Abstract
Nuclear hormone receptors exert transcriptional activation of target genes upon hormone induction via interactions with the basal transcription machinery. This interaction is mediated by cofactors which physically bind to receptors, thereby acting as coactivators or corepressors leading to activation or repression, respectively. Here we report the screening for and cloning of a peroxisome proliferator receptor-interacting protein, the rat homolog of TIF2. By sequence comparison with the related coactivator SRC-1, we identified three short conserved motifs (NR boxes) in both proteins which are the putative binding sites of TIF2 to nuclear hormone receptors. We demonstrate here by generation of amino acid exchanges within the NR boxes that all three boxes located in the receptor interaction domain of TIF2 are necessary and sufficient for interaction. The three boxes individually can bind to hormone receptors but display preferences in binding for certain receptors. In addition, we show that the interaction domain of TIF2 can compete with other AF-2-dependent cofactors for binding to receptors. Finally, we demonstrate cooperative binding of two TIF2 molecules to a heterodimeric nuclear receptor complex even in the presence of only one cognate ligand, indicating an allosteric effect on the heterodimeric partner upon coactivator binding.
Nuclear hormone receptors are transcription factors which activate transcription of their target genes upon ligand binding. The nuclear receptors can be divided into two subgroups. One subgroup includes the receptors for steroid hormones, e.g., glucocorticoid receptor (GR), estrogen receptor (ER), and progesterone receptor. These receptors are associated with heat shock protein 90 in the non-ligand-bound form and bind after induction by hormone to palindromic recognition sites as homodimers. The members of the other subgroup do not interact with heat shock protein 90 and bind preferentially to direct repeats as heterodimers with retinoid X receptor (RXR) (28). To this group belong, among others, the receptors for thyroid hormone (TR), retinoids (retinoic acid receptor [RAR] and RXR), vitamin D3, and peroxisome proliferators (PPAR). Nuclear hormone receptors are composed of three domains: an N-terminal domain containing a transactivation function (AF-1), a DNA binding domain (DBD) harboring two zinc finger motifs, and the C-terminal ligand binding domain (LBD), which contains the dimerization interface and a second activation domain (AD), AF-2, responsible for ligand-induced activation (2, 4, 12, 13).
Some receptors negatively influence transcription as repressors in the absence of hormone by association with corepressors (9, 20, 23, 31, 33, 34, 47, 48). In the presence of hormone, the corepressors are displaced, and by the subsequent recruitment of coactivators, nuclear receptors exert their positive effects on gene transcription. It has been shown that the binding of coactivators depends on the integrity of helices 11 and 12, located at the C-terminal ends of the receptors (15, 21). A deletion of helix 12, also described as AF-2 AD, τc, or Tau 4, generates a dominant negative receptor with high affinity to corepressors (2, 11, 32). After ligand binding, the LBDs of nuclear receptors undergo a conformational change. Mainly affected by this structural rearrangement is helix 12, which in the non-ligand-bound state protrudes from the LBD into the aqueous phase but which in the ligand-bound conformation is tightly associated with the ligand binding pocket and helices 3 and 4. This modifying effect on the receptor structure exerted by the ligand is a very conserved mechanism within the group of ligand-dependent nuclear hormone receptors and is probably a prerequisite for coactivator interaction.
Various AF-2 AD-dependent coactivators have been identified by yeast two-hybrid screening or far-Western expression cloning. One group of coactivators, including SRC-1/NCoA-1 (29, 38), TIF2/GRIP1/NCoA-2 (19, 38, 40), and p/CIP/RAC3/ACTR/AIB1 (1, 8, 27, 38), consists of proteins of about 160 kDa. They share an N-terminal basic helix-loop-helix region, regions of high similarity to PAS A and PAS B domains of PAS/basic helix-loop-helix factors (15a), and a C-terminal glutamine-rich region. It was shown that all three members can coactivate nuclear hormone receptors in yeast and in mammalian cells in a hormone- and AF-2 AD-dependent manner. Several members possess a transactivation function active in yeast as well as in mammalian cells (18, 29, 38, 40). It was demonstrated that SRC-1 can interact with TBP and TFIIB (36). Furthermore, interaction with CBP/p300 as well as with P/CAF was proven, indicating a complex of mutually interacting factors. Besides interactions with downstream targets, a histone acetylase function was demonstrated for the related proteins SRC-1 and ACTR and previously also for CBP and p300 (3, 8, 35). A possible mechanism of coactivation by the SRC-1-related proteins may therefore be a targeted change of chromatin structure, a process thought to be involved in regulation of gene expression by nuclear hormone receptors (39, 45). This possibility is strengthened by the finding that the corepressors SMRT and N-CoR form complexes with Sin3 and proteins which are involved in histone deacetylation, a possible mechanism for gene repression (17, 44).
Recently the short motif LXXLL was identified, within nearly all cofactors, to be indispensable for the interaction with hormone receptors. This motif was first described for TIF1 and designated the NR box (16, 24). Each of the three SRC-1-related coactivators contains several of these motifs. The central interaction domain (IAD) contains three boxes in all three related proteins, whereas only SRC-1 contains an additional motif at its very C terminus.
Here we report the cloning of TIF2 from rats and the characterization of its interaction with different nuclear hormone receptors. To gain further insight into receptor-coactivator assembly, we isolated the nuclear receptor IAD of TIF2. We investigated whether each of the three NR boxes is necessary for interaction with nuclear hormone receptors. We demonstrated synergistic binding of all three modules to different receptors as well as specificities of different NR boxes for certain receptors. Additionally, we provide evidence that NR box-containing IADs of different AF-2-dependent cofactors are able to compete for binding to the nuclear hormone receptor, and furthermore, by analysis of the interaction with heterodimeric DNA-bound receptors, we delineated a novel mechanism of allosteric activation.
MATERIALS AND METHODS
Cloning and mutagenesis of the nuclear receptor IAD.
First, pBKCMV HA was constructed by insertion of a double-stranded oligonucleotide (5′ATCCGCCGCCACCATGGATTACCCATACGACGTCCCAGACTACGCTCAGATCTCCGAATTCC3′) encoding a Kozak translation start site in front of the hemagglutinin (HA) epitope into the BamHI/XhoI-linearized pBKCMV vector (Stratagene). pBKCMV HA TIF2 was constructed by insertion of the fused cDNA fragments into the EcoRI site of pBKCMV HA.
The IAD was amplified by PCR with the construct pBKCMV HA TIF2 as the template. The following primers were used: 5′ primer, 5′TCAGAATTCACAACTGGACAAGCACAGGCC3′, and 3′ primer, 5′CTGTCCAACCGCTCGAGTTTGGGGG3′ Mutated variants of the different modules were constructed by two independent PCRs with the flanking primers indicated above and sets of the following mutagenesis primers: NR box 1ss, 5′GGGCAAACCAAACTCCTACAGGCGGCCACCACCAAGTCCG3′; NR box 1as, 5′CGGACTTGGTGGTGGCCGCCTGTAGGAGTTTGGTTTGCCC3′; NR box 2ss, 5′GCATAAGATTTTGCACAGAGCCGCACAGGACAGCAGCTCCCC3′; NR box 2as, 5′GGGGAGCTGCTGTCCTGTGCGGCTCTGTGCAAAATCTTATGC3′; NR box 3ss, 5′CGCACTACTGCGCTATGCGGCCGACAAAGATGATACTAAAG3′; and NR box 3as, 5′CTTTAGTATCATCTTTGTCGGCCGCATAGCGCAGTAGTGCG3′. The corresponding PCR products were isolated and fused by an additional PCR with the flanking primers. The products of these PCRs were isolated, EcoRI/XhoI digested, and cloned into pBKCMV HA digested with the same enzymes. Representive clones of each generated mutant were analyzed by sequencing. One positive clone of each mutant was amplified in bacteria and, after preparation of plasmid DNA, used for construction of the different vectors.
Plasmid constructs.
The constructs with a GAL4 DBD (amino acids [aa] 1 to 147) fusion were derived from the plasmid pGBT9 or pAS2/2-1 (Clontech). GAL-PPARα LBD (aa 166 to 468), which was used for interaction studies and also served as a bait for the two-hybrid screening, was generated by inserting a PCR fragment into EcoRI/SalI-linearized pGBT9. In addition, all GAL4-receptor LBD fusion proteins, i.e., those with PPARαΔc (aa 166 to 455), rRXRβ (aa 153 to 451), human TRα (hTRα) (aa 122 to 410), and hGR (aa 485 to 777), were expressed in pAS2-1 after cloning of the corresponding PCR fragments into the EcoRI/SalI-linearized vector. All GAL4 AD (GAD) (aa 768 to 881) fusion constructs were derived from the plasmid pGAD-GH (Clontech). The GAD fusions containing the IAD variants were constructed by EcoRI/XhoI digestion of the corresponding pBKCMV HA constructs and cloning into EcoRI/SalI-linearized pGAD-GH.
