Abstract
Paclitaxel is a potent anti-cancer drug that is mainly produced through semi-synthesis, which still requires plant materials as precursors. The content of paclitaxel and 10-deacetyl baccatin III (10-DAB) in Taxus yunnanensis has been found to differ from that of other Taxus species, but there is little research on the mechanism underlying the variation in paclitaxel content in T. yunnanensis of different provenances. In this experiment, the contents of taxoids and precursors in twigs between a high paclitaxel-yielding individual (TG) and a low paclitaxel-yielding individual (TD) of T. yunnanensis were compared, and comparative analyses of transcriptomes as well as chloroplast genomes were performed. High-performance liquid chromatography (HPLC) detection showed that 10-DAB and baccatin III contents in TG were 18 and 47 times those in TD, respectively. Transcriptomic analysis results indicated that genes encoding key enzymes in the paclitaxel biosynthesis pathway, such as taxane 10-β-hydroxylase (T10βH), 10-deacetylbaccatin III 10-O-acetyltransferase (DBAT), and debenzoyl paclitaxel N-benzoyl transferase (DBTNBT), exhibited higher expression levels in TG. Additionally, qRT-PCR showed that the relative expression level of T10βH and DBAT in TG were 29 and 13 times those in TD, respectively. In addition, six putative transcription factors were identified that may be involved in paclitaxel biosynthesis from transcriptome data. Comparative analysis of plastid genomes showed that the TD chloroplast contained a duplicate of rps12, leading to a longer plastid genome length in TD relative to TG. Fifteen mutation hotspot regions were identified between the two plastid genomes that can serve as candidate DNA barcodes for identifying high-paclitaxel-yield individuals. This experiment provides insight into the difference in paclitaxel accumulation among different provenances of T. yunnanensis individuals.
Keywords: Taxus yunnanensis, High paclitaxel yield, Transcriptome analysis, Chloroplast genome, Gene expression, qRT-PCR
1. Introduction
Paclitaxel was first isolated from the bark of Taxus brevifolia in 1967 and has been used as an anti-cancer drug for the treatment of various cancers such as breast cancer, lung cancer, and ovarian cancer [1]. However, owing to destructive harvesting methods at production sites, the complex purification process, and low yield, the supply of paclitaxel remains far from meeting market demand [2]. To increase the yield of paclitaxel, a paclitaxel total synthesis method has been reported [3]. However, its complex synthesis process leads to a decrease in yield, an increase in cost, and the production of various toxic products, making this method difficult to commercialize [4]. After elucidating the total synthesis method for paclitaxel, a semi-synthesis method was also published, starting from a high-content paclitaxel intermediate, such as 10-deacetyl baccatin III (10-DAB), to chemically synthesize paclitaxel [5]. Later, Chen et al. [6]isolated endophytic fungi that can produce paclitaxel from the branches and bark of T. yunnanensis. However, the semi-synthesis [7] relies on plant-supplied precursors, while biosynthesis [8] is limited by the recalcitrant behavior of Taxus spp. plant cell cultures in vitro [9]. Previous research shows that the differential expression of key genes leads to variations in the content of paclitaxel and paclitaxel intermediates among different species [10], so identifying high-paclitaxel-yield individuals is an important direction for optimizing paclitaxel production.
The biosynthetic pathway of paclitaxel involves 19 catalytic reactions and has three main stages. The first stage is the synthesis of the taxane skeleton. As a diterpenoid compound, paclitaxel is synthesized from isoprene precursors [11]. Three units of isopentenyl pyrophosphate (IPP) and one unit of dimethylallyl pyrophosphate (DMAPP) condense to form geranylgeranyl pyrophosphate (GGPP) via the methyl erythritol phosphate (MEP) pathway [12]. Then, catalyzed by taxadiene synthase (TS), GGPP cyclizes to form taxadiene, creating the tricyclic diterpene skeleton structure of paclitaxel [13]. Notably, while this step is slow but still not the rate-limiting step in paclitaxel biosynthesis [14]. Afterwards, taxane is hydroxylated, acylated and ketonized to form baccatin III [15]. During this process, an increase in the expression of 10-deacetylbaccatin III-10-β-O-acetyltransferase (DBAT) increases accumulation of baccatin III [16]. Finally, the C-13 side chain of baccatin III is assembled as the last step in paclitaxel biosynthesis [17] (Fig. 1).
Fig. 1.
Paclitaxel biosynthetic pathway from geranylgeranyl diphosphate. Steps shown with dotted arrows are not yet fully elucidated. Enzyme abbreviations: TS, taxadiene synthase; T5αH, taxane-5α-hydroxylase; TAT, taxane-5α-ol-O-acetyltransferase; T10βH, taxane-10β-hydroxylase; T13αH, taxane-13α-hydroxylase; T2αH, taxane 2α-hydroxylase; T7βH, taxane 7β-hydroxylase; TBT, taxane-2α-O-benzoyl transferase; DBAT, 10-deacetylbaccatinIII-10-O-acetyltransferase; PAM, phenylalanine aminomutase; PCL, phenylalanine-CoA ligase; BAPT, C-13 phenylpropanoyl-CoA transferase; T2′αH, taxane2′α-hydroxylase; DBTNBT, debenzoyl paclitaxel N-benzoyl transferase [2].
RNA-Seq, a high-precision and cost-effective DNA sequencing technology, is being increasingly used for assessing the complexity of transcriptomes [18]. Currently, RNA-Seq is widely used to investigate the expression of paclitaxel pathways in different species and plant tissues [19]. Li et al. studied the regulation of gene expression in the T. chinensis paclitaxel synthesis pathway under application of methyl-jasmonate and found that the activation of a series of transcription factor (TF) families, such as MYB, bHLH, ERF, AP2, and MYC, can enhance the expression of related genes in the paclitaxel synthesis pathway [20]. Zhou et al. measured the paclitaxel content in different Taxus species and found that differential expression of genes that are involved in the paclitaxel biosynthesis pathway, may provide a potential explanation for interspecific differences in paclitaxel accumulation [21]. Meng et al. found that different TF families, such as NAC, WRKY and bZIP, participated in the response of T. yunnanensis to low temperature stress through transcriptome analysis of T. yunnanensis under different temperatures [22].
T. yunnanensis is mainly distributed in Yunnan Province, China. As a slow-growing tree species, it has been extensively studied owing to its valuable secondary metabolite paclitaxel [23,24]. Since Zhang et al. [25] reported the chemical composition of T. yunnanensis in 1990, more than 100 taxane diterpenes have been isolated from various parts of T. yunnanensis. Among them, the most representative compound, paclitaxel, and its biosynthetic precursor 10-DAB have been characterized as having higher content in T. yunnanensis than in other Taxus spp [26]. Previous studies have found that the paclitaxel content of T. yunnanensis of different provenances varies significantly [27], but there are currently no reports on the molecular mechanism underlying the differences in paclitaxel or 10-DAB content among T. yunnanensis individuals. Thus, in a preliminary experiment, two excellent clonal lines of T. yunnanensis with high taxanes content were selected based on their morphological characteristics and content of taxanes, following the screening of T. yunnanensis from 25 natural distribution areas in Yunnan and T. yunnanensis artificial forests [28]. The present study used RNA-Seq technology and chloroplast genome sequencing to analyze two T. yunnanensis trees of different provenances. This study investigates the expression patterns of key genes involved in taxane compound metabolism and the differences in chloroplast genomes between two T. yunnanensis individuals of different provenances grown in the same environment.
