Abstract
Clinical progress in multiple myeloma (MM), an incurable plasma cell (PC) neoplasia, has been driven by therapies which have limited applications beyond MM/PC neoplasias and do not target specific oncogenic mutations in MM. Instead, these agents target pathways critical for PC biology yet largely dispensable for malignant or normal cells of most other lineages. We systematically characterized the lineage-preferential molecular dependencies of MM through genome-scale CRISPR studies in 19 MM versus hundreds of non-MM lines. This identified 116 genes whose disruption more significantly affects MM cell fitness compared to other malignancies. These genes, some known, others not previously linked to MM, encode transcription factors, chromatin modifiers, endoplasmic reticulum components, metabolic regulators or signaling molecules. Most of these genes are not among the top amplified, overexpressed or mutated in MM. Functional genomics approaches thus define new therapeutic targets in MM not readily identifiable by standard genomic, transcriptional, or epigenetic profiling analyses.
Keywords: CRISPR editing, genome-scale functional genomics, myeloma, essential genes
Introduction
Multiple myeloma (MM), a plasma cell (PC) neoplasia and the second most common hematologic malignancy in the Western world, remains incurable despite major therapeutic progress during the last two decades. Much of this progress was achieved through use of proteasome inhibitors, thalidomide and its derivatives, anti-CD38 monoclonal antibodies and more recently BCMA-targeting therapies. These agents have limited therapeutic applications outside MM, do not target specific oncogenic mutations in MM cells, but perturb pathways which are critical for PC biology yet largely dispensable for most other normal or malignant cell types1,2. By contrast, established or investigational therapeutics that target mutated gene products and pathways of MM3 generally yield short-lived clinical responses. Identification of genes essential for malignant or normal PCs, but dispensable for most other cell types, normal or malignant, could uncover putative therapeutic targets for MM. Therefore, we performed a systematic characterization of the molecular vulnerabilities of MM cells, compared to other types of neoplastic cells, through genome-scale CRISPR gene-editing screens. We hypothesized that these functional screens would not only “re-identify” known MM/PC dependencies but also pinpoint additional genes whose preferential role in MM might not be readily predicted from patterns of molecular alterations in MM cells, including mutations, DNA copy number changes, structural rearrangements or overexpression.
Results
MM-preferential dependencies identified by CRISPR screens
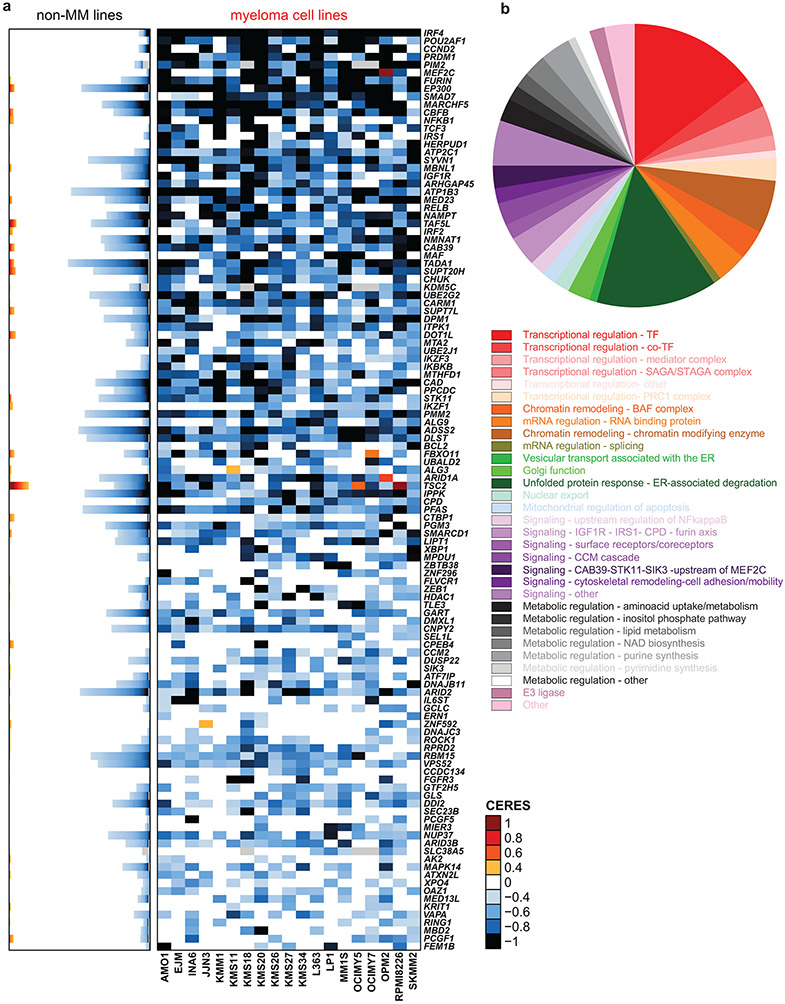
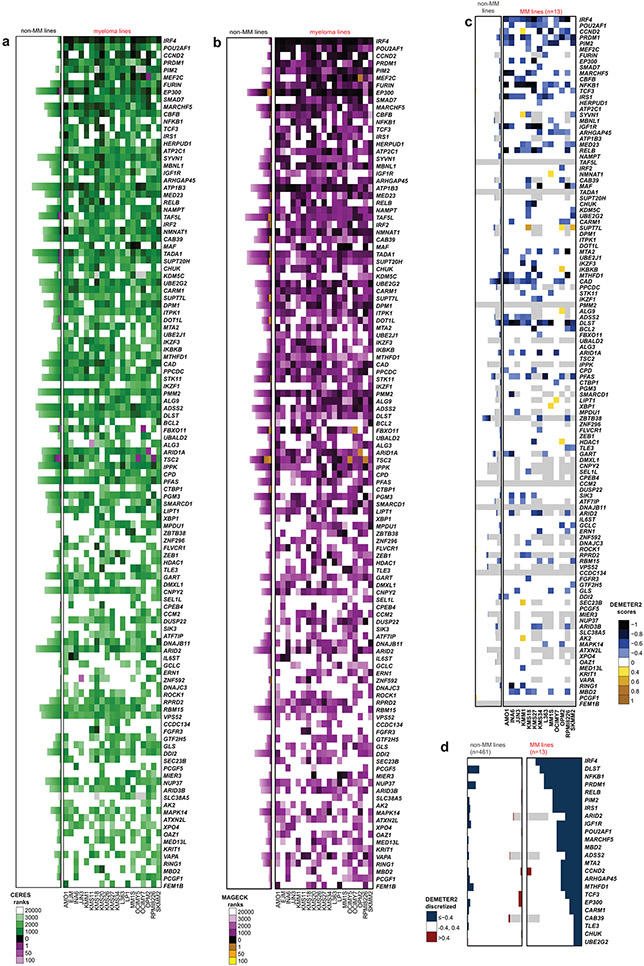
We sought to identify genes whose loss-of-function (LOF) more efficiently and consistently inhibits growth/survival of MM compared to non-MM cells. CRISPR/Cas9-based gene-editing screens were performed in 19 MM and 770 non-MM lines (see Methods and 4). Guide RNAs for genes more essential for MM are predicted to be eliminated more profoundly in MM than non-MM cells. We compared the patterns of gene essentiality in MM vs. non-MM lines using CERES scores (Fig. 1a, Extended Data Fig. 1a-b and Supplementary Table 1), the ranks of genes according to their CERES scores in a given cell line (Extended Data Fig. 2a) or MaGECK ranks (Extended Data Fig. 2b). These comparisons, based on criteria outlined in Methods, identified genes with statistically significant differences in quantitative metrics of essentiality in MM vs. non-MM lines, while filtering out those genes with a similar frequency of essentiality in MM vs. non-MM, including “core essential” genes required across all cancer cell lines. These analyses identified 116 MM-preferential dependencies (Fig. 1a, Extended Data Figs. 1 and 2a, Supplementary Table 1). In retrospective analyses of sequential releases of data from the Dependency Map (DepMap) program, which included increasing numbers of cell lines, the identity of MM-preferential dependencies was largely stable, with 72 genes identified in 5 consecutive releases. Additional cell lines in the later datasets allowed identification of >30 additional preferential dependencies (Extended Data Fig. 1a). These analyses were not influenced by the computational correction (e.g., in CERES score calculation) of the gene-independent copy number effects of CRISPR gene-editing, because MAGeCK analyses without such correction provided concordant results for these MM-preferential dependencies (Extended Data Fig. 2b, Supplementary Fig.1). Collectively, the use of multiple analytical methods and versions of the DepMap data offers greater confidence in the identification of MM-preferential dependencies. While some of these genes can also be identified by shRNA-based screens, including IRF45, PIM2, PRDM1, POU2AF1, NFKB1, RELB, IGF1R, IRS1, EP300, or TCF3 (Extended Data Fig. 2c,d), many others were identified only by gene editing studies.
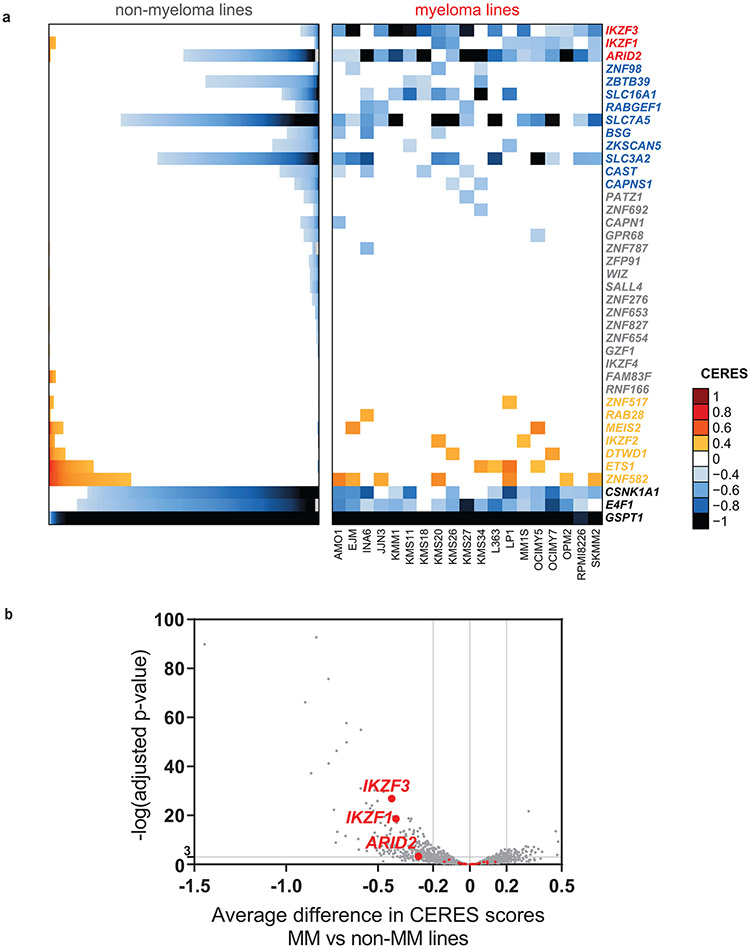
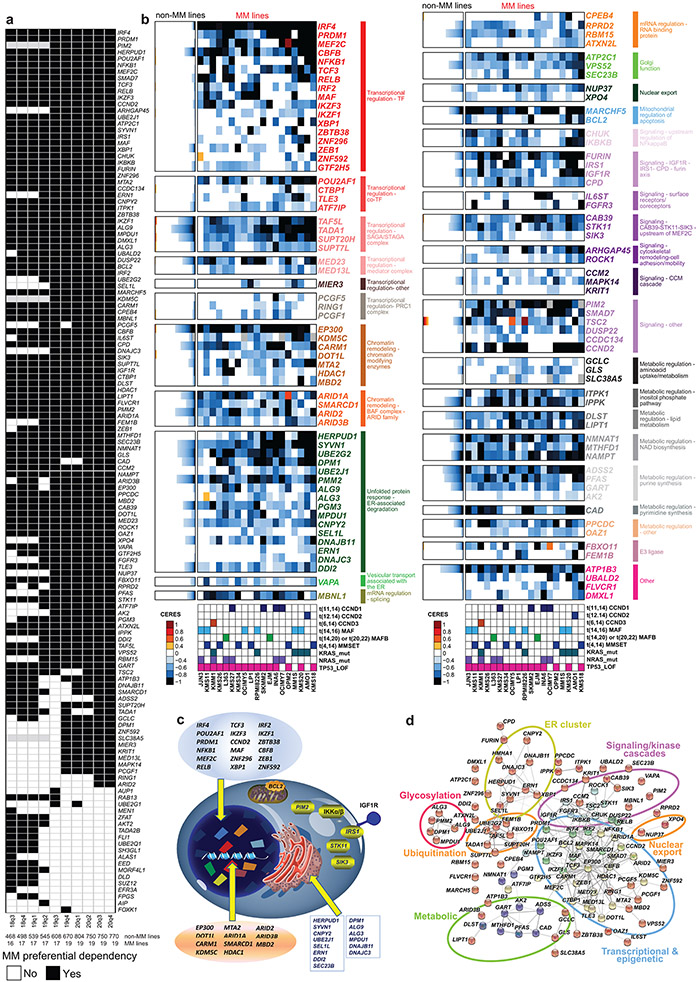
Figure 1 ∣. Myeloma–preferential dependencies identified by genome-scale CRISPR-based gene-editing screens.
a, Color-coded heatmaps depict CERES scores, as a quantitative metric of dependence of human tumor cell lines to each gene in CRISPR/Cas9 gene-editing screens (AVANA sgRNA library). CERES scores for MM lines (n=19) are depicted as a matrix (right side of graph) of cell lines (in columns) and genes (in rows). For non-MM lines (n=770), data are depicted for each gene (row) in stacked bar graphs, which visualize the CERES score of each gene in descending order (from left to right). Black or dark blue signifies negative CERES scores compatible with pronounced sgRNA depletion of a given gene for a specific cell line. MM-preferential dependencies were identified based on average CERES scores in MM cell lines ≤−0.2; difference in average CERES scores in MM vs. non-MM lines ≤−0.2; two-sided limma t-test with adjusted p-value (FDR) <0.05 for comparison of CERES scores; and additional criteria outlined in Methods. b, Pie chart of the distribution of MM-preferential dependencies to different functional groups, pathways, or biological functions.
Approximately one third of the genes preferentially essential for MM encode transcriptional and epigenetic regulators (Fig. 1b, Extended Data Fig. 1b-d). These include regulators of plasma cell biology (e.g., IRF4, PRDM1, XBP1, IKZF1, IKZF3), members of the NF-κB pathway (e.g., NFKB1, RELB), or other genes involved in MM pathogenesis (e.g., MAF). Several transcription factors with underappreciated roles in MM, including MEF2C, CBFB, TCF3, IRF2, ZBTB38, ZNF296, and ZNF592, as well as transcriptional cofactors such as POU2AF1, CTBP1, TLE3 and ATF7IP were also identified. Disruption of several epigenetic enzymes had a more pronounced effect on MM compared to non-MM cell lines, including EP300, KDM5C, CARM1, DOT1L, and HDAC1; as well as members of the BAF (SWI/SNF) complex (ARID1A, SMARCD1, ARID2); STAGA complex (TAF5L, TADA1, SUPT20H, SUPT7L); Mediator complex (MED23, MED13L) and PRC1 (PCGF5, RING1 and PCGF1). MBNL1, a regulator of alternative splicing of pre-mRNAs, and several RNA binding proteins (CPEB4, RPRD2, RBM15 and ATXN2L) were also more essential in MM cell lines.
Consistent with the highly secretory nature of plasma cells, a large group of MM-preferential dependencies are involved in endoplasmic reticulum (ER) function (Fig. 1b), including genes encoding ER membrane protein complexes mediating dislocation of misfolded proteins from the luminal side of the ER to the cytosol (e.g., HERPUD1 and SEL1L); ER-specific E2 ubiquitin conjugating enzymes (e.g., UBE2J1, UBE2G2) or the E3 ligase SYVN1; enzymes required for N-glycan-dependent surveillance of quality control for luminal ER glycoproteins (e.g., DPM1, PMM2, ALG3, ALG9, PGM3, MPDU1) chaperones for misfolded ER proteins (e.g., DNAJB11, DNAJBC3); the ER stress-sensor IRE1a (ERN1) and the target of its RNA processing activity, XBP1. Other molecules involved in ER stress sensing and response (e.g. CNPY2 6, DDI2); or involved in transport of proteins from ER to the Golgi network (e.g. ATP2C1, SEC23B) are also preferentially essential for MM cells (Extended Data Fig. 1b,c).
Several genes preferentially essential for MM cells encode proteins participating in proliferative/anti-apoptotic signaling cascades (Fig. 1b) including, the serine/threonine kinase PIM2; IKBKB (IKK-β) and CHUK (IKK-α), which are upstream of NF-κB transcription factors; members of the IGF1R signaling cascade, including IGF1R itself, its downstream effector IRS1 and the peptidases FURIN and CPD (carboxypeptidase D) which regulate the cleavage of the IGF1R polypeptide to its mature form7; as well as IL6ST (gp130; a coreceptor for IL-6 and other cytokines). FGFR3 is also a MM-preferential dependency, likely reflecting MM cell lines with t(4;14) chromosomal translocationthat results in FGFR3 overexpression, versus the highly infrequent nature of FGFR3 essentiality in other malignancies. Notably, STK11, a tumor suppressor in lung cancer, its positive regulator CAB398 as well as SIK3, a downstream target of STK11 and an upstream regulator of MEF2C in other systems9, are preferentially required for MM cells. Additional signaling-related MM-preferential dependencies include the negative regulator of TGF-β signaling SMAD7; ARHGAP45 (HMHA1) and ROCK1, which are involved in regulation of cell adhesion and motility; as well as the CCM signaling complex members CCM2, KRIT1 (CCM1), and their downstream interactor MAPK14. Finally, other genes preferentially essential for MM cells include those encoding the mitochondrial regulator of apoptosis BCL2 and the mitochondrial E3 ligase MARCH5 (MARCHF5); the E3 ligases FBXO11 and FEM1B; and the nuclear transport proteins NUP37 and XPO4.
