Graphical abstract
Keywords: Methicillin-resistant Staphylococcus aureus, Sophora flavescens, Lavandulylated flavonoids, Sophoraflavanone G, Kurarinone
Highlights
-
•
Sophoraflavanone G and kurarinone exhibit potent antibacterial activity and do not develop drug resistance.
-
•
Sophoraflavanone G and kurarinone disrupt the integrity of cell membranes and prevent the synthesis of biofilms.
-
•
Sophoraflavanone G and kurarinone can disrupt the energy metabolism process of methicillin-resistant Staphylococcus aureus.
-
•
Sophoraflavanone G and kurarinone can improve wound infection and promote wound healing.
-
•
Sophoraflavanone G and kurarinone may be potential candidates for the development of new antibiotics.
Abstract
Introduction
The continuous emergence and rapid spread of multidrug-resistant bacteria have accelerated the demand for the discovery of alternative antibiotics. Natural plants contain a variety of antibacterial components, which is an important source for the discovery of antimicrobial agents.
Objective
To explore the antimicrobial activities and related mechanisms of two lavandulylated flavonoids, sophoraflavanone G and kurarinone in Sophora flavescens against methicillin-resistant Staphylococcus aureus.
Methods
The effects of sophoraflavanone G and kurarinone on methicillin-resistant Staphylococcus aureus were comprehensively investigated by a combination of proteomics and metabolomics studies. Bacterial morphology was observed by scanning electron microscopy. Membrane fluidity, membrane potential, and membrane integrity were determined using the fluorescent probes Laurdan, DiSC3(5), and propidium iodide, respectively. Adenosine triphosphate and reactive oxygen species levels were determined using the adenosine triphosphate kit and reactive oxygen species kit, respectively. The affinity activity of sophoraflavanone G to the cell membrane was determined by isothermal titration calorimetry assays.
Results
Sophoraflavanone G and kurarinone showed significant antibacterial activity and anti-multidrug resistance properties. Mechanistic studies mainly showed that they could target the bacterial membrane and cause the destruction of the membrane integrity and biosynthesis. They could inhibit cell wall synthesis, induce hydrolysis and prevent bacteria from synthesizing biofilms. In addition, they can interfere with the energy metabolism of methicillin-resistant Staphylococcus aureus and disrupt the normal physiological activities of the bacteria. In vivo studies have shown that they can significantly improve wound infection and promote wound healing.
Conclusion
Kurarinone and sophoraflavanone G showed promising antimicrobial properties against methicillin-resistant Staphylococcus aureus, suggesting that they may be potential candidates for the development of new antibiotic agents against multidrug-resistant bacteria.
Introduction
Staphylococcus aureus is a prominent zoonotic pathogen and is considered a major cause of nosocomial and community-acquired infections [1]. Recent studies have shown that epidemic strains have continued to emerge since the discovery of methicillin-resistant Staphylococcus aureus (MRSA), which has become a terrible threat in the clinic due to its high mortality [2]. Although vancomycin, linezolid, daptomycin, tigecycline, iclaprim, 5th-generation cephalosporins, and other new antibiotics are widely used in the clinical treatment of drug-resistant bacterial infections, with the increase of misuse, bacteria resistant to the above agents have emerged in recent years [3], [4], [5]. Nowadays, the treatment of multidrug-resistant bacteria is facing a crisis. Therefore, novel effective antibacterial agents are urgently needed to combat the growing problem of antimicrobial resistance.
Natural plants have always been a valuable source of drugs, whether for the discovery of leading compounds or direct use as therapeutic agents, such as traditional Chinese medicine, Indian medicine, Persian medicine, and African medicine [6], [7], [8]. Plants do not have an immune system like mammals, so they have evolved to produce various active molecules to fight against microbial invasion [9], [10]. Recently, studies have found that active molecules from Chinese herbal medicine have varying degrees of antibacterial activity. In traditional Chinese medicine, there are many cases of combating microbic infections: Coptis chinensis Franch. can be used to treat cholera [11]. Artemisinin, an active molecule isolated from Artemisia annua Linn., has saved the lives of millions of people suffering from malaria [12], [13]. Compared to the existing antibiotics, antimicrobial compounds from traditional Chinese medicine have better bactericidal activity and can reverse multidrug resistance in bacteria [14], [15]. For example, Scutellaria baicalensis extracts may be effective against inflammation and Salmonella enteritidis infection [16]. Flavonoids and polyphenols in Psoralea corylifolia and Glycyrrhiza show promising antibacterial activity against pathogenic bacteria [17], [18]. What's more, the active ingredients in Chinese herbal medicine also exert a synergistic antibacterial effect with antibiotics and reduce the occurrence of multidrug resistance [19], [20].
The root of Sophora flavescens, also known as Kushen (Radix Sophorae Flavescentis), is commonly used to treat skin diseases and bacterial diarrhea in China and other Asian countries[21], [22]. Previous studies have found that S. flavescens contains abundant alkaloids and flavonoids, which showed excellent antibacterial activity [23], [24]. Sophoraflavanone G (SFG) and kurarinone (KE) are two representative lavandulylated flavonoids with the highest content in S. flavescens. Both showed potent antibacterial activity with a minimum inhibitory concentration (MIC) or fractional inhibitory concentration (FIC) <10 μg/mL and have a synergistic effect with common antibiotics [24], [25], [26], [27]. They have been reported to inhibit bacterial growth by reducing the fluidity of cell membranes [28]. However, their specific mechanisms of action remain unclear.
In this study, to comprehensively elucidate the antibacterial effect and related mechanism of SFG and KE on MRSA, the metabolomics and proteomics profiles of MRSA treated with SFG or KE were invested. Potential targets for SFG or KE on MRSA were identified and validated. Mechanistic studies revealed that both SFG and KE exhibited potent antibacterial activity through a distinct mode of action. Finally, the antibacterial activity of these two flavonoids was further demonstrated in a mouse model of MRSA wound infection. Our findings suggest that flavonoids with unique lavandulyl structures may be an untapped resource for the discovery of novel antimicrobial agents to address the growing prevalence of antimicrobial resistance.
Materials and methods
Chemicals and reagents
Sophoraflavanone G (purity ≥ 98 %, CAS No. 97938-30-2, Cat. No. B30760), kurarinone (purity ≥ 98 %, CAS No. 34981-26-5, Cat. No. B24034) and 3,3-dipropylthiadicarbocyanine iodide [DiSC3(5), purity ≥ 98 %, CAS No. 53213-94-8, Cat. No. S47975] were provided by Yuanye Technology Co. Ltd (Tianjin, China). Glutaraldehyde (CAS No. 111-30-8, Cat. No. G105906) and propidium iodide (CAS No. 25535-16-4, Cat. No. P266304) were purchased from Shanghai Aladdin (Shanghai, China). Phosphatidylglycerol (purity ≥ 99 %, CAS No. 322647-44-9, Cat. No. 841148P), phosphatidyl-ethanolamine (purity ≥ 99 %, CAS No. 97281-52-2, Cat. No. P6636), cardiolipin (purity ≥ 97 %, CAS No. 383907-10-6, Cat. No. C1649), trifluoroacetic acid (purity ≥ 99 %, CAS No. 76-05-1, Cat. No. 302031), formic acid (purity ≥ 99 %, CAS No. 64-18-6, Cat. No. F0507), iodoacetamide (purity ≥ 99 %, CAS No. 144-48-9, Cat. No. I6125), dithiothreitol (purity ≥ 99 %, CAS No. 16096-97-2, Cat. No. D9760), carbamide (purity ≥ 99 %, CAS No. 57-13-6, Cat. No. U5378), protease inhibitor (Cat. No. P2714), ethylene diamine tetraacetic acid (purity ≥ 97 %, CAS No. 67-42-5, Cat. No. E3889) and triethylammonium bicarbonate (purity ≥ 99 %, CAS No. 15751-58-9, Cat. No. T7408) were purchased from Sigma-Aldrich (St. Louis, USA). The reactive oxygen species kit (Cat. No. E004-1-1) and bacterial total RNA extraction kit (Cat. No. N069) were provided by Nanjing Jiancheng Bioengineering Institute (Nanjing, China). ATP assay kit (Cat. No. S0026) and BCA Protein Assay Kit (Cat. No. P0012S) were obtained from Beyotime Biotechnology (Nanjing, China). cDNA synthesis kit (Cat. No. AH321-01) and PCR fluorescence quantitative kit (Cat. No. AUQ-01) were provided by TransGen Biotech (Beijing, China). PCR primers were provided by Sangon Biotech (Shanghai, China). MRSA standard strain USA300 (ATCC® BAA-1717), S. aureus ATCC 29213, and S. aureus ATCC 25923 were purchased from the American type culture collection.
Antibacterial and resistance development test
SFG or KE was dissolved in Mueller-Hinton broth (MHB) containing 0.1 % dimethyl sulfoxide at a concentration of 62.5 μg/mL and serially diluted twofold with MHB. The SFG or KE solutions were then added to 100 μL of bacterial suspensions containing 1.0 × 106 CFU/mL bacterial cells. After incubation for 18 h, the MICs of SFG and KE were evaluated by the microdilution broth method [29]. The inhibition rate was calculated by measuring the OD600 values. The minimum concentration of the tested drug that inhibited 90 % bacterial growth was determined as MIC90.
The resistance development test was performed by a serial passage method as previously reported [30]. Penicillin and oxacillin were used as the control drugs. On the first day, 100 μL of bacterial suspensions (MRSA USA300) containing 1.0 × 104 CFU/mL bacterial cells were inoculated into a 96-well microtiter plate with different concentrations of SFG, KE, or control drugs. After overnight incubation, MIC90s were determined as described above. The MRSA culture with sublethal concentrations (OD600 > 0.1) of the tested drugs was diluted 1:1,000 in MHB and inoculated into a 96-well microtiter plate for the next MIC90 determination. The experiment was repeated up to 20 serial passages.
Time-Killing curves analysis
The time-kill kinetics of SFG and KE against MRSA (USA300), S. aureus ATCC 29213, and S. aureus ATCC 25923 were performed according to a previously reported procedure [31]. Briefly, bacteria were cultured in MHB at 37 °C overnight, then 50 μL of bacterial inoculum (5.0 × 105 CFU/mL) was mixed with 50 μL of the solution containing SFG or KE at different concentrations (1/2 ×, 1 ×, and 2 × MIC) in a 96-well microtiter plate and incubated 37 °C. At selected time points (0 h, 2 h, 4 h, 6 h, 8 h, 12 h, 24 h), the culture solution in each well was diluted with an appropriate multiple and plated onto Luria-Bertani (LB) agar and then incubated at 37 °C overnight. Colonies in each spot were counted and the time-killing curves were constructed by plotting mean colony counts (log CFU/ml, taking into account the dilution factors) versus time.
Biofilm inhibition assay
Antibiofilm activity was performed by crystal violet staining assay [32]. MRSA (USA300) was incubated in MHB at 37 °C and diluted to a final concentration of 1.0 × 105 CFU/mL. Bacteria were inoculated into a 96-well microtiter plate and cultured for 24 h. Bacterial cells were collected after centrifugation at 5000g for 5 min and rinsed twice with sterile PBS. Then 200 μL of MHB containing different concentrations of SFG or KE (1/16 ×, 1/8 ×, 1/4 ×, and 1/2 × MIC) were mixed with bacteria and cultured at 37 °C for 18 h. The MHB without SFG or KE was used as a control. After incubation, the culture was discarded and the bacteria were rinsed with PBS. The bacteria were then stained with 0.1 % crystal violet solution for 30 min. After staining, each well was rinsed three times with sterile PBS and decolored with 95 % ethanol solution for 30 min. The optical density was determined at 595 nm to calculate the relative percentage of absorbance compared to the control.