2. Materials and methods
2.1. High- and low-paclitaxel-yield individuals
High- and low-paclitaxel-yield individuals of T. yunnanensis (TG and TD, respectively) were planted in a greenhouse at the Laboratory of Forest Plant Cultivation and Utilization, Yunnan Academy of Forestry & Grassland Science (25°8′E, 102°44′N). Samples were obtained from three-year-old cultivated TG and TD individuals that had been planted in close proximity, and the growth conditions were maintained at 25–30 °C with a light/dark cycle of 12/12 h and 80–85% relative humidity [28]. Three replicates of twigs were collected from TG and TD plants, respectively, for RNA extraction.
2.2. Determinations of paclitaxel content
Fresh twigs were cut from the T. yunnanensis trees and dried in a 65 °C oven for 6 h before being ground into a powder. The powder was filtered through a 40-mesh sieve to remove large particles. Five grams of the filtered powder was added to 100 mL, 50 mL, and 50 mL of methanol solution for three separate reflux extractions. The combined extract volume was then evaporated to 100 mL using a rotary evaporator. The extract was then extracted four times with an equal volume of hexane. The methanol extract was then evaporated using a rotary evaporator and reconstituted to 25 mL with methanol. The extract was then filtered through a 0.22-μm filter for high-performance liquid chromatography (HPLC) analysis.
HPLC analysis was conducted utilizing the Agilent 1100 HPLC system. Chromatographic separation was achieved employing the Agilent Hypersil 5 ODS C18 column (250 × 4.0 mm, 5 μm). The reference substances, 10-DAB, baccatin III, 10-deacetylpaclitaxel, 7-xylosyl-10-deacetylpaclitaxel, cephalomannine, and paclitaxel were purchased from Sigma-Aldrich China Inc. (Shanghai, China). The injection volume was 5 μL, and detection occurred at a wavelength of 227 nm, with gradient elution at a flow rate of 1.0 mL/min, maintained at 30 °C. The mobile phase consisted of acetonitrile: water (30:70 [v: v]) from 0 to 28 min, followed by acetonitrile: water (54:46 [v: v]) after 28 min. Precise amounts of 10-DAB, baccatin III, 10-deacetylpaclitaxel, 7-xylosyl-10-deacetylpaclitaxel, cephalomannine, and paclitaxel reference standards were weighed and sequentially prepared into five different concentrations to establish the standard curve as indicated: 10-DAB (0.1377, 0.2755, 0.4133, 0.551, 0.6888 mg/mL), baccatin III (0.1330, 0.2660, 0.3990, 0.5320, 0.6650 mg/mL), 7-xylosyl-10-deacetylpaclitaxel (0.1910, 0.3820, 0.5730, 0.7640, 0.9550 mg/mL), 10-deacetylpaclitaxel (0.2050, 0.4100, 0.6150, 0.8200, 1.0250 mg/mL), Cephalomannine (0.1717, 0.3434, 0.5151, 0.6868, 0.8585 mg/mL), paclitaxel (0.1188, 0.3564, 0.5940, 0.8316, 1.1880 mg/mL) [29].
2.3. RNA extraction
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to its manual. Contaminating DNA was removed by adding DNase I to the mixture. Clean RNA was detected by 1% agarose gel electrophoresis. The quality of total RNA was confirmed using the RNA 6000 Nano LabChip Kit (Agilent, Santa Clara, CA, USA), with an RNA integrity number >7.0 required.
2.4. Construction and sequencing of cDNA library
The sequencing library was constructed using the NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA). Oligo(dT) magnetic beads were used to enrich for mRNAs with a poly-A tail from the total RNA. The RNA was fragmented into pieces approximately 300 bp in length using ion disruption. The first strand of cDNA was synthesized from RNA using 6-base random primers and reverse transcriptase. The second strand of cDNA was synthesized using the first-strand cDNA as the template. After library construction, PCR amplification was used to enrich library fragments. The library size was selected to be 380 bp based on fragment size, and the library quality was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The constructed library was sequenced by Personal Biotechnology Co., Ltd (Shanghai, China) using the Illumina HiSeq 2000 sequencing platform (Illumina, San Diego, CA, USA) for paired-end (PE) sequencing. The quality of obtained raw reads was checked with the FastQC tool. To acquire valid sequencing data, the raw data were filtered using Cutadapt software (v2.3). In this step, the reads with adapters or ambiguous bases (‘N’ > 5% of a read), and low-quality reads (with more than 10% Q < 20) bases were discarded.
2.5. Functional annotation and enrichment analysis
Cutadapt was used to remove adapter sequences and low-quality reads. All assembled unigenes were searched for in various databases, including the non-redundant (Nr) protein database, Gene Ontology (GO), SwissProt, Kyoto Encyclopedia of Genes and Genomes (KEGG), and eggNOG database with an E value < 0.00001 threshold. Local Perl scripts were used to perform GO and KEGG enrichment analysis on differentially expressed genes (DEGs).
2.6. Differentially expressed unigene analysis
Clean reads of each sample were mapped to the reference sequence (GCA_018340775.1). The expression level of each transcript in different samples was according to fragments per kilobase of transcript per million mapped reads (FPKM), normalizing for gene length and sequencing depth. The differential expression fold change between different samples was calculated based on the FPKM values of gene expression. DESeq was used for differential screening analysis, with a threshold set both at p-value <0.05 and log2(fold change) > 1 to screen for DEGs.
2.7. Phylogenetic and structural analyses
The phylogenetic tree of WRKY, bHLH, and ERF was constructed using the seven TFs confirmed to be involved in the paclitaxel biosynthesis pathway, including TcJAMYC1/2/3, TcERF12/15, and TcWRKY1/8/47 [[30], [31], [32]] as reference sequences, by MEGA11. The tree-building method was neighbor-joining (NJ), with 1000 bootstrap replicates and the Poisson correction model.
2.8. Real-time PCR validation
Quantitative real-time PCR (qRT-PCR) was performed on TG and TD samples using the SYBR Premix Ex Taq Kit (TaKaRa, Dalian, China) and a DNA sequence detection system (ABI PRIM 7700; Thermo Fisher Scientific, Waltham, MA, USA). The actin sequence was used as an internal reference gene, and relative fold differences in expression were calculated using the comparative cycle threshold method (2−ΔΔCt). Expression analysis was performed with three biological replicates. The primer sequences are list in Table S1.