Molecular alterations of MM-preferential dependencies
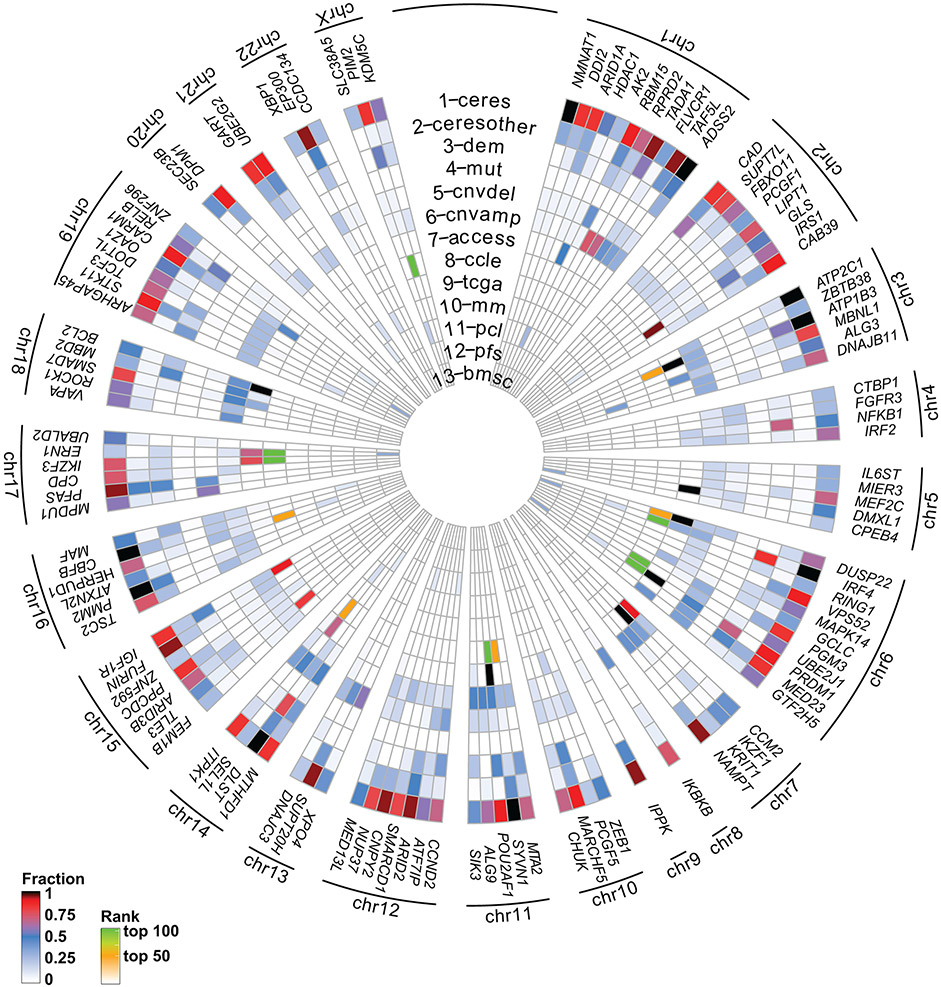
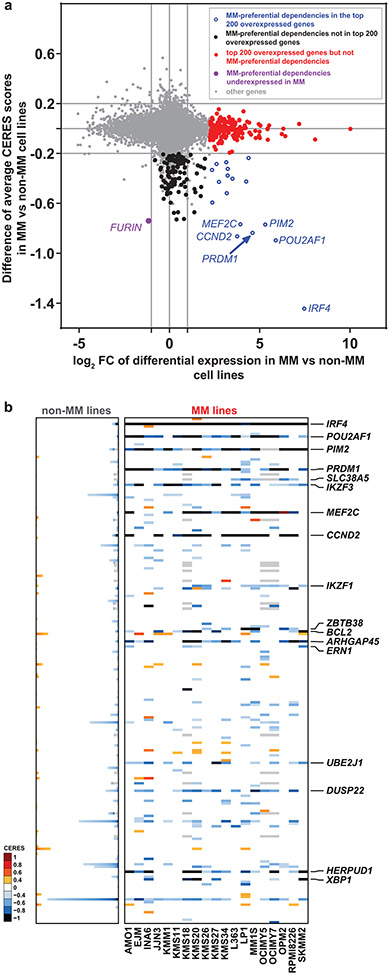
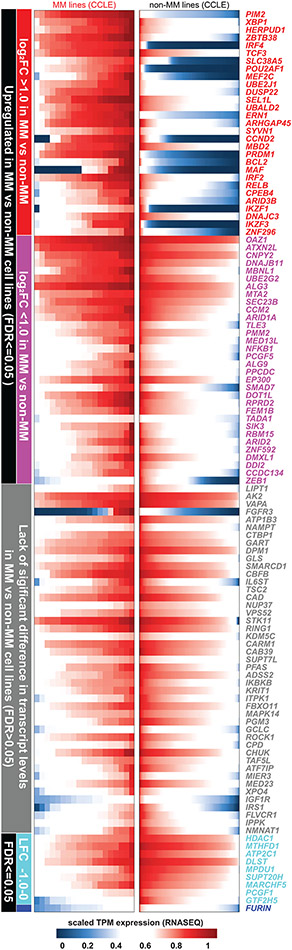
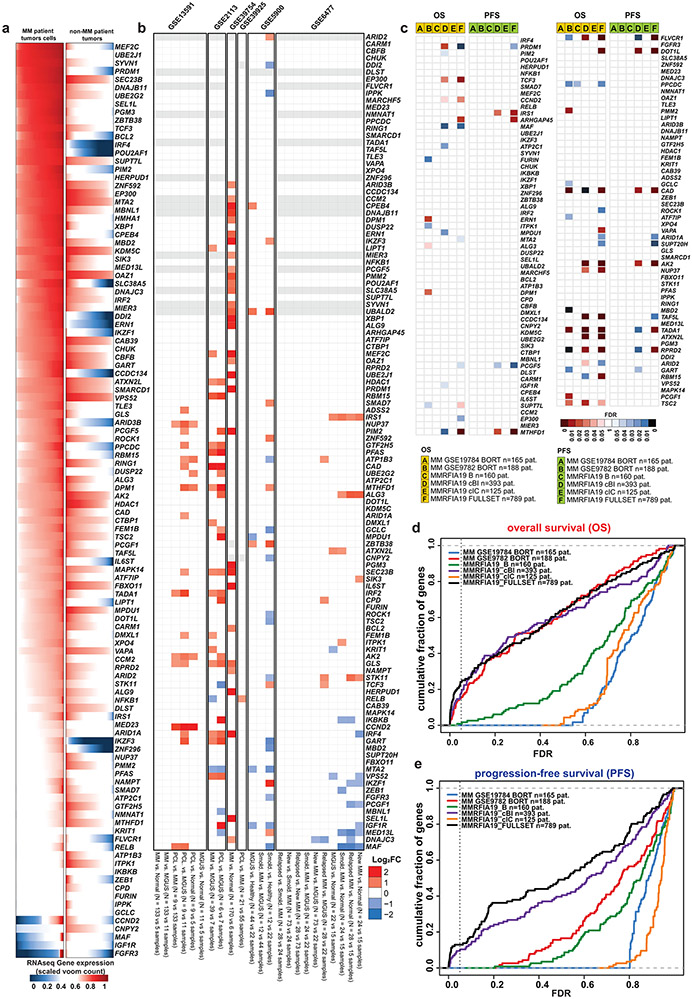
We examined whether there are recurrent molecular alterations in the genes preferentially essential for MM cells (as summarized in Fig. 2). Among 834 genes overexpressed (log2FC>1.0, FDR<0.05) in MM vs. non-MM cells lines (Cancer Cell Line Encyclopedia [CCLE], Fig. 3a), only 4% (29) are among the 116 MM-preferential dependencies. Notably, 6 of these genes have the greatest difference in essentiality scores in MM vs. non-MM cells. These include the lineage-defining transcription factors IRF4 and PRDM1, as well as POU2AF1, PIM2, MEF2C and CCND2. However, only a minority of MM-preferential dependencies are in the top 100–200 overexpressed genes when ranked by log2FC (Fig. 3b) or FDR (Fig. 2, circle 8) in MM vs. other tumor types and some MM-preferential dependencies are less highly expressed in MM lines (Fig. 3a, Extended Data Fig. 3). Similar observations were made when examining transcript levels for these genes in MM vs. non-MM patient tumor samples (Extended Data Fig. 4a). Most MM-preferential dependencies are not overexpressed in MM vs. normal PCs or more highly expressed in later vs. earlier stages of myelomagenesis (Extended Data Fig. 4b) and do not consistently correlate with adverse patient outcome (Extended Data Fig. 4c), even under relaxed statistical criteria (Extended Data Fig. 4d,e). Moreover, most MM-preferential dependencies were not among the top overexpressed transcripts in MM cells co-cultured with mesenchymal bone marrow stromal cells (BMSCs) (Fig. 2, circle 13), an interaction which attenuates MM cell responses to diverse therapies10-12.
Figure 2 ∣. Integrated molecular profiling analyses for MM-preferential dependencies.
CIRCOS plot summarizing results of integrated molecular analyses for MM-preferential dependencies (more details in Figs. 3, 4 and Extended Data Fig. 3-6) to examine whether most of them are among the top genes with most frequent molecular alterations (for example, mutations, DNA copy number gains or differential expression) in MM cells. Concentric circles depict for each gene: (1-2) fraction of MM (1; “ceres”) or non-MM (2; “ceresother”) lines with CERES scores ≤−0.4; (3) fraction of MM lines with DEMETER scores ≤−0.4 (“dem”); (4-6) fraction of MM cell lines with non-synonymous mutations (4; “mut”; see Extended Data Fig. 5), CNV loss (5; “cnvdel”) or CNV gain (6; “cnvamp”) (see Extended Data Fig. 5); (7) fraction of MM cell lines with a super-accessible chromatin region annotated by closest proximity to the gene of interest (“access”). Circles 8-12 summarize whether expression of a gene is higher in (8) MM vs. non-MM cell lines of CCLE (“ccle”; see Fig. 3 and Extended Data Fig. 3); (9) tumor samples from patients with MM (CoMMpass study) vs. non-MM patients (TCGA) (“tcga”; see Extended Data Fig. 4a); (10) MM patient samples vs. normal PCs (“mm”; see Extended Data Fig. 4b); (11) PCL (or advanced MM) vs. early/newly diagnosed MM (“pcl”; see Extended Fig. 4b); (12) patients with shorter PFS (“pfs”; see Extended Data Fig. 4c); and (13) when MM cells are co-cultured with bone marrow stromal cells (BMSCs) (“bmsc”) in dataset GSE20540. For circles 8 and 9, transcripts with log2FC>1.0 and FDR <0.05 are in green or orange, if they rank (based on FDR), respectively, in the top 1-50 or 51-100 most upregulated genes (white depicts genes that did not satisfy all these criteria). Each of the circles 10-13 integrates several individual comparisons (see Methods) and depicts (based on the color-coded scale) the fraction of these comparisons with upregulation by log2FC ≥1.0 and FDR ≤0.05 and ranking (based on FDR) in the top 100 most upregulated genes.
Figure 3 ∣. Most MM-preferential dependencies do not rank among the top overexpressed genes in MM vs non-MM cell lines.
a, Scatter plot depicting for each gene the log2FC of differential expression in MM (n=25) vs. non-MM (n=991) cell lines in CCLE (x-axis) vs. average differences in CERES score (y-axis, see Supplementary Table 1) in MM vs non-MM lines in CRISPR gene editing screens (N = 17,436 genes with matching gene symbols between CCLE and CERES data). The plot highlights genes that are (i) preferentially essential and in the top N = 200 overexpressed genes (log2FC>1.0, two-sided limma t-test, FDR<0.05, ranking based on log2FC) in MM (blue circles); (ii) the top N = 200 overexpressed genes that are not preferentially essential in MM (red dots); (iii) a MM-preferential dependency that is under-expressed (log2FC<−1.0, FDR<0.05) in MM vs. non-MM cell lines in CCLE (purple dot); (iv) other MM preferential-dependencies that are not in the top N = 200 overexpressed genes (black dots) and (v) other genes (gray dots). b, Heat-maps for MM (N = 19 cell lines) (right; matrix) and non-MM (N = 770 cell lines) (left; stacked bars) depict CERES scores of the top N = 200 most upregulated genes in MM vs. non-MM cell lines (CCLE) for which both transcript and CERES data are available (significantly upregulated genes were ranked according to log2FC of differential expression, distinctly from the FDR-based ranking of differentially expressed genes for Fig. 2). Gene symbols are depicted for the minority of top upregulated genes that represent MM-preferential dependencies. Gene expression data for a, was accessed from the initial CCLE portal, with concordant observations based on subsequent releases of these data through DepMap portal.
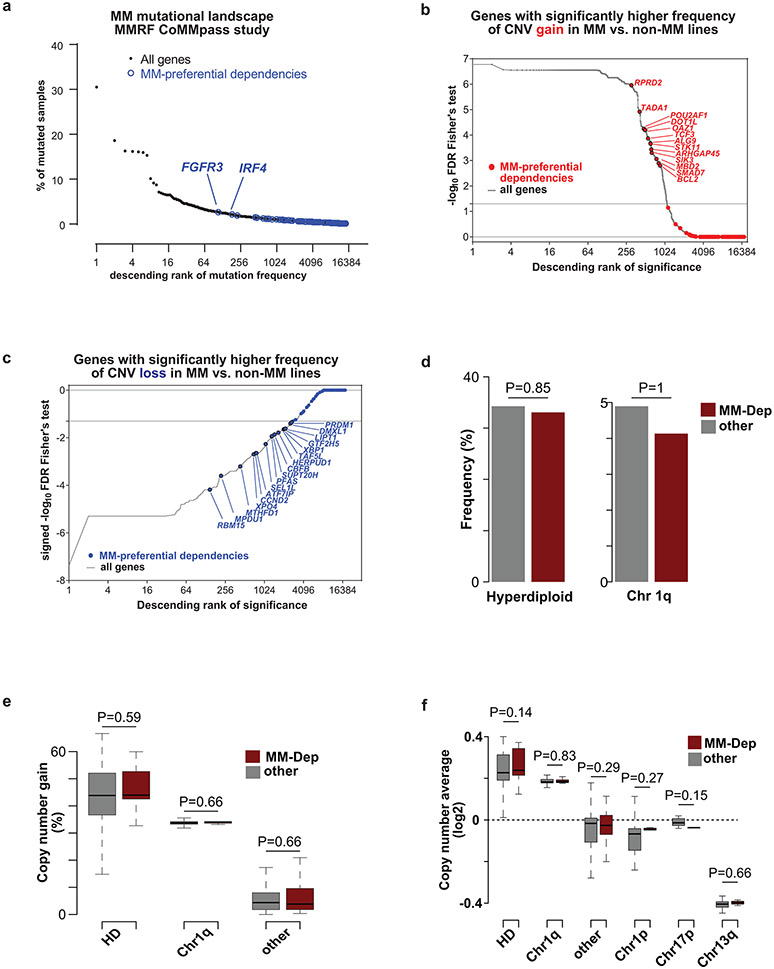
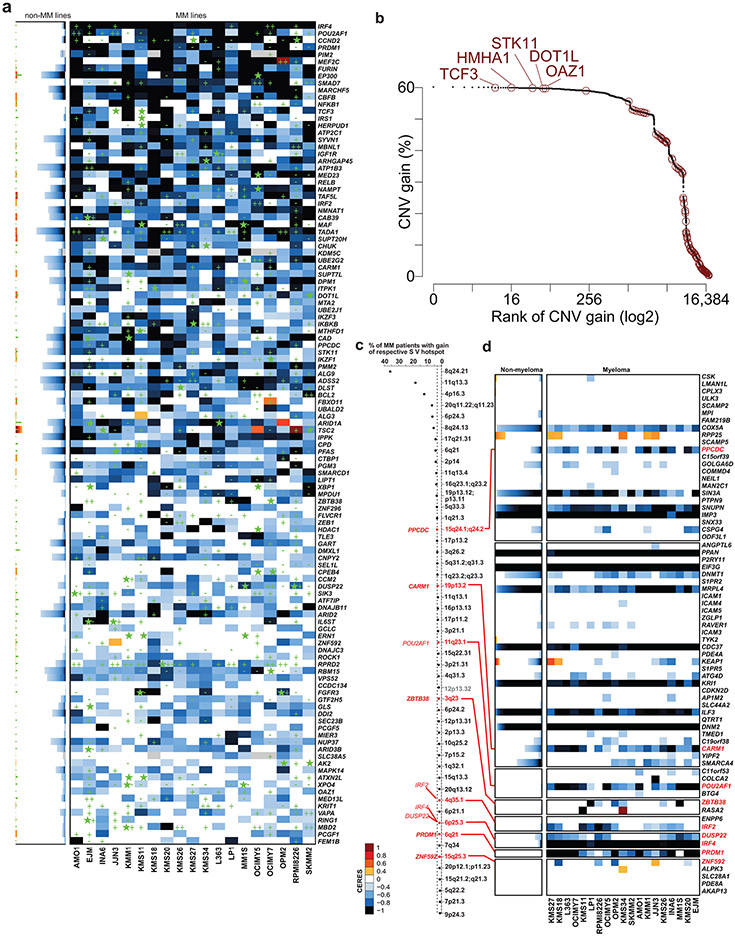
Only 10/116 MM-preferential dependency genes were mutated in more than one of the MM cell lines (Extended Data Fig. 5a). Only two MM-preferential dependencies (FGFR3 and IRF4, mutated in 2% of patients) are among the top 200 most frequently mutated genes in patients with newly diagnosed MM (Fig. 4a). Furthermore, the large majority of MM-preferential dependencies did not have higher frequency of DNA copy number variation (CNV) gains, while some had a higher rate of CNV losses, in MM vs. non-MM cell lines (CCLE; Fig. 4b,c). In patient-derived MM samples, MM-preferential dependencies are not enriched within regions of frequent large CNV gains (e.g., hyperdiploid chromosomes or chromosome 1q) or losses (Fig. 4d,e). In patient-derived MM samples, MM-preferential dependency genes did not exhibit a higher frequency of CNV gains (Fig. 4e) or DNA copy number (Fig. 4f). Furthermore, only 5 MM-preferential dependencies are among the top 200 genes with the highest frequency of CNV gains in patient samples (Extended Data Fig. 5b). Regarding structural variants (SVs) that result in focal CNVs and complex somatic events13, 45 regions, which harbor in total 475 genes evaluated in our CRISPR screens, were recently identified13 as hotspots for SVs that cause gain of chromosomal material. Of these 45 regions, 8 contain 9 of the 116 MM preferential dependencies, namely IRF4 (and its neighboring DUSP22), POU2AF1, IRF2, PPCDC, CARM1, ZBTB38, PRDM1 and ZNF592 (Extended Data Fig. 5c,d). Notably, two MM preferential dependencies (MPDU1, PFAS) are located in a SV loss hotspot for MM (specifically within 17p 13). Overall, a limited number of MM-preferential dependencies may be located in regions with structural rearrangements or copy number alterations, but most MM-preferential dependencies do not rank among the top genes in terms of the frequency of these events in MM or their enrichment in MM compared with non-MM.