Cell membrane integrity analysis
The effect of SFG or KE on MRSA membranes was tested by propidium iodide (PI) according to the previous reports [33]. SFG or KE was added into the bacterial culture medium respectively at a final concentration of 1/2 ×, 1 ×, 2, and 4 × MIC, and cultured for 2 h (37 °C, 180 rpm). Bacterial cells were collected after centrifugation at 5000g for 5 min and rinsed by sterile PBS. 10 μL PI solution (10 μg/ mL) was added into the suspension and the solution was placed at 37 °C for 30 min. The bacterial cells were collected after centrifugation at 5000g for 5 min and the fluorescence was determined using an automatic microplate reader.
Antibacterial activity of the mixtures of SFG or KE with different phospholipids
Phosphatidylglycerol (PG), phosphatidylethanolamine (PE), and cardiolipin (CL) were each dissolved in 20 % DMSO respectively to get an initial concentration of 64 μg/mL and then diluted twofold to a final concentration of 1 μg/mL. The MIC90s of SFG or KE mixed with different phospholipids (1 to 64 µg/mL) were determined by the microdilution broth method described previously [29].
Membrane fluidity analysis
The effect of SFG or KE on the membrane fluidity of MRSA was tested by the Laurdan fluorescence probe method [34]. SFG or KE was added to the bacterial culture medium at a final concentration of 1 × MIC and cultured for 2 h (37 °C, 180 rpm). The bacterial cells were collected after centrifugation at 5000g for 5 min and rinsed with sterile PBS. Then, 10 nmol/L laurdan solution was added and the solution was placed in the dark at 37 °C for 30 min. After centrifugation at 5000g for 5 min, fluorescence was determined using an automated microplate reader.
Determination of membrane potential
The effect of SFG or KE on the membrane potential was determined by the DiSC3(5) fluorescent probe method according to the previous reports [18]. SFG or KE was added to the bacterial culture medium at the final concentration of 1/2 ×, 1 ×, 2 ×, and 4 × MIC, respectively, and then cultured for 2 h (37 °C, 180 rpm). The bacterial cells were collected after centrifugation at 5000g for 5 min and rinsed with sterile PBS. Then, 10 μL of 10 μg/mL DiSC3(5) solution was added and the solution was placed in the dark at 37 °C for 30 min. After centrifugation at 5000g for 5 min, fluorescence was measured using an automatic microplate reader at an excitation wavelength of 622 nm and an emission wavelength of 670 nm.
Determination of ATP
SFG or KE was added to the bacterial culture medium at the final concentration of 1/2 ×, 1 ×, 2 ×, and 4 × MIC, respectively, and then cultured for 2 h (37 °C, 180 rpm). The culture medium was centrifuged at 5000g for 5 min. The bacterial cells were collected after centrifugation at 5000g for 5 min and rinsed with sterile PBS. The supernatant was then collected for the determination of extracellular ATP levels. Meanwhile, the bacterial sediments were lysed with lysozyme for the determination of intracellular ATP levels [18]. ATP levels were determined using the ATP kit according to the manufacturer's instructions.
Reactive oxygen species detection
The effect of SFG or KE on ROS production of MRSA was determined using the DCFH-DA fluorescent probe according to the previous reports [18]. Bacteria in the logarithmic phase were diluted to OD600 = 0.6 in fresh medium, and then 10 μL of 10 × DCFH-DA solution was added. The solution was placed in the dark at 37 °C for 30 min, and then SFG or KE was added to the bacterial culture medium to the final concentrations of 1/2 ×, 1 ×, 2 ×, and 4 × MIC, respectively, and then cultured for 2 h (37 °C, 180 rpm). The bacterial cells were collected after centrifugation at 5000g for 5 min and rinsed with sterile PBS. Fluorescence was determined using an automated microplate reader.
Scanning electron microscopy analysis
MRSA was cultured in an LB broth medium for 18 h. SFG or KE was then added to the culture medium to a final concentration of 1 × MIC each and then cultured for 2 h (37 °C, 180 rpm). Bacterial cells were collected after centrifugation at 5000g for 5 min and rinsed with sterile PBS. The bacteria were mixed with 2.5 % glutaraldehyde solution for 2 h, then rinsed three times with sterile PBS, followed by gradient dehydration with ethanol (30 %, 50 %, 70 %, 90%, and 100 %). After dehydration, the bacterial cells were freeze-dried and coated with a gold spray [35]. The morphological changes of the bacterial cells were analyzed using a scanning electron microscope (Zeiss, Oberkochen, Germany).
Isothermal titration calorimetry analysis
To evaluate the interaction between 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) sodium salt (POPG) and SFG, the calorimetric experiment was performed on MicroCal PEAQ-ITC automated system (Malvern Panalytical, Shanghai, China) [18]. POPG was dissolved in HEPES and then injected into the cells mixed with SFG. The injection was repeated 20 times with an equilibration interval of 200 s. Data were managed using MicroCal PEAQ-ITC analysis software to calculate stoichiometry, equilibrium dissociation constant, and enthalpy changes.
Label-Free quantitative proteomics
Bacterial cells were mixed with 400 µL of lysis buffer on ice, homogenized by sonication, and boiled for 5 min. The cell lysis solution was centrifuged at 12,000g for 10 min and the supernatants were collected. The protein concentration of each sample was measured using a BCA protein assay kit, followed by trypsin digestion according to previous studies [36]. The samples were separated by an ultra-performance liquid chromatography (UPLC) system, then injected into an NSI ion source for ionization and analyzed by Q Exactive™ HF-X mass spectrometry. The ion source voltage was set to 2.1 kV, and the peptide parent ions and their secondary fragments were detected and analyzed using the high-resolution mode. The data acquisition mode is used as a data-dependent scan program. After the first-level scan, the parent ions of the top ten peptide segments with the highest signal intensity were selected to enter the HCD collision cell in sequence at 28 % fragmentation energy, and the second-level mass spectrum analysis was also performed sequentially.
Untargeted Metabolomics Methods[37].
Protein in the samples was removed by mixing 400 µL acetonitrile/methanol (1:1, v/v) with 100 µL aliquots at 4 °C. The mixture was centrifuged at 12,000 rpm for 15 min. The supernatant was mixed with 5 μL internal standard (dichlorophenylalanine, 1 mg/mL) for LC-MS analysis. Samples were separated on the UPLC system (WatersTM, USA) using an ACQUITY UPLC HSS T3 column (2.1 × 100 mm, 1.8 μm). Mass spectrometry detection was performed in a Q-Exactive Orbitrap mass spectrometer (Thermo ScientificTM, USA). Mass conditions were set as follows: ESI+: Heater temperature, 300 °C; Sheath gas flow, 45 arb; Aux gas flow, 15 arb; Sweep gas flow, 1 arb; Spray voltage, 3.0 kV; Capillary temperature, 350 °C; S-Lens RF level, 30 %. ESI−: Heater temp 300 °C, Sheath gas flow rate, 45 arb; Aux gas flow rate, 15 arb; Sweep gas flow rate, 1 arb; Spray voltage, 3.2 kV; Capillary temp, 350 °C; S-Lens RF level, 60 %.
RT-PCR analysis
MRSA was cultured to the logarithmic stage, and SFG or KE at the concentration of 1 × MIC was added and cultured at 180 rpm for 2 h. Bacteria were collected by centrifugation, and bacterial RNA was extracted using the total bacterial RNA extraction kit. The mixture of the configured reverse transcription reaction system solution (10 μL 2 × ES reaction mix, 1 μL oligo(dT)18, 1 μL RI enzyme mix, 1 μL gDNA remover) was prepared to synthesize the first chain according to the cDNA one-chain synthesis kit procedure (65 °C, 5 min; 0 °C, 2 min; 42 °C, 15 min; 85 °C, 5 s) (primers for each gene are shown in Table S6). TOP Green premix (10 μL 2 × TOP Green Mix, 0.5 μL forward primer, 0.5 μL reverse primer, 4 μL cDNA, 5 μL ddH2O) was used for quantitative fluorescence analysis. Relative gene expression levels were measured by the 2−△△Ct method [35].
Ethics statement
All experiments involving animals were conducted according to the ethical policies and procedures approved by the ethics committee of the Nanjing University of Chinese Medicine, China (Approval No. 202207A059).
Animal studies
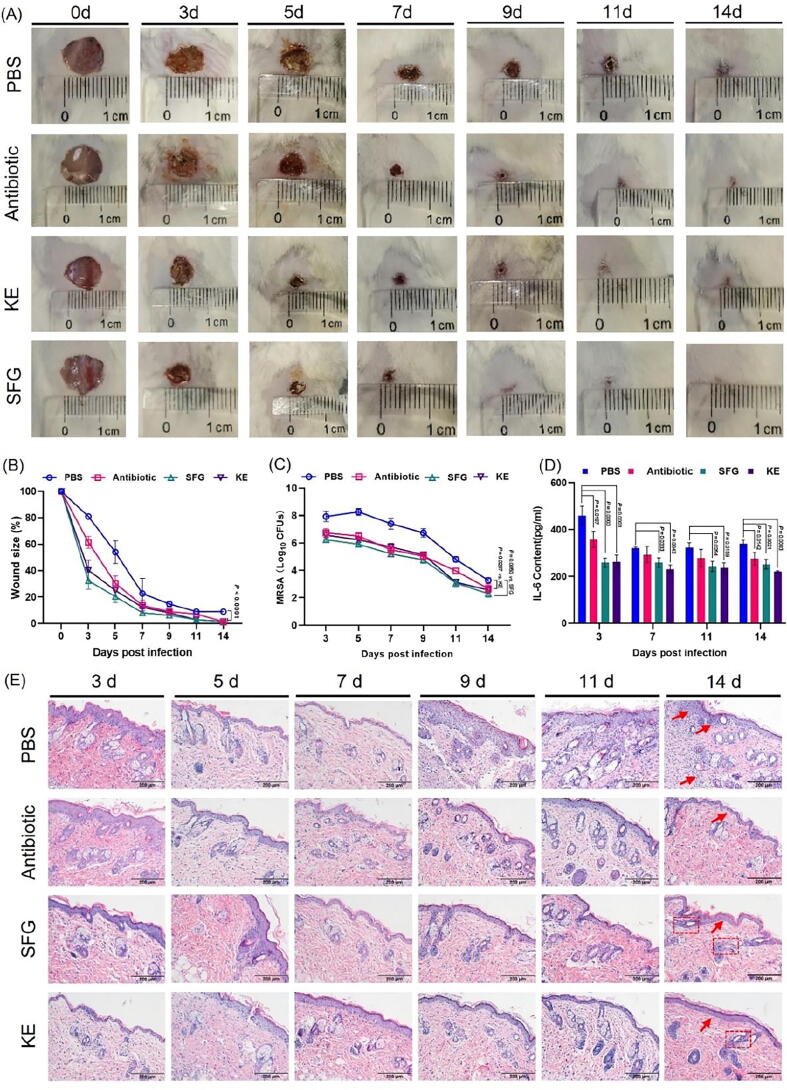
Male ICR mice aged 6–8 weeks (body weight 23.0 ± 0.5 g) were provided by Qinglongshan Animal Breeding Farm (Nanjing, China, animal license number SYXK2018-0049). Animals were housed in a facility at 23 ± 2 °C, with 12 h/12 h light/dark cycle conditions and 50 ± 5 % relative humidity. All animals had free access to distilled water and were fed standard rodent chow. After one week of adaptive feeding, mice were randomly divided into four groups: PBS group (n = 18), antibiotic group (n = 18), SFG group (n = 18), and KE group (n = 18). All mice were anesthetized with isoflurane. The fur on the back was removed and 10 mm diameter wounds were made with surgical scissors. Each wound was inoculated with 100 µL of bacterial suspension (1.0 × 107 CFU). One hour after infection, PBS, SFG (3.9 mg•kg−1), KE (7.8 mg•kg−1), or vancomycin (1.0 mg•kg−1) was applied to the wound. Wound size was determined by photographing the wound after treatment on days 3, 5, 7, 9, 11, and 14. Meanwhile, three mice in each group were sacrificed. Serum was collected for measurement of IL-6 levels. Wound tissue was excised and homogenized in PBS for bacterial enumeration. Meanwhile, the wound tissue was stained with hematoxylin and eosin to evaluate the pathological changes of wound skin tissue.
