen A.J., Naumenko N., Puttonen K., Lehtonen Š., Gubert Olivé M., Shakirzyanova A., Leskelä S., Sarajärvi T., Viitanen M., et al. PSEN1 Mutant iPSC-Derived Model Reveals Severe Astrocyte Pathology in Alzheimer's Disease. Stem Cell Rep. 2017;9:1885–1897. doi: 10.1016/j.stemcr.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song D., Wang D., Yang Q., Yan T., Wang Z., Yan Y., Zhao J., Xie Z., Liu Y., Ke Z., et al. The lateralization of left hippocampal CA3 during the retrieval of spatial working memory. Nat. Commun. 2020;11:2901. doi: 10.1038/s41467-020-16698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen T., Ma J., Liu Y., Chen Z., Xiao N., Lu Y., Fu Y., Yang C., Li M., Wu S., et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res. 2022;50 doi: 10.1093/nar/gkab1081. D1522-d1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., Li K., Zhang G., Jin Z., He F., et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky869. D1211-d1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambert M.P., Barlow A.K., Chromy B.A., Edwards C., Freed R., Liosatos M., Morgan T.E., Rozovsky I., Trommer B., Viola K.L., et al. Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reimand J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A., Wadi L., Meyer M., Wong J., Xu C., et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019;14:482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plaisier S.B., Taschereau R., Wong J.A., Graeber T.G. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S., et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51 doi: 10.1093/nar/gkac1000. D638-d646. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The original mass spectra and the protein sequence databases have been deposited at ProteomeXchange (PXD040802) (https://www.proteomexchange.org/) and are publicly accessible at https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD040802.
-
•
Human postmortem proteomic data (ROSMAP: syn17015098, MSBB: syn20801227and Banner: syn20818199) were not generate by this work and can be obtained from synapse website upon request. Customized code was uploaded to github (https://github.com/liukf10/5xFAD-PCx-proteomic-data-analysis).
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.