Abstract
The blue whale, Balaenoptera musculus, is the largest animal known to have ever existed, making it an important case study in longevity and resistance to cancer. To further this and other blue whale-related research, we report a reference-quality, long-read-based genome assembly of this fascinating species. We assembled the genome from PacBio long reads and utilized Illumina/10×, optical maps, and Hi-C data for scaffolding, polishing, and manual curation. We also provided long read RNA-seq data to facilitate the annotation of the assembly by NCBI and Ensembl. Additionally, we annotated both haplotypes using TOGA and measured the genome size by flow cytometry. We then compared the blue whale genome with other cetaceans and artiodactyls, including vaquita (Phocoena sinus), the world's smallest cetacean, to investigate blue whale's unique biological traits. We found a dramatic amplification of several genes in the blue whale genome resulting from a recent burst in segmental duplications, though the possible connection between this amplification and giant body size requires further study. We also discovered sites in the insulin-like growth factor-1 gene correlated with body size in cetaceans. Finally, using our assembly to examine the heterozygosity and historical demography of Pacific and Atlantic blue whale populations, we found that the genomes of both populations are highly heterozygous and that their genetic isolation dates to the last interglacial period. Taken together, these results indicate how a high-quality, annotated blue whale genome will serve as an important resource for biology, evolution, and conservation research.
Keywords: cetaceans, body size, evolution, conservation, developmental biology, genetic diversity, animal genomes, segmental duplications
Introduction
The blue whale appears to be the largest animal to have ever existed (Sears and Perrin 2008). An adult can reach up to 110 feet (∼33 m) and weigh 330,000 pounds (150 metric tons) (Sears and Perrin 2008). Genomic studies of giant animals are of interest to several subfields of biomedical science. Understanding developmental mechanisms that control body size may have applications in regenerative medicine and animal husbandry. Large mammals tend to have long lives and have developed mechanisms that make them resistant to cancer, in spite of having orders of magnitude more cells (and thus more cell divisions). This puzzling phenomenon is known as Peto's Paradox (Caulin and Maley 2011; Tollis et al. 2017). Previously sequenced genomes of large animals, including other large whales, yielded some clues with regard to their possible mechanisms of cancer resistance. These include, for example, mutations and duplications of known tumor suppressors and other potentially relevant genes involved in processes such as DNA repair and apoptosis (Keane et al. 2015; Sulak et al. 2016; Vazquez et al. 2018; Tollis et al. 2019), and control over the abundance of microsatellite repeats (Park et al. 2016).
Mammalian genomes contain relatively large regions of duplicated sequence, known as segmental duplications (SDs), many of which contain genes. Copy number differences in genes inside SD loci have been reported as associated with longevity and increased body size in cetaceans (Tollis et al. 2019; Tejada-Martinez et al. 2021) and elephants (Sulak et al. 2016; Vazquez et al. 2018). In order to identify SD events that may affect mammalian development, body size, longevity, or susceptibility to cancer, we have compared the blue whale to the vaquita (Phocoena sinus), the world's smallest cetacean. Vaquitas, weighing up to 54.5 kg, differ from the blue whale by nearly 3,000-fold in body size. There is also a substantial difference in life span, with vaquitas living about 20 yr and blue whales living 80 to 90 yr (Hohn et al. 1996; Sears and Perrin 2008). The vaquita genome was sequenced and assembled using long-read data, enabling comparative analysis of repetitive DNA. We also computed SDs from long-read genomes assembled for bottlenose dolphin (Tursiops truncatus), the only other cetacean for which such data were available at the time of writing, and cattle (Bos taurus), for comparison as a distantly related artiodactyl (Rosen et al. 2020; Vertebrate Genomes Project 2020b; Morin et al. 2021).
The blue whale is an endangered species and included on the IUCN Red List, on Appendix I of the Convention on International Trade in Endangered Species of Wild Fauna and Flora, and on the Convention on the Conservation of Migratory Species of Wild Animals (Cooke 2018). Genomes of endangered species facilitate studies of their population structure, diversity, and demographic history, thereby aiding conservation efforts (Ming et al. 2019; Zhou et al. 2019; Morin et al. 2021; Robinson et al. 2022).
Here we report a reference-quality genome assembly and several analyses that may shed light on important aspects of blue whale biology. Our findings include a recent burst in segmental duplication activity, which resulted in amplification of several genes. We also discovered that the insulin-like growth factor 1 (IGF1) gene has several sites whose alleles appear to be associated with large body size in cetaceans. Finally, we show that the genomes of both Pacific and Atlantic blue whales are highly heterozygous, suggesting large, genetically diverse populations that became genetically isolated from each other during the last interglacial period.
Results
A Reference-Quality Genome Assembly
We generated a reference-quality genome assembly using methods developed by the Vertebrate Genome Project (VGP). The assembly is based on PacBio long reads, with Illumina/10×, Bionano optical maps, and Dovetail Hi-C used for scaffolding and polishing (Rhie et al. 2021). Manual curation resulted in multiple corrections of the computationally generated assembly, introducing 29 breaks and 64 joins between contigs within scaffolds (supplementary fig. S1, Supplementary Material online). A total of 99.8% of the assembled sequence could be assigned to 23 chromosomal-level scaffolds, which reflects the expected karyotype (21 autosomes plus X and Y). The Y chromosome could only be partially assembled (supplementary figs. S2 and S3, Supplementary Material online). To further improve the assembly, we applied additional false duplication removal and base accuracy polishing steps (supplementary methods, Supplementary Material online). Our assembly quality metrics are shown in Table 1. We also assembled the mitogenome, using the mitoVGP pipeline (Formenti et al. 2021) (supplementary methods, Supplementary Material online).
Table 1.
Assembly quality metrics
| Quality category | Quality metric | Computation method | Value |
|---|---|---|---|
| Input data | Sequencing technology | … | PacBio CLR |
| Coverage | … | 51X | |
| Scaffolding and polishing | … | 10X, optical maps, Hi-C | |
| Contiguity | Contig (NG50)a | VGP asm_stats.sh | 5,559,837 |
| Scaffold (NG50)a | VGP asm_stats.sh | 107,421,550 | |
| Num gaps per Gb | VGP asm_stats.sh | 329 | |
| CC ratio | Manual calculation | 42.3 | |
| Structural accuracy | False duplications | BUSCO % Duplicatedb | 1.3 |
| Curation improvements | N/A | Yes | |
| Base accuracy | Base pair QV | MERQURYc | 39.4 |
| k-mer completeness, primary | MERQURYc | 94.2 | |
| k-mer completeness, diploid | MERQURYc | 98.3 | |
| % of genes with frameshift mutations | GNOMON | 10% | |
| Haplotype phasing | Phased block (N50) | N50 of alt haplotype | 275,582 |
| Functional completeness | Genes, primary haplotype | BUSCO % Completeb | 88.1 |
| Genes, diploid | BUSCO % Completeb | 89.2 |
aAssuming genome size of 2,695,100,000, per flow cytometry estimate.
bBUSCO4, Cetartiodactyla lineage, ODB10 database. Running BUSCO in genome mode produced different scores compared to those shown in Fig. 1, which were generated by NCBI based on RefSeq annotations.
cUsing an independent Illumina sequencing dataset from the same blue whale, which has not been used in any of the assemblies reported in this table (Vertebrate Genomes Project 2020a).
Our assembly has been annotated by NCBI (Thibaud-Nissen et al. 2013; National Center for Biotechnology Information (US) 2020) and Ensembl (Ensembl 2020). Additionally, we annotated both primary and alternate pseudohaplotype by projecting human and mouse genes using TOGA (Hiller Lab 2020a, 2020b; Kirilenko et al. 2023). We also predicted GO terms for all protein coding genes identified by the NCBI Eukaryotic Genome Annotation Pipeline using Phylo-PFP (Jain and Kihara 2019) as described in supplementary methods, Supplementary Material online: see supplementary file blue_whale.tar.gz in the Open Science Framework (OSF) repository (Jain and Kihara 2022).
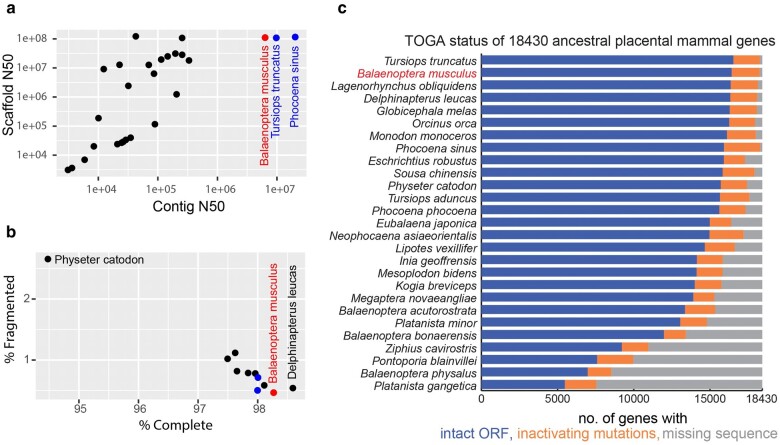
Our blue whale genome assembly is one of the highest quality of any cetacean species. Figure 1 shows data for cetacean assemblies available in the NCBI Assembly database (National Center for Biotechnology Information (US) 2022a). The blue whale and the other 2 VGP assemblies (vaquita Phocoena sinus and bottlenose dolphin Tursiops truncatus) stand out by being at least an order of magnitude more contiguous at the contig level (Fig. 1a), among the most complete and least fragmented (Fig. 1b), and having the least missing sequence and artifact inactivating mutations (Fig. 1c).
Fig. 1.
Assembly quality metrics. Blue whale (Balaenoptera musculus) data are shown in red; the 2 other VGP assemblies, vaquita (Phocoena sinus) and bottlenose dolphin (Tursiops truncatus), are in blue. a) Assembly contig and scaffold N50 metrics. Contigs are segments of contiguous, i.e. gapless sequence. Scaffolds are sets of contigs that have been ordered and oriented using long-range mapping data such as optical maps and Hi-C with gaps between contigs. N50 is a measure of average length, e.g. 50% of all bases are contained in contigs of length N50 or longer. b) % of complete and fragmented universal single copy BUSCO orthologs found in an annotated genome. Universal single copy orthologs are genes that are present in a single copy in all or most genomes within a phylogenetic group. A high % complete score is an indication that a genome assembly is not missing a large amount of gene-coding sequence (Simão et al. 2015; Manni et al. 2021). C) TOGA status of 18,430 ancestral placental mammal genes. Note: For 2 species, different assemblies were used in panel C compared to panel A: GCA_004363415.1 instead of GCA_002189225.1 for Eschrichtius robustus and GCA_008795845.1 instead of GCA_023338255.1 for Balaenoptera physalus.
Another blue whale genome assembly, based on 10× synthetic long-read technology has recently been published (Yuan et al. 2021). It is somewhat less contiguous than our own 10×-based assembly (supplementary table S1, Supplementary Material online). However, it seems to contain fewer false duplications, as evidenced by a lower BUSCO % duplicated score, 1.7 versus 2.3 in our 10×-based assembly.
Genome Size and Assembly Completeness
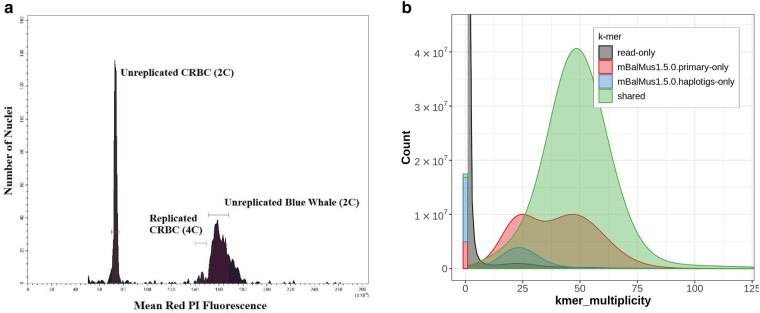
We estimated the genome size of the blue whale using flow cytometry, which quantifies the amount of propidium iodide (Hare and Johnston 2011) incorporated into the major groove of the nuclear DNA of a sample and a co-prepared standard. Comparing fluorescence intensity of blue whale cell nuclei to chicken red blood cell (CRBC) controls resulted in the genome size estimate of 1C = 2,695 ± 11.5 Mb (∼2.7 Gb) (Fig. 2a).
Fig. 2.
Blue whale genome size. a) Genome size estimation by flow cytometry. CRBC were used as the standard. b) K-mer spectra plot generated by the Merqury software (Rhie et al. 2020).
Genome assemblies tend to be somewhat smaller than genome size estimates due to the missing satellite DNA (Gordon et al. 2016). A short reads-based k-mer spectra plot produced by the Merqury software (Rhie et al. 2020) shows that only a small number of haploid k-mers are missing in the unique fraction of the genome (Fig. 2b). K-mers derived from sequencing reads of homozygous regions have an average multiplicity of around 50× coverage and match sequences of either the primary or both assembled haplotypes (“shared”). K-mers derived from reads of heterozygous regions have an average multiplicity of around 25 and match either the primary or alternate haplotype. Some of the k-mers that match neither haplotype (“read-only”) also have multiplicity of around 25, suggesting that the assembly is missing a small fraction of heterozygous k-mers, likely from the alternate haplotype. Overall, we conclude that the nonrepetitive fraction of our genome appears almost complete.
No Evidence of Convergent Evolution for Single Nucleotide Body Size QTLs in Cetacea and Cattle
A meta-analysis of genome-wide association studies (GWAS) in cattle (Bos taurus), which are land relatives of cetaceans, identified 163 loci associated with body size (Bouwman et al. 2018). We used whole-genome alignments to map these loci to the blue whale and vaquita. A total of 52 loci mapped to the former and 53 to the latter, the remainder being in regions not well mapped between species. The blue whale being the largest cetacean and the vaquita the smallest, we hypothesized that blue whale genomic sequences would predominantly match alleles associated with large body size in cattle, while vaquita would match the small-body-size alleles. However this was not the case, with blue whale and vaquita matching the same bovine alleles. For example, a total of 22 loci mapped from cattle to cetaceans had nucleotides corresponding to the large-body-size allele in the latter. Sixteen of these mapped to both blue whale and vaquita and had the same nucleotide in both, while 3 mapped to each genome uniquely. Thus, both the blue whale and vaquita genome had 19 loci with a nucleotide corresponding to increased body size, which contradicts the hypothesis that loci governing size are conserved between cattle and cetaceans.