2.9. Chloroplast genomic comparison
Chloroplast DNA extraction and assembly annotation were performed according to previously described methods [33]. In brief, paired-end reads were sequenced by the Illumina HiSeq system (Illumina). High-quality clean reads were generated, and adaptors were trimmed. Aligning, assembly, and annotation were conducted using CLC de novo assembler (CLC Bio, Aarhus, Denmark), BLAST, GeSeq, and Geneious v 11.0.5 (Biomatters Ltd, Auckland, New Zealand). To detect the degree of sequence differences between the chloroplasts of the two Taxus plants, the sequences were visually compared using the online tool mVISTA. Codon usage in the chloroplast genome was assessed using CodonW v 1.4.2. In addition, single sequence repeat (SSR) sites in the chloroplast genome were identified using the online tool misa.
2.10. Identification of divergence hotspots
In order to identify mutation hotspot regions, the protein coding genes, non-coding regions, and intron regions of the two Taxus plastomes were extracted with local Python scripts and aligned with MAFFT v 7.221. Then, alignments of more than 200 bp in length were used to evaluate nucleotide diversity (Pi) using DnaSP v 5.0.
3. Results
3.1. Differences in paclitaxel content between TG and TD
Contents of paclitaxel and other taxoids in twigs of the collected samples are shown in Fig. 2. All contents of taxoids were higher in TG than in TD, except for 7-xylosyl-10-deacetylpaclitaxel. Among those taxoids, 10-DAB, baccatin III, 10-deacetylpaclitaxel, cephalomannine, and paclitaxel accumulated significantly more in TG, but the content of 7-xylosyl-10-deacetylpaclitaxel did not significantly differ between TG and TD (Fig. 2).
Fig. 2.
The contents of six taxoids in high- and low-paclitaxel-yielding Taxus yunnanensis (TG and TD, respectively). Statistical significance was assessed at a p < 0.01 threshold.
3.2. Transcriptomes of different Taxus samples
Transcriptome sequencing was performed on the TG and TD samples using the Illumina HiSeq 2000 high-throughput sequencing platform, obtaining a total of 93,498,780 raw reads. After removing adapter sequences and low-quality reads, a total of 86,761,854 clean reads were obtained, accounting for 92.7% of raw reads; 89.30% and 93.03% of clean reads were mapped to the reference transcriptome, respectively. The percentages of N, Q20, and Q30 bases for the TG and TD sequences after data filtering were 0.000445% and 0.000421%, 98.2% and 97.47%, and 94.76% and 92.91%, respectively. The data quality is summarized in Table 1. A total of 54,689 unigenes were obtained with a library length of 380 bp. Among them, 19,587 unigenes were annotated in the GO database, accounting for 35.18% of all unigenes; 16,296 unigenes were annotated in KEGG, accounting for 29.79% of all unigenes; 39,822 unigenes were annotated in eggNOG, accounting for 72.81% of all unigenes; 33,296 unigenes were annotated in the SwissProt database, accounting for 60.88% of all unigenes (Table 1).
Table 1.
Comparison of the quality statistics for filtered reads between high- and low-paclitaxel-yielding Taxus yunnanensis (TG and TD, respectively).
| Sample | Reads No. | Bases(bp) | Q30(bp) | N(%) | Q20(%) | Q30(%) |
|---|---|---|---|---|---|---|
| TG | 47721860 | 7158279000 | 6783238535 | 0.000445 | 98.2 | 94.76 |
| TD | 45776920 | 6866538000 | 6380253689 | 0.000421 | 97.47 | 92.91 |
3.3. Identification and analysis of DEGs between TG and TD
In this experiment, a total of 1286 DEGs were found, including 598 up-regulated genes and 688 down-regulated genes. The paclitaxel biosynthesis pathway is an important part of terpenoid biosynthesis and is a currently well-characterized biosynthesis pathway. It includes the supply of precursors, diterpenoid taxane core synthesis, hydroxylations, acylations, baccatin III formation, and C13-side chain assembly intermediate reaction steps (Fig. 1). To obtain further clarity into the relationship between DEGs and paclitaxel biosynthesis, GO enrichment analysis was performed (Fig. S2). Four GO terms directly related to paclitaxel synthesis were identified (Table S2): paclitaxel biosynthetic process (GO: 0042617), paclitaxel metabolic process (GO: 0042616), taxane 10-beta-hydroxylase activity (GO: 0050597), and taxadiene 5-alpha-hydroxylase activity (GO: 0050604). Among them, paclitaxel biosynthetic process, paclitaxel metabolic process, and taxane 10-beta-hydroxylase activity were all significantly down-regulated in TD relative to TG. KEGG analysis of DEGs revealed that there were significant differences in 17 KEGG pathways between TG and TD (Fig. 3A). Almost all of the differences were related to metabolism pathways: one pathway related to transport and catabolism, two pathways related to metabolism of other amino acids, one pathway related to metabolism of cofactors and vitamins, four pathways related to lipid metabolism, one pathway related to glycan biosynthesis and metabolism, one pathway related to energy metabolism, two pathways related to carbohydrate metabolism, two pathways related to biosynthesis of other secondary metabolites, and three pathways related to amino acid metabolism (Fig. 3B).
Fig. 3.
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis: (A) KEGG classification of unigenes; (B) KEGG enrichment analysis of differentially expressed genes (DEGs). The significant p-value of each KEGG term between the high- and low-paclitaxel-yielding Taxus yunnanensis (TG and TD, respectively) is shown in the heatmap.
3.4. Variation in the precursors of paclitaxel biosynthesis between TG and TD
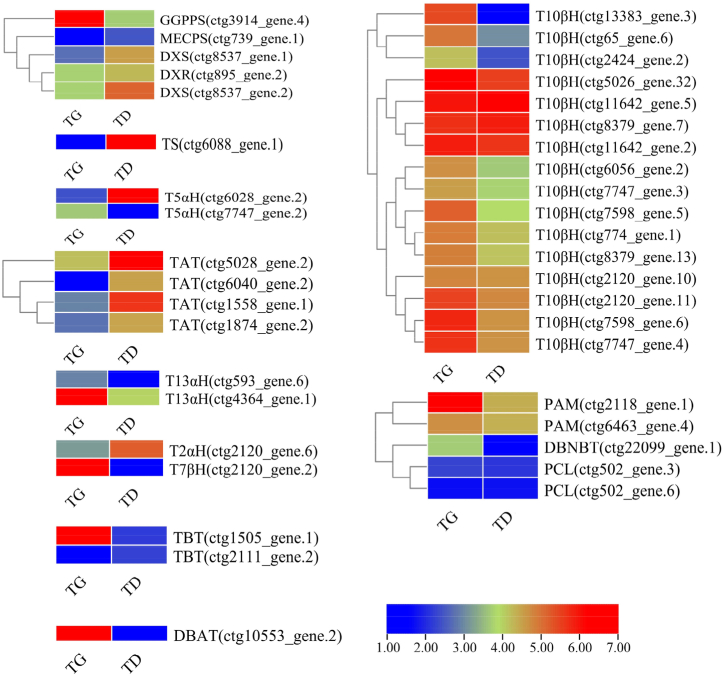
The MEP pathway provides the key precursor GGPP for the paclitaxel biosynthesis pathway. According to the transcriptome data obtained in this experiment, five genes related to the MEP pathway were found: one DXR-encoding unigene, two DXS-encoding unigenes, one MECPS-encoding unigene, and one GGPPS-encoding unigene. Although the expression levels of MEP pathway-related genes found in this experiment were higher in TD than in TG, except for GGPPS, there were no significant differences in the expression levels of all related genes (Fig. 4).
