Abstract
Background: Meningiomas are among the most common intracranial tumors. In these tumors, volumetric assessment is not only important for planning therapeutic intervention but also for follow-up examination.However, a highly accurate automated volumetric method for meningiomas using single-modality magnetic resonance imaging (MRI) has not yet been reported. Here, we aimed to develop a deep learning-based automated volumetry method for meningiomas in MRI and investigate its accuracy and potential clinical applications. Methods: For deep learning, we used MRI images of patients with meningioma who were referred to Osaka University Hospital between January 2007 and October 2020. Imaging data of eligible patients were divided into three non-overlapping groups: training, validation, and testing. The model was trained and tested using the leave-oneout cross-validation method. Dice index (DI) and root mean squared percentage error (RMSPE) were measured to evaluate the model accuracy. Result: A total of 178 patients (64.6 ± 12.3 years [standard deviation]; 147 women) were evaluated. Comparison of the deep learning model and manual segmentation revealed a mean DI of 0.923 ± 0.051 for tumor lesions. For total tumor volume, RMSPE was 9.5 ± 1.2%, and Mann–Whitney U test did not show a significant difference between manual and algorithm-based measurement of the tumor volume (p = 0.96). Conclusion: The automatic tumor volumetry algorithm developed in this study provides a potential volume-based imaging biomarker for tumor evaluation in the field of neuroradiological imaging, which will contribute to the optimization and personalization of treatment for central nervous system tumors in the near future
Keywords: Algorithm, Biomarkers, Brain neoplasms, Deep learning, Magnetic resonance imaging, Meningeal neoplasms, Meningioma
1. Introduction
Meningiomas are among the most common intracranial tumors. These tumors account for approximately 30% of primary intracranial tumors and are often discovered incidentally.1,2 Radiological and autopsy studies have reported an incidence of approximately 1.2 cases of incidental meningiomas per 100,000 people per year.3,4 The World Health Organization (WHO) classifies meningiomas as benign (grade I), atypical (grade II), or malignant (grade III), with most of them being diagnosed as Grade I.5 The WHO grades of meningiomas diagnosed based on histopathologic findings, are related to imaging features and grade II or III meningiomas grow at a comparatively faster rate.6, 7, 8 Enlarged meningiomas have been reported in 70% of cases, and increased meningioma volume is known to be associated with its symptoms.9, 10, 11, 12 The primary treatment strategy for asymptomatic meningioma is observation, wherein some patients require regular follow-up with imaging studies to help determine the timing of therapeutic intervention.13
Manual segmentation by physicians is commonly used to measure tumor volume; however, differences between inter- and intra-observers may make it difficult to achieve a precise follow-up and study.14,15 To the best of our knowledge, no previous studies have reported an automated volumetric method for meningiomas using single-modality magnetic resonance imaging (MRI) with high accuracy.
Therefore, in this study, we used deep learning techniques with contrast-enhanced T1 (CET1)-weighted MRI to develop and evaluate a supportive algorithm for automated volumetry of meningiomas.
2. Materials and methods
This retrospective study was approved by the institutional review board of our institution. Patient informed consent was waived because this study consisted of a retrospective analysis of existing records.
2.1. Datasets
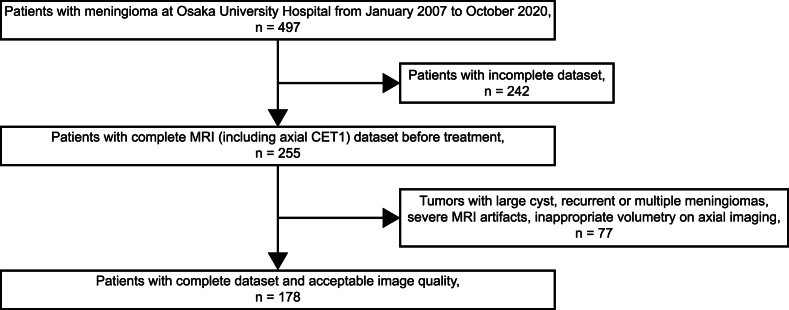
A total of 497 patients who were referred to Osaka University Hospital for meningioma treatment and underwent MRI between January 2007 and October 2020 were included. MRI Digital Imaging and Communications in Medicine data from different scanners, including data from a referral center, were used. Among the 255 patients with a completely available MRI dataset before treatment, those with axial CET1-weighted MRI sequence were further included. If a patient underwent multiple MRI scans, we only considered the final images. Furthermore, 77 patients were excluded from further assessment for the following reasons (Fig. 1): tumor with a large cyst, previous treatment, image size other than 256 × 256 or 512 × 512 pixels, recurrent or multiple meningiomas, and severe MRI artifacts. Patients with inappropriate volumetry on axial imaging, such as remarkably small meningiomas located at high convexity, were excluded.