Segmental Duplications
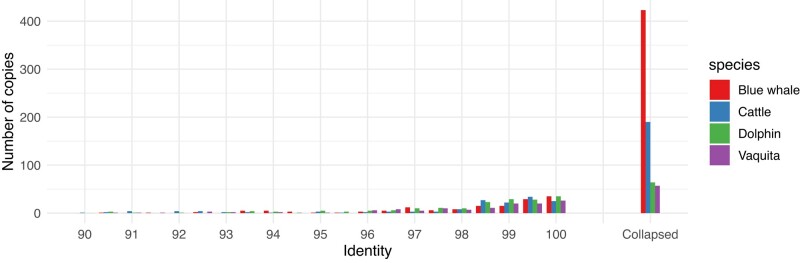
Previous reports of cetacean genomes have associated expansion of certain gene families through SDs with aquatic adaptation (Yim et al. 2014) and potential resistance to cancer (Tollis et al. 2019). To explore how gene expansions may relate to traits unique to the blue whale, we annotated SDs in blue whale, vaquita, and bottlenose dolphin, along with cattle as an outgroup (Fig. 3; supplementary fig. S4, Supplementary Material online). The standard definition of SD is duplicated sequences with at least 90% identity over 1 kb. Typical SD annotations exclude mobile elements and simple repeats (Bailey et al. 2001). To account for incomplete repeat masking, we also exclude sequences duplicated over 20 times in a genome. The approach to SD annotation used here was developed for vertebrate genomes and previously reported in (Toh et al. 2022). It quantifies SD using whole-genome self-alignments (Numanagic et al. 2018), and annotates duplicated gene copy numbers by excess read depth and whole-genome multimapping. We further distinguish between resolved and collapsed duplications. Resolved duplications are 2 or more distinct but highly similar segments, identified by a genome self-alignment. Collapsed duplications, a technical rather than biological term, arise when a genome assembly algorithm is unable to resolve multiple copies of a segment and the resulting assembly contains fewer copies than the genome. They are identified by excess read depth, as reads originating from multiple copies of a segment in the genome map to a single “collapsed” interval in the assembly.
Fig. 3.
Gene duplications in blue whale, vaquita, dolphin, and cattle. Duplications that are resolved in the assembly are stratified by duplication identity. The identity of collapsed duplications may not be directly discovered from read alignments.
Contrary to patterns of recent bursts of SD in primate (Bailey and Eichler 2006) as well as muroid genomes (Cheung et al. 2003; Thybert et al. 2018; Toh et al. 2022), SDs represent a smaller fraction of genomes in cetartiodactyla. Using the estimated mutation rate of 0.045% bp−1 My−1 (Jackson et al. 2009), both the blue whale and vaquita had relatively quiescent genomes until ∼20 M years ago. We detected a total of 20.2 Mb resolved duplications in blue whale, 30.2 Mb in vaquita, 43.6 Mb in bottlenose dolphin, and 73.2 Mb in cattle. We also identified 50.1 to 94.2 Mb of collapsed duplications in the blue whale, 15.5 to 20.8 Mb in vaquita, 29.4 to 35.0 Mb in the dolphin, and 17.6 to 22.7 Mb in cattle. Therefore, in spite of having fewer duplications resolved in the genome assembly, the blue whale shows an overall increase in duplicated bases compared to the other 3 genomes. The relatively few bases of resolved SD sequence in the blue whale assembly may reflect the sequencing depth used: 50× PacBio read coverage versus 121× in vaquita, 70× in dolphin, and ∼106× in cattle. Overall, between 2.1% to 4.8% of cetartiodactyl genomes are duplicated regions, compared to 4% to 14% in rodent and primate genomes.
SDs in the blue whale are gene rich, amounting to a roughly 7.1× burst in gene duplications relative to vaquita and dolphin, and 3.0× relative to cattle. This difference is largely driven by dramatic expansions of a limited number of genes in the blue whale. For example, the 10 most highly amplified genes account for 331 gene copies out of 700 (47%) total duplicated genes. The majority of copies of these genes could not be resolved by our genome assembly and are therefore contained in collapsed duplications (Fig. 3). The blue whale has 46 genes that have ≥4 copies, at least some of which are collapsed, compared to 8 in cattle, 9 in dolphin, and 6 in vaquita, after filtering for genes with single exons, genes annotated in pericentric DNA, and genes overlapping nuclear DNA of mitochondrial origin (supplementary table S2, Supplementary Material online).
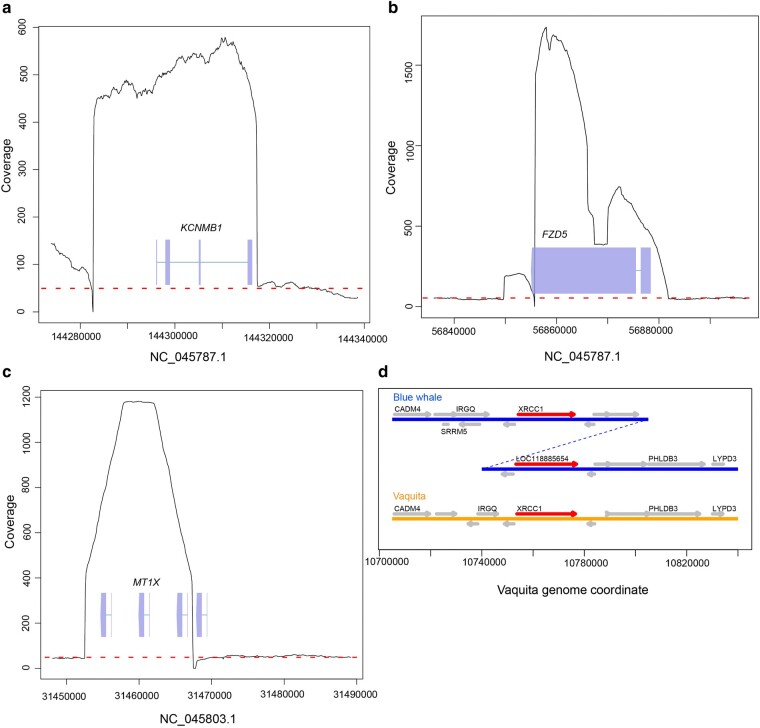
Using algorithms developed for assembling missing segments in collapsed duplications (Vollger et al. 2019), we were able to resolve additional copies of several genes, including KCNMB1, MYH8, DPEP2, and NXF2. The assembly sizes ranged from 10 to 30 Kb, largely reflecting the longest sequence overlapping the resolved copy. The pairwise similarity of the resolved sequences ranged from 86.2% to 99.4%, indicating high sequence diversity in the missing copies of each gene (supplementary table S3, Supplementary Material online).
In total, we detected 580 duplicated genes: 234 in the blue whale, 167 in the vaquita, 211 in dolphin, and 205 in cattle (supplementary table S4, Supplementary Material online). Some examples of genes duplicated in the blue whale genome are listed in Table 2 and shown in Fig. 4 and supplementary figs. S5 and S6, Supplementary Material online.
Table 2.
Examples of genes duplicated in the blue whale
| Gene (human ortholog) | Description (human ortholog) | Number of copies | Comments | |||
|---|---|---|---|---|---|---|
| Blue whale | Vaquita | Bottlenose dolphin | Cattle | |||
| C2orf78 | Chromosome 2 Open Reading Frame 78 | 150 | 3 | 4 | 1 | Uncharacterized protein. Allelic and copy number variants and a genomic rearrangement linked to cancers (Park et al. 2012; Jaratlerdsiri et al. 2017; Xicola et al. 2018). Apparent copy number in the blue whale may be affected by proximity to a centromere. |
| MT1A/MT1X | Metallothionein | 53 | 7 | 1 | 1 | Sequesters toxic metals, linked to longevity and cancer (Goulding et al. 1995; Pedersen et al. 2009; Sigel et al. 2015; Pabis et al. 2021). |
| MTRNR2L5 | MT-RNR2 like 5 (pseudogene) | 41 | 5 | 12 | 14 | Humanin-like: a member of a large family of micropeptides encoded by nuclear paralogs of the mitochondrial MT-RNR2 gene. |
| DPEP2 | Dipeptidase 2 | 32 | 3 | 3 | 1 | Linked to body size (Deciphering Developmental Disorders Study 2015), obesity and diabetes (Liu et al. 2013; Heemskerk et al. 2015; Sárvári et al. 2021), Alzheimer's disease (Carlyle et al. 2022), cancer, immunity, and inflammation (Yang et al. 2019; Rao et al. 2021; Huang et al. 2022; Han et al. 2023) |
| FZD5 | Frizzled 5 | 17 | 1 | 1 | 1 | A member of the WNT signaling pathway, involved in development, regeneration, and cancer (Bhanot et al. 1996; Yang-Snyder et al. 1996; He et al. 1997; Shulman et al. 1998; Ishikawa et al. 2001; Huang and Klein 2004; Burns et al. 2008; Thiele et al. 2018; Sun et al. 2020; Dong et al. 2021) |
| DDX24 | DEAD-box helicase 24 | 14 | 1 | 1 | 1 | An RNA helicase involved in RNA virus-host interactions and cancer (Ma et al. 2008: 24; Ma et al. 2013: 24; Shi et al. 2016: 24) |
| NCAM1 (also known as CD56) | Neural cell adhesion molecule 1 | 15 | 1 | 1 | 1 | Nervous and immune systems, blood cancers (Atz et al. 2007: 1; Alegretti et al. 2011: 56; Vukojevic et al. 2020; Jennings et al. 2022: 1) |
| KCNMB1 | Potassium calcium-activated channel subfamily M regulatory beta subunit 1 | 12 | 1 | 1 | 1 | Linked to hypertension (Sentí et al. 2005: 1; Kelley-Hedgepeth et al. 2008: 1; Grimm et al. 2009: 1) |
| XRCC1 | X-ray repair cross-complementing protein 1 | 2 | 1 | 1 | 1 | Plays an important role in DNA damage repair, associated with several human cancers (Tebbs et al. 1999; Schneider et al. 2008; Hanssen-Bauer et al. 2012; Wang and Ai 2014; Ghosh et al. 2016; Meng et al. 2017; Wu et al. 2020; Demin et al. 2021; Gong et al. 2021; Tang and Çağlayan 2021). |
| CDK20 | Cyclin-dependent kinase 20 | 4 | 1 | 1 | 1 | A key regulator of the cell cycle, involved in mammalian development and cancer (Snouffer et al. 2017; Lai et al. 2020). |
| CHRNB1 | Cholinergic receptor nicotinic beta 1 subunit | 2 | 2 | 3 | 1 | Linked to body size traits in sheep, BMI in humans. and congenital myasthenic syndromes that affect neuromuscular transmission (Müller et al. 2007; Kominakis et al. 2017; Zhao et al. 2021). The second copy has pseudogenized in vaquita. |
This is a subset of genes listed in supplementary table S4, Supplementary Material online, chosen for large copy numbers in blue whale, copy number differences between blue whale and vaquita, and/or potential relevance to body size and longevity traits.
Fig. 4.
Examples of duplicated genes. a to c) Sequencing read coverage plots of the collapsed duplications containing KCNMB1, FZD5, and MT1X genes. Average coverage is shown in panels (a) to (c) in the dashed red line. MT1X duplication is partially resolved, as evidenced by the four resolved copies of the gene, shown as boxes. d) Genomic region containing XRCC1 in blue whale and vaquita. XRCC1 genes are highlighted in red and labeled by the gene name. The second XRCC1 locus in the blue whale is labeled by its locus number, LOC118885654. This locus also has an increased read coverage, suggesting an unresolved third copy; see supplementary fig. S5, Supplementary Material online.
In order to identify duplication events potentially linked to body size and longevity, we selected 283 genes whose predicted copy numbers differ at least 2-fold between blue whale and vaquita (supplementary table S5, Supplementary Material online). To prioritize variants, we identified 8,649 candidate genes linked to body size, development, longevity, and susceptibility to cancer (supplementary table S6, Supplementary Material online) from published studies in whales (Tollis et al. 2019; Lagunas-Rangel 2021), dogs (Ostrander et al. 2017), cattle (Bouwman et al. 2018), and sheep (Kominakis et al. 2017), and automated literature mining engines (Kuusisto et al. 2020; Raja et al. 2020). Intersecting these 2 lists identified 133 genes of potential interest (supplementary table S7, Supplementary Material online). These included KCNMB1, previously reported by (Tollis et al. 2019), for which our workflow detected a collapsed duplication with 12 copies (Fig. 4a). Details of several other genes on this list linked to longevity (MT1X), body size (CHRNB1, DPEP2), development (FZD5, CDK20), cancer (C2orf78, FZD5, DDX24, NCAM1, MT1X, XRCC1, CDK20), obesity and diabetes (DPEP2), and the immune system (NCAM1) are available in Table 2 and Wiki pages on OSF (Bukhman et al. 2021b).
IGF1 Alleles Potentially Linked to Body Size in Cetaceans
Next, to further explore possible genetic bases for the large body size of the blue whale, we assessed variants of the IGF1, which had been reported to have large effects on body size in dogs (Ostrander et al. 2017). Domestic dogs present a unique system for the study of genetic determinants of mammalian body size due to the more than 40-fold variation in size between different breeds of the same species. Alleles of a single nucleotide within an intron of the IGF1 gene were found to be associated with body size not only in domestic dogs, but in canines generally (Plassais et al. 2022). Therefore, we decided to investigate IGF1 sequences in artiodactyls, including both cetaceans and their land relatives, in an attempt to discover variants that may be associated with body size in this mammalian order. To this end, we aligned whole-gene sequences and their flanking regions and carried out statistical analyses on the columns of the alignment matrix to identify sites with significant associations between adult body sizes and nucleotides across species.
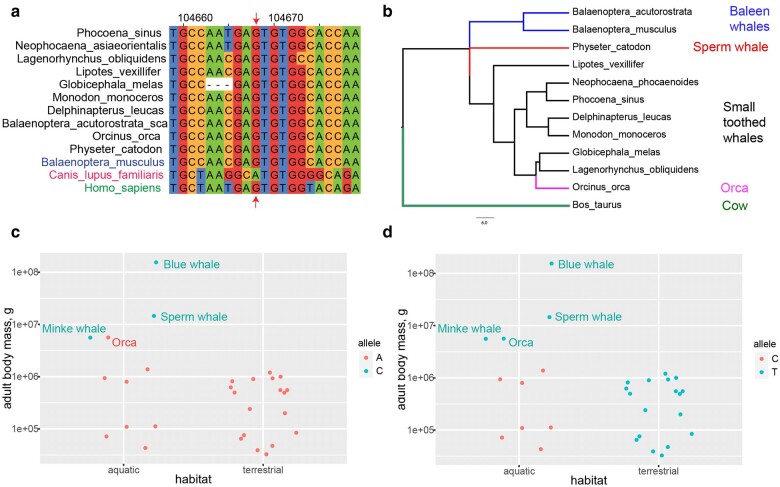
We first examined the SNP (rs22397284) in canine IGF1 reported by (Plassais et al. 2022). This site appears to be specific to canines, with a different sequence appearing in artiodactyls, humans, and other mammals (Fig. 5a and Fig. 3 of Plassais et al. (2022)). Where canines have the CA or CG dinucleotide for large and small body size respectively, artiodactyls and other mammals have AG regardless of body size.
