Abstract
PURPOSE
Although immune checkpoint inhibitors (ICIs) have improved outcomes in certain patients with cancer, they can also cause life-threatening immunotoxicities. Predicting immunotoxicity risks alongside response could provide a personalized risk-benefit profile, inform therapeutic decision making, and improve clinical trial cohort selection. We aimed to build a machine learning (ML) framework using routine electronic health record (EHR) data to predict hepatitis, colitis, pneumonitis, and 1-year overall survival.
METHODS
Real-world EHR data of more than 2,200 patients treated with ICI through December 31, 2018, were used to develop predictive models. Using a prediction time point of ICI initiation, a 1-year prediction time window was applied to create binary labels for the four outcomes for each patient. Feature engineering involved aggregating laboratory measurements over appropriate time windows (60-365 days). Patients were randomly partitioned into training (80%) and test (20%) sets. Random forest classifiers were developed using a rigorous model development framework.
RESULTS
The patient cohort had a median age of 63 years and was 61.8% male. Patients predominantly had melanoma (37.8%), lung cancer (27.3%), or genitourinary cancer (16.4%). They were treated with PD-1 (60.4%), PD-L1 (9.0%), and CTLA-4 (19.7%) ICIs. Our models demonstrate reasonably strong performance, with AUCs of 0.739, 0.729, 0.755, and 0.752 for the pneumonitis, hepatitis, colitis, and 1-year overall survival models, respectively. Each model relies on an outcome-specific feature set, though some features are shared among models.
CONCLUSION
To our knowledge, this is the first ML solution that assesses individual ICI risk-benefit profiles based predominantly on routine structured EHR data. As such, use of our ML solution will not require additional data collection or documentation in the clinic.
Predicting #immunotherapy OS and toxicities across tumors using EHR data @GEHealthCare @VUMChealth.
INTRODUCTION
New cancer immunotherapies extend lives in previously rapidly fatal cancers.1 Although some patients experience remarkable response to immunotherapy,2,3 other patients have severe immune-related adverse events (irAEs).2,4,5 These irAEs can occur at any point, affecting 20%-30% of patients receiving immune checkpoint inhibitor (ICI) monotherapy and more than 50% receiving ICI combinations.2,4,5 Notably, effectiveness is associated with toxicity, suggesting that patients most likely to benefit from immunotherapy are also most at risk.6-8
CONTEXT
Key Objective
To build a machine learning (ML) solution that can assess a patient's immune checkpoint inhibitor (ICI) risk-benefit profile based primarily on routinely collected, structured electronic health record (EHR) data.
Knowledge Generated
We developed random forest classification models for four prediction outcomes—pneumonitis, colitis, hepatitis, and 1-year overall survival—after ICI initiation. The models primarily used structured EHR data before ICI treatment, and they demonstrate reasonably strong performance with area under the receiver operator curve between 0.729 and 0.755 for the four outcomes.
Relevance
The authors present a ML algorithm to predict 1 year survival and risk of pneumonitis, hepatitis and colitis during the 1 year after initiation of ICI treatment. With further validation across other data sets, this tool could help clinicians assess the risk/benefit tradeoff of initiating ICI therapy.
There are no practical reliable methods for predicting efficacy/effectiveness and toxicity to maximize safety and benefit in clinical care and clinical trials. Instead, current practice is limited to routine monitoring and management of significant and potentially fatal toxicities.2,4,5,9
Predictive toxicity-effectiveness modeling using machine learning (ML) could provide clinical decision support. However, most existing methods for predicting toxicity or effectiveness cannot be practically implemented clinically and require measurement of irAE-associated biomarkers. Many of these, such as cytokines, immune cell subsets (eg, CD4+ lymphocytes), and polygenic signatures, are novel, not collected in clinical routine, and largely unstudied in larger cohorts.10-15 Other studies used more practical data but either used small cohorts, data that were not human-verified, did not consider multiple toxicities, were limited in drug or disease setting, or did not address both toxicity and efficacy/effectiveness.16-18 Thus, there is a need for comprehensive tools to accurately predict irAEs alongside effectiveness from routinely collected data.
We formed an academic-industry partnership between Vanderbilt University Medical Center (VUMC) and GE HealthCare to develop a practical, comprehensive toxicity-efficacy tool. Here, we report on the development of random forest classification models for effectiveness and each of the three most common severe irAEs—autoimmune hepatitis, colitis, and pneumonitis. We demonstrate that this multimodel framework can reliably predict toxicities and effectiveness in a test data set. In contrast to past studies,19-21 we developed and validated our algorithms using a large cohort of more than 2,200 ICI-treated individuals from an National Cancer Institute–designated Comprehensive Cancer Center with human-verified outcomes data. Our models have potential utility in clinical decision support guiding therapy selection and cessation, such as avoiding combination therapies for patients at higher predicted risk for toxicities.
METHODS
This study was conducted under the approval of the VUMC Institutional Review Board (study: 211814) and granted consent exemption for study participants.
Study Population
Patients (N = 2,710) were identified using drug keywords and pharmaceutical codes at Vanderbilt-Ingram Cancer Center (Nashville, TN). The following patients were excluded: no cancer diagnosis, last known alive age older than 90 years, no ICI treatment receipt, or ICI receipt only on clinical trial (n = 480). Patients receiving confirmed ICI before December 31, 2018, and with requisite modeling data were included. For hepatitis, patients were excluded if they did not have laboratory measurements of the four liver enzymes within 1 year of ICI initiation (n = 274). We excluded patients with an indefinite colitis or pneumonitis diagnosis as determined by expert curators or a clinician (n = 95 and n = 151, respectively). In these cases, the patients' treating physicians did not give and laboratory or imaging did not yield a definitive diagnosis of colitis or pneumonitis or lack thereof. For 1-year overall survival (OS), patients lost to follow-up within 1 year of ICI initiation with no death date were excluded (n = 213). Unanticipated systematic reasons why some patients were lost to follow-up could have introduced bias into the models; additional details about OS data completeness and provenance are included in the Data Supplement. Patients were randomly partitioned into training (80%) and test (20%) sets for development and validation, respectively (n = 1,564 and 392 for hepatitis; n = 1,708 and 427 for colitis; n = 1,663 and 416 for pneumonitis; n = 1,614 and 403 for OS; Fig 1).
FIG 1.
