Abstract
G-protein-coupled receptors (GPCRs) transduce the signals for a wide range of hormonal and sensory stimuli by activating a heterotrimeric guanine nucleotide-binding protein (G protein). The analysis of loss-of-function and constitutively active receptor mutants has helped to reveal the functional properties of GPCRs and their role in human diseases. Here we describe the identification of a new class of mutants, dominant-negative mutants, for the yeast G-protein-coupled α-factor receptor (Ste2p). Sixteen dominant-negative receptor mutants were isolated based on their ability to inhibit the response to mating pheromone in cells that also express wild-type receptors. Detailed analysis of two of the strongest mutant receptors showed that, unlike other GPCR interfering mutants, they were properly localized at the plasma membrane and did not alter the stability or localization of wild-type receptors. Furthermore, their dominant-negative effect was inversely proportional to the relative amount of wild-type receptors and was reversed by overexpressing the G-protein subunits, suggesting that these mutants compete with the wild-type receptors for the G protein. Interestingly, the dominant-negative mutations are all located at the extracellular ends of the transmembrane segments, defining a novel region of the receptor that is important for receptor signaling. Altogether, our results identify residues of the α-factor receptor specifically involved in ligand binding and receptor activation and define a new mechanism by which GPCRs can be inactivated that has important implications for the evaluation of receptor mutations in other G-protein-coupled receptors.
G-protein-coupled receptors (GPCRs) comprise a large family of receptors that are found in a wide range of eukaryotic organisms from yeasts to humans (4, 10). These receptors respond to diverse stimuli including hormones, neurotransmitters, and other chemical messengers (48). GPCRs transduce their signal by stimulating the α subunit of a heterotrimeric guanine nucleotide binding protein (G protein) to bind GTP (4, 16). This releases the α subunit from the βγ subunits, and then either the α subunit or the βγ subunits go on to promote signaling depending on the specific pathway (28).
GPCRs are structurally similar in that they contain seven transmembrane domains (TMDs) connected by intracellular and extracellular loops. Although many techniques have been applied to study receptor function, much of our knowledge on the mechanisms of GPCR activation comes from the characterization of mutant receptors. Loss-of-function and supersensitive mutants have helped to identify receptor regions needed for ligand binding, G-protein activation, and down-regulation of signaling (4, 49). Furthermore, the study of constitutively active receptor mutations has played a key role in the development of current models for receptor activation (26). Naturally occurring GPCR mutations have also been implicated in a number of human diseases (8, 25, 42). Interestingly, the analysis of different mutant receptors indicates that GPCRs utilize common structural domains for similar functions. In particular, the third intracellular loop has an essential role in G-protein activation in a wide range of GPCRs.
The genetic approaches possible in the yeast Saccharomyces cerevisiae have been used to examine the relationship between structure and function of the G-protein-coupled mating pheromone receptors. The α-factor and a-factor pheromones induce conjugation in yeast by binding to receptors with seven TMDs that activate a G-protein signal pathway that is highly conserved with mammalian signaling pathways (24). In fact, some human GPCRs can activate the pheromone signal pathway when they are expressed in yeast (19, 29). The analysis of loss-of-function, supersensitive, and constitutively active α-factor receptor mutants has begun to reveal the mechanisms for activation and regulation of this receptor. For example, the analysis of constitutively active mutants indicates that movement in the transmembrane segments plays a key role in α-factor receptor activation (22). Constitutive mutations and loss-of-function mutations implicate the third intracellular loop in G-protein activation (7, 34, 44). Mutagenesis studies also indicate that the cytoplasmic C terminus is not needed for G-protein activation but is involved in down-regulation of receptors by endocytosis (17) and desensitization of receptors by phosphorylation (6). In addition, studies with chimeric receptors suggest that the specificity for α-factor binding is determined by discontinuous segments of the α-factor receptor that include the transmembrane and extracellular regions (36, 37). Although some of the important domains of the α-factor receptor have been identified in these studies, the molecular mechanism of receptor signaling remains to be determined.
Dominant-negative (DN) mutants represent an important class of mutation in which a mutant receptor interferes with the function of the wild-type (WT) version of the receptor. Since the inhibitory phenotype in DN mutants implies loss of some but not all functions of the protein, these mutants have been used to great advantage in other receptor systems. For example, in the case of receptor tyrosine kinases, DN mutants have been used to assign particular functions to specific structural features or to study the effects of blocking receptor signaling (18). In view of the large number of mutations reported for GPCRs, it is intriguing that there are few examples of dominant GPCR mutations (42, 43). Furthermore, in cases where it has been examined, dominant mutations in GPCRs seem to affect primarily the targeting of receptors to the plasma membrane and not directly the function of the WT receptors. Therefore, we sought to determine if the analysis of DN mutants could be applied to GPCRs by taking advantage of the genetic accessibility of the yeast S. cerevisiae. In this report, we describe the identification of DN mutations in the α-factor pheromone receptor. Interestingly, our results indicate that these DN mutants interfere with the activity of the WT receptors by competing for the G protein. In addition, these mutations identify a new domain on the extracellular side of the TMDs that is important for receptor function.
MATERIALS AND METHODS
Strains and plasmids.
Yeast strains are described in Table 1. Cells were grown in media as described by Sherman (40). Cells carrying plasmids were grown in synthetic medium containing adenine and amino acid additives but lacking uracil or leucine to select for plasmid maintenance. The STE2 gene was inserted into the high-copy-number vector YEplac195 (2μm URA3) (12) to create pJK75 and into the low-copy-number vector YCplac33 (12) to create pDB02 (22). To construct the hemagglutinin (HA) epitope-tagged version of STE2, pJK75 was digested with BclI, the overhanging ends were made blunt with mung bean nuclease, and then the plasmid was digested with SacI. Three tandem copies of the HA epitope sequence (50) were prepared from plasmid SKp/x3HA by digestion with PstI, treatment with mung bean nuclease, and then digestion with SacI. After ligation, the reading frames were joined so that the three copies of the epitope sequence were present at the 3′ end of the STE2 coding sequence. The STE2-HA gene was subcloned into the integrating vector YIplac211 (12) and then used to integrate a copy of STE2-HA in the genome to create strain YLG122-2. To construct pMD82 (YEp-Gαβγ) carrying GPA1, STE4, and STE18 under control of their own promoters, fragments from nucleotides −717 to 1981 of GPA1, nucleotides −1168 to 1667 of STE4, and nucleotides −1155 to 1628 of STE18 were amplified by PCR. Oligonucleotide pairs used in the PCR contained flanking restriction sites for PstI-SalI, SalI-BamHI, and BamHI-KpnI, respectively, to clone the GPA1, STE4, and STE18 genes in tandem into the high-copy-number vector YEplac181 (2μm LEU2) (12).
TABLE 1.
Yeast strains used
| Strain | Genotype |
|---|---|
| DJ211-5-3 | MATa ade2-1o bar1-1 cry1 his4-580a leu2 lys2o trp1a tyr1o ura3 SUP4-3ts |
| JKY25 | MATa ade2-1o his4-580a lys2o trp1a tyr1o leu2 ura3 SUP4-3ts bar1-1 mfa2::FUS1-lacZ |
| JKY99-1 | MATa ade2-1o bar1-1 cry1 his4-580a leu2 lys2o trp1a tyr1o ura3 SUP4-3ts end4::LEU2 |
| JKY117-3 | MATa trp1::hisG ura3 ste5-3ts gpa1::LEU2 |
| JKY7434-2 | Isogenic to DJ211-5-3 except MATa/MATa/MATa/MATa TYR1/tyr1/tyr1/tyr1 STE2/STE2/STE2/STE2 |
| JKY7435-2 | Isogenic to JKY7434-2 except STE2/STE2/STE2/ste2::LEU2 |
| JKY7436-1 | Isogenic to JKY7434-2 except STE2/STE2/ste2::LEU2/ste2::LEU2 |
| JKY7437-1 | Isogenic to JKY7434-2 except STE2/ste2::LEU2/ste2::LEU2/ste2::LEU2 |
| PT2α | MATα hom3 ilv1 can |
| YLG122-2 | MATa ade2-1o his4-580a lys2o trp1a tyr1o leu2 ura3 SUP4-3ts bar1-1 mfa2::FUS1-lacZ STE2-HA |
| YLG123 | MATa ade2-1o his4-580a lys2o trp1a tyr1o leu2 ura3 SUP4-3ts bar1-1 mfa2::FUS1-lacZ ste2::LEU2 |
Genetic screen for DN receptor mutants.
