Abstract
Objective
This review provides a comprehensive overview of the existing research on the seminal microbiome and its association with male infertility, while also highlighting areas that warrant further investigation.
Methods
A narrative review was conducted, encompassing all relevant studies published between 1980-2023 on the male reproductive tract microbiome in humans. This review considered studies utilizing culture-based, polymerase chain reaction (PCR)-based, and next-generation sequencing (NGS)-based methodologies to analyze the microbiome. Data extraction encompassed sample types (semen or testicular tissue), study designs, participant characteristics, employed techniques, and critical findings.
Results
We included 37 studies comprising 9,310 participants. Among these, 16 studies used culture-based methods, 16 utilized NGS, and five employed a combination of methods for microorganism identification. Notably, none of the studies assessed fungi or viruses. All NGS-based studies identified the presence of bacteria in all semen samples. Two notable characteristics of the seminal microbiome were observed: substantial variability in species composition among individuals and the formation of microbial communities with a dominant species. Studies examining the testicular microbiome revealed that the testicular compartment is not sterile. Interestingly, sexually active couples shared 56% of predominant genera, and among couples with positive cultures in both partners, 61% of them shared at least one genital pathogen. In couples with infertility of known causes, there was an overlap in bacterial composition between the seminal and vaginal microbiomes, featuring an increased prevalence of Staphylococcus and Streptococcus genera. Furthermore, the seminal microbiome had discernible effects on reproductive outcomes. However, bacteria in IVF culture media did not seem to impact pregnancy rates.
Conclusion
Existing literature underscores that various genera of bacteria colonize the male reproductive tract. These organisms do not exist independently; instead, they play a pivotal role in regulating functions and maintaining hemostasis. Future research should prioritize longitudinal and prospective studies and investigations into the influence of infertility causes and commonly prescribed medication to enhance our understanding of the seminal microbiota’s role in reproductive health.
Keywords: male infertility, microbiome, microbiota composition, semen, spermatozoa, testicular microbiome, testis
Introduction
Contrary to earlier perceptions that primarily portrayed bacteria as pathogenic adversaries, contemporary insights reveal a fascinating truth: the human body is teeming with more bacteria than human cells (1). This revelation aligns with the recognition that nearly all organs and systems host a companion microbiota composed of bacteria, fungi, and viruses that coexist harmoniously with human hosts (2). These organisms do not lead solitary lives; instead, they play a pivotal role in regulating bodily functions and maintaining hemostasis. Perturbations in the microbiota, termed dysbiosis, which can encompass imbalances in microbial community composition, loss of beneficial symbionts, proliferation of pathobionts or opportunistic organisms, and disruptions in inter-microbial competition and diversity, have been implicated in the onset or exacerbation of various diseases (3).
The term ‘microbiome’ refers to diverse microorganisms inhabiting specific organs, systems, or biofluids. Next-generation sequencing (NGS) technology has ushered in a new era of understanding the human microbiome, enabling the detection of previously unknown commensal and pathogenic microorganisms (4). Leveraging this high-throughput technique, the ‘Human Microbiome Project’ has characterized microbiomes in various bodily organs and has reported that the urogenital tract microbiome constitutes approximately 9% of the total human microbiota (5, 6). Notably, dysbiosis of the female reproductive microbiome has been associated with reduced pregnancy rates and adverse pregnancy outcomes (7).
Despite considerable progress in elucidating the human microbiome, the characterization of the male genital tract microbiome remains in its early stages. Most studies concerning the male reproductive microbiota center on the seminal microbiome. Semen comprises secretions from the testicles, epididymis, prostate, seminal vesicles, bulbourethral glands, and periurethral glands, providing a conducive environment for microbial growth due to its nutrient content (8, 9). Therefore, the seminal microbiome serves as a representative of the entire male genital system.
A male factor is identified in up to 50% of infertile couples, and urogenital tract infection represents a potential etiological contributor (10, 11). Additionally, as many as 25% of men with abnormal semen analysis results are categorized as having idiopathic infertility due to the absence of discernible causes using current diagnostic tools (7, 12). Beyond the conventional mechanisms by which pathogenic bacteria can adversely affect male fertility, such as impairing sperm motility and capacitation and inducing oxidative stress and apoptosis (13–21), some researchers propose that dysbiosis of the seminal microbiome may also exert adverse effects on male fertility through as-yet-unclear pathways (2).
To consolidate the evidence concerning the seminal microbiome and its association with male infertility, we conducted a comprehensive narrative review of all studies about the male reproductive tract microbiome in humans from 1980 to 2023. Our review encompassed research that employed various methodologies, including culture-based, polymerase chain reaction (PCR)-based, and NGS-based techniques to investigate the microbiome in human semen or testicular tissue samples. We systematically collected information on sample types (semen or testicular tissue), study designs, participant demographics, employed methodologies, and key findings. To assess the quality of the included studies, we utilized the ‘Study Quality Assessment Tool for Before-After (Pre-Post) Studies with no Control Group’ developed by the National Heart, Lung, and Blood Institute. The eligible studies were categorized into three tiers based on their quality: high, medium, or low (www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools). Our review also explores the influence of environmental factors on the seminal microbiome. Finally, we examine the evolving clinical practices stemming from this emerging knowledge and recommend topics for future research to address the existing knowledge gaps.
How to assess the microbiome
The initial investigations into the bacterial content of semen relied on culture-based techniques primarily targeting well-known pathogenic bacteria, like Staphylococcus, Enterococcus, Escherichia, and Ureaplasma (22–24). Consequently, these earlier endeavors yielded limited insights into the resident seminal microbiota, particularly concerning anaerobes and fastidious bacteria, which are challenging to cultivate (5). Subsequently, PCR-based studies made strides in identifying a broader spectrum of bacteria genera. However, they still failed to provide a comprehensive overview of the seminal microbiome (25). This limitation arose from the requirement to predetermine the genera of bacteria under investigation, rendering the technique less effective for polymicrobial specimens and frequently resulting in data that were challenging to interpret (26).
The emergence of NGS technology marked a remarkable breakthrough in exploring the human microbiome. This method directly sequences microbial DNA or RNA within samples, eliminating the reliance on traditional culture-based approaches (4). Two primary NGS techniques employed for microbiome characterization are amplicon sequencing and shotgun metagenomic sequencing (27).
Amplicon sequencing involves amplifying a specific region of DNA through PCR and then sequencing the resultant product. Typically, this entails targeting one or more hypervariable regions of the bacterial 16S ribosomal RNA (rRNA) gene (4). The hypervariable regions, being highly conserved and ubiquitous among bacteria, offer a suitable basis for analysis (28). Nevertheless, due to practical constraints related to time and cost, only a subset of these variable regions is generally chosen for sequencing. This approach introduces potential bias since no single region effectively distinguishes all bacteria species, and sequencing specific hypervariable regions may yield varying results.
In contrast, shotgun metagenomic sequencing (SMS) comprehensively assesses all the DNA within a given sample. This method involves DNA extraction and random fragmentation, followed by the ligation of barcodes and adapters to each fragment, facilitating sample identification and DNA sequencing. Subsequently, the obtaining reads are meticulously cleaned and aligned with a reference database to identify taxa and assess functional potential (28). Unlike amplicon sequencing, SMS metagenomic sequencing enables the detection of fungi, parasites, and DNA viruses (29). Furthermore, SMS has superior resolution and sensitivity in detecting species-level changes and predicting functional potential (28).
Seminal microflora of healthy men
A limited number of studies employing NGS have investigated the seminal microbiome of healthy men, often including them as part of a control group ( Table 1 ). Notably, two distinct features have emerged regarding the seminal microbiome: a wide variation in species composition among individuals and the formation of microbial communities dominated by particular species (2, 22).
Table 1.
Characteristics of studies examining the seminal microbiome in healthy men.
| Author, year, (country) | Design | Patients | Sample type | Technique | Main Phyla/Genera | Other findings | Study quality |
|---|---|---|---|---|---|---|---|
| Veneruso et al., 2023 (30) (Italy) |
Cross-sectional | 7 men with normal SA | Semen | Sequencing V4 – V6 hypervariable region of 16S rRNA gene | Proteobacteria were the most abundant phylum; Achromobacte, Staphylococcus, Gardnerella, and Serratia were the most abundant genera |
High | |
| Yao et al., 2022 (31) (China) |
Cross-sectional | 20 men with normal SA | Semen | Sequencing V3 - V4 hypervariable region of 16S rRNA gene |
Streptococcus, Lactobacillus, Burkholderia-Caballeronia-Paraburkholderia, Staphylococcus, Gardnerella
Lactobacillus-enriched group predominated in men with normal SA Streptococcus-enriched group predominated in men with leukocytospermia |
High | |
| Bukharin et al., 2022 (32) (Russia) |
Cross-sectional | 30 healthy men | Semen | Culture + Sequencing of 16S rRNA gene) | Staphylococcus, Corynebacterium, Enterococcus, Neisseria, Veillonella; | Fair | |
| Lundy et al., 2021 (33) (USA) |
Cross-sectional | 12 men with proven paternity | Semen Urine Rectal swab |
Sequencing V3 - V4 hypervariable region of 16S rRNA gene) | Urine and semen contained an abundance of Gardnerella and Corynebacterium compared to the rectum Decreased Veillonella, Prevotella and increased Pseudomonas, Pseudoxanthomonas, and Acidovorax in semen compared to urine |
Collinsella and Staphylococcus depleted in semen following vasectomy; Finegoldia in uncircumcised men |
High |
| Pagliuca et al., 2021 (16) (Italy) |
Cross-sectional | 16 men with normal SA | Semen | Culture positive if concentration > 10³cfu/m and PCR |
Staphylococcus coagulase negative, Enterococcus faecalis, Streptococcus anginosus, Streptococcus agalactiae, Staphylococcus aureus | High | |
| Okwelogu et al., 2021 (34) (Nigeria) |
Cohort | 11 men with normal basic SA | Semen | Sequencing V4 hypervariable region of 16S rRNA gene) | Lactobacillus, Gardnerella, Veillonella, Corynebacterium, Escherichia, Haemophilus, Prevotella | Couples shared 56% of the predominant genera; Couples with clinical pregnancy after IVF had increased abundance of Lactobacillus jensenii, and Faecalibacterium, whereas Proteobacteria, Prevotella,and Bacteroidetes were decreased |
High |
| Yang et al., 2020 (35) (China) |
Cross-sectional | 58 healthy controls | Semen | Sequencing V1 and V2 hypervariable region of 16S rRNA gene) | Pelomonas, Propionibacterium, Boseagenosp, Bosea, Afipia, Sphingomonas, and Vogesella | Fair | |
| Baud et al., 2019 (36) (Switzerland) |
Cross-sectional | 26 men with normal SA | Semen | Sequencing V1 and V2 hypervariable regions of 16S rRNA gene) | Overall: Actinobacteria, Bacteroidetes, Firmicute, and Proteobacteria phyla;
Staphylococcus genus was significantly more abundant in the normal SA group; Lactobacillus genus was enriched in samples with normal morphology |
Three broad microbiota profiles identified: Prevotella-dominant, Lactobacillus-dominant, and Polymicrobial |
High |
| Alfano et al., 2018 (37) (Italy) |
Cross-sectional | 5 men with normal spermatogenesis who underwent orchiectomy | Testicular tissue | Sequencing V3 to V5 hypervariable regions of 16S rRNA gene) | Normal germline: Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria | High | |
| Zeyad et al., 2018 (38) (Germany) |
Cross-sectional | 55 non-bacteriospermic men | Semen | Culture: positive if concentration > 10³cfu/ml |
S. aureus (9%), E. coli (7%), S epidermidis (6%), S haemolyticus (5%), E. faecalis (5%), and S. agalactiae (2%) |
No significant DFI differences in men with bacteriospermia; Decreased fertilization in men with bacteriospermia (p<0.05) |
High |
| Chen et al., 2018 (39) (China) |
Cross-sectional | 5 fertile semen donors | Semen | Sequencing V4 hypervariable regions of 16S rRNA gene | Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria were the predominant phyla Lactobacillus, Prevotella, Proteus, Pseudomonas, and Veillonella were the dominant genera |
Alicyclobacillus, Amaricoccus, Anaeromyxobacter, Aquicella, Arsenicicoccus were decreased when compared to men with azoospermia | High |
| Monteiro et al., 2017 (40) (Portugal) |
Cross-sectional | 29 men with normal basic SA | Semen (pooled by subgroups) |
Sequencing V3 to V6 hypervariable regions of 16S rRNA gene) | Overall: Enterococcus, Staphylococcus, Anaerococcus, Corynebacterium, Peptoniphilus, and Propionibacterium |
Corynebacterium, Haemophilus, and Streptococcus
at considerable abundancies (>1%) |
High |
| Vilvanathan et al., 2016 (41) (India) |
Cross-sectional | 47 men with normal sperm count | Semen | Culture: positive if concentration > 10³cfu/ml | Overall: E. faecalis (30%), Coagulase negative Staphylococcus (23.3%), Staphylococcus aureus (20%), E. coli (10%), Klebsiella pneumoniae (6.6%), Proteus sp (6.6%), and Citrobacter sp (3.3%) |
High | |
| Fraczek et al., 2016 (42) (Poland) |
Cross-sectional | 30 normozoospermic men without bacteriospermia or leucocytospermia | Semen | Culture: positive if concentration > 10⁴cfu/ml | Coagulase-negative Staphylococcus (22.9%), Streptococcus spp (12.3%), Enterococcus spp (13.8%), Mycoplasma spp (4.6%);
Gram+ aerobic (16.5%): Corynebacterium glucuronolyticum-seminale, C. striatum, and C. propinquum Gram-negative aerobic (3.7%): Escherichia coli, and Proteus mirabilis) Gram+ anaerobic (6.4%): Propionibacterium acnes, P. propionicum, P. avidum, and Bifidobacterium sp. Gram negative anaerobic (13.8%): Bacteroides urealyticum, Prevotella melaninogenica, P. intermedia, and Fusobacterium varium; One sample: Candida albicans |
High | |
| Weng et al., 2014 (27) (China) |
Cross-sectional | 10 men with abnormal semen volume | Semen | Sequencing V4 hypervariable region of 16S rRNA gene | Overall: Lactobacillus, Pseudomonas, Prevotella, Gardnerella, Rhodanobacter, Streptococcus, Finegoldia and Haemophilus
Normal SA group: Lactobacillus, Pseudomonas, Gardnerella, Prevotella Rhodanobacter, and Streptococcus, |
High | |
| Hou et al., 2013 (43) (China) |
Cross-sectional | 19 healthy sperm donors | Semen | Sequencing V1 and V2 hypervariable regions of 16S rRNA gene | Overall: Ralstonia, Lactobacillus, Corynebacterium, Streptococcus, Staphylococcus, Prevotella, Finegoldia, and Anaerococcus |
Fair | |
| Kjaergaard et al., 1997 (44) (Denmark) |
Cross-sectional | 115 normozoospermic men | Semen | Culture positive if concentration >10³cfu/m and PCR |
Commensals: Ureaplasma urealyticum, Gardnerella vaginalis, Enterococcus faecalis, and Enterobacteriaceae |
No association between semen quality and semen microorganisms | High |
SA, semen analysis; DFI, Sperm DNA Fragmentation Index.