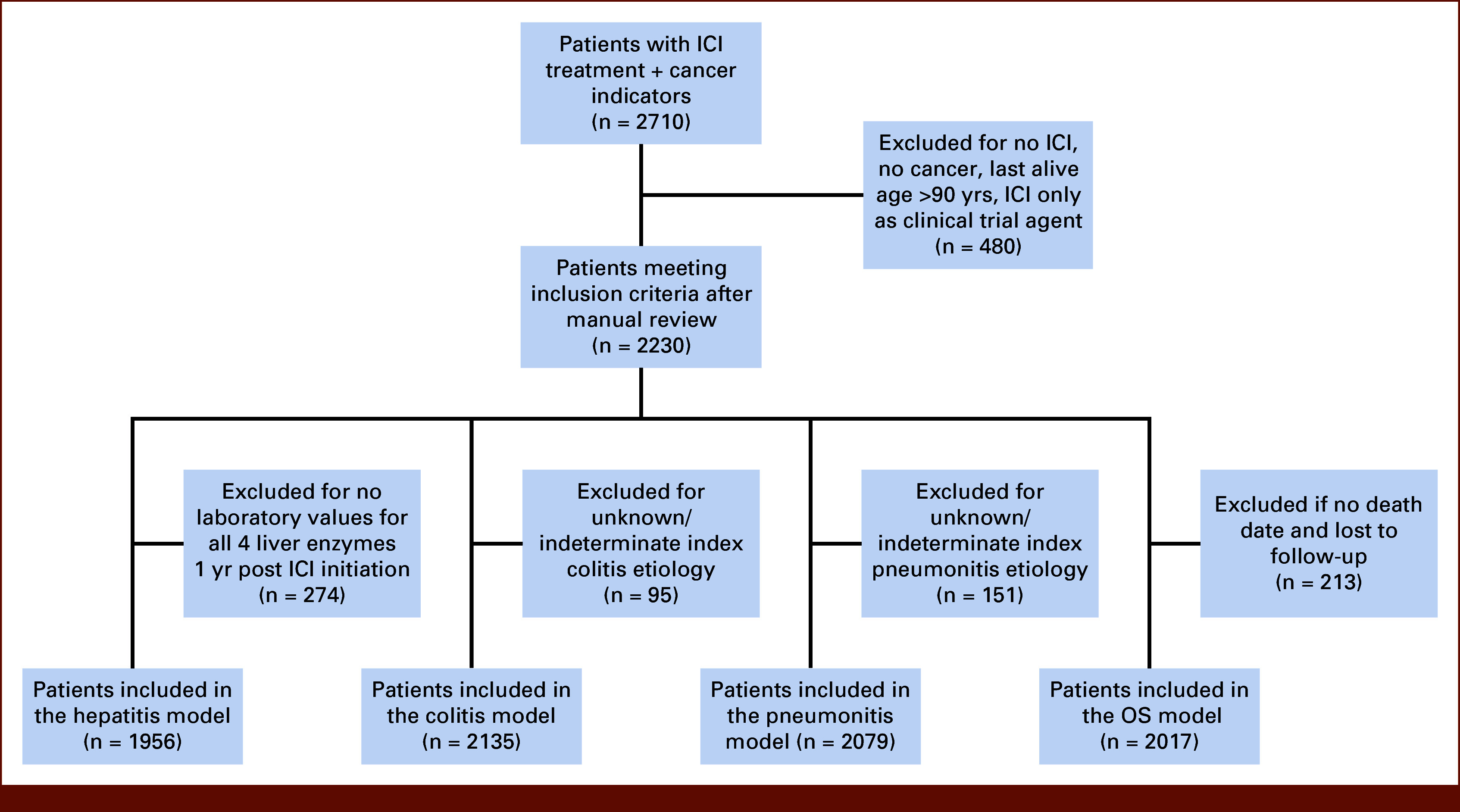
Flow diagram. ICI, immune checkpoint inhibitor; OS, overall survival.
Manual Curation of Unstructured EHR Data
Automated electronic health record (EHR) extraction methods often yield conflicting and incomplete information. To ensure robustness of predictive models, human data verification was used to reduce noise and improve label quality. We developed manual curation databases for clinicopathologic variables, pneumonitis, and colitis using Research Electronic Data Capture (REDCap).22,23 Standard curation protocols were used to extract and structure data from clinician notes and other EHR natural language elements. We manually curated variables for treatment start/stop dates (including treatment pauses), cancer diagnosis, biomarkers, and medication for each patient. For pneumonitis, curators globally searched each EHR for pneumonitis, followed by assignment of positive, negative, or unknown status for each new pneumonitis event. All unknown status cases were adjudicated by a physician (T.J.O). For colitis, because of the frequency of keywords (eg, diarrhea) needed to identify ICI-related colitis in EHR notes, we created an automated pipeline to facilitate chart review, which has been previously described.24 For positive colitis or pneumonitis cases, curators extracted occurrence dates, etiology, any ICI hold dates, and details of additional toxicity-related treatment. See curator training, QC process descriptions, and REDCap codebooks in the Data Supplement.
Design of the Predictive Models
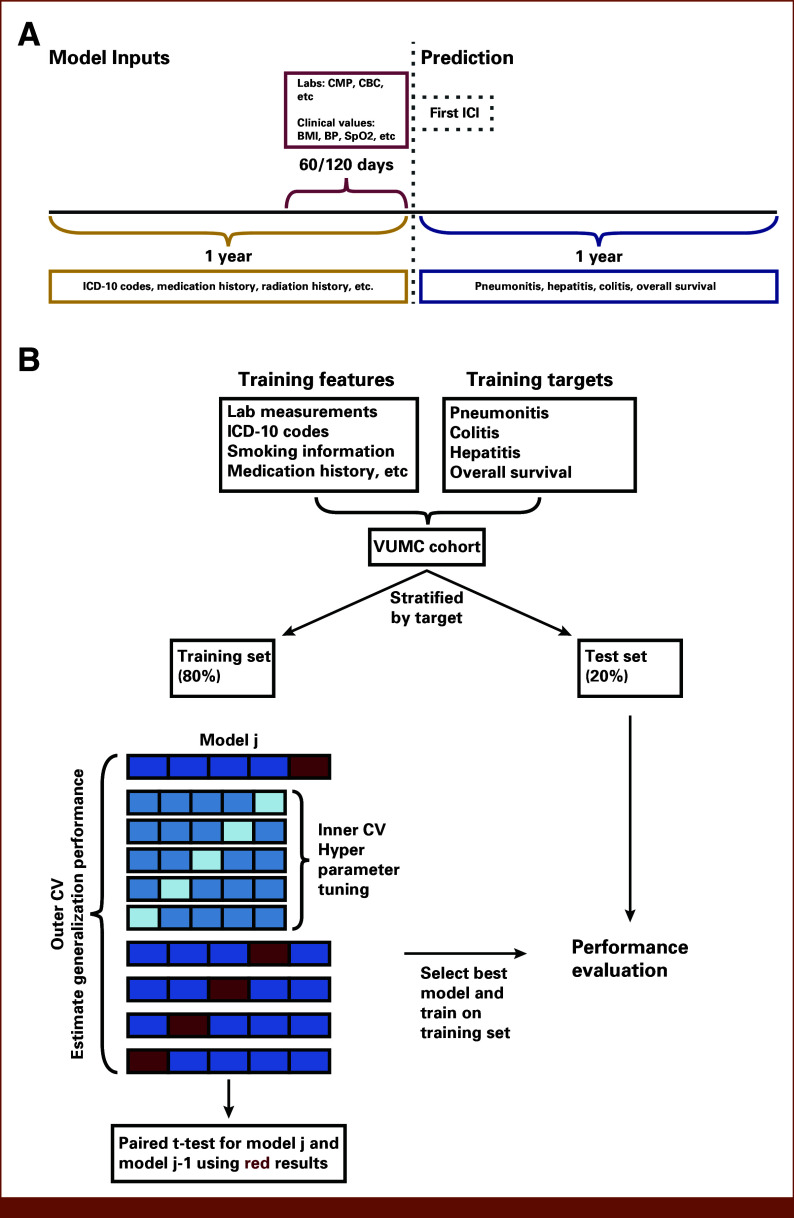
Figure 2A depicts the model design. For each patient, the prediction time point was set as ICI initiation date. Patient data were aggregated to create the predictive features for the models. A 60- or 120-day aggregation time window was applied to generate features from laboratory and clinical measurement data. A 1-year window was used for all other data sources, such as diagnosis codes, medications, smoking history, and procedures.