The plasmids used for in vitro transcription-translation were created by cloning of PCR-amplified fragments of the following yeast two-hybrid constructs into pBKCMV HA linearized with BamHI/XhoI: GAL4-PPARα (aa 166 to 468), GAL4-PPARαΔc (aa 166 to 455), and GAL4-hTRα (aa 122 to 410). The 5′ PCR primers for GAL4 introduce a Kozak translation start site. pT7-hTRβ (aa 1 to 410) was received from Stefan Nilsson; pGEM3Z (Promega), containing rRXRα (aa 1 to 467) has been described previously; and rRXRαΔN (aa 112 to 467) was a gift from D. Feltkamp. Construction of the wild-type IAD of TIF2 (IAD wt) and TIF2 in pBKCMV HA is described above. The SRC-1 construct was a gift from B. W. O’Malley. pEF-RIP140 was a gift from M. G. Parker. The corresponding pBKCMV HA clone was generated by ligation of the PCR product containing the coding region of RIP140 into BglII/XhoI-digested pBKCMV HA. Proteins were synthesized in vitro by using the T3 or T7 RNA polymerase-based rabbit reticulocyte lysate coupled transcription-translation kit (TNT; Promega).
The following glutathione S-transferase (GST) constructs were generated by insertion of fragments of the corresponding pBKCMV HA or yeast two-hybrid constructs into EcoRI/SalI-digested pGEX4T-1 (Pharmacia): hTRα (aa 122 to 410), PPARα (aa 166 to 468), rRXRα (aa 112 to 467), and all IAD variants.
Screening and cloning.
To identify PPARα-interacting proteins, a mouse embryo cDNA library (Clontech) in the vector pGAD10 containing the GAD was transfected into the yeast reporter strain HF7c [MATa ura-52 his3-00 lys2-801 ade2-101 trp1-901 leu2-3,112 gal4-542 gal80-538 LYS::GAL1-HIS3 URA3::(GAL4 17-mer)3-CYC-lacZ] carrying pGBT9-PPARα LBD. More than 3 × 106 transformants were plated onto selective SD medium lacking histidine, leucine, and tryptophan and grown for 3 to 5 days at 30°C. Growing colonies were restreaked onto fresh selective plates. The library DNA from all His+ colonies was isolated by electroporation of total yeast DNA into Escherichia coli HB101. The transformed bacteria were identified by selection on synthetic M9 medium lacking leucine. After classification by PCR and restriction analysis, the different cDNA inserts were sequenced by using the GAD10 5′ primer. About 50% of the interacting clones represent the isoforms of RXR. Additionally, the isolated DNAs represent interacting parts of SMRT, N-CoR, an unknown protein, and two clones with high homologies to hTIF2. These two clones represent aa 320 to 1119 and aa 402 to 1464 plus 3′ the untranslated region of murine TIF2 (mTIF2), respectively. We used 5′ and 3′ regions of the clone containing aa 320 to 1119 as radioactively labeled probes to screen a rat liver cDNA library cloned in the λ-ZAP system (Stratagene) for isolation of the full-length TIF2 sequence. A total of 2 × 106 phage were plated and analyzed after filter lift with the radioactive probes. Eighteen different positive clones were isolated by two subsequent screenings and analyzed by sequencing. The full-length rTIF2 sequence was generated by fusing parts of three different overlapping clones by ligation and finally cloning into the EcoRI site of pBKCMV HA.
Yeast two-hybrid interaction assay.
A two-hybrid mating assay was used to examine interactions between various GAL4-nuclear receptor fusion proteins and different variants of GAD-NR box fusion proteins. GAD plasmids were introduced into the reporter strain Y187 (MATα ura3-52 his3-200 ade2-101 trp1-901 leu2-3, 112, gal4Δ met− gal80ΔURA3::GAL1-lacZ) and selected on plates lacking tryptophan. Growing colonies were mated with HF7c (MATa) carrying the various GAL4 constructs (pGBT9 or pAS2 derivatives) for 12 to 16 h in liquid YPD medium. Selection for the presence of both Leu and Trp plasmids on plates lacking tryptophan and leucine was carried out. The positive clones were used in quantitative liquid β-galactosidase assays.
Expression and purification of GST-tagged proteins.
E. coli BL218(DE3) carrying the pGEX fusion constructs was grown in Luria-Bertani medium containing 0.5% Casamino Acids and 0.5% glucose at 37°C and were induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 to 3 h at 30°C. Bacteria were harvested by centrifugation and lysed in resuspension buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 0.5 mg of lysozyme per ml, 10 mM MgCl2, 1 mM MnCl2, 10 μg of DNase I per ml, 10 μg of RNase A per ml) for 30 min with rotation at 4°C. After centrifugation at 10,000 × g for 30 min at 4°C, the lysates were frozen or immediately used for the binding reaction (pull-down assay). To produce pure GST fusion protein for gel shift assays, supernatants from ca. 500 ml of culture were mixed with 0.5 ml of glutathione-Sepharose 4B (Pharmacia) for 2 h at 4°C. The beads were washed three times with phosphate-buffered saline, eluted with 4 volumes of 10 mM glutathione in 50 mM Tris (pH 8.0), and stored at −70°C. GST fusion protein concentrations were determined by the Bradford dye-binding procedure (Bio-Rad Laboratories). To produce pure NR box protein for GST competition experiments, the beads were washed three times with buffer containing 50 mM Tris-Cl (pH 7.5) and 150 mM NaCl, washed once with cleavage buffer containing in addition 2.5 mM CaCl2, and then cleaved with thrombin in a small volume of cleavage buffer containing 1 μl of thrombin (1 U/μl; Sigma) per 10 ml of bacterial culture for 2 h at room temperature.
In vitro protein-protein interaction assay (GST pull-down assay).
Approximately 5 μg of GST fusion protein was bound to glutathione-Sepharose 4B beads (Pharmacia). The beads were incubated for 2 h with 2 μl of [35S]methionine-labeled protein generated in rabbit reticulocyte lysate by using the TNT coupled in vitro transcription-translation system (Promega) in the presence of 1 μl of ligand in dimethyl sulfoxide (DMSO) (final concentration, between 1 and 100 μM) or DMSO alone in a total volume of 200 μl of incubation buffer (50 mM KPi [pH 7.4], 100 mM NaCl, 1 mM MgCl2, 10% glycerol, 0.1% Tween 20, 1.5% bovine serum albumin [BSA]) with rotation at 4°C. Beads were separated by centrifugation and washed three times for 15 min each with incubation buffer without BSA. Washed beads were resuspended in 50 μl of 1 × sodium dodecyl sulfate sample buffer, and an aliquot was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Before autoradiography, gels were stained with Coomassie blue to control for the stability of the GST fusion proteins and equal loading.
GST pull-down competition assay.
Approximately 1 μg of GST-TR bound to glutathione-Sepharose 4B beads was incubated with 1 μl of NR box protein (about 4 μg/μl) for 1 h at 4°C in the presence of the corresponding hormone. The in vitro-translated protein was then added, and the protocol for the above-described GST pull-down method was followed.
Electrophoretic mobility shift assays.
TRβ and RXRαΔN were synthesized in rabbit reticulocyte lysate by using the TNT coupled in vitro transcription-translation system (Promega). Double-stranded oligonucleotides (1 μg) (synthetic DR4-TRE 5′TCGATCAGGTCATTTCAGGTCAGAG3′) were end labeled with [γ-32P]ATP. Binding reactions were performed in a total volume of 20 μl including 1× reaction buffer [5% glycerol, 5 mM dithiothreitol, 5 mM EDTA, 250 mM KCl, 100 mM HEPES (pH 7.5), 1 μg of poly(dI-dC), 25 mM MgCl2, 1 mg of BSA per ml, 0.05% Triton X-100], 0.5 ng of labeled probe, 2 μl of each in vitro-translated receptor protein, and, where indicated, 1 μl of ligands in DMSO. Finally, the purified GST fusion protein (10 to 500 ng/reaction) was added as indicated in Results. In the case of the pure proteins, 500 ng of IAD wt and box 2 and 2,500 ng of box 3 were added. The binding reaction was allowed to proceed for 20 min on ice before the reaction mixtures were loaded on a 4% nondenaturing polyacrylamide gel containing 5% glycerol. After electrophoresis for 2 h in 0.5× Tris-borate-EDTA at 4°C, the gels were dried and autoradiographed.
