Abstract
Background
Panax ginseng, one of the valuable perennial medicinal plants, stores numerous pharmacological substrates in its storage roots. Given its perennial growth habit, organ regeneration occurs each year, and cambium stem cell activity is necessary for secondary growth and storage root formation. Cytokinin (CK) is a phytohormone involved in the maintenance of meristematic cells for the development of storage organs; however, its physiological role in storage-root secondary growth remains unknown.
Methods
Exogenous CK was repeatedly applied to P. ginseng, and morphological and histological changes were observed. RNA-seq analysis was used to elucidate the transcriptional network of CK that regulates P. ginseng growth and development. The HISTIDINE KINASE 3 (PgHK3) and RESPONSE REGULATOR 2 (PgRR2) genes were cloned in P. ginseng and functionally analyzed in Arabidopsis as a two-component system involved in CK signaling.
Results
Phenotypic and histological analyses showed that CK increased cambium activity and dormant axillary bud formation in P. ginseng, thus promoting storage-root secondary growth and bud formation. The evolutionarily conserved two-component signaling pathways in P. ginseng were sufficient to restore CK signaling in the Arabidopsis ahk2/3 double mutant and rescue its growth defects. Finally, RNA-seq analysis of CK-treated P. ginseng roots revealed that plant-type cell wall biogenesis-related genes are tightly connected with mitotic cell division, cytokinesis, and auxin signaling to regulate CK-mediated P. ginseng development.
Conclusion
Overall, we identified the CK signaling-related two-component systems and their physiological role in P. ginseng. This scientific information has the potential to significantly improve the field-cultivation and biotechnology-based breeding of ginseng.
Keywords: cytokinin, Panax ginseng, phytohormones, secondary growth, storage root
1. Introduction
P. ginseng, also known as Korean ginseng, has long been used as an important herbal remedy in Asia, particularly in Korea, China, and Japan, to treat a range of diseases [1,2]. The roots of P. ginseng plants contain a variety of pharmacologically active ginsenosides, which have a wide range of therapeutic effects on disease and aging, owing to their anti-allergenic, antidiabetic, anticancer, anti-aging, and immunity- and vitality-boosting properties [[2], [3], [4]]. Even though P. ginseng roots have been used as a major therapeutic herb for thousands of years, few genetic and physiological elements controlling the formation and development of its storage roots have been identified [5]. Progress has recently been made in understanding the physiological properties and the primary and secondary growth of P. ginseng storage roots. However, the implementation of functional genomic approaches in P. ginseng is challenging because of its allotetraploid character (2n = 4x = 48) and its relatively large genome size (3.2 Gb) with high quantities of repetitive DNA [6,7]. To shed light on the mechanism of secondary growth of storage roots, the main endogenous signaling components governing the growth and developmental features of P. ginseng should be characterized.
The maintenance of cambium stem cells directly affects the regulation of plant secondary growth. Previous studies have shown that a variety of plant hormones, including auxin, cytokinin (CK), and gibberellin (GA), as well as their crosstalk, affect the division and differentiation of cambium stem cells [[8], [9], [10], [11], [12], [13]]. Recent research shows that increasing cambium activity through GA and nitrate treatments improves the development of storage parenchyma cells [8,13]. Furthermore, CK, a key phytohormone controlling meristem activity and cell division, regulates the development of vascular tissue and storage organ formation by increasing sink strength [11,12]. The two-component phosphorelay system plays a central role in initiating CK signaling pathways in plants. The extracellular CHASE domain of plasma membrane-anchored histidine kinases (HKs) recognizes active CKs directly [[14], [15], [16]]. This signaling cue is subsequently transferred to the conserved aspartic acid (Asp) residue in the receiver domain of type-B RESPONSE REGULATORS (RRs) via the cytoplasmic HISTIDINE-CONTAINING PHOSPHOTRANSFER PROTEINS (HPTs). Upon phosphorylation-induced activation, the type-B RRs regulate the expression of CK-responsive genes, including type-A RRs, thus acting as negative regulators of CK signaling pathways [17]. A CK-responsive two-component signaling (TCS) reporter was designed using the target binding sequence of type-B RRs. These CK signaling components also act as a crosstalk node within the plant, coordinating a wide range of physiological responses to environmental cues.
In this study, we first defined the functionality of a two-component circuit associated with CK signaling in P. ginseng, and investigated the effects of exogenous CK on the secondary growth of storage roots. Next, we confirmed the evolutionary conservation of HK-triggered CK signaling pathways in the P. ginseng genome. Functional characterization of PgHK3 and PgRR2 in the model plant Arabidopsis thaliana indicated that the encoded proteins comprise a two-component phosphorelay system that regulates CK signal transduction in P. ginseng. Exogenous CK treatment markedly enhanced root mass, root diameter, and bud number. Histological microscopy analysis revealed that CK treatment enhances cambium stem cell activity to promote radial secondary growth of storage roots. Furthermore, transcriptome analysis provided additional evidence that CK stimulates cell division, cell cycle, cytokinesis, and auxin responses, all of which are closely linked to plant secondary cell wall biogenesis. We show that the CK signaling pathway is functionally conserved and promotes secondary growth and development in P. ginseng. Overall, our results shed light on the link between CK and secondary growth in P. ginseng.
2. Materials and methods
2.1. Plant material and growth conditions
Arabidopsis Col-0 and ahk2ahk3 was used as the wild type and genetic backgrounds for the transgenic lines overexpressing PgHK3-HA and PgHK3HQ-HA. The seeds were germinated in a medium containing 1/2 MS, 1% sucrose, and 0.8% agar (pH 5.7) at 22°C under long-day conditions (16 h light/8 h dark). 2-Year-old of Panax ginseng (cv. Sunwon and Yeonpung) plants were provided by the Rural Development Administration and was grown under the same condition of a plant growth room. For observation of vascular cambium activity upon CK, the Sunwon and Yeonpung cultivars were treated with 6-benzylaminopurine (BAP) and Kinetin, respectively, for 8-10 weeks after shoot emergence. BAP and Kinetin were dissolved in DMSO and treated with concentrations of 0.1 mM, 0.25 mM, 0.5 mM, and 1 mM, respectively, and DMSO was used as a mock control. CKs or Mock was applied to the soil at 25 ml per plant on a weekly basis using the overhead flooding method.