Fig. 4.
The expression patterns of the genes in the paclitaxel biosynthetic pathway in high- and low-paclitaxel-yielding Taxus yunnanensis (TG and TD, respectively).
3.5. Variation in the paclitaxel biosynthesis pathway between TG and TD
The paclitaxel biosynthesis pathway has been elucidated in recent years. In the present study, a total of 40 genes related to the paclitaxel biosynthesis pathway were found (Table S3), including one TS-encoding unigene, two T5αH-encoding unigenes, four TAT-encoding unigenes, two T13αH-encoding unigenes, sixteen T10βH-encoding unigenes, one T7βH-encoding unigene, one T2αH-encoding unigene, two TBT-encoding unigenes, one DBAT-encoding unigene, one DBTNBT-encoding unigene, two PAM-encoding unigenes, and two PCL-encoding unigenes. Among them, the unigenes encoding TAT, T10βH, T2αH, DBAT, and DBTNBT exhibited different expression patterns in TG and TD. Most of the T10βH unigenes and the DBAT and DBTNBT unigenes had higher expression levels in TG than in TD, while the expression levels of TAT and T2αH were lower in TG than in TD (Fig. 4).
3.6. Identification and phylogenetic analysis of TF families between TD and TG
TFs can activate the co-expression of multiple genes in secondary metabolic pathways, which effectively regulates the production of secondary metabolites. Therefore, many TFs play an important role in the biosynthesis of paclitaxel, such as bHLH, MYB, and AP2/ERF TFs. A total of 57 TF families were identified in this experiment, and 6585 genes were annotated as putative TFs, which included 133 WRKYs, 126 MYBs, 234 ERFs, 295 bHLHs, and 21 AP2s (Table S4). Sequences of the TFs determined to participate in the paclitaxel biosynthesis pathway were used to construct a phylogenetic tree. After removing data with poor homology, a total of six TFs were finally used for phylogenetic analysis, including two WRKYs (Fig. S3), three bHLHs (Fig. S4), and one ERF (Fig. S5). Except for one WRKY and one AP2 with expression in TG that was lower than that in TD, the other TFs had higher expression levels in TG.
3.7. Validation of DEGs by qPCR
To validate the expression levels of key genes in different paclitaxel biosynthesis pathways, eight genes in paclitaxel biosynthesis pathway were randomly selected for qRT-PCR: TS, T5αH, T13αH, T2αH, T7βH, TBT, T10βH, and DBAT (Fig. 5). Among them, except for TS and T2αH, which had higher expression levels in TD, the remaining genes involved in paclitaxel synthesis had more transcript accumulation in TG than in TD. The accumulations of T10βH and DBAT in TG were particularly significant, being 29-fold and 13-fold higher than those in TD, respectively.
Fig. 5.
Differential expression of the genes involved in the paclitaxel biosynthesis pathway as measured by qPCR. Significant differences are indicated by “*” (p < 0.01).
3.8. General features of TG and TD chloroplast genomes
The sequence lengths of the chloroplast genomes of TG and TD were 128,356 bp and 129,190 bp, respectively, and their total GC contents were 34.69% and 34.65%, respectively. The two plastomes encoded 116 unique genes. The TG and TD plastomes encoded 83 and 82 genes, respectively, as well as 30 tRNA genes each and 4 rRNA genes each. There was a duplicate of rps12 in TG. The petL gene was lost in TG; rps16 was absent in TD (Table 2).
Table 2.
Comparison of plastome features between high- and low-paclitaxel-yielding Taxus yunnanensis (TG and TD, respectively).
| Taxon | Total GC content (%) | Total length (bp) | Total genes | Protein coding genes | rRNA genes | tRNA genes |
|---|---|---|---|---|---|---|
| TG | 34.69 | 128,356 | 117 | 83 | 4 | 30 |
| TD | 34.65 | 129,190 | 116 | 82 | 4 | 30 |
In order to analyze the codon usage of TG and TD, the sequences of 82 protein-coding genes were extracted for analysis. The concatenated extracted sequence lengths were 73,432 bp and 73,950 bp, respectively, containing 42,785 and 43,063 codons, respectively. In addition, the relative synonymous codon usage (RSCU) values of all codons ranged from 0.32 to 2.23 in the two plastomes. It is worth noting that there were 60 codons with RSCU values greater than 1 in the two chloroplast genomes; however, Arg only had an RSCU value greater than 1 in TG, and Ser only had an RSCU value greater than 1 in TD (Fig. 6).
Fig. 6.
The relative synonymous codon usage (RSCU) values of all concatenated protein-coding genes for high- and low-paclitaxel-yielding Taxus yunnanensis (TG and TD, respectively).
The number of SSRs in TG and TD were 42 and 54, respectively, and there were 11 and 21 SSRs located in their non-coding regions, respectively. While psbE-petG, rps12-2-trnV-GAC, and rpl16-intron1 only appeared in TG, atpA-atpF, rpl36-rps11, psbE-petL, rpl16-rps3, rps7-trnV-GAC, and rps 19 only appeared in TD. Among all SSRs, mononucleotide repeats were the most common type, with 19 and 20 in TG and TD, respectively, followed by dinucleotide repeats, with 6 and 13, respectively (Table S5).
3.9. Plastome hotspot identification
The chloroplast genomes of TG and TD were compared using mVISTA (Fig. 7). Both tRNA and rRNA genes were relatively conserved in TG and TD, and the coding regions were more conserved than the non-coding regions. Based on the degree of divergence between sequences, nucleotide diversity was calculated in the coding and non-coding regions of the two chloroplast genomes. In the coding region, three highly variable hotspots were found that could be utilized as potential DNA barcodes, namely infA, rpl23, and rps3, each of which exhibited high nucleotide diversity (Pi > 0.002). In the non-coding region, a total of 12 highly variable hotspots were found: atpE-rbcL, clpP-infA, petN-trnD-GUC, rbcL-accD, rpl32-trnL-UAG, rpoB-trnC-GCA, rps12-rps8, rrn16-trnI-GAU-1, trnI-CAU-ycf2, trnI-GAU-2-rrn23, trnL-UAA-2-trnF-GAA, ycf1-chlN. Each of these exhibited rather high nucleotide diversity (Pi > 0.1) (Fig. 8).
Fig. 7.
Sequence identity plots for high- and low-paclitaxel-yielding Taxus yunnanensis (TG and TD, respectively).
Fig. 8.