RESULTS
Nuclear hormone receptors may stimulate basal transcription upon activation by their corresponding hormones. This effect may be enhanced by so-called coactivators, which interact with the hormone-bound receptors and are thought to constitute a bridge between the hormone receptor and transcription initiation complex. To get further insights into receptor-coactivator assembly and function, we screened a mouse embryo library for interacting proteins by using the yeast two-hybrid system. For this screening we used the LBD of PPARα fused to the GAL4 DBD as a bait and identified various PPAR-interacting proteins. Two of these PPAR-interacting proteins represented fragments highly homologous to the coactivator hTIF2 (40). The first one encoded aa 321 to 1119 of mTIF2, a clone also identified in a yeast two-hybrid screening with the LBD of GR as a bait (19). The second clone encoded the C-terminal end of mTIF2 corresponding to aa 402 to 1464 and additionally represented the full 3′ untranslated region. These cDNAs were used in a screening of a rat liver library for isolation of the rat homolog of hTIF2 (Fig. 1A).
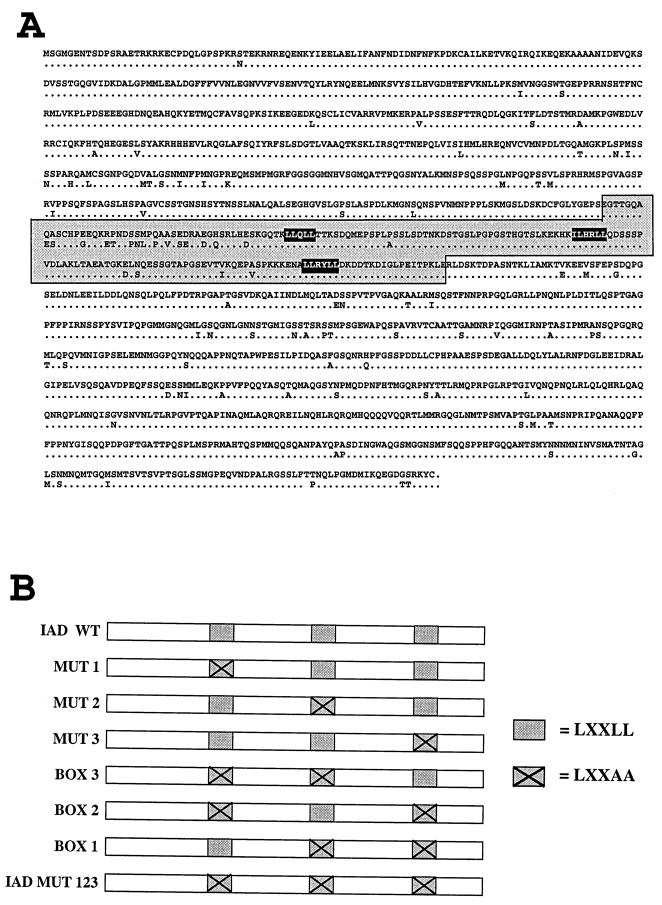
FIG. 1.
(A) The upper sequence shows the amino acid sequence of TIF2. Differences in hTIF2 are shown below this sequence. IAD wt is marked by a grey box. The three NR boxes located within the IAD are marked by black boxes. The three NR boxes, beginning with the N-terminal one, are NR box 1, NR box 2, and NR box 3, respectively. (B) Different variants of the IAD wt were constructed by replacement of two Leu residues by Ala residues within the LXXLL motif as indicated. By mutation of only one box within the otherwise intact IAD, we generated variants named Mut 1, 2, and 3. Mutations of two boxes led to variants harboring only one functional box, named boxes 1, 2, and 3. The variant carrying mutations of all three NR boxes was named IAD Mut 123.
The overlapping region of both screened mTIF2 clones, aa 402 to 1119, presumably includes the IAD responsible for receptor binding. By comparison of this putative TIF2 IAD with the RAR IAD of SRC-1 (45a) we identified three small motifs as the only highly conserved amino acid residues in both corresponding parts of TIF2 and SRC-1. These motifs comprise an LXXLL sequence, and its importance for receptor interaction is consistent with the finding of one single such module in TIF1 (24). In contrast to SRC-1, which contains an additional fourth motif at its C terminus (16, 29), TIF2 harbors only these three elements in its predicted receptor IAD, which we therefore narrowed down to aa 594 to 766 (Fig. 1). During preparation of this paper, publications from other groups appeared describing these motifs as NR boxes. NR boxes are found in a large variety of LBD/AF-2-interacting proteins. (16, 24).
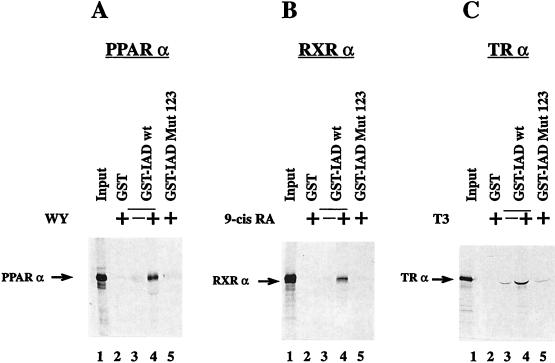
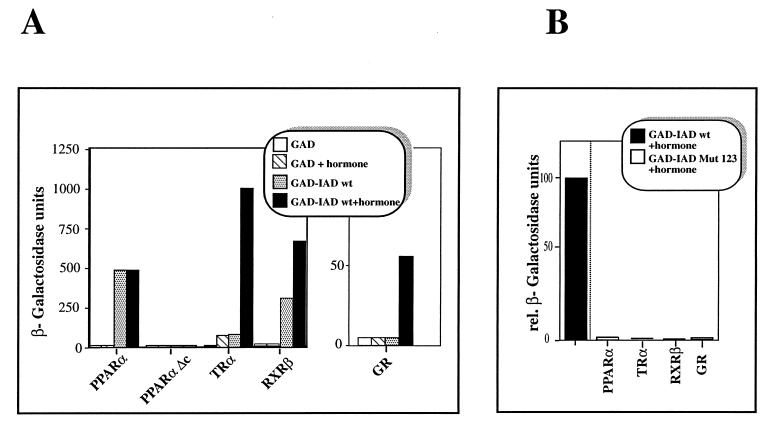
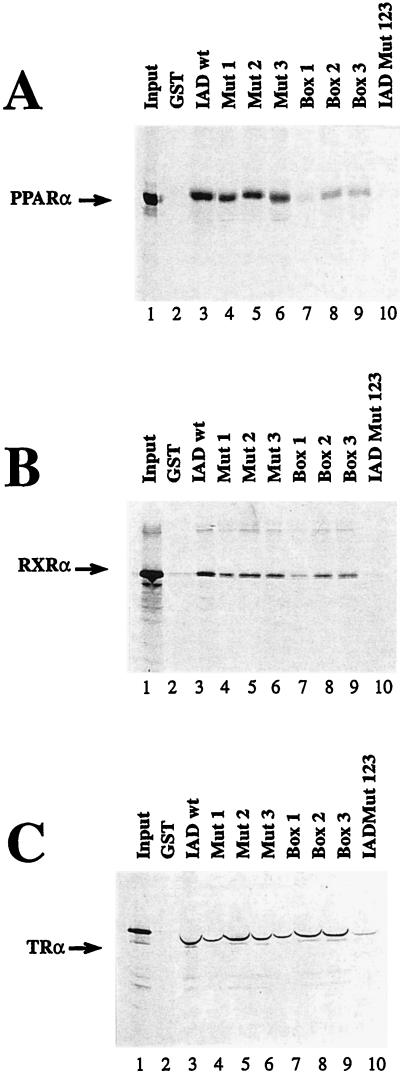
We wondered whether these motifs are necessary and sufficient for receptor binding or whether other parts within the receptor IADs of coactivators also contribute to receptor interaction. We therefore tested a variant of the IAD-containing mutated NR boxes. All three motifs were altered to LXXAA, a change influencing putative hydrophobic interactions. This mutant (IAD Mut 123) (Fig. 1B) was compared with IAD wt for interaction with hormone receptors in in vitro GST pull-down experiments (Fig. 2) as well as in in vivo yeast two-hybrid assays (Fig. 3). In the GST pull-down assay IAD wt, fused to GST, interacts with each tested in vitro-translated receptor, i.e., PPARα (Fig. 2A), RXRα (Fig. 2B), and TRα (Fig. 2C). The interactions with PPARα and RXRα strongly depend on the presence of hormone; hormone-independent interactions with these receptors were not detectable in this assay (compare lanes 3 and 4 in Fig. 2). A hormone-independent interaction with TRα was found, but it was quite insignificant compared with the ligand-induced interaction (Fig. 2C, lanes 3 and 4). In contrast to IAD wt, the variant IAD Mut 123 failed to interact with the LBD of any receptor in the presence of hormone (Fig. 2, lane 5) indicating the requirement for a correct NR box sequence for receptor interaction in vitro. In yeast, IAD wt also interacted with all tested nuclear hormone receptors (Fig. 3). It is known that PPARα ligands have no or very low effects in yeast systems. All other tested receptors exhibited a much stronger IAD wt interaction in yeast in the presence of the respective hormone. However, in contrast to our in vitro results, relatively strong interactions in the absence of the cognate ligands were also seen for TR and RXR. Only the binding to GR was restricted to occurring in the presence of hormone. Apparently, the IAD of TIF2 has a certain capacity to bind some unliganded receptors in yeast, possibly due to by the presence of endogenous ligands or improper folding of the nuclear receptors, leading to a quasi-hormone-bound structure (Fig. 3A).
