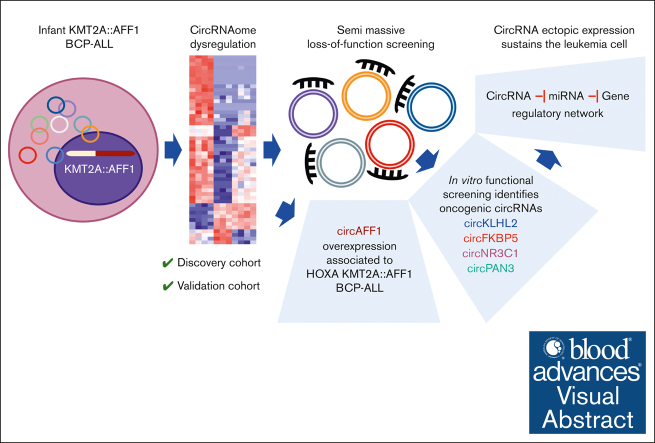
Key Points
-
•
New data on circRNAome dysregulation in KMT2A::AFF1 ALL indicate aberrant transcripts from KMT2A signature genes and many new disease loci.
-
•
Functional study disclosed the oncogenic role in KMT2A::AFF1 ALL of circFKBP5, circKLHL2, circNR3C1, and circPAN3.
Visual Abstract
Abstract
Circular RNAs (circRNAs) are emerging molecular players in leukemogenesis and promising therapeutic targets. In KMT2A::AFF1 (MLL::AF4)-rearranged leukemia, an aggressive disease compared with other pediatric B-cell precursor (BCP) acute lymphoblastic leukemia (ALL), data about circRNAs are limited. Here, we disclose the circRNA landscape of infant patients with KMT2A::AFF1 translocated BCP-ALL showing dysregulated, mostly ectopically expressed, circRNAs in leukemia cells. Most of these circRNAs, apart from circHIPK3 and circZNF609, previously associated with oncogenic behavior in ALL, are still uncharacterized. An in vitro loss-of-function screening identified an oncogenic role of circFKBP5, circKLHL2, circNR3C1, and circPAN3 in KMT2A::AFF1 ALL, whose silencing affected cell proliferation and apoptosis. Further study in an extended cohort disclosed a significantly correlated expression of these oncogenic circRNAs and their putative involvement in common regulatory networks. Moreover, it showed that circAFF1 upregulation occurs in a subset of cases with HOXA KMT2A::AFF1 ALL. Collectively, functional analyses and patient data reveal oncogenic circRNA upregulation as a relevant mechanism that sustains the malignant cell phenotype in KMT2A::AFF1 ALL.
Introduction
Circular RNAs (circRNAs), transcripts in which by backsplicing the splice donor site is covalently bound to an upstream acceptor site, are key molecular players whose dysregulation and functions affect almost all cancer hallmarks1,2 and attract high interest in studies aiming to discover new disease mechanisms and therapeutic targets.3, 4, 5, 6
The prominent function of circRNAs is their ability to act as efficient microRNA (miRNA) sponges, regulating axes involving miRNAs.7, 8, 9 CircRNAs can interact with a variety of RNA-binding proteins10, 11, 12 and even generate functional peptides13, 14, 15, 16 not coded by linear transcripts.
CircRNAs are pervasively expressed in the hematopoietic compartment,1,17,18 and we and others have shown the dysregulation and oncogenic potential of circRNAs in acute leukemias.18, 19, 20, 21, 22
In addition, recent discoveries have opened new perspectives for the circRNA role in leukemias with KMT2A/MLL rearrangements (KMT2Are).1,2,23,24 In leukemia with KMT2A::MLLT3 (MLL::AF9) translocation, oncogenic fusion circRNAs exhibited transforming activity linked to an enhancement of the oncogenic potential of the fusion protein.25 Furthermore, preliminary targeted investigation of specific circRNAs in B-cell precursor acute lymphoblastic leukemia (BCP-ALL) with KMT2Are18 and of the expression dysregulation of circRNAs expressed by the translocation partner genes in KMT2A::MLLT1 ALL26 provided evidence that MLL and several translocation partner genes express circRNAs in hematopoietic cells and that KMT2Are, beyond generating fusion circRNAs, can affect circRNA expression in general.26
Infant KMT2A::AFF1 leukemia is an aggressive disease with short event-free survival compared with other pediatric BCP-ALLs,27 and its unique features, including extensive spread beyond the hematopoietic compartment, resistance to therapies, tendency to relapse, and myeloid switch,28 motivated a thorough investigation of the biology underlying this malignancy ever since the first clinical report of a large set of patients carrying this translocation published in the early 80s.29 The low frequency of somatic mutations in KMT2A::AFF130 indicated the fusion protein as the main disease driver, even if KMT2A::AFF1–induced leukemogenesis was difficult to model.31, 32, 33 Several studies disclosed the impact of KMT2A::AFF1 on gene expression, defining 2 different types of KMT2A::AFF1 signatures34,35 and identifying the chimera direct targets.36, 37, 38 Less is known about the transcripts expressed by KMT2A fusion genes. In KMT2A::AFF1 ALL, all whole transcriptome studies focused on linear transcripts only, and data about circRNA expression and roles in KMT2A::AFF1ALL are very limited. We recently detected fusion circRNAs in the RS4;11 cell line and in patients harboring KMT2A::AFF1 translocation.39 A circAFF1 isoform contributing to release miR-128-3p inhibition on KMT2A::AFF1 protein expression has been implicated in leukemogenesis.40
This study defining the KMT2A::AFF1 ALL genomic landscape of circRNA expression was instrumental in identifying, through extensive functional investigation, multiple oncogenic circRNAs whose upregulation in malignant cells play an active role in the disease.
Materials and methods
Patients and samples
Leukemia cells from bone marrow samples at the diagnosis of 4 infant patients with BCP-ALL carrying KMT2A::AFF1re and 2 additional patient-derived xenograft samples were obtained (supplemental Methods).
CD34+ hematopoietic stem cells from cord blood samples of 3 healthy donors (HDs) and CD19+ healthy B cells/B lineage cells sorted using a fluorescence-activated cell sorter from peripheral blood mononuclear cells of 4 HDs18 were used as normal counterparts (GSE213990).
An enlarged cohort of pediatric patients with BCP-ALL at diagnosis, including cases with KMT2A::AFF1 translocation (N = 45) and wild-type KMT2A (N = 13), was collected to assess the expression of circRNAs under investigation by real-time quantitative polymerase chain reaction (qRT-PCR), in comparison with bone marrow samples of HDs (N = 5). All samples were collected with informed consent, according to the Declaration of Helsinki.
RNA isolation and sequencing
RNA was isolated using TRIzol (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions. RNA quality was assessed with Agilent 2100 Bioanalyzer (Santa Clara, CA) keeping only samples with RNA integrity number > 7. RNA was quantified using the Qubit Assay (Thermo Fisher Scientific).
RNA libraries were prepared with the TruSeq Stranded Total RNA Ribo-Zero Gold kit and sequenced with an Illumina HiSeq2000 (San Diego, CA) at an average sequencing depth of 90 million reads per sample (paired-end reads 100-150 bp).
CircRNA detection and quantification from RNA-seq data
CircRNA identification and quantification and calculation of the circular-to-linear proportion (CLP) from RNA sequencing (RNA-seq) data were obtained with CirComPara version 0.6.341,42 as described in supplemental Methods. Ensembl GRCh38 human genome and annotation version 93 was used for all analyses.
CircRNA expression was normalized with the regularized logarithm method and corrected using “sva.”43 Differential expression (DE) was assessed by DESeq2 (version 1.22.2)44 with the parametric model, including Wald significance tests and no independent filtering, using an adjusted (Benjamini Hochberg) P value ≤ .05 as the significance threshold.
Cell lines
RS4;11 and SEM cell lines harboring KMT2A::AFF1 (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) were cultured in RPMI 1640 (Gibco, Life Technologies, CA) medium supplemented with 10% fetal bovine serum (Gibco), 1% L-glutamine and 1% penicillin/streptomycin and maintained at 500 000/mL concentration at 37°C in a 5% CO2 incubator.
CircRNA silencing
Silencer select small interfering RNAs (siRNAs) were designed and synthesized by Thermo Fisher Scientific, specifically targeting the backsplice site of each circRNA, and tested to exclude off-targets among known human transcripts. Silencer Select Negative Control (Thermo Fisher Scientific) no.1 siRNA was used as the negative control (sirNEG).
