Abstract
Protein kinase C (PKC) functions are essential for synaptic plasticity, learning, and memory. However, the roles of specific members of the PKC family in synaptic function, learning, and memory are poorly understood. Here, we investigated the role of individual PKC homologs for synaptic plasticity in Caenorhabditis elegans and found a differential role for pkc-2 and tpa-1, but not pkc-1 and pkc-3 in associative olfactory learning and memory. More specifically we show that PKC-2 is essential for associative learning and TPA-1 for short-term associative memory (STAM). Using endogenous labeling and cell-specific rescues, we show that TPA-1 and PKC-2 are required in AVA for their functions. Previous studies demonstrated that olfactory learning and memory in C. elegans are tied to proper synaptic content and trafficking of AMPA-type ionotropic glutamate receptor homolog GLR-1 in the AVA command interneurons. Therefore, we quantified synaptic content, transport, and delivery of GLR-1 in AVA and showed that loss of pkc-2 and tpa-1 leads to decreased transport and delivery but only a subtle decrease in GLR-1 levels at synapses. AVA-specific expression of both PKC-2 and TPA-1 rescued these defects. Finally, genetic epistasis showed that PKC-2 and TPA-1 likely act in the same pathway to control GLR-1 transport and delivery, while regulating different aspects of olfactory learning and STAM. Thus, our data tie together cell-specific functions of 2 PKCs to neuronal and behavioral outcomes in C. elegans, enabling comparative approaches to understand the evolutionarily conserved role of PKC in synaptic plasticity, learning, and memory.
Keywords: AMPA receptors, associative memory, C. elegans, PKC, synaptic plasticity, synaptic trafficking
Significance Statement
The protein kinase C family of kinases are essential signaling elements of synaptic plasticity. However, how different members interact to contribute to learning and memory and how this relates to cellular functions is technically challenging in vertebrate models. Here, using Caenorhabditis elegans, we describe an approach combining behavior and cellular analysis for TPA-1 and PKC-2, one novel and one classical PKC isoform. Our findings show that changes in learning and memory due to the loss of TPA-1 and PKC-2 can be related to changes in the dynamics of synaptic AMPA receptors. Thus, our approach using C. elegans offers the opportunity to better understand how the molecular functions of PKCs relate to their effect on learning and memory.
Introduction
Members of the protein kinase C family of serine/threonine kinases play a prominent role in neuronal function and synaptic plasticity across animal models. They are divided into three subgroups, which include the classical PKC (cPKC; a, b1, b2, and g), the novel PKC (nPKC; d, e, h, q, and m), and the atypical PKC (aPKC; z, i, and l) subfamilies (Stabel and Parker, 1991; Newton, 1995; Sun and Alkon, 2014). PKCs are widely found in the CNS, and many isoforms are simultaneously translated into the same cells with variations in their expression levels. Furthermore, it is thought that besides tissue and cellular location, signal timing is also a crucial determinant of their activation pattern (Sun and Alkon, 2014). The classical cPKCs (PKCa, b, and g) require calcium and diacylglycerol (DAG) for activation, the novel nPKCs require only DAG, and the atypical PKCs do not require either calcium or DAG. The classical and novel PKCs have a conserved catalytic domain and regulatory domains whereas the atypical PKCs lack calcium binding and part of the catalytic domain. All the PKCs have an N-terminal motif that acts as an autoinhibitory domain that can be cleaved leading to the formation of a persistently active kinase (PKM). The variation in domains and activation conditions of the different PKCs are thought to be important for their function in synaptic transmission and plasticity (Sun and Alkon, 2014).
Long-lasting changes in excitatory synaptic transmission, including long-term potentiation (LTP) and long-term depression (LTD), are cellular hallmarks of learning and memory (Malenka and Nicoll, 1999; Malinow et al., 2000; Man et al., 2000; Xia et al., 2000; Sheng and Lee, 2001; Redondo and Morris, 2011; Choquet and Triller, 2013; Huganir and Nicoll, 2013). Stable insertion or removal of the α-amino-3-hydroxy-5-methylsisoazole-4-propionic acid receptor (AMPAR) into or from postsynaptic sites (Hayashi et al., 2000; Chung et al., 2003; Park et al., 2004; Boehm et al., 2006; Hanus et al., 2014) is considered to be the main molecular mechanisms behind LTP and LTD. Synaptic plasticity combines calcium and protein kinase signaling, implicating phosphorylation of residues in AMPARs as key modulators of their insertion and removal at postsynaptic sites (Roche et al., 1996; Lisman and Zhabotinsky, 2001; Passafaro et al., 2001; Lisman et al., 2002; Esteban et al., 2003; Boehm et al., 2006; Lu et al., 2010; Sathler et al., 2021). In particular, PKC phosphorylation of the highly conserved GluA1 C-terminal tail is critical for hippocampal LTP (Boehm et al., 2006) and cerebellar LTD (Chung et al., 2003).
Although good molecular models for the activation and downstream phosphorylation of the targets exist for the single PKC isoforms, the models become unreliable when investigating how the different isoforms contribute to different processes of synaptic plasticity in vertebrate experimental models. For each PKC, there are several genes and splice forms that partially overlap and compensate for each other’s function (Sossin, 2007); thus, direct synaptic and molecular observation of an individual PKC family member is difficult in complex vertebrate brains.
The transparent genetic model Caenorhabditis elegans, with a complete synaptic connectome and only four PKC genes and few isoforms, offers the opportunity to better dissect the role of different PKCs in excitatory synaptic function as well as short and long-term memory. Excellent prior studies have contributed to our understanding of the molecular function of PKCs (Sano et al., 1995; Islas-Trejo et al., 1997) as well as attributing roles of different PKC isoforms to certain behaviors (Okochi et al., 2005; Kindt et al., 2007; Hyde et al., 2011; Land and Rubin, 2017). Thus, PKC-1 function is important for thermotaxis learning and memory and mechanosensory adaptation (Hyde et al., 2011) and PKC-2 for thermotaxis as well as salt (NaCl) learning and memory (Edwards et al., 2012; Land and Rubin, 2017; Hiroki and Iino, 2022), and finally TPA-1 has been implicated in salt chemotaxis learning (Hiroki and Iino, 2022). However, little is known about the specific roles of pkc-1, 2, and 3 or tpa-1 in associative olfactory short- and long-term learning and memory. In C. elegans, AMPA (GLR-1 to GLR-4) and NMDA (NMR-1 and NMR-2) subtypes of receptors are essential for learning and memory (Rose et al., 2003; Kano et al., 2008b) and critical for olfactory associative memory (Kano et al., 2008a; Stetak et al., 2009; Vukojevic et al., 2012; Hadziselimovic et al., 2014). Synaptic GLR-1 levels, insertion, and transport are regulated by calcium signaling, CaMKII, and MAPK (Hoerndli et al., 2015b, 2022; Doser et al., 2020). However, the role of PKC in regulating AMPAR trafficking in C. elegans has not been studied.
Therefore, we first investigated the endogenous expression of the different PKC genes using clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-driven tagging followed by analyzing the role of specific isoforms in olfactory learning and memory. We found that the PKC-2 function is essential for associative olfactory learning while the TPA-1 function is critical for short-term associative memory (STAM). In addition, PKC-1 seems to be important only in olfaction in AWA. Neither tpa-1 nor pkc-2 were required for long-term associative olfactory memory. Second, we analyzed the synaptic AMPA receptor levels and transport in tpa-1(lf) and pkc-2(lf) mutants, which show a decrease in transport and total synaptic levels. Thus, our study defines the different roles of individual PKCs in chemosensation and olfactory associative memory in C. elegans and links their activity to GLR-1 receptor dynamics.
Materials and Methods
Strains
C. elegans strains were maintained on nematode growth media (NGM) and fed with the E. coli strain OP50 at 20°C (Brenner, 2003). Strains used in these experiments contained alleles presented in Table 1.
Table 1.
List of genetic alleles, the corresponding gene, the effect of the mutation, and reported functional change along with original references mentioning the allele
| Gene | Allele | Mutation | Functional change | References |
|---|---|---|---|---|
| glr-1 | ky176 | Premature stop | Truncated, unfunctional receptor | Maricq et al. (1995) |
| pkc-1 | nj3 | W218 stop | Premature stop | WormBase, Okochi et al. (2005) |
| pkc-1 | ok563 | 1.6 kbp deletion | Premature stop | WormBase, Hyde et al., (2011) |
| pkc-2 | ok238 | 2.3 kbp deletion with insertion of ttttcag | Premature stop | Kim and Horvitz (1990) and Islas-Trejo et al. (1997) |
| pkc-2 | utr58 | YPET::AID::FLAG insertion | C-terminal tag in isoforms b, c, and f | This study |
| pkc-3 | utr59 | YPET::AID::FLAG insertion | C-terminal tag | This study |
| tpa-1 | k501 | Substitution | P544/407S | Tabuse et al. (1989) and Ziegler et al. (2009) |
| tpa-1 | utr71 | YPET::AID::FLAG insertion | C-terminal tag | This study |
akIs201 V, rig-3p::SEP::mCherry::glr-1;csfEx21, flp-18p::PKC2b::3′UTR let858 + egl-20p::nls::DsRed;csfEx;, pkc-1(nj1), utrEx70[odr-10p::pkc-1 wt; sur-5p::dsRed; myo-2p::mCherry], pkc-2(ok328), utrEx46[WRM0639dD11;sur-5dsRed], pkc-2(ok328), utrEx131[rig-3p::pkc-2b+;myo-2p::mCherry], tpa-1(k501), utrEx133[rig-3p::tpa-1+;myo-2p::mCherry].
Generation of transgenic strains
Transgenic strains were created by microinjection using egl-20p::nls::DsRed, sur-5p::dsRed, or myo-2p::mCherry as injection markers. Fluorescent marker insertions using SEC (self-excising cassette) CRISPR were performed as described (Dickinson et al., 2015). The sgRNA corresponding sequence for pkc-2 (aattaaaaataatatgatca), pkc-3 (tgggactcgggggatggtgg), or tpa-1 (cgacgacttcacgaacgaga) was fused into pDD162 vector using q5 in vitro mutagenesis kit (New England Biolabs).
CRISPR repair constructs were generated by inserting 500–800 bp long, left and right homology arms of the target gene flanking the protospacer adjacent motif site into SpeI/AvrII sites of the pJW1584 vector. The fluorescent protein insertion for both pkc-2 and tpa-1 was on the C-terminal end prior to the stop codon. Following insertion, the SEC cassettes were eliminated using a heat shock as described (Dickinson et al., 2015), and worms were twice backcrossed to the N2 strain. For the AVA expression constructs, pkc-2 or tpa-1 cDNA and the 3′UTR of the genes were fused to a 7.7 kb long rig-3 promoter using NEBuilder HiFi DNA assembly master mix (New England Biolabs). For pkc-1 rescue in AWA, the 1.2 kb odr-10 promoter was fused to the 4.6 kb genomic region containing the PKC-1 gene.
Confocal microscopy
The gene expression patterns were recorded with a Zeiss LSM 880 confocal microscope using 514 nm (YPET) and 633 nm (DiD) excitation lasers with a 25×/0.8 oil or a 63×/1.4 objective. DiD (DiIC18(5); 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine,4-chlorobenzenesulfonate salt) is a lipophilic carbocyanine dye similar to DiI but with longer absorption and emission wavelengths). Tilled Z-stacked images were processed and merged using Zeiss Zen (edition black). Imaging was also carried out on a spinning disc confocal microscope (Olympus IX83) equipped with 488 and 561 nm excitation lasers (Andor ILE Laser Combiner). Images were captured using an Andor iXon Ultra EMCCD camera through either a 10×/0.40 or a 100×/1.40 oil objective (Olympus). Devices were controlled remotely for image acquisition using MetaMorph 7.10.1 (Molecular Devices).