FIG 2.
Design of the predictive models and model development framework. (A) For each patient, ICI initiation date was set as the prediction time point (dotted line). A 60- or 120-day time window (red curly brace) was applied to aggregate laboratory and clinical measurements data (top red box), indicated in the pre-ICI period; a 1-year time window (gold curly brace) before ICI initiation (gray dotted box) was applied to other data types (eg, diagnosis codes, drugs; bottom left gold box). The features were used to train ML models to predict four binary outcomes (bottom right blue box). The binary outcome label indicates the occurrence of at least one episode (for toxicities) within 1 year of ICI initiation (blue curly brace). (B) For each outcome, patients were randomly partitioned into training (80%) and test (20%) sets using stratified split on the basis of the binary target. Nested CV was used in model development, hyperparameter optimization was performed in the inner loop, and generalization performance was estimated in the outer loop. Each experiment included a baseline and an alternative model. The alternative model was selected if it demonstrated statistically significant superiority over the baseline model based on a paired t-test using the outer loop AUC results. BP, blood pressure; CMP, complete metabolic panel; CV, cross-validation; ICD-10, International Classification of Diseases; ICI, immune checkpoint inhibitor; ML, machine learning; SpO2, oxygen saturation.
The four prediction outcomes consisted of three of the most common irAEs and OS as an effectiveness proxy. Binary outcome labels were created using a 1-year time window since most toxicity events occur by this time point. The goal was to build four binary classification models—one for each outcome—using ML.
Feature Engineering
Model features were created from both structured and curator-structured natural language EHR data. The REDCap data tables were processed into model development-compatible formats. VUMC stores structured EHR data using the Observational Medical Outcomes Partnership (OMOP) Common Data Model.25 We used OMOP data tables to derive features describing demographics, laboratory (eg, albumin) and clinical measurements (eg, BMI), diagnoses, drugs, procedures, visits, and observations from structured EHR data. In a multistep data transformation process, concept tables were used to map OMOP table concept codes to concept names (Fig 3). Condition codes were derived from EHR data encoded in different diagnosis coding systems (eg, International Classification of Diseases [ICD]-9, ICD-10, SNOMED CT [Systemized Nomenclature of Medicine—Clinical Terms]); all codes were transformed to ICD-10. The ICD-10 codes used in the features are defined in the Data Supplement (Table S1). Laboratory measurement units were harmonized for consistency. Sex variables recorded at VUMC are based on legal sex; therefore, this variable will not represent a unified variable of either sex assigned at birth or gender identity. Medication data were synthesized from both curator-structured data tables and structured EHR variable data. Intermediate tables were constructed for time-dependent data. The final data table was assembled by aggregating laboratory and clinical measurements within a 60- or 120-day time window and applying a 1-year time window to all other data types. The feature engineering procedure resulted in approximately 10,000 features.
FIG 3.
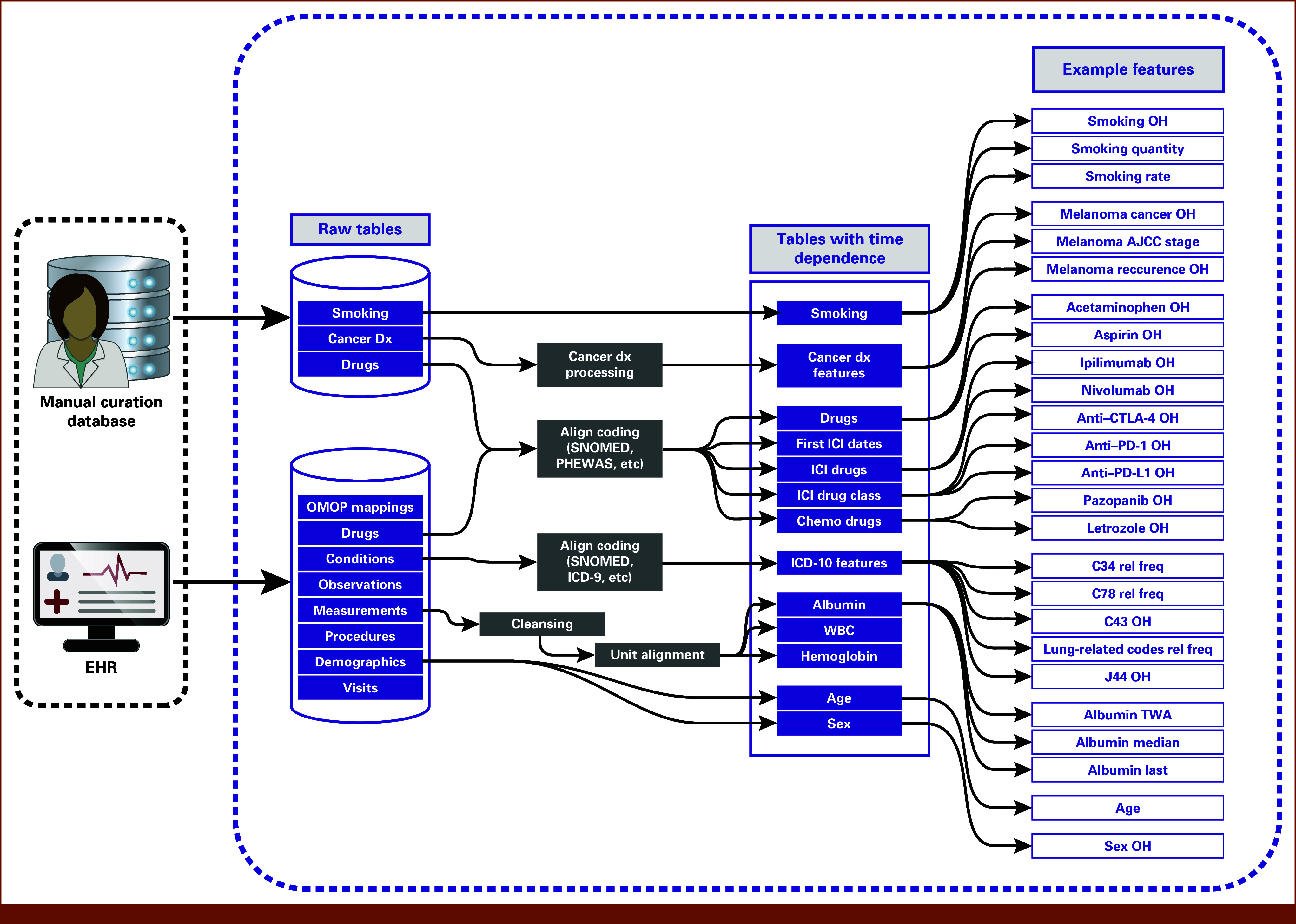
Feature engineering pipeline. Clinician notes and other unstructured EHR data were transformed into structured tables by curators. Data tables constructed directly from structured EHR variables included demographic, laboratory (eg, albumin levels) and clinical measurement (eg, BMI), condition, drug, procedure, visit, and observation data. Condition codes and drug codes were aligned into common coding systems. Laboratory measurements underwent cleansing procedures, and units were harmonized. Intermediate tables were constructed containing data for each patient at various time points. Features were created by aggregating data using different time windows, example features displayed on the right. The ICD-10 codes used in the features are defined in the Data Supplement (Table S1). AJCC, American Joint Committee on Cancer; BMI, body mass index; dx, diagnosis; EHR, electronic health record; ICD, International Classification of Diseases; ICI, immune checkpoint inhibitor; OH, one-hot encoding; OMOP, Observational Medical Outcomes Partnership; PHEWAS, phenome-wide association studies; rel freq, relative frequency; SNOMED, Systemized Nomenclature of Medicine; TWA, time-weighted average.