Statistical data analysis
The data are presented as Mean ± standard deviation (SD) and plotted using GraphPad Prism 9.0 software (San Diego, USA). Statistical significance was analyzed by one-way ANOVA. Statistical significance was defined as the probability (P value) < 0.05.
Results
SFG and KE are potent antibiotic candidates
Different side-chain substitutions have a major influence on the bioactivity of flavonoids. Although the antibacterial activity of flavonoids has been reported previously, the effect of side-chain substitution on bacteriostatic activity remains unclear. Since the presence of the lavandulyl side-chain greatly increases the lipophilicity of flavonoids [38], we speculate whether the lavandulylation modulates the antibacterial activity of flavonoids by targeting the bacterial membrane. We found that SFG and KE, two of the most abundant lavandulylated flavonoids in S. flavescens (Fig. S1A and B), exhibited robust antibacterial activity. The MIC90 value of SFG against MRSA (USA300), MSSA ATCC 25923, and MSSA ATCC 29213 was 3.9 μg/mL. The MIC90 value of KE against MRSA (USA300) and MSSA ATCC 29213 was 7.8 μg/mL, and against MSSA ATCC 25923 was 3.9 μg/mL. In contrast, the non-lavandulyl substituted flavonoid, naringenin, showed no significant antibacterial activity (Table S1), suggesting that the lavandulyl group is critical for antibacterial activity. Then we evaluated the time-killing kinetics of SFG and KE. The results showed that both SFG and KE could promptly reduce the viable bacteria below the detection limit within 8–12 h. The results indicated that SFG and KE had robust in vitro pharmacodynamic potentials (Fig. S1E and F). In addition, the potential development of drug resistance in MRSA after long-term exposure to SFG or KE was assessed. As shown in Fig. S1C and D, the MIC90 of SFG and KE remained stable after 20 serial subcultures, indicating that MRSA has a relatively high sensitivity to SFG and KE.
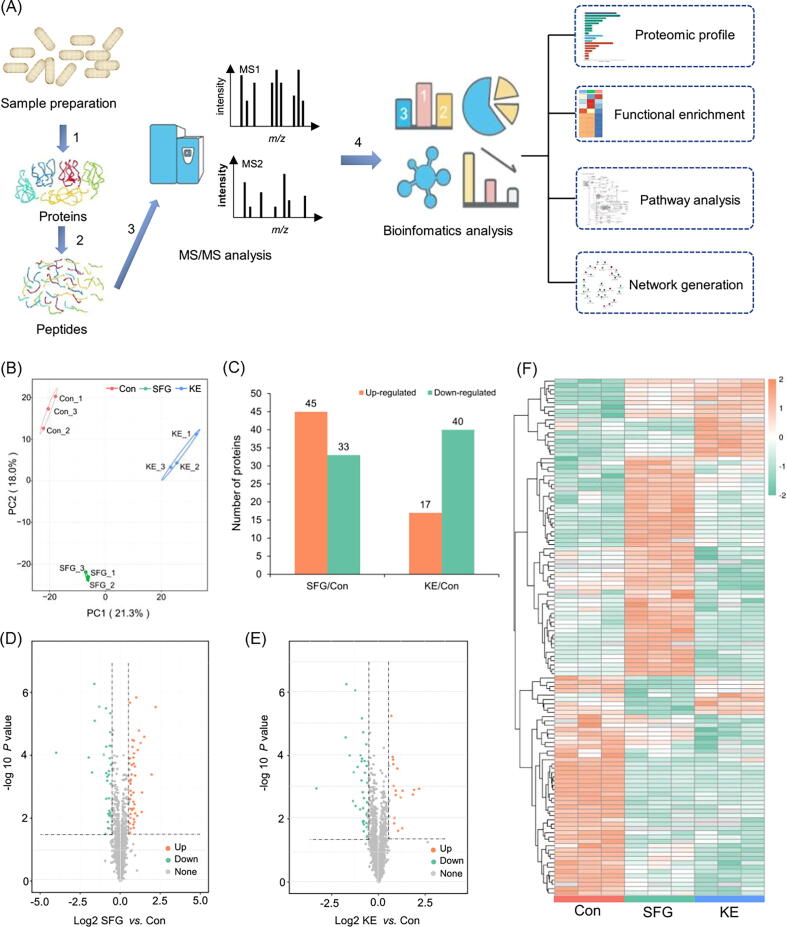
Fig. 1.
Proteomic profiling of MRSA treated with SFG or KE. (A) Flowchart illustrating the proteomic procedures for protein identification. MRSA proteins were extracted (step 1) and trypsinized (step 2). One aliquot of peptides was separated by high-performance liquid chromatography and analyzed by MS/MS (step 3). A separate aliquot was subjected to affinity enrichment and bioinformatics analysis (step 4). (B) Principal component analysis of the proteome resulted in a clear separation of three groups. The three replicates of the same species clustered tightly. (C) Differentially expressed proteins in MRSA treated with SFG and KE. (D, E) Volcano plots showing differentially expressed proteins in SFG and KE treated MRSA (in comparison with the untreated control). Dots highlighted in red (FC > 1.5/1) and green (FC < 1/1.5) indicate proteins whose expression was significantly altered (P < 0.05). (F) Heat-map and hierarchical clustering analysis of the proteins differentially abundant in MRSA cells and SFG- and KE-treated MRSA cells (P < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Proteomic profiling of MRSA treated with SFG or KE
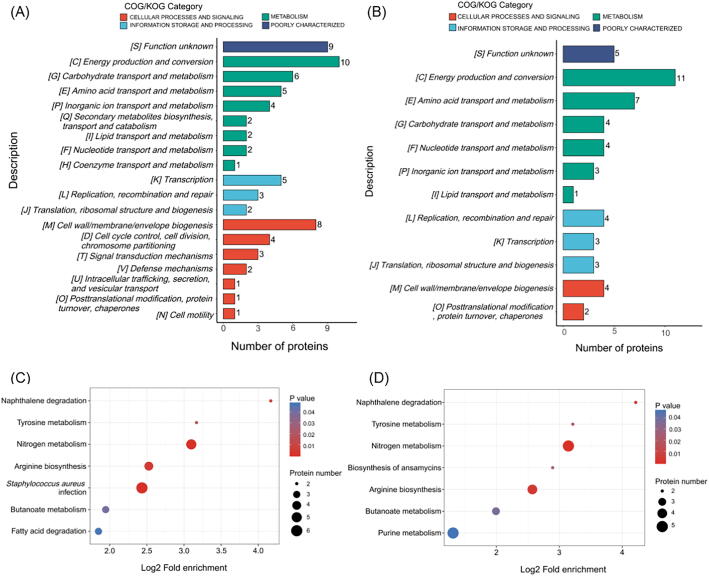
To comprehensively understand the antibacterial mechanism and potential targets of SFG and KE on MRSA. In this study, a label-free quantitative proteomics technique was used to perform proteomics studies on MRSA treated with SFG or KE (Fig. 1A). We identified 21,098 peptides by spectrograph analysis, of which 14,634 were specific peptides. Most of the identified peptides are distributed within 7–20 amino acids, which follows the general rule of mass spectrometry fragmentation mode and the requirement of quality control (Fig. S2A and B). PCA analysis showed that three biological replicates in each group had good repeatability, and there was a large difference among the three groups (Fig. 1B). The differentially expressed proteins (DEPs) are considered with the fold-change value above 1.5 and below 1/1.5 (Table S2 and Table S3, ratio = SFG/Con or KE/Con, P value < 0.05). We identified 45 up-regulated proteins and 33 down-regulated proteins in the SFG group and 17 up-regulated proteins and 40 down-regulated proteins in the KE group compared to the control group (Fig. 1C-E). Hierarchical cluster analysis showed a significant difference between the DEPs in the control and treatment groups (Fig. 1F). Gene ontology (GO) annotation showed that biological process was highly enriched in cellular and metabolic processes and most DEPs were involved in catalytic activity and binding (Fig. S2C and D). Clusters of orthologous groups (COG) analysis revealed that the DEPs were mainly involved in energy production and conversion, transport and metabolism of carbohydrates and amino acids, cell wall/membrane/envelope biogenesis, and cell division (Fig. 2A and B). Furthermore, KEGG pathway enrichment analysis revealed that these DEPs are involved in the following pathways: S. aureus infection; alanine, aspartate, and glutamate metabolism; nitrogen metabolism; arginine biosynthesis; tyrosine metabolism; butanoate metabolism; naphthalene degradation; fatty acid degradation and purine metabolism (Fig. 2C and D).
Fig. 2.
Functional enrichment analysis of differentially expressed proteins in MRSA treated with SFG or KE. (A, B)COG/KOG category of identified DEPs in MRSA treated with SFG (A) or KE (B). (C, D) KEGG pathway-based enrichment of identified DEPs in MRSA treated with SFG (C) or KE (D). Heatmap colors represent log2-fold changes in expression levels.