In one of the initial NGS-based studies, Hou et al. sequenced the V1-V2 regions of 16S rRNA genes, revealing that even in healthy sperm donors, semen harbors a more diverse bacteria population than sperm itself (43). Their findings highlighted Ralstonia, Lactobacillus, Corynebacterium, Streptococcus, and Staphylococcus as the most prevalent bacteria in seminal fluid; these bacteria are organized into six distinct communities based on species composition and structure.
Weng et al. employed sequencing of the V4 hypervariable region of the 16S rRNA gene to examine 36 semen samples with normal basic semen analysis parameters. Their study identified Lactobacillus, Pseudomonas, Gardnerella, Prevotella, and Rhodanobacter as the most common genera (27). Additionally, the bacterial communities formed three main clusters: Pseudomonas-predominant, Lactobacillus-predominant, and Prevotella-predominant, with Lactobacillus-predominant group being the most frequent in the normal samples.
Similarly, Baud et al., utilizing sequencing of the V1 and V2 hypervariable regions of the 16S rRNA gene, examined 26 samples from men undergoing fertility evaluation who had normal basic semen analysis parameters. Their findings revealed three distinct microbiota communities: a Lactobacillus-predominant group, a Prevotella-predominant group, and a polymicrobial group (36). Staphylococcus was associated with normal semen analysis parameters, while the Lactobacillus genus was enriched in samples with normal morphology.
Another study reported that Lactobacillus, Gardnerella, Veillonella, Corynebacterium, and Escherichia were the most prevalent genera in the semen of men with normal basic semen analysis results (34). Bukharin et al. analyzed the seminal microbiome composition in 30 healthy men, identifying Staphylococcus, Corynebacterium, Enterococcus, Neisseria, and Veillonella as the most prevalent genera (32). Moreover, Yao et al. examined semen samples of 20 men with normal basic semen analysis parameters, revealing the main genera as Streptococcus, Lactobacillus, Burkholderia-Caballeronia-Paraburkholderia, Staphylococcus and Gardnerella (31). Interestingly, a Lactobacillus-enriched group predominated among these men.
Conversely, Monteiro et al. reported a low prevalence of Lactobacillus and a high prevalence of Enterococcus in semen samples of men with normal basic semen analysis parameters, employing sequencing of the V3 to V6 hypervariable regions of the 16S rRNA gene (40). However, it is essential to note that all the samples used in this study were derived from leftovers of assisted reproduction procedures, making it possible that some cases might have involved male factor infertility. Correspondingly, Yang et al., using sequencing of the V1 and V2 hypervariable regions of the 16S rRNA gene, demonstrated that Pseudomonas, Propionibacterium, Boseagenosp, Bosea, and Afipia were the most prevalent genera in healthy men with normal basic semen analysis parameters (35). Intriguingly, these authors observed an increased abundance of Lactobacillus in men with abnormal semen analysis results.
Given the diverse microfluidic components of semen and the microbiome’s complexity, it is estimated that approximately 30% of microorganisms in the semen originate from the urethra microbiome (35). Furthermore, specific genera, such as Pseudomonas, Pseudoxanthomonas, and Acidovorax, are overrepresented in the seminal microbiome compared to the urethral microbiome, suggesting their origin from upstream anatomic compartments (33). Thus, the seminal microbiome represents a composite of the microbiomes of the testicular, epididymal, prostatic, vesicular, and urethral regions (33) ( Figure 1 ). Higher microbiota diversity in the gut, skin, and oral cavity is often considered beneficial for human health (45). Interestingly, data from studies regarding the male genital tract microbiome is heterogeneous. Some authors have suggested that greater microbiota diversity harms sperm health (32, 46, 47), while others have found that reduced seminal biodiversity is associated with poor semen quality (30, 39).
Figure 1.
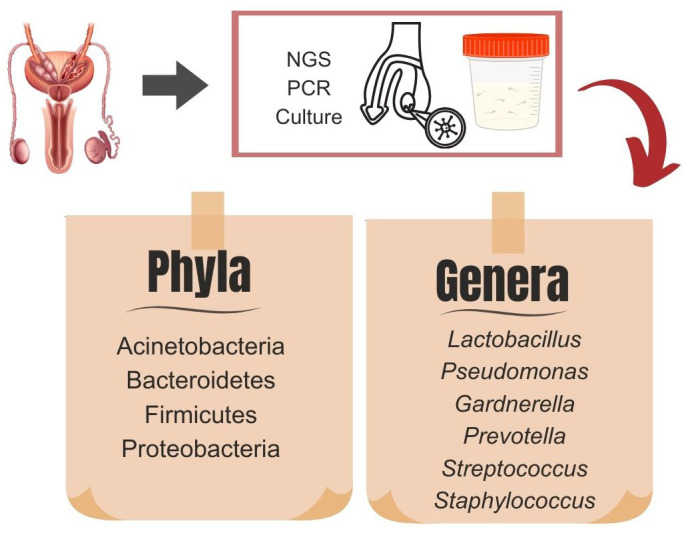
Dominant microbiota phyla and genera in testicular tissue samples and semen analysis obtained by existing diagnostic methods: next-generation sequencing (NGS), culture, and polymerase chain reaction (PCR).
Seminal microflora of men with altered semen quality
Most culture-based studies examining cohorts of infertile couples have failed to establish a conclusive link between the presence of bacteria in semen and abnormal semen analysis parameters (33, 41, 48–51). However, Ricci et al. reported reduced sperm motility in samples testing positive for microorganisms compared to negative samples (52). They also observed a negative association between E. faecalis and semen quality. Likewise, Zeyad et al. identified a negative impact of bacterial presence on sperm concentration and motility (38). Along these lines, Pagliuca et al. showed a significant correlation between infected status assessed by culture and PCR with semen volume, sperm concentration, and motility (16). Below, we summarize findings from studies using NGS to explore the microbiome of men with abnormal semen analysis parameters ( Figure 2 ; Table 2 ).
Figure 2.
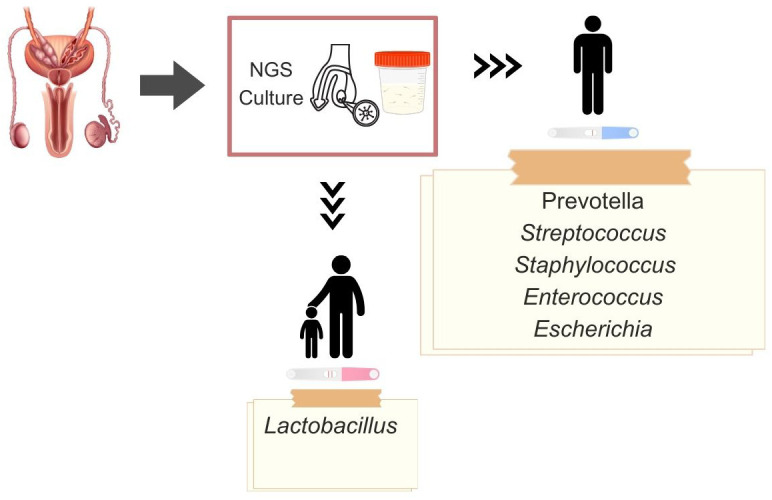
Dominant microbiota genera in fertile and infertile men, obtained by existing diagnostic methods: next-generation sequencing (NGS), culture, and polymerase chain reaction (PCR).
Table 2.
Characteristics of studies investigating semen/sperm microbiome in infertile men.
