Abstract
The Clock gene plays an essential role in the manifestation of circadian rhythms (≅24 h) in mice and is a member of the basic helix-loop-helix (bHLH) PER-ARNT-SIM (PAS) superfamily of transcription factors. Here we report the characterization of a novel Drosophila bHLH-PAS protein that is highly homologous to mammalian CLOCK. (Similar findings were recently described by Allada et al. Cell 93:791–804, 1998, and Darlington et al., Science 280:1599–1603, 1998.) Transcripts from this putative Clock ortholog (designated dClock) undergo daily rhythms in abundance that are antiphase to the cycling observed for the RNA products from the Drosophila melanogaster circadian clock genes period (per) and timeless (tim). Furthermore, dClock RNA cycling is abolished and the levels are at trough values in the absence of either PER or TIM, suggesting that these two proteins can function as transcriptional activators, a possibility which is in stark contrast to their previously characterized role in transcriptional autoinhibition. Finally, the temporal regulation of dClock expression is quickly perturbed by shifts in light-dark cycles, indicating that this molecular rhythm is closely connected to the photic entrainment pathway. The isolation of a Drosophila homolog of Clock together with the recent discovery of mammalian homologs of per indicate that there is high structural conservation in the integral components underlying circadian oscillators in Drosophila and mammals. Nevertheless, because mammalian Clock mRNA is constitutively expressed, our findings are a further example of striking differences in the regulation of putative circadian clock orthologs in different species.
Many organisms ranging from bacteria to humans display circadian (≅24 h) rhythms in a wide variety of biochemical, physiological, and behavioral phenomena (for a review, see reference 19). These rhythms are regulated by endogenous biochemical oscillators or clocks that can be entrained (synchronized) by environmental stimuli, most notably the daily changes in light intensity and temperature. It is likely that the most significant evolutionarily conserved role of circadian clocks is to enable organisms to organize their physiology and behavior to occur at biologically advantageous times in a day.
A major goal in the study of circadian rhythms is to understand the molecular underpinnings of the pacemakers that govern these rhythms. Genetic approaches using model organisms have played a pivotal role in identifying genes that are critical for the manifestation of a variety of behavioral rhythms. The best characterized examples of such genes are period (per) (24) and timeless (tim) (42) in Drosophila melanogaster, frequency (frq) (31) in Neurospora crassa, and Clock (51) in mice. Although the in vivo biochemical roles of the protein products encoded by these genes are not clear, they all appear to have a primary function in the regulation of transcription (for a recent review, see reference 38).
For example, in D. melanogaster, daily cycles in per and tim mRNAs are largely governed by a transcriptional feedback loop, likely negative, that depends on the presence of both PER and TIM, suggesting that a shared mechanism participates in the autoregulation of per and tim transcription (17, 18, 43, 47, 53). The observation that PER and TIM physically interact to form a functional complex that enters the nucleus in a temporally gated manner, an event that is accompanied by decreases in the levels of per and tim transcripts, likely explains this reciprocal autoregulation (8, 13, 25, 37, 54). Likewise, the Neurospora FRQ protein also participates in a negative transcriptional feedback loop that is dependent on the nuclear localization of FRQ and leads to daily oscillations in the abundance of frq transcripts (3, 28). In addition, PER, TIM, and FRQ undergo temporally regulated changes in abundance as well as phosphorylation (10, 12, 22, 30, 32, 53, 54). Current understanding posits that the protein and RNA products of per, tim, and frq comprise transcription-translation-based autoinhibitory loops that are central to the oscillatory mechanisms in these species (reviewed in reference 38). Although PER, TIM, and FRQ behave as negative elements in circadian transcriptional loops, they do not share significant sequence similarities (4, 31, 33). Furthermore, it does not appear that these proteins bind DNA, suggesting indirect modes of action for their roles in transcriptional autoinhibition. Specifically, it has been proposed that PER might inhibit gene expression by forming nonfunctional heterodimers with transcription factors (reviewed in reference 38).
The predicted involvement of CLOCK in transcriptional regulation is based on the demonstration that it is a member of the basic helix-loop-helix (bHLH) PER-ARNT-SIM (PAS) superfamily of transcription factors (23). The structure of bHLH-PAS proteins is highly conserved (for a recent review, see reference 5); the bHLH domain which mediates protein dimerization and DNA-binding is near the amino terminus and is closely followed by another protein dimerization motif called the PAS domain (named for the first three proteins identified with this motif: D. melanogaster PER, mammalian aryl hydrocarbon receptor nuclear translocator [ARNT], and D. melanogaster SIM). In bHLH-PAS-containing proteins, the PAS domain is ∼250 to 300 amino acids long and contains two well-conserved repeats of approximately 50 amino acids, designated PAS-A and PAS-B. Finally, in many cases, the carboxyl termini of these proteins have glutamine-rich regions associated with transcriptional activation. In this context, it is noteworthy that a mutation that causes a deletion within the glutamine-rich region of the carboxy terminus of mCLOCK results in mice with altered activity rhythms (23). Thus, in contrast to PER, TIM, and FRQ, which function as negative regulators, CLOCK is predicted to have a role in the stimulation of transcription. It is anticipated that a significant role for transcriptional activators in circadian feedback loops is to stimulate the rhythmic production of elements associated with negative regulation, such as PER, TIM, and FRQ. Indeed, a recent study has shown that a transcription factor termed WC-2 is necessary for expression of frq and the manifestation of overt circadian rhythms in Neurospora (6). Presumably, after an appropriately timed delay following the production of the negative regulators, these factors complete the transcriptional feedback loop by somehow inhibiting the activities of the transcriptional activators. The emerging picture is that transcriptional feedback loops composed of positive and negative regulatory elements that alternate in their functioning are core features of circadian oscillatory mechanisms.
In addition to the inferred functional conservation of transcriptional feedback loops in circadian oscillatory mechanisms, the recent discovery of per homologs in mice (mper-1 and mper-2) (1, 44, 45, 48, 49) indicates that the Drosophila and mammalian clocks also share components that are structurally conserved, raising the possibility of a Drosophila Clock homolog. Moreover, in D. melanogaster, a circadian transcriptional enhancer that drives high-amplitude per mRNA cycling contains an E-box element that is necessary for high-level expression, suggesting the involvement of a bHLH-containing transcription factor in the regulation of per expression (16). In this report, we characterize a putative D. melanogaster ortholog of mammalian Clock (dClock). Similar findings were recently reported by two independent groups (2, 9). We show that dClock transcript levels are regulated in a circadian fashion in sharp contrast to the constitutive expression of mammalian Clock (48, 49). Taken together with recent findings, our data indicate that putative circadian clock orthologs in different species can be subject to very different regulatory schemes. Surprisingly, our data also indicate that PER and TIM can function as positive elements in the regulation of gene expression.