Metabolomic profiling of MRSA treated with SFG or KE
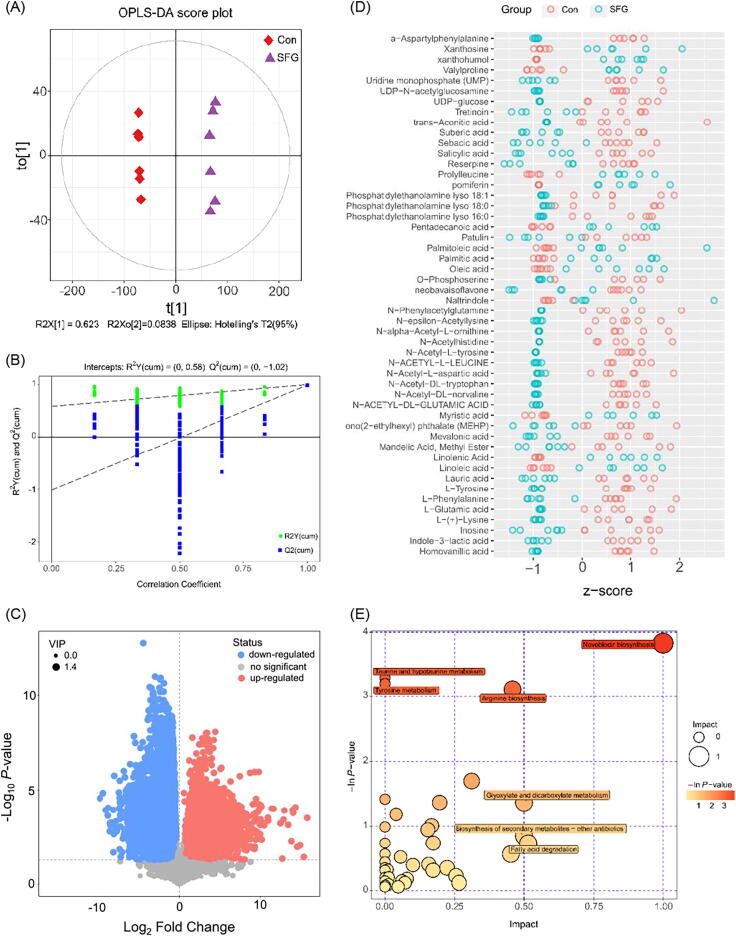
The application of metabolomics can fully understand how bacterial metabolic activities are affected by external influences. Metabolomics and proteomics analyses complement and confirm each other, allowing us to gain a more comprehensive understanding of how antibacterial agents affect bacteria and their potential mechanisms. In the present study, the metabolomics approach based on UPLC-Q-TOF/MS system and multivariate statistical analysis is applied to analyze the perturbation of the metabolite spectrum in MRSA cells treated with SFG or KE. Differentiated metabolites (DMs) were selected according to the OPLS-DA model which provided convincing parameters with high R2Y and Q2Y values. Both the control and treatment groups showed clear aggregation behavior. All replicates in each group were grouped into the same cluster, confirming the observed differences in metabolic profiles between the control and treatment groups (Fig. 3A and B, Fig. S3A and B). A total of 192 significantly altered metabolites were defined in the SFG group, of which 148 metabolites were downregulated and 44 metabolites were upregulated. In the KE group, 189 significantly altered metabolites were identified, of which 132 metabolites were downregulated and 57 metabolites were upregulated (VIP > 1, P-value < 0.05) (Fig. 3 C and D, Fig. S3C and D, Fig. S4). KEGG enrichment analysis showed that the DMs were mainly involved in the amino acid and fatty acid metabolism pathway (Fig. 3E and Fig. S3 E).
Fig. 3.
Metabolites profiling of MRSA treated with SFG at negative mode. (A) Score scatter plot of the OPLS-DA model of normalized LC − MS data from samples in Con and SFG group. (B) Permutation test of the OPLS-DA model. (C): Volcano plot of significantly differently metabolites in MRSA treated with SFG at negative mode; significantly upregulated and downregulated metabolites are shown in red and blue, respectively. Non-significantly different metabolites are shown in grey. (D) The Z-score plot of the differentially expressed metabolites. (E) KEGG enrichment analysis of metabolic pathways. The most significantly enriched pathways are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Effect of SFG and KE on the biofilm and cell wall synthesis
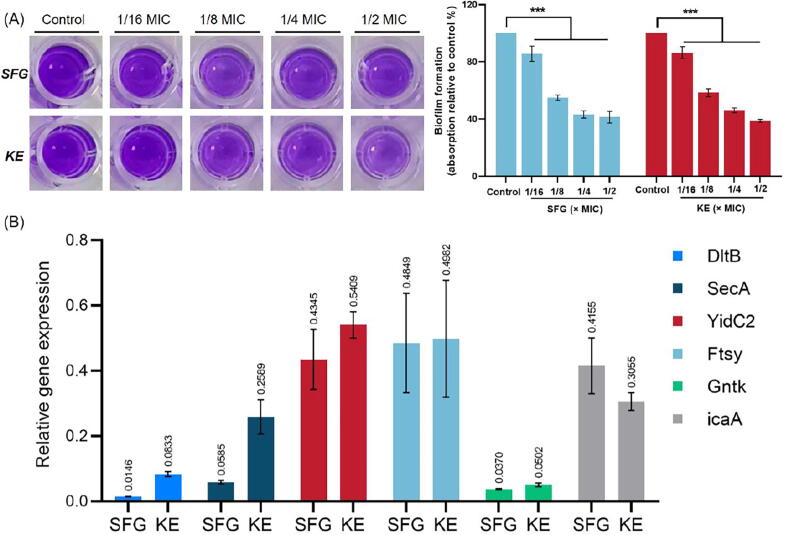
Bacterial biofilms are highly structured microbial communities surrounded by a self-produced protective extracellular matrix and are associated with multidrug resistance. Bacterial biofilms have superior resistance to antimicrobial agents, making MRSA and MSSA infections difficult to eradicate [39]. In this study, we evaluated the biofilm formation of MRSA (USA300) treated with SFG or KE at different concentrations. As shown in Fig. 4A, both SFG and KE exhibited significant inhibition of the MRSA biofilm in a dose-dependent manner (P < 0.001). At 1/2 × MIC concentration, the average inhibition rates of SFG and KE on the biofilm were 58.68 % and 61.24 %, respectively. We also assessed the expression of the icaA gene, which encodes for N-acetylglucosaminyltransferase and is essential for biofilm formation [40]. The results of the qRT-PCR analyses showed a down-regulation of the icaA gene in MRSA after SFG or KE treatment (Fig. 4B). In general, these results showed that SFG and KE have a robust inhibitory effect on the MRSA biofilm formation.
Fig. 4.
SFG and KE exert antibiofilm effects and regulate the expression of genes involved in the cell wall biosynthesis, protein transport, and glucose metabolism. (A) Biofilms treated with SFG or KE at respective MICs for 18 h and stained with 1 % crystal violet. Biofilm formation was quantified by measuring sample absorbance at a wavelength of 595 nm and percentages were calculated with the untreated biofilm as the basis. (B) The qRT-PCR analysis of genes involved in the cell wall biosynthesis, protein transport, and glucose metabolism. Values are presented as Mean ± SD. P < 0.05 was considered statistically significant, ***P < 0.001 vs. control group. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Bacteria respond to environmental stress by increasing cell wall synthesis. The cell wall of Gram-positive bacteria consists mainly of teichoic acids (TAs) and peptidoglycan, which can effectively separate cells from the extracellular environment and is essential for maintaining the structural integrity of the bacterial cell [41], [42]. The D-alanylation of TAs regulates the interaction between the cell wall and the bacteriostatic agent, which may confer resistance to the antibacterial agent. The D-alanylation of TAs was mainly mediated by the DTL pathway [43]. In the present study, the expression of DltB, a key regulator of the DTL pathway, was significantly downregulated at both protein and transcript levels after treatment with SFG or KE (Fig. 4B, Table S2 and S3). Consistent with this finding, prolonged exposure to SFG and KE did not induce drug resistance in MRSA. This may be explained by the downregulation of DltB, which interferes with the synthesis of TAs and hinders cell wall biosynthesis, resulting in the direct action of SFG or KE on the cell membrane. Meanwhile, during bacterial proliferation, cells secrete autolysin (Atl) to hydrolyze their peptidoglycan layer and facilitate cell division. Atl is an autolytic enzyme secreted by S. aureus that has the bifunctional active domains of amidase and aminosinase [44], [45]. As shown in Table S2, the expression of Atl was significantly upregulated in the treatment group, indicating that SFG or KE may lead to the overexpression of Atl and excessive hydrolysis of the peptidoglycan layer. Taken together, these results suggested that SFG and KE could inhibit the function of DltB and induce the overexpression of Atl, leading to the disruption of the cell wall’s biosynthesis and inducing cell wall degradation.
Efficacy of SFG and KE on MRSA cell membrane
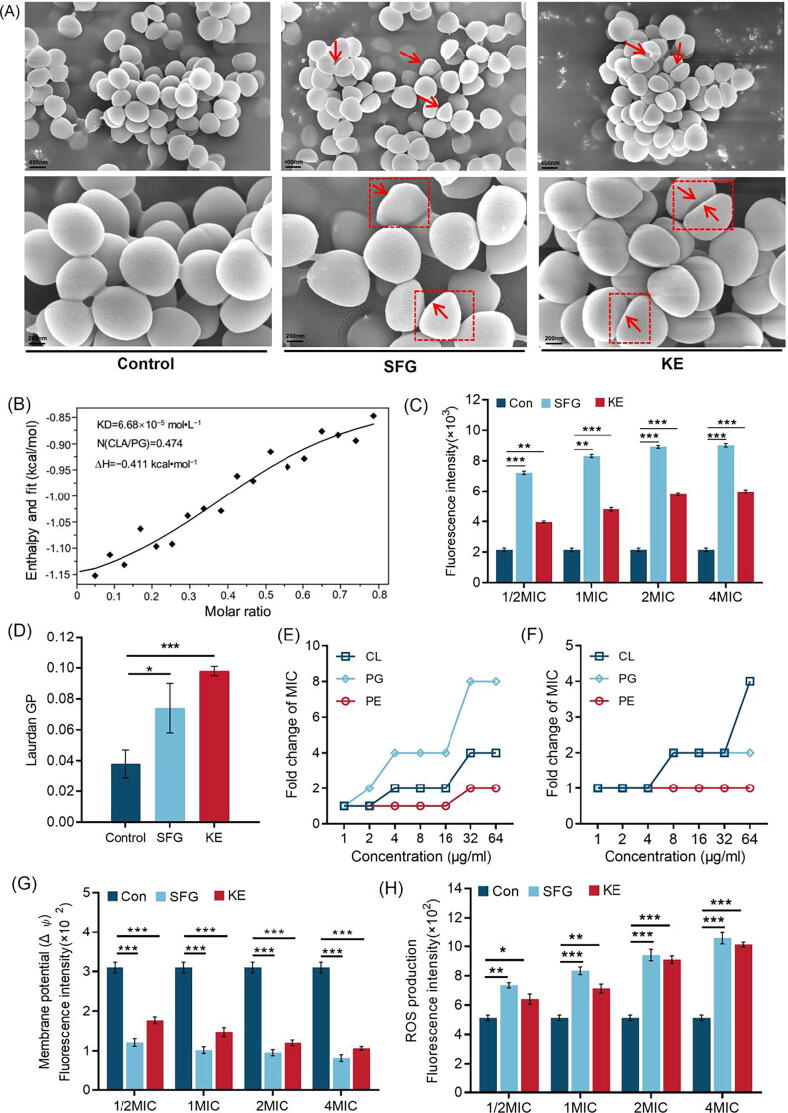
When the integrity of the cell wall is destroyed, the cell membrane is exposed to the external environment and interacts directly with the bacteriostatic agent, resulting in the disruption of the normal function of the cell membrane. We first observed the morphological changes of MRSA after treatment with SFG or KE. Both SFG and KE had a clear effect on the morphology of MRSA cells. Bacterial cells in the control group showed a complete and smooth membrane with normal morphology. However, MRSA treated with SFG or KE was characterized by irregular depressions and surface collapse compared to normal MRSA (Fig. 5A). Furthermore, the affinity between SFG and POPG was evaluated by using an ITC analysis (Fig. 5B), where the equilibrium dissociation constant (KD), number of binding sites (N), and molar binding enthalpy (ΔH) is 6.68 × 10−5 mol L−1, 0.474 and − 0.411 kJ mol−1, respectively. We then assessed membrane permeability by using the DNA-binding dye PI [46]. By measuring the fluorescence intensity, we can assess the integrity of the cell membrane and the membrane permeability capacity of the SFG or KE. As shown in Fig. 5C, the fluorescence value increased with the concentration, indicating that the degree of cell membrane damage was positively correlated with the SFG or KE in a dose-dependent manner (P < 0.01). In addition, we detected the membrane fluidity with the fluorescent probe Laurdan and observed a remarkable change in membrane fluidity after treatment with SFG or KE (Fig. 5D). These results indicate that SFG and KE have high affinity for bacterial cell membranes and can interact with cell membranes, causing changes in cell membrane fluidity and permeability.