2.2. Plasmid construction
For transient expression assay, PgHK3 and PgRR2 were cloned into the 35S–C4PPDK promoter containing plant expression vector fused with the GFP-tag (Hwang and Sheen 2001). To generate overexpressed transgenic plants, PgHK3 or PgHK3-H442Q was cloned into the pCB302ES vector containing human influenza hemagglutinin (HA). PgHK3-H442Q was constructed by substituting glutamine for histidine at position 442 in the amino acid sequence of PgHK3. These constructs were transformed into Agrobacterium tumefaciens GV3101, and the Arabidopsis plants were transformed using the floral dipping method.
2.3. Protoplast transient expression assay
For the reporter assay, 2 × 104 of protoplasts were co-transfected with 20 μg of plasmid DNA composed of reporter (pTCS:LUC) and effector (PgHK3-GFP , PgHKH442Q-GFP, PgrRR2-GFP). The transfected protoplasts were incubated for 12 h under continuous light in CK-treated or untreated conditions.
2.4. Histochemical staining
The plant samples including stem and hypocotyl of 5-week-old Arabidopsis, main storage roots and dormancy buds of P. ginseng were fixed with Fix solution (0.2 M sodiumphosphate, 3% formaldehyde, 2.5% glutaraldehyde) before embedding in paraffin. Samples prepared to a thickness of 7-10 um with a HisoCore MULTICUT microtom (Leica, Wetzlar, Germany) were stained as a previously reported [8,13]. The prepared samples were observed using a slide scanner VS200 system and a BX53 microscope (OLYMPUS).
2.5. RNA-seq analysis and quantitative reverse transcription qRT-PCR
Total RNA was extracted from the main storage root samples (a 2 cm sample taken from the top region of the storage roots of P. ginseng cv. Yeonpung) applied to mock (DMSO, the mock treated RNA samples are the same ones used as a control in our previous work [8]) and CK (Kinetin) treatments once a week for 4 weeks. Total RNAs were prepared for RNA-seq libraries with three biological replicates, using TruSeq Stranded mRNA Library Prep Kit (Illumina, Inc., San Diego, CA). Following PCR enrichment and library purification, the libraries were sequenced on the Illumina HiSeq 4000 platform to produce 100-bp paired-end reads. The RNA-seq raw data are available in the NCBI Short Read Archive (accession number; SAMN12273127). Each sample's clean paired-end reads were aligned to the reference sequences for ginseng using Bowtie2 [18]. For each transcript, read counts and TPM (trimmed mean of M value-normalized transcripts per million) values were calculated using the RSEM 1.3.0 program [18]. The differential gene expression analysis's negative binomial dispersion across conditions was computed using EdgeR version 3.16.5 [19]. If a gene displayed a false discovery rate (FDR)-adjusted P < 0.05, it was found to be significantly differentially expressed [20].
qRT-PCR was performed for the validation of RNA-seq analysis. Topscript™ RT DRyMIX (enzynomics) was used for cDNA synthesis and then qRT-PCR was carried out with a SYBR qPCR MASTER MIX (TOYOBO). PgActin (Pg_S0354.5) was used as an internal control for qRT-PCR validation. All primer sequences are presented in Table S1.
2.6. Network analysis
Functional annotation of differentially expressed genes was performed using the BLAST tool with an e-value cutoff of 1E−5 against the Arabidopsis thaliana protein database. DAVID was used to perform a Gene Ontology (GO) term enrichment analysis, and enriched GO terms were identified using the Fisher Exact test (P < 0.05) [21]. According to our earlier research [22], enriched GO genes were further examined using Gene Set Enrichment Analysis (GSEA). Network analysis was carried out with Cytoscape's GeneMANIA program [23,24]. The gene subset that contributed the most to the enrichment score is represented by the red line in the enrichment graphic (ES). The correlation between a gene and the plant phenotype is quantified by the ranking list metric in the graphic. In the ranking list, positive values indicate genes up-regulated in mock-treated control samples with red color gradient, and negative values indicate genes down-regulated in the mock-treated root samples with blue color.
3. Results
3.1. Exogenous CK treatment promotes bud formation and root secondary growth by increasing cambium activity in P. ginseng
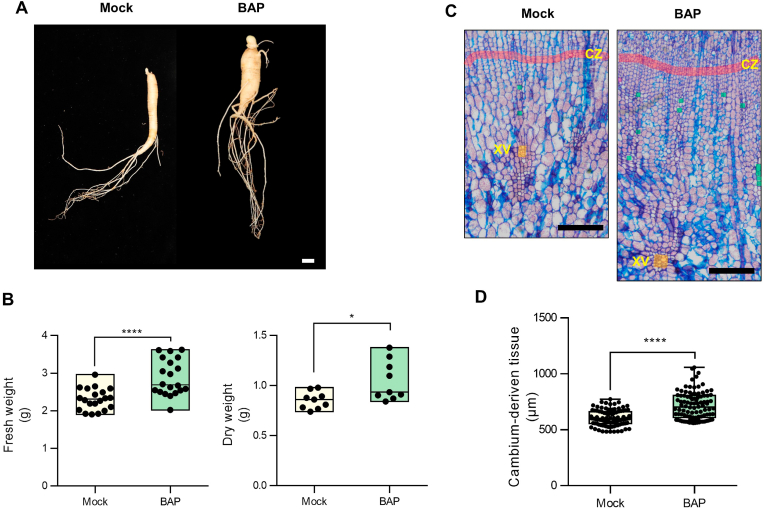
The stimulation of cell division and cambium stem cell activity is largely responsible for secondary growth in plant storage organs. CK is one of the most important growth factors that increase meristem activity and promote cell division [[10], [11], [12],25]. We initially examined the effect of CK on root development in P. ginseng to determine the relationship between CK responses and storage organ formation. Exogenous application of CK (BAP) enhanced biomass by promoting the secondary growth of the main storage roots of P. ginseng cv. Sunwon. The fresh and dried weight of P. ginseng roots increased by approximately 30% following CK-treatment (Fig. 1 A and B). We previously showed that cambium-driven storage parenchyma formation contributes to the secondary growth of storage roots in P. ginseng [8,13]. To determine whether the increased root biomass was a result of storage parenchyma cell growth, the paraffin-embedded sections of mock- and CK-treated P. ginseng root samples were subjected to safranine-astra blue combination staining. The results showed that the number of cambium-driven storage parenchyma cells and the activity of cambium cells were considerably increased in the CK-treated root samples compared with the mock treatment (Fig. 1C). Additionally, the CK-mediated acceleration of cambium cell division and production of storage parenchyma cells increased the area of the newly deposited parenchyma tissue in storage roots (Fig. 1D). These results indicate that CK plays a critical role in facilitating the secondary growth of storage roots in P. ginseng.