Fig. 5.
IGF1 sites potentially associated with body size. a) Dog site rs22397284 in the context of a multiple alignment with cetaceans and the human genome. The sequences here are reverse complements of those shown in (Plassais et al. 2022), Fig. 3. rs22397284 is marked by an arrow. b) Phylogenetic tree of 11 cetaceans considered in this analysis, generated by Timetree. The 3 clades discussed in the text and the Orca are shown in different colors. c) An example type 1 site, blue whale chromosome 10 position 85,169,891. d) An example type 2 site, blue whale chromosome 10 position 85,160,822. See Table 3 for site types.
Our multiple alignment included whole-gene IGF1 sequences of 11 cetacean and 18 land artiodactyl species annotated by RefSeq (Thibaud-Nissen et al. 2013). The 11 cetaceans fell into 3 phylogenetic clades (Fig. 5b): first, large baleen whales, blue and minke, Balaenoptera musculus and B. acutorostrata; second, a giant toothed whale, sperm whale Physeter catodon; third, smaller toothed whales, dolphins, and porpoises. The latter also includes the orca, Orcinus orca, the largest dolphin, as large as the minke whale.
Fitting linear models of adult body weight versus nucleotide to all columns of the IGF1 multiple alignment matrix uncovered 2 types of potentially interesting sites. The first type is where 3 large whales, blue, minke, and sperm, have a different allele from all other artiodactyls (Fig. 5c). These sites are statistically significant when all artiodactyls are compared without regard to their habitat (linear model P-values of 2.5 × 10−5 and the false discovery rates (FDR) of 0.1 estimated using a permutation technique: see Materials and Methods). There are 2 such sites in the intergenic region upstream of the gene, 4 in the second intron, and 1 in the third or fourth intron, depending on the IGF1 isoform (Table 3). The orca has the same allele as its smaller relatives at these sites. The second type includes sites where the four largest whales, blue, minke, sperm, and orca, have a different nucleotide compared to all smaller cetaceans. These sites appear as statistically significant when the habitat is taken into account (linear model P-values of 3.2 × 10−5 and FDR of 0.09). There are four such sites in the second intron and one in the third intron (Table 3). Interestingly, all land artiodactyls have the same variant as the large whales (Fig. 5d). Therefore, the large whales probably have the ancestral allele, while the small ones have evolved an alternative. Although the orca is more closely related to smaller dolphins, it still has the allele characteristic of baleen and sperm whales. A smaller potential ancestor of the orca is known from the fossil record (Capellini 1883; Bianucci 1996). It is possible that the orca retained the ancestral large-body allele due to incomplete lineage sorting. Additionally, the orca has a unique SINE:CHR2A_Ttr insertion in intron 2, not found in any other species in this analysis. This might be a unique adaptation compensating for the orca's lack of sites of the first type. Finally, several sites appeared statistically significant but were clearly artifacts, located in low-complexity regions where the alignment is ambiguous. These sites are not shown in Table 3. All statistically significant sites and notes are available in supplementary data, Supplementary Material online file on OSF (IGF1/hits.xlsx in (Bukhman et al. 2021b)).
Table 3.
IGF1 sites potentially associated with body size. Sites within the IGF1 gene and flanking genomic regions that are significantly associated with body size in cetaceans
| Site typea | Small cetacean allele | Large cetacean allele | Blue whale chromosome 10 coordinate | Genomic feature(s) | Multiple alignment column |
|---|---|---|---|---|---|
| 1 | C | T | 85,170,164 | Intergenic region 5′ of IGF1 | 24,474 |
| 1 | A | C | 85,169,891 | Intergenic region 5′ of IGF1; LINE element L2c | 24,791 |
| 2 | C | T | 85,160,822 | Intron 2 | 35,626 |
| 2 | G | A | 85,160,446 | Intron 2 | 36,017 |
| 1 | G | C | 85,150,447 | Intron 2 | 48,867 |
| 1 | A | G | 85,140,848 | Intron 2; LINE element L2b | 62,044 |
| 1 | C | T | 85,140,276 | Intron 2 | 62,965 |
| 2 | C | T | 85,132,164 | Intron 2; LINE element L2d | 78,042 |
| 2 | C | T | 85,126,807 | Intron 2 | 87,228 |
| 1 | G | A | 85,118,402 | Intron 2 | 99,043 |
| 2 | A | G | 85,112,779 | Intron 3 | 106,724 |
| 1 | C | T | 85,108,387 | Intron 3-4 | 113,202 |
Historical Demography
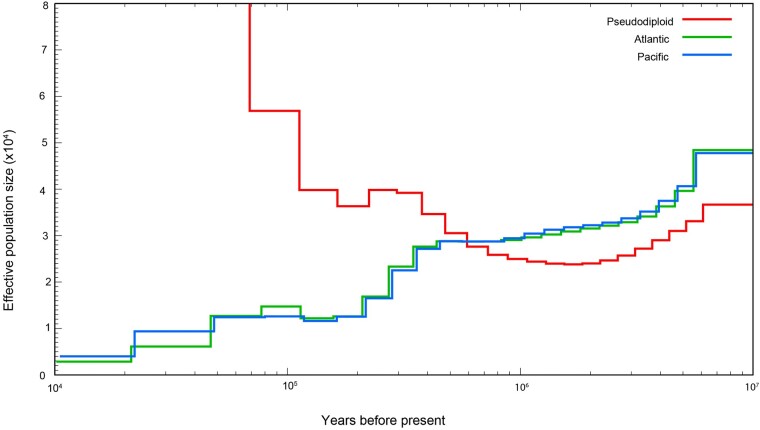
Next, to gain insight into the history of blue whale populations, we used the pairwise sequential Markovian coalescent (PSMC) model (Li and Durbin 2011) for an analysis of historical demography. This analysis utilized reference-guided genome assemblies of Illumina sequencing data generated using our blue whale genome as the reference, with repeats masked after read alignment. We assembled Pacific blue whale Illumina sequencing reads generated by us and previously published Atlantic blue whale reads by (Árnason et al. 2018). The assemblies for the Pacific and Atlantic blue whales had 43.3× and 33.6× average depth of coverage, respectively. The PSMC plot of historical demography for the Atlantic sample was similar to the previously published analysis (Árnason et al. 2018), despite our use of a faster mutation rate (see supplementary fig. S8, Supplementary Material online for PSMC plots with alternate mutation rates). (Árnason et al. 2018) used the bowhead genome for reference-guided assembly of the blue whale and other rorqual genomes, with the maximum coalescent parameter (t) set to 20, whereas we used the de novo blue whale reference genome (repeat masked) and t = 15. We tested the effect of changing the maximum coalescent time to t = 20, and found that it had almost no effect on the plot (supplementary fig. S9, Supplementary Material online), but did reduce the number of recombination events to <10 in the last few atomic intervals in the PSMC, so we used t = 15 for all analyses reported here.
The historical demography plot of the Pacific blue whale matched that of the Atlantic blue whale until approximately 125 kyr ago, around the end of the Saalian ice age (Fig. 6). At that point, the Atlantic population appears to have grown slightly larger than the Pacific population before both ocean population sizes decreased approaching the last glacial maximum (LGM) that occurred ∼20 kyr ago; however, the apparent increase could also be due to gene flow from other blue whale populations, e.g. in the Southern Ocean. The pseudodiploid PSMC plot shows an exponential increase indicative of population division, possibly due to cessation of gene flow between the 2 ocean basins as the Eemian warm period took hold. This pattern has also been found in sperm whales from these 2 ocean basins (Morin et al. 2018).
Fig. 6.
Historical demography of Pacific and Atlantic blue whales from PSMC analysis of genomes. The pseudodiploid plot represents coalescence between the 2 genomes, where the rapid increase starting approximately 125 kyr ago indicates cessation of gene flow (coalescence) between the populations. Generation time = 30.8 yr; Autosomal mutation rate (µA) = 1.58E-08 substitutions/bp/generation.
Heterozygosity
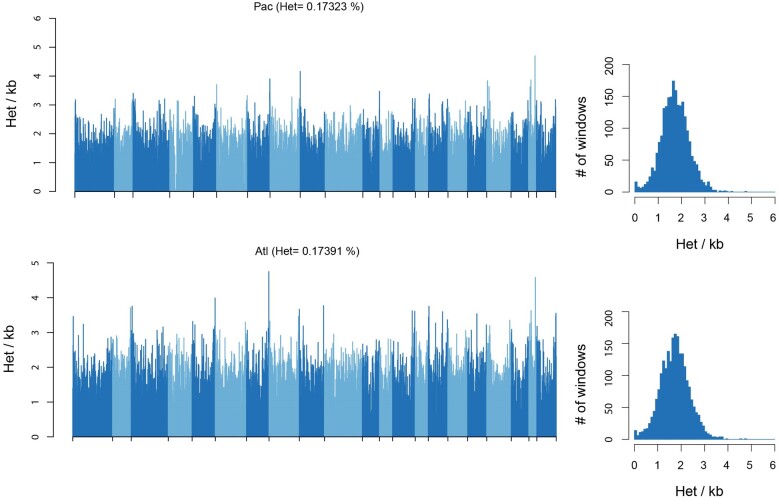
The distribution of heterozygosity across the genome was determined using previously described analysis pipelines (Robinson et al. 2019). Briefly, genotypes were called and filtered from the genome assembly (above) using Genome Analysis Toolkit (GATK) (McKenna et al. 2010), filtering out loci with <1/3× or >2× mean depth of coverage, and heterozygosity was calculated as the number of heterozygous sites divided by the total number of called genotypes in nonoverlapping 1 Mb windows across each scaffold. We found high and evenly distributed levels of heterozygosity indicative of large, outbred populations (Fig. 7). See (Robinson et al. 2019) for a discussion of the effect of different demographic histories on heterozygosity.
Fig. 7.
Distributions of heterozygosity across the genomes of the North Pacific and North Atlantic blue whales. (left) Barplot shows per-site heterozygosity in nonoverlapping 1-Mb windows across 22 scaffolds >10 Mb in length. Scaffolds are shown in alternating shades. (right) Histogram of the count of per-window heterozygosity levels.
Runs of Homozygosity (ROH) Analysis
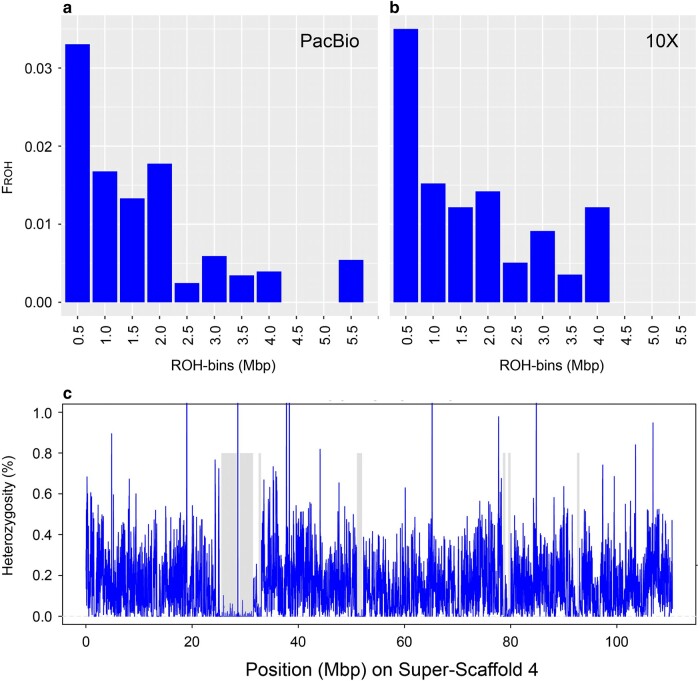
Runs of homozygosity (ROH) are indicative of the frequency and relative timing of inbreeding events and were frequently used to assess the impact of inbreeding on different mammalian and cetacean populations (Foote et al. 2021; Wolf et al. 2022; de Jong et al. 2023). ROH was identified by scanning the previously called genotypes using a nonoverlapping sliding window approach as implemented in DARWINDOW (de Jong 2021). We visualized heterozygosity distribution for all 23 super-scaffolds and manually altered heterozygosity thresholds, window size, and minimal window number until all visible drops in heterozygosity were marked as ROH. Using a window size of 20 kb, a minimal window number of 25, and a heterozygosity threshold of 2.5%, 108 runs of 500 kb or longer were found in both assemblies. Over 70% of these ROH were shorter than 1 Mb, indicating that most ROH were probably fragmented over time by frequent outcrossing (McQuillan et al. 2008).
Compared to the long-read assembly, a slightly larger proportion of the linked-short-read assembly was covered in ROH, as indicated by a higher inbreeding coefficient, Froh (500 Kb), of 0.107 compared to 0.102 of the long-read assembly (Fig. 8a and b), respectively. The longest run of 5.5 Mb was located on super-scaffold 4 of the long-read assembly (Fig. 8c). In the linked-short-read assembly, this ROH is probably fragmented as indicated by a higher number of short (500 Kb) and medium-sized (2.5 to 4 Mb) ROHs.
Fig. 8.
Comparison of inbreeding factors (FROH) based on the genome coverage of ROH between (a) the long-read assembly and (b) the linked-short-read assembly. ROH were identified with DARWINDOW, using a sliding-window-based approach, and sorted into respective length-bins. In both assemblies, 108 ROH over 500 kb were found; however, they appear to be more continuous in the long-read assembly as indicated by the longest ROH located on Super-Scaffold 4 (chromosome 8) of the respective assembly. A visual representation of this ROH is given in (c) and depicts the heterozygosity distribution of 20 kb windows over the scaffold in blue while identified ROH were marked as gray bars.
Discussion
Genome Assembly
We have generated a reference-quality blue whale genome assembly based on PacBio long-read sequencing for contigs, and Illumina/10× linked reads, Bionano optical maps, and Dovetail Hi-C data for scaffolding and polishing. This is one of the first cetacean genomes generated in the context of the VGP, aiming at producing reference-quality genomes of all vertebrate species. We have shown that our assembly, along with 2 other VGP cetacean assemblies, are the highest quality cetacean genomes among those available from the NCBI Assembly database (National Center for Biotechnology Information (US) 2022a).
Segmental Duplications
Previous studies of cetacean genomes have identified expansions of duplicated genes or gene families using multimapped short reads (Tollis et al. 2019) and analysis of short-read assemblies (Yim et al. 2014). High-quality genomes from long-read assemblies have been shown to resolve more duplicated sequences than short-read assemblies (Gordon et al. 2016), indicating this study enables comparative analysis of repetitive DNA and gene copy numbers in cetacean genomes. We applied a computational pipeline recently developed to analyze duplicated genes in rodentia (Toh et al. 2022) to assess if there are duplications that could be associated with size differences in cetacea. The vaquita, the world's smallest cetacean, provides a comparison point to the blue whale in terms of body size, while cattle is a terrestrial relative of both species.