RS4;11 and SEM cell lines were transfected using Amaxa 4D-Nucleofector instrument (Lonza, Davis, NC) and Mirus electroporation solution (Ingenio Electroporation Solution) with an optimized siRNA concentration of 30pMol (siRNA sequence are reported in supplemental Table 1).
SEM (EN-138 electroporation program) and RS4;11 (DS-130 program) cell lines were transfected using 16-well strips, with a cell (400 000 cells per well)-siRNA-Mirus final volume of 20 μL. After electroporation, we seeded 1 000 000 cells per mL. At least 3 independent experiments were performed for all tests.
Cell viability, proliferation, and apoptosis assays
Cell lines were collected 24 hours and 48 hours after transfection and subjected to different in vitro functional assays to measure cell proliferation (5-ethynyl-2'-deoxyuridine incorporation) and apoptosis rate (annexin V–propidium iodide [PI]) as described in supplemental Methods.
RNA extraction, RNase R treatment, RT-PCR, and qRT-PCR
RNA was extracted from transfected cell lines using TRIzol (Thermo Fisher Scientific) and quantified using Qubit Assay (Thermo Fisher Scientific). For the validation of circRNAs presence in cell lines, RNA was treated with RNase R (Epicenter Biotechnologies, Madison, WI) to enhance circRNA abundance at a ratio of 2 U/μg. A total of 1 μg of RNA was retrotranscribed using Random hexamers (Invitrogen and Thermo Fisher Scientific) and SuperScript II Reverse Transcriptase (Invitrogen).
To confirm the circRNA silencing efficiency, we performed the qRT-PCR using Platinum Sybr Green qPCR SuperMix-UDG (Invitrogen) carried out for 40 cycles and divergent primers to detect the backsplice site of each circRNA molecule (supplemental Table 2).
qRT-PCR with convergent primers (supplemental Table 3) was also performed to monitor the linear transcript expression. The 2(-ΔΔCt) method was applied to obtain circRNA or linear transcript expression using sirNEG results as calibrators. GAPDH was used as a reference gene. Statistical analysis of experimental results was performed using Prism 8 (GraphPad Software Inc, La Jolla, CA).
CircRNA expression in patients was quantified using quantitative reverse transcription polymerase chain reaction (qRT-PCR) with divergent primers (supplemental Table 2), as previously described, without RNase R treatment. Bone marrow samples from HDs were used as a calibrator to calculate 2(-ΔΔCt).
CircRNA functional predictions
The CRAFT tool45 was applied to predict miRNA binding sites of circRNAs and to retrieve validated miRNA target genes (supplemental Methods). Functional enrichments have been calculated with the ClusterProfiler 4.0 package.
Gene expression profiling of patients harboring KMT2A::AFF1
Gene expression profiling data already available in the Laboratory of Onco-Hematology at University of Padova (GSE77416; pediatric KMT2A::AFF1 ALL samples from GSE13204) were processed as described in supplemental Methods.
Results
CircRNA expression in patients harboring KMT2A::AFF1
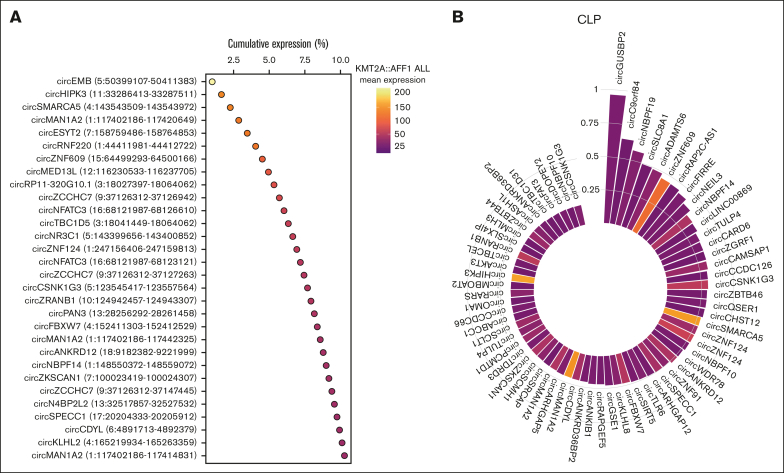
The first aim of this study was to disclose the circRNAome of KMT2A::AFF1 BCP-ALL. Analysis with CirComPara42 of high-depth ribodepleted RNA-seq data in a discovery cohort of infant patients with KMT2A::AFF1 (N = 4) identified 7596 circRNAs expressed from 3297 loci.
The 100 most expressed circRNAs account for about 18% of the expression in KMT2A::AFF1 samples, whereas 1050 isoforms cover 50% of circRNA expression (supplemental Figure 1). Among the most expressed circRNAs in KMT2A::AFF1 leukemia (Figure 1A), there are both circRNAs with well-characterized functions in other diseases, such as the oncogenic circHIPK346 and circZNF609,47 and less-characterized molecules, such as circRNF220 and circNR3C1.48,49
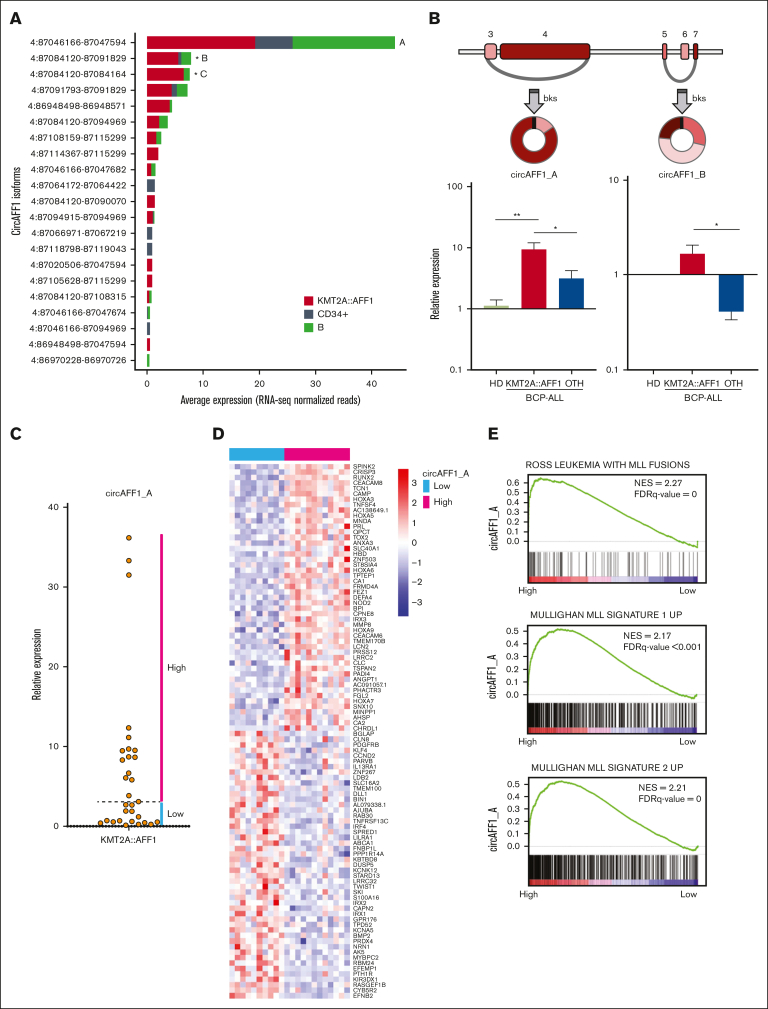
Figure 1.
CircRNAome of KMT2A::AFF1 ALL. (A) Cumulative expression of the 30 circRNAs most abundantly expressed in the samples from patients with KMT2A::AFF1 ALL; (B) CLP (bar height) and expression (bar color, as in panel A) in KMT2A::AFF1 ALL of the circRNAs with highest CLP (at least 0.15) and in the top 5% most expressed in patients.
In line with previous reports,18, 19, 20 about half of circRNA host genes expressed multiple circular isoforms. Eleven genes can generate at least 15 circRNAs expressed in the discovery sample set, with the highest number of isoforms detected for UBAP2 (27 circRNAs), ZCCHC7 (21), and ANKRD36 (20) (supplemental Figure 2A). A single predominant circRNA more highly expressed than the other isoforms was observed for 5 of these genes, including ANKRD36, PAN3, and AKT3, whereas 3, such as BIRC6 and ANKRD36C, presented 2 circRNAs with similarly high expression and the other circRNAs with medium to low expression (supplemental Figure 2B). The remaining genes presented multiple isoforms with comparably high expression.