Transport imaging and analysis
All transport imaging was conducted on strains containing akIs201 in the glr-1 null background (ky176). One-day-old hermaphrodite adults from these strains were mounted on a 10% agarose pad with 1.6 µl of a mixture containing equal measures of polystyrene beads (Polybead, Polysciences) and 30 mM muscimol (MP Biomedicals). The worm was positioned so that the AVA interneurons were in close proximity to the coverslip through which the AVA neurites would be imaged. Once the AVAs were located using the 100× objective and a 561 nm excitation laser, a proximal section of the AVA immediately after the second chiasma of AVAR and AVAL was photobleached using a 0.5 W output and a 1 s pulse time of 3 W, 488 nm coherent solid-state laser (Genesis MX MTM). The photobleaching laser was targeted to a defined portion of AVA using a Mosaic II digital mirror device (Andor Mosaic 3) controlled through MetaMorph. Immediately following photobleaching, a 500-frame image stream was collected in a single z-plane with the 561 nm excitation laser with a 100 ms exposure time. Kymographs were generated using the kymograph tool in MetaMorph with a 20-pixel line width as previously reported (Hoerndli et al., 2013). Transport quantification, stops, and velocities were analyzed using the ImageJ plugin KymoAnalyzer (Neumann et al., 2017).
Fluorescence recovery after photobleaching FRAP
Strains containing akIs201 were mounted for imaging as described above. For photobleaching and image acquisition, a proximal section of the AVA ∼80 µm immediately after the second chiasma of AVAR and AVAL was localized. This imaging position was memorized and recorded using MetaMorph’s stage position memory function. Then, an image stack was acquired using the 561 nm and then the 488 nm excitation laser (500 ms exposure) around the AVA process (20 images were taken every 0.25 µm starting 2.5 µm below to 2.5 µm above the process). A ∼80 µm proximal and distal section of the AVA flanking the imaging region was then photobleached using a 3 W, 488 nm coherent solid-state laser (Genesis MX MTM) set to 0.5 W output and a 1 s pulse time. Immediately following using the recorded stage position, the imaging region was photobleaching using the same condition, and after photobleaching, two image stacks (with 561 nm and then 488 nm excitation) were acquired for the 0 min time point. This was repeated at 2, 4, 8, and 16 min following the photobleaching of the imaging region. Image stacks from all time points were converted to maximum projections using MetaMorph’s stack arithmetic function. The average fluorescence in the imaging region at each time point was analyzed using the region measurement tool in ImageJ 1.51 s (Java 1.8). The background fluorescence (i.e., outside of the AVA) from each maximum projection was then subtracted from the average fluorescence of the imaging region. The resulting fluorescence from the maximum projections 0 min following photobleaching was subtracted from the fluorescence values of all subsequent time points. These values were then compared with those from images taken before photobleaching to determine the percent of signal recovery for each channel within the imaging region.
Behavioral assays
Chemotaxis of the different mutant worms was tested using 1% or 0.1% benzaldehyde (benzald.), isoamyl alcohol (IAA), or diacetyl (DA) as described previously (Bargmann et al., 1993). Briefly, 50–200 1-d-old hermaphrodite worms were placed in the center of a 10 cm chemotaxis plate (CTX) [5 mM KH2PO4/K2HPO4 (pH 6.0), 1 mM CaCl2, 1 mM MgSO4, 2% agar] with 1 µl of 10% sodium azide and either odorant in EtOH or EtOH alone on each side of the plate. Worms were allowed to move freely for 1 h at room temperature, and the chemotaxis index was defined as follows: CI = (number of animals attractant − number of animals EtOH)/total number of animals) as described (Bargmann et al., 1993).
Worm motility rates upon different treatments were tested as described (Sawin et al., 2000; Mohri et al., 2005). Briefly, worms were grown under uncrowded conditions with or without food on CTX plates for 1 h. Individual worms were then transferred onto fresh 6 cm assay plates seeded with OP50 bacteria, and then 2 min after transfer, the number of body bends of the animals from each strain was counted for 1 min.
For spontaneous reversals, worms were transferred on fresh NGM plates seeded with OP50 and allowed to recover for 2 min, and spontaneous reversals were monitored for 5 min under a dissecting microscope.
The nose touch assay was performed as described in Kaplan and Horvitz (Kaplan and Horvitz, 1993), using well-fed young adult worms tested by placing an eyelash in front of a moving worm allowing spontaneous collision of the animal to the eyelash.
Short-term and long-term associative memory (STAM and LTAM, respectively). Assays were conducted as described previously (Hadziselimovic et al., 2014). Briefly, animals were synchronized using the egg preparation method (Stiernagle, 2006) and then grown on 10 cm NGM plates on OP50. After collecting 1-d-old hermaphrodite adult animals and washing them three times in CTX buffer, animals were subjected to different treatment conditions corresponding to either naive, 1× conditioning, STAM, 2× conditioning, or LTAM, as described below. For all conditions, the chemotaxis of the animals after treatment was tested the following way: 50–200 worms were placed in the center of 10 cm chemotaxis plates [CTX 5 mM KH2PO4/K2HPO4 (pH 6.0), 1 mM CaCl2, 1 mM MgSO4, 2% agar] with 1 µl of 10% sodium azide and either 0.1% DA in EtOH or EtOH alone on each side of the plate. After 1 h at room temperature with closed lids, animals immobilized at the 0.1% DA, at the EtOH, and outside of the spots were counted. The chemotaxis index was calculated as follows: CI = (number of animals at 0.1%DA − number of animals at EtOH)/total number of animals) as described (Bargmann et al., 1993). Counting of animals on the plates was done blind to genotype. The chemotaxis assay was conducted in triplicate for each genotype for each condition and repeated a minimum of three times. Data from all chemotaxis assays for each condition and genotype were combined to generate the average CI per genotype and condition. For STAM, 50–200 worms were conditioned for 1 h without food in the presence of 2 µl DA on the top of the lid of a 10 cm CTX plate. Animals were then either tested immediately (1× conditioning) or left for 1 h on NGM with OP50 and tested for STAM (1 h memory). For LTAM, the 1 h conditioning was repeated twice with a 30 min interval in between conditioning sessions where worms were able to roam on plates with food. After the second conditioning, worms were either tested immediately for chemotaxis (conditioned) or put on NGM plates with OP50 for 24 h before LTAM was tested (24 h memory). For PMA treatment, worms prior to conditioning were incubated for 15 min in M9 supplemented with the given concentration of PMA dissolved in DMSO. The DMSO concentration was set to 0.5% for all conditions. In addition to the preincubation step, PMA was applied in all washing steps during the conditioning procedure.
Image presentation and data analysis
All images were acquired under nonsaturating conditions. Quantification of GLR-1 transport, FRAP, and calcium imaging is described in the appropriate sections above. Representative images shown were chosen and processed following analysis only to the extent necessary to appreciate the corresponding quantifications. Image processing Red, Green and Blue (RGB) colors, cropping, adjustment of brightness, and contrast) was performed in Photoshop (21.1.1). For FRAP images, the mCherry signal of SEP::mCherry::GLR-1 is shown in magenta for colorblind vision. This was achieved in Photoshop by duplicating the 561 nm information to create the red and blue channels of the RGB image which was then merged to create the magenta image as published previously (Hoerndli et al., 2013). All images in each panel were identically processed.
Experimental design and statistical analyses
All experiments were performed using 1-d-old adult hermaphrodite C. elegans animals as determined by a single row of eggs and picking as precisely identifiable L4 stage larvae 24 h before imaging and behavior experiments. All mutant strains were backcrossed with N2 wild-type animals at least two times. All imaging reagents such as SEP::mCherry::GLR-1 and GCAMP6f were crossed into strains carrying genetic mutations in the same manner, verifying the presence of the knock-out glr-1 allele ky176 and genetic mutations using PCR genotyping on at least two generations. Primer sequences are available on request.
For statistical analysis, all datasets were screened for outliers using a Thompson tau test. For datasets including only two experimental groups, statistical significance was tested using a two-tailed Student’s t test. For datasets comparing more than two experimental groups, a one-way Brown–Forsythe ANOVA with a Dunnett’s correction for multiple comparisons was used, and if the datasets were not distributed in a Gaussian manner, a one-way ANOVA with Kruskal–Wallis test and Dunn’s correction was used. FRAP differences between groups were determined using an extra-sum-of-squares F statistic testing the hypothesis of one fit for both control and mutant. Behavioral assays were analyzed using mixed model two-way ANOVA with Bonferroni’s correction for multiple testing. All statistical analyses were performed using Prism software (versions 8 and 9). In all cases, post hoc tests were performed and were reported only if the initial F test was significant. The details of every statistical test, for each panel of every figure, are reported in Table 2.
Table 2.
Summary of all statistical tests performed
| Dataset | Model | Condition tested | Levels | Test result | df | p-value |
|---|---|---|---|---|---|---|
| Figure 2A | ANOVAa | Genotype | Wild type, pkc-1(lf):Ex[odr-10::pkc1+] | F = 7.39 | 1|87 | 0.0079 |