A major finding of this work is the presence of large copy number expansions of a number of genes in the blue whale, while relatively few such expansions are observed in vaquita, bottlenose dolphin, and cattle. These expansions are recent in evolutionary time, making the gene copies highly similar and difficult to resolve, even when using long-read sequencing technology. Development of giant body size is also relatively recent (Slater et al. 2017; Bianucci et al. 2019), suggesting a possible link with gene expansions. However, the exact connection of these genes to body size, if any, is difficult to ascertain in the context of this study.
The majority (102/151) of duplicated gene families discovered in the blue whale were resolved by the assembly, however the majority of duplicated gene counts were identified as collapsed duplications from excess read depth (571/719), with some genes having both resolved and collapsed copies. Even with the high contiguity enabled by long-read assemblies, certain duplication architectures, in particular long tandem arrays of duplications, currently require specialized methods and data to resolve (Nurk et al. 2022).
The long-read assembly of the blue whale provides further insight into segmentally duplicated genes over a previously reported short-read analysis (Tollis et al. 2019). For example, read-depth with short reads previously identified the expansion of KCNMB1 in blue whales (Tollis et al. 2019). Our long-read segmental duplication assembly methods were able to resolve individual copies with 0.4% to 7% divergence, indicating that the KCNMB1 locus contains ancient (>20 Mya) duplication events. Furthermore, the long-read assembly provides additional substrate for read-depth analysis, as several additional genes of potentially high impact on body size or longevity were found to be amplified in blue whales, including vastly expanded MT1X and FZD5, additional copies of XRCC1, CDK20, and CHRNB1, and others.
Although this work represents a significant advance over short-read-based genome assemblies, some issues remain. We have not been able to resolve individual copies of highly amplified genes. In many cases, it was also not possible to ascertain whether duplicated gene copies form tandem arrays or are interspersed throughout the genome. Additionally, excess copies of duplicated genes were occasionally missed by our algorithm, although we were able to infer their existence from TOGA and NCBI annotations. Regarding gene copy number comparisons between species, since our workflow depends on the availability of long-read sequencing data, we have been constrained to comparing only 3 cetaceans. Such a comparison lacks statistical power and the differentially duplicated genes that we have identified should be treated as preliminary findings, rather than definitive discoveries. Finally, the biological significance of gene duplication events may vary, with greater number of copies not necessarily meaning a more important effect. Thus, in some cases, such as amylase enzymes in humans and commensal species, additional copies of a gene are fully functional and represent adaptations to factors such as diet change (Arendt et al. 2014; Pajic et al. 2019; Toh et al. 2022), in other cases, such as hemoglobin in some species of Antarctic notothenioid fishes, they are nonfunctional and ultimately lost (Bista et al. 2023). In yet other cases, such as certain TP53 and LIF retrogenes in elephants (Sulak et al. 2016; Vazquez et al. 2018), some nonfunctional duplicates are refunctionalized again, and sometimes, e.g. in case of an enzyme-turned-antifreeze protein in notothenioid fishes (Bista et al. 2023), the new function is completely different from the original. Ongoing developments in genome sequencing, assembly, and segmental duplication analysis algorithms, as well as long-read sequencing of other cetacean species, will enable better resolution of SDs and more rigorous cross-species comparisons in the near future.
IGF1 Sites Potentially Associated With Body Size
Alternative alleles of IGF1 have been shown to have large effects on body size in domestic dogs (Ostrander et al. 2017). One site, located in an intron, was associated with body size in canines more generally (Plassais et al. 2022). We identified several sites within the IGF1 gene that, while being conserved among terrestrial artiodactyls, appear to be associated with body size in cetaceans. In one set of such sites, large whales have a different allele compared to land artiodactyls and smaller cetaceans. In another, large whales have the same allele as land artiodactyls while the smaller ones have evolved an alternative. Additionally, orca, the giant dolphin, has a unique mobile element insertion not found in any other artiodactyls. It is notable that, similar to the canine site identified by (Plassais et al. 2022), all of the sites identified by our analysis are located in noncoding regions in and adjacent to the gene. The canine site is part of an lncRNA encoded on the opposite strand and possibly affecting the expression of the IGF1 gene, rather than the sequence of the protein that it encodes. The cetacean sites identified in this study might also affect gene regulation, which could be elucidated by future research.
Although we have controlled for FDR and manually excluded sites in low-complexity regions that were obvious alignment artifacts, these results should still be treated with caution due to the large size of the whole-gene alignment, >100,000 bp, and relatively small number of species with high-quality long-read-based assemblies. Vigorous efforts are underway to generate reference-quality genomes of all cetacean species (Morin et al. 2020), which will increase statistical power for similar analyses in the near future.
Historical Demography and Genetic Diversity
Our assembly of the North Pacific blue whale genome enabled an analysis of its population history using PSMC and a comparison to the existing Atlantic blue whale genomic data (Árnason et al. 2018) to provide insight into demography and improve granularity of blue whale taxonomy. Four subspecies of blue whales are currently recognized (Committee on Taxonomy 2020), 3 of which are found in the Southern Hemisphere and northern Indian Ocean. The fourth and nominate subspecies include blue whales in the North Atlantic and North Pacific, which have not been formally compared. Outstanding questions about subspecies designation and genetic diversity thus persist. Our PSMC analysis strongly suggests that blue whales in the North Atlantic and eastern North Pacific began to diverge 100 to 200 kyr years ago, becoming completely genetically isolated around the time of the last interglacial period. These results suggest that blue whale taxonomy should be revisited to determine whether, similar to fin and humpback whales in these ocean basins (Jackson et al. 2014; Archer et al. 2019), blue whale populations should be divided into Pacific and Atlantic subspecies in the Northern Hemisphere.
As blue whales in the North Pacific and North Atlantic diverged, the effective population sizes of both ocean basins initially remained stable before declining at ∼50 kyr ago, approaching the LGM (Fig. 6). While Antarctic blue whales use both the South Pacific and South Atlantic (Stafford et al. 2004; Branch et al. 2007a; Samaran et al. 2019; Thomisch et al. 2019), no other populations of blue whales have been confirmed to regularly use the South Atlantic. In contrast, at least 2 populations of blue whales can be found in the South Pacific year-round (Hucke-Gaete et al. 2004; Branch et al. 2007b; Galletti Vernazzani et al. 2012; Balcazar et al. 2015; Barlow et al. 2018). While less is known about the population in the western South Pacific, both morphological and genetic studies have shown similarities between the eastern South Pacific and eastern North Pacific blue whales (Gilpatrick and Perryman 2008; Durban et al. 2016; Leduc et al. 2017), suggesting they likely diverged recently. The inferred slightly larger population size followed by the sharper decline in the North Atlantic approximately 50 to 60 kyr ago (Fig. 6) may be an artifact resulting from episodic gene flow between the North Atlantic and Antarctic subspecies, ending during the approach of LGM, while the effective size in the eastern North and South Pacific declined as a metapopulation with ongoing gene flow. These demographic inferences provide insight into global gene flow and divergence patterns, with implications for taxonomy and response to climate change.
The disagreement between the pseudodiploid PSMC plot and the 2 individual PSMC plots prior to the cessation of coalescence (>100 kyr ago) also suggests that there were more complex population dynamics over time (Fig. 6), likely affected by changing population structure and levels of mixing, potentially via populations in the Southern Hemisphere. Additional genomic studies of samples from other blue whale subspecies and populations, including analysis of admixture (Foote et al. 2019) are required to further elucidate global blue whale evolutionary and demographic history.
Our analyses of heterozygosity patterns across the genome indicate that the blue whale genome is highly heterozygous and contains mostly short ROH. Even though extrapolations from a single genome to a population are challenging, we assume that this indicates a large and outbred population in the Northern Pacific, especially when compared to well-known examples of threatened or extinct populations like the mountain gorilla, the Scottish killer whale, or the Malay Peninsula rhinoceros (van der Valk et al. 2019; Foote et al. 2021; von Seth et al. 2021). Although blue whales were hunted to less than 1% of their global abundance in the late 19th to mid-20th centuries, they are believed to be recovering in all major ocean basins (Branch et al. 2007b; Pike et al. 2009; Monnahan et al. 2014). Given the separation into multiple subspecies and populations, it is probable that some populations have suffered a loss of genetic diversity and potential increase in inbreeding. While the distribution of heterozygosity across the genome of the North Pacific and North Atlantic blue whales suggests maintenance of high levels of genomic diversity, population-level genetic studies are required to assess population structure and regional inbreeding levels, especially in regions that were more heavily impacted by commercial whaling (Clapham and Baker 2018).
Conclusion
Our high-quality chromosome-scale genome assembly of the blue whale provides more and notably more accurate genetic information for evolutionary, conservation, and other studies than previously available. Here, we used the assembly to investigate and compare SDs and other genomic features potentially important to the development of large body size, as well as to more precisely determine Pacific and Atlantic blue whale population history and genetic diversity. The assembly will serve as a valuable resource to the scientific community, enabling future comparative genomics studies to further our understanding of large animal longevity and associated resistance to cancer, and helping conserve this magnificent species.
Materials and Methods
Genome Size Estimation by Flow Cytometry
The genome size of the blue whale was estimated using nuclei isolated from blue whale fibroblast cell culture BW-04. The DNA amount per nucleus was estimated using flow cytometry following methods described in (Johnston et al. 2019). In brief, whole cells from the blue whale cell culture were placed into 1 mL of ice-cold Galbraith buffer in a 2 mL Kontes Dounce grinder along with a single drop of commercially prepared chicken red blood cells (CBRCs). The nuclei of sample and standard were released by grinding the mixture using 12 strokes of the A pestle at a rate of 12 strokes in approximately 9 s. Nuclei released by the grinding were passed through a 40 µm nylon mesh, stained with 25 µL/mg propidium iodide, and held in the dark and cold for 3 h. The relative red (PI) fluorescence of the 2C nuclei from the sample and standard were scored to channel number using a Beckman Coulter CytoFLEX flow cytometer. The 1C amount of DNA in the blue whale was determined as the ratio of the 2C red fluorescent mean 2C peak positions of the sample and standard times the 1C amount of DNA in the standard. Because the commercial CRBCs are supplied with no information on the sex or strain of the chicken, the genome size of the CBRCs was scored by multiple co-preparations of CBRC and lab strains standards, Drosophila virilis (1C = 328 Mb) and Callosobruchus maculatus (1C = 1,205 Mb). The resulting estimate of the CRBC genome size was 1C = 1,250 Mb, consistent with published values for a white leghorn rooster.
Construction and Sequencing of High Molecular Weight Genomic DNA Libraries Using PacBio Technology
HMW DNA was prepared according to the Pacific Biosciences 20 kb gDNA library protocol (Pacific Biosciences). The quality and quantity of the finished libraries were assessed using an Agilent Fragment Analyzer and Qubit® dsDNA HS Assay Kit, respectively. Libraries were sequenced on one Pacific Biosciences Sequel 1 M SMRT Cell for a 10-h movie.
VGP Genome Assembly Workflow
The blue whale VGP genome was assembled using version 1.5 of the standard VGP assembly workflow described in (Rhie et al. 2021). Briefly, PacBio genomic reads were assembled using Falcon. The assembly was then partially phased using Falcon-unzip, bases polished using Arrow, and false duplications purged using purge_haplotigs. This was followed by successive scaffolding with: 10× linked-read data using 2 rounds of scaff10x; Bionano optical maps using Solve; and Dovetail Hi-C data using Salsa2. The scaffolded assembly was polished using Arrow using the CLR long reads and 2 rounds of freebayes using the 10× linked reads. Assembly assessment, decontamination, manual curation, and assignment of chromosomal-scale scaffolds was performed as previously described (Howe et al. 2021) using gEVAL (Chow et al. 2016).
Segmental Duplications Analysis
Segmental duplications and gene copy numbers were defined using a combination of self-alignments and read depth defined in the Segmental Duplication Annotation pipeline (Chaisson 2023). The boundaries of collapsed duplications identified from excess read depth are annotated using a hidden Markov model (HMM) trained on single-molecule sequencing read depth. The copy number of a collapsed duplication is calculated from the average depth in each collapsed region, for collapses greater than copy number 6, or from the HMM for collapses at or below this copy number. Genes in collapsed duplications were identified as genes that fully overlap intervals identified as collapse.
This pipeline identifies duplicated genes by first mapping human Gencode v29 sequences using minimap2 -x splice (version 2.17-r941), extracting the sequence of the entire gene body, and re-mapping using minimap2 allowing for multiple alignments of a gene. Genes with a single exon were excluded. In order to identify genes that conform to the definition of a segmental duplication, duplicated gene copies are identified as sequences with at least 90% identity and 90% similar gene length.
The SDA method was used to assemble missing collapsed duplications (Vollger and Chaisson 2022). A coverage of 40 was specified, and the method was used with the “–collapse” option.
Identification of Genes Linked to Body Size and Related Traits
We identified genes linked to body size, development, longevity, and susceptibility to cancer by manual review of published studies in whales (Tollis et al. 2019; Lagunas-Rangel 2021), dogs (Ostrander et al. 2017), cattle (Bouwman et al. 2018), and sheep (Kominakis et al. 2017). We also used KinderMiner (Kuusisto et al. 2020) and SKiM (Raja et al. 2020) literature mining systems to identify genes linked to relevant search terms, including “developmental clock”, “body size”, “dwarfism”, “gigantism”, “longevity”, “cancer”, “growth”, and “overgrowth”. We also searched a curated gene-disease database for genes linked to dwarfism. See our OSF repository (Bukhman et al. 2021a) for a more detailed description in wiki/1_body_size_and_development and the R script 1_body_size_and_development/body_size_and_development_genes.R. See also (Toh et al. 2022) for a more detailed description of our literature mining methodology.
IGF1 Multiple Alignment and Statistical Analysis of the Alignment Matrix
We made use of NCBI e-utilities (National Center for Biotechnology Information (US) 2010; Winter 2017; Bukhman 2022a) to retrieve genomic sequences of entire IGF1 genes plus 10 Kb of flanking sequence on both sides. The retrieved sequences are available on OSF (Bukhman 2022b). The alignment of 29 artiodactyl, dog, and human genomes was performed using clustalw2 with the following command: clustalw2 -MAXSEQLEN = 200,000 -ALIGN -TREE -QUICKTREE -TYPE = DNA -KTUPLE = 4 -TOPDIAGS = 2 -WINDOW = 4 -INFILE = IGF1_genomic_seqs.fna. The resulting alignment is available on OSF (Bukhman 2022c: 1). The rows of the alignment were manually sorted by habitat and adult body mass using Jalview (Waterhouse et al. 2009). The sorted alignment is available on OSF (Bukhman 2022d). We fitted 2 linear models for each column of the alignment matrix, modeling the logarithm of adult body size as a function of the nucleotide alone or the nucleotide and the habitat. The latter was defined as “aquatic” for cetacea or “terrestrial” for land-dwelling artiodactyls. The R markdown notebook of this analysis is available on GitHub (Bukhman 2022e). Adult body mass values were retrieved from PANTHERIA and other sources (Jones et al. 2009; Treseus 2022; Wikipedia contributors 2022a; Wikipedia contributors 2022b; Wikipedia contributors 2022c; Wikipedia contributors 2022d; Castillo-Ruiz).
