Abstract
Previously, we found that the cause of autosomal dominant selective tooth agenesis in one family is a missense mutation resulting in an arginine-to-proline substitution in the homeodomain of MSX1. To determine whether the tooth agenesis phenotype may result from haploinsufficiency or a dominant-negative mechanism, we have performed biochemical and functional analyses of the mutant protein Msx1(R31P). We show that Msx1(R31P) has perturbed structure and reduced thermostability compared with wild-type Msx1. As a consequence, the biochemical activities of Msx1(R31P) are severely impaired, since it exhibits little or no ability to interact with DNA or other protein factors or to function in transcriptional repression. We also show that Msx1(R31P) is inactive in vivo, since it does not display the activities of wild-type Msx1 in assays of ectopic expression in the limb. Furthermore, Msx1(R31P) does not antagonize the activity of wild-type Msx1 in any of these assays. Because Msx1(R31P) appears to be inactive and does not affect the action of wild-type Msx1, we propose that the phenotype of affected individuals with selective tooth agenesis is due to haploinsufficiency.
Tooth agenesis, or missing teeth, is one of the most common developmental anomalies in humans (12). Agenesis of one or more teeth is reported to occur in as many as 9% of the population, excluding third molar (wisdom tooth) agenesis, which is more prevalent (12). Inherited tooth agenesis is likely to be caused by an impairment of one or more of the molecular processes that regulate tooth formation. As with many other organs, tooth development involves sequential and reciprocal signaling processes between epithelial and mesenchymal cell layers that are orchestrated by a hierarchy of genes encoding secreted growth factors, extracellular matrix components, and transcriptional regulators (26–29). Because the regulatory genes required for tooth formation are common components of signaling cascades involved in development of other embryonic structures and because of its relative simplicity, the tooth is an excellent model for studying the molecular processes that underlie organogenesis.
Among the transcriptional regulatory genes required for tooth formation, the Msx1 homeobox gene is highly expressed in the dental mesenchyme (17, 19, 20) and is essential for tooth development, since targeted gene disruption results in arrested tooth formation at an early stage in Msx1(−/−) mice (6, 23). In addition to its expression in the tooth primordia, Msx1 expression is prominent in regions of epithelial-mesenchymal interactions in several other embryonic structures, including other craniofacial structures and the limb (reviewed in reference 7). These findings have led to the hypothesis that Msx1 is an important component in the signaling events that occur between epithelial and mesenchymal tissues.
Previously, we reported that a missense mutation in the human MSX1 gene causes selective tooth agenesis of secondary dentition in one family (30). This mutation results in a protein, MSX1(R31P), that contains an arginine-to-proline substitution at position 31 within the homeodomain. Since this trait is autosomal dominant, the resulting phenotype may be due to haploinsufficiency, a dominant-negative activity, or a novel activity of MSX1(R31P). To distinguish among these possibilities, we have now investigated the consequences of the R31P substitution on the structure of the resulting protein, as well as on its biochemical and biological activities. We present evidence that the missense mutation in MSX1 is likely to cause selective tooth agenesis through haploinsufficiency. Our findings highlight the importance of dosage for mediating the biological actions of MSX1, as well as the significance of a detailed understanding of the consequences of missense mutations for interpreting the molecular bases of genetic disease.
MATERIALS AND METHODS
Plasmid construction and protein expression.
Most studies were performed with the murine Msx1 cDNA, which shares 94% similarity with human MSX1 (100% identity within the homeodomain) (7). The Msx1 and Msx1(R31A) plasmids used for in vitro transcription and translation [pGEM7zf(+)-Msx1(1–297) and pGEM7zf(+)-Msx1(1–297):R196A] and the expression plasmids used for transient transfection [pCB6+-Msx1(1–297) and pCB6+-Msx1(1–297):R196A] were described previously (5, 33, 34). Note that Msx1(R31A) refers to Msx1-D from a previous report (33). To construct Msx1(R31P), we introduced, by PCR-mediated site-directed mutagenesis, a substitution to replace arginine 196 (homeodomain position 31) with proline. The product was subcloned into the BamHI and HindIII sites of plasmids pGEM7zf(+) (Promega) and pCB6+ for use in in vitro transcription and translation and mammalian expression, respectively. The Msx1HD plasmid, used for production of the recombinant homeodomain polypeptide, was described previously [pDS56-Msx1(157–233)] (3). Msx1HD(R31P) and Msx1HD(R31A) were obtained by PCR amplification of the respective sequences encoding amino acids 157 to 233 and subcloned into the BamHI and HindIII sites of plasmid pDS56. The recombinant homeodomain polypeptides, made as hexahistidine fusion proteins, were expressed in Escherichia coli and purified by nickel affinity chromatography as described previously (3). Note that this purification procedure renders virtually homogeneous protein preparations (Fig. 1A). Procedures for DNA binding, glutathione S-transferase (GST) interaction, and transient-transfection assays have been described elsewhere (3, 5, 33, 34). The complete sequences of all Msx1, Msx1(R31P), and Msx1(R31A) constructs were verified by using Sequenase version 2.0 (U.S. Biochemicals). Msx1(R31P) and Msx1(R31A) are comparable to Msx1 in all respects, except for the relevant substitutions.
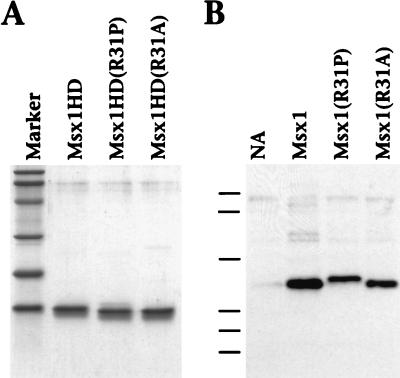
FIG. 1.