Comparative analysis of the Pi values between high- and low-paclitaxel-yielding Taxus yunnanensis (TG and TD, respectively): (A) protein-coding genes; (B) non-coding and intron sequences.
4. Discussion
As the main natural source of paclitaxel, Taxus spp. have become a critical research focus owing to the huge economic value of paclitaxel [10,21,34]. In the present study, the contents of 10-DAB, baccatin III, 10-deacetylpaclitaxel, 7-xylosyl-10-deacetylpaclitaxel, cephalomannine. and paclitaxel in two different T. yunnanensis individuals, TG and TD, were measured. It was found that except for the content of 7-xylosyl-10-deacetylpaclitaxel, the contents of all other assayed substances were higher in TG than in TD (Fig. 2). Previous studies have measured the paclitaxel content in the twigs of T. yunnanensis, finding a range of 0.00172–0.0084% [35,36]. In the present study, paclitaxel content in TG (0.011%) was significantly higher than TD (0.001%), and in TD twigs was lower than the previously reported content range. The paclitaxel biosynthesis intermediate 10-DAB is an important precursor in the semi-synthesis method, and its content in TG (0.6266%) was 18 times that in TD (0.0362%). At the same time, the content of 10-DAB in TG was higher than that reported in other Taxus spp. such as T. yunnanensis previously (0.005%), T. media (0.07%), T. cuspidata (0.003%), and T. chinensis (0.006%) [36,37]. In addition, 10-deacetylpaclitaxel, 7-xylosyl-10-deacetylpaclitaxel, and cephalomannine can also be converted to paclitaxel via a few steps [[38], [39], [40], [41], [42]]. The contents of 10-deacetylpaclitaxel and 7-xylosyl-10-deacetylpaclitaxel detected in TG (0.016% and 0.0031%) and TD (0.026% and 0.00323%) are lower than those in other Taxus spp., such as T. cuspidata (0.0144–0.0288% and 0.009%), T. chinensis (0.0072–0.043% and 0.042–0.111%), and T. media (0.0216–0.0288% and 0.009%) [43,44], but the content of cephalomannine in TG (0.1306%) twigs was significantly higher than that in twigs of TD (0.04695%) and other Taxus spp., such as T. media (0.033–0.055%), T. cuspidata (0.033–0.066%) [43,44]. To reveal the mechanisms underlying the differences in paclitaxel accumulation between the two different provenances of T. yunnanensis, further RNA-Seq and qRT-PCR analyses were performed.
The MEP pathway, as an important pathway for the supply of paclitaxel biosynthesis precursors, can increase the accumulation of paclitaxel biosynthesis precursors by upregulating their expression [45]. GGPPS, as an enzyme regulating taxoid production in the MEP pathway, plays an important role in the MEP pathway. The expression level of GGPPS in TG is twice that of TD, suggesting a more abundant precursor supply in TG. In addition, there are also some important enzymes in the paclitaxel biosynthesis pathway that significantly differ in expression levels. TAT and T13αH are catalytic enzymes on the branch pathways of paclitaxel biosynthesis, respectively. T13αH catalyzes taxadiene-5α-ol into taxadiene-5α-13α-diol, while TAT is responsible for acylation of taxa 4(20),11(12)-dien-5α-ol, which is then hydroxylated by hydroxylases such as T10αH [11,46] (Fig. 1). In this experiment, the expression levels of two T13αH genes were higher in TG, and the expression levels of four TAT genes were higher in TD (Fig. 4). However, regarding the subsequent expression levels of hydroxylases, the expression levels of 16 T10αH genes were higher in TG. DBAT is another key enzyme that catalyzes 10-deacetylbaccatin III into baccatin III [47]. DBAT catalysis is a rate-limiting step in paclitaxel biosynthesis [48], and its expression level can affect the content of baccatin III, the last diterpene intermediate before paclitaxel [49]. One DBAT-related gene was found, and it was highly expressed in TG, which indicates that the conversion of 10-DAB to paclitaxel is higher in TG [16]. These results were also corroborated by relative content measurements by qPCR (Fig. 5). The secondary metabolite accumulation in plants is influenced by a combination of environmental and genetic factors. In the present study, however, the two experimental lines were grown under the same environmental conditions, and the results are thus attributable to only the genetic differences between TG and TD [29]. Based on the HPLC data and gene expression analysis, it can be inferred that the reason for the higher paclitaxel content in TG twigs is the high expression levels of key genes involved in paclitaxel biosynthesis, GGPPS and DBAT, which leads to high accumulation of baccatin III in the twigs of TG. Although gene expression levels are associated with the accumulation of secondary metabolites in plants, transcriptomic analysis often involves sampling at specific time points rather than continuously monitoring gene expression across the entire plant over time. Meanwhile, the accumulation of secondary metabolites in plants changes along with gene expression levels over time. In a study measuring taxane content and expression levels of genes related to the paclitaxel biosynthetic pathway in T. baccata cell suspension cultures, it was found that the expression levels of genes related to the paclitaxel biosynthetic pathway increased with experiment duration, while changes in taxane content showed no clear pattern [50]. Therefore, time-series experiments should be conducted in the future to provide more convincing evidence of the differences between TG and TD.
TFs play a key role in regulating the production of secondary metabolites [51]. Multiple studies have reported a series of TFs that can increase the expression of paclitaxel synthesis genes [32,52,53]. For example, TcWRKY1 can specifically bind to the W-box element (TTGAC(C/T)) in the DBAT promoter and activate the expression of DBAT [54]. TcJamyc1/2/4 and TcERF12/15 have also been found to regulate the paclitaxel synthesis pathway. The phylogenetic tree constructed in the present study revealed six genes that are closely related to the TFs that regulate the paclitaxel biosynthesis pathway, including two WRKYs, three bHLHs, and one ERF. Among them, one WRKY and one AP2s had different expression patterns between TG and TD, providing more candidate regulatory factors for the paclitaxel biosynthesis pathway.