FIG. 2.
The in vitro interaction of the IAD of TIF2 with nuclear receptors is ligand dependent. PPARα (A), RXRα (B), and TRα (C) were in vitro translated in the presence of [35S]methionine and analyzed with the indicated GST fusion proteins bound to glutathione-Sepharose beads in a pull-down assay. The input (lanes 1) always represents 50% of the amount of labeled protein used in the pull-down assay. The interaction with GST-IAD was analyzed as indicated in the absence or presence of the respective ligands, WY-14,643, 9-cis-retinoic acid (9-cis RA), and T3 (lanes 3 and 4). The interactions with GST (lanes 2) and GST-IAD Mut 123 (lanes 5) were analyzed only in the presence of the corresponding hormones.
FIG. 3.
(A) The binding of the IAD of TIF2 to different hormone receptors depends on the presence of hormone. IAD wt of TIF2 fused to the GAD was tested in a yeast two-hybrid assay for interaction with the LBDs of PPARα (aa 166 to 468), the C-terminally deleted mutant PPARαΔc lacking helix 12 (aa 166 to 455), TRα (aa 122 to 410), GR (aa 485 to 777), and RXRβ (aa 153 to 451) fused to the GAL4 DBD in the absence or presence of the corresponding ligands as indicated. As a control, the interaction of the GAD with the LBDs was tested. (B) The integrity of the NR boxes is crucial for the interaction of the IAD with nuclear receptors in yeast. The variant IAD Mut 123 was fused to the GAD and analyzed for interaction with the LBDs of different nuclear receptors fused to the GAL4 DBD in the presence of hormone. The given values are relative (rel.) β-galactosidase units in comparison to the measured interaction with IAD wt and the respective receptor in the presence of ligand. For hormone induction 10 μM WY (PPARα), 1.5 μM T3 (TRα), 20 μM TA (GR), or 10 μM 9-cis-retinoic acid (RXRβ) was used.
Also in the yeast two-hybrid assay, the mutated IAD failed to interact with any receptor even in the presence of hormone, strengthening the notion of the importance of the three NR boxes for receptor interaction as demonstrated by the GST pull-down experiments (Fig. 3B).
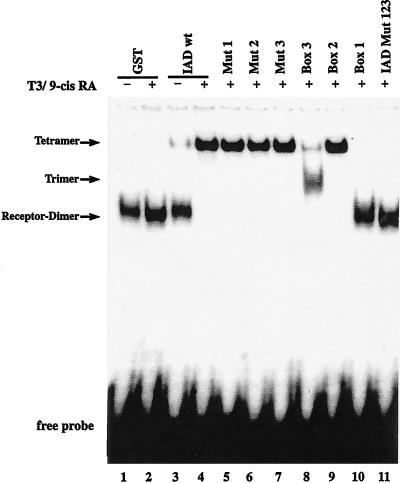
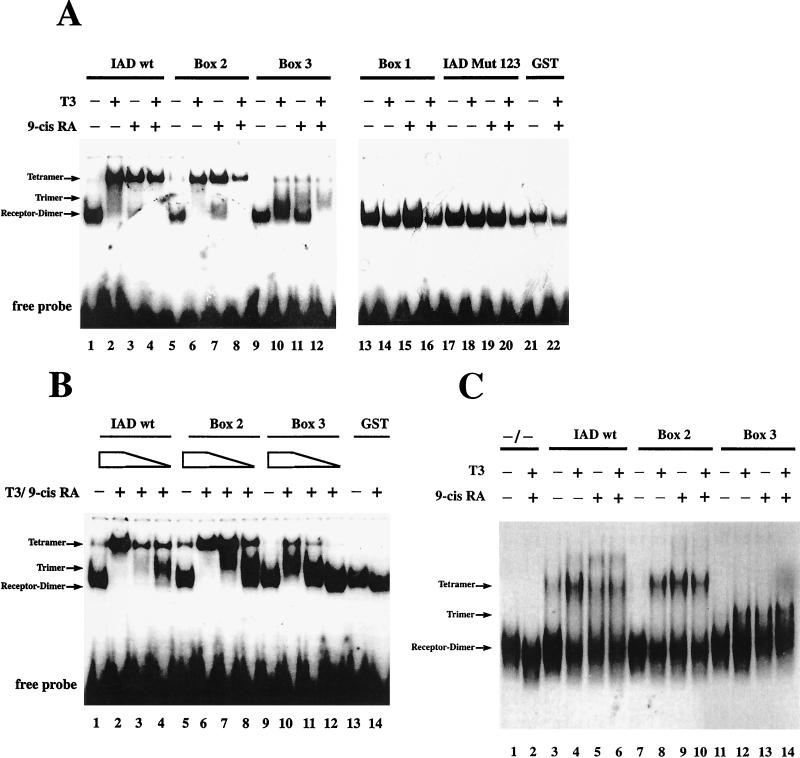
The nuclear receptors TR and PPAR exert their positive effects on gene transcription upon heterodimerization with RXR. To investigate whether TIF2 also binds a heterodimeric complex attached to DNA, we used full-length TR and RXR bound to a radioactively labeled DR4 in a gel retardation (band shift) assay. Whereas the E. coli-expressed IAD of TIF2 failed to interact with the heterodimer in the absence of hormone, it completely supershifted the dimer, by forming a ternary complex, in the presence of both corresponding hormones (Fig. 4, lanes 3 and 4). Consistent with the two-hybrid and pull-down assay results, in this assay also there was no receptor interaction of the variant IAD Mut 123 (Fig. 4, lane 11).
FIG. 4.
Point mutations of the NR boxes inhibit TIF2 binding to TR-RXR heterodimers differently. Electrophoretic mobility shift assays were performed with in vitro-translated TRβ and RXRβ and bacterially expressed GST-tagged TIF2 IAD variants. The interactions of GST (lanes 1 and 2) and GST-IAD wt (lanes 3 and 4) with the heterodimer were analyzed in the absence or presence of both T3 (100 μM) and 9-cis-retinoic acid (9-cis RA) (100 μM), as indicated. The interactions with the mutated variants were analyzed in the presence of both hormones.
An additional, NR box-independent interaction function within the IAD was not detectable by any of the approaches used. Hence, the LXXLL motifs within the IAD of TIF2 are necessary and sufficient for hormone-dependent interaction with nuclear receptors.
Whereas the leucine residues of the LXXLL motif are conserved among all NR box-containing proteins, the amino acid residues between the leucine residues as well as the adjacent sequences are heterogeneous. These residues also differ among the three motifs within TIF2. We wondered whether these differences may contribute to variations in receptor interaction among the three NR boxes. By analysis of mutations within the LXXLL motif, we investigated whether every single box is necessary for binding to receptors. Furthermore, we examined whether there is a specificity of these NR boxes for certain receptors. To test the influence of each motif, we mutated each box and analyzed it in the context of the entire IAD (Mut 1, 2, and 3) (Fig. 1B). In addition, by fusing two mutated boxes, we generated variants of the IAD containing only one functional motif (box 1, 2, and 3) (Fig. 1B). The resulting eight different variants (Fig. 1B) were fused to GST and subsequently analyzed in vitro in GST pull-down assays for interaction with in vitro-translated PPARα, RXRα, and TRα (Fig. 5). Equal amounts of protein were loaded as determined by Coomassie blue staining prior to autoradiography (data not shown). A single mutation of any motif did not show significant influence on receptor interaction. We assume that due to the large excess of GST fusion protein, even weak interactions are sufficient to be detected in GST pull-down assays. Mutation of two modules within the IAD strongly affects the interaction in the pull-down experiment. For PPARα, NR box 2 binds best (Fig. 5A, lane 8), and motif 3 still has capacity for binding (Fig. 5A, lane 9), whereas NR box 1 alone only shows very weak interaction (Fig. 5A, lane 7). The interactions with RXRα and TRα yielded about the same results (Fig. 5B and C). However, the interactions seen with the variants carrying two mutations seem to be stronger than those with PPARα, indicating a higher capacity of each individual NR box for binding to those receptors.