Fig. 1.
Exogenous CK treatment enhances root biomass by promoting secondary growth in P. ginseng. (A) Phenotype of 2-year-old P. ginseng (Sunwon) plants treated with mock control (Mock) or 0.5 mM BAP for 10 weeks. Scale bar = 2 cm. (B) Measurement of the fresh weight and dried weight of CK-treated P. ginseng (Sunwon) roots (n = 20). (C) Representative stained cross-sections image of P. ginseng (Sunwon) roots treated with mock, or 0.5 mM BAP. Red line and yellow regions indicate cambial cell layer zone (CZ) and xylem vessel (XV), respectively (n = 9). Scale bar = 100 μm (D) Measurement of cambium-driven tissue length in (C) (n = 100). Scale bar = 100 μm. Error bars represent standard error. The significance of the difference was analyzed by t-test method (*P < 0.05, ****P > 0.0001).
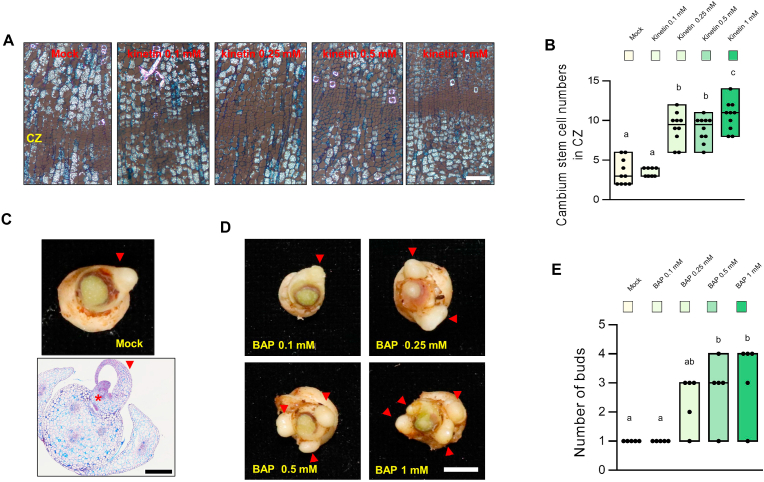
Next, we investigated whether the increase in cambial activity was a general effect of CK in ginseng cultivars and whether it depended on the concentration of exogenous CK. To test this, we treated Kinetin, another synthetic CK, with a dosage dependent manner in the Yeonpung cultivar. The results showed that cambium activity and cell division promoted with the increase in CK (Kinetin) concentration (Fig. 2 A and B). Storage roots treated with 0.1 mM CK or mock treatment contained approximately 3–6 cambium cells, whereas those treated with 0.25 mM or higher concentrations of CK contained approximately 10 dividing cells (Fig. 2B). These results support that the CK enhances vascular cambium activity during storage root development in P. ginseng. Furthermore, we observed a correlation between new bud formation and exogenous CK concentration in P. ginseng. A single dormant bud is generally observed on the head of a storage root (Fig. 2C, top). Histological analysis of the newly developed dormant bud revealed the establishment of a developing shoot and floral primordium (Fig. 2C, bottom). However, the number of emerging buds increased with the increase in CK concentration (Fig. 2D), and up to four dormant buds were observed on the head of storage roots treated with 0.5 mM or higher concentrations of CK (Fig. 2C and D). These results demonstrate that CK plays a positive role in the secondary growth of P. ginseng roots by enhancing the activity of cambium cells and promoting the generation of perennial dormant buds.
Fig. 2.
Exogenous CK treatment increased the cambial stem cell activity and dormancy bud formation. (A) Phenotype of the cambial stem cells of storage roots of 2-year-old P. ginseng (Yeonpung) treated with Kinetin (0.1 mM, 0.25 mM, 0.5mM, 1mM) and Mock for 10 weeks. Scale bar = 100 μm. (B) The numbers of cambial stem cells in the cambial cell layer zone (CZ). Error bars represent standard error (n = 10). Different lowercase letters indicate statistically significant differences P < 0.05; one-way ANOVA, followed by Tukey's multiple range test. (C, D) Representative images of bud samples with mock (Sunwon) (C) and treated with BAP (0.1 mM, 0.25 mM, 0.5 mM and 1 mM/ D) for 10 weeks. Red arrows are buds. (upper panel) and cross-section image (lower panel). Scale bar = 0.5. (E) The numbers of buds which are presented in (D). Error bars represent standard error (n = 5). Different lowercase letters indicate statistically significant differences P < 0.05; one-way, followed by Tukey's multiple range test.