MATERIALS AND METHODS
Fly strains and collections.
The wild-type Canton-S (CS) flies and the mutant per01 flies used in this study were descendants of stocks originally maintained in the laboratory of M. Rosbash (Brandeis University, Waltham, Mass.) and were previously described (see, e.g., reference 10). The tim0 flies were descendants of stocks originally maintained in the laboratory of A. Sehgal (University of Pennsylvania Medical School, Philadelphia) and were previously described (42). All flies were grown and maintained in vials containing standard agar-cornmeal-sugar-yeast-tegosept medium. Vials containing ∼100 young (2-to-6-day-old) adult flies were placed in incubators (Precision Scientific) at 25°C, exposed to at least 2 cycles of 12 h of light and 12 h of darkness (12-h LD) (where zeitgeber time 0 [ZT0] is lights on and ZT12 is lights off) and were subsequently maintained in the dark (DD). At selected times during the 12-h LD and the DD periods, flies were collected by freezing. In the 12-h LD shift experiment (see Fig. 4), four groups of flies were exposed to 3 days of 12-h LD. On the third day of 12-h LD, two groups of flies were subjected to a shift in the relative timing of the 12-h LD cycle. One group was removed at ZT11.9 and placed in another incubator where the lights-on period was continued from ZT12 to ZT16. At ZT16, the lights were turned off and the flies were maintained in the dark for the standard 12-h period followed by the next lights-on period at ZT4 (ZTs are given relative to the original 12-h LD entraining condition). Thus, on the third day of 12-h LD, the light period was extended by 4 h, followed by a standard 12-h LD cycle. The other experimental group was removed at ZT20 and placed in another incubator where it was exposed to light for the standard 12-h period followed by a lights-off period beginning at ZT8 (relative to the original 12-h LD entraining condition). Thus, on the third day of 12-h LD, the light period was initiated 4 h earlier, followed by a standard 12-h LD cycle. The two other groups of flies were maintained in the original 12-h LD condition used during entrainment and served as controls for each of the two groups of flies that were subjected to 12-h LD shifts.
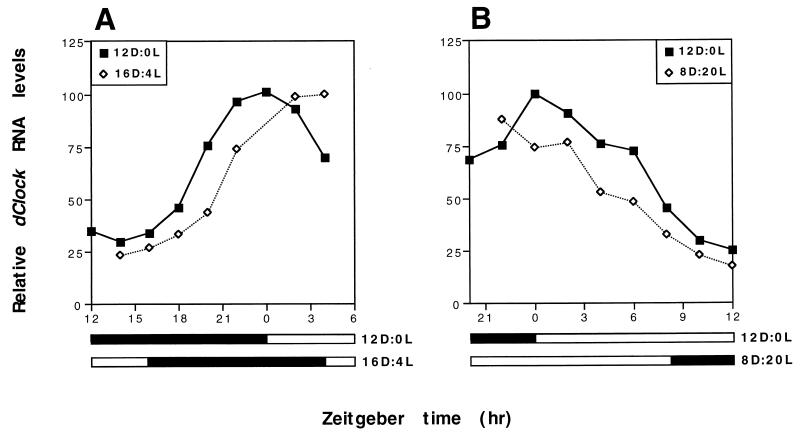
FIG. 4.
dClock RNA levels in flies exposed to shifts in the timing of a 12-h LD cycle. Four groups of wild-type D. melanogaster were entrained by a standard 12-h LD cycle for 3 days. Subsequently, two groups of flies were transferred from the original 12-h LD condition and treated with either a backward (A) or forward (B) shift of 4 h in the timing of the 12-h LD cycle; a group of flies maintained in standard 12-h LD conditions served as the control for each group of flies that was exposed to a 12-h LD shift (see Materials and Methods). (A) The 4-h backward shift was initiated by extending the light period for four extra hours (between ZT12 and ZT16) and beginning the dark period at ZT16 (16D:4L) relative to the original 12-h LD cycle (12D:0L). (B) The 4-h forward shift was initiated by beginning the light period 4 h earlier at ZT20 (8D:20L) relative to the original 12-h LD cycle (12D:0L). Peak levels of dClock mRNA under standard 12-h LD conditions were set to 100. Levels of dClock transcripts were determined by RNase protection assays. Relative RNA levels refers to ratios of dClock to RP49 RNA. Closed bar, darkness; open bar, light. The experiment was done twice with similar results, and one example is shown.
5′ RACE.
To obtain DNA sequences upstream of an uncharacterized Drosophila melanogaster expressed sequence tag (EST) (GenBank accession no. AA698290) whose deduced translation product shows very high homology to the PAS-B motif of mCLOCK (data not shown and see Fig. 1) we used the 5′ rapid amplification of cDNA ends (5′ RACE) system from Gibco/BRL (version 2.0). 5′ RACE was performed according to manufacturer’s recommended procedure on total head RNA isolated from wild-type CS flies collected between ZT15 and 16. Total RNA was extracted from ∼30 μl of fly heads by using TriReagent (Sigma) as previously described (29). 5′ RACE-PCR products were directly subcloned into the pGEM-T Easy vector (Promega), and the inserts were sequenced (described below). The initial round of 5′ RACE experiments was done with three nested primers based on the EST sequence available in the database (GenBank accession no. AA698290). The primers used in the initial 5′ RACE-PCR experiment were 5′ CATTCCAGTATCGGAGGT 3′, 5′ CGTTGTCGTAGTGGTTGTTACCCTC 3′, and 5′ CGATACGGCTCCAAGACTTCCTAT 3′. This yielded several prominent PCR products ranging in size, and the largest (∼0.6 kb) was selected for sequencing. As expected from the size of the 0.6-kb PCR fragment, the predicted size of a CLOCK homolog, and a putative PAS-B starting point in the 5′ RACE experiment, DNA sequence analysis (described below) indicated that this 5′ RACE-PCR product did not contain the translation start site (data not shown and see Fig. 1). In a second round of 5′ RACE experiments, a new set of two nested primers based on the EST sequence were used: 5′ CGTTCAGAACGATGATGGCTAAG 3′ and 5′ GTACTTCTCGATTCCGTTTGCCCA 3′. Two prominent PCR products of approximately 1.0 and 1.3 kb were selected for sequencing. Sequence analysis indicated that the 5′ terminus of the ∼1.0-kb PCR product was slightly upstream of a region encoding the PAS-A domain, whereas the ∼1.3-kb fragment terminated upstream of the predicted translation start site (see below and data not shown).