Fig. 1.
Flowchart depicting the patient selection process.
All scans were performed for clinical indications. MRI acquisitions were performed using diverse scanners from our institution and the referring institutions. The examinations were mainly performed using MRI units (Achieva, Philips Medical Systems, Best, the Netherlands and SIGNA, GE Healthcare, Milwaukee, WI, USA). Each patient underwent imaging with a CET1-weighted MRI sequence after the administration of gadolinium.
2.2. Algorithm development methods
All meningioma segmentations were conducted manually by two experienced neurosurgeons (R.H. and S.Y.) using Horos (www.horosproject.org; Horos Project) for macOS and Onis 2.5 Professional (www.onis-viewer.com; DigitalCore). The segmentation data were converted into a binarized image, which was used as the ground truth. The input MRI data of the deep learning network included only slices that contained the tumor. Images of size 256 × 256 pixels were converted to 512 × 512 pixels.
Convolutional neural networks are often used for deep-learning-based segmentation. We performed transfer learning by fine-tuning a DeepLabV3 model that was pretrained by ResNet-101 to fully automate the process of meningioma segmentation. The networks were created in PyTorch using a Windows desktop with a 32-GB RAM and NVIDIA RTX 2080. For training, we used the Adam optimizer with a batch size of four. The learning rate was initialized to exp (−2). Because tumors are 3D objects, three neighboring 2D slices were stacked as the inputs after the input images were converted to grayscale. Thus, the 2D network could detect a small range of 3D contexts each time. Image augmentation was employed to improve the generalizability of the network. Training was terminated if the validation loss did not improve after 20 epochs. The eligible data were divided into training (90%) and test (10%) datasets. The model was trained and tested using leave-one-out 10-fold cross-validation (Supplementary Fig. 1).
2.3. Statistical analysis
For quantitative analysis, the Dice index (DI) and root mean squared percentage error (RMSPE) of the tumor volume were measured. We defined pixels with tumors as true and those without tumors as false.
The DI is a proven statistical validation metric used to evaluate the performance and spatial overlap between two sets of segmentations of the same anatomy. The DI is represented as a percentage using the below formula, with 100% being a perfect voxel-wise match between results.
DI, Dice index; TP, true positive; FP, false positive; FN, false negative.
RMSPE is defined by the below formula and is the error between the manually measured volume of each tumor and the volume calculated using artificial intelligence. Continuous variables are expressed as means and standard deviations. The Mann–Whitney U test was used to compare the two groups. JMP Pro 16.0.0 (SAS Institute, Cary, NC, USA) was used for statistical analyses.
RMSPE, root mean squared percentage error; Vi, manually measured volume; , automatically measured volume
3. Results
The training dataset included 178 patients (147 females and 31 males) with meningiomas. The characteristics of all patients are listed in Table 1. The mean age of the patients was 64.6 ± 12.3 years. Tumor locations included the convexity (n = 66), parasagittal (n = 34), falx (n = 26), cerebellopontine angle (n = 16), and other locations (n = 36). Peritumoral brain edema was observed in 65 patients.
Table 1.
Patient and tumor characteristics.
