Abstract
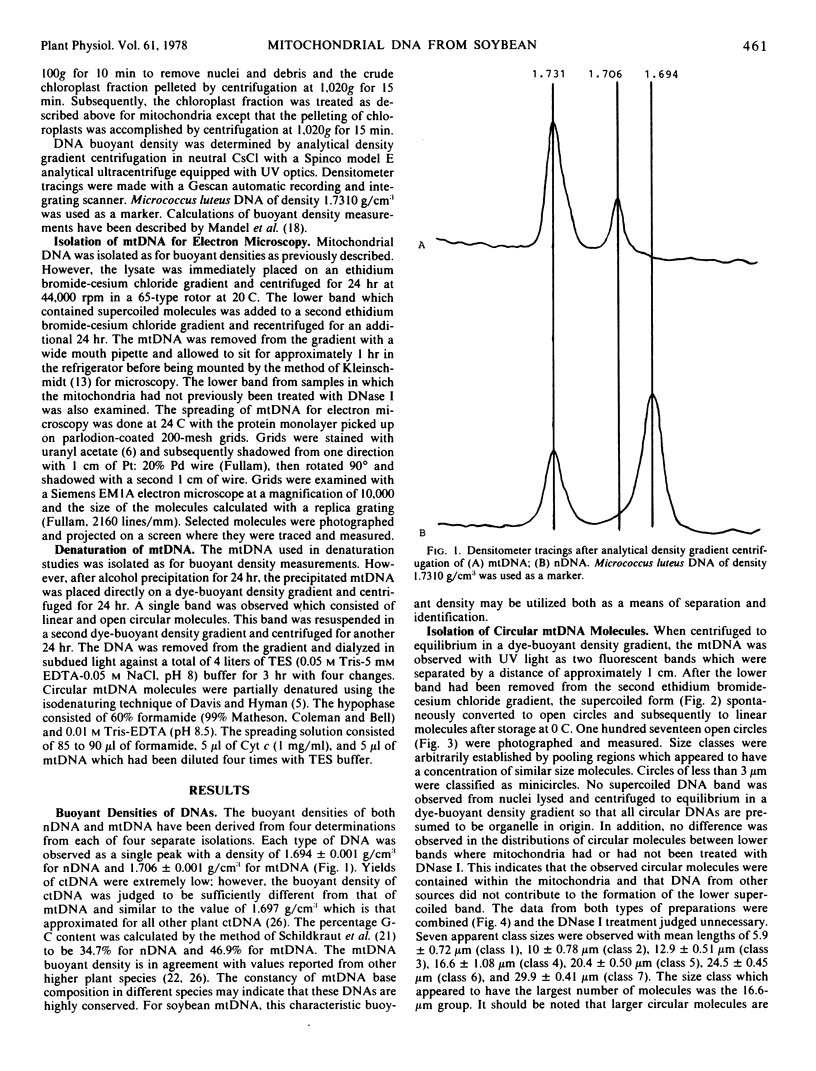
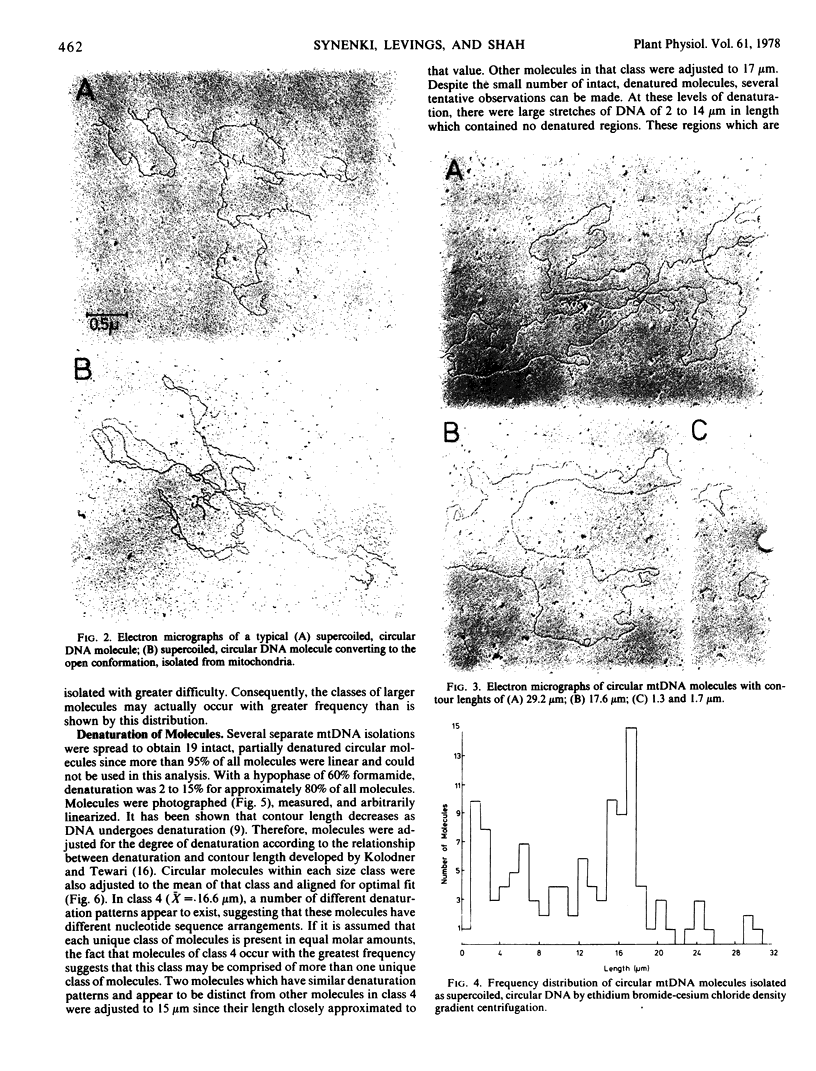
Mitochondrial DNA (mtDNA) of soybean (Glycine max L.) was isolated and its buoyant density was contrasted with that of nuclear (nDNA) and chloroplast (ctDNA) DNA. Each of the three DNAs banded at a single, characteristic buoyant density when centrifuged to equilibrium in a CsCl gradient. Buoyant densities were 1.694 g/cm3 for nDNA and 1.706 g/cm3 for mtDNA. These values correspond to G-C contents of 34.7 and 46.9%, respectively. Covalently closed, circular mtDNA molecules were isolated from soybean hypocotyls by ethidium bromide-cesium chloride density gradient centrifugation. Considerable variation in mtDNA circle size was observed by electron microscopy. There were seven apparent size classes with mean lengths of 5.9 μm (class 1), 10 μm (class 2), 12.9 μm (class 3), 16.6 μm (class 4), 20.4 μm (class 5), 24.5 μm (class 6), and 29.9 μm (class 7). In addition, minicircles were observed in all preparations. Partially denatured, circular mtDNA molecules with at least one representative from six of the seven observed size classes were mapped. In class 4, there appear to be at least three distinct denaturation patterns, indicating heterogeneity within this class. It is proposed that the mitochondrial genome of soybean is distributed among the different size circular molecules, several copies of the genome are contained within these classes and that the majority of the various size molecules may be a result of recombination events between circular molecules.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernard U., Pühler A., Mayer F., Küntzel H. Denaturation map of the circular mitochondrial genome of Neurospora crassa. Biochim Biophys Acta. 1975 Aug 21;402(2):270–278. doi: 10.1016/0005-2787(75)90047-7. [DOI] [PubMed] [Google Scholar]
- Clowes R. C. Molecular structure of bacterial plasmids. Bacteriol Rev. 1972 Sep;36(3):361–405. doi: 10.1128/br.36.3.361-405.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Kopecko D. J. Structural evolution of bacterial plasmids: role of translocating genetic elements and DNA sequence insertions. Fed Proc. 1976 Jul;35(9):2031–2036. [PubMed] [Google Scholar]