Beyond the absolute expression (AE) of circRNAs, we also considered the CLP, indicating the relative abundance of each circRNA with respect to the overlapping host gene transcripts. Figure 1B shows the group of 67 circRNAs with the highest CLP in KMT2A::AFF1 leukemia and AE. CircGUSBP2 accounted for almost all the expression of its host gene, circC90orf84 (alias SHOC1) and circNBPF19 were more abundant than their linear counterparts (CLP > 0.5). CircZNF609, circSMARCA5, circMAN1A2, and circHIPK3 were among the circRNAs with the highest CLP and particularly high AE.
Dysregulation of circRNA absolute and relative expression level in KMT2A::AFF1 leukemia
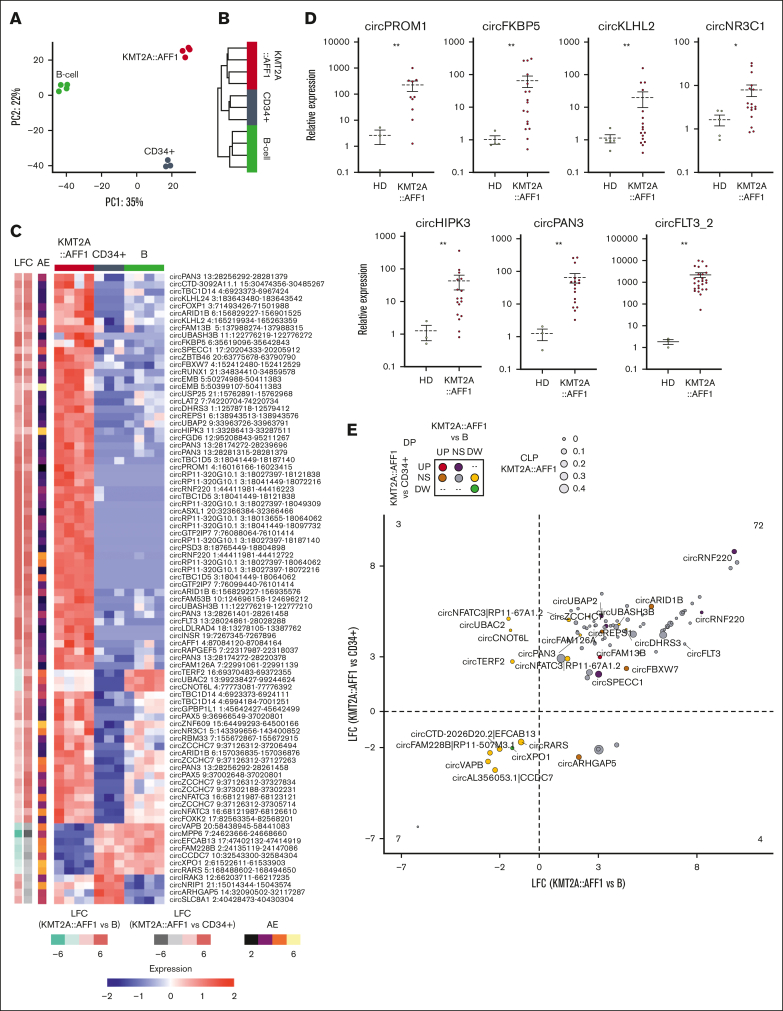
Next, to identify dysregulated circRNAs in KMT2A::AFF1 possibly specific to leukemia, the circRNAome of KMT2A::AFF1 leukemia was compared with CD34+ hematopoietic stem cells and B cells from HDs. For this comparison, 8607 expressed circRNAs, derived from 3602 loci, were used.
Unsupervised analyses of circRNA expression by principal component analysis (Figure 2A) and hierarchical clustering (Figure 2B) clearly separated KMT2A::AFF1 ALL samples from the 2 healthy cell populations as well as CD34+ cells from B cells. In terms of circRNAome, KMT2A::AFF1 ALL is closer to CD34+ cells than to B cells. In line, DE analysis identified 206 circRNAs that significantly varied (109 upregulated and 97 downregulated) in KMT2A::AFF1 ALL when compared with CD34+ samples and 1116 (714 upregulated and 402 downregulated) when compared with B cells.
Figure 2.
The KMT2A::AFF1 circRNAome is highly dysregulated. (A) Principal component (PC) analysis and (B) hierarchical clustering using Euclidean distance of samples from patients with KMT2A::AFF1 ALL and CD34+ cells and B-cell populations from HDs, based on circRNA expression profiles. (C) Heat map of standardized expression of the 86 circRNAs differentially expressed when comparing KMT2A::AFF1 ALL vs B cells and CD34+ cells from HDs; (left) the log2 fold change (LFC) in the 2 comparisons and the average AE in all samples are shown. (D) CircRNA expression quantification by RQ-PCR in an extended patient cohort (relative expression calculated as 2-ΔΔCT values using glyceraldehyde-3-phosphate dehydrogenase as reference and bone marrow samples from at least 3 HDs as calibrator; Mann-Whitney U test P values: ∗P < .05 and ∗∗ P < .01); N = 10 MLL:AF4 BCP-ALL for circPROM1; N = 17 KMT2A::AFF1 BCP-ALL for circFKBP5, circKLHL2, circNR3C1, circHIPK3, and circPAN3; and N = 27 MLL:AF4 BCP-ALL for circFLT3_2. (E) Relation between dysregulation of circRNA expression and proportion in KMT2A::AFF1 ALL for the same 86 DE circRNAs in panel C. The dot size represents the CLP value in patients with KMT2A::AFF1, the dot color indicates DP variation categories in the 2 comparisons; names of circRNAs DP in at least 1 comparison (KMT2A::AFF1 vs B cells and/or KMT2A::AFF1 vs CD34+ cells) are shown, and the number of circRNAs in each quadrant is indicated. UP, upregulated; NS, unvaried; DW, decreased.
Of particular interest, a group of 86 circRNAs was DE in KMT2A::AFF1 ALL compared with both healthy cell populations (Figure 2C; supplemental Table 4). Seven of these circRNAs were expressed in leukemia cells at an intermediate level compared with the 2 control populations: circTERF2, circUBAC2, and circCNOT6L being more expressed in cells from patients with KMT2A::AFF1 than in CD34+ cells but less than that in B cells. The opposite was observed for circIRAK3, circNRIP1, circARHGAP5, and circSLC8A1 (more expressed in cells from patients with KMT2A::AFF1 than in B cells but less than in CD34+ cells). The remaining 79 circRNAs were clearly aberrantly expressed. CircMPP6, circRARS, circXPO1, circEFCAB13, circFAM228B, circVAPB, and circCCDC7 were depleted in malignant cells.
Of importance, 72 circRNAs ectopically expressed in KMT2A::AFF1 ALL represented candidate oncogenic circRNAs. These circRNAs were derived from 44 genes, 12 with multiple circular isoforms upregulated in malignant cells, among which were TBC1D5 (8 circular isoforms) and PAN3 (6 circular isoforms). Two circRNF220 isoforms, circPROM1 and circFLT3, almost or completely absent in the control cell populations, were instead highly expressed in KMT2A::AFF1 ALL. In addition, the most expressed and upregulated circRNAs included circKLHL2, circTBC1D14, circHIPK3, circSPECC1, circFKBP5, circNR3C1, circZNF609, circNR3C1, and circFLT3 (Figure 2C). Quantification by qRT-PCR of 7 of the most upregulated circRNAs in a validation cohort of pediatric patients with KMT2A::AFF1 ALL confirmed their significant upregulation in these patients (Figure 2D).