FIG. 5.
The in vitro interaction of TIF2 with different nuclear receptors depends on the integrity of the NR boxes. PPARα (A), RXRα (B), and TRα (C) were in vitro translated in the presence of [35S]methionine and analyzed with the indicated GST fusion proteins bound to glutathione-Sepharose beads in a pull-down assay for interaction with different IAD variants as indicated. The interaction was analyzed in the presence of the corresponding ligand. The experimental conditions were otherwise similar to those described in the legend to Fig. 3.
To gain further insight into the involvement of different hormones in coactivator-receptor interaction on a heterodimeric complex, we performed band shift assays with TRβ-RXRα heterodimers on a DR4 element and the different variants of the TIF2 IAD. IAD wt bound to the receptor in the presence of both corresponding hormones, as described above (Fig. 4, lanes 3 and 4). The variants containing only one mutated box exhibited the same binding pattern as IAD wt (Fig. 4, lanes 5 to 7). In contrast, the variants carrying only one functional box acted differently (Fig. 4, lanes 8 to 10). Box B1 was not able to interact when tested alone (Fig. 4, lane 10). In contrast, box 2 behaved similarly to IAD wt and bound strongly to the heterodimer (Fig. 4, lane 9). Interestingly, the capacity of box 3 to upshift the receptor dimer was apparently lower than that of box 2 but still detectable. The supershift with IAD wt and box 2 was formed only weakly by box 3 (Fig. 4, lane 8). A lower band appeared to migrate between the receptor dimer and the supershift. This intermediate band probably represented a trimer consisting of the receptor dimer and one molecule of TIF2 IAD, whereas the upper shift resulting from addition of IAD wt or box 2 represented a tetrameric association between the two receptors and two molecules of TIF2 IAD. Furthermore, we conclude that there are differences in affinities between the three boxes for binding to heterodimeric, DNA-bound receptors. Whereas box 2 showed nearly wild-type affinity, box 3 interacted only weakly. NR box 1 alone has no capacity for interaction, but it contributes in concert with NR box 3 to the resulting interaction (Fig. 4, lane 6). The variant Mut 2 contains functional NR boxes 1 and 3. The supershift with Mut 2 differs from that of NR box 3 alone (Fig. 4, lane 8) indicating a cooperative effect of NR box 1 in conjunction with NR box 3. The single elements thus have different affinities to the heterodimer, in line with the results from the GST pull-down experiment.
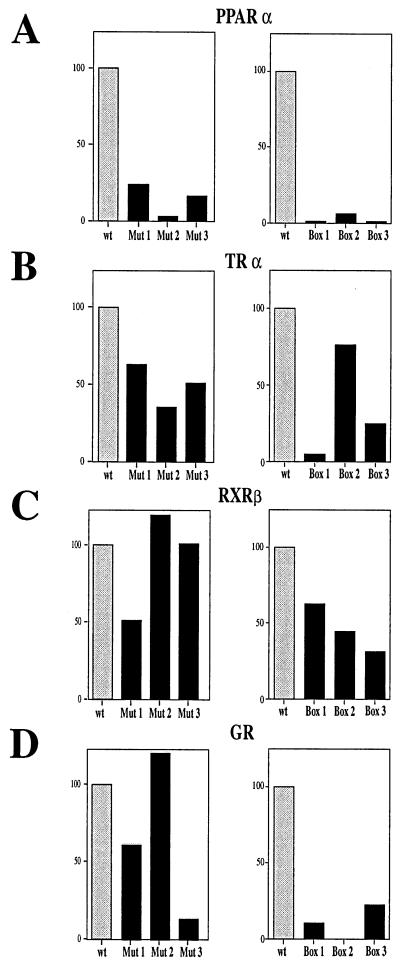
Because of the high molar excess of bacterially expressed protein compared to the in vitro-translated receptors, slight differences in interaction affinities are difficult to detect in GST pull-down and band shift experiments. To study differences in interactions at an approximately 1:1 ratio of the proteins, we analyzed the IAD variants in the yeast two-hybrid assay. These experiments were carried out by using the LBDs of different nuclear receptors fused to the GAL4 DBD and with the variants of the IAD fused to the GAD. In comparison to IAD wt, every IAD variant harboring a single mutated box strongly affected the binding to PPARα (Fig. 6). Mutation of NR box 2 had the largest influence on the interaction with PPARα, reducing the relative β-galactosidase activity to about 2% of that with IAD wt (Fig. 6A). In line with this, only NR box 2 was capable of interacting with PPARα LBD when it was tested as a single functional motif. The addition of all three single interaction capacities of each NR box does not result in the 100% interaction measured with IAD wt. Therefore, binding of the three motifs to PPARα seems to involve significant cooperative interactions.
FIG. 6.
The three NR boxes within the IAD of TIF2 bind preferentially to different receptors. Mutated variants of the IAD of TIF2 were fused to the GAD and tested in the presence of the corresponding hormones in a yeast two-hybrid assay for interaction with the LBDs of PPARα (A), TRα (B), GR (C), and RXRβ (D). (Left panels) Effects of point mutations of single modules (Mut 1, Mut 2, and Mut 3) within the IAD compared to IAD wt, defined as 100%. (Right panels) Effects of variants carrying only single modules (box 1, box 2, and box 3) within the IAD compared to IAD wt, defined as 100%.
For TRα also, NR box 2 seem to be the most important motif, followed by NR box 3, whereas NR box 1 has a low capacity for interaction. In that way, the different NR boxes interacted similarly with TRα as with PPARα. However, the variants containing mutations in single boxes have weaker effects than they have on the interaction with PPARα. In line with this, box 3 had a comparatively high capacity for interaction, and box 2 interacted with TRα with about 75% of the affinity of IAD wt (Fig. 6B).
In the case of RXRβ, differences in NR box interactions compared to those with PPARα and TRα were seen. For instance, NR box 1 appears to be the most important motif. Moreover, all three NR boxes were relatively equally efficient in interaction with RXRβ compared with all other tested receptors (Fig. 6C).
Strikingly, the interaction results with GR differ from those obtained with PPARα, TRα, and RXRβ. NR box 2 seems to have no effect on GR interaction, whereas in the case of PPARα and TRα, this motif is the most important one. Accordingly, NR boxes 1 and 3 together synergistically interact with the same efficiency as IAD wt, obviously not due to additive single effects, but in a cooperative manner (Fig. 6D).
In conclusion, there are binding specificities for the different motifs within the interaction domain of TIF2. NR box 1 has a relative preference for binding to RXRβ, NR box 2 has a preference for binding to PPARα and TRα, and NR box 3 has a preference for binding to GR. Moreover, NR boxes bind to GR and PPARα in a cooperative manner, whereas the interaction with TRα and RXRβ is based on additive effects of single elements, indicating different binding mechanisms in coactivator interactions with different receptors.
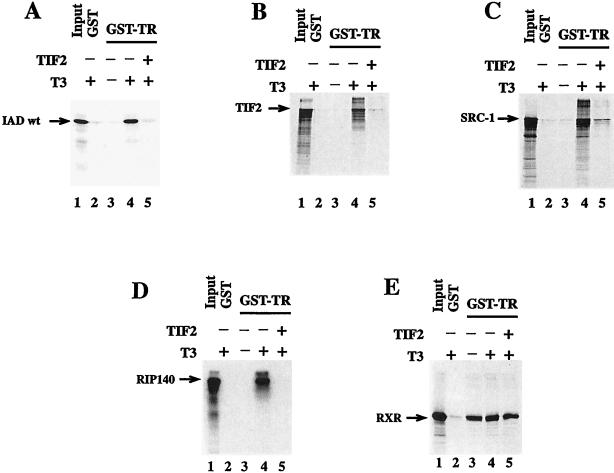
The motif LXXLL, which is responsible for protein-protein interaction, is present in different LBD/AF-2-interacting proteins. In this paper we have demonstrated certain preferences of different NR boxes for different receptors, but we also state that IAD wt binds in a promiscuous manner to every receptor we tested. Accordingly, we questioned whether TIF2 interferes with the binding of other NR box-containing proteins to nuclear receptors. To test this, we expressed IAD wt as a GST fusion protein in E. coli. After purification and cleavage with thrombin, this protein was used in a competition experiment in a GST pull-down assay. In this assay we used GST-TR and the in vitro-translated interacting proteins RIP140 (7, 25), SRC-1, and TIF2, as well as IAD wt of TIF2 as a positive control. In addition, we examined whether the GST-TR binding of RXRα, the heterodimeric partner of TRα, is inhibited by an excess of IAD wt protein (Fig. 7). The binding of all interacting proteins was dependent on the presence of T3. In contrast, the binding of RXRα to TRα was hormone independent (Fig. 7E, lanes 3 and 4). The addition of purified IAD wt protein (Fig. 7A, lane 5) abolished the binding of in vitro-translated IAD wt, as expected because of the excess of E. coli-expressed protein. The interaction with TIF2 also was abolished, indicating that there are no other interaction motifs in TIF2 within the regions N and C terminal to the IAD (Fig. 7B, lane 5). The interaction of the TIF2-related protein SRC-1 was also inhibited. Even the binding of cofactor RIP140, which contains at least nine different NR boxes (7, 16, 25), was efficiently competed by the IAD of TIF2 (Fig. 7C and D, lanes 5). In contrast, the interaction of RXRα with TRα was not influenced by the excess of IAD wt protein (Fig. 7E, lane 5). The same results were obtained when E. coli-expressed RIP140 instead of the TIF2 IAD was used as a competitor (38a).