Expression of Msx1 proteins. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis demonstrates the purity of the recombinant Msx1 homeodomain polypeptides Msx1HD, Msx1HD(R31P), and Msx1HD(R31A). Each protein (2.5 μg) was separated on a 15% polyacrylamide gel and visualized by staining with Coomassie brilliant blue. (B) Western blot assay demonstrates the equivalent expression of the indicated Msx1 proteins in infected CEFs. Cell lysates were prepared from CEFs that were not infected (NA) or that were infected with a retrovirus expressing Msx1, Msx1(R31P), or Msx1(R31A). The lysates were separated on a 10% polyacrylamide gel, and the Msx1 proteins, which were Myc tagged, were detected with a monoclonal antibody against the Myc epitope. Note that the R31P substitution results in a more slowly migrating protein. Molecular mass standards (shown in panel A by the marker lane and in panel B by dashes) are phosphorylase B (100 kDa), bovine serum albumin (77 kDa), ovalbumin (48.2 kDa), carbonic anhydrase (33.8 kDa), soybean trypsin inhibitor (28.6 kDa), and lysozyme (20.5 kDa).
Retroviral infection.
For construction of retroviral expression vectors, a Myc epitope was introduced at the 5′ end of the coding region of Msx1, Msx1(R31P), and Msx1(R31A) by PCR amplification. We have previously found that the N-terminal Myc tag, which facilitates detection, does not affect the activity of Msx1 in various assays (data not shown). The resulting PCR products were first cloned into the BamHI and HindIII sites of the SLAX13 shuttle vector (14) and then subcloned as ClaI fragments into the corresponding site of the replication-competent avian retroviral vectors RCASBP(A) and RCASBP(B) (see Fig. 7A) (9, 14). For comparison, chicken Msx1(GMsx1) and the corresponding Msx1R31P mutant (without the Myc epitope) were subcloned into the NcoI and HindIII sites of SLAX13 and subcloned into RCASBP(A). A control retrovirus expressing human alkaline phosphatase (AP) in RCASBP(B) was described in reference 9. For production of high-titer virus (∼108 to 109 CFU/ml), supernatants from virus-infected chicken embryo fibroblasts (CEFs) were concentrated as described previously (9). Note that the retroviruses express similar levels of Msx1, Msx1(R31P), and Msx1(R31A) (Fig. 1B), and all three proteins were localized to the nucleus (data not shown). Virus was injected into stage 10 or stage 17 chicken embryos in the area fated to become the right wing as described previously (11). Embryos were staged according to the method of Hamburger and Hamilton (13). Wings were dissected at stages 36 to 39, stained with Alcian blue or green, and cleared with KOH-glycerol as described previously (11). Dissected wings were imaged by video capture; the bone lengths were measured, and other parameters of the phenotype (feather germ formation and altered morphology) were scored. The ratios of infected (right wing) to uninfected (left wing) bone lengths from the same embryos were calculated for the humerus, radius, ulna, and the longest digit (digit III). The mean bone length indices of Msx1-infected embryos were compared to the 99% confidence interval for the mean bone length indices of control [AP- and Msx1(R31P)-infected] embryos.
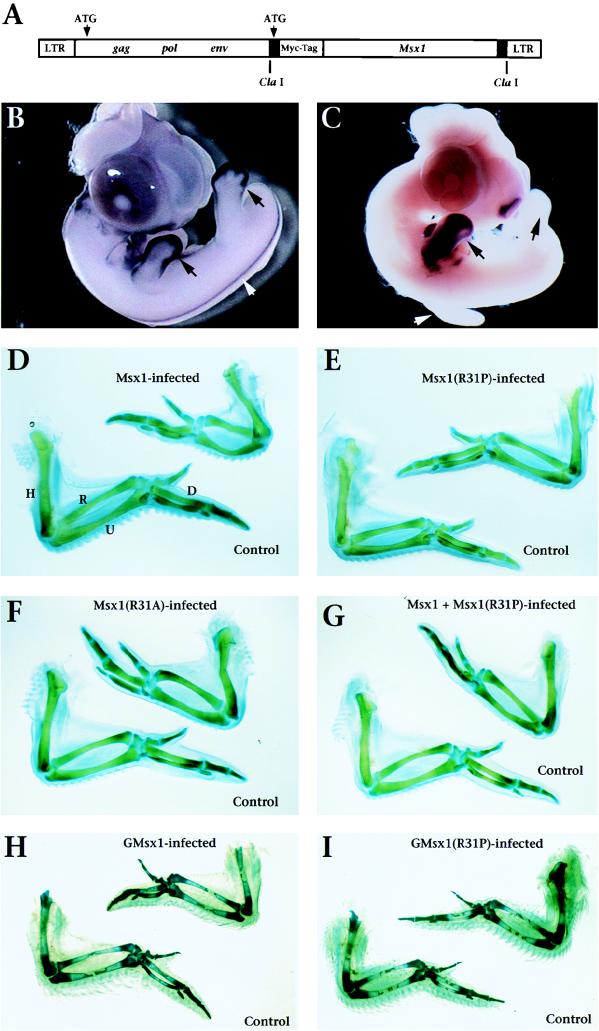
FIG. 7.