CircRNA expression can be dysregulated because of epigenetic and transcriptional aberrancies that are expected to affect linear transcripts as well or because of more specific mechanisms of circRNA biogenesis, including backsplicing regulation and modulation of circRNA stability. In this regard, the examination of the CLP across conditions was informative. To understand how much the circRNAome dysregulation in KMT2A::AFF1 ALL was independent from the linear gene expression disturbances, we tested for significantly different CLPs in leukemia compared with those in CD34+ cells and B cells, identifying 82 and 1281 circRNAs with significantly varied CLPs (differential proportion [DP]), respectively. Of note, 10 (5%) and 255 (23%) of DE circRNAs when comparing KMT2A::AFF1 ALL with CD34+ and CD19+ B cells also had DP in the same comparison. A concordant behavior of expression and proportion (for instance, both absolute and relative expression levels increased) of most of these circRNAs indicated that the expression variation of the circRNAs does not simply reflect the variation of the linear counterpart (supplemental Figure 3A-B; supplemental Table 5). Two circRNAs were both DP and DE in KMT2A::AFF1 ALL compared with normal populations: circFAM13B was upregulated and had an increased proportion in malignant cells, whereas both absolute and relative circXPO1 expression levels were reduced (Figure 2E). Several circRNAs, including 2 circRNF220 isoforms, circSPECC1 and circARID1B, and circFBXW7, upregulated in leukemia cells, also accounted for a significantly increased expression proportion in KMT2A::AFF1 compared with at least 1 normal population. In contrast, 80% of the 72 circRNAs with significantly upregulated expression in KMT2A::AFF1 ALL did not present a significantly varied CLP in either comparison, indicating that the observed circRNA upregulation was prevalently concordant with the increase in linear counterpart expression.
Are dysregulated circRNAs expressed by known KMT2Are signature genes?
Functional enrichment showed that host genes of the circRNAs dysregulated in KMT2A::AFF1 ALL samples in comparison with both normal populations are significantly enriched in leukemia-associated genes (DISGenet signatures of acute myeloid leukemia with ryelodysplasia-related changes: ASXL1, FLT3, and RUNX1; primary myelofibrosis: ASXL1, FLT3, RARS, NR3C1, RUNX1, and FKBP5; and precursor B-cell lymphoblastic leukemia-lymphoma: FLT3, INSR, PAX5, NR3C1, and RUNX1).
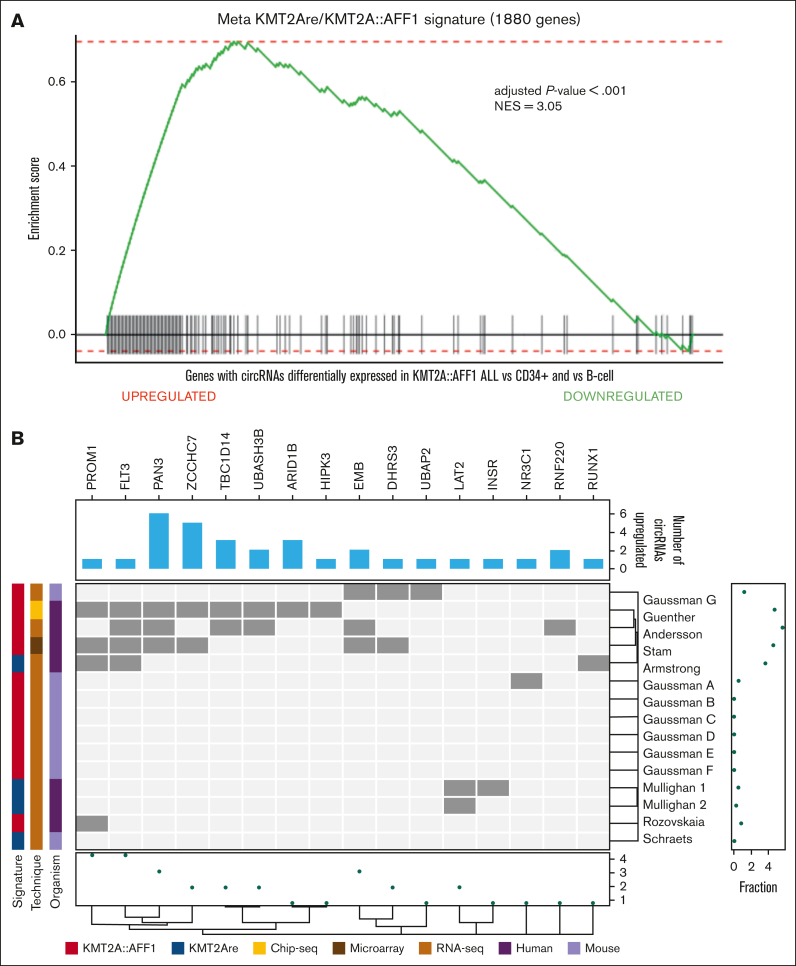
Next, we assessed if these leukemia-associated genes are part of the known KMT2A::AFF1 signature defined by the linear transcriptome studies based on microarrays and RNA-seq or DNA-binding by CHIP-seq data.
We collected 15 human or murine50, 51, 52, 53, 54, 55 signatures from public databases of genes upregulated in association with KMT2A rearrangements in general or specifically with the KMT2A::AFF1 fusion, and assembled a KMT2Are/KMT2A::AFF1 meta-signature of 1881 genes (supplemental Table 6). Custom gene set enrichment analysis (GSEA) using this meta-signature demonstrated a significant enrichment of genes with circRNAs upregulated in KMT2A::AFF1 ALL (Figure 3A). In fact, 121 (6.4%) signature genes expressed 1 or more circRNAs DE in KMT2A::AFF1 ALL, most of them being upregulated in 1 or both comparisons (supplemental Figure 4).
Figure 3.
Relation between circRNAs dysregulated in KMT2A::AFF1 ALL and known KMT2Are signature genes. (A) Custom GSEA of the host genes expressing the 86 circRNAs most dysregulated in KMT2A::AFF1 using custom assembled KMT2Are/KMT2A::AFF1 meta-signature of 1881 genes previously associated with the disease. (B) Bar plot of the number of circRNAs dysregulated in KMT2A::AFF1 expressed from genes belonging to the KMT2Are/KMT2A::AFF1 meta-signature. The heat map shows the genes belonging to each original signature (dark gray); the columns are clustered according to the Dice distance, and the rows according to the Sokal and Michener distance; the marginal dot plots display the number signatures that contain each gene (bottom) and the fraction of genes of each signature that host at least 1 circRNA (right); information about the original signatures considered is given by the left bars. NES, normalized enrichment score.
Up to 6 (PAN3) upregulated circRNAs were displayed by 16 host genes previously included in at least 1 signature (Figure 3B). Most DE circRNAs we detected were not expressed by genes previously associated with KMT2A rearrangements and represent novel findings of loci potentially associated with this disease.
More than 80% of the signature gene circRNAs had a poorly altered CLP in KMT2A::AFF1 ALL, including circHIPK3, circNR3C1, circRNF220, and circTBC1D14, with AE markedly increased in leukemia cells, whose previously reported gene upregulation could be linked to the circRNA overexpression.
CircAFF1 upregulation is linked to HOXA KMT2A::AFF1 ALL
Using RNA-seq analysis in the discovery cohort, we detected 21 circular isoforms of the AFF1 gene (Figure 4A). The most expressed circRNA formed by backsplicing of exons 3 and 4 (4:87046166-87047594, named circAFF1_A), previously shown to be upregulated in KMT2A::AFF1 ALL,40 was not significantly increased in leukemia cells according to our RNA-seq data. Instead, we detected a significant upregulation of a circular isoform formed by backsplicing of exon 5 with 7 (4:87084120-87091829, circAFF1_B) and a very short one (circularized exon 5; 4:87084120-87084164, circAFF1_C). Further quantification by qRT-PCR in an extended pediatric patient cohort (32 KMT2A::AFF1 ALL and 12 BCP-LL non–KMT2A-rearranged and bone marrow samples from 5 HDs) showed a higher expression of circAFF1_B in KMT2A::AFF1 ALL than in non-KMT2Are BCP-ALL, however, without a dramatic increase compared with the normal counterpart (Figure 4B). The average expression of circAFF1_A was significantly higher in KMT2A::AFF1 ALL than in controls and non-KMT2Are cases (Figure 4B). Notably, in the KMT2A::AFF1 ALL cohort, circAFF1_A expression was heterogeneous (Figure 4C), and stratifying the cases based on circAFF1_A expression, we detected a different expression profile in the 2 groups. The genes more upregulated in cases with high circAFF1_A expression (greater than the median; Figure 4D) were significantly enriched, according to GSEA based on C2CGP MSigDB, in genes characterizing the KMT2A signatures, including HOX cluster genes (Figure 4E). On the contrary, patients with less circAFF1_A expression showed an upregulation of IRX1 and IRX2 genes (Figure 4D).
Figure 4.