As the genome of a semiautonomous organelle, chloroplast genomes have been explored for the development of many molecular markers that have been used for provenances- and species-level identification. For example, SSRs were used to distinguish the red alga Gracilaria tenuistipitata between two different provenances [55]. Thus, it is of value to use chloroplast DNA molecular markers to identify molecular genetic differences between high- and low-paclitaxel-yield individuals in order to further understand their differentiation. The chloroplast genomes of two T. yunnanensis individuals were sequenced, assembled and compared, which revealed some obvious differences (Table 2). The length of the TG chloroplast genome was 834 bp less than that of the TD chloroplast genome. The total length of chloroplast genome introns in TG was significantly higher than that in TD, while the total length of non-protein coding sequences in the chloroplast genome was lower in TG than in TD. A duplicate of rps12 was found in TG. Moreover, rps12 is usually necessary for trans-splicing of PPR4 and EMB2654 transcripts in maize and Arabidopsis [56], but there is currently no research on PPR4 in T. yunnanensis. Secondly, petL was absent in TG but not TD, but Andreas et al. [57] found that petL deletion does not necessarily hinder the growth of higher plants through knockout of petL in tobacco. The loss of rps16 in the TD chloroplast genome may be linked to its nuclear translocation during the course of evolution. The loss of multiple chloroplast-encoded rps16 genes has been recorded in various seed plants [58]. For example, rps16 is missing in the plastomes in Penthorum chinense, Trachelium caeruleum. and Pelargonium × hortorum [[59], [60], [61]]. Additionally, rps16 has also been observed to be completely absent in a wide range of taxa from ferns to angiosperms [[62], [63], [64], [65], [66], [67], [68]]. With the emergence of DNA barcode technology, using short DNA sequences to distinguish species and taxa has become common [69]. Although rcbL, matK, and trnH-psbA are commonly used as species-level identification barcodes in plants, these DNA barcodes are not completely reliable [[70], [71], [72], [73], [74]]. In the present study, there were no nucleotide substitutions between TG and TD in rcbL and matK. Based on the observed sequence variation, a total of 15 highly variable regions (infA, rpl23, rps3, atpE-rbcL, clpP-infA, petN-trnD-GUC, rbcL-accD, rpl32-trnL-UAG, rpoB-trnC-GCA, rps12-rps8, rrn16-trnI-GAU-1, trnI-CAU-ycf2, trnI-GAU-2-rrn23, trnL-UAA-2-trnF-GAA, ycf1-chlN) can be used as potential DNA barcodes to distinguish between TG and TD (Fig. 8). Further testing must be conducted to determine whether it can be used as a reliable DNA barcode to distinguish between TG and TD T. yunnanensis.
In the present study, HPLC technology was utilized to determine the taxane compounds in twigs from two different T. yunnanensis individuals, and significant differences were found between them. Through analysis of the chloroplast genome, potential DNA barcodes were identified to distinguish the experimental samples. However, these results are preliminary. In future experiments, more accurate quantification methods, such as HPLC-MS/MS, will be used to determine the taxane compounds, and further experiments will be conducted to validate these potential DNA barcodes.
5. Conclusions
TG accumulated a greater quantity of both paclitaxel and taxanes compared to TD. Correspondingly, the genes associated with the paclitaxel biosynthesis pathway in TG exhibited distinct expression patterns in TD. These differentially expressed genes may elucidate the factors explaining the variation in paclitaxel content among T. yunnanensis individuals from different provenances. Analysis of the chloroplast genome revealed multiple highly variable regions that could serve as potential DNA barcodes for distinguishing high-paclitaxel-yielding individuals. These experimental results contribute to a deeper understanding of the differences in the accumulation of taxoids among T. yunnanensis individuals from different provenances.
Funding
This work was supported by Kunming International (foreign) Science and Technology Cooperation Base (GHJD-2020031); Yunnan Province “Ten Thousand Plan” Industrial Technology Leading Talents (Zhang Jinfeng); the Reserve Talents for Young and Middle-aged Academic and Technical Leaders of the Yunnan Province (202205AC160044).
Data availability statement
Data associated with this study has been deposited at GenBank and Sequence Read Archive (Accession number: SRR25203150 for high-paclitaxel-yield individual, SRR25203151 for low-paclitaxel-yield individual), National Center for Biotechnology Information database.
CRediT authorship contribution statement
Dong Wang: Writing – original draft, Data curation. Jiansheng Wei: Methodology, Data curation. Xiaolong Yuan: Investigation, Formal analysis. Zhonghua Chen: Resources, Methodology. Lei Wang: Resources. Yunfen Geng: Investigation, Conceptualization. Jinfeng Zhang: Resources, Funding acquisition. Yi Wang: Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27223.
Contributor Information
Yunfen Geng, Email: gengyunfen@yafg.ac.cn.
Yi Wang, Email: wangyi@yafg.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Weaver B.A. How Taxol/paclitaxel kills cancer cells. Mol. Biol. Cell. 2014;25(18):2677–2681. doi: 10.1091/mbc.E14-04-0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mutanda I., et al. Recent advances in metabolic engineering, protein engineering, and transcriptome-guided insights toward synthetic production of taxol. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.632269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicolaou K.C., et al. Total synthesis of taxol. Nature. 1994;367(6464):630–634. doi: 10.1038/367630a0. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed Khalil A., et al. Recent developments and anticancer therapeutics of paclitaxel: an update. Curr. Pharmaceut. Des. 2022;28(41):3363–3373. doi: 10.2174/1381612829666221102155212. [DOI] [PubMed] [Google Scholar]
- 5.Baloglu E., Kingston D.G. A new semisynthesis of paclitaxel from baccatin III. J. Nat. Prod. 1999;62(7):1068–1071. doi: 10.1021/np990040k. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y., et al. Screening endophytic fungus to produce taxol from Taxus yunnanensis. Biotechnology. 2003;13(2):10–11. 10.16519/j.cnki.1004-311x.2003.02.007. [Google Scholar]
- 7.Kingston D.G., et al. Synthesis of taxol from baccatin III via an oxazoline intermediate. Tetrahedron Lett. 1994;35(26):4483–4484. doi: 10.1016/s0040-4039(00)60706-2. [DOI] [Google Scholar]
- 8.Kolewe M.E., Gaurav V., Roberts S.C. Pharmaceutically active natural product synthesis and supply via plant cell culture technology. Mol. Pharm. 2008;5(2):243–256. doi: 10.1021/mp7001494. [DOI] [PubMed] [Google Scholar]