Ectopic expression of Msx1, but not Msx1(R31P), alters chicken embryonic limb morphology. (A) The replication-competent retroviral vector RCASBP (9, 14) provides a vehicle with which to introduce Msx1, Msx1(R31P), or Msx1(R31A) into the limb and to drive their expression. Note that the murine genes are Myc tagged, but the chicken genes are not (see Materials and Methods). The retroviruses that contain the murine Msx1 sequences allow for detection of the exogenous genes as distinguished from the endogenous chicken gene (GMsx1) by in situ hybridization. (B) Whole-mount in situ hybridization shows expression of endogenous GMsx1 at stage 26 in the embryonic limb buds, where it is confined primarily to the distal mesenchyme (progress zone; arrows). At this stage, GMsx1 is also expressed in the dorsal neural tube (white arrowhead) and branchial arches (not shown). (C) Whole-mount in situ hybridization shows ectopic expression of murine Msx1 in the forelimb bud at stage 26 following retroviral infection at stage 17. Exogenous Msx1 is expressed throughout the forelimb, but not the hindlimb, on the infected side (black arrows) and not in the forelimb of the uninfected (control) side (white arrowhead) or in other regions of endogenous GMsx1 expression. A similar pattern of ectopic expression was observed for GMsx1 and GMsx1(R31P) (data not shown). Whole-mount in situ hybridization was performed with a chicken (B) or murine (C) Msx1 antisense riboprobe as described in reference 34. (D to F) Ectopic expression of Msx1 (D) or Msx1(R31A) (F), but not Msx1(R31P) (E), in the forelimb produces a smaller wing on the infected side (upper right) relative to the wing on the uninfected side (control, lower left). (G) Ectopic expression of Msx1 together with Msx1(R31P) produces a smaller wing comparable to that seen with ectopic expression of Msx1 alone. (H to I) Ectopic expression of GMsx1 (H), but not GMsx1(R31P) (I), in the forelimb produces a smaller wing and smaller feathers on the infected side relative to the uninfected one (control). In panels D to I, infection was achieved by injection of high-titer virus (108 CFU/ml) into the region of the prospective forelimb (right side) at stage 10 (H and I) or stage 17 (D to G); embryos were staged according to the method of Hamburger and Hamilton (13). Infection was generally restricted to the site of injection (Fig. 7C); however, blood-borne virus sometimes led to ectopic expression in the heart (not shown). Two retroviral vectors encoding alternative viral envelope proteins were used [RCASBP(A) and RCASBP(B)]. Ectopic expression of Msx1 by using either retroviral vector produced equivalent results. In panel G, coinfection of Msx1 and Msx1(R31P) was achieved with a 1:1 mixture of Msx1-RCASBP(A) and Msx1(R31P)-RCASBP(B). Panels D to I show representative wings (stages 36 to 39) with the ventral surface facing up. Data analysis is provided in Table 1. R, radius; U, ulna; H, humerus; D, digit III.
CD.
Circular dichroism (CD) measurements were performed with an Aviv model 62 DS spectropolarimeter fitted with a thermally regulated cell holder in 0.1-cm (far-UV spectra) or 1.0-cm (near-UV spectra) rectangular cuvettes. The midpoint temperatures (Tms) of the unfolding transitions were determined by fitting the change in ellipticity at 208 nm to the equations k = exp{[ΔH/(RT)] [(T/Tm) − 1]}, y = k/(1 + k), and f = [(u − l)y] + 1, where k is the equilibrium constant of folding at any temperature (T), Tm is the midpoint temperature of the folding transition, and f is the fraction folded at any temperature. T and Tm are in degrees kelvin, converted to degrees celsius in Fig. 3. R is the gas constant, and ΔH is the enthalpy of folding, u represents the ellipticity values where the protein is completely folded, and l is the ellipticity value where the protein is completely unfolded. To calculate the Tm values, initial values of ΔH, u, l, and Tm are estimated. The CD data are then fit to f by using the Levenberg-Marquardt algorithm (21) implemented in SigmaPlot. The percentage of α-helicity was calculated from the ellipticity at 222 nm with the equation %helix = 100[θ(Observed) − θ(Coil)]/[θ(Helix) − θ(Coil)], where θ(Helix) = −40,000 (1 − 2.5/n) + 100T and θ(Coil) = 640 − 45T. θ(Helix) and θ(Coil) are the values for 100% α-helix and 100% random coil respectively, expressed in degrees times centimeter squared per decimole, T is the temperature in degrees celsius, and n is the number of residues in the chain (24).
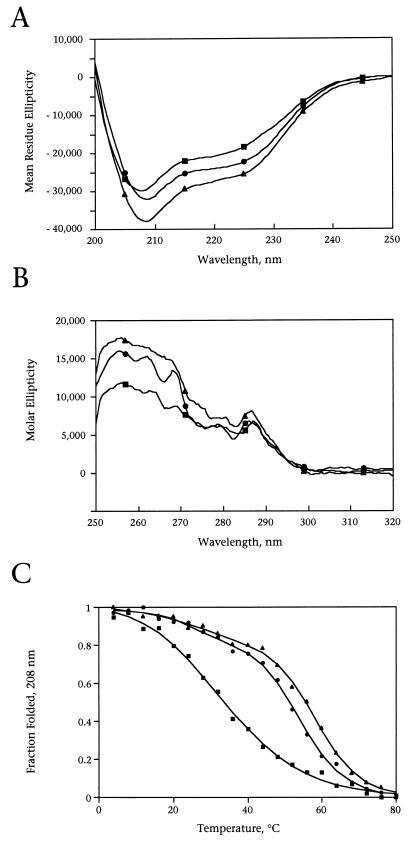
FIG. 3.
CD analysis demonstrates altered structure and reduced stability of Msx1HD(R31P) relative to Msx1HD and Msx1HD(R31A). CD spectra were collected by using the purified Msx1 homeodomain polypeptides (Fig. 1A) Msx1HD (•), Msx1HD(R31P) (■), and Msx1HD(R31A) (▴). (A) Far-UV CD spectra (200 to 250 nm) show that the α-helical content of Msx1HD(R31P) (56%) is less than that of Msx1HD (65%), whereas Msx1HD(R31A) (71%) has greater α-helical content. This is consistent with the known helix-disrupting propensity of proline and the helix-promoting propensity of alanine (22). (B) Near-UV CD spectra (250 to 320 nm) show that Msx1HD(R31P) has reduced absorbance in the characteristic tyrosine and phenylalanine regions (260 and 280 nm, respectively), whereas the tryptophan peak (290 nm) is similar for all three proteins. (C) Tm curves show that Msx1HD(R31P) has a lower Tm (33°C) than Msx1HD (53°C) or Msx1HD(R31A) (58°C). The fraction of folded protein was calculated from the far-UV CD spectra at 208 nm, taken between 0 and 80°C; a similar profile was obtained at 222 nm (data not shown). In panels A, B, and C, protein concentrations were determined by A280 (ɛ280 = 7,000 cm−1/M−1) in the presence of 6 M guanidine-HCl (8) and were adjusted to 0.06 mg/ml (A and C) or 0.6 mg/ml (B). All data were collected in 10 mM potassium phosphate buffer (pH 7.0) in triplicate with a step size of 0.25 nm. In panels A and B, spectra were recorded at 20°C; for clarity, only five data points are shown. CD analysis was performed three times with two independent protein preparations; representative data are shown.