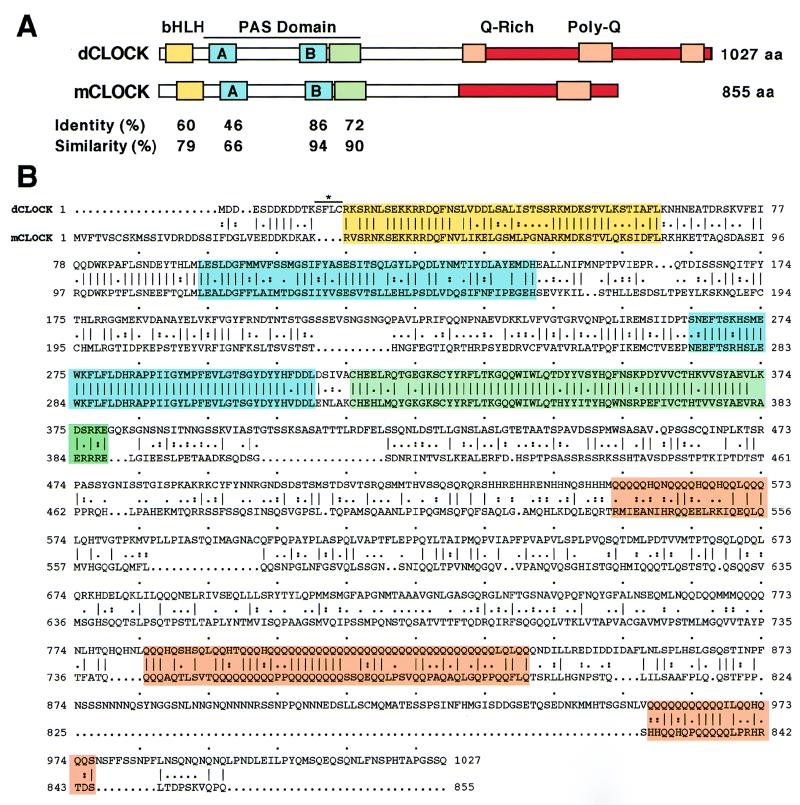
FIG. 1.
Comparison of the predicted protein sequences of dCLOCK and mCLOCK. (A) A schematic representation of dCLOCK and mCLOCK with homologous regions highlighted by different colors is shown. Yellow, amino-terminal region containing bHLH; blue, A and B repeats of PAS; green, region in PAS domain that is immediately carboxy terminal to PAS-B; orange, polyglutamine stretch; red, glutamine-rich region. The boundaries of the bHLH-PAS repeats, glutamine-rich region, and polyglutamine stretch were according to King et al. (23). The boundary of the region immediately carboxy terminal to the PAS-B repeat is based on Saez and Young (37). (B) Pairwise alignment of dCLOCK and mCLOCK amino acids was done with the Genetics Computer Group program GAP. Amino acid identity is indicated by a vertical line and similarity is indicated by dots. Overall, the amino acid sequences of dCLOCK and mCLOCK are 50% identical and 67% similar (data not shown). The mCLOCK sequence is from King et al. (23). A line above the four amino acids that differ between our sequence and that recently published by Darlington et al. (9) is shown.
DNA sequencing and analysis.
Five independent inserts derived from 5′ RACE-PCR experiments (i.e., one isolate of the 0.6-kb fragment and two independent isolates of both the 1.0- and 1.3-kb fragments; see above) and a cDNA clone containing an EST that showed high homology to mCLOCK (GenBank accession no. AA698290; obtained from Genome Systems Inc., St. Louis, Mo.) were sequenced in both directions. DNA sequences were determined by the DNA synthesis and sequencing laboratory at the University of Medicine and Dentistry of New Jersey with an ABI 377 Sequencer with the enzyme Amplitaq polymerase FS. The predicted open reading frame (ORF) for dCLOCK (see Fig. 1) was constructed by combining sequences derived from the five independent 5′ RACE-PCR products in addition to that derived from sequencing the cDNA clone obtained from Genome Systems Inc. Protein searches of nonredundant databases were done with ADVANCED BLAST. The GAP program (Genetics Computer Group) was used for sequence alignments.
RNase protection assay.
For each time point, total RNA was extracted from ∼10 μl of fly heads by using TriReagent (Sigma) as previously described (29). The abundance of dClock, tim, and per transcripts was determined by RNase protection assays (18) performed with the modifications described by Zeng et al. (53). The radiolabeled antisense probes used to determine the levels of per and tim RNA were as previously described (46). To measure dClock RNA levels, we used PCR to generate a DNA fragment that contained nucleotides 3 to 293 of the Drosophila EST clone showing high homology to mCLOCK (numbering according to GenBank accession no. AA698290). The oligonucleotides used in the PCR were (nucleotides corresponding to the EST are underlined) 5′ AATTGGATCCGAAGTTTTTGTTTCTGGATCACCGTG 3′ and 5′ AATTCTCGAGCAGAACCTCGGCATAGCTGACAAC 3′. The PCR product was subcloned into the pGEM-T Easy vector (Promega) and linearized with BamHI, and antisense radiolabeled probe was produced in vitro by using SP6 RNA polymerase as previously described (18). As a control for RNA loading in each lane, a ribosomal protein probe (RP49) was included in each protection assay (18). Protected bands were quantified with a PhosphorImager from Molecular Dynamics, and values were normalized relative to those of RP49 (18).
Nucleotide sequence accession number.
The cDNA sequence for dClock has been submitted to GenBank under the accession no. AF06997.
RESULTS
Identification of a novel Drosophila bHLH-PAS protein with high homology to mammalian CLOCK.
In a search for D. melanogaster cDNAs with homology to mammalian Clock, we noticed an uncharacterized EST in the database that encodes a conceptual translation product with very high sequence identity to the PAS-B domain of mouse and mammalian CLOCK (GenBank accession no. AA698290). A cDNA clone containing this EST was obtained (Genome Systems Inc.) and as expected because the synthesis of this cDNA was primed with oligo(dT), sequencing verified that a poly(A) tail is present at the 3′ terminus of the insert. The remaining portion of the ORF encoded by this gene was cloned by using 5′ RACE PCR (see Materials and Methods). In total, we sequenced 3,727 bp of cDNA. Analysis of the cDNA sequence indicated a single large ORF of 3,081 bp. The presumptive 5′ untranslated region has at least two ATG codons that are quickly followed by in-frame stop codons (data not shown). The predicted initiating AUG codon for the large ORF lies within a favorable Kozak consensus sequence for translation start sites (−3 AAAATGG +4). A sequence of 264 bp follows the stop codon at the end of the large ORF and as expected it contains a consensus sequence for poly(A) addition (ATAAA) that is closely followed by a stretch of 18 adenosine residues at its 3′ terminus.