Prediction Outcomes
All outcomes were modeled as binary variables indicating occurrence (for death) or occurrence of at least one episode (for toxicities) within 1 year of ICI initiation. Although curators recorded EHR-identified toxicity etiologies, we modeled all-cause pneumonitis as the proxy variable instead of ICI-induced pneumonitis alone. This was decided because EHR documentation of etiology was inconsistent, and, even when recorded, clinical determination of pneumonitis etiology is imprecise and subjective. Although use of all-cause pneumonitis may sometimes lead to risk identification for non–ICI-caused pneumonitis, such as radiation-induced pneumonitis occurring during ICI therapy but shortly after radiation therapy, many pneumonitis cases would have had to have been excluded from the training and test data sets. Colitis prediction was limited to episodes that met a standardized definition of ICI-related etiology (eg, were clinically determined to be ICI-related, who had immune-related colitis on biopsy, whose colitis responded to ICI cessation, immune suppressive therapy and/or rebounded on ICI restart, etc). Hepatitis was defined, guided by clinical expertise, as any laboratory measurement in the 1-year window where any of the four liver enzymes, namely AST, ALT, alkaline phosphatase (ALP), and total bilirubin, exceeded three times the upper limit of normal. Similar to our use of all-cause pneumonitis, the specific etiology of hepatitis during ICI therapy can be difficult to ascertain and does not affect resulting clinical decisions. The hepatitis model may be limited by the inclusion of bilirubin, which can indicate progression. Additionally, although isolated bilirubin is not traditionally associated with hepatitis,26 bilirubin levels more than three times the upper limit of normal would often lead clinicians to discontinue therapy as they would in the case of typically defined hepatitis. OS served as a proxy variable for ICI-related effectiveness; death dates were obtained from either the EHR or from a third-party vendor (Redsson, Toledo, OH).
Model Development and Evaluation
For each outcome, we created development and test data sets using stratified random assignment to ensure equal proportions of patients with the outcome in the training and test data sets, which yielded different sets of patients in each outcome model. The characteristics of the data and prediction tasks shaped modeling framework choice. Because our data set is tabular and its scale does not meet the typical volume required for models with high complexity (usually deep learning models), we considered traditional ML approaches (ie, logistic regression, random forest algorithms). Furthermore, many studies using similar data apply traditional ML algorithms for prediction tasks.9,10 Our data set exceeded 2,200 patients while the number of engineered features exceeded 10,000. Because of the curse of dimensionality,27 the number of patients necessary to achieve accurate generalization of ML models grows exponentially with the dimensionality of the feature space, limiting us to a relatively small number of features (<100) for each model. Finally, we aimed to build models that are easy to deploy in clinical practice. This led us to start from a minimal feature set and experiment with adding new features. For each model, clinically validated features showing strong association with the binary target in the training set were incorporated into the minimal feature set. Subsequently, data scientists and clinicians collaboratively formulated hypotheses regarding additional features that may increase the predictive power of the models (Data Supplement). Feature selection was performed on the basis of validation performance in our setting. The immense number of possible feature combinations causes a high risk of overfitting in model selection.28 To mitigate this risk, we conducted hypothesis tests to make modeling decisions. Nested cross-validation (CV) was used to optimize the hyperparameters of the ML algorithms (Data Supplement, Table S2) in the inner loop (10-fold CV) and estimated the generalization performance in the outer loop (10-fold CV with three repeats). Each experiment incorporated a baseline model and an alternative model, where the latter included additional features or used a different ML algorithm. A paired t-test using the outer loop AUC results was conducted with the null hypothesis of no performance difference between models. If the null hypothesis was experimentally rejected at an a priori α of .05, the alternative model was selected. A moderate number of experiments were performed to reduce erroneous inferences from the multiple testing problem.29
For each outcome, a binary classification model was developed following the procedure above. Each model was evaluated on the corresponding 20% test set (Fig 2B). Predictive performance was measured using AUC, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV); 1,000-fold bootstrap was performed to calculate 95% CI (Data Supplement).
RESULTS
Cohort Characteristics
The study cohort included 2,230 patients (Table 1). The median age of patients was 63 years; 61.8% were male. The cohort primarily included three cancer types: melanoma (37.8%), lung cancer (27.3%), and genitourinary cancer (16.4%). Most patients (60.4%) received PD-1 ICI treatment. PD-L1 and CTLA-4 treatment were administered to 9.0% and 19.7% of the patients, respectively. 60.1% had chemotherapy before ICI. Slightly more than half of the patients (52.4%) had confirmed smoking history.
TABLE 1.
Characteristics of 2,230 Included Patients
| Patient Characteristic | No. (%) |
|---|---|
| Sex | |
| Female | 852 (38.2) |
| Male | 1,378 (61.8) |
| Cancer type | |
| Melanoma | 842 (37.8) |
| Lung | 608 (27.3) |
| Genitourinary | 365 (16.4) |
| Other | 490 (22.0) |
| Drug class | |
| PD-1 | 1,348 (60.4) |
| PD-L1 | 201 (9.0) |
| CTLA-4 | 440 (19.7) |
| PD-1 + CTLA-4 | 236 (10.6) |
| PD-L1 + CTLA-4 | 5 (0.2) |
| Traditional chemotherapy before ICI | 1,340 (60.1) |
| Smoking history | |
| Current or former smoker | 1,169 (52.4) |
| Never-smoker | 774 (34.7) |
| Unknown | 287 (12.9) |
| Age at first ICI, years, median (IQR) | 63 (54-70) |
NOTE. Details on the categorization of cancer diagnoses into cancer types can be found in the Data Supplement.