CircAFF1_A and circAFF1_B isoform structure and expression in the extended patient cohort. (A) Expression levels of the circAFF1 isoforms in samples from patients with KMT2A::AFF1 ALL and CD34+ cells and B cells populations from HDs, according to RNA-seq analysis (significant DE is indicated by ∗). (B) Schematic representation of circAFF1_A and circAFF1_B structure (exons number according to ENST00000395146 transcript; colored boxes represent exons; gray segments, introns; and arches, circRNAs;) and bar graphs of expression levels quantified by qRT-PCR in bone marrow samples from HDs (N = 3), in pediatric patients with BCP-ALL, 31 with (KMT2A::AFF1, N = 31 for circAFF1_A and N = 17 for circAFF1_B) and 13 without (OTH) KMT2A::AFF1 rearrangement (mean ± standard error of the mean is shown; Mann-Whitney U test P value ∗P < .05 and ∗∗P < .01). (C) circAFF1_A level stratify patients into high- and low-expressors groups. (D) Patients with high and low circAFF1_A expression level are characterized by different genes expression profiles according to microarrays analysis (the top 50 ranked genes differentially expressed are shown in the heat map); (E) MLL signatures resulted positively enriched among genes differentially expressed between patients with high and low circAFF1_A expression. bks, backsplice site; FDR, false discovery rate.
Collectively, these data indicate that circAFF1_A expression is associated with the previously identified gene expression KMT2A subgroups (ie, HOXA and IRX subtype).
Functional screening identified the oncogenic role of circFKBP5, circKLHL2, circNR3C1, and circPAN3 in KMT2A::AFF1 ALL in vitro
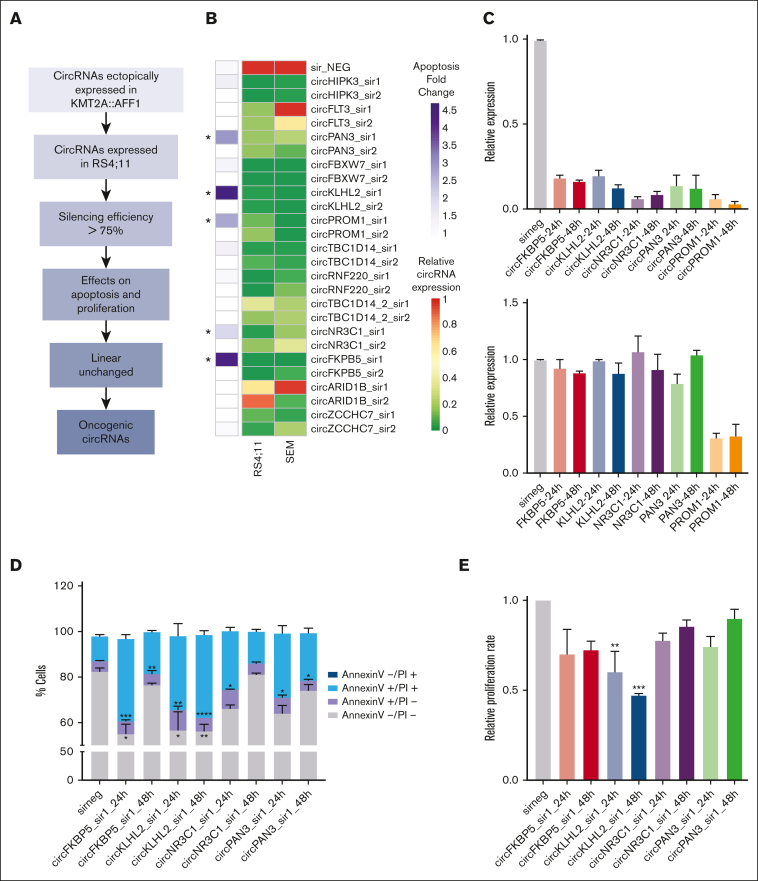
Next, we conducted an extensive functional screening using a loss-of-function approach in vitro to identify oncogenic circRNAs in KMT2A::AFF1 cells (Figure 5A). We prioritized 27 circRNAs to be functionally studied, for which high expression in KMT2A::AFF1 ALL was confirmed by additional RNA-seq data of patient-derived xenograft samples and the RS4;11 cell line (supplemental Figure 5), including the 7 (circPROM1, circFLT3, circNR3C1, circPAN3, circKLHL2, circFKBP5, and circHIPK3) previously validated in the extended cohort (Figure 2).
Figure 5.
Functional screening of circRNAs using a loss-of-function approach in vitro. (A) Workflow of the strategy used for circRNA prioritization and functional screening. (B) Results of the semimassive siRNA screening: the heat map on the left shows the average relative apoptotic rate with respect to sirNEG (measured as relative percentage of annexin and propidium iodide (PI) marked, ie, the sum of annexin-positive/PI-positive and annexin-positive/PI-negative; ∗indicates LFC > 1.5) upon silencing of 13 circRNAs with high siRNA efficiency (right columns; at least 75% of circRNA expression reduction in SEM and RS4;11 cell lines; relative circRNA expression measured by qRT-PCR and expressed as 2-ΔΔCt using sir_NEG as calibrator and GAPDH as reference gene). (C) Relative circular RNA (top) and linear transcript (bottom) expression in the silenced samples compared to the sirNEG (gray bar). (D) Percentage of annexin-PI marked cells and (E) relative cell proliferation rate according to EdU assay upon circRNA silencing at 24 hours and 48 hours. For each bar, data are reported as the mean ± standard error of the mean of at least 3 independent replicates in RS4;11.
We designed a custom siRNA panel, including 2 siRNAs targeting each of the selected circRNA molecules, specific for the backsplicing region and without any predicted human off-target transcripts, plus sirNEG siRNAs (supplemental Table 1). Thirteen circRNAs were silenced with an efficiency higher than 75% in both RS4;11 and SEM cell lines 24 hours after transfection by at least 1 of the siRNAs tested (Figure 5B).
A first functional test revealed an increased percentage of apoptotic cells after silencing of circFKBP5, circKLHL2, circNR3C1, circPAN3, and circPROM1 (Figure 5B). Silencing of circFKBP5, circKLHL2, circNR3C1, and circPAN3 (Figure 5C, top) did not alter the expression of the linear counterpart (Figure 5C, bottom). Further in vitro experiments were performed considering these 4 circRNAs, using, for each of them, the siRNA with the best silencing performance.
For all the 4 tested circRNAs, the annexin-PI assay from at least 3 independent replicates confirmed an increased ratio of apoptotic cells upon in vitro silencing in KMT2A::AFF1 specimens with respect to sirNEGs, both 24 hours and 48 hours (Figure 5D). The effect was particularly marked for circFKBP5 and circKLHL2. Moreover, the EdU-incorporation assay highlighted a tendency toward reduced cell proliferation after knockdown of these 4 circRNAs, with a reduction of ≤50% compared with that in the control (Figure 5E). A high effect was observed upon circKLHL2 silencing, with a significant reduction at both time points (24 hours P = .0032 and 48 hours P = .0003). Results supported an oncogenic role of circFKBP5, circKLHL2, circNR3C1, and circPAN3 ectopically expressed in KMT2A::AFF1 ALL.