Fig. 5.
SFG and KE exert antibacterial effects through the disruption on cell membrane. (A) Bacterial morphology affected by SFG or KE in the control group and treatment group. (B) Isothermal titration calorimetry (ITC) analysis of the interaction between POPG and SFG. Thermodynamic parameters were calculated, including equilibrium dissociation constant (KD = 6.68 × 10−5 mol•L−1), number of binding sites (N = 0.474), molar binding enthalpy (ΔH = − 0.411 kcal•mol−1). (C) Increased membrane permeability after treatment of SFG or KE at 1/2–4 MIC. (D) The fluidity of the cell membrane was decreased after treatment with SFG or KE. (E, F) The exogenous addition of phosphatidylglycerol (PG), phosphatidylethanolamine (PE), and cardiolipin (CL) abolished the antibacterial activity of SFG (E) or KE (F) against MRSA. The concentrations of phospholipids are in the range of 1 to 64 µg mL−1. (G) The membrane potential of MRSA after treatment with SFG or KE. (H) The effect of SFG or KE on the ROS accumulation in MRSA. Values are presented as Mean ± SD. P < 0.05 was considered statistically significant, *P < 0.05, **P < 0.01, ***P < 0.001 vs. control (Con) group.
The changes in membrane permeability and fluidity could disrupt cellular homeostasis and cause perturbations in membrane potential (Δψ) and osmotic pressure. We measured Δψ using the fluorescent DiSC3(5) method [18]. DiSC3(5) is a membrane potential-sensitive probe that aggregates in the phospholipid bilayer and causes self-quenching of the dyes. When membrane depolarization occurs and the membrane potential changes, DiSC3(5) releases the immersed solution causing changes in fluorescence intensity [47]. As shown in Fig. 5G, the fluorescence intensity decreased significantly after treatment with SFG or KE, indicating that the Δψ of the MRSA cell membrane was depolarized. Betaine and choline are two common osmotic protective substances that can maintain the osmotic pressure balance of microbial cells. The metabolic disruption of these two substances could lead to an alteration in the osmotic pressure [48]. In this study, metabolomics and proteomics results showed that the expression of proline/betaine transporter (ProP) was decreased after SFG or KE treatment. Meanwhile, the levels of betaine and choline in MRSA were also significantly reduced (Table S2-S5). These results indicated that SFG and KE could induce a significant change in the Δψ and osmotic pressure of MRSA cells.
Subsequently, disruption of membrane homeostasis can lead to oxidative stress and ROS accumulation. In the present study, ROS production was measured using a DCFH-DA fluorescent probe [18]. Like many bactericidal antibiotics, SFG or KE treatment induced the excessive accumulation of ROS in a dose-dependent manner (Fig. 5H). These results indicated that SFG and KE caused significant damage to the cell membrane of MRSA, and may induce an oxidative stress response to produce excessive ROS, which further leads to cell death.
Interestingly, proteomics and metabolomics results suggested that SFG and KE also have profound effects on cell membrane biosynthesis. The CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase (Pgs1), which is mainly involved in the biosynthesis of anionic phospholipids was significantly downregulated in MRSA treated with SFG or KE, suggesting that the synthesis of phosphatidylglycerol and cardiolipin may be inhibited (Table S3). In addition, we found that the ratio of medium-chain fatty acids to long-chain fatty acids was significantly altered in MRSA treated with SFG or KE (Fig. S5), which may cause disturbances in cell membrane synthesis and cell membrane fluidity [49]. To further verify that SFG and KE have significant effects on cell membrane synthesis, we evaluated the antibacterial activity of SFG or KE mixed with different phospholipids. The results showed that the exogenous addition of phospholipids could effectively reduce the inhibitory activity of SFG and KE on MRSA in a dose-dependent manner (Fig. 5E and F). In addition, the qRT-PCR results indicated that SFG and KE strongly downregulated the expression of cell membrane homeostasis-related genes YidC2, SecA, and FtsY (Fig. 4B). These results suggest that SFG and KE have a high affinity for bacterial cell membranes and can interfere with cell membrane biosynthesis and homeostasis, which may be the reason for their robust activity against MRSA.
Efficacy of SFG and KE in the energy production and conversion
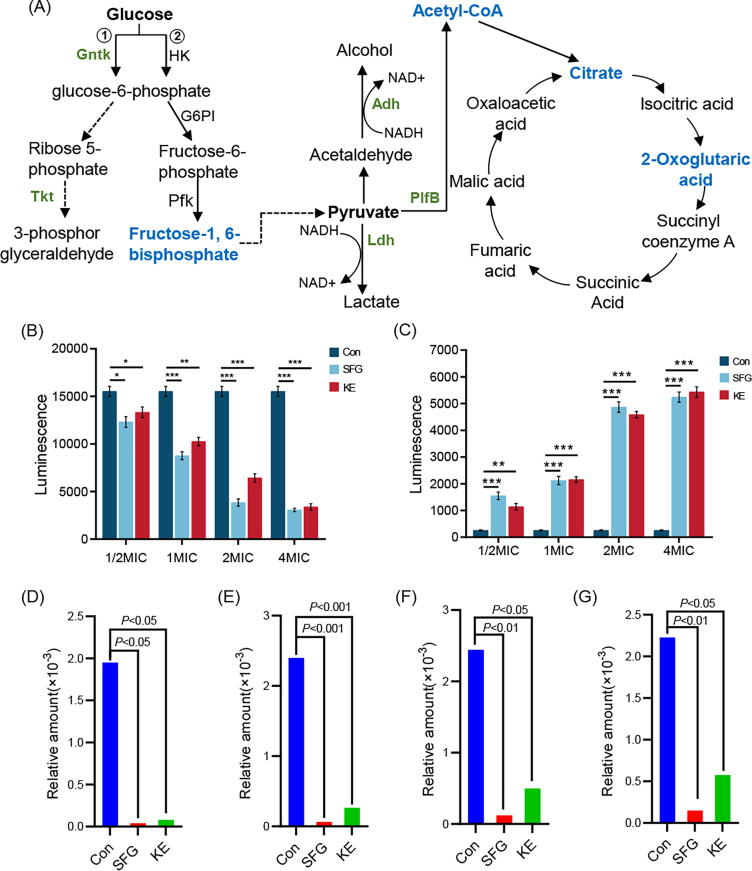
When the cell membrane is damaged, membrane permeability changes can induce the leakage of cellular soluble substances such as ATP, nucleic acids, proteins, etc. [50], [51], [52]. In this study, we found that the extracellular ATP was gradually increased with the increased concentrations of SFG or KE (Fig. 6B and C). When ATP leakage occurs, cells must increase their energy metabolism to produce more ATP to support their basic activities. MRSA produces ATP mainly through oxidative phosphorylation of glucose in which the Embden-Meyerhof pathway (EMP) is one of the main approaches for microbial energy production and acquisition. Microorganisms metabolize glucose to produce ATP and pyruvate. Fructose 1,6-diphosphate (FDP) is a key intermediate in the EMP process, and the metabolism of FDP is closely related to this process. Pyruvate produced by the EMP pathway is catalyzed by formate C-acetyltransferase (plfB, EC.3.1.54) to produce acetyl-CoA, which then enters the tricarboxylic acid (TCA) cycle and releases a large amount of energy. Continuous operation of the EMP requires sufficient NAD+ as a hydrogen acceptor to participate in the dehydrogenation of glyceraldehyde 3-phosphate to 1, 3-bisphosphoglycerate. There are two main sources of NAD+ in microorganisms: one is the dehydrogenation and oxidation of NADH2 by the respiratory chain under aerobic conditions to produce NAD+. The other is the reduction of pyruvate as a substrate to form lactic acid or ethanol under anaerobic conditions to produce NAD+, which is catalyzed by lactate dehydrogenase (Ldh) and ethanol dehydrogenase (ADH), respectively [53]. Proteomic and metabolomic results showed that the levels of Ldh, Adh, PlfB, FDP, and TCA cycle products (acetyl-CoA, citric acid, and 2-oxoglutaric acid) in MRSA were significantly reduced after SFG and KE treatment (Fig. 6A and D-G), suggesting that SFG and KE could inhibit the EMP of MRSA and thus reduce the production of ATP. In conclusion, under the stress of SFG or KE, the membrane permeability of MRSA was changed, causing a substantial leakage of ATP and the energy metabolism of MRSA was disturbed, which eventually led to cell damage and death.
Fig. 6.
Altered proteins and metabolites in the energy production and conversion pathway. (A) Diagram of glycolysis and TCA cycle pathway. Green: proteins with decreased levels in the MRSA treated with SFG or KE. Blue: metabolites with a decreased level in the MRSA treated with SFG or KE. (B) The intracellular ATP levels of MRSA treated with SFG or KE. (C) The extracellular ATP levels of MRSA treated with SFG or KE. (D-G) Relative amount of fructose 1,6-diphosphate (D), acetyl-CoA (E), citrate (F), and 2-oxoglutaric acid (G) in all groups. Values are presented as Mean ± SD. P < 0.05 was considered statistically significant, *P < 0.05, **P < 0.01, ***P < 0.001 vs. control (Con) group. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Efficacy of SFG and KE on the nitrogen and amino acid metabolism
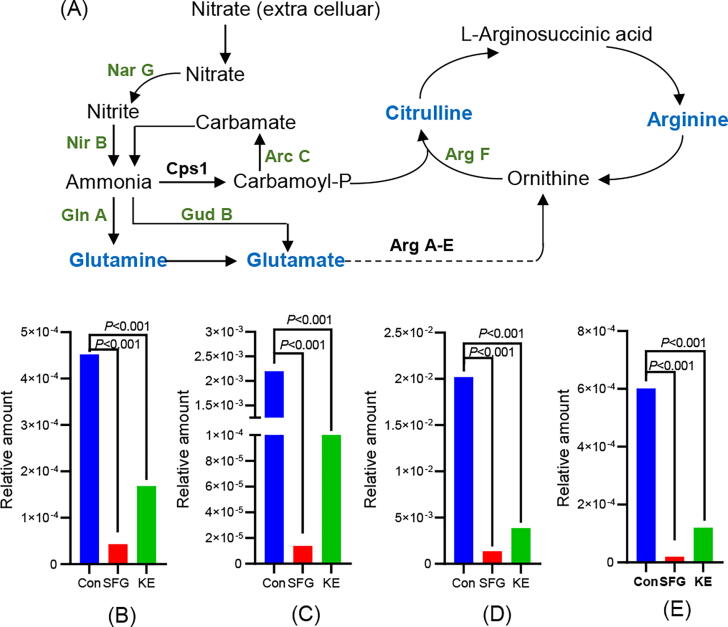
Nitrate is a major source of nitrogen nutrition for bacteria, which can be used to produce amino acids essential for survival. In addition, when the carbohydrate metabolism of the bacteria is deprived, the cell can produce energy by reducing nitrate [54], [55]. In this study, we found that SFG and KE had significant effects on the nitrogen and amino acid metabolism of MRSA. The key catalytic enzymes Nar G and Nir B involved in the reduction of nitrate to ammonia were significantly downregulated (P < 0.01). Inhibition of this process may reduce intracellular levels of ammonia, which is a key component in the synthesis of glutamine and glutamate. In addition, inhibition of glutamate synthesis will further affect arginine synthesis. Proteomic results showed that the expression of glutamate dehydrogenase (GudB), glutamine synthetase (GlnA), carbamate kinase (ArcC), and ornithine carbamoyltransferase (ArgF) were significantly downregulated (Fig. 7A, Table S2 and S3). Correspondingly, metabolomic results showed a significant reduction in glutamine, glutamate, citrulline, and arginine levels in MRSA treated with SFG and KE (Fig. 7B-E) (P < 0.01). In conclusion, SFG and KE may interfere with the utilization of nitrate by MRSA and inhibit the synthesis of amino acids, leading to the disruption of the energy metabolism of MRSA.