3.2. Transcriptome analysis of secondary growth in response to CK in P. ginseng
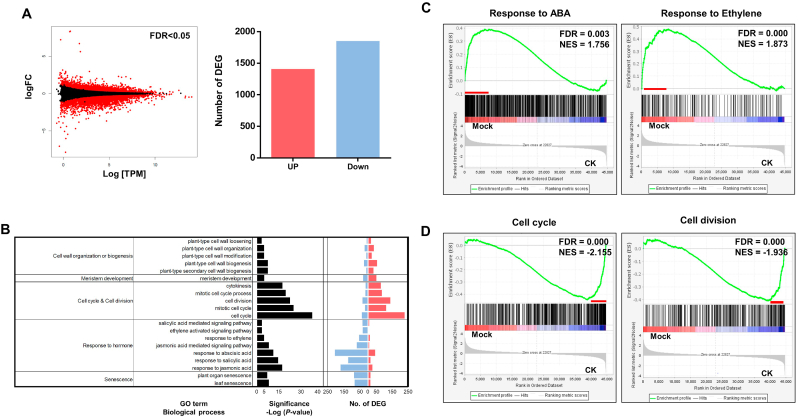
To elucidate the effect of CK on root secondary growth and bud development in P. ginseng, we identified the genes differentially expressed between mock- and CK-treated P. ginseng roots using RNA-seq analysis (Fig. 3A). We selected over 3,200 differentially expressed genes (DEGs) in the CK (Kinetin)-treated root samples. Gene Ontology (GO) enrichment analysis of these DEGs revealed significant enrichment of the following functional categories: ‘Cell wall organization or biogenesis’’ (p = 4.42 × 10−5, plant-type cell wall loosening; p = 5.07 × 10−6, plant cell wall modification; p = 6.03 × 10−5, plant-type cell wall biogenesis; p = 7.78 × 10−6, plant-type cell wall organization; p = 4.56 × 10−5, plant-type secondary cell wall organization), ‘Cell cycle & Cell division’ (p = 1.44 × 10−4, cytokinesis; p = 2.64 × 10−10 cell cycle, p = 7.79 × 10−8, cell division; p = 9.29 × 10−5, mitotic cell cycle process; p = 2.98 × 10−5, mitotic cell cycle), ‘Response to hormone’ (p = 1.28 × 10−14, response to jasmonic acid; p = 6.42 × 10−9, jasmonic acid-mediated signaling pathway; p = 1.80 × 10−12, response to abscisic acid; p = 3.40 × 10−19, response to salicylic acid; p = 3.95 × 10−4, salicylic acid-mediated signaling pathway), and ‘Senescence’ (p = 4.23 × 10−5, leaf senescence; p = 1.45 × 10−7, plant organ senescence) (Fig. 3B). These data indicate that CK-mediated storage root and dormant bud formation in P. ginseng are significantly connected with mitotic cell division, cell wall organization, hormonal regulation, and meristematic cell development. Gene set enrichment analysis (GSEA) revealed that CK treatment compromised responses related to stress hormones such as abscisic acid (ABA) and ethylene (false discovery rate [FDR] = 0.003; Fig. 3C and Fig. S1). Cell cycle- and cell division-related gene sets with growth hormone auxin responses, on the other hand, were significantly enriched in CK-treated P. ginseng roots (Fig. 3D and Figs S2A–D and S3). Finally, we validated the expression patterns of selected DEGs, including those related to ABA, ethylene, cell cycle, cell division, and secondary cell wall biogenesis, by real-time qRT-PCR (Fig. S4). These findings support the notion that exogenous CK-induced activation of cambium stem cells is strongly linked with the signaling pathways related to cell division, cell cycle, cytokinesis, and plant-type cell wall biogenesis through the suppression of stress-related hormone regulation.
Fig. 3.
Transcriptome profiling of P. ginseng roots treated with or without CK. (A) MA plot shows differential expression between mock-and CK (Kinetin)-treated P. ginseng (Yeonpung) root samples. Red dots and bar graphs indicate the either up-(1,409) and down-(1,884) regulated genes with q < 0.05 and ≥ |1.5|-fold change. (B) Gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs) identified by comparison of control and CK (Kinetin)-treated P. ginseng (Yeonpung) root samples. GO terms in biological process, with EASE score < 0.01 were selected (left panel). The number of up-regulated genes (red) and down-regulated genes (blue) categorized under the enriched GO terms are shown in the right panel. Enrichment plots for (C) response to ABA (GO:0010427, FDR = 0.0031986109, NES = 1.7566888) and ethylene-activated signaling pathway (GO:0009873, FDR = 0.00061904767, NES = 1.8739792), (D) cell cycle (GO:0007049, FDR = 0.000, NES = −2.1552036) and cell division (GO:0051301, FDR = 0.000, NES = −1.9369985).
3.3. Plant-type secondary cell wall biogenesis-related gene sets play a central role in the CK-mediated secondary growth of P. ginseng roots
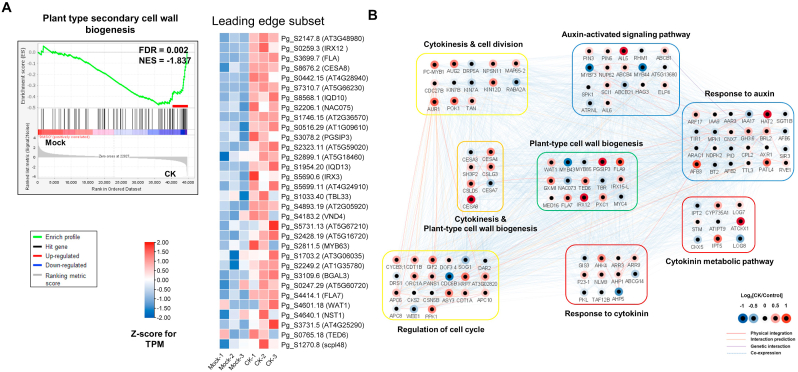
Recent studies showed that the secondary growth of P. ginseng storage roots is highly correlated with active cell division in the cambial zone and with cell wall biogenesis [8,13]. Consistently, the GO term ‘plant-type secondary cell wall biogenesis’ was significantly enriched in the CK-treated P. ginseng roots (FDR = 0.002) (Fig. 4A). Through GSEA, 31 CK-induced genes were identified as the leading-edge subset related to ‘plant-type secondary cell wall biogenesis’ (Fig. 4A). Among these, 9 genes were significantly induced by CK treatment, with more than 1.5-fold change in expression (q < 0.05) (Fig. 4B and Table S2). These genes, including GXM1, PGSIP3, and IRXs, have been demonstrated to play a crucial role in secondary cell wall biogenesis and secondary root growth [8,26].
Fig. 4.
Functional enrichment of cell wall biogenesis in P. ginseng root in response to CK and the transcriptional network. (A) Enrichment plot for the plant-type secondary cell wall biogenesis (GO:0009834), and an expression heatmap (FDR = 0.0025, NES = −1.837). (B) Transcriptional network genes related to cell wall biogenesis.
Next, we focused on the identification of the signaling network associated with CK-mediated plant-type cell wall biogenesis. The transcriptional network of plant-type cell wall biogenesis was analyzed based on A. thaliana and its protein–protein interaction network using the STRING database (Fig. 4B). The effects of CK treatment on cell wall biogenesis interacted strongly with those on cytokinesis, cell division, regulation of cell cycle, response to auxin, auxin-activated signaling pathway, and response to CK (Fig. 4B). Interestingly, genes related to ‘auxin-related signaling pathway’ and ‘response to auxin’ directly interacted with those related to ‘plant secondary cell wall biogenesis’ and ‘responses to cytokinin’ in the network (Fig. 4B). These results suggest that the transcriptional regulatory network affecting CK-mediated secondary growth is achieved through the regulation of ‘cell division’, ‘cell wall biogenesis’, and ‘auxin signaling’. Therefore, our results provide highly reliable biological information on the transcriptional regulatory network affecting secondary growth in response to CK in P. ginseng.