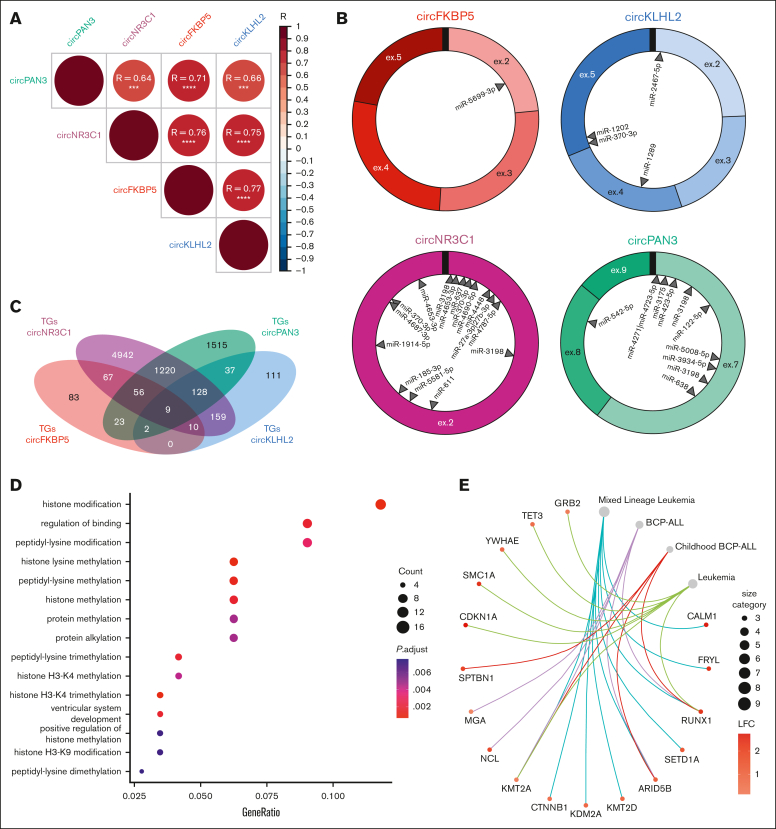
Next, the high correlation between the expression profiles of these 4 oncogenic circRNAs in the enlarged cohort of patients with KMT2A::AFF1 at diagnosis (Figure 6A; supplemental Figure 6) motivated a further investigation of their possible overlapping networks of action, potentially involving common miRNAs and target genes. For each of the 4 circRNAs, binding sites were predicted for 1 up to 14 miRNAs, for a total of 21 miRNAs (Figure 6B). CircNR3C1 shared binding sites for miR-370-3p with circKLHL2 and for miR-3198 with circPAN3. Despite the limited number of predicted binding miRNAs shared between the 4 circRNAs, we found 205 validated miRNA target genes potentially commonly regulated by at least 3 of these 4 circRNAs, mainly through different miRNAs (Figure 6B-C). Upon GSEA, these genes were significantly (false discovery rate < 0.05) associated with histone modification and methylation, key biological processes affected by the KMT2A::AFF1 chimera complex (Figure 6D) and the KMT2A wild-type protein (Figure 6E; supplemental Table 7). According to KMT2A::AFF1 CHIP-seq data available for SEM and RS4;11 cell lines,56 only 14 of the genes targeted by miRNAs potentially sequestered by the circRNAs under consideration are direct KMT2A::AFF1 chimera targets (Figure 6E), including ARID5B and RUNX1, which are strongly associated with this leukemia. Overall, these data suggest that the identified ectopically expressed oncogenic circRNAs could operate as a reinforcement of the KMT2A::AFF1 chimera activity, although prevalently by regulatory action on genes not under the direct control of the fusion protein.
Figure 6.
Correlated expression and putative functions of oncogenic circFKBP5, circKLHL2, circNR3C1, and circPAN3 ectopically expressed in KMT2A::AFF1 ALL. CircFKBP5, circKLHL2, circNR3C1, and circPAN3 (A) pairwise expression correlation in a cohort of 17 pediatric patients with KMT2A::AFF1 ALL; (B) miRNA-binding sites predicted by CRAFT; (C) venn diagram of the overlap of validated target genes (TGs) of miRNAs potentially bound by circFKBP5, circKLHL2, circNR3C1 and circPAN3; overrepresentation of (D) biological processes; and (E) diseases among genes commonly linked, through miRNAs, to at least 3 of the 4 oncogenic circRNAs identified and with positive LFC comparisons of KMT2A::AFF1 ALL samples with CD34+ cells from HDs.
Discussion
In this study, we described for the first time, to our knowledge, the circRNAome of pediatric KMT2A::AFF1 translocated leukemia, bringing to light new players that sustain the molecular mechanisms contributing to leukemogenesis.
Through deep analysis of RNA-seq data with cutting-edge bioinformatic methods, we observed that the circRNA landscape is dysregulated in patients with KMT2A::AFF1 ALL, with circRNAs aberrantly expressed in malignant cells compared with the normal counterpart. Most of the dysregulated circRNAs were ectopically expressed in patients, thus being candidate oncogenic molecules.
Interestingly, several circRNAs are aberrantly overexpressed and derived from host genes associated with KMT2A::AFF1 gene signatures, including PROM1, FLT3, PAN3, ZCCHC7, TBC1D14, HIPK3, and RNF220.57,58 Nevertheless, most of the altered circRNAs we detected did not derive from genes previously associated with KMT2Are, in general, or with the KMT2A::AFF1 fusions, in particular, and indicated several new loci, including FKBP5 and KLHL2, with a potential role in this disease. An orthogonal analysis of the CLP in KMT2A::AFF1 ALL showed that most (80%) of the circRNAs overexpressed in leukemia cells followed the expression profiles of their linear counterparts, whereas we also found genes in which the circRNAs accounted for a significantly increased expression proportion in KMT2A::AFF1 than in healthy cells. Collectively, these observations support the added value of the study of circRNA expression and potential associations to the molecular mechanism of this disease. The observed excess of upregulated circRNAs in leukemia cells can be at least partly explained by the activation of transcriptional programs in KMT2A::AFF1 ALL because several of them are expressed from known chimera target genes. Nevertheless, the CLP increase in malignant cells indicates that disturbances of posttranscriptional processes, including RNA splicing dysregulation, can play a role.
Several circular isoforms of the AFF1 gene were expressed in the samples, including circAFF1_A previously shown to be upregulated in KMT2A::AFF1 ALL.40 Here, we disclosed that patients can be classified according to the expression level of circAFF1_A and that the cases with the highest expression of this circRNA present a specific gene expression profile, with a marked upregulation of KMT2A signature genes, particularly HOX cluster genes. A low circAFF1_A expression was instead associated with upregulation of the IRX1 and IRX2 genes. Of novelty, our examination of a sizable cohort disclosed that the features associated with high expression of circAFF1_A are not a general characteristic of KMT2A::AFF1 ALL but specific to a patient subgroup. Several studies have reported that patients with t(4;11) can be grouped based on HOXA gene expression and that the missing expression of these genes is correlated with high IRX1 or IRX2.34,59 Indeed, we demonstrated that high expression of circAFF1 is associated with a high HOXA expression, AFF1 overexpression, and a KMT2A-leukemia related signature, emphasizing the explanation of its role to regulate KMT2A::AFF1 expression.40 Differently, in patients with activation of the IRX homeobox protein, either due to the absence of AFF1::KMT2A expression or to its repression, circAFF1-mediated regulation can be lacking.
We did not see upregulation in KMT2A::AFF1 ALL of the circRNAs expressed from KMT2A, in line with our previous data in KMT2A::MLLT1 (MLL::ENL) cases26 and the recent interesting finding that a circKMT2A isoform overexpressed before disease onset plays a role in the acquisition of KMT2Are.60
Among the circRNAs most aberrantly expressed in KMT2A::AFF1 ALL, circHIPK361, 62, 63, 64 and circZNF60947,65 had been previously described as being deregulated in other tumors and developmental processes. Regarding pediatric leukemias, we showed the upregulation of circHIPK3 in different subtypes of BCP-ALL18 and a possible oncogenic role of circZNF609 in T-cell ALL.20 Other circRNAs upregulated in KMT2A::AFF1 ALL are lacking functional information to our knowledge, particularly circFKBP5. CircKLHL2 was shown to be upregulated in multiple myeloma.66 CircNR3C1 involvement in tumorigenic processes was described in different contexts but not in leukemias. An oncogenic role of circPAN3 in acute myeloid leukemia has been reported, linking its overexpression to doxorubicin resistance.9 Beyond circPAN3, it is worth noting that other circRNAs previously associated with acute myeloid leukemia,67 such as circFBXW721 and circRNF220,68 are overexpressed in KMT2A::AFF1 ALL.
Because most of the identified upregulated circRNAs were not previously characterized, we set up a functional screening using specific circRNA silencing, through which we identified new circRNAs with oncogenic potential in KMT2A::AFF1 ALL in vitro. Starting with 27 circRNAs, for about half of them, we obtained an efficient silencing in 2 cell lines. The specific silencing of circFKBP5, circKLHL2, circNR3C1, and circPAN3 had an effect on the cell apoptosis rate. More in-depth functional investigation provided robust data about the oncogenic roles of these circRNAs in KMT2A::AFF1 ALL in vitro. The silencing of each of these 4 circRNA, particularly circFKBP5, circKLHL2, significantly increased the ratio of apoptotic cells, indicating that their overexpression sustains the survival of KMT2A::AFF1 ALL cells. In addition, reduced cell proliferation upon the silencing of these circRNAs was observed, with the highest effect linked to circKLHL2 silencing. Overall, these results support an oncogenic role of each of these 4 circRNAs in KMT2A::AFF1 ALL, which is the most important highlight of our study.