Figure 4 ∣. Landscape of single nucleotide variants and DNA copy number variants for MM-preferentially essential genes.
a, Frequency of non-synonymous single nucleotide variants (SNVs) in N=940 samples from MM patients (CoMMpass study, IA17 release). MM-preferential dependencies (as defined in Fig. 1a, Supplementary Table 1) are highlighted in blue. b,c, Ranking of MM-preferential dependencies and other genes in terms of statistical significance (FDR, two-sided Fisher’s test) of the frequency of CNV gains (b) or losses (c) in MM (n=33) vs. non-MM (n=1721) lines of CCLE panel (based on data and annotation from DepMap 22Q1 release, concordant observations with other releases). d, Frequency of MM-preferential dependencies (MM-dep; red) and other (gray) genes that fall in sites of common CNV gains, including hyperdiploid (HD) chromosomes (e.g., 3, 5, 7, 9, 11, 15, 19, 21) in MM. e, Frequency of CNV gains in CoMMpass samples for MM-preferential dependencies and all other genes stratified by hyperdiploid (HD) chromosomes, chromosome 1q, and other. f, Average DNA copy number in CoMMpass samples for MM preferential dependencies vs. other genes stratified by HD, chromosome 1q, other, chromosome 1p, chromosome 17p and chromosome 13q. P-values are from two-sided Fisher’s exact test (d) or two-sided Mann-Whitney U-test (e-f). Panels e-f evaluated N=932 patient samples for 19054 genes with DNA copy number data available in the CoMMpass study (IA15 release).
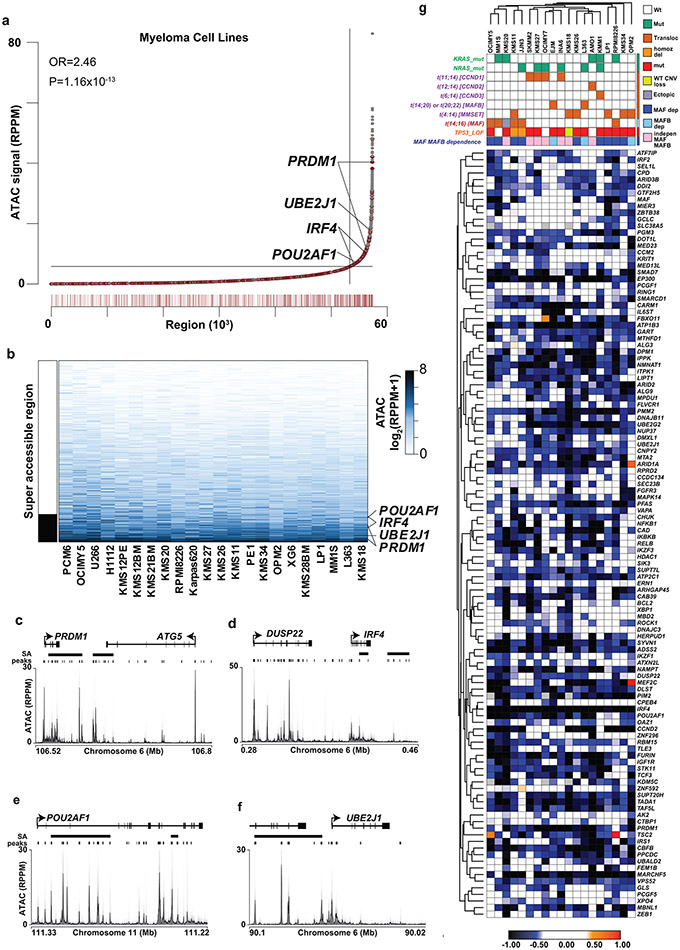
Chromatin regions such as “super-enhancers”, defined by dense transcription factor binding, H3K27 acetylation, and chromatin accessibility, facilitate gene expression critical for cell identity14. To determine if such gene regulatory features defined MM-preferential dependencies, we examined chromatin accessibility (ATAC-Seq) in 12 MM cell lines with a focus on MM-preferentially essential genes. This identified on average 5-6 chromatin accessible regions within 100 kb of the MM-preferential dependency genes and these were modestly enriched at super-accessible regions (Extended Data Fig. 6a) that were largely consistent across the 22 MM cell lines analyzed (Extended Data Fig. 6b). Examples of these chromatin accessible regions can be found at PRDM1, UBE2J1, and IRF4; in regions of the DUSP22 gene that may regulate nearby IRF4; as well as in POU2AF1 (Extended Data Fig. 6c-f). While 55/116 MM-preferential dependencies were ≤100 kb from a highly accessible region, there were over 4,000 super-accessible regions covering over 3,400 genes, and therefore MM-preferential dependencies could not be readily identified by chromatin accessibility alone.
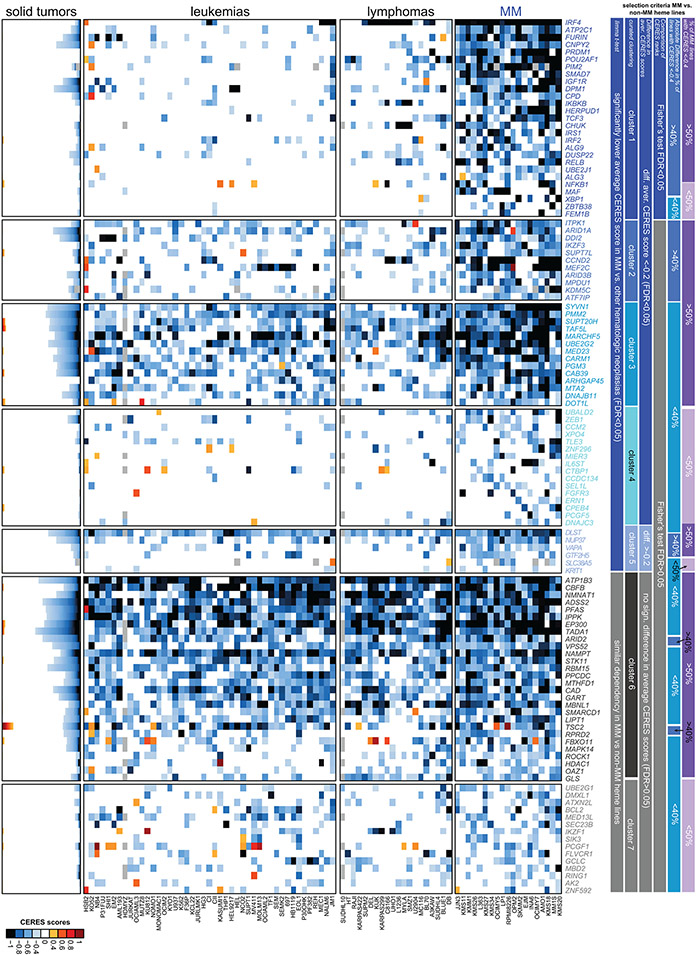
Collectively, these data (Fig. 2) indicate that many MM-preferential dependencies identified by CRISPR gene-editing screens are not among the top recurrently mutated, amplified or aberrantly expressed genes in MM. This observation is concordant with data on preferential dependencies in other malignancies, such as ER+ breast, renal, or colon cancer, melanoma, and acute myeloid leukemia (Supplementary Figures 2-6).
MM encompasses several subgroups defined by molecular features, such as chromosomal translocations involving immunoglobulin gene enhancers or mutations/DNA copy number events in key oncogenes or tumor suppressors. MM lines with translocations targeting CCND1, CCND2, CCND3, MAF, and FGFR3; or with KRAS or NRAS mutations tend to be dependent on these respective genes. Hierarchical clustering of MM cell lines according to the essentiality scores for preferential dependencies revealed that 4 of 5 MM lines with MAF rearrangement and another line with ectopic MAF overexpression were in the same branch of the dendrogram, while 4 lines with CCND1 rearrangement were in adjacent branches. Overall, however, clustering of lines based on their essentiality scores for MM-preferential dependencies as a group does not distinguish molecular subtypes, perhaps reflecting the limited numbers of lines from each MM subtype (Extended Data Fig. 6g) and a need for gene editing studies in larger panels of MM lines in order to better define subtype-specific MM dependencies.
MM dependencies: shared or distinct roles in other cancers
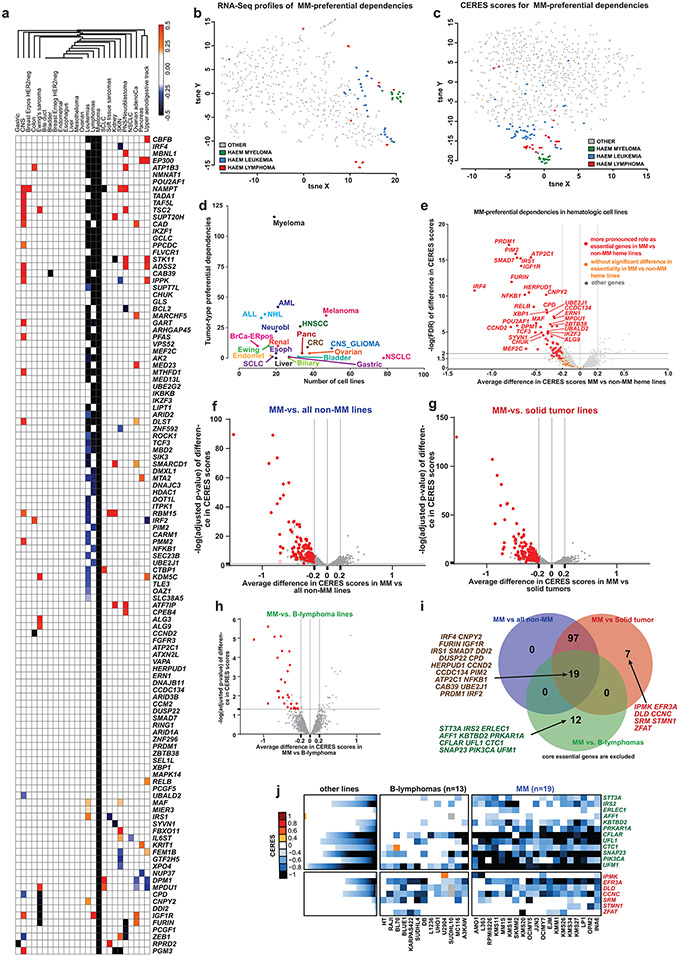
The 116 genes we identified are preferentially, but not necessarily exclusively, important for MM cells. Several of them are recurrently essential in other neoplasias, e.g. EP300, MARCHF5, CBFB, MBNL1, DOT1L or FURIN in leukemia (Extended Data Figs. 7 and 8a-e), while IRF4 is important for lymphoma and a subset of melanoma lines (Extended Data Fig. 8a). Notably, though, the essentiality scores for the MM-preferential dependencies define a tight cluster of MM lines distinct from all non-MM, including other hematologic, cell lines in the t-distributed stochastic neighbor embedding (t-SNE) clustering analysis (Extended Data Fig. 8b,c). Applying in other neoplasias across the DepMap dataset the criteria we used to define preferential dependencies for MM, we identified genes previously known to be essential for tumors of different lineages including CTNNB1 for colorectal or ESR1, FOXA1 and SPDEF for ER+ breast cancer (Supplementary Figs.2-6). However, in general, non-MM tumor types had fewer preferentially essential genes, even those with gene editing screens in higher numbers of cell lines than MM. (Extended Data Fig. 8a,d).
The high number of MM-preferential dependencies might not reflect solely biological differences between malignant hematopoietic vs. solid tumor cells but a specific set of vulnerabilities associated with PC biology. Consistent with this notion, 68 MM-preferential dependencies were more essential to MM cells than to non-MM blood cancer cell lines (difference in average CERES scores ≤−0.2, FDR<0.05; Extended Data Fig. 8e). Gene editing with a sub-genome scale library that included single guide RNAs (sgRNAs) for 89 MM-preferential dependencies was performed in two cell lines representing Waldenström’s macroglobulinemia (WM), a lymphoplasmacytic lymphoma, which is related to MM but also has several distinct biological and genetic features. Disruption of 28 MM-preferential dependency genes had no effect on either WM cell line and 40 additional MM-preferential dependencies were not essential in one of the WM lines (Supplementary Fig. 7a-c), highlighting the distinct pattern of genetic vulnerabilities of MM, even when compared to a closely related malignancy.
Some genes do not meet all criteria for designation as preferential dependencies in MM when compared to all other non-MM (heme or solid) tumor lines but are more essential in MM vs. B-cell lymphoma; or in MM vs. solid tumor lines (Extended Data Fig. 8f-j). Several of these genes function in similar pathways as some MM-preferentially essential genes, such as the ER-associated degradation (ERAD)-related genes ERLEC1, STT3A, UFL1, and UFM1 which are more essential in MM vs. B-cell lymphomas (Extended Data Fig. 8i,j). IKZF1, IKZF3, or BCL2 which can be therapeutically targeted, are more essential for MM compared with all non-MM lines, but have a similar importance for B cell lymphomas. Conversely, some genes such as PIK3CA are more essential for MM vs. B-cell lymphomas, but are similarly critical for all other non-MM cell lines (Extended Data Fig. 8i,j) and have not yet proved to be clinically actionable in MM. These examples highlight that defining differential dependencies for MM cells may provide distinct information depending on the comparator group, e.g., all non-MM tumor cells or specific hematologic malignancies: the latter comparisons inform about potential biological differences in MM vs. the respective neoplasias and warrant studies in larger cell line panels.
Further highlighting their distinct roles in MM, several MM-preferential dependencies function as tumor suppressors in others diseases, e.g., FBXO1115 or PRDM116 in lymphoma; or STK11, CAB39, and TSC2 in many cancers17. This seemingly paradoxical observation may relate to the biology of PCs and the functional relationships of these genes with other MM-preferential dependencies. For instance, CERES scores for TSC2 and several other negative regulators of mTORC1 signaling (e.g., DDIT4, DEPDC5, NPRL2) exhibit positive correlation in MM and other lines (Supplementary Fig. 7d-g), concordant with the TSC1/2 complex as negative mTORC1 regulator in MM cells. RHEB, direct downstream target of TSC1/2 and positive regulator of mTORC1, has higher CERES scores in MM vs. other cell lines (Supplementary Fig. 7h,i). Therefore, disruption of the TSC1/2 complex leading to hyperactive mTORC1 can drive growth of other cell types but also leads to increased ER stress18, to which MM cells are particularly susceptible. Recent studies in leukemia9 reported that STK11 activates SIK3 and SIK2, which in turn activate MEF2C, another gene preferentially required by MM (Supplementary Fig. 7j,k). These examples suggest that cell lineage is critical for interpretation of gene essentiality screens.
Preferential dependencies previously implicated in MM
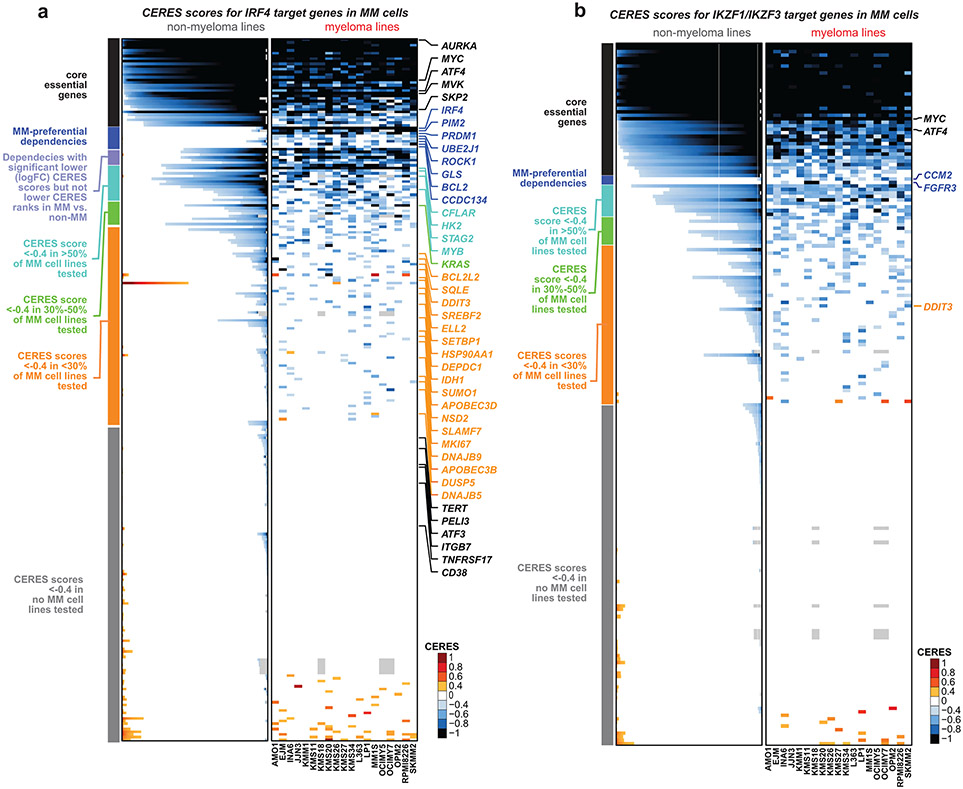
Several MM-preferential dependencies (IRF4 5, MAF, CCND2, IKZF3, IKZF1, NFKB) have known roles in MM, but limited, if any, prior formal evaluation of their preferential essentiality in MM compared with other cancers. We examined the patterns of essentiality of genes targeted by IRF45 (Supplementary Fig. 7l, Extended Data Fig. 9a) or IKZF1 and IKZF319 (Extended Data Fig. 9b) in MM. Each of these TFs regulates in MM cells genes which, represent distinct clusters, including genes essential across all tumor types; genes with recurrent proliferative, anti-apoptotic, or oncogenic roles across many cancers (e.g., regulation KRAS by IRF4); as well as genes that individually are not required for growth of MM or other cancer cell lines. Notably, several putative IRF4 targets (e.g., PRDM1, PIM2, BCL2, UBE2J1 and CCDC134) are themselves MM-preferential dependencies (Extended Data Fig. 9a, based on data from Fig. 1-2), which may explain why IRF4 disruption is so disadvantageous to MM cell fitness.