Abbreviation: ICI, immune checkpoint inhibitor.
Model Performance
For each outcome, we conducted multiple experiments and selected features using the model development framework, with statistically significantly superior validation performance driving model selection. Despite exploration of alternative ML algorithms including logistic regression, random forest was experimentally chosen for each outcome. Using the 20% test cohort, we evaluated overall model performance and model performance in the three largest cancer subgroups using AUC, sensitivity, specificity, PPV, and NPV (Table 2). Categorization of cancer diagnoses into subgroups was performed based on the keywords provided in the Data Supplement (Table S3). Except for AUC, all metrics require binarized predictions; predicted probabilities were dichotomized by optimizing sensitivity and specificity in the training set.30
TABLE 2.
Performance Results for the Four Models Predicting Three Toxicity Outcomes and OS in the 20% Test Set and in the Three Largest Cancer Diagnosis Groups
| Outcome Modeled | Diagnosis (N) | AUC (95% bootstrap CI) | Sensitivity (95% bootstrap CI) | Specificity (95% bootstrap CI) | PPV (95% bootstrap CI) | NPV (95% bootstrap CI) | IR, Half the Patients on the Basis of Model Predictions, % | IR, Random Selection of Patients, % |
|---|---|---|---|---|---|---|---|---|
| Pneumonitis 1y | All cancer types (n = 416) | 0.739 (0.638 to 0.823) | 0.711 (0.561 to 0.845) | 0.701 (0.653 to 0.746) | 0.193 (0.129 to 0.261) | 0.960 (0.937 to 0.981) | 3.8 | 9.1% |
| Melanoma (n = 166) | 0.590 (0.353 to 0.841) | 0.444 (0.125 to 0.800) | 0.917 (0.874 to 0.956) | 0.235 (0.059 to 0.462) | 0.966 (0.936 to 0.993) | 6.0 | 5.4% | |
| Lung cancer (n = 111) | 0.723 (0.600 to 0.840) | 0.952 (0.846 to 1.000) | 0.256 (0.172 to 0.346) | 0.230 (0.146 to 0.317) | 0.958 (0.867 to 1.000) | 10.7 | 18.9 | |
| GU cancer (n = 67) | 0.845 (0.686 to 0.960) | 0.857 (0.500 to 1.000) | 0.617 (0.484 to 0.741) | 0.207 (0.069 to 0.357) | 0.974 (0.914 to 1.000) | 0.0 | 10.4 | |
| Hepatitis 1y | All (n = 392) | 0.729 (0.655 to 0.804) | 0.455 (0.333 to 0.574) | 0.859 (0.822 to 0.899) | 0.395 (0.284 to 0.508) | 0.886 (0.851 to 0.921) | 8.2 | 16.8 |
| Melanoma (n = 136) | 0.567 (0.373 to 0.777) | 0.214 (0.000 to 0.455) | 0.893 (0.838 to 0.943) | 0.188 (0.000 to 0.400) | 0.908 (0.850 to 0.958) | 8.8 | 10.3 | |
| Lung cancer (n = 107) | 0.884 (0.792 to 0.965) | 0.500 (0.200 to 0.818) | 0.907 (0.845 to 0.959) | 0.357 (0.125 to 0.636) | 0.946 (0.896 to 0.989) | 0.0 | 9.3 | |
| GU cancer (n = 53) | 0.639 (0.376 to 0.863) | 0.444 (0.111 to 0.777) | 0.795 (0.674 to 0.909) | 0.308 (0.077 to 0.583) | 0.875 (0.769 to 0.958) | 15.4 | 17.0 | |
| Colitis 1y | All (n = 427) | 0.755 (0.638 to 0.856) | 0.750 (0.581 to 0.909) | 0.608 (0.563 to 0.654) | 0.134 (0.086 to 0.182) | 0.968 (0.943 to 0.988) | 3.3 | 7.5 |
| Melanoma (n = 161) | 0.747 (0.631 to 0.850) | 0.957 (0.857 to 1.000) | 0.174 (0.112 to 0.238) | 0.162 (0.100 to 0.223) | 0.960 (0.864 to 1.000) | 5.0 | 14.3 | |
| Lung cancer (n = 112) | 0.853 (0.717 to 0.964) | 0.750 (0.000 to 1.000) | 0.833 (0.764 to 0.900) | 0.143 (0.000 to 0.308) | 0.989 (0.958 to 1.000) | 0.0 | 3.6 | |
| GU cancer (n = 68) | 0.560 (0.447 to 0.672) | 0.000 (0.000 to 0.000) | 0.806 (0.712 to 0.894) | 0.000 (0.000 to 0.000) | 0.982 (0.934 to 0.983) | 0.0 | 1.5 | |
| OS 1y | All (n = 403) | 0.752 (0.706 to 0.796) | 0.817 (0.764 to 0.865) | 0.558 (0.487 to 0.627) | 0.664 (0.609 to 0.719) | 0.741 (0.673 to 0.806) | 67.8 | 51.6 |
| Melanoma (n = 158) | 0.794 (0.716 to 0.861) | 0.917 (0.861 to 0.868) | 0.500 (0.362 to 0.639) | 0.798 (0.723 to 0.866) | 0.735 (0.585 to 0.882) | 83.5 | 68.4 | |
| Lung cancer (n = 115) | 0.681 (0.580 to 0.781) | 0.674 (0.533 to 0.814) | 0.611 (0.500 to 0.714) | 0.509 (0.387 to 0.630) | 0.759 (0.642 to 0.8680 | 50.0 | 37.4 | |
| GU cancer (n = 63) | 0.741 (0.600 to 0.858) | 0.742 (0.576 to 0.889) | 0.625 (0.448 to 0.814) | 0.657 (0.487 to 0.814) | 0.714 (0.542 to 0.880) | 65.6 | 49.2 |
NOTE. Detailed interpretation of the results can be found in the Data Supplement.
Abbreviations: 1y, 1-year prediction time window; GU, genitourinary; IR, incidence rate; NPV, negative predictive value; OS, overall survival; PPV, positive predictive value.
All models performed reasonably well,31 achieving an AUC between 0.72 and 0.76 when considering all test patients (Table 2). Lower limits of the 95% bootstrap CI were above 0.63 for all models, demonstrating statistically significant superiority to random chance. The toxicity models' performances vary among cancer subgroups, whereas the effectiveness model exhibits good performance throughout. All models perform strongly in patients with lung cancer. Pneumonitis and hepatitis models show diminished performance for patients with melanoma as does the colitis model for patients with genitourinary cancer.