| ANOVAa | Treatment | Naive, cond., 1 h delay | F = 156.35 | 2|87 | <1E−15 | |
| ANOVAa | Genotype::treatment | F = 1.884 | 2|87 | 0.158 | ||
| Figure 2D | Two-tailed t test | Genotype | Wild type, pkc-1(lf) | t = 0.3998 | 12 | 0.6963 |
| Figure 3A | ANOVAb | F = 38.497 | 11|81 | <1E−15 | ||
| Figure 3B | ANOVAb | F = 10.612 | 11|96 | 1.693E−12 | ||
| Figure 3C | ANOVAa | Genotype | Wild type, pkc-2(lf) | F = 1.5721 | 1|119 | 0.21236 |
| ANOVAa | Treatment | Fed/food, fed/empty, starved/food | F = 261.345 | 2|119 | <1E−15 | |
| Figure 3D | ANOVAa | Genotype | Wild type, tpa-1(lf) | F = 0.2157 | 1|113 | 0.64323 |
| ANOVAa | Treatment | Fed/food, fed/empty, starved/food | F = 135.3511 | 2|113 | <1E−15 | |
| Figure 4A | ANOVAa | Treatment | Naive, cond., 1 h delay | F = 662.8892 | 2|99 | <1E−15 |
| ANOVAa | Genotype | Wild type, pkc-2(lf), pkc-2(lf);Ex[rig-3P::pkc-2+] | F = 67.4533 | 2|99 | <1E−15 | |
| ANOVAa | Treatment::genotype | F = 6.6336 | 4|99 | 9.0049E−05 | ||
| Figure 4B | ANOVAa | Treatment | Naive, cond, 1 h delay | F = 832.2073 | 2|98 | <1E−15 |
| ANOVAa | Genotype | Wild type, tpa-1(lf), tpa-1(lf);Ex[rig-3P::tpa-1+] | F = 16.2933 | 2|98 | 7.7809E−07 | |
| ANOVAa | Treatment::genotype | F = 11.4409 | 4|98 | 1.1624E−07 | ||
| Figure 4C | ANOVAa | Treatment | Naive, cond., 1 h delay | F = 1,034.7553 | 2|130 | <1E−15 |
| ANOVAa | Genotype | Wild type, tpa−1(lf), pkc-2(lf), tpa-1(lf);pkc-2(lf) | F = 42.1245 | 3|130 | <1E−15 | |
| ANOVAa | Treatment::genotype | F = 8.0675 | 6|130 | 2.0498E−07 | ||
| Figure 4D | ANOVAa | Treatment | Naive, cond, 24 h delay | F = 243.2827 | 2|97 | <1E−15 |
| ANOVAa | Genotype | Wild type, pkc-2(lf) | F = 4.3385 | 1|97 | 0.03989 | |
| ANOVAa | Treatment::genotype | F = 0.2521 | 2|97 | 0.77666 | ||
| Figure 4E | ANOVAa | Treatment | Naive, cond., 24 h delay | F = 326.904 | 2|92 | <1E−15 |
| ANOVAa | Genotype | Wild type, tpa-1(lf) | F = 1.2902 | 1|92 | 0.25896 | |
| ANOVAa | Treatment::genotype | F = 0.7107 | 2|92 | 0.49396 | ||
| Figure 4F | ANOVAa | Treatment | Naive, cond., 24 h delay | F = 577.2622 | 2|63 | <1E−15 |
| ANOVAa | Genotype | Wild type, pkc-2(lf);tpa-1(lf) | F = 0.1851 | 1|63 | 0.66846 | |
| ANOVAa | Treatment::genotype | F = 2.4587 | 2|63 | 0.093723 | ||
| Figure 5A | ANOVAa | Treatment | None, 0.001 µg/ml PMA, 0.01 µg/ml PMA, 0.1 µg/ml PMA, 1 µg/ml PMA | F = 8.279 | 2|119 | 2.0736E−06 |
| ANOVAa | Genotype | Naive, cond. | F = 244.79 | 1|119 | <1E−15 | |
| ANOVAa | Treatment::genotype | F = 5.96 | 2|119 | 1.688E−07 | ||
| Figure 5B | ANOVAa | Treatment | None, 0.001 µg/ml PMA, 0.01 µg/ml PMA, 0.1 µg/ml PMA, 1 µg/ml PMA | F = 2.25287 | 4|68 | 0.016788 |
| ANOVAa | Genotype | Naive, cond. | F = 372.1359 | 1|68 | <1E−15 | |
| ANOVAa | Treatment::genotype | F = 2.18363 | 4|68 | 0.080013 | ||
| Figure 5C | ANOVAa | Treatment | None, 0.001 µg/ml PMA, 0.01 µg/ml PMA, 0.1 µg/ml PMA, 1 µg/ml PMA | F = 5.49726 | 4|124 | 0.00041567 |
| ANOVAa | Genotype | Naive, cond. | F = 728.77 | 1|124 | <1E−15 | |
| ANOVAa | Treatment::genotype | F = 6.1731 | 4|124 | 0.00014561 | ||
| Figure 5D | ANOVAb | F = 85.913 | 5|66 | <1E−15 | ||
| Figure 5E | ANOVAa | Treatment | Naive, conditioned, 1 h delay + none, 1 h delay + 0.01 µg/ml PMA, 1 h delay + 0.1 µg/ml PMA, 1 h delay + 1 µg/ml PMA | F = 23.6468 | 5|105 | 0.00000408 |
| ANOVAa | Genotype | Wild type, pkc-2(lf) | F = 68.3975 | 1|105 | <1E−15 | |
| ANOVAa | Treatment::genotype | F = 3.78 | 5|105 | 0.0034572 | ||
| Figure 5F | ANOVAa | Treatment | Naive, conditioned, 1 h delay + none, 1 h delay + 0.001 µg/ml PMA, 1 h delay + 0.01 µg/ml PMA, 1 h delay + 0.1 µg/ml PMA, 1 h delay + 1 µg/ml PMA | F = 5.49726 | 6|159 | 0.000126 |
| ANOVAa | Genotype | Wild type, tpa-1(lf) | F = 728.77 | 1|159 | <1E−15 | |
| ANOVAa | Treatment::genotype | F = 6.1731 | 6|159 | 0.0001055 | ||
| Figure 5G | ANOVAa | Treatment | Naive, conditioned, 1 h delay + none, 1 h delay + 0.1 µg/ml PMA, 1 h delay + 1 µg/ml PMA | F = 146.2974 | 4|110 | <1E−15 |
| ANOVAa | Genotype | Wild type, pkc-2(lf);tpa-1(lf) | F = 103.93275 | 1|110 | <1E−15 | |
| ANOVAa | Treatment::genotype | F = 12.36205 | 4|110 | 2.4499E−08 | ||
| Figure 6C | Kruskal–Wallis test | H = 13.25 | 6|127 | 0.0211 | ||
| Figure 6D | ANOVAb | F = 4.236 | 5|121 | 0.0014 | ||
| Figure 6E | Fisher’s exact test | Genotype | Wild type, pkc-1(lf) | 0.02355 | ||
| Figure 6F | Two-tailed t test | Genotype | Wild type, pkc-1(lf) | t = 7.62793 | 9 | 0.00000149 |
| Figure 7C | Kruskal–Wallis test | H = 35.56 | 6|108 | <0.0001 | ||
| Figure 7D | Extra-sum of squares | One curve fits both | Control versus pkc-2(lf) | F = 49.34 | 15 | <0.0001 |
| Extra-sum of squares | One curve fits both | Control versus tpa-1(lf) | F = 2.549 | 15 | 0.074 | |
| Extra-sum of squares | One curve fits both | Control versus pkc-2(lf);tpa-1(lf) | F = 5.671 | 15 | 0.0034 | |
| Figure 7E | Kruskal–Wallis test | H = 39.87 | 4|6,512 | <0.0001 | ||
| Figure 8C | Extra-sum of squares | One curve fits both | Control versus pkc-2(lf) | F = 0.2575 | 3|96 | 0.8558 |
| Extra-sum of squares | One curve fits both | Control versus tpa-1(lf) | F = 3.065 | 2|88 | 0.0517 | |
| Extra-sum of squares | One curve fits both | Control versus pkc-2(lf);tpa-1(lf) | F = 5.742 | 2|91 | 0.0045 | |
| Figure 8D | Extra-sum of squares | One curve fits both | Control versus pkc-2(lf) | F = 0.4809 | 2|98 | 0.6197 |
| Extra-sum of squares | One curve fits both | Control versus tpa-1(lf) | F = 7.769 | 2|92 | 0.0008 | |
| Extra-sum of squares | One curve fits both | Control versus pkc-2(lf);tpa-1(lf) | F = 3.899 | 2|89 | 0.0238 |
Figure panels to which the dataset corresponds to the statistical model used, conditions tested and levels to which the test was applied to, test statistic, degrees of freedom (df), and uncorrected exact p-values.
Two-way ANOVA.
One-way ANOVA.
Results
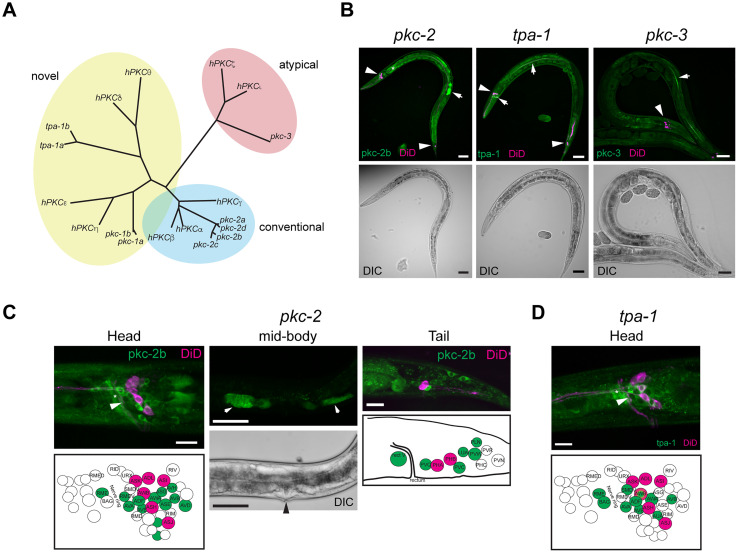
In vertebrates, the PKC family is encoded by nine genes (Callender and Newton, 2017) while the C. elegans genome contains only four vertebrate homologs (pkc-1, pkc-2, pkc-3, and tpa-1; Tabuse, 2002). A homology analysis using Clustal Omega shows that pkc-2 is more closely related to the classical PKC isozymes (Fig. 1A), whereas pkc-1 and tpa-1 belong to the novel PKC isozymes, and pkc-3 is related to the atypical PKC isozyme group (Fig. 1A). pkc-1 expression has been reported previously, including in command interneurons, and rescue experiments have shown pkc-1 to be important for mechanosensory transduction and neuropeptide release (Okochi et al., 2005; Kindt et al., 2007; Hyde et al., 2011; Jia and Sieburth, 2021). pkc-2, tpa-1, and pkc-3 expression, however, has not been documented in adult worms, so first we performed endogenous CRISPR-mediated YPET tagging of these three genes to observe their expression pattern in the nervous system of C. elegans (Fig. 1B–D, Materials and Methods Table 1). PKC-2b expression was observed in many head and tail neurons including command interneurons (RME, RMDV, AVA, ADF, AVE, AVD, AVB, AVH, ASG, ASE, AWA, PVC, PVQ, PVW, LUA, and PLN). PKC-3 was not detectably expressed in any neurons (Fig. 1B), and TPA-1 was exclusively expressed in neurons of the head (Fig. 1D; RME, SMDV, AVA, ADF, AVE, AWA, AWB, and AUA), which include the command interneurons and thus mostly overlapped in expression with PKC-2 (AVA, AVE, and AVB neurons).
Figure 1.
Conservation and expression of different PKC isoforms in C. elegans. A, Clustal Omega analysis of the similarities between vertebrate PKC and C. elegans PKC gene families. B, Low-magnification whole-animal representative images of the lipophilic fluorescent dye DiD (similar to DiL; see Materials and Methods for details) and YPET expression using YPET knock-in pkc-2, tpa-1, and pkc-3. Scale bar = 50 µm. The arrowheads point to the DiD staining, and the arrows point to the YPET signal in the head, tail, or gut. C, D, Higher-magnification images of images of DiD (similar to DiL; see Materials and Methods for details) and PKC-2 and TPA-1::YPET expression. Scale bars = 10 µm (C, left panel, and D) or 50 µm (C, middle panel). The arrowheads point to the AVA cells in the worm head region, the arrows point to the spermatheca, and the arrowhead on the differential interference contrast image points to the vulva position.
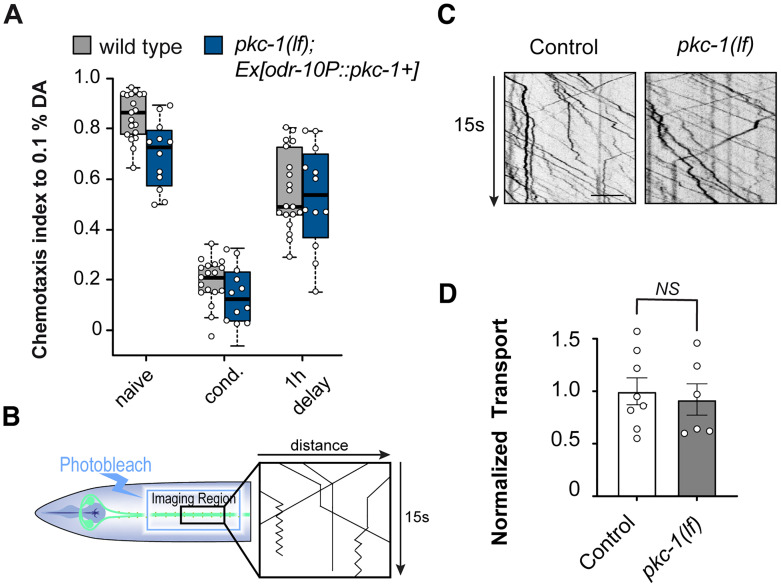
Interestingly, previous studies (Hoerndli et al., 2009; Stetak et al., 2009) have documented that some of the PKC-expressing interneurons (in particular, AVA, AVE, AVD, and AVB) are important for short- and long-term olfactory associative learning and memory in C. elegans suggesting that besides chemosensation, the different PKCs could play various roles in learning and memory. Previously, PKC-1 was shown to play a role in olfaction mediated by the AWA and AWC sensory neurons in addition to its well-defined role in thermotaxis (Okochi et al., 2005). To test the putative role of PKC-1 in learning or memory, we analyzed the effect of PKC-1 loss in worms rescued with PKC-1 expression in AWA-mediated olfaction using the odr-10 promoter–driven pkc-1 gene. We tested pkc-1(lf);Ex[odr-10p::pkc-1+] for their STAM to DA, comparing naive, 1 h DA starved conditioned (cond.) and animals conditioned but left to rest on food for one hour (1 h delay; Fig. 2A). Expression of PKC-1 in sensory neurons led to normal chemotaxis to 0.1% DA with no apparent defect in either learning or STAM (Fig. 2A). Previous findings have shown that PKC-1 is necessary for command interneurons for nose touch reversal but did not affect GLR-1 levels (Hyde et al., 2011). We have found in the past that steady-state GLR-1::GFP levels are difficult to interpret and that observing the transport of GLR-1 in AVA sometimes reveals additional aspects of signaling mechanisms (Hoerndli et al., 2013, 2015a). Thus, we tested whether loss of PKC-1 affected GLR-1 transport in AVA, using photobleaching of GLR-1::GFP in AVA (Fig. 2B, Materials and Methods) but saw no significant difference with control (Fig. 2C,D).