3.4. Identification of CK signaling-related two-component system in P. ginseng
The CK responses in the secondary growth of storage roots and transcriptome analysis indicated that conserved CK signaling pathways would be functionally regulated in P. ginseng. Canonical CK signal transduction acts in concert with a two-component system [14,16,27]. Phylogenetic analysis revealed putative CK receptors and type-B RRs in the P. ginseng genome (Figs S5 and S6). We identified four P. ginseng HKs related to Arabidopsis CK receptors (AHK2, AHK3, AHK4), and cloned PgHK3 (Gene ID: KG ISO 082030), which is closely related to AHK3 (Fig. S5A) [28]. Analysis of the primary structure of PgHK3 revealed that functional domains, including the CK-binding CHASE domain and the histidine kinase and receptor domains, are evolutionarily conserved (Fig. S5B). PgRR2 (Gene ID: KG ISO 060520.78810) was also identified, and the evolutionary conservation of functional core amino acid sequences and domains was validated (Fig. S6A and B) [28]. The nuclear-localized PgRR2 enhanced proTCS::LUC activity, thus acting as a CK signaling reporter in a dose-dependent manner (Fig. S6C and D). These findings demonstrate that PgHK3 and PgRR2 are involved in the CK response signaling pathway, and that the two-component system-mediated CK signaling is well-conserved in P. ginseng.
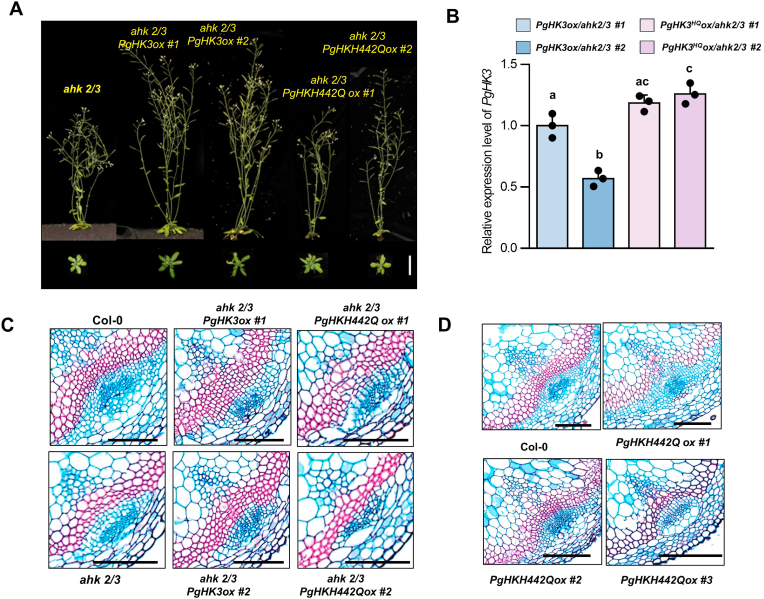
To investigate the physiological role of PgHK3 in CK signaling, we performed a complementation assay using the Arabidopsis ahk2/3 double knockout mutant, which exhibits CK-deficient growth defects. We overexpressed PgHK3 and its loss-of-function variant, PgHK3H442Q, in the ahk2/3 double mutant under the control of the constitutively active 35S promoter (Fig. 5A and B). Compared with the control ahk2/3 double mutant, transgenic lines overexpressing 35S::PgHK3 in the ahk2/3 background displayed larger shoots and secondary xylem development in the inflorescence stems. However, overexpression of PgHK3H442Q failed to restore the shoot architecture and vascular tissue secondary growth of ahk2/3 (Fig. 5A and C). Next, we generated 35S::PgHK3H442Q transgenic lines in the Col-0 background. Two lines showing moderate expression levels of PgHK3H442Q exhibited similar growth characteristics, whereas plants of 35S::PgHK3H442Q line #3 showed strong transgene expression and exhibited severely retarded shoot growth and inflorescence stem development (Fig. S7A and B). The secondary growth of inflorescence stems was consistently defective in 35S::PgHK3H442Q line #3 plants (Fig. 5D). Taken together, our findings support that PgHK3 acts as a CK receptor to control plant growth and development. Also, our data demonstrate that P. ginseng exhibits an evolutionarily well-conserved CK signaling pathway for modulating storage root development, which is regulated by a two-component circuit common to land plants (Fig. 6).
Fig. 5.
Complementation of the Arabidopsis ahk2/3 loss-of-function mutant via PgHK3 overexpression. (A) Phenotype of the CK-insensitive dwarf phenotype of the ahk2/3 double mutant and overexpression of PgHK3 and PgHK3-H442Q genes in the ahk2/3 background. Scale bar = 1cm. (B) Relative expression level of PgHK3, PgHK3-H442Q in 7-day-old- transgenic plants. Different lowercase letters indicate statistically significant differences P < 0.05; one-way ANOVA, followed by Tukey's multiple range test. Cross-section images of (C) stem of the transgenic plants. (D)The vascular bundle phenotype of PgHKH442Q overexoression line in Col-0 background. Scale bar = 100 μm.