| CASES (n = 178) | ||
|---|---|---|
| SEX | Female | 147 (82.6%) |
| Male | 31 (17.4%) | |
| AGE (YEARS) | Mean ± SD | 64.6 ± 12.3 |
| Range | 23–89 | |
| LOCATION | Convexity | 66 (37.1%) |
| Parasagittal | 34 (19.1%) | |
| Falx | 26 (14.6%) | |
| CP-angle | 16 (9.0%) | |
| Other | 36 (20.2%) | |
| TUMOR VOLUME (CM3) | Mean ± SD | 20.5 ± 26.0 |
| Range | 0.9–138.8 | |
| PERITUMORAL BRAIN EDEMA | Yes | 65 (36.5%) |
| No | 113 (63.5%) |
3.1. Segmentation and volumetry performance of the network
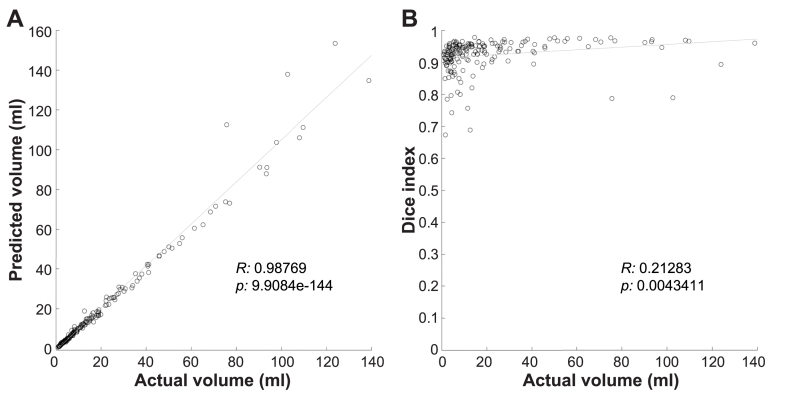
Among all patients, manual segmentation volume of tumors averaged 20.5 ± 26.0 cm3 (range 0.9–138.8 cm3). The accuracy and consistency of the developed algorithm were further evaluated using the DI to assess the 2D accuracy of the individual images. The mean DI for the tumor region was 0.923 ± 0.051. The actual tumor volume was significantly correlated with the predicted volume (R: 0.9877, p < 0.001; Pearson correlation analysis). The tumor volume showed a weak correlation with the DI (R: 0.21283, p: 0.0043411; Pearson correlation analysis) (Fig. 2). Furthermore, we compared the DI in two groups of tumors: those below and above the median volume of all tumors in this study. This analysis revealed a significantly lower DI for tumors with a volume below the median compared with those above the median (DI = 0.91 ± 0.051 and 0.94 ± 0.049; p < 0.0001; Wilcoxon rank sum test).
Fig. 2.
(A)Plot of actual vs. predicted tumor volume. Tumor volume was significantly correlated with the predicted volume (R: 0.9877, p: 9.9084e-144; Pearson correlation analysis). (B) Plot of tumor volume vs. Dice index (DI). Tumor volume showed a weak correlation with the DI (R: 0.21283, p: 0.0043411; Pearson correlation analysis).
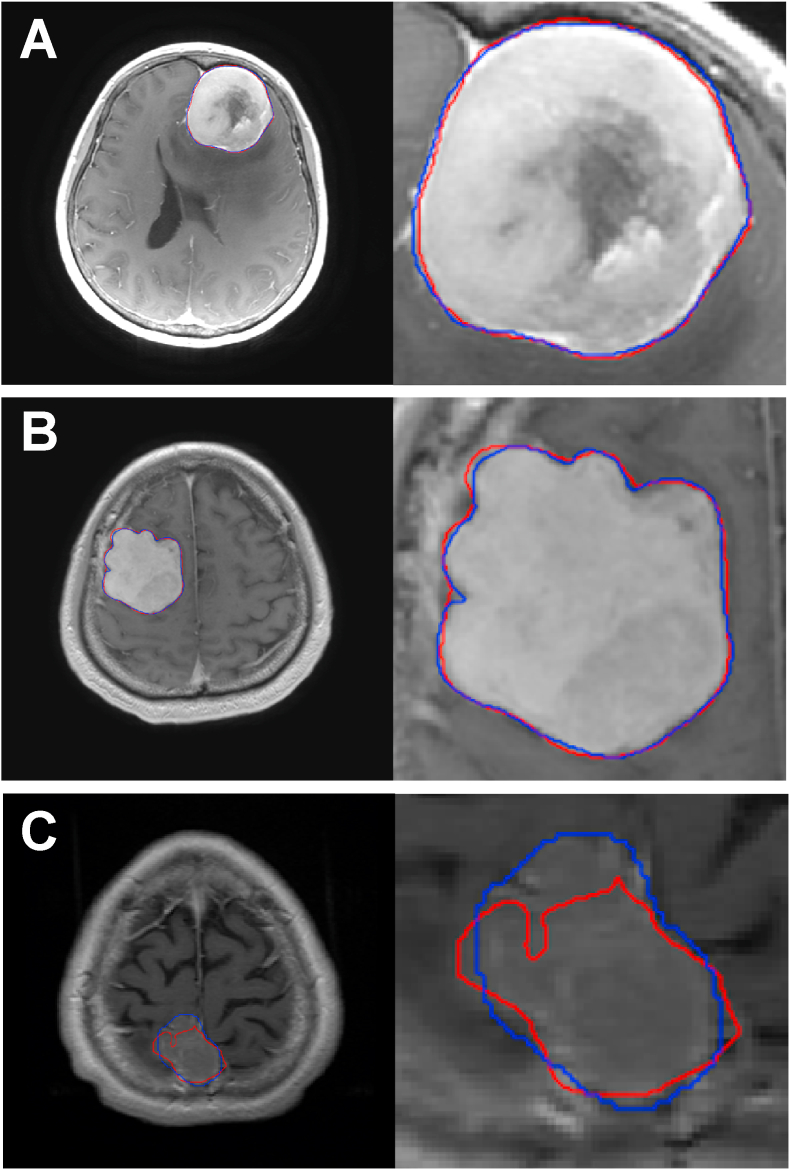
Representative cases are shown in Fig. 3. Cases with a clear contrast effect, but with the tumor in contact with the skull base (Fig. 3A) and cases with a large tumor and no large surrounding vessels to be contrasted (Fig. 3B), but with irregularly shaped tumor, were automatically segmented with a high DI. In contrast, cases with small tumors and poor contrast effects (Fig. 3C) were poorly segmented with a low DI.
Fig. 3.
Representative cases are shown. The left column shows a contrast-enhanced T1 image with the outline of a detected tumor. The right column image is a magnified image of the tumor and its surrounding area. The blue contour shows manual segmentation, while the red contour represents automated segmentation using the algorithm.A (case no. 30) is a case of left frontal base meningioma with a Dice index (DI) of 0.975, which is representative of highly accurate segmentation of the region bordering the superior sagittal sinus and falx cerebri.B (case no. 212) is a case of right convexity meningioma. Although the tumor was irregularly shaped, it was segmented with high accuracy (DI: 0.973).C (case no. 109) is a case of right parasagittal meningioma with a DI of 0.688 and poor segmentation. The tumor was small, and the contrast effect with the surrounding parenchyma was poor.