The single large ORF encodes a protein product of 1,027 amino acid residues with a predicted molecular mass of 116.1 kDa. A BLAST analysis (ADVANCED BLAST) against a nonredundant amino acid database (National Center for Biotechnology Information) performed with the entire 1,027 amino acid sequence identified mammalian CLOCK (23) and MOP4 (20) (also referred to as NPAS2 [55]) as the proteins most highly homologous to the predicted Drosophila protein (designated dCLOCK) (data not shown). Although the function of MOP4 is not known, it is a bHLH-PAS protein that shows very high homology to CLOCK (23). Alignment of dCLOCK, mouse CLOCK (mCLOCK), and human MOP4 (hMOP4) showed that extensive homology is largely limited to four regions (Fig. 1 and data not shown): (i) an amino terminal region that includes a bHLH domain; (ii) PAS-A repeats; (iii) PAS-B repeats; and (iv) a region immediately carboxy terminal to the PAS-B repeat which was shown to function as a cytoplasmic localization determinant in D. melanogaster PER (37). All three proteins share the same putative basic region of EKKRR (Fig. 1) (20), which is found in only a few of the characterized bHLH-PAS proteins, suggesting that the E-box DNA elements with which these proteins presumably interact contain similar half-sites. The most significant homology was found in the PAS-B repeat; over a stretch of 46 amino acids this region in dCLOCK (amino acids 264 to 309) shows a remarkable 86 to 90% identity and 94 to 96% similarity with mammalian CLOCK and MOP4 (Fig. 1A and data not shown). However, it is noteworthy that dCLOCK and mCLOCK but not MOP4 have very prominent polyglutamine stretches in their carboxy terminal regions (Fig. 1) (20, 23, 55). Polyglutamine stretches in the carboxy terminal regions of many proteins function to activate transcription.
The sequence analysis clearly indicates that the Drosophila cDNA we characterized encodes a novel bHLH-PAS protein that is very homologous to mammalian CLOCK (overall homology is 50% identity and 67% similarity). Around the time that we submitted our manuscript, two independent groups reported the molecular isolation of dClock (2, 9). Conceptual translation of our large ORF (in the original version of our manuscript we designated it dPAS1 [GenBank accession no. AF06997]) yields a protein sequence that is identical to that reported by Darlington et al. (9) except that our sequence contains four extra amino acids (Fig. 1). Because we detected cDNA sequences encoding these four amino acids in only one of two independent isolates derived from reverse transcriptase PCR (see Materials and Methods), it is possible that they are a cloning artifact. Furthermore, the dCLOCK sequence shown in Allada et al. (2) is also missing these four amino acids and otherwise differs from our sequence and that of Darlington et al. (9) only in that the longest polyglutamine repeat in Allada et al. (2) is 25 rather than 33 residues in length. The discrepancy in the length of the longest polyglutamine stretch is presumably due to the polymorphic nature of this region (2).
dClock transcripts undergo daily cycles in abundance that are antiphase to per and tim mRNAs.
If dClock is an integral component of the Drosophila circadian timekeeping mechanism, then it might be expected to show rhythmic expression. To determine whether the abundance of dClock RNA changes during the course of a day, we exposed wild-type CS flies to three 12-h LD cycles (where zeitgeber time 0 [ZT0] is when lights were turned on and ZT12 is when lights were turned off). At different times during the 12-h LD cycles, flies were collected, total RNA was isolated from heads, and the levels of dClock transcripts were determined by RNase protection (see Materials and Methods). Heads were used because it is the anatomical location of the best characterized circadian clock in D. melanogaster (11, 15). Furthermore, the dClock cDNA we characterized was derived from D. melanogaster head and sensory organs (see Materials and Methods and data not shown). The results indicate that in light-dark conditions, dClock RNA displays robust cycling with an approximately fivefold peak-trough amplitude (n = 5), reaching peak abundance within several hours of the dark-light transition at ZT0 and trough values around the light-dark transition at ZT12 (Fig. 2).
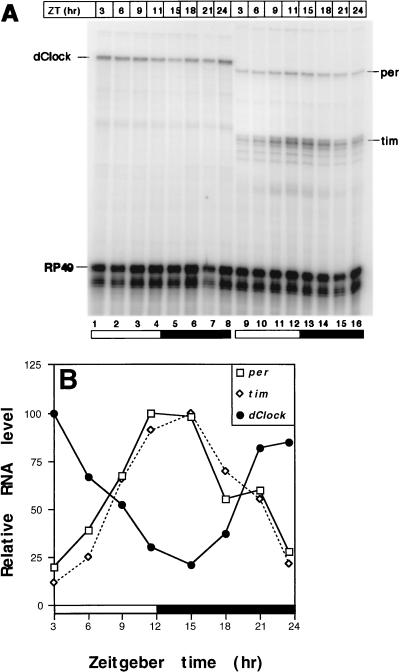
FIG. 2.
Daily cycling of dClock mRNA in heads from wild-type D. melanogaster. (A) Autoradiogram of dClock, per, tim, and RP49 transcripts in wild-type flies during a 12-h LD cycle. Levels of dClock, per, tim, and RP49 transcripts were determined by RNase protection assays (see Materials and Methods). (B) Quantitation of results shown in panel A. Time course of dClock, per, and tim transcript levels in wild-type flies during 12-h LD conditions. Peak levels for each mRNA were set to 100. Relative RNA level refers to ratio of dClock, per, or tim transcripts to RP49 RNA. Closed bar, darkness; open bar, light. Similar results were obtained in five independent experiments, and a representative example is shown.
Intriguingly, the time course of dClock transcript cycling in adult heads is strikingly antiphase to that observed for per and tim RNAs (Fig. 2). The circadian cycling of four D. melanogaster transcripts has been well characterized (i.e., per [18], tim [43], Crg-1 [35], and Dreg-5 [50]), and they all have similar phases reaching peak values in the early night. To the best of our knowledge, dClock is the first documented case of a D. melanogaster clock-regulated RNA that peaks in the early day, similar in phase to transcripts from the Neurospora circadian clock gene frequency (frq) (3). Our data indicate that circadian oscillators in the Drosophila head can drive phase-specific cycling of different transcripts. Recently, Darlington et al. (9) also reported that the levels of dClock transcripts oscillate in a 12-h LD cycle. However, they describe the oscillation as bimodal with a major peak at ZT23 and a minor peak at ZT5. In five independent experiments, we never observed a bimodal distribution (Fig. 2 through 4 and data not shown). Furthermore, they detected at least two variants of dClock transcripts (9): (i) a major form that encodes full-length dCLOCK (variant A) and (ii) a minor form that is produced by the use of an alternative splice acceptor site (variant B), generating a coding region that goes out of frame after the bHLH domain. The possibility that the differences in the RNA curve observed in our study and that reported by Darlington et al. (9) are due to the detection of different dClock mRNA variants is unlikely because the riboprobe we used in our RNase protection assays is directed to a region that is present in both variants (i.e., to a region that encodes the PAS-B domain; see Materials and Methods). Moreover, Darlington et al. (9) showed that both splice variants cycled in phase with a bimodal distribution, and therefore detecting either variant should have yielded qualitatively similar results. At present it is not clear why the curves describing the dClock mRNA cycles in the two studies differ.