Fig. 7.
Altered proteins and metabolites in the nitrogen metabolism and arginine biosynthesis pathway. (A) Diagram of nitrogen metabolism and arginine biosynthesis pathway. Green: proteins with decreased levels in the MRSA treated with SFG or KE. Blue: metabolites with a decreased level in the MRSA treated with SFG or KE. (B-E) The relative amount of arginine (B), glutamine (C), glutamate (D), and citrulline (E) in all groups. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Efficacy of SFG and KE on the mouse skin wound infection model
To further explore the potential therapeutic effects of SFG and KE on bacterial infectious diseases, we established a mice wound infection model. As shown in Fig. 8A and B, after MRSA infection, untreated wounds heal slowly and show purulent wounds on day 3. However, SFG and KE significantly promoted wound closure and inhibited purulent secretion. Wound size was remarkably reduced under the treatment of SFG or KE (P < 0.001). Meanwhile, the number of bacteria in the infected areas was reduced almost tenfold (Fig. 8C, P < 0.01 vs. SFG, P < 0.05 vs. KE). In addition, SFG and KE dramatically suppressed the levels of pro-inflammatory cytokine IL-6 throughout the experimental period (Fig. 8D, P < 0.05). H&E staining showed that the wound in the untreated group had more inflammatory cells and necrotic tissue. However, SFG and KE could reduce inflammatory cell infiltration and promote neovascularization (Fig. 8E). In conclusion, these results suggested that SFG and KE could effectively treat skin infections caused by MRSA.
Fig. 8.
SFG and KE exerted promising therapeutic potential in wound infection models. (A) Representative photographs of the wounds under the treatment of SFG, KE, or vancomycin. The morphology of the wounds under the treatment of SFG or KE displayed comparable trends compared with antibiotics. (B) Skin wound size was reduced under the treatment of SFG, KE, or vancomycin on days 0, 3, 5, 7, 9,11, and 14 post-infections. (C) SFG and KE reduced the bacterial loads in skin wound infections. (D) SFG and KE treatment reduced the serum IL-6 level in wound infection mice. (E) Representative wound tissue histopathology on days 0, 3, 5, 7, 9,11, and 14 post-infections. Values are presented as Mean ± SD. P < 0.05 was considered statistically significant.
Discussion
Although bioactive molecules from natural plants are used for a variety of purposes, their applications in combating clinical pathogens and bacterial resistance issues are still largely overlooked. S. flavescens has been used in traditional Chinese medicine since ancient times as an herbal remedy for skin diseases, and clinical application has demonstrated its promising antibacterial activity. Previous studies have mainly focused on the bacteriostatic properties of quinolizidine alkaloids in S. flavescens [56]. Here, we investigated the antibacterial activity and related mechanism of two flavonoids, SFG and KE derived from S. flavescens. Based on the antibacterial assays, we found that the lavandulylation of the phenolic backbone is a prerequisite for bacteriostatic activity. SFG and KE showed significant inhibitory activity against common MRSA (USA300) and MSSA (ATCC 25923 and ATCC 29213), and can rapidly inhibit bacterial growth in a short time. In addition, continuous treatment with SFG or KE did not induce resistance in MRSA compared to β-lactam antibiotics. Finally, we investigated the potential therapeutic effects of SFG and KE in a rat wound infection model. In vitro and in vivo studies revealed that SFG and KE have a robust bacteriostatic activity which can be potential candidates for the development of new antibiotic agents against bacteria-associated infections.
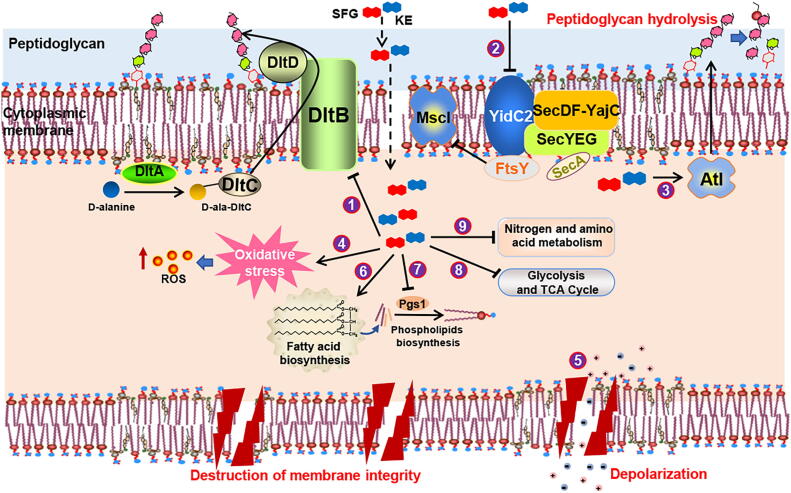
To comprehensively understand the in-depth biological mechanism of SFG and KE against MRSA, we used proteomics and metabolomics methods to describe the major changes in the biological processes of MRSA after SFG or KE treatment. The results showed that SFG and KE interfered with several key proteins and metabolites involved in the crucial biological pathways of MRSA including the biofilm and cell wall synthesis, cell membrane synthesis, energy production, and amino acid and fatty acid metabolism. Thus, unlike commonly used antibiotics, we speculate that SFG and KE may interfere with multiple biological processes of MRSA to rapidly inhibit MRSA growth (Fig. 9).
Fig. 9.
Schematic representation of the multifarious antibacterial mode of action for SFG and KE. (1) The downregulation of dltB inhibits D-alanylation of teichoic acids and leading to the disruption of the cell wall’s biosynthesis. (2) The downregulation of YidC2, SecA, FtsY, and MscL has evidenced that SFG and KE affected cell membrane biosynthesis and homeostasis. (3) SFG and KE activated bacterial autolysin, resulting in hydrolysis of peptidoglycan and destruction of the cell wall. (4) SFG and KE induced oxidative stress and increased ROS levels. (5) SFG and KE induced cell depolarization and alteration of osmotic pressure. (6) and (7) Changes in the synthesis of fatty acids and the ratio of fatty acids affect the synthesis of phospholipids and thus inhibit the synthesis of cell membranes. (8) Proteins involved in energy metabolism were inhibited by SFG and KE, resulting in inhibition of energy production and conversion. (9) SFG and KE might interfere with MRSA's utilization of nitrate and inhibit the synthesis of amino acids, resulting in the disorder of MRSA's energy metabolism.
Bacterial biofilm is one of the major factors that cause bacterial resistance, promoting wound inflammation and delaying wound healing [57]. In the present study, an apparent biofilm inhibition was observed at the sub-MIC concentration of SFG and KE. Polysaccharide intercellular adhesin (PIA)is the major component of the biofilm matrix of Gram-positive bacteria and has the greatest influence on biofilm formation. Studies have shown that S. aureus possesses the ica A-D operon, in which icaA encodes for N-acetylglucosaminyltransferase, an important enzyme for the synthesis of PIA [58], [59]. In the present study, the expression of icaA in MRSA was significantly downregulated after SFG or KE treatment. Thus, the anti-biofilm effect may be achieved by inhibiting the synthesis of PIA.
The cell wall is an important protection for bacteria and is essential for maintaining the shape and structural integrity of bacterial cells. TAs and peptidoglycan are the major components of the cell wall. TAs are modified by D-alanylation and transported across membranes to bind to peptidoglycan (wall teichoic acid) or anchor to the cell membrane (lipoteichoic acid). It is reported that DltB and DltD are reported to be involved in the migration of the D-alanyl moiety across the membrane [60]. Autolysin is a proteolytic enzyme produced by bacteria that can degrade bacterial cell walls and is associated with bacterial cell division, biofilm formation, surface adhesion, genetic receptivity, and cell wall renewal [61]. Cell wall thickening is an important mechanism of antibiotic resistance in S. aureus. Increased peptidoglycan maturation is the main element of MRSA cell wall thickening. Weakening of Atl expression reduces the enzymatic hydrolysis of peptidoglycan, resulting in the accumulation of the peptidoglycan layer and excessive peptidoglycan synthesis [62], [63]. In this study, the proteomics and qRT-PCR results showed that SFG and KE could significantly inhibit the expression of DltB and increase the expression of Atl, indicating that they could block the transport of teichoic acids and disrupt cell wall synthesis while causing excessive hydrolysis of the cell wall. This undermines the protection of the bacteria, which is a prerequisite for their potent antibacterial activity.
The integrity and function of the cell membrane are critical for bacterial survival. Therefore, antibacterial agents that target bacterial membranes have the promising therapeutic potential [64], [65]. For example, Rhodomyrtosone B, a natural product derived from Rhodomyrtus tomentosa, exerted antibacterial activity by disrupting membrane permeability [66]. Due to the presence of the lavandulyled side chain, SFG, and KE showed strong lipophilicity. Therefore, they have a high affinity with cell membranes. In this study, we found that SFG and KE could cause the increase in membrane permeability, the change of membrane potential, and the disruption of membrane integrity of MRSA. This may be related to the alteration of large conductance mechanosensitive channel protein (MscL) and YidC2. MscL is an important channel protein in the bacterial cell membrane that can protect bacterial cells from low osmotic pressure downshock. Previous studies have shown that the deletion of the signal recognition particle (SRP) and its receptor FtsY has a significant effect on the MscL function [67], [68]. The YidC protein is critical for membrane biogenesis and has been shown to interact with the Sec pathway. Inhibition of SecA affects the transmembrane transport of microbial proteins such as the insertion of the membrane protein MscL [69], [70]. In this study, proteomic results showed that MscL was downregulated after SFG and KE treatment, and qPCR results showed that YidC2 was downregulated along with SecA and FtsY after SFG or KE treatment. These results suggest that SFG and KE alter the cell homeostasis of MRSA by inhibiting the function of MscL, resulting in changes in cell osmotic pressure and disruption of cell membrane integrity.
In addition, SFG and KE also showed a profound effect on cell membrane synthesis. The cell membrane of bacteria is mainly composed of phospholipids, and it has been reported that the exogenous addition of phospholipids, such as cardiolipin, phosphatidylglycerol, and phosphatidylethanolamine, which constitute bacterial cell membranes, could effectively reduce the inhibitory activity of isobavachalcone against S. aureus [18]. In the present study, we also found that the MIC of SFG and KE increased with the increase in the amount of phospholipid addition. Phospholipids are synthesized from fatty acids, and the disturbance of fatty acid metabolism will affect the synthesis of phospholipids and then affect the composition and fluidity of cell membranes. Here, we found that the ratio of medium-chain fatty acids to long-chain fatty acids was significantly changed in MRSA treated with SFG or KE, which might cause disturbances in cell membrane synthesis and cell membrane fluidity.