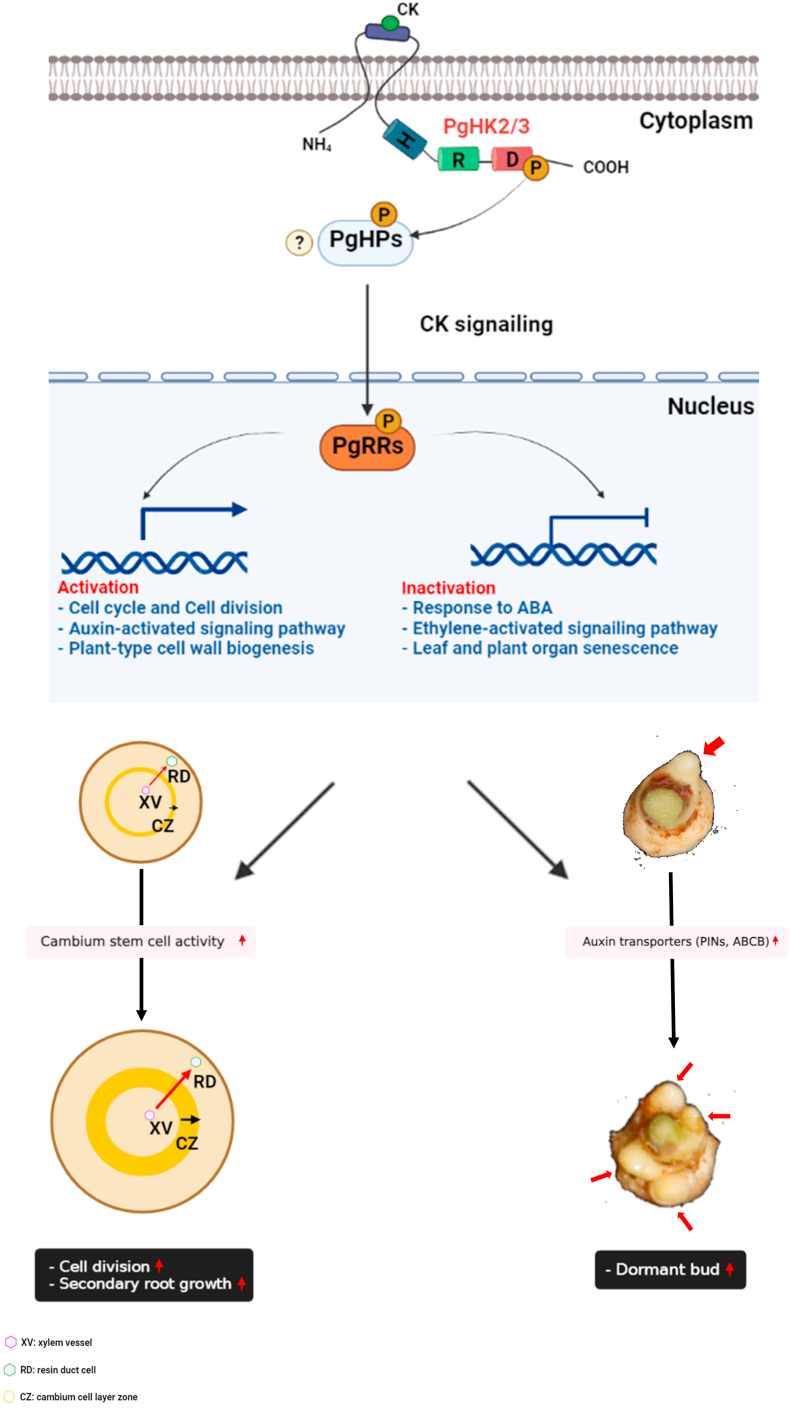
Fig. 6.
A schematic model for CK signaling pathway and transcriptional network for modulating secondary growth of P. ginseng storage root (The model was created with BioRender.com).
4. Discussion
4.1. CK signaling pathways are evolutionary conserved and facilitate the secondary growth of storage roots in P. ginseng
P. ginseng was domesticated as a medicinal root herb, primarily because of its ability to store beneficial pharmacological compounds in the storage roots [1,29]. The secondary growth of P. ginseng storage organs is crucial for high crop yield [8,30]. Plant hormones and environment factors modulate cambium stem cell activity to control plant secondary growth. The homeostasis of cambium stem cells, which is primarily controlled by the integrated signaling networks of plant hormones such as auxin and CK, is directly related to the regulation of secondary growth [[10], [11], [12]]. Previous studies demonstrated that numerous plant hormones and their interactions regulate the division and development of cambium stem cells [12,[30], [31], [32]]. GA treatment increases the division and differentiation of storage parenchyma cells by increasing cambial activity in P. ginseng [8]. Furthermore, CK plays an important role in controlling cambial stem cell maintenance and vascular tissue differentiation in plants [[9], [10], [11], [12],15]. However, given the complex structure and polyploid nature of its genome, the functional genomic analysis of P. ginseng has been challenging [6,7]. In this study, we performed a functional genomic analysis of exogenous CK-induced physiological changes in P. ginseng, and examined their genetic regulation by integrating in silico RNA-seq analysis. Our findings suggest that the CK-induced secondary growth of P. ginseng storage roots is more closely related to cambium stem cell proliferation than to cell growth and elongation. The molecular basis of root meristem regulation in Arabidopsis implies that CK signaling pathways are crucially involved in the proliferation of meristematic stem cells [11,25,[33], [34], [35]]. Mitotic cell division and procambial cell number are greatly decreased in mutants lacking CK signaling, including wooden leg (wol) and the ahk2/ahk3/ahk4 triple mutant [36,37]. Consistent with these results, CK treatment promoted the secondary growth of P. ginseng storage roots in this study by increasing the division of cambium stem cells (Fig. 1, Fig. 2). Additionally, our results showed that PgHK3 and PgRR, homologs of the Arabidopsis CK receptors HK3 and RR2, respectively, are evolutionarily conserved (Fig. S5 and S6), and the growth defects of the ahk2/3 double mutant could be complemented by the overexpression of PgHK3 (Fig. 5 and Fig. S7). In addition, exogenous CK treatment enhanced storage root growth and development in P. ginseng by promoting cambium cell division and increasing dormant bud formation (Fig. 1, Fig. 2). These results demonstrate that CK signaling is well-conserved in P. ginseng at the genomic level and has the potential to increase storage root yield.
4.2. Secondary growth is closely related to cell wall biogenesis-related networks and cell division
In this study, transcriptome profiling of CK-treated P. ginseng roots revealed how genetic regulators and signaling networks control the growth and development of a storage organ in P. ginseng. Our transcriptome analysis results showed that CK induces secondary growth and development in P. ginseng by promoting cambium cell division. CKs have been generally regarded as important regulators of cell division. Transcriptome and GO enrichment analyses revealed that 'cell cycle', 'cell division', 'cell movement', 'response to auxin', 'response to cytokinin', and 'plant-type secondary cell wall biogenesis' were mainly involved in CK-mediated secondary growth regulation (Fig. 4). Previous studies showed that the plant cell wall is closely related to the functions of 'cell cycle/division' and 'cell growth' [8,13]. Transcriptome profiling of CK-treated P. ginseng roots supported the possibility that CK-induced secondary growth is synergistically linked to secondary cell wall biosynthesis-related pathways. The reconstruction of the cell wall, which is closely related to cell division, is closely associated with secondary cell division. Our results suggest that secondary cell wall production in plants is linked to cell cycle, cell division, cytokinesis, CK response, and auxin response (Fig. 4B). These results demonstrate that transcriptional networks regulate root secondary growth in P. ginseng. Furthermore, local auxin accumulation in the cambium organizer cells during plant secondary growth was significantly affected by the cell wall growth-related genes and auxin-activated signaling factors, including WAT1, MYB, IRX, ABCB, and PINs. WAT1, MYB, and IRXs are essential for secondary cell wall formation during xylem development [[38], [39], [40]]. Auxin transporters, such as ABCB and PIN, are also important for the lignification of secondary cell walls through auxin distribution [41]. Therefore, our findings provide an advanced understanding of the transcriptional network controlling secondary growth and development in response to CK in P. ginseng storage roots.