MM-preferential dependencies also include targets for anti-MM therapies, including the thalidomide derivative targets IKZF3 and IKZF120,21 and a recently identified CRBN neo-substrate ARID222, but not other CRBN neosubstrates23-32 (Fig. 5). Genes required for MM cell fitness also include those encoding molecules mediating the anti-MM activity of proteasome inhibitors such as members of the NF-κB pathway33 and regulators of ER-associated protein degradation. These results are concordant with the fact that the clinical effects of thalidomide derivatives and proteasome inhibitors are mostly limited to PC malignancies. HDAC1 and BCL2 are also MM-preferential dependencies, consistent with the activity of inhibitors against these targets in clinical trials. IGF1R and members/regulators of its pathway (for example, IRS1, FURIN7) are also MM-preferential dependencies, consistent with the greater preclinical activity of IGF1R inhibitors against MM compared with other cell types 34,35. These data suggest that other genes identified from this study may have therapeutic relevance.
Fig. 5 ∣. CERES scores for reported substrates or targets for thalidomide derivatives.
a, Heatmaps depict CERES scores for known/proposed substrates or targets of thalidomide derivatives. Results as depicted as a matrix for N=19 MM cell lines (right side of graph) and stacked bar plots for N=770 non-MM cell lines (with format and color-coding similar to other figures, e.g., Fig. 1a). Gene symbols (for N=39 genes) are highlighted in red for MM-preferential dependencies whose protein products are known (IKZF1, IKZF3) or recently proposed (ARID2) neo-substrates for thalidomide derivatives; black for “core essential” genes; blue for genes that are not “core essential” or MM-preferentially essential and have CERES scores <−0.4 in ≥2 MM lines tested; and gray or orange for other known or reported CRBN neo-substrates / targets of thalidomide derivatives.
b, Dot plot depicting for each gene the −log10FDR (Limma t-test) for comparison of CERES scores in MM (N =19 cell lines) vs non-MM (N = 768 cell lines) (y-axis) vs. the difference in average CERES scores in MM vs. non-MM cell lines (x-axis) (N=18,119 genes, also see Supplementary Table 1). Genes whose protein products are known or proposed targets/neosubstrates of thalidomide or its derivatives are highlighted in red dots and those genes (IKZF3, IKZF1 and ARID2) that also meet the criteria for MM-preferential dependencies are highlighted by their symbols.
In vitro studies supporting CRISPR screen results
MM lines harboring doxycycline (Doxy)-inducible SpCas9 were transduced with sgRNAs directed against MM-preferential dependencies including PIM2, MEF2C, TCF3 and DOT1L. Doxy treatment led to significant depletion of MM cells transduced with these sgRNAs (Supplementary Fig. 8a,b) compared with control sgRNAs for olfactory receptor genes, which are not expressed in MM36. As an orthogonal validation of the gene disruption screening results, treatment of MM lines with antagonists for the methyltransferase CARM1 (PRMT4)37,38, the CBFB transcription factor39, the SIK kinases (including SIK3) or PIM kinases (including PIM2) decreased the relative viability of MM cells (Supplementary Fig. 8c-e). Additional validation of genome-scale CRISPR studies was offered by data from pharmacological screens (Supplementary Fig. 8f,g). Inhibitors against the products of several genes preferentially essential for MM were more active against MM lines compared to lines from solid tumors or other hematologic malignancies. These included “positive controls” such as lenalidomide (targeting IZKF1, IZKF3), bortezomib (targeting ER function or NF-κB); as well as inhibitors for BCL-2, IKK1/IKK2 (CHUK/IKBKB), IGF1R, HDAC1 and NAMPT (Supplementary Fig. 8f,g).
POU2AF1, an essential transcriptional cofactor for MM cells
The roles for many MM-preferentially essential genes were previously only modestly explored. One example is POU2AF1, encoding the OCA-B transcriptional cofactor. Prior studies in MM suggested that POU2AF1 regulates expression of BCMA (TNFRSF17)40; TCR-engineered T cells recognizing POU2AF1 peptides can have therapeutic applications41; while elevated POU2AF1 protein levels correlates with adverse prognosis42. However, the role of POU2AF1 as a dependency in MM has been understudied. POU2AF1 was the most preferentially essential gene encoding a transcriptional co-factor in MM (Fig. 1) and was also essential for several MM cell lines in shRNA studies (Extended Data Fig. 2c,d). Depletion of POU2AF1 protein levels through Doxy-inducible CRISPR interference (Extended Data Fig. 10a) decreased MM cell growth (Fig. 6a), while CRISPR-based activation of POU2AF1 (Extended Data Fig. 10b) stimulated growth of LP-1 MM cells (Fig. 6b and Extended Data Fig. 10c). POU2AF1 overexpression also triggered upregulation (Fig. 6c) of other MM-preferential dependencies (e.g., PRDM1, SUPT7L, UBE2G2), TSC1, KRAS and other genes implicated in the pathogenesis of MM or other cancers (e.g., FGFR3, RUNX243, SMO, MEF2D, and PCGF2); as well as downregulation of CDKN1C (Fig. 6c), encoding a cyclin-dependent kinase inhibitor. POU2AF1 overexpression also led to downregulation of MHC class II molecules (Fig. 6d) and their transcriptional activator CIITA (Fig. 6c), suggesting potential roles of POU2AF1 in immune evasion.
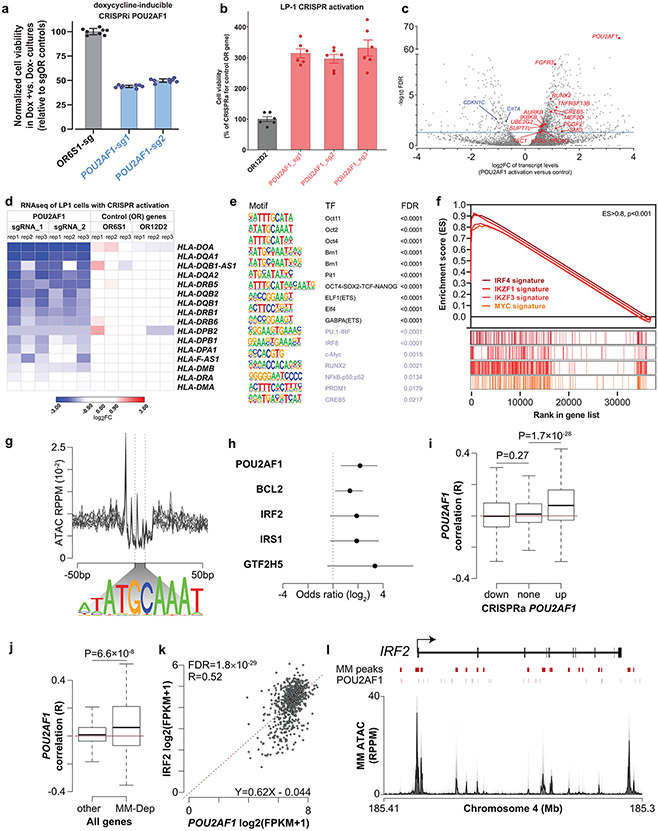
Fig. 6: ∣. Biological role of POU2AF1 in MM cells.
a,b, Relative number of viable cells after Doxy-inducible CRISPR interference (CRISPRi) (KMS-11 cells, 11 days after sgRNA transduction) (a) or CRISPR activation (CRISPRa) (LP-1 cells, 19 days after sgRNA transduction) (b) of POU2AF1 vs. control OR genes. CTG assays, N = 8 (a) or N = 6 (b) independent replicate cell cultures per condition; mean±SEM, one-way analysis of variance (ANOVA) and Tukey’s post-hoc test (detailed results in source data), P < 0.001 for each POU2AF1 sgRNA vs. OR gene sgRNA). c–f, Transcriptional signature of POU2AF1 overexpression in LP1 MM cells: volcano plot of transcripts differentially expressed in LP1 cells with CRISPR activation of POU2AF1 vs. OR controls (blue line denotes FDR = 0.05) (c); HLA class II transcript levels with POU2AF1 activation vs. control (d); TF DNA-binding motifs enriched in sites of chromatin binding of POU2AF1, where top ten most statistically significant motifs (in black) include POU2AF1 partner Oct2 (POU2F2), whereas others include motifs for TFs relevant to MM, such as Myc, PU.1-IRF, NF-κB, PRDM1 and CREB5, which is overexpressed with POU2AF1 activation (e); GSEA plots examining the transcriptional signature of POU2AF1 activation identify enrichment for genes previously determined as targets of IRF4, IKZF3, IKZF1 or Myc (P < 0.001, for each plot) (f). g–l, POU2AF1 binding motifs are enriched in chromatin accessible regions near select MM-preferential dependencies: ATAC-seq signal at POU2AF1 binding motifs in 12 MM DepMap cell lines (top), with the POU2AF1 consensus binding motif shown (bottom) (g); MM-preferential dependencies with significant enrichment of POU2AF1 binding motifs in chromatin accessible regions (odds ratio of enrichment lines denoting 95% confidence intervals shown, Fisher’s exact test) (h); correlation of transcript levels in N = 768 newly diagnosed primary MM specimens (CoMMpass study, IA15 release) for POU2AF1 expression with genes downregulated (down), not significantly changed (none) or upregulated (up) by CRISPR activation of POU2AF1 (i); correlation of POU2AF1 expression with transcript levels of MM-preferential dependencies (MM-Dep; N = 116 genes) or all other N = 55,092 genes (two-sided t-test for i and j; box plots denote median, lower/upper quartiles, with whiskers extending up to 1.5 times the interquartile range of the box) (j); gene expression correlation between POU2AF1 (x axis) and IRF2 (y axis) in N = 768 patient samples (CoMMpass study IA15 release), with significance determined by edgeR and FDR corrected), and gene expression measured in fragments per kilobase per million reads (FPKM) (k); genome plot of IRF2 showing MM chromatin accessible regions (MM peaks), POU2AF1 consensus binding motifs (POU2AF1) with motifs overlapping accessible chromatin (red), and a composite ATAC profile of 12 MM lines (l).
ATAC-Seq indicated that chromatin surrounding the POU2AF1 locus was highly accessible in MM cells (Extended Data Fig. 6e), concordant with its consistent expression (Extended Data Fig. 3 and 4a). Motif analysis of ChIP-Seq data for POU2AF1 (GSE79480) identified overlap with DNA-binding motifs for POU family TFs such as OCT2 (POU2F2), the binding partner of POU2AF1, members of the ETS family and other TFs with roles in MM including c-MYC, IRF4, NF-κB, PRDM1 and RUNX2 (Fig. 6e), suggesting that POU2AF1 may act as a cofactor for these factors. In further support of this notion, gene set enrichment analyses (GSEA) showed that the transcriptional signature of POU2AF1 overexpression is enriched for genes regulated by MM TFs such IRF4, IKZF1, IKZF3 and MYC (Fig. 6f). Motifs associated with POU2AF1 binding are also enriched near the transcriptional start site of several MM-preferential dependencies including POU2AF1 itself, BCL2, IRF2 and IRS1 (Fig. 6g,h). Genes correlating with POU2AF1 expression in MM cells across 768 patients with newly diagnosed MM were enriched among the genes upregulated by CRISPR activation of POU2AF1 in the LP1 MM cell line, suggesting that many are bona fide POU2AF1 targets (Fig. 6i). POU2AF1 expression was also correlated with expression of the 116 MM-preferential dependencies in MM patient samples (Fig. 6j), as exemplified by IRF2 (Fig. 6k), with multiple POU2AF1 binding sites in the accessible chromatin regions of this gene (Fig. 6l). POU2AF1, like IRF4, may be critical for MM cell fitness due to its ability to stimulate expression of other genes essential for MM proliferation and survival.
Endoplasmic reticulum genes preferentially essential for MM
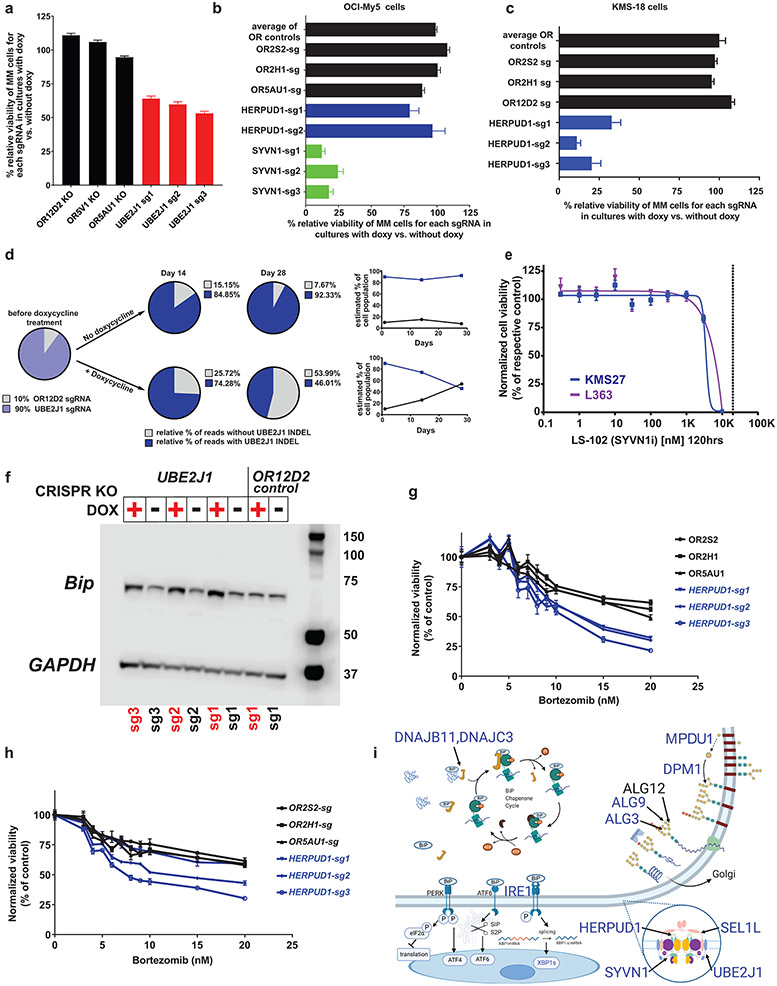
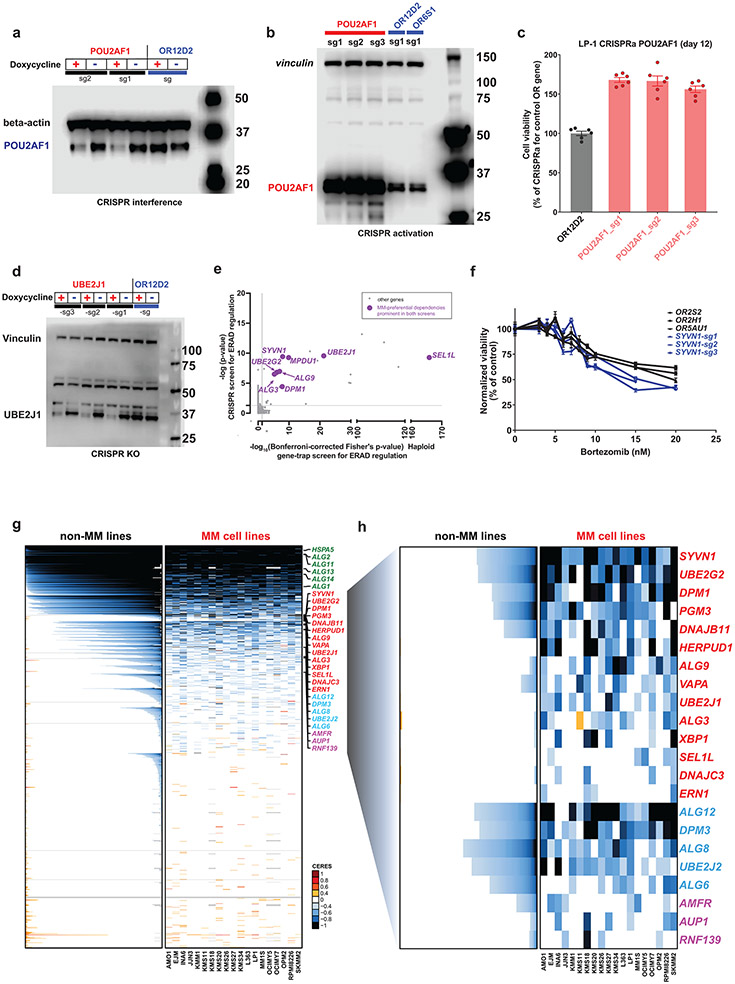
ER-associated degradation (ERAD) for unfolded proteins represents an important biological vulnerability for MM cells, given the proteostatic stress associated with immunoglobulin production2,44. Multiple genes preferentially essential for MM encode previously underappreciated components of the ERAD system (Fig. 1 and Extended Data Fig. 1b,d). Doxy-inducible CRISPR knockout (KO) of UBE2J1 (Fig. 7a and Extended Data Fig. 10d), SYVN1 (Fig. 7b) or HERPUD1 (Fig. 7c) validated observations from gene editing screens in the respective cell lines (Fig. 1). Accordingly, HERPUD1 knockout affected viability of KMS18 cells, but not OCI-My5 cells (Fig. 7b). Moreover, in a competition assay of KMS18 cells harboring Doxy-inducible SpCas9 and sgRNA against UBE2J1 or an olfactory receptor (OR2D12) negative control, UBE2J1 KO cells were outcompeted by control cells only in the presence of Doxy (Fig. 7d). While there are no small molecule inhibitors for UBE2J1 or HERPUD1, LS-102, an inhibitor of SYVN1, inhibited growth of MM cell lines at micromolar concentrations (Fig. 7e). Consistent with the role of UBE2J1 in ERAD, UBE2J1 KO led to induction of the heat shock protein BiP, a marker of ER stress (Fig. 7f). In a reanalysis of a retroviral gene-trap mutagenesis screen and a gene-editing screen for genes involved in ERAD regulation in KBM7 haploid cells45, UBE2J1 was one of the top hits, together with its partners in the ER dislocon (HERPUD1 and SYVN), that facilitates translocation of misfolded proteins from the ER lumen to the cytoplasm (Extended Data Fig. 10e). Given that proteasome inhibitors induce ER stress in MM, we examined whether disruption of ER-associated genes preferentially important for MM could enhance response to proteasome inhibitors. In support of this notion, inducible KO of HEPRUD1 further decreased viability of MM cell lines treated with bortezomib (Fig. 7g,h), while KO of SYVN1 had a more modest effect (Extended Data Fig. 10f). The variable impact that perturbation of different ER-associated genes on MM cell response to proteasome inhibition may reflect diverse roles of these proteins in ER function. Collectively, these data support an important role in MM cells for a series of ER-associated genes (Fig. 7i) which may represent additional targets to enhance efficacy of proteasome inhibitors.