3.2. Comparison of automated and manual volumetry
The volume RMSPE was measured to assess the 3D and clinical utility of individual cases. The mean RMSPE for tumor volume was 9.5 ± 1.2%, and the Mann–Whitney U test did not show a significant difference between the tumor volume measured manually and that measured with our algorithm (p = 0.96).
4. Discussion
In this study, we developed a deep learning-based method for the automated volumetry of meningiomas. The DI and RMSPE showed that the automatically segmented tumor regions correlated well with manual segmentation, which was highly accurate compared to other previous studies based on brain lesion segmentation using deep learning.16,17
Artificial intelligence has been demonstrated to be reaching a point where it can be used reliably in clinical applications.15, 16, 17 Glioma is a clinically important brain tumor due to its nature and has been a research target for automated tumor segmentation.16,17 Meningioma segmentation has also been a research focus, with several automated approaches being reported since tracking of tumor volume plays an important role in determining therapeutic interventions for meningiomas.18,19 Recent studies have reported the accuracy of meningioma tumor volume measurement using deep learning methods with multiple MRI sequences, including T1-weighted image (T1WI) and T2WI as well as CET1 and fluid-attenuated inversion recovery (FLAIR) images.20,21 The segmentation accuracy of CE tumor regions has reached a DI of up to 0.91 ± 0.08.21 In order to obtain a high accuracy, only CET1-weighted images were used in our study. The reasons for the high accuracy of this algorithm are that a large number of examples were used, transfer learning was well adapted, and noise was added during augmentation to minimize the overfitting of the algorithm and generalizability.
Automatic detection is highly reproducible and objective as it avoids observer differences in the definition of tumor volume.14,15 Moreover, conventional diameter measurements tend to underestimate tumor growth, whereas volume measurements based on deep learning can reliably detect tumor growth by volume, thereby supporting decision-making for treatment strategies and providing a precise understanding of the disease state.14 Thus, the reliable automatic volume measurement shown in this study can replace the time-consuming manual segmentation and deal with large amounts of data.