| Author, year, (country) | Design | Patients | Sample type | Technique | Main Phyla/Genera | Other findings |
Study quality |
|---|---|---|---|---|---|---|---|
| Veneruso et al., 2023 (30) (Italy) |
Cross-sectional | 13 men with abnormal SA | Semen | Sequencing V4 – V6 hypervariable region of 16S rRNA gene | Proteobacteria were the most abundant phylum; Lactobacillus, Escherichia, Shigella, and Serratia were the most abundant genera |
The genera Mannheimia, Escherichia_Shigella, and Varibaculum
were significantly increased in men with abnormal SA when compared to men with normal SA |
High |
| Yao et al., 2022 (31) (China) |
Cross-sectional | 13 men with asthenozoospermia, 22 men with leukocytospermia, and 32 men with asthenozoospermia and leukocytospermia; |
Semen | Sequencing V3 - V4 hypervariable region of 16S rRNA gene | Overall: Streptococcus, Lactobacillus, Burkholderia-Caballeronia-Paraburkholderia, Staphylococcus, and Gardnerella;
Lactobacillus-enriched group predominated in men with asthenozoospermia, whereas Streptococcus-enriched group predominated in men with leukocytospermia |
Diversity increased in men with leukocytospermia; Bacteroides were increased in men with leukocytospermia |
High |
| Bukharin et al., 2022 (32) (Russia) |
Cross-sectional | 42 infertile men with abnormal SA | Semen | Culture + Sequencing of 16S rRNA gene | Staphylococcus, Corynebacterium, Enterococcus, Streptococcus, and Escherichia | Fair | |
| Molina et al., 2021 (53) (Spain) |
Cross-sectional | 7 azoospermic men (13 samples), 3 men with high SDF (9 samples), and 1 man with severe OAT (2 samples) |
Testicular tissue | Sequencing V3 and V4 hypervariable regions of 16S rRNA gene | Blautia, Cellulosibacter, Clostridium XIVa, Clostridium XIVb, Clostridium XVIII, Collinsella, Prevotella, Prolixibacter, Robinsoniella, and Wandonia. | 50-70% contamination | High |
| Lundy et al., 2021 (33) (USA) |
Cross-sectional | 25 men with primary idiopathic infertility | Semen, Urine, and Rectal swab |
Sequencing V3 - V4 hypervariable region of 16S rRNA gene | Infertile group: Increased Aerococcus, Prevotella, Pseudomonas, and decreased Collinsella
Infertile group + varicocele: Bacteroids, Peptoniphilus |
Rectum of infertile men: decreased Anaerococcus and increased Lachnospiraceae, Collinsella, and Coprococcus;
Urine of infertile men: increased Anaerococcus; SAM cycle strongly over-represented in the urine and semen of infertile men |
High |
| Pagliuca et al., 2021 (16) (Italy) |
Cross-sectional | 37 men with abnormal SA | Semen | Culture positive if concentration > 10³cfu/m and PCR |
Staphylococcus coagulase negative, Haemophilus haemolyticus, Enterococcus faecalis, Haemophilus parainfluenzae, Gardnerella vaginalis | Bacteria were found more frequently in men with abnormal SA when compared to those with normal SA (70% vs 31%) | High |
| Okwelogu et al., 2021 (34) (Nigeria) |
Cohort | 36 male partners of infertile couples: 7 men with oligozoospermia, 7 men with azoospermia, 10 men with asthenozoospermia, and 1 man with teratozoospermia | Semen | Sequencing V4 hypervariable region of 16S rRNA gene | Oligozoospermia: Prevotella, Escherichia, Lactobacillus, Shuttleworthia, Serratia, Megasphaera, Gardnerella, Sneathia, Porphyromonas;
Azoospermia: Lactobacillus, Enterococcus, Corynebacterium, Veillonella, Gardnerella, Ureaplasma, and Prevotella |
Leukocytospermia: Increased Bacteroides and Prevotella;
Decreased Lactobacillus reuteri group, and Faecalibacterium |
High |
| Campisciano et al., 2020 (54) (Italy) |
Cohort | 47 male partners of infertile couples: 22 men with explained infertility, and 25 with unexplained infertility | Semen | Sequencing V3 hypervariable region of 16S rRNA gene | Overall: Prevotella
Explained Infertility group: Increased Prevotella (p. bivia and Staphylococcus); Unexplained Infertility group: Increased Lactobacillus gasseri |
Prevotella had a higher relative abundance in HPV-positive semen samples (25% vs. 17%) | High |
| Yang et al., 2020 (35) (China) |
Cross-sectional | 8 men with azoospermia, 58 men with asthenozoospermia, and 22 men with oligoasthenozoospermia |
Semen | Sequencing V1 and V2 hypervariable region of 16S rRNA gene | Men with asthenozoospermia had increased abundance of Sneathia, Ralstonia, Ureaplasma, Bacteroides, and Chryseobacterium Men with oligoasthenozoospermia had an increased abundance of Ralstonia, Oscillospira, Parabacteroides, Lachnospira, and Phascolarctobacterium |
Fair | |
| Baud et al., 2019 (36) (Switzerland) |
Cross-sectional | 68 men with abnormal SA | Semen | Sequencing V1 and V2 hypervariable regions of 16S rRNA gene | Prevotella genus was significantly enriched in the abnormal SA group | Three broad microbiota profiles identified: Prevotella-dominant, Lactobacillus-dominant, and Polymicrobial |
High |
| Ndiokwere et al., 2019 (55) (Nigeria) |
Cross-sectional | 22 semen samples from men undergoing fertility evaluation | Semen | Sequencing V4 hypervariable region of 16S rRNA gene | Serratia, Lactobacillus, Corynebacterium, Staphylococcus, and Prevotella | Species: Serratia marcescens Lactobacillus iners, Serratia entomophila, Haemophilus parainfluenzae, and Corynebacterium tuberculostearicum | Fair |
| Zeyad et al., 2018 (38) (Germany) |
Cross-sectional | 29 men with bacteriospermia | Semen | Culture: positive if concentration > 10³cfu/ml |
S. aureus (9%), E. coli (7%), S. epidermidis (6%), S. haemolyticus (5%), E. faecalis (5%), and S. agalactiae (2%) |
Bacteriospermia 34.5% of samples; Bacteriospermia associated with reduced sperm concentration and motility; Bacteriospermia not associated with increased DFI; Bacteriospermia associated with decreased fertilization |
High |
| Ricci et al., 2018 (52) (Italy) | Cross-sectional | 285 male partners of infertile couples | Semen | Culture positive if concentration > 10³cfu/m | Bacteriospermia in 29.1% of specimens; Staphylococcus aureus (0.7%), Enterococcus fecalis (11.6%), Streptococcus agalactiae (4.6%), Escherichia coli (6.7%), Streptococcus anginosus (0.3%), S. haemolyticus (2%), and U. urealyiticum (2%) |
Bacteriospermia associated with a decrease in total motility and progressive motility; Enterococcus fecalis associated with reduced sperm motility and morphology |
High |
| Chen et al., 2018 (39) (China) |
Cross | 6 men with OA; 6 men with iNOA |
Semen | Sequencing V4 hypervariable regions of 16S rRNA gene | Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria were the predominant phyla Lactobacillus, Prevotella, Proteus, Pseudomonas, and Veillonella were the dominant genera |
Solibacillus, Campylobacter, Campyiobacteraceae and Plesiomonas were reduced in the OA group; Sneathia and Lysobacter were reduced in iNOA group |
High |
| Alfano et al., 2018 (37) (Italy) |
Cross-sectional | 10 men with iNOA: 5 with positive sperm retrieval, and 5 with negative sperm retrieval |
Testicular tissue | Sequencing V3 to V5 hypervariable regions of 16S rRNA gene |
Actinobacteria
and Firmicutes |
Increased number of bacteria in the testis of iNOA men; Positive sperm retrievals: Actinobacteria and Firmicutes Negative sperm retrievals: Actinobacteria |
High |
| Zeyad et al., 2017 (56) (Germany) |
Cross-sectional | 36 men with bacteriospermia | Semen | Culture: positive if concentration > 10³cfu/ml |
Staphylococcus sp (15%; aureus, epidermidis, haemolyticus, xylosus); Escherichia coli (5%); Streptococcus spp (6%: agalactie, pneumoniae); Enterococcus faecalis (4%), and Klebsiella pneumoniae (1.6%) |
Bacteriospermia associated with reduced sperm concentration and motility; Neither morphology nor DFI was significantly impacted by bacteriospermia |
High |
| Monteiro et al., 2017 (40) (Portugal) |
Cross-sectional | 27 men with AT, 35 men with OAT, And 27 men with hyperviscosity |
Semen (pooled by subgroups) |
Sequencing V3 to V6 hypervariable regions of 16S rRNA gene | Overall: Enterococcus, Staphylococcus, Anaerococcus, Corynebacterium, Peptoniphilus, and Propionibacterium; OAT and Hyperviscosity groups: Cyanobacteria and Fusobacteria |
Lower prevalence of Lactobacillus and Propionibacterium; Higher prevalence of Pseudomonas, Klebsiella, Aerococcus, Actinobaculum, and Neisseria in OAT and hyperviscosity groups |
High |
| Vilvanathan et al., 2016 (41) (India) |
Cross-sectional | 37 men with oligozoospermia and 1 individual with azoospermia |
Semen | Culture: positive if concentration > 10³cfu/ml | Bacteriospermia in 35% of specimens; Overall: E. faecalis (30%), Coagulase-negative Staphylococcus (23.3%), Staphylococcus aureus (20%), E. coli (10%), Klebsiella pneumoniae (6.6%), Proteus sp (6.6%), and Citrobacter sp (3.3%) |
Presence of asymptomatic bacteriospermia not associated with abnormal semen parameters; Altered semen quality among different bacterial species lacked significant associations |
Fair |
| Mashaly et al., 2016 (57) (Egypt) | Cross-sectional | 60 infertile men: 30 without leukocytospermia (G1), and 30 with leukocytospermia (G2) | Semen | Culture: positive if concentration > 10.000 cfu/ml | G1: Corynebacterium (26.7%), Corynebacterium + E. coli (3.3%), Staphylococcus aureus (13.3%), Haemolytic streptococci + E.coli (3.3%); G2: Corynebacterium (10%), Corynebacterium + E.coli (0%), Staphylococcus aureus (10%), Haemolytic streptococci + E.coli (0%) |
Bacteriospermia in 33% of specimens; 20% Corynebacteria;
Sperm motility considerably lower in positive culture with Corynebacteria; Nonsignificant difference in sperm concentration and morphology between patients with Corynebacteria positive or negative cultures |
High |
| Ruggeri et al., 2016 (58) (Italy) | Cross-sectional | 246 male partners of infertile couples: 212 negative semen culture; 15 positive semen culture; 19 mixed flora | Semen | Not specified |
Enterococcus faecalis most common in both men (2.8%) and women (3.6%); Escherichia coli: men (0.8%) vs. women (3.2%); Ureaplasma urealyticum: 3.2% (men) |
High | |
| Fraczek et al., 2016 (42) (Poland) |
Cross-sectional | 30 normozoospermic men with isolated bacteriospermia; 22 normozoospermic with bacteriospermia and leukocytospermia; 19 normozoospermic with isolated leukocytospermia; |
Semen | Culture: positive if concentration > 10⁴cfu/ml | Coagulase-negative: Staphylococcus (22.9%), Streptococcus spp (12.3%), Enterococcus spp (13.8%), Mycoplasma spp (4.6%), Gram+ aerobic (16.5%), Corynebacterium glucuronolyticum-seminale, C. striatum, and C. propinquum; Gram negative aerobic (3.7%): Escherichia coli, and Proteus mirabilis); Gram+ anaeroibic (6.4%): Propionibacterium acnes, P. propionicum, P. avidum, Bifidobacterium sp.) Gram negative anaerobic (13.8%): Bacteroides ureolyticum, Prevotella melaninogenica, P. intermedia, and Fusobacterium varium One sample: Candida albicans |
Reduced sperm concentration in all groups compared to the control group; Significant sdeterioration of motility in the isolated leucocytospermia group; Necrozoospermia significantly higher in the combined bacteriospermia + leucocytospermia group; Teratozoospermia significantly higher in the isolated bacteriospermia group |
Fair |
| Mändar et al., 2015 (59) (Estonia) |
Cross-sectional | 23 infertile men | Semen | Sequencing V6 hypervariable region of 16S rRNA gene |
Lactobacillus, Flavobacterium, Prevotela, Porphyromonas, and Gardnerella;
The mean proportion of proteobacteria was higher in leukocytospermic men |
After intercourse, the seminal microbiome shifted the vaginal microbiome | High |
| Weng et al., 2014 (27) (China) |
Cross-sectional | 10 men with abnormal semen volume, 13 men with oligozoospermia, 12 men with asthenozoospermia, 44 men with teratozoospermia, 10 men with antisperm antibodies, And 18 men with; leukocytospermia |
Semen | Sequencing V4 hypervariable region of 16S rRNA gene | Abnormal SA group: Lactobacillus, Prevotella, Pseudomonas, Haemophilus, Finegoldia, Rhodanobacter, Corynebacterium and Streptococcus | High | |