PER and TIM are necessary for high-level expression of dClock.
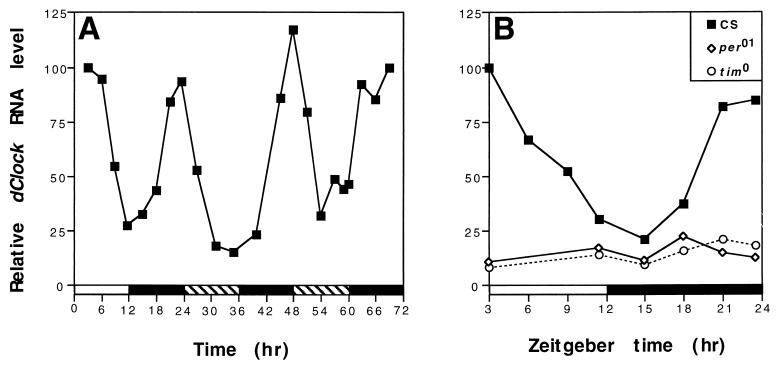
The cycling of dClock mRNA in 12-h LD (Fig. 2) might be an exogenous response to the daily changes in light and dark or reflect bona fide circadian regulation. To test whether the daily cycling of dClock mRNA is driven by an endogenous circadian clock, we measured its time course in 12-h LD-entrained flies that were maintained in constant DD. The level of dClock RNA continued to undergo daily oscillations for at least 2 days in constant DD (Fig. 3A), strongly suggesting that this molecular cycle is clock regulated. Furthermore, a phase relationship is maintained in DD similar to that observed during 12-h LD with no indication of bimodal behavior. To confirm that dClock RNA is regulated in a circadian manner, we used two arrhythmic D. melanogaster mutants that do not produce functional PER (per01) (24) or TIM (tim0) (42). As expected, the levels of dClock RNA were constant throughout a daily cycle in both mutants (Fig. 3B). Surprisingly, however, the abundance of dClock transcripts was similar to or lower than that observed for trough values in wild-type flies. Although the biochemical roles of PER and TIM are not clear, numerous lines of evidence indicate that these two proteins interact in the cytoplasm to form a PER-TIM complex that enters the nucleus, where it functions to inhibit per and tim transcription (see Discussion). Moreover, a role for the PER-TIM complex in the negative regulation of other genes is indicated by the observation that in the per01 (and where analyzed in the tim0) mutant the levels of the circadianly regulated Crg-1 (35) and Dreg-5 (50) transcripts are close to their respective peak levels in wild-type flies. In stark contrast, the ability to accumulate high levels of dClock transcripts requires PER and TIM, indicating a novel role for these two proteins in the stimulation of gene expression.
FIG. 3.
Circadian regulation of dClock mRNA cycling. (A) Levels of dClock mRNA in wild-type flies during the last 12-h LD cycle and the first 2 days in constant dark conditions. Peak levels of dClock mRNA at ZT3 were set to 100. Time refers to hours since last dark-to-light transition at ZT0. (B) PER and TIM are required for high-level expression of dClock. Levels of dClock mRNA in wild-type CS, per01, and tim0 flies during 12-h LD. Peak levels of dClock mRNA in wild-type flies was set to 100. Relative RNA levels refers to ratios of dClock to RP49 RNA. Closed bar, darkness; open bar, light; striped bar, subjective day. Each experiment was done at least three times with similar results, and representative examples are shown.
Shifts in the timing of light-dark cycles elicit rapid changes in the temporal regulation of dClock RNA levels.
A hallmark feature of circadian rhythms is that they are reset by changes in the timing of light-dark cycles (reviewed in reference 19). This adaptive property enables circadian rhythms to maintain phase synchrony with the entraining agent. To determine how quickly the dClock RNA rhythm responds to shifts in 12-h LD cycles, four groups of flies were exposed for several days to a standard 12-h LD cycle (dark period began at ZT12 and light period began at ZT0 [abbreviated hereafter in the form 12D:0L]). Subsequently, two groups of flies were transferred from the original 12-h LD condition to a 12-h LD cycle that was either delayed (16D:4L) or advanced (8D:20L) by 4 h (Fig. 4) (see Materials and Methods). Exposure of flies to photic stimuli in the early (e.g., 16D:4L) and late (e.g., 8D:20L) night results in output behavioral rhythms that are phase delayed or phase advanced, respectively (references 26, 30, 32, 34, and 41 and data not shown).
Both a backward (Fig. 4A) and a forward (Fig. 4B) shift in the timing of the 12-h LD cycle evoked rapid effects on the temporal regulation of dClock RNA abundance, indicating that this molecular rhythm is quickly entrained by photic stimuli. Extending the light period from ZT12 to ZT16 (16D:4L) was accompanied by a delay of several hours in the accumulation profile of dClock RNA (Fig. 4A). Conversely, beginning the light period prematurely (at ZT20 [8D:20L]) led to a more rapid decline in the levels of dClock RNA (Fig. 4B). The effects of the 12-h LD shifts on the temporal regulation of dClock transcript levels are consistent with the direction of the phase shifts in the clock-controlled locomotor activity rhythm in D. melanogaster exposed to similar photic treatments. Likewise, the per-tim transcriptional-translational feedback loop is also perturbed by light pulses in a manner that is consistent with the direction of the phase shifts in behavioral rhythms (25, 30, 54). However, in stark contrast to dClock expression, photic stimuli in the early night delay the declining phase in per and tim mRNA levels, whereas similar light treatments in the late night result in an earlier rise in the levels of these mRNAs (references 25 and 30 and data not shown). The results clearly indicate that photic signals elicit opposite effects on the expression of dClock compared to per and tim.
Recent studies have shown that the primary clock-specific response to light in D. melanogaster is the photic-induced degradation of TIM (22, 32, 54). A model that can account for the time-of-day-specific response of the clock to light (i.e., phase delays in the early night and phase advances in the late night) has been proposed (22, 25, 32, 54). Essentially, photic stimuli in the early night are accompanied by delays in the nuclear entry time of the PER-TIM complex. Conversely, exposure of flies to light in the late night elicits the rapid degradation of nuclear TIM and PER leading to the premature release of the autoinhibitory mechanism and consequently an earlier accumulation of per and tim transcripts. In the 16D:4L shift (Fig. 4A), the accumulation and nuclear entry of PER and TIM is retarded whereas in the 8D:20L shift, the levels of nuclear PER and TIM undergo more rapid decreases (references 22, 25, 30, 32 and data not shown). Thus, results from the 12-h LD shift experiments suggest that PER, TIM, or the PER-TIM complex must be present in the nucleus to stimulate expression of dClock. This contention is in agreement with the observation that high levels of dClock transcripts occur at times in a daily cycle when PER and TIM are present in the nucleus. Nevertheless, we cannot exclude the formal possibility that cytoplasmic PER and/or TIM decreases the stability of dClock mRNA and that once they localize to the nucleus they no longer modulate dClock expression. Finally, because the response of dClock expression to changes in 12-h LD cycles is rapid, it argues against the possibility that there is a very extended delay between the activity of PER and TIM and the regulation of dClock mRNA levels.