The deep learning method developed in this study is also clinically useful because it can be implemented using a single MRI modality. Furthermore, MR images from different scanners, including those from a referral center, were used to prove that the method could provide accurate volume measurements. Because meningiomas have less structural complexity than gliomas and other types of tumors, the segmentation accuracy should be higher. However, segmentation of meningiomas may be rendered difficult due to the following tumor features1: significant edema around the tumor,2 infiltration into the skull base or bone invasion,3 wide dural tail sign,4 large cysts,5 necrosis, and6 CE vascular structures adjacent to the tumor. Therefore, more data are needed on the accurate segmentation of tumors with these characteristics.
The current study has several limitations. First, this was a single-center, retrospective study. In the future, its validity will be necessary to be evaluated under the imaging conditions at other facilities. Second, the accuracy of detecting the presence of tumors was not evaluated because the training was performed only on slices where the tumor was present. In addition, to increase specificity, training on a mixture of datasets with and without meningiomas is required. However, in clinical follow-up situations, assessing the increase in meningioma volume may be more important than detecting the presence of meningioma. Third, CET1-weighted images were used in this study to develop a method that minimizes the variability inherent in manual volume measurements. In clinical practice, excellent agreement between meningioma size and growth is obtained from T1 3D-gadolinium and 2D-T2WI, suggesting that the use of non-contrast images may be sufficient for the follow-up of untreated meningiomas.22 However, our dataset did not include non-contrast T1 and T2 annotation data, limiting our ability to extend our findings to these imaging modalities. Therefore, we are planning a study to create a similar algorithm using T2WI or FLAIR images.
5. Conclusions
Our study results suggest that automatic volume calculation using deep learning can be easily used for follow-up and treatment planning of meningiomas with high reproducibility and accuracy. Additionally, the automatic tumor volumetry algorithm developed in this study provides a potential volume-based imaging biomarker for tumor evaluation in the field of neuroradiological imaging, which will contribute to the optimization and personalization of treatment for central nervous system tumors in the near future.
Funding
This investigation was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant number 22K07279).
CRediT authorship contribution statement
Takamitsu Iwata: Writing – original draft. Ryuichi Hirayama: Writing – review & editing. Shuhei Yamada: Data curation, Funding acquisition. Noriyuki Kijima: Data curation. Yoshiko Okita: Data curation. Naoki Kagawa: Data curation. Haruhiko Kishima: Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Abbreviations
- CET1
Contrast-enhanced T1
- DI
Dice index
- FLAIR
fluid-attenuated inversion recovery
- MRI
Magnetic resonance imaging
- RMSPE
Root mean squared percentage error
- T1WI
T1-weighted image
- WHO
World Health Organization
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.wnsx.2024.100353.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
A diagram illustrating the process of 10-fold cross-validation was utilized in this study. The dataset was divided into ten subsets, with nine of these subsets being used for training, and one subset for test. The mean Dice index was calculated based on the ten partitioned test results, and this value was utilized as the accuracy of the model, serving as a performance indicator for the cross-validation framework.
References
- 1.Dolecek T.A., Propp J.M., Stroup N.E., Kruchko C. CBTRUS statistical Report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro Oncol. 2012;14(suppl 5):v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom Q.T., Patil N., Cioffi G., Waite K., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro Oncol. 2020;22(Supplement_1):iv1–iv96. doi: 10.1093/neuonc/noaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuratsu J ichi, Kochi M., Ushio Y. Incidence and clinical features of asymptomatic meningiomas. J Neurosurg. 2000;92(5):766–770. doi: 10.3171/jns.2000.92.5.0766. [DOI] [PubMed] [Google Scholar]
- 4.Nakasu S., Hirano A., Shimura T., Llena J.F. Incidental meningiomas in autopsy study. Surg Neurol. 1987;27(4):319–322. doi: 10.1016/0090-3019(87)90005-X. [DOI] [PubMed] [Google Scholar]