Figure 7: ∣. Biological role of UBE2J1 and other ER-associated MM-preferential dependencies.
a–d, Doxy-inducible CRISPR KO of ER-associated MM preferential dependencies or control OR genes in KMS-18 (a, c and d) or OCI-My5 (b) MM cells. Cells were cultured with or without Doxy (14 days in a–c; 14 or 28 days in d). In a–c, cell viability was evaluated by CTG (mean ± SEM), one-way ANOVA and Tukey’s post-hoc tests (see source data) at P < 0.001 for each ER gene sgRNA (except HERPUD1 in b) vs. each of the OR sgRNAs; 80, 32 and 40 independent replicate cell cultures/sgRNA in a–c, respectively. In d, KMS18 cells with Doxy-inducible SpCas9 and transduced with sgRNA against UBE2J1 or OR2D12 were mixed at a 9:1 ratio, respectively, in a competition assay. INDEL analyses (at days 14 and 28) calculated the relative percentage of cells with CRISPR-induced frameshift mutations of UBE2J1. e, In vitro treatment with SYVN1 inhibitor LS-102 (5 days; vertical dotted line represents reported in vitro half maximal inhibitory concentration (IC50) for inhibition of this target). CTG; mean; biological replicates N = 30 independent replicate cell cultures for drug-free controls in both lines, n = 3 or 4 independent replicate cell cultures, respectively, in L363 and KMS27 MM cells for each drug dose; nonlinear curve fitting with variable slope (four parameters). f, Immunobloting for BiP, a marker of ER stress, in KMS18 cells with Doxy-inducible CRISPR KO of UBE2J1 or control OR gene, cultured with versus without Doxy. g,h, In vitro bortezomib treatment (24 h) of KMS18 (g) or OPM-2 (h) cells with Doxy-inducible CRISPR KO of HERPUD1 or control OR genes. (CTG; mean ± SEM; n = 8 independent replicate cell cultures for drug-free controls and n = 4 independent replicate cell cultures per drug dose for each KO; two-way ANOVA (P < 0.001); detailed results of Tukey posthoc tests in source data). i, Schematic figure of ER-associated dependencies. MM-preferential ER dependencies (blue symbols) involve ER membrane protein complexes mediating dislocation of misfolded ER proteins to cytosol (e.g. HERPUD1, SEL1L) and associated ER-specific E2/E3 enzymes (SYVN1, UBE2J1, UBE2G2); enzymes (e.g. DPM1, ALG3, ALG9) required for N-glycan-dependent surveillance of quality control for luminal ER glycoproteins; chaperones (e.g. DNAJB11, DNAJBC3) for BiP complexes with misfolded proteins; and the known ER stress-sensor IRE1a (ERN1) and its downstream transcription factor XBP1.
The patterns of essentiality of all ER-associated genes in MM vs other cancers (Extended Data Fig. 10g) reveal that a minority are “core essential” genes (Extended Data Fig. 10g, top of the graph); and a large proportion are essential for few, if any, cancer cell lines (Extended Data Fig. 10g, lower part of graph). Additionally, we identified ER-associated genes which do not meet all criteria for MM-preferential dependencies, are not broadly essential across all cancers, but are essential for many MM cell lines (Extended Data Fig. 10g,h). These latter genes encode for ER proteins involved in dislocation of misfolded ER proteins to the cytosol (AUP1, AMFR and RNF139); or in N-glycan-dependent quality control for luminal ER glycoproteins (ALG12, ALG6, ALG8): these additional ER-associated genes may also represent candidate therapeutic targets for MM.
In vivo studies confirm role of MM-preferential dependencies
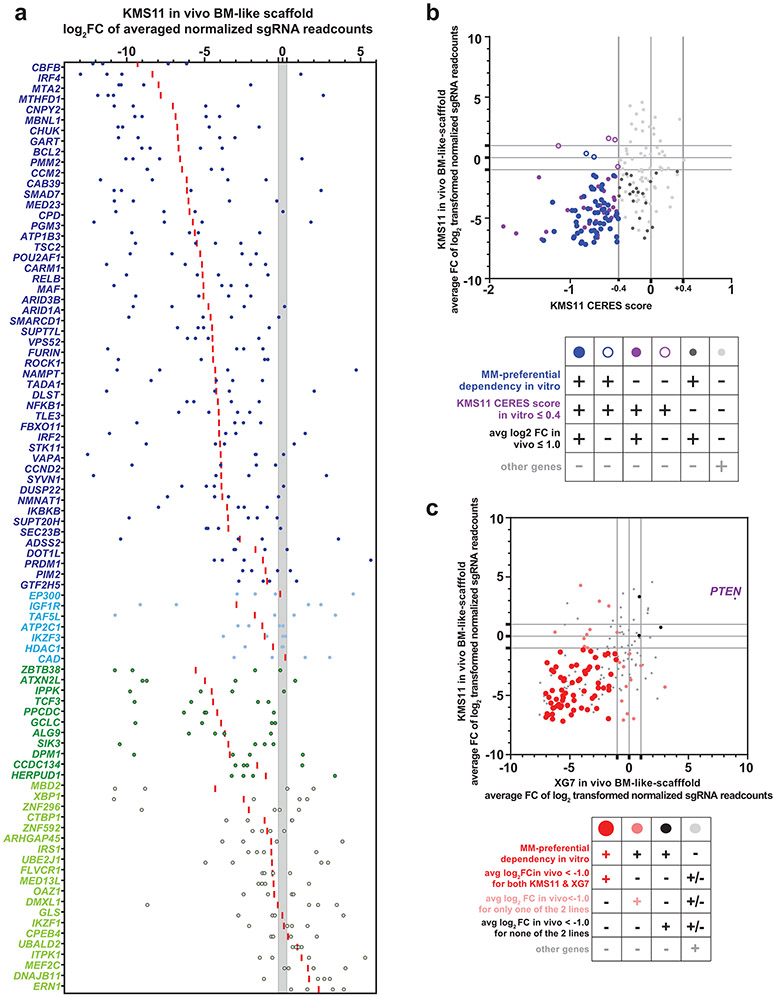
We examined if MM-preferential dependencies identified in vitro were also essential for MM cells grown in vivo within a bone marrow (BM)-like scaffold system engineered to simulate the human marrow microenvironment46 and enhance MM growth. Bicalcium phosphate scaffolds were populated ex vivo with primary human mesenchymal BM stromal cells under conditions favoring osteogenic differentiation (46 and Supplementary Fig. 8h). Scaffolds were subcutaneously implanted into NOD-scid gamma (NSG) mice and injected with KMS11 or XG7 SpCas9+ MM cell lines transduced with a focused sgRNA library targeting 89 MM-preferentially essential genes, genes with broad roles across many tumor types and controls. Analysis of sgRNA distribution of tumors recovered from the mice revealed that a large majority of MM-preferential dependencies identified in vitro were also essential for MM cells in vivo. For example, among the 57 MM-preferential dependencies with CERES scores ≤ −0.4 in KMS11 cells in vitro, their large majority exhibited depletion of their cognate sgRNAs in vivo (average log2FC≤−1.0 and depletion of 3-4 of 4 sgRNAs; Fig. 8a,b). These included genes encoding TFs/cofactors (e.g., IRF4, PRDM1, POU2AF1, RELB, MAF); epigenetic regulators (e.g., CARM1); kinases upstream of NF-κB (CHUK, IKBKB); and ER regulators. Core-essential genes and broad-spectrum oncogenes essential in vitro (e.g., MYC, CFLAR, CDK7 on both lines; KRAS in XG7) remained essential in vivo; while PTEN KO cells were enriched consistent with the tumor suppressive role of this gene (Fig. 8c). Overall, the large majority of MM-preferential dependencies examined were essential for MM cell growth in vivo of either KMS11 or XG7 cells; and most were essential for both lines (Fig. 8c). Knockout of several genes had a greater effect in vivo than in vitro. For instance, BCL2, the ER-associated genes HEPRUD1, ALG9, and DPM1; and the TFTCF3 (a gene examined with individual sgRNAs in another MM line in vitro; Supplementary Fig. 8) had in vitro CERES scores in the range of or greater than −0.40 in KMS11 cells, but sgRNAs for these genes were depleted in the in vivo setting (Fig. 8a). These observations indicate that most MM-preferential dependencies identified in vitro are also required when MM cells interact in vivo with a highly supportive microenvironment.
Figure 8 ∣. In vivo studies to validate the role of key examples of MM-preferential dependencies identified in vitro.
a, Results from study of KMS11 cells in the “humanized” BM-like scaffold-based in vivo model using a single-gene CRISPR KO system. The graph depicts, for each gene N=88 MM-preferential dependencies with in vitro CERES scores of <0.4 in KMS11 cells), the log2FC of averaged read counts for each of their sgRNAs (blue dots for individual values; red bar for average). The region highlighted in gray delineates the upper and lower limit of the 95% CIs for log2FC of averaged read counts for sgRNAs of OR genes as controls. Genes for which their sgRNA log2fold change are outside the 95% CIs for the OR gene sgRNAs were considered to have depletion or enrichment. Gene symbols for MM-preferential dependencies with CERES scores <−0.4 in KMS11 in vitro are indicated in dark blue vs. light blue if these genes did vs. did not exhibit, respectively, depletion of 3-4 out of 4 sgRNAs per gene in vivo. MM-preferential dependencies with CERES scores >−0.4 in KMS11 in vitro are indicated in dark green vs. light green, if these genes did vs. did not exhibit, respectively, depletion of 3-4 sgRNAs per gene in vivo. b, Average log2FC of read counts for sgRNAs of N=184 genes (4 sgRNAs/gene) in KMS11 cells in the in vivo humanized BM-like scaffold-based model (N=5 mice) (y-axis) and their respective CERES score in KMS11 cell line in vitro (x-axis). c, Scatterplot of average log2FC of read counts for sgRNAs of genes examined through sub-genome scale focused CRISPR KO study of the KMS11 cells (N=5 mice) (y-axis) vs. XG-7 cells (N=8 mice) (x-axis) in the in vivo humanized BM-like scaffold-based model.
Discussion
Recent advances in MM treatment have relied on therapeutics that are primarily effective against PC neoplasias. This preferential anti-MM activity could not have been readily predicted by the genomic characterization of MM cells, as these agents do not target mutated oncogenes or the malignant state of MM PCs, but rather pathways critical for PC biology. This was originally recognized for thalidomide derivatives and proteasome inhibitors1 and also applies for subsequently developed therapies targeting the preferentially high expression of CD38, BCMA or GPRC5D on PCs, malignant and normal. Notably, some of the most successful anticancer therapies also target both malignant and normal cells of lineages dispensable for survival of adult patients, sparing other tissues and avoiding major life-threatening complications. Such lineage-specific therapies include rituximab for lymphomas, hormonal therapies for prostate or breast cancer or radioactive iodine for thyroid carcinoma. The profound impact of lineage-specific treatments in MM and beyond prompted us to functionally ascertain, through genome-scale CRISPR screens, genes that are preferentially essential for MM compared with the overwhelming majority of neoplasias from other lineages.
Reassuringly, several MM-preferential dependencies identified in this study are known regulators of MM biology (e.g., IRF4) or targets/mediators for therapies with preferential clinical activity against MM/PC neoplasias. Among diverse proposed mediators of anti-MM activity of thalidomide derivatives, IKZF3, IKZF1 and ARID2 emerged as MM-preferential dependencies. Prior work primarily centered on IKZF1 as the critical target of thalidomide derivatives, but our present study identifies more pronounced and recurrent MM cell dependence on IKZF3. ARID2 is a CRBN neosubstrate with pomalidomide, but not lenalidomide, treatment22. Our observations suggest that additional emphasis is warranted on IKZF3, ARID2, and their downstream effects.
In terms of the pronounced activity of proteasome inhibitors (PIs) against PC neoplasias (vs. limited activity against most other tumor types), the precise mechanistic contribution of NF-κB inhibition vs. ER stress had remained an unanswered question. Our study points to a contribution of both pathways, because MM cells are, compared to other neoplasias, preferentially dependent on both NF-κB pathway genes and ER regulators. The latter include molecules with previously underappreciated roles in MM, including the ER-resident E2 ligase UBE2J1 and E3 ligase SYVN1; or their ER-to-cytosol retrotranslocation partners SEL1lL and HERPUD1 which contribute to the quality control system for misfolded proteins in the ER. These proteins and their respective complexes may represent therapeutic targets in MM.
The identification of BCL2, HDAC1, and PIM2 as MM-preferential dependencies is also notable, given that BCL2 inhibitors have promising clinical activity in a subset of MM patients47; and broad-spectrum inhibitors of class I HDACs48 or PIM kinases49 have exhibited activity in clinical studies in MM, but only limited clinical efficacy in other settings.
By this logic, other MM-preferential dependencies could represent putative therapeutic targets. Many transcriptional/epigenetic regulators identified in this study have received limited attention as therapeutic targets in MM. Others (e.g., DOT1L50 or CARM151) have been targeted therapeutically in preclinical MM studies which, however, did not comprehensively compare the role of these targets in MM vs. other cancers. A translational implication of our study is that selective direct inhibitors of the expression of MM-preferential dependencies or function of their product(s) merit preclinical and clinical evaluation in MM, without excluding possible applications in other neoplasias. Our data do not imply MM-“exclusive” essentiality for these genes, as several are also recurrent/preferential dependencies for other malignancies. However, a large fraction of MM-preferential dependencies do not exhibit a similar role in other hematologic neoplasias and some were even reported as tumor suppressors in other lymphoid malignancies (e.g., FBXO1152 and PRDM116) or solid tumors (e.g., STK11 and TSC2). Examining other neoplasias, beyond MM, for their respective preferential dependencies, identified some known examples of dependencies related to the respective cell of origin, but overall fewer genes per disease compared to those identified for MM. This may reflect the highly distinct molecular network that is essential for the MM cells and their identity as PCs, specifically their status as highly secretory cells, which require high levels of ER function, as well as distinct transcriptional, epigenetic and signaling vulnerabilities, compared to most other tumor types. Collectively, MM-preferential dependencies cannot be attributed exclusively to biological differences between blood cancers vs. solid tumors but may reflect the major underlying differences in the molecular network of MM cells compared to all other cancers.
The identified MM-preferential dependencies vary in terms of the fraction of MM lines dependent on each gene or the magnitude of essentiality scores. Future studies in larger panels of MM lines may reveal molecular determinants of these differences, e.g., if any of these genes are preferential to individual MM subtypes, defined by either genomic or CRISPR-based functional criteria. Some MM-preferential dependencies defined by CRISPR are also apparent in shRNA studies, but others are not, perhaps reflecting a more pronounced and less variable suppression of gene function by CRISPR-based gene-editing. Time-course studies may provide important additional insights on the kinetics and the cytostatic vs. cytocidal impact of CRISPR KO of MM-preferential dependencies and whether during the course of a CRISPR screen tumor cells “re-wire” to accommodate the loss of such genes.
Our in vivo studies validated the large majority of MM-preferential dependencies identified in vitro. Additional genes may conceivably be preferentially essential for MM cells in vivo but not in vitro. Interaction with the BM milieu may alter the patterns of dependencies in MM cells, as evidenced by our observation that some genes were more essential for growth in vivo than growth in vitro. Future studies will likely define microenvironment-related in vivo dependencies (e.g., growth factor receptors or cell adhesion molecules critical for cell-cell interactions) in models that faithfully simulate the support of the local BM milieu on MM cells and ideally involve local production by human stromal cells of cytokine/growth factors since many murine cytokines do not react with the human receptors. Our xenograft studies in immunocompromised mice could not examine the impact of MM-preferential dependencies on immune recognition. Notably, activation of POU2AF1, one of the top MM-preferential dependencies, was associated with decreased expression of MHC class II molecules, while other MM-preferential dependencies (e.g., MPDU1, ARID1A) influence tumor cell responses to natural killer cells53,54. Therefore, at least some MM-preferential dependencies could have pleiotropic roles beyond the cell autonomous regulation of MM cell survival and proliferation.