DISCUSSION
In this study, we characterized a novel D. melanogaster bHLH-PAS protein that shows very high homology to mammalian CLOCK. Similar findings were recently reported by Allada et al. (2) and Darlington et al. (9). Together, these results provide further evidence that the components of mammalian and Drosophila circadian clocks are highly conserved. Nevertheless, our data highlight some dramatic differences in the regulation and possible biochemical activities of putative clock orthologs.
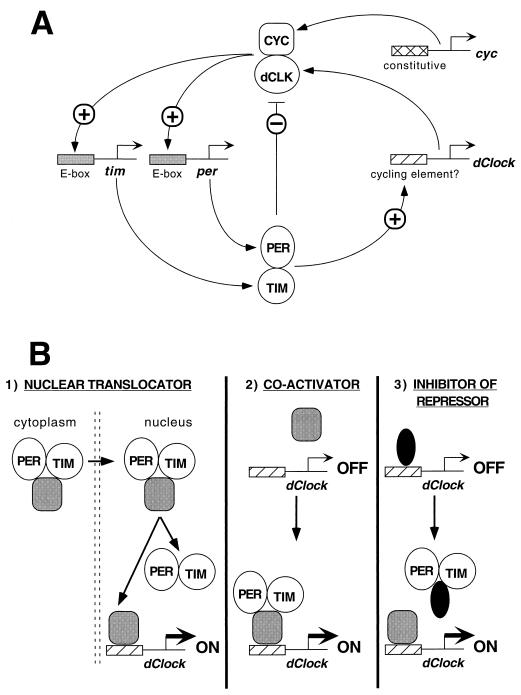
What might be the role of dCLOCK in pacemaker function? Because it is a member of the bHLH-PAS superfamily and contains a polyglutamine repeat, it is almost certain that dCLOCK functions as a transcriptional activator. Indeed, a recent study showed that in Drosophila tissue culture cells, expression of dClock can drive the expression of a reporter gene flanked on the 5′ end with E-box elements present in either per or tim (9). Moreover, a mutation in dClock (Jrk) that eliminates much of the putative activation domain of this transcription factor results in flies with low per and tim transcription, likely explaining the arrhythmic phenotype of the homozygous mutant (2). The putative in vivo partner of dCLOCK is likely the Drosophila homolog of the bHLH-PAS protein BMAL1, termed CYC (cycle) (2, 9). Importantly, a mutation in cyc also leads to low transcription of per and tim and to an arrhythmic mutant phenotype (36). A similar situation has been shown in the mammalian system where mCLOCK interacts with BMAL1 or its close relative MOP3 to activate transcription through E-box elements such as those found in per from either Drosophila or mammals (14, 21). Based on these recent findings, it is postulated that in Drosophila, dCLOCK interacts with CYC to function as a positive element in a circadian transcriptional loop by stimulating the expression of per and tim (2, 9) (Fig. 5A). To generate the negative element in the transcriptional feedback loop, PER, TIM, and/or the PER-TIM complex (the latter possibility is shown in Fig. 5A) inhibits the activity of the dCLOCK-CYC complex (9). Daily fluctuations in the subcellular distribution and levels of PER and TIM are thought to contribute to a properly timed delay in the ability of these two proteins to inhibit dCLOCK-CYC activity resulting in a transcriptional feedback loop that is regulated in a circadian manner.
FIG. 5.
Model of how PER and TIM might regulate dClock expression and function in the transcriptional feedback loop. (A) dCLOCK (dCLK) and CYC interact to form a heterodimer that binds E-box elements on per and tim 5′ regulatory sequences leading to transcriptional activation (2, 9). Increases in the levels of per and tim transcripts are subsequently followed by a rise in the amounts of PER and TIM proteins (not shown). After attaining a critical intracellular concentration, PER and TIM interact to form a complex that enters the nucleus (8, 13, 25, 37, 54), where PER, TIM, and/or the PER-TIM complex (the latter case is shown) inhibit the transcriptional activity of dCLOCK-CYC (9). In addition, nucleus-localized PER, TIM and/or the PER-TIM complex stimulate the rhythmic expression of dClock. Unlike per, tim, and dClock, cyc is expressed constitutively (36). (B) Three examples of how PER and TIM might lead to the stimulation of dClock transcription. See Discussion for more details. Gray oval, hypothetical transcription complex that stimulates dClock expression; black oval, hypothetical transcription complex that inhibits dClock expression.
Although the biochemical modes of action for PER and TIM in clock function are not established, evidence accumulated over many years has generated very strong evidence for the participation of these two proteins in the inhibition of transcription. These include the observations that (i) high-level expression of per from a transgene containing a constitutive promoter is accompanied by the low abundance of noncycling per RNA from the wild-type chromosomal copy (53); (ii) in the per01 mutant, the levels of the circadianly regulated Crg-1 and Dreg-5 transcripts are close to their respective peak levels in wild-type flies (35, 50); (iii) light pulses in the early and late night delay the declining phase and advance the accumulating phase of per mRNA (25), respectively (in sharp contrast to dClock expression [Fig. 4]); (iv) failure to turn off the lights in a standard 12-h LD cycle results in a delay of the PER-TIM dependent autoinhibition (30); (v) in mammalian tissue culture cells, forced expression of Drosophila PER inhibits the transcriptional stimulatory activity of ARNT and AHR, two mammalian bHLH-PAS proteins that form a functional heterodimer (27); and most recently (vi) the demonstration that coexpression of PER and TIM can inhibit dCLOCK-dependent gene expression in tissue culture cells (9). Presumably, PER and/or TIM inhibits its own transcription by interacting with one or more components of a dCLOCK-CYC-containing complex blocking its function (9). Because PER has a PAS domain but no bHLH region, it is reasonable to expect that PER is the direct factor that interacts with either dCLOCK or CYC yielding nonfunctional heterodimers. However, although TIM does not have a PAS domain, it can nevertheless interact with the PAS domain of PER (13), suggesting that TIM can also bind bHLH-PAS containing transcription factors.