Figure 2.
pkc-1(lf) does not affect STAM nor GLR-1 transport. A, Attraction of C. elegans animals to 0.1% DA, as naive untrained animals, after 1 h of starvation conditioning in the presence of pure DA, and 1 h after conditioning (STAM), in controls and pkc-1(lf) expressing a wild-type copy of PKC-1 under the odr-10p promoter. (F(2|87) = 1.884, p = 0.158). B, Diagram of the procedure used to record GFP-tagged GLR-1 transport events in the AVA and generate a kymograph. The proximal region of AVAR and AVAL after the second chiasma is imaged with a 20–25-µm-long stretch photobleached using 1 s of 0.5 W, 488 nm solid-state laser. A total of 500 frames at 10 frames per second (fps) are recorded and then transformed into a kymograph using the kymograph function of MetaMorph (see Materials and Methods for details). C, Representative kymographs of GFP-tagged GLR-1 transport in control and pkc-1(lf) animals. D, Quantification of the total number of transport events in all 50 s kymographs normalized to daily controls. N = 10 animals per condition, unpaired two-tailed t test (t(10) = 0.3998, p = 0.6963), NS = not significant. The error bars are SEM. See Table 2 for statistical details.
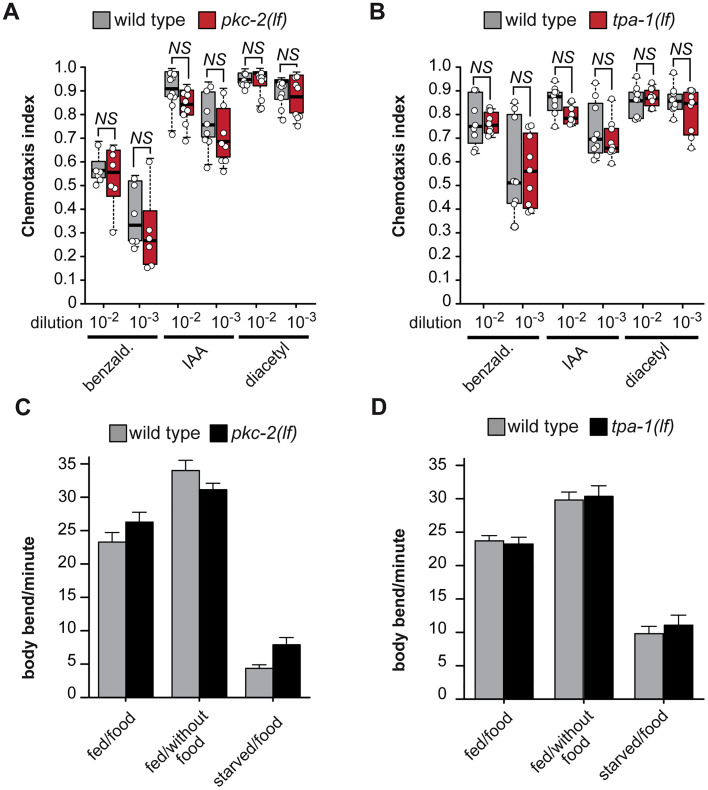
Next, we assessed the chemotaxis sensory abilities of pkc-2(lf) and tpa-1(lf) mutant animals. Neither pkc-2(lf) nor tpa-1(lf) mutants showed any defects in chemotaxis to benzaldehyde, IAA, or DA compared with wild-type control (Fig. 3A,B). Besides testing the olfaction, we also investigated whether loss-of-function mutations of pkc-2 or tpa-1 affect animal behavior such as body bends on or off food and their ability to adapt without food (Fig. 3C,D). Both pkc-2(lf) and tpa-1(lf) mutants moved at the same speed as controls on or off food and adapted to longer food deprivation.
Figure 3.
tpa-1(lf) and pkc-2(lf) mutants do not have obvious olfactory or motor deficiencies. A, B, Attraction of naive C. elegans animals to different concentrations of benzaldehyde (benzald.), IAA, or DA was tested (see Materials and Methods for details) in wild-type, (A) pkc-2(lf), or (B) tpa-1(lf) animals. The ANOVA F tests for panels A (F(11|81) = 38.497, p < 1E−15) and B (F(11|96) = 0.10612, p = 1.693E−12) showed significant differences in sample means, but the post hoc testing showed no difference between mutant and corresponding control. NS = not significant. D, E, Motility of wild-type, (C) pkc-2(lf), or (D) tpa-1(lf) well-fed (fed) or starved (starved) young adult animals was tested on empty NGM plates (without food) or on OP50 bacteria (food). Locomotor behavior and response to starvation were assessed by counting body bends for 1 min. N = 10. The ANOVA F tests for panels C (F(1|119) = 1.5721, p = 0.2136) and D (F(1|113) = 0.2157, p = 0.6432) were not significant. See Table 2 for statistical details.
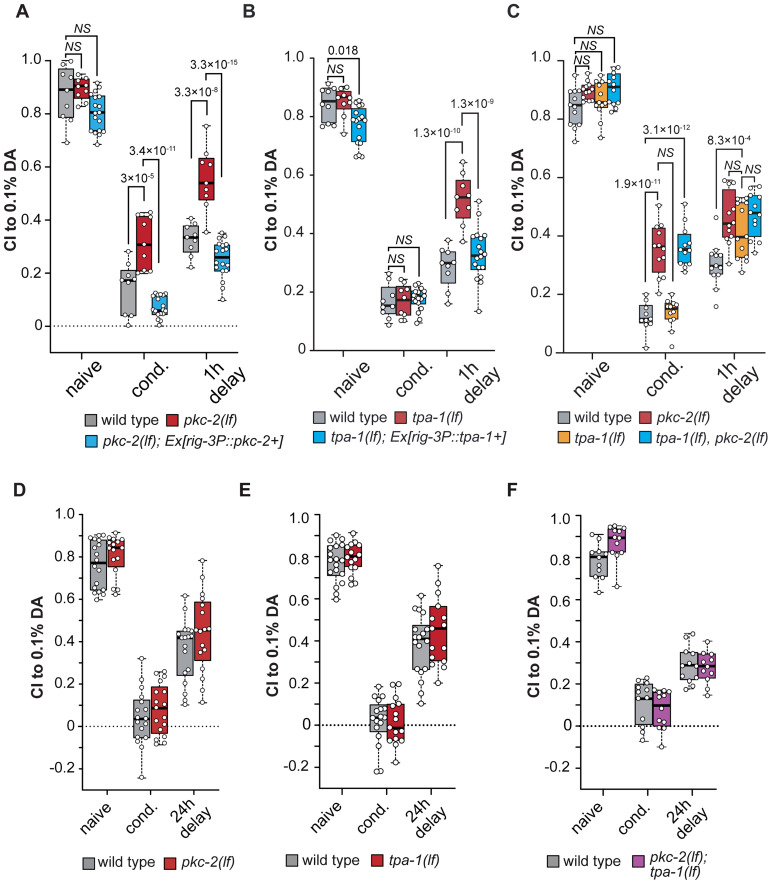
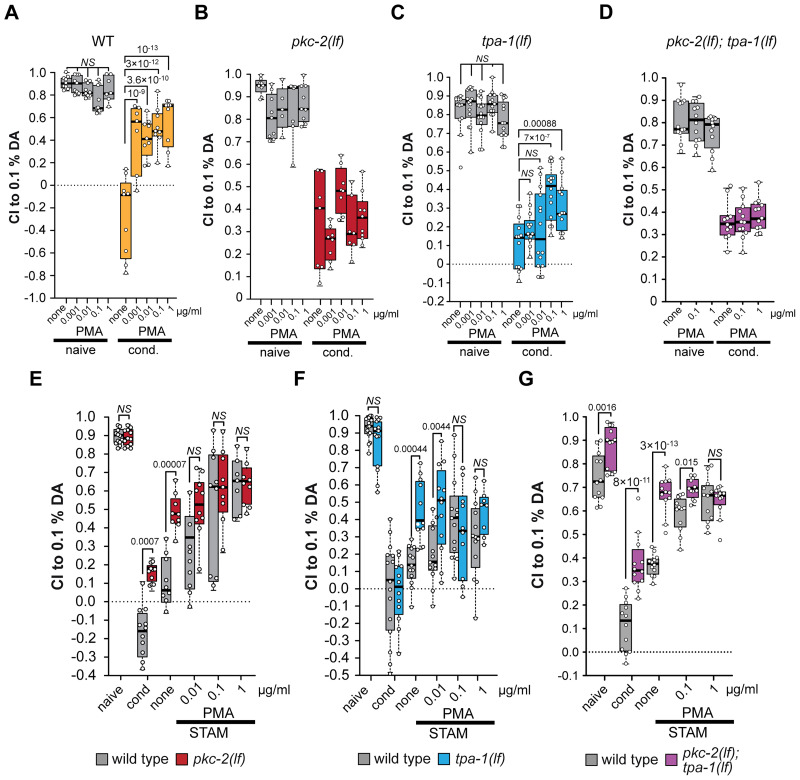
We then analyzed pkc-2(lf) animals for their STAM to DA, comparing naive, 1 h DA starved conditioned (cond.) and animals conditioned but left to rest on food for 1 h (1 h delay; Fig. 4A). Our analysis revealed that pkc-2(lf) mutants showed a defect in aversive associative learning (red box plots) compared with controls (gray box plots), and defect in short-term memory compared with wild-type control animals. Given the strong learning defect observed in 4A for pkc-2(lf), it is possible that the memory defect is a result of defective learning rather than a memory defect. This phenotype was dependent on the expression of PKC-2b in AVA as the expression of PKC-2b using the rig-3p promoter rescued both learning and memory defects of pkc-2(lf) mutants (Fig. 4A, blue box plots). Indeed, overexpression of PKC-2b in AVA led to a statistically significant, slightly better associative learning and memory compared with wild-type controls (see Table 2 for details). In comparison, tpa-1(lf) mutants did not show a defect in learning compared with controls (Fig. 4B, gray and red box plots) while tpa-1(lf) showed a sizeable defect memory compared with controls, which was rescued by expression of TPA-1 under the rig-3p promoter (Fig. 4B, blue box plots). These experiments suggest that whereas pkc-2 is important for associative learning, tpa-1 is important for short-term memory, both requiring expression in AVA for function. To determine whether pkc-2 and tpa-1 can complement each other in associative learning and memory, we also analyzed how pkc-2(lf);tpa-1(lf) double mutants would perform during the DA associative learning and memory (Fig. 4C). The single tpa-1(lf) mutants (yellow box plot) did not exhibit a learning defect but both the pkc-2(lf) and the pkc-2(lf);tpa-1(lf) did (red and blue box plots) compared with controls (gray box plots). However, the learning defect for pkc-2(lf);tpa-1(lf) was not worse than pkc-2 alone). Furthermore, pkc-2(lf) and tpa-1(lf) single mutants both showed a defect in short-term memory, and the pkc-2(lf);tpa-1(lf) was not different from the single mutants. These experiments suggest that although pkc-2 and tpa-1 regulate different stages of short-term associative learning and memory, they both likely act in the same signaling pathway since their effects on learning or memory are not additive. Based on the genetic epistasis test, pkc-2 probably acts upstream of tpa-1. To further investigate the role of pkc-2 and tpa-1 in associative learning and memory, we also tested long-term memory as described previously (Vukojevic et al., 2012). Briefly, animals were conditioned twice for 1 h with a 30 min rest on food in between and then transferred to plates with food for 24 h before testing their attraction to 0.1% DA (Fig. 4D,E). In this assay, neither pkc-2(lf) nor tpa-1(lf) single mutants (red box plots) showed a learning or memory defect compared with controls (gray box plots). This analysis thus indicates that pkc-2 and tpa-1 are important for short- but not long-term associative learning and memory. Additional analyses confirmed that pkc-2(lf);tpa-1(lf) double mutants (magenta box plots) also did not show defects in long-term learning or memory (Fig. 4F).
Figure 4.
PKC-2 and TPA-1 have differential roles in associative learning and memory in C. elegans. A–C, Attraction of C. elegans animals to 0.1% DA, as naive untrained animals, after 1 h of starvation conditioning in the presence of pure DA, and 1 h after conditioning (STAM), in various mutants and cell-specific rescues. Statistics were obtained using a two-way ANOVA with Bonferroni’s correction comparing all samples in the same treatment condition. See Table 2 for details. D–F, Attraction of C. elegans animals to 0.1% DA, before conditioning, after conditioning for long-term associative memory, and 24 h after conditioning (see Materials and Methods for details). Statistical comparisons between samples were done using a two-way ANOVA, showing no significant differences between samples in the same conditions on panels D (F(2|97) = 0.25211, p = 0.77666), E (F(2|92) = 0.71073, p = 0.49396), and F (F(2|63) = 2.45865, p = 0.093723). See Table 2 for statistical details.