FIG. 7.
TIF2 competes with other cofactors for binding to nuclear receptors. The in vitro interactions between GST-hTRα and IAD wt of TIF2 (A), full-length TIF2 (B), hSRC-1 (aa 1 to 1061) (C), RIP140 (aa 1 to 1158) (D), and RXRα (aa 112 to 467) (E) were analyzed in a GST pull-down assay in the absence or presence of 1 μM T3 as indicated. The experimental conditions were otherwise similar to those described in the legend to Fig. 3, except that ca. 1 μg of IAD protein was added in lanes 5. The sizes of the major translation products are 20 kDa (IAD), 160 kDa (TIF2), 114 kDa (SRC-1), 127 kDa (RIP140), and 43 kDa (RXR). Lanes 1 represent 50% input.
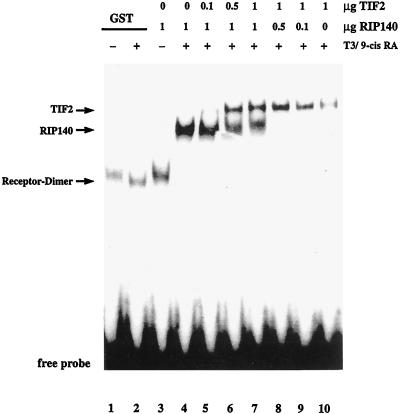
The finding that the IAD of TIF2 is able to compete with other coactivators for binding to hormone-bound TRα raises the question of whether two different interacting proteins can bind to one receptor at the same time. In a band shift assay we investigated whether TIF2 can compete for binding of RIP140 to the TRβ-RXRα heterodimer. Both E. coli-expressed interacting proteins bound hormone dependently to the receptors, resulting in distinguishable supershifts (Fig. 8, lanes 4 and 10). An increase of TIF2 protein with a given RIP140 concentration in the reaction mixture led to the appearance of the TIF2-receptor supershift, whereas the supershift formed by RIP140 decreased (Fig. 8, lanes 5 to 9). A 1:1 ratio of the proteins led to similar intensities of the respective supershifted bands (Fig. 8, lane 7). Accordingly, both TIF2 and RIP140 appear to bind to the receptor heterodimer with about the same affinity. A binding of both interacting proteins at the same time should result in a further upshift of the supershift. This could not been observed, indicating either that both proteins bind to the same site of the heterodimer or that binding of one interacting protein inhibits binding of the other one by steric hindrance or by changing the receptor conformation.
FIG. 8.
TIF2 competes with RIP140 for binding to a TR-RXR heterodimer. Electrophoretic mobility shift assays were performed as described in the legend to Fig. 4. For analysis of competitive interaction with the heterodimer, bacterially expressed GST-TIF2 and/or His-RIP140 was used. The amounts of protein and the presence or absence of hormones is indicated. 9-cis RA, 9-cis-retinoic acid.
The fact that an IAD of one protein interferes with the binding of other receptor-interacting proteins supports the existence of a general mechanism of NR box binding to nuclear receptors. Furthermore, the finding that the binding of RXRα to TRα is not affected by NR boxes clearly indicates the existence of at least two different and independent mechanisms of protein-protein interaction: an NR box-dependent interaction between a receptor and an interacting protein and a heterodimeric interaction between two receptors.
Every receptor that we tested is able to interact with TIF2 in a hormone-dependent manner. For a given heterodimer, we would assume that each receptor can bind one molecule of TIF2 in the presence of its cognate ligand. In the presence of both ligands, a tetrameric supershift should appear, whereas in the presence of only one hormone, we would expect an intermediate shift formed by a trimer. Alternatively, there is the possibility that the binding of a second TIF2 molecule is prevented by steric hindrance. In this case only one supershift should be formed in the presence of one or both hormones. As shown above, an intermediate shift was detectable with the IAD variant box 3 in the presence of both hormones, strongly supporting the idea of tetramer formation. To test whether an intermediate shift is reproducibly formed and to further understand the mechanisms by which the different supershifts are formed, we used different hormone combinations in conjunction with different IAD variants in a gel retardation experiment (Fig. 9A). Whereas IAD Mut 123 and box 1 are not able to interact even in the presence of both hormones (Fig. 9A, lanes 13 to 20), IAD wt, box 2, and box 3 engaged in hormone-dependent interactions. Surprisingly, IAD wt interacted in the presence of both hormones as well as in the presence of only one cognate ligand. Box B2 exhibited the same interaction pattern as IAD wt, whereas box 3 only weakly formed the supershift. Instead of the supershift, the intermediate shift arose in addition to the unbound heterodimer in the presence of single hormones (Fig. 9A, lanes 10 and 11). After addition of both hormones, the heterodimer is completely upshifted to the intermediate shift (Fig. 9A, lane 12). Apparently the binding to each receptor is relatively weak in the presence of one hormone only; the presence of two activated receptors leads to additive effects and thereby to the complete upshift, forming a trimeric complex. To get further insight into the formation of tetramers, we used limiting amounts of IAD wt and variant IAD protein in the band shift assay (Fig. 9B). The lowest concentration of IAD wt also causes the intermediate shift (Fig. 9B, lane 4). According to their respective different affinities to the receptors, box 2 and box 3 at certain concentrations also formed this monomeric shift, representing a single TIF2 IAD molecule bound to the receptor heterodimer (Fig. 9B, lanes 7, 8, 10, and 11). For IAD wt and box 2, but not for box 3, the upper, tetrameric shift was favored even when only little trimer formation was detectable (Fig. 9B, lanes 3 and 7). Additionally, at high concentrations of IAD wt and box 2 protein, the hormone-independent interaction led directly to the tetrameric shift and not, as expected in the case of noncooperative binding, first to a trimeric shift (Fig. 9B, lanes 1 and 5). We therefore conclude that the receptor interaction of IAD wt and box 2 occurs in a cooperative manner. Binding of one monomer leads to subsequent binding of the second one, forming a tetrameric complex. In contrast to these results obtained with IAD wt and box 2, interactions with box 3 favored the trimeric shift compared to the tetrameric supershift (Fig. 9B, lanes 10 and 11). We conclude that in contrast to NR box 2, which interacts cooperatively with both receptors, NR box 3 does not contribute to the cooperative binding of IAD wt. Therefore, the potency for cooperative binding of TIF2 IAD to the receptor heterodimer is mediated by NR box 2.
FIG. 9.
(A) Even one hormone is sufficient to form a tetrameric complex. Electrophoretic mobility shift assays were performed as described in the legend to Fig. 4. For analysis of interaction with the heterodimer, the bacterially expressed GST fusion proteins indicated above the lanes were used. The interaction with TR-RXR was tested in the absence or presence of one or both cognate hormones as indicated. 9-cis RA, 9-cis-retinoic acid. (B) The NR box modules differently affect the formation of the supershift. For analysis of interaction with the heterodimer, the bacterially expressed GST fusion proteins indicated above the lanes were used at 5 μg (lanes 1 and 2, 5 and 6, 9 and 10, and 13 and 14), 0.5 μg (lanes 3, 7, and 11), and 0.05 μg (lanes 4, 8, and 12). The influence of the presence or absence of hormone was analyzed as indicated. (C) The GST moiety of the fusion protein does not affect the interaction. For analysis of the interaction of the pure TIF2 moiety, this part was generated by thrombin cleavage of the corresponding GST fusion protein and used in electrophoretic mobility shift assays. The proteins indicated above the lanes were used. As a control, no purified protein was added in lanes 1 and 2. The interaction with TR-RXR was tested in the absence or presence of one or both cognate ligands as indicated.