RESULTS AND DISCUSSION
Functional domains of Msx1.
The murine Msx1 gene encodes a highly conserved DNA binding protein that functions as a transcriptional repressor through its interactions with general transcription factors, such as the TATA binding protein (TBP), and other homeoproteins, including members of the Dlx family (4, 5, 31, 33, 34). Of several conserved functional domains of Msx1, the homeodomain in particular is essential for DNA binding, transcriptional repression, interactions with TBP and Dlx, and protein stability (Fig. 2) (3, 4, 8, 25, 33, 34). Therefore, the R31P substitution may affect any, or all, of these biochemical activities of Msx1.
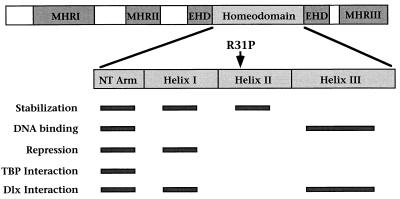
FIG. 2.
Functional domains of Msx1. Schematic diagram of Msx1 showing the regions conserved among Msx proteins: the Msx homology regions I to III (MHRI to -III), the extended homeodomain (EHD), and the homeodomain (7). MHRI and MHRII contribute to transcriptional repression, whereas MHRIII promotes protein stability (4). Contributions made by the homeodomain subdivisions (the N-terminal arm [NT Arm] and helices I, II, and III) to protein stability, DNA binding specificity, transcriptional repression, and protein interactions are indicated by bars (8, 25, 33, 34). Note that the R31P substitution occurs in helix II, which is primarily important for protein stability.
In considering the possible consequences of the R31P substitution, we noted that this mutation occurs within helix II of the homeodomain, which makes an important contribution to protein stability (Fig. 2) (8, 25). Moreover, proline residues are rarely found at position 31 in homeodomain sequences (2), which is not surprising given the known propensity of prolines to disrupt α-helices (22). Therefore, the R31P substitution may affect the activity of Msx1 as a consequence of introducing a proline residue within helix II, which may be distinct from effects due to the loss of the basic arginine side chain. To distinguish between these possibilities, we have compared the activities of Msx1(R31P) to that of Msx1 and also to that of an Msx1 polypeptide containing a substitution of arginine 31 by alanine [Msx1(R31A)], since alanine is a neutral amino acid that is known to promote, rather than destabilize, α-helix formation (22).
Msx1HD(R31P) has altered structure and reduced stability relative to Msx1HD and Msx1HD(R31A).
To determine whether the R31P substitution affects the structure of Msx1, we performed CD analysis using homeodomain polypeptides corresponding to the wild-type sequence [Msx1HD], the R31P substitution [Msx1HD(R31P)], or the R31A substitution [Msx1HD(R31A)] (Fig. 1A). CD analysis in the far-UV range provides a quantitative measurement of the α-helical content of proteins (1), which is particularly useful for homeodomains, since they are primarily α-helical in structure (10). CD analysis in the near-UV range measures the aromatic amino acid side chain conformations and provides an indirect measurement of protein conformation (1). As shown by their far-UV CD spectra, all three homeodomain polypeptides form α-helices at 20°C (Fig. 3A). However, Msx1HD(R31P) has reduced α-helical content (56%) relative to Msx1HD (65%), whereas Msx1HD(R31A) has increased α-helical content (71%). The near-UV CD spectra show that the protein conformation of Msx1HD(R31P) is altered relative to those of Msx1HD and Msx1HD(R31A) (Fig. 3B). In particular, the characteristic tryptophan peak (at 290 nm) is similar for all three polypeptides, whereas the tyrosine (280 nm) and phenylalanine (260 nm) peaks are reduced for Msx1HD(R31P) relative to those for Msx1HD and Msx1HD(R31A) (Fig. 3B). The single tryptophan residue in the Msx1 homeodomain is located in helix III, whereas tyrosine and phenylalanine residues are found in helices I and II, indicating a local unfolding in the vicinity of the proline substitution.
To examine directly whether the R31P substitution affects protein stability, we determined the Tms of Msx1HD, Msx1HD(R31P), and Msx1HD(R31A) (Fig. 3C). The Tm refers to the temperature at which 50% of the protein is folded and is calculated from the far-UV CD spectra taken between 0 and 80°C. This analysis revealed that Msx1(R31P) is significantly less stable (Tm = 33°C) than Msx1HD and Msx1HD(R31A) (Tm = 53 and 58°C, respectively) (Fig. 3C). We note that the Tm of Msx1HD(R31P) is lower than the physiological temperature, suggesting that a considerable fraction of the Msx1HD(R31P) protein may be in a partially unfolded state in vivo.
Msx1R31P has reduced activity in biochemical functions that require the homeodomain.
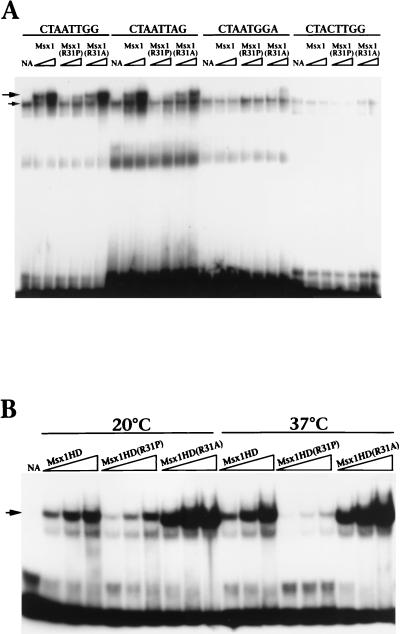
We next examined the consequences of the R31P substitution on biochemical activities of Msx1 mediated by the homeodomain. To compare the DNA binding activities of Msx1, Msx1(R31P), and Msx1(R31A), we performed gel retardation assays using full-length proteins obtained by in vitro translation (Fig. 4A). As shown in Fig. 4A and as previously described (3), Msx1 interacts with its consensus DNA site (e.g., CTAATTGG) and certain variations of this site (e.g., CTAATTAG), but not with sites that have substitutions of critical nucleotides within the consensus site (e.g., CTAATGGA and CTACTTGG) (Fig. 4A). Whereas the binding profile of Msx1(R31A) is essentially identical to that of Msx1, Msx1(R31P) does not interact significantly with any of these DNA sites (Fig. 4A) or with several other DNA sites tested (data not shown).