| Sellami et al., 2014 (24) (Tunisia) | Cross-sectional | 85 infertile men | Semen | Culture positive if concentration > 10⁴cfu/m, and PCR | Bacteriospermia in 7% of specimens; Culture: Group B Streptococcus (3.5%), Enterococcus spp (1.1%), Staphylococcus aureus (1.1%), and Corynebacterium spp (1.1%); PCR: C. trachomatis (15.2%), N gonorrhea (5.8%), U. urealyticum (5.8%), M. genitalium (5.8%), U. parvum (5.8%), and M. hominis (5.8%) |
C. trachomatis associated with decreased sperm quality and increased apoptosis | High |
| Hou et al., 2013 (43) (China) |
Cross-sectional | 10 men with asthenozoospermia, 23 men with oligoasthenozoospermia, and 25 with oligozoospermia or azoospermia |
Semen | Sequencing V1 and V2 hypervariable regions of 16S rRNA gene) | Overall: Ralstonia, Lactobacillus, Corynebacterium, Streptococcus, Staphylococcus, Prevotella, Finegoldia, and Anaerococcus; No differences among the groups |
Anaerococcus had a negative association with sperm quality | Fair |
| Aghazarian et al., 2013 (50) (Iran) |
Cross-sectional | 171 men undergoing infertility evaluation | Semen | Not specified | Bacteriospermia in 36.2% of specimens; Ureaplasma urealyticum + Gardnerella vaginalis (25.8%), Ureaplasma urealyticum (19.4%), G. vaginalis (16.1%), Enterococcus faecalis (9.7%), E. coli + E. faecalis (1.6%) |
No significant association between bacteriospermia and leukocytospermia; No significant differences in semen parameters in men with bacteriospermia |
High |
| Domes et al., 2012 (51) (Canada) |
Retrospective cohort | 4935 samples from infertile men | Semen | Culture positive if concentration > 10³cfu/m |
Bacteriospermia in 15% of specimens; Staphylococcus aureus (5%), Enterococcus fecalis (56%), Escherichia coli (16%), Group B streptococcus (13%), Klebsiella pneumoniae (2.2%), Proteus mirabilis (1.7%), Citrobacter koseri (1.5%), and Morganella morganii (1.3%) |
Bacteriospermia associated with an increase in DFI; Elevated seminal leukocytes dominant factor associated with deterioration in semen parameters |
High |
| Isaiah et al., 2011 (60) (Nigeria) | Cross-sectional | 140 infertile men | Semen | Culture | Bacteriospermia in 65.7% of specimens; Staphylococcus aureus (28.3%), Staphylococcus saprohyticus (13%), Pseudomonas aerouginosa (6.5%), Escherichia coli (19.6%), Proteus mirabilis (10.8%), Staphylococcus spp (10.8%), and Proteus vulgaris (10.8%) |
Staphylococcus saprohyticus and Escherichia coli associated with altered sperm motility and morphology; Significant (p<0.001) relationship between bacteriospermia, leukocytes, and total sperm count |
High |
| Moretti et al., 2009 (61) (Italy) | Cross-sectional | 236 men with bacteriospermia | Semen | Culture: positive if concentration > 10⁴cfu/ml if gram + and > 10⁵cfu/ml if gram |
E. faecalis Bacteriospermia in 33.2% of specimens; (32.1%), E.coli (20.3%), Streptococcus agalactiae (13.4%), U. urealyiticum (11.8%), Staphylococcus epidermidis (9.7%), Streptococcus anginosus (9.3%), and Morganella morganii (3.2%) |
Sperm concentration lower than in controls; progressive motility lower than controls except for samples positive for S. agalactiae and S. anginosus | High |
| Gdoura et al., 2008 (62) (Tunisia) |
Cross-sectional | 166 men undergoing infertility evaluation | Semen | Culture and PCR | Overall: Chlamydia trachomatis (41.4%), Ureaplasma urealyticum (15.5%), and Mycoplasma hominis (10.3%) Culture: E. coli (1.7%), Streptococcus agalactiae (0.9%), Citrobacter diversus (0.9%), Enterococcus faecalis (0.9%), and Gardnerella vaginalis (0.9%) |
Bacteriospermia 56.9%; bacteria in 56% of semen samples by PCR; Bacteria in 5.2% semen samples by culture |
High |
| Virecoulon et al., 2005 (45) (France) |
Cross-sectional | 534 male partners of infertile couples | Semen | Culture: positive if concentration > 10³cfu/ml | Gardnerella vaginalis (26.1%), coagulase-negative staphylococci (15.7%), Streptococcus anginosus (14.2%), Ureaplasma urealyticum (15.5%), Enterobacteriaceae (E. coli, Proteus mirabilis), Corynebacterium spp, and Lactobacillus spp | Sterile in 28.8%; polymicrobial flora in 49.3%; No relationship between the bacterial flora and leukocytospermia; Low titers of U. urealyticum in semen were not associated with a disturbance of the ecosystem |
High |
| Levy et al., 1999 (24) (France) | Cross-sectional | 92 male partners of infertile couples | Semen | Culture positive if concentration > 10⁴cfu/m, and PCR | Culture: Ureaplama urealyticum (13%) PCR: Chlamydia trachomatis (11%) |
No relation between the presence of microorganisms in semen and serum antibodies | High |
| Debata et al., 1999 (63) (India) | Cross-sectional | 197 infertile men | Semen | Culture | Ureaplasma. urealyticum (43%), Mycoplasma hominis (17%) | No association between Ureaplasma and sperm count; Bacteriospermia associated with altered sperm morphology |
High |
| Kjaergaard et al., 1997 (44) (Denmark) |
Cross-sectional | 60 men with mild/moderate oligozoospermia and 26 men with severe oligozoospermia | Semen | Culture positive if concentration > 10³cfu/m, and PCR | Mild/moderate oligozoospermia: Commensals, Ureaplasm. Urealyticum, Gardnerella vaginalis, Enterococcus faecalis, Enterobacteriaceae, and Mycoplasma;
Severe oligozoospermia: Commensals, Ureaplasm. Urealyticum, Enterococcus faecalis, Gardnerella vaginalis, Enterobacteriaceae, and Mycoplasma |
No association between semen quality and microorganisms | High |
| Bussen et al., 1997 (49) (Italy) | Cross-sectional | 88 male partners of infertile couples Group 1: 28 negative culture + 14 positive culture for microorganisms that colonize skin (considered control group); Group 2: 46 positive cultures |
Semen | >100 colonies per plate | Bacteriospermia in 68% of specimens; S. epidermidis (33%): considered to be commensal S. aureus (9%); E. coli (8%); Enterobacter spp. (7%); Group B streptococcus (8%); Corynebacteria (8%) |
No differences in sperm concentration, count, sperm morphology, and fertilization rates between groups | High |
| Shalika et al., 1996 (64) (USA) | Cross-seccional | 342 male partners of infertile couples | Semen | Culture | Bacteriospermia in 32% of specimens; Culture: S. aureus (3%), Enterococcus spp (23%), Ureaplasma spp (11%), E. coli (3%), Proteus mirabillis (0.5%), and Streptococcus spp (2%) |
Enterococcus spp did not adversely affect IVF pregnancy rate; E. coli, S aureus, and Ureaplasma urealyticum potentially affecting IVF pregnancy rates |
High |
| Eggert-Kruse et al., 1995 (48) (Germany) |
Cross-sectional | 126 male partners of infertile couples | Semen | Culture: positive if concentration > 106cfu/ml |
Peptococcus sp (38.1%), Peptostreptococcus sp (32.5%), Veillonella spp (27.8%), Lactobacillus spp (20.6%), Bacterioides spp (7.9%: B. disiens, B.capillosus, B. ruminicola, B. bivius), Propionibacterium spp (7.1%), Fusobacterium spp (3.2%: F. varium, F. mortiferum, F. nucleatum), Gardnerella vaginalis (3.1%), and Actinomyces spp (1.6%: A. meyeri, A. viscosus); Gram-negative non-identified anaerobic rods (5.6%); Anaerobic bacteria not identified (11.9%) Mycoplasma hominis (6.1%); Ureaplasma urealyticum (21.2%) |
99% of samples colonized with anaerobic; 71% potentially pathogenic species; Potentially pathogenic aerobic microorganisms more frequent in oligozoospermia group; Bacteroides spp and Fusobacterium spp more frequent in the asthenozoospermia and teratozoospermia groups (not statistically significant) |
Fair |
AT, asthenoteratozoospermia; OA, Obstructive Azoospermia; OAT, oligoasthenoteratozoospermia; H, hyperviscosity; iNOA, idiopathic non-obstructive azoospermia; SA, semen analysis; SDF, Sperm DNA Fragmentation; DFI, sperm DNA fragmentation index; PCR, polymerase chain reaction; cfu, colony forming units; HPV, human papillomavirus; SAM, S-adenosyl-L-methionine.
Oligozoospermia
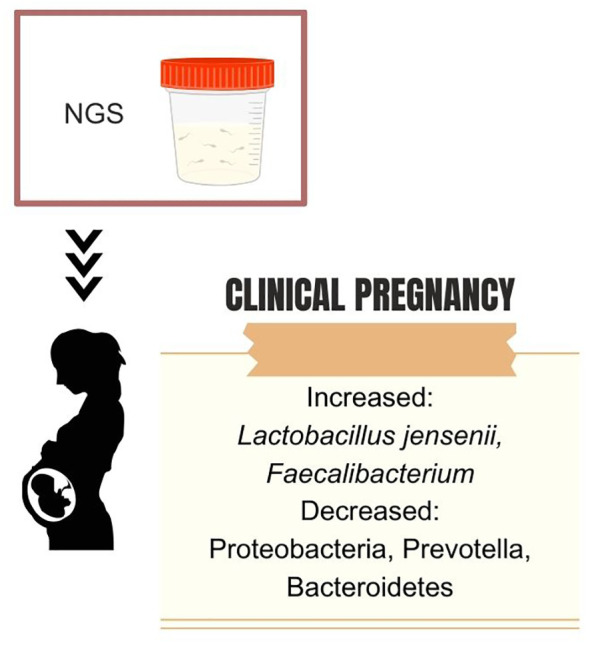
Oligozoospermia, characterized by a sperm concentration below the WHO reference limit (e.g., 16 x 106 sperm/mL) (9, 65), was associated with specific bacterial genera in the study of Okwelogu and colleagues (34). Prevotella, Escherichia, Lactobacillus, Shuttleworthia, and Serratia were the most abundant genera in oligozoospermic men (34). This observation was corroborated by Lundy and colleagues, who described an inverse association between seminal abundance of Prevotella and sperm concentration (33).
Asthenozoospermia
Bacterial presence in semen significantly affects motility (66), a critical component of a basic semen analysis assessment. Asthenozoospermia is typically defined as having less than 30% progressive spermatozoa or less than 42% total motility (9, 65, 67). Yang and colleagues showed that men with asthenozoospermia exhibited an increased abundance of Sneathia, Ralstonia, Ureaplasma, Bacteroides, and Chryseobacterium (35). Moreover, in men with oligoasthenozoospermia, the genera Ralstonia, Oscillospira, Parabacteroides, Lachnospira, and Phascolarctobacterium were more abundant. Notably, the authors reported an increased prevalence of Lactobacillus in men with astheno- or oligoasthenozoospermia compared to controls with normal basic semen analysis parameters, suggesting Lactobacillus as a potential bacterial biomarker for asthenozoospermia (receiver operating characteristics value of 0.841) (35). Similarly, Yao and colleagues found a Lactobacillus-enriched seminal microbial community prevailing in men with asthenozoospermia (31). In semen samples from men undergoing in vitro fertilization (IVF), Štšepetova and colleagues noted negative associations between sperm motility and the phyla Bacteroidetes and Proteobacteria, as well as the classes Alphaproteobacteria and Sphingobacteria (68). In contrast, another study found that the seminal abundance of Pseudomonas, a proteobacteria, was directly associated with total motile sperm count (33).
Oligoasthenoteratozoospermia
Oligoasthenoteratozoospermia (OAT), characterized by abnormalities in the three primary semen analysis parameters (i.e., sperm concentration, motility, and morphology) (9, 67), is indicative of severe impairment of spermatogenesis and is linked to reduced chances of natural pregnancy (69). Monteiro and colleagues associated OAT with the presence of Cyanobacteria and Fusobacteria, an increased prevalence of Pseudomonas, Klebsiella, Aerococcus, Actinobaculum, and Neisseria, as well as a decreased prevalence of Lactobacillus and Propionibacterium (40).
Azoospermia
Azoospermia is the lack of spermatozoa in the ejaculate (70). Examining men undergoing IVF, Okwelogu and colleagues found Lactobacillus, Enterococcus, Corynebacterium, Veillonella, and Gardnerella were the most abundant genera in azoospermic men (34). However, the authors did not specify the cause of azoospermia.
Semen quality in general
Several studies investigated the microbiome in men with abnormalities in any basic semen analysis parameters, often referred to as low-quality semen. Hou et al. found no significant differences in the seminal bacterial composition between healthy semen donors and infertile men with abnormal basic semen analysis parameters (43). However, they did observe a negative association between semen quality and the presence of Anaerococcus. In contrast, Weng et al. demonstrated that a Prevotella-predominant bacterial community was associated with low-quality semen (27). Similarly, Baud et al. found that the Prevotella genus was significantly enriched in the semen of men with abnormal semen analysis parameters (36).
Furthermore, Lundy et al. reported an increased prevalence of Aerococcus and decreased Collinsella in semen samples from infertile men compared to fertile controls (33). In this study, male infertility was defined as the presence of altered basic semen parameters and an inability to father a child after 12 months of trying. Along these lines, Bukharin et al., also studying infertile men, demonstrated that Staphylococcus, Corynebacterium, Enterococcus, Streptococcus, and Escherichia were the most prevalent genera in the semen (32).