Plasmid DNA was mutagenized by treatment with hydroxylamine (23) or by PCR. For PCR mutagenesis, primers were used to amplify the MluI-AatII or AatII-PstI fragments of the STE2 gene under essentially standard conditions except that one of the four deoxynucleoside triphosphates was at a concentration (40 μM) five times lower than that of the other three (200 μM). The cycle profile was 1 min at 94°C, 1 min at 50°C, and 2 min at 75°C for 35 cycles. Pools of mutagenized DNA fragments were subsequently cloned into pJK75 and then transformed into yeast strain JKY7436-1. In total, approximately 60,000 colonies were screened for resistance to α-factor-induced cell division arrest by replica plating cells onto medium containing a concentration of α-factor sufficient to arrest WT cells (≥3 × 10−8 M). The STE2 plasmids were recovered from yeast cells that grew in the presence of α-factor and were transformed into Escherichia coli for purification and analysis. The plasmids were also transformed back into a WT yeast strain to confirm that the resistance to α-factor was due to a mutation on the plasmid and not to a chromosomal mutation. DNA sequence analysis was carried out by using a dideoxy DNA sequencing kit from United States Biochemical. To confirm that the observed mutations accounted for DN receptor activity, a 1,219-bp MluI-AatII fragment, a 276-bp AatII-ClaI fragment, or a 329-bp ClaI-PstI fragment containing the mutation was subcloned into pJK75, and then the plasmids carrying specific mutations were tested for the ability to promote resistance to α-factor-induced cell division arrest in yeast.
α-Factor receptor analysis.
Logarithmic-phase cells adjusted to 107/ml were collected directly or treated with α-factor (5 × 10−7 M, final concentration) for the appropriate time, poisoned with 10 mM NaN3 and 10 mM KF to halt endocytosis, and collected. Western immunoblot analysis was carried out by lysing approximately 2.5 × 108 cells with glass beads in 250 μl of lysis buffer (2% sodium dodecyl sulfate, 100 mM Tris [pH 6.8], 8 M urea). Protein concentration was determined by using a Bio-Rad protein assay kit; equal amounts of extract were separated by electrophoresis on a sodium dodecyl sulfate–9% polyacrylamide gel, transferred to nitrocellulose, and then probed with rabbit anti-Ste2p antibodies (21) or with monoclonal anti-HA antibody 12CA5 (Boehringer Mannheim). Immunoreactive proteins were detected by enhanced chemiluminescence (ECL), using an Amersham ECL kit. α-Factor binding assays were carried out as described previously (22, 34). Fractionations of membranes on density gradients were performed essentially as described previously (34) except that Renocal-76 (Bracco Diagnostics) was used in place of Renografin-76, which is no longer commercially available. Mouse anti-Pma1p monoclonal antibody (kindly provided by J. Aris), rabbit polyclonal anti-HDEL (kindly provided by N. Dean), and rabbit anti-G6PDH (anti-glucose-6-phosphate dehydrogenase; Sigma) were used as markers for plasma membrane, internal membranes and cytosol, and cytosol, respectively. The distribution of the marker proteins was consistently restricted to fractions 10 to 12 for Pma1p, fractions 2 to 6 and 12 to 14 for proteins with the HDEL motif, and fractions 13 and 14 for G6PDH.
α-Factor-induced responses.
Induction of FUS1-lacZ was assayed in cells grown overnight to log phase in selective medium, diluted to 4 × 106 cells/ml, and then incubated with the indicated concentrations of α-factor for 2 h. β-Galactosidase assays were performed in duplicate, using the colorimetric substrate o-nitrophenyl-β-d-galactopyranoside (ONPG) as described previously (22). The average value of at least two independent experiments was reported for each assay, and the standard deviation was always less than 10%. Recovery from α-factor-induced cell division arrest was assayed by treating log-phase cells with α-factor (10−7 M); then samples were withdrawn at the indicated time intervals and fixed with formaldehyde, and at least 200 cells were examined microscopically to determine the percentage of cells with buds. To assay resistance to α-factor-induced cell division arrest, cells were adjusted to 106/ml, 10-fold dilutions were made, and then 5-μl aliquots of each dilution were placed on petri plates containing the indicated concentration of α-factor. The growth of the cells was recorded after 2 days for strains incubated at 30°C and 4 days for cells incubated at 23°C. Halo assays for α-factor-induced cell division arrest were performed by spreading approximately 3 × 105 cells from an overnight culture onto the appropriate solid media. α-Factor was added to sterile filter disks and incubated at 30°C for 2 days. Similar results were observed in at least two independent assays. In addition, similar results were always observed in both the spot assays and halo assays for cell division arrest.
RESULTS
Isolation of DN mutants.
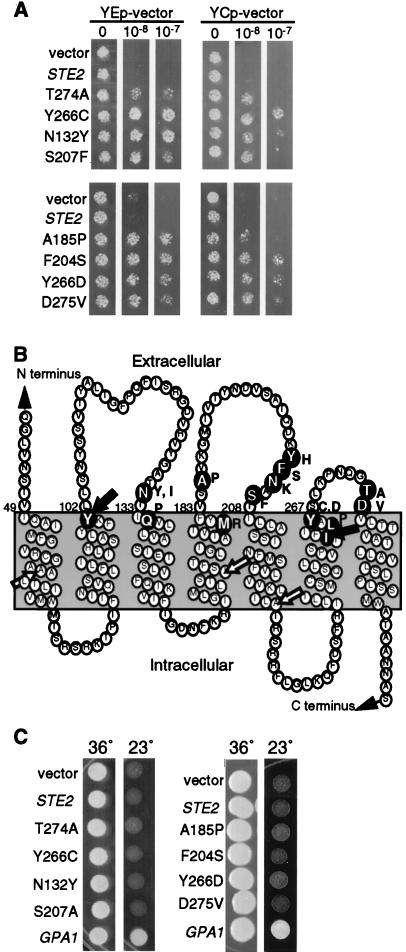
To search for DN mutations in the α-factor receptor gene (STE2), we took advantage of the fact that α-factor pheromone causes MATa S. cerevisiae cells to arrest cell division. Receptor mutants that interfered with pheromone signaling could therefore be isolated based on the ability to promote resistance to α-factor-induced cell division arrest. In practice, a plasmid carrying the STE2 gene was mutagenized with hydroxylamine and introduced into cells carrying a WT STE2 gene in the chromosome, and then cells that could grow on petri plates containing α-factor were identified (see Materials and Methods). Plasmids were recovered from the mutant cells and retransformed into a fresh culture of yeast in order to demonstrate that the resistance to α-factor was due to a mutation in the STE2 gene on the plasmid and not to a chromosomal mutation. Five DN STE2 plasmids were isolated from a screen of about 20,000 colonies. DNA sequence analysis showed that they represented four different mutations; Thr274 to Ala (T274A), Tyr266 to Cys (Y266C), Asn132 to Tyr (N132Y), and Ser207 to Phe (S207F). Subsequent analysis of the DN mutant receptors showed that they differed in the ability to promote resistance to α-factor-induced cell division arrest (Fig. 1A). From this set, the Y266C and N132Y mutants had the strongest DN effect, followed by S207F and the weakest DN mutant, T274A.
FIG. 1.
Identification of DN α-factor receptor mutants that interfere with the mating pathway upstream of the Gβγ subunits. (A) Yeast strain JKY25 carrying a copy of the WT receptor gene (STE2) in the genome and the indicated STE2 allele on multicopy plasmid YEplac195 or on the low-copy-number plasmid YCplac33 were tested for resistance to α-factor-induced cell division arrest. Cells were placed on solid medium containing the indicated concentration of α-factor and photographed after a 2-day incubation at 30°C. (B) A model of the transmembrane topology of the receptor protein which shows that DN mutations are located at the extracellular ends of transmembrane segments. The amino acid changes caused by the DN mutations are indicated as dark circles with white letters. The substituted amino acid is indicated on the right. The positions of DN linker insertions at residues 101 and 261 are indicated with black arrows; the positions of recessive linker insertion mutations at residues 62, 169, and 229 are indicated with white arrows. (C) The gpa1::LEU2 ste5-3ts strain JKY117-3 carrying the indicated STE2 allele on multicopy plasmid vector YEplac195 was incubated at 23 or 36°C for 3 days. The gpa1::LEU2 mutation deletes the gene encoding the Gα subunit, resulting in constitutive signaling from the Gβγ subunits at 23°C but not at 36°C, where signaling is blocked by the ste5-3ts mutation.