In addition to interfering with the function of the cell membrane, SFG and KE also affect the basic energy metabolism of bacteria. ATP is the energy base for cellular activities in which bacteria produce ATP through fermentation and aerobic and anaerobic respiration [71]. We found that SFG and KE significantly inhibited the glycolysis pathway of MRSA and reduced the level of acetyl-CoA, a key precursor of the TCA cycle. Meanwhile, SFG and KE might interfere with the utilization of nitrate by MRSA and inhibit the synthesis of amino acids, leading to the disruption of the energy metabolism of MRSA. Thus, we speculated that SFG and KE disrupted the utilization of carbon and nitrogen sources in MRSA, preventing the cells from producing enough energy to survive.
In conclusion, the current research showed that SFG and KE exerted robust antibacterial activities against MRSA. We found that their antibacterial activity is mainly achieved through cell membrane disruption. SFG and KE can first prevent the synthesis of bacterial biofilms and cell walls, thus removing the protection of the bacteria. Then, they can directly disrupt the cell membrane by altering the homeostasis and integrity of the cell membrane. In addition, they can disrupt the normal physiological activities of bacteria by interfering with energy metabolism. Taken together, the combination of these effects ultimately leads to the death of the bacteria.
Although we have demonstrated the possible mechanism of SFG and KE involved in killing bacteria. The determinants for the antibacterial activity of SFG and KE still need further studies. In addition, lavandulylated flavonoids are widely distributed in natural plants, especially in Leguminosae plants. Therefore, the discovery of leading compounds for antibacterial drugs with similar structures and better activity in more plants needs further exploration.
Conclusions
In conclusion, this study demonstrated that lavandulylated flavonoids from natural plants have promising antibacterial activities against MRSA. Both SFG and KE could be potential candidates for the development of new antibiotic agents against bacterial-associated infections.
CRediT authorship contribution statement
Zebin Weng: Conceptualization, Methodology, Writing – original draft, Project administration. Fei Zeng: Methodology, Investigation. Minxin Wang: Writing – review & editing. Sheng Guo: Writing – review & editing, Resources, Investigation. Zhijuan Tang: Software, Formal analysis, Investigation. Kiyoshi Itagaki: Writing – review & editing, Resources. Yajuan Lin: Investigation. Xinchun Shen: Writing – review & editing, Resources. Yaqi Cao: Visualization, Software. Jin-ao Duan: Project administration, Funding acquisition, Supervision. Fang Wang: Project administration, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the Innovation Team and Talent Support Program of the National Administration of Traditional Chinese Medicine (ZYYCXTD-D-202005), Major increase and decrease spending projects of the central government (2060302), the Supporting project of the National Natural Youth Fund of Nanjing University of Chinese Medicine (NZY81903940) and Traditional Chinese and Western Medicine Clinical Medicine Brand Construction Project of Jiangsu Higher Education Institutions (Phase II).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.04.017.
Contributor Information
Jin-ao Duan, Email: dja@njucm.edu.cn.
Fang Wang, Email: wangfang8875@163.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Lee A.S., de Lencastre H., Garau J., Kluytmans J., Malhotra-Kumar S., Peschel A., et al. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primers. 2018;4:18033. doi: 10.1038/nrdp.2018.33. [DOI] [PubMed] [Google Scholar]
- 2.Turner N.A., Sharma-Kuinkel B.K., Maskarinec S.A., Eichenberger E.M., Shah P.P., Carugati M., et al. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat Rev Microbiol. 2019;17:203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rehm SJ, Tice A. Staphylococcus aureus: methicillin-susceptible S. aureus to methicillin-resistant S. aureus and vancomycin-resistant S. aureus. Clin Infect Dis 2010;51 Suppl 2:S176-182. [DOI] [PubMed]
- 4.McGuinness W.A., Malachowa N., DeLeo F.R. Vancomycin resistance in Staphylococcus aureus. Yale J Biol Med. 2017;90:269–281. [PMC free article] [PubMed] [Google Scholar]
- 5.Wroblewska M.M., Rudnicka J., Marchel H., Luczak M. Multidrug-resistant bacteria isolated from patients hospitalised in Intensive Care Units. Int J Antimicrob Agents. 2006;27:285–289. doi: 10.1016/j.ijantimicag.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 6.Cragg G.M., Newman D.J. Natural products: A continuing source of novel drug leads. Biochim Biophys Acta. 2013;1830:3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen T., Samanta S.K. Medicinal plants, human health and biodiversity: A broad review. Adv Biochem Eng Biotechnol. 2015;147:59–110. doi: 10.1007/10_2014_273. [DOI] [PubMed] [Google Scholar]
- 8.Feyzabadi Z., Ghorbani F., Vazani Y., Zarshenas M.M. A critical review on phytochemistry, pharmacology of Viola odorata L. and related multipotential products in traditional Persian medicine. Phytother Res. 2017;31:1669–1675. doi: 10.1002/ptr.5909. [DOI] [PubMed] [Google Scholar]
- 9.Zipfel C., Oldroyd G.E. Plant signalling in symbiosis and immunity. Nature. 2017;543:328–336. doi: 10.1038/nature22009. [DOI] [PubMed] [Google Scholar]
- 10.Cheng Y.T., Zhang L., He S.Y. Plant-microbe interactions facing environmental challenge. Cell Host Microbe. 2019;26:183–192. doi: 10.1016/j.chom.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mekseepralard C., Kamkaen N., Wilkinson J.M. Antimicrobial and antioxidant activities of traditional Thai herbal remedies for aphthous ulcers. Phytother Res. 2010;24:1514–1519. doi: 10.1002/ptr.3158. [DOI] [PubMed] [Google Scholar]
- 12.Talman A.M., Clain J., Duval R., Ménard R., Ariey F. Artemisinin bioactivity and resistance in Malaria Parasites. Trends Parasitol. 2019;35:953–963. doi: 10.1016/j.pt.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Ma N., Zhang Z., Liao F., Jiang T., Tu Y. The birth of artemisinin. Pharmacol Ther. 2020;216 doi: 10.1016/j.pharmthera.2020.107658. [DOI] [PubMed] [Google Scholar]
- 14.Wang G., Li L., Wang X., Li X., Zhang Y., Yu J., et al. Hypericin enhances β-lactam antibiotics activity by inhibiting sarA expression in methicillin-resistant Staphylococcus aureus. Acta Pharm Sin B. 2019;9:1174–1182. doi: 10.1016/j.apsb.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y., Wei L., Wang Y., Ding L., Guo Y., Sun X., et al. Inhibitory effect of the traditional Chinese medicine Ephedra sinica granules on Streptococcus pneumoniae Pneumolysin. Biol Pharm Bull. 2020;43:994–999. doi: 10.1248/bpb.b20-00034. [DOI] [PubMed] [Google Scholar]
- 16.Varmuzova K., Matulova M.E., Gerzova L., Cejkova D., Gardan-Salmon D., Panhéleux M., et al. Curcuma and Scutellaria plant extracts protect chickens against inflammation and Salmonella Enteritidis infection. Poult Sci. 2015;94:2049–2058. doi: 10.3382/ps/pev190. [DOI] [PubMed] [Google Scholar]
- 17.Villinski J.R., Bergeron C., Cannistra J.C., Gloer J.B., Coleman C.M., Ferreira D., et al. Pyrano-isoflavans from Glycyrrhiza uralensis with antibacterial activity against Streptococcus mutans and Porphyromonas gingivalis. J Nat Prod. 2014;77:521–526. doi: 10.1021/np400788r. [DOI] [PubMed] [Google Scholar]
- 18.Song M., Liu Y., Li T., Liu X., Hao Z., Ding S., et al. Plant natural flavonoids against multidrug resistant pathogens. Adv Sci (Weinh) 2021;8:e2100749. doi: 10.1002/advs.202100749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian X., Wang P., Li T., Huang X., Guo W., Yang Y., et al. Self-assembled natural phytochemicals for synergistically antibacterial application from the enlightenment of traditional Chinese medicine combination. Acta Pharm Sin B. 2020;10:1784–1795. doi: 10.1016/j.apsb.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian M., Tang S., Wu C., Wang Y., He T., Chen T., et al. Synergy between baicalein and penicillins against penicillinase-producing Staphylococcus aureus. Int J Med Microbiol. 2015;305:501–504. doi: 10.1016/j.ijmm.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 21.He X., Fang J., Huang L., Wang J., Huang X. Sophora flavescens ait.: Traditional usage, phytochemistry and pharmacology of an important traditional Chinese medicine. J Ethnopharmacol. 2015;172:10–29. doi: 10.1016/j.jep.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y., Chen X.J., He L., Yan X.L., Li Q.R., Zhang X., et al. Systematic elucidation of the bioactive alkaloids and potential mechanism from Sophora flavescens for the treatment of eczema via network pharmacology. J Ethnopharmacol. 2023;301 doi: 10.1016/j.jep.2022.115799. [DOI] [PubMed] [Google Scholar]
- 23.Sohn H.Y., Son K.H., Kwon C.S., Kwon G.S., Kang S.S. Antimicrobial and cytotoxic activity of 18 prenylated flavonoids isolated from medicinal plants: Morus alba L., Morus mongolica Schneider, Broussnetia papyrifera (L.) Vent, Sophora flavescens Ait and Echinosophora koreensis Nakai. Phytomedicine. 2004;11:666–672. doi: 10.1016/j.phymed.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Li P., Chai W.C., Wang Z.Y., Tang K.J., Chen J.Y., Venter H., et al. Bioactivity-guided isolation of compounds from Sophora flavescens with antibacterial activity against Acinetobacter baumannii. Nat Prod Res. 2021:1–9. doi: 10.1080/14786419.2021.1983570. [DOI] [PubMed] [Google Scholar]
- 25.Sakagami Y., Mimura M., Kajimura K., Yokoyama H., Linuma M., Tanaka T., et al. Anti-MRS activity of sophoraflavanone G and synergism with other antibacterial agents. Lett Appl Microbiol. 1998;27:98–100. doi: 10.1046/j.1472-765x.1998.00386.x. [DOI] [PubMed] [Google Scholar]
- 26.Cha J.D., Moon S.E., Kim J.Y., Jung E.K., Lee Y.S. Antibacterial activity of sophoraflavanone G isolated from the roots of Sophora flavescens against methicillin-resistant Staphylococcus aureus. Phytother Res. 2009;23:1326–1331. doi: 10.1002/ptr.2540. [DOI] [PubMed] [Google Scholar]
- 27.Sun Z.L., Sun S.C., He J.M., Lan J.E., Gibbons S., Mu Q. Synergism of sophoraflavanone G with norfloxacin against effluxing antibiotic-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106098. [DOI] [PubMed] [Google Scholar]
- 28.Tsuchiya H., Iinuma M. Reduction of membrane fluidity by antibacterial sophoraflavanone G isolated from Sophora exigua. Phytomedicine. 2000;7:161–165. doi: 10.1016/S0944-7113(00)80089-6. [DOI] [PubMed] [Google Scholar]
- 29.Humphries R, Bobenchik AM, Hindler JA, Schuetz AN. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, m100, 31st edition. J Clin Microbiol 2021;59:e0021321. [DOI] [PMC free article] [PubMed]