In our data, several circRNAs were altered in malignant cells, and at least 4 of them were proven to have oncogenic properties by functional investigation. In our view, the attribution of leukemogenic potential to single circRNA molecules can be overly simplistic and somewhat fail to account for the pleiotropic roles of these molecules and for the possible interplay of circRNAs concurrently dysregulated in malignant cells. Along this line and because of the observation of a significant correlation between oncogenic circRNA expression in the extended cohort of KMT2A::AFF1 ALL cases, we explored the possible regulatory networks controlled by the oncogenic circFKBP5, circKLHL2, circNR3C1, and circPAN3 in this malignancy. Integrative analysis of the miRNA binding potential of these circRNAs and of the expression of validated target miRNA genes disclosed a set of genes for which upregulation could be sustained by circRNA overexpression. These genes showed a notable enrichment of leukemia-associated genes, most of which are not under direct control of the KMT2A::AFF1 chimera complex. These observations, together with the results of our functional studies in vitro, suggest oncogenic circRNA upregulation as an additional mechanism that sustains the malignant cell phenotype in KMT2A::AFF1 ALL. In the future, more in-depth functional studies, including in vivo experiments, will be needed to provide a clearer vision of the leukemogenic potential of circRNAs in this neoplasm. Nevertheless, our data are laying new bases for the development of innovative RNA-based therapies.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Acknowledgments
This work was supported by grants from Fondazione AIRC per la Ricerca sul Cancro (IG 2017 #20052) (S. Bortoluzzi), Fondazione Cariparo (FCR 31/05) (S. Bresolin and B.B.), and Fondazione Umberto Veronesi (E.G. and A.C.).
Authorship
Contribution: S. Bortoluzzi, S. Bresolin, and G.t.K. conceived the study; S. Bortoluzzi and S. Bresolin supervised the study development; E.B., P.G., A.M.V., G.C., A.B., F.L., B.B., and L.H.M. provided patient and control samples; C.T.P., G.L., A.C. and V.S. performed experiments; A.D.M., S. Bresolin, and S. Bortoluzzi did bioinformatic analysis, E.G. and A.B. provided bioinformatics methods. C.T.P., A.D.M., S. Bresolin and S.Bortoluzzi prepared tables and figures; A.D.M., S. Bresolin and S. Bortoluzzi wrote the manuscript; and all authors revised and approved the manuscript.
Footnotes
C.T.P. and A.D.M. contributed equally to this study.
S. Bresolin and S. Bortoluzzi contributed equally to this study.
All the RNA-seq data of the study, including samples of leukemia cells of patients with B-cell precursor acute lymphoblastic leukemia harboring MLL::AF4 rearrangements, patient-derived xenografts and the RS4;11 cell line, and CD34+ hematopoietic stem cells (GSE213172) and CD19+ B cells of healthy donors (GSE110159), are deposited in the Gene Exoression Omnibus database (accession nmber GSE213990).
The full-text version of this article contains a data supplement.
Supplementary Material
References
- 1.Bonizzato A, Gaffo E, Te Kronnie G, Bortoluzzi S. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016;6(10):e483. doi: 10.1038/bcj.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visci G, Tolomeo D, Agostini A, Traversa D, Macchia G, Storlazzi CT. CircRNAs and Fusion-circRNAs in cancer: new players in an old game. Cell Signal. 2020;75 doi: 10.1016/j.cellsig.2020.109747. [DOI] [PubMed] [Google Scholar]
- 3.Dahl M, Daugaard I, Andersen MS, et al. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab Invest. 2018;98:1657–1669. doi: 10.1038/s41374-018-0108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng WL, Mohidin TBM, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018:1–11. doi: 10.1080/15476286.2018.1486659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou X, Du J. CircRNAs: novel therapeutic targets in multiple myeloma. Mol Biol Rep. 2022 doi: 10.1007/s11033-022-07668-8. [DOI] [PubMed] [Google Scholar]
- 6.Sharma AR, Banerjee S, Bhattacharya M, Saha A, Lee SS, Chakraborty C. Recent progress of circular RNAs in different types of human cancer: Technological landscape, clinical opportunities and challenges (review) Int J Oncol. 2022;60 doi: 10.3892/ijo.2022.5346. [DOI] [PubMed] [Google Scholar]
- 7.Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu J, Han Q, Gu Y, et al. Circular RNA PVT1 expression and its roles in acute lymphoblastic leukemia. Epigenomics. 2018;10:723–732. doi: 10.2217/epi-2017-0142. [DOI] [PubMed] [Google Scholar]
- 9.Shang J, Chen W-M, Wang Z-H, et al. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp Hematol. 2019;70:42–54.e3. doi: 10.1016/j.exphem.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by circPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji X, Ding W, Xu T, et al. MicroRNA-31-5p attenuates doxorubicin-induced cardiotoxicity via quaking and circular RNA Pan3. J Mol Cell Cardiol. 2020;140:56–67. doi: 10.1016/j.yjmcc.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. Mol Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicolet BP, Engels S, Aglialoro F, et al. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 2018;46:8168–8180. doi: 10.1093/nar/gky721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaffo E, Boldrin E, Dal Molin A, et al. Circular RNA differential expression in blood cell populations and exploration of circRNA deregulation in pediatric acute lymphoblastic leukemia. Sci Rep. 2019;9(1):14670. doi: 10.1038/s41598-019-50864-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dal Molin A, Hofmans M, Gaffo E, et al. CircRNAs dysregulated in juvenile myelomonocytic leukemia: CircMCTP1 stands out. Front Cell Dev Biol. 2021;8:613540. doi: 10.3389/fcell.2020.613540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buratin A, Paganin M, Gaffo E, et al. Large-scale circular RNA deregulation in T-ALL: unlocking unique ectopic expression of molecular subtypes. Blood Adv. 2020;4:5902–5914. doi: 10.1182/bloodadvances.2020002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papaioannou D, Volinia S, Nicolet D, et al. Clinical and functional significance of circular RNAs in cytogenetically normal AML. Blood Adv. 2020;4:239–251. doi: 10.1182/bloodadvances.2019000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lux S, Blätte TJ, Gillissen B, et al. Deregulated expression of circular RNAs in acute myeloid leukemia. Blood Adv. 2021;5:1490–1503. doi: 10.1182/bloodadvances.2020003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agraz-Doblas A, Bueno C, Bashford-Rogers R, et al. Unraveling the cellular origin and clinical prognostic markers of infant B-cell acute lymphoblastic leukemia using genome-wide analysis. Haematologica. 2019;104:1176–1188. doi: 10.3324/haematol.2018.206375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghetti M, Vannini I, Storlazzi CT, Martinelli G, Simonetti G. Linear and circular PVT1 in hematological malignancies and immune response: two faces of the same coin. Mol Cancer. 2020;19:69. doi: 10.1186/s12943-020-01187-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 26.Dal Molin A, Bresolin S, Gaffo E, et al. CircRNAs are here to stay: a perspective on the MLL recombinome. Front Genet. 2019:10. doi: 10.3389/fgene.2019.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan AKN, Chen C-W. Rewiring the epigenetic networks in MLL-rearranged leukemias: epigenetic dysregulation and pharmacological interventions. Front Cell Dev Biol. 2019;7 doi: 10.3389/fcell.2019.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Sluis IM, de Lorenzo P, Kotecha RS, et al. Blinatumomab added to chemotherapy in infant lymphoblastic leukemia. N Engl J Med. 2023;388:1572–1581. doi: 10.1056/NEJMoa2214171. [DOI] [PubMed] [Google Scholar]
- 29.Chromosomal abnormalities and their clinical significance in acute lymphoblastic leukemia. Third International Workshop on Chromosomes in Leukemia. Cancer Res. 1983;43:868–873. [PubMed] [Google Scholar]
- 30.Fujita TC, Sousa-Pereira N, Amarante MK, Watanabe MAE. Acute lymphoid leukemia etiopathogenesis. Mol Biol Rep. 2021;48:817–822. doi: 10.1007/s11033-020-06073-3. [DOI] [PubMed] [Google Scholar]