FIG. 4.
Msx1R31P has reduced DNA binding activity, compared with that of Msx1 or Msx1(R31A), and is temperature sensitive. (A) A gel retardation assay was performed at 20°C with proteins obtained by in vitro transcription and translation (1 or 2 μl, indicated by the triangle) and with the DNA sites shown; these sites were described in reference 3. The lanes labeled NA contain the unprogrammed reticulocyte lysate (2 μl). The upper arrow indicates the position of the specific Msx1-DNA complex; the lower arrow indicates a nonspecific complex present in the unprogrammed lysate. (B) The gel retardation assay was performed with recombinant protein (5, 10, or 20 ng, indicated by the triangle) and the Msx1 consensus DNA site (CTAATTGG) (3). Protein-DNA complexes were formed at 20 or at 37°C, as shown. The lanes labeled NA contain no added protein; the arrow indicates the position of the Msx1HD-DNA complex. Note that Msx1HD(R31A) binds to DNA more avidly than Msx1HD, which is presumably a consequence of its enhanced stability (Fig. 3). For panels A and B, assays were performed a minimum of three times; representative data are shown.
To examine the relationship between loss of DNA binding and reduced stability of Msx1(R31P), we compared the binding activities of Msx1HD, Msx1HD(R31P), and Msx1HD(R31A) at temperatures below (20°C) and above (37°C) the Tm of Msx1HD(R31P) (Fig. 4B). We used the recombinant homeodomain polypeptides, since Msx1HD(R31P) interacts better with DNA than full-length Msx1(R31P) (compare Fig. 4A and B). We found that the DNA binding activities of Msx1HD and Msx1HD(R31A) were not altered at the temperatures examined (Fig. 4B). In contrast, the DNA binding activity of Msx1HD(R31P) was significantly reduced at 37°C compared to that at 20°C (Fig. 4B). Taken together, these findings indicate that the impaired DNA binding activity of Msx1(R31P) is a consequence of its reduced stability rather than the loss of the arginine side chain.
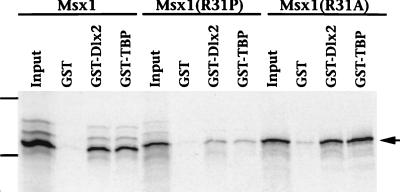
The Msx1 homeodomain also mediates functional interactions with various protein factors, including Dlx2 and TBP (Fig. 2) (33, 34). Therefore, we tested the ability of Msx1(R31P) to associate with these proteins in GST interaction assays (Fig. 5). In contrast to the strong interaction observed with Msx1 and Msx1(R31A), Msx1(R31P) exhibited a weak interaction with Dlx2 and TBP (Fig. 5). Since residues in helix II do not contribute directly to these protein-protein interactions (Fig. 2) (33, 4), we infer that the reduced ability of Msx1(R31P) to bind to Dlx2 and TBP is also due to its structural perturbation and/or reduced stability.
FIG. 5.
Msx1(R31P) interacts inefficiently with Dlx2 and TBP. The GST interaction assay was performed with 5 μg of GST, GST-Dlx2, or GST-TBP and 35S-labeled Msx1, Msx1(R31P), or Msx1(R31A), as shown. Immobilized proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by autoradiography (arrow). The input lane contains 20% (1 μl) of the total 35S-labeled protein (5 μl) used in the interaction assays. The dashes show the positions of the molecular mass standards (bovine serum albumin, 77 kDa; carbonic anhydrase, 33.8 kDa). Assays were performed a minimum of three times; representative data are shown.
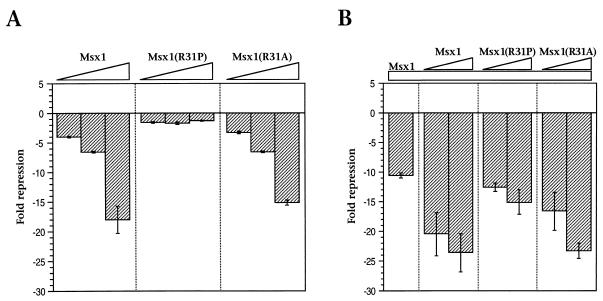
The biological actions of Msx1 are presumed to be mediated through its function as a transcriptional repressor, and the homeodomain is known to be essential for this activity (4, 5, 31, 33). Therefore, we examined whether Msx1(R31P) is capable of functioning as a transcriptional repressor in transient-transfection assays (Fig. 6). As we have found previously (15, 33), Msx1 and Msx1(R31A) exhibited potent repressor activity on an Msx1-responsive reporter in transfection assays (Fig. 6A). In contrast, Msx1(R31P) had no transcriptional repressor activity through this Msx1-responsive reporter (Fig. 6A) or through various other reporters (data not shown).
FIG. 6.
Msx1(R31P) lacks transcriptional repressor activity and does not influence the transcriptional repressor activity of Msx1. Transient-transfection assays were performed with C2C12 cells by using expression plasmids encoding the indicated proteins and a luciferase reporter plasmid containing an Msx1-responsive element (the WIP element [15]). In panel A, the amounts of each expression plasmid were 62.5, 125, or 250 ng (indicated by triangles). In panel B, each sample contained 125 ng of the Msx1 expression plasmid (indicated by the bar) alone or in combination with 125 or 250 ng of the Msx1, Msx1(R31P), and Msx1(R31A) expression plasmids (indicated by triangles). Data are represented as fold luciferase activity; a representative assay is shown with error bars indicating the difference between duplicates. Assays were performed a minimum of three times; representative data are shown.