Interestingly, the PKC isoforms that demonstrated an impact on short-term associative learning belong to the conventional and novel PKC isozymes, which depend on DAG for activation and have been shown to be activated by PMA treatment (Leach and Blumberg, 1985). Therefore, we speculated that some of the observed effects may be redundant between the PMA-sensitive PKC isoforms. Thus, hyperactivation of the remaining isoform with PMA may compensate for the loss of the other isoform. To test this hypothesis, we treated wild-type, pkc-2(lf),tpa-1(lf) single or pkc-2(lf);tpa-1(lf) double mutant worms in M9 with 0.5% DMSO alone or with various concentrations of PMA for 15 min prior to conditioning or immediately after conditioning (Fig. 5A–D). The short-term learning and memory performance was tested after PMA treatment as indicated (Fig. 5E–G). PMA treatment in wild-type animals prior to training strongly interfered with learning (Fig. 5A) and PMA treatment following training also impaired memory (Fig. 5E–G). These results suggest that not only loss of the PKC activity but also overactivation of the phorbol ester–sensitive PKCs impairs learning and memory. Therefore, the precise amount or timing of PKC activity is likely required for proper learning and memory formation. In contrast to the wild-type, pkc-2(lf) learning was not affected by PMA treatment, and the original pkc-2(lf) defect was not rescued by PMA (Fig. 5B). In addition, PMA treatment of the pkc-2(lf) mutant worms after conditioning did not affect memory (Fig. 5D). PMA treatment of the tpa-1(lf) mutant animals strongly impaired learning, while we observed no effect on memory compared with DMSO treatment (Fig. 5C,F). Interestingly, tpa-1(lf) mutant worms seem to be less sensitive to PMA and the learning defect can be observed only at higher concentrations. This finding further supports the idea that a tight control of PKC activity is needed for learning and strong hyperactivation of the general kinase activity beyond a threshold leads to learning impairment. In the absence of TPA-1, hyperactivation of PKC-2 requires higher PMA concentration as in wild-type animals where both PKC isoforms are present. On the other hand, PMA treatment prior training of pkc-2(lf);tpa-1(lf) double mutant animals had no additional effect on learning suggesting that PKC-2 is the dominant isoform responsible for short-term learning. In addition, pkc-2(lf);tpa-1(lf) double mutants behaved similarly to pkc-2(lf), and we have not detected any additive phenotype in the double mutant worms (Fig. 5G). Altogether, these results further support the idea that PKC-2 acts upstream of TPA-1 in associative short-term memory. In line with our hypothesis, they also show that overactivation of the PMA-sensitive PKCs (likely PKC-2) in the absence of TPA-1 has a major effect on learning but is not crucial for memory. Altogether, this set of experiments further supports our initial findings demonstrating that PKC-2 is predominantly regulating learning while TPA-1 is more involved in short-term memory maintenance.
Figure 5.
External activation of PKCs with the phorbol ester PMA does not rescue memory association. A–D, The effect of PMA on associative learning was tested by measuring the attraction of PMA pretreated (15 min prior conditioning) (A) wild-type, (B) pkc-2(lf), (C) tpa-1(lf) single, or (D) pkc-2(lf);tpa-1(lf) double mutant worms to 0.1% DA, as untrained animals (naive) and following 1 h of starvation conditioning in the presence of pure DA (cond.). Statistics were obtained using a two-way ANOVA with Bonferroni’s correction comparing all samples in the same treatment condition. E–G, The effect of PMA on associative memory was tested by measuring attraction of (E) pkc-2(lf), (F) tpa-1(lf) mutant, or (G) pkc-2(lf);tpa-1(lf) double mutant C. elegans animals to 0.1% DA, before conditioning (naive), directly after conditioning (cond) or 1 h after conditioning (1 h delay). Specifically, the effect of PMA treatment on memory was tested by soaking the worms in vehicle or different concentrations of PMA as indicated for 15 min directly following the 1 h of starvation conditioning in the presence of pure DA (see Materials and Methods for details). Statistical comparisons between samples were performed using a two-way ANOVA with post hoc t test with Bonferroni’s correction. See Table 2 for statistical details.
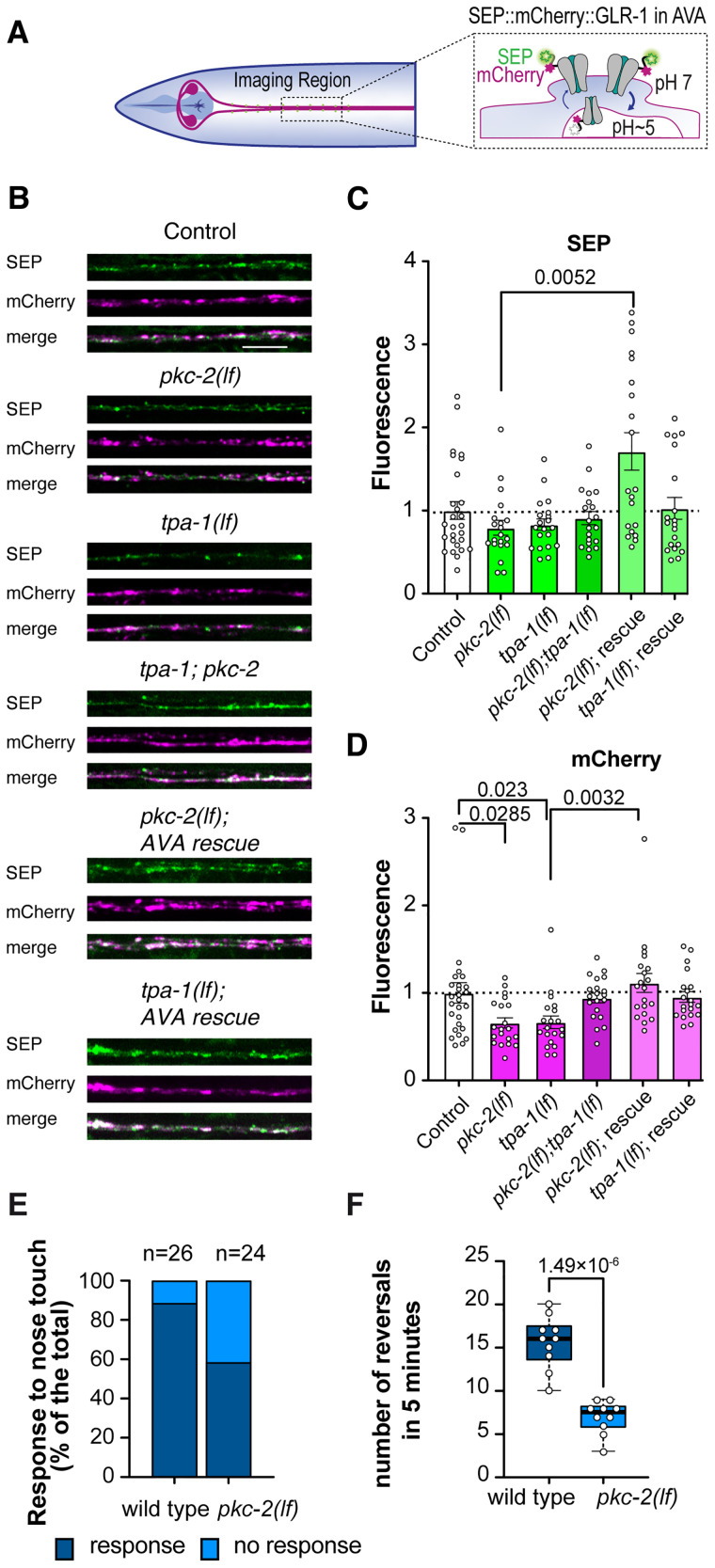
Associative olfactory learning to DA in C. elegans requires the expression of the glutamate receptor subunit GLR-1 in the AVA command interneurons (Stetak et al., 2009; Hadziselimovic et al., 2014). GLR-1 is the closest homolog to vertebrate GluA1 and is mostly functional as GLR-1/2 hetero-tetramers like GluA1/2 in vertebrates (Hart et al., 1995; Maricq et al., 1995; Mellem et al., 2002). In addition, it is well known that one of the primary targets of PKC phosphorylation during LTP and LTD in vertebrate hippocampal neurons is GluA1, leading to either increased or decreased expression of synaptic surface GluA1 (Sossin, 2007). To determine if pkc-2 and tpa-1 play a similar role in C. elegans GLR-1 surface expression, we generated single and double mutants of the loss-of-function alleles to observe their effect on synaptic SEP::mCherry::GLR-1 levels (Hoerndli et al., 2013, 2015b) in a complete loss-of-function glr-1(ky176) genetic background. The superecliptic pHluorin (SEP) fluorescence enables the quantification of synaptic surface GLR-1 receptors since the pH-sensitive SEP signal is quenched in acidic endosomes, and the mCherry fluorescence quantifies the total number of receptors at synapses (Hoerndli et al., 2013, 2015b; Doser et al., 2020; Luth et al., 2021). As previously described, we monitored the synaptic GLR-1 signal in the proximal region of AVA (Fig. 6) and quantified the SEP and mCherry signal at punctae in maximum projections containing stacks of 21 confocal images (see Materials and Methods section). Neither pkc-2(lf), nor tpa-1(lf), nor the double mutants showed a significant decrease in synaptic surface GLR-1(Fig. 6B,C). However, both single mutants demonstrated a significant decrease in the total number of GLR-1 receptors at synapses (Fig. 6B,D), with PKC-2b but not TPA-1 expression in AVA using the flp-18p promoter as previously described (Hoerndli et al., 2013) rescuing these defects. Interestingly, PKC-2 overexpression in AVA alone led to a very high level of SEP signal suggesting an increase beyond control of synaptic surface GLR-1 (1.7 ± SEM compared with 1.0 ± SEM for controls). These results suggest that loss of pkc-2 or tpa-1 leads to a decreased total pool of synaptic GLR-1 receptors but not a significant change in the number of receptors at the cell surface. Changes in the total endosomal levels of GLR-1 have been previously associated with decreased responsiveness of AVA to sensory input such as nose touch (Maricq et al., 1995) or in spontaneous reversal frequency of animals (Brockie et al., 2001; Juo et al., 2007). Thus, we tested whether the decrease in total GLR-1 we observed in pkc-2(lf) resulted in more than a deficit in associative learning. Figure 6, E and F, shows that pkc-2(lf) mutants had decreased responsiveness to nose touch as well as a significant decrease in spontaneous reversal compared with wild-type controls. These results further support a necessary role for PKC-2 regulation of GLR-1 function in AVA.
Figure 6.
Synaptic GLR-1 levels are decreased in pkc-2(lf) and tpa-1(lf). A, Diagram indicating the imaging region and how SEP::mCherry::GLR-1 fluorescence depends on subcellular localization. B, Representative maximal projection images of SEP, mCherry, and merge of both channels from SEP::mCherry::GLR-1 expression in AVA, in various mutants and rig-3p or flp-18p driven PKC-2a and TPA-1 rescues. Scale bar = 5 µm. C, D, Quantification of the synaptic fluorescence for each channel (see Materials and Methods for details). Statistics were obtained using a one-way ANOVA with Dunn’s multiple testing correction on the comparison of all mutants to the control strain. N > 20 animals per condition. The error bars are SEM. E, Behavioral response to nose touch in wild-type controls and pkc-2(lf) as percent response. Fisher’s exact test(26,24), p = 0.02355. F, Behavioral analysis of spontaneous reversals in 5 min comparing wild-type controls to pkc-2(lf). Two-tailed t test (t(10)= 7.62793, p = 1.49E-06). See Table 2 for statistical details.