MM cell behavior is shaped by their intrinsic “PC biology” and their superimposed “cancer biology”1: comprehensive understanding and therapeutic targeting of both aspects is warranted1. By comparing dependencies in MM vs. all other malignancies, our study addresses this former aspect of “PC biology” of MM and yields many previously underappreciated targets which do not require genomic perturbations in order to serve as essential genes and candidate therapeutic vulnerabilities for MM. Indeed, the large majority of MM-preferential dependencies are not among the top genes in terms of the frequency of mutations or DNA copy number gains in MM, are not necessarily located in highly accessible regions of chromatin and are not among the top differentially expressed genes in MM vs. other neoplasias. Conversely, most genes overexpressed in MM cells (compared with other tumor types) are not essential for MM cells. Collectively, our study identifies MM-preferential dependencies, most of which would not be readily identified as MM driver genes with highly recurrent genomic perturbations, and thus is complementing the long-standing efforts to define therapeutic targets for the “cancer biology” aspect of MM.
For nearly two decades, research in MM and other malignancies focused on profiling of tumor cell lines and patient samples for alterations in their genome, transcriptome, epigenome, and proteome, with the hope that molecules with the most recurrent or pronounced dysregulation could represent attractive therapeutic targets. Our study highlights that CRISPR-based functional genomics approaches4,55,56, by directing assessing the impact of gene perturbation on tumor cell fitness, can identify genes critical for tumor cells from a particular cell lineage and define promising therapeutic targets not readily identifiable based on alterations in the tumor genome, transcriptome, or epigenome.
Methods
This research complies with all relevant ethical regulations. In vivo studies were performed according to a protocol approved by the Dana-Farber Cancer Institute Animal Care and Use Committee.
Cell lines.
Details about the cell lines examined in the genome-scale CRISPR-Cas9 gene editing studies are available at https://depmap.org/portal/. Information about lines used in additional experiments is included in Supplementary Table 2. Cell line identity was validated by short tandem repeat analysis and cultures were regularly tested for Mycoplasma.
CRISPR-based genome-scale screens
Genome-scale CRISPR-Cas9 screens were performed in human MM and other cell lines stably transduced with lentiviral vector pXPR-311Cas9, selected with blasticidin and then infected with a lentiviral library of 76,106 sgRNAs (AVANA) targeting 17,670 genes protein coding (~4 sgRNAs/gene) and including 995 nontargeting control sgRNAs. Cells were selected in puromycin and blasticidin for 7 days and then passaged without selection (with target representation of 500 cells per sgRNA) for 21 days. Genomic DNA was purified from endpoint cell pellets, sgRNA barcodes were PCR amplified with sufficient gDNA to maintain representation, and PCR products were sequenced using Illumina protocols as described55,57. Data processing and quality control was performed as in previous studies4,55,56,58. CERES scores, a metric of relative essentiality of an individual gene in a given cell line, were calculated as in55 to correct for gene-independent DNA copy number effects of CRISPR gene editing. The CERES scores for all cell lines in this study are available at https://depmap.org. Unless noted, figures represent data reported in the 20Q4v2 release and exhibit very high degree of concordance with results from other releases (e.g., in Extended Data Fig. 1). Essentiality was also evaluated by converting the CERES scores into ranks of CERES scores (also referred to as “CERES ranks”) for each gene within each cell line; or the MAGeCK algorithm59 to assess sgRNA depletion or enrichment without correction for DNA copy number. Dependency data based on RNA interference were derived from Achilles Heel shRNA screens and Novartis’ Project DRIVE60, and were reprocessed using the DEMETER2 algorithm to calculate gene dependencies56.
Computational methods to identify preferential dependencies:
To identify candidate tumor type-preferential dependencies, we examined genes with significant difference and lower (more essential) average CERES scores in MM vs. non-MM cell lines; in similar comparisons of a different tumor type vs. all others; or comparing MM lines vs. e.g., solid tumors. Statistical significance was assessed using empirical-Bayes moderated t-statistics using Limma software with an adjusted p-value of <0.05 and a difference in CERES score of <−0.2 between cell types was considered significant. To identify a refined list of candidate MM-preferential dependencies, we focused on genes which satisfied the following criteria: 1) adjusted p-value (FDR) <0.05 in Limma tests comparing CERES scores in MM vs. non-MM cell lines; 2) average CERES score difference of ≤−0.2 between MM vs. non-MM cell lines; 3) average CERES scores of ≤−0.2 in MM cell lines; 4) at least 15% MM cell lines with CERES score ≤−0.4; 5) the fraction of non-MM cell lines with ≤−0.4 CERES score is ≤0.8 (to filter-out broadly essential / “core essential” genes); 6) adjusted p-value (FDR) <0.05 in Fisher’s test comparing ranks of CERES scores in MM vs. non-MM cell lines; 7) log2(TPM+1) of ≥1.0 in at least 30% of MM cell lines tested (TPM: transcripts per million). For genes in the X chromosome, CERES-based correction for their copy number status was not applied in early versions of DepMap data. Such genes are indicated in gray for the respective DepMap releases (Extended Data Fig. 1a). We also compared MM vs. non-MM cell lines, using the same statistical tests as for CERES ranks, in terms of the distribution of DNA copy number-uncorrected ranks based on the MAGeCK algorithm59 of sgRNA depletion.
Molecular profiling and other datasets:
Transcriptional profiles, DNA copy number status and mutational landscapes of human MM and non-MM cell lines examined were accessed from the Cancer Cell Line Encyclopedia (CCLE) portal (https://portals.broadinstitute.org/ccle/data, data versions from 2017-2018) or the Dependency Map portal (https://depmap.org/portal). Transcriptional profiles, mutational and CNV data on MM tumor cells from patients and clinical data on progression-free survival (PFS) and overall survival (OS) of the CoMMpass study were accessed from the MMRF Researcher Gateway (https://research.themmrf.org/, data releases IA8-IA19): PFS and OS data were evaluated (e.g., Extended Data 4c-e) for patients receiving bortezomib plus IMID (immunomodulatory thalidomide derivative (IMID) (cBI group), bortezomib without IMID (B group), IMID plus carfilzomib (cIC group) and all patients (full set) of the datataset. Gene expression profiles on patient tumors with non-MM malignancies (e.g., in Extended Data Fig. 4a or Supplementary Fig.2-6) were derived from The Cancer Genome Atlas (TCGA) and accessed from https://gdac.broadinstitute.org/ (version 2016012800), https://portal.gdc.cancer.gov/. TCGA and MMRF CoMMpass data can also be retrieved from the UCSC Xena platform61. For evaluation of gene expression, after a library size normalization and voom transformation62, the Limma moderated t-test was applied between samples of MM and TCGA (excluding acute myeloid leukemia [LAML]) to identify genes with FDR ≤0.05 and log2FC <−1.0 or above ≥1.0. The patterns of transcript expression for MM-preferential dependencies were also examined in publicly available datasets of samples representing different stages of MM or settings with distinct differences in the clinical or biological behavior of MM (GSE2113, GSE5900, GSE6477, GSE13591, GSE39754, GSE39925, GSE66293); or MM patients receiving bortezomib-based or other treatments (GSE19748, GSE9782) or MM cells interacting with BMSCs (GSE20540). IRF4 target genes were identified previously5 (in datasets GSE8958, GSE9067, and GSE9367), and genes downregulated by IKZF1 or IKZF3 loss-of-function were derived in prior studies (GSE113031)19. ATAC-Seq data of MM lines (from GSE121912) were analyzed to determine accessible regions of chromatin with MACS2 (v2.1.0.20151222)63,64 using default parameters and a q-value of 0.01. Regions that overlapped ENCODE blacklisted regions were removed65. ATAC-seq data was normalized for reads per peak million (RPPM) for visualization using the following formula: RPPM = reads x (106 / total reads in autosomal peaks. Super accessible regions were determined using the GenomicRanges (v1.36.1) and GenomicAlignments (v1.20.1) packages in R (v3.6.3) where regions within 12.5 kb were linked together excluding those within 2.5 kb of a transcription start site. Regions were ranked by accessibility (RPPM) and regions that were past the inflection point were considered super accessible regions. Genome-wide chromatin immunoprecipitation analyses (ChIP-Seq) for POU2AF1 (OCA-B) were accessed from GSE79480. Functional genomic data of retroviral gene-trap mutagenesis screen and a gene-editing screen for genes involved in ERAD regulation in KBM7 haploid cells were derived from45. The GDSC1 and GDSC2 datasets of pharmacological screens were derived from the Genomics of Drug Sensitivity Project (66 and https://www.cancerrxgene.org/). The direct (physical) and indirect (functional) associations of the MM-preferential dependencies (Extended Data Fig. 1d), based on computational prediction, knowledge transfer between organisms, interactions aggregated from other (primary) databases or other resources integrated were visualized using the STRING database (String-DB, https://string-db.org/ v11.0)67.
Cloning of individual sgRNAs:
sgRNAs for CRISPR-KO, CRISPRi and CRISPRa were packaged in pLVX-hyg-sgRNA1, pXPR-502 (RRID:Addgene_96923) and pXPR-050 (RRID:Addgene_96925) as described. Briefly, target sgRNA oligos (Supplementary Table 2) were mixed with Guide-it Oligo annealing buffer (Takara Bio 632630), denatured at 95°C or 2 min and cooled to 25°C over 15 min. Annealed oligos were ligated into gel-purified vectors using DNA Ligation Mighty Mix (Takara Bio USA, 6023) at 16 °C for ~30 minutes, transformed into Stellar™ Competent Cells (Takara Bio USA), with resulting colonies picked, expanded with DNA isolated using the QIAprep Spin Miniprep Kit (Qiagen, 27106), screened for inserts and the resulting plasmids sequenced.
Addback studies:
In-frame fusion of sequences encoding HA-FKBP12F36V in pLEX_305-N-dTAG (Addgene, #91797) to the complementary DNA (cDNA) encoding IRF4 to yield HA-FKBP12F36V-IRF4 cDNA was performed by Gateway recombination (Invitrogen). Individual sgRNAs against intron–exon junctions (IEJs) of IRF4 were designed using the Broad Institute sgRNA design portal (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). All sgRNA sequences were synthesized by CustomArray Inc (Bothell, WA) and cloned into a pHKO9 vector (as described in https://media.addgene.org/cms/filer_public/4f/ab/4fabc269-56e2-4ba5-92bd-09dc89c1e862/zhang_lenticrisprv2_and_lentiguide_oligo_cloning_protocol_1.pdf). Production of lentiviral particles for IRF4 fusion constructs and individual sgRNAs, and lentiviral transduction were performed based on published protocols68,69. The viability of cell populations transduced with HA-FKBP12F36V-IRF4, sgRNAs against intron-exon junctions of IRF4 or both was assessed 3 days after hygromycin selection for the last of the transductions (for the sgRNAs against IEJs of IRF4) using CellTiter-Glo (CTG, Promega).
Tumor cell viability assays:
In vitro anti-MM activity for small molecular-weight inhibitors.
CellTiter-Glo (CTG) assays were performed for studies with pan-PIM inhibitors LGB-321(AdooQ BioScience # A14420-5) or SGI-1776; the CBFB inhibitor (CBFBi) Ro5-3335 (Fisher Scientific # 469410), the CARM1 inhibitor (CARM1i) Merck 217531 (EMD Millipore #217531), the SYVN1 inhibitor LS-102 (Fisher Scientific #NC1398267), and SIK inhibitor HG-9-91-01 (MedChemexpress, HY-15776-5MG). Cell lines were seeded with inhibitor for 3-5 days as indicated. CTG assays at indicated timepoints were measured by a BioTek Synergy 2 plate reader (BioTek, Winooski, VA).
Assessment of cell viability after CRISPR gene editing, activation or interference.
Lenti-X-293T cells were transduced using lipofectamine with packaging plasmids psPAX2 (RRID:Addgene 12260) (5 μg) and MD2.G (RRID:Addgene_12259) (2.5 μg) and plasmids encoding individual sgRNAs packaged in the pXPR_502 or pLVX-hyg-sgRNA1 (5μg). Virus was collected after 24 hours and 1mL applied to 1 x 106 target cells.
For CRISPR gene editing studies, viability of KMS18 or OCI-My5 cells harboring Tet-inducible SpCas9 construct and transduced with sgRNAs for genes of interest (details in Supplementary Table 3) were seeded (100 cells/well) into 384-well plates, in 10% Tet-negative FBS media (50 μL) with or without Doxycycline (2 μg/mL) and add another 50 μL media with or without Doxycycline (2 μg/mL) at day 7. CellTiter-Glo reagent was added to each well at day 14, and plates were read using a microplate reader. For CRISPR interference studies, KMS11 cells with Tet-inducible dCAS9_KRAB construct and transduced with sgRNAs (details in Supplementary Table 3) were seeded (0.3x106 cells/well) into 24-well plates, in 10% Tet-negative FBS media 1 mL with or without Doxycycline (2 μg/mL), and seeded in 384 well plates. Media were changed every 3-4 days with cell viability checked by CellTiter-Glo at day 11.
For CRISPR activation, LP1 dCAS9-VP64 cells were plated in 1 mL of complete RPMI1640 medium per well in a 24-well plate. Cells were incubated in cell medium containing polybrene (4 μg/mL; Santa Cruz Biotechnology) and 1%HEPES (1M), and same amount of viral prep, were centrifuged at 1500 g for 2 h and incubated overnight at 37°C 5%CO2. Media were changed next day and, after another 48 h, selection with puromycin 2 μg/mL) for seven days. After 12 and 19 days from transduction, cells were detached from flask by trypsin, allowed to recover at 1mL of complete medium. Then, 50 μL aliquots were seeded in 384-well plate and were assessed using CellTiter-Glo.
Competition assay evaluated by INDEL analysis:
Competition assays with gene edited cells were performed as previous studies70. KMS18 cells stably transduced with Doxy-inducible SpCas9 were transduced with pLVX-hyg-sgRNA1 plasmid harboring specific gRNAs (Supplementary Table 3) and selected in Hygromycin B (350 μg/mL). OR12D2 KO cells and UBE2J1 KO cells were mixed at a 1:9 ratio and maintained with or without Doxycycline at 2 μg/mL (replenished every three or four days). Cells were collected at day 14 or 28. Genomic DNA was extracted from cell pellets and targeted lesion of sgRNA sequence were amplified by PCR and analyzed by next generation sequencing (MGH DNA core; https://dnacore.mgh.harvard.edu/new-cgi-bin/site/pages/crispr_sequencing_main.jsp). Indel analysis and estimation of % of cells with frameshift mutations was performed with CRISPRESSO (http://crispresso.pinellolab.org).
Immunoblotting.
Similar to prior studies70, cells (3 x 106 per condition) were collected and lysed using RIPA buffer (ThermoFisher) with protease/phosphatase inhibitor cocktail (Cell Signaling Technology) by incubating on ice for 10 min. Lysates were collected by centrifugation (15,000 g for 10 min at 4 °C) and lysate concentration was determined using BCA (ThermoFisher). Protein samples were resuspended in Bolt LDS sample buffer (NuPage, Invitrogen) with sample-reducing agent (NuPage), heated to 70°C for 10 min and 10-20μg/sample loaded on 4-12% Bis-Tris gels (NuPage) and run at 125V for 70 min using MOPS running buffers. Gels were transferred onto PVDF membranes using SDS-based transfer buffer (NuPage), blocked in 5% skim milk in TBS-T for 1h and probed with primary antibodies overnight at 4 °C. Secondary antibodies in 1% skim milk in TBS-T were applied to the membranes for 1.5 h at room temperature prior to incubation in ECL (ThermoFisher #34075) substrate. Information on antibodies used in these studies is included in Supplementary Table 2. Immunoblots were visualized using a C-DiGit®Blot Scanner (LI-COR Biotechnology, Lincoln, NE).
RNA-sequencing.
Triplicate cultures of LP1 cells transduced with sgRNAs for CRISPR activation of POU2AF1 or control genes were pelleted and frozen at −80°C. RNA was extracted by RNeasy Plus Mini Kit (Qiagen 74134) and ERCC RNA Spike-In Mix (Thermo Fisher 4456740) was added at the first step of extraction. RNA sequencing was performed by the Molecular Biology Core Facilities (MBCF, DFCI). results are available (GSE186997). RNA-seq raw data processing and generation of gene read counts was performed with STAR71. Analysis performed with edgeR Bioconductor package involved ERCC-based normalization, a generalized linear model and, for the likelihood ratio test, pooling of the coefficients of each sgRNA within the control or POU2AF1 activation groups. Gene set enrichment analysis (GSEA) was performed using the pre-ranked option (e.g., ranking according to -log10FDR x log2FC) with custom sets representing genes suppressed by loss of function of IRF4, IKZF1, IKZF35,19 or genes upregulated with MYC amplification (e.g., Kim_MYC_Amplification_Targets_UP), using default settings (through Gene Pattern, https://www.genepattern.org/).
Subgenome-scale CRISPR editing studies in vitro and in vivo
A library of 1372 oligonucleotides for sgRNAs was designed to include typically 4 guides per gene for each of 184 genes, including 89 MM-preferential dependencies; broad-spectrum oncogenes; select tumor suppressor genes (e.g. PTEN); and genes with limited in vitro essentiality in MM cells, including some with significantly higher expression in MM vs. non-MM lines, and 155 olfactory receptor (OR) genes “DNA cutting” control sgRNAs. These oligonucleotides were synthesized in pooled format (CustomArray), PCR-amplified and gel-purified using a Qiagen gel extraction kit and used as template for a second PCR reaction with the flanking sequence to attach to the lentiGuide-Puro vector. After gel purification, 0.1 pmol of PCR product, 90 ng of lentiguide-puro and Gibson assembly kit with water were incubated for 30 min at 16°C. Next, 1200 ng of the resulting plasmid DNA was transformed into 300 μL of ElectroMAX Stbl4 electrocompetent cells by electroporation and put into 2.5 mL of SOC medium before being shaken 1 hr at 37°C. After incubation, cells were plated in 3 mL of medium on a total of 3 bioassay plates and incubated for 16 hr at 37°C. After 16 hrs of incubation, cells are collected with 30 mL of cold LB each by biospreader and pelleted at 4°C at 6000 g for 15min. Plasmid DNA was extracted using the QIAGEN Plasmid Plus Maxi Kit. Lenti-X-293T cells were plated in T175 culture flasks in DMEM with 10%FBS and incubated overnight. The next day, cells were transduced using lipofectamine with the library plasmids (30 μg) and MD2.G encoding VSV-G (12.5 μg). Viral supernatants were collected after 24h and stored at −80°C prior to use.