Yet our data indicate that PER and TIM are required for the high-level expression of dClock (Fig. 3B). This finding is consistent with the observation that dClock RNA levels begin to increase around the time when the PER-TIM complex begins to accumulate in the nucleus (ZT18-19) (8, 25) and decrease during the declining phase in the accumulation of nuclear PER and TIM (ZT2-8) (10, 22, 32, 53, 54). Although mutations that eliminate functional PER and TIM both lead to low levels of dClock RNA (Fig. 3B), it is not clear that both clock proteins are actively required in the nucleus to effect a transcriptional response. For example, the nuclear localization of PER and TIM are dependent on each other (37, 52). A role for PER and/or TIM in the stimulation of gene expression is further supported by the effects of changing the timing of light-dark cycles on the time course of dClock RNA (Fig. 4). Shifting the timing of a light-dark cycle such that the nuclear entry time of the PER-TIM complex is delayed (25) resulted in a concomitant delay in the accumulation of dClock mRNA abundance (Fig. 4A). Consistently, exposure of flies to light in the late night elicits the rapid degradation of TIM and premature disappearance of PER from the nucleus (22, 25, 32, 54), events that are accompanied by an earlier decline in the levels of dCLOCK transcripts (Fig. 4B). These results suggest that the presence of PER and/or TIM in the nucleus is necessary for the high-level expression of dClock. Taken together, our findings indicate that PER, TIM, or the PER-TIM complex also functions as a positive element in a circadian feedback loop. They also suggest an explanation for the finding that in the per01 mutant, the levels of per mRNA are approximately 50% of the peak value observed in wild-type flies (18). If PER was functioning solely as a negative regulator of its own transcription, a prediction is that in the absence of PER, its mRNA levels should be similar to the wild-type peak values. Notwithstanding this prediction, our findings suggest that the levels of dCLOCK are much lower in the mutant background (Fig. 3B). Thus, PER and TIM are acting as both negative regulators of dCLOCK protein activity (9) (Fig. 5A) and as positive regulators of dClock expression (Figs. 3B, 4, and 5A). We postulate that the combination of no PER-TIM-mediated inhibition but lower dCLOCK levels in per01 flies results in a dCLOCK-CYC complex with an effective activity that is ca. 50% lower than that present during the transcription of peak amounts of per and tim in wild-type flies.
How might the apparently contradictory findings showing that PER and TIM can act as both positive and negative elements in the regulation of gene expression be reconciled? A speculative proposal is that PER or TIM do not function to directly activate transcription but that during their daily subcellular movement they shuttle a transcription factor(s) from the cytoplasm to the nucleus where it can interact with specific DNA elements on dClock to elicit an appropriate transcriptional response (Fig. 5B). A second possibility is that PER and/or TIM can act as both a dominant negative inhibitor (Fig. 5A) and a coactivator (Fig. 5B) depending on the transcriptional complex it interacts with. Finally, in keeping with its previously suggested biochemical function, PER and/or TIM might abrogate the activity of a transcriptional complex that inhibits dClock expression (Fig. 5B), a mechanism that could phenocopy a stimulatory response. This postulated mechanism is likely more difficult to resolve when considering feedback loops. Our findings are also consistent with posttranscriptional effects of PER and TIM, e.g., increasing the stability of dClock transcripts. Future studies to understand better the biochemical functions of PER and TIM in the control of transcription in vivo will be required. For example, because PER interacts with TIM via its PAS domain (13), it is not clear if PER must first disengage from TIM prior to interacting with other PAS-containing proteins. In any event, our results demonstrate for the first time that PER and TIM are required for high-level gene expression. It will be of interest to determine how widespread a role PER and TIM play in the stimulation of gene expression.
The observation that dClock mRNA levels undergo daily cycles (9) (Figs. 2 through 4) suggests that it might be a state variable in a circadian timekeeping mechanism (a state variable is a clock component whose rhythmic changes in abundance or activity, rather than mere presence in the cell, is a necessary element of the timekeeping mechanism [3]). Although it is not clear whether the cycling of dClock mRNA is reflected at the protein level, the rhythmic expression of dClock suggests that regulation of dCLOCK abundance is important for clock function. Indeed, that the Drosophila timekeeping mechanism is very sensitive to the levels of dCLOCK is strongly suggested by the recent finding showing that reductions in the gene dosage of dClock from two to one copy resulted in flies manifesting activity rhythms that are approximately 1.5 to 2 h longer than those observed in wild-type controls (2). At this level of analysis, the Drosophila circadian pacemaker is more sensitive to the dosage of dClock than to that of the two previously characterized state variables in this species, per and tim (2). Based on the constitutive expression of cyc RNA (reference 36 and data not shown), fluctuations in the abundance of dCLOCK could contribute to the production of a transcriptional complex that regulates gene expression in a circadian manner (Fig. 5A). However, fluctuations in the levels of one or both components of the dCLOCK-CYC complex are not necessary to generate cyclical transcriptional activity because temporal changes in the activity of this complex could be regulated by oscillations in the abundance of PER and/or TIM. A more complicated scenario in which other bHLH-PAS proteins that can compete for binding with either dCLOCK or CYC are expressed in a temporally regulated manner can also be envisaged.
Despite the high structural conservation of circadian clock components in different species, there is remarkable diversity in the regulation and possible biochemical function of putative clock orthologs. For example, although a circadian pacemaker in the brains of silkmoths is dependent on PER (40), the PER and TIM proteins in this organism are apparently exclusively cytoplasmic (39), in stark contrast to the situation in D. melanogaster, in which the temporal regulation of PER and TIM nuclear entry is necessary for clock functioning (8, 37, 52). In addition, light elicits rapid increases in the levels of mouse per1 transcripts (1, 44, 45), whereas early changes in the levels of per (or tim) RNA are not observed in D. melanogaster (22, 25, 30). The photosensitivity of mper-1 is very reminiscent of the rapid increases in frq RNA levels by light pulses (7). Although a TIM homolog in mammals has not been described and the functional relationship of mper-1 and mper-2 (and possibly other mper homologs) to Drosophila per is not clear, these results suggest that putative circadian clock orthologs in different species might be regulated in very different manners. Here we show a striking example whereby dClock transcripts show daily rhythms in contrast to the constitutive expression of mammalian Clock (48, 49). Thus, the emerging picture seems to encompass the combination of the following two extreme possibilities: (i) clock components that do not share structural similarity, such as the PER and FRQ proteins, can nevertheless be similarly regulated and appear to have very similar roles in clock function; and (ii) putative clock orthologs, such as D. melanogaster and silkmoth PER, share extensive structural features yet apparently have different biochemical modes of action. Differences in how clock orthologs function within molecular loops and how they are regulated by input pathways are likely to reflect species-specific adaptations that confer biological advantages.
ACKNOWLEDGMENTS
This work was partially supported by an NIH grant to I.E.
K.B. and C.L. contributed equally to this study.