Sites of DN mutations.
Because there are so few examples of dominant GPCR mutations, it was of special interest to predict the effects of the structural changes caused by the DN mutations. Since the positions of the mutations did not appear to be clustered in the linear sequence of the receptor protein, sites of the altered residues were examined relative to the membrane topology of the receptor. A model for transmembrane topology of Ste2p was developed in which the seven TMD segments were predicted by hydropathy analysis (5) and drawn to be 21 residues long (Fig. 1B). Where possible, an aromatic residue was aligned at the lipid interface because studies have shown that aromatic amino acid side chains partition into the lipid head group and are often found at this position in other membrane proteins (51). This model for the topology of the receptor is generally consistent with other models such as that developed by the Viseur program (http://www.lctn.u-nancy.fr/viseur/viseur.html). When mapped onto this model, the DN mutations were all located near the extracellular ends of TMDs. The N132Y, S207F, Y266C, and T274A mutations were found at the ends of TMD3, -5, -6 and -7, respectively.
Given the clustering of the DN mutations at the extracellular ends of TMDs, a region not targeted in previous mutagenesis studies, it was of interest to examine the significance of their unique topological position. As a further test of this unique distribution, we screened for additional DN mutations and identified two more, Q135P and A185P, that mapped to the extracellular ends of TMD3 and -4, respectively. In addition, a separate screen carried out by using PCR to introduce mutations in STE2 identified eight more mutations that also mapped to the ends of the transmembrane segments (Fig. 1B, N132I, M180R, Y203H, F204S, N205K, L264P, Y266D, and D275V). To demonstrate that not all receptor mutations are dominant, we tested a set of five linker insertion mutants that are all defective in α-factor binding and signaling (20). Interestingly, two of the linker insertion mutations that mapped to extracellular ends of TMD2 (insertion at codon 101) and TMD6 (insertion at codon 261) (black arrows in Fig. 1B) displayed DN effects, whereas the three linker mutations that were recessive mapped to sites expected to be intracellular or within the TMDs (white arrows in Fig. 1B). Similarly, loss-of-function mutations within the third intracellular loop of Ste2p did not have DN properties (data not shown) (44).
Gene dosage relationship of DN mutants.
The genetic screen to identify DN mutant genes made use of a multicopy YEp plasmid vector (YEplac195) that results in about 10-fold overproduction of receptor protein. To determine whether overproduction of mutant receptors was required to observe their negative effects on signaling, the DN receptor genes were subcloned onto a YCp plasmid vector (YCplac33) that is usually present in a single copy per cell. Figure 1A shows a comparison of the effects of eight of the DN mutant alleles when carried on YEp and YCp vectors. Interestingly, overproduction of mutant receptors was not required because cells carrying the mutant genes on a low-copy-number vector were also resistant to α-factor. Overproduction did, however, enhance the DN effects of the mutant receptors.
To examine the gene dosage relationship between the WT and DN receptor genes more closely, we analyzed the effects of four representative DN mutants in a series of tetraploid yeast strains that carry one, two, three, or four copies of the WT receptor gene (STE2) in the genome. As shown in Table 2, cells with one genomic copy of STE2 were more resistant to α-factor if they carried a DN mutant gene on a low-copy-number YCp plasmid. However, the interfering effects of the DN mutants carried on YCp plasmids were less pronounced in cells containing multiple copies of STE2 in the genome, and their effects were not detectable in cells with four genomic copies of STE2. Similarly, the effects of the DN mutants could also be reversed in haploid cells by overproducing the WT receptors with a multicopy YEp-STE2 plasmid vector (data not shown). On the other hand, tetraploid cells carrying the DN mutant genes on multicopy YEp plasmids showed greater resistance to α-factor than did cells carrying the YCp plasmid versions. Furthermore, the DN mutant genes carried on multicopy plasmids conferred resistance to α-factor in cells with multiple genomic copies of STE2 (Table 2). This type of gene dosage relationship indicates that there is a stoichiometric relationship between the activities of the DN and WT receptors.
TABLE 2.
Gene dosage relationship between DN and WT receptor (STE2) genesa
| Vector | STE2 allele on plasmid | Relative growth
|
|||
|---|---|---|---|---|---|
| 1b | 2 | 3 | 4 | ||
| YCp | STE2 | − | − | − | − |
| T274A | +/− | − | − | − | |
| Y266C | + | + | +/− | − | |
| N132Y | + | +/− | − | − | |
| S207F | +/− | − | − | − | |
| YEp | STE2 | − | − | − | − |
| T274A | + | +/− | +/− | +/− | |
| Y266C | +++ | +++ | ++ | ++ | |
| N132Y | ++ | ++ | + | + | |
| S207F | ++ | + | + | + | |
Tetraploid yeast strains varying in STE2 dosage (see Table 1) and carrying different alleles of STE2 on multicopy plasmid YEplac195 or low-copy-number plasmid YCplac33 were plated on solid media in the absence or presence of 10−7 M α-factor. The cells were scored for growth after a 2-day incubation at 30°C. Relative cell growth in the presence of α-factor is indicated as − (0%), +/− (<1%), + (≥1%), ++ (≥10%), or +++ (100%).
Number of copies of STE2 in genome.
DN mutants antagonize an early step in the pheromone pathway.
The gene dosage studies suggested that the DN mutants interfere with WT receptor function. However, since some dominant forms of GPCRs have been reported to interfere with downstream components of the signaling pathway (9, 33), we tested the abilities of our mutants to antagonize signaling initiated by free Gβγ subunits. To this end, the DN mutants were assayed for the ability to counteract the constitutive cell division arrest caused by deletion of the GPA1 gene that encodes the Gα subunit (gpa1::LEU2). These gpa1::LEU2 cells also carried a temperature-sensitive mutation in a gene that acts further downstream in the pathway, ste5-3ts, so that cells could be propagated at the nonpermissive temperature (36°C) and then shifted down to the permissive temperature (23°C) to assay the effects of Gβγ signaling. As expected, gpa1::LEU2 cells carrying a control plasmid arrested division, as evidenced by the absence of growth at 23°C (Fig. 1C). In contrast, a GPA1 plasmid rescued the ability of the gpa1::LEU2 cells to grow. Interestingly, cells containing DN mutant plasmids failed to grow at 23°C, indicating that the mutant receptors interfere with an early step in the pheromone pathway, such as the receptor or Gα subunit, and do not interfere with signaling promoted by the free Gβγ subunits.
Effects of DN mutants on short-term responses to α-factor.
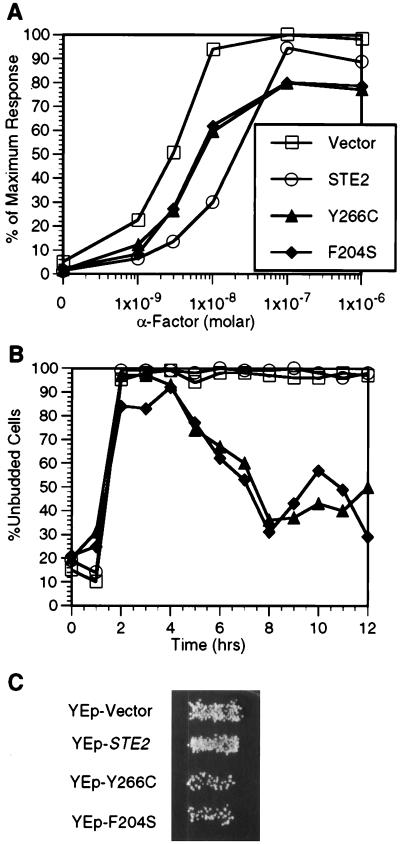
To analyze the effects of the DN mutants in detail, we selected two of the strongest mutants, Y266C and F204S, for further study. First, we examined their abilities to interfere with the induction of the FUS1-lacZ reporter gene by WT receptors (Fig. 2A). The FUS1-lacZ reporter gene is highly induced by pheromone (46) and serves for use in a short-term assay (2 h) that is more sensitive to α-factor than the long-term cell division arrest assays (2 days) described above. In these assays, cells containing the empty vector or a plasmid carrying the WT STE2 gene presented slightly different dose-response curves. Our results reproducibly showed that cells overproducing WT receptors were, for reasons that are unclear, somewhat less efficient at inducing the reporter gene when treated with low doses of α-factor. Despite this difference, the cells containing either the empty vector or the WT STE2 plasmid induced the reporter gene to similar maximum levels. In contrast, the maximum levels of the FUS1-lacZ reporter gene were consistently lower (80% of maximum) in cells producing the Y266C or F204S DN receptor, indicating that the DN mutants interfere with the induction of the reporter gene.