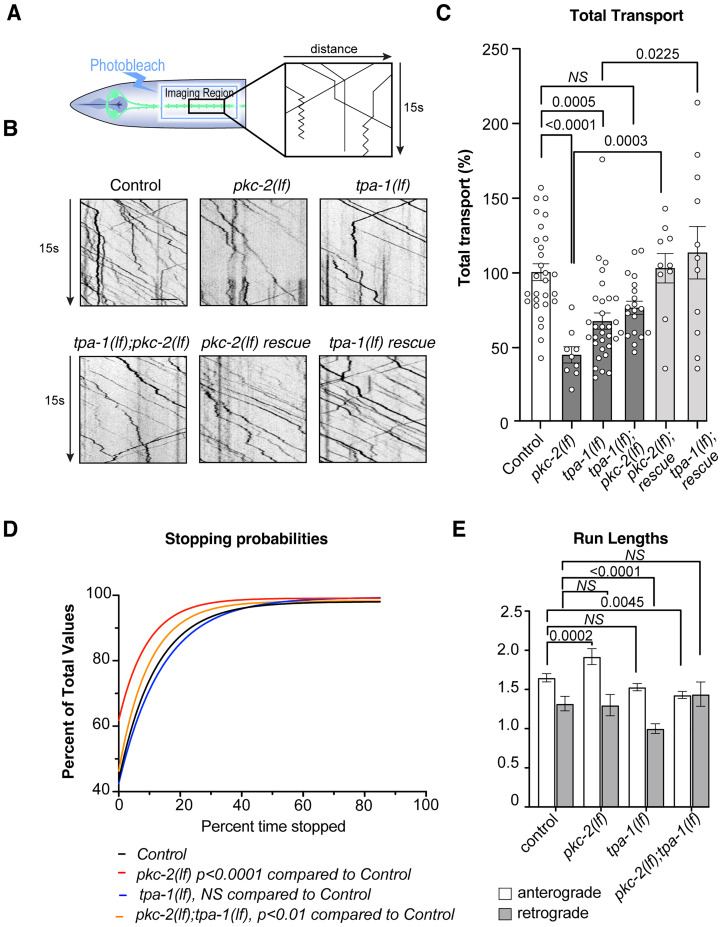
Previous studies have shown that the total number of synaptic receptors at synapses is dependent on the delivery of these GLR-1 receptors from long-distance transport (Hoerndli et al., 2013). To determine if pkc-2 and tpa-1 are necessary for long-distance GLR-1 transport, we combined photobleaching and continuous imaging of mCherry GLR-1 as described previously (Fig. 7A; Doser et al., 2020). This approach revealed that both pkc-2(lf) and tpa-1(lf) mutants had decreased GLR-1 transport compared with controls and the double pkc-2;tpa-1 mutants were not different from the single mutants (Fig. 7B,C). Furthermore, the transport defect observed in the single mutants could be rescued by postsynaptic AVA-specific expression of PKC-2 and TPA-1. To better understand which aspect of GLR-1 transport pkc-2 and tpa-1 could regulate, we quantified the percentage of the time a transport vesicle remained stopped during the imaging. We have previously shown that transport stops or pauses correlate with the delivery of receptors to synapses and are regulated by calcium signaling (Doser et al., 2020). Using a cumulative distribution analysis of the percent time a vesicle remained stopped (Fig. 7D) showed a significant shift toward lower stopping in pkc-2(lf) mutants but not in tpa-1(lf) mutants. In addition, pkc-2(lf) showed significantly increased anterograde run lengths (Fig. 7E) consistent with the results in Fig. 7D. tpa-1(lf) on the other hand led to a decreased retrograde run length (Fig. 7E). As was seen with stopping, pkc-2(lf) and tpa-1(lf) have slightly different effects on GLR-1 transport perhaps explaining why the pkc-2(lf);tpa-1(lf) doubles show an intermediate or less severe transport phenotype. Finally, neither pkc-2(lf) nor tpa-1(lf) significantly changed the ratio of anterograde/retrograde transport events (data not shown). Overall, GLR-1 transport analysis revealed fewer transport vesicles in both pkc-2(lf) and tpa-1(lf), with pkc-2(lf) showing a decrease in synaptic stopping.
Figure 7.
GLR-1 transport in pkc-2(lf) and tpa-1(lf) is decreased. A, Diagram depicting photobleaching and imaging region and generation of kymographs from vesicle movement and imaging of GFP-tagged GLR-1 in AVA. B, Kymographs representing GFP-tagged GLR-1 transport in control, mutants, double mutants, rig-3p or flp-18p pkc-2a, and tpa-1 rescues. Scale bar = 5 µm. C, Quantification of the total number of transport events per transport acquisition (50 s) normalized to daily controls in all the mutants and rescues shown in (A). Statistics were obtained using a one-way ANOVA to controls with Dunn’s multiple-comparison correction. D, Stopping probabilities of transport events as cumulative distributions in controls and different mutants shown. Statistics were obtained using an extra-sum of squares to test if one curve fit to both control and mutant data points, pkc-2(lf) (F(18,18) = 49.34, p < 0.0001), tpa-1(lf) (F(18,18) = 2.549, p = 0.074), and pkc-2(lf);tpa-1(lf) (F(18,18) = 5.671, p = 0.034). E, Average anterograde (white bars) and retrograde (gray bars) run lengths, in controls and mutants. Statistical comparisons were done between samples of anterograde or retrograde run lengths using a nonparametric Kruskal–Wallis z test. The error bars are SEM. See Table 2 for statistical details.
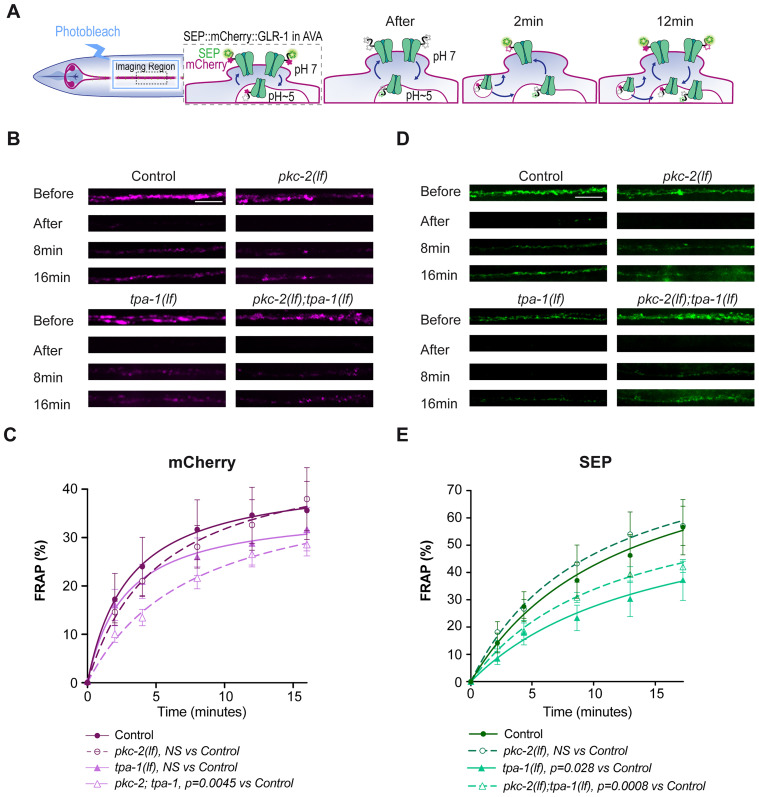
Although transport analysis is strongly correlated with the number of synaptic GLR-1 receptors (Doser et al., 2020), it is possible that due to the short time window and lower sensitivity, it is not completely representative of the mechanism of delivery and insertion of GLR-1 at synapses. To better determine the requirements for pkc-2 and tpa-1 in synaptic GLR-1 delivery and insertion, we turned to FRAP analysis of SEP::mCherry::GLR-1 in the same dendritic AVA region imaged for transport and puncta distribution (Fig. 8). The area of interest was first imaged, and then a region of ∼20 µm was photobleached with 1 s illumination of 0.5 W of a 488 nm laser. A full confocal stack before, immediately after, and then 2, 4, 8, and 16 min after photobleaching was obtained for both SEP and mCherry fluorescence. The fluorescence along the process was quantified as a percent of the initial fluorescence before photobleaching. Using this approach, we first measured the FRAP of the mCherry signal (Fig. 8B,C). mCherry FRAP in single pkc-2(lf) and tpa-1(lf) mutants was not significantly different from controls although the dynamics showed a difference, either with an initial slow recovery rate for pkc-2(lf) or faster plateau reached for tpa-1(lf). On the other hand, the double pkc-2;tpa-1 mutant showed significantly slower recovery suggesting that PKC-2 and TPA-1 function can partially complement each other in terms of endosomal delivery. SEP FRAP analysis revealed that tpa-1(lf) and the pkc-2;tpa-1 double had a significantly reduced FRAP recovery (Fig. 8D,E). Altogether, the total GLR-1 FRAP results support the decreased transport and decreased synaptic pool of GLR-1 observed in Figures 6 and 7. Likewise, the decreased membrane-associated GLR-1 FRAP observed for tpa-1(lf) and pkc-2;tpa-1 indicates that these genes likely affect synaptic exocytosis. Altogether, our single-cell in vivo analysis of synaptic GLR-1 levels and dynamics provides a further understanding of the roles of PKC-2 and TPA-1 in short-term learning and memory in C. elegans.
Figure 8.
tpa-1(lf) but not pkc-2(lf) lead to decreased SEP FRAP of GLR-1. A, Diagram depicting the FRAP process in AVA using SEP::mCherry::GLR-1 and how fluorescence changes at different time points and subcellular localization. B, D, Representative maximum projection images of mCherry and SEP signals before, immediately after, and 8 and 16 min after FRAP. Scale bar = 5 µm. C, E, Quantification of the percentage mCherry or SEP recovery compared with before photobleaching at different time points. All curves are exponential fits to the data points shown. An extra-sum of squares statistic test was used to compare the recovery curves of each mutant to the control. N > 10 animals per condition. The error bars are SEM. See Table 2 for statistical details.
Discussion
Previous studies have shown that the PKC family plays a prominent role in synaptic signaling and plasticity (Sossin, 2007; Gould and Newton, 2008; Sun and Alkon, 2014). However, the complexity and the number of genes and isoforms in this family have made it difficult to assign precise individual roles in synaptic plasticity using classic genetic approaches or molecular replacement approaches in mammalian rodent models. Here, we made use of the limited number of PKC family members in C. elegans, containing a single gene for each subfamily, to investigate their individual roles in olfaction and short- and long-term associative memory. We identified pkc-2 and tpa-1, but not pkc-1 or pkc-3, as important contributors to olfactory associative learning and short-term memory in C. elegans. More specifically, our results indicated pkc-2 is necessary for negative associative learning with DA leading to defects in STAM and that tpa-1 is necessary for STAM but not associative learning. In addition, we show that their function is necessary in AVA for learning and short-term memory and acts broadly in the same signaling pathway albeit probably at different steps. Furthermore, we demonstrated that GLR-1 transport and synaptic delivery are also modulated by the pkc-2 and tpa-1 functions in AVA. Since GLR-1 transport, function, and synaptic levels have been associated with synaptic plasticity, this further strengthens the functional necessity of pkc-2 and tpa-1 for memory in C. elegans.
Some of the limitations of this study lie in the fact that PKC-2 and TPA-1 diverge in terms of their amino acid sequence from their closest vertebrate homologs (PKC-2 is 62–67% identical to human cPKC family members while TPA-1 shows only 48% identity to human PKCd) and in the simplicity of the neuronal anatomy of C. elegans circuits. However, although these differences exist, it is important to mention that reductive approaches in C. elegans have often been the key to understanding the conserved functions of molecular players involved in synaptic plasticity and function, as well as memory (Bono and Villu Maricq, 2005; Ikeda et al., 2008; Lau et al., 2013; Jin et al., 2016).