- 5.Louis D.N., Perry A., Reifenberger G., et al. The 2016 World Health Organization Classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 6.Hale A.T., Wang L., Strother M.K., Chambless L.B. Differentiating meningioma grade by imaging features on magnetic resonance imaging. J Clin Neurosci. 2018;48:71–75. doi: 10.1016/j.jocn.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 7.Violaris K., Katsarides V., Karakyriou M., Sakellariou P. Surgical Outcome of treating Grades II and III meningiomas: a Report of 32 cases. Neurosci J. 2013;2013:1–4. doi: 10.1155/2013/706481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jääskeläinen J., Haltia M., Laasonen E., Wahlström T., Valtonen S. The growth rate of intracranial meningiomas and its relation to histology. An analysis of 43 patients. Surg Neurol. 1985;24(2):165–172. doi: 10.1016/0090-3019(85)90180-6. [DOI] [PubMed] [Google Scholar]
- 9.Nakasu S., Nakasu Y. Natural history of meningiomas: review with Meta-analyses. Neurol Med -Chir. 2020;60(3):109–120. doi: 10.2176/nmc.ra.2019-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada S., Kinoshita M., Nakagawa T., et al. The Impact of 5-year tumor Doubling time to predict the Subsequent Long-Term natural history of asymptomatic meningiomas. World Neurosurg. 2021;151:e943–e949. doi: 10.1016/j.wneu.2021.05.023. [DOI] [PubMed] [Google Scholar]
- 11.Kauke M., Safi A.F., Stavrinou P., Krischek B., Goldbrunner R., Timmer M. Does meningioma volume correlate with clinical disease Manifestation Irrespective of histopathologic tumor Grade? J Craniofac Surg. 2019;30(8):e799–e802. doi: 10.1097/SCS.0000000000005845. [DOI] [PubMed] [Google Scholar]
- 12.Yamada S., Kijima N., Nakagawa T., et al. How Much tumor volume is Responsible for Development of clinical symptoms in patients with convexity, parasagittal, and falx meningiomas? Front Neurol. 2021;12 doi: 10.3389/fneur.2021.769656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldbrunner R., Minniti G., Preusser M., et al. EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol. 2016;17(9):e383–e391. doi: 10.1016/S1470-2045(16)30321-7. [DOI] [PubMed] [Google Scholar]
- 14.Chang V., Narang J., Schultz L., et al. Computer-aided volumetric analysis as a sensitive tool for the management of incidental meningiomas. Acta Neurochir. 2012;154(4):589–597. doi: 10.1007/s00701-012-1273-9. [DOI] [PubMed] [Google Scholar]
- 15.Akkus Z., Galimzianova A., Hoogi A., Rubin D.L., Erickson B.J. Deep learning for brain MRI segmentation: state of the Art and future Directions. J Digit Imaging. 2017;30(4):449–459. doi: 10.1007/s10278-017-9983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan Y., Rahmat R., Price S.J. Deep learning for glioblastoma segmentation using preoperative magnetic resonance imaging identifies volumetric features associated with survival. Acta Neurochir. 2020;162(12):3067–3080. doi: 10.1007/s00701-020-04483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naser M.A., Deen M.J. Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Comput Biol Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103758. [DOI] [PubMed] [Google Scholar]
- 18.Yao A., Pain M., Balchandani P., Shrivastava R.K. Can MRI predict meningioma consistency?: a correlation with tumor pathology and systematic review. Neurosurg Rev. 2018;41(3):745–753. doi: 10.1007/s10143-016-0801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimol E ben, Joskowicz L., Eliahou R., Shoshan Y. Computer-based radiological longitudinal evaluation of meningiomas following stereotactic radiosurgery. Int J Comput Assist Radiol Surg. 2018;13(2):215–228. doi: 10.1007/s11548-017-1673-7. [DOI] [PubMed] [Google Scholar]
- 20.Laukamp K.R., Thiele F., Shakirin G., et al. Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI. Eur Radiol. 2019;29(1):124–132. doi: 10.1007/s00330-018-5595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laukamp K.R., Pennig L., Thiele F., et al. Automated meningioma segmentation in multiparametric MRI. Clin Neuroradiol. 2021;31(2):357–366. doi: 10.1007/s00062-020-00884-4. [DOI] [PubMed] [Google Scholar]
- 22.Boto J., Guatta R., Fitsiori A., Hofmeister J., Meling T.R., Vargas M.I. Is contrast Medium Really needed for follow-up MRI of untreated intracranial meningiomas? Am J Neuroradiol. 2021;42(8):1421–1428. doi: 10.3174/ajnr.A7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A diagram illustrating the process of 10-fold cross-validation was utilized in this study. The dataset was divided into ten subsets, with nine of these subsets being used for training, and one subset for test. The mean Dice index was calculated based on the ten partitioned test results, and this value was utilized as the accuracy of the model, serving as a performance indicator for the cross-validation framework.