SpCas9-expressing cell lines (KMS11, XG-7, RPCI-WM and BCWM1) were incubated for 16 hrs in cell medium containing 8 μg/mL polybrene, 10mM HEPES (pH 7.4) and viral prep (6 mL) diluted to achieve transfection rate of 0.3. After the end of the incubation with the viral preps, cells were washed and incubated for an additional two days. Transduced cells were treated with puromycin (2 μg/mL) for up to 7 days after 3 days from transduction. The RPCI-WM and BCWM1 cell lines transduced with this focused sgRNA library were cultured (3 replicates per cell line) in vitro for 3 weeks. At the end of this incubation, tumor cells were collected, and PCR amplification and next-generation sequencing of the samples were performed53,70, to quantify the abundance of sgRNAs. The KMS11 and XG7 cell lines transduced with this focused sgRNA library were introduced in vivo into bicalcium phosphate (BCP) particles: the latter had been loaded with human primary mesenchymal bone marrow stromal cells, cultured ex vivo under conditions favoring osteogenic differentiation of these stromal cells46 and implanted subcutaneously (two scaffolds per mouse) into 8-week-old NSG female mice. Seven weeks after scaffold implantation, 1.5 million KMS11-SpCas9 or XG-7-SpCas9 cells transduced with the focused sgRNA library were injected directly into the scaffolds (5 mice for KMS11 and 8 for XG7 study). Without exceeding the maximal tumor burden (20mm of diameter in any direction) permitted by DFCI IACUC, tumors were removed, and processed for DNA isolation (Blood & Cell Culture DNA Maxi Kit #13362), pooling of material from the same mouse, PCR amplification and next-generation sequencing53,70, to quantify the abundance sgRNAs for genes of interest (vs sgRNAs for control OR genes). Read counts normalized according to the OR control sgRNAs were analyzed, with averaging of read counts examined both before (e.g., Fig. 8a) and after (e.g., Fig. 8b,c) log2 transformation, yielding concordant conclusions regarding the patterns of depletion for sgRNAs targeting MM-preferential dependencies.
Statistics and Reproducibility:
To identify and further characterize genes preferentially essential for MM, this study involved multiple essentiality metrics and criteria for the identification of these genes; corroboration of results across multiple iterations of genome-scale screens; functional characterization of many of these genes; integration of their molecular features across multiple datasets; and alternative methods of analyses of data (information on additional approaches for data analyses not included in this study are available through the corresponding author). Details on sample size(s) and statistical test(s) are provided in the respective sections. Statistical tests were two-sided (except rank aggregation analyses), and distribution of individual data points was assumed to be normal, but this was not formally tested. No statistical methods were used to predetermine sample sizes, but in this study these sample sizes (e.g., numbers of replicates in CRISPR experiments) were similar to those reported in prior publications4. Animal studies were performed according to a protocol approved by the Dana-Farber Cancer Institute Animal Care and Use Committee and did not involve treatment administration; thus randomization was not pertinent. For other experiments, data collection and analysis were not performed in a manner blinded to the conditions of the experiment. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Figure 1 ∣. MM-preferential dependencies in genome-scale CRISPR-based gene editing screens.
a, Summary matrix of results for identification of MM-preferential dependencies in genome-scale CRISPR-based gene-editing screens from different releases of the Dependency Map program. The criteria used to identify MM preferential dependencies in the 20Q4v2 Dependency Map data were also applied in earlier releases (18Q3 to 20Q3). The matrix summarizes results for all genes that met these criteria in at least one of the releases. Black or white indicate, respectively, that a gene did vs. did not meet criteria for MM preferential dependency in the respective data release (gray signifies that CERES scores were not calculated for a given gene in the data release). b, MM-preferential dependencies clustered according to molecular pathways represented in this group of genes. Color-coded heatmaps for CERES scores following the format of Fig. 1a. Genes are clustered based on their related functional groups, pathways, or biological functions, based on aggregate information from the literature. c-d, Molecular pathways enriched for MM-preferential dependencies. c, Schematic representation of functional groups represented in the MM-preferential dependencies, such as transcription factors/co-factors, other regulators of transcriptional responses and chromatic signaling; kinases serving as upstream regulators of these pathways (e.g., kinases activating NF-κB); or endoplasmic reticulum/Golgi regulators. d, Visualization of the direct (physical) and indirect (functional) associations of the MM-preferential dependencies, based on computational prediction, knowledge transfer between organisms, interactions aggregated from other (primary) databases or other resources integrated and visualized by the online STRING database (https://string-db.org/, v11.0)67.
Extended Data Figure 2 ∣. Additional metrics of essentiality for MM preferential dependencies.
a-b, Ranks of CERES scores or DNA copy number-uncorrected ranks of sgRNA depletion for MM-preferential dependencies. Essentiality metrics are depicted in color-coded heatmaps similar in format to Fig. 1a, with results presented for MM lines as a matrix (each line in a separate column) while results for non-MM lines are stacked separately for each gene from lowest to highest essentiality (from left to right in each row). For each cell line, the top 3000 genes with the lowest CERES scores (in a) or with most pronounced sgRNA depletion based on MAGeCK rank aggregation (in b) are depicted in green or purple, respectively. For each cell line, the top 100 genes with highest CERES scores (in a) or highest MAGeCK ranks for sgRNA enrichment (in b) are depicted in purple and yellow/orange, respectively, according to the respective color-coded scales.
c-d, Patterns of depletion for shRNAs targeting genes defined by CRISPR as MM-preferential dependencies. c, DEMETER2 scores are depicted as a matrix for MM (n=13 cell lines; right) and as separate stacked plots for non-MM (n=461 cell lines; left), according to the color-coded scale (black/ blue for shRNA depletion; yellow/orange/brown for shRNA enrichment; white for DEMETER2 scores between −0.4 and +0.4; and gray for genes not examined in the shRNA screen of the respective cell line). d, DEMETER2 scores for key examples of MM-preferential dependencies are depicted (in rows) for both non-MM (left) and MM lines (right) as stacked bar graphs, according to the color-coded scale.
Extended Data Figure 3 ∣. Patterns of expression of MM-preferential dependencies in MM vs non-MM cell lines.
RNA-Seq data (CCLE dataset) for MM-preferential dependencies in MM vs. non-MM cell lines. Transcript levels (log2(TPM+1)) for each gene (row) across MM and non-MM cell lines are scaled by maximum value resulting in a value range between 0 and 1 and presented as stacked bar plots. Color bars on the side of the graph denote different clusters of genes, defined based on analyses of Fig. 3 (based on 2-sided limma t-test FDR and log2FC of differential expression of each gene in MM vs non-MM lines).
Extended Data Figure 4 ∣. Patterns of transcript levels for MM-preferential dependencies in different biological or clinical contexts.
a, RNA-Seq data for MM-preferential dependencies in patient-derived tumor samples for MM vs. non-MM. Transcript levels (presented as stacked plots) for each gene (row) across MM (n=591 samples; MMRF CoMMpass study, IA8 release) and non-MM (n=11060 samples, TCGA; accessed from GDAC). Raw counts were voom normalized, negative voom values were set to zero, scaled by maximum value for each gene, resulting in a value range between 0 and 1. Concordant observations also obtained with other versions of MMRF and TCGA datasets. b, Comparative analyses of transcript levels for MM-preferential dependencies in different stages of myelomagenesis or settings with distinct differences in clinical or biological aggressiveness of MM. Heatmap summarizes results from comparisons performed between groups of samples within each of the gene expression profiling datasets indicated in the figure. Red and blue denote statistically significant (FDR<0.05, Limma t-test, log2FC > 1.0 or < −1.0) up- or down-regulation, respectively, for a gene in a given group of samples vs. its indicated comparator group. Genes in gray do not have perfect match probes in the respective array. White indicates no statistically significant difference for a given comparison. Number of samples per group is indicated next to each comparison. c-e, Transcript levels of most MM-preferential dependencies do not consistently correlate with adverse clinical outcome. c, Overall survival (OS) or progression free survival (PFS) were examined for MM patients at high vs. intermediate vs. low tertiles of expression of each MM-preferential dependency in each dataset indicated in the graph (see Methods). Red and blue denote statistically significant (at FDR<0.05, two-sided log-rank test) correlation of transcript levels for a given gene with adverse or favorable, respectively, clinical outcome (white indicates FDR>0.05). d-e, Cumulative plots summarizing results of c, in terms of OS (d), or PFS (e), between MM patients with high vs. intermediate vs. low tertile of expression of each gene in each dataset indicated in the graph. For each potential FDR value (x-axis), the y-axis depicts, separately for OS or PFS in each dataset, the cumulative fraction of MM-preferential dependencies exhibiting FDR levels equal or lower to those depicted in each respective position of the x-axis. For all evaluated datasets, <25% of MM-preferential dependencies exhibit FDR<0.05 for the correlation of transcript levels with PFS or OS. Number of patient samples in c-e is indicated for each dataset.
Extended Data Figure 5 ∣. Genomic landscape of MM-preferentially essential genes.
a, Mutational and DNA copy number data for MM-preferential dependencies in MM vs. non-MM cell lines is included in heatmaps of CERES scores (similar to the format of Fig. 1a). Green stars represent non-synonymous mutations; while CNV gains and losses are depicted by “+” or “−”, respectively. In stacked plots for non-MM cell lines, green stars are also stacked and are not linked with the CERES scores in respective lines. b, Rank of genes with most frequent CNV gains in MM patient tumor samples (N = 932 samples; N = 18,057 genes with CERES data (20Q4v2) and CNV data in CoMMpass study, IA15 release): MM-preferential dependencies are highlighted in red and their gene symbols are labeled for those MM-preferential dependencies ranked in the top 200 genes (genes are ranked on the x-axis on a log2 scale). c, Top hotspots for gain of structural variants (SVs) ranked based on their frequency in MM patient tumor samples (CoMMpass study), derived from analyses of Rustad et al13. MM-preferential dependencies residing in 8 of these hotspots are highlighted, and those in bold have not been previously proposed as candidate drivers of the respective hotspots. Gray denotes hotspots which contain no genes evaluated in the genome-scale CRISPR screens. d, Heat maps for CERES scores in MM vs. non-MM lines of genes in each of the 8 SV gain hotspots of panel c that contain MM-preferential dependencies.
Extended Data Figure 6 ∣. Overlap or proximity of chromatin accessible regions with MM-preferential dependencies.
a, Plot of stitched regions of chromatin accessibility with average ATAC-seq signal (RPPM) across 22 MM cell lines shown in gray. Black lines denote the inflection point that denotes super-accessible (SA) regions. Regions within 100 kb of MM-preferential dependencies are denoted by red tick marks on the bottom and the odds ratio (OR) and P-value (two-sided Fisher’s exact test) of enrichment of MM-preferential dependencies found near super-accessible regions are shown. b, Heatmap of chromatin accessible regions within 100 kb of MM-preferential dependencies across 22 MM cell lines. c-f, Genomic plots of ATAC-seq for select examples of MM-dependencies (PRDM1, IRF4, POU2AF1, UBE2J1) that overlap with super-accessible (SA) regions. Each cell line is shown in a transparent gray and the average is shown in black. Note the proximity of IRF4 and DUSP22 and the multiple prominent areas of accessible chromatin within intronic regions of DUSP22. g, Hierarchical clustering of MM cell lines based on their CERES scores for MM preferential dependencies. MM cell lines are annotated for their status for genomic events, such as translocations targeting CCND1, CCND2, CCND3, MAF, MAFB, MMSET/NSD2, mutations for KRAS or NRAS, loss-of-function for TP53; or the functional status of their dependence (based in CRISPR data) on either MAF or MAFB.
Extended Data Figure 7 ∣. Comparative analysis of CERES scores for MM preferential dependencies in MM vs other hematologic malignancies vs. solid tumors.
Results are presented in a manner similar to Fig. 1, with stacked bar plots for solid tumors (left); separate matrices for cell lines from non-MM hematologic malignancies (middle) vs. MM (right). Genes are included in 7 different clusters determined based on the criteria included in the color-coded bars on the right-hand side of the graph (FDR of comparison of CERES scores and difference in average CERES scores in MM vs. non-MM hematologic cell lines; Fisher’s test FDR for comparison of CERES ranks; absolute difference in % of MM vs. non-MM hematologic cell lines with CERES scores ≤ −0.4; and % of MM lines with CERES score ≤ −0.4). Results highlight that several MM-preferential dependencies are shared between MM and other hematologic malignancies, but many others are preferentially essential only for MM cell lines, a statement also supported by results of Extended Data Fig. 8.
Extended Data Figure 8 ∣. MM-preferential dependencies with distinct vs. overlapping roles in MM vs. other hematologic neoplasias or solid tumors.
a, Heat map for MM-preferential dependencies, summarizing their potential roles as preferential dependencies for other malignancies. Color coding indicates the difference in average CERES score for each gene in a given tumor type vs. all others: black/blue or red/orange denote FDR<0.05 and lower or higher, respectively, average CERES scores for a given gene in the respective neoplasia vs. all other cancer types. White denotes FDR>0.05. b-c, t-SNE plots of cell lines (depicted as dots), from MM, leukemias, lymphomas or other neoplasias, clustered according to RNA-Seq profiles b, or CERES scores c, for MM-preferential dependencies. RPKM data in b from CCLE [2018] for lines with matching 20Q4v2 CERES scores (N=15, 33, 16, 505 lines, respectively). In c, N=19, 44, 20, 706 lines, respectively (20Q4v2). d, Numbers of CRISPR-defined preferential dependencies (y-axis; identified based on the same criteria applied for MM) vs. number of lines for each indicated tumor type (x-axis). e, Volcano plot of -log10FDR (Limma t-test) for comparison of CERES scores in MM vs non-MM hematopoietic cell lines (y-axis) vs. difference in average CERES scores in MM vs. non-MM cell hematopoietic lines (x-axis). MM-preferential dependencies (identified in this study by comparison of MM vs. all non-MM cell lines) are depicted in red and orange, respectively, if they did vs. did not exhibit significantly lower CERES scores in MM compared with non-MM hematopoietic lines. f-h, Dependencies with differential role in MM vs. solid tumors or vs. B-cell lymphomas. Volcano plots for comparisons of CERES scores in MM lines (N=19) vs. f, all non-MM cell lines, from both hematologic malignancies and solid tumors (N=768; also see Supplementary Table 1); g, only solid tumor cell lines (N=701); h, B cell lymphoma lines (N=13) (x-axis: difference in average CERES scores between respective groups; y-axis: -log10FDR, Limma t-test). Red dots in each plot indicate genes satisfying criteria for more pronounced essentiality in MM compared with the respective groups of cell lines; i, Venn diagram highlighting genes with differential role in MM vs. solid tumors or B-cell lymphomas, based on panels f-h. j, CERES scores for genes that do not meet criteria for MM-preferential dependencies (comparison of MM vs. all other non-MM lines), but are more essential in MM vs. solid tumors or B-cell lymphomas, based on panels f-h.
Extended Data Figure 9 ∣. Pattern of essentiality for genes downstream of IRF4 or IKZF1/IKZF3.
CERES scores for genes previously defined as a, IRF4 target genes in MM cells5 or b, genes that are downregulated by loss-of-function of IKZF1 or IKZF3 (GSE113031) are depicted in a heatmap format (similar to Fig. 1) and in clusters of genes which (i) can be considered “core essential” genes (e.g., CERES <−0.4 in ≥90% of cell lines across cancers); (ii) meet all criteria for MM-preferential dependencies vs. other genes that have CERES scores <−0.4 in (iii) >50% of MM cell lines tested, (iv) 30-50% of MM cell lines tested; (v) <30% of MM cell lines tested; or (vi) none of the MM cell lines tested.
Extended Data Figure 10 ∣. Molecular and functional studies of POU2AF1 and ER-associated dependencies.
a-b, Immunoblotting analyses to confirm that protein levels of POU2AF1 are decreased with Doxy-inducible CRISPR interference (a, KMS11 cells) and increased with CRISPR activation (b, LP-1 cells) compared to cells with sgRNAs for control OR genes. Beta-actin a, or vinculin b, were probed as loading controls in the same respective membrane concurrently with POU2AF1. c, Relative numbers of viable LP-1 cells with CRISPR-based activation of POU2AF1 vs. a control OR gene (day 12 after end of transduction with sgRNAs for POU2AF1; results qualitatively concordant with those at later time-point in Fig. 6b). CTG assay, mean +/− SEM results; n=6 independent replicate cell cultures per condition; one-way ANOVA and Tukey’s post-hoc test (detailed results included in Source Data), p<0.001 for each POU2AF1 sgRNA vs. OR12D2 sgRNA). d, Immunoblotting for UBE2J1 after doxy-inducible CRISPR-based KO of UBE2J1 (or a control OR gene). Vinculin was probed as loading control concurrently with the staining for UBE2J1. Each experiment in a-d was performed once. e, UBE2J1, its dislocon complex partners SEL1L, SYVN1, and other ER-related MM preferential dependencies are among the top “hits” in two genome-scale screens (using retroviral gene-trap mutagenesis and CRISPR gene-editing)45 for genes involved in ERAD regulation (in KBM7 haploid cells). f, In vitro bortezomib treatment (24 h) of KMS18 cells with Doxy-inducible CRISPR KO of SYVN1 or control OR genes. (CTG; mean +/− SEM; n=8 independent replicate cell cultures for drug-free control and n=4 independent replicate cell cultures per drug dose for each KO; 2-way analyses of variance (p<0.001); detailed results of Tukey post-hoc tests included in Source Data). g-h, Patterns of CERES scores in MM (n=19) and non-MM (n=770) lines for g, ER/ERAD/Golgi-related genes and h, select ER genes. Results are presented similar to format of Fig. 1. Highlighted gene symbols include MM-preferential dependencies (red); examples of core essential genes (green); and genes which do not meet all criteria for MM-preferential dependencies but are recurrently essential for MM cell lines and are linked with the function of the ER glycoprotein quality control system (blue) and the ER translocon system (purple).