We also assessed the models' capabilities as an eligibility criterion for a clinical trial cohort of patients with a low risk of toxicity or high likelihood of ICI effectiveness (two rightmost columns in Table 2). We imagined selection of patients at the 50th percentile or lower likelihood of experiencing a toxicity or death. Random selection would yield comparable toxicity incidences and OS rates as those observed in the complete data set. In Table 2, we compare the incidence rates on the basis of random selection versus prioritization of 50% by selecting the patients with the lowest model-predicted toxicity probabilities and highest model-predicted OS probabilities. The results show that model-driven prioritization reduces the incidence rate for each toxicity by over half and elevates the OS incidence rate from 52% to 68%.
The modeling strategy selected a different feature subset for each outcome; these features cover a wide variety of patient information, including ICD-10 codes, procedures, smoking history, medication history, demographics data, and laboratory and clinical measurements (Table 3). Most features are EHR structured data; only two (smoking status, number of ICI drugs) are derived from unstructured notes. We used a 60- or 120-day time window before treatment initiation measurement aggregation, using basic functions such as minimum, maximum, last value, and time-weighted average. Other data types were aggregated over a 1-year time window before treatment initiation, primarily using relative frequency and one-hot encoding. Smoking indicator was a binary variable distinguishing ever-smokers and never-smokers. Feature contributions for each model are depicted in the Data Supplement (Fig S1). Feature sensitivity analysis is detailed in the Data Supplement (Fig S2).
TABLE 3.
Features Used in Each of the Models
| Outcome Modeled | Feature | Data Type | Time Window |
|---|---|---|---|
| Pneumonitis 1y | Rel freq of C34, C78, R91, J, R05, R06, R07, R09 codesa | ICD-10 condition codes | 1 year |
| Rel freq of C34 code | ICD-10 condition codes | ||
| Rel freq of C78 code | ICD-10 condition codes | ||
| OH of J44 code indicator | ICD-10 condition codes | ||
| OH of smoking indicator | Curation | Smoking history | |
| OH of smoking indicator and OH (C34 or C78 codes) indicator interaction | Curation and ICD-10 condition codes | Smoking history and 1 year | |
| TWA of oxygen saturation in blood values | Laboratory measurements | 120 days | |
| TWA of body mass index values | Body measurements | ||
| Hepatitis 1y | Min, max, last value, TWA of aspartate aminotransferase (AST) values | Laboratory measurements | 60 days |
| Min, max, last value, TWA of ALT values | |||
| Min, max, last value, TWA of alkaline phosphatase (ALP) values | |||
| Min, max, last value, TWA of total bilirubin values | |||
| Colitis 1y | Rel freq of K50, K51, K52, K57, K58 codes | ICD-10 condition codes | 1 year |
| OH of chemotherapy administration indicator | Procedures | ||
| OH of C43 code indicator | ICD-10 condition codes | ||
| OH of anti–CTLA-4 drug indicator | Drugs | ||
| OH of anti–PD-1 drug indicator | Drugs | ||
| Freq of ICI drugs | Drugs | ||
| Rel freqs of hemoglobin values below and above normal range | Laboratory measurements | 120 days | |
| Rel freqs of albumin values below and above normal range | |||
| Rel freqs of red blood cell values below and above normal range | |||
| Rel freqs of absolute lymphocytes count values below and above normal range | |||
| Rel freqs of white blood cell values below and above normal range | |||
| OS 1y | Freq of chemotherapy instances | Procedures | 1 year |
| Last value, TWA of albumin values | Laboratory measurements | 120 days | |
| Last value of neutrophils % values | |||
| Last value of lymphocytes % values | |||
| Last value of lactate dehydrogenase (LDH) | |||
| Last value, time-weighted average of alkaline phosphatase values | |||
| Average difference of alanine transaminase values | |||
| Average difference of packed cell volume % values | |||
| Last value of oxygen saturation in blood | |||
| Frequency of C78 codes | ICD-10 condition codes | 1 year | |
| Age in years | Demographics | At first ICI |
Abbreviations: freq, frequency; ICD, International Classification of Diseases; ICI, immune checkpoint inhibitor; OH, one-hot encoding; OS, overall survival; rel freq, relative frequency; TWA, time-weighted average.
Codes collapsed to a single indicator due to clinical reasoning (all codes lung-related). Details on feature types and descriptions of the ICD-10 codes can be found in the Data Supplement. Feature contributions are shown in Data Supplement (Fig S1).
This study relied on patients who had data available before initiation of ICI. However, not every feature was consistently populated across all patients. Missing data were imputed using the median of the feature in the training folds.
DISCUSSION
Here, we describe ML models leveraging routinely collected clinical data to accurately forecast effectiveness and toxicities of cancer immunotherapy. To our knowledge, this approach is the first capable of immunotherapy risks and benefits assessment using only routinely collected EHR data. Most ML-based immunotherapy solutions focus on a single prediction outcome, usually effectiveness, and are restricted to one or few cancer indications and a single ICI drug.16,17 Although studies have been conducted that focus on immunotherapy irAEs,19,20 this research marks the first to address three significant toxicities.
A primary advantage of our models is that inputs are readily available in clinical practice. Only two features—smoking status and number of ICI drugs—were drawn from manually curated data. In the pneumonitis model, Smoking:C34/C78 was the third and Smoking was the eighth most important feature as depicted in the Data Supplement (Fig S1). ICI drugs, freq was the fourth most important feature in the colitis model. These features were incorporated as no alternative feature sets that excluded these variables exhibited statistically significant superior performance. They are easily attainable by treating physicians, their assistants, or clinical trial staff and could be readily entered into the model. We intentionally designed models using practically available data because many published models are not available for clinical use and/or do not have clinical utility because of onerous additional testing or data ascertainment (eg, using nonstandard biomarkers).
Because of broad availability of the input features, the models presented here have potential for wide deployment and adoption. One envisaged use case is to assist prescreening of eligible clinical trials patients. The OS and toxicity models may inform novel immunotherapy trial cohort selection on the basis of patient safety profiles. Toxicity prediction may also assist patient management in routine clinical care through early detection of high-grade toxicities that cause significant morbidity and mortality. Improved awareness of likely toxicities will allow additional monitoring visits and tests for higher-risk patients, facilitating early detection and management of irAEs. Other scenarios include assigning patients at high risk of toxicity to less intensive therapy (eg, monotherapy instead of combination, particularly if they had favorable OS profiles) or conducting early intervention or prevention studies with immunomodulators to prevent toxicities. In clinical practice, attention should be paid to tradeoffs in sensitivity and PPV. In some scenarios a poor PPV may not be a barrier because the intervention in response to an elevated risk for a patient is feasible and reasonable, whereas in other scenarios, clinicians may prefer a strong NPV to optimize clinical trial enrollment. Care should be taken in applying these models clinically, considering the implications of both false positives and false negatives.