Leukocytospermia
Leukocytospermia, characterized by the presence of >1.0 million leukocytes per mL of semen (71), is classically associated with genitourinary tract infection since bacteriospermia can trigger the recruitment of seminal leukocytes (10). However, other conditions, such as exposure to vaginal products during intercourse, smoking, genitourinary procedures, and autoimmunity, may increase the number of leukocytes in semen (51). In most studies employing standard culture techniques, the presence of bacteria in the semen of asymptomatic men was not associated with an increase of seminal leukocytes (45, 50–52), even when leukocytospermia was defined with low cutoff values (e.g., 0.2 x 106 leukocytes/mL) (62). Despite that, some authors have reported associations between leukocytospermia and bacteriospermia (38, 44, 60, 61). For instance, Yao et al., using NGS to assess the seminal microbiome, reported that a Streptococcus-enriched bacterial community predominated in men with leukocytospermia (31). The authors also found an increased prevalence of Bacteroidetes associated with leukocytospermia. Additionally, Štšepetova et al. showed that in men undergoing IVF, Staphylococcus sp. was associated with leukocytospermia (68). However, Lundy and colleagues found an inverse association between Aerococcus abundance and leukocytospermia. Nevertheless, when comparing the seminal microbiome between infertile men with and without leukocytospermia, no differences were observed in measures of bacterial diversity (33).
Oxidative stress and sperm DNA fragmentation
Oxidative stress and sperm DNA fragmentation are commonly observed in infertile men and may result from the activation of seminal leukocytes (19, 72–75). When using NGS to evaluate the seminal microbiome of men with elevated oxidative stress (oxidation-reduction potential >1.34 mV/106 sperm/ml), Lundy et al. reported modest differences in three taxa (Serratia, Streptococcus and Curvibacter) (33). Conversely, using culture-based methods, Zeyad and colleagues did not find differences in sperm DNA fragmentation levels between men with or without bacteriospermia (38). However, in a large study including nearly 5,000 infertile men, Domes et al. identified a negative association between culture-positive semen and sperm DNA integrity (41). In line with this, when assessing only healthy men with normal basic semen analysis parameters, Fraczek and colleagues reported increased sperm DNA damage in those with positive semen culture but no increase in oxidative stress markers (42).
Inflammatory markers
Bacteria in the genitourinary tract may lead to inflammatory responses mediated by various cytokines produced by leukocytes (72). Hence, it is reasonable to assume that the seminal microbiome can influence the production of these inflammatory mediators. Bukharin et al. demonstrated that Staphylococcus isolated from the semen of healthy men degraded IL-10 and IL-17 more intensely than those from the semen of infertile men (32). Additionally, Enterococcus from infertile men reduced IL-1 levels, and Corynebacterium from these individuals reduced TNF-α levels to a greater extent than those isolated from healthy subjects (32). These findings suggest that the seminal microbiome can influence the host’s inflammatory response, at least locally. By contrast, culture-based studies did not establish an association between the presence of bacteria in semen and seminal levels of inflammatory markers (50, 76).
Epididymal and testicular microflora
Evaluating the epididymal or testicular microbiome requires harvesting biofluids or tissue samples from these organs. In this context, Alfano et al. conducted a study using testicular samples from men with idiopathic non-obstructive azoospermia (iNOA) and normozoospermic men who underwent orchiectomy (37). Employing NGS to sequence the V3 to V5 hypervariable regions of the 16S rRNA gene with NGS, the authors made a groundbreaking discovery, revealing that the testicular compartment is not sterile. In the testicular tissue of men with normal spermatogenesis, they identified the phyla Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria. However, despite the increased presence of bacterial DNA in testicular samples from men with iNOA, only the phyla Actinobacteria and Firmicutes were identified in these samples.
Furthermore, the testicular microbiome of NOA men with complete germline cell aplasia exhibited reduced bacterial richness and diversity, with a dominance of the Actinobacteria phylum and the absence of Clostridia. Similarly, Molina et al. utilized testicular samples from sperm retrieval procedures in men with azoospermia, severe oligoasthenozoospermia, or high DNA fragmentation to study the testicular microbiome (53). The authors also observed low levels of bacteria and identified ten genera specific to the testicles, including Blautia (phylum Firmicutes), Cellulosibacter (Firmicutes), Clostridium XIVa (Firmicutes), Clostridium XIVb (Firmicutes), Clostridium XVIII (Firmicutes), Collinsella (Actinobacteria), Prevotella (Bacteroidetes), Prolixibacter (Bacteroidetes), Robinsoniella (Firmicutes), and Wandonia (Bacteroidetes). Notably, despite stringent antiseptic measures, contamination accounted for 50–70% of all detected bacterial reads, suggesting that sperm retrieval from the testes is not performed under sterile conditions (53).
Using a different strategy, Lundy et al. demonstrated that Collinsella and Staphylococcus were prevalent in semen samples from healthy fertile men and depleted in samples from men who underwent vasectomy, implying that these two genera are constituents of the testicular or epididymal microbiome (33). To date, no studies have examined the epididymal microbiome.
Other factors that can affect the seminal microbiome
Diet (gut microbiome)
High-fat (HFD) and high-sugar “Western” diets have been associated with obesity, metabolic disorders, and alterations in gut microbiota composition in both humans and animals (3). However, the impact of HFD-induced dysbiosis on reproductive function remains unclear. In a study by Zhang et al., significant differences in the bacterial composition of the gut microbiota were observed between the HFD and normal diet groups (77). The HFD was associated with a decreased abundance of Bacteroidetes and Verrucomicrobia and an increased abundance of Firmicutes and Proteobacteria (3). Notably, the HFD resulted in reduced sperm concentration and motility, along with a decrease in spermatocyte and round spermatid numbers. Analysis of the gut microbiota in the HFD group revealed an increased abundance of Bacteroides, Prevotella, Rikenella, and Lactobacillus. The authors also analyzed fecal samples from healthy semen donors and infertile men with asthenozoospermia, oligozoospermia, and teratozoospermia, demonstrating a similar strong negative correlation between sperm motility and the combined abundance of Bacteroides and Prevotella. Moreover, Prevotella copri, a dominant species within Prevotella, was implicated in spermatogenic defects. These findings suggest a potential role of HFD-induced gut microbiota dysbiosis in impairing spermatogenesis and sperm motility.
Sexual habits
A culture-based study demonstrated that men who never had sexual intercourse exhibited lower total seminal bacterial concentration and diversity than sexually active men (78). Nelson et al. applied sequencing of the V1-V9 sub-regions of 16 S rRNA alleles to evaluate the coronal sulcus microbiome from eighteen healthy 14–17 year-old teens (79). The authors reported that some taxa associated with bacterial vaginitis including Mycoplasma, Ureaplasma, and Sneathia were detected only in participants with sexual experience, mainly vaginal intercourse and fellatio. Moreover, studying men who have sex with men, Liu et al. observed that bacteria in the semen of these men overlapped with those previously described in the vagina, including Streptococcus, Corynebacterium, Staphylococcus, Prevotella and Mycoplasma (80). These finding suggest that partnered sexual activity influences on the composition of the seminal microbiome. Unfortunately, there is no data in the current literature regarding the association of specific modalities of sexual activity to changes in the seminal microbiome.
Sexually transmitted infections
Human papillomavirus (HPV) infection has been associated with reduced semen quality (17, 79, 81). The mechanisms underlying this association are unclear but may include apoptosis of sperm cells, sperm DNA damage, and the production of antisperm antibodies. Additionally, HPV-positive semen samples exhibited higher Moraxellaceae, Streptococcus, and Peptostreptococcus abundances than HPV-negative semen samples (80). Notwithstanding these observations, the authors of the above study did not perform semen analysis to assess the impact of these alterations on semen quality.
Furthermore, Human Immunodeficiency Virus (HIV) has been shown to induce changes in the seminal microbiome. Liu et al. demonstrated that men with HIV infection had decreased semen microbiome diversity and richness, which were restored after six months of antiretroviral therapy (82). The semen bacterial load was associated with pro-inflammatory semen cytokines and semen viral load, suggesting a role of the semen microbiome in HIV sexual transmission (82).
Impact of seminal microbiome on the female genital tract
The seminal microbiome influences the microflora of the female genital tract. A well-established example of such influence is the association between bacterial vaginosis, a dysbiotic condition, and frequent vaginal intercourse (83). A study examining risk factors for bacterial vaginosis in women with and without HIV infection demonstrated that the presence of spermatozoa in Gram-stained vaginal smear samples, which serves as a biological marker of recent exposure to semen, was the only common factor in both groups (84).
Furthermore, the production of H2O2 by certain vaginal lactobacilli is essential for maintaining a healthy vaginal environment (85). Having more than two sexual partners during the past year has been identified as a risk factor for the absence of H2O2-producing lactobacilli among women with bacterial vaginosis (86). Several studies that simultaneously assessed the seminal and vaginal microbiomes of sexual partners have confirmed this finding. For instance, Okwelogu et al. found that couples shared 56% of predominant genera, suggesting that the composition of the reproductive tract microbiota, whether healthy or dysbiotic, could influence the microbial composition of their sexual partners (34). Similarly, Campisciano et al. demonstrated that couples with infertility of known causes exhibited an overlap in the bacterial composition of their seminal and vaginal microbiomes, including an increased prevalence of Staphylococcus and Streptococcus genera (54). The authors also noted a higher abundance of Lactobacillus gasseri in the semen of couples with unexplained infertility than those with explained infertility. Using PCR and culture-based techniques, Borovkova et al. found that up to seven new species could be introduced, and the same number removed from vaginal microflora after intercourse (87). Additionally, using culture-based methods, Ricci et al. found that 61% of couples with positive cultures in both partners shared at least one genital pathogen (52).
Impact of seminal microbiome on the reproductive outcomes
Natural pregnancy
Early studies utilizing culture-based methods failed to identify differences in the microbial patterns in the ejaculate of men from couples who achieved natural pregnancy compared to those who did not (49, 64, 76). In a study by Eggert-Kruse et al., anaerobic and “potentially pathogenic” bacteria were cultured in 94.7% and 84.2% of the fertile men, respectively. Furthermore, there was no association between microbial colonization and natural pregnancy after a 6-month follow-up (76).
Assisted reproduction technology
Semen and vaginal cultures are typically carried out before assisted reproduction technology (ART). However, interpreting positive cultures in asymptomatic patients can be challenging due to the possibility of contamination. Nonetheless, specific pathogens, such as E. faecalis, U. urealyticum, M. hominis, G. vaginalis, and E. coli, were more prevalent in the genital tract of couples that had experienced IVF failure (52). Additionally, Zeyad et al. reported a weak negative correlation between bacteriospermia and fertilization rates in couples undergoing IVF (r=−0.239, p<0.05) (38). Interestingly, sperm preparation techniques like swim-up and density gradient (88), commonly used to process semen for ART, can reduce bacteria counts in asymptomatic infertile men, but total clearance is rarely achieved (89). Thus, it seems evident that ART is commonly performed in a non-sterile environment despite taking precautions to prevent sample and equipment contamination (90).
NGS studies corroborate this idea and have reported associations between specific types of seminal microbiome and in vitro fertilization/intracytoplasmic sperm injection (IVF/ICSI) outcomes ( Figure 3 ). Štšepetova et al. investigated the microbiome of raw semen, processed semen, incubated sperm, and IVF culture media from 50 couples undergoing IVF (68). The authors utilized sequencing of the V2 and V3 hypervariable region of the 16S rRNA gene and real-time PCR. They observed decreasing bacterial reads count as semen samples underwent processing (i.e., raw > washed >incubated). The most abundant genera of bacteria in raw semen were Lactobacillus, Incertae sedis XI, Staphylococcus, and Prevotella. Processed semen samples exhibited a more heterogeneous microbial composition. Higher counts of Alphaproteobacteria and Gammaproteobacteria in washed sperm, as well as Corynebacterium sp. in raw semen samples, were associated with reduced embryo quality. Conversely, couples with increased embryo quality had a higher mean proportion of the Enterobacteriaceae group in raw semen ( Figure 4 ). Bacterial reads were detected in IVF culture media in 8% of the samples via NGS and more than 70% by the real-time PCR method, with Lactobacillus and Phyllocterium being the most frequent genera.
Figure 3.
Dominant seminal microbiota genera associated with pregnancy success after assisted reproductive technology.
Figure 4.