FDRs were estimated by randomly permuting species labels of the body mass values. All values were permuted in the first version of the analysis, which did not account for the habitat. Values were permuted separately within terrestrial and aquatic groups for the second version of the analysis. FDRs for different P-value thresholds were estimated by dividing the average number of hits in permuted datasets by the number of hits in the original dataset. Each analysis included 2 runs of 1,000 random permutations each, in order to verify the stability of the FDR estimates. R scripts, P-values of alignment positions, and FDR estimates are available on OSF (Bukhman et al. 2021b) in folder IGF1/body-size-loci_try2_artiodactyls-etc/4_permut-fdr.
PSMC and Genome Heterozygosity
We used PSMC (Li and Durbin 2011) for analysis of historical demography. Illumina 150 bp (Pacific) or 100 bp (Atlantic) paired-end reads were trimmed with the BBduk function of BBTools (Bushnell) for average quality (q ≥ 20), 3′ ends trimmed to q ≥ 15, and minimum length (≥40 nucleotides). Trimmed reads were aligned to a previously generated reference blue whale mitogenome, GenBank accession MF409242.1 (Árnason et al. 2018), using BWA mem (Li and Durbin 2009), and the unmapped reads exported as reads representing only the nuclear genome. Nuclear reads were aligned to the reference genome primary pseudohaplotype, NCBI Assembly accession GCF_009873245.2, also using BWA mem. Duplicate reads were removed using Picard Tools v2.20 (Broad Institute) and the resulting genome alignments from one library (Atlantic sample) and 2 combined libraries (Pacific sample) were assessed for average depth of coverage using ANGSD (Korneliussen et al. 2014). From each assembly, the diploid genome was extracted using SAMtools (Li et al. 2009) following (Morin et al. 2018), with minimum and maximum depth of coverage set to 1/3× and 2× the average depth of coverage for each assembly, and PSMC run with generation time of 30.8 yr (Taylor et al. 2007), and an autosomal mutation rate (µA) of 1.58E-08 substitutions/bp/generation, based on mean rate of 5.14E-10 substitutions/bp/year for mysticetes (Jackson et al. 2009), with 100 bootstrap resamplings on all PSMC analyses. The PSMC parameters were left as the default values used for humans (Li and Durbin 2011), and we verified that, after 20 rounds of iterations, at least ∼10 recombinations were inferred to occur in the intervals each parameter spans: P = (4 + 25*2 + 4 + 6) (Li and Durbin 2011). After the generation of the PSMC results, data were plotted with multiple mutation rates to test the effect of mutation rate uncertainty.
A pseudodiploid genome was made by randomly sampling an allele at each site from each of the genome assemblies from the 2 ocean basins using seqtk (Li 2022), followed by PSMC analysis using the same parameters as above, as previously described (Morin et al. 2018; Foote et al. 2019). The pseudodiploid PSMC analysis provides information on changes in the rate of coalescence between 2 individual genomes through time, and, therefore, the timing of changes in population structure relative to the changes in effective population size inferred by the single-genome PSMC analyses (Cahill et al. 2016; Chikhi et al. 2018).
The distribution of heterozygosity across the genome was determined using previously described analysis pipelines (Robinson et al. 2019). Briefly, genotypes were called and filtered from the genome assembly (above) using GATK (McKenna et al. 2010), filtering out loci with <1/3× or >2× coverage, and heterozygosity was calculated as the number of heterozygous sites divided by the total number of called genotypes in nonoverlapping 1Mb windows across each scaffold.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors would like to thank Cene S. Bryant for assisting in field tissue sample collection from free ranging blue whales; the San Diego Zoo Wildlife Alliance for karyotyping support; the University of Wisconsin Biotechnology Center DNA Sequencing Facility (Research Resource Identifier—RRID:SCR_017759) for providing Pacific Biosciences library prep and sequencing services; the University of Minnesota Genomics Center for preparing the 10× Chromium sequencing library; the Washington University at St. Louis McDonnell Genome Institute (MGI) for performing the optical mapping experiment and scaffolding the 10×-based genome assembly; Dovetail Genomics for performing the Hi-C experiment and scaffolding the 10×-based genome assembly; Michael Collins for setting up software tools and the computing environment at Morgridge Institute for Research; Woori Kwak of C&K Genomics for submitting data to NCBI; Francoise Thibaud-Nissen for leading the genome annotation effort at NCBI; Amy Freitag and Alicia Williams for helping with editing the text; Marlys Houck, John Fogg, Cecile Ane, Jacqueline A. Robinson, and Francoise Thibaud-Nissen for stimulating discussions and advice.
Contributor Information
Yury V Bukhman, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA.
Phillip A Morin, Southwest Fisheries Science Center, National Oceanic and Atmospheric Administration (NOAA), La Jolla, CA 92037, USA.
Susanne Meyer, Neuroscience Research Institute, University of California, Santa Barbara, CA, USA.
Li-Fang Chu, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA; Department of Comparative Biology and Experimental Medicine, University of Calgary, Calgary, Canada.
Jeff K Jacobsen, V.E. Enterprises, Arcata, CA, USA.
Jessica Antosiewicz-Bourget, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA.
Daniel Mamott, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA.
Maylie Gonzales, Neuroscience Research Institute, University of California, Santa Barbara, CA, USA.
Cara Argus, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA.
Jennifer Bolin, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA.
Mark E Berres, University of Wisconsin Biotechnology Center, Bioinformatics Resource Center, University of Wisconsin - Madison, Madison, WI 53706, USA.
Olivier Fedrigo, Vertebrate Genome Lab, The Rockefeller University, New York, NY 10065, USA.
John Steill, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA.
Scott A Swanson, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA.
Peng Jiang, Center for Gene Regulation in Health and Disease (GRHD), Cleveland State University, Cleveland, OH, USA; Department of Biological, Geological and Environmental Sciences, Cleveland State University, Cleveland, OH, USA; Center for RNA Science and Therapeutics, School of Medicine, Case Western Reserve University, Cleveland, OH, USA.
Arang Rhie, Genome Informatics Section, National Human Genome Research Institute, Bethesda, MD 20892, USA.
Giulio Formenti, Laboratory of Neurogenetics of Language, The Rockefeller University/HHMI, New York, NY 10065, USA.
Adam M Phillippy, Genome Informatics Section, National Human Genome Research Institute, Bethesda, MD 20892, USA.
Robert S Harris, Department of Biology, Pennsylvania State University, University Park, PA 16802, USA.
Jonathan M D Wood, Tree of Life, Wellcome Sanger Institute, Cambridge CB10 1SA, UK.
Kerstin Howe, Tree of Life, Wellcome Sanger Institute, Cambridge CB10 1SA, UK.
Bogdan M Kirilenko, LOEWE Centre for Translational Biodiversity Genomics, 60325 Frankfurt, Germany; Senckenberg Research Institute, 60325 Frankfurt, Germany; Institute of Cell Biology and Neuroscience, Faculty of Biosciences, Goethe University Frankfurt, 60438 Frankfurt, Germany.
Chetan Munegowda, LOEWE Centre for Translational Biodiversity Genomics, 60325 Frankfurt, Germany; Senckenberg Research Institute, 60325 Frankfurt, Germany; Institute of Cell Biology and Neuroscience, Faculty of Biosciences, Goethe University Frankfurt, 60438 Frankfurt, Germany.
Michael Hiller, LOEWE Centre for Translational Biodiversity Genomics, 60325 Frankfurt, Germany; Senckenberg Research Institute, 60325 Frankfurt, Germany; Institute of Cell Biology and Neuroscience, Faculty of Biosciences, Goethe University Frankfurt, 60438 Frankfurt, Germany.
Aashish Jain, Department of Computer Science, Purdue University, West Lafayette, IN 47907, USA.
Daisuke Kihara, Department of Computer Science, Purdue University, West Lafayette, IN 47907, USA; Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA.
J Spencer Johnston, Department of Entomology, Texas A&M University, College Station, TX 77843, USA.
Alexander Ionkov, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA.
Kalpana Raja, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA.
Huishi Toh, Neuroscience Research Institute, University of California, Santa Barbara, CA, USA.
Aimee Lang, Southwest Fisheries Science Center, National Oceanic and Atmospheric Administration (NOAA), La Jolla, CA 92037, USA.
Magnus Wolf, Institute for Evolution and Biodiversity (IEB), University of Muenster, 48149, Muenster, Germany; Senckenberg Biodiversity and Climate Research Centre (BiK-F), Frankfurt am Main, Germany.
Erich D Jarvis, Vertebrate Genome Lab, The Rockefeller University, New York, NY 10065, USA; Laboratory of Neurogenetics of Language, The Rockefeller University/HHMI, New York, NY 10065, USA.
James A Thomson, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA; Department of Molecular, Cellular and Developmental Biology, University of California Santa Barbara, Santa Barbara, CA 93106, USA; Department of Cell and Regenerative Biology, University of Wisconsin School of Medicine and Public Health, Madison, WI 53726, USA.
Mark J P Chaisson, Department of Quantitative and Computational Biology, University of Southern California, Los Angeles, Los Angeles, CA 90089, USA.
Ron Stewart, Regenerative Biology, Morgridge Institute for Research, Madison, WI 53715, USA.
Authors’ Contributions
J.A.T. and R.S. conceived of the project. S.M. and J.K.J. procured blue whale skin biopsy. S.M., L.F.C., J.A.B., and D.M. worked on blue whale cell cultures. S.M. and M.G. performed karyotyping. C.A. and J.B. performed Illumina sequencing. M.E.B. worked on PacBio sequencing of the genome. O.F. worked on PacBio Iso-seq sequencing of the transcriptome. J.S., S.A.S., and P.J. processed and analyzed Illumina data. A.R., G.F., A.M.P., and Y.V.B. worked on genome assemblies. R.S.H. and Y.V.B. worked on the Y chromosome. J.W. and K.H. curated the VGP genome assembly. Y.V.B. did assembly quality assessments and comparisons with other cetacean genomes. B.K., C.M., and M.H. generated TOGA projections of human and mouse genes to blue whale and vaquita genomes. A.J. and D.K. annotated blue whale proteins with GO terms. J.S.J., Y.V.B., A.R., A.I., and M.J.P.C. worked on genome size analyses. M.J.P.C. projected cattle GWAS sites onto blue whale and vaquita genomes. K.R. and R.S. carried out text mining analyses. M.J.P.C., Y.V.B., H.T., L.F.C., and R.S. performed segmental duplication analyses and evaluated biological significance of duplicated genes. Y.V.B. and R.S. worked on IGF1. P.A.M. and A.L. performed genome heterozygosity and historical demography analyses. M.W. performed the ROH analysis. Y.V.B., E.D.J., J.A.T., M.J.P.C., P.A.M., and R.S. coordinated and provided guidance. Y.V.B., M.J.P.C., and P.A.M. co-wrote the manuscript, with input from all other co-authors. All of the authors read and approved the final manuscript.
Funding
This study was supported by Morgridge Institute for Research departmental funds. R.S. and J.A.T. acknowledge a grant from Marv Conney. M.J.P.C. is funded by National Science Foundation (NSF) CAREER grant 2046753. M.H. acknowledges support from the LOEWE-Centre for Translational Biodiversity Genomics (TBG) funded by the Hessen State Ministry of Higher Education, Research and the Arts (HMWK). This work was supported, in part, by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (AR, AMP). O.F. and E.D.J. were supported by the Howard Hughes Medical Institute and the Rockefeller University.
Conflict of Interest
The authors declare no competing interests.
Data Availability
Raw sequencing and mapping data and various versions of genome assemblies are available from VGP GenomeArk (Vertebrate Genomes Project 2022a: 1; Vertebrate Genomes Project 2022b). Primary pseudohaplotype genome assembly and annotation are available from NCBI Genome, id 7017 (National Center for Biotechnology Information (US) 2022b). Primary pseudohaplotype assembly is mBalMus1.pri.v3, RefSeq accession GCF_009873245.2, and GenBank accession GCA_009873245.3. Alternate pseudohaplotype assembly is mBalMus1.alt.v2, GenBank accession GCA_008658375.2. Mitochondrion assembly generated in this study has GenBank accession CM018075.1. Fibroblast transcriptomics data have been deposited to the NCBI SRA archive. Iso-seq accession SRX6360705, Illumina RNA-seq accession SRX7696402. Additional supplementary materials and datasets are available on OSF (Bukhman et al. 2019). Whole genome annotations are available from NCBI RefSeq (National Center for Biotechnology Information (US) 2020), Ensembl Rapid Release (Ensembl 2020), and TOGA (both haplotypes) (Hiller Lab 2020a; Hiller Lab 2020b).
Ethics Approval and Consent to Participate
Blue whale skin tissue sample was collected under NMFS-SWFSC permit # 19091-01. Blue whale cell line development and use by the Thomson Laboratory was performed under NMFS permits # 14097-06 and # 20646. No human subjects participated in this study.
Consent for Publication
Not applicable.
Code Availability
All software used in this study is in the public domain, with the sole exception of Dovetail HiRise. VGP software repository is available on GitHub (Vertebrate Genomes Project 2022c). Y.V.B.'s R scripts and R Markdown notebooks are likewise available on GitHub (Bukhman 2022f). References to other software repositories are given in the Methods.