4.3. CK promotes bud formation and delays senescence
CKs are involved in diverse aspects of plant growth and development [34,42]. CKs stimulate shoot development and cell division in plants as well as shoot formation from undifferentiated callus cells [14,25]. Thus, CKs play an important role in organogenesis, leaf senescence, shoot meristem formation, and apical dominance [15,25,35,43,44]. CKs regulate shoot branch growth by activating axillary bud formation, but this mechanism is not yet fully understood [25,31]. In Arabidopsis, shoot branching is also affected by auxin homeostasis [25,31,32]. Interestingly, several auxin transporters, such as PIN3, PIN4, and PIN7, are targets of CK signaling during shoot branching [31]. PINs play a major role in shoot branching, and numerous studies indicate that CK-controlled polar auxin transport is critically involved in shoot development [45,46]. Interestingly, in this study, the number of dormant buds in P. ginseng increased upon exogenous CK treatment (Fig. 2C and D). Our transcriptome analysis also supported the relationship between CK and auxin transporters during shoot production and secondary growth in P. ginseng. Exogenous CK treatment induces the expression of PIN3, PIN6, and ABCB (Fig. 4B and Table S3). In addition, among the endogenous developmental signals, CK has a particularly important effect on the longevity of plant organs [43,44]. It has been well established that increasing CK biosynthesis during senescence can significantly delay plant organ senescence and boost crop yields [25,44]. Consistently, our transcriptome analysis showed that 'response to ethylene', 'plant organ senescence', and 'leaf senescence' were significantly enriched in mock-treated P. ginseng roots (Fig. 3 and Fig. S2D and E). These senescence related GO terms were significantly downregulated in CK-treated P. ginseng root samples, suggesting that CK could regulate senescence-related genes and hormones in P. ginseng to delay senescence.
Elucidating the molecular basis of CK signaling and its connections with other signal transduction pathways will enhance our understanding of the mechanisms governing the growth and development of perennials. Although it remains to be found which downstream signaling pathways interact with the upstream CK signaling pathway to govern storage root secondary growth, our results provide strong evidence in support of the role of CK-induced cell division in P. ginseng root secondary growth. Overall, our results suggest that understanding the function of CK in P. ginseng will not only improve root crop productivity but will also be valuable for future research.
Author contributions
H.C and H.R designed the experiments and supervised this study. K.R.G, Y.K, J.H, J.L, S.H and W.B carried out P. ginseng plant growth experiments and histological sectioning analysis. Y.K and J.G generated transgenic Arabidopsis plants and analysis. K.R.G and J.H analyzed RNA-Seq data and bioinformatic analysis. K.R.G, Y.L, J.H, H.C and H.R wrote the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development (No. PJ01482004), National Research Foundation (NRF-2021R1I1A3050947) and (NRF-RS-2023-00209134).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2023.11.002.
Contributor Information
Hyunwoo Cho, Email: hwcho@chungbuk.ac.kr.
Hojin Ryu, Email: hjryu96@chungbuk.ac.kr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hu SY. The genus Panax (ginseng) in Chinese medicine. Econ. Bot. 1976;30:11–28. [Google Scholar]
- 2.Mok DW, Mok MC. Ginsengs: a review of safety and efficacy. Nutr Clin Pract. 2000;3(2):90–101. [Google Scholar]
- 3.Mok D.W., Mok M.C. Cytokininschemistry, activity, and function. CRC press; 1994. [Google Scholar]
- 4.Kang S., Min H. Ginseng, the'immunity boost': the effects of Panax ginseng on immune system. J Ginseng Res. 2012;36(4):354. doi: 10.5142/jgr.2012.36.4.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi HI, Waminal NE, Park HM, Kim NH, Choi BS, Park M, Choi D, Lim YP, Kwon SJ, Park BS, et al. Major repeat components covering one-third of the ginseng (Panax ginseng C.A. Meyer) genome and evidence for allotetraploidy. Plant J. 2014;77(6):906–916. doi: 10.1111/tpj.12441. [DOI] [PubMed] [Google Scholar]
- 6.Hong CP, Lee SJ, Park JY, Plaha P, Park YS, Lee YK, Choi JE, Kim KY, Lee JH, Lee J, et al. Construction of a BAC library of Korean ginseng and initial analysis of BAC-end sequences. Mol Genet Genomics. 2004;271(6):709–716. doi: 10.1007/s00438-004-1021-9. [DOI] [PubMed] [Google Scholar]
- 7.Jang W, Kim N-H, Lee J, Waminal NE, Lee S-C, Jayakodi M, Choi H-I, Park JY, Lee J-E, Yang T-J. A glimpse of Panax ginseng genome structure revealed from ten BAC clone sequences obtained by SMRT sequencing platform. Plant Breed. Biotech. 2017;5(1):25–35. [Google Scholar]
- 8.Hong CP, Kim J, Lee J, Yoo SI, Bae W, Geem KR, et al. Gibberellin signaling promotes the secondary growth of storage roots in Panax ginseng. Int J Mol Sci. 2021;22(16) doi: 10.3390/ijms22168694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Rybel B., Adibi M., Breda AS, Wendrich JR, Smit ME, Novák O, Yamaguchi N, Yoshida S, Van Isterdael G, Palovaara J. Palovaara. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science. 2014;345(6197) doi: 10.1126/science.1255215. [DOI] [PubMed] [Google Scholar]
- 10.Nieminen K., Immanen J, Laxell M, Kauppinen L, Tarkowski P, Dolezal K, Tähtiharju S, Elo A, Decourteix M, Ljung K. Cytokinin signaling regulates cambial development in poplar. P Natl Acad Sci USA. 2008;105(50):20032–20037. doi: 10.1073/pnas.0805617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto-Kitano M., Kusumoto T, Tarkowski P, Kinoshita-Tsujimura K, Vaclavikova K, Miyawaki K, Kakimoto T. Cytokinins are central regulators of cambial activity. P Natl Acad Sci USA. 2008;105(50):20027–20031. doi: 10.1073/pnas.0805619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye L.L., Wang X, Lyu M, Siligato R, Eswaran G, Vainio L, Blomster T, Zhang J, Mahonen AP. Cytokinins initiate secondary growth in the Arabidopsis root through a set of LBD genes. Curr Biol. 2021;31(15):3365–+. doi: 10.1016/j.cub.2021.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geem K.R., Kim J, Bae W, Jee MG, Yu J, Jang I, Lee DY, Hong CP, Shim D, Ryu H. Nitrate enhances the secondary growth of storage roots in Panax ginseng. Journal of Ginseng Research. 2023;47(3):469–478. doi: 10.1016/j.jgr.2022.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K, Ryu H, Cho YH, Scacchi E, Sabatini S, Hwang I. Cytokinin-facilitated proteolysis of ARABIDOPSIS RESPONSE REGULATOR 2 attenuates signaling output in two-component circuitry. Plant J. 2012;69(6):934–945. doi: 10.1111/j.1365-313X.2011.04843.x. [DOI] [PubMed] [Google Scholar]