Previously, different isoforms of PKC have been associated with short and long-term memory in Aplysia, Drosophila, and mice (Sossin, 2007; Sun and Alkon, 2014). Based on sequence homology, PKC-2 can be considered part of the cPKC family and TPA-1 part of the nPKC family or δ PKCs. Classical PKCs were found to be important for short-term plasticity and memory in Aplysia, Drosophila, and mice. Our results indicate that PKC-2 is required for olfactory associative learning in C. elegans and has a cell-specific effect on GLR-1 transport and synaptic delivery (Figs. 7, 8) in addition to the previously implicated role in thermotaxis memory (Land and Rubin, 2017). The effect of pkc-2(lf) is strongest on GLR-1 transport but does not affect FRAP of SEP or mCherry GLR-1, indicating a broader role of PKC-2 on transport rather than a local GLR-1 trafficking effect. On the other hand, loss of TPA-1 leads to a substantial decrease in associative olfactory short-term memory without influencing learning in C. elegans. This is associated with a strong effect on both transport and local synaptic trafficking of GLR-1 (Fig. 7, 8). The loss of PKC-2 or TPA-1 does not seem to affect long-term associative olfactory learning and memory (Fig. 4D,E), suggesting that in long-term olfactory memory, the conditions of learning and retrieval are different enough to permit compensation by other signaling pathways or PKCs. PKM and PKCl functions have been implicated as necessary in long-term memory and LTP in vertebrate models (Matt and Hell, 2013; Sacktor and Hell, 2017). Based on sequence homology, only pkc-3 would correspond to these atypical forms of PKC in C. elegans, but the insertion of YPET in the genomic sequence of pkc-3 does not show a detectable neuronal expression. The truncation of classical PKC can lead to PKM forms that enable sustained PKC signaling associated with synaptic plasticity (Matt and Hell, 2013; Sacktor and Hell, 2017). At this point, we do not know if there is cleavage of the regulatory domain of TPA-1 or PKC-2 that could lead to a PKM form. The tpa-1b isoform seems to be missing the N-terminal and perhaps regulatory domains of TPA-1 which could make it akin to the PKM form; however, the tpa-1(k501) allele affecting both isoforms did not show a long-term memory phenotype in our olfactory associative assays.
Previous studies using cell culture and hippocampal slices as well as studies in Aplysia and Drosophila have shown an important role for PKCs in regulating the synaptic trafficking of AMPA receptors (Sossin, 2007). In addition, to local synaptic recycling, AMPA receptors are also delivered to synapses using microtubule-dependent motors, and recent studies have shown that this transport is dynamically regulated by neuronal activity (Hangen et al., 2018; Doser et al., 2020; Hoerndli et al., 2022). However, the role of PKCs in AMPA receptor transport has not been studied. Here, we show that TPA-1 and PKC-2 modulate ∼50% of AMPA receptor transport, and genetic studies suggest that both proteins are acting in the same pathway regulating AMPA transport numbers (Fig. 4). However, whereas PKC-2 seems to be important in the synaptic stopping of AMPARs, TPA-1 does not (Figs. 7, 8). Sequence alignment of the C-terminal of C. elegans GLR-1, mouse, and human GluA1 starting from after the last residue of the last transmembrane domain shows conservation of several serines including one that could be the equivalent of S831 phosphorylated by PKCs in mouse (data not shown). However, additional approaches using hybrid receptors and CRISPR-based mutations would be required to test this more thoroughly.
Synaptic trafficking of AMPARs relies on endosomal pools of receptors and is well documented as one of the fundamental mechanisms implicated in synaptic plasticity and ultimately learning and memory (Groc and Choquet, 2020). In addition, recent studies have shown that S831A or S831D mutations of GluA1 lead to changes in GluA1 transport velocity, stopping, and synaptic delivery (Hangen et al., 2018). Previous studies have shown that PKC phosphorylation at S831in GluA1 and S880 in GluA2 increases association with PICK1, leading to endocytosis of the AMPARs (Chung et al., 2000, 2003; Matsuda et al., 2000; Xia et al., 2000; Hanley, 2008).
Here, we show that the transport of AMPARs in C. elegans is important for the synaptic trafficking of AMPARs and that decreases in transport associated with loss of PKC-2 or TPA-1 have consequences on associative learning and memory. Our results indicate a relatively strong effect on the transport of GLR-1 in pkc-2(lf) and tpa-1(lf) mutants, which is rescued by postsynaptic expression of both PKC-2b and TPA-1. However, these transport defects do not result in strong constitutive changes in synaptic GLR-1 surface receptors indicating that at a steady state, 50% transport is sufficient for normal cycling of receptors (Figs. 6, 7). In addition, although pkc-2(lf) and tpa-1(lf) both lead to decreased transport the double mutant is not significantly different from controls which is consistent with the effects observed for the total pool of receptors where the double mutants are less affected than single mutants. The stopping and transport run length analyses in Figure 7, D and E, suggest that loss of pkc-2 or tpa-1 does not affect transport in the same way. So perhaps, their effect on the steady-state endosomal pool might interfere with each other in the double mutants. FRAP experiments suggest that dynamics of SEP::GLR-1 recovery are statistically affected by loss of TPA-1 but not PKC-2. This is not consistent with the fact that average synaptic SEP::GLR-1 is not affected in tpa-1(lf) mutants (Fig. 6C). These results suggest that either the imaging of SEP::mCherry::GLR-1 is not accurately reporting dynamics of endogenous receptors, missing the GLR-1/2 complexes, or reporting correctly but suggesting that other mechanisms in addition to exocytosis such as endocytosis of receptors may be affected. Additional experiments using S to A mutations at the S831 residue equivalent in GLR-1 C-terminus, as well as synapse-specific markers for olfactory inputs to AVA, will be necessary to achieve a mechanistic understanding of how PKC regulation of GLR-1 trafficking is necessary for associative learning and memory. Since our results suggest that loss of PKC-2 and TPA-1 function leads to a decrease in transport and delivery, this would suggest that their function in olfactory associative learning in C. elegans is a positive regulation of AMPAR transport and delivery, contrary to the known role of PKCs in AMPAR trafficking in LTD (Chung et al., 2000, 2003; Xia et al., 2000) but in line with a role of PKCs in LTP (Sossin, 2007).
Overall, this study has identified PKC-2 function in command interneurons for short-term but not long-term olfactory associative learning and demonstrated that TPA-1 is necessary for short-term olfactory associative memory. A detailed analysis of GLR-1 dynamics suggests that although GLR-1 dynamics do require PKC-2 and TPA-1 function in AVA, these two gene products may have additional effects related to AMPAR trafficking or function that will need to be investigated in the future. Altogether, this study lays the groundwork for linking PKC activation and olfactory associative memory and creates a foundation for future studies in C. elegans to elucidate in depth the exact molecular role of specific PKC genes in synaptic plasticity, learning, and memory.
References
- Bargmann CI, Hartwieg E, Horvitz HR (1993) Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74:515–527. 10.1016/0092-8674(93)80053-H [DOI] [PubMed] [Google Scholar]
- Boehm J, Kang MG, Johnson RC, Esteban J, Huganir RL, Malinow R (2006) Synaptic incorporation of AMPA receptors during LTP is controlled by a PKC phosphorylation site on GluR1. Neuron 51:213–225. 10.1016/j.neuron.2006.06.013 [DOI] [PubMed] [Google Scholar]
- Bono MD, Villu Maricq A (2005) Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci 28:451–501. 10.1146/annurev.neuro.27.070203.144259 [DOI] [PubMed] [Google Scholar]
- Brenner S (2003) Nature's gift to science (Nobel lecture). Chembiochem 4:683–687. 10.1002/cbic.200300625 [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Mellem JE, Hills T, Madsen DM, Maricq AV (2001) The C. elegans glutamate receptor subunit NMR-1 is required for slow NMDA-activated currents that regulate reversal frequency during locomotion. Neuron 31:617–630. 10.1016/S0896-6273(01)00394-4 [DOI] [PubMed] [Google Scholar]
- Callender JA, Newton AC (2017) Conventional protein kinase C in the brain: 40 years later. Neuronal Signal 1:NS20160005. 10.1042/ns20160005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet D, Triller A (2013) The dynamic synapse. Neuron 80:691–703. 10.1016/j.neuron.2013.10.013 [DOI] [PubMed] [Google Scholar]
- Chung HJ, Steinberg JP, Huganir RL, Linden DJ (2003) Requirement of AMPA receptor GluR2 phosphorylation for cerebellar long-term depression. Science 300:1751–1755. 10.1126/science.1082915 [DOI] [PubMed] [Google Scholar]
- Chung HJ, Xia J, Scannevin RH, Zhang X, Huganir RL (2000) Phosphorylation of the AMPA receptor subunit GluR2 differentially regulates its interaction with PDZ domain-containing proteins. J Neurosci 20:7258–7267. 10.1523/JNEUROSCI.20-19-07258.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B (2015) Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200:1035–1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doser RL, Amberg GC, Hoerndli FJ (2020) Reactive oxygen species modulate activity-dependent AMPA receptor transport in C. elegans. J Neurosci 40:7405–7420. 10.1523/JNEUROSCI.0902-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Johnson JR, Rankin K, Jenkins RE, Maguire C, Morgan A, Burgoyne RD, Barclay JW (2012) PKC-2 phosphorylation of UNC-18 Ser322 in AFD neurons regulates temperature dependency of locomotion. J Neurosci 32:7042–7051. 10.1523/JNEUROSCI.4029-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban JA, Shi SH, Wilson C, Nuriya M, Huganir RL, Malinow R (2003) PKA phosphorylation of AMPA receptor subunits controls synaptic trafficking underlying plasticity. Nat Neurosci 6:136–143. 10.1038/nn997 [DOI] [PubMed] [Google Scholar]
- Gould CM, Newton AC (2008) The life and death of protein kinase C. Current Drug Targets 9:614–625. 10.2174/138945008785132411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groc L, Choquet D (2020) Linking glutamate receptor movements and synapse function science. New York, NY: American Association for the Advancement of Science. [DOI] [PubMed] [Google Scholar]
- Hadziselimovic N, et al. (2014) Forgetting is regulated via Musashi-mediated translational control of the Arp2/3 complex. Cell 156:1153–1166. 10.1016/j.cell.2014.01.054 [DOI] [PubMed] [Google Scholar]
- Hangen E, Cordelieres FP, Petersen JD, Choquet D, Coussen F (2018) Neuronal activity and intracellular calcium levels regulate intracellular transport of newly synthesized AMPAR. Cell Rep 24:1001–1012e1003. 10.1016/j.celrep.2018.06.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JG (2008) PICK1: a multi-talented modulator of AMPA receptor trafficking. Pharmacol Ther 118:152–160. 10.1016/j.pharmthera.2008.02.002 [DOI] [PubMed] [Google Scholar]
- Hanus C, Kochen L, Tom Dieck S, Racine V, Sibarita JB, Schuman EM, Ehlers MD (2014) Synaptic control of secretory trafficking in dendrites. Cell Rep 7:1771–1778. 10.1016/j.celrep.2014.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM (1995) Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature 378:82–85. 10.1038/378082a0 [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Shi SH, Esteban JA, Piccini A, Poncer JC, Malinow R (2000) Driving AMPA receptors into synapses by LTP and CaMKII: requirement for GluR1 and PDZ domain interaction. Science 287:2262–2267. 10.1126/science.287.5461.2262 [DOI] [PubMed] [Google Scholar]