References
- Alegretti AP, Bittar CM, Bittencourt R, Piccoli AK, Schneider L, Silla LM, Bó SD, Xavier RM. The expression of CD56 antigen is associated with poor prognosis in patients with acute myeloid leukemia. Rev Bras Hematol Hemoter. 2011:33(3):202–206. 10.5581/1516-8484.20110054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer FI, Brownell RL Jr, Hancock-Hanser BL, Morin PA, Robertson KM, Sherman KK, Calambokidis J, Urbán R J, Rosel PE, Mizroch SA, et al. Revision of fin whale Balaenoptera physalus (Linnaeus, 1758) subspecies using genetics. J Mammal. 2019:100(5):1653–1670. 10.1093/jmammal/gyz121. [DOI] [Google Scholar]
- Arendt M, Fall T, Lindblad-Toh K, Axelsson E. Amylase activity is associated with AMY2B copy numbers in dog: implications for dog domestication, diet and diabetes. Anim Genet. 2014:45(5):716–722. 10.1111/age.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Árnason Ú, Lammers F, Kumar V, Nilsson MA, Janke A. Whole-genome sequencing of the blue whale and other rorquals finds signatures for introgressive gene flow. Sci Adv. 2018:4(4):eaap9873. 10.1126/sciadv.aap9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atz ME, Rollins B, Vawter MP. NCAM1 association study of bipolar disorder and schizophrenia: polymorphisms and alternatively spliced isoforms lead to similarities and differences. Psychiatr Genet. 2007:17(2):55–67. 10.1097/YPG.0b013e328012d850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006:7(7):552–564. 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001:11(6):1005–1017. 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcazar NE, Tripovich JS, Klinck H, Nieukirk SL, Mellinger DK, Dziak RP, Rogers TL. Calls reveal population structure of blue whales across the southeast Indian Ocean and the southwest Pacific Ocean. J Mammal. 2015:96(6):1184–1193. 10.1093/jmammal/gyv126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DR, Torres LG, Hodge KB, Steel D, Baker CS, Chandler TE, Bott N, Constantine R, Double MC, Gill P, et al. Documentation of a New Zealand blue whale population based on multiple lines of evidence. Endanger Species Res. 2018:36:27–40. 10.3354/esr00891. [DOI] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996:382(6588):225–230. 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Bianucci G. The odontoceti (Mammalia, Cetacea) from Italian pliocene. Systematics and phylogenesis of delphinidae. Palaeontogr Ital. 1996:83:73–167. Available from: https://www.researchgate.net/publication/292704391_The_Odontoceti_Mammalia_Cetacea_from_Italian_Pliocene_Systematics_and_phylogenesis_of_Delphinidae. [Google Scholar]
- Bianucci G, Marx FG, Collareta A, Di Stefano A, Landini W, Morigi C, Varola A. Rise of the titans: baleen whales became giants earlier than thought. Biol Lett. 2019:15(5):20190175. 10.1098/rsbl.2019.0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bista I, Wood JMD, Desvignes T, McCarthy SA, Matschiner M, Ning Z, Tracey A, Torrance J, Sims Y, Chow W, et al. Genomics of cold adaptations in the Antarctic notothenioid fish radiation. Nat Commun. 2023:14(1):3412. 10.1038/s41467-023-38567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman AC, Daetwyler HD, Chamberlain AJ, Ponce CH, Sargolzaei M, Schenkel FS, Sahana G, Govignon-Gion A, Boitard S, Dolezal M, et al. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat Genet. 2018:50(3):362–367. 10.1038/s41588-018-0056-5. [DOI] [PubMed] [Google Scholar]
- Branch TA, Abubaker EMN, Mkango S, Butterworth DS. Separating southern blue whale subspecies based on length frequencies of sexually mature females. Mar Mammal Sci. 2007a:23(4):803–833. 10.1111/j.1748-7692.2007.00137.x. [DOI] [Google Scholar]
- Branch TA, Stafford KM, Palacios DM, Allison C, Banniter JL, Burton CLK, Cabrera E, Carlson CA, Galletti Vernazzani B, Gill PC, et al. Past and present distribution, densities and movements of blue whales Balaenoptera musculus in the Southern Hemisphere and Northern Indian Ocean. Mammal Rev. 2007b:37(2):116–175. 10.1111/j.1365-2907.2007.00106.x. [DOI] [Google Scholar]
- Broad Institute . Picard Tools. Available from: http://broadinstitute.github.io/picard/.
- Bukhman YV. retrieve_gene_seqs.R. 2022a. Available from: https://github.com/stewart-lab/blue_whale/blob/c83d6f2f13eacd359a727a8d292801674effdb76/IGF1/body-size-loci_try2_artriodactyls-etc/retrieve_gene_seqs.R.
- Bukhman YV. IGF1_genomic_seqs.fna. 2022b. Available from: https://osf.io/h4dus.
- Bukhman YV. IGF1_genomic_seqs.aln. 2022c. Available from: https://osf.io/h8pjv.
- Bukhman YV. IGF1_genomic_seqs_sorted.aln. 2022d. Available from https://osf.io/c8aj5.
- Bukhman YV. IGF1_vs_body-size_2.Rmd. 2022e. Available from: https://github.com/stewart-lab/blue_whale/blob/c83d6f2f13eacd359a727a8d292801674effdb76/IGF1/body-size-loci_try2_artriodactyls-etc/IGF1_vs_body-size_2.Rmd.
- Bukhman YV. General · stewart-lab/blue_whale. GitHub [Internet]. 2022f. Available from: https://github.com/stewart-lab/blue_whale.
- Bukhman YV, Chaisson M, Raja K, Chu L-F, Toh H, Stewart R. Gene lists. 2021a. Available from: https://osf.io/rc2pf/.
- Bukhman YV, Morin PA, Meyer S, Chu L-F, Jacobsen JK, Antosiewicz-Bourget J, Mamott D, Gonzales M, Argus C, Bolin J, et al. Blue whale genome paper supplementary materials. 2019. Available from: https://osf.io/584jv/. [DOI] [PMC free article] [PubMed]
- Bukhman YV, Toh H, Chu L-F, Chaisson M, Stewart R, Raja K. Interesting genes. 2021b. Available from: https://osf.io/9kmx2/.
- Burns CJ, Zhang J, Brown EC, Van Bibber AM, Van Es J, Clevers H, Ishikawa T, Taketo MM, Vetter ML, Fuhrmann S. Investigation of Frizzled-5 during embryonic neural development in mouse. Dev Dyn. 2008:237:1614–1626. 10.1002/dvdy.21565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. BBMap. SourceForge [Internet]. Available from: https://sourceforge.net/projects/bbmap/.
- Cahill JA, Soares AE, Green RE, Shapiro B. Inferring species divergence times using pairwise sequential Markovian coalescent modelling and low-coverage genomic data. Philos Trans R Soc Lond B Biol Sci. 2016:371(1699):20150138. 10.1098/rstb.2015.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini Giovanni. Di un orca fossile scoperta a Cetona in Toscana. Memorie dell'Accademia delle Scienze dell'Instituto di Bologna. 1883:4(4):1–25. Available from: https://books.google.com/books/about/Di_un_orca_fossile_scoperta_a_Cetona_in.html?id=qDAQMwEACAAJ. [Google Scholar]
- Carlyle BC, Kitchen RR, Mattingly Z, Celia AM, Trombetta BA, Das S, Hyman BT, Kivisäkk P, Arnold SE. Technical performance evaluation of Olink proximity extension assay for blood-based biomarker discovery in longitudinal studies of Alzheimer's disease. Front Neurol. 2022:13:889647. 10.3389/fneur.2022.889647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Ruiz A. Lama pacos (alpaca). Anim. Divers. Web [Internet]. Available from: https://animaldiversity.org/accounts/Lama_pacos/.
- Caulin AF, Maley CC. Peto's Paradox: evolution's prescription for cancer prevention. Trends Ecol Evol. 2011:26(4):175–182. 10.1016/j.tree.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJP. ChaissonLab/SegDupAnnotation, release Blue whale genome v2. GitHub [Internet]. 2023. Available from: https://github.com/ChaissonLab/SegDupAnnotation/tree/vBW3.
- Cheung J, Wilson MD, Zhang J, Khaja R, MacDonald JR, Heng HH, Koop BF, Scherer SW. Recent segmental and gene duplications in the mouse genome. Genome Biol. 2003:4(8):R47. 10.1186/gb-2003-4-8-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi L, Rodriguez W, Grusea S, Santos P, Boitard S, Mazet O. The IICR (inverse instantaneous coalescence rate) as a summary of genomic diversity: insights into demographic inference and model choice. Heredity (Edinb). 2018:120(1):13–24. 10.1038/s41437-017-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, Sealy I, Torrance J, Howe K. gEVAL—a web-based browser for evaluating genome assemblies. Bioinformatics. 2016:32(16):2508–2510. 10.1093/bioinformatics/btw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham PJ, Baker CS. Modern whaling. In: Würsig BThewissen JGM, Kovacs KM, editors. Encyclopedia of marine mammals. 3rd ed. San Diego (CA): Academic Press/Elsevier; 2018. p. 1070–1074. USA [Google Scholar]
- Committee on Taxonomy . List of marine mammal species and subspecies. 2020. Available from: www.marinemammalscience.org
- Cooke JG. Balaenoptera musculus (errata version published in 2019). The IUCN Red List of Threatened Species 2018: e.T2477A156923585. [accessed 2024 Mar 01]. 10.2305/IUCN.UK.2018-2.RLTS.T2477A156923585.en. [DOI]
- Deciphering Developmental Disorders Study . Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015:519(7542):223–228. 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong MJ. Darwindow. 2021. Available from: https://github.com/mennodejong1986/Darwindow.
- de Jong MJ, Niamir A, Wolf M, Kitchener AC, Lecomte N, Seryodkin IV, Fain SR, Hagen SB, Saarma U, Janke A. Range-wide whole-genome resequencing of the brown bear reveals drivers of intraspecies divergence. Commun Biol. 2023:6(1):153. 10.1038/s42003-023-04514-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demin AA, Hirota K, Tsuda M, Adamowicz M, Hailstone R, Brazina J, Gittens W, Kalasova I, Shao Z, Zha S, et al. XRCC1 prevents toxic PARP1 trapping during DNA base excision repair. Mol Cell. 2021:81(14):3018–3030.e5. 10.1016/j.molcel.2021.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Na L, Zhou K, Wang Z, Sun Y, Zheng Q, Gao J, Zhao C, Wang W. FZD5 prevents epithelial-mesenchymal transition in gastric cancer. Cell Commun Signal. 2021:19(1):21. 10.1186/s12964-021-00708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durban JW, Moore MJ, Chiang G, Hickmott LS, Bocconcelli A, Howes G, Bahamonde PA, Perryman WL, LeRoi DJ. Photogrammetry of blue whales with an unmanned hexacopter. Mar Mammal Sci. 2016:32(4):1510–1515. 10.1111/mms.12328. [DOI] [Google Scholar]
- Ensembl . Balaenoptera_musculus_GCA_009873245.3—Ensembl 109. 2020. Available from: https://rapid.ensembl.org/Balaenoptera_musculus_GCA_009873245.3/Info/Index.
- Foote AD, Hooper R, Alexander A, Baird RW, Baker CS, Ballance L, Barlow J, Brownlow A, Collins T, Constantine R, et al. Runs of homozygosity in killer whale genomes provide a global record of demographic histories. Mol Ecol. 2021:30(23):6162–6177. 10.1111/mec.16137. [DOI] [PubMed] [Google Scholar]
- Foote AD, Martin MD, Louis M, Pacheco G, Robertson KM, Sinding M-HS, Amaral AR, Baird RW, Baker CS, Ballance L, et al. Killer whale genomes reveal a complex history of recurrent admixture and vicariance. Mol Ecol. 2019:28(14):3427–3444. 10.1111/mec.15099. [DOI] [PubMed] [Google Scholar]
- Formenti G, Rhie A, Balacco J, Haase B, Mountcastle J, Fedrigo O, Brown S, Capodiferro MR, Al-Ajli FO, Ambrosini R, et al. Complete vertebrate mitogenomes reveal widespread repeats and gene duplications. Genome Biol. 2021:22(1):120. 10.1186/s13059-021-02336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti Vernazzani B, Carlson CA, Cabrera E, Brownell RL Jr. Chilean blue whales off Isla Grande de Chiloe, 2004-2010: distribution, site-fidelity and behaviour. J Cetacean Res Manag. 2012:12(3):353–360. 10.47536/jcrm.v12i3.567. [DOI] [Google Scholar]
- Ghosh S, Ghosh S, Bankura B, Saha ML, Maji S, Ghatak S, Pattanayak AK, Sadhukhan S, Guha M, Nachimuthu SK, et al. Association of DNA repair and xenobiotic pathway gene polymorphisms with genetic susceptibility to gastric cancer patients in West Bengal, India. Tumour Biol. 2016:37(7):9139–9149. 10.1007/s13277-015-4780-5. [DOI] [PubMed] [Google Scholar]
- Gilpatrick JW Jr, Perryman WL. Geographic variation in external morphology of North Pacific and Southern Hemisphere blue whales (Balaenoptera musculus). J Cetacean Res Manag. 2008:10(1):9–21. 10.47536/jcrm.v10i1.654. [DOI] [Google Scholar]
- Gong Z, Platek ME, Till C, Goodman PJ, Tangen CM, Platz EA, Neuhouser ML, Thompson IM, Santella RM, Ambrosone CB. Associations between polymorphisms in genes related to oxidative stress and DNA repair, interactions with serum antioxidants, and prostate cancer risk: results from the prostate cancer prevention trial. Front Oncol. 2021:11:808715. 10.3389/fonc.2021.808715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Huddleston J, Chaisson MJP, Hill CM, Kronenberg ZN, Munson KM, Malig M, Raja A, Fiddes I, Hillier LW, et al. Long-read sequence assembly of the gorilla genome. Science. 2016:352(6281):aae0344. 10.1126/science.aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding H, Jasani B, Pereira H, Reid A, Galea M, Bell JA, Elston CW, Robertson JF, Blamey RW, Nicholson RA, et al. Metallothionein expression in human breast cancer. Br J Cancer. 1995:72(4):968–972. 10.1038/bjc.1995.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm PR, Irsik DL, Settles DC, Holtzclaw JD, Sansom SC. Hypertension of Kcnmb1−/− is linked to deficient K secretion and aldosteronism. Proc Natl Acad Sci U S A. 2009:106(28):11800–11805. 10.1073/pnas.0904635106. [DOI] [Google Scholar]
- Han T, Liu Y, Wu J, Bai Y, Zhou J, Hu C, Zhang W, Guo J, Wang Q, Hu D. An immune indicator based on BTK and DPEP2 identifies hot and cold tumors and clinical treatment outcomes in lung adenocarcinoma. Sci Rep. 2023:13(1):5153. 10.1038/s41598-023-32276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen-Bauer A, Solvang-Garten K, Akbari M, Otterlei M. X-ray repair cross complementing protein 1 in base excision repair. Int J Mol Sci. 2012:13(12):17210–17229. 10.3390/ijms131217210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare EE, Johnston JS. Genome size determination using flow cytometry of propidium iodide-stained nuclei. Methods Mol Biol. 2011:772:3–12. 10.1007/978-1-61779-228-1_1. [DOI] [PubMed] [Google Scholar]
- He X, Saint-Jeannet JP, Wang Y, Nathans J, Dawid I, Varmus H. A member of the Frizzled protein family mediating axis induction by wnt-5A. Science. 1997:275(5306):1652–1654. 10.1126/science.275.5306.1652. [DOI] [PubMed] [Google Scholar]
- Heemskerk MM, Giera M, Bouazzaoui FE, Lips MA, Pijl H, Van Dijk KW, Van Harmelen V. Increased PUFA content and 5-lipoxygenase pathway expression are associated with subcutaneous adipose tissue inflammation in obese women with type 2 diabetes. Nutrients. 2015:7(9):7676–7690. 10.3390/nu7095362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller Lab . Hiller Lab UCSC Genome Browser on Blue whale (GCF_009873245.2_mBalMus1.pri.v3/HLbalMus2). 2020a. Available from: https://genome.senckenberg.de/cgi-bin/hgTracks? db=HLbalMus2.