Although Msx1(R31P) is not transcriptionally active, it may influence the repressor activity of Msx1. To test this possibility, we performed mixing experiments by using a constant amount of the Msx1 expression plasmid and increasing amounts of additional Msx1, Msx1(R31P), or Msx1(R31A) (Fig. 6B). A comparable level of repression was observed with equivalent amounts of additional Msx1 or Msx1(R31A) (Fig. 6B). In contrast, Msx1(R31P) did not modify the repressor activity of Msx1 significantly (Fig. 6B). In other mixing experiments, we have observed that Msx1(R31P) neither increased nor decreased the DNA binding activity of wild-type Msx1 in gel retardation assays (data not shown). Taken together, these biochemical studies demonstrate that Msx1(R31P) (i) has little or no activity on its own, (ii) does not perturb the actions of wild-type Msx1, and (iii) has no apparent novel activities.
Ectopic expression of Msx1, but not Msx1(R31P), alters chicken embryonic limb morphology.
To address whether these in vitro studies may also reflect the biological properties of Msx1(R31P), we misexpressed Msx1, Msx1(R31P), or Msx1(R31A) in embryonic chicken limb buds by using retroviral infection (Fig. 7). Clearly, there are differences between tooth and limb development that are exemplified by the expression pattern and function of Msx1 in these tissues. In particular, Msx1 expression is more dynamic in the developing tooth than in the limb, and the functional consequences of targeted gene disruption of Msx1 and the R31P mutation are evident in the tooth, but not in the limb (7, 23, 30). On the other hand, limb development is similar to tooth formation in that both require sequential and reciprocal signaling processes between epithelial and mesenchymal cell layers, and Msx1 expression in the limb and tooth mesenchyme is an important component in these signaling events (7). Therefore, the chicken limb bud assay provides a means of comparing the biological activities of Msx1 and Msx1(R31P), whose functions in the tooth may be more specialized.
Within the developing chicken limb bud, expression of endogenous Msx1 (GMsx1) is restricted to the outer margin of the mesoderm, termed the progress zone, which is in close proximity to the overlying ectoderm (Fig. 7B). We infected the prospective right forelimb with the Msx1-expressing retroviruses at the onset of limb bud outgrowth at stage 17 (13) and examined the consequences of this ectopic expression at later stages of limb development (stages 24 to 37) (Fig. 7). Ectopic expression of Msx1 throughout the wing (Fig. 7C) resulted in a reduction in the size of the wing on the infected side compared with the control, uninfected, wing (Fig. 7D and Table 1). In particular, the skeletal elements (humerus, radius, ulna, and digits) were reduced by an average of 10 to 20% in infected wings compared with uninfected, control wings (Table 1). We observed a reduction in the size and number of the feather buds in the infected wings compared with the control wings (Table 1). For the Msx1-infected wings, the mean infected/uninfected (right/left) ratio of length of the various skeletal elements was statistically different from the same ratio for control wings (P < 0.01 for all skeletal elements measured). A similar phenotype was produced by infecting the prospective forelimb field with retroviruses expressing GMsx1 at an earlier developmental stage (stage 10) (Fig. 7H). Since these wings were examined at a later stage (stage 39), the feather bud defect is more evident. These alterations of the skeletal elements and feather buds are likely to be a consequence of changes in the expression patterns of Msx1-responsive genes, as well as direct effects of Msx1 on cellular proliferation and differentiation (1a).
TABLE 1.
Effect of retrovirus infection on chicken wing cartilage pattern and feather germ formation
| Protein expressed | No. of embryos infected | No. of embryos with phenotype | Proportion with:
|
Cartilage length indexc
|
||||
|---|---|---|---|---|---|---|---|---|
| Feather germ defecta | Altered morphologyb | Humerus | Radius | Ulna | Digit III | |||
| Msx1 | 54 | 49 | 0.61 | 0.49 | 0.89 ± 0.01 | 0.88 ± 0.01 | 0.87 ± 0.01 | 0.83 ± 0.03 |
| Msx1(R31P) | 52 | 4d | 0.08 | 0 | 1.00 ± 0.01 | 1.01 ± 0.01 | 1.00 ± 0.01 | 1.01 ± 0.01 |
| Msx1(R31A) | 50 | 45 | 0.82 | 0.60 | 0.93 ± 0.01 | 0.93 ± 0.01 | 0.91 ± 0.01 | 0.86 ± 0.01 |
| APe | 6 | 0 | 0 | 0 | 1.00 ± 0.01 | 1.03 ± 0.01 | 1.00 ± 0.01 | 0.99 ± 0.02 |
| Msx1 + AP | 11 | 10 | 0.80 | 0.60 | 0.89 ± 0.03 | 0.86 ± 0.04 | 0.86 ± 0.04 | 0.85 ± 0.05 |
| Msx1 + Msx1(R31P) | 8 | 8 | 0.75 | 0.50 | 0.92 ± 0.02 | 0.90 ± 0.03 | 0.91 ± 0.03 | 0.83 ± 0.04 |
Feather germs were significantly reduced in size or absent along the posterior wing margin of the right infected wing compared with the contralateral uninfected (control) wing.
The angle made by the ulna and digit III of infected wings was often ≥180°, compared to the angle made by the ulna and digit III of the uninfected contralateral (control) wing, which was between >90° and <180°. Compare the wings shown in Fig. 7D as an example.
The cartilage length index is the ratio of the length of the infected right element to the length of the uninfected contralateral element and is expressed as the mean ± standard error.
Wing size of the Msx1(R31P)-infected embryos was never observed to change, but in a small number of embryos, there was some difference in the size of feather germs on the right infected wing compared with the contralateral uninfected (control) wing.
AP, human AP.