To rule out the possibility that the GST moiety of the fusion proteins is involved in oligomerization and thereby might be at least partially responsible for the observed differences in complex formation, we performed band shift experiments with thrombin-cleaved IAD variants, omitting the GST portion (Fig. 9C). This experiment was carried out to investigate complex formation of the pure variants in the presence of different hormones. As already observed with the GST fusion proteins (Fig. 9A) the pure IAD wt and box 2 form a supershift in the presence of the single hormones (Fig. 9C, lanes 4, 5, 8, and 9) as well as in the presence of both ligands (Fig. 9C, lanes 6 and 10). The supershifts formed by IAD wt and by box 2 migrate at the same position. Furthermore, the results with box 3 are consistent with the data shown in Fig. 9A. This variant forms an intermediate shift with an increased mobility compared to the supershifts induced by IAD wt and box 2. This interaction is detectable in the presence of both ligands as well as in the presence of only one ligand (Fig. 9C, lanes 12, 13, and 14). The pure box 1 and Mut 123, as well as their GST-fused counterparts, are unable to interact with the heterodimer (data not shown). In summary, the differences in interaction observed for the variants are due only to the integrity of different NR boxes present in these variants. The GST moiety has no influence on the differences in interaction. IAD wt and box 2 give rise to a tetrameric shift with the heterodimer, whereas only one box 3 molecule interacts with the receptors, forming a trimer.
We have demonstrated that in the presence of one hormone, only the tetrameric shift is the dominant one in the case of IAD wt as well as box 2. The trimeric shift is favored by box 3 under these conditions (Fig. 9B). When we compare these results with the data from the band shift experiments analyzing effects of different hormones eliciting cooperative binding (Fig. 9A), we conclude that the binding of the TIF2 IAD to one receptor strongly facilitates the interaction of the TIF2 IAD with the other receptor even in the absence of its cognate hormone. We therefore suggest that the binding of one molecule of TIF2 to one heterodimeric partner alters the structure of the other heterodimeric protein in such a way that it may now bind to TIF2 without its corresponding hormone (Fig. 10). This effect is mediated mainly by NR box 2.
FIG. 10.
Model of coactivator-induced allosteric conformational change. In the unliganded state, both receptors within the DNA-bound heterodimer are in a conformation unable to associate with coactivators such as TIF2. (A) Upon addition of both hormones, both structures are altered, allowing TIF2 interaction. (B) Addition of a single ligand leads in a first step to a conformational change and cofactor association with the corresponding receptor. This interaction causes a conformational change in the unliganded partner receptor, allowing the subsequent binding of a second cofactor molecule and formation of a tetrameric complex.
DISCUSSION
In this work we have studied the roles of different NR boxes within the coactivator TIF2. We tested the involvement of these elements in protein-protein interactions with different hormone receptors in several in vivo and in vitro assays in an effort to understand the individual properties of the different interacting motifs and how these properties may lead to certain binding preferences. With our approach of examining the different NR boxes in the context of the entire IAD of TIF2, we took into account the possibility that adjacent amino acid residues might contribute to the binding effects and also that different surface accessibilities might have an influence on the interactions. By generating point mutations in the individual NR boxes, we analyzed binding characteristics of the different mutated motifs separately or in combination with one another in comparison with those of the intact IAD.
We could thus determine the relative binding preferences of the different modules to different receptors. NR box 1 seems to play the major role in RXR binding, NR box 2 is most important for TR and PPAR interactions, and NR box 3 has its strongest impact on GR interaction. Conversely, NR box 2 exhibits no interaction with GR. Possibly steroid receptors in general interact most efficiently with NR box 3, whereas heterodimeric partners of RXR are targets of NR box 2. Arguing against this model, it was shown with NR box peptides in a yeast assay that the corresponding NR box 2 of SRC-1 binds best to ERα, followed by NR box 1 and NR box 3 (16). This may reflect differences between SRC-1 and TIF2 or between ER and GR, but the contrasting results may also relate to differences between the experimental approaches.
Considering (i) these differences in interaction preferences between the three NR boxes and (ii) the high conservation of the amino acid residues of the NR boxes between SRC-1, TIF2, and ACTR, we propose that at least the amino acids between the leucine residues but presumably also the N- and C-terminal surroundings of the LXXLL motifs are important for specific receptor interactions. For high-interaction capacity, it is apparently significant that of the two amino acids between the hydrohobic residues, one is polar and the other one is basic. Additionally, adjacent to the LXXLL motif, a basic N-terminal region seems to be important for efficient interactions. The flanking C-terminal region often contains polar and acidic amino acids, and their importance for receptor interaction is consistent with the findings that a deletion of the glutamic acid and asparagine residues of the C-terminal region of a RIP140 NR box leads to a strongly decreased binding to ERα (16). It remains to be shown whether these regions are involved in direct protein-protein interactions and therefore play a role in the binding specificities of the different boxes. In addition, the distance between the boxes is well conserved among SRC-1, TIF2, and ACTR. We have provided evidence for binding of more than one box to any tested receptor, and it is therefore most likely that a definite distance between the boxes is a steric prerequisite for binding of more than one box to one receptor at the same time.
The NR box seems to be the major interacting motif existing in TIF2 and other receptor-interacting proteins. So far, no other motif responsible for LBD/AF-2-dependent cofactor-receptor interaction has been described. NR boxes have been found in many of these cofactors, with only the exceptions of SUG1 and ARA70, which do not contain a classical LXXLL motif (41, 46). However, these proteins also carry NR box-like sequences, possibly serving as interaction motifs. These facts raise two interesting questions. First, why do some interacting proteins harbor more than one motif, and second, why are there so many NR box-containing interacting factors? We showed that for wild-type interaction, depending on the receptor in question, two or all three boxes within TIF2 are necessary. Furthermore, we demonstrated that different motifs show certain preferences for different receptors. The reason for the obvious redundancy in NR box-containing proteins is not yet clear. We have shown here by competition experiments that receptor binding of NR box-containing factors seems to occur according to a general mechanism. The IAD of TIF2 competes with both SRC-1 and RIP140 for binding to nuclear receptors. In contrast to the SRC-1-related coactivators, RIP140 has not been shown to significantly enhance gene transcription mediated by nuclear hormone receptors. In coexpression experiments with mammalian cells, we showed that RIP140 caused a repression of SRC-1-coactivated transcription, suggesting an antagonistic function of both proteins on transcriptional activation (38a). Coactivated transcription probably depends on the relative amounts of different coactivators with distinct binding affinities for certain receptors. This decides which coactivator interacts and which downstream targets will join the receptor-coactivator complex.
In this paper we have described in some detail the IAD and the single interaction motifs of the coactivator TIF2. In contrast, the corresponding interaction interfaces on the receptor are still unknown. Based on our competition experiments, we conclude that neither SRC-1 nor RIP140, which harbors at least nine NR boxes, interacts with a receptor target different from that for TIF2, strongly arguing for a common interaction interface within nuclear receptors responsible for the interaction with NR boxes. As we have shown, the interaction of TIF2 with PPAR is dependent on the presence of helix 12. Helix 12, sometimes also denoted AF-2 AD or τ4, is a conserved region present in all members of the steroid and nonsteroid nuclear receptor family (2, 4, 12, 13). Structural analysis of hormone-bound RAR and unliganded RXR indicated that the main receptor conformational change occurring upon ligand binding relates to the orientation of helix 12. Without hormone, helix 12 protrudes from the LBD, whereas it is tightly folded against helix 4 and the ligand binding pocket in the hormone-bound receptor (5, 30, 42). Coincidentally with this structural change, corepressors dissociate and coactivators bind to the receptors. Here we have demonstrated the binding of three NR boxes within the IAD of TIF2 to one receptor. For the tested receptors we observed at least additive effects when binding of all three boxes occurred, providing evidence for binding of all boxes at the same time. As we know from the dimerization interface between the subunits in ERα homodimers, which contains LXXLL motifs, the interaction engages a region of about 12 aa (6). Although the involved LXXLL motifs in ERα are not conserved within the nuclear hormone receptor family and are not of the type contacting the LBD/AF-2 motif, it is nevertheless not unlikely that ERα homodimerization might occur via a type of interaction similar to that between cofactor and receptor. Assuming that a binding analogous to that between the ERα subunits in the homodimer also occurs between the NR box in TIF2 and a receptor, a region of at least 12aa residues should be involved. Simultaneous binding of two or three NR boxes to one receptor would necessarily involve more regions in the receptor than exclusively helix 12. We therefore suggest that binding of ligand to the receptor LBD results in a conformational change creating a coactivator-binding interface consisting of helix 12 as well as other regions, probably helix 3 and/or helix 4.