FIG. 2.
Effects of DN α-factor receptors in a WT STE2 strain. (A) WT STE2 strain JKY25 carrying the indicated STE2 allele on a multicopy plasmid (YEplac195) was incubated in the absence or presence of the indicated concentration of α-factor for 2 h and then assayed for β-galactosidase activity to measure induction of the FUS1-lacZ gene. The results were normalized to a value of 100% for WT STE2 cells treated with 10−6 M α-factor, which corresponded to a value of 121 U of activity. (B) The cells used for panel A were assayed for maintenance of cell division arrest in the unbudded stage at the indicated time after treatment with 10−7 M α-factor. (C) Mating ability of cells used for panel A with MATα strain PT2α. Diploid cells that were formed after mating were detected by examining growth on a selective plate as shown.
These results of the FUS1-lacZ induction assays indicate that the signaling activity of WT receptors, although inhibited, is not strongly blocked by the DN mutants at short times after α-factor treatment. We therefore tested the effects of the DN receptors on α-factor-induced cell division arrest at early time points. As expected, control cells carrying a YEp vector or a YEp-STE2 plasmid arrested in G1 as unbudded cells and maintained the arrested state for over 12 h (Fig. 2B). In contrast, cells carrying YEp-DN mutant plasmids started to recover and resumed cell division after 4 to 5 h, as evidenced by the percentage of cells that formed buds. Thus, cells carrying the DN mutants transiently arrested division but were not able to maintain the response to α-factor.
To examine the overall effect of the DN mutants in the pheromone pathway, we tested their abilities to interfere with mating. Mating assays carried out on petri plates clearly showed that the DN mutants inhibited mating (Fig. 2C). Quantitation of diploid formation in 4-h mating reactions showed that cells carrying the Y266C mutant mated at 16% and cells carrying the F204S mutant mated at 14% of WT efficiency. Taken together with the reporter gene induction and cell division arrest assays, these results indicate that the DN receptors strongly interfere with the ability of WT receptors to sustain pheromone signal transduction.
Signaling activity of DN mutants in the absence of WT receptors.
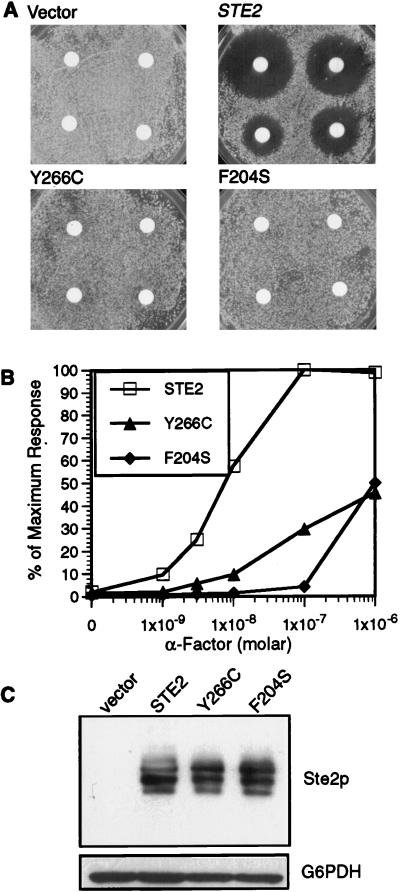
To better understand the signaling properties of the mutant receptors, we introduced them into a yeast strain in which the genomic copy of STE2 was deleted (ste2::LEU2). Cells carrying the DN receptor genes as the only receptor gene were then assayed for induction of cell division arrest. This analysis was carried out by placing filter disks containing α-factor onto a lawn of cells spread on a solid medium agar plate. Diffusion of the α-factor caused a zone of growth inhibition (halo) for cells carrying a WT STE2 plasmid but not the vector control (Fig. 3A). ste2::LEU2 cells carrying the Y266C or F204S plasmid also failed to undergo detectable cell division arrest. More interestingly, analysis of the ability to induce the FUS1-lacZ gene demonstrated that the mutant receptors are strongly defective for signaling. As shown in Fig. 3B, the Y266C mutant weakly induced the reporter gene, reaching only 40% of the maximum WT levels, after treatment with α-factor. The F204S mutant also showed a strong signaling defect, as these cells required at least 100-fold more α-factor than did WT cells to induce significant levels of FUS1-lacZ. Comparison of Y266C and F204S with the other DN receptor mutants in the ste2::LEU2 strain showed that their signaling defects were proportional to their ability to act as DN mutants in an STE2 strain (data not shown).
FIG. 3.
Activity of DN receptors in the absence of WT receptors. The ste2::LEU2 strain YLG123 carrying the indicated STE2 allele on a multicopy YEplac195 plasmid was assayed for responses to α-factor. (A) Halo assay for cell division arrest. Filter disks containing different amounts of α-factor (0.6, 0.3, 0.1 and 0.06 nmol, proceeding clockwise from top left) were placed on a lawn of the indicated cells and then incubated for 2 days. (B) Induction of the FUS1-lacZ reporter gene. Cells were incubated with α-factor for 2 h, and then β-galactosidase activity was assayed. Values were normalized to the level for WT STE2 cells treated with 10−6 M α-factor, which was 126 U of activity. (C) Western blot analysis of cells carrying the indicated receptor gene on the YCplac195 vector. Equal amounts (10 μg) of protein from lysates of exponentially growing cells were resolved by gel electrophoresis, transferred to nitrocellulose, and probed with anti-Ste2p or with anti-G6PDH to show that equal protein amounts were loaded in all lanes.
We then examined the production and ligand binding properties of the mutant receptors. Western blot analysis showed that the DN and WT receptor proteins were produced at similar levels and displayed similar heterogeneity due to glycosylation, indicating that the signaling defect of the DN mutants was due to impaired receptor function and not due to decreased receptor production (Fig. 3C). The ligand binding properties of the receptors were examined in equilibrium binding assays with 35S-labeled α-factor. As summarized in Table 3, ste2::LEU2 cells containing a low-copy-number YCp-STE2 plasmid bound α-factor with properties consistent with those previously reported for WT cells (Kd = 8.3 nM and 5,440 receptors/cell) (22, 34). When the ste2::LEU2 cells contained a plasmid encoding Y266C receptors, the binding affinity was about threefold lower and the number of cell surface binding sites was also diminished. In the case of F204S cells, a more dramatic defect in ligand binding was apparent since binding of α-factor was not detectable under the conditions that we used. This defect in ligand binding can account for the signaling defects of F204S cells. However, the Y266C cells still retain a significant binding affinity, suggesting that they may also be defective in transducing the α-factor signal.
TABLE 3.
α-Factor binding properties of cells containing mutant DN receptors
| Chromosomal STE2 allele | Plasmid STE2 allele | Kd (nM) | Receptors/cell |
|---|---|---|---|
| ste2Δ | STE2 | 8.3 | 5,440 |
| ste2Δ | Y266C | 21.5 | 1,530 |
| ste2Δ | F204S | NDB | NDB |
| STE2 | STE2 | 8.1 | 10,800 |
| STE2 | Y266C | 10.1 | 6,720 |
| STE2 | F204S | 9.1 | 6,700 |
Yeast strains YLG123 (ste2Δ) and JKY25 (STE2) containing different alleles of STE2 in the YCplac33 vector were assayed for binding to 35S-labeled α-factor in equilibrium binding assays. Values for Kd and receptors/cell were calculated by Scatchard plot analysis and are the means of two or three independent experiments. NDB, no detectable binding under our experimental conditions.