REFERENCES
- 1.Albrecht U, Sun Z S, Eichele G, Lee C C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 2.Allada R, White N E, So W V, Hall J C, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 3.Aronson B D, Johnson K A, Loros J J, Dunlap J C. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 4.Citri Y, Colot H V, Jacquier A C, Yu Q, Hall J C, Baltimore D, Rosbash M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature. 1987;326:42–47. doi: 10.1038/326042a0. [DOI] [PubMed] [Google Scholar]
- 5.Crews S T. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- 6.Crosthwaite S K, Dunlap J C, Loros J J. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 7.Crosthwaite S K, Loros J J, Dunlap J C. Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell. 1995;81:1003–1012. doi: 10.1016/s0092-8674(05)80005-4. [DOI] [PubMed] [Google Scholar]
- 8.Curtin K D, Huang Z J, Rosbash M. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron. 1995;14:365–372. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 9.Darlington T K, Wager-Smith K, Ceriani M F, Staknis D, Gekakis N, Steeves T D L, Weitz C J, Takahashi J S, Kay S A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 10.Edery I, Zwiebel L J, Dembinska M E, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewer J, Frisch B, Hamblen-Coyle M J, Rosbash M, Hall J C. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garceau N Y, Liu Y, Loros J J, Dunlap J C. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- 13.Gekakis N, Saez L, Delahaye-Brown A M, Myers M P, Sehgal A, Young M W, Weitz C J. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 14.Gekakis N, Staknis D, Nguyen H B, Davis F C, Wilsbacher L D, King D P, Takahashi J S, Weitz C J. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 15.Handler A M, Konopka R J. Transplantation of a circadian pacemaker in Drosophila. Nature. 1979;279:236–238. doi: 10.1038/279236a0. [DOI] [PubMed] [Google Scholar]
- 16.Hao H, Allen D L, Hardin P E. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardin P E, Hall J C, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci USA. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardin P E, Hall J C, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 19.Hastings J W, Rusak B, Boulos Z. Circadian rhythms: the physiology of biological timing. In: Prosser C L, editor. Neural and integrative animal physiology. New York, N.Y: Wiley-Liss, Inc.; 1991. pp. 435–546. [Google Scholar]
- 20.Hogenesch J B, Chan W K, Jackiw V H, Brown R C, Gu Y Z, Pray-Grant M, Perdew G H, Bradfield C A. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 21.Hogenesch J B, Gu Y Z, Jain S, Bradfield C A. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 23.King D P, Zhao Y, Sangoram A M, Wilsbacher L D, Tamaka M, Antoch M P, Steeves T D L, Vitaterna M H, Kornhauser J M, Lowry P L, Turek F W, Takahashi J S. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konopka R J, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- 26.Levine J D, Casey C I, Kalderon D D, Jackson F R. Altered circadian pacemaker functions and cyclic AMP rhythms in the Drosophila learning mutant dunce. Neuron. 1994;13:967–974. doi: 10.1016/0896-6273(94)90262-3. [DOI] [PubMed] [Google Scholar]
- 27.Lindebro M C, Poellinger L, Whitelaw M L. Protein-protein interaction via PAS domains: role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo C, Loros J J, Dunlap J C. Nuclear localization is required for function of the essential clock protein FRQ. EMBO J. 1998;17:1228–1235. doi: 10.1093/emboj/17.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majercak J, Kalderon D, Edery I. Drosophila melanogaster deficient in protein kinase A manifest behavior-specific arrhythmia but normal clock function. Mol Cell Biol. 1997;17:5915–5922. doi: 10.1128/mcb.17.10.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marrus S B, Hongkui Z, Rosbash M. Effect of constant light and circadian entrainment of perS flies: evidence for light-mediated delay of the negative feedback loop in Drosophila. EMBO J. 1996;15:6877–6886. [PMC free article] [PubMed] [Google Scholar]
- 31.McClung C R, Fox B A, Dunlap J C. The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period. Nature. 1989;339:558–562. doi: 10.1038/339558a0. [DOI] [PubMed] [Google Scholar]
- 32.Myers M P, Wager-Smith K, Rothenfluh-Hilfiker A, Young M W. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 33.Myers M P, Wager-Smith K, Wesley C S, Young M W, Sehgal A. Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science. 1995;270:805–808. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- 34.Qiu J, Hardin P E. per mRNA cycling is locked to lights-off under photoperiodic conditions that support circadian feedback loop function. Mol Cell Biol. 1996;16:4182–4188. doi: 10.1128/mcb.16.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouyer F, Rachidi M, Pikielny C, Rosbash M. A new gene encoding a putative transcription factor regulated by the Drosophila circadian clock. EMBO J. 1997;16:3944–3954. doi: 10.1093/emboj/16.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutila J E, Suri V, Le M, So W V, Rosbash M, Hall J C. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 37.Saez L, Young M W. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 38.Sassone-Corsi P. Molecular clocks: mastering time by gene regulation. Nature. 1998;392:871–874. doi: 10.1038/31821. [DOI] [PubMed] [Google Scholar]
- 39.Sauman I, Reppert S M. Circadian clock neurons in the silkmoth Antheraea pernyi: novel mechanisms of Period protein regulation. Neuron. 1996;17:889–900. doi: 10.1016/s0896-6273(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 40.Sauman I, Tsai T, Roca A L, Reppert S M. Period protein is necessary for circadian control of egg hatching behavior in the silkmoth Antheraea pernyi. Neuron. 1996;17:901–909. doi: 10.1016/s0896-6273(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 41.Saunders D S, Gillanders S W, Lewis R D. Light pulse phase response curves for the locomoter activity rhythm in period mutants. J Insect Physiol. 1994;40:957–968. [Google Scholar]
- 42.Sehgal A, Price J L, Man B, Young M W. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 43.Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers M P, Young M W. Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 44.Shearman L P, Zylka M J, Weaver D R, Kolakowski L F, Jr, Reppert S M. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 45.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros J J, Dunlap J C, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 46.Sidote D, Majercak J, Parikh V, Edery I. Differential effects of light and heat on the Drosophila circadian clock proteins PER and TIM. Mol Cell Biol. 1998;18:2004–2013. doi: 10.1128/mcb.18.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.So W V, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Z S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 49.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 50.Van Gelder R N, Krasnow M A. A novel circadianly expressed Drosophila melanogaster gene dependent on the period gene for its rhythmic expression. EMBO J. 1996;15:1625–1631. [PMC free article] [PubMed] [Google Scholar]
- 51.Vitaterna M H, King D P, Chang A M, Kornhauser J M, Lowrey P L, McDonald J D, Dove W F, Pinto L H, Turek F W, Takahashi J S. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vosshall L B, Price J L, Sehgal A, Saez L, Young M W. Block in nuclear localization of period protein by a second clock mutation, timeless. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- 53.Zeng H, Hardin P E, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng H, Qian Z, Myers M P, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y D, Barnard M, Tian H, Li X, Ring H Z, Francke U, Shelton J, Richardson J, Russell D W, McKnight S L. Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc Natl Acad Sci USA. 1997;94:713–718. doi: 10.1073/pnas.94.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]