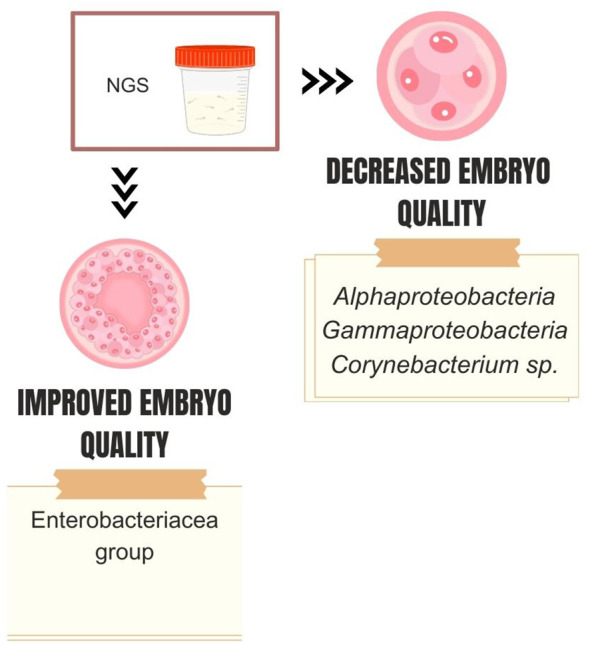
Dominant seminal microbiota genera associated with embryo quality.
Nevertheless, bacteria in IVF culture media do not seem to influence pregnancy rates. Utilizing sequencing of the V4 region of the 16S rRNA, Okwelogu et al. demonstrated that semen samples from couples with clinical pregnancy after ICSI exhibited increased colonization by Lactobacillus jensenii group and Faecalibacterium, along with a decreased prevalence of Proteobacteria, Prevotella, Bacteroidetes taxa compared to those with adverse outcomes (34). Conversely, a study applying sequencing of variable regions 3 and 4 of the 16S rRNA found no differences in seminal microbiome composition and diversity between male partners of couples that had or did not have a successful pregnancy after intrauterine insemination (91). On the female side, a recent analysis of the endometrial microbiome of women undergoing IVF demonstrated that 73.9% of the endometrial samples assessed with NGS were colonialized by one or more microbes, further highlighting the fact that human reproduction often happens in the presence of a bacterial microbiota (92). Figure 5 summarizes the main bacterial phyla or genera associated with fertility status and outcomes.
Figure 5.
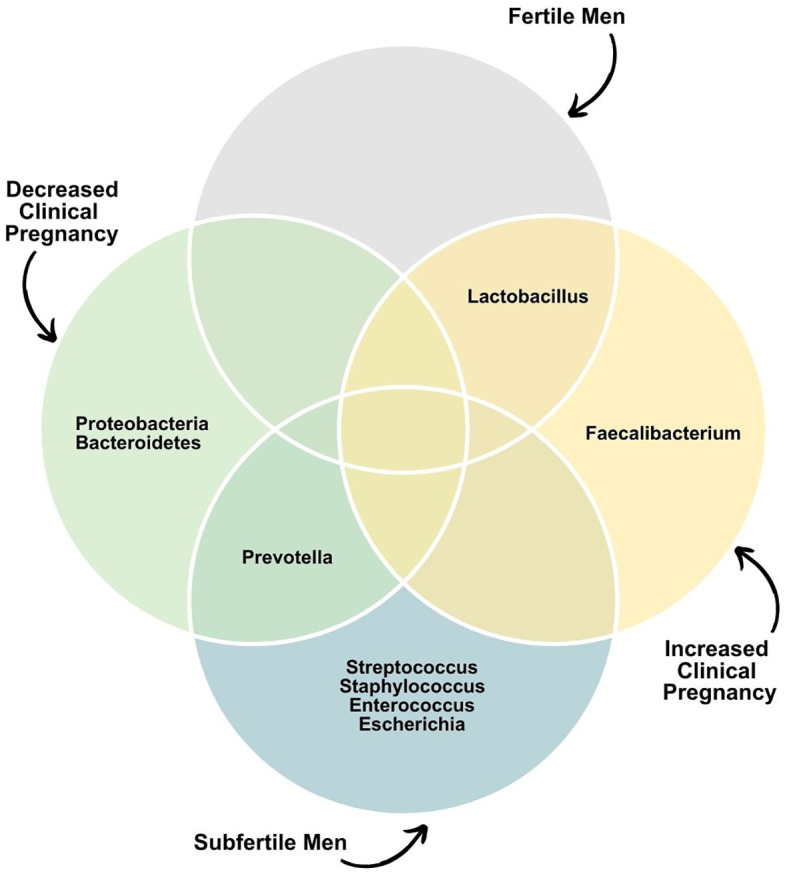
Dominant seminal microbiota phyla or genera associated with fertility status and outcomes.
Future directions and recommendations for semen microbiome studies
The emerging field of seminal microbiota research has illuminated the intricate microbial communities residing in the male reproductive tract. While significant progress has been made in characterizing these microorganisms and their potential functions of these microorganisms, several critical areas detailed below merit further exploration (93).
1. Standardized Protocols: To ensure the reliability and reproducibility of results, it is imperative to establish standardized protocols for sample collection, handling, DNA extraction, and NGS analysis. Future research should formulate guidelines and best practices to mitigate technical variations and biases that may arise during these processes. This will facilitate robust comparisons between studies and enable the integration and comparison of findings across different research groups.
2. Optimal Variable Regions: The choice of variable regions within the 16S rRNA gene for sequencing can impact the accuracy and resolution of results. Future studies should aim to pinpoint the most informative variable regions specific to the seminal microbiota. This will help establish a standardized sequencing approach, enhancing comparability across studies and facilitating meta-analyses.
3. Shotgun Metagenomics: While most research has focused on bacterial communities, the seminal microbiota likely includes viruses and fungi. Future investigations should leverage shotgun metagenomics approaches to comprehensively identify and characterize these components. This will provide a complete understanding of the microbial landscape and its potential role in male infertility and semen abnormalities.
4. Contamination Mitigation: Contamination poses a significant challenge in microbiota research. Future studies should prioritize stringent measures to prevent and detect contamination at each experimental stage. This includes incorporating negative controls during sample collection, DNA extraction, library preparation, and sequencing. Stringent quality control measures will enhance the reliability of results and minimize the impact of potential contamination on data interpretation.
5. Pathogenic strain determination: It is known that some bacteria species can have pathogenic and non-pathogenic strains. Thus, it is of utmost importance to differentiate the strains that have the potential to cause harm from those that are commensals. This subtyping can be done using NGS (64), and coupled with data from databases such as the National Center for Biotechnology Information Pathogen Detection, this approach can better identify “friends and foes”.
6. Functional role of seminal microbiota. Few studies have delved into the role played by the male genital tract microbiome and who it interacts with spermatogenesis, but it is plausible that this microbiota regulates the immune microenvironment of testis, playing a role in providing nutrients, regulating the testicular immune microenvironment, and modulating signal transduction (88, 89). To advance in this field, further studies should focus not only on describing the components of the seminal microbiome, but also to assess their function in this system by using in vitro and ex vivo experimental systems for studying host–microbiome interactions, similar to what has been used to study gut and respiratory microbiomes (90).
7. Multi-site Investigation: Given that the seminal microbiome likely originates from multiple sites within the reproductive tract, simultaneous assessment of the microbiome composition of each of these organs (i.e., testis, epididymis, vas deferens, prostate, seminal vesicles, urethra, and penis) may provide deeper insights into their relevance to male infertility conditions, enabling more tailored treatment.
8. Longitudinal Studies: Longitudinal research is crucial for capturing dynamic changes in the seminal microbiota over time and understanding its potential impact on male fertility. Future research should prioritize longitudinal study designs to explore temporal variations in the microbiota composition and function within individuals and across different stages of reproductive health. This will elucidate the role of seminal microbiota in physiological and pathological conditions and its contribution to infertility and semen alterations.
9. Prospective Studies: Prospective studies are necessary to establish a direct link between seminal microbiota and reproductive outcomes. These studies should involve monitoring the male reproductive tract microbiota in men attempting natural conception or undergoing ART. Researchers might unravel the seminal microbiome’s potential impact on fertility and ART success by correlating microbiota profiles with pregnancy rates, embryo development, and other ART outcomes.
10. Impact of Male Infertility Causes: Male infertility can stem from various causes, including genetic factors, hormonal imbalances, infections, and structural abnormalities. Future research should investigate the specific influence of different infertility causes on the seminal microbiome. This will shed light on whether distinct microbial signatures are linked to specific infertility etiologies and guide the development of targeted therapeutic strategies tailored to individual patients.
11. Effects of Commonly Prescribed Drugs: Several drugs and treatments are commonly prescribed for male infertility management, such as vitamin supplements, antibiotics, and hormonal therapy. Future research should explore whether and how these interventions impact the seminal microbiome. Understanding the effects of these therapeutic agents on microbial communities will provide insights into their potential contributions to fertility outcomes and guide the development of more personalized treatment regimens.
Conclusions
The exploration of the seminal microbiome has opened a fascinating realm of research, shedding light on the intricate microbial communities residing within the male reproductive tract. This emerging field has revealed a complex interplay between these microorganisms and male fertility, semen quality, and their potential influence on female reproductive health. The evidence compiled from various studies using culture-based and NGS techniques has provided valuable insights into these microbial communities’ composition, dynamics, and potential functions. One of the key takeaways from this review is the pressing need for standardized protocols and best practices in sample collection, DNA extraction, and NGS analysis. By establishing rigorous methodologies, the scientific community can ensure the reliability and reproducibility of results, fostering more robust comparisons between studies and facilitating meta-analyses. Additionally, the choice of variable regions within the 16S rRNA gene for sequencing and the application of shotgun metagenomics for a comprehensive assessment of viruses and fungi within the seminal microbiota are vital considerations for future research. Prospective and longitudinal studies and investigations into the impacts of various male infertility causes and commonly prescribed drugs hold promise for unraveling the intricate relationships between the seminal microbiome and male reproductive outcomes. This knowledge enhances our understanding of male fertility and paves the way for personalized interventions and treatments tailored to individual patients. Exploring the seminal microbiome represents a dynamic and rapidly evolving field poised to advance our comprehension of male reproductive health and potentially revolutionize clinical approaches to male infertility and semen alterations.