DN receptors do not affect the subcellular localization of WT receptor proteins.
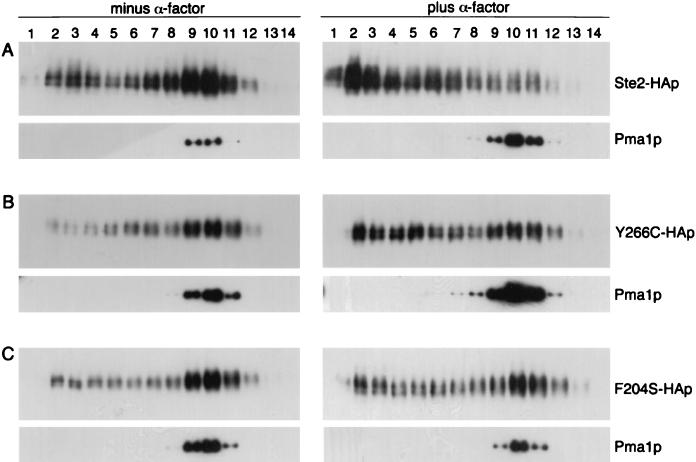
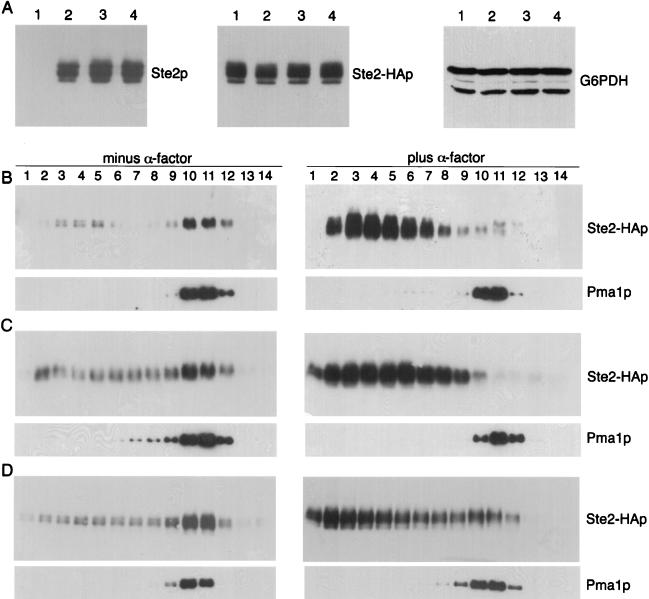
Previous reports about dominant GPCR mutations have shown that some mutant receptor proteins were mislocalized and also caused mislocalization of WT receptors (42). Therefore, it was of interest to investigate the subcellular localization of the DN receptors. For this analysis, cell extracts were separated by density gradient centrifugation under conditions that resolve the heavier plasma membrane fractions from the lighter membrane fractions consisting of endoplasmic reticulum, Golgi, and vacuolar membranes (34). Western blot analysis of the gradient fractions showed that WT receptors (Ste2p) were found primarily in the denser fractions (fractions 10 to 12) that also contained the plasma membrane ATPase, Pma1p (Fig. 4A). Similar sedimentation profiles were observed for the Y266C and F204S mutant receptors, indicating that they are properly localized to the plasma membrane (Fig. 4B and C). In all these experiments, the plasma membrane-containing fractions were well resolved from those containing internal membranes (fractions 2 to 6) and cytosol (fractions 13 to 14) (see Materials and Methods). When cells were treated with α-factor for 12 min, WT Ste2p shifted to the lighter fractions, consistent with the receptors undergoing endocytosis and transfer to the vacuole as reported previously (34). In contrast, significant amounts of the Y266C and F204S proteins were still detected at the plasma membrane after 12 min of α-factor treatment. The difference was the most dramatic for the F204S mutant, which was still primarily detected in the plasma membrane fractions after α-factor treatment. The greater stability of the DN receptors at the plasma membrane is very likely due to their defects in ligand binding and signal transduction, since the binding of α-factor specifically triggers receptors to undergo endocytosis.
FIG. 4.
Plasma membrane localization of DN receptors. YLG123 (ste2::LEU2) cells containing HA-tagged versions of STE2 (A), STE2-Y266C (B), and STE2-F204S (C) on YCplac33 were collected from exponentially growing cultures (left) or after treatment for 12 min with 5 × 10−7 M α-factor (right). Membranes were resolved on Renocal-76 density gradients, and the same amount of each fraction was subjected to immunoblotting with anti-HA monoclonal antibody to detect Ste2 proteins. As a control for plasma membrane localization, the fractions were also probed with anti-Pma1p monoclonal antibody.
Although the DN α-factor receptors were properly localized at the plasma membrane, they were defective in endocytosis, and therefore it was of interest to examine whether they could alter the distribution or internalization properties of WT receptors. To specifically detect the WT receptors, we generated a strain in which the HA epitope tag sequence was inserted into the chromosomal copy of the receptor gene (STE2-HA). As shown in Fig. 5A (middle panel), the presence of the DN receptors did not alter the steady-state levels of WT Ste2-HAp receptor protein in the cells. To determine if the WT Ste2-HAp was properly localized, cell extracts were fractionated on density gradients and analyzed by Western blotting. Cells carrying STE2-HA in the genome and an untagged WT receptor gene on a multicopy YEp-STE2 plasmid showed the expected plasma membrane localization of Ste2-HAp in the absence of α-factor and then its shift to lighter fractions after α-factor treatment (Fig. 5B). Similar results were also observed for cells carrying the Y266C and F204S mutant receptors on multicopy plasmids (Fig. 5C and D). This indicates that the presence of the DN mutant receptors does not alter the plasma membrane localization of the WT receptors or their ability to undergo endocytosis. In agreement with these results, we observed that the DN receptors have no effect on the ability of WT receptors to bind ligand (Table 3). Altogether, the results of these experiments show that the DN mutants do not interfere with the localization or stability of WT receptors, but rather interfere with the function of WT receptors at a step after ligand binding.
FIG. 5.
Production and plasma membrane localization of WT receptors is not altered by DN receptors. (A) Western blot analysis of strain YLG122-2 carrying an HA epitope-tagged version of WT STE2 in the genome and plasmid YEplac195 (lane 1), YEp-STE2 (lane 2), YEp-Y266C (lane 3), or YEp-F204S (lane 4). Total Ste2p was detected with rabbit anti-Ste2p under conditions that detected only the overproduced Ste2p encoded in the plasmids. Epitope-tagged Ste2-HAp was detected with anti-HA monoclonal antibody. Samples were also probed with anti-G6PDH antibodies to show that equal protein amounts were loaded in all lanes. (B to D) Analysis of the membrane localization of Ste2-HAp in YLG122-2 cells carrying the WT STE2 (B), STE2-Y266C (C), or STE2-F204S (D) gene in YEplac195. Cells were collected when exponentially growing (left) or after treatment with 5 × 10−7 M α-factor for 2 h (right), and then the extracts were resolved on density gradients as described in the legend to Fig. 4. Levels of WT Ste2-HAp were specifically monitored by immunoblot analysis of the gradient fractions with anti-HA monoclonal antibody. Localization of the control plasma membrane marker Pma1p is shown for each experiment.
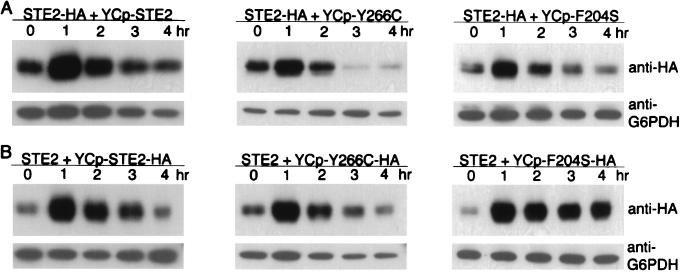
Stability of DN receptors after α-factor treatment.