Recently, allosteric effects between heterodimeric nuclear receptor partners have been suggested. It was shown that heterodimerization with RXR leads to an increase in solubility and stability of the partner (26). In studies of RAR-RXR dimers, it was demonstrated also that the ligand binding affinity of RXR is dramatically reduced after heterodimerization (14, 22). A novel mechanism of receptor activation upon heterodimerization has been described for the OR1-RXR heterodimer. The orphan receptor OR1 is activated not only by the presence of ligand but also by conformational changes as a result of heterodimerization with RXR, even in the absence of OR1 ligand (43). Receptor ligands also play an important role in allosteric activation or inhibition. For instance, formation of TR-RXR heterodimers is facilitated by the presence of T3 (10). The ligand binding affinity of RXR, which is inhibited upon heterodimerization with RAR and TR, is restored after ligand activation of RAR and further inhibited by T3 (14). Apparently heterodimerization leads in both cases to structural rearrangements. Binding of ligand causes conformational changes in the cognate receptor (37), generating an allosteric rearrangement in the partner receptor. This allosteric conformational change can result in hormone-independent activation of the partner receptor of RXR, as recently shown for RXR-RAR and RXR-PPAR heterodimers. This allosteric effect, described as the phantom ligand effect, is thought to alter the structure of the heterodimeric partner into a quasiactivated conformation which allows coactivator binding (32a).
Here we suggest a new model also involving coactivators in allosteric effects. We have shown by GST pull-down experiments that the IAD of TIF2 binds to receptors strictly depending on the presence of ligand. Addition of both ligands to a heterodimeric complex leads to conformational changes in the cognate receptors that allow cofactor association (Fig. 10A). This is consistent with the findings in our GST pull-down assays revealing that each of the receptors interacts with the IAD in the presence of ligand. However, addition of a single ligand also leads to the association of two TIF2 molecules, indicating that the unbound receptor also interacts with TIF2. We suggest that in the presence of a single ligand recognizing one of the subunits of the DNA-bound TR-RXR heterodimer, the induced interaction with one molecule of TIF2 subsequently leads to the binding of an additional molecule of TIF2 to the partner receptor even in the absence of its cognate ligand. (Fig. 10B). We therefore propose that TIF2 interaction with one receptor causes conformational changes within the heterodimeric partner receptor via the dimerization interface. As discussed above, a prerequisite for receptor-cofactor interaction is an intact helix 12, which is rearranged after ligand binding. Therefore, we believe that the conformational changes in the heterodimeric partner caused by coactivator binding to the first receptor lead to a quasi-ligand-induced structure allowing the second cofactor molecule to interact with the second receptor even in the absence of its cognate ligand.
ACKNOWLEDGMENTS
We thank M. G. Parker, B. W. O’Malley, and S. Nilsson for generously providing constructs. We are grateful to Dorothee Feltkamp and Franziska Wiebel for providing material, and we thank all members of the orphan receptor group for helpful discussions, especially AnneMarie Witte for excellent technical support.
J.L. receives a grant from the European Community (MCFA). E.T. obtained a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft. This work was supported by a grant from the Swedish Cancer Society.
ADDENDUM
During the process of review of this paper, similar results regarding the binding preferences of different NR boxes within TIF2 and SRC-1 to nuclear receptors were reported independently (12a, 21a, 40a).
REFERENCES
- 1.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 2.Baniahmad A, Leng X, Burris T P, Tsai S Y, Tsai M J, O’Malley B W. The tau 4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Barettino D, Vivanco Ruiz M M, Stunnenberg H G. Characterization of the ligand-dependent transactivation domain of thyroid hormone receptor. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature (London) 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 6.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engström O, Öhman L, Greene G L, Gustafsson J Å, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature (London) 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 7.Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H W, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakartani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 9.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 10.Collingwood T N, Butler A, Tone Y, Clifton-Bligh R J, Parker M G, Chatterjee V K. Thyroid hormone-mediated enhancement of heterodimer formation between thyroid hormone receptor beta and retinoid X receptor. J Biol Chem. 1997;272:13060–13065. doi: 10.1074/jbc.272.20.13060. [DOI] [PubMed] [Google Scholar]
- 11.Damm K, Heyman R A, Umesono K, Evans R M. Functional inhibition of retinoic acid response by dominant negative retinoic acid receptor mutants. Proc Natl Acad Sci USA. 1993;90:2989–2993. doi: 10.1073/pnas.90.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Nuclear receptor-binding sites of coactivator glucocorticoid receptor interacting protein 1 (GRIP1) and steroid coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 13.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forman B M, Umesono K, Chen J, Evans R M. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 15.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 15a.Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 16.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 17.Heinzel T, Lavinsky R M, Mullen T M, Söderström M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 18.Hong H, Kohli K, Garabedian M J, Stallcup M R. GRIP1, a transcriptional coactivator for teh AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hörlein A J, Näär A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Söderström M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature (London) 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 21.Horwitz K B, Jackson T A, Rain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 21a.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the estrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld M G, Heyman R A, Glass C K. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature (London) 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 23.Kurokawa R, Söderström M, Hörlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature (London) 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 24.Le Douarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1-alpha and TIF1-beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 25.L’Horset F, Dauvois S, Heery D M, Cavailles V, Parker M G. RIP140 interacts with multiple nuclear receptors by means of two distinct sites. Mol Cell Biol. 1996;16:6029–6036. doi: 10.1128/mcb.16.11.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Schwabe J W, Banayo E, Evans R M. Coexpression of nuclear receptor partners increases their solubility and biological activities. Proc Natl Acad Sci USA. 1997;94:2278–2283. doi: 10.1073/pnas.94.6.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF-2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 29.Oñate S A, Tsai S Y, Tsai M J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 30.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding bound to all-trans retinoic acid. Nature (London) 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 31.Sande S, Privalsky M L. Identification of TRACs (T-3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 32.Schulman I G, Juguilon H, Evans R M. Activation and repression by nuclear hormone receptors: hormone modulates an equilibrium between active and repressive states. Mol Cell Biol. 1996;16:3807–3813. doi: 10.1128/mcb.16.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Schulman I G, Li C, Schwabe J W, Evans R M. The phantom ligand effect: allosteric control of transcription by the retinoic X receptor. Genes Dev. 1997;11:299–308. doi: 10.1101/gad.11.3.299. [DOI] [PubMed] [Google Scholar]
- 33.Seol W, Mahon M J, Lee Y K, Moore D D. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- 34.Shibata H, Nawaz Z, Tsai S Y, Omalley B W, Tsai M J. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT) Mol Endocrinol. 1997;11:714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- 35.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J X, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature (London) 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 36.Takeshita A, Yen P M, Misiti S, Cardona G R, Liu Y, Chin W W. Molecular cloning and properties of a full-length putative thyroid hormone receptor coactivator. Endocrinology. 1996;137:3594–3597. doi: 10.1210/endo.137.8.8754792. [DOI] [PubMed] [Google Scholar]
- 37.Toney J H, Wu L, Summerfield A E, Sanyal G, Forman B M, Zhu J, Samuels H H. Conformational changes in chicken thyroid hormone receptor alpha 1 induced by binding to ligand or to DNA. Biochemistry. 1993;32:2–6. doi: 10.1021/bi00052a001. [DOI] [PubMed] [Google Scholar]
- 38.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 38a.Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson J Å. A regulatory role for RIP140 in nuclear receptor activation. Mol Endocrinol. 1998;12:864–881. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- 39.Truss M, Bartsch J, Schelbert A, Hache R J, Beato M. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 1995;14:1737–1751. doi: 10.1002/j.1460-2075.1995.tb07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 40a.Voegel J J, Heine M J S, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transcription through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.vom Baur E, Zechel C, Heery D, Heine M J, Garnier J M, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature (London) 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 43.Wiebel F F, Gustafsson J A. Heterodimeric interaction between retinoid X receptor alpha and orphan nuclear receptor OR1 reveals dimerization-induced activation as a novel mechanism of nuclear receptor activation. Mol Cell Biol. 1997;17:3977–3986. doi: 10.1128/mcb.17.7.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 45.Wong J, Shi Y B, Wolffe A P. A role for nucleosome assembly in both silencing and activation of the Xenopus TR beta A gene by the thyroid hormone receptor. Genes Dev. 1995;9:2696–2711. doi: 10.1101/gad.9.21.2696. [DOI] [PubMed] [Google Scholar]
- 45a.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor activator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zamir I, Harding H P, Atkins G B, Horlein A, Glass C K, Rosenfeld M G, Lazar M A. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zamir I, Zhang J, Lazar M A. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]