Direct comparison of our models to those in the literature is challenging because of prediction outcome definition variation, data types used, and data set size differences. Nevertheless, our models' performances are similar to those previously reported.16,20 OS model performance is comparable with summary AUC (sAUC) values reported in a large-scale meta-analysis that pooled results of studies on US Food and Drug Administration–approved PD-L1 immunohistochemistry tests.10 A significant limitation of the previous study is that sAUC was calculated for objective response rate rather than 1-year OS, on the basis of prior ICI trials reporting it as a well-correlated surrogate for 1-year OS.32 Our 1-year OS model on the whole test cohort shows a significant gain in performance (AUC = 0.75 v sAUC of 0.65; PPV of 0.66 v 0.34) versus this previous study.
Additional work will be needed to ensure that these models align with clinical knowledge and have practical clinical utility. The work presented in this article represents the first step: these models were developed in close collaboration with clinical subject matter experts, as tying appropriate potential interventions to the model predictions will be critical for success. In the future, we plan to extend this work to longitudinal predictions. We are in the process of validating models with external data sets from other health systems and countries. EHR-based models could provide clinical decision support to oncologists in tumor board settings. It remains to be seen whether additional data sources, such as tumor and germline genomic data, can further improve models as these data become increasingly available.
ACKNOWLEDGMENT
We thank the data curation team, including authors M.L.L.-N, M.E.H., N.M.J., C.N.M., M.B., Y.M., and M.M.; and contributors (alphabetical) Ryan Ahmed, Mary Banasiewicz, Kimberly Fields, Adara Holland, Steven Houtschilt, Alexandria Manis, Lilyanna McArthur, Lori Michalowski, Ndidiamaka Odinkemelu, Rhonda Potter, Joyce Raisan, Andrew Sayce, Timothy Schurz, Alysse Sephel, Marina Shannon, Morgan Shannon, Jarrod Smith, Susan Sommers, Samuel Trump, Stephanie Winchell, and Annie Zwaschka. We also thank Lara Franck, Tahsin Reasat, Peter Louis, Gergely Horváth, Levente Török, and the Vanderbilt Coordinating Center for their contributions, as well as Julia Casey, Mohamed Fouda, Reed Omary, Jennifer Pietenpol, and Kimryn Rathmell for valuable advice.
PRIOR PRESENTATION
Presented in part at IO360, New York, NY, February 7–10, 2023, and at SITC, San Diego, CA, November 1-5, 2023
SUPPORT
Supported by GE HealthCare. REDCap is funded by UL1 TR000445 from NCATS/NIH, the recipient is Gordon Bernard.
AUTHOR CONTRIBUTIONS
Conception and design: Levente Lippenszky, Kathleen F. Mittendorf, Michele L. LeNoue-Newton, Pablo Napan-Molina, Balázs Laczi, Eszter Csernai, David S. Smith, Ben H. Park, Christine M. Micheel, Daniel Fabbri, Jan Wolber, Travis J. Osterman
Collection and assembly of data: Levente Lippenszky, Kathleen F. Mittendorf, Michele L. LeNoue-Newton, Pablo Napan-Molina, Protiva Rahman, Cheng Ye, Neha M. Jain, Marilyn E. Holt, Christina N. Maxwell, Madeleine Ball, Yufang Ma, Margaret B. Mitchell, Christine M. Micheel, Jan Wolber
Data analysis and interpretation: Levente Lippenszky, Kathleen F. Mittendorf, Zoltán Kiss, Michele L. LeNoue-Newton, Pablo Napan-Molina, Eszter Csernai, Douglas B. Johnson, David S. Smith, Ben H. Park, Christine M. Micheel, Daniel Fabbri, Jan Wolber
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Levente Lippenszky
Employment: GE Healthcare
Patents, Royalties, Other Intellectual Property: Inventor on pending patent applications as part of full-time employment with GE Healthcare
Travel, Accommodations, Expenses: GE Healthcare
Kathleen F. Mittendorf
Employment: Zoom Care, Lavender Spectrum Health, Hands On Medicine, Folx Health, Folx Health
Leadership: Lavender Spectrum Health
Stock and Other Ownership Interests: Lavender Spectrum Health
Research Funding: GE Healthcare (Inst)
Patents, Royalties, Other Intellectual Property: Patent pending - algorithms for predicting immunotoxicity and efficacy of immune therapy in cancer
Uncompensated Relationships: Lavender Spectrum Health
Zoltán Kiss
Employment: GE Healthcare
Michele L. LeNoue-Newton
Employment: DaVita
Stock and Other Ownership Interests: DaVita, General Electric
Research Funding: GE Healthcare (Inst)
Patents, Royalties, Other Intellectual Property: Pending patent application 63/335,215 filed in relation to work conducted on predictive modeling
Pablo Napan-Molina
Patents, Royalties, Other Intellectual Property: Patent pending about toxicity modeling methodologies
Eszter Csernai
Employment: GE HealthCare
Patents, Royalties, Other Intellectual Property: GE HealthCare patent pending
Marilyn E. Holt
Employment: HCA/Sarah Cannon, McKesson
Margaret B. Mitchell
Employment: Mass General Brigham
Travel, Accommodations, Expenses: Mass General Brigham
Douglas B. Johnson
Consulting or Advisory Role: Bristol Myers Squibb, Merck, Novartis, Pfizer, Mosaic ImmunoEngineering, Targovax, Mallinckrodt
Research Funding: Incyte, Bristol Myers Squibb
Patents, Royalties, Other Intellectual Property: Intellectual property and patents pending surrounding use of MHC-II and response to immune therapy
David S. Smith
Employment: Vanderbilt University Medical Center
Research Funding: GE Healthcare (Inst)
Ben H. Park
Leadership: Loxo
Stock and Other Ownership Interests: Loxo, Celcuity
Consulting or Advisory Role: Horizon Discovery, Loxo, Casdin Capital, Jackson Laboratory for Genomic Medicine, Celcuity, Sermonix Pharmaceuticals, Hologic, EQRx, Guardant Health, Janssen, Caris Life Sciences, AstraZeneca
Research Funding: Abbvie, Pfizer, GE Healthcare, Lilly
Patents, Royalties, Other Intellectual Property: Royalties paid through inventions at Johns Hopkins University by Horizon Discovery LTD
Travel, Accommodations, Expenses: Lilly, Loxo
Uncompensated Relationships: Tempus
Christine M. Micheel
Research Funding: GenomOncology (Inst), GE Healthcare (Inst)
Daniel Fabbri
Employment: Vanderbilt University Medical Center
Leadership: Maize Analytics
Stock and Other Ownership Interests: Imprivata, Prediction Health, Kythera labs, Innova Health
Research Funding: AACR Project GENIE (Inst), GE Healthcare (Inst)
Patents, Royalties, Other Intellectual Property: Royalty from University of Michigan for patent technology for data privacy and security. Related to Maize Analytics
Jan Wolber
Employment: GE Healthcare
Stock and Other Ownership Interests: GE Healthcare
Patents, Royalties, Other Intellectual Property: Inventor on various patents and patent applications as part of full-time employment with GE Healthcare
Travel, Accommodations, Expenses: GE Healthcare
Travis J. Osterman
Stock and Other Ownership Interests: Faculty Coaching
Honoraria: Amazon Web Services
Consulting or Advisory Role: eHealth, AstraZeneca, Outcomes Insights, Biodesix, MDoutlook, GenomOncology, Cota Healthcare, Cota Healthcare, Flagship Biosciences, Microsoft, Dedham Group, Oncollege
Research Funding: GE Healthcare, Microsoft, IBM
Travel, Accommodations, Expenses: GE Healthcare, Amazon Web Services
No other potential conflicts of interest were reported.