The observations that the DN mutants were more stable at the surface than WT receptors in the presence of α-factor (Fig. 4) and that the strength of their interfering effect is proportional to their levels of expression (Fig. 1 and Table 2) suggested that the difference in stability of the mutant receptors could cause their interfering effects to become predominant with time of exposure to α-factor, as was observed in Fig. 2B. To test this idea, cells carrying epitope-tagged STE2-HA in the genome and a WT or mutant version of the receptor gene on a low-copy-number plasmid were assayed for the level of Ste2-HAp at various times after addition of α-factor (Fig. 6A). Western blot analysis showed that there was an initial increase in WT receptor protein, probably due to α-factor-induced expression of the STE2 gene (14). After this initial increase, the levels of protein diminished over time and after 3 to 4 h of treatment with α-factor reached levels similar to those found in the absence of pheromone. The decrease in receptor protein is consistent with the observations that ligand-bound receptors are rapidly endocytosed and degraded in the vacuole (17, 34). New receptor synthesis to replace the old receptors presumably slows down after several hours, as the division-arrested cells do not continue to grow in size at the same rate. Similar changes in the levels of WT receptors were also observed in cells coexpressing DN receptors except that the decrease in WT receptor protein was more pronounced, particularly in cells coexpressing Y266C mutant receptors. The more rapid decrease in WT receptors may be due to inhibition of pheromone signaling by DN receptors that would consequently result in lower induction of the receptor expression. Interestingly, the reciprocal experiments showed that HA-tagged DN receptors were significantly more stable than the WT receptors. The Y266C receptors took longer to decrease to unstimulated levels, and the F204S receptors remained at the induced levels for over 4 h (Fig. 6B). The results of these analyses show that DN mutant receptors are more stable than the WT receptors in the presence of α-factor and are consistent with the idea of cells becoming more resistant to α-factor as the ratio of DN to WT receptor protein increases.
FIG. 6.
Receptor stability in the presence of α-factor. (A) Western blot analysis of the levels of WT Ste2-HAp in strain YLG122-2 that also carried the indicated STE2 allele on low-copy-number vector YCplac33. Cells were incubated with 5 × 10−7 M α-factor for 0, 1, 2, 3, or 4 h as indicated above each lane and then processed for Western blot analysis with anti-HA antibody to specifically detect the chromosomally encoded WT Ste2-HAp receptor protein. (B) Western blot analysis of DN Ste2p at various times after treatment with α-factor. Strain JKY25 containing a WT untagged chromosomal STE2 gene and the indicated HA-tagged versions of STE2 on YCplac33 were treated with α-factor as indicated for panel A and analyzed by Western blotting with anti-HA antibody to specifically detect the HA-tagged Ste2 receptors encoded on the plasmid.
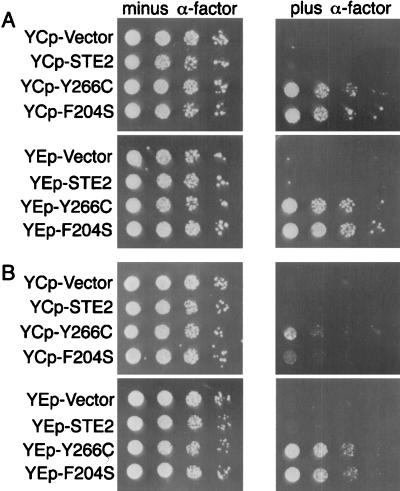
To examine further the role of endocytosis in the DN effect, we tested the abilities of mutant receptors to interfere with signaling in an end4Δ strain that is defective in α-factor-stimulated receptor endocytosis (31). This analysis was carried out by spotting dilutions of cells on plates containing α-factor and then observing the abilities of the DN receptors to interfere with the maintenance of cell division arrest. In the case of WT cells, cells carrying the DN receptor genes on a low-copy-number YCp plasmid vector were able to interfere with signaling, as evidenced by the ability of the cells to form colonies (Fig. 7A). The interfering effects were improved when the DN receptor genes were carried on multicopy YEp plasmid vectors. In contrast, end4Δ cells containing the DN receptors on low-copy-number plasmids did not display significant ability to grow in the presence of α-factor (Fig. 7B). The end4Δ cells carrying the DN receptor genes on multicopy YEp plasmids were able to grow in the presence of α-factor, but to a much lower extent than in the END4+ cell background. Thus, the weaker effects of the DN mutant receptors in the end4Δ cells also suggest that the greater stability of the DN mutants in the presence of α-factor may account for the stronger effects of the DN mutants in long-term assays such as cell division arrest than in short-term assays such as reporter gene induction.
FIG. 7.
Interfering effects of DN receptors in an endocytosis-defective end4Δ strain. Growth of JKY25 (END4+) (A) or JKY99-1 (end4::LEU2) (B) cells carrying the indicated STE2 alleles on low-copy-number vector YCplac33 (upper panels) or multicopy vector YEplac195 (lower panels). Serial dilutions of cells were spotted on plates in the absence or presence of 10−7 M α-factor as indicated.
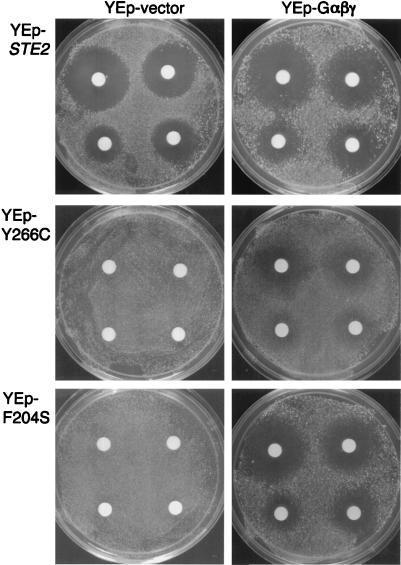
Interfering effects of DN mutants can be reversed by overproducing G protein.
The results described above suggested the possibility that the DN receptors compete with WT receptors for a factor required for signaling. In particular, it seemed likely that they could compete for the G protein. If this possibility is true, we reasoned that it should be possible to diminish the interfering effects of the DN receptors by overproducing the G-protein subunits. To test this hypothesis, a multicopy plasmid carrying the GPA1 (Gα), STE4 (Gβ), and STE18 (Gγ) genes (YEp-Gαβγ) was constructed (see Materials and Methods) and introduced into cells expressing both WT and DN receptors. Measurement of the response to α-factor in a halo assay (Fig. 8) showed that indeed the cells carrying the YEp-Gαβγ plasmid were now able to maintain cell division arrest. Overproduction of the G-protein subunits in this manner enabled cells containing the Y266C receptors to display significant ability to maintain cell division arrest. In the case of cells containing F204S receptors, the degree of cell division arrest was nearly the same as that observed for WT cells. By examining the ability of dilutions of cells to grow on plates containing α-factor, we estimated that the YEp-Gαβγ plasmid improved cell division arrest over 100-fold for Y266C cells and over 1,000 for F204S cells relative to the corresponding cells carrying the empty YEp vector (data not shown). As a control, we showed that G-protein overproduction did not improve signaling from DN receptors in a strain lacking WT receptors (data not shown). Taken together, all of these results strongly suggest that the DN receptors compete with WT receptors for G protein.
FIG. 8.
Effects of DN receptors in cells overproducing G protein, determined by assays of pheromone-induced growth arrest (halo assays) for JKY25 cells carrying the indicated STE2 allele on the multicopy vector YEplac195 (2μm URA3). Also, as indicated at the top, the cells contained either the control vector YEplac181 (2μm LEU2) or the same vector containing the GPA1 (Gα), STE4 (Gβ), and STE18 (Gγ) genes (YEp-Gαβγ). Halo assays were performed as described in the legend to Fig. 3. The amounts of α-factor applied to the disks on each plate were 0.6, 0.3, 0.1, and 0.06 nmol, proceeding clockwise from top left.