- 30.Friedman L., Alder J.D., Silverman J.A. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaskovich M.A.T., Hansford K.A., Gong Y., Butler M.S., Muldoon C., Huang J.X., et al. Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria. Nat Commun. 2018;9:22. doi: 10.1038/s41467-017-02123-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W., Lin F., Hung A., Barlow A., Sani M.A., Paolini R., et al. Enhancing proline-rich antimicrobial peptide action by homodimerization: Influence of bifunctional linker. Chem Sci. 2022;13:2226–2237. doi: 10.1039/d1sc05662j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho J., Choi H., Lee J., Kim M.S., Sohn H.Y., Lee D.G. The antifungal activity and membrane-disruptive action of dioscin extracted from Dioscorea nipponica. Biochim Biophys Acta. 2013;1828:1153–1158. doi: 10.1016/j.bbamem.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Verstraeten S., Catteau L., Boukricha L., Quetin-Leclercq J., Mingeot-Leclercq M.P. Effect of ursolic and oleanolic acids on lipid membranes: Studies on MRSA and models of membranes. Antibiotics (Basel) 2021:10. doi: 10.3390/antibiotics10111381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S., Tang H., Wang Y., Nie B., Yang H., Yuan W., et al. Antibacterial and antibiofilm effects of flufenamic acid against methicillin-resistant Staphylococcus aureus. Pharmacol Res. 2020;160 doi: 10.1016/j.phrs.2020.105067. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y., Li Y., Yang K., Wei G., Huang A. Antimicrobial peptide BCp12 inhibits Staphylococcus aureus growth by altering lysine malonylation levels in the arginine synthesis pathway. J Agric Food Chem. 2022;70:403–414. doi: 10.1021/acs.jafc.1c05894. [DOI] [PubMed] [Google Scholar]
- 37.Lin Y., Huang L., Zhang X., Yang J., Chen X., Li F., et al. Multi-omics analysis reveals anti-Staphylococcus aureus activity of actinomycin D originating from Streptomyces parvulus. Int J Mol Sci. 2021;22 doi: 10.3390/ijms222212231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boozari M., Soltani S., Iranshahi M. Biologically active prenylated flavonoids from the genus Sophora and their structure-activity relationship-a review. Phytother Res. 2019;33:546–560. doi: 10.1002/ptr.6265. [DOI] [PubMed] [Google Scholar]
- 39.Craft K.M., Nguyen J.M., Berg L.J., Townsend S.D. Methicillin-resistant Staphylococcus aureus (MRSA): Antibiotic-resistance and the biofilm phenotype. Medchemcomm. 2019;10:1231–1241. doi: 10.1039/c9md00044e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdel-Shafi S., El-Serwy H., El-Zawahry Y., Zaki M., Sitohy B., Sitohy M. The association between icaA and icaB genes, antibiotic resistance and biofilm formation in clinical isolates of Staphylococci spp. Antibiotics (Basel) 2022:11. doi: 10.3390/antibiotics11030389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berni F., Enotarpi J., Voskuilen T., Li S., van der Marel G.A., Codée J.D.C. Synthetic carbohydrate-based cell wall components from Staphylococcus aureus. Drug Discov Today Technol. 2020;38:35–43. doi: 10.1016/j.ddtec.2021.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Peacock S.J., Paterson G.K. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem. 2015;84:577–601. doi: 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- 43.Pandin C., Caroff M., Condemine G. Antimicrobial peptide resistance genes in the plant pathogen Dickeya dadantii. Appl Environ Microbiol. 2016;82:6423–6430. doi: 10.1128/AEM.01757-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porayath C., Suresh M.K., Biswas R., Nair B.G., Mishra N., Pal S. Autolysin mediated adherence of Staphylococcus aureus with fibronectin, gelatin and heparin. Int J Biol Macromol. 2018;110:179–184. doi: 10.1016/j.ijbiomac.2018.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nega M., Tribelli P.M., Hipp K., Stahl M., Götz F. New insights in the coordinated amidase and glucosaminidase activity of the major autolysin (Atl) in Staphylococcus aureus. Commun Biol. 2020;3:695. doi: 10.1038/s42003-020-01405-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buzón-Durán L., Alonso-Calleja C., Riesco-Peláez F., Capita R. Effect of sub-inhibitory concentrations of biocides on the architecture and viability of MRSA biofilms. Food Microbiol. 2017;65:294–301. doi: 10.1016/j.fm.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Taggar R., Singh S., Bhalla V., Bhattacharyya M.S., Sahoo D.K. Deciphering the antibacterial role of peptide from Bacillus subtilis subsp. spizizenii Ba49 against Staphylococcus aureus. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.708712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breisch J., Bendel M., Averhoff B. The choline dehydrogenase BetA of Acinetobacter baumannii: A flavoprotein responsible for osmotic stress protection. Environ Microbiol. 2022;24:1052–1061. doi: 10.1111/1462-2920.15741. [DOI] [PubMed] [Google Scholar]
- 49.Mostofian B., Zhuang T., Cheng X., Nickels J.D. Branched-chain fatty acid content modulates structure, fluidity, and phase in model microbial cell membranes. J Phys Chem B. 2019;123:5814–5821. doi: 10.1021/acs.jpcb.9b04326. [DOI] [PubMed] [Google Scholar]
- 50.Li N., Luo M., Fu Y.J., Zu Y.G., Wang W., Zhang L., et al. Effect of corilagin on membrane permeability of Escherichia coli, Staphylococcus aureus and Candida albicans. Phytother Res. 2013;27:1517–1523. doi: 10.1002/ptr.4891. [DOI] [PubMed] [Google Scholar]
- 51.Varma G.Y.N., Kummari G., Paik P., Kalle A.M. Celecoxib potentiates antibiotic uptake by altering membrane potential and permeability in Staphylococcus aureus. J Antimicrob Chemother. 2019;74:3462–3472. doi: 10.1093/jac/dkz391. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z., Yang Q., Wang X., Li R., Qiao H., Ma P., et al. Antibacterial activity of xanthan-oligosaccharide against Staphylococcus aureus via targeting biofilm and cell membrane. Int J Biol Macromol. 2020;153:539–544. doi: 10.1016/j.ijbiomac.2020.03.044. [DOI] [PubMed] [Google Scholar]
- 53.Liu L., Zeng X., Zheng J., Zou Y., Qiu S., Dai Y. AHL-mediated quorum sensing to regulate bacterial substance and energy metabolism: A review. Microbiol Res. 2022;262 doi: 10.1016/j.micres.2022.127102. [DOI] [PubMed] [Google Scholar]
- 54.Schlievert P.M., Strandberg K.L., Lin Y.C., Peterson M.L., Leung D.Y. Secreted virulence factor comparison between methicillin-resistant and methicillin-sensitive Staphylococcus aureus, and its relevance to atopic dermatitis. J Allergy Clin Immunol. 2010;125:39–49. doi: 10.1016/j.jaci.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balasubramanian D., Harper L., Shopsin B., Torres V.J. Staphylococcus aureus pathogenesis in diverse host environments. Pathog Dis. 2017:75. doi: 10.1093/femspd/ftx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang S.Y., Li W., Nie H., Liao M., Qiu B., Yang Y.L., et al. Five new alkaloids from the roots of Sophora flavescens. Chem Biodivers. 2018;15:e1700577. doi: 10.1002/cbdv.201700577. [DOI] [PubMed] [Google Scholar]
- 57.Vijayakumar K., Bharathidasan V., Manigandan V., Jeyapragash D. Quebrachitol inhibits biofilm formation and virulence production against methicillin-resistant Staphylococcus aureus. Microb Pathog. 2020;149 doi: 10.1016/j.micpath.2020.104286. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen H.T.T., Nguyen T.H., Otto M. The staphylococcal exopolysaccharide PIA - biosynthesis and role in biofilm formation, colonization, and infection. Comput Struct Biotechnol J. 2020;18:3324–3334. doi: 10.1016/j.csbj.2020.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoang T.M., Zhou C., Lindgren J.K., Galac M.R., Corey B., Endres J.E., et al. Transcriptional regulation of icaADBC by both icaR and tcaR in Staphylococcus epidermidis. J Bacteriol. 2019;201 doi: 10.1128/JB.00524-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood BM, Santa Maria JP, Jr., Matano LM, Vickery CR, Walker S. A partial reconstitution implicates DltD in catalyzing lipoteichoic acid d-alanylation. J Biol Chem 2018;293:17985-17996. [DOI] [PMC free article] [PubMed]
- 61.Kalali Y., Haghighat S., Mahdavi M. Passive immunotherapy with specific IgG fraction against autolysin: Analogous protectivity in the MRSA infection with antibiotic therapy. Immunol Lett. 2019;212:125–131. doi: 10.1016/j.imlet.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 62.Wittekind M., Schuch R. Cell wall hydrolases and antibiotics: Exploiting synergy to create efficacious new antimicrobial treatments. Curr Opin Microbiol. 2016;33:18–24. doi: 10.1016/j.mib.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 63.Cui J., Zhang H., Mo Z., Yu M., Liang Z. Cell wall thickness and the molecular mechanism of heterogeneous vancomycin-intermediate Staphylococcus aureus. Lett Appl Microbiol. 2021;72:604–609. doi: 10.1111/lam.13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo Y., Song G., Sun M., Wang J., Wang Y. Prevalence and therapies of antibiotic-resistance in Staphylococcus aureus. Front Cell Infect Microbiol. 2020;10:107. doi: 10.3389/fcimb.2020.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bowman C., Bumah V.V., Niesman I.R., Cortez P., Enwemeka C.S. Structural membrane changes induced by pulsed blue light on methicillin-resistant Staphylococcus aureus (MRSA) J Photochem Photobiol B. 2021;216 doi: 10.1016/j.jphotobiol.2021.112150. [DOI] [PubMed] [Google Scholar]
- 66.Zhao L.Y., Liu H.X., Wang L., Xu Z.F., Tan H.B., Qiu S.X. Rhodomyrtosone B, a membrane-targeting anti-MRSA natural acylgphloroglucinol from Rhodomyrtus tomentosa. J Ethnopharmacol. 2019;228:50–57. doi: 10.1016/j.jep.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 67.Facey S.J., Neugebauer S.A., Krauss S., Kuhn A. The mechanosensitive channel protein Mscl is targeted by the SRP to the novel YidC membrane insertion pathway of Escherichia coli. J Mol Biol. 2007;365:995–1004. doi: 10.1016/j.jmb.2006.10.083. [DOI] [PubMed] [Google Scholar]
- 68.Pop O.I., Soprova Z., Koningstein G., Scheffers D.J., van Ulsen P., Wickström D., et al. YidC is required for the assembly of the Mscl homopentameric pore. Febs J. 2009;276:4891–4899. doi: 10.1111/j.1742-4658.2009.07188.x. [DOI] [PubMed] [Google Scholar]
- 69.Shanmugam S.K., Backes N., Chen Y., Belardo A., Phillips G.J., Dalbey R.E. New insights into amino-terminal translocation as revealed by the use of YidC and Sec depletion strains. J Mol Biol. 2019;431:1025–1037. doi: 10.1016/j.jmb.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 70.Tsirigotaki A., De Geyter J., Šoštaric N., Economou A., Karamanou S. Protein export through the bacterial Sec pathway. Nat Rev Microbiol. 2017;15:21–36. doi: 10.1038/nrmicro.2016.161. [DOI] [PubMed] [Google Scholar]
- 71.Baeza N., Mercade E. Relationship between membrane vesicles, extracellular ATP and biofilm formation in Antarctic Gram-Negative bacteria. Microb Ecol. 2021;81:645–656. doi: 10.1007/s00248-020-01614-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.