- Hiroki S, Iino Y (2022) The redundancy and diversity between two novel PKC isotypes that regulate learning in Caenorhabditis elegans. Proc Natl Acad Sci U S A 119. 10.1073/pnas.2106974119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerndli FJ, Brockie PJ, Wang R, Mellem JE, Kallarackal A, Doser RL, Pierce DM, Madsen DM, Maricq AV (2022) MAPK signaling and a mobile scaffold complex regulate AMPA receptor transport to modulate synaptic strength. Cell Rep 38:110577. 10.1016/j.celrep.2022.110577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerndli FJ, Kallarackal AJ, Maricq AV (2015a) Mobile AMPARs are required for synaptic plasticity. Channels 9:230–232. 10.1080/19336950.2015.1080931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerndli FJ, Maxfield DA, Brockie PJ, Mellem JE, Jensen E, Wang R, Madsen DM, Maricq AV (2013) Kinesin-1 regulates synaptic strength by mediating the delivery, removal, and redistribution of AMPA receptors. Neuron 80:1421–1437. 10.1016/j.neuron.2013.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerndli FJ, Walser M, Hoier EF, De Quervain D, Papassotiropoulos A, Hajnal A (2009) A conserved function of C. elegans CASY-1 calsyntenin in associative learning. PLoS ONE 4. 10.1371/journal.pone.0004880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerndli FJ, Wang R, Mellem JE, Kallarackal A, Brockie PJ, Thacker C, Madsen DM, Maricq AV (2015b) Neuronal activity and CaMKII regulate kinesin-mediated transport of synaptic AMPARs. Neuron 86:457–474. 10.1016/j.neuron.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huganir RL, Nicoll RA (2013) AMPARs and synaptic plasticity: the last 25 years. Neuron 80:704–717. 10.1016/j.neuron.2013.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde R, Corkins ME, Somers GA, Hart AC (2011) PKC-1 acts with the ERK MAPK signaling pathway to regulate Caenorhabditis elegans mechanosensory response. Genes Brain Behav 10:286–298. 10.1111/j.1601-183X.2010.00667.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda DD, Duan Y, Matsuki M, Kunitomo H, Hutter H, Hedgecock EM, Iino Y (2008) CASY-1, an ortholog of calsyntenins/alcadeins, is essential for learning in Caenorhabditis elegans. Proc Natl Acad Sci U S A 105:5260–5265. 10.1073/pnas.0711894105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas-Trejo A, Land M, Tcherepanova I, Freedman JH, Rubin CS (1997) Structure and expression of the Caenorhabditis elegans protein kinase C2 gene. Origins and regulated expression of a family of Ca2+-activated protein kinase C isoforms. J Biol Chem 272:6629–6640. 10.1074/jbc.272.10.6629 [DOI] [PubMed] [Google Scholar]
- Jia Q, Sieburth D (2021) Mitochondrial hydrogen peroxide positively regulates neuropeptide secretion during diet-induced activation of the oxidative stress response. Nat Commun 12:1–22. 10.1038/s41467-020-20314-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Pokala N, Bargmann CI (2016) Distinct circuits for the formation and retrieval of an imprinted olfactory memory. Cell 164:632–643. 10.1016/j.cell.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juo P, Harbaugh T, Garriga G, Kaplan JM (2007) CDK-5 regulates the abundance of GLR-1 glutamate receptors in the ventral cord of Caenorhabditis elegans. Mol Biol Cell 18:3883–3893. 10.1091/mbc.e06-09-0818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano T, Brockie PJ, Sassa T, Fujimoto H, Kawahara Y, Iino Y, Mellem JE, Madsen DM, Hosono R, Maricq AV (2008a) Memory in Caenorhabditis elegans is mediated by NMDA-type ionotropic glutamate receptors. Curr Biol 18:1010–1015. 10.1016/j.cub.2008.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano T, Brockie PJ, Sassa T, Fujimoto H, Kawahara Y, Iino Y, Mellem JE, Madsen DM, Hosono R, Maricq AV (2008b) Memory in Caenorhabditis elegans is mediated by NMDA-type ionotropic glutamate receptors. Curr Biol 18:1010–1015. 10.1016/j.cub.2008.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JM, Horvitz HR (1993) A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc Natl Acad Sci U S A 90:2227–2231. 10.1073/pnas.90.6.2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Horvitz HR (1990) The Caenorhabditis elegans gene lin-10 is broadly expressed while required specifically for the determination of vulval cell fates. Genes Dev 4:357–371. 10.1101/gad.4.3.357 [DOI] [PubMed] [Google Scholar]
- Kindt KS, Quast KB, Giles AC, De S, Hendrey D, Nicastro I, Rankin CH, Schafer WR (2007) Dopamine mediates context-dependent modulation of sensory plasticity in C. elegans. Neuron 55:662–676. 10.1016/j.neuron.2007.07.023 [DOI] [PubMed] [Google Scholar]
- Land M, Rubin CS (2017) A calcium- and diacylglycerol-stimulated protein kinase C (PKC), Caenorhabditis elegans PKC-2, links thermal signals to learned behavior by acting in sensory neurons and intestinal cells. Mol Cell Biol 37:e00192–00117. 10.1128/MCB.00192-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau HL, Timbers TA, Mahmoud R, Rankin CH (2013) Genetic dissection of memory for associative and non-associative learning in Caenorhabditis elegans. Genes BrainBehav 12:210–223. 10.1111/j.1601-183X.2012.00863.x [DOI] [PubMed] [Google Scholar]
- Leach KL, Blumberg PM (1985) Modulation of protein kinase C activity and [3H]phorbol 12,13-dibutyrate binding by various tumor promoters in mouse brain cytosol. Cancer Res 45:1958–1963. [PubMed] [Google Scholar]
- Lisman J, Schulman H, Cline H (2002) The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci 3:175–190. 10.1038/nrn753 [DOI] [PubMed] [Google Scholar]
- Lisman JE, Zhabotinsky AM (2001) PP1 switch that potentiates transmission by organizing an AMPA receptor anchoring assembly. Neuron 31:191–201. 10.1016/S0896-6273(01)00364-6 [DOI] [PubMed] [Google Scholar]
- Lu W, Isozaki K, Roche KW, Nicoll RA, Lua W, Isozaki K, Roche KW, Nicoll RA (2010) Synaptic targeting of AMPA receptors is regulated by a CaMKII site in the first intracellular loop of GluA1. PNAS 107:22266–22271. 10.1073/pnas.1016289107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luth ES, Hodul M, Rennich BJ, Riccio C, Hofer J, Markoja K, Juo P (2021) VER/VEGF receptors regulate AMPA receptor surface levels and glutamatergic behavior. PLoS Genet 17:e1009375–e1009375. 10.1371/journal.pgen.1009375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Nicoll RA (1999) Long-term potentiation–a decade of progress? Science 285:1870–1874. 10.1126/science.285.5435.1870 [DOI] [PubMed] [Google Scholar]
- Malinow R, Mainen ZF, Hayashi Y (2000) LTP mechanisms: from silence to four-lane traffic. Curr Opin Neurobiol 10:352–357. 10.1016/S0959-4388(00)00099-4 [DOI] [PubMed] [Google Scholar]
- Man HY, Ju W, Ahmadian G, Wang YT (2000) Intracellular trafficking of AMPA receptors in synaptic plasticity. Cell Mol Life Sci 57:1526–1534. 10.1007/PL00000637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricq AV, Peckol E, Driscoll M, Bargmann CI (1995) Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature 378:78–81. 10.1038/378078a0 [DOI] [PubMed] [Google Scholar]
- Matsuda S, Launey T, Mikawa S, Hirai H (2000) Disruption of AMPA receptor GluR2 clusters following long-term depression induction in cerebellar Purkinje neurons. EMBO J 19:2765–2774. 10.1093/emboj/19.12.2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt L, Hell JW (2013) PKClambda: a new player in LTP coming to the rescue of PKCzeta’s faltering role in LTP? EMBO J 32:1348–1349. 10.1038/emboj.2013.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellem JE, Brockie PJ, Zheng Y, Madsen DM, Maricq AV (2002) Decoding of polymodal sensory stimuli by postsynaptic GLutamate receptors in C. elegans. Neuron 36:933–944. 10.1016/S0896-6273(02)01088-7 [DOI] [PubMed] [Google Scholar]
- Mohri A, Kodama E, Kimura KD, Koike M, Mizuno T, Mori I (2005) Genetic control of temperature preference in the nematode Caenorhabditis elegans. Genetics 169:1437–1450. 10.1534/genetics.104.036111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S, Chassefeyre R, Campbell GE, Encalada SE (2017) KymoAnalyzer: a software tool for the quantitative analysis of intracellular transport in neurons. Traffic 18:71–88. 10.1111/tra.12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC (1995) Protein kinase C: structure, function, and regulation. J Biol Chem 270:28495–28498. 10.1074/jbc.270.48.28495 [DOI] [PubMed] [Google Scholar]
- Okochi Y, Kimura KD, Ohta A, Mori I (2005) Diverse regulation of sensory signaling by C. elegans nPKC-epsilon/eta TTX-4. The EMBO J 24:2127–2137. 10.1038/sj.emboj.7600697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD (2004) Recycling endosomes supply AMPA receptors for LTP. Science 305:1972–1975. 10.1126/science.1102026 [DOI] [PubMed] [Google Scholar]
- Passafaro M, Piech V, Sheng M (2001) Subunit-specific temporal and spatial patterns of AMPA receptor exocytosis in hippocampal neurons. Nat Neurosci 4:917–926. 10.1038/nn0901-917 [DOI] [PubMed] [Google Scholar]
- Redondo RL, Morris RG (2011) Making memories last: the synaptic tagging and capture hypothesis. Nat Rev Neurosci 12:17–30. 10.1038/nrn2963 [DOI] [PubMed] [Google Scholar]
- Roche KW, O’Brien RJ, Mammen AL, Bernhardt J, Huganir RL (1996) Characterization of multiple phosphorylation sites on the AMPA receptor GluR1 subunit. Neuron 16:1179–1188. 10.1016/S0896-6273(00)80144-0 [DOI] [PubMed] [Google Scholar]
- Rose JK, Kaun KR, Chen SH, Rankin CH (2003) GLR-1, a non-NMDA glutamate receptor homolog, is critical for long-term memory in Caenorhabditis elegans. J Neurosci 23:9595–9599. 10.1523/JNEUROSCI.23-29-09595.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacktor TC, Hell JW (2017) The genetics of PKMζ and memory maintenance science signaling. American Association for the Advancement of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Tabuse Y, Nishiwaki K, Miwa J (1995) The tpa-1 gene of Caenorhabditis elegans encodes two proteins similar to Ca(2+)-independent protein kinase Cs: evidence by complete genomic and complementary DNA sequences of the tpa-1 gene. J Mol Biol 251:477–485. 10.1006/jmbi.1995.0449 [DOI] [PubMed] [Google Scholar]
- Sathler MF, Khatri L, Roberts JP, Schmidt IG, Zaytseva A, Kubrusly RCC, Ziff EB, Kim S (2021) Phosphorylation of the AMPA receptor subunit GluA1 regulates clathrin-mediated receptor internalization. J Cell Sci 134. 10.1242/jcs.257972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin ER, Ranganathan R, Horvitz HR (2000) C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26:619–631. 10.1016/s0896-6273(00)81199-x [DOI] [PubMed] [Google Scholar]
- Sheng M, Lee SH (2001) AMPA receptor trafficking and the control of synaptic transmission. Cell 105:825–828. 10.1016/S0092-8674(01)00406-8 [DOI] [PubMed] [Google Scholar]
- Sossin WS (2007) Isoform specificity of protein kinase Cs in synaptic plasticity. Learn Mem 14:236–246. 10.1101/lm.469707 [DOI] [PubMed] [Google Scholar]
- Stabel S, Parker PJ (1991) Protein kinase C. Pharmacol Ther 51:71–95. 10.1016/0163-7258(91)90042-K [DOI] [PubMed] [Google Scholar]
- Stetak A, Horndli F, Maricq AV, van den Heuvel S, Hajnal A (2009) Neuron-specific regulation of associative learning and memory by MAGI-1 in C. elegans. PLoS One 4:e6019. 10.1371/journal.pone.0006019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T (2006) Maintenance of C. elegans (Feb11, 2006). The C. elegans research community, WormBook. Available at: http://www.wormbook.org. 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun MK, Alkon DL (2014) The “memory kinases”: roles of PKC isoforms in signal processing and memory formation. Prog Mol Biol Transl Sci 122:31–59. 10.1016/B978-0-12-420170-5.00002-7 [DOI] [PubMed] [Google Scholar]
- Tabuse Y (2002) Protein kinase C isotypes in C. elegans. J Biochem 132:519–522. 10.1093/oxfordjournals.jbchem.a003251 [DOI] [PubMed] [Google Scholar]
- Tabuse Y, Nishiwaki K, Miwa J (1989) Mutations in a protein kinase C homolog confer phorbol ester resistance on Caenorhabditis elegans. Science 243:1713–1716. 10.1126/science.2538925 [DOI] [PubMed] [Google Scholar]
- Vukojevic V, Gschwind L, Vogler C, Demougin P, de Quervain DJ, Papassotiropoulos A, Stetak A (2012) A role for α-adducin (ADD-1) in nematode and human memory. EMBO J 31:1453–1466. 10.1038/emboj.2012.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Chung HJ, Wihler C, Huganir RL, Linden DJ (2000) Cerebellar long-term depression requires PKC-regulated interactions between GluR2/3 and PDZ domain-containing proteins. Neuron 28:499–510. 10.1016/S0896-6273(00)00128-8 [DOI] [PubMed] [Google Scholar]
- Ziegler K, Kurz CL, Cypowyj S, Couillault C, Pophillat M, Pujol N, Ewbank JJ (2009) Antifungal innate immunity in C. elegans: PKCdelta links G protein signaling and a conserved p38 MAPK cascade. Cell Host Microbe 5:341–352. 10.1016/j.chom.2009.03.006 [DOI] [PubMed] [Google Scholar]