- 31.Metzler M, Forster A, Pannell R, et al. A conditional model of MLL-AF4 B-cell tumourigenesis using invertor technology. Oncogene. 2006;25:3093–3103. doi: 10.1038/sj.onc.1209636. [DOI] [PubMed] [Google Scholar]
- 32.Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamai H, Miyake K, Takatori M, et al. Activated K-Ras protein accelerates human MLL/AF4-induced leukemo-lymphomogenicity in a transgenic mouse model. Leukemia. 2011;25:888–891. doi: 10.1038/leu.2011.15. [DOI] [PubMed] [Google Scholar]
- 34.Trentin L, Giordan M, Dingermann T, et al. Two independent gene signatures in pediatric t(4;11) acute lymphoblastic leukemia patients. Eur J Haematol. 2009;83:406–419. doi: 10.1111/j.1600-0609.2009.01305.x. [DOI] [PubMed] [Google Scholar]
- 35.Stam RW, Schneider P, Hagelstein JAP, et al. Gene expression profiling-based dissection of MLL translocated and MLL germline acute lymphoblastic leukemia in infants. Blood. 2010;115:2835–2844. doi: 10.1182/blood-2009-07-233049. [DOI] [PubMed] [Google Scholar]
- 36.Kerry J, Godfrey L, Repapi E, et al. MLL-AF4 spreading identifies binding sites that are distinct from super-enhancers and that govern sensitivity to DOT1L inhibition in leukemia. Cell Rep. 2017;18:482–495. doi: 10.1016/j.celrep.2016.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prange KHM, Mandoli A, Kuznetsova T, et al. MLL-AF9 and MLL-AF4 oncofusion proteins bind a distinct enhancer repertoire and target the RUNX1 program in 11q23 acute myeloid leukemia. Oncogene. 2017;36:3346–3356. doi: 10.1038/onc.2016.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice S, Jackson T, Crump NT, et al. A human fetal liver-derived infant MLL-AF4 acute lymphoblastic leukemia model reveals a distinct fetal gene expression program. Nat Commun. 2021;12:6905. doi: 10.1038/s41467-021-27270-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dal Molin A, Tretti Parenzan C, Gaffo E, et al. Discovery of fusion circular RNAs in leukemia with KMT2A::AFF1 rearrangements by the new software CircFusion. Brief Bioinform. 2023;24(1):bbac589. doi: 10.1093/bib/bbac589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang W, Fang K, Chen T-Q, et al. CircRNA circAF4 functions as an oncogene to regulate MLL-AF4 fusion protein expression and inhibit MLL leukemia progression. J Hematol Oncol. 2019;12:103. doi: 10.1186/s13045-019-0800-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaffo E, Bonizzato A, Kronnie G, Bortoluzzi S. CirComPara: a multi-method comparative bioinformatics pipeline to detect and study circRNAs from RNA-seq data. Noncoding RNA. 2017;3:8. doi: 10.3390/ncrna3010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaffo E, Buratin A, Dal Molin A, Bortoluzzi S. Sensitive, reliable and robust circRNA detection from RNA-seq with CirComPara2. Brief Bioinform. 2022;23 doi: 10.1093/bib/bbab418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dal Molin A, Gaffo E, Difilippo V, et al. CRAFT a bioinformatics software for custom prediction of circular RNA functions. Brief Bioinform. 2022:23. doi: 10.1093/bib/bbab601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature Commun. 2016;7(1) doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossi F, Legnini I, Megiorni F, et al. Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene. 2019;38:3843–3854. doi: 10.1038/s41388-019-0699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie F, Xiao X, Tao D, et al. CircNR3C1 suppresses bladder cancer progression through acting as endogenous blocker of BRD4/C-myc complex. Mol Ther Nucleic Acids. 2020 doi: 10.1016/j.omtn.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng F, Wang M, Li Y, et al. CircNR3C1 inhibits proliferation of bladder cancer cells by sponging miR-27a-3p and downregulating cyclin D1 expression. Cancer Lett. 2019;460:139–151. doi: 10.1016/j.canlet.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 50.Mullighan CG, Kennedy A, Zhou X, et al. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007;21:2000–2009. doi: 10.1038/sj.leu.2404808. [DOI] [PubMed] [Google Scholar]
- 51.Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 52.Rozovskaia T, Ravid-Amir O, Tillib S, et al. Expression profiles of acute lymphoblastic and myeloblastic leukemias with ALL-1 rearrangements. Proc Natl Acad Sci U S A. 2003;100:7853–7858. doi: 10.1073/pnas.1132115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schraets D, Lehmann T, Dingermann T, Marschalek R. MLL-mediated transcriptional gene regulation investigated by gene expression profiling. Oncogene. 2003;22:3655–3668. doi: 10.1038/sj.onc.1206438. [DOI] [PubMed] [Google Scholar]
- 54.Gaussmann A, Wenger T, Eberle I, et al. Combined effects of the two reciprocal t(4;11) fusion proteins MLL·AF4 and AF4·MLL confer resistance to apoptosis, cell cycling capacity and growth transformation. Oncogene. 2007;26:3352–3363. doi: 10.1038/sj.onc.1210125. [DOI] [PubMed] [Google Scholar]
- 55.Lin S, Luo RT, Ptasinska A, et al. Instructive role of MLL-fusion proteins revealed by a model of t(4;11) pro-B acute lymphoblastic leukemia. Cancer Cell. 2016;30:737–749. doi: 10.1016/j.ccell.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 56.Wilkinson AC, Ballabio E, Geng H, et al. RUNX1 is a key target in t(4;11) leukemias that contributes to gene activation through an AF4-MLL complex interaction. Cell Rep. 2013;3:116–127. doi: 10.1016/j.celrep.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bueno C, Ayllón V, Montes R, et al. FLT3 activation cooperates with MLL-AF4 fusion protein to abrogate the hematopoietic specification of human ESCs. Blood. 2013;121:3867–3878. doi: 10.1182/blood-2012-11-470146. S1–3. [DOI] [PubMed] [Google Scholar]
- 58.Godfrey L, Crump NT, O’Byrne S, et al. H3K79me2/3 controls enhancer-promoter interactions and activation of the pan-cancer stem cell marker PROM1/CD133 in MLL-AF4 leukemia cells. Leukemia. 2021;35:90–106. doi: 10.1038/s41375-020-0808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isobe T, Takagi M, Sato-Otsubo A, et al. Multi-omics analysis defines highly refractory RAS burdened immature subgroup of infant acute lymphoblastic leukemia. Nat Commun. 2022;13:4501. doi: 10.1038/s41467-022-32266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conn VM, Gabryelska M, Toubia J, et al. Circular RNAs drive oncogenic chromosomal translocations within the MLL recombinome in leukemia. Cancer Cell. 2023;41(7):1309–1326. doi: 10.1016/j.ccell.2023.05.002. [DOI] [PubMed] [Google Scholar]
- 61.Wen Y, Li B, He M, et al. CircHIPK3 promotes proliferation and migration and invasion via regulation of miR-637/HDAC4 signaling in osteosarcoma cells. Oncol Rep. 2021;45:169–179. doi: 10.3892/or.2020.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hong W, Zhang Y, Ding J, et al. CircHIPK3 acts as competing endogenous RNA and promotes non-small-cell lung cancer progression through the miR-107/BDNF signaling pathway. Biomed Res Int. 2020;2020:6075902. doi: 10.1155/2020/6075902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen G, Shi Y, Liu M, Sun J. CircHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175. doi: 10.1038/s41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin C, Zhao W, Zhang Z, Liu W. Silencing circular RNA circZNF609 restrains growth, migration and invasion by up-regulating microRNA-186-5p in prostate cancer. Artif Cells Nanomed Biotechnol. 2019;47:3350–3358. doi: 10.1080/21691401.2019.1648281. [DOI] [PubMed] [Google Scholar]
- 66.Zhou F, Wang D, Wei W, et al. Comprehensive profiling of circular RNA expressions reveals potential diagnostic and prognostic biomarkers in multiple myeloma. BMC Cancer. 2020;20:40. doi: 10.1186/s12885-020-6515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rahmati A, Mafi A, Soleymani F, et al. Circular RNAs: pivotal role in the leukemogenesis and novel indicators for the diagnosis and prognosis of acute myeloid leukemia. Front Oncol. 2023;13:1149187. doi: 10.3389/fonc.2023.1149187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X, Liu X, Cai M, et al. CircRNF220, not its linear cognate gene RNF220, regulates cell growth and is associated with relapse in pediatric acute myeloid leukemia. Mol Cancer. 2021;20:139. doi: 10.1186/s12943-021-01395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.