In contrast to the defects observed with retroviruses expressing Msx1 or Msx1(R31A), infection with the Msx1(R31P)-expressing retrovirus in the prospective forelimb did not produce any significant alteration in the size or shape of the infected wings (Fig. 7D to F, H, and I and Table 1). In fact, the wings infected with either the mouse or chicken Msx1(R31P)-expressing retroviruses were indistinguishable from the uninfected wings, suggesting that Msx1(R31P) is inactive in vivo as well as in vitro.
To examine whether Msx1(R31P) affects the activity of Msx1 in vivo, we infected the prospective forelimb with a 1:1 mixture of Msx1- and Msx1(R31P)-expressing retroviruses. In control experiments, we verified the efficacy of coinfection by using a 1:1 mixture of the Msx1- and AP-expressing retroviruses, which produced the Msx1 phenotype, as well as AP activity throughout the limb (Table 1 and data not shown). Coinfection with the Msx1- and Msx1(R31P)-expressing retroviruses produced a wing phenotype that is indistinguishable from that produced by infection with Msx1 alone (compare Fig. 7D and G and Table 1). These findings indicate that Msx1(R31P) does not affect the activity of Msx1 in vivo.
Conclusions.
In summary, we have found that Msx1(R31P) is partially or completely inactive in vitro and in vivo because of a perturbation of structure and decreased stability that results from the introduction of a proline residue within helix II of the homeodomain. Furthermore, Msx1(R31P) does not appear to influence the activity of wild-type Msx1, nor does it display any novel activities in the assays performed. We therefore propose that the phenotype in affected individuals with selective tooth agenesis is due to haploinsufficiency.
These findings raise several interesting questions regarding the mode of action of MSX1 and its particular importance in tooth morphogenesis. Msx1 is expressed throughout the tooth mesenchyme (17, 19, 20) as well as other embryonic regions (reviewed in reference 7), and complete loss of Msx1 function in mice results in a failure of tooth development (6, 23). Yet the missense mutation of MSX1 in humans selectively affects certain teeth, particularly the second premolars and third molars (30). Apparently, the reduced dosage of MSX1 in other embryonic regions, and even in other teeth, is tolerated, suggesting that morphogenesis of the affected teeth requires a greater dosage of MSX1. This idea is supported by the clinical observation that while individuals affected with selective tooth agenesis always fail to develop second premolars and third molars, flanking teeth are more variably affected (30). Alternatively, tooth morphogenesis in the affected family may be particularly susceptible to a reduced MSX1 dosage because of the specific effects of genetic background. It is noteworthy that mice heterozygous for Msx1 exhibit no abnormalities in tooth development (23). Although the absence of this phenotype may simply reflect the fact that mice lack premolars, it would be of interest to examine tooth morphogenesis in heterozygous mice in different genetic backgrounds.
Interestingly, a missense mutation in human MSX2, which is the cause of Boston-type craniosynostosis, also results in the substitution of a single residue within the homeodomain (16). In this case, substitution of a proline for a histidine renders the protein more stable than wild-type MSX2, and the resulting phenotype is believed to occur from a gain-of-function activity (18, 32). In combination with the present study, these findings highlight the significance of structural integrity and protein stability as a means of regulating the activities of proteins such as MSX1. Furthermore, this study demonstrates the importance of a detailed analysis of the biochemical and biological consequences of missense mutations for understanding the molecular bases of genetic disease.
ACKNOWLEDGMENTS
G.H., H.V., and A.J.B. contributed equally to this work.
We are indebted to Cliff Tabin (Harvard Medical School) for generous assistance with the chick retroviral expression studies and for many helpful discussions and Lee Niswander (Memorial Sloan Kettering) for the gift of the GMsx1 probe. We thank Michael Shen and Aaron Shakin (CABM) for helpful comments on the manuscript and Qing Li for assistance with the CD analysis.
This work was supported by NIH grant HD29446-06 to C.A.-S.; HHMI funding to C.E.S., J.G.S., and H.V.; predoctoral grants from the American Heart Association to G.H. and H.Z.; and a grant from the American Association of Orthodontists Foundation to H.V. C.A.-S. is a recipient of an NSF Young Investigator award.
REFERENCES
- 1.Alder A J, Greenfield N J, Fasman G D. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 1973;27:675–735. doi: 10.1016/s0076-6879(73)27030-1. [DOI] [PubMed] [Google Scholar]
- 1a.Bendall, A. J., G. Hu, and C. Abate-Shen. Unpublished data.