- Hiller Lab . Hiller Lab UCSC Genome Browser on Blue whale (mBalMus1.alt.v2/HLbalMus2B). 2020b. Available from: https://genome.senckenberg.de/cgi-bin/hgTracks? db=HLbalMus2B.
- Hohn AA, Read AJ, Fernandez S, Vidal O, Findley LT. Life history of the vaquita, phocoena sinus (phocoenidae, cetacea). J Zool. 1996:239(2):235–251. 10.1111/j.1469-7998.1996.tb05450.x. [DOI] [Google Scholar]
- Howe K, Chow W, Collins J, Pelan S, Pointon D-L, Sims Y, Torrance J, Tracey A, Wood J. Significantly improving the quality of genome assemblies through curation. Gigascience. 2021:10(1):giaa153. 10.1093/gigascience/giaa153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H-C, Klein PS. The Frizzled family: receptors for multiple signal transduction pathways. Genome Biol. 2004:5(7):234. 10.1186/gb-2004-5-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Zhang J, Gong L, Wang X, Zhang B, Liu D. Characterization of the fatty acid metabolism-related genes in lung adenocarcinoma to guide clinical therapy. BMC Pulm Med. 2022:22(1):486. 10.1186/s12890-022-02286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucke-Gaete R, Osman LP, Moreno CA, Findlay KP, Ljungblad DK. Discovery of a blue whale feeding and nursing ground in southern Chile. Proc Biol Sci. 2004:271 Suppl 4(Suppl 4):S170–S173. 10.1098/rsbl.2003.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Tamai Y, Zorn AM, Yoshida H, Seldin MF, Nishikawa S, Taketo MM. Mouse Wnt receptor gene Fzd5 is essential for yolk sac and placental angiogenesis. Development. 2001:128(1):25–33. 10.1242/dev.128.1.25. [DOI] [PubMed] [Google Scholar]
- Jackson JA, Baker CS, Vant M, Steel DJ, Medrano-González L, Palumbi SR. Big and slow: phylogenetic estimates of molecular evolution in baleen whales (suborder mysticeti). Mol Biol Evol. 2009:26(11):2427–2440. 10.1093/molbev/msp169. [DOI] [PubMed] [Google Scholar]
- Jackson JA, Steel DJ, Beerli P, Congdon BC, Olavarria C, Leslie MS, Pomilla C, Rosenbaum H, Baker CS. Global diversity and oceanic divergence of humpback whales (Megaptera novaeangliae). Proc Biol Sci. 2014:281(1786):20133222. 10.1098/rspb.2013.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Kihara D. Phylo-PFP: improved automated protein function prediction using phylogenetic distance of distantly related sequences. Bioinformatics. 2019:35(5):753–759. 10.1093/bioinformatics/bty704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Kihara D. GO term predictions. 2022. Available from: https://osf.io/w9fk6/.
- Jaratlerdsiri W, Chan EKF, Petersen DC, Yang C, Croucher PI, Bornman MSR, Sheth P, Hayes VM. Next generation mapping reveals novel large genomic rearrangements in prostate cancer. Oncotarget. 2017:8(14):23588–23602. 10.18632/oncotarget.15802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MJ, Kagiava A, Vendredy L, Spaulding EL, Stavrou M, Hathazi D, Grüneboom A, De Winter V, Gess B, Schara U, et al. NCAM1 and GDF15 are biomarkers of Charcot-Marie-Tooth disease in patients and mice. Brain:awac. 2022:145(11):3999–4015. 10.1093/brain/awac055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JS, Bernardini A, Hjelmen CE. Genome size estimation and quantitative cytogenetics in insects. Insect genomics. New York (NY): Humana Press; 2019. p. 15–26. [DOI] [PubMed] [Google Scholar]
- Jones KE, Bielby J, Cardillo M, Fritz SA, O’Dell J, Orme CDL, Safi K, Sechrest W, Boakes EH, Carbone C, et al. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology. 2009:90(9):2648–2648. 10.1890/08-1494.1. [DOI] [Google Scholar]
- Keane M, Semeiks J, Webb AE, Li YI, Quesada V, Craig T, Madsen LB, van Dam S, Brawand D, Marques PI, et al. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 2015:10(1):112–122. 10.1016/j.celrep.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley-Hedgepeth A, Peter I, Kip KE, Montefusco MC, Kogan S, Cox D, Ordovas JM, Levy D, Reis SE, Mendelsohn ME, et al. The protective effect of KCNMB1 E65K against hypertension is restricted to blood pressure treatment with β-blockade. J Hum Hypertens. 2008:22(7):512–515. 10.1038/jhh.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilenko BM, Munegowda C, Osipova E, Jebb D, Sharma V, Blumer M, Morales AE, Ahmed A-W, Kontopoulos D-G, Hilgers L, et al. Integrating gene annotation with orthology inference at scale. Science. 2023:380(6643):eabn3107. 10.1126/science.abn3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominakis A, Hager-Theodorides AL, Zoidis E, Saridaki A, Antonakos G, Tsiamis G. Combined GWAS and ‘guilt by association’-based prioritization analysis identifies functional candidate genes for body size in sheep. Genet Sel Evol. 2017:49(1):41. 10.1186/s12711-017-0316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics. 2014:15(1):356. 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusisto F, Ng D, Steill J, Ross I, Livny M, Thomson J, Page D, Stewart R. KinderMiner web: a simple web tool for ranking pairwise associations in biomedical applications. F1000Res. 2020:9:832. 10.12688/f1000research.25523.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunas-Rangel FA. Deciphering the whale's secrets to have a long life. Exp Gerontol. 2021:151:111425. 10.1016/j.exger.2021.111425. [DOI] [PubMed] [Google Scholar]
- Lai L, Shin GY, Qiu H. The role of cell cycle regulators in cell survival-dual functions of cyclin-dependent kinase 20 and p21Cip1/Waf1. Int J Mol Sci. 2020:21(22):8504. 10.3390/ijms21228504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc RG, Archer FI, Lang AR, Martien KK, Hancock-Hanser B, Torres-Florez JP, Hucke-Gaete R, Rosenbaum HC, van Waerebeek K, Brownell RL, et al. Genetic variation in blue whales in the eastern pacific: implication for taxonomy and use of common wintering grounds. Mol Ecol. 2017:26(3):740–751. 10.1111/mec.13940. [DOI] [PubMed] [Google Scholar]
- Li H. lh3/seqtk. 2022. Available from: https://github.com/lh3/seqtk.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009:25(14):1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011:475(7357):493–496. 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup . The sequence alignment/map format and SAMtools. Bioinformatics. 2009:25(16):2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L-F, Shen W-J, Ueno M, Patel S, Azhar S, Kraemer FB. Age-related modulation of the effects of obesity on gene expression profiles of mouse bone marrow and epididymal adipocytes. PLoS One. 2013:8(8):e72367. 10.1371/journal.pone.0072367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Moore R, Xu X, Barber GN. DDX24 negatively regulates cytosolic RNA-mediated innate immune signaling. PLoS Pathog. 2013:9(10):e1003721. 10.1371/journal.ppat.1003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Rong L, Zhou Y, Roy BB, Lu J, Abrahamyan L, Mouland AJ, Pan Q, Liang C. The requirement of the DEAD-box protein DDX24 for the packaging of human immunodeficiency virus type 1 RNA. Virology. 2008:375(1):253–264. 10.1016/j.virol.2008.01.025. [DOI] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 2021:38(10):4647–4654. 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010:20(9):1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan R, Leutenegger A-L, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, Smolej-Narancic N, Janicijevic B, Polasek O, Tenesa A, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008:83(3):359–372. 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Wang S, Tang W, Wu S, Gao N, Zhang C, Cao X, Li X, Zhang Z, Aschner M, et al. XRCC1 mediated the development of cervival cancer through a novel Sp1/Krox-20 swich. Oncotarget. 2017:8(49):86217–86226. 10.18632/oncotarget.21040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming Y, Jian J, Yu X, Wang J, Liu W. The genome resources for conservation of Indo-Pacific humpback dolphin, Sousa chinensis. Sci Data. 2019:6(1):68. 10.1038/s41597-019-0078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnahan CC, Branch TA, Punt AE. Do ship strikes threaten the recovery of endangered eastern North Pacific blue whales? Mar Mammal Sci. 2014:31(1):279–297. 10.1111/mms.12157. [DOI] [Google Scholar]
- Morin PA, Alexander A, Blaxter M, Caballero S, Fedrigo O, Fontaine MC, Foote AD, Kuraku S, Maloney B, McCarthy ML, et al. Building genomic infrastructure: sequencing platinum-standard reference-quality genomes of all cetacean species. Mar Mammal Sci. 2020:36(4):1356–1366. 10.1111/mms.12721. [DOI] [Google Scholar]
- Morin PA, Archer FI, Avila CD, Balacco JR, Bukhman YV, Chow W, Fedrigo O, Formenti G, Fronczek JA, Fungtammasan A, et al. Reference genome and demographic history of the most endangered marine mammal, the vaquita. Mol Ecol Resour. 2021:21(4):1008–1020. 10.1111/1755-0998.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PA, Foote AD, Baker CS, Hancock-Hanser BL, Kaschner K, Mate BR, Mesnick SL, Pease VL, Rosel PE, Alexander A. Demography or selection on linked cultural traits or genes? Investigating the driver of low mtDNA diversity in the sperm whale using complementary mitochondrial and nuclear genome analyses. Mol Ecol. 2018:27(11):2604–2619. 10.1111/mec.14698. [DOI] [PubMed] [Google Scholar]
- Müller JS, Mihaylova V, Abicht A, Lochmüller H. Congenital myasthenic syndromes: spotlight on genetic defects of neuromuscular transmission. Expert Rev Mol Med. 2007:9(22):1–20. 10.1017/S1462399407000427. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information (US) . Balaenoptera musculus genome assembly mBalMus1.pri.v3. Natl Cent Biotechnol Inf US [Internet]. 2020. Available from: https://identifiers.org/refseq.gcf:GCF_009873245.2.
- National Center for Biotechnology Information (US) . Assembly—NCBI. 2022a. Available from: https://www.ncbi.nlm.nih.gov/assembly.
- National Center for Biotechnology Information (US) . Balaenoptera musculus Genome (ID 7017). 2022b. Available from: https://www.ncbi.nlm.nih.gov/genome/7017.
- National Center for Biotechnology Information (US) . Entrez programming utilities help. Bethesda (MD): National Center for Biotechnology Information (US); 2010. Available from: https://www.ncbi.nlm.nih.gov/books/NBK25501/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numanagic I, Gökkaya AS, Zhang L, Berger B, Alkan C, Hach F. Fast characterization of segmental duplications in genome assemblies. Bioinforma Oxf Engl. 2018:34(17):i706–i714. 10.1093/bioinformatics/bty586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, et al. The complete sequence of a human genome. Science. 2022:376(6588):44–53. 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Wayne RK, Freedman AH, Davis BW. Demographic history, selection and functional diversity of the canine genome. Nat Rev Genet. 2017:18(12):705–720. 10.1038/nrg.2017.67. [DOI] [PubMed] [Google Scholar]
- Pabis K, Chiari Y, Sala C, Straka E, Giacconi R, Provinciali M, Li X, Brown-Borg H, Nowikovsky K, Valencak TG, et al. Elevated metallothionein expression in long-lived species mediates the influence of cadmium accumulation on aging. Geroscience. 2021:43(4):1975–1993. 10.1007/s11357-021-00393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic P, Pavlidis P, Dean K, Neznanova L, Romano R-A, Garneau D, Daugherity E, Globig A, Ruhl S, Gokcumen O. Independent amylase gene copy number bursts correlate with dietary preferences in mammals. Elife. 2019:8:e44628. 10.7554/eLife.44628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, An Y-R, An C-M, Kang J-H, Kim EM, Kim H, Cho S, Kim J. Evolutionary constraints over microsatellite abundance in larger mammals as a potential mechanism against carcinogenic burden. Sci Rep. 2016:6:25246. 10.1038/srep25246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park NI, Rogan PK, Tarnowski HE, Knoll JHM. Structural and genic characterization of stable genomic regions in breast cancer: relevance to chemotherapy. Mol Oncol. 2012:6(3):347–359. 10.1016/j.molonc.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen MØ, Larsen A, Stoltenberg M, Penkowa M. The role of metallothionein in oncogenesis and cancer prognosis. Prog Histochem Cytochem. 2009:44(1):29–64. 10.1016/j.proghi.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Pike DG, Víkingsson GA, Gunnlaugsson T, Øien N. A note on the distribution and abundance of blue whales (Balaenoptera musculus) in the Central and Northeast North Atlantic. NAMMCO Sci Publ. 2009:7:19–29. 10.7557/3.2703. [DOI] [Google Scholar]
- Plassais J, vonHoldt BM, Parker HG, Carmagnini A, Dubos N, Papa I, Bevant K, Derrien T, Hennelly LM, Whitaker DT, et al. Natural and human-driven selection of a single non-coding body size variant in ancient and modern canids. Curr Biol. 2022:32(4):889–897.e9. 10.1016/j.cub.2021.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja K, Steill J, Ross I, Tsoi LC, Kuusisto F, Ni Z, Livny M, Thomson J, Stewart R. SKiM—A generalized literature-based discovery system for uncovering novel biomedical knowledge from PubMed. bioRxiv 343012. 10.1101/2020.10.16.343012, 17 October 2020, preprint: not peer reviewed. [DOI]
- Rao CV, Xu C, Farooqui M, Zhang Y, Asch AS, Yamada HY. Survival-critical genes associated with copy number alterations in lung adenocarcinoma. Cancers (Basel). 2021:13(11):2586. 10.3390/cancers13112586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J, et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021:592(7856):737–746. 10.1038/s41586-021-03451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020:21(1):245. 10.1186/s13059-020-02134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JA, Kyriazis CC, Nigenda-Morales SF, Beichman AC, Rojas-Bracho L, Robertson KM, Fontaine MC, Wayne RK, Lohmueller KE, Taylor BL, et al. The critically endangered vaquita is not doomed to extinction by inbreeding depression. Science. 2022:376(6593):635–639. 10.1126/science.abm1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JA, Raikkonen J, Vucetich LM, Vucetich JA, Peterson RO, Lohmueller KE, Wayne RK. Genomic signatures of extensive inbreeding in Isle Royale wolves, a population on the threshold of extinction. Sci Adv. 2019:5(5):eaau0757. 10.1126/sciadv.aau0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen BD, Bickhart DM, Schnabel RD, Koren S, Elsik CG, Tseng E, Rowan TN, Low WY, Zimin A, Couldrey C, et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience. 2020:9(3):giaa021. 10.1093/gigascience/giaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaran F, Berne A, Leroy EC, Moreira S, Stafford KM, Maia M, Royer J. Antarctic blue whales (Balaenoptera musculus intermedia) recorded at the Equator in the Atlantic Ocean. Mar Mammal Sci. 2019:35(2):641–648. 10.1111/mms.12559. [DOI] [Google Scholar]
- Sárvári AK, Van Hauwaert EL, Markussen LK, Gammelmark E, Marcher A-B, Ebbesen MF, Nielsen R, Brewer JR, Madsen JGS, Mandrup S. Plasticity of epididymal adipose tissue in response to diet-induced obesity at single-nucleus resolution. Cell Metab. 2021:33(2):437–453.e5. 10.1016/j.cmet.2020.12.004. [DOI] [PubMed] [Google Scholar]
- Schneider J, Classen V, Helmig S. XRCC1 polymorphism and lung cancer risk. Expert Rev Mol Diagn. 2008:8(6):761–780. 10.1586/14737159.8.6.761. [DOI] [PubMed] [Google Scholar]
- Sears R, Perrin W. Blue whale balaenoptera musculus. In: Würsig BPerrin WFThewissen JGMWursig BPerrin WF, Thewissen JGMH, editors. Encyclopedia of marine mammals. San Diego, UNITED STATES: Elsevier Science & Technology; 2008. p. 121–124. Available from: http://ebookcentral.proquest.com/lib/wisc/detail.action?docID=405935. [Google Scholar]
- Sentí M, Fernández-Fernández JM, Tomás M, Vázquez E, Elosua R, Marrugat J, Valverde MA. Protective effect of the KCNMB1 E65K genetic polymorphism against diastolic hypertension in aging women and its relevance to cardiovascular risk. Circ Res. 2005:97(12):1360–1365. 10.1161/01.RES.0000196557.93717.95. [DOI] [PubMed] [Google Scholar]
- Shi D, Dai C, Qin J, Gu W. Negative regulation of the p300-p53 interplay by DDX24. Oncogene. 2016:35(4):528–536. 10.1038/onc.2015.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman JM, Perrimon N, Axelrod JD. Frizzled signaling and the developmental control of cell polarity. Trends Genet. 1998:14(11):452–458. 10.1016/S0168-9525(98)01584-4. [DOI] [PubMed] [Google Scholar]
- Sigel A, Freisinger E, Sigel RKO eds. 2015. Metallothioneins and Related Chelators. De Gruyter. Available from: https://www.degruyter.com/document/doi/10.1515/9783110436273/html.