Author contributions
FL: Writing – original draft. MV: Writing – original draft. FC: Funding acquisition, Writing – review & editing. AC: Funding acquisition, Writing – review & editing. CA: Funding acquisition, Writing – review & editing. SE: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by an unrestricted research grant from the Department of Neuroscience, Reproductive Science and Odontostomatology, University of Naples, Federico II, Naples, Italy.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AG declared a shared affiliation with the author(s) FC, AC, CA to the handling editor at the time of review.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. (2016) 164(3):337–40. doi: 10.1016/j.cell.2016.01.013 [DOI] [PubMed] [Google Scholar]
- 2. Altmäe S, Franasiak JM, Mändar R. The seminal microbiome in health and disease. Nat Rev Urol. (2019) 16(12):703–21. doi: 10.1038/s41585-019-0250-y [DOI] [PubMed] [Google Scholar]
- 3. Zhang F, Aschenbrenner D, Yoo JY, Zuo T. The gut mycobiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. Lancet Microbe (2022) 3(12):E969–E983. doi: 10.1016/S2666-5247(22)00203-8 [DOI] [PubMed] [Google Scholar]
- 4. Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect (2008) 14(10):908–34. doi: 10.1111/j.1469-0691.2008.02070.x [DOI] [PubMed] [Google Scholar]
- 5. Methé AB, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower C. A framework for human microbiome research. Nature. (2012) 486(7402):215–21. doi: 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. The NIH human microbiome project. Genome Res (2009) 19(12):2317–23. doi: 10.1101/gr.096651.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. García-Velasco JA, Menabrito M, Catalán IB. What fertility specialists should know about the vaginal microbiome: a review. Reprod BioMed Online. (2017) 35(1):103–12. doi: 10.1016/j.rbmo.2017.04.005 [DOI] [PubMed] [Google Scholar]
- 8. Drabovich AP, Saraon P, Jarvi K, Diamandis EP. Seminal plasma as a diagnostic fluid for male reproductive system disorders. Nat Rev Urol. (2014) 11(5):278–88. doi: 10.1038/nrurol.2014.74 [DOI] [PubMed] [Google Scholar]
- 9. Esteves SC. Evolution of the World Health Organization semen analysis manual: where are we? Nat Rev Urol (2022) 19(7):439–46. doi: 10.1038/s41585-022-00593-2 [DOI] [PubMed] [Google Scholar]
- 10. Esteves SC. Who cares about oligozoospermia when we have ICSI. Reprod BioMed Online. (2022) 44(5):769–75. doi: 10.1016/j.rbmo.2021.11.026 [DOI] [PubMed] [Google Scholar]
- 11. Esteves SC, Humaidan P. Towards infertility care on equal terms: a prime time for male infertility. Reprod BioMed Online. (2023) 47(1):11–4. doi: 10.1016/j.rbmo.2023.04.003 [DOI] [PubMed] [Google Scholar]
- 12. Esteves SC, Achermann APP, Simoni M, Santi D, Casarini L. Male infertility and gonadotropin treatment: What can we learn from real-world data? Best Pract Res Clin Obstet Gynaecol (2023) 86:102310. doi: 10.1016/j.bpobgyn.2022.102310 [DOI] [PubMed] [Google Scholar]
- 13. Villegas J, Schulz M, Soto L, Sanchez R. Bacteria induce expression of apoptosis in human spermatozoa. Apoptosis. (2005) 10(1):105–10. doi: 10.1007/s10495-005-6065-8 [DOI] [PubMed] [Google Scholar]
- 14. Gimenes F, Souza RP, Bento JC, Teixeira JJ, Maria-Engler SS, Bonini MG, et al. Male infertility: a public health issue caused by sexually transmitted pathogens. Nat Rev Urol. (2014) 11(12):672–87. doi: 10.1038/nrurol.2014.285 [DOI] [PubMed] [Google Scholar]
- 15. Gallegos G, Ramos B, Santiso R, Goyanes V, Gosálvez J, Fernández JL. Sperm DNA fragmentation in infertile men with genitourinary infection by Chlamydia trachomatis and Mycoplasma. Fertil Steril. (2008) 90(2):328–34. doi: 10.1016/j.fertnstert.2007.06.035 [DOI] [PubMed] [Google Scholar]
- 16. Pagliuca C, Cariati F, Bagnulo F, Scaglione E, Carotenuto C, Farina F, et al. Microbiological evaluation and sperm DNA fragmentation in semen samples of patients undergoing fertility investigation. Genes (Basel) (2021) 12(5). doi: 10.3390/genes12050654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carbone L, Conforti A, La Marca A, Cariati F, Vallone R, Raffone A, et al. The negative impact of most relevant infections on fertility and assisted reproduction technology. Minerva Obstet Gynecol. (2022) 74(1):83–106. doi: 10.23736/S2724-606X.21.04870-3 [DOI] [PubMed] [Google Scholar]
- 18. Esteves SC, Lombardo F, Garrido N, Alvarez J, Zini A, Colpi GM, et al. SARS-CoV-2 pandemic and repercussions for male infertility patients: A proposal for the individualized provision of andrological services. Andrology. (2021) 9(1):10–8. doi: 10.1111/andr.12809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Esteves SC, Santi D, Simoni M. An update on clinical and surgical interventions to reduce sperm DNA fragmentation in infertile men. Andrology. (2020) 8(1):53–81. doi: 10.1111/andr.12724 [DOI] [PubMed] [Google Scholar]
- 20. Hallak J, Esteves SC. Concise practice recommendations for the provision of andrological services and assisted reproductive technology for male infertility patients during the SARS-CoV-2 in Brazil. Int Braz J Urol. (2020) 46(6):1082–9. doi: 10.1590/s1677-5538.ibju.2020.06.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hallak J, Teixeira TA, Bernardes FS, Carneiro F, Duarte SAS, Pariz JR, et al. SARS-CoV-2 and its relationship with the genitourinary tract: Implications for male reproductive health in the context of COVID-19 pandemic. Andrology. (2021) 9(1):73–9. doi: 10.1111/andr.12896 [DOI] [PubMed] [Google Scholar]
- 22. Farahani L, Tharakan T, Yap T, Ramsay JW, Jayasena CN, Minhas S. The semen microbiome and its impact on sperm function and male fertility: A systematic review and meta-analysis. Andrology. (2021) 9(1):115–44. doi: 10.1111/andr.12886 [DOI] [PubMed] [Google Scholar]
- 23. Sellami H, Znazen A, Sellami A, Mnif H, Louati N, Ben Zarrouk S, et al. Molecular detection of Chlamydia trachomatis and other sexually transmitted bacteria in semen of male partners of infertile couples in Tunisia: the effect on semen parameters and spermatozoa apoptosis markers. PloS One (2014) 9(7):e98903. doi: 10.1371/journal.pone.0098903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levy R, Layani-Milon MP, Giscard D'Estaing S, Najioullah F, Lornage J, Aymard M, et al. Screening for Chlamydia trachomatis and Ureaplasma urealyticum infection in semen from asymptomatic male partners of infertile couples prior to in vitro fertilization. Int J Androl. (1999) 22(2):113–8. doi: 10.1046/j.1365-2605.1999.00157.x [DOI] [PubMed] [Google Scholar]
- 25. Wensel CR, Pluznick JL, Salzberg SL, Sears CL. Next-generation sequencing: insights to advance clinical investigations of the microbiome. J Clin Invest. (2022) 132(7). doi: 10.1172/JCI154944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol (2000) 38(10):3623–30. doi: 10.1128/JCM.38.10.3623-3630.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weng SL, Chiu CM, Lin FM, Huang WC, Liang C, Yang T, et al. Bacterial communities in semen from men of infertile couples: metagenomic sequencing reveals relationships of seminal microbiota to semen quality. PloS One (2014) 9(10):e110152. doi: 10.1371/journal.pone.0110152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods (2007) 69(2):330–9. doi: 10.1016/j.mimet.2007.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Poretsky R, Rodriguez RL, Luo C, Tsementzi D, Konstantinidis KT. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PloS One (2014) 9(4):e93827. doi: 10.1371/journal.pone.0093827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Veneruso I, Cariati F, Alviggi C, Pastore L, Tomaiuolo R, D'Argenio V. Metagenomics reveals specific microbial features in males with semen alterations. Genes (Basel). (2023) 14(6). doi: 10.3390/genes14061228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yao Y, Qiu XJ, Wang DS, Luo JK, Tang T, Li YH, et al. Semen microbiota in normal and leukocytospermic males. Asian J Androl. (2022) 24(4):398–405. doi: 10.4103/aja202172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bukharin OV, Perunova NB, Ivanova EV, Chaynikova IN, Bekpergenova AV, Bondarenko TA, et al. Semen microbiota and cytokines of healthy and infertile men. Asian J Androl. (2022) 24(4):353–8. doi: 10.4103/aja202169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lundy SD, Sangwan N, Parekh NV, Selvam MKP, Gupta S, McCaffrey P, et al. Functional and taxonomic dysbiosis of the gut, urine, and semen microbiomes in male infertility. Eur Urol. (2021) 79(6):826–36. doi: 10.1016/j.eururo.2021.01.014 [DOI] [PubMed] [Google Scholar]
- 34. Okwelogu SI, Ikechebelu JI, Agbakoba NR, Anukam KC. Microbiome compositions from infertile couples seeking in vitro fertilization, using 16S rRNA gene sequencing methods: Any correlation to clinical outcomes? Front Cell Infect Microbiol (2021) 11:709372. doi: 10.3389/fcimb.2021.709372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang H, Zhang J, Xue Z, Zhao C, Lei L, Wen Y, et al. Potential pathogenic bacteria in seminal microbiota of patients with different types of dysspermatism. Sci Rep (2020) 10(1):6876. doi: 10.1038/s41598-020-63787-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baud D, Pattaroni C, Vulliemoz N, Castella V, Marsland BJ, Stojanov M. Sperm microbiota and its impact on semen parameters. Front Microbiol (2019) 10:234. doi: 10.3389/fmicb.2019.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alfano M, Ferrarese R, Locatelli I, Ventimiglia E, Ippolito S, Gallina P, et al. Testicular microbiome in azoospermic men-first evidence of the impact of an altered microenvironment. Hum Reprod (2018) 33(7):1212–7. doi: 10.1093/humrep/dey116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zeyad A, Hamad M, Amor H, Hammadeh ME. Relationships between bacteriospermia, DNA integrity, nuclear protamine alteration, sperm quality and ICSI outcome. Reprod Biol (2018) 18(1):115–21. doi: 10.1016/j.repbio.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 39. Chen H, Luo T, Chen T, Wang G. Seminal bacterial composition in patients with obstructive and non-obstructive azoospermia. Exp Ther Med (2018) 15(3):2884–90. doi: 10.3892/etm.2018.5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Monteiro C, Marques PI, Cavadas B, Damião I, Almeida V, Barros N, et al. Characterization of microbiota in male infertility cases uncovers differences in seminal hyperviscosity and oligoasthenoteratozoospermia possibly correlated with increased prevalence of infectious bacteria. Am J Reprod Immunol (2018) 79(6):e12838. doi: 10.1111/aji.12838 [DOI] [PubMed] [Google Scholar]
- 41. Vilvanathan S, Kandasamy B, Jayachandran AL, Sathiyanarayanan S, Tanjore Singaravelu V, Krishnamurthy V, et al. Bacteriospermia and its impact on basic semen parameters among infertile men. Interdiscip Perspect Infect Dis (2016) 2016:2614692. doi: 10.1155/2016/2614692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fraczek M, Hryhorowicz M, Gill K, Zarzycka M, Gaczarzewicz D, Jedrzejczak P, et al. The effect of bacteriospermia and leukocytospermia on conventional and nonconventional semen parameters in healthy young normozoospermic males. J Reprod Immunol (2016) 118:18–27. doi: 10.1016/j.jri.2016.08.006 [DOI] [PubMed] [Google Scholar]
- 43. Hou D, Zhou X, Zhong X, Settles ML, Herring J, Wang L, et al. Microbiota of the seminal fluid from healthy and infertile men. Fertil Steril. (2013) 100(5):1261–9. doi: 10.1016/j.fertnstert.2013.07.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kjaergaard N, Kristensen B, Hansen ES, Farholt S, Schønheyder HC, Uldbjerg N, et al. Microbiology of semen specimens from males attending a fertility clinic. Apmis. (1997) 105(7):566–70. doi: 10.1111/j.1699-0463.1997.tb05054.x [DOI] [PubMed] [Google Scholar]
- 45. Virecoulon F, Wallet F, Fruchart-Flamenbaum A, Rigot JM, Peers MC, Mitchell V, et al. Bacterial flora of the low male genital tract in patients consulting for infertility. Andrologia. (2005) 37(5):160–5. doi: 10.1111/j.1439-0272.2005.00673.x [DOI] [PubMed] [Google Scholar]
- 46. Kermes K, Punab M, Lõivukene K, Mändar R. Anaerobic seminal fluid micro-flora in chronic prostatitis/chronic pelvic pain syndrome patients. Anaerobe. (2003) 9(3):117–23. doi: 10.1016/S1075-9964(03)00085-4 [DOI] [PubMed] [Google Scholar]