- Cohen S. N. Transposable genetic elements and plasmid evolution. Nature. 1976 Oct 28;263(5580):731–738. doi: 10.1038/263731a0. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Guerineau M., Grandchamp C., Slonimski P. P. Circular DNA of a yeast episome with two inverted repeats: structural analysis by a restriction enzyme and electron microscopy. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3030–3034. doi: 10.1073/pnas.73.9.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerineau M., Slonimski P. P., Avner P. R. Yeast episome: oligomycin resistance associated with a small covalently closed non-mitochondrial circular DNA. Biochem Biophys Res Commun. 1974 Nov 27;61(2):462–469. doi: 10.1016/0006-291x(74)90979-6. [DOI] [PubMed] [Google Scholar]
- Hollenberg C. P., Borst P., van Bruggen E. F. Mitochondrial DNA. V. A 25 micron closed circular duplex DNA molecule in wild-type yeast mitochondria. Stucture and genetic complexity. Biochim Biophys Acta. 1970 May 21;209(1):1–15. [PubMed] [Google Scholar]
- Hollenberg C. P., Degelmann A., Kustermann-Kuhn B., Royer H. D. Characterization of 2-mum DNA of Saccharomyces cerevisiae by restriction fragment analysis and integration in an Escherichia coli plasmid. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2072–2076. doi: 10.1073/pnas.73.6.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourcade D., Dressler D., Wolfson J. The nucleolus and the rolling circle. Cold Spring Harb Symp Quant Biol. 1974;38:537–550. doi: 10.1101/sqb.1974.038.01.058. [DOI] [PubMed] [Google Scholar]
- Kasamatsu H., Vinograd J. Replication of circular DNA in eukaryotic cells. Annu Rev Biochem. 1974;43(0):695–719. doi: 10.1146/annurev.bi.43.070174.003403. [DOI] [PubMed] [Google Scholar]
- Kolodner R. D., Tewari K. K. Denaturation mapping studies on the circular chloroplast deoxyribonucleic acid from pea leaves. J Biol Chem. 1975 Jul 10;250(13):4888–4895. [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. Physicochemical characterization of mitochondrial DNA from pea leaves. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1830–1834. doi: 10.1073/pnas.69.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. The molecular size and conformation of the chloroplast DNA from higher plants. Biochim Biophys Acta. 1975 Sep 1;402(3):372–390. doi: 10.1016/0005-2787(75)90273-7. [DOI] [PubMed] [Google Scholar]