DISCUSSION
To examine the mechanisms of receptor activation, we carried out a genetic screen to isolate DN mutations in the α-factor receptor. The rationale for this approach is that since the interfering phenotype of a dominant mutant usually results from loss of some, but not all, functions of the protein, they should be useful for identifying important domains of GPCRs. The genetic screen for DN mutants yielded 16 different mutations in the α-factor receptor, based on their abilities to interfere with α-factor-induced cell division arrest. The DN mutants appeared to specifically interfere with the pheromone signal pathway because they also diminished mating efficiency and the ability of pheromone to induce a FUS1-lacZ reporter gene. The interfering effects on signaling caused by the DN mutants could be reversed by overproducing the WT receptors or overproducing the G-protein subunits, indicating that the mutant receptors were not simply inhibiting the function of membrane signaling molecules in a nonspecific manner. In addition, the DN mutants affect an early step in the signal pathway because they did not suppress the constitutive signaling caused by free Gβγ subunits in a strain lacking the Gα subunit. Thus, these DN mutants represent a new class of mutation in the α-factor receptor that interfere with the signaling activity of WT α-factor receptors.
The DN receptor mutants identified in this study differ from other interfering forms of GPCRs that have been reported. For example, production of the opposite pheromone receptor (a-factor receptor) in MATa yeast suppresses the constitutive signaling caused by free Gβγ subunits, indicating that the interference occurred at a later stage of the pathway (9). Similarly, expression of an alternatively spliced form of the follicle-stimulating hormone receptor interferes with a post-G-protein component of its signaling pathway (33). The DN α-factor receptor mutants also differ from some dominant forms of rhodopsin that cause retinitis pigmentosa (42), the gonadotropin-releasing hormone receptor (13), and the calcium-sensing receptor (1), which apparently cause mislocalization of the corresponding WT receptors. In the case of the DN α-factor receptor mutants, our analysis of receptor distribution in fractionated extracts showed that WT receptors were properly localized to the plasma membrane. In addition, WT receptors were properly internalized by endocytosis in response to α-factor. Therefore, the α-factor receptor mutants identified in this study represent a novel mechanism of interference for a dominant GPCR mutant.
A likely explanation for the mechanism of action of the DN α-factor mutants is that they may present an altered conformation that confers the ability to interfere with the WT receptors. This interference could happen either by interacting directly with the WT receptors and blocking their function or by sequestering the G protein away from WT receptors. Although several reports have suggested that dimerization may occur in GPCRs (15, 20, 27), our results do not support the idea of an interfering interaction between the DN mutants with WT receptors since they did not affect the ligand binding properties of WT receptors and did not alter the ability of α-factor to stimulate endocytosis of WT receptors. Instead, our data are consistent with a model in which DN receptors compete with WT receptors for the G proteins. In support of this, we observed that overexpression of the G-protein subunits reversed the effects of DN receptors. Also consistent with this competition model is the observation that the greater plasma membrane stability of the DN receptors in the presence of α-factor correlated with their effects being more pronounced in long-term assays than in short-term assays. The delayed internalization kinetics of DN receptors (Fig. 4 and unpublished data) increases the ratio of DN to WT receptors over time and would thus enable DN receptors to better compete for a factor in the plasma membrane (i.e., G protein). Altogether, these results suggest that the DN receptors are capable of associating with G proteins but are unable to activate them.
The unique topological distribution of the DN mutations is of great interest because the altered residues are all situated toward the extracellular face and thus cannot be in direct contact with the G protein on the intracellular side. It is also interesting that the DN mutations are clustered toward the extracellular ends of the TMDs and are not detected throughout the extracellular loops. This localization contrasts with the occurrence of loss-of-function mutations (7, 20, 34, 44) and constitutive mutations (7, 22) in many different regions of the α-factor receptor. Assuming that the α-factor receptor forms a seven-helix bundle, as proposed for other members of the GPCR family (2), the amino acid residues affected by the DN mutations should all lie in close spatial proximity. Thus, it appears that these mutations define a specific domain of the receptor. Three of the DN mutations result from introduction of proline residues that are expected to perturb the conformation of this domain (Q135P, A185P, and L264P). The other substitutions may also alter the structure of this domain because, in many cases, the introduced residue is less polar (N132Y, N132I, Q135P, S207F, Y266C, T274A, and D275V) or more polar (Y203H, F204S, and N205K) than the WT residue. One of the DN mutations that we identified, Y266C, was previously detected as a second-site suppressor of an E143K mutation in TMD3 of the α-factor receptor (41), and it was proposed that this suppression might occur as a result of altered packing of the TMDs. A direct role of this domain in ligand binding is suggested by the lower affinity for α-factor of the Y266C and F204S mutants.
This disruption of ligand binding activity can account for the signaling defect in the F204S mutant. However, the Y266C mutant retains significant ligand binding affinity and therefore must also be defective in the conformational changes that lead to G-protein activation. This idea is further suggested by the delay in ligand-stimulated endocytosis of Y266C receptors in comparison to WT receptors, even after treatment with saturating doses of α-factor (Fig. 5 and data not shown). Interestingly, structural studies on rhodopsin indicate that the region on the extracellular face is in an ordered configuration (47), and studies on other GPCRs have also suggested that residues near the extracellular ends of TMDs play an important role in receptor conformation (35). For example, the introduction of metal ion binding sites into the extracellular ends of the TMDs for both the tachykinin receptor and rhodopsin caused a metal-dependent inhibition of receptor activity (11, 39). Thus, the domain of the receptor containing the DN mutations plays a key role in the conformational changes that lead to G-protein activation. An interesting possibility is that the DN mutations lock the α-factor receptor in an inactive conformation that is still capable of associating with a G protein. This possibility is consistent with the idea that WT α-factor receptors may associate with G proteins in the absence of ligand. That unoccupied WT receptors may bind G proteins and sequester them away from other receptors has been suggested to explain the ability of WT receptors to interfere with the signaling activity of certain hypersensitive, constitutive, or chimeric mutant pheromone receptors and with mammalian GPCRs expressed in yeast (3, 21, 22, 30, 32, 38, 45).
Altogether, the results of this study show, to our knowledge, the first clear evidence of DN mutant GPCRs that interfere with the function of WT receptors. The discovery of DN mutants of the α-factor receptor also suggests that similar DN mutations can occur in other GPCRs and that they may help to reveal the functional properties of other receptors in this family. Furthermore, in view of the fact that loss-of-function mutations and constitutively activate mutations in GPCRs are known to cause human diseases (8, 25, 42, 43), DN receptor mutations should also be considered as a possible cause of human disease.
ACKNOWLEDGMENTS
We thank our colleagues for their helpful comments on the manuscript. We thank Neta Dean for plasmids and antibodies, John Aris for antibodies, and Duane Jenness for advice on density gradient fractionation experiments.
M.D. was supported in part by NIH training grant 5T32CAO9176. This work was supported by NIH grant GM55107 awarded to J.B.K.