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015:31(19):3210–3212. 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Slater GJ, Goldbogen JA, Pyenson ND. Independent evolution of baleen whale gigantism linked to plio-pleistocene ocean dynamics. Proc Biol Sci. 2017:284(1855):20170546. 10.1098/rspb.2017.0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snouffer A, Brown D, Lee H, Walsh J, Lupu F, Norman R, Lechtreck K, Ko HW, Eggenschwiler J. Cell Cycle-Related Kinase (CCRK) regulates ciliogenesis and Hedgehog signaling in mice. PLoS Genet. 2017:13(8):e1006912. 10.1371/journal.pgen.1006912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford KM, Bohnenstiehl DR, Tolstoy M, Chapp E, Mellinger DK, Moore SE. Antarctic-type blue whale calls recorded at low latitudes in the Indian and eastern Pacific Oceans. Deep Sea Res Part I Oceanogr Res Pap. 2004:51(10):1337–1346. 10.1016/j.dsr.2004.05.007. [DOI] [Google Scholar]
- Sulak M, Fong L, Mika K, Chigurupati S, Yon L, Mongan NP, Emes RD, Lynch VJ. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. Elife. 2016:5:e11994. 10.7554/eLife.11994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wang Z, Na L, Dong D, Wang W, Zhao C. FZD5 contributes to TNBC proliferation, DNA damage repair and stemness. Cell Death Dis. 2020:11(12):1060. 10.1038/s41419-020-03282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Çağlayan M. The scaffold protein XRCC1 stabilizes the formation of polβ/gap DNA and ligase IIIα/nick DNA complexes in base excision repair. J Biol Chem. 2021:297(3):101025. 10.1016/j.jbc.2021.101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Chivers SJ, Larese J, Perrin WF. Generation length and percent mature estimates for IUCN assessments of cetaceans. Administrative Report. La Jolla (CA): Southwest Fisheries Science Center; 2007. Available from https://www.researchgate.net/publication/254460229_Generation_length_and_percent_mature_estimates_for_IUCN_assessments_of_cetaceans.
- Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol. 1999:208(2):513–529. 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- Tejada-Martinez D, de Magalhães JP, Opazo JC. Positive selection and gene duplications in tumour suppressor genes reveal clues about how cetaceans resist cancer. Proc Biol Sci. 2021:288(1945):20202592. 10.1098/rspb.2020.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud-Nissen F, Souvorov A, Murphy T, DiCuccio M, Kitts P. Eukaryotic genome annotation pipeline. Bethesda (MD): National Center for Biotechnology Information (US); 2013. Available from: https://www.ncbi.nlm.nih.gov/books/NBK169439/. [Google Scholar]
- Thiele S, Zimmer A, Göbel A, Rachner TD, Rother S, Fuessel S, Froehner M, Wirth MP, Muders MH, Baretton GB, et al. Role of WNT5A receptors FZD5 and RYK in prostate cancer cells. Oncotarget. 2018:9(43):27293–27304. 10.18632/oncotarget.25551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomisch K, Boebel O, Bachmann J, Filun D, Neumann S, Spiesecke S, Van Opzeeland I. Temporal patterns in the acoustic presence of baleen whale species in a presumed breeding area off Namibia. Mar Ecol Prog Ser. 2019:620:201–214. 10.3354/meps12952. [DOI] [Google Scholar]
- Thybert D, Roller M, Navarro FCP, Fiddes I, Streeter I, Feig C, Martin-Galvez D, Kolmogorov M, Janoušek V, Akanni W, et al. Repeat associated mechanisms of genome evolution and function revealed by the Mus caroli and Mus pahari genomes. Genome Res. 2018:28(4):448–459. 10.1101/gr.234096.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Yang C, Formenti G, Raja K, Yan L, Tracey A, Chow W, Howe K, Bergeron LA, Zhang G, et al. A haplotype-resolved genome assembly of the Nile rat facilitates exploration of the genetic basis of diabetes. BMC Biol. 2022:20(1):245. 10.1186/s12915-022-01427-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Boddy AM, Maley CC. Peto's paradox: how has evolution solved the problem of cancer prevention? BMC Biol. 2017:15(1):60. 10.1186/s12915-017-0401-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Robbins J, Webb AE, Kuderna LFK, Caulin AF, Garcia JD, Bèrubè M, Pourmand N, Marques-Bonet T, O’Connell MJ, et al. Return to the sea, get huge, beat cancer: an analysis of cetacean genomes including an assembly for the humpback whale (Megaptera novaeangliae). Mol Biol Evol. 2019:36(8):1746–1763. 10.1093/molbev/msz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treseus . Husbandry—Treseus. 2022. Available from: https://treseus.com/en/cattle-raising/cattle-raising-husbandry/.
- van der Valk T, Díez-del-Molino D, Marques-Bonet T, Guschanski K, Dalén L. Historical genomes reveal the genomic consequences of recent population decline in eastern gorillas. Curr Biol. 2019:29(1):165–170.e6. 10.1016/j.cub.2018.11.055. [DOI] [PubMed] [Google Scholar]
- Vazquez JM, Sulak M, Chigurupati S, Lynch VJ. A zombie LIF gene in elephants is upregulated by TP53 to induce apoptosis in response to DNA damage. Cell Rep. 2018:24(7):1765–1776. 10.1016/j.celrep.2018.07.042. [DOI] [PubMed] [Google Scholar]
- Vertebrate Genomes Project . GenomeArk, blue whale Illumina genomic sequencing data. 2020a. Available from: https://genomeark.s3.amazonaws.com/index.html? prefix=species/Balaenoptera_musculus/mBalMus1/genomic_data/illumina/.
- Vertebrate Genomes Project . Tursiops truncatus (common bottlenose dolphin) genome assembly mTurTru1.mat.Y. 2020b. Available from: https://identifiers.org/ncbi/insdc.gca:GCA_011762595.1.
- Vertebrate Genomes Project . GenomeArk, Balaenoptera musculus, mBalMus1. 2022a. Available from: https://genomeark.s3.amazonaws.com/index.html? prefix=species/Balaenoptera_musculus/mBalMus1/.
- Vertebrate Genomes Project . 2022b. GenomeArk, Balaenoptera musculus. GenomeArk [Internet]. Available from: https://www.genomeark.org/vgp-curated-assembly/Balaenoptera_musculus.html.
- Vertebrate Genomes Project . 2022c. Vertebrate Genomes Project. GitHub [Internet]. Available from: https://github.com/VGP.
- Vollger MR, Chaisson M. 2022. mrvollger/SDA: v0.1.0. Available from: https://zenodo.org/record/5879923.
- Vollger MR, Dishuck PC, Sorensen M, Welch AE, Dang V, Dougherty ML, Graves-Lindsay TA, Wilson RK, Chaisson MJP, Eichler EE. Long-read sequence and assembly of segmental duplications. Nat Methods. 2019:16(1):88–94. 10.1038/s41592-018-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Seth J, Dussex N, Díez-del-Molino D, van der Valk T, Kutschera VE, Kierczak M, Steiner CC, Liu S, Gilbert MTP, Sinding M-HS, et al. Genomic insights into the conservation status of the world's last remaining Sumatran rhinoceros populations. Nat Commun. 2021:12(1):2393. 10.1038/s41467-021-22386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukojevic V, Mastrandreas P, Arnold A, Peter F, Kolassa I-T, Wilker S, Elbert T, de Quervain DJ-F, Papassotiropoulos A, Stetak A. Evolutionary conserved role of neural cell adhesion molecule-1 in memory. Transl Psychiatry. 2020:10(1):1–13. 10.1038/s41398-020-00899-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ai Z. Association of XRCC1 polymorphisms with thyroid cancer risk. Tumour Biol. 2014:35(5):4791–4797. 10.1007/s13277-014-1629-2. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009:25(9):1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikipedia contributors . 2022a. Elk. Wikipedia Free Encycl. [Internet]. Available from: https://en.wikipedia.org/w/index.php? title=Elk&oldid=1098803917.
- Wikipedia contributors . 2022b. Hereford cattle. Wikipedia Free Encycl. [Internet]. Available from: https://en.wikipedia.org/w/index.php? title=Hereford_cattle&oldid=1080987824.
- Wikipedia contributors . 2022c. Wild yak. Wikipedia [Internet]. Available from: https://en.wikipedia.org/w/index.php? title=Wild_yak&oldid=1094738040.
- Wikipedia contributors . 2022d. Yangtze finless porpoise. Wikipedia [Internet]. Available from: https://en.wikipedia.org/w/index.php? title=Yangtze_finless_porpoise&oldid=1092461136.
- Winter DJ. Rentrez: an R package for the NCBI eUtils API. R J. 2017:9(2):520–526. 10.32614/RJ-2017-058. [DOI] [Google Scholar]
- Wolf M, de Jong M, Halldórsson SD, Árnason Ú, Janke A. Genomic impact of whaling in north Atlantic fin whales. Mol Biol Evol. 2022:39(5):msac094. 10.1093/molbev/msac094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Miao X, Zhang Y, Li D, Zou Q, Yuan Y, Liu R, Yang Z. XRCC1 is a promising predictive biomarker and facilitates chemo-resistance in gallbladder cancer. Front Mol Biosci. 2020:7:70. 10.3389/fmolb.2020.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicola RM, Manojlovic Z, Augustus GJ, Kupfer SS, Emmadi R, Alagiozian-Angelova V, Triche T Jr, Salhia B, Carpten J, Llor X, et al. Lack of APC somatic mutation is associated with early-onset colorectal cancer in African Americans. Carcinogenesis. 2018:39(11):1331–1341. 10.1093/carcin/bgy122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang-Snyder J, Miller JR, Brown JD, Lai CJ, Moon RT. A frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr Biol. 1996:6(10):1302–1306. 10.1016/S0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- Yang X, Yue Y, Xiong S. Dpep2 emerging as a modulator of macrophage inflammation confers protection against CVB3-induced viral myocarditis. Front Cell Infect Microbiol. 2019:9:57. 10.3389/fcimb.2019.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H-S, Cho YS, Guang X, Kang SG, Jeong J-Y, Cha S-S, Oh H-M, Jae-Hak L, Yang EC, Kwon KK, et al. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 2014:46(1):88–92. 10.1038/ng.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zhang Y, Zhang P, Liu C, Wang J, Gao H, Hoelzel AR, Seim I, Lv M, Lin M, et al. Comparative genomics provides insights into the aquatic adaptations of mammals. Proc Natl Acad Sci U S A. 2021:118(37):e2106080118. 10.1073/pnas.2106080118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Luo H, Huang X, Wei C, Di J, Tian Y, Fu X, Li B, Liu GE, Fang L, et al. Integration of a single-step genome-wide association study with a multi-tissue transcriptome analysis provides novel insights into the genetic basis of wool and weight traits in sheep. Genet Sel Evol. 2021:53(1):56. 10.1186/s12711-021-00649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Xiao S, Lin G, Chen D, Cen W, Xue T, Liu Z, Zhong J, Chen Y, Xiao Y, et al. Chromosome genome assembly and annotation of the yellowbelly pufferfish with PacBio and Hi-C sequencing data. Sci Data. 2019:6(1):267. 10.1038/s41597-019-0279-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing and mapping data and various versions of genome assemblies are available from VGP GenomeArk (Vertebrate Genomes Project 2022a: 1; Vertebrate Genomes Project 2022b). Primary pseudohaplotype genome assembly and annotation are available from NCBI Genome, id 7017 (National Center for Biotechnology Information (US) 2022b). Primary pseudohaplotype assembly is mBalMus1.pri.v3, RefSeq accession GCF_009873245.2, and GenBank accession GCA_009873245.3. Alternate pseudohaplotype assembly is mBalMus1.alt.v2, GenBank accession GCA_008658375.2. Mitochondrion assembly generated in this study has GenBank accession CM018075.1. Fibroblast transcriptomics data have been deposited to the NCBI SRA archive. Iso-seq accession SRX6360705, Illumina RNA-seq accession SRX7696402. Additional supplementary materials and datasets are available on OSF (Bukhman et al. 2019). Whole genome annotations are available from NCBI RefSeq (National Center for Biotechnology Information (US) 2020), Ensembl Rapid Release (Ensembl 2020), and TOGA (both haplotypes) (Hiller Lab 2020a; Hiller Lab 2020b).