- 9.Bestoso F., et al. In vitro cell cultures obtained from different explants of Corylus avellanaproduce Taxol and taxanes. BMC Biotechnol. 2006;6(1) doi: 10.1186/1472-6750-6-45. 10.1186/1472-6750-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu C., et al. Omic analysis of the endangered Taxaceae species Pseudotaxus chienii revealed the differences in taxol biosynthesis pathway between Pseudotaxus and Taxus yunnanensis trees. BMC Plant Biol. 2021;21(1):104. doi: 10.1186/s12870-021-02883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howat S., et al. Paclitaxel: biosynthesis, production and future prospects. N. Biotech. 2014;31(3):242–245. doi: 10.1016/j.nbt.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Eisenreich W., et al. Studies on the biosynthesis of taxol: the taxane carbon skeleton is not of mevalonoid origin. Proc. Natl. Acad. Sci. U. S. A. 1996;93(13):6431–6436. doi: 10.1073/pnas.93.13.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wildung M.R., Croteau R. A cDNA clone for taxadiene synthase, the diterpene cyclase that catalyzes the committed step of taxol biosynthesis. J. Biol. Chem. 1996;271(16):9201–9204. doi: 10.1074/jbc.271.16.9201. [DOI] [PubMed] [Google Scholar]
- 14.Williams D.C., et al. Heterologous expression and characterization of a "Pseudomature" form of taxadiene synthase involved in paclitaxel (Taxol) biosynthesis and evaluation of a potential intermediate and inhibitors of the multistep diterpene cyclization reaction. Arch. Biochem. Biophys. 2000;379(1):137–146. doi: 10.1006/abbi.2000.1865. [DOI] [PubMed] [Google Scholar]
- 15.Croteau R., et al. Taxol biosynthesis and molecular genetics. Phytochemistry Rev. 2006;5(1):75–97. doi: 10.1007/s11101-005-3748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nasiri J., et al. Seasonal-based temporal changes fluctuate expression patterns of TXS, DBAT, BAPT and DBTNBT genes alongside production of associated taxanes in Taxus baccata. Plant Cell Rep. 2016;35(5):1103–1119. doi: 10.1007/s00299-016-1941-y. [DOI] [PubMed] [Google Scholar]
- 17.Jiménez-Barbero J., Amat-Guerri F., Snyder J.P. The solid state, solution and tubulin-bound conformations of agents that promote microtubule stabilization. Curr. Med. Chem. Anticancer Agents. 2002;2(1):91–122. doi: 10.2174/1568011023354416. [DOI] [PubMed] [Google Scholar]
- 18.Li C., et al. De novo assembly and characterization of fruit transcriptome in Litchi chinensis Sonn and analysis of differentially regulated genes in fruit in response to shading. BMC Genom. 2013;14:552. doi: 10.1186/1471-2164-14-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao da C., et al. The first insight into the tissue specific taxus transcriptome via Illumina second generation sequencing. PLoS One. 2011;6(6) doi: 10.1371/journal.pone.0021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S.T., et al. Transcriptional profile of Taxus chinensis cells in response to methyl jasmonate. BMC Genom. 2012;13:295. doi: 10.1186/1471-2164-13-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou T., et al. Transcriptome analyses provide insights into the expression pattern and sequence similarity of several taxol biosynthesis-related genes in three Taxus species. BMC Plant Biol. 2019;19(1):33. doi: 10.1186/s12870-019-1645-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng D., et al. Transcriptomic response of Chinese yew (taxus chinensis) to cold stress. Front. Plant Sci. 2017;8:468. doi: 10.3389/fpls.2017.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miao Y.C., et al. Phylogeography and genetic effects of habitat fragmentation on endangered Taxus yunnanensis in southwest China as revealed by microsatellite data. Plant Biol. (Stuttg.) 2014;16(2):365–374. doi: 10.1111/plb.12059. [DOI] [PubMed] [Google Scholar]
- 24.Ohtsuki K., et al. Biochemical characterization of novel lignans isolated from the wood of Taxus yunnanensis as effective stimulators for glycogen synthase kinase-3β and the phosphorylation of basic brain proteins by the kinase in vitro. Biol. Pharm. Bull. 2012;35(3):385–393. doi: 10.1248/bpb.35.385. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z., Jia Z. Taxanes from taxus yunnanensis. Phytochemistry. 1990;29(11):3673–3675. doi: 10.1016/0031-9422(90)85303-w. [DOI] [Google Scholar]
- 26.Glasenapp-Breiling M., et al. Progress in the Chemistry of Organic Natural Products/Fortschritte der Chemie organischer Naturstoffe. 2002. The chemistry of taxol and related taxoids; pp. 53–225. [DOI] [PubMed] [Google Scholar]
- 27.Ge G.B., et al. Rapid qualitative and quantitative determination of seven valuable taxanes from various Taxus species by UFLC-ESI-MS and UFLC-DAD. Planta Med. 2010;76(15):1773–1777. doi: 10.1055/s-0030-1249959. [DOI] [PubMed] [Google Scholar]
- 28.Wang L., et al. The clone selection of high taxane content of taxus wallichiana var. wallichiana. Forest Resour. Manag. 2013;(6):154–156. doi: 10.13466/j.cnki.lyzygl.2013.06.034. [DOI] [Google Scholar]
- 29.Mubeen S., et al. Comparative transcriptome analysis revealed the tissue-specific accumulations of taxanes among three experimental lines of taxus yunnanensis. J. Agric. Food Chem. 2018;66(40):10410–10420. doi: 10.1021/acs.jafc.8b03502. [DOI] [PubMed] [Google Scholar]
- 30.Lenka S.K., et al. Jasmonate-responsive expression of paclitaxel biosynthesis genes in Taxus cuspidata cultured cells is negatively regulated by the bHLH transcription factors TcJAMYC1, TcJAMYC2, and TcJAMYC4. Front. Plant Sci. 2015;6:115. doi: 10.3389/fpls.2015.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M., et al. Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis. Plant Mol. Biol. 2015;89(4–5):463–473. doi: 10.1007/s11103-015-0382-2. [DOI] [PubMed] [Google Scholar]
- 32.Zhang M., et al. Transcriptome-wide identification and screening of WRKY factors involved in the regulation of taxol biosynthesis in Taxus chinensis. Sci. Rep. 2018;8(1):5197. doi: 10.1038/s41598-018-23558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng Y., et al. The complete chloroplast genome sequence of Taxus yunnanensis. Mitochondrial DNA B Resour. 2020;5(3):2756–2757. doi: 10.1080/23802359.2020.1788442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He C.T., et al. Transcriptome profiling reveals specific patterns of paclitaxel synthesis in a new Taxus yunnanensis cultivar. Plant Physiol. Biochem. 2018;122:10–18. doi: 10.1016/j.plaphy.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 35.Zhu X. Content of paclitaxel and its related compounds in different parts of Taxus yunnanensis. Fitoterapia. 1996;67(2):149–151. [Google Scholar]
- 36.Yu C., et al. Comparative metabolomics reveals the metabolic variations between two endangered Taxus species (T. fuana and T. yunnanensis) in the Himalayas. BMC Plant Biol. 2018;18(1):197. doi: 10.1186/s12870-018-1412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S., et al. Determination of paclitaxel and other six taxoids in Taxus species by high-performance liquid chromatography–tandem mass spectrometry. J. Pharmaceut. Biomed. Anal. 2009;49(1):81–89. doi: 10.1016/j.jpba.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Denis J.N., et al. Highly efficient, practical approach to natural taxol. J. Am. Chem. Soc. 1988;110(17):5917–5919. doi: 10.1021/ja00225a063. [DOI] [Google Scholar]
- 39.Gunatilaka A.L., Chordia M.D., Kingston D.G. Efficient conversion of cephalomannine to paclitaxel and 3'-N-acyl-3'-N-debenzoylpaclitaxel analogs. J. Org. Chem. 1997;62:3775–3778. [Google Scholar]
- 40.Nikolakakis A., et al. Taxus canadensis abundant taxane: conversion to paclitaxel and rearrangements. Bioorg. Med. Chem. 2000;8(6):1269–1280. doi: 10.1016/s0968-0896(00)00056-0. [DOI] [PubMed] [Google Scholar]
- 41.Rao K.V. Semi‐synthesis of paclitaxel from naturally occurring glycosidic precursors. J. Heterocycl. Chem. 1997;34(2):675–680. doi: 10.1002/jhet.5570340255. [DOI] [Google Scholar]