- Locker J., Rabinowitz M., Getz G. S. Electron microscopic and renaturation kinetic analysis of mitochondrial DNA of cytoplasmic petite mutants of Saccharomyces cerevisiae. J Mol Biol. 1974 Sep 15;88(2):489–507. doi: 10.1016/0022-2836(74)90497-5. [DOI] [PubMed] [Google Scholar]
- Nass S. The significance of the structural and functional similarities of bacteria and mitochondria. Int Rev Cytol. 1969;25:55–129. doi: 10.1016/s0074-7696(08)60201-6. [DOI] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962 Jun;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- Smith C. A., Vinograd J. Small polydisperse circular DNA of HeLa cells. J Mol Biol. 1972 Aug 21;69(2):163–178. doi: 10.1016/0022-2836(72)90222-7. [DOI] [PubMed] [Google Scholar]
- Stanfield S., Helinski D. R. Small circular DNA in Drosophila melanogaster. Cell. 1976 Oct;9(2):333–345. doi: 10.1016/0092-8674(76)90123-9. [DOI] [PubMed] [Google Scholar]
- Vedel F., Quetier F. Physico-chemical characterization of mitochondrial DNA from potato tubers. Biochim Biophys Acta. 1974 Apr 10;340(4):374–387. doi: 10.1016/0005-2787(74)90059-8. [DOI] [PubMed] [Google Scholar]
- Wells R., Ingle J. The constancy of the buoyant density of chloroplast and mitochondrial deoxyribonucleic acids in a range of higher plants. Plant Physiol. 1970 Jul;46(1):178–179. doi: 10.1104/pp.46.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstonholme D. R., Kirschner R. G., Gross N. J. Heart denaturation studies of rat liver mitrochondrial DNA. A denaturation map and changes in molecular configurations. J Cell Biol. 1972 May;53(2):393–406. doi: 10.1083/jcb.53.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong F. Y., Wildman S. G. Simple procedure for isolation of satellite DNA's from tobacco leaves in high yield and demonstration of minicircles. Biochim Biophys Acta. 1972 Jan 18;259(1):5–12. doi: 10.1016/0005-2787(72)90468-6. [DOI] [PubMed] [Google Scholar]