- 47. Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. (2012) 486(7402):207–14. doi: 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eggert-Kruse W, Rohr G, Ströck W, Pohl S, Schwalbach B, Runnebaum B. Anaerobes in ejaculates of subfertile men. Hum Reprod Update. (1995) 1(5):462–78. doi: 10.1093/humupd/1.5.462 [DOI] [PubMed] [Google Scholar]
- 49. Bussen S, Zimmermann M, Schleyer M, Steck T. Relationship of bacteriological characteristics to semen indices and its influence on fertilization and pregnancy rates after IVF. Acta Obstet Gynecol Scand (1997) 76(10):964–8. doi: 10.3109/00016349709034910 [DOI] [PubMed] [Google Scholar]
- 50. Aghazarian A, Stancik I, Pflüger H, Lackner J. Influence of pathogens and moderate leukocytes on seminal interleukin (IL)-6, IL-8, and sperm parameters. Int Urol Nephrol. (2013) 45(2):359–65. doi: 10.1007/s11255-013-0400-8 [DOI] [PubMed] [Google Scholar]
- 51. Domes T, Lo KC, Grober ED, Mullen JB, Mazzulli T, Jarvi K. The incidence and effect of bacteriospermia and elevated seminal leukocytes on semen parameters. Fertil Steril. (2012) 97(5):1050–5. doi: 10.1016/j.fertnstert.2012.01.124 [DOI] [PubMed] [Google Scholar]
- 52. Ricci S, De Giorgi S, Lazzeri E, Luddi A, Rossi S, Piomboni P, et al. Impact of asymptomatic genital tract infections on in vitro Fertilization (IVF) outcome. PloS One (2018) 13(11):e0207684. doi: 10.1371/journal.pone.0207684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Molina NM, Plaza-Díaz J, Vilchez-Vargas R, Sola-Leyva A, Vargas E, Mendoza-Tesarik R, et al. Assessing the testicular sperm microbiome: a low-biomass site with abundant contamination. Reprod BioMed Online. (2021) 43(3):523–31. doi: 10.1016/j.rbmo.2021.06.021 [DOI] [PubMed] [Google Scholar]
- 54. Campisciano G, Iebba V, Zito G, Luppi S, Martinelli M, Fischer L, et al. Lactobacillus iners and gasseri, Prevotella bivia and HPV Belong to the Microbiological Signature Negatively Affecting Human Reproduction. Microorganisms (2020) 9(1). doi: 10.3390/microorganisms9010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ndiokwere C, Olise N, Nmewurum V, Omoregie R, Agbakoba N, Anukam K. 16S rRNA Metagenomics of Seminal Fluids from Medical Microbiology Laboratory in a Tertiary Hospital, Southern Nigeria (2019) 29:86–109. [Google Scholar]
- 56. Zeyad A, Hamad MF, Hammadeh ME. The effects of bacterial infection on human sperm nuclear protamine P1/P2 ratio and DNA integrity. Andrologia (2017) 50(2). doi: 10.1111/and.12841 [DOI] [PubMed] [Google Scholar]
- 57. Mashaly M, Masallat DT, Elkholy AA, Abdel-Hamid IA, Mostafa T. Seminal Corynebacterium strains in infertile men with and without leucocytospermia. Andrologia (2016) 48(3):355–9. doi: 10.1111/and.12457 [DOI] [PubMed] [Google Scholar]
- 58. Ruggeri M, Cannas S, Cubeddu M, Molicotti P, Piras GL, Dessole S, et al. Bacterial agents as a cause of infertility in humans. New Microbiol (2016) 39(3):206–209. [PubMed] [Google Scholar]
- 59. Mändar A, Punab MF, Borovkova ME, Lapp ME, Kiiker ME, Korrovits ME, et al. Complementary seminovaginal microbiome in couples. Res Microbiol (2015) 166(5):440–7. doi: 10.1016/j.resmic.2015.03.009 [DOI] [PubMed] [Google Scholar]
- 60. Isaiah IN, Nche BT, Nwagu IG, Nnanna II. Current studies on bacterospermia the leading cause of male infertility: a protégé and potential threat towards mans extinction. N Am J Med Sci (2011) 3(12):562–4. doi: 10.4297/najms.2011.3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moretti E, Capitani S, Figura N, Pammolli A, Federico MG, Giannerini V, et al. The presence of bacteria species in semen and sperm quality. J Assist Reprod Genet (2009) 26(1):47–56. doi: 10.1007/s10815-008-9283-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gdoura R, Kchaou W, Znazen A, Chakroun N, Fourati M, Ammar-Keskes L, et al. Screening for bacterial pathogens in semen samples from infertile men with and without leukocytospermia. Andrologia. (2008) 40(4):209–18. doi: 10.1111/j.1439-0272.2008.00845.x [DOI] [PubMed] [Google Scholar]
- 63. Debata NK, Venkatesh V, Misra RN, Lapp Y, Chander VC, Ohri RK, et al. Ureaplasmas urealyticum and human infertility: effect on spermatozoa morphology. Med J Armed Forces India (1999) 55(3):193–6. doi: 10.1016/S0377-1237(17)30439-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shalika S, Dugan K, Smith RD, Padilla SL. The effect of positive semen bacterial and Ureaplasma cultures on in-vitro fertilization success. Hum Reprod (1996) 11(12):2789–92. doi: 10.1093/oxfordjournals.humrep.a019211 [DOI] [PubMed] [Google Scholar]
- 65. WHO . Laboratory Manual for the Examination and Processing of Human Semen, 6th ed. (2021). Geneva:World Health Organization. [Google Scholar]
- 66. Wang H, Chen T, Chen Y, Luo T, Tan B, Chen H, et al. Evaluation of the inhibitory effects of vaginal microorganisms on sperm motility in vitro. Exp Ther Med (2020) 19(1):535–44. doi: 10.3892/etm.2019.8237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Esteves SC. Clinical relevance of routine semen analysis and controversies surrounding the 2010 World Health Organization criteria for semen examination. Int Braz J Urol. (2014) 40(4):443–53. doi: 10.1590/S1677-5538.IBJU.2014.04.02 [DOI] [PubMed] [Google Scholar]
- 68. Štšepetova J, Baranova J, Simm J, Parm Ü, Rööp T, Sokmann S, et al. The complex microbiome from native semen to embryo culture environment in human in vitro fertilization procedure. Reprod Biol Endocrinol (2020) 18(1):3. doi: 10.1186/s12958-019-0562-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. van der Steeg JW, Steures P, Eijkemans MJ, JD FH, Hompes PG, Kremer JA, et al. Role of semen analysis in subfertile couples. Fertil Steril. (2011) 95(3):1013–9. doi: 10.1016/j.fertnstert.2010.02.024 [DOI] [PubMed] [Google Scholar]
- 70. Andrade DL, Viana MC, Esteves SC. Differential diagnosis of azoospermia in men with infertility. J Clin Med (2021) 10(14):5–24. doi: 10.3390/jcm10143144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hamada A, Esteves SC, Nizza M, Agarwal A. Unexplained male infertility: diagnosis and management. Int Braz J Urol. (2012) 38(5):576–94. doi: 10.1590/S1677-55382012000500002 [DOI] [PubMed] [Google Scholar]
- 72. Pasqualotto FF, Sharma RK, Nelson DR, Thomas AJ, Agarwal A. Relationship between oxidative stress, semen characteristics, and clinical diagnosis in men undergoing infertility investigation. Fertil Steril. (2000) 73(3):459–64. doi: 10.1016/S0015-0282(99)00567-1 [DOI] [PubMed] [Google Scholar]
- 73. Gosálvez J, Coppola L, Fernández JL, López-Fernández C, Góngora A, Faundez R, et al. Multi-centre assessment of nitroblue tetrazolium reactivity in human semen as a potential marker of oxidative stress. Reprod BioMed Online. (2017) 34(5):513–21. doi: 10.1016/j.rbmo.2017.01.014 [DOI] [PubMed] [Google Scholar]
- 74. Esteves SC, López-Fernández C, Martínez MG, Silva EA, Gosálvez J. Reliability of the sperm chromatin dispersion assay to evaluate sperm deoxyribonucleic acid damage in men with infertility. Fertil Steril. (2022) 117(1):64–73. doi: 10.1016/j.fertnstert.2021.08.045 [DOI] [PubMed] [Google Scholar]
- 75. Esteves SC, Zini A, Coward RM, Evenson DP, Gosálvez J, Lewis SEM, et al. Sperm DNA fragmentation testing: Summary evidence and clinical practice recommendations. Andrologia. (2021) 53(2):e13874. doi: 10.1111/and.13874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Eggert-Kruse W, Kiefer I, Beck C, Demirakca T, Strowitzki T. Role for tumor necrosis factor alpha (TNF-alpha) and interleukin 1-beta (IL-1beta) determination in seminal plasma during infertility investigation. Fertil Steril. (2007) 87(4):810–23. doi: 10.1016/j.fertnstert.2006.08.103 [DOI] [PubMed] [Google Scholar]
- 77. Ding N, Zhang X, Zhang XD, Jing J, Liu SS, Mu YP, et al. Impairment of spermatogenesis and sperm motility by the high-fat diet-induced dysbiosis of gut microbes. Gut. (2020) 69(9):1608–19. doi: 10.1136/gutjnl-2019-319127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mändar R, Türk S, Korrovits P, Ausmees K, Punab M. Impact of sexual debut on culturable human seminal microbiota. Andrology. (2018) 6(3):510–2. doi: 10.1111/andr.12482 [DOI] [PubMed] [Google Scholar]
- 79. Tramontano L, Sciorio R, Bellaminutti S, Esteves SC, Petignat P. Exploring the potential impact of human papillomavirus on infertility and assisted reproductive technology outcomes. Reprod Biol (2023) 23(2):100753. doi: 10.1016/j.repbio.2023.100753 [DOI] [PubMed] [Google Scholar]
- 80. Tuominen H, Rautava J, Kero K, Syrjänen S, Collado MC, Rautava S. HPV infection and bacterial microbiota in the semen from healthy men. BMC Infect Dis (2021) 21(1):373. doi: 10.1186/s12879-021-06029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Souho T, Benlemlih M, Bennani B. Human papillomavirus infection and fertility alteration: a systematic review. PloS One (2015) 10(5):e0126936. doi: 10.1371/journal.pone.0126936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu CM, Osborne BJ, Hungate BA, Shahabi K, Huibner S, Lester R, et al. The semen microbiome and its relationship with local immunology and viral load in HIV infection. PloS Pathog (2014) 10(7):e1004262. doi: 10.1371/journal.ppat.1004262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fethers KA, Fairley CK, Hocking JS, Gurrin LC, Bradshaw CS. Sexual risk factors and bacterial vaginosis: a systematic review and meta-analysis. Clin Infect Dis (2008) 47(11):1426–35. doi: 10.1086/592974 [DOI] [PubMed] [Google Scholar]
- 84. Gallo MF, Warner L, King CC, Sobel JD, Klein RS, Cu-Uvin S, et al. Association between semen exposure and incident bacterial vaginosis. Infect Dis Obstet Gynecol. (2011) 2011:842652. doi: 10.1155/2011/842652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Agnew KJ, Hillier SL. The effect of treatment regimens for vaginitis and cervicitis on vaginal colonization by lactobacilli. Sex Transm Dis (1995) 22(5):269–73. doi: 10.1097/00007435-199509000-00001 [DOI] [PubMed] [Google Scholar]
- 86. Beigi RH, Wiesenfeld HC, Hillier SL, Straw T, Krohn MA. Factors associated with absence of H2O2-producing Lactobacillus among women with bacterial vaginosis. J Infect Dis (2005) 191(6):924–9. doi: 10.1086/428288 [DOI] [PubMed] [Google Scholar]
- 87. Borovkova N, Korrovits P, Ausmees K, Türk S, Jõers K, Punab M, et al. Influence of sexual intercourse on genital tract microbiota in infertile couples. Anaerobe. (2011) 17(6):414–8. doi: 10.1016/j.anaerobe.2011.04.015 [DOI] [PubMed] [Google Scholar]
- 88. Esteves SC, Roque M, Bedoschi G, Haahr T, Humaidan P. Intracytoplasmic sperm injection for male infertility and consequences for offspring. Nat Rev Urol. (2018) 15(9):535–62. doi: 10.1038/s41585-018-0051-8 [DOI] [PubMed] [Google Scholar]
- 89. Abeysundara PK, Dissanayake D, Wijesinghe PS, Perera R, Nishad A. Efficacy of two sperm preparation techniques in reducing non-specific bacterial species from human semen. J Hum Reprod Sci (2013) 6(2):152–7. doi: 10.4103/0974-1208.117169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Esteves SC, Bento FC. Implementation of air quality control in reproductive laboratories in full compliance with the Brazilian Cells and Germinative Tissue Directive. Reprod BioMed Online. (2013) 26(1):9–21. doi: 10.1016/j.rbmo.2012.10.010 [DOI] [PubMed] [Google Scholar]
- 91. Amato V, Papaleo E, Pasciuta R, Viganò P, Ferrarese R, Clementi N, et al. Differential composition of vaginal microbiome, but not of seminal microbiome, is associated with successful intrauterine insemination in couples with idiopathic infertility: A prospective observational study. Open Forum Infect Dis (2020) 7(1):ofz525. doi: 10.1093/ofid/ofz525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cariati F, Carotenuto C, Bagnulo F, Pacella D, Marrone V, Paolillo R, et al. Endometrial microbiota profile in in-vitro fertilization (IVF) patients by culturomics-based analysis. Front Endocrinol (Lausanne). (2023) 14:1204729. doi: 10.3389/fendo.2023.1204729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zuber A, Peric A, Pluchino N, Baud D, Stojanov M. Human male genital tract microbiota. Int J Mol Sci (2023) 24(8). doi: 10.3390/ijms24086939 [DOI] [PMC free article] [PubMed] [Google Scholar]