- 42.Rao K.V., et al. Synthesis and evaluation of some 10-mono-and 2', 10-diesters of 10-deacetylpaclitaxel. J. Med. Chem. 1995;38(17):3411–3414. doi: 10.1021/jm00017a026. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y.-F., et al. Natural taxanes: developments since 1828. Chem. Rev. 2011;111(12):7652–7709. doi: 10.1021/cr100147u. [DOI] [PubMed] [Google Scholar]
- 44.Nižnanský Ľ., et al. Natural taxanes: from plant composition to human pharmacology and toxicity. Int. J. Mol. Sci. 2022;23(24) doi: 10.3390/ijms232415619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ketchum R.E., et al. The kinetics of taxoid accumulation in cell suspension cultures of Taxus following elicitation with methyl jasmonate. Biotechnol. Bioeng. 1999;62(1):97–105. doi: 10.1002/(sici)1097-0290(19990105)62:1<97::aid-bit11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 46.Walker K., et al. Partial purification and characterization of acetyl coenzyme A: taxa-4(20),11(12)-dien-5 alpha-ol O-acetyl transferase that catalyzes the first acylation step of taxol biosynthesis. Arch. Biochem. Biophys. 1999;364(2):273–279. doi: 10.1006/abbi.1999.1125. [DOI] [PubMed] [Google Scholar]
- 47.You L.F., et al. Activity essential residue analysis of taxoid 10β-O-acetyl transferase for enzymatic synthesis of baccatin. Appl. Biochem. Biotechnol. 2018;186(4):949–959. doi: 10.1007/s12010-018-2789-0. [DOI] [PubMed] [Google Scholar]
- 48.Onrubia M., et al. The relationship between TXS, DBAT, BAPT and DBTNBT gene expression and taxane production during the development of Taxus baccata plantlets. Plant Sci. 2011;181(3):282–287. doi: 10.1016/j.plantsci.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Wheeler A.L., et al. Taxol biosynthesis: differential transformations of taxadien-5 alpha-ol and its acetate ester by cytochrome P450 hydroxylases from Taxus suspension cells. Arch. Biochem. Biophys. 2001;390(2):265–278. doi: 10.1006/abbi.2001.2377. [DOI] [PubMed] [Google Scholar]
- 50.Perez-Matas E., et al. Insights into the control of taxane metabolism: molecular, cellular, and metabolic changes induced by elicitation in Taxus baccata cell suspensions. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.942433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broun P. Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 2004;7(2):202–209. doi: 10.1016/j.pbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 52.Nims E., et al. Expression profiling of genes involved in paclitaxel biosynthesis for targeted metabolic engineering. Metab. Eng. 2006;8(5):385–394. doi: 10.1016/j.ymben.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 53.Wang T., et al. Transcriptome sequencing reveals regulatory mechanisms of taxol synthesis in Taxus wallichiana var. Mairei. Int. J. Genomics. 2019 doi: 10.1155/2019/1596895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuang X., et al. Iso-Seq analysis of the Taxus cuspidata transcriptome reveals the complexity of Taxol biosynthesis. BMC Plant Biol. 2019;19(1):210. doi: 10.1186/s12870-019-1809-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song S.L., et al. Development of chloroplast simple sequence repeats (cpSSRs) for the intraspecific study of Gracilaria tenuistipitata (Gracilariales, Rhodophyta) from different populations. BMC Res. Notes. 2014;7:77. doi: 10.1186/1756-0500-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee K., et al. The coordinated action of PPR4 and EMB2654 on each intron half mediates trans-splicing of rps12 transcripts in plant chloroplasts. Plant J. 2019;100(6):1193–1207. doi: 10.1111/tpj.14509. [DOI] [PubMed] [Google Scholar]
- 57.Fiebig A., Stegemann S., Bock R. Rapid evolution of RNA editing sites in a small non-essential plastid gene. Nucleic Acids Res. 2004;32(12):3615–3622. doi: 10.1093/nar/gkh695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jansen R.K., Ruhlman T.A. Genomics of Chloroplasts and Mitochondria. 2012. Plastid genomes of seed plants; pp. 103–126. [DOI] [Google Scholar]
- 59.Dong W., et al. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of saxifragales. Genome Biol. Evol. 2013;5(5):989–997. doi: 10.1093/gbe/evt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haberle R.C., et al. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 2008;66(4):350–361. doi: 10.1007/s00239-008-9086-4. [DOI] [PubMed] [Google Scholar]
- 61.Chumley T.W., et al. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006;23(11):2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- 62.Gao L., et al. Plastome sequences of Lygodium japonicum and Marsilea crenata reveal the genome organization transformation from basal ferns to core leptosporangiates. Genome Biol. Evol. 2013;5(7):1403–1407. doi: 10.1093/gbe/evt099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Magee A.M., et al. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 2010;20(12):1700–1710. doi: 10.1101/gr.111955.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sabir J., et al. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol. J. 2014;12(6):743–754. doi: 10.1111/pbi.12179. [DOI] [PubMed] [Google Scholar]
- 65.Saski C., et al. Complete chloroplast genome sequence of Gycine max and comparative analyses with other legume genomes. Plant Mol. Biol. 2005;59(2):309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- 66.Tangphatsornruang S., et al. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 2010;17(1):11–22. doi: 10.1093/dnares/dsp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsudzuki J., et al. Chloroplast DNA of black pine retains a residual inverted repeat lacking rRNA genes: nucleotide sequences of trnQ, trnK, psbA, trnI and trnH and the absence of rps16. Mol. Gen. Genet. 1992;232(2):206–214. doi: 10.1007/bf00279998. [DOI] [PubMed] [Google Scholar]
- 68.Ueda M., et al. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 2008;25(8):1566–1575. doi: 10.1093/molbev/msn102. [DOI] [PubMed] [Google Scholar]
- 69.Li X., et al. Plant DNA barcoding: from gene to genome. Biol. Rev. Camb. Phil. Soc. 2015;90(1):157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
- 70.Coissac E., et al. From barcodes to genomes: extending the concept of DNA barcoding. Mol. Ecol. 2016;25(7):1423–1428. doi: 10.1111/mec.13549. [DOI] [PubMed] [Google Scholar]
- 71.Hollingsworth P.M., Graham S.W., Little D.P. Choosing and using a plant DNA barcode. PLoS One. 2011;6(5) doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hollingsworth P.M. Refining the DNA barcode for land plants. Proc. Natl. Acad. Sci. U. S. A. 2011;108(49):19451–19452. doi: 10.1073/pnas.1116812108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hollingsworth P.M., et al. Telling plant species apart with DNA: from barcodes to genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371(1702) doi: 10.1098/rstb.2015.0338. 10.1098/rstb.2015.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hollingsworth PM F.L., Spouge J.L., Hajibabaei M., Ratnasingham S., van der Bank M., et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. U. S. A. 2009;106(31):12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at GenBank and Sequence Read Archive (Accession number: SRR25203150 for high-paclitaxel-yield individual, SRR25203151 for low-paclitaxel-yield individual), National Center for Biotechnology Information database.