Supplementary Material
Acknowledgements
This work was supported by grants NIH R01 CA050947 (C.S.M.), CA179483 (C.S.M.), CA196664 (C.S.M. and R.W.J.G.), R01CA276156 (C.S.M.), CA180475 (J.D.L. and C.S.M.), CA192844 (L.H.B.), U01CA225730 (C.S.M.) and U01CA176058 (W.C.H); and by the de Gunzburg Myeloma Research Fund (C.S.M.), Leukemia and Lymphoma Society (LLS) Translational Research Program (C.S.M.), LLS Quest for Cure Program (C.S.M. and R.W.J.G.), LLS Scholar Award (C.S.M.), LLS Specialized Center for Research (J.D.L.), LLS Special Fellow Award (D.D.R.), Multiple Myeloma Research Foundation (MMRF) Answer Fund (C.S.M., J.D.L. and L.H.B.), MMRF Translational Network of Excellence (C.S.M.), MMRF Epigenetics Program Project grant (J.D.L. and C.S.M.), Shawna Ashlee Corman Investigatorship in Multiple Myeloma Research (C.S.M.), Cobb Family Myeloma Research Fund (C.S.M.), Paula and Rodger Riney Foundation (L.H.B.), Chambers Family Advanced Myeloma Research Fund (C.S.M.), International Waldenstrom’s Macroglobulinemia Foundation (C.S.M.), Department of Defense grant W81XWH-15-1-0012 (A.C. and C.S.M.), International Myeloma Foundation (G.M.M.), American-Australian Association (G.M.M.), Associazione Italiana per la Ricerca sul Cancro (S.G.), Claudia Adams Barr Program in Innovative Basic Cancer Research at Dana-Farber Cancer Institute (M.S., E.D. and C.S.M.), Dutch Cancer Society grant VU2011-5127 (R.W.J.G.) and Ludwig Center at Harvard (C.S.M.). Q.L.S. was supported by award number T32GM007753 from the National Institute of General Medical Sciences. B.G.B. was supported by an MMRF Research Fellow Award and an ASH Research Scholar Award. V.A.G. was supported by the American Cancer Society grant #IRG-14-188-01 to the Winship Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. The authors thank S. Sarkar, M. A. Bariteau and M. Borah for technical assistance in experiments related to this study and J. Sorrell for administrative coordination. The authors apologize in advance for the inability to reference in our paper all possible studies in the literature that are directly relevant to our findings.
Footnotes
Competing interests
R.S. serves research funding from FIMECS and honoraria from Janssen Pharma. G.M.M. is an employee of Aculeus Therapeutics. N.V.D. is an employee of Genentech, a member of the Roche Group. A.J.A. has consulted for Anji Pharmaceuticals, Arrakis Therapeutics, AstraZeneca, Boehringer Ingelheim, Oncorus, Inc., Merck & Co., Inc., Mirati Therapeutics, Nimbus Therapeutics, Plexium, Revolution Medicines, Reactive Biosciences, Riva Therapeutics, Servier Pharmaceuticals, Syros Pharmaceuticals and T-knife Therapeutics. A.J.A. holds equity in Riva Therapeutics. A.J.A. has research funding from Bristol Myers Squibb, Deerfield, Inc., Eli Lilly, Mirati Therapeutics, Novartis, Novo Ventures, Revolution Medicines and Syros Pharmaceuticals. C.J.O. has received research support from Gilead, Scorpion Therapeutics and eFFECTOR Therapeutics. J.M.D. is a consultant and holds equity in Jumble Therapeutics. B.L.E. has received research funding from Celgene, Deerfield, Novartis and Calico and consulting fees from GRAIL. He is a member of the scientific advisory board and shareholder for Neomorph Inc., TenSixteen Bio, Skyhawk Therapeutics and Exo Therapeutics. J.E.B. is a Scientific Founder of Syros Pharmaceuticals, SHAPE Pharmaceuticals, Acetylon Pharmaceuticals, Tensha Therapeutics (now Roche) and C4 Therapeutics and is the inventor on intellectual property licensed to these entities. J.E.B. has recently served as an executive and shareholder in Novartis AG. N.K. is currently an employee of Kymera Therapeutics, Inc. N.S.G. is a founder, science advisory board member (SAB) and equity holder in Syros, C4, Allorion, Lighthorse, Voronoi, Inception, Matchpoint, CobroVentures, GSK, Shenandoah (board member), Larkspur (board member) and Soltego (board member). The Gray lab receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Jansen, Kinogen, Arbella, Deerfield, Springworks, Interline and Sanofi. J.J.K. has received research funding from Amgen, Genentech and Janssen. J.M.M. has received research funding from the Dependency Map Consortium. W.C.H. is a consultant for Thermo Fisher, Solasta Ventures, MPM Capital, KSQ Therapeutics, Frontier Medicines, Tyra Biosciences, Function Oncology, Jubilant Therapeutics, Riva Therapeutics, RAPPTA Therapeutics, Calyx and Serinus Biosciences. L.H.B. is a consultant for and receives honoraria and research funding from AstraZeneca, and is a consultant for Genentech and Abbvie. J.D.L. receives research funding from Ipsen/ Epizyme and is consultant for AstraZeneca. A.T. is a consultant for Cedilla Therapeutics, Foghorn Therapeutics and The Center for Protein Degradation (Deerfield), and is a SAB member and holds equity in Turbine Simulated Cell Technologies. F.V. receives research support from the Dependency Map Consortium, Bristol Myers Squibb, Novo Ventures and Riva Therapeutics, and has shares and is a consultant for Riva Therapeutics. C.S.M. serves on the Scientific Advisory Board of Adicet Bio and discloses consultant/honoraria from Genentech, Fate Therapeutics, Ionis Pharmaceuticals, FIMECS, Secura Bio and Oncopeptides, and research funding from Janssen/Johnson & Johnson, EMD Serono, Arch Oncology, Karyopharm, Sanofi, Nurix, BMS, H3 Biomedicine/Eisai, Springworks, Abcuro and Novartis. The remaining authors declare no competing interests.
Data Availability.
RNA–seq data that support the findings of this study are publicly available in the Gene Expression Omnibus (GEO) under accession code GSE186997. Previously published data that were re-analyzed here are available under accession codes GSE2113, GSE5900, GSE6477, GSE13591, GSE39754, GSE39925, GSE66293, GSE20540, GSE8958, GSE9067, GSE9367, GSE19748, GSE9782, GSE113031, GSE121912 and GSE79480. Molecular profiling data were derived for CCLE lines from the CCLE portal (https://portals.broadinstitute.org/ccle/data) or the Dependency Map portal (https://depmap.org/portal); for MM patient samples from MMRF Researcher Gateway (https://research.themmrf.org/); and for non-MM patient samples from the TCGA Research Network (https://portal.gdc.cancer.gov/, http://cancergenome.nih.gov/). Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code Availability.
The analyses of our study involved standard workflows and sequential use of available code, e.g., through R-packages from CRAN (https://cran.r-project.org/) and Bioconductor (https://www.bioconductor.org) and R build-in functions for graphing, statistical tests and data analyses, e.g., moderated t-test with the limma Bioconductor R Package, Fisher’s exact test with the fisher.test R build-in function, survival analysis log-rank test the survival CRAN R package, gene expression analysis with edgeR Bioconductor R package. For processing large data matrices we used the data.table CRAN R Package. For data visualization, e.g., generation of heatmaps with the ComplexHeatmap Bioconductor R Package, the circos plot with circlize Bioconductor R Package and for dimensionality reduction visualization the tsne CRAN R Package. We further used for data analysis and visualization the statistical and graphing software GraphPad Prism 9, heatmaps with Morpheus (https://software.broadinstitute.org/morpheus/), and network analysis with StringDB (https://string-db.org/). RNA-seq raw data processing and generation of gene read counts with STAR71.
Resource Availability.
Requests for resources and reagents should be directed to the Lead Contact, Constantine Mitsiades (Constantine_Mitsiades@dfci.harvard.edu).
REFERENCES
- 1.Boise LH, Kaufman JL, Bahlis NJ, Lonial S & Lee KP The Tao of myeloma. Blood 124, 1873–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barwick BG, Gupta VA, Vertino PM & Boise LH Cell of Origin and Genetic Alterations in the Pathogenesis of Multiple Myeloma. Front Immunol 10, 1121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrulis M. et al. Targeting the BRAF V600E mutation in multiple myeloma. Cancer Discov 3, 862–9 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Tsherniak A. et al. Defining a Cancer Dependency Map. Cell 170, 564–576 e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaffer AL et al. IRF4 addiction in multiple myeloma. Nature 454, 226–31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong F. et al. CNPY2 is a key initiator of the PERK-CHOP pathway of the unfolded protein response. Nat Struct Mol Biol 24, 834–839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khatib AM et al. Inhibition of proprotein convertases is associated with loss of growth and tumorigenicity of HT-29 human colon carcinoma cells: importance of insulin-like growth factor-1 (IGF-1) receptor processing in IGF-1-mediated functions. J Biol Chem 276, 30686–93 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Hawley SA et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol 2, 28 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarumoto Y. et al. LKB1, Salt-Inducible Kinases, and MEF2C Are Linked Dependencies in Acute Myeloid Leukemia. Mol Cell 69, 1017–1027 e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMillin DW et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med 16, 483–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMillin DW et al. Compartment-Specific Bioluminescence Imaging platform for the high-throughput evaluation of antitumor immune function. Blood 119, e131–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMillin DW, Negri JM & Mitsiades CS The role of tumour-stromal interactions in modifying drug response: challenges and opportunities. Nat Rev Drug Discov 12, 217–28 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Rustad EH et al. Revealing the impact of structural variants in multiple myeloma. Blood Cancer Discov 1, 258–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whyte WA et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan S. et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature 481, 90–3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasqualucci L. et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med 203, 311–7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henske EP, Jozwiak S, Kingswood JC, Sampson JR & Thiele EA Tuberous sclerosis complex. Nat Rev Dis Primers 2, 16035 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Ozcan U. et al. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell 29, 541–51 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedele PL et al. IMiDs prime myeloma cells for daratumumab-mediated cytotoxicity through loss of Ikaros and Aiolos. Blood 132, 2166–2178 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Kronke J. et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu G. et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto J. et al. ARID2 is a pomalidomide-dependent CRL4(CRBN) substrate in multiple myeloma cells. Nat Chem Biol 16, 1208–1217 (2020). [DOI] [PubMed] [Google Scholar]
- 23.An J. et al. pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4(CRBN) ubiquitin ligase. Nat Commun 8, 15398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donovan KA et al. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray Syndrome. Elife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eichner R et al. Immunomodulatory drugs disrupt the cereblon-CD147-MCT1 axis to exert antitumor activity and teratogenicity. Nat Med 22, 735–43 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Hideshima T. et al. p53-related protein kinase confers poor prognosis and represents a novel therapeutic target in multiple myeloma. Blood 129, 1308–1319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer ES et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millrine D, Tei M, Gemechu Y & Kishimoto T Rabex-5 is a lenalidomide target molecule that negatively regulates TLR-induced type 1 IFN production. Proc Natl Acad Sci U S A 113, 10625–30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang J. et al. A calcium- and calpain-dependent pathway determines the response to lenalidomide in myelodysplastic syndromes. Nat Med 22, 727–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matyskiela ME et al. A Cereblon Modulator (CC-220) with Improved Degradation of Ikaros and Aiolos. J Med Chem 61, 535–542 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Sievers QL et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 362(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heider M. et al. The IMiD target CRBN determines HSP90 activity toward transmembrane proteins essential in multiple myeloma. Mol Cell 81, 1170–1186 e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsiades N. et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci U S A 99, 14374–9 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsiades CS et al. Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell 5, 221–30 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Barretina J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dashevsky O. et al. Use of Olfactory Receptor Genes As Controls for Genome-Scale CRISPR Functional Genomic Studies to Define Treatment Resistance Mechanisms. Blood 136, 36–36 (2020).32430502 [Google Scholar]
- 37.Ferreira de Freitas R. et al. Discovery of a Potent and Selective Coactivator Associated Arginine Methyltransferase 1 (CARM1) Inhibitor by Virtual Screening. J Med Chem 59, 6838–47 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Shen Y. et al. Discovery of a Potent, Selective, and Cell-Active Dual Inhibitor of Protein Arginine Methyltransferase 4 and Protein Arginine Methyltransferase 6. J Med Chem 59, 9124–9139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunningham L. et al. Identification of benzodiazepine Ro5-3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFbeta interaction. Proc Natl Acad Sci U S A 109, 14592–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao C. et al. POU2AF1, an amplification target at 11q23, promotes growth of multiple myeloma cells by directly regulating expression of a B-cell maturation factor, TNFRSF17. Oncogene 27, 63–75 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Jahn L. et al. TCR-based therapy for multiple myeloma and other B-cell malignancies targeting intracellular transcription factor BOB1. Blood 129, 1284–1295 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Toman I. et al. Expression and prognostic significance of Oct2 and Bob1 in multiple myeloma: implications for targeted therapeutics. Leuk Lymphoma 52, 659–67 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Gowda PS et al. Runx2 Suppression by miR-342 and miR-363 Inhibits Multiple Myeloma Progression. Mol Cancer Res 16, 1138–1148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obeng EA et al. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 107, 4907–16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timms RT et al. Genetic dissection of mammalian ERAD through comparative haploid and CRISPR forward genetic screens. Nat Commun 7, 11786 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groen RW et al. Reconstructing the human hematopoietic niche in immunodeficient mice: opportunities for studying primary multiple myeloma. Blood 120, e9–e16 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Kumar SK et al. Venetoclax or placebo in combination with bortezomib and dexamethasone in patients with relapsed or refractory multiple myeloma (BELLINI): a randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol 21, 1630–1642 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Panobinostat approved for multiple myeloma. Cancer Discov 5, OF4 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Raab MS et al. The first-in-human study of the pan-PIM kinase inhibitor PIM447 in patients with relapsed and/or refractory multiple myeloma. Leukemia 33, 2924–2933 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Ishiguro K. et al. DOT1L inhibition blocks multiple myeloma cell proliferation by suppressing IRF4-MYC signaling. Haematologica (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drew AE et al. Identification of a CARM1 Inhibitor with Potent In Vitro and In Vivo Activity in Preclinical Models of Multiple Myeloma. Sci Rep 7, 17993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagel S. et al. Identification of a tumor suppressor network in T-cell leukemia. Leuk Lymphoma 58, 2196–2207 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Sheffer M. et al. Genome-scale screens identify factors regulating tumor cell responses to natural killer cells. Nat Genet (2021). [DOI] [PubMed] [Google Scholar]
- 54.Dufva O. et al. Single-cell functional genomics of natural killer cell evasion in blood cancers (* co-senior authors). bioRxiv, 2022.08.22.504722 (2022). [Google Scholar]
Methods-Only References
- 55.Meyers RM et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McFarland JM et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun 9, 4610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dempster JM et al. Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. bioRxiv, 720243 (2019). [Google Scholar]
- 58.Cowley GS et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data 1, 140035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McDonald ER 3rd et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell 170, 577–592 e10 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Goldman MJ et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 38, 675–678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Law CW, Chen Y, Shi W & Smyth GK voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barwick BG et al. Multiple myeloma immunoglobulin λ translocations portend poor prognosis. Nature Communications, 340877–340877 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amemiya HM, Kundaje A & Boyle AP The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci Rep 9, 9354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang W. et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 41, D955–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szklarczyk D. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45, D362–D368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erb MA et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 543, 270–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nabet B. et al. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol 14, 431–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shirasaki R. et al. Functional Genomics Identify Distinct and Overlapping Genes Mediating Resistance to Different Classes of Heterobifunctional Degraders of Oncoproteins. Cell Rep 34, 108532 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA–seq data that support the findings of this study are publicly available in the Gene Expression Omnibus (GEO) under accession code GSE186997. Previously published data that were re-analyzed here are available under accession codes GSE2113, GSE5900, GSE6477, GSE13591, GSE39754, GSE39925, GSE66293, GSE20540, GSE8958, GSE9067, GSE9367, GSE19748, GSE9782, GSE113031, GSE121912 and GSE79480. Molecular profiling data were derived for CCLE lines from the CCLE portal (https://portals.broadinstitute.org/ccle/data) or the Dependency Map portal (https://depmap.org/portal); for MM patient samples from MMRF Researcher Gateway (https://research.themmrf.org/); and for non-MM patient samples from the TCGA Research Network (https://portal.gdc.cancer.gov/, http://cancergenome.nih.gov/). Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
The analyses of our study involved standard workflows and sequential use of available code, e.g., through R-packages from CRAN (https://cran.r-project.org/) and Bioconductor (https://www.bioconductor.org) and R build-in functions for graphing, statistical tests and data analyses, e.g., moderated t-test with the limma Bioconductor R Package, Fisher’s exact test with the fisher.test R build-in function, survival analysis log-rank test the survival CRAN R package, gene expression analysis with edgeR Bioconductor R package. For processing large data matrices we used the data.table CRAN R Package. For data visualization, e.g., generation of heatmaps with the ComplexHeatmap Bioconductor R Package, the circos plot with circlize Bioconductor R Package and for dimensionality reduction visualization the tsne CRAN R Package. We further used for data analysis and visualization the statistical and graphing software GraphPad Prism 9, heatmaps with Morpheus (https://software.broadinstitute.org/morpheus/), and network analysis with StringDB (https://string-db.org/). RNA-seq raw data processing and generation of gene read counts with STAR71.
Requests for resources and reagents should be directed to the Lead Contact, Constantine Mitsiades (Constantine_Mitsiades@dfci.harvard.edu).