- 15.Schmulling T. New insights into the functions of cytokinins in plant development. J Plant Growth Regul. 2002;21(1):40–49. doi: 10.1007/s003440010046. [DOI] [PubMed] [Google Scholar]
- 16.To J.P.C., Kieber J.J. Cytokinin signaling: two-components and more. Trends in Plant Science. 2008;13(2):85–92. doi: 10.1016/j.tplants.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Ramírez-Carvajal GA, Morse AM, Dervinis C, Davis JM. The cytokinin type-B response regulator PtRR13 is a negative regulator of adventitious root development in Populus. Plant Physiol. 2009;150(2):759–771. doi: 10.1104/pp.109.137505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. P Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26(22):2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P., Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azizi P., Rafii MY, Maziah M, Abdullah SNA, Hanafi MM, Latif MA, Rashid AA, Sahebi M. Understanding the shoot apical meristem regulation: a study of the phytohormones, auxin and cytokinin, in rice. Mechanisms of Development. 2015;135:1–15. doi: 10.1016/j.mod.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Zhong R, Cui D, Ye ZH. Secondary cell wall biosynthesis. New Phytol. 2019;221(4):1703–1723. doi: 10.1111/nph.15537. [DOI] [PubMed] [Google Scholar]
- 27.Kakimoto T. Perception and signal transduction of cytokinins. Annu Rev Plant Biol. 2003;54:605–627. doi: 10.1146/annurev.arplant.54.031902.134802. [DOI] [PubMed] [Google Scholar]
- 28.Jo I.-H., Lee J, Hong CE, Lee DJ, Bae W, Park S-G, Ahn YJ, Kim YC, Kim JU, Lee JW. Isoform sequencing provides a more comprehensive view of the Panax ginseng transcriptome. Genes. 2017;8(9):228. doi: 10.3390/genes8090228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu XL, Xi QY, Yang L, Li HY, Jiang QY, Shu G, Wang SB, Gao P, Zhu XT, Zhang YL. The effect of dietary Panax ginseng polysaccharide extract on the immune responses in white shrimp, Litopenaeus vannamei. Fish Shellfish Immun. 2011;30(2):495–500. doi: 10.1016/j.fsi.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Agusti J., Herold S, Schwarz M, Sanchez P, Ljung K, Dun EA, Brewer PB, Beveridge CA, Sieberer T, Sehr EM. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. P Natl Acad Sci USA. 2011;108(50):20242–20247. doi: 10.1073/pnas.1111902108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waldie T, Leyser O. Cytokinin targets auxin transport to promote shoot branching. Plant Physiol. 2018;177(2):803–818. doi: 10.1104/pp.17.01691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willige B.C., Isono E, Richter R, Zourelidou M, Schwechheimer C. Gibberellin regulates PIN-FORMED abundance and is required for auxin transport–dependent growth and development in Arabidopsis thaliana. The Plant Cell. 2011;23(6):2184–2195. doi: 10.1105/tpc.111.086355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto-Kitano M., Kusumoto T, Tarkowski P, Kinoshita-Tsujimura K, Václavíková K, Miyawaki K, Kakimoto T. Cytokinins are central regulators of cambial activity. P Natl Acad Sci USA. 2008;105(50):20027–20031. doi: 10.1073/pnas.0805619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mok MC. Cytokinins and plant development. Cytokinins: chemistry, activity. and function. 1994:155–166. [Google Scholar]
- 35.Kurakawa T., Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445(7128):652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 36.Ishida K, Yamashino T, Yokoyama A, Mizuno T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008;49(1):47–57. doi: 10.1093/pcp/pcm165. [DOI] [PubMed] [Google Scholar]
- 37.Higuchi M., Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S. Planta functions of the Arabidopsis cytokinin receptor family. P Natl Acad Sci USA. 2004;101(23):8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J, Kim H, Park SG, Hwang H, Yoo SI, Bae W, Kim E, Kim J, Lee HY, Heo TY, et al. Brassinosteroid-BZR1/2-WAT1 module determines the high level of auxin signalling in vascular cambium during wood formation. New Phytol. 2021;230(4):1503–1516. doi: 10.1111/nph.17265. [DOI] [PubMed] [Google Scholar]
- 39.Coomey JH, Sibout R, Sibout R, Hazen SP, Hazen SP. Grass secondary cell walls, Brachypodium distachyon as a model for discovery. New Phytol. 2020;227(6):1649–1667. doi: 10.1111/nph.16603. [DOI] [PubMed] [Google Scholar]
- 40.Hong J, Ryu H. Identification of WAT1-like genes in Panax ginseng and functional analysis in secondary growth. J Plant Biotechnol. 2022;49(3):171–177. [Google Scholar]
- 41.Kaneda M, Schuetz M, Lin BSP, Chanis C, Hamberger B, Western TL, Ehlting J, Samuels AL. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J Exp Bot. 2011;62(6):2063–2077. doi: 10.1093/jxb/erq416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kieber JJ, Schaller GE. Cytokinin signaling in plant development. Development. 2018;145(4):dev149344. doi: 10.1242/dev.149344. [DOI] [PubMed] [Google Scholar]
- 43.Gan S.S., Amasino R.M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270(5244):1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- 44.Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. P Natl Acad Sci USA. 2006;103(3):814–819. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol. 2006;16(6):553–563. doi: 10.1016/j.cub.2006.01.058. [DOI] [PubMed] [Google Scholar]
- 46.Domagalska MA, Leyser O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 2011;12(4):211–221. doi: 10.1038/nrm3088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.