- 2.Burglin T R. A comprehensive classification of homeobox genes. In: Duboule D, editor. Guidebook to the homeobox genes. Oxford, United Kingdom: Oxford University Press; 1994. pp. 27–71. [Google Scholar]
- 3.Catron K M, Iler N, Abate C. Nucleotides flanking a conserved TAAT core dictate the DNA binding specificity of three murine homeodomain proteins. Mol Cell Biol. 1993;13:2354–2365. doi: 10.1128/mcb.13.4.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catron K M, Wang H, Hu G, Shen M M, Abate-Shen C. Comparison of Msx1 and Msx2 suggests a molecular basis for functional redundancy. Mech Dev. 1996;55:185–199. doi: 10.1016/0925-4773(96)00503-5. [DOI] [PubMed] [Google Scholar]
- 5.Catron K M, Zhang H, Marshall S C, Inostroza J A, Wilson J M, Abate C. Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. Mol Cell Biol. 1995;15:861–871. doi: 10.1128/mcb.15.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y P, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- 7.Davidson D. The function and evolution of Msx genes: pointers and paradoxes. Trends Genet. 1995;11:405–411. doi: 10.1016/s0168-9525(00)89124-6. [DOI] [PubMed] [Google Scholar]
- 8.Ebu Isaac V, Sciavolino P, Abate C. Multiple amino acids determine the DNA binding specificity of the Msx1 homeodomain. Biochemistry. 1995;34:7127–7134. doi: 10.1021/bi00021a026. [DOI] [PubMed] [Google Scholar]
- 9.Fekete D M, Cepko C L. Replication-competent retroviral vectors encoding alkaline phosphatase reveal spatial restriction of viral gene expression/transduction in the chick embryo. Mol Cell Biol. 1993;13:2604–2613. doi: 10.1128/mcb.13.4.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gehring W J, Qian Y Q, Billeter M, Furukubo-Tokunaga K, Schier A F, Resendez-Perez D, Affolter M, Otting G, Wüthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 11.Goff D J, Tabin C J. Analysis of Hoxd-13 and Hoxd-11 misexpression in chick limb bud reveals that Hox genes affect bone condensation and growth. Development. 1997;124:627–636. doi: 10.1242/dev.124.3.627. [DOI] [PubMed] [Google Scholar]
- 12.Graber L W. Congenital absence of teeth: a review with emphasis on inheritance patterns. J Am Dent Assoc. 1978;96:266–275. doi: 10.14219/jada.archive.1978.0054. [DOI] [PubMed] [Google Scholar]
- 13.Hamburger V, Hamilton H L. A series of normal stages in the development of the chick embryo. J Morphol. 1992;88:49–92. [PubMed] [Google Scholar]
- 14.Hughes S H, Greenhouse J J, Petropoulos C J, Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iler N, Rowitch D H, Echelard Y, McMahon A, Abate-Shen C. A single homeodomain binding site confers spatial restriction of Wnt-1 expression in the developing brain. Mech Dev. 1995;53:87–96. doi: 10.1016/0925-4773(95)00427-0. [DOI] [PubMed] [Google Scholar]
- 16.Jabs E W, Müller U, Li X, Ma L, Luo W, Haworth I S, Klisak I, Sparkes R, Warman M L, Mulliken J B, Snead M L, Maxson R. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75:443–450. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- 17.Jowett A K, Vainio S, Ferguson M W, Sharpe P T, Thesleff I. Epithelial-mesenchymal interactions are required for msx 1 and msx 2 gene expression in the developing murine molar tooth. Development. 1993;117:461–470. doi: 10.1242/dev.117.2.461. [DOI] [PubMed] [Google Scholar]
- 18.Ma L, Golden S, Maxson R. The molecular basis of Boston type craniosynostosis: the Pro148>His mutation in the N-terminal arm of the Msx2 homeodomain stabilizes DNA binding without altering nucleotide sequence preferences. Hum Mol Genet. 1996;12:1915–1920. doi: 10.1093/hmg/5.12.1915. [DOI] [PubMed] [Google Scholar]
- 19.MacKenzie A, Ferguson M W J, Sharpe P T. Hox-7 expression during murine craniofacial development. Development. 1991;113:601–611. doi: 10.1242/dev.113.2.601. [DOI] [PubMed] [Google Scholar]
- 20.MacKenzie A, Leeming G L, Jowett A K, Ferguson M W J, Sharpe P T. The homeobox gene Hox 7.1 has specific regional and temporal expression patterns during early murine craniofacial embryogenesis, especially tooth development in vivo and in vitro. Development. 1991;111:269–285. doi: 10.1242/dev.111.2.269. [DOI] [PubMed] [Google Scholar]
- 21.Marquardt D W. An algorithm for least-squares estimation of non-linear parameters. J Soc Ind Appl Math. 1963;11:431–441. [Google Scholar]
- 22.Richardson J S, Richardson D C. Principles and patterns of protein conformation. In: Fasman G D, editor. Prediction of protein structure and principles of protein conformation. New York, N.Y: Plenum Press; 1989. pp. 1–99. [Google Scholar]
- 23.Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–355. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 24.Scholtz J M, Qian H, York E J, Stewart J M, Baldwin R L. Parameters of helix-coil transition theory for alanine based peptides of varying chain length in water. Biopolymers. 1991;31:1463–1470. doi: 10.1002/bip.360311304. [DOI] [PubMed] [Google Scholar]
- 25.Shang Z, Ebu Isaac V, Li H, Patel L, Catron K M, Curran T, Montelione G T, Abate C. Design of a “minimAl” homeodomain: the N-terminal arm modulates DNA binding affinity and stabilizes homeodomain structure. Proc Natl Acad Sci USA. 1994;91:8373–8377. doi: 10.1073/pnas.91.18.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharpe P T. Homeobox genes and orofacial development. Connect Tissue Res. 1995;32:17–25. doi: 10.3109/03008209509013701. [DOI] [PubMed] [Google Scholar]
- 27.Thesleff I, Nieminen P. Tooth morphogenesis and cell differentiation. Curr Opin Cell Biol. 1996;8:844–850. doi: 10.1016/s0955-0674(96)80086-x. [DOI] [PubMed] [Google Scholar]
- 28.Thesleff I, Sahlberg C. Growth factors as inductive signals regulating tooth morphogenesis. Semin Cell Dev Biol. 1996;7:185–193. [Google Scholar]
- 29.Thesleff I, Sharpe P. Signalling networks regulating dental development. Mech Dev. 1997;67:111–123. doi: 10.1016/s0925-4773(97)00115-9. [DOI] [PubMed] [Google Scholar]
- 30.Vastardis H, Karimbux N, Guthua S W, Seidman J G, Seidman C E. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- 31.Woloshin P, Song K, Degnin C, Killary A M, Goldhamer D J, Sassoon D, Thayer M J. MSX1 inhibits MyoD expression in fibroblast × 10T1/2 cell hybrids. Cell. 1995;82:611–620. doi: 10.1016/0092-8674(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 32.Wu L, Wu H, Sangiorgi F, Wu N, Bell J R, Lyons G E, Maxson R. Miz1, a novel zinc finger transcription factor that interacts with Msx2 and enhances its affinity for DNA. Mech Dev. 1997;65:3–17. doi: 10.1016/s0925-4773(97)00032-4. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Catron K M, Abate-Shen C. A role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TBP interaction and transcriptional repression. Proc Natl Acad Sci USA. 1996;93:1764–1769. doi: 10.1073/pnas.93.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen M M, Abate-Shen C. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]