REFERENCES
- 1.Bai M, Janicic N, Trivedi S, Quinn S J, Cole D E, Brown E M, Hendy G N. Markedly reduced activity of mutant calcium-sensing receptor with an inserted Alu element from a kindred with familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism. J Clin Investig. 1997;99:1917–1925. doi: 10.1172/JCI119359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin J M. The probable arrangement of the helices in G protein-coupled receptors. EMBO J. 1993;12:1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boone C, Davis N G, Sprague G F., Jr Mutations that alter the third cytoplasmic loop of the a-factor receptor lead to a constitutive and hypersensitive phenotype. Proc Natl Acad Sci USA. 1993;90:9921–9925. doi: 10.1073/pnas.90.21.9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourne H R. How receptors talk to trimeric G proteins. Curr Opin Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 5.Burkholder A C, Hartwell L H. The yeast α-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985;13:8463–8475. doi: 10.1093/nar/13.23.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Q, Konopka J B. Regulation of the G-protein-coupled α-factor pheromone receptor by phosphorylation. Mol Cell Biol. 1996;16:247–257. doi: 10.1128/mcb.16.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark C D, Palzkill T, Botstein D. Systematic mutagenesis of the yeast mating pheromone receptor third intracellular loop. J Biol Chem. 1994;269:8831–8841. [PubMed] [Google Scholar]
- 8.Coughlin S. Expanding horizons for receptors coupled to G proteins: diversity and disease. Curr Opin Cell Biol. 1994;6:191–197. doi: 10.1016/0955-0674(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 9.Couve A, Hirsch J P. Loss of sustained Fus3p kinase activity and the G1 arrest response in cells expressing an inappropriate pheromone receptor. Mol Cell Biol. 1996;16:4478–4485. doi: 10.1128/mcb.16.8.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dohlman H G, Thorner J, Caron M G, Lefkowitz R J. Model systems for the study of 7-transmembrane-segment receptors. Annu Rev Biochem. 1991;60:653–688. doi: 10.1146/annurev.bi.60.070191.003253. [DOI] [PubMed] [Google Scholar]
- 11.Elling C E, Nielsen S M, Schwartz T W. Conversion of antagonist-binding site to metal-ion site in the tachykinin NK-1 receptor. Nature. 1995;374:74–77. doi: 10.1038/374074a0. [DOI] [PubMed] [Google Scholar]
- 12.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 13.Grosse R, Schoneberg T, Schultz G, Gudermann T. Inhibition of gonadotropin-releasing hormone receptor signaling by expression of a splice variant of the human receptor. Mol Endocrinol. 1997;11:1305–1318. doi: 10.1210/mend.11.9.9966. [DOI] [PubMed] [Google Scholar]
- 14.Hartig A, Holly J, Saari G, MacKay V L. Multiple regulation of STE2, a mating-type-specific gene of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:2106–2114. doi: 10.1128/mcb.6.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hebert T E, Moffett S, Morello J P, Loisel T P, Bichet D G, Barret C, Bouvier M. A peptide derived from a β2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J Biol Chem. 1996;271:16384–16392. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 16.Hepler J R, Gilman A G. G proteins. Trends Biochem Sci. 1992;17:382–387. doi: 10.1016/0968-0004(92)90005-t. [DOI] [PubMed] [Google Scholar]
- 17.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 18.Kashles O, Yarden Y, Fischer R, Ullrich A, Schlessinger J. A dominant negative mutation suppresses the function of normal epidermal growth factor receptors by heterodimerization. Mol Cell Biol. 1991;11:1454–1463. doi: 10.1128/mcb.11.3.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King K, Dohlman H G, Thorner J, Caron M G, Lefkowitz R J. Control of yeast mating signal transduction by a mammalian β2-adrenergic receptor and the Gs α-subunit. Science. 1990;250:121–123. doi: 10.1126/science.2171146. [DOI] [PubMed] [Google Scholar]
- 20.Konopka J B, Jenness D D. Genetic fine-structural analysis of the Saccharomyces cerevisiae α-pheromone receptor. Cell Regul. 1991;2:439–452. doi: 10.1091/mbc.2.6.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konopka J B, Jenness D D, Hartwell L H. The C terminus of the Saccharomyces cerevisiae α-pheromone receptor mediates an adaptive response to pheromone. Cell. 1988;54:609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- 22.Konopka J B, Margarit M, Dube P. Mutation of pro-258 in transmembrane domain 6 constitutively activates the G protein-coupled α-factor receptor. Proc Natl Acad Sci USA. 1996;93:6764–6769. doi: 10.1073/pnas.93.13.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence C W. Classical mutagenesis techniques. Methods Enzymol. 1991;194:273–281. doi: 10.1016/0076-6879(91)94021-4. [DOI] [PubMed] [Google Scholar]
- 24.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 25.Lefkowitz R J. Turned on to ill effect. Nature. 1993;365:603–604. doi: 10.1038/365603a0. [DOI] [PubMed] [Google Scholar]
- 26.Lefkowitz R J, Cotecchia S, Samama P, Costa T. Constitutive activity of receptors coupled to guanine nucleotide regulatory proteins. Trends Pharmacol Sci. 1993;14:303–307. doi: 10.1016/0165-6147(93)90048-O. [DOI] [PubMed] [Google Scholar]
- 27.Maggio R, Vogel Z, Wess J. Reconstitution of functional muscarinic receptors by co-expression of amino- and carboxyl-terminal receptor fragments. FEBS Lett. 1993;319:195–200. doi: 10.1016/0014-5793(93)80066-4. [DOI] [PubMed] [Google Scholar]
- 28.Neer E J. Heterotrimeric G proteins: organizers of transmembrane signals. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 29.Price L A, Kajkowski E M, Hadcock J R, Ozenberger B A, Pausch M H. Functional coupling of a mammalian somatostatin receptor to the yeast pheromone response pathway. Mol Cell Biol. 1995;15:6188–6195. doi: 10.1128/mcb.15.11.6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price L A, Strnad J, Pausch M, Hadcock J R. Pharmacological characterization of the rat A2A-adenosine receptor functionally coupled to the yeast pheromone response pathway. Mol Pharmacol. 1996;50:829–837. [PubMed] [Google Scholar]
- 31.Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reneke J E, Blumer K J, Courchesne W E, Thorner J. The carboxyl terminal domain α-factor receptor is a regulatory domain. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- 33.Sairam M R, Jiang L G, Yarney T A, Khan H. Follitropin signal transduction: alternative splicing of the FSH receptor gene produces a domainant negative form of receptor which inhibits hormone action. Biochem Biophys Res Commun. 1996;226:717–722. doi: 10.1006/bbrc.1996.1419. [DOI] [PubMed] [Google Scholar]
- 34.Schandel K A, Jenness D D. Direct evidence for ligand-induced internalization of the yeast α-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz T W, Rosenkilde M M. Is there a ’lock’ for all agonist ’keys’ in 7TM receptors? Trends Pharmacol Sci. 1996;17:213–216. doi: 10.1016/0165-6147(96)10017-1. [DOI] [PubMed] [Google Scholar]
- 36.Sen M, Marsh L. Noncontiguous domains of the α-factor receptor of yeasts confer ligand specificity. J Biol Chem. 1994;269:968–973. [PubMed] [Google Scholar]
- 37.Sen M, Shah A, Marsh L. Two types of α-factor receptor determinants for pheromone specificity in the mating-incompatible yeasts S. cerevisiae and S. kluyveri. Curr Genet. 1997;31:235–240. doi: 10.1007/s002940050200. [DOI] [PubMed] [Google Scholar]
- 38.Shah A, Marsh L. Role of SST2 in modulating G protein-coupled receptor signaling. Biochem Biophys Res Commun. 1996;226:242–246. doi: 10.1006/bbrc.1996.1340. [DOI] [PubMed] [Google Scholar]
- 39.Sheikh S P, Zvyaga T A, Lichtarge O, Sakmar T P, Bourne H R. Rhodopsin activation blocked by metal-ion-binding sites linking transmembrane helices C and F. Nature. 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 40.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 41.Sommers C M, Dumont M E. Genetic interactions among the transmembrane domains of the G protein-coupled receptor encoded by the yeast STE2 gene. J Mol Biol. 1997;266:559–575. doi: 10.1006/jmbi.1996.0816. [DOI] [PubMed] [Google Scholar]
- 42.Spiegel A M. Defects in G protein-coupled signal transduction in human disease. Annu Rev Physiol. 1995;58:143–170. doi: 10.1146/annurev.ph.58.030196.001043. [DOI] [PubMed] [Google Scholar]
- 43.Spiegel A M. Mutations in G proteins and G protein-coupled receptors in endocrine disease. J Clin Endocrinol Metab. 1996;81:2434–2442. doi: 10.1210/jcem.81.7.8675557. [DOI] [PubMed] [Google Scholar]
- 44.Stefan C J, Blumer K J. The third cytoplasmic loop of a yeast G-protein-coupled receptor controls pathway activation, ligand discrimination, and receptor internalization. Mol Cell Biol. 1994;14:3339–3349. doi: 10.1128/mcb.14.5.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stefan C J, Overton M C, Blumer K J. Mechanisms governing the activation and trafficking of yeast G protein-coupled receptors. Mol Biol Cell. 1998;9:885–899. doi: 10.1091/mbc.9.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trueheart J, Boeke J D, Fink G R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unger V M, Hargrave P A, Baldwin J M, Schertler G F. Arrangement of rhodopsin transmembrane α-helices. Nature. 1997;389:203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- 48.Watson S, Arkinstall S. The G-protein linked receptor factsbook. London, England: Academic Press, Ltd.; 1994. [Google Scholar]
- 49.Wess J. G-protein-coupled receptors: molecular mechanisms involved in receptor activation and selectivity of G-protein recognition. FASEB J. 1997;11:346–354. [PubMed] [Google Scholar]
- 50.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 51.Wimley W C, White S H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;10:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]