REFERENCES
- 1.Lin EP, Hsu CY, Berry L, et al. : Analysis of cancer survival associated with immune checkpoint inhibitors after statistical adjustment: A systematic review and meta-analyses. JAMA Netw Open 5:e2227211, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martins F, Sofiya L, Sykiotis GP, et al. : Adverse effects of immune-checkpoint inhibitors: Epidemiology, management and surveillance. Nat Rev Clin Oncol 16:563-580, 2019 [DOI] [PubMed] [Google Scholar]
- 3.Waldman AD, Fritz JM, Lenardo MJ: A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat Rev Immunol 20:651-668, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson DB, Nebhan CA, Moslehi JJ, et al. : Immune-checkpoint inhibitors: Long-term implications of toxicity. Nat Rev Clin Oncol 19:254-267, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gumusay O, Callan J, Rugo HS: Immunotherapy toxicity: Identification and management. Breast Cancer Res Treat 192:1-17, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maher VE, Fernandes LL, Weinstock C, et al. : Analysis of the association between adverse events and outcome in patients receiving a programmed death protein 1 or programmed death ligand 1 antibody. J Clin Oncol 37:2730-2737, 2019 [DOI] [PubMed] [Google Scholar]
- 7.Eggermont AMM, Kicinski M, Blank CU, et al. : Association between immune-related adverse events and recurrence-free survival among patients with stage III melanoma randomized to receive pembrolizumab or placebo: A secondary analysis of a randomized clinical trial. JAMA Oncol 6:519-527, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akamatsu H, Murakami E, Oyanagi J, et al. : Immune-related adverse events by immune checkpoint inhibitors significantly predict durable efficacy even in responders with advanced non-small cell lung cancer. Oncologist 25:e679-e83, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider BJ, Naidoo J, Santomasso BD, et al. : Management of immune-related adverse events in patients treated with immune checkpoint inhibitor therapy: ASCO guideline update. J Clin Oncol 39:4073-4126, 2021 [DOI] [PubMed] [Google Scholar]
- 10.Lu S, Stein JE, Rimm DL, et al. : Comparison of biomarker modalities for predicting response to PD-1/PD-L1 checkpoint blockade: A systematic review and meta-analysis. JAMA Oncol 5:1195-1204, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei F, Azuma K, Nakahara Y, et al. : Machine learning for prediction of immunotherapeutic outcome in non-small-cell lung cancer based on circulating cytokine signatures. J Immunother Cancer 11:e006788, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong L, Gong J, Sun X, et al. : Identification and prediction of immune checkpoint inhibitors-related pneumonitis by machine learning. Front Immunol 14:1138489, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman RS, Jung S, Balko JM, et al. : Biomarkers of immune checkpoint inhibitor response and toxicity: Challenges and opportunities. Immunol Rev 318:157-166, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He LN, Li H, Du W, et al. : Machine learning-based risk model incorporating tumor immune and stromal contexture predicts cancer prognosis and immunotherapy efficacy. iScience 26:107058, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng W, Wang J, Yang J, et al. : Identification of immune activation-related gene signature for predicting prognosis and immunotherapy efficacy in lung adenocarcinoma. Front Immunol 14:1217590, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pires da Silva I, Ahmed T, McQuade JL, et al. : Clinical models to define response and survival with anti-PD-1 antibodies alone or combined with ipilimumab in metastatic melanoma. J Clin Oncol 40:1068-1080, 2022 [DOI] [PubMed] [Google Scholar]
- 17.Wu Y, Zhu W, Wang J, et al. : Using machine learning for mortality prediction and risk stratification in atezolizumab-treated cancer patients: Integrative analysis of eight clinical trials. Cancer Med 12:3744-3757, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dionese M, Basso U, Pierantoni F, et al. : Prognostic role of systemic inflammation indexes in metastatic urothelial carcinoma treated with immunotherapy. Future Sci OA 9:FSO878, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim W, Cho YA, Kim DC, et al. : Factors associated with thyroid-related adverse events in patients receiving PD-1 or PD-L1 inhibitors using machine learning models. Cancers (Basel) 13:5465, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou JG, Wong AH, Wang H, et al. : Elucidation of the application of blood test biomarkers to predict immune-related adverse events in atezolizumab-treated NSCLC patients using machine learning methods. Front Immunol 13:862752, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benzekry S, Grangeon M, Karlsen M, et al. : Machine learning for prediction of immunotherapy efficacy in non-small cell lung cancer from simple clinical and biological data. Cancers (Basel) 13:6210, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris PA, Taylor R, Thielke R, et al. : Research Electronic Data Capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42:377-381, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris PA, Taylor R, Minor BL, et al. : The REDCap consortium: Building an international community of software platform partners. J Biomed Inform 95:103208, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman P, Ye C, Mittendorf KF, et al. : Accelerated curation of checkpoint inhibitor-induced colitis cases from electronic health records. JAMIA Open 6:ooad017, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.OHDSI CDM Working Group : OMOP common data model. https://ohdsi.github.io/CommonDataModel/
- 26.Shroff H, Maddur H: Isolated elevated bilirubin. Clin Liver Dis (Hoboken) 15:153-156, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keogh E, Mueen A: Curse of dimensionality, in Sammut C, Webb GI (eds): Encyclopedia of Machine Learning. Boston, MA, Springer US, 2010, pp 257-258 [Google Scholar]
- 28.Cawley GC, Talbot NLC: On over-fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Res 11:2079-2107, 2010 [Google Scholar]
- 29.Streiner DL: Best (but oft-forgotten) practices: The multiple problems of multiplicity-whether and how to correct for many statistical tests. Am J Clin Nutr 102:721-728, 2015 [DOI] [PubMed] [Google Scholar]
- 30.Chowell D, Yoo SK, Valero C, et al. : Improved prediction of immune checkpoint blockade efficacy across multiple cancer types. Nat Biotechnol 40:499-506, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosmer DW, Lemeshow S, Sturdivant RX: Applied Logistic Regression Electronic Resource (ed 3). Hoboken, NJ, Wiley, 2013 [Google Scholar]
- 32.Kok PS, Yoon WH, Lord S, et al. : Tumor response end points as surrogates for overall survival in immune checkpoint inhibitor trials: A systematic review and meta-analysis. JCO Precis Oncol